
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Genome Ed. , 11 January 2023
Sec. Genome Editing in Plants
Volume 4 - 2022 | https://doi.org/10.3389/fgeed.2022.1085023
This article is part of the Research Topic Plant Breeding Innovations - CRISPR as a Powerful Weapon for Agricultural Crops View all 5 articles
Targeted mutagenesis via programmable nucleases including the clustered regulatory interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) (CRISPR/Cas9) system has been broadly utilized to generate genome-edited organisms including flowering plants. To date, specific expression of Cas9 protein and guide RNA (gRNA) in reproductive cells or tissues is considered one of the most effective genome-editing approaches for heritable targeted mutagenesis. In this report, we review recent advances in genome editing methods for reproductive cells or tissues, which have roles in transmitting genetic material to the next-generation, such as egg cells, pollen grains, zygotes, immature zygotic embryos, and shoot apical meristems (SAMs). Specific expression of Cas9 proteins in initiating cells efficiently induces targeted mutagenesis via Agrobacterium-mediated in planta transformation. In addition, genome editing by direct delivery of CRISPR/Cas9 components into pollen grains, zygotes, cells of embryos and SAMs has been successfully established to generate genome-edited plant lines. Notably, DNA-free genome editing by the delivery of Cas9-gRNA ribonucleoproteins (RNPs) is not associated with any legislative concerns about genetically modified organisms. In summary, the genome editing methods for reproductive cells or tissues have enormous potential for not only basic studies for plant reproduction but also applied sciences toward molecular plant breeding.
Technology involving targeted mutagenesis using programmable nucleases, such as zinc-finger nucleases (ZFNs) (Urnov et al., 2010), transcription activator-like effector nucleases (TALENs) (Cermak et al., 2011), and RNA-guided endonucleases (RGENs), has been rapidly developing and has enormous potential to accelerate basic and applied sciences. The programmable nucleases produce double-strand breaks (DSBs) at target sites in genomic DNA, and these DSBs can be repaired by two independent pathways: non-homologous end-joining (NHEJ) and homology-directed repair (HDR) (Roth and Wilson, 1986; Puchta et al., 1993; Moore and Haber, 1996; Jasin and Rothstein, 2013).
In RGENs, the clustered regulatory interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) (CRISPR/Cas9) system has paved the way for the development of rapid and cost-effective procedures to create new mutant populations in plants (Belhaj et al., 2013; Voytas 2013). In general, the CRISPR/Cas9 expression cassette and selectable marker are integrated into plasmid DNA, and the constructs are delivered into plant cells via Agrobacterium tumefaciens-mediated transformation or particle bombardment (Kumar and Jain, 2015; Luo et al., 2016). Plant lines that have integrated the constructs into genomic DNA are selected by the selectable marker and genome-edited plant lines can be screened by the sequencing of target sites. However, constitutive expression of CRISPR/Cas9 in the plant life cycle generates a large proportion of non-heritable mutations in somatic cells (Feng et al., 2014), and increases the likelihood of DNA cleavage at non-specific loci, so-called off-target modifications, in plant genome editing (Lawrenson et al., 2015). To induce heritable mutations and reduce off-target modifications, a genome editing system through CRISPR/Cas9 expression under a reproductive cell- or tissue-specific promoter has been developed, which we will summarize later in this review.
In animals, to produce genetically heritable traits of interest, in vitro transcribed RNAs encoding Cas9 and gRNA are directly delivered into eggs or zygotes, resulting in the highly efficient production of genetically modified animals (Hwang et al., 2013; Wang et al., 2013). In angiosperms, although female gamete, zygote, and embryo exist in the embryo sac deeply embedded in ovular tissue (Russell 1992; Raghavan 2003), such reproductive cells/tissues isolated from flowers have been successfully used as targets for the direct delivery of CRISPR/Cas9 vectors or preassembled Cas9 protein-guide RNA (gRNA) ribonucleoproteins (RNPs). Moreover, shoot apical meristems (SAMs) including a subepidermal cell layer, L2, from which germ cells later develop during floral organogenesis have also been target tissues for an inheritable genome editing approach. In this mini review, we summarize the current approaches of genome editing using plant reproductive cells/tissues, such as egg cell, pollen grain, zygote, embryo, and SAM, based on the frequencies of targeted mutagenesis and off-target mutations.
In general, Agrobacterium-mediated in planta transformation has been applied to introduce the CRISPR/Cas9 expression cassette into Arabidopsis. Ubiquitously expressed Cas9 protein and gRNA generate targeted gene modifications with high efficiency; however, only the gene modification generated in reproductive cells can be transmitted to the next-generation (Feng et al., 2014). To efficiently induce inheritable targeted mutations, specific promoters for the germline (Elongation Factor-1α(EF1α) promoter; Osakabe et al., 2016) and egg cell (EC promoter; Wang et al., 2015; Zheng et al., 2020, and DD45 promoter; Mao et al., 2016) have been successfully used for the exogenous expression of Cas9-gRNA complexes in reproductive cells of Arabidopsis. Moreover, the RIBOSOMAL PROTEIN S5A (RPS5A) promoter, which is constitutively active at the beginning of the process of egg cell formation, was shown to be efficient for driving the expression of Cas9 in Arabidopsis female germ cells (Figure 1A; Tsutsui and Higashiyama, 2017). In addition to preferential Cas9 expression in female gametes, the SPOROCYTELESS (SPL) genomic expression cassette, which is specifically expressed in sporogenous cells and microsporocytes, has been used for germline-specific Cas9 expression in male Arabidopsis gametocytes (Mao et al., 2016). Furthermore, the YAO promoter, which is preferentially active in the embryo sac, embryo, endosperm, pollen and SAM, has been used for the expression of Cas9 (Yan et al., 2015). These approaches efficiently and preferentially generate progeny with a high diversity of mutations at the targeted locus.
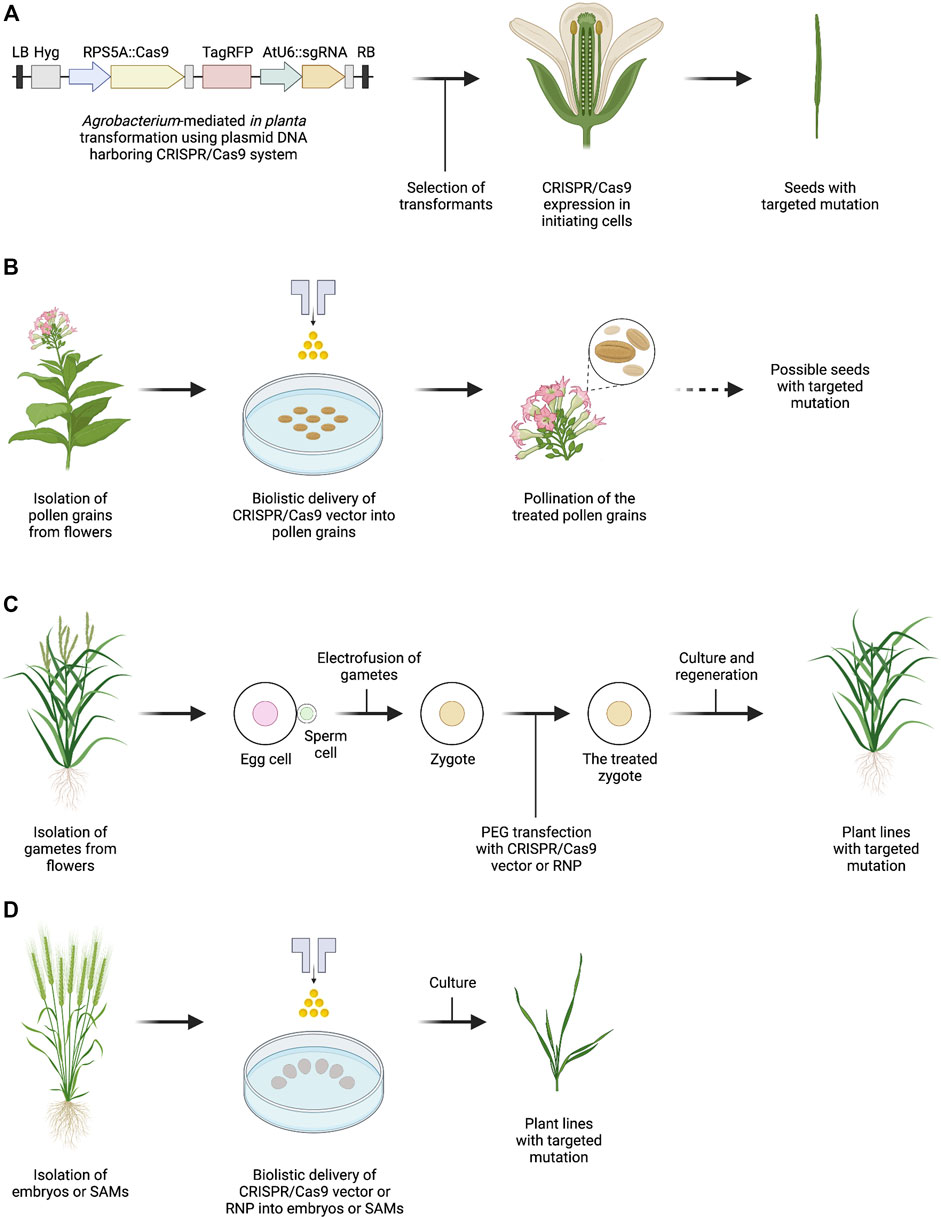
FIGURE 1. Outline of genome editing methods for plant reproductive cells or tissues. (A) Agrobacterium-mediated in planta transformation with CRISPR/Cas9 vector in which promoters for Cas9 expression are specifically expressed in Arabidopsis initiating cells. Vector information is taken with reference to Tsutsui and Higashiyama (2017). (B) Biolistic delivery of CRISPR/Cas9 vector into tobacco pollen grains. The dotted arrow indicates a procedure that has not been experimentally tested. (C) PEG-Ca2+-mediated transfection of rice zygotes with CRISPR/Cas9 vector or Cas9-gRNA RNP. (D) Biolistic delivery of CRISPR/Cas9 vector or Cas9-gRNA RNP into cells of wheat embryos and SAMs. The figure was created with BioRender.com.
In addition to NHEJ-based genome editing, a cell/tissue-specific promoter for initiating cells has been applied for Cas9 expression to induce heritable gene targeting (GT) in Arabidopsis. In this strategy, parental lines expressing Cas9 under the egg cell- and early embryo-specific DD45 promoter were used in combination with the delivery of HDR donor DNA to increase genome editing activity, resulting in high efficiency of GT of ca. 5.3%–9.1% (Miki et al., 2018). Wolter et al. (2018) also demonstrated that the use of Cas9 under a ubiquitin promoter leads to seeds harboring GT events with a low frequency, whereas the use of Cas9 controlled under an egg cell-specific promoter was the most efficient approach, achieving a frequency of around 1% of the seeds. These results indicate that the use of a reproductive cell- or tissue-specific promoter is an effective genome editing approach to achieve heritable targeted mutagenesis via either NHEJ or HDR by Agrobacterium-mediated in planta transformation.
Particle bombardment can be used to deliver macromolecules into various tissues such as immature zygotic embryos, leaf disks, and calli, and is not limited by plant-host range (Altpeter et al., 2005). Pollen grains are structurally simple tissue containing male germ cells, and are easily isolated from anthers. Therefore, the direct delivery of CRISPR/Cas9 vector into Nicotiana benthamiana pollens via biolistic delivery triggers genome editing of the pollen grains, and the bombarded pollen enables the elongation of pollen tubes and delivery of sperm cells into the embryo sac (Table 1; Nagahara et al., 2021). Although biolistic delivery conditions and seed detection methods should be optimized, this delivery approach using pollen grains may be broadly applicable to obtaining progeny with targeted mutations (Figure 1B). Furthermore, procedures for delivering exogenous materials into pollen grains have been demonstrated with various approaches (Eapen et al., 2011; Zhao et al., 2017; Bhowmik et al., 2018). Recently, Lei et al. (2021) has reported genome editing using pollen-specific Cas9 expression via Agrobacterium vacuum infiltration in cotton. In addition, magnetic nanoparticles have been reported as a novel physiological procedure for transforming pollen grains (Zhao et al., 2017), although pollen magnetofection can only be applied in cotton pollen (Vejlupkova et al., 2020).
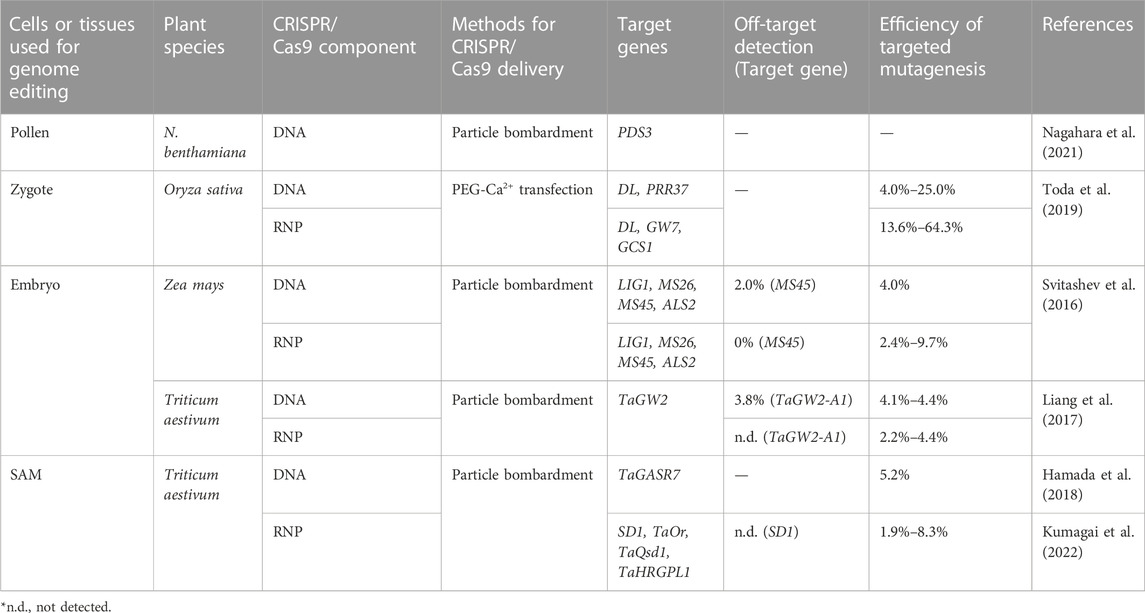
TABLE 1. Genome editing by direct delivery of CRISPR/Cas9 components into plant reproductive cells or tissues.
In animals, to produce genetically heritable traits of interest, in vitro transcribed Cas9 mRNA and sgRNA or preassembled Cas9 protein-sgRNA complexes are delivered into zygotes by direct injection, resulting in the production of bi-allelic mutants with high efficiency (Gratz et al., 2013; Hwang et al., 2013; Wang et al., 2013). In angiosperms, a genome editing system via direct delivery of CRISPR/Cas9 vectors or Cas9-gRNA RNPs into rice zygotes has recently been developed (Figure 1C; Toda et al., 2019). CRISPR/Cas9 vectors or Cas9-gRNA RNPs were transfected into rice zygotes produced by in vitro fertilization (IVF) of isolated gametes via polyethylene glycol-calcium (PEG-Ca2+)-mediated transfection (Koiso et al., 2017). Thereafter, the treated zygotes were cultured in the absence of selection agents, resulting in the regeneration of rice plants with targeted mutations, at frequencies in the range of ca. 4%–64% (Table 1; Toda et al., 2019). In addition to rice, IVF systems have been established in maize (Kranz and Lörz, 1993) and wheat (Maryenti et al., 2019), suggesting that a zygote-based genome editing approach would be applicable to other crop species.
Although particle bombardment delivery of CRISPR/Cas9 expression cassette into immature zygotic embryos has showed successful genome editing, Mendelian segregation distortion was observed in progeny plants (Svitashev et al., 2015). One possibility is that constitutive expression of CRISPR/Cas9 lead to somatic mutations, resulting in chimeric plants (Feng et al., 2014; Svitashev et al., 2015). Therefore, to overcome the issue, genome editing approach by direct delivery of Cas9-gRNA RNPs into cells of embryos has been developed in maize, and the frequencies of targeted mutagenesis were in the range of 2.4%–9.7% (Table 1; Svitashev et al., 2016). Similarly, 2.2%–4.4% of regenerated plants contained target mutations were obtained in wheat (Figure 1D; Table 1; Liang et al., 2017).
In addition to embryos, in planta transformation using biolistic delivery of CRISPR/Cas9 vector to wheat SAMs, which maintain the potential to develop into flower organs, has been reported as an in planta particle bombardment (iPB) method, with targeted mutations in 5.2% of the bombarded plants (Figure 1D; Table 1; Hamada et al., 2018). Notably, the iPB method is a non-culture method that does not require callus culture and regeneration procedures. Furthermore, a system of directly delivering Cas9-gRNA RNPs into wheat SAMs has recently been established, and no mutations were found at the potential off-target sites (Table 1; Kumagai et al., 2022).
In animals, genome editing approaches have been established using germline cells, zygotes, and embryos to obtain genome-edited organisms by inducing heritable genetic changes (Cooper et al., 2017; Lea and Niakan 2019; Koslova et al., 2020). In this mini review, we described genome editing approaches using plant reproductive cells or tissues toward efficient and precise genome editing. In Arabidopsis, specific and sufficient expression of Cas9 proteins in initiating cells, such as germ cells, egg cells, and SAMs, is crucial for efficient targeted mutagenesis through Agrobacterium-mediated in planta transformation (reviewed in Osakabe and Osakabe, 2017).
In addition to Agrobacterium-mediated methods, genome editing via direct delivery of CRISPR/Cas9 components into plant cells or tissues has been developed. Notably, DNA-free genome editing, which can avoid the introduction of foreign DNA sequences into genomic DNA, has been achieved by the direct delivery of Cas9-gRNA RNP into somatic protoplasts via PEG-Ca2+-mediated transfection, such as in tobacco, Arabidopsis, lettuce, rice (Woo et al., 2015), Petunia (Subburaj et al., 2016), grapevine, apple (Malnoy et al., 2016), potato (Andersson et al., 2018), and tomato (Liu et al., 2022). Although a somatic protoplast-based genome editing can use abundant isolated cells for transfection, it remains a major challenge to apply it generally in a wide range of plant species due to difficulties in plant regeneration and obtaining a low frequency of genome-edited plants.
Agrobacterium-mediated transformation- and somatic protoplast-based genome editing has not been applicable to some plant species or cultivars; in contrast, the new system for directly delivering macromolecules to reproductive cells or tissues described here has the potential to be applied for producing genome-edited lines in a wide range of species or cultivars. Genome editing approaches by direct delivery of Cas9-gRNA RNPs into rice zygotes (via PEG-Ca2+-mediated transfection; Toda et al., 2019), cells of maize and wheat embryo cells (via particle bombardment; e.g., Svitashev et al., 2016; Liang et al., 2017), and cells of wheat SAMs (via particle bombardment; Kumagai et al., 2022) have been successfully established. Because foreign DNA sequences cause legislative concerns about genetically modified organisms (Jones 2015), the production of genome-edited rice, wheat, and maize via Cas9-gRNA RNPs is highly desirable for gene functional studies as well as for application to molecular plant breeding (Gu et al., 2021) and can reduce the frequency of off-target changes (Woo et al., 2015; Svitashev et al., 2016; Liang et al., 2017).
DSBs are mainly repaired via NHEJ pathways in the present methods, except for HDR-mediated gene editing in maize embryo cells (Svitashev et al., 2015). Thus, gene targeting via the HDR pathway can in principle be applied and function when Cas9-gRNA RNPs and donor DNA are delivered in reproductive cells/tissues. Although further optimization of procedures for preparation and delivery of RNPs-donor DNA components is required toward production of genome-edited lines possessing the donor DNA at the targeted genome site, these approaches using plant reproductive cells/tissues in various plant species or cultivars have the potential to accelerate a range of different research. This includes basic research, for example, functional analysis of genes of interest involved in reproductive and developmental events, such as gamete differentiation, fertilization, embryogenesis, and endosperm development in flowering plants, as well as applied sciences toward molecular plant breeding.
ET conceived the review. ET wrote a draft of the manuscript and prepared the figure and table. NK, TH, and TO edited and finalized the manuscript. All authors read and approved the final manuscript.
This work was supported, in part, by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (MAFF commissioned project study, Grant No. JPJ008723) and the Japan Society for the Promotion of Science (Grant-in-Aid for JSPS Fellows, Grant No. 21J01093, and Grant-in-Aid for Early-Career Scientists, Grant No. 21K15126 to ET).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Altpeter, F., Baisakh, N., Beachy, R., Bock, R., Capell, T., Christou, P., et al. (2005). Particle bombardment and the genetic enhancement of crops: Myths and realities. Mol. Breed. 15, 305–327. doi:10.1007/s11032-004-8001-y
Andersson, M., Turesson, H., Olsson, N., Fält, A. S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 164, 378–384. doi:10.1111/ppl.12731
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., and Nekrasov, V. (2013). Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/cas system. Plant Methods 9, 39. doi:10.1186/1746-4811-9-39
Bhowmik, P., Ellison, E., Polley, B., Bollina, V., Kulkarni, M., Ghanbarnia, K., et al. (2018). Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Sci. Rep. 8, 6502. doi:10.1038/s41598-018-24690-8
Cermak, T., Doyle, E. L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., et al. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82. doi:10.1093/nar/gkr218
Cooper, C. A., Challagulla, A., Jenkins, K. A., Wise, T. G., O’Neil, T. E., Morris, K. R., et al. (2017). Generation of gene edited birds in one generation using sperm transfection assisted gene editing (STAGE). Transgenic Res. 26, 331–347. doi:10.1007/s11248-016-0003-0
Eapen, S. (2011). Pollen grains as a target for introduction of foreign genes into plants: An assessment. Physiol. Mol. Biol. Plants 17, 1–8. doi:10.1007/s12298-010-0042-6
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 4632–4637. doi:10.1073/pnas.1400822111
Gratz, S. J., Cummings, A. M., Nguyen, J. N., Hamm, D. C., Donohue, L. K., Harrison, M. M., et al. (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035. doi:10.1534/genetics.113.152710
Gu, X., Liu, L., and Zhang, H. (2021). Transgene-free genome editing in plants. Front. Genome Ed. 3, 805317. doi:10.3389/fgeed.2021.805317
Hamada, H., Liu, Y., Nagira, Y., Miki, R., Taoka, N., and Imai, R. (2018). Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat. Sci. Rep. 8, 14422. doi:10.1038/s41598-018-32714-6
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229. doi:10.1038/nbt.2501
Jasin, M., and Rothstein, R. (2013). Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5, 012740. doi:10.1101/cshperspect.a012740
Jones, H. D. (2015). Regulatory uncertainty over genome editing. Nat. Plants 1, 14011. doi:10.1038/nplants.2014.11
Koiso, N., Toda, E., Ichikawa, M., Kato, N., and Okamoto, T. (2017). Development of gene expression system in egg cells and zygotes isolated from rice and maize. Plant Direct 1, 00010. doi:10.1002/pld3.10
Koslová, A., Trefil, P., Mucksová, J., Reinišová, M., Plachý, J., Kalina, J., et al. (2020). Precise CRISPR/Cas9 editing of the NHE1 gene renders chickens resistant to the J subgroup of avian leukosis virus. Proc. Natl. Acad. Sci. U.S.A. 117, 2108–2112. doi:10.1073/pnas.19138.27117
Kranz, E., and Lörz, H. (1993). In vitro fertilization with isolated, single gametes results in zygotic embryogenesis and fertile maize plants. Plant Cell 5, 739–746. doi:10.1105/tpc.5.7.739
Kumagai, Y., Liu, Y., Hamada, H., Luo, W., Zhu, J., Kuroki, M., et al. (2022). Introduction of a second ‘Green Revolution’ mutation into wheat via in planta CRISPR/Cas9 delivery. Plant Physiol. 188, 1838–1842. doi:10.1093/plphys/kiab570
Kumar, V., and Jain, M. (2015). The CRISPR–cas system for plant genome editing: Advances and opportunities. J. Exp. Bot. 66, 47–57. doi:10.1093/jxb/eru429
Lawrenson, T., Shorinola, O., Stacey, N., Li, C., Østergaard, L., Patron, N., et al. (2015). Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16, 258. doi:10.1186/s13059-015-0826-7
Lea, R., and Niakan, K. (2019). Human germline genome editing. Nat. Cell Biol. 21, 1479–1489. doi:10.1038/s41556-019-0424-0
Lei, J., Dai, P., Li, J., Yang, M., Li, X., Zhang, W., et al. (2021). Tissue-specific CRISPR/Cas9 system of cotton pollen with GhPLIMP2b and GhMYB24 promoters. J. Plant Biol. 64, 13–21. doi:10.1007/s12374-020-09272-4
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261. doi:10.1038/ncomms14261
Liu, Y., Andersson, M., Granell, A., Cardi, T., Hofvander, P., and Nicolia, A. (2022). Establishment of a DNA-free genome editing and protoplast regeneration method in cultivated tomato (Solanum lycopersicum). Plant Cell Rep. 41, 1843–1852. doi:10.1007/s00299-022-02893-8
Luo, M., Gilbert, B., and Ayliffe, M. (2016). Applications of CRISPR/Cas9 technology for targeted mutagenesis, gene replacement and stacking of genes in higher plants. Plant Cell Rep. 35, 1439–1450. doi:10.1007/s00299-016-1989-8
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904. doi:10.3389/fpls.2016.01904
Mao, Y., Zhang, Z., Feng, Z., Wei, P., Zhang, H., Botella, J. R., et al. (2016). Development of germ-line-specific CRISPR-cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 14, 519–532. doi:10.1111/pbi.12468
Maryenti, T., Kato, N., Ichikawa, M., and Okamoto, T. (2019). Establishment of an in vitro fertilization system in wheat (Triticum aestivum L. Plant Cell Physiol. 60, 835–843. doi:10.1093/pcp/pcy250
Miki, D., Zhang, W., Zeng, W., Feng, Z., and Zhu, J.-K. (2018). CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat. Commun. 9, 1967. doi:10.1038/s41467-018-04416-0
Moore, J. K., and Haber, J. E. (1996). Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell Biol. 16, 2164–2173. doi:10.1128/mcb.16.5.2164
Nagahara, S., Higashiyama, T., and Mizuta, Y. (2021). Detection of a biolistic delivery of fluorescent markers and CRISPR/Cas9 to the pollen tube. Plant Reprod. 34, 191–205. doi:10.1007/S00497-021-00418-Z
Osakabe, Y., and Osakabe, K. (2017). Genome editing to improve abiotic stress responses in plants. Prog. Mol. Biol. Transl. Sci. 149, 99–109. doi:10.1016/bs.pmbts.2017.03.007
Osakabe, Y., Watanabe, T., Sugano, S. S., Ueta, R., Ishihara, R., Shinozaki, K., et al. (2016). Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 6, 26685. doi:10.1038/srep26685
Puchta, H., Dujon, B., and Hohn, B. (1993). Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 21, 5034–5040. doi:10.1093/nar/21.22.5034
Raghavan, V. (2003). Some reflections on double fertilization, from its discovery to the present. New Phytol. 159, 565–583. doi:10.1046/j.1469-8137.2003.00846.x
Roth, D. B., and Wilson, J. H. (1986). Nonhomologous recombination in mammalian cells: Role for short sequence homologies in the joining reaction. Mol. Cell Biol. 6, 4295–4304. doi:10.1128/mcb.6.12.4295
Russell, S. D. (1992). Double fertilization. Int. Rev. Cytol. 40, 357–388. doi:10.1016/S0074-7696(08)61102-X
Subburaj, S., Chung, S. J., Lee, C., Ryu, S.-M., Kim, D. H., Kim, J.-S., et al. (2016). Site-directed mutagenesis in Petunia x hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 35, 1535–1544. doi:10.1007/s00299-016-1937-7
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., and Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. doi:10.1104/pp.15.00793
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., Cigan, A. M., and Mark CigAn, A. (2016). Genome editing in maize directed by CRISPR-cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274. doi:10.1038/ncomms13274
Toda, E., Koiso, N., Takebayashi, A., Ichikawa, M., Kiba, T., Osakabe, K., et al. (2019). An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants 5, 363–368. doi:10.1038/s41477-019-0386-z
Tsutsui, H., and Higashiyama, T. (2017). pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol. 58, 46–56. doi:10.1093/pcp/pcw191
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S., and Gregory, P. D. (2010). Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646. doi:10.1038/nrg2842
Vejlupkova, Z., Warman, C., Sharma, R., Scheller, H. V., Mortimer, J. C., and Fowler, J. E. (2020). No evidence for transient transformation via pollen magnetofection in several monocot species. Nat. Plants 6, 1323–1324. doi:10.1038/s41477-020-00798-6
Voytas, D. F. (2013). Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. doi:10.1146/annurev-arplant-042811-105552
Wang, H., Yang, H., Shivalila, C. S., Dawlaty, M. M., Cheng, A. W., Zhang, F., et al. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. doi:10.1016/j.cell.2013.04.025
Wang, Z. P., Xing, H. L., Dong, L., Zhang, H. Y., Han, C. Y., Wang, X. C., et al. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144. doi:10.1186/s13059-015-0715-0
Wolter, F., Klemm, J., and Puchta, H. (2018). Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 94, 735–746. doi:10.1111/tpj.13893
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-Free genome editing in plants with preassembled CRISPR-cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi:10.1038/nbt.3389
Yan, L., Wei, S., Wu, Y., Hu, R., Li, H., Yang, W., et al. (2015). High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8, 1820–1823. doi:10.1016/j.molp.2015.10.004
Zhao, X., Meng, Z., Wang, Y., Chen, W., Sun, C., Cui, B., et al. (2017). Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat. Plants 3, 956–964. doi:10.1038/s41477-017-0063-z
Keywords: CRISPR/Cas9, embryo, initiating cell, targeted mutagenesis, plant, pollen grain, shoot apical meristem, zygote
Citation: Toda E, Kato N, Higashiyama T and Okamoto T (2023) Genome editing approaches using reproductive cells/tissues in flowering plants. Front. Genome Ed. 4:1085023. doi: 10.3389/fgeed.2022.1085023
Received: 31 October 2022; Accepted: 30 December 2022;
Published: 11 January 2023.
Edited by:
Jochen Kumlehn, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), GermanyReviewed by:
Piero Barone, Corteva Agriscience™, United StatesCopyright © 2023 Toda, Kato, Higashiyama and Okamoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erika Toda, ZXRvZGFAZy5lY2MudS10b2t5by5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.