
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genome Ed., 07 January 2025
Sec. Genome Editing in Plants
Volume 6 - 2024 | https://doi.org/10.3389/fgeed.2024.1488024
This article is part of the Research TopicGene Editing: Approaches and Applications for Trait Enhancement in Legume and Oilseeds CropsView all articles
 Charli Kaushal1†
Charli Kaushal1† Mahak Sachdev2†
Mahak Sachdev2† Mansi Parekh2
Mansi Parekh2 Harini Gowrishankar2
Harini Gowrishankar2 Mukesh Jain3
Mukesh Jain3 Subramanian Sankaranarayanan1*
Subramanian Sankaranarayanan1* Bhuvan Pathak2*
Bhuvan Pathak2*Plant-derived oils provide 20%–35% of dietary calories and are a primary source of essential omega-6 (linoleic) and omega-3 (α-linolenic) fatty acids. While traditional breeding has significantly increased yields in key oilseed crops like soybean, sunflower, canola, peanut, and cottonseed, overall gains have plateaued over the past few decades. Oilseed crops also experience substantial yield losses in both prime and marginal agricultural areas due to biotic and abiotic stresses and shifting agro-climates. Recent genomic, transcriptomic, and metabolomics research has expanded our understanding of the genetic and physiological control of fatty acid biosynthesis and composition. Many oilseed species have inherent stress-combating mechanisms, including transcription factor regulation. Advances in genome editing tools like CRISPR/Cas9 offer precise genetic modifications, targeting transcription factors and binding sites to enhance desirable traits, such as the nutritional profile and chemical composition of fatty acids. This review explores the application of genome editing in oilseed improvement, covering recent progress, challenges, and future potential to boost yield and oil content. These advancements could play a transformative role in developing resilient, nutritious crop varieties essential for sustainable food security in a changing climate.
Oil seeds account for more than 207.8 M Ha of the world’s cultivated area and production is estimated to be 666.7 million tons (FAO, 2023/24). Soybean is the major oil crop with a production of 425.4 million tons, followed by rapeseed at 87.2 million tons, sunflower seed at 50.4 million tons, cottonseed at 44.6 million tons, and groundnut at 50.3 million tons (USDA, 2023/24).
The oil crops accumulate the high-energy triacylglycerols (TAGs) consisting of three glycerol-bound esterified fatty acids (FAs) as the primary source of stored carbon. Plant-derived oils typically account for 20%–35% of dietary calorie intake and play a critical role in human nutrition. While the human body can synthesize omega-9 FA (oleic acid), it relies on dietary intake for essential omega-6 (linoleic) and omega-3 (α-linolenic) FAs (Saini and Keum, 2018). The unique omega-3, -6 and -9 FAs profiles of the plant-derived oils are linked to improved cardiovascular health, reduced cancer risk, and decreased inflammation (Tian et al., 2023). Seeds of various oilseed crops such as peanut (Arachis hypogaea), canola or rapeseed (Brassica napus), Ethiopian mustard (Brassica carinata), Chinese mustard (Brassica juncea), false flax (Camelina sativa), soybean (Glycine max), sunflower (Helianthus annus) and cottonseed (Gossypium hirsutum) are rich in ‘common’ FAs, including palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1Δ9), linoleic acid (C18:2Δ9,12), and α-linolenic acid (C18:3Δ9,12,15) (Attia et al., 2021). Seed oils also contain structurally diverse FAs with unusual features, such as atypical carbon chain lengths (fewer than 16 or more than 18 carbons), varying degrees of unsaturation, double-bond positions, cis/trans orientations, acetylenic bonds, or unique side-chain decorations (e.g., epoxy, hydroxy, cyclopropene, and furan rings). These ‘unusual’ FAs hold high value as nutraceuticals, biofuels, and industrial chemicals (Cahoon and Li-Beisson, 2020).
Achieving environmentally sustainable food security for the growing world population needs pragmatic emphases on improving crop yields and nutritional quality. Changing climate conditions continue to exacerbate the effects of temperature, drought and salinity stress across marginal agricultural landscapes. The abiotic stress conditions also increase the crops’ susceptibility to existing pathogens and have been implicated in increased virulence of previously mild strains (Wu et al., 2020). These biotic and abiotic stresses significantly impact agricultural productivity, especially in staple oilseed crops like soybean, sunflower, canola, peanut, and cottonseed, causing substantial yield losses. Despite notable gains in yield through traditional breeding, oilseed crop productivity has plateaued in recent decades. Both prime and marginal agricultural areas experience considerable yield losses in oilseeds due to combined stressors, amplified by climate change. For example, season-long high temperatures of 32°C significantly reduce biomass and yield in peanuts (Prasad et al., 2003), while yield models suggest a 3.1% reduction in soybean yield for every 1°C increase in average temperature (Zhao et al., 2017).
Recent advances in genomic, transcriptomic, and metabolomics research have unveiled a wealth of information about the genetic and physiological regulation of FA biosynthesis in oilseed crops. Large-scale genome sequencing of key crops such as canola, groundnuts, soybean, flax, and sunflower offers valuable data on traits governing yield, oil composition, and stress resilience. This genomic information significantly enhances our ability to develop crop varieties better suited to withstand environmental stresses and improve productivity (Varshney et al., 2021; Bohra et al., 2022).
Traditionally, studies on lipid metabolism in oilseeds have focused on enhancing FA biosynthesis by either overexpressing or disabling specific genes (Ruiz-Lopez et al., 2014). Transgenic approaches for improving traits in oilseed crops face challenges like low consumer acceptance, stringent regulation protocols, and the labor-intensive process of integrating transgenes into elite varieties (Meesapyodsuk et al., 2018; Na et al., 2018; Shah et al., 2018; Kim et al., 2019). Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated protein (Cas) provide a molecular toolkit for precise editing of genetic information (Mohanta et al., 2017). CRISPR/Cas-mediated editing approaches offer targeted bi-allelic/homozygous mutations in multiple alleles. By targeting transcription factor (TF) binding sites, CRISPR/Cas toolkits enable redesign of the desired traits, such as the nutritional composition of FAs. Additionally, editing pathogen effector binding sites can suppress disease susceptibility genes, boosting disease resistance and yield (Greenwood et al., 2023). Nuanced variations of CRISPR/Cas, such as CRISPR/Cas-mediated transcriptional activation (CRISPRa) (Selma et al., 2019; Pan et al., 2021; Xiong et al., 2021; Ren et al., 2022) and CRISPR-interference (CRISPRi) (Tang et al., 2017; Ming et al., 2020), hold immense potential in generating crop varieties that are disease resistant, stress-tolerant and nutritionally balanced, particularly benefiting developing regions.
Complex allopolyploid genomes of oilseed crops, analyzed through large transcriptomic datasets and machine learning models, can guide CRISPR/Cas-mediated high-throughput workflows and reveal post-editing effects for optimized phenotyping (O’brien et al., 2021). This review explores genome editing’s role in tackling current challenges and advancing oilseed crop improvement. By synthesizing progress, highlighting challenges, and proposing future directions, it offers insights into developing resilient, nutritious oilseed varieties essential for sustainable food security in a changing climate.
Oilseed crops contain over 200 FAs, with seeds storing up to 60% of their dry weight as TAGs during maturation (Ohlrogge and Jaworski, 1997; Kazaz et al., 2022; Zhou et al., 2023). FAs are classified as saturated (no double bonds) or unsaturated (one or more double bonds, e.g., Δ12,15). Genes encoding FA biosynthesis proteins are identified and functionally annotated across several plant species. Despite varied oil compositions, the processes of production, modification, TAG assembly, and FA storage are generally conserved in oilseed crops (Zhou et al., 2023). FA biosynthesis occurs in plastids and mitochondria, involving enzymes like acetyl-CoA carboxylase (ACCase) and fatty acid synthase (FAS) (Ohlrogge et al., 1979).
ACCase catalyzes the ATP-dependent carboxylation of acetyl-CoA to malonyl-CoA. Acyl-carrier protein (ACP), an essential co-factor, accepts the malonyl group to form malonyl-ACP, which provides the thiol group for FA synthesis, forming thioesters with the elongating acyl chain (Lessire and Stumpe, 1983; Clough et al., 1992; Sasaki and Nagano, 2004; Park and Kim, 2022). Through a series of enzymatic steps, ACP-bound β-ketoacyl intermediates elongate by two-carbon units until the desired acyl length is achieved, aided by ketoacyl synthase (KS), reductase (KR), dehydratase (DH), and enoyl reductase (ER) enzymes (Harwood, 1996; Park and Kim, 2022). Finally, thioesterase (TE) terminates synthesis, producing free FAs like palmitic and stearic acids for export to the cytoplasm (Jones et al., 1995; Salas and Ohlrogge, 2002; Park and Kim, 2022) (Figure 1).
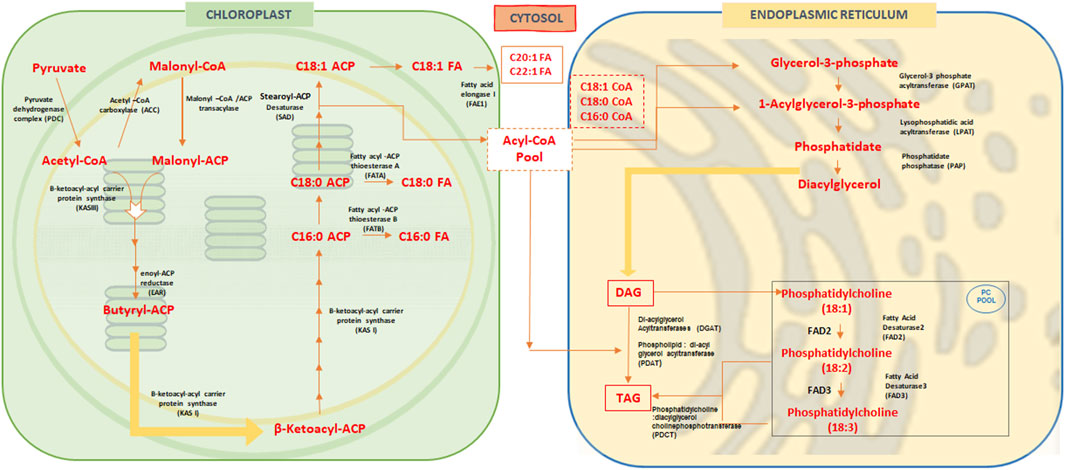
Figure 1. Overview of Fatty acid biosynthesis pathway in oilseed crops. In oilseed or higher plants, fatty acid biosynthesis occurs in the plastids, while polyunsaturated fatty acids (PUFAs) are synthesized in the endoplasmic reticulum (ER). In the chloroplasts, acetyl-CoA is first converted to malonyl-CoA by acetyl-CoA carboxylase. Malonyl-CoA is then converted to malonyl-ACP (acyl carrier protein), and two carbons are added after each cycle to the malonyl-acyl chain, synthesizing β-ketoacyl-ACP. This is followed by the formation of palmitic acid (C16:0), which is then elongated to stearic acid (C18:0) and subsequently desaturated to oleic acid (C18:1) by stearoyl-ACP desaturase (SAD). These fatty acids are transported to the cytosol, where further elongation occurs, converting them to C20:1 and C22:1 fatty acids by fatty acid elongase 1 (FAE1). Some fatty acids, such as C18:1-CoA, C18:0-CoA, and C16:0-CoA, are transported to the ER for desaturation. The acyl chains derived from these fatty acids are attached to glycerol-3-phosphate to form glycolipids. Through the action of enzymes like glycerol-3-phosphate acyltransferase (GPAT) and lysophosphatidic acid acyltransferase (LPAT), diacylglycerols (DAGs) are synthesized. The acyl chain C18:1 is then transferred to phosphatidylcholine (PC) and desaturated at the Δ-12 position to form C18:2 (linoleic acid) by FAD2 (fatty acid desaturase 2). This is further desaturated at the Δ-15 position to produce C18:3 (alpha-linolenic acid) by FAD3 (fatty acid desaturase 3). These acyl chains on phosphatidylcholine are subsequently used for the synthesis of triacylglycerols (TAGs).
Stearic acid is desaturated in plastids to oleic acid (18:1Δ9) by stearoyl-ACP desaturase (SAD). Free FAs exit plastids via ABC transporter proteins (Jouhet et al., 2007) and are activated into acyl-CoAs by acyl-CoA synthetases for cytosolic processes (Schnurr et al., 2002). In the cytosol, acyl-CoAs bind to acyl-CoA-binding proteins (ACBPs), increasing solubility for transport, desaturation, and elongation in the endoplasmic reticulum (ER) (Fox et al., 2000; Bates et al., 2013; Vanhercke et al., 2019). Fatty acid export (FAX) family proteins in the ER, such as FAX2 and FAX4, aid FA transport during seed oil accumulation (Li et al., 2020), with desaturase enzymes like FAD2 and FAD3 converting oleic acid to linoleic and alpha-linolenic acids, respectively (Lemieux et al., 1990; Dar et al., 2017; Park and Kim, 2022).
Plant TAG biosynthesis from FAs occurs via both acyl-CoA-dependent and independent pathways. TAG biosynthesis intersects with membrane lipid production and affects the oil composition in seeds (Bates and Browse, 2012; Bates et al., 2013; Bates, 2016; Bates, 2022; Parchuri et al., 2024; Zhou et al., 2023). Acyltransferase enzymes in the ER transfer acyl-CoAs to glycerol-3-phosphate (G3P) to form TAG (Li-Beisson et al., 2013; Park and Kim, 2022). Glycerol-3-phosphate acyltransferase (GPAT) adds acyl-CoA to the G3P at the sn-1 position to produce lysophosphatidic acid (LPA) (Shockey et al., 2016; Park and Kim, 2022). Lysophosphatidic acid acyltransferase (LPAT) then transfers to acyl-CoA to the sn-2 position of LPA to form phosphatidic acid (PA). Phosphate phosphatase (PAP) dephosphorylates PA to produce diacylglycerol (DAG) (Carman and Han, 2006; Park and Kim, 2022). Lastly, diacylglycerol acyltransferase (DGAT) adds acyl-CoA to the sn-3 position of DAG to create TAG (Zou et al., 1999; Park and Kim, 2022) (Figure 1).
Empirical evidence suggests that phosphatidylcholine (PC) provides polyunsaturated fatty acids (PUFAs) in TAGs (Park and Kim, 2022). Lysophosphatidylcholine acyltransferases (LPCAT) catalysis releases PUFAs from PC thereby augmenting the acyl-CoA pool (Lager et al., 2013). In the ER, TAG is synthesized using acyl-CoA as an acyl donor and DAG as an acceptor, catalyzed by the enzyme acyl-CoA:diacylglycerol acyltransferase (Zou et al., 1999; Kim et al., 2005; Shockey et al., 2016; Park and Kim, 2022). Nonetheless, an acyl-CoA-independent mechanism for TAG biosynthesis has also been described in plants and in the yeast (Saccharomyces cerevisiae). Phospholipid:diacylglycerol acyltransferase (PDAT) converts PUFAs in PC to DAG, directly producing TAG (Dahlqvist et al., 2000; Park and Kim, 2022).
Recent advances in genomics, transcriptomics, proteomics, and metabolomics, have provided significant insights into the regulatory networks involved in fatty acid biosynthesis during oilseed maturation and enrichment (Liu N. et al., 2022; Turquetti-Moraes et al., 2022; Bu et al., 2023; Li et al., 2024). Through these omics approaches, understanding of oilseed crop genomes provide comparative insights into the evolution of genetic mechanisms controlling lipid biosynthesis, accumulation, and species-specific variations in lipid profiles. Constructing gene regulation networks can help identify highly connected gene hubs related to oil biosynthesis (Zuo et al., 2024).
In peanut (Arachis hypogaea L.), one of the world’s major oilseed crops, salt stress poses a significant challenge to growth, particularly in regions prone to saline soils. Transcriptome analysis of peanut seedlings under salt stress (250 mM NaCl for 4 days, S4) and subsequent recovery (3 days post-transfer to standard conditions, R3) revealed 1,742 differentially expressed genes (DEGs) under stress and 390 during recovery (Sui et al., 2018). Among these, genes encoding ω-3 fatty acid desaturase, crucial for converting linoleic acid (18:2) to alpha-linolenic acid (18:3), were downregulated under salt stress and upregulated during recovery, shedding light on the genetic basis of salt tolerance and fatty acid biosynthesis in peanuts.
In other oilseed crops, such as Camellia oleifera and olives, integrative omics approaches have elucidated the molecular mechanisms of fatty acid accumulation and oil quality. A combination of transcriptome sequencing (RNA-seq) and isobaric tags for relative and absolute quantification (iTRAQ) during seed maturation revealed a reduction in flavonoids and a corresponding increase in fatty acid accumulation (Ye et al., 2021). A meta-analysis of RNA-seq data identified key genes associated with oil quality in olives by comparing three growth stages (S1, S2, and S3). The analysis revealed 155, 473, and 241 DEGs across these comparisons, which were classified into four groups based on their roles in oil biosynthesis and quality determinants. Pathways like galactose metabolism and FA biosynthesis were highlighted, showcasing the olive’s efficiency in FA production (Asadi et al., 2023). Transcriptome and proteome analyses in peony seeds have identified key genes involved in unsaturated FA biosynthesis, with FAD genes playing a crucial role in alpha-linolenic acid (ALA) accumulation. An integrated analysis of transcriptomic and proteomic data identified 38,482 unigenes and 2,841 proteins across nine developmental stages. These were grouped into three developmental clusters, revealing 211 genes and 35 proteins linked to fatty acid metabolism, with a particular emphasis on genes involved in unsaturated fatty acid (UFA) and ALA biosynthesis. Notably, eight FAD genes exhibited peak expression 53 days after pollination, indicating their crucial role in ALA accumulation (Wang X. et al., 2019; Wang et al., 2019). Similarly, in peanuts, iTRAQ proteomics identified fatty acid biosynthesis 2 (FAB2) as a critical enzyme in oleic acid biosynthesis during early seed development in high-oleate cultivars (Liu et al., 2018).
Metabolomic analyses across different stages of seed development and maturation, combined with transcriptomic profiles, have deepened our understanding of lipid biosynthesis. By integrating transcriptome and metabolome profiles at different stages of seed maturation, a comprehensive understanding of the molecular mechanisms underlying lipid biosynthesis can be achieved. Such studies have shown that changes in metabolomes correspond to gene expression associated with lipid biosynthesis, aiding in the identification of TFs involved in these processes. This approach can identify critical time intervals for gene engineering-based breeding strategies (Tan et al., 2019; Jia C. et al., 2024). Thus, integrating genomics, transcriptomics, proteomics, and metabolomics data offers a holistic view of the molecular mechanisms that govern seed oil quality.
Oilseeds are important sources of seed-specific proteins, TAGs, and other energy reserves that accumulate during embryogenesis. Fatty acid biosynthesis is a complex, multi-level process regulated at the transcriptional level by various transcription factors (Table 1). The seed maturation and embryogenesis processes are also regulated by a network of transcription factors (Verma et al., 2022). Key B3 domain-containing transcription factors, such as ABI3, FUS3, and LEC2, are essential in regulating seed maturation, transitioning from cell division to storage accumulation (Figure 2A). In dicotyledonous plants, seed development involves two main phases: morphogenesis, with cell division and expansion, followed by seed maturation, marked by storage component accumulation and preparation for desiccation tolerance (Pathak et al., 2020).
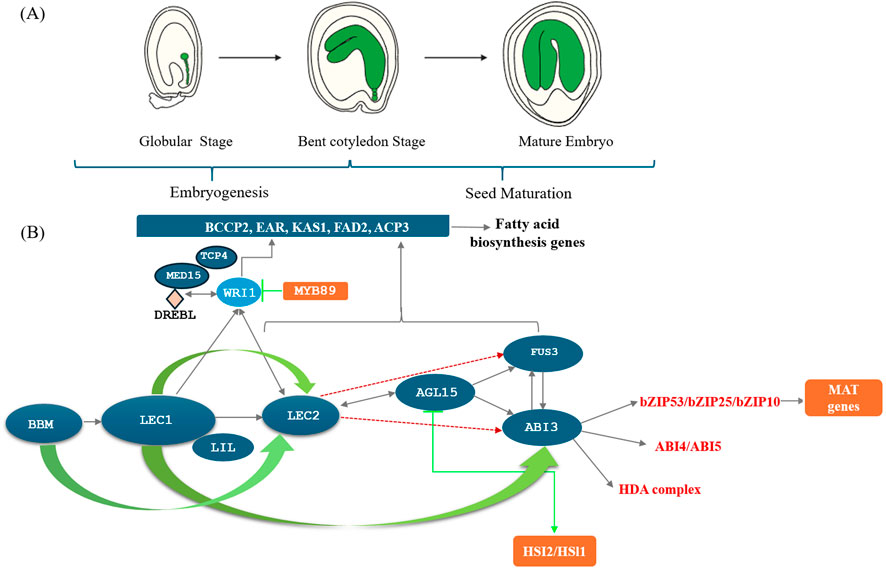
Figure 2. Networking of Transcription factors during oilseed maturation and Transcriptional regulation of seed development in Arabidopsis thaliana. (A) Stages of seed development in Arabidopsis thaliana: This figure shows broad stages of embryo development in Arabidopsis. Stage1: Globular stage, Stage 2: Bent cotyledon stage, Stage 3: Mature embryo. (B) Transcription Factor Network during Oilseed Maturation: Seed development is broadly categorized into two stages in seeds of A. thaliana and other crop plants. Stage 1- Embryogenesis, Stage 2-Seed maturation involving globular stage, bent cotyledon stage and mature embryo. Seed specific transcription factors include B3-domain TF (B3-domain containing transcription factors); LEC (Leafy cotyledons) - LEC1, LEC2 and LIL/FUS3; NF-Y TF (Nuclear factor–Y)-NF-YB/NF-YC; AP2/ERF (APETALA2/ETHYLENE RESPONSE FACTOR) –WRI1/BBM (Baby Boom TF), ARF(Auxin Response Factors),WRKY TF, MYB TF (myeloblastosis TF); bZIP TF (Basic leucine zipper transcription factor), ABA (Abscisic acid insensitive) -ABI3/4/5; AGL TF (Agamous-Like TF) -AGL15/18 play important role during different stages of seed development. LEC1 and BBM (AP2-ERF TF) initiate the embryo formation or embryogenesis followed by coordinated expression of vital transcription factors like LAFL-LEC2, ABI3, FUS3 and LIL/LEC1 network initiating embryo maturation. The BABY BOOM (BBM) transcription factor regulates embryo formation and seed development and is required for embryo initiation and maintenance. The figure is created with bioRender.com.
The AP2/ERF domain transcription factor Baby Boom (BBM), along with AINTEGUMENTA-LIKE (AIL), plays a critical role in regulating zygotic embryo development and the cellularization of the endosperm (Chen et al., 2022). CRISPR bbm-cr mutants showed their importance in embryo patterning (Chen et al., 2022).
LEAFY COTYLEDON1 (LEC1) acts as a molecular signal, transmitting cues from the endosperm to the embryo, thereby initiating various developmental programs essential for embryo maturation (Song et al., 2021). Mutations disrupting LEC1 lead to reduced desiccation tolerance and lipid accumulation in soybean seeds (Pelletier et al., 2017; Jo et al., 2019). These mutations also lead to chromatin de-repression and the subsequent activation of LAFL genes, which are pivotal in initiating embryogenesis within the embryo of Arabidopsis seeds (Song et al., 2021). LEC2, activated by LEC1, plays a crucial role in signaling early embryogenesis, establishing an ideal cellular environment that supports both morphogenesis and subsequent seed maturation (Figures 2A, B). This gene is integral not only in the early stages of embryo formation but also during the later phases of seed development (Stone et al., 2001).
LEC2, along with FUS3 and ABI3, operates as a central regulator of seed maturation, with each factor interdependently influencing the others. Notably, LEC2 can activate both LEC1 and FUS3 to kickstart the maturation process (Stone et al., 2008; Liu B. et al., 2021). Moreover, LEC2 can directly induce AGL15, a positive regulator of LAFL, and regulate the expression of gibberellic acid-related genes crucial for seed maturation (Stone et al., 2008) (Figure 2). LEC2 also exerts control over WRINKLED1 (WRI1), a master regulator of fatty acid synthesis, thereby playing a key role in fatty acid accumulation in oilseeds (Baud et al., 2007). Conversely, repressors such as TCP4 and MED15 act as repressors of WRI1, thereby inhibiting seed oil accumulation (Kong et al., 2020) (Figures 2A, B).
Seed maturation prepares the seed for germination, though this readiness is held in check until the seed encounters a suitable environment. ABI3, a seed-specific transcription factor, is instrumental in preparing seeds for dormancy, desiccation tolerance, and long-term viability (Delmas et al., 2013; Bedi et al., 2016; Tian et al., 2020). The intricate network of transcription factors that regulate seed embryogenesis, maturation (including lipid and protein accumulation), and dormancy represents a potential target for gene-editing technologies. Such interventions could enhance or biofortify oil content or yield cultivated plant varieties with improved tolerance to abiotic and biotic stresses.
Leafy Cotyledon1 (LEC1), a subunit of the NF-Y transcription factor, plays a pivotal role in embryogenesis and seed maturation (Boulard et al., 2018). LEC1 plays a critical role during seed maturation by coordinating the expression of various seed maturation genes. LEC1 and its interactions with AFL transcription factors (ABI3, FUS3, and LEC2) form a regulatory network directing embryo development (Verma et al., 2022). LEC1 expression in the endosperm can activate maturation genes even without embryo-expressed LEC1, reinforcing the endosperm’s role in seed maturation (Song et al., 2021) (Table 1; Figure 2B).
The AFL transcription factors play specific roles in activating genes responsible for seed storage protein synthesis, further reinforcing their importance in seed maturation (Yang T. et al., 2023). In Arabidopsis, LEC1 mutant plants exhibit underdeveloped seeds characterized by short embryo axes, underdeveloped cotyledons with anthocyanin accumulation, and desiccation intolerance. Conversely, overexpression of LEC1 leads to an upregulation of fatty acid biosynthetic genes, resulting in increased fatty acid levels. Additionally, LEC1 enhances the expression of FUS3, ABI3, and LEC2, which regulate genes involved in photosynthesis and other developmental processes during early and late seed maturation (Jo et al., 2019).
LEC1’s role in the regulatory hierarchy of seed maturation is further emphasized by its interactions with other transcription factors. It interacts sequentially with different transcription factors, such as LEC2 and ABI3, through the B2 domain to activate specific seed development genes, although it does not interact with FUS3, which lacks the B2 domain (Boulard et al., 2018) (Figure 2A). The interaction between LEC1 and LEC2 is highly specific, with an amino acid residue (D86) being crucial for this interaction (Boulard et al., 2018). The translocation of the LEC2/LEC1/NF-YC2 complex from the cytoplasm to the nucleus is critical for activating LEC2 and regulating downstream genes (Boulard et al., 2018). LEC2, another central embryonic regulator with a B3 domain, is vital for establishing the cellular environment necessary for embryo formation and later stages of seed development (Stone et al., 2001).
Although LEC2 is primarily expressed during early embryogenesis, it remains essential for normal development throughout morphogenesis and maturation (Liu Q. et al., 2021). LEC2 specifically binds to the conserved RY element (5′TAGAC-3′) in the promoter region of almost all seed-specific genes in plants. DNA microarray experiments during Arabidopsis seed development revealed that ectopic induction of LEC2 in seedlings led to a rapid accumulation of RNAs encoding seed-specific proteins, such as 2S and 12S storage proteins, oleosin, and steroleosin, all typically expressed during seed maturation. These genes were significantly enriched with the RY motif within 1 kb upstream of the transcription start site, and DNA binding assays confirmed LEC2’s specific binding to this element (Braybrook et al., 2006).
Mutant lines lacking LEC2 in Arabidopsis showed altered embryo morphology, changes in seed protein profiles, and desiccation intolerance (Stone et al., 2001) LEC2 functions upstream in the regulatory hierarchy, upregulating LEC1, FUS3, and ABI3 during embryo maturation (Table 1; Figures 2A, B). It also regulates the WRINKLED1 (WRI1) transcription factor, which controls TAG biosynthesis (Baud et al., 2007). Loss-of-function mutants of LEC1, LEC2, and FUS3 exhibit defects in embryo formation or partial conversion of cotyledons into leaves. Additionally, LEC2 is reported to trigger metabolic steps that increase TAG accumulation in vegetative tissues upon induction of senescence in Arabidopsis leaves (Kim et al., 2015).
WRINKLED1 (WRI1) is an APETALA2 (AP2)/EREBP (Ethylene Response Element Binding Protein) transcription factor that plays a critical role in seed oil biosynthesis during seed maturation (Kong et al., 2020). Loss-of-function mutants show reduced seed weight (by approximately 25%–40%) and fatty acid content (by around 45%–55%) (Baud et al., 2007). Specifically, there was an increase in the levels of end products of fatty acid synthesis and desaturation reactions, such as C18:3 and C22:1, while the levels of C18:1, C18:2, and C20:1 were decreased (Baud et al., 2007). Earlier studies in 1998 showed that the WRI1 mutation could reduce seed oil content by about 80%. Homozygous wrinkled mutants were found to have impaired ability to incorporate sucrose and glucose into triacylglycerol, though they incorporated pyruvate and acetate at an increased rate (Focks and Benning, 1998). Comparative genetic studies suggest that the expression of genes involved in late glycolysis and fatty acid synthesis is reduced in WRI1 mutants (Ruuska et al., 2002).
Orthologous WRI1 genes have been identified in various dicot and monocot plants, including B. napus, C. sativa, G. max, Z. mays, and H. annuus. The WRI1-binding cis-element, known as the AW-box (CnTnG7[CG]; where, ‘n’ represents any nucleotide), has been identified in the promoters of genes regulated by WRI1 (Maeo et al., 2009).
A notable variant of WRI1, AtWRI1W74R, has been reported to enhance oil synthesis in Arabidopsis seeds. This variant demonstrated about a 60% increase in TAG content when transiently expressed in N. benthamiana (Qiao et al., 2022). The W74R variant exhibits a tenfold increase in binding affinity to AW-box sequences and is highly conserved across WRI1 orthologs in diverse plant species. When this W74R mutant was introduced into other plants, such as B. napus, C. sativa, Z. mays, and G. max, consistently resulted in enhanced TAG accumulation upon transient expression in Nicotiana benthamiana (Qiao et al., 2022).
The structure and interaction of the WRI1 protein with cis-elements in gene promoters have been characterized in Arabidopsis, using the dsDNA of the AW-box isolated from the promoter of one of WRI1’s target genes (Table 1). The WRI1 protein structure comprises two similar AP2 domains (each consisting of two antiparallel β-sheets flanked by an α-helix) linked by a V-shaped arrangement of two α-helices (Qiao et al., 2022).
Another significant regulator of fatty acid biosynthesis in Arabidopsis is the TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP4). TCP4 acts as a repressor of AtWRI1, thereby reducing seed oil accumulation (Figures 2A, B). Arabidopsis mutants with reduced TCP4 activity accumulated more seed oil than the wild type, whereas Arabidopsis plants overexpressing TCP4 (expressing rTCP4) showed reduced expression of AtWRI1 target genes (Kong et al., 2020).
Abscisic Acid Insensitive 3 (ABI3) is crucial for seed maturation, desiccation tolerance, entry into the quiescent stage, and seed longevity. ABI3 mutants, exhibit defects in desiccation tolerance and storage product accumulation. Although loss-of-function mutants of ABI3 do not show defects in embryo formation, the seeds remain green and fail to develop desiccation tolerance (Nambara et al., 1995; Delmas et al., 2013).
The expression of ABI3, regulated by hormone abscisic acid, is essential for de-greening of seeds in oilseed crops, which is necessary for seed maturation and seed oil quality (Delmas et al., 2013). Recent global mapping of ABI3 DNA binding sites using DNA chip immunoprecipitation-tiling array assays has provided further insights (Mönke et al., 2012; Lim et al., 2013; Bedi et al., 2016). Transcriptome analysis comparing abi3 three to five mutants with wild-type Arabidopsis seeds revealed that genes regulated by FUS3 and ABI3 often contain at least one RY-element CATGCA sequence (Table 1) (Wang and Perry, 2013; Tian et al., 2020). Genes with overlapping binding sites for both ABI3 and FUS3 are notably overrepresented in lipid biosynthesis, seed-oil biosynthesis (77.04%), and seed dormancy (Figures 2A, B). Approximately 43 genes are directly bound by both FUS3 and ABI3 and are also induced in response to these factors, predominantly those involved in lipid storage. Additionally, FUS3 has been shown to induce the expression of miRNA156, which prevents premature maturation of Arabidopsis seeds (Wang and Perry, 2013; Tian et al., 2020).
ABI3 directly activates or induces the expression of genes and transcription factors related to desiccation tolerance, such as PLATZ1, PLATZ2, and AGAMOUS LIKE67 (AGL67) (Tian et al., 2020). It also induces LEA genes like LEA14 and genes involved in seed longevity, such as SEED IMBIBITION PROTEIN2 and raffinose synthase. Other ABI3-regulated genes include NYC1 and STAY-GREEN2 (SGR2), which are involved in chlorophyll breakdown (Tian et al., 2020). Both ABI3 and FUS3 directly induce WRI1 (Wang and Perry, 2013; Tian et al., 2020). ABI3 also directly induces the FAD3 gene, which is involved in the synthesis of ALA in plants (Tian et al., 2020). ABI3 contains B1 and B2 domains for interaction with bZIPs and LEC1 proteins, as well as a B3 DNA binding domain (Tian et al., 2020). Additionally, ABI3 induces other seed-specific factors, such as CRUCIFERIN1, CRUCIFERIN3, and various LEA genes, including LEA76, LEA6, DEHYDRIN LEA, and LEA-LIKE (Bedi et al., 2016).
The basic leucine zipper (bZIP) family of plant transcription factors is one of the largest and most diverse, playing crucial roles in regulating various synthesis pathways related to plant growth, development, and responses to biotic and abiotic stresses (Yu et al., 2020). In Arabidopsis, approximately 78 bZIP transcription factors have been identified. These factors typically bind to palindromic or pseudo-palindromic hexamer sequences with a core motif of 5′-ACGT-3' (Dröge-Laser et al., 2018).
Recent research has highlighted the role of bZIP52 in seed oil accumulation. Overexpression of bZIP52 in Arabidopsis led to a reduction in seed oil content, while CRISPR/Cas9-mediated knockout of bZIP52 resulted in increased seed oil accumulation (Yang Y. et al., 2023). This suggests that bZIP52 functions as a repressor of fatty acid biosynthesis. Specifically, bZIP52 interacts with the AP2 domain of AtWRI1 (residues 58–240), preventing its binding to the AW-Box sequence in the promoters of fatty acid synthesis genes. This interaction indicates that the AtWRI1–bZIP52 module is a key regulator of seed oil biosynthesis (Yang Y. et al., 2023).
Additionally, the regulation of seed maturation in Arabidopsis has been shown to involve the bZIP53 transcription factor and its heterodimerizing partners, bZIP10 and bZIP25 (Jain et al., 2017) (Table 1) (Figures 2A, B). Another bZIP67 TF was found to be induced by LEC1 during embryogenesis along with the transcriptional complex comprising LIL and NF-YC2 (Mu et al., 2008; Mendes et al., 2013; Tian et al., 2024). This complex activates ALA production by regulating the FAD3 gene. The binding of bZIP67 to cis-elements in the FAD3 gene promoter enhances expression, but this binding and subsequent activation depends on the assembly of LIL and NF-YC2 (Mendes et al., 2013; Tian et al., 2024). Null mutants of FUS3 and ABI3 showed no direct requirement for the enhanced activation of FAD3 achieved with the bZIP67-LIL-NF-YC2 complex (Mendes et al., 2013). However, LEC1 triggers high levels of C18:3 during Arabidopsis seed maturation, which, in conjunction with the induction of FUS3, either alone or cooperatively with LEC2, leads to the activation of bZIP67, LIL, and NF-YC2. This forms a transcription complex that binds to G-boxes in the FAD3 gene promoter (Mendes et al., 2013).
In Arabidopsis, the R2R3-MYB transcription factor MYB89 functions as a negative regulator of seed oil accumulation (Li et al., 2017). Predominantly expressed during seed development, MYB89 plays a crucial role in seed maturation (Dubos et al., 2010). Mutations in MYB89 lead to a significant increase in total fatty acid levels during seed maturation. This increase is associated with the upregulation of the conversion of acetyl-CoA to malonyl-CoA, which maintains the flux of fatty acids (Li et al., 2024). Additionally, the expression of the BCCP1 and BIO2 genes is also elevated. In MYB89 mutants, the expression of PLA2 and ROD1 genes is also increased (Table 1) (Figures 2A, B). The PLA2 gene hydrolyzes the sn-2 position of phospholipids, preferentially targeting linoleoyl acyl chains. The ROD1 gene encodes Phosphatidylcholine-diacylglycerol choline phosphotransferase, which facilitates the transfer of C18:1 to phosphatidylcholine for desaturation and the reverse transfer of C18:2 and C18:3 in the TAG biosynthesis pathway (Li et al., 2017).
MYB89 directly represses WRI, ENO1, BCCP1, KASI, KCS11, and PLA2
Epigenetics regulates gene expression through heritable but reversible mechanisms, affecting both mitotic and meiotic cells (Ding et al., 2022). Key processes include DNA methylation, histone modification, and RNA-directed DNA methylation (RdDM), which can modify gene expression without altering DNA sequences (Kumar and Rani, 2023).
In oilseed crops, seed maturation and development are tightly regulated by epigenetic mechanisms, with chromatin modifiers repressing seed-specific genes. This epigenetic regulation of seed-specific and fatty acid biosynthesis genes is complex and critical for proper seed development, affecting oil seed content and quality (Zhang and Ogas, 2009). For instance, DNA methylation and histone modifications can change gene accessibility, impacting transcription and seed oil accumulation. This intricate interaction underscores epigenetics’ role in optimizing seed oil production and crop performance, although it’s molecular mechanism is yet to be fully understood.
Histone acetyltransferase General Control Non-Repressed Protein 5 (GCN5) plays a crucial role in regulating the FAD3 gene in Arabidopsis. The enzyme FAD3, a microsomal ω-3 fatty acid desaturase, is primarily responsible for synthesizing C18:3 in seeds (Browse et al., 1993). According to Wang et al. (2016), GCN5 influences FAD3 post-transcriptionally through acetylation of histone H3 at lysines 9 and 14 in the upstream regulatory region. This acetylation is essential for the proper expression of FAD3. Mutations in GCN5 lead to a decreased ALA to LA ratio in Arabidopsis seeds, highlighting its role in fatty acid composition. The constitutive expression of the FAD3 gene in gcn5 mutant seeds restores the ALA/LA ratio to normal levels. Conversely, overexpression of GCN5 enhances the ALA/LA ratio, indicating that GCN5 positively regulates ALA synthesis (Wang et al., 2016).
Chromatin immunoprecipitation sequencing (ChIP-Seq) and ChIP-qPCR analyses have shown that in gcn5 mutant seeds, there is a downregulation of several fatty acid biosynthesis-related genes, including FAD3, LACS2, LPP3, and PLAIIIβ. Within the FAD3 promoter, three specific regions have been identified as sites of acetylation by H3K9 and H3K14: approximately 200–400 bp upstream of the transcription start site (P1 region), 0–200 bp upstream (P2 region), and within the first exon (P3 region). These regions exhibit significantly reduced acetylation in the gcn5 mutant compared to wild-type seeds. Overall, GCN5 is crucial for maintaining the ALA/LA ratio in Arabidopsis seeds by regulating the expression of FAD3.
HSI2/VAL1 interacts with the AGL15 gene through a specific DNA sequence and histone modifications, which are crucial for the shift from mature seed to germinating seedling stages (Figures 2A, B). This interaction silences the expression of LAFL genes, which are key regulators of seed maturation, by repressing AGL15—a positive regulator of these genes. This repression is further assisted by other silencing factors, such as MSI1, a polycomb DNA packaging protein (Chen N. et al., 2018). Recent studies have shown that Histone Deacetylase 19 (HDA19) can directly bind to the chromatin of seed maturation genes. HDA19 interacts with HSL1 to repress seed maturation during germination, and the homozygous double mutant of both genes is embryonically lethal (Zhou et al., 2013). Additionally, the DOG1 (Delay of Germination 1) gene, a regulator of seed dormancy, is also controlled by HSI2 and HSL1. These proteins bind to RY elements in the promoter region of DOG1, recruiting polycomb proteins CLF and LHP1. This recruitment leads to histone methylation (H3K27me3), which represses DOG1 and thus breaks seed dormancy (Chen et al., 2020).
PICKLE (PKL) is a chromatin-remodeler protein that plays a crucial role in the regulation of the ABI3 and ABI5 genes, both of which are essential for conferring osmotolerance and resistance to germination under high-stress conditions. PKL is responsible for the epigenetic repression of these genes during germination, particularly in the presence of abscisic acid (ABA). In pkl mutants, ABI3 and ABI5 are less associated with repressed chromatin, leading to a failure in proper gene silencing (Figures 2A, B). PKL restricts the transcriptional activity of embryogenesis genes by modifying chromatin, thereby maintaining the appropriate expression levels required for seed germination under stress (Perruc et al., 2007).
Chromo Domain-Like 1 (CHR1), also known as CHR5, is an epigenetic regulator that plays a contrasting role to PKL during embryogenesis. CHR1 activates the embryo development program by downregulating the expression of key embryogenesis genes such as LEC1, ABI3, and FUS3, while promoting the accumulation of storage seed proteins (SSPs). CHR1 expression is prominent during late embryogenesis, where it antagonizes the function of PKL by reducing nucleosome occupancy near the transcription start sites (TSS) of ABI3 and FUS3 genes, thereby facilitating their activation (Shen et al., 2015).
These two proteins, PKL and CHR1, function antagonistically to regulate the balance between gene repression and activation during seed germination and embryogenesis, ensuring proper seed development.
Transcription factors are essential in regulating gene expression during seed formation stages in oilseed crops, impacting fatty acid biosynthesis, elongation, desaturation, and export. In soybean (G. max), a key leguminous oilseed crop, these regulatory mechanisms are well-demonstrated. For instance, Chen L. et al. (2018) showed that GmWRI1a, a soybean WRI1 ortholog, is highly expressed in maturing seeds and promotes fatty acid accumulation by binding to the AW-box sequence in fatty acid metabolism gene promoters. Additionally, transcription factors LEC1 and ABI3, crucial in seed development, work within a network involving AREB3 and bZIP67 to regulate gene expression during seed maturation. Although LEC1 and ABI3 do not directly interact, AREB3 and bZIP67 form heterodimers that link with the LEC1-NF-YC dimer, merging LEC1 and ABI3 into a functional complex, which finely tunes seed maturation gene expression (Jo et al., 2019).
Studies show that overexpression of Dof-type transcription factors GmDOF4 and GmDOF11 in soybean seeds increases lipid content by activating key metabolic genes like acetyl-CoA carboxylase and long-chain acyl-CoA synthetase. These factors also repress protein synthesis by binding to the CRA1 promoter (Wang et al., 2007). GmDOF4’s ortholog, GmDOF4.2, is linked to drought tolerance in N. tabacum (Zhai et al., 2023).
In sesame, another oilseed crop often grown in drought-prone areas, the genome has 132 AP2/ERF genes. Dossa et al. (2016) reported that 23 DREB genes are upregulated in response to drought, indicating a role in stress tolerance. In oil biosynthesis, the DREB-type transcription factor GmDREBL activates the WRINKLED promoter, along with downstream oil biosynthesis genes. GmDREBL expression is regulated by GmABI3 and GmABI5, suggesting that gene editing of GmDREBL could enhance oil content and improve stress resistance in oilseed crops (Zhang et al., 2016).
Genome editing (GE) is a precise technique employed to alter the genomes of various organisms, including yeast, bacteria, plants, and animals. Among the available tools for this purpose—such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs)—CRISPR/Cas9 has emerged as a particularly effective and economical method for genetic modification, significantly enhancing crop yields (Negi et al., 2022; Cardi et al., 2023; Gawande et al., 2024). Prior to CRISPR/Cas9, methods like ZFNs were both costly and time-consuming (Cardi et al., 2023). Since its initial adaptation from bacterial immune systems for targeted DNA cleavage (Jinek et al., 2012), CRISPR/Cas9 has become central to genome engineering. Unlike meganucleases, ZFNs, and TALENs, which recognize target sequences through protein-DNA interactions, the CRISPR/Cas system employs Watson–Crick base pairing for DNA targeting. This system uses the Cas9 endonuclease in conjunction with a single-guide RNA (sgRNA) that includes a 20-base pair complementary sequence, guiding precise genome modifications through repair processes like nonhomologous end-joining (NHEJ) or homology-directed repair (HDR) (Jinek et al., 2012; Makarova et al., 2015; Cardi et al., 2023; Pan and Qi, 2023).
The CRISPR toolbox has since expanded to include various Cas nucleases with unique features and PAM recognition sites, which are employed for plant genome editing (Pramanik et al., 2021; Gawande et al., 2024). Cas9 from Streptococcus pyogenes (SpCas9) remains the most widely used due to its well-understood mechanism, which involves the Cas9-sgRNA complex searching for target DNA sequences with the protospacer adjacent motif (PAM). This complex binds to the target DNA, inducing a double-strand break (DSB) facilitated by the RuvC and HNH nuclease domains. Additionally, the development of Cas variants—such as nickase (nCas9) and dead Cas (dCas9)—has enabled the creation of CRISPR tools with diverse editing capabilities, including gene knockout, base editing, and epigenetic modifications (Gao, 2021; Pramanik et al., 2021; Wang and Doudna, 2023; Gawande et al., 2024; Shelake et al., 2024). Recently, a transposon-associated TnpB protein has been successfully employed as an alternative to Cas9 for genome editing in plants (Karmakar et al., 2024). Novel tools like CRISPR-TSKO also offer tissue-specific genome editing, allowing for spatially and temporally controlled genetic alterations (Decaestecker et al., 2019).
Genome editing techniques have emerged as a superior method for enhancing oil quality in oil-producing crops, surpassing conventional breeding and genetic engineering approaches. Traditional breeding has long served as the foundation for crop improvement, focusing on selecting traits like yield, pest resistance, and adaptability. However, while breeding has achieved significant successes, it has limitations in precision and speed, especially with complex traits like oil composition and stress resilience. Recent advancements in transcriptional engineering and genome editing, such as CRISPR/Cas9, complement breeding by allowing specific, targeted modifications at the genetic level (Cardi et al., 2023; Gawande et al., 2024). These tools enable breeders to precisely regulate genes involved in traits like fatty acid biosynthesis and stress responses, accelerating the development of high-performing, resilient oilseed varieties. By integrating these methods, breeders can enhance traditional crop lines with desirable genetic traits more efficiently, meeting the increasing demand for improved nutritional profiles, higher yields, and robust environmental tolerance.
Traditional methods involving transgenics face limitations such as unintended genetic changes, reduced efficacy, lengthy processes, and the challenges of transgene insertion (Wang and Doudna, 2023). In contrast, the successful application of CRISPR/Cas9 in oil-producing crops has been well-documented (Table 2). Techniques like transcriptome analysis, map-based cloning, gene silencing, and pathway analysis have been instrumental in functionally validating target genes. In polyploid oilseed crops such as rapeseed, canola, camelina, peanut, and cottonseed, CRISPR technology enables the simultaneous editing of multiple genes (Yuan et al., 2019; Zhang et al., 2019; 2023; Chen et al., 2021; Neelakandan et al., 2022). This approach also facilitates the generation of stable knock-out mutants that can be backcrossed to develop transgene-free oilseed crops, significantly accelerating genetic improvements in these plants (Table 2).
Peanut, an allotetraploid (AABB) crop with 2n = 4x = 40 chromosomes, is an important economic resource due to its high-quality oil yield (Husted, 1936; Bertioli et al., 2019; Leal-Bertioli et al., 2021). According to the United States Department of Agriculture (USDA), global peanut production in 2024 reached 51.31 million metric tons (https://ipad.fas.usda.gov). The quality of peanut oil is determined by the ratio of saturated fatty acids (SFAs) to making up the remaining 80% (Wang et al., 2015; Shasidhar et al., 2017; Tang et al., 2022).
In 2018, Wen et al. successfully used transcription activator-like effector nucleases (TALENs) to modify the FAD2 gene in peanuts, resulting in mutant varieties with significantly higher oleic acid content (90.45%) in their oil. This modification also led to a decrease in linoleic acid levels by 3%–19% in fad2 T2 transgenic seeds compared to the wild type, marking the first successful instance of genome editing in peanuts and showcasing the potential of TALEN-induced mutagenesis in peanut breeding (Table 2) (Wen et al., 2018). Additionally, 18 AhFatB genes identified in the A. hypogaea genome, which are crucial for glycerolipid content and play essential roles in seed and flower development, were studied. The CRISPR/Cas9 system revealed mutations at Arahy.4E7QKU, resulting in low palmitic acid and high oleic acid phenotypes (Tang et al., 2022) (Table 2). Similar results were observed when CRISPR/Cas9 identified three mutations—G448A in ahFAD2A, 441_442insA, and G451T in ahFAD2B—with G451T being a novel mutation and G448A and 441_442insA previously identified in high oleate variants (Yuan et al., 2019) (Table 2) (Figure 3).

Figure 3. Schematic representation of the different CRISPR-edited oilseed crops. (A) BnFAD2, BnGTR2, BnLPAT2/5, BnTT2, BnaFAE1, SFAR genes responsible for oil quality improvement by CRISPR in Brassica napus; (B) EgFAD2, EgPAT genes responsible for oil quality improvement by CRISPR in Elaeis guineensis; (C) GmFAD2-1A, GmFAD2-1B, GmFAD2-2B, GmFAD2-2C, GmFAD2-2D, GmFATB1, GmFAD2-2A, FAD2-2 genes responsible for oil quality improvement by CRISPR in Glycine max; (D) GhFAD2 gene responsible for oil quality improvement by CRISPR in Gossypium hirsutum; (E) AhFAD2, AhFATB genes responsible for oil quality improvement by CRISPR in Arachis hypogaea. The figure is created with BioRender.com.
Further advancements in peanut genome editing were demonstrated through base editing using CRISPR/nCas9 technology (Neelakandan et al., 2022). Researchers constructed two expression vectors, pDW3873 and pDW3876, by fusing Cas9 to different deaminases to induce point mutations in the promoter and coding sequences of the AhFAD2 genes. These constructs were tested for their ability to replace cytosine with thymine or other bases in the targeted editing window. The pDW3873 vector showed higher efficiency, indicating its potential as a superior base editor in peanuts. This technique could significantly shorten the time required to introgress beneficial mutations into elite peanut varieties, offering substantial benefits to breeders, farmers, and consumers alike.
In addition, a recent study by Huai et al. (2024) investigated reducing very long-chain fatty acids (VLCFAs) in peanut kernels using CRISPR-Cas9 technology. Natural high levels of VLCFAs in peanuts are detrimental to human health, being linked to cardiovascular diseases. Previous studies identified AhKCS1 and AhKCS28 as putative regulators of VLCFA content in peanut kernels. The biosynthesis of VLCFAs in plants is regulated by β-ketoacyl-CoA synthase (KCS) (Wang et al., 2017). In a prior study, 30 AhKCS genes were identified in peanut genomes, with AhKCS1 and AhKCS28 found to be key regulators of VLCFA content. The VLCFA content in available peanut germplasm ranges from 4.3% to 9.8%, with no sequence variation observed within or surrounding the AhKCS1 and AhKCS28 genes, suggesting that gene editing is the only viable method for further reduction. Consequently, AhKCS1 and AhKCS28 were genetically disrupted using the CRISPR/Cas9 system, producing novel peanut mutants with significantly reduced VLCFA levels. The results demonstrated up to a 100% reduction in VLCFA content in the double mutants, underscoring the efficacy of CRISPR-Cas9 in modifying peanut fatty acid profiles and highlighting its potential in enhancing crop genetics for global food security and human health (Huai et al., 2024).
Soybean, a polyploid (2n = 4x = 40) legume and essential oil crop (Ren et al., 2021; Yuan and Song, 2023), is cultivated on about 120 million hectares worldwide (Berbenets et al., 2020), with global production nearing 352 million tons (Voora et al., 2020; Sokolovskaya and Valevskaya, 2023). Enhancing oleic acid content and improving the oleic-to-linoleic (O/L) ratio in soybean is a major breading goal (Monteros et al., 2008). Using CRISPR/Cas9, researchers have edited five FAD2 genes (GmFAD2-1B, GmFAD2-1A, GmFAD2-2B, GmFAD2-2C, and GmFAD2-2D) in soybean varieties (JN38, T6098, and T7010), resulting in higher oleic acid levels (54.07%) and lower linoleic acid levels (26.17%) (Zhang et al., 2023) (Table 2), showing that targeting multiple FAD2-2 genes can enhance soybean oil’s nutritional quality. Al Amin et al. (2019) also used CRISPR-Cas9 to modify the FAD2-2 gene, achieving 65.58% oleic acid and 16.08% linoleic acid (Table 2).
Do et al. (2019) used CRISPR/Cas9 with two gRNAs to target separate exons of GmFAD2-1A and GmFAD2-1B, achieving CRISPR-edited DNA in 77.8% of T0 plants, with edits inherited in T1 progeny. This resulted in a 1.3%–2.7% decrease in linoleic acid and over 80% oleic acid content. Wu et al. (2020) observed similar results in T2 and T3 generations. Additionally, Ma et al. (2021) used CRISPR/Cas9 to create mutants of the FATB genes, GmFATB1a and GmFATB1b, which resulted in reduced stearic and palmitic acids and a 1.3%–3.6% increase in linoleic acid (Table 2) (Figure 3).
Rapeseed (B. napus, allotetraploid, 2n = 4x = 38), is a major global source of edible oils and is valued for its nutritional profile and biodiesel potential (Dupont et al., 1989; Ohlrogge, 1994; Thelen and Ohlrogge, 2002; Sun et al., 2017; Liu Y. et al., 2022; Shi et al., 2022). According to USDA data, global rapeseed production in 2023/2024 reached 89.33 million metric tons (https://ipad.fas.usda.gov).
CRISPR/Cas9-mediated editing of the BnFAD2 and BnFAE1 genes has enhanced the nutritional quality of the rapeseed cultivar CY2, which contains roughly 50% oil content and 40% erucic acid (Shi et al., 2022). These edits increased oleic acid content by 70%–80% and reduced erucic acid levels in lines with mutations at all target sites (Shi et al., 2022) (Table 2). Similarly, Okuzaki et al. (2018) reported that CRISPR/Cas9-modified FAD2_Aa mutants achieved oleic acid concentrations of 73%–80% and lower linoleic acid levels of 16%–9%, with corroborating results from Huang et al. (2020) (Table 2).
Targeted knockout of BnaFAE1 has significantly improved the nutritional quality of B. napus seed oil, especially in BnaA08.fae1 and BnaC03.fae1 double mutants, which resulted in a ∼66% increase in oleic acid content (Liu Y. et al., 2022) (Table 2). Xie et al. (2020) utilized CRISPR/Cas9 to target Transparent Testa (TT) genes in B. napus, leading to reduced flavonoid levels, increased oil content, and enhanced fatty acid composition with higher linoleic (C18:2) and linolenic acids (C18:3) (Table 2). CRISPR-derived Bna.gtr2 mutants, including the double mutant BnaC03.gtr2 and its three paralogs, positively regulated seed glucosinolate accumulation, resulting in smaller seeds with higher oil content (Tan et al., 2022) (Table 2). Zhang et al. (2019) used CRISPR/Cas9 to target Lysophosphatidic Acid Acyltransferase (LPTA) genes, BnLPAT2 and BnLPAT5, leading to increased oil body size and decreased oil content, highlighting their roles in oil biosynthesis (Table 2) (Figure 3).
Cotton (allotetraploid, 2n = 4x = 52) is the world’s fifth-largest source of vegetable oil and serves as a significant cash crop for natural textiles (Chen et al., 2021). According to USDA, global cotton production for 2023/2024 is projected to reach 113.66 million bales (https://fas.usda.gov/data/production/commodity/2631000). The oil extracted from cottonseed contains 26% palmitic acid, 15% oleic acid, and 58% linoleic acid (Liu et al., 2002; Chen et al., 2021). In a recent study, Chen et al. (2021) used CRISPR/Cas9 to delete the GhFAD2 gene, resulting in mutants with elevated oleic acid content, marking the first successful production of high-oleic cotton using this technology (Chen et al., 2021) (Table 2) (Figure 3). Additionally, another recent study Jia et al. (2024) identified the BEL1-LIKE HOMEODOMAIN (BLH) gene GhBLH1, which positively regulates cotton fiber cell elongation. Their research revealed that GhBLH1 enhances linolenic acid accumulation by activating the GhFAD7A-1 gene, also leading to longer fiber cells. This finding highlights the role of long-chain and very-long-chain fatty acids in fiber cell elongation and underscores the importance of linolenic acid in cotton fiber growth. The molecular mechanism uncovered in this study involves the binding of the POX domain of GhBLH1 to the TGGA cis-element in the GhFAD7A-1 promoter, leading to increased transcription and subsequently, longer fiber cells. Conversely, knockout of GhFAD7A-1 using CRISPR/Cas9 results in shorter fibers, while its overexpression promotes fiber elongation. These studies exemplify how targeted genetic modifications can influence key traits in crops, providing insights into the genetic and molecular controls of oil and fiber production in cotton.
Camelina sativa, an allohexaploid oilseed from the Brassicaceae family, is increasingly valued for its diverse applications (Nguyen et al., 2013; Morineau et al., 2017; Lee et al., 2021; Lin et al., 2022). However, its high linolenic acid content (30%–40%) limits its use in biofuels, lubricants, and food products (Nguyen et al., 2013; Iskandarov et al., 2014; Morineau et al., 2017; Lee et al., 2021).
CRISPR/Cas9 gene editing has been used to modify FAD2 genes in Camelina, increasing oleic acid levels from 16% to over 50% while reducing linoleic acid (16% to <4%) and linolenic acid (35% to <10%) (Table 2) (Morineau et al., 2017). Similar results were achieved with a CRISPR-Cas9 triple CsFAD2 knockout, which showed an 80% increase in oleic acid compared to the wild type (Lee et al., 2021) (Table 2). Additionally, modifying triacylglycerol synthesis genes CsDGAT1 or CsPDAT1 in Camelina seeds has led to changes in oil content and fatty acid composition (Aznar-Moreno and Durrett, 2017) (Table 2).
In a recent study, Cai et al. (2024) utilized CRISPR/Cas9 to disrupt the Transparent Testa 8 (TT8) transcription factor genes in Camelina, resulting in a yellow seed phenotype due to reduced flavonoid accumulation (up to 44%) and a disrupted seed coat mucilage layer. Their study identified and edited three TT8 genes, one from each Camelina subgenome, creating independent CsTT8 lines with frameshift mutations. This genetic modification led to significant upregulation of lipid-related transcription factors (LEC1, LEC2, FUS3, and WRI1) and their targets, enhancing fatty acid synthesis. Consequently, total fatty acid (TFA) content increased from 32.4% to as high as 38.0% of seed weight, and TAG yield rose by over 21%, with no significant changes in starch or protein levels compared to the parental line (Table 2).
These findings underscore the potential of CRISPR/Cas9 to create high-oil Camelina lines, contributing to future sustainable biofuel production and the development of high-yield lipid-derived bioproducts.
Pennycress is a homozygous diploid species (2n = 2x = 14) (Navarrete et al., 2022; Nunn et al., 2022) that is native to Eurasia and has since become widely distributed globally (Best, 1978; Sharma et al., 2007; Jarvis et al., 2021). In their research, Jarvis et al. (2021) utilized CRISPR/Cas9 technology to enhance the oleic acid content in pennycress seed oil by targeting the fatty acid desaturase 2 (FAD2) and reduced oleate desaturation 1 (ROD1) genes. The resulting double knockdown mutants demonstrated a significant increase in oleic acid production, highlighting a promising strategy for improving commercial yields.
Elaeis guineensis is a diploid species with 16 chromosomes (Low et al., 2024). According to the USDA, global palm oil production for the 2023/2024 season is projected to reach 77.28 million metric tons, with Indonesia being the world’s largest producer, accounting for 57% of this output (https://fas.usda.gov/data/production/commodity/4243000). In Malaysia, the oil palm is a crucial commodity crop that contributes to 35% of global oil and fats production (Bahariah et al., 2021; Parveez et al., 2021; Bahariah et al., 2023; Norfaezah et al., 2024; Ramesh et al., 2024). In their study, Bahariah et al. (2023) employed the CRISPR/Cas9 technology to create knockout mutants with enhanced oleic acid content by targeting the EgFAD2 and palmitoyl-acyl carrier protein thioesterase (EgPAT) genes in oil palm. The Cas9/sgRNA genome-editing approach effectively knocked out both genes, resulting in the generation of single- and double-knockout mutants (Bahariah et al., 2021; Bahariah et al., 2023) (Table 2) (Figure 3).
Transcriptional engineering represents a revolutionary approach to enhancing the value of oilseed crops, focusing on both improving oil quality and increasing yield. Advances in gene editing technologies, particularly CRISPR/Cas9, along with synthetic biology tools, have enabled precise modifications of the transcriptional networks governing oil biosynthesis and storage. By combining transcriptional engineering with precision agriculture practices, it is possible to develop crop varieties specifically tailored to distinct environmental conditions and farming practices, thereby enhancing resource use efficiency, reducing environmental impact, and improving crop resilience. Furthermore, developing oilseed crops that can withstand extreme weather conditions and fluctuating climates is critical due to the detrimental impact of climate change on crop yield. Moreover, engineering crops to enhance their nutritional profiles, such as increasing the content of essential fatty acids and other beneficial compounds, can provide significant health benefits and help address nutritional deficiencies in various populations.
The successful application of CRISPR/Cas technology on rapeseed, soybean, cotton, camelina, pennycress, and palm, has demonstrated remarkable potential for enhancing oil quality and nutritional value. By targeting specific genes involved in fatty acid biosynthesis and composition, researchers have been able to develop crop variants with improved oil profiles. By precisely modulating the expression of genes involved in stress tolerance mechanisms, such as heat shock proteins (HSPs), researchers can develop crop varieties with improved resilience to heat stress and other abiotic stresses. Future research in this field should prioritize the simultaneous improvement of multiple traits, including yield, oil quality, and stress resistance. This holistic approach necessitates the use of sophisticated gene stacking techniques and advanced regulatory network analyses to achieve comprehensive enhancements. Additionally, synthetic biology can introduce new biosynthetic pathways into oilseed crops, allowing for the production of novel oils and value-added compounds. This innovation could create new markets and applications for oilseed-derived products, expanding the economic potential of these crops. Moreover, the development of transgene-free oilseed crops through CRISPR-mediated genome editing offers a promising avenue for ensuring the safety and acceptance of genetically modified organisms in agriculture. Despite the progress made in utilizing CRISPR/Cas9 for crop improvement, challenges remain, particularly in oilseed crops with complex genomes and resistance to regeneration like cotton and peanut, which pose technical barriers to efficient genome editing.
Comparative evaluations reveal that while some crops respond robustly to transcriptional interventions, others require additional optimization to overcome genetic complexity and regeneration difficulties. For instance, in Brassica napus (rapeseed), CRISPR-based transcriptional modifications have notably increased oleic acid content and decreased undesirable fatty acids like erucic acid. This progress addresses both nutritional quality and regulatory standards, making rapeseed oil more appealing for food and industrial applications (Shi et al., 2022). In contrast, Gossypium hirsutum (cotton) presents greater complexity, as its polyploid genome has proven more challenging for stable transcriptional edits. However, recent breakthroughs have shown potential in enhancing oleic acid levels and fiber quality simultaneously, suggesting that multiplexed editing strategies could unlock further gains in cotton (Chen et al., 2021).
Glycine max (soybean) has also benefited from transcriptional engineering with relatively straightforward genome editing, leading to increased oleic acid and reduced linoleic acid content, improving oil stability and shelf life (Do et al., 2019). Soybean’s amenability to editing has made it a model for other legumes and oilseed crops, showcasing how regulatory transcription factors, when edited, can achieve specific oil profile enhancements. By contrast, Arachis hypogaea (peanut) has posed unique challenges due to its allotetraploid genome structure, which complicates the precise targeting of transcriptional elements. Despite these hurdles, targeted editing of fatty acid desaturase genes in peanuts has produced high-oleic varieties with better oil quality, although achieving stable, heritable modifications remains an ongoing challenge (Tang et al., 2022).
In Camelina sativa, a relatively new candidate for industrial oils and biofuels, transcriptional modulation has led to significant increases in oleic acid and reductions in polyunsaturated fatty acids, making it suitable for biofuel production (Morineau et al., 2017). Camelina’s simpler genome structure and compatibility with genetic modification approaches position it as a promising oilseed for future applications. Thlaspi arvense (pennycress), while less studied, has demonstrated potential as a biofuel feedstock through CRISPR-mediated modulation of fatty acid pathways, increasing oleic acid while reducing undesirable fatty acids, though further research is needed to fully harness its agronomic potential (Jarvis et al., 2021).
These comparative insights emphasize that while transcriptional technologies offer universal benefits for oilseed crop improvement, species-specific challenges, such as polyploidy and regeneration efficiency, require tailored approaches (Pramanik et al., 2021). Moving forward, a balanced approach that combines genome editing with advances in transcriptomics and synthetic biology will allow for more comprehensive enhancements across diverse oilseed crops. This trajectory not only meets the growing demand for nutritionally rich oils but also ensures the sustainability and economic viability of oilseed production under changing climatic conditions.
However, ongoing research efforts aim to overcome these challenges and unlock the full potential of genome editing technologies for enhancing the resilience and productivity of oilseed crops in the face of climate change.
Furthermore, as ‘omics’ technologies unravel the intricacies of lipid metabolism in oilseeds, a deeper understanding of regulatory networks emerges. Integration of genomic, transcriptomic, proteomic, and metabolomic data offers invaluable insights into enhancing seed oil quality, pivotal for future crop enhancement endeavours. Harnessing transcriptional and epigenetic regulation mechanisms presents further avenues for crop improvement through targeted breeding and genetic engineering.
In conclusion, by embracing cutting-edge biotechnologies and addressing the associated challenges, we can pave the way for a sustainable and prosperous future for oil crop improvement. Ongoing research and innovations in transcriptional engineering will contribute to the next green revolution, ensuring food security and environmental sustainability for future generations. This comprehensive approach will not only meet the escalating global demand for vegetable oils but also ensure the economic viability and ecological sustainability of oilseed crop production.
CK: Writing–original draft. MS: Writing–original draft. Mansi Parekh: Writing–original draft. HG: Writing–original draft. MJ: Conceptualization, Writing–review and editing. SS: Conceptualization, Funding acquisition, Supervision, Writing–original draft, Writing–review and editing. BP: Conceptualization, Funding acquisition, Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work is supported by Gujarat State Biotechnology Mission, Govt. of Gujarat’s research grant to BP and SS under the Research Support Scheme 2022-23, project # GSBTM/JD (R&D)/626/22-23/00018354.
BP acknowledges Ahmedabad University for the start-up funds. SS acknowledges DBT for the Re-entry fellowship grant, SERB and the Indian Institute of Technology Gandhinagar for start-up grant. CK and MS acknowledge RA fellowships from GSBTM.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abe, K., Araki, E., Suzuki, Y., Toki, S., and Saika, H. (2018). Production of high oleic/low linoleic rice by genome editing. Plant Physiology Biochem. 131, 58–62. doi:10.1016/j.plaphy.2018.04.033
Al Amin, N., Ahmad, N., Wu, N., Pu, X., Ma, T., Du, Y., et al. (2019). CRISPR-Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max.L). BMC Biotechnol. 19, 9. doi:10.1186/s12896-019-0501-2
Asadi, A. A., Shariati, V., Mousavi, S., Mariotti, R., and Hosseini Mazinani, M. (2023). Meta-analysis of transcriptome reveals key genes relating to oil quality in olive. BMC Genomics 24, 566. doi:10.1186/s12864-023-09673-y
Attia, Z., Pogoda, C. S., Reinert, S., Kane, N. C., and Hulke, B. S. (2021). Breeding for sustainable oilseed crop yield and quality in a changing climate. Theor. Appl. Genet. 134, 1817–1827. doi:10.1007/s00122-021-03770-w
Aznar-Moreno, J. A., and Durrett, T. P. (2017). Simultaneous targeting of multiple gene homeologs to alter seed oil production in camelina sativa. Plant Cell Physiol. 58, 1260–1267. doi:10.1093/pcp/pcx058
Bahariah, B., Masani, M. Y. A., Fizree, M. P. M. A. A., Rasid, O. A., and Parveez, G. K. A. (2023). Multiplex CRISPR/Cas9 gene-editing platform in oil palm targeting mutations in EgFAD2 and EgPAT genes. J. Genet. Eng. Biotechnol. 21, 3. doi:10.1186/s43141-022-00459-5
Bahariah, B., Masani, M. Y. A., Rasid, O. A., and Parveez, G. K. A. (2021). Multiplex CRISPR/Cas9-mediated genome editing of the FAD2 gene in rice: a model genome editing system for oil palm. J. Genet. Eng. Biotechnol. 19, 86. doi:10.1186/s43141-021-00185-4
Bates, P. D. (2016). Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1861, 1214–1225. doi:10.1016/j.bbalip.2016.03.021
Bates, P. D. (2022). “The plant lipid metabolic network for assembly of diverse triacylglycerol molecular species,” in Advances in botanical research. doi:10.1016/bs.abr.2021.07.003
Bates, P. D., and Browse, J. (2012). The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front. Plant Sci. 3, 147. doi:10.3389/fpls.2012.00147
Bates, P. D., Stymne, S., and Ohlrogge, J. (2013). Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16, 358–364. doi:10.1016/j.pbi.2013.02.015
Baud, S., Mendoza, M. S., To, A., Harscoët, E., Lepiniec, L., and Dubreucq, B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50, 825–838. doi:10.1111/j.1365-313X.2007.03092.x
Bedi, S., Sengupta, S., Ray, A., and Nag Chaudhuri, R. (2016). ABI3 mediates dehydration stress recovery response in Arabidopsis thaliana by regulating expression of downstream genes. Plant Sci. 250, 125–140. doi:10.1016/j.plantsci.2016.06.006
Berbenets, O., Pugach, A., and Lebedenko, O. (2020). Economic aspects of extruded soybeans’ usage in livestock and poultry. Bulg. J. Agric. Sci. 26, 1034–1040.
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884. doi:10.1038/s41588-019-0405-z
Best, K. F. (1978). Studies on the flowering of thlaspi arvense L. IV. Genetic and ecological differences between early- and late-flowering strains. Bot. Gaz. 139, 190–195. doi:10.1086/336986
Bohra, A., Kilian, B., Sivasankar, S., Caccamo, M., Mba, C., McCouch, S. R., et al. (2022). Reap the crop wild relatives for breeding future crops. Trends Biotechnol. 40, 412–431. doi:10.1016/j.tibtech.2021.08.009
Boulard, C., Thévenin, J., Tranquet, O., Laporte, V., Lepiniec, L., and Dubreucq, B. (2018). LEC1 (NF-YB9) directly interacts with LEC2 to control gene expression in seed. Biochim. Biophys. Acta Gene Regul. Mech. 1861, 443–450. doi:10.1016/j.bbagrm.2018.03.005
Braybrook, S. A., Stone, S. L., Park, S., Bui, A. Q., Le, B. H., Fischer, R. L., et al. (2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. U. S. A. 103, 3468–3473. doi:10.1073/pnas.0511331103
Browse, J., McConn, M., James, D., and Miquel, M. (1993). Mutants of Arabidopsis deficient in the synthesis of α-linolenate: biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 268, 16345–16351. doi:10.1016/s0021-9258(19)85427-3
Bu, M., Fan, W., Li, R., He, B., and Cui, P. (2023). Lipid metabolism and improvement in oilseed crops: recent advances in multi-omics studies. Metabolites 13, 1170. doi:10.3390/metabo13121170
Cahoon, E. B., and Li-Beisson, Y. (2020). Plant unusual fatty acids: learning from the less common. Curr. Opin. Plant Biol. 55, 66–73. doi:10.1016/j.pbi.2020.03.007
Cai, Y., Liang, Y., Shi, H., Cui, J., Prakash, S., Zhang, J., et al. (2024). Creating yellow seed Camelina sativa with enhanced oil accumulation by CRISPR-mediated disruption of Transparent Testa 8. Plant Biotechnol. J. 22, 2773–2784. doi:10.1111/pbi.14403
Cardi, T., Murovec, J., Bakhsh, A., Boniecka, J., Bruegmann, T., Bull, S. E., et al. (2023). CRISPR/Cas-mediated plant genome editing: outstanding challenges a decade after implementation. Trends Plant Sci. 28, 1144–1165. doi:10.1016/j.tplants.2023.05.012
Carman, G. M., and Han, G. S. (2006). Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem. Sci. 31, 694–699. doi:10.1016/j.tibs.2006.10.003
Chen, B., Maas, L., Figueiredo, D., Zhong, Y., Reis, R., Li, M., et al. (2022). BABY BOOM regulates early embryo and endosperm development. Proc. Natl. Acad. Sci. U. S. A. 119, e2201761119. doi:10.1073/pnas.2201761119
Chen, L., Zheng, Y., Dong, Z., Meng, F., Sun, X., Fan, X., et al. (2018a). Soybean (Glycine max) WRINKLED1 transcription factor, GmWRI1a, positively regulates seed oil accumulation. Mol. Genet. Genomics 293, 401–415. doi:10.1007/s00438-017-1393-2
Chen, M., Xuan, L., Wang, Z., Zhou, L., Li, Z., Du, X., et al. (2014). TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol. 165, 905–916. doi:10.1104/pp.114.235507
Chen, N., Veerappan, V., Abdelmageed, H., Kang, M., and Allen, R. D. (2018b). HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in arabidopsis. Plant Cell 30, 600–619. doi:10.1105/tpc.17.00655
Chen, N., Wang, H., Abdelmageed, H., Veerappan, V., Tadege, M., and Allen, R. D. (2020). HSI2/VAL1 and HSL1/VAL2 function redundantly to repress DOG1 expression in Arabidopsis seeds and seedlings. New Phytol. 227, 840–856. doi:10.1111/nph.16559
Chen, Y., Fu, M., Li, H., Wang, L., Liu, R., Liu, Z., et al. (2021). High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) generated by knockout of GhFAD2 genes with CRISPR/Cas9 system. Plant Biotechnol. J. 19, 424–426. doi:10.1111/pbi.13507
Clough, R. C., Matthis, A. L., Barnum, S. R., and Jaworski, J. G. (1992). Purification and characterization of 3-ketoacyl-acyl carrier protein synthase III from spinach: a condensing enzyme utilizing acetyl-coenzyme A to initiate fatty acid synthesis. J. Biol. Chem. 267, 20992–20998. doi:10.1016/s0021-9258(19)36787-0
Dahlqvist, A., Ståhl, U., Lenman, M., Banas, A., Lee, M., Sandager, L., et al. (2000). Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. U. S. A. 97, 6487–6492. doi:10.1073/pnas.120067297
Dar, A. A., Choudhury, A. R., Kancharla, P. K., and Arumugam, N. (2017). The FAD2 gene in plants: occurrence, regulation, and role. Front. Plant Sci. 8, 1789. doi:10.3389/fpls.2017.01789
Decaestecker, W., Buono, R. A., Pfeiffer, M. L., Vangheluwe, N., Jourquin, J., Karimi, M., et al. (2019). CRISPR-Tsko: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 31, 2868–2887. doi:10.1105/tpc.19.00454
Delmas, F., Sankaranarayanan, S., Deb, S., Widdup, E., Bournonville, C., Bollier, N., et al. (2013). ABI3 controls embryo degreening through Mendel’s i locus. Proc. Natl. Acad. Sci. U. S. A. 110, E3888–E3894. doi:10.1073/pnas.1308114110
Ding, X., Jia, X., Xiang, Y., and Jiang, W. (2022). Histone modification and chromatin remodeling during the seed life cycle. Front. Plant Sci. 13, 865361. doi:10.3389/fpls.2022.865361
Do, P. T., Nguyen, C. X., Bui, H. T., Tran, L. T. N., Stacey, G., Gillman, J. D., et al. (2019). Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol. 19, 311. doi:10.1186/s12870-019-1906-8
Dossa, K., Wei, X., Li, D., Fonceka, D., Zhang, Y., Wang, L., et al. (2016). Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biol. 16, 171. doi:10.1186/s12870-016-0859-4
Dröge-Laser, W., Snoek, B. L., Snel, B., and Weiste, C. (2018). The Arabidopsis bZIP transcription factor family — an update. Curr. Opin. Plant Biol. 45, 36–49. doi:10.1016/j.pbi.2018.05.001
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi:10.1016/j.tplants.2010.06.005
Dupont, J., White, P. J., Johnston, K. M., Alexander Heggtveit, D. H., McDonald, B. E., Grundy, S. M., et al. (1989). Food safety and health effects of canola oil. J. Am. Coll. Nutr. 8, 360–375. doi:10.1080/07315724.1989.10720311
Focks, N., and Benning, C. (1998). wrinkled1: a novel, low-seed-oil mutant of arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism 1. Plant Physiol. 118, 91–101. doi:10.1104/pp.118.1.91
Fox, S. R., Hill, L. M., Rawsthorne, S., and Hills, M. J. (2000). Inhibition of the glucose-6-phosphate transporter in oilseed rape (Brassica napus L.) plastids by acyl-CoA thioesters reduces fatty acid synthesis. Biochem. J. 352, 525. doi:10.1042/0264-6021:3520525
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635. doi:10.1016/j.cell.2021.01.005
Gawande, N. D., Bhalla, H., Watts, A., Shelake, R. M., and Sankaranarayanan, S. (2024). Application of genome editing in plant reproductive biology: recent advances and challenges. Plant Reprod. 37, 441–462. doi:10.1007/s00497-024-00506-w
Greenwood, J. R., Zhang, X., and Rathjen, J. P. (2023). Precision genome editing of crops for improved disease resistance. Curr. Biol. 33, R650–R657. doi:10.1016/j.cub.2023.04.058
Harwood, J. L. (1996). Recent advances in the biosynthesis of plant fatty acids. Biochimica Biophysica Acta - Lipids Lipid Metabolism 1301, 7–56. doi:10.1016/0005-2760(95)00242-1
Huai, D., Xue, X., Wu, J., Pandey, M. K., Liu, N., Huang, L., et al. (2024). Enhancing peanut nutritional quality by editing AhKCS genes lacking natural variation. Plant Biotechnol. J. 22, 3015–3017. doi:10.1111/pbi.14423
Huang, H., Cui, T., Zhang, L., Yang, Q., Yang, Y., Xie, K., et al. (2020). Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9-mediated gene editing in Brassica napus. Theor. Appl. Genet. 133, 2401–2411. doi:10.1007/s00122-020-03607-y
Husted, L. (1936). Cytological Studies an the Peanut, Arachis. II Chromosome number, morphology and behavior, and their application to the problem of the origin of the cultivated forms. Cytol. (Tokyo) 7, 396–423. doi:10.1508/cytologia.7.396
Iskandarov, U., Kim, H. J., and Cahoon, E. B. (2014). “Camelina: an emerging oilseed platform for advanced biofuels and bio-based materials,” in Plants and BioEnergy. doi:10.1007/978-1-4614-9329-7_8
Jain, P., Shah, K., Sharma, N., Kaur, R., Singh, J., Vinson, C., et al. (2017). A-ZIP53, a dominant negative reveals the molecular mechanism of heterodimerization between bZIP53, bZIP10 and bZIP25 involved in Arabidopsis seed maturation. Sci. Rep. 7, 14343. doi:10.1038/s41598-017-14167-5
Jarvis, B. A., Romsdahl, T. B., McGinn, M. G., Nazarenus, T. J., Cahoon, E. B., Chapman, K. D., et al. (2021). CRISPR/Cas9-Induced fad2 and rod1 mutations stacked with fae1 confer high oleic acid seed oil in pennycress (thlaspi arvense L.). Front. Plant Sci. 12, 652319. doi:10.3389/fpls.2021.652319
Jia, C., Lai, Q., Zhu, Y., Feng, J., Dan, X., Zhang, Y., et al. (2024a). Intergrative metabolomic and transcriptomic analyses reveal the potential regulatory mechanism of unique dihydroxy fatty acid biosynthesis in the seeds of an industrial oilseed crop Orychophragmus violaceus. BMC Genomics 25, 29. doi:10.1186/s12864-023-09906-0
Jia, T., Wang, H., Cui, S., Li, Z., Shen, Y., Li, H., et al. (2024b). Cotton BLH1 and KNOX6 antagonistically modulate fiber elongation via regulation of linolenic acid biosynthesis. Plant Commun. 5, 100887. doi:10.1016/j.xplc.2024.100887
Jiang, W. Z., Henry, I. M., Lynagh, P. G., Comai, L., Cahoon, E. B., and Weeks, D. P. (2017). Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15, 648–657. doi:10.1111/pbi.12663
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 1979, 816–821. doi:10.1126/science.1225829
Jo, L., Pelletier, J. M., and Harada, J. J. (2019). Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J. Integr. Plant Biol. 61, 564–580. doi:10.1111/jipb.12806
Jones, A., Davies, H. M., and Voelker, T. A. (1995). Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell 7, 359–371. doi:10.1105/tpc.7.3.359
Jouhet, J., Maréchal, E., and Block, M. A. (2007). Glycerolipid transfer for the building of membranes in plant cells. Prog. Lipid Res. 46, 37–55. doi:10.1016/j.plipres.2006.06.002
Karmakar, S., Panda, D., Panda, S., Dash, M., Saha, R., Das, P., et al. (2024). A miniature alternative to Cas9 and Cas12: transposon-associated TnpB mediates targeted genome editing in plants. Plant Biotechnol. J. 22, 2950–2953. doi:10.1111/pbi.14416
Karunarathna, N. L., Wang, H., Harloff, H. J., Jiang, L., and Jung, C. (2020). Elevating seed oil content in a polyploid crop by induced mutations in seed fatty acid reducer genes. Plant Biotechnol. J. 18, 2251–2266. doi:10.1111/pbi.13381
Kazaz, S., Miray, R., Lepiniec, L., and Baud, S. (2022). Plant monounsaturated fatty acids: diversity, biosynthesis, functions and uses. Prog. Lipid Res. 85, 101138. doi:10.1016/j.plipres.2021.101138
Kim, H. U., Lee, K. R., Jung, S. J., Shin, H. A., Go, Y. S., Suh, M. C., et al. (2015). Senescence-inducible LEC2 enhances triacylglycerol accumulation in leaves without negatively affecting plant growth. Plant Biotechnol. J. 13, 1346–1359. doi:10.1111/pbi.12354
Kim, H. U., Li, Y., and Huang, A. H. C. (2005). Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in arabidopsis. Plant Cell 17, 1073–1089. doi:10.1105/tpc.104.030403
Kim, R. J., Kim, H. U., and Suh, M. C. (2019). Development of camelina enhanced with drought stress resistance and seed oil production by co-overexpression of MYB96A and DGAT1C. Ind. Crops Prod. 138, 111475. doi:10.1016/j.indcrop.2019.111475
Kong, Q., Yang, Y., Guo, L., Yuan, L., and Ma, W. (2020). Molecular basis of plant oil biosynthesis: insights gained from studying the WRINKLED1 transcription factor. Front. Plant Sci. 11, 24. doi:10.3389/fpls.2020.00024
Kuczynski, C., McCorkle, S., Keereetaweep, J., Shanklin, J., and Schwender, J. (2022). An expanded role for the transcription factor WRINKLED1 in the biosynthesis of triacylglycerols during seed development. Front. Plant Sci. 13, 955589. doi:10.3389/fpls.2022.955589
Kumar, M., and Rani, K. (2023). Epigenomics in stress tolerance of plants under the climate change. Mol. Biol. Rep. 50, 6201–6216. doi:10.1007/s11033-023-08539-6
Kumar, V., Jha, P., and Van Staden, J. (2020). LEAFY COTYLEDONs (LECs): master regulators in plant embryo development. Plant Cell Tissue Organ Cult. 140, 475–487. doi:10.1007/s11240-019-01752-x
Lager, I., Yilmaz, J. L., Zhou, X. R., Jasieniecka, K., Kazachkov, M., Wang, P., et al. (2013). Plant Acyl-CoA:lysophosphatidylcholine acyltransferases (LPCATs) have different specificities in their forward and reverse reactions. J. Biol. Chem. 288, 36902–36914. doi:10.1074/jbc.M113.521815
Leal-Bertioli, S. C. M., Nascimento, E. F. M. B., Chavarro, C. M. F., Custódio, A. R., Hopkins, M. S., Moretzsohn, M. C., et al. (2021). Spontaneous generation of diversity in Arachis neopolyploids (Arachis ipaënsis × Arachis duranensis)4x replays the early stages of peanut evolution. G3 Genes, Genomes, Genet. 11, jkab289. doi:10.1093/g3journal/jkab289
Lee, H. G., Kim, H., Suh, M. C., Kim, H. U., and Seo, P. J. (2018). The MYB96 transcription factor regulates triacylglycerol accumulation by activating DGAT1 and PDAT1 expression in arabidopsis seeds. Plant Cell Physiol. 59, 1432–1442. doi:10.1093/pcp/pcy073
Lee, K. R., Jeon, I., Yu, H., Kim, S. G., Kim, H. S., Ahn, S. J., et al. (2021). Increasing monounsaturated fatty acid contents in hexaploid camelina sativa seed oil by FAD2 gene knockout using CRISPR-cas9. Front. Plant Sci. 12, 702930. doi:10.3389/fpls.2021.702930
Lemieux, B., Miquel, M., Somerville, C., and Browse, J. (1990). Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor. Appl. Genet. 80, 234–240. doi:10.1007/BF00224392
Lessire, R., and Stumpe, P. K. (1983). Nature of the fatty acid synthetase systems in parenchymal and epidermal cells of Allium porrum L. Leaves. Plant Physiol. 73, 614–618. doi:10.1104/pp.73.3.614
Li, D., Jin, C., Duan, S., Zhu, Y., Qi, S., Liu, K., et al. (2017). MYB89 transcription factor represses seed Oil accumulation. Plant Physiol. 173, 1211–1225. doi:10.1104/pp.16.01634
Li, H., Che, R., Zhu, J., Yang, X., Li, J., Fernie, A. R., et al. (2024). Multi-omics-driven advances in the understanding of triacylglycerol biosynthesis in oil seeds. Plant J. 117, 999–1017. doi:10.1111/tpj.16545
Li, N., Meng, H., Li, S., Zhang, Z., Zhao, X., Wang, S., et al. (2020). Two plastid fatty acid exporters contribute to seed oil accumulation in Arabidopsis. Plant Physiol. 182, 1910–1919. doi:10.1104/PP.19.01344
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2013). Acyl-lipid metabolism. Arab. Book 11, e0161. doi:10.1199/tab.0161
Lim, S., Park, J., Lee, N., Jeong, J., Toh, S., Watanabe, A., et al. (2013). ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25, 4863–4878. doi:10.1105/tpc.113.118604
Lin, P., Wang, K., Wang, Y., Hu, Z., Yan, C., Huang, H., et al. (2022). The genome of oil-Camellia and population genomics analysis provide insights into seed oil domestication. Genome Biol. 23, 14. doi:10.1186/s13059-021-02599-2
Liu, B., Sun, G., Liu, C., and Liu, S. (2021a). Leafy Cotyledon 2: a regulatory factor of plant growth and seed development. Genes (Basel) 12, 1896. doi:10.3390/genes12121896
Liu, H., Li, H., Gu, J., Deng, L., Ren, L., Hong, Y., et al. (2018). Identification of the candidate proteins related to oleic acid accumulation during peanut (Arachis hypogaea L.) seed development through comparative proteome analysis. Int. J. Mol. Sci. 19, 1235. doi:10.3390/ijms19041235
Liu, N., Liu, J., Fan, S., Liu, H., Zhou, X. R., Hua, W., et al. (2022a). An integrated omics analysis reveals the gene expression profiles of maize, castor bean, and rapeseed for seed oil biosynthesis. BMC Plant Biol. 22, 153. doi:10.1186/s12870-022-03495-y
Liu, Q., Singh, S. P., and Green, A. G. (2002). High-stearic and high-oleic cottonseed oils produced by hairpin RNA-mediated post-transcriptional gene silencing. Plant Physiol. 129, 1732–1743. doi:10.1104/pp.001933
Liu, Q., Yang, F., Zhang, J., Liu, H., Rahman, S., Islam, S., et al. (2021b). Application of crispr/cas9 in crop quality improvement. Int. J. Mol. Sci. 22, 4206. doi:10.3390/ijms22084206
Liu, Y., Du, Z., Lin, S., Li, H., Lu, S., Guo, L., et al. (2022b). CRISPR/Cas9-Targeted mutagenesis of BnaFAE1 genes confers low-erucic acid in Brassica napus. Front. Plant Sci. 13, 848723. doi:10.3389/fpls.2022.848723
Low, E. T. L., Chan, K. L., Zaki, N. M., Taranenko, E., Ordway, J. M., Wischmeyer, C., et al. (2024). Chromosome-scale Elaeis guineensis and E. oleifera assemblies: comparative genomics of oil palm and other Arecaceae. G3 (Bethesda). G3, jkae135. doi:10.1093/g3journal/jkae135
Ma, J., Sun, S., Whelan, J., and Shou, H. (2021). CRISPR/Cas9-mediated knockout of GmFATB1 significantly reduced the amount of saturated fatty acids in soybean seeds. Int. J. Mol. Sci. 22, 3877. doi:10.3390/ijms22083877
Maeo, K., Tokuda, T., Ayame, A., Mitsui, N., Kawai, T., Tsukagoshi, H., et al. (2009). An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 60, 476–487. doi:10.1111/j.1365-313X.2009.03967.x
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi:10.1038/nrmicro3569
Meesapyodsuk, D., Ye, S., Chen, Y., Chen, Y., Chapman, R. G., and Qiu, X. (2018). An engineered oilseed crop produces oil enriched in two very long chain polyunsaturated fatty acids with potential health-promoting properties. Metab. Eng. 49, 192–200. doi:10.1016/j.ymben.2018.08.009
Mendes, A., Kelly, A. A., van Erp, H., Shaw, E., Powers, S. J., Kurup, S., et al. (2013). bZIP67 regulates the omega-3 fatty acid content of arabidopsis seed oil by activating fatty acid DESATURASE3. Plant Cell 25, 3104–3116. doi:10.1105/tpc.113.116343
Ming, M., Ren, Q., Pan, C., He, Y., Zhang, Y., Liu, S., et al. (2020). CRISPR–Cas12b enables efficient plant genome engineering. Nat. Plants 6, 202–208. doi:10.1038/s41477-020-0614-6
Mohanta, T. K., Bashir, T., Hashem, A., Abd Allah, E. F., and Bae, H. (2017). Genome editing tools in plants. Genes (Basel) 8, 399. doi:10.3390/genes8120399
Mönke, G., Seifert, M., Keilwagen, J., Mohr, M., Grosse, I., Hähnel, U., et al. (2012). Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 40, 8240–8254. doi:10.1093/nar/gks594
Monteros, M. J., Burton, J. W., and Boerma, H. R. (2008). Molecular mapping and confirmation of QTLs associated with oleic acid content in N00-3350 soybean. Crop Sci. 48, 2223–2234. doi:10.2135/cropsci2008.05.0287
Morineau, C., Bellec, Y., Tellier, F., Gissot, L., Kelemen, Z., Nogué, F., et al. (2017). Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 15, 729–739. doi:10.1111/pbi.12671
Mu, J., Tan, H., Zheng, Q., Fu, Y., Liang, Y., Zhang, J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148, 1042–1054. doi:10.1104/pp.108.126342
Na, G., Aryal, N., Fatihi, A., Kang, J., and Lu, C. (2018). Seed-specific suppression of ADP-glucose pyrophosphorylase in Camelina sativa increases seed size and weight. Biotechnol. Biofuels 11, 330. doi:10.1186/s13068-018-1334-2
Nambara, E., Keith, K., McCourt, P., and Naito, S. (1995). A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121, 629–636. doi:10.1242/dev.121.3.629
Navarrete, T. G., Arias, C., Mukundi, E., Alonso, A. P., and Grotewold, E. (2022). Natural variation and improved genome annotation of the emerging biofuel crop field pennycress (Thlaspi arvense). G3 Genes, Genomes, Genet. 12, jkac084. doi:10.1093/g3journal/jkac084
Neelakandan, A. K., Wright, D. A., Traore, S. M., Ma, X., Subedi, B., Veeramasu, S., et al. (2022). Application of CRISPR/Cas9 system for efficient gene editing in peanut. Plants 11, 1361. doi:10.3390/plants11101361
Negi, C., Vasistha, N. K., Singh, D., Vyas, P., and Dhaliwal, H. S. (2022). Application of CRISPR-mediated gene editing for crop improvement. Mol. Biotechnol. 64, 1198–1217. doi:10.1007/s12033-022-00507-y
Nguyen, H. T., Silva, J. E., Podicheti, R., Macrander, J., Yang, W., Nazarenus, T. J., et al. (2013). Camelina seed transcriptome: a tool for meal and oil improvement and translational research. Plant Biotechnol. J. 11, 759–769. doi:10.1111/pbi.12068
Norfaezah, J., Masani, M. Y. A., Fizree, M. P. M. A. A., Bahariah, B., Shaharuddin, N. A., Ho, C. L., et al. (2024). DNA-free CRISPR/Cas9 genome editing system for oil palm protoplasts using multiple ribonucleoproteins (RNPs) complexes. Ind. Crops Prod. 208, 117795. doi:10.1016/j.indcrop.2023.117795
Nunn, A., Rodríguez-Arévalo, I., Tandukar, Z., Frels, K., Contreras-Garrido, A., Carbonell-Bejerano, P., et al. (2022). Chromosome-level Thlaspi arvense genome provides new tools for translational research and for a newly domesticated cash cover crop of the cooler climates. Plant Biotechnol. J. 20, 944–963. doi:10.1111/pbi.13775
O’brien, A. R., Burgio, G., and Bauer, D. C. (2021). Domain-specific introduction to machine learning terminology, pitfalls and opportunities in CRISPR-based gene editing. Brief. Bioinform 22, 308–314. doi:10.1093/bib/bbz145
Ohlrogge, J. B. (1994). Design of new plant products: engineering of fatty acid metabolism. Plant Physiol. 104, 821–826. doi:10.1104/pp.104.3.821
Ohlrogge, J. B., and Jaworski, J. G. (1997). Regulation of fatty acid synthesis. Annu. Rev. Plant Biol. 48, 109–136. doi:10.1146/annurev.arplant.48.1.109
Ohlrogge, J. B., Kuhn, D. N., and Stumpf, P. K. (1979). Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proc. Natl. Acad. Sci. U. S. A. 76, 1194–1198. doi:10.1073/pnas.76.3.1194
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiology Biochem. 131, 63–69. doi:10.1016/j.plaphy.2018.04.025
Pan, C., and Qi, Y. (2023). CRISPR-Combo–mediated orthogonal genome editing and transcriptional activation for plant breeding. Nat. Protoc. 18, 1760–1794. doi:10.1038/s41596-023-00823-w
Pan, C., Wu, X., Markel, K., Malzahn, A. A., Kundagrami, N., Sretenovic, S., et al. (2021). CRISPR–Act3.0 for highly efficient multiplexed gene activation in plants. Nat. Plants 7, 942–953. doi:10.1038/s41477-021-00953-7
Parchuri, P., Bhandari, S., Azeez, A., Chen, G., Johnson, K., Shockey, J., et al. (2024). Identification of triacylglycerol remodeling mechanism to synthesize unusual fatty acid containing oils. Nat. Commun. 15, 3547. doi:10.1038/s41467-024-47995-x
Park, M. E., and Kim, H. U. (2022). Applications and prospects of genome editing in plant fatty acid and triacylglycerol biosynthesis. Front. Plant Sci. 13, 969844. doi:10.3389/fpls.2022.969844
Parveez, G. K. A., Tarmizi, A. H. A., Sundram, S., Loh, S. K., Ong-Abdullah, M., Palam, K. D. P., et al. (2021). Oil palm economic performance in Malaysia and R&D progress in 2020. J. Oil Palm. Res. 33. doi:10.21894/jopr.2021.0026
Pathak, A. K., Singla, R., Juneja, M., and Tuli, R. (2020). Shortlisting genes important in seed maturation by principal component analysis of gene expression data. bioRxiv 2020 (06), 158832. doi:10.1101/2020.06.19.158832
Pelletier, J. M., Kwong, R. W., Park, S., Le, B. H., Baden, R., Cagliari, A., et al. (2017). LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. U. S. A. 114, E6710-E6719. doi:10.1073/pnas.1707957114
Perruc, E., Kinoshita, N., and Lopez-Molina, L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52, 927–936. doi:10.1111/j.1365-313X.2007.03288.x
Pramanik, D., Shelake, R. M., Park, J., Kim, M. J., Hwang, I., Park, Y., et al. (2021). CRISPR/Cas9-mediated generation of pathogen-resistant tomato against tomato yellow leaf curl virus and powdery mildew. Int. J. Mol. Sci. 22, 1878. doi:10.3390/ijms22041878
Prasad, P. V. V., Boote, K. J., Allen, L. H., and Thomas, J. M. G. (2003). Super-optimal temperatures are detrimental to peanut (Arachis hypogaea L.) reproductive processes and yield at both ambient and elevated carbon dioxide. Glob. Chang. Biol. 9, 1775–1787. doi:10.1046/j.1365-2486.2003.00708.x
Qiao, Z., Kong, Q., Tee, W. T., Lim, A. R. Q., Teo, M. X., Olieric, V., et al. (2022). Molecular basis of the key regulator WRINKLED1 in plant oil biosynthesis. Sci. Adv. 8, eabq1211. doi:10.1126/sciadv.abq1211
Ramesh, S. V., Rajesh, M. K., Das, A., and Hebbar, K. B. (2024). CRISPR/Cas9 –based genome editing to expedite the genetic improvement of palms: challenges and prospects. Front. Plant Sci. 15, 1385037. doi:10.3389/fpls.2024.1385037
Ren, C., Li, H., Liu, Y., Li, S., and Liang, Z. (2022). Highly efficient activation of endogenous gene in grape using CRISPR/dCas9-based transcriptional activators. Hortic. Res. 9, uhab037. doi:10.1093/hr/uhab037
Ren, D., Yang, H., Zhou, L., Yang, Y., Liu, W., Hao, X., et al. (2021). The Land-Water-Food-Environment nexus in the context of China’s soybean import. Adv. Water Resour. 151, 103892. doi:10.1016/j.advwatres.2021.103892
Ruiz-Lopez, N., Haslam, R. P., Napier, J. A., and Sayanova, O. (2014). Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop. Plant J. 77, 198–208. doi:10.1111/tpj.12378
Ruuska, S. A., Girke, T., Benning, C., and Ohlrogge, J. B. (2002). Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14, 1191–1206. doi:10.1105/tpc.000877
Saini, R. K., and Keum, Y. S. (2018). Omega-3 and omega-6 polyunsaturated fatty acids: dietary sources, metabolism, and significance — a review. Life Sci. 203, 255–267. doi:10.1016/J.LFS.2018.04.049
Salas, J. J., and Ohlrogge, J. B. (2002). Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch. Biochem. Biophys. 403, 25–34. doi:10.1016/S0003-9861(02)00017-6
Sandgrind, S., Li, X., Ivarson, E., Wang, E. S., Guan, R., Kanagarajan, S., et al. (2023). Improved fatty acid composition of field cress (Lepidium campestre) by CRISPR/Cas9-mediated genome editing. Front. Plant Sci. 14, 1076704. doi:10.3389/fpls.2023.1076704
Sasaki, Y., and Nagano, Y. (2004). Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 68, 1175–1184. doi:10.1271/bbb.68.1175
Sasnauskas, G., Manakova, E., Lapėnas, K., Kauneckaitė, K., and Siksnys, V. (2018). DNA recognition by Arabidopsis transcription factors ABI3 and NGA1. FEBS J. 285, 4041–4059. doi:10.1111/febs.14649
Schnurr, J. A., Shockey, J. M., De Boer, G. J., and Browse, J. A. (2002). Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol. 129, 1700–1709. doi:10.1104/pp.003251
Selma, S., Bernabé-Orts, J. M., Vazquez-Vilar, M., Diego-Martin, B., Ajenjo, M., Garcia-Carpintero, V., et al. (2019). Strong gene activation in plants with genome-wide specificity using a new orthogonal CRISPR/Cas9-based programmable transcriptional activator. Plant Biotechnol. J. 17, 1703–1705. doi:10.1111/pbi.13138
Shah, S., Karunarathna, N. L., Jung, C., and Emrani, N. (2018). An APETALA1 ortholog affects plant architecture and seed yield component in oilseed rape (Brassica napus L.). BMC Plant Biol. 18, 380. doi:10.1186/s12870-018-1606-9
Sharma, N., Cram, D., Huebert, T., Zhou, N., and Parkin, I. A. P. (2007). Exploiting the wild crucifer Thlaspi arvense to identify conserved and novel genes expressed during a plant’s response to cold stress. Plant Mol. Biol. 63, 171–184. doi:10.1007/s11103-006-9080-4
Shasidhar, Y., Vishwakarma, M. K., Pandey, M. K., Janila, P., Variath, M. T., Manohar, S. S., et al. (2017). Molecular mapping of oil content and fatty acids using dense genetic maps in groundnut (Arachis hypogaea L.). Front. Plant Sci. 8, 794. doi:10.3389/fpls.2017.00794
Shelake, R. M., Pramanik, D., and Kim, J. Y. (2024). CRISPR base editor-based targeted random mutagenesis (BE-TRM) toolbox for directed evolution. BMB Rep. 57, 30–39. doi:10.5483/BMBRep.2023-0086
Shen, Y., Devic, M., Lepiniec, L., and Zhou, D. X. (2015). Chromodomain, Helicase and DNA-binding CHD1 protein, CHR5, are involved in establishing active chromatin state of seed maturation genes. Plant Biotechnol. J. 13, 811–820. doi:10.1111/pbi.12315
Shi, J., Ni, X., Huang, J., Fu, Y., Wang, T., Yu, H., et al. (2022). CRISPR/Cas9-Mediated gene editing of BnFAD2 and BnFAE1 modifies fatty acid profiles in Brassica napus. Genes (Basel) 13, 1681. doi:10.3390/genes13101681
Shockey, J., Regmi, A., Cotton, K., Adhikari, N., Browse, J., and Bates, P. D. (2016). Identification of arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. 170, 163–179. doi:10.1104/pp.15.01563
Sokolovskaya, O., and Valevskaya, L. (2023). Research of soybean seed quality indicators. Food Sci. Technol. 17. doi:10.15673/fst.v17i2.2596
Song, J., Xie, X., Chen, C., Shu, J., Thapa, R. K., Nguyen, V., et al. (2021). LEAFY COTYLEDON1 expression in the endosperm enables embryo maturation in Arabidopsis. Nat. Commun. 12, 3963. doi:10.1038/s41467-021-24234-1
Stone, S. L., Braybrook, S. A., Paula, S. L., Kwong, L. W., Meuser, J., Pelletier, J., et al. (2008). Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc. Natl. Acad. Sci. U. S. A. 105, 3151–3156. doi:10.1073/pnas.0712364105
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U. S. A. 98, 11806–11811. doi:10.1073/pnas.201413498
Sui, N., Wang, Y., Liu, S., Yang, Z., Wang, F., and Wan, S. (2018). Transcriptomic and physiological evidence for the relationship between unsaturated fatty acid and salt stress in peanut. Front. Plant Sci. 9, 7. doi:10.3389/fpls.2018.00007
Sun, F., Fan, G., Hu, Q., Zhou, Y., Guan, M., Tong, C., et al. (2017). The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 92, 452–468. doi:10.1111/tpj.13669
Tan, H., Zhang, J., Qi, X., Shi, X., Zhou, J., Wang, X., et al. (2019). Correlation analysis of the transcriptome and metabolome reveals the regulatory network for lipid synthesis in developing Brassica napus embryos. Plant Mol. Biol. 99, 31–44. doi:10.1007/s11103-018-0800-3
Tan, Z., Xie, Z., Dai, L., Zhang, Y., Zhao, H., Tang, S., et al. (2022). Genome- and transcriptome-wide association studies reveal the genetic basis and the breeding history of seed glucosinolate content in Brassica napus. Plant Biotechnol. J. 20, 211–225. doi:10.1111/pbi.13707
Tang, X., Lowder, L. G., Zhang, T., Malzahn, A. A., Zheng, X., Voytas, D. F., et al. (2017). A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 3, 17018. doi:10.1038/nplants.2017.18
Tang, Y., Huang, J., Ji, H., Pan, L., Hu, C., Qiu, X., et al. (2022). Identification of AhFatB genes through genome-wide analysis and knockout of AhFatB reduces the content of saturated fatty acids in peanut (Arichis hypogaea L.). Plant Sci. 319, 111247. doi:10.1016/j.plantsci.2022.111247
Thelen, J. J., and Ohlrogge, J. J. (2002). Metabolic engineering of fatty acid biosynthesis in plants. Metab. Eng. 4, 12–21. doi:10.1006/mben.2001.0204
Tian, M., Bai, Y., Tian, H., and Zhao, X. (2023). The chemical composition and health-promoting benefits of vegetable oils—a review. Molecules 28, 6393. doi:10.3390/molecules28176393
Tian, R., Wang, F., Zheng, Q., Niza, V. M. A. G. E., Downie, A. B., and Perry, S. E. (2020). Direct and indirect targets of the arabidopsis seed transcription factor abscisic acid insensitive3. Plant J. 103, 1679–1694. doi:10.1111/tpj.14854
Tian, Y., Song, K., Li, B., Song, Y., Zhang, X., Li, H., et al. (2024). Genome-wide identification and expression analysis of NF-Y gene family in tobacco (Nicotiana tabacum L.). Sci. Rep. 14 (1), 5257. doi:10.1038/s41598-024-55799-8
Turquetti-Moraes, D. K., Moharana, K. C., Almeida-Silva, F., Pedrosa-Silva, F., and Venancio, T. M. (2022). Integrating omics approaches to discover and prioritize candidate genes involved in oil biosynthesis in soybean. Gene 808, 145976. doi:10.1016/j.gene.2021.145976
Vanhercke, T., Dyer, J. M., Mullen, R. T., Kilaru, A., Rahman, M. M., Petrie, J. R., et al. (2019). Metabolic engineering for enhanced oil in biomass. Prog. Lipid Res. 74, 103–129. doi:10.1016/j.plipres.2019.02.002
Varshney, R. K., Bohra, A., Roorkiwal, M., Barmukh, R., Cowling, W. A., Chitikineni, A., et al. (2021). Fast-forward breeding for a food-secure world. Trends Genet. 37, 1124–1136. doi:10.1016/j.tig.2021.08.002
Verma, S., Attuluri, V. P. S., and Robert, H. S. (2022). Transcriptional control of Arabidopsis seed development. Planta 255, 90. doi:10.1007/s00425-022-03870-x
Voora, V., Larrea, C., and Bermudez, S. (2020) “Global market report: soybeans,”, I. The International Institute for Sustainable Development. “JSTOR”. https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=Global+market+report%3A+soybeans%2C%E2%80%9D%2C+I.+The+International+Institute+for+Sustainable+Development.&btnG=.
Wang, F., and Perry, S. E. (2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161, 1251–1264. doi:10.1104/pp.112.212282
Wang, H. W., Zhang, B., Hao, Y. J., Huang, J., Tian, A. G., Liao, Y., et al. (2007). The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 52, 716–729. doi:10.1111/j.1365-313X.2007.03268.x
Wang, J. Y., and Doudna, J. A. (2023). CRISPR technology: a decade of genome editing is only the beginning. Science 1979, eadd8643. doi:10.1126/science.add8643
Wang, M. L., Khera, P., Pandey, M. K., Wang, H., Qiao, L., Feng, S., et al. (2015). Genetic mapping of QTLs controlling fatty acids provided insights into the genetic control of fatty acid synthesis pathway in peanut (Arachis hypogaea L.). PLoS One 10, e0119454. doi:10.1371/journal.pone.0119454
Wang, T., Xing, J., Liu, X., Liu, Z., Yao, Y., Hu, Z., et al. (2016). Histone acetyltransferase general control non-repressed protein 5 (GCN5) affects the fatty acid composition of Arabidopsis thaliana seeds by acetylating fatty acid desaturase3 (FAD3). Plant J. 88, 794–808. doi:10.1111/tpj.13300
Wang, X., Guan, Y., Zhang, D., Dong, X., Tian, L., and Qing Qu, L. (2017). A β-ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiol. 173, 944–955. doi:10.1104/pp.16.01527
Wang, X., Liang, H., Guo, D., Guo, L., Duan, X., Jia, Q., et al. (2019a). Integrated analysis of transcriptomic and proteomic data from tree peony (P. ostii) seeds reveals key developmental stages and candidate genes related to oil biosynthesis and fatty acid metabolism. Hortic. Res. 6, 111. doi:10.1038/s41438-019-0194-7
Wang, Z., Yang, M., Sun, Y., Yang, Q., Wei, L., Shao, Y., et al. (2019b). Overexpressing Sesamum indicum L.’s DGAT1 increases the seed oil content of transgenic soybean. Mol. Breed. 39, 101. doi:10.1007/s11032-019-1016-1
Wen, S., Liu, H., Li, X., Chen, X., Hong, Y., Li, H., et al. (2018). TALEN-mediated targeted mutagenesis of fatty acid desaturase 2 (FAD2) in peanut (Arachis hypogaea L.) promotes the accumulation of oleic acid. Plant Mol. Biol. 97, 177–185. doi:10.1007/s11103-018-0731-z
Wu, N., Lu, Q., Wang, P., Zhang, Q., Zhang, J., Qu, J., et al. (2020). Construction and analysis of GmFAD2-1A and GmFAD2-2A soybean fatty acid desaturase mutants based on CRISPR/Cas9 technology. Int. J. Mol. Sci. 21, 1104. doi:10.3390/ijms21031104
Xie, T., Chen, X., Guo, T., Rong, H., Chen, Z., Sun, Q., et al. (2020). Targeted knockout of BnTT2 homologues for yellow-seeded Brassica napus with reduced flavonoids and improved fatty acid composition. J. Agric. Food Chem. 68, 5676–5690. doi:10.1021/acs.jafc.0c01126
Xiong, X., Liang, J., Li, Z., Gong, B. Q., and Li, J. F. (2021). Multiplex and optimization of dCas9-TV-mediated gene activation in plants. J. Integr. Plant Biol. 63, 634–645. doi:10.1111/jipb.13023
Yamamoto, A., Kagaya, Y., Usui, H., Hobo, T., Takeda, S., and Hattori, T. (2010). Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol. 51, 2031–2046. doi:10.1093/pcp/pcq162
Yang, T., Wu, X., Wang, W., and Wu, Y. (2023a). Regulation of seed storage protein synthesis in monocot and dicot plants: a comparative review. Mol. Plant 16, 145–167. doi:10.1016/j.molp.2022.12.004
Yang, Y., Kong, Q., Tee, W. T., Li, Y., Low, P. M., Patra, B., et al. (2023b). Transcription factor bZIP52 modulates Arabidopsis seed oil biosynthesis through interaction with WRINKLED1. Plant Physiol. 192, 2628–2639. doi:10.1093/plphys/kiad270
Ye, Z., Yu, J., Yan, W., Zhang, J., Yang, D., Yao, G., et al. (2021). Integrative iTRAQ-based proteomic and transcriptomic analysis reveals the accumulation patterns of key metabolites associated with oil quality during seed ripening of Camellia oleifera. Hortic. Res. 8, 157. doi:10.1038/s41438-021-00591-2
Yu, Y., Qian, Y., Jiang, M., Xu, J., Yang, J., Zhang, T., et al. (2020). Regulation mechanisms of plant basic leucine zippers to various abiotic stresses. Front. Plant Sci. 11, 1258. doi:10.3389/fpls.2020.01258
Yuan, J., and Song, Q. (2023). Polyploidy and diploidization in soybean. Mol. Breed. 43, 51. doi:10.1007/s11032-023-01396-y
Yuan, M., Zhu, J., Gong, L., He, L., Lee, C., Han, S., et al. (2019). Mutagenesis of FAD2 genes in peanut with CRISPR/Cas9 based gene editing. BMC Biotechnol. 19, 24. doi:10.1186/s12896-019-0516-8
Zhai, Y., Chen, J., He, J., Zhang, J., Sha, W., Yu, H., et al. (2023). Isolation, characterization and functional validation of a soybean transcription factor, GmDof4.2 improves drought tolerance in transgenic tobacco. Plant Cell Tissue Organ Cult. 152, 357–367. doi:10.1007/s11240-022-02412-3
Zhang, H., and Ogas, J. (2009). An epigenetic perspective on developmental regulation of seed genes. Mol. Plant 2, 610–627. doi:10.1093/mp/ssp027
Zhang, K., Nie, L., Cheng, Q., Yin, Y., Chen, K., Qi, F., et al. (2019). Effective editing for lysophosphatidic acid acyltransferase 2/5 in allotetraploid rapeseed (Brassica napus L.) using CRISPR-Cas9 system. Biotechnol. Biofuels 12, 225. doi:10.1186/s13068-019-1567-8
Zhang, Q., Liu, L., Xiao, Z., Sun, Y., Xi, Y., Sun, T., et al. (2023). Construction and functional evaluation of CRISPR/Cas9 multiple knockout vectors of the FAD2 gene family. Agronomy 13, 1737. doi:10.3390/agronomy13071737
Zhang, Y. Q., Lu, X., Zhao, F. Y., Li, Q. T., Niu, S. L., Wei, W., et al. (2016). Soybean GmDREBL increases lipid content in seeds of transgenic arabidopsis. Sci. Rep. 6, 34307. doi:10.1038/srep34307
Zhao, C., Liu, B., Piao, S., Wang, X., Lobell, D. B., Huang, Y., et al. (2017). Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. U. S. A. 114, 9326–9331. doi:10.1073/pnas.1701762114
Zheng, Y., Ren, N., Wang, H., Stromberg, A. J., and Perry, S. E. (2009). Global identification of targets of the arabidopsis MADS domain protein AGAMOUS-like15. Plant Cell 21, 2563–2577. doi:10.1105/tpc.109.068890
Zhou, X. R., Liu, Q., and Singh, S. (2023). Engineering nutritionally improved edible plant oils. Annu. Rev. Food Sci. Technol. 14, 247–269. doi:10.1146/annurev-food-052720-104852
Zhou, Y., Tan, B., Luo, M., Li, Y., Liu, C., Chen, C., et al. (2013). HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25, 134–148. doi:10.1105/tpc.112.096313
Zou, J., Wei, Y., Jako, C., Kumar, A., Selvaraj, G., and Taylor, D. C. (1999). The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J. 19, 645–653. doi:10.1046/j.1365-313X.1999.00555.x
Keywords: transcription factors, genome editing, CRISPR/Cas9, oilseed crops, fatty acid biosynthesis, seed maturation, omics
Citation: Kaushal C, Sachdev M, Parekh M, Gowrishankar H, Jain M, Sankaranarayanan S and Pathak B (2025) Transcriptional engineering for value enhancement of oilseed crops: a forward perspective. Front. Genome Ed. 6:1488024. doi: 10.3389/fgeed.2024.1488024
Received: 29 August 2024; Accepted: 16 December 2024;
Published: 07 January 2025.
Edited by:
Milind Ratnaparkhe, ICAR Indian Institute of Soybean Research, IndiaReviewed by:
Sadhana Singh, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), IndiaCopyright © 2025 Kaushal, Sachdev, Parekh, Gowrishankar, Jain, Sankaranarayanan and Pathak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subramanian Sankaranarayanan, cy5zYW5rYXJAaWl0Z24uYWMuaW4=; Bhuvan Pathak, Ymh1dmFuLnBhdGhha0BhaGR1bmkuZWR1Lmlu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.