
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genome Ed. , 20 November 2020
Sec. Genome Editing in Plants
Volume 2 - 2020 | https://doi.org/10.3389/fgeed.2020.605768
This article is part of the Research Topic Targeted Genome Editing in Crops View all 12 articles
 Limin Hu
Limin Hu Olalekan Amoo
Olalekan Amoo Qianqian Liu
Qianqian Liu Shengli Cai
Shengli Cai Miaoshan Zhu
Miaoshan Zhu Xiaoxiao Shen
Xiaoxiao Shen Kaidi Yu
Kaidi Yu Yungu Zhai
Yungu Zhai Yang Yang
Yang Yang Lei Xu
Lei Xu Chuchuan Fan*
Chuchuan Fan* Yongming Zhou
Yongming ZhouRapeseed is one of the world's most important sources of oilseed crops. Single nucleotide substitution is the basis of most genetic variation underpinning important agronomic traits. Therefore, genome-wide and target-specific base editing will greatly facilitate precision plant molecular breeding. In this study, four CBE systems (BnPBE, BnA3A-PBE, BnA3A1-PBE, and BnPBGE14) were modified to achieve cytidine base editing at five target genes in rapeseed. The results indicated that genome editing is achievable in three CBEs systems, among which BnA3A1-PBE had the highest base-editing efficiency (average 29.8% and up to 50.5%) compared to all previous CBEs reported in rapeseed. The editing efficiency of BnA3A1-PBE is ~8.0% and fourfold higher, than those of BnA3A-PBE (averaging 27.6%) and BnPBE (averaging 6.5%), respectively. Moreover, BnA3A1-PBE and BnA3A-PBE could significantly increase the proportion of both the homozygous and biallelic genotypes, and also broaden the editing window compared to BnPBE. The cytidine substitution which occurred at the target sites of both BnaA06.RGA and BnaALS were stably inherited and conferred expected gain-of-function phenotype in the T1 generation (i.e., dwarf phenotype or herbicide resistance for weed control, respectively). Moreover, new alleles or epialleles with expected phenotype were also produced, which served as an important resource for crop improvement. Thus, the improved CBE system in the present study, BnA3A1-PBE, represents a powerful base editor for both gene function studies and molecular breeding in rapeseed.
Rapeseed (Brassica napus L., AACC, 2n = 38) is one of the most important sources of oilseed crops in the world, accounting for ~16% of the entire global vegetable oil production (Woodfield et al., 2017). Achieving high yields through genetic improvements has always been the major goal in rapeseed production. The constant creation and use of novel genetic variation are important to both genetic research and plant trait improvement. In order to optimize the agronomic traits of crops, breeders applied various methods such as chemical compounds and irradiation to produce heritable mutations. However, these traditional techniques are not target-specific and require genome-scale screening, which is time-and-labor-consuming (Russell et al., 1958; Sega, 1984). As an allotetraploid species, rapeseed has a complicated genome in which most genes have several homologous copies (Chalhoub et al., 2014). Thus, obtaining mutations at all homologous copies is challenging by traditional mutagenesis. With the rapid progress in molecular biology, genome-editing technologies have proven to be a powerful tool to address this issue.
In recent years, CRISPR/Cas9 systems have been proven to be very efficient in improving agronomic traits, especially yield-related traits of rapeseed through genome editing (Braatz et al., 2017; Yang et al., 2018; Zhai et al., 2020). The traditional CRISPR/Cas9 system prefers to generate small insertions and deletions (indels) and is best suited to create knockout mutations. This makes the traditional CRISPR/Cas9 system ineffective when precise base substitutions are needed. However, many desired agronomic traits involve only single nucleotide variants within genes, such as the reported cytidine (C) to thymidine (T) replacement at particular sites of BnaALS, BnaRGA, and BnaA3.IAA7 that conferred gain-of-function mutations with valuable benefits for agricultural applications in rapeseed (Liu et al., 2010; Li et al., 2015, 2019). Therefore, precise base editing has great potential for the production of desired alleles and trait improvement. Using this method, desirable traits can be introgressed into elite lines without compromise, and the resulting lines with targeted improvement will be utilized for practical production.
Recently, base editors, including cytidine base editors (CBEs) and adenine base editors (ABEs), enable precise base alterations in the genome without inducing DNA double-stranded breaks (DSBs) (Komor et al., 2016; Nishida et al., 2016; Gaudelli et al., 2017). CBEs using a Cas9 variant fused with cytidine deaminase have enabled C-to-T conversion without requiring DSBs formation and homology-directed repair in mammalian cells (Komor et al., 2016). Currently, the most commonly used CBE, named BE3, consists of the rat cytidine deaminase APOBEC1 (rAPOBEC1) and uracil DNA glycosylase inhibitor (UGI) fused to Cas9 nickase (nCas9) (Li et al., 2017; Lu and Zhu, 2017; Ren et al., 2017; Zong et al., 2017). The BE3 system typically allows C-T substitution within a small editing window from C4 to C8 of the protospacer (Komor et al., 2016; Li et al., 2017). Several studies have reported the successful applications of CBE in several crop species including rice, maize, wheat, tomato, cotton, and rapeseed (Li et al., 2017; Zong et al., 2017, 2018; Qin et al., 2019; Wu et al., 2020). In addition, many other base editing systems have been developed in plants to improve gene editing accuracy and efficiency. A Petromyzon marinus cytidine deaminase (PmCDA1)-based CBE has resulted in efficient editing in rice and tomato (Shimatani et al., 2017). Moreover, a separate study has shown that PmCDA1 has higher base editing activity than rAPOBEC1 in rice (Tang et al., 2018). Zong et al. (2018) further improved CBEs by using the more effective human APOBEC3A (named A3A-PBE) which worked efficiently in wheat, rice and potato with a 17-nucleotide editing window, independent of sequence context.
During the preparation of our manuscript, Wu et al. (2020) reported the successful application of cytosine base-editing in rapeseed using rat cytidine deaminase APOBEC1. The editing efficiency was 1.8%, which is relatively lower when compared to other crops, and only one copy of BnaALS gene was edited (Wu et al., 2020). Cheng et al. (2020) successfully used A3A-PBE system to target ALS, RGA, and IAA7 genes with an averaging editing efficiency of 23.6%, which also needs further improvement in the editing efficiency. In addition, they provided very limited information on the editing feature of the A3A-PBE system (Cheng et al., 2020). Therefore, further studies are required to establish more effective CBE systems in rapeseed based on the commonly used cytidine deaminases like rAPOBEC1, PmCDA1, and APOBEC3A.
In this study, we modified four CBE systems to achieve cytidine base editing at different genome sites in rapeseed. Five important genes with well-known functions, including BnaCLV3, BnaRGA, BnaA3.IAA7, BnaDA1, and BnaALS, were selected for precise base editing to improve agronomic traits in rapeseed (Liu et al., 2010; Li et al., 2015, 2019; Wang et al., 2017; Yang et al., 2018). Our results indicated that BnA3A1-PBE represents the best CBE editor in rapeseed at present, with the highest base-editing efficiency (up to 50.5%) and higher proportion of both homozygous and biallelic genotypes. The cytidine substitution that occurred at the target sites of BnaRGA and BnaALS were stably inherited and conferred expected phenotype in the T1 generation, indicating its powerful application prospect in rapeseed improvement.
To construct BnPBE and BnA3A-PBE vectors, cytidine deaminase (rAPOBEC1 or APOBEC3A), nCas9 and UGI units were amplified from pnCas9-PBE or A3A-PBE template plasmid (Zong et al., 2017), while the 35S promoter and ccdB units were amplified from PYLCRISPRCas9P35s-H (Ma et al., 2015). The resulting polymerase chain reaction (PCR) products were inserted into the PmeI/BamHI sites of binary vector PYLCRISPRCas9P35s-H through a Pro Ligation-Free Cloning Kit (Applied Biological Materials Inc, Canda, Cat.No.E086/E087). Furthermore, the cereal plant APOBEC3A sequences were codon-optimized for dicotyledon plant and synthesized commercially (Nanjing, China, GenScript) to create BnA3A1-PBE. The multiple sgRNA constructs were generated following a previous protocol used in combining sgRNAs to PYLCRISPRCas9P35s-H (Yang et al., 2018). Then, the multiple sgRNAs were amplified from the generating vector and the resulting PCR product was inserted into the AscI sites of BnPBE, BnA3A-PBE and BnA3A1-PBE, using the Pro Ligation-Free Cloning Kit. To construct BnPBGE14 vector, nCas9 and PmCDA1 (Shimatani et al., 2017) were codon-optimized for dicotyledon plant and then synthesized commercially to replace Cas9 in PYLCRISPRCas9P35s-H. The multiple target sequences were synthesized and ligated to the BsaI sites of PYLCRISPRCas9P35s-H. Primers used for vector construction are listed in Supplementary Table 1.
Following verification of the fused constructs via sequencing, the CBE expressing binary vectors were transformed into an elite cultivar (J9707) via the Agrobacterium tumefaciens-mediated hypocotyl method (Zhou and Fowke, 2002). Hygromycinselection (25 mg/L) was used to screen the transgenic plants.
Genomic DNA was extracted from the T0 transgenic and wild type rapeseed plants using the CTAB method. The positive transgenic plants were screened by PCR using the specific primer pairs PB-L/PB-R (Supplementary Table 1). Then, the targeted mutations were determined in transgenic plants using the high-throughput tracking of mutations (Hi-TOM) platform (Liu et al., 2019). The sequencing analysis was conducted following the approach previously described by Zhai et al. (2020). The targets specific primer sets are listed in Supplementary Table 1.
Putative off-target sites, which contained 2–4-nucleotide mismatches relative to the BnaCLV3 and BnaRGA target sites, were identified using Cas-OFFinder and CRISPR-P software (Bae et al., 2014; Liu et al., 2017). These potential off-target sites were detected in all edited T0 transgenic plants using targeted deep sequencing. For each target gene, mixed genomic DNA from all T0 editing plants was used as the template, and DNA of wild type plant was included as a control. All PCR products were purified and mixed in equal amounts (50 ng for each) as one sample. The DNA library construction, sequencing using the Illumina HiSeq 3000 system and data analysis were conducted according to the methods previously described by Yang et al. (2018). The independent sequence reads of each off-target site were aligned to the genomic wild type sequence, which covered each off-target site as a reference.
The T1 mutants and wild type plants grown in the greenhouse (23°C, 16 h light/20°C, 8 h dark) were treated with commercial sulfonylureas at 1, 2, and 4 times field-recommended concentration (200, 400, and 800 mg/L). Representative pictures were taken 3 weeks after treatment.
In this study, we adopted the base editor units (cytidine deaminase, nCas9 and UGI) from the PBE and A3A-PBE plasmid (Zong et al., 2018) to replace Cas9 in the pYLCRISPR/Cas9P35S-H binary vector (Yang et al., 2018), leading to the BnPBE and BnA3A-PBE systems, respectively. A codon-optimized APOBEC3A for Brassica plants was synthesized to optimize A3A-PBE, resulting in the creation of BnA3A1-PBE (Figure 1). PmCDA1 and nCas9 sequences (Shimatani et al., 2017) were codon-optimized for Brassica plants and synthesized to replace Cas9 in the pYLCRISPR/Cas9P35S-H binary vector, leading to the BnPBGE14 system (Figure 1). Thus, four CBE systems were modified to test cytidine base editing in B. napus. Three of them, including BnPBE, BnA3A-PBE, and BnA3A1-PBE, used the 35S promoter and the AtU3/AtU6 promoters to express the base editor unit and sgRNAs, respectively; while, BnPBGE14 use a 35S promoter to express both the base editor unit and sgRNAs in one ORF, in which multiple sgRNAs were further released using the tRNA-processing system-based strategy (Figure 1).
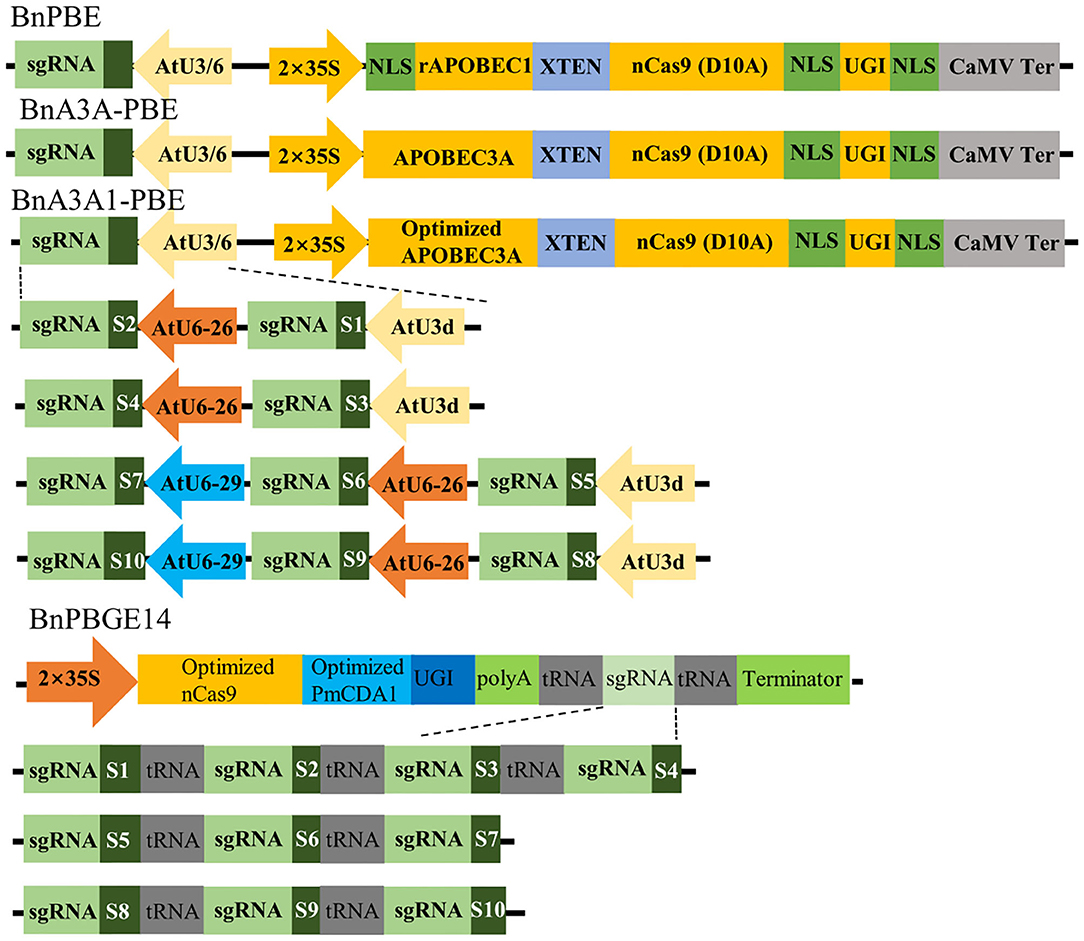
Figure 1. Schematic representation of four CBE systems and 10 sgRNAs used in this study. The main difference among BnPBE, BnA3A-PBE, BnA3A1-PBE, and BnPBGE1 is the sequence of cytidine deaminase. Ten sgRNAs were tested in each CBE system to compare the editing efficiency of the different systems.
To investigate the feasibility and efficacy of these CBE systems in rapeseed, we designed 10 sgRNAs for five endogenous genes: sgRNA1 (S1) and S2 for BnaCLV3, S3 for BnaRGA, S4 for BnaA3.IAA, S5 to S7 for BnaDA1, and S8 to S10 for BnaALS. Then, four constructs for each CBE system were generated and introduced separately into the rapeseed variety J9707 through Agrobacterium-mediated transformation. An average of 78 independent T0 transgenic lines were generated for each of the 15 CRISPR constructs (Table 1, Supplementary Table 2).
Base-editing of the generated plants was assessed by Hi-TOM through sequencing of the sgRNA target sites (Liu et al., 2019). The observed mutations at the S1 and S2 sites in this study were considered as one due to the overlapping of the sgRNA sequences. It showed that three of CBEs (BnPBE, BnA3A-PBE, and BnA3A1-PBE) were active at all sgRNAs, except BnPBE showed no editing at the S4-to-S7 sites (Figure 2). And the BnPBGE14 was inactive at all sgRNAs (Supplementary Table 2). BnA3A1-PBE had the highest base-editing efficiency, with an average editing efficiency of 29.8%, which is ~8.0% and fourfold higher than those of BnA3A-PBE (averaging 27.6%) and BnPBE (averaging 6.5%), respectively (Figure 2; Table 1). The C-to-T substitution efficiencies reached up to 50.5% in BnA3A1-PBE (Figure 2; Table 1), which is comparable with the efficiency in other crops (Qin et al., 2019). Recently, there were two reports about the successful application of different CBE systems in rapeseed [i.e., a PBE system (1.8% editing efficiency) reported by Wu et al. (2020) and an A3A-PBE system (averaging 23.6% editing efficiency) reported by Cheng et al. (2020)]. The performance of BnA3A1-PBE was much better than these two reported CBE systems, and thus represents the best CBE editor in rapeseed at present.
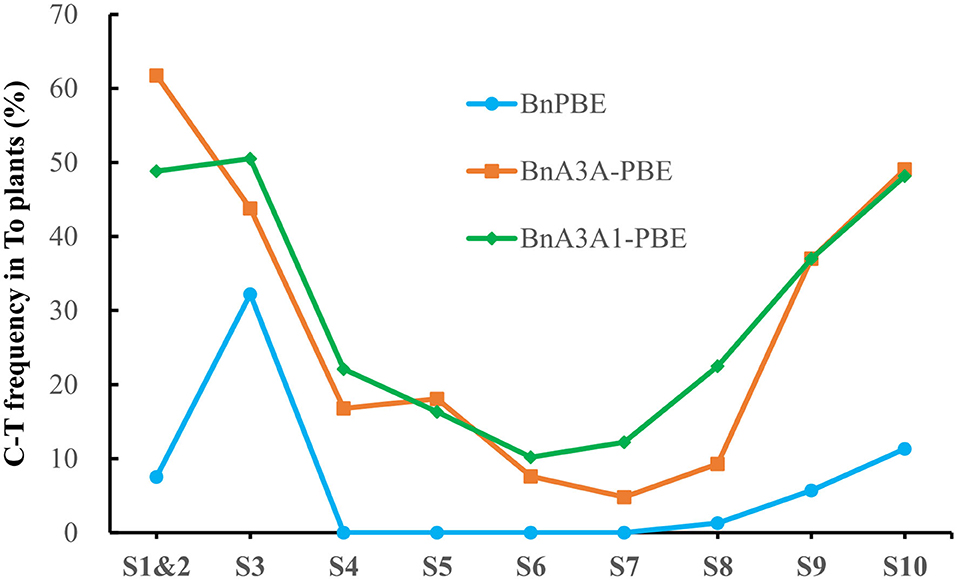
Figure 2. Frequencies of C-to-T conversion using three active CBE systems at 10 sgRNAs in T0 plants.
The overall base-editing efficiencies of the three CBE systems showed a similar trend at all sgRNAs [i.e., a higher editing efficiency at S1–S3, S9, and S10 and a lower editing efficiency at S4–S7 (Figure 2)]. In accordance with previous results, the difference in editing efficiency at these sgRNAs might be due to their nucleotide composition, GC content or promoter activities. Based on the fact that base editing with rAPOBEC1 is limited to a narrow deamination window and is inefficient in the GC contexts (Komor et al., 2016; Zong et al., 2018), this might be the reason why BnPBE has no editing activity at the S4–S7 sites.
Analyses of the base-editing efficiencies at every protospacer position across different sgRNAs revealed that the deamination window for BnA3A1-PBE spanned 17 nucleotides from protospacer positions 2–18, compared with 2–16 for BnA3A-PBE, and 3–8 for BnPBE (Figure 3), which is consistent with previous reports (Zong et al., 2018). Furthermore, we found that the on-target editing products were different among the active three CBE systems: BnA3A-PBE and BnA3A1-PBE preferred to substitute more C into T simultaneously compared with BnPBE. For example, at the S10 site, BnPBE created four types of mutations with one to three substitutions (C6, C6C7 or C6C7C8), while BnA3A-PBE and BnA3A1-PBE created six types of mutations, where simultaneous editing of three or four Cs (C6C7C8 and C6C7C8C10) occurred more frequently (Supplementary Table 3). These results suggested that there was an obvious difference in the main mutation genotypes between rat-APOBEC1-based BnPBE and human-APOBEC3A-based BnA3A-PBE or BnA3A1-PBE. Thus, BnA3A-PBE and BnA3A1-PBE could increase the production of novel alleles with diverse genetic variations because of their broad editing window. Whereas, BnPBE could reduce the possibility of introducing undesired mutations at specific sites because of the narrow editing window. However, BnPBE has a lower editing efficiency than BnA3A-PBE and BnA3A1-PBE (Figures 2, 3). Therefore, it is critical to fully understand the characteristics of these editing systems for better utilization.
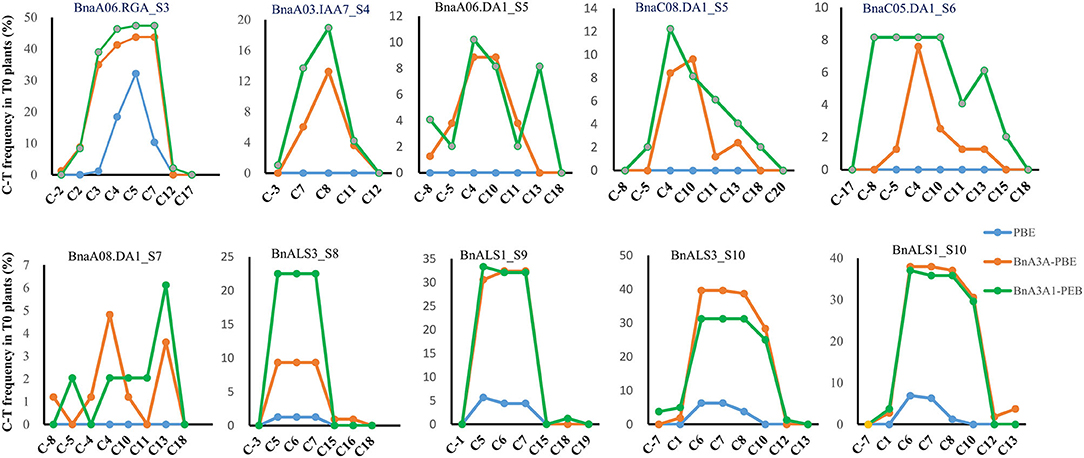
Figure 3. Frequencies of single C-to-T conversions by BnPBE, BnA3A-PBE, and BnA3A1-PBE at 10 target sites in T0 plants. Substitution efficiency was calculated by the ratio of the edited plants in the total transgenic positive plants.
By analyzing the ratio of different mutation genotypes in base-edited plants, the three active CBE systems produced mutants with a similar trend [i.e, heterozygous (Hetero) > homozygous (Homo) > biallelic (Bi) > chimerism (Ch) (Figure 4)]. Compared with BnPBE, BnA3A-PBE, and BnA3A1-PBE could significantly reduce the proportion of heterozygous genotypes and increase the proportion of both homozygous and biallelic genotypes (Figure 4).
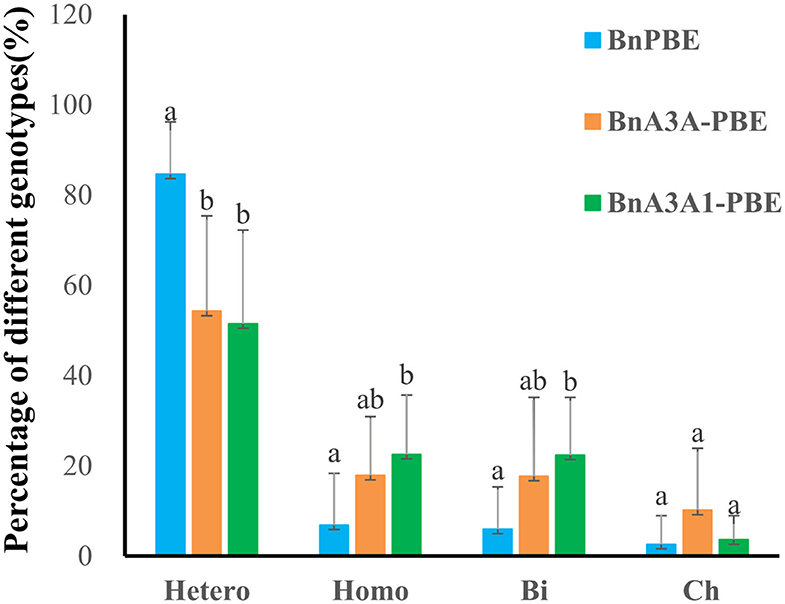
Figure 4. Percentage of four genotypes of base mutations in 10 targets using three CBE systems. Uppercase letters indicate a significant difference at the 0.05 probability level among the three CBE systems based on a multiple comparison test. Hetero, heterozygous; Homo, homozygous; Bi, biallelic; Ch, chimerism.
We compared the base-editing efficiencies of these sgRNA sites that could target both the A and C subgenomes of rapeseed in this study. The data showed that all the CBEs had uniform editing rate between the two subgenomes for six sgRNAs, whereas an obvious bias of base editing at S8 and S9 was observed in the C-subgenome (26.7% on average) compared to that in the A-subgenome (11.0% on average). More than 80% of base editing in BnPBE occurred at only C subgenomes, whereas more than 56.7 and 61.7% of base editing occurred simultaneously at both subgenomes for BnA3A-PBE and BnA3A1-PBE, respectively (Table 1).
The overall unexpected nucleotide changes and indels in the putative editing window occurred with much lower frequencies than C-to-T base editing, and BnA3A-PBE and BnA3A1-PBE yielded relatively higher frequencies of these undesired edits than that those observed in BnPBE (Table 1). This showed that the frequency of undesired edits was positively correlated with the editing efficiency of CBE systems.
To detect any potential off-target effects of the CBE systems reported here, we selected the target sites with the highest editing efficiency corresponding to the three active CBE systems to detect the off-target efficiency. There were 23 and four potential off-target sites identified for BnaCLV3 and BnaRGA sites, respectively, with up to 4-nucleotide mismatches (Bae et al., 2014; Liu et al., 2017). High-throughput sequencing of the PCR products of these potential off-target sites revealed that no significant difference was observed in the off-target ratio between the base-edited and wild-type plants (Supplementary Table 4). These results revealed that the three active CBE systems have a high specificity for targeted mutagenesis in rapeseed, which is consistent with previous reports in animals and plants (Kim et al., 2017; Qin et al., 2019; Cheng et al., 2020).
The S3 target site was fully matched with a functional copy of BnaA06.RGA (BnaA06g34810D) which encodes a DELLA protein, serving as a Gibberellins (GA) signaling repressor. A C-to-T substitution conferred a mutation (P91L) in its TVHYNP motif and resulted in a dwarf phenotype (Liu et al., 2010). Sequencing analysis revealed that 108 (41.2%) of the T0 plants contained a C-to-T substitution which occurred at C2, C3, C4, C5, C7, and C12 from the protospacer position at the S3 site (Table 1, Supplementary Table 3). A total of 14 different mutation genotypes were detected from the 108 edited lines in the T0 generation, and the homozygous substitutions at the conserved P91 in three different lines showed obvious dwarf phenotypes (Figure 5A).
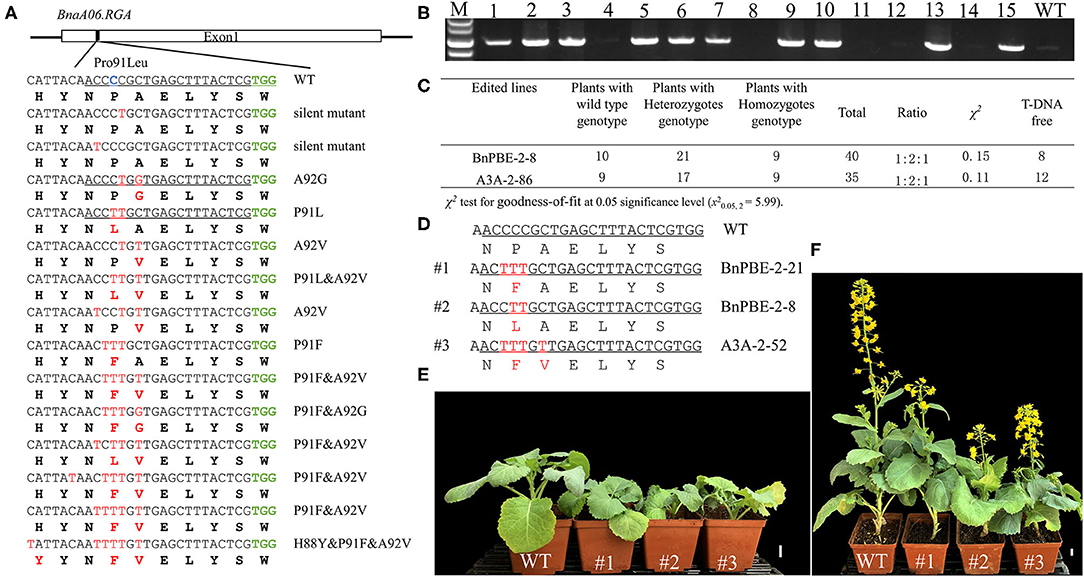
Figure 5. P91 substitution in BnaA06.RGA T0 mutants confer a dwarf phenotype. (A) Diverse editing events at the P91 site of BnaA06.RGA. Target sequence, expected substitution, PAM sequence and DNA or protein modifications are indicated by green, blue, and red text, respectively. (B) PCR analysis to detect the exogenous T-DNA in the T1 generation. (C) The genotypic segregation ratio of two edited lines in the T1 generation and the total number of plants with T-DNA free. The mutation genotypes of three edited lines (D) and their plant height were compared with wild type (WT) at seedling (E) and maturation stage (F). Scale bar, 2 cm.
To obtain stable homozygous mutants and test whether the base-editing mutants are inherited, two independent heterozygous lines (BnPBE3-2-8 and BnA3A-PBE-2-86) were self-pollinated. Then, their respective T1 progeny were genotyped via Hi-TOM sequencing at the S3 site. In the T1 progeny from the BnPBE3-2-8 and BnA3A-PBE-2-86 lines, the segregation ratio observed for the heterozygous, homozygous and wild type genotype was ~1:2:1 (χ2 = 0.15 and 0.11, P > 0.05; Figure 5C). These results indicated that the produced base substitution was successfully transmitted from T0 to T1 generation, with an expected monogenic segregation pattern. Furthermore, the PCR assay was performed to detect exogenous T-DNA using the PB-L/R primer pairs (Figure 5B). Twenty edited mutant plants, including seven homozygous mutants without exogenous T-DNA were obtained in the T1 generation (Figure 5C).
Indeed, several T1 plants with the expected P91L or novel P91F&A92V substitutions showed a decreased plant height compared with wild-type plants (Figures 5D–F). The significant reduction in height was due to a lower first branch position and shorter internodes compared with wild-type plants. Besides, we found that the heterozygous mutants also show a significant reduction in plant height. Previous report showed that the target substitution in BnaC09.RGA conserved domain generated dwarf phenotype (Cheng et al., 2020). Altogether, we can conclude that both the functional copies of BnaA06.RGA and BnaC09.RGA can achieve gain-of-function mutations at the conserved P91 through CBE system. The utilization of these semi-dwarf mutants produced in the study could improve the lodging resistance in rapeseed breeding.
In the edited plants, we were excited to obtain expected substitutions at the conserved P197 site of the acetolactate synthase gene (BnaALS) targeted by S10, which probably confer resistance to sulfonylurea herbicides (Li et al., 2015). Sequencing results revealed that 101 (29.0%) of the T0 plants contained a C-to-T substitution at the S10 site, which occurred at C1, C6, C7, C8, and C10 from the protospacer position (Table 1). Among these mutant plants, 57 harbored missense mutations in both functional copies of BnaALS, among which 21 and 23 had missense mutations in BnaC01g25380D (BnaALS1) and BnaA01g20380D (BnaALS3), respectively. Diverse editing events were detected at the target sites of BnaALS1 and BnaALS3 (Figure 6A). The expected amino acid substitution (P197S or P197L) was rarely detected in these mutants, whereas most of the editing lines carried P197F and P197F&R198C substitutions (Supplementary Table 3).

Figure 6. P197F substitution in BnaALS T0 mutants confer resistance to sulfonylureas herbicides. (A) Diverse co-editing events at the P197 site of BnaALS. The BnaALS-P197 site is conserved in the (A,C) subgenomes and 197 was numbered according to the corresponding sequence of Arabidopsis. The PAM were underlined in black line (B) Representative P197F substitution in BnaALS T0 mutants and their transmission to T1 generation. Hetero, heterozygous; Homo, homozygous; Bi, biallelic; WT, wild type. (C,D) Phenotypes of base-edited plants with different alleles and wild type before and after being tested with 200, 400, and 800 mg/L tribenuron-methyl herbicide in the T1 generation. “aaCC,” “AAcc,” and “aacc” represent homozygous mutations of the target gene in BnaALS3, BnaALS1 and both copies, respectively. Scale bar, 2 cm.
To test whether the observed mutations are stably inherited and obtain stable homozygous mutants, four independent T0 editing lines of BnaALS were self-pollinated to produce T1 progeny. The target mutations of progenies from these T0 lines were verified by Hi-TOM sequencing analysis of the target sites. As expected, the observed base substitutions were transmitted to the T1 generation, and different single and double mutants were obtained (Figure 6B).
To determine whether these P197 missense mutations in the BnaALS conferred sulfonylureas herbicide resistance, the T1 mutants with homozygous P197-substitutions at a single (AAcc, aaCC) or double (aacc) copies of BnaALS gene were treated with various field application levels of tribenuron-methyl at the four-leaf stage. The mutants carrying the P197F edited alleles grew better at 1, 2, and 4 times field-recommended rates (200, 400, and 800 mg/L) over three weeks compared with wild type (Figures 6C,D), the resistance of aacc mutant was the best, followed by AAcc and aaCC mutants at the field application levels of 200 to 800 mg/L sulfonylurea herbicides. Thus, both copies of BnaALS gene likely confer herbicide resistance with a similar effect and work in an additive manner (Figure 6D), which is different from the report that BnaALS3 confer better herbicide resistance than BnaALS1 (Cheng et al., 2020). Thus, the P197F substitution represents a novel allele which confers herbicide resistance in rapeseed.
There are five homologous copies of BnaIAA7 in the B. napus genome, and the S4 was designed to fully target four out of the five gene copies, with BnaC05.IAA7 having one base mismatched (Supplementary Figure 1A). A G-to-A mutation changed the glycine at the 84th position to glutamic acid (G84E) in BnaA3.IAA7, which contributes to reducing the length of internodes and branch angles in rapeseed (Li et al., 2019). As expected, this substitution caused the conversion of the conserved Gly84 to glutamic acid (Supplementary Figure 1B). However, all the edited plants carry a G-to-A mutation at C8, which results in the insertion of a stop codon at the 85th position (Supplementary Figure 1B). Our results suggested that base editing can also be utilized to create knockout mutations by insertion of a stop codon which results in premature termination.
In the B. napus genome, there are four copies of BnaDA1 gene. S5 was designed to target both BnaA06.DA1 and BnaC08.DA1, while S6 and S7 were designed to target BnaC05.DA1 and BnaA08.DA1, respectively. We designed these targets intending to obtain base substitutions at the conserved Arg358 of the BnaDA1 (DA means big in Chinese) targeted by S5–S7. The A358K conversion of BnaDA1 probably contributes to the improvement of seed weight in rapeseed (Wang et al., 2017). S8 and S9 were designed to target BnaALS3 and BnaALS1, respectively. The alanine at position 122 of ALS is converted to valine, which endow mutant resistance to imidazolinone herbicide (Li et al., 2008; Sala et al., 2008; Han et al., 2012). In the T0 edited plants, only 11 edited plants contained the intended R358K conversion at S5–S7 and only 1 edited plant contained the intended A122V conversion at S8–S9 since the target bases (C13 or C15) are located outside of the hot spot of the deamination window (Figure 3; Supplementary Table 5). Thus, it is imperative to develop an engineered SpCas9 variant that recognizes not only the NGG protospacer adjacent motif (PAM), to ensure that the desired target bases are located within the hot spot of the editing window. The seed weight of the 11 edited plants containing the intended R358K conversion at S5–S7 will be tested in the next generation since there were limited seeds to conduct a field experiment in the T1 plants.
Streptococcus pyogenes Cas9 (SpCas9) recognizes a very simple NGG PAM, making it the most commonly used CRISPR-Cas9 system. The canonical NGG PAM limits its targeting scope in a genome, especially for applications that require precise Cas9 positioning such as base editing (Wang et al., 2019). Recently, the engineered SpCas9 variant, SpCas9-NG, which recognize NG PAMs are more efficient than the xCas9 variant (Hu et al., 2018; Nishimasu et al., 2018), Moreover, SpCas9-NG coupled with the activation-induced cytidine deaminase (AID) can mediate the C-to-T conversion at target sites with NG PAMs in human cells (Nishimasu et al., 2018). Endo et al. (2019) reported that the SpCas9-NG can efficiently mutagenize endogenous target sites with NG PAMs in rice and Arabidopsis genomes. For B. napus genome, the 17-nt editing window of BnA3A1-PBE theoretically increases up to 15.3% and 1.3-fold the number of genomic cytidines and guanidine available for base editing when compared to BnA3A-PBE and BnPBE, respectively (Figure 7). When combined with SpCas9, xCas9, and other variants with NG PAM, BnA3A1-PBE theoretically targets 93% of the cytidines and guanidine in the rapeseed genome (Figure 7), which makes it as an ideal editing system for further improvement in future research.
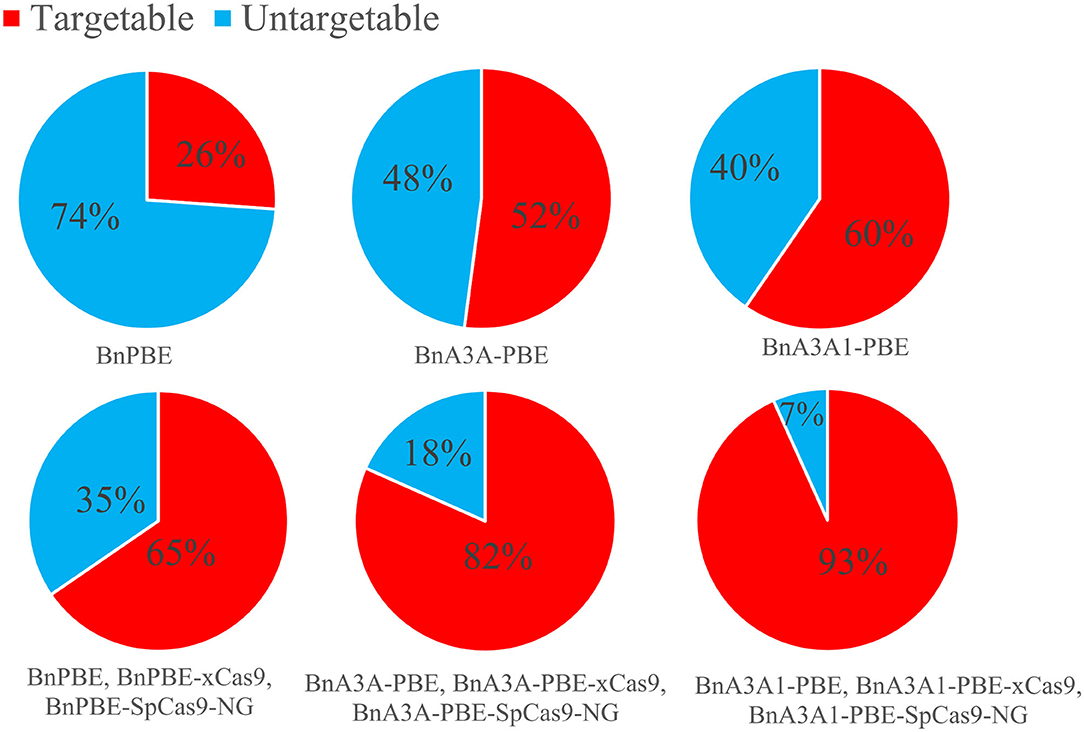
Figure 7. Computational analysis of the rapeseed genome revealed that Cytidines (NGG PAM) and guanidines (CCN PAM) can potentially be edited by BnPBE, BnA3A-PBE, and BnA3A1-PBE. The above three CBEs together with the various Cas9 variants (SpCas9 and xCas9) promise to increase the scope of base editing of targeted cytidines and guanidines in rapeseed genome.
In conclusion, the three CBEs described here can efficiently and specifically perform precise C-to-T substitutions across a broad range of endogenous genomic loci in rapeseed. The improved BnA3A1-PBE performed efficiently as a base editor with higher editing efficiency, a more broadened editing window coupled with a higher proportion of homozygous and biallelic genotypes compared with BnA3A-PBE and BnPBE systems. When compared with the two latest studies which reported the successful application of cytosine base-editing in rapeseed (Wu et al., 2020, Cheng et al., 2020), BnA3A1-PBE has a high editing efficiency, which is ~16-fold than PBE (Wu et al., 2020), and 26% higher than those of A3A-PBE (Cheng et al., 2020). Thus, BnA3A1-PBE represents a powerful base editor for both gene function study and molecular breeding in rapeseed.
Sequencing data has been deposited in NCBI Sequence Read Archive (SRA) database under the accession no. PRJNA663415.
LH, CF, and YZho: conceived and designed the experiments. LH, QL, OA, SC, MZ, XS, KY, YZha, YY, and LX: performed the experiments. LH and CF: wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (Nos. 31371240 and 31671279). This study was also supported by Grants 2015CB150200 from the National Key Basic Research Program of China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Caixia Gao of the Institute of Genetics and Developmental Biology, CAS, for her kind gift of the plasmid A3A-PBE for this research.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeed.2020.605768/full#supplementary-material
Supplementary Figure 1. The desired substitution accompany with a stop codon occurred at the S4 target site. (A) The target sequences are shown with the PAM underlined in red line. (B) Diverse co-editing events at the BnaIAA7-Gly84 site, all of these new alleles introduce an early stop codon.
Supplementary Table 1. The primers used in this study.
Supplementary Table 2. Detail information on the numbers of T0 plants in BnPBG14 system.
Supplementary Table 3. Detail information of the mutation of all edited T0 plants in this study.
Supplementary Table 4. Summary of deep sequencing data for most likely off-target sites of S1, S2, and S3.
Supplementary Table 5. The edited lines contain the intended C13 to T13 conversion at S5-S7.
Bae, S., Park, J., and Kim, J. S. (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. doi: 10.1093/bioinformatics/btu048
Braatz, J., Harloff, H. J., Mascher, M., Stein, N., Himmelbach, A., and Jung, C. (2017). CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174, 935–942. doi: 10.1104/pp.17.00426
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A. P., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Cheng, H., Hao, M., Ding, B., Mei, D., Wang, W., Wang, H., et al. (2020). Base editing with high efficiency in allotetraploid oilseed rape by A3A-PBE base-editing system. Plant Biotechnol. J. doi: 10.1111/pbi.13444. [Epub ahead of print].
Endo, M., Mikami, M., Endo, A., Kaya, H., Itoh, T., Nishimasu, H., et al. (2019). Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM. Nat. Plants. 5, 14–17. doi: 10.1038/s41477-018-0321-8
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi: 10.1038/nature24644
Han, H., Yu, Q., Purba, E., Li, M., Walsh, M., Friesen, S., et al. (2012). A novel amino acid substitution Ala-122-Tyr in ALS confers high-level and broad resistance across ALS-inhibiting herbicides. Pest Manag. Sci. 68, 1164–1170. doi: 10.1002/ps.3278
Hu, J. H., Miller, S. M., Geurts, M. H., Tang, W., Chen, L., Sun, N., et al. (2018). Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. doi: 10.1038/nature26155
Kim, D., Lim, K., Kim, S. T., Yoon, S. H., Kim, K., Ryu, S. M., et al. (2017). Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 35, 475–480. doi: 10.1038/nbt.3852
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. doi: 10.1038/nature17946
Li, D., Barclay, I., Jose, K., Stefanova, K., and Appels, R. (2008). A mutation at the Ala122 position of acetohydroxyacid synthase (AHAS) located on chromosome 6D of wheat: improved resistance to imidazolinone and a faster assay for marker-assisted selection. Mol. Breeding 22, 217–225. doi: 10.1007/s11032-008-9168-4
Li, H., Li, J., Song, J., Zhao, B., Guo, C., Wang, B., et al. (2019). An auxin signaling gene BnaA3.IAA7 contributes to improved plant architecture and yield heterosis in rapeseed. New Phytol. 222, 837–851. doi: 10.1111/nph.15632
Li, H., Li, J., Zhao, B., Wang, J., Yi, L., Liu, C., et al. (2015). Generation and characterization of tribenuron-methyl herbicide-resistant rapeseed (Brasscia napus) for hybrid seed production using chemically induced male sterility. Theor. Appl. Genet. 128, 107–118. doi: 10.1007/s00122-014-2415-7
Li, J., Sun, Y., Du, J., Zhao, Y., and Xia, L. (2017). Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant. 10, 526–529. doi: 10.1016/j.molp.2016.12.001
Liu, C., Wang, J., Huang, T., Wang, F., Yuan, F., Cheng, X., et al. (2010). A missense mutation in the VHYNP motif of a DELLA protein causes a semi-dwarf mutant phenotype in Brassica napus. Theor. Appl. Genet. 121, 249–258. doi: 10.1007/s00122-010-1306-9
Liu, H., Ding, Y., Zhou, Y., Jin, W., Xie, K., and Chen, L. L. (2017). CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant. 10, 530–532. doi: 10.1016/j.molp.2017.01.003
Liu, Q., Wang, C., Jiao, X., Zhang, H., Song, L., Li, Y., et al. (2019). Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7. doi: 10.1007/s11427-018-9402-9
Lu, Y., and Zhu, J. (2017). Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant. 10, 523–525. doi: 10.1016/j.molp.2016.11.013
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Nishida, K., Arazoe, T., Yachie, N., Banno, S., Kakimoto, M., Tabata, M., et al. (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353:aaf8729. doi: 10.1126/science.aaf8729
Nishimasu, H., Shi, X., Ishiguro, S., Gao, L., Hirano, S., Okazaki, S., et al. (2018). Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262. doi: 10.1126/science.aas9129
Qin, L., Li, J., Wang, Q., Xu, Z., Sun, L., Alariqi, M., et al. (2019). High efficient and precise base editing of C· G to T· A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. J. 18, 45–56. doi: 10.1111/pbi.13168
Ren, B., Yan, F., Kuang, Y., Li, N., Zhang, D., Lin, H., et al. (2017). A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice. Sci. China Life Sci. 60, 516–519. doi: 10.1007/s11427-016-0406-x
Russell, W. L., Russell, L. B., and Kelly, E. (1958). Radiation dose rate and mutation frequency. Science 128, 1546–1550. doi: 10.1126/science.128.3338.1546
Sala, C., Bulos, M., Echarte, M., Whitt, S. R., and Ascenzi, R. (2008). Molecular and biochemical characterization of an induced mutation conferring imidazolinone resistance in sunflower. Theor. Appl. Genet. 118, 105–112. doi: 10.1007/s00122-008-0880-6
Sega, G. A. (1984). A review of the genetic effects of ethyl methanesulfonate. Mutat. Res. 134, 113–142. doi: 10.1016/0165-1110(84)90007-1
Shimatani, Z., Kashojiya, S., Takayama, M., Terada, R., Arazoe, T., Ishii, H., et al. (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443. doi: 10.1038/nbt.3833
Tang, X., Ren, Q., Yang, L., Bao, Y., Zhong, Z., He, Y., et al. (2018). Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol. J. 17, 1431–1445. doi: 10.1111/pbi.13068
Wang, J., Tang, M., Chen, S., Zhen, X., Mo, H., Li, S., et al. (2017). Down-regulation of BnDA1, whose gene locus is associated with the seeds weight, improves the seeds weight and organ size in Brassica napus. Plant Biotechnol. J. 15, 1024–1033. doi: 10.1111/pbi.12696
Wang, M., Wang, Z., Mao, Y., Lu, Y., Yang, R., Tao, X., et al. (2019). Optimizing base editors for improved efficiency and expanded editing scope in rice. Plant Biotechnol. J. 17, 1697–1699. doi: 10.1111/pbi.13124
Woodfield, H. K., Sturtevant, D., Borisjuk, L., Munz, E., Guschina, I. A., Chapman, K., et al. (2017). Spatial and temporal mapping of key lipid species in Brassica napus seeds. Plant Physiol. 173, 1998–2009. doi: 10.1104/pp.16.01705
Wu, J., Chen, C., Xian, G., Liu, D., Lin, L., Yin, S., et al. (2020). Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnol. J. doi: 10.1111/pbi.13368
Yang, Y., Zhu, K., Li, H., Han, S., Meng, Q., Khan, S. U., et al. (2018). Precise editing of CLAVATA genes in Brassica napus, L. regulates multilocular silique development. Plant Biotechnol. J. 16, 1322–1335. doi: 10.1111/pbi.12872
Zhai, Y., Yu, K., Cai, S., Hu, L., Amoo, O., Xu, L., et al. (2020). Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 18, 1153–1168. doi: 10.1111/pbi.13281
Zhou, Y., and Fowke, L. C. (2002). Control of petal and pollen development by the plant cyclin-dependent kinase inhibitor ICK1 in transgenic Brassica plants. Planta 215, 248–257. doi: 10.1007/s00425-002-0752-2
Zong, Y., Song, Q., Li, C., Jin, S., Zhang, D., Wang, Y., et al. (2018). Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 36, 950–953. doi: 10.1038/nbt.4261
Keywords: Brassica napus, base-editing, cytidine deaminase, gain-of-function mutation, crop improvement
Citation: Hu L, Amoo O, Liu Q, Cai S, Zhu M, Shen X, Yu K, Zhai Y, Yang Y, Xu L, Fan C and Zhou Y (2020) Precision Genome Engineering Through Cytidine Base Editing in Rapeseed (Brassica napus. L). Front. Genome Ed. 2:605768. doi: 10.3389/fgeed.2020.605768
Received: 13 September 2020; Accepted: 27 October 2020;
Published: 20 November 2020.
Edited by:
Lanqin Xia, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Hongliang Zhu, China Agricultural University, ChinaCopyright © 2020 Hu, Amoo, Liu, Cai, Zhu, Shen, Yu, Zhai, Yang, Xu, Fan and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuchuan Fan, ZmFuY2h1Y2h1YW5AbWFpbC5oemF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.