
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Genome Ed., 08 January 2025
Sec. Genome Editing in Plants
Volume 6 - 2024 | https://doi.org/10.3389/fgeed.2024.1506468
This article is part of the Research TopicDevelopments in Genetic Transformation and Genome Editing in Horticultural Crops and Their PathogensView all articles
Virus-induced genome editing (VIGE) technologies have been developed to address the limitations to plant genome editing, which heavily relies on genetic transformation and regeneration. However, the application of VIGE in plants is hampered by the challenge posed by the size of the commonly used gene editing nucleases, Cas9 and Cas12a. To overcome this challenge, we employed intein-mediated protein splicing to divide the SaCas9 transcript into two segments (Split-v1) and three segments (Split-v3). The Split-v1 system demonstrated genome editing efficiencies in transgenic plants comparable to those achieved with wild-type SaCas9, with efficiencies ranging from 70.2% to 96.1%. Additionally, we constructed barley stripe mosaic virus (BSMV)-based vectors to co-express Split-v1 SaCas9 and gRNAs targeting LcHRC, LcGW2, and LcTB1 in sheepgrass (Leymus chinensis), a Gramineae forage species known for its recalcitrance to genetic transformation. Infected leaves of sheepgrass exhibited genome editing efficiencies ranging from 10.40% to 37.03%. These results demonstrate the potential of intein-mediated split nuclease systems to broaden the applicability of VIGE in challenging plant species.
The CRISPR/Cas technology of genome editing is a powerful tool for making targeted changes in plants. However, due to its reliance on genetic transformation and plant regeneration, this technology has been restricted to a handful of species, and even within a species, some varieties are recalcitrant (Zhan et al., 2021). In recent years, virus-induced genome editing (VIGE) technologies have been developed to overcome the limitations to in vitro tissue culture and regeneration (Ellison et al., 2020; Liu and Zhang, 2020; Ma et al., 2020; Li et al., 2021; Liu et al., 2024). In VIGE, the modified viral vectors used to express genome editing components were delivered into the plant by agroinfiltration or viral homogenate inoculation, which spread throughout the plant, including the infected meristematic region, and generate heritable gene-edited seeds (Ellison et al., 2020; Li et al., 2021; Lei et al., 2022; Kim et al., 2023). Furthermore, VIGE avoids the stable integration of genome editing components such as expression cassettes of Cas and gRNA (usually seen in biolistics or Agrobacterium-mediated transformation) and reduces off-target effects and insertional mutations (Ali et al., 2018). However, because the length of the foreign gene insertion is known to correlate negatively with the stability of the plant virus vector, the size of the two most commonly used gene editing nucleases, Cas9 and Cas12a, poses a challenge for their efficient expression using plant virus vectors, which limits the broad application of VIGE in plants.
Employing a split-protein system has been proven to be an effective method for reducing the size of a gene transcription unit, as demonstrated by several studies (Wright et al., 2015; Kaya et al., 2017). However, the identification of suitable split sites is significantly limited by structural constraints (Wright et al., 2015). Crucially, this strategy has a profound effect on the efficiency of genome editing activities (Costa Carrió, 2015; Payacán Ortiz, 2022). Inteins, which are internal protein elements, facilitate post-translational protein splicing by self-excision from precursor proteins and the ligation of adjacent protein sequences. Recent evidence has shown that intein-mediated splits of SpCas9 and Cas12j2 are effective in the protoplasts of Arabidopsis and rice, respectively (Yuan et al., 2022; Sun Y. et al., 2024). However, there have been no reports yet on the application of intein-mediated Cas nuclease splitting in stable transgenic plants or its potential applicability to VIGE.
In this study, we utilized intein-mediated splicing to split Staphylococcus aureus Cas9 (SaCas9), which comprises only 1,053 amino acids, making it smaller than SpCas9 (1,386 amino acids), yet it retains a higher efficiency of editing. Results show that an intein-mediated split SaCas9 system attains editing efficiencies comparable to that of the wild-type SaCas9 in transgenic plants. We also demonstrate that this intein-mediated split SaCas9 was successfully applied in VIGE for sheepgrass, a Gramineae forage challenging for genetic transformation.
The full-length SaCas9 was cloned from the SaCas9-gRNA vector reported in Qin et al. (2019). Fragments P1, P2, P3, and P4, fused with inteinN or inteinC, were synthesized by Shenggong Bioengineering (Shanghai). The full-length SaCas9 or fused fragments were cloned into the pCXUN backbone for stable rice transformation or into RNAγ1 or RNAγ2 vectors for VIGE in sheepgrass. The RNAγ1 and RNAγ2 vectors were constructed following the method described by Cheuk and Houde (2018). All subcloning procedures were carried out using the pEASY-Uni Seamless Cloning and Assembly Kit (TransGen Biotech). The plasmid maps, including full-length SaCas9, split-v1, split-v2, and split-v3, are shown in Supplementary Figure S1. The gRNAs targeting OsNYC1, OsNYC4, and OsPDS were synthesized using overlapping PCR. Taking OsNYC1 as an example, PCR amplification was performed using the SaCas9-gRNA template with the primer pairs U3pmeF/OsNYC1-T1F and OsNYC1-T1F/U3pmeR, producing two PCR products. These products were mixed in a 1:1 ratio and used as the template for a subsequent amplification with U3pmeF/U3pmeR. The resulting PCR product was then cloned into the Pme I restriction site of vectors containing either full-length or split SaCas9. For the gRNAs targeting the LcHRC, LcGW2, and LcTB1 genes, taking LcHRC as an example, the gRNA scaffold was obtained by amplifying the SaCas9-gRNA template using the primers γ1-LcHRC-T1F/γ1-gRNAR. This scaffold was then seamlessly cloned into the Apa I restriction site of the RNAγ1 backbone using the Assembly Kit. All primers used in this study are provided in Supplementary Table S1.
The stable transformation of rice was conducted following previously published protocols (Hiei et al., 1994; Saha et al., 2006). In brief, the vectors described above were transformed into the Agrobacterium tumefaciens strain EHA105 by electroporation. Dehulled seeds of the japonica rice (Oryza sativa L.) variety Zhonghua 11 were surface sterilized and cultured on the N6D solid medium to induce callus formation. Rice calli were pre-cultured and inoculated with the A. tumefaciens strain EHA105 carrying the recombinant vector. After 3 days of co-cultivation, the calli were washed thoroughly with sterilized water to remove residual bacteria and transferred to the N6-S medium for selection under antibiotic pressure for 2 weeks. Resistant calli were then transferred to the RE-III medium for a 2-week cultivation period and subsequently sub-cultured onto the fresh RE-III medium every 2 weeks to promote regeneration. Regenerated plants were successfully obtained, with a minimum of 18 independent transgenic lines generated for each vector.
The Western blot and immunoprecipitation experimental methods were primarily conducted following previously published protocols (Saha et al., 2006). In brief, the protein extract from wild-type and transgenic rice plants was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane (Merck Millipore, IPVH00010). For Western blot and immunoprecipitation, SaCas9 was assayed using the mouse anti-SaCas9 monoclonal antibody [6H4] (EpiGentek, A-9001). The loading control was probed using the anti-actin antibody (Abmart, M20009), with a goat anti-mouse antibody (Abcam, ab6789) utilized as the secondary antibody.
The constructed plasmids were transformed into A. tumefaciens EHA105 strains for agroinfiltration of wild-type or overexpressed RNAα and RNAβ Nicotiana benthamiana leaves. Equal volumes of Agrobacterium strains harboring individual plasmids were mixed to a final OD600 of 0.3 and infiltrated into leaves of 3- to 4-week-old N. benthamiana plants, as described previously (Yuan et al., 2011). Virus-infected leaves were harvested after 7 days postinfection (dpi) and ground in 10 mM phosphate buffer each containing 0.5% of celite 545 (Roth) on ice. The virus was used to inoculate the fully emerged third leaves of 2- to 3-week-old sheepgrass using the finger-rub method. Each treatment included a minimum of four replicates.
The mutation types of the transgenic rice plant were assayed by Sanger sequencing of the PCR product and analyzed by CRISPR-GE DSDecodeM software (http://skl.scau.edu.cn/.). For next-generation sequencing (NGS), mutations were analyzed by the online tool high-throughput tracking of mutations 2.0 (Hi-TOM 2.0) (Sun T. et al., 2024). In brief, the target region was amplified from genomic DNA using site-specific primers containing barcode sequences. The resulting amplicons were submitted for next-generation sequencing at the State Key Laboratory of Rice Biology and Breeding, utilizing the Hi-TOM platform (China National Rice Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou).
Statistical analysis was performed using GraphPad Prism version 8 for Windows (https://www.graphpad.com/), while the figures were further processed with Adobe Illustrator 2020 and Adobe Photoshop CC 2019 (https://www.adobe.com/).
To test whether intein-mediated protein splicing is an optional approach to reduce the size of SaCas9 transcription unit in plants and its potential use in VIGE, we analyzed the sequence of SaCas9, and results showed that there are 105 potential sites that can be split by intein derived from NpuDnaE (Supplementary Figure S2). In this study, the SaCas9 protein was first split into a 533 aa N-terminal fragment (P1) and a 520 aa C-terminal fragment (P2), with the split site located at HNH endonuclease domains that cleave the DNA strands complementary to the guide RNA (split-v1), and these two fragments were fused with N-terminal and C-terminal of NpuDnaE intein, respectively (Figure 1 and Supplementary note S1). The fused coding sequence was driven by maize ubiquitin and the double 35 S promoter, respectively (Figure 1B and Supplementary note S1). The full-length SaCas9 and split SaCas9 without fusion with intein (split-v2) were designed as control groups for comparison (Figure 1B and Supplementary note S1). Subsequently, three gRNAs driven by an OsU3 promoter targeted to the OsPDS, OsNYC1, and OsNYC4 genes were constructed and cloned into the vector containing the full-length or split SaCas9 expression cassette, respectively (Figure 1B and Supplementary note S1). Sequence analysis of the transgenic rice plants revealed that the editing efficiency of split-v1 is comparable to that of full-length SaCas9 across all three target genes. Specifically, the editing efficiencies of split-v1 in OsNYC1, OsNYC4, and OsPDS were 72.2%, 96.1%, and 81%, respectively, while the corresponding efficiencies for full-length SaCas9 were 75%, 92%, and 80%. For the OsNYC4 and OsPDS targets, the editing efficiency of split-v1 was slightly higher than that of full-length SaCas9; however, the proportion of homologous and biallelic mutations was lower, with split-v1 achieving 69.2% and 47.7% for OsNYC4 and OsPDS, respectively, compared to 76% and 65% for full-length SaCas9 (Figures 1D, E). However, split-v2 showed no editing activity (Figrure 1D). These findings are in line with the Western blot and immunoprecipitation results: the full-length SaCas9 was detected in split-v1 transgenic plants but not in those with split-v2. Possibly due to antibody characteristics or the lower abundance of the target protein, full-length SaCas9 could not be detected via Western blot, but it can be detected using the immunoprecipitation method (Figure 2A).
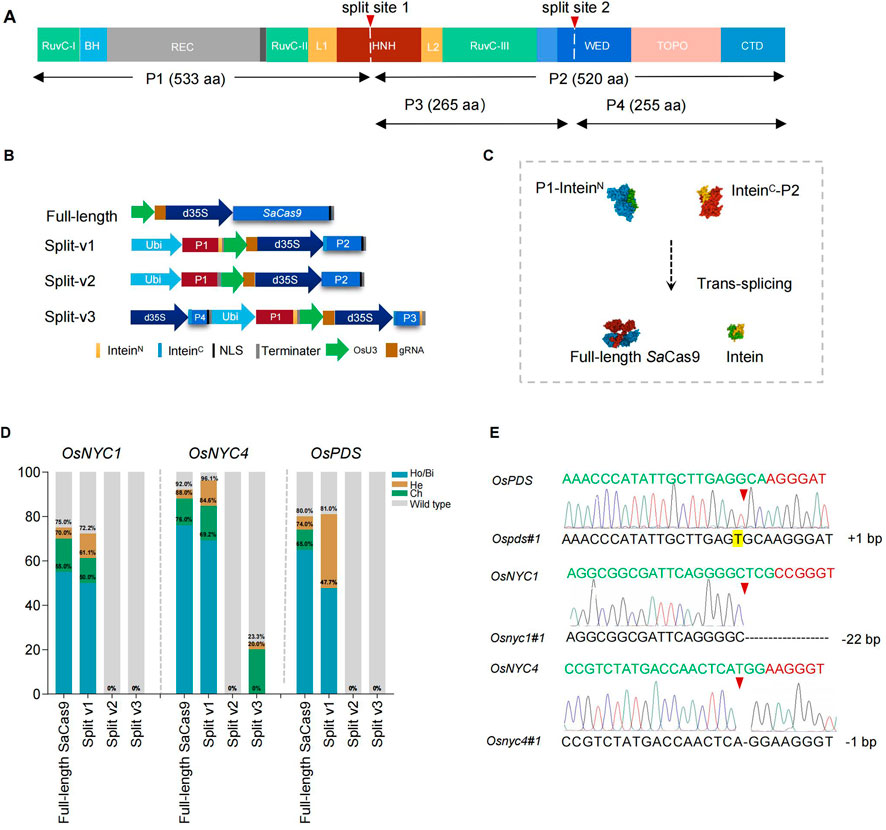
Figure 1. Intein-mediated split SaCas9 for genome editing in plants. (A) Domain organization and split sites of SaCas9, with the white dashed line indicating the split site. (B) Schematic representation of the split SaCas9 and gRNA expression cassettes. (C) Trans-splicing reaction mechanism of split SaCas9 mediated by NpuDnaE intein. (D) Mutation types and editing efficiency induced by full-length and split SaCas9 in transgenic rice plants. Ho/Bi indicates homozygous/biallelic mutations; He, heterozygous mutations; Ch, chimeric mutations. (E) Sanger sequencing results of regenerated rice plants, with red arrows indicating the edited site. The PAM sequence is highlighted in red, and the protospacer target, in green. Insertions are shaded, and black dashes represent deletions.

Figure 2. Analysis of SaCas9 trans-splicing in transgenic plants and phenotypes of Ospds, Osnycl, and Osnyc4 mutants. (A) Western blot and immunoprecipitation analysis of trans-splicing in SaCas9 protein. Full-length SaCas9 protein is detectable in both full-length SaCas9 and Split-v1 transgenic plants but not in Split-v2. The split SaCas9 protein is detected in both Split-v1 and Split-v2 transgenic plants but not in full-length SaCas9. (B) Phenotypes of the Osnycl and Osnyc4 mutants, which exhibit a stay-green phenotype in dark-induced leaf senescence compared to the wild type. (C) Phenotypes of the wild-type and Ospds mutant, with photobleaching observed in the Ospds mutant.
To explore the possibility of splitting SaCas9 into three parts, we divided P2 into P3 and P4 and fused them with inteinN or inteinC (split-v3), respectively (Figures 1A, B). The results demonstrated a significant decrease in efficiency, with only the vector targeted to OsNYC4 showing an editing efficiency of 23.3%, and among these edited plants, no homozygous/biallelic lines were found (Figure 1D). The lower efficiency observed in split-v3 may be attributed to the misassembly of P1, P3, and P4, which could potentially be improved using different inteins.
The phenotype of randomly selected Ospds, Osnyc1, and Osnyc4 mutants confirmed the sequencing results, with photobleaching observed in the Ospds mutant (Figure 2B) and a stay-green phenotype in dark-induced leaf senescence of Osnyc1 and Osnyc4 mutants (Figure 2C).
To test whether split SaCas9 can be utilized in VIGE, we modified the barley stripe mosaic virus (BSMV) vector as previously described (Cheuk and Houde, 2018). The modified vectors have the four-component BSMV system and have been demonstrated to allow overexpression of cDNAs of up to 2,100 nucleotides. In this study, we utilized RNAγ1 to express a gRNA targeting the LcHRC gene of sheepgrass (Leymus chinensis). Specifically, we constructed three vector combinations: in the first combination (Comb1), RNAγ1 expressed P1-inteinN and inteinC-P2; in the second combination (Comb2), RNAγ1 expressed P1-inteinN, while RNAγ2 expressed inteinC-P2; and in the third combination (Comb3), RNAγ2 expressed both P1-inteinN and inteinC-P2 (Figure 3). These constructs were then introduced into N. benthamiana plants for further analysis. After 7 days, the homogenate containing BSMV was harvested and used to infect sheepgrass (Figures 3A, B). After 14 days, we sequenced the LcHRC gene of the infected leaves and new leaves using next-generation sequencing. The results show that no editing events were detected in the sheepgrass. We hypothesize that the lack of genome editing activity may be attributed to the excessive amount of Agrobacterium components simultaneously injected into N. benthamiana. To test this, we separately introduced three vector combinations into N. benthamiana plants overexpressing RNAα and RNAβ and subsequently used the resulting virus to infect sheepgrass leaves. Next-generation sequencing revealed detectable genome editing activity in the Comb3-infected leaves, with an average editing efficiency reaching 30.95% (Figures 3B–D). In contrast, genome editing activity was undetectable in the leaves infected with Comb1 and Comb2, as well as in the control group, where RNAγ1 was used to express the gRNA and RNAγ2 was used to express the full-length SaCas9. To evaluate the efficiency of this approach and its applicability as a high-throughput gRNA screening platform in sheepgrass, additional target sites were designed for validation. These included LcHRC-T1 targeting the LcHRC gene; LcGW2-A-T1, LcGW2-B-T1, LcGW2-A-T2, and LcGW2-B-T2 targeting the LcGW2 gene; and LcTB1-A-T1 and LcTB1-B-T1 targeting the LcTB1 gene. All target sites exhibited detectable editing activity, with genome editing efficiencies ranging from 10.40% to 37.03%. Surprisingly, genome editing activity was not detectable in the new leaves of plants that had not been infected with the virus. This may be due to the size of the split SaCas9 still having some impact on the pathogenicity or mobility of the virus. Overcoming this limitation can be achieved by 1) optimizing viral vectors to enhance the ability of viruses to express exogenous genes and 2) improving the gene editing efficiency of reported smaller nucleases like CasΦ (Pausch et al., 2020; Liu et al., 2022; Li et al., 2023; Wang et al., 2023; Sun Y. et al., 2024; Zhao et al., 2024), Cas12f1 (Bigelyte et al., 2021; Wu et al., 2021; Kim et al., 2022), Casλ (Al-Shayeb et al., 2022), and TnpB (Karvelis et al., 2021) in plants and employing inteins to facilitate splicing and achieve smaller transcription units.
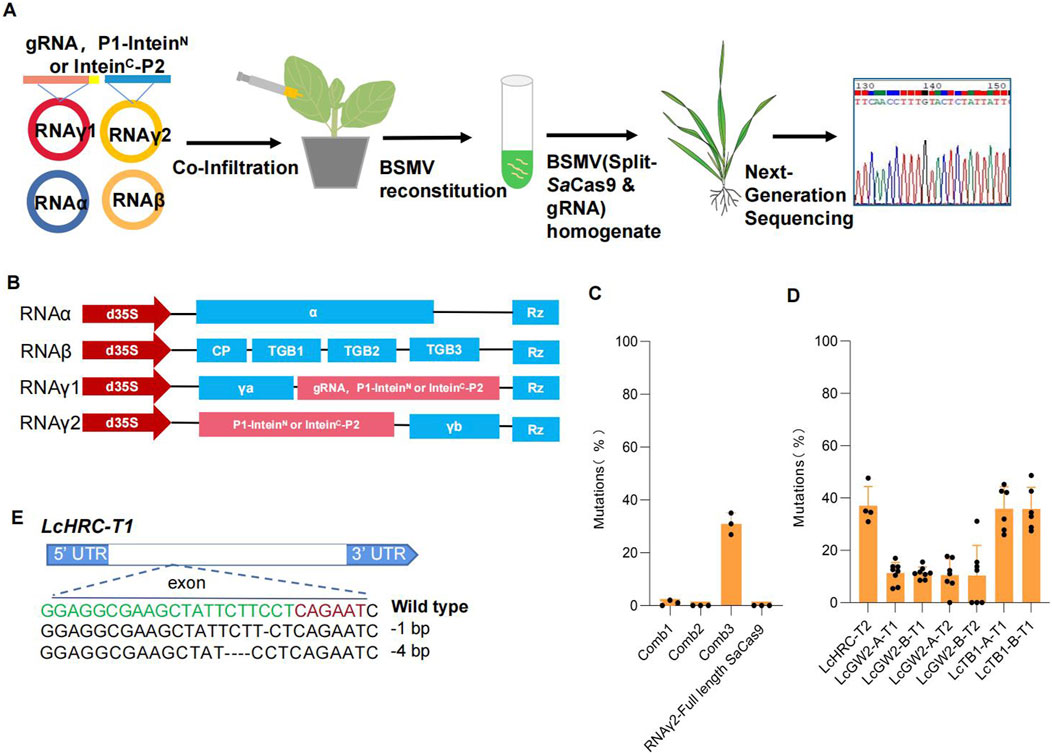
Figure 3. Validation of intein-mediated split-SaCas9 in VIGE for genome editing of sheepgrass. (A) Schematic illustration of BSMV-mediated genome editing in sheepgrass. (B) Schematic illustration of the vectors used by BSMV-mediated genome editing. (C) Genome editing efficiency of different vector combinations targeting the LCHRC gene. (D) Genome editing efficiency mediated by BSMV at seven endogenous loci in LCHRC, LCGW2, and LCTB1 genes. For (C, D), each point represents a biological replicate from an independent experiment (n > 3). Data are presented as mean values ±SD. (E) Alignment of indel mutations of the LCHRC gene mediated by VIGE. PAM is shown in red, and the protospacer target, in green; black dashes denote deletions.
We have demonstrated that intein-mediated split SaCas9 functions effectively in transgenic plants. Additionally, we validated that intein-mediated split SaCas9 can be successfully applied in the VIGE in sheepgrass, a type of Gramineae forage that is difficult to be genetically transformed. This strategy creates new opportunities not only for VIGE but also for virus-mediated base editing and prime editing in plants.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
DH: writing–original draft, data curation, formal analysis, and project administration. LH: formal analysis, writing–original draft, data curation, and methodology. YL: formal analysis, writing–original draft, data curation, methodology, and investigation. XD: formal analysis, writing–original draft, data curation, methodology, and investigation. XC: writing–original draft, data curation, formal analysis, investigation, and methodology. SB: writing–original draft, data curation, and project administration. LZ: writing–original draft, data curation, and formal Analysis. DL: data curation, formal analysis, methodology, and writing–review and editing. YS: writing–original draft, writing–review and editing, data curation, formal analysis, methodology, conceptualization, funding acquisition, investigation, project administration, resources, software, supervision, validation, and visualization.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation of China (32160111, 32370431, and 31770261), the Central Government Guiding Special Funds for the Development of Local Science and Technology (2020ZY0005), the Inner Mongolia Natural Science Foundation (2020MS03027 and 2021ZD04), and program for Young Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region (NJYT22105).
The authors thank Prof. Dawei Li (China Agriculture University) for providing the BSMV vectors. This work was funded by the National Natural Science Foundation of China (32160111, 32370431, and 31770261), the Central Government Guiding Special Funds for the Development of Local Science and Technology (2020ZY0005), the Inner Mongolia Natural Science Foundation (2020MS03027 and 2021ZD04), and the program for Young Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region (NJYT22105).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeed.2024.1506468/full#supplementary-material
Ali, Z., Eid, A., Ali, S., and Mahfouz, M. M. (2018). Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res. 244, 333–337. doi:10.1016/j.virusres.2017.10.009
Al-Shayeb, B., Skopintsev, P., Soczek, K. M., Stahl, E. C., Li, Z., Groover, E., et al. (2022). Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors. Cell 185, 4574–4586.e16. doi:10.1016/j.cell.2022.10.020
Bigelyte, G., Young, J. K., Karvelis, T., Budre, K., Zedaveinyte, R., Djukanovic, V., et al. (2021). Miniature type VF CRISPR-Cas nucleases enable targeted DNA modification in cells. Nat. Commun. 12, 6191. doi:10.1038/s41467-021-26469-4
Cheuk, A., and Houde, M. (2018). A new barley stripe mosaic virus allows large protein overexpression for rapid function analysis. Plant Physiol. 176, 1919–1931. doi:10.1104/pp.17.01412
Costa Carrió, P. (2015). Creation of a split-cas9 system for effective plant genome editing. Master’s thesis. Barcelona, Spain: Universitat Politècnica de Catalunya.
Ellison, E. E., Nagalakshmi, U., Gamo, M. E., Huang, P.-J., Dinesh-Kumar, S., and Voytas, D. F. (2020). Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. plants 6, 620–624. doi:10.1038/s41477-020-0670-y
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi:10.1046/j.1365-313x.1994.6020271.x
Karvelis, T., Druteika, G., Bigelyte, G., Budre, K., Zedaveinyte, R., Silanskas, A., et al. (2021). Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696. doi:10.1038/s41586-021-04058-1
Kaya, H., Ishibashi, K., and Toki, S. (2017). A split Staphylococcus aureus Cas9 as a compact genome-editing tool in plants. Plant Cell Physiology 58, 643–649. doi:10.1093/pcp/pcx034
Kim, D. Y., Lee, J. M., Moon, S. B., Chin, H. J., Park, S., Lim, Y., et al. (2022). Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 40, 94–102. doi:10.1038/s41587-021-01009-z
Kim, H., Oh, Y., Park, E., Kang, M., Choi, Y., and Kim, S.-G. (2023). “Heritable virus-induced genome editing (VIGE) in Nicotiana attenuata,” in Base editors: methods and protocols (Springer), 203–218.
Lei, J., Li, Y., Dai, P., Liu, C., Zhao, Y., You, Y., et al. (2022). Efficient virus-mediated genome editing in cotton using the CRISPR/Cas9 system. Front. Plant Sci. 13, 1032799. doi:10.3389/fpls.2022.1032799
Li, T., Hu, J., Sun, Y., Li, B., Zhang, D., Li, W., et al. (2021). Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. plant 14, 1787–1798. doi:10.1016/j.molp.2021.07.010
Li, Z., Zhong, Z., Wu, Z., Pausch, P., Al-Shayeb, B., Amerasekera, J., et al. (2023). Genome editing in plants using the compact editor CasΦ. Proc. Natl. Acad. Sci. 120, e2216822120. doi:10.1073/pnas.2216822120
Liu, D., Ellison, E. E., Myers, E. A., Donahue, L. I., Xuan, S., Swanson, R., et al. (2024). Heritable gene editing in tomato through viral delivery of isopentenyl transferase and single-guide RNAs to latent axillary meristematic cells. Proc. Natl. Acad. Sci. 121, e2406486121. doi:10.1073/pnas.2406486121
Liu, H., and Zhang, B. (2020). Virus-based CRISPR/Cas9 genome editing in plants. Trends Genet. 36, 810–813. doi:10.1016/j.tig.2020.08.002
Liu, S., Sretenovic, S., Fan, T., Cheng, Y., Li, G., Qi, A., et al. (2022). Hypercompact CRISPR–Cas12j2 (CasΦ) enables genome editing, gene activation, and epigenome editing in plants. Plant Commun. 3, 100453. doi:10.1016/j.xplc.2022.100453
Ma, X., Zhang, X., Liu, H., and Li, Z. (2020). Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants 6, 773–779. doi:10.1038/s41477-020-0704-5
Pausch, P., Al-Shayeb, B., Bisom-Rapp, E., Tsuchida, C. A., Li, Z., Cress, B. F., et al. (2020). CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 369, 333–337. doi:10.1126/science.abb1400
Payacán Ortiz, C. V. (2022). A non-integrative CRISPR/Cas9 genome-editing approach for use in vegetable crop breeding. Master’s thesis. Coventry, Warwickshire, United Kingdom: University of Warwick.
Qin, R., Li, J., Li, H., Zhang, Y., Liu, X., Miao, Y., et al. (2019). Developing a highly efficient and wildly adaptive CRISPR-SaCas9 toolset for plant genome editing. Plant Biotechnol. J. 17 (4), 706. doi:10.1111/pbi.13047
Saha, P., Majumder, P., Dutta, I., Ray, T., Roy, S., and Das, S. (2006). Transgenic rice expressing Allium sativum leaf lectin with enhanced resistance against sap-sucking insect pests. Planta 223, 1329–1343. doi:10.1007/s00425-005-0182-z
Sun, T., Liu, Q., Chen, X., Hu, F., and Wang, K. (2024a). Hi-TOM 2.0: an improved platform for high-throughput mutation detection. Sci. China Life Sci. 67, 1532–1534. doi:10.1007/s11427-024-2555-x
Sun, Y., Hu, J., Hu, Z., Zhou, H., Gao, Y., Liu, Y., et al. (2024b). Engineered and split an efficient hypercompact CRISPR-CasΦ genome editor in plants. Plant Commun. 100881. doi:10.1016/j.xplc.2024.100881
Wang, Y., Qi, T., Liu, J., Yang, Y., Wang, Z., Wang, Y., et al. (2023). A highly specific CRISPR-Cas12j nuclease enables allele-specific genome editing. Sci. Adv. 9, eabo6405. doi:10.1126/sciadv.abo6405
Wright, A. V., Sternberg, S. H., Taylor, D. W., Staahl, B. T., Bardales, J. A., Kornfeld, J. E., et al. (2015). Rational design of a split-Cas9 enzyme complex. Proc. Natl. Acad. Sci. 112, 2984–2989. doi:10.1073/pnas.1501698112
Wu, Z., Zhang, Y., Yu, H., Pan, D., Wang, Y., Wang, Y., et al. (2021). Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat. Chem. Biol. 17, 1132–1138. doi:10.1038/s41589-021-00868-6
Yuan, C., Li, C., Yan, L., Jackson, A. O., Liu, Z., Han, C., et al. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS one 6, e26468. doi:10.1371/journal.pone.0026468
Yuan, G., Lu, H., De, K., Hassan, M. M., Liu, Y., Li, Y., et al. (2022). An intein-mediated split–nCas9 system for base editing in plants. ACS Synth. Biol. 11, 2513–2517. doi:10.1021/acssynbio.1c00507
Zhan, X., Lu, Y., Zhu, J. K., and Botella, J. R. (2021). Genome editing for plant research and crop improvement. J. Integr. plant Biol. 63, 3–33. doi:10.1111/jipb.13063
Keywords: CRISPR/Cas, intein, SaCas9, VIGE, plants
Citation: Hu D, Hu L, Lu Y, Dong X, Cao X, Bai S, Zhang L, Li D and Sun Y (2025) Intein-mediated split SaCas9 for genome editing in plants. Front. Genome Ed. 6:1506468. doi: 10.3389/fgeed.2024.1506468
Received: 05 October 2024; Accepted: 13 December 2024;
Published: 08 January 2025.
Edited by:
Prasenjit Saha, Planet 13 Holdings, Inc., United StatesReviewed by:
Rui Zhang, Chinese Academy of Sciences (CAS), ChinaCopyright © 2025 Hu, Hu, Lu, Dong, Cao, Bai, Zhang, Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongwei Sun, c3VueW9uZ3dlaUBpbXUuZWR1LmNu; Dongming Li, bGlkb25nbWluZzAxMThAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.