
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Genome Ed., 30 August 2023
Sec. Genome Editing in Plants
Volume 5 - 2023 | https://doi.org/10.3389/fgeed.2023.1247702
This article is part of the Research TopicWomen in Genome EditingView all 3 articles
In this study, we generated and compared three cytidine base editors (CBEs) tailor-made for potato (Solanum tuberosum), which conferred up to 43% C-to-T conversion of all alleles in the protoplast pool. Earlier, gene-edited potato plants were successfully generated by polyethylene glycol-mediated CRISPR/Cas9 transformation of protoplasts followed by explant regeneration. In one study, a 3–4-fold increase in editing efficiency was obtained by replacing the standard Arabidopsis thaliana AtU6-1 promotor with endogenous potato StU6 promotors driving the expression of the gRNA. Here, we used this optimized construct (SpCas9/StU6-1::gRNA1, target gRNA sequence GGTC4C5TTGGAGC12AAAAC17TGG) for the generation of CBEs tailor-made for potato and tested for C-to-T base editing in the granule-bound starch synthase 1 gene in the cultivar Desiree. First, the Streptococcus pyogenes Cas9 was converted into a (D10A) nickase (nCas9). Next, one of three cytosine deaminases from human hAPOBEC3A (A3A), rat (evo_rAPOBEC1) (rA1), or sea lamprey (evo_PmCDA1) (CDA1) was C-terminally fused to nCas9 and a uracil-DNA glycosylase inhibitor, with each module interspaced with flexible linkers. The CBEs were overall highly efficient, with A3A having the best overall base editing activity, with an average 34.5%, 34.5%, and 27% C-to-T conversion at C4, C5, and C12, respectively, whereas CDA1 showed an average base editing activity of 34.5%, 34%, and 14.25% C-to-T conversion at C4, C5, and C12, respectively. rA1 exhibited an average base editing activity of 18.75% and 19% at C4 and C5 and was the only base editor to show no C-to-T conversion at C12.
The CRISPR–Cas9 editing system/complex consists, in its basic form, of a guide RNA (gRNA) and a Streptococcus pyogenes nuclease SpCas9 enzyme, which generate a targeted double-stranded DNA break, leading to the formation of insertions and/or deletions (indels) via the activation of the non-homologous end joining (NHEJ) DNA repair pathway frequently resulting in frameshift of the reading frame and loss of gene function (LOF) (Jinek et al., 2012a). Basic CRISPR–SpCas9-mediated gene editing has been further developed into cytidine base editors (CBEs), where single targeted cytosines are converted into thymines (C-to-T) (Komor et al., 2016) and later expanded to include targeted adenine-to-guanine (A-to-G) adenine base editors (ABEs) (Gaudelli et al., 2017) and C-to-G base editors (Kurt et al., 2021). Base editing (BE) was first and mainly employed in mammalian systems (Komor et al., 2016; Nishida et al., 2016; Gaudelli et al., 2017; Komor et al., 2017; Koblan et al., 2018; Thuronyi et al., 2019; Koblan et al., 2021; Kurt et al., 2021) but have since been adjusted to plants, including crops such as rice (Shimatani et al., 2017; Zong et al., 2017; Zong et al., 2018; Jin et al., 2019; Li C. et al., 2020; Hua et al., 2020; Xiong et al., 2022), wheat (Zong et al., 2017; Zong et al., 2018), maize (Zong et al., 2017) potato (Zong et al., 2018; Veillet et al., 2019a; Veillet et al., 2019b; Veillet et al., 2020a; Veillet et al., 2020b), and tomato (Shimatani et al., 2017; Veillet et al., 2019b; Veillet et al., 2020b). BE has been introduced and tested in potato protoplasts using Agrobacterium-mediated delivery of integrative constructs followed by editing analysis of regenerated explants (Zong et al., 2018; Veillet et al., 2019a; Veillet et al., 2019b; Veillet et al., 2020a; Veillet et al., 2020b) and using PEG-mediated delivery of non-integrative constructs into potato protoplasts (Zong et al., 2018). Both approaches included targeting Granular-bound starch synthase (StGBSS), where Agrobacterium-mediated delivery generally conferred high C-to-T conversion, some indel formation, and undesired C-to-A and C-to-G conversions in the explants examined (Veillet et al. (2019a), and delivery to protoplasts, in one instance, conferred an average of up to 18%–20% of C-to-T editing (Zong et al., 2018). Prime editing (PE) is a recent additional editing tool that allows controlled editing directly into the target site through the use of a reverse transcriptase and a specialized prime editing guide RNA (pegRNA), which confers the targeting and editing specificity and the binding capability to the nickase (nCas9) of the prime editing complex (Anzalone et al., 2019). PE has interesting potential within clinical applications (Surun et al., 2020; Geurts et al., 2021; Happi Mbakam et al., 2022a; Happi Mbakam et al., 2022b; Tremblay et al., 2022) and in crop breeding (Jiang et al., 2020b; Li H. Y. et al., 2020; Lin et al., 2020). Implementation of PE in plants on a wider scale, however, has proven difficult, perhaps due to the mode of action and the complex pegRNA structure (Zhao et al., 2023), underpinning the continued relevance of base editing. However, the applicability of base editing on a wider scale is constrained by moderate targeting specificity and efficiencies, which, to some degree, may be alleviated by design and efficiency optimizations of the BE construct at hand. Here, we further developed a non-integrative CRISPR/SpCas9 construct, optimized and custom-made for potato protoplasts via replacement of the standard AtU6-1 promotor with a native potato StU6-1 promotor, to generate and compare three CBE constructs with different origins of the deaminase. When targeted to the granule-bound starch synthase (GBSS) 1 gene and tested on protoplasts of the cultivar Desiree, the three BEs generally conferred high C-to-T base editing efficiencies with, in one instance, 43% C-to-T conversion of a single cytosine.
Potato (Solanum tuberosum) cultivar Desiree plantlets were grown and maintained in vitro on medium A, as described in the work of Nicolia et al. (2015) and Nicolia et al. (2021). The potato plants were grown in a Fitotron growth cabinet model SGC 120 from Weiss Technik with a diurnal rhythm of 16/8 h, 24°C/20°C, 70% humidity, at a light intensity of 65 μE.
The basic construct, SpCas9/StU6-1::sgRNA1, comprising the 35SPPDK::SpCas9 cassette, driving the expression of the codon-optimized Streptococcus pyogenes Cas9 nuclease (SpCas9) originally from the plasmid pHBT-pcoCas9 (Li et al., 2015) (Addgene plasmid #52254) and the StU6-1 promoter::sgRNA-1 cassette (StU6-1 promoter (NCBI accession no. Z17290)) described in the work of Johansen et al. (2019) was used as the basis for generation of the base editing constructs. Each of the cytosine deaminases, hAPOBEC3A (A3A) (Zong et al., 2018) (Addgene #119768) from human, evo_rAPOBEC1 (rA1) (Thuronyi et al., 2019) (Addgene #122611) from rat, and evo_PmCDA1 (CDA1) (Thuronyi et al., 2019) (Addgene #122608) from sea lamprey (Petromyzon marinus), were codon-optimized for potato and purchased from GenScript (https://www.genscript.com), delivered in the pUC57 vector. The plasmid SpCas9/StU6-1::sgRNA1 cassette was used first for the generation of the nCas9 nickase (D10A) using site-directed mutagenesis, which then served as the basis for the generation of the base editing constructs via Gibson construct assembly with either of the cytosine deaminases using the NEBuilder HiFi DNA Assembly Master Mix (New England BioLabs) according to the manufacturer’s recommendations. Final constructs consisted of the 35SPPDK::nCas9 cassette, the StU6-1 promoter::sgRNA1, and one of three deaminases A3A, rA1, and CDA1, a uracil glycosylase inhibitor (UGI), interconnected by flexible serine–glycine (SG)-extended XTEN linker (SGGSSGGSSGSETPGTSESATPESSGGSSGGS) and serine–glycine–glycine–serine (SGGS) linkers.
Codon-optimized nucleotide sequences including assembly overhangs are provided in Supplementary Information.
The D10A mutation was introduced into SpCas9/StU6-1::sgRNA1 by site-directed mutagenesis PCR using 12.5 pmol of primer 567 and 12.5 pmol of primer 568, 12.5 μL 2 X CloneAmpTMHiFi PCR Premix from Takara, and 100 ng of template (SpCas9/StU6-1::sgRNA1) in a total reaction volume of 25 μL. PCR cycle parameters were 98°C 3 min, followed by 15 cycles of 98°C for 10 s, 55°C for 15 s, and 72°C for 5 min.
Primers were ordered from TAG Copenhagen A/S (https://www.tagc.com) and are listed in Supplementary Table S1. For working applications, 5 pmol/μL dilutions in Milli-Q water were prepared.
2 μL of Gibson assembly mix (Base editor construct assembly) was transformed into and multiplied in E. coli. Plasmids were extracted using the E.Z.N.A.(R) Plasmid DNA Mini Kit I (D6943-02) from Omega Bio-tek according to the manufacturer’s instructions and sequenced by EZ-sequencing services provided by Macrogen to ascertain the correct sequence.
Following confirmation of the correct sequence, plasmids were amplified in E. coli and isolated by CTAB large-scale-prep plasmid phenol extraction and then diluted to a concentration of 1 μg/μL to be used for protoplast transformation.
Media used for isolation and transformation include medium B, plasmolysis solution, medium C, wash solution, sucrose solution, transformation buffer 1, transformation buffer 2, PEG solution, and medium E, with recipes outlined in the work of Nicolia et al. (2021). Protoplast isolation was carried out as described in the work of Nicolia et al. (2015) and Nicolia et al. (2021). The intactness and purity of isolated protoplasts were checked by light microscopy and diluted to a concentration of ca. 1.6 × 103 protoplasts/μL in transformation buffer 2. Then, 110 µL protoplasts (ca. 1.6 × 103 protoplasts/μL) in transformation buffer 2 were gently mixed with 10 µL (1 μg/μL) of base editing plasmid, and 110 µL 25% PEG solution was added, gently mixed, and incubated for 3 min at RT. Transfection was stopped by adding 6 mL of wash solution and then spun at 500 RPM for 5 min (minimum acceleration and deceleration), RT, the wash solution was carefully removed, and 1 mL of ½ medium E (diluted with 0.4 M sorbitol) was added. Protoplasts were then incubated in the dark for 2 days at 60 RPM, RT, which yielded optimal editing when using the original construct (Johansen et al., 2019). Following incubation, the protoplasts were harvested by spinning for 3 min at 4000 RPM, and the pellet was re-dissolved in 50 µL of Milli-Q water, frozen in N2, heated for 15 min at 96°C, and stored at −20°C. The protoplast slurry was thawed, placed on ice, and then, vortexed prior to entering as a template in PCR amplifications.
PCR amplification of the target region of GBSS1 was performed using 6.25 pmol of primer 472 and 6.25 pmol of primer 384, 12.5 µL 2 X CloneAmpTMHiFi PCR Premix from Takara, and 1 µL of protoplast slurry (ca. 1.6 × 103 protoplasts/μL) in a total volume of 25 µL. PCR cycle parameters were 2 min at 98°C, followed by 40 cycles of 10 s at 98°C, 15 s at 64°C, and 30 s at 72°C, followed by 2 min at 72°C. PCR products were purified using the BioLine ISOLATE II PCR & Gel Kit or NucleoSpin Gel and the PCR Clean-up Mini kit from Macherey-Nagel according to the manufacturer’s recommendations. PCR products were then sent for sequencing (Sequencing directly on PCR products).
PCR amplification of the GBSS1 target region was performed using 6.25 pmol of primer 475 and 6.25 pmol of primer FAM481 (5’ end labeled with fluorescein amidite (FAM)), 12.5 µL 2 X CloneAmpTMHiFi PCR Premix from Takara, and 1 µL of protoplast slurry in a total volume of 25 µL. PCR cycle parameters were 2 min at 98°C, followed by 40 cycles of 10 s at 98°C, 15 s at 64°C, and 30 s at 72°C, followed by 2 min at 72°C. PCR amplicons were wrapped in aluminum foil and stored at −20°C until being subjected to indel detection amplicon analysis (IDAA) analysis at COBO Technologies Aps, Denmark, where the fluorescently labeled fragments were run on a sequenator 3500xL Genetic Analyzer (Applied Biosystems) and separated according to size by capillary electrophoresis, with a separation resolution down to fragments differing ±1 bp in length as described in the work of Yang et al. (2015).
StyI digestions were performed in a total volume of 10 μL containing 80 ng of PCR fragment DNA, 1 μL 10 x CutSmart Buffer (New England BioLabs), and 2 U StyI enzyme (New England BioLabs) and incubated at 37°C for 3 h. Then, 4 U of StyI enzyme was additionally added and incubated for 1 h. BsrI digestion was performed in a total volume of 10 μL containing 80 ng of PCR fragment DNA, 1 μL 10 x NEBuffer 3.1 (New England BioLabs), and 2 U BsrI enzyme (New England BioLabs) and incubated at 65°C for 2 h.
Editing was also analyzed by Sanger sequencing, using the EZ-seq sequencing services provided by Macrogen, directly on PCR products using 20 ng of purified PCR product (PCR amplification and product purification) and 25 pmol of primer 589. It should be noted that for direct sequencing on PCR amplicons of the protoplast cell pool, discernable/readable sequence chromatograms were only obtained when using ca. 20 ng of purified PCR product as opposed to the 50–75 ng recommended by Macrogen EZ-seq.
Editing efficiency was determined by analyzing sequence chromatograms using the EditR software (Kluesner et al., 2018). IDAA chromatograms were obtained using the online software VIKING (https://viking-suite.com/).
Earlier, we used CRISPR/Cas9 for knockout of the GBSS 1 target gene in potato (Solanum tuberosum) (cultivar Desiree and Wotan), where the CRISPR/Cas9 components were transiently expressed from plasmids delivered by polyethylene glycol (PEG)-mediated transformation to protoplasts and explants regenerated from single edited protoplast cells (Johansen et al., 2019). Here, the target region of StGBSS1 (5’ UTR, exon 1, intron 1, including length and single-nucleotide polymorphisms (SNPs)) in the potato cultivars Desiree and Wotan were sequenced and mapped, providing the allele-specific foundation for gRNA and diagnostic PCR primer designs for targeting and editing scoring of the CBE editors A3A, rA1, and CDA1 (Figure 1) in the present study.
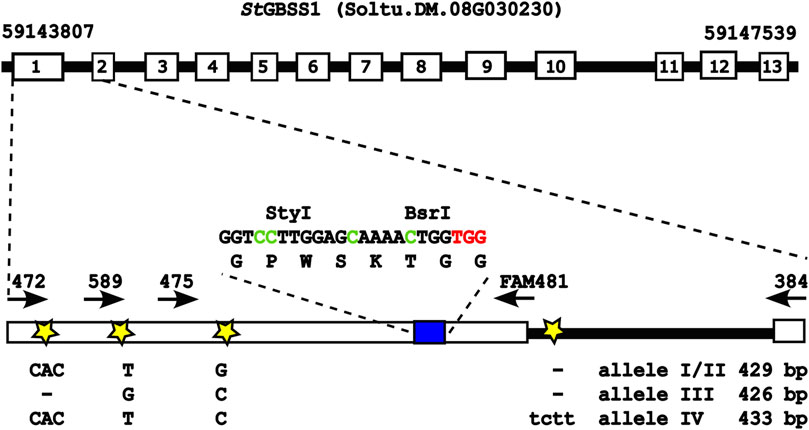
FIGURE 1. GBSS 1 target gene (cultivar Desiree). Exon 1 of the GBSS 1 target gene with both length and SNPs between the four alleles in the cultivar Desiree is indicated. The target gRNA sequence GGTC4C5TTGGAGC12AAAAC17TGG (blue box), with target cytosines (Cs) in green, PAM in red, and the diagnostic restrictions sites StyI and BsrI, is indicated. White numbered boxes depict exons, while stars indicate SNPs or size polymorphisms between the four alleles. Arrows indicate diagnostic PCR primers for editing scoring, amplifying the target region inside or outside the length polymorphisms. The figure is based on and adapted from the work of Johansen et al. (2019).
Nickase and cytidine base editing activities were tested by the transient expression of the SpCas9/StU6-1::sgRNA1 nickase construct or the C-to-T base editors A3A, rA1, and CDA1 using PEG transformation of isolated potato protoplasts (cell pool) of cultivar Desiree, which were then cultured for 2 days as described in the work of Nicolia et al. (2015) and Nicolia et al. (2021) and outlined in Materials and methods, after which the target region was PCR-amplified, and each PCR amplicon was analyzed by both IDAA and amplicon sequencing, including EditR analysis, for potential nuclease-induced indels and C-to-T base editing activity. First, the SpCas9 in the construct SpCas9/StU6-1::sgRNA1 (Johansen et al., 2019) was converted into a nickase (nCas9) by changing the aspartic acid (Asp10) into alanine (Ala10) (D10A) (Jinek et al., 2012b) through the use of site-directed mutagenesis. The absence of nuclease activity from nCas9 was confirmed by full digestion of the BsrI restriction site situated 3 bp upstream of the protospacer adjacent motif (PAM) and confirmed by IDAA (Yang et al., 2015; Bennett et al., 2020), which displayed a PCR amplicon with unchanged length (see Supplementary Information) and sanger sequencing of PCR products (data not shown). Effect of placement of the three deaminases, A3A (human hAPOBEC3A), rA1 (rat evo_rAPOBEC1), and CDA1 (sea lamprey Petromyzon marinus, evo_PmCDA1), and the use of different linkers between fusion partners have been investigated earlier (Nishida et al., 2016; Zong et al., 2018; Tan et al., 2019; Thuronyi et al., 2019; Choi et al., 2021; Huang et al., 2021). In the present study, the deaminase was fused to the N-terminal of nCas9 because the three deaminases have been proven to be functionally active in this design and in order to enable comparison between the three CBEs. The CBEs, A3A, rA1, and CDA1, were initially scored for editing activity by checking for destruction of the StyI restriction site (C4C5WWGG) 11 bp upstream of the PAM site (TGG), where conversion of either or both of the two cytosines C4 and C5 would lead to resistance to StyI digestion (see Supplementary Information). C-to-T editing efficiencies of A3A, rA1, and CDA1 were confirmed and scored by direct sequencing and quantified using the EditR software (Kluesner et al., 2018), with A3A having the best overall activity with an average 34.5%, 34.5%, and 27% C-to-T conversion at C4, C5, and C12 in the target (gRNA) sequence (GGTC4C5TTGGAGC12AAAAC17TGG), respectively, whereas CDA1 showed an average C-to-T conversion of 34.5%, 34%, and 14.25% at C4, C5, and C12, respectively. rA1 showed an average C-to-T conversion of 18.75% and 19% at C4 and C5 and was the only base editor to show no C-to-T conversion at C12. C17 conversion was not observed for any of the three base editors. All three base editors showed stable conversion rates with at least 21% C-to-T conversion for C4 and C5 (a single exception being rA1 replicate 2), and A3A and CDA1 showed an average 34% conversion rate for C4 and C5. The highest C-to-T conversion was observed for A3A replicate 4, which showed 39%, 43%, and 36% for C4, C5, and C12, respectively (Figures 2B–D). Neither indel formation, as evidenced by IDAA and direct sequencing results (Supplementary Information), nor unintended C-to-A or C-to-G changes, as evidenced by direct sequencing results (Figure 2B and Supplementary Information) and EditR analysis (Figure 2C and Supplementary Information), were encountered in the present study, which, however, was confined to the protoplast pool. Direct sequencing identified two allele-specific SNPs (Figure 1 and Supplementary Information), indicating amplification of the four alleles. Detailed information regarding constructs, protoplast isolation, PEG-mediated transformation and incubation, restriction enzyme, IDAA analyses, and direct sequencing on the protoplast cell pool is provided in Materials and methods and Supplementary Information.
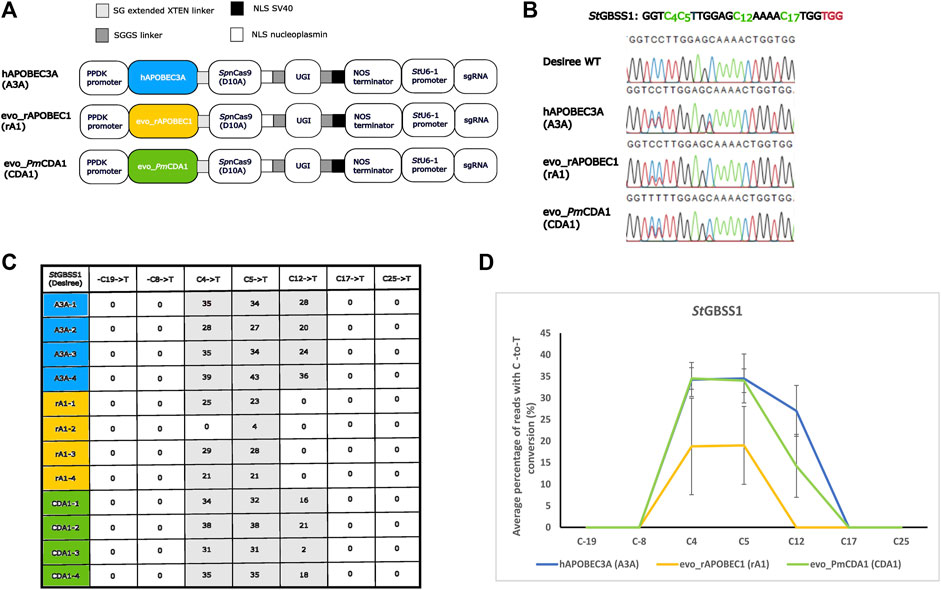
FIGURE 2. C-to-T conversion derived from cytosine base editors A3A, rA1, and CDA1 optimized for potato. (A) Three cytosine base editor constructs comprising a cassette driving expression of the gRNA (target conferring part, GGTC4C5TTGGAGC12AAAAC17TGG) from the StU6-1 promotor (Johansen et al., 2019), the SpCas9 nickase (nCas9) in fusion with one of the deaminases hAPOBEC3A (A3A) from human, evo_rAPOBEC1 (rA1) from rat, or sea lamprey evo_PmCDA1 (CDA1), followed by a uracil-DNA glycosylase inhibitor (UGI), interspaced by long flexible linkers are depicted. NLS, nuclear localization signal; serine (S) and glycine (G) linkers: SG and SGGS. (B) Chromatograms from direct sequencing of PCR products from the protoplast cell pool transformed with the base editing constructs with A3A showing 39% (C4), 43% (C5), and 36% (C12) editing (sample A3A replicate 4), rA1 showing 29% (C4) and 28% (C5) (sample rA1 replicate 3), and CDA1 showing 38% (C4), 38% (C5), and 21% (C12) (sample CDA1 replicate 2) C-to-T conversion in the target region (exon 1 of GBSS1 (cultivar Desiree)) when compared to the WT sequence. Target Cs in the gRNA and adjacent PAM site are shown in green and red, respectively. (C) C-to-T conversion (%) of Cs within (C4, C5, C12, and C17) and most closely adjacent (−C19, −C8 and C25) to the gRNA of protoplasts transformed with A3A, rA1, and CDA1 as evidenced by EditR analysis. Numbers 1–4 indicate replicates. (D) Average percentage of reads with C-to-T conversion (%) rates of protoplasts transformed with A3A, rA1, and CDA1 as shown for Cs within (C4, C5, C12, and C17) and most closely adjacent (−C19, −C8 and C25) to the gRNA. Data are shown as mean ± sd of four biological replicates.
The use of CRISPR-based precise gene editing, including base and prime editing, for crop improvement has recently been reviewed (Butt et al., 2020; Gurel et al., 2020; Mishra et al., 2020), with a particular focus on potato protoplasts, e.g., provided in the work of Hofvander et al. (2022).
Here, we further developed a CRISPR/SpCas9 construct optimized for potato, where replacement of the standard Arabidopsis thaliana AtU6-1 promotor driving the expression of the gRNA, with the endogenous potato StU6-1 promotor, resulted in a 3–4-fold increase in editing efficiencies at the protoplast cell pool level (Johansen et al., 2019), into CBE constructs. We used three different CBE constructs, in which either of three deaminases, A3A (human hAPOBEC3A), rA1 (rat evo_rAPOBEC1), and CDA1 (sea lamprey Petromyzon marinus, evo_PmCDA1), were C-terminally fused to a SpCas9 nickase (nCas9) and uracil-DNA glycosylase inhibitor (UGI), which were combined with the native potato StU6-1::gRNA-1 cassette expressing the gRNA. Each CBE was targeted to exon 1 of the GBSS1 gene and transformed into protoplasts of potato cultivar Desiree with their base editing conversions scored. All three constructs displayed high C-to-T conversion activities, peaking at C4 and C5 in the target (GGTC4C5TTGGAGC12AAAAC17TGGTGG) sequence, with A3A, CDA1, and rA1 displaying average C4 and C5 C-to-T conversions of 34.5% & 34.5%, 34.5% & 34%, and 18.75% & 19%, respectively. A3A and CDA1 displayed 27% and 14.25% C-to-T conversion at C12, and rA1 showed no C12 C-to-T conversion, which is in agreement with the fact that the rAPOBEC1 deaminase, from which rA1 is derived, has previously been reported to be inefficient in a GC context (Zong et al., 2018). The importance of the sequential location of Cs and different sequence preferences for different deaminases have been highlighted in other studies (Tan et al., 2019; Tan et al., 2020; Huang et al., 2021).
With the exception of a single replicate, all three CBEs conferred >= 21% C-to-T conversion of C4 and C5 in the target sequence (see Figure 2C rA1-2), with an average 34% C-to-T conversion for C4 and C5 for both the A3A and CDA1, which, to our knowledge, are the highest average C-to-T conversions obtained when employing PEG-mediated delivery into potato protoplasts. In comparison, Zong et al. (2018) obtained, in one instance, an average of up to 18%–20% C-to-T editing when targeting the StGBSS1 gene in potato protoplasts (Zong et al., 2018). Protoplasts transformed with non-integrative constructs will, unlike agrobacterium-transformed plants that may display chimerism (Faize et al., 2010), generate single-protoplast-cell-derived genetically uniform explants and enable a potential replacement of plasmid with ribonucleoprotein (RNP), thereby excluding the presence of DNA in the entire editing process.
The averaged significantly higher editing efficiency obtained in the present study may be attributed to the use of the native potato StU6-1 promoter, driving the gRNA, which appeared rate limiting in Johansen et al.’s (2019) study, although differences in CBE construct architecture and composition or methodology may also be contributing factors. Zong et al., 2018 pioneered the implementation of C-to-T base editing in plants using the human APOBEC3A-based and the rat APOBEC1-based cytidine deaminase construct (Zong et al., 2018). The APOBEC3A- and APOBEC1-based CBEs were delivered into cells of potato, rice, and wheat by PEG-mediated transformation of protoplasts, Agrobacterium-mediated transformation of callus, or biolistic delivery into immature embryo cells. In most experiments, the human APOBEC3A-based CBE outperformed the rat APOBEC1-based CBE, a tendency which was confirmed for the A3A and rA1 CBEs generated and tested in the present study.
Distribution of C-to-T conversion across the target sequence, i.e., editing frequencies at C4, C5, C12 and C17, seemed to be somewhat similar to what has been reported for other base editing constructs (Huang et al., 2021). However, careful construct design/architecture, e.g., adjustments of flexible linker lengths, may elevate a desired target position accuracy (Tan et al., 2019; Tan et al., 2020). In addition, the development of base editors with alternative PAM specificities expands the freedom to operate and may potentially affect precision (Veillet et al., 2020a; Veillet et al., 2020b). The base editing efficiencies presented here were obtained via transient non-integrative PEG transformation of the protoplast cell pool level, where the A3A CBE, in one instance, conferred 43% C-to-T conversion of C5.
CBEs have, in some settings, been reported to additionally generate C-to-G or C-to-A conversions, although at lower frequencies than the targeted C-to-T conversions (Komor et al., 2017) and indels, whereas in one study, 75% of explants transformed with an agrobacterium-mediated integrative CBE construct were found to contain indels (Veillet et al., 2019b). Similar undesired conversions or indel formation, e.g., as evidenced by direct sequencing, EditR analysis, and IDAA, were, within the resolution of the analytic methods applied, not encountered in the present study, which, however, was confined to the protoplast cell pool.
PE enables controlled generation of small insertions, deletions, or base substitutions as part of the prime editing guide RNA (pegRNA) and was originally described as a tool for correcting DNA in humans in relation to disease (Anzalone et al., 2019). PE has also been applied in plants, such as rice (Li H. Y. et al., 2020; Lin et al., 2020) and maize (Jiang et al., 2020a), with moderate success, highlighting the importance of testing a range of pegRNAs. Recent implementation of PE in the model plant Physcomitrium patens and tetraploid potato also pinpointed limitations of the technology, which need to be overcome before PE may become a versatile efficient tool in precision plant breeding (Perroud et al., 2022). The PE repertoire has, as in the case of the base editor repertoire, been expanded with alternative PAM specificities (Kweon et al., 2021).
The construct design and protocols for scoring C-to-T base editing presented in this study may readily be converted into A-to-G base editors (ABEs), probably with comparable efficiencies. Thus, for now, and with the editing efficacies obtained in this study, BE still remains a competitive relevant tool in the toolbox for precise plant breeding.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Independent Research Fund Denmark (1032-00399B) (2021) “Green CRISPR production platform for new starch qualities”; Novo Nordisk foundation (NNF19OC0057208); Grønt Udviklings og Demonstrations program (GUDP) (2020) “KRISPS Kartofler med Resistens og Innovativ Stivelse som Platform for Synergi egenskaber i kartoffel vha. DNA-fri CRISPR“; Kartoffelafgiftfonden (2022) “Reduceret miljø og klima aftryk em grøn og økonomisk bæredygtighed” (34009-19-1531); Kartoffelafgiftfonden (2021) “Samtidig introduktion af flere egenskaber gennem præcis forædling af skimmel resistens i stivelses kartofler” are acknowledged for funding support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeed.2023.1247702/full#supplementary-material
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 (7785), 149–157. doi:10.1038/s41586-019-1711-4
Bennett, E. P., Petersen, B. L., Johansen, I. E., Niu, Y. Y., Yang, Z., Chamberlain, C. A., et al. (2020). INDEL detection, the 'achilles heel' of precise genome editing A survey of methods for accurate profiling of gene editing induced indels. Nucleic Acids Res. 48 (21), 11958–11981. doi:10.1093/nar/gkaa975
Butt, H., Zaidi, S. S. E. A., Hassan, N., and Mahfouz, M. (2020). CRISPR-based directed evolution for crop improvement. Trends Biotechnol. 38 (3), 236–240. doi:10.1016/j.tibtech.2019.08.001
Choi, M., Yun, J. Y., Kim, J. H., Kim, J. S., and Kim, S. T. (2021). The efficacy of CRISPR-mediated cytosine base editing with the RPS5a promoter in Arabidopsis thaliana. Sci. Rep. 11 (1), 8087. doi:10.1038/s41598-021-87669-y
Faize, M., Faize, L., and Burgos, L. (2010). Using quantitative real-time PCR to detect chimeras in transgenic tobacco and apricot and to monitor their dissociation. Bmc Biotechnol. 10, 53. doi:10.1186/1472-6750-10-53
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A.T to G.C in genomic DNA without DNA cleavage. Nature 551 (7681), 464–471. doi:10.1038/nature24644
Geurts, M. H., de Poel, E., Pleguezuelos-Manzano, C., Oka, R., Carrillo, L., Andersson-Rolf, A., et al. (2021). Evaluating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids. Life Sci. Alliance 4 (10), e202000940. doi:10.26508/lsa.202000940
Gurel, F., Zhang, Y. X., Sretenovic, S., and Qi, Y. P. (2020). CRISPR-Cas nucleases and base editors for plant genome editing. Abiotech 1 (1), 74–87. doi:10.1007/s42994-019-00010-0
Happi Mbakam, C., Rousseau, J., Lu, Y., Bigot, A., Mamchaoui, K., Mouly, V., et al. (2022a). Prime editing optimized RTT permits the correction of the c.8713C>T mutation in DMD gene. Mol. Ther. Nucleic Acids 30, 272–285. doi:10.1016/j.omtn.2022.09.022
Happi Mbakam, C., Rousseau, J., Tremblay, G., Yameogo, P., and Tremblay, J. P. (2022b). Prime editing permits the introduction of specific mutations in the gene responsible for duchenne muscular dystrophy. Int. J. Mol. Sci. 23 (11), 6160. doi:10.3390/ijms23116160
Hofvander, P., Andreasson, E., and Andersson, M. (2022). Potato trait development going fast-forward with genome editing. Trends Genet. 38 (3), 218–221. doi:10.1016/j.tig.2021.10.004
Hua, K., Tao, X. P., Liang, W. Y., Zhang, Z. X., Gou, R. Y., and Zhu, J. K. (2020). Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotechnol. J. 18 (3), 770–778. doi:10.1111/pbi.13244
Huang, T. P., Newby, G. A., and Liu, D. V. R. (2021). Publisher Correction Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat. Protoc. 16 (12), 5740. doi:10.1038/s41596-021-00525-1
Jiang, Y. Y., Chai, Y. P., Lu, M. H., Han, X. L., Lin, Q. P., Zhang, Y., et al. (2020b). Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 21 (1), 257. doi:10.1186/s13059-020-02170-5
Jiang, Y. Y., Chai, Y. P., Lu, M. H., Han, X. L., Lin, Q., Zhang, Y., et al. (2020a). Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 21 (1), 257. doi:10.1186/s13059-020-02170-5
Jin, S., Zong, Y., Gao, Q., Zhu, Z., Wang, Y., Qin, P., et al. (2019). Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364 (6437), 292–295. doi:10.1126/science.aaw7166
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012a). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821. doi:10.1126/science.1225829
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012b). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821. doi:10.1126/science.1225829
Johansen, I. E., Liu, Y., Jorgensen, B., Bennett, E. P., Andreasson, E., Nielsen, K. L., et al. (2019). High efficacy full allelic CRISPR/Cas9 gene editing in tetraploid potato. Sci. Rep. 9 (1), 17715. doi:10.1038/s41598-019-54126-w
Kluesner, M. G., Nedveck, D. A., Lahr, W. S., Garbe, J. R., Abrahante, J. E., Webber, B. R., et al. (2018). EditR A method to quantify base editing from sanger sequencing. CRISPR J. 1 (3), 239–250. doi:10.1089/crispr.2018.0014
Koblan, L. W., Doman, J. L., Wilson, C., Levy, J. M., Tay, T., Newby, G. A., et al. (2018). Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36 (9), 843–846. doi:10.1038/nbt.4172
Koblan, L. W., Erdos, M. R., Wilson, C., Cabral, W. A., Levy, J. M., Xiong, Z. M., et al. (2021). In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature 589 (7843), 608–614. doi:10.1038/s41586-020-03086-7
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533 (7603), 420–424. doi:10.1038/nature17946
Komor, A. C., Zhao, K. T., Packer, M. S., Gaudelli, N. M., Waterbury, A. L., Koblan, L. W., et al. (2017). Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:a base editors with higher efficiency and product purity. Sci. Adv. 3 (8), eaao4774. doi:10.1126/sciadv.aao4774
Kurt, I. C., Zhou, R., Iyer, S., Garcia, S. P., Miller, B. R., Langner, L. M., et al. (2021). CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 39 (1), 41–46. doi:10.1038/s41587-020-0609-x
Kweon, J., Yoon, J. K., Jang, A. H., Shin, H. R., See, J. E., Jang, G., et al. (2021). Engineered prime editors with PAM flexibility. Mol. Ther. 29 (6), 2001–2007. doi:10.1016/j.ymthe.2021.02.022
Li, C., Zhang, R., Meng, X., Chen, S., Zong, Y., Lu, C., et al. (2020a). Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 38 (7), 875–882. doi:10.1038/s41587-019-0393-7
Li, H. Y., Li, J. Y., Chen, J. L., Yan, L., and Xia, L. Q. (2020b). Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 13 (5), 671–674. doi:10.1016/j.molp.2020.03.011
Li, J. F., Zhang, D., and Sheen, J. (2015). Targeted plant genome editing via the CRISPR/Cas9 technology. Methods Mol. Biol. 1284, 239–255. doi:10.1007/978-1-4939-2444-8_12
Lin, Q. P., Zong, Y., Xue, C. X., Wang, S. X., Jin, S., Zhu, Z. X., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38 (5), 582–585. doi:10.1038/s41587-020-0455-x
Mishra, R., Joshi, R. K., and Zhao, K. J. (2020). Base editing in crops Current advances, limitations and future implications. Plant Biotechnol. J. 18 (1), 20–31. doi:10.1111/pbi.13225
Nicolia, A., Falt, A. S., Hofvander, P., and Andersson, M. (2021). Protoplast-based method for genome editing in tetraploid potato. Crop Breed. 2264, 177–186. doi:10.1007/978-1-0716-1201-9_12
Nicolia, A., Proux-Wera, E., Ahman, I., Onkokesung, N., Andersson, M., Andreasson, E., et al. (2015). Targeted gene mutation in tetraploid potato through transient TALEN expression in protoplasts. J. Biotechnol. 204, 17–24. doi:10.1016/j.jbiotec.2015.03.021
Nishida, K., Arazoe, T., Yachie, N., Banno, S., Kakimoto, M., Tabata, M., et al. (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353 (6305), aaf8729. doi:10.1126/science.aaf8729
Perroud, P. F., Guyon-Debast, A., Veillet, F., Kermarrec, M. P., Chauvin, L., Chauvin, J. E., et al. (2022). Prime Editing in the model plant Physcomitrium patens and its potential in the tetraploid potato. Plant Sci. 316, 111162. doi:10.1016/j.plantsci.2021.111162
Shimatani, Z., Kashojiya, S., Takayama, M., Terada, R., Arazoe, T., Ishii, H., et al. (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35 (5), 441–443. doi:10.1038/nbt.3833
Surun, D., Schneider, A., Mircetic, J., Neumann, K., Lansing, F., Paszkowski-Rogacz, M., et al. (2020). Efficient generation and correction of mutations in human iPS cells utilizing mRNAs of CRISPR base editors and prime editors. Genes (Basel) 11 (5), 511. doi:10.3390/genes11050511
Tan, J. J., Zhang, F., Karcher, D., and Bock, R. (2019). Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat. Commun. 10, 439. doi:10.1038/s41467-018-08034-8
Tan, J. J., Zhang, F., Karcher, D., and Bock, R. (2020). Expanding the genome-targeting scope and the site selectivity of high-precision base editors. Nat. Commun. 11 (1), 629. doi:10.1038/s41467-020-14465-z
Thuronyi, B. W., Koblan, L. W., Levy, J. M., Yeh, W. H., Zheng, C., Newby, G. A., et al. (2019). Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 37 (9), 1070–1079. doi:10.1038/s41587-019-0193-0
Tremblay, G., Rousseau, J., Mbakam, C. H., and Tremblay, J. P. (2022). Insertion of the Icelandic mutation (A673T) by prime editing A potential preventive treatment for familial and sporadic alzheimer's disease. CRISPR J. 5 (1), 109–122. doi:10.1089/crispr.2021.0085
Veillet, F., Chauvin, L., Kermarrec, M. P., Sevestre, F., Merrer, M., Terret, Z., et al. (2019a). The Solanum tuberosum GBSSI gene A target for assessing gene and base editing in tetraploid potato. Plant Cell Rep. 38 (9), 1065–1080. doi:10.1007/s00299-019-02426-w
Veillet, F., Kermarrec, M. P., Chauvin, L., Chauvin, J. E., and Nogue, F. (2020a). CRISPR-induced indels and base editing using theStaphylococcus aureusCas9 in potato. Plos One 15 (8), e0235942. doi:10.1371/journal.pone.0235942
Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M. P., Guyon-Debast, A., Chauvin, J. E., et al. (2019b). Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 20 (2), 402. doi:10.3390/ijms20020402
Veillet, F., Perrot, L., Guyon-Debast, A., Kermarrec, M. P., Chauvin, L., Chauvin, J. E., et al. (2020b). Expanding the CRISPR toolbox in P. patens using SpCas9-NG variant and application for gene and base editing in solanaceae crops. Int. J. Mol. Sci. 21 (3), 1024. doi:10.3390/ijms21031024
Xiong, X., Li, Z., Liang, J., Liu, K., Li, C., and Li, J. F. (2022). A cytosine base editor toolkit with varying activity windows and target scopes for versatile gene manipulation in plants. Nucleic Acids Res. 50 (6), 3565–3580. doi:10.1093/nar/gkac166
Yang, Z., Steentoft, C., Hauge, C., Hansen, L., Thomsen, A. L., Niola, F., et al. (2015). Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res. 43 (9), e59. doi:10.1093/nar/gkv126
Zhao, Z., Shang, P., Mohanraju, P., and Geijsen, N. (2023). Prime editing Advances and therapeutic applications. Trends Biotechnol. 41, 1000–1012. doi:10.1016/j.tibtech.2023.03.004
Zong, Y., Song, Q., Li, C., Jin, S., Zhang, D., Wang, Y., et al. (2018). Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 36, 950–953. doi:10.1038/nbt.4261
Keywords: cytosine base editor, U6 promotor, native promotor, protoplast, potato, genome editing
Citation: Westberg I, Carlsen FM, Johansen IE and Petersen BL (2023) Cytosine base editors optimized for genome editing in potato protoplasts. Front. Genome Ed. 5:1247702. doi: 10.3389/fgeed.2023.1247702
Received: 26 June 2023; Accepted: 03 August 2023;
Published: 30 August 2023.
Edited by:
Humira Sonah, Laval University, CanadaReviewed by:
Sanskriti Vats, National Agri-Food Biotechnology Institute, IndiaCopyright © 2023 Westberg, Carlsen, Johansen and Petersen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bent Larsen Petersen, YmxwQHBsZW4ua3UuZGs=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.