
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Genet., 11 March 2025
Sec. Human and Medical Genomics
Volume 16 - 2025 | https://doi.org/10.3389/fgene.2025.1581299
This article is part of the Research TopicUnderstanding Molecular Mechanisms to Facilitate the Development of Biomarkers for Therapeutic Intervention in Gastrointestinal Diseases and SepsisView all 11 articles
Editorial on the Research Topic
Understanding molecular mechanisms to facilitate the development of biomarkers for therapeutic intervention in gastrointestinal diseases and sepsis
Gastrointestinal (GI) disorders include a range of pathological conditions with varying severities and outcomes that impact the integrity and function of the GI tract. These conditions include indigestion, the inflammatory bowel diseases (IBDs) ulcerative colitis (UC) and Crohn’s disease (CD), and malignant tumors. The resulting dysfunction of the intestinal barrier, leading to impaired permeability, allows for the translocation of luminal contents, including intact microbes and microbial products. This situation can cause severe sepsis and potentially fatal outcomes if timely intervention is not provided. Advancements in omics technology have facilitated the identification and evaluation of molecular biomarkers for disease diagnosis. These encompass genomic (e.g., single nucleotide polymorphisms), transcriptomic (including non-coding RNAs), epigenetic (e.g., DNA methylation), proteomic, metabolomic, and microbiome biomarkers (Dalal et al., 2020; Sahoo et al., 2024a; Sahoo et al., 2022b; Figure 1). These biomarkers hold significant clinical potential for improving diagnosis, prognosis, and treatment strategies in patients with GI disorders and sepsis.
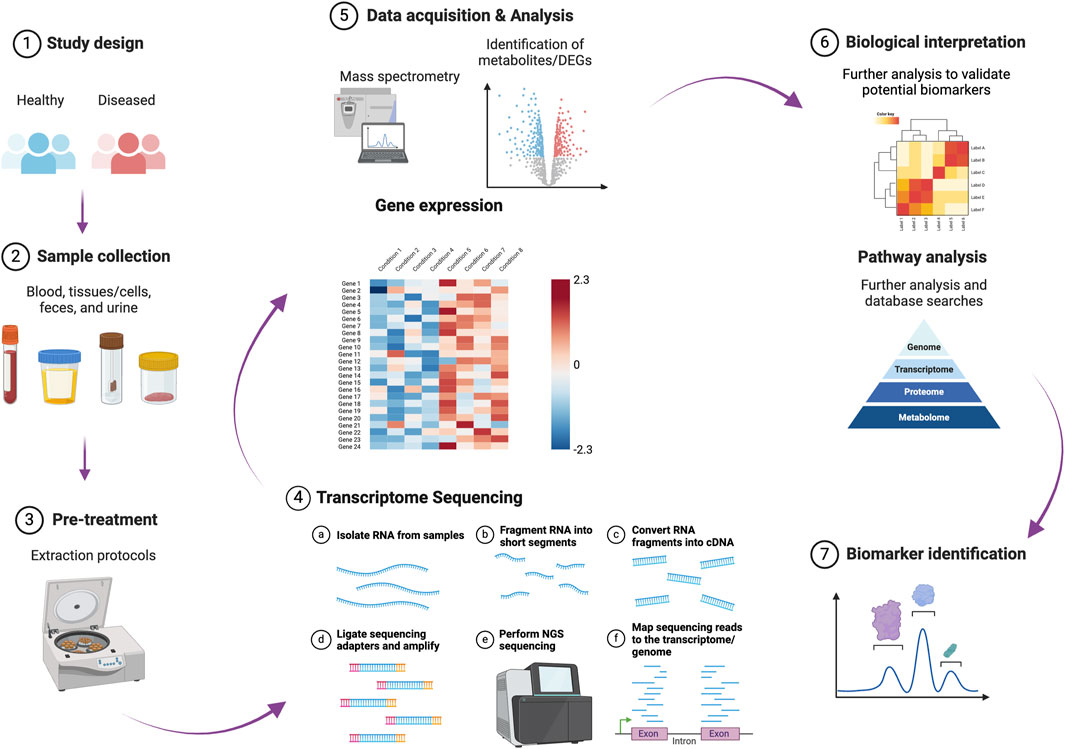
Figure 1. A schematic representation of the different phases involved in identifying and validating molecular biomarkers for disease diagnosis and prognosis. This figure was created in BioRender (https://BioRender.com/d22b503). DEGs: differentially expressed genes.
For diagnosing sepsis, in addition to lactate as a widely utilized biomarker, other surrogate markers, such as C-reactive protein (CRP) and procalcitonin, which are produced in response to infection and inflammation, may assist in identifying patients at risk of developing severe sepsis before significant organ dysfunction occurs (Faix, 2013). As oxidative stress (OS) plays a key role in the progression of sepsis and septic shock to multiple organ failure (Sahoo et al., 2024b; Wong et al., 2025), OS markers associated with sepsis, specifically, superoxide dismutase, soluble endoglin, asymmetric dimethylarginine, and neopterin, merit further clinical investigation (Helan et al., 2022). Several emerging biomarkers such as microRNA-486-5p, circular RNAs (circRNAs), HOXA distal transcript antisense RNA (a lncRNA located on chromosome 7q15.2), protein C (a vitamin K-dependent glycoprotein), prokineticin 2, and triiodothyronine also hold potential for enhancing early detection and prognostic assessment of sepsis with high sensitivity and specificity (He et al., 2024).
Mendelian randomization (MR) examines the causal effects of modifiable exposures, such as potential risk factors, on health by utilizing genetic variants linked to those specific exposures (Richmond and Smith, 2022). MR analysis by Zhang et al. showed that CMPF (3-carboxy-4-methyl-5-propyl-2-furanpropanoate) has an association with 28-day all-cause mortality in clinical cases of sepsis. The metabolic pathway of alpha-linolenic acid and linoleic acid was identified as a crucial factor in the development and progression of sepsis. The study by Ye et al. demonstrated that levels of soluble suppression of tumorigenicity 2 (sST2) in the blood have significant clinical diagnostic and prognostic implications in sepsis. Moreover, sST2 showed a comparable predictive capability to the SOFA (Sequential Organ Failure Assessment) and APACHE II (Acute Physiology and Chronic Health Evaluation II) scores and had a greater predictive capability than lactic acid levels in assessing the prognosis of patients with sepsis. Jin et al. used single-cell RNA sequencing (scRNA-seq) and identified several diagnostic markers for sepsis, such as PIM1 (proviral integration site for Moloney murine leukemia virus kinase 1), HIST1H1C (Histone Cluster 1 H1 Family Member C), and IGSF6 (Immunoglobulin Superfamily Member 6). The involvement of PIM1 in modulating the immune-inflammatory response during sepsis was verified through experimental validation, indicating that PIM1 is a promising novel therapeutic target.
The endoscopic evaluation of patients suspected of IBD with the collection of mucosal biopsies for histopathological confirmation continues to be the gold standard for establishing an IBD diagnosis, assessing treatment efficacy, and identifying post-operative recurrence; however, it is associated with high costs and invasiveness. Biomarkers enable non-invasive disease evaluation, with C-reactive protein and fecal calprotectin being the most frequently utilized biomarkers in current clinical practice. The T>C substitution SNPs that affect the functionality of the DLG5 (discs large homolog 5) protein, along with the lack of CARD15/NOD2 (caspase recruitment domain family number 15/nucleotide-binding oligomerization domain-containing protein 2) SNPs associated with CD pathogenesis, could serve as genomic biomarkers (Dudzińska et al., 2018). Several potential microbiome biomarkers (microbial markers) have been reported. For example, Faecalibacterium prausnitzii and its phylogroups and elevated Escherichia coli counts serve as potential biomarkers for CD diagnosis. Akkermansia muciniphila is identified for pediatric CD diagnosis. Additionally, reductions in Firmicutes (Clostridiales) levels correlate with IBD severity, while increased abundance of Lactobacilllaceae and Enterococcaceae families, as well as the genera Lactobacillus, Enterococcus, and Eggerthella, are noted in UC patients (Wang et al., 2024). In dogs with CIE, there is a notable dysbiotic profile in both luminal and mucosal intestinal bacteria, marked by a reduction in Clostridium and Bacteroides and an increase in Enterobacteriaceae (Sahoo et al., 2022a).
The regulation of gene expression mediated by microRNA (miRNA) is essential for the appropriate development and functioning of the intestine (Shanahan et al., 2021). Numerous studies have effectively identified unique miRNA profiles that indicate the upregulation or downregulation of one or more miRNAs in intestinal biopsy samples from patients with IBD (James, 2020) and dogs affected with chronic inflammatory enteropathy (CIE) (Sahoo et al., 2024a). The colonic mucosa of patients with active UC was shown to overexpress specific miRNAs, including miR-21, miR-150, and miR-155, and have a reduction in miRNAs like miR-143 and miR-145 when compared to healthy controls (Alghoul et al., 2022). Comparing the colonic mucosa of patients with active CD and healthy controls, there was an upregulation of miR-196 and a downregulation of miR-7 (Alghoul et al., 2022). The differential expression of miRNAs in saliva, blood, and colon tissue samples was analyzed in UC and CD patients (Schaefer et al., 2015). This research highlighted multiple miRNAs (specifically, miR-21, miR-31, miR-142-3p, miR-142-5p) with expression levels exhibiting significant changes across all three sample types when comparing IBD patients to non-IBD controls (Schaefer et al., 2015). Recent studies indicate that DNA methylation of specific genes contributes to the pathogenesis of IBD, implying their potential utility as clinical biomarkers (Cooke et al., 2012; Nimmo et al., 2012). A comprehensive analysis of methylation patterns across the genome, performed on rectal mucosal biopsies, revealed specific differential gene signatures, including Fanconi anemia complementation group (FANCC), thyroid hormone receptor-associated protein 2 (THRAP2), globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 (GBGT1), tumor necrosis factor ligand superfamily member 4 (TNFSF4), TNF superfamily member 12 (TNFSF12), docking protein 2 (DOK2), and fucosyltransferase 7 (FUT7). These genes exhibited notable differences in methylation levels in specimens from patients with CD or UC compared to healthy individuals (Cooke et al., 2012). Response to infliximab treatment in patients with IBD resulted in notable decreases in macrophage-derived cluster of differentiation 14 (CD14) and CD86 levels, along with the chemokine CCL2 (Magnusson et al., 2015), highlighting their potential as surrogate biomarkers to monitor IBD patients during treatment.
The research by Song et al. employing MR suggests that interleukin-13 (IL-13) contributes to the pathophysiology of IBD (CD and UC). Whereas macrophage migration inhibitory factor appears to be specifically related to CD, stem cell factor is more likely to play a role in the progression of IBD (CD and UC). Another MR study by Qian et al. showed that patients with gastric cancer exhibit decreased blood levels of tryptophan, nonadecanoate (19:0), and erythritol. Zhu et al. employed MR and demonstrated that the genes GPBAR1 (G protein-coupled bile acid receptor 1), IL1RL1 (Interleukin 1 receptor-like 1), PRKCB (Protein Kinase C Beta), and PNMT (Phenylethanolamine N-Methyltransferase) are linked to an increased risk of IBD. Whereas IL1RL1 was shown to have a protective effect against the risk of CD, GPX1 (Glutathione peroxidase 1), GPBAR1, and PNMT are implicated in the risk of UC. In a scRNA-seq study by Keever-Keigher et al., common expression patterns were observed in GI disorders, including an extensive upregulation of MTRNR2L8 (MT-RNR2 Like 8) across various cell types. The increase of XIST (X Inactive Specific Transcript) expression across different cell types in individuals with UC and an elevated expression of Th2 (T helper 2)-associated genes in eosinophilic disorders is also noteworthy.
Colorectal cancer (CRC) is a common GI neoplasia. To facilitate the early detection of CRC, a minimally invasive and reproducible technique known as liquid biopsy (LB) has been established. This method isolates cancer-derived components from the patients’ peripheral blood and/or other body fluids, including circulating tumor cells (CTC), miRNA, long non-coding RNAs (lncRNAs), and circulating tumor DNA (ctDNA) (Zhang et al., 2023). Heat-shock protein 27 (Hsp27) has been identified as expressed explicitly in well-differentiated CRC and is linked to other significant CRC biomarkers, such as epidermal growth factor receptor (EGFR), tumor necrosis factors, protein kinase B (AKT), and human epidermal growth factor receptor 2 (ERBB2) (Gan et al., 2014). Glutathione S-transferase pi1 (GSTP1) and KTR8 were overexpressed in both well-differentiated and poorly differentiated CRCs. In contrast, triosephosphate isomerase (TPI), tubulin beta chain (TUBB), and fatty acid-binding protein (FABP1) were upregulated exclusively in well-differentiated CRCs. Human leukocyte antigen A (HLA-A) was observed to be increased in poorly differentiated CRC (Gan et al., 2014). Circular RNAs (circRNAs) such as hsa_circ_001978, hsa_circ_103627, hsa_circ_105039, and circ_0124554 may also serve as indicators for a diagnosis of CRC. Levels of serum miR-21, miR-29a, and miR-125b can potentially differentiate patients with early colorectal neoplasia from healthy ones (Yamada et al., 2015). Serum miRNAs, such as miR-21, miR-92a, miR-182S, and miR-223, along with other miRNAs like miR-17–5p, miR-18a–5p, miR-18b–5p, miR-103a–3p, miR-127–3p, miR-151a–5p, and miR-181a–5p may have clinical utility as biomarkers in the non-invasive screening for CRC (Zhang et al., 2023). CircRNAs have the potential to synergize with various proteins or RNAs, demonstrated by circ_0000523 and methyltransferase-like 3 (METTL3), to enhance the accuracy of a CRC diagnosis (Wang et al., 2022).
Carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) currently serve as the primary serum tumor markers for assessing the prognosis of CRC. The research conducted by Dai et al. indicates that both the overall survival rate and the disease-free survival rate in patients with CRC progressively decline with an increasing number of positive tumor markers before and after surgical intervention. Nomograms utilizing pre-and postoperative CEA and CA19-9 demonstrate high accuracy in predicting survival and recurrence for stage I-III CRC patients following radical surgery, significantly outperforming the American Joint Committee on Cancer (AJCC) 8th Tumor-Node-Metastasis (TNM) stage. The research by Rad et al. highlights the significant impact of machine learning algorithms on predicting CRC recurrence, specifically by examining the least number of serial CEA measurements necessary for accurate prediction of recurrence. The research conducted by Pan et al. involved a retrospective analysis of clinical data from 36,708 patients who underwent gastroscopy and colonoscopy between 2005 and 2022. Conventional adenomas (CAs), serrated polyps (SPs), non-adenomatous polyps (NAPs), and CRC were all associated with an increased risk of Helicobacter pylori infection and older age. The presence of moderate to severe intestinal metaplasia was associated with an increased risk of NAP and CAs. The risk of CRC was found to be increased with low-grade intraepithelial neoplasia, whereas gastric cancer was linked to high-grade intraepithelial neoplasia. A correlation was also observed between advanced gastric pathology and an increased risk of CRC.
In individual patients without clinical signs, biomarkers or biomarker panels have the potential to serve as a significant resource for screening to identify cancer at an early stage or recognize precancerous conditions. For symptomatic patients, these biomarkers can help differentiate cancerous from benign states. Furthermore, in cancer patients undergoing treatment such as surgical procedures, radiation therapy, and/or chemotherapy, surrogate disease biomarkers are valuable tools for evaluating the success of tumor elimination (complete resection and remission) and the potential for disease recurrence. Identifying and validating optimal biomarkers and biomarker panels is a crucial step, as it offers considerable potential for enhancing personalized medicine and overall clinical outcomes. However, further research into the specificity of molecular biomarkers for sepsis, IBD, and CRC is necessary before they can be utilized as diagnostic tools in clinical practice.
DS: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. RH: Conceptualization, Resources, Writing–original draft, Writing–review and editing. AP: Conceptualization, Resources, Writing–original draft, Writing–review and editing.
The author(s) declare that no financial support was received for the research and/or publication of this article.
DKS acknowledges the support of the Department of Veterinary Clinical Sciences (VCS) Core Lab, College of Veterinary Medicine, Iowa State University, Ames, Iowa, United States.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alghoul, Z., Yang, C., and Merlin, D. (2022). The current status of molecular biomarkers for inflammatory bowel disease. Biomedicines 10, 1492. doi:10.3390/BIOMEDICINES10071492
Cooke, J., Zhang, H., Greger, L., Silva, A. L., Massey, D., Dawson, C., et al. (2012). Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm. Bowel Dis. 18, 2128–2137. doi:10.1002/IBD.22942
Dalal, N., Jalandra, R., Sharma, M., Prakash, H., Makharia, G. K., Solanki, P. R., et al. (2020). Omics technologies for improved diagnosis and treatment of colorectal cancer: technical advancement and major perspectives. Biomed. and Pharmacother. 131, 110648. doi:10.1016/J.BIOPHA.2020.110648
Dudzińska, E., Gryzinska, M., and Kocki, J. (2018). Single nucleotide polymorphisms in selected genes in inflammatory bowel disease. Biomed. Res. Int. 2018, 6914346. doi:10.1155/2018/6914346
Faix, J. D. (2013). Biomarkers of sepsis. Crit. Rev. Clin. Lab. Sci. 50, 23–36. doi:10.3109/10408363.2013.764490
Gan, Y., Chen, D., and Li, X. (2014). Proteomic analysis reveals novel proteins associated with progression and differentiation of colorectal carcinoma. J. Cancer Res. Ther. 10, 89–96. doi:10.4103/0973-1482.131396
He, R. R., Yue, G. L., Dong, M. L., Wang, J. Q., and Cheng, C. (2024). Sepsis biomarkers: advancements and clinical applications—a narrative review. Int. J. Mol. Sci. 25, 9010–9025. doi:10.3390/IJMS25169010
Helan, M., Malaska, J., Tomandl, J., Jarkovsky, J., Helanova, K., Benesova, K., et al. (2022). Kinetics of biomarkers of oxidative stress in septic shock: a pilot study. Antioxidants 11, 640. doi:10.3390/ANTIOX11040640
Magnusson, M. K., Strid, H., Isaksson, S., Bajor, A., Lasson, A., Ung, K. A., et al. (2015). Response to infliximab therapy in ulcerative colitis is associated with decreased monocyte activation, reduced CCL2 expression and downregulation of Tenascin C. J. Crohns Colitis 9, 56–65. doi:10.1093/ECCO-JCC/JJU008
Nimmo, E. R., Prendergast, J. G., Aldhous, M. C., Kennedy, N. A., Henderson, P., Drummond, H. E., et al. (2012). Genome-wide methylation profiling in Crohn’s disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm. Bowel Dis. 18, 889–899. doi:10.1002/IBD.21912
Richmond, R. C., and Smith, G. D. (2022). Mendelian randomization: concepts and scope. Cold Spring Harb. Perspect Med. 12, a040501. doi:10.1101/CSHPERSPECT.A040501
Sahoo, D. K., Allenspach, K., Mochel, J. P., Parker, V., Rudinsky, A. J., Winston, J. A., et al. (2022a). Synbiotic-IgY therapy modulates the mucosal microbiome and inflammatory indices in dogs with chronic inflammatory enteropathy: a randomized, double-blind, placebo-controlled study. Veterinary Sci. 10, 25. doi:10.3390/VETSCI10010025
Sahoo, D. K., Borcherding, D. C., Chandra, L., Jergens, A. E., Atherly, T., Bourgois-Mochel, A., et al. (2022b). Differential transcriptomic profiles following stimulation with lipopolysaccharide in intestinal organoids from dogs with inflammatory bowel disease and intestinal mast cell tumor. Cancers (Basel) 14, 3525. doi:10.3390/cancers14143525
Sahoo, D. K., Heilmann, R. M., Ackermann, M., Parker, V., Rudinsky, A., Winston, J. A., et al. (2024a). Tu1739 MicroRNAs as potential biomarkers for diagnosis and monitoring chronic inflammatory enteropathy in dogs. Gastroenterology 166, S-1401–1401. doi:10.1016/S0016-5085(24)03676-X
Sahoo, D. K., Wong, D., Patani, A., Paital, B., Yadav, V. K., Patel, A., et al. (2024b). Exploring the role of antioxidants in sepsis-associated oxidative stress: a comprehensive review. Front. Cell Infect. Microbiol. 14, 1348713. doi:10.3389/fcimb.2024.1348713
Schaefer, J. S., Attumi, T., Opekun, A. R., Abraham, B., Hou, J., Shelby, H., et al. (2015). MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 16, 5. doi:10.1186/S12865-015-0069-0
Shanahan, M. T., Kanke, M., Oyesola, O. O., Hung, Y. H., Koch-Laskowski, K., Singh, A. P., et al. (2021). Multiomic analysis defines the first microRNA atlas across all small intestinal epithelial lineages and reveals novel markers of almost all major cell types. Am. J. Physiol. Gastrointest. Liver Physiol. 321, G668–G681. doi:10.1152/ajpgi.00222.2021
Wang, X., Peng, J., Cai, P., Xia, Y., Yi, C., Shang, A., et al. (2024). The emerging role of the gut microbiota and its application in inflammatory bowel disease. Biomed. and Pharmacother. 179, 117302. doi:10.1016/J.BIOPHA.2024.117302
Wang, Y., Zhang, B., Zhu, Y., Zhang, Y., Li, L., Shen, T., et al. (2022). hsa_circ_0000523/miR-let-7b/METTL3 axis regulates proliferation, apoptosis and metastasis in the HCT116 human colorectal cancer cell line. Oncol. Lett. 23, 186. doi:10.3892/OL.2022.13306
Wong, D., Sahoo, D. K., Faivre, C., Kopper, J., Dersh, K., Beachler, T., et al. (2025). Oxidative stress in critically ill neonatal foals. J. Vet. Intern Med. 39, e17297. doi:10.1111/JVIM.17297
Yamada, A., Horimatsu, T., Okugawa, Y., Nishida, N., Honjo, H., Ida, H., et al. (2015). Serum MIR-21, MIR-29a, and MIR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin. Cancer Res. 21, 4234–4242. doi:10.1158/1078-0432.CCR-14-2793
Keywords: gastrointestinal disorders, inflammatory bowel diseases, ulcerative colitis, molecular biomarkers, sepsis, colorectal cancer, disease diagnosis
Citation: Sahoo DK, Heilmann RM and Patel A (2025) Editorial: Understanding molecular mechanisms to facilitate the development of biomarkers for therapeutic intervention in gastrointestinal diseases and sepsis. Front. Genet. 16:1581299. doi: 10.3389/fgene.2025.1581299
Received: 21 February 2025; Accepted: 25 February 2025;
Published: 11 March 2025.
Edited and reviewed by:
Jared C. Roach, Institute for Systems Biology (ISB), United StatesCopyright © 2025 Sahoo, Heilmann and Patel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dipak Kumar Sahoo, ZHNhaG9vQGlhc3RhdGUuZWR1, ZGlwYWtzYWhvbzExQGdtYWlsLmNvbQ==; Romy M. Heilmann, cm9teS5oZWlsbWFubkBrbGVpbnRpZXJrbGluaWsudW5pLWxlaXB6aWcuZGU=; Ashish Patel, dW5pLmFzaGlzaEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.