
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 06 March 2025
Sec. Human and Medical Genomics
Volume 16 - 2025 | https://doi.org/10.3389/fgene.2025.1551171
 Riley H. Tough1,2*
Riley H. Tough1,2* Paul J. McLaren1,2*
Paul J. McLaren1,2*A previous study investigated a genomic region on chromosome 1 associated with reduced human immunodeficiency virus type 1 (HIV) set-point viral load, implicating CHD1L as a novel HIV inhibitory factor. However, given that regulatory variants can influence expression of multiple nearby genes, further work is necessary to determine the impact of genetic variants on other genes in the region. This study evaluates the potential for genetic regulation of PRKAB2, a gene located upstream of CHD1L and encoding the β2 regulatory subunit of the AMPK complex, and for downstream impacts on HIV pathogenesis. Using genotype and gene expression data from the Gene Expression Omnibus repository and Genotype-Tissue Expression database, we observed cell-type-specific correlations between CHD1L and PRKAB2 expression, with a strong positive association in whole blood and negative correlation in monocytes. Notably, we found that individuals with HIV set-point viral load associated variants exhibited significantly reduced PRKAB2 expression in imputed whole blood models and ex vivo monocytes. Functional analyses using PRKAB2−/− induced pluripotent stem cells suggest that PRKAB2 loss-of-function may influence CHD1L expression, and genes regulating cytokine activity, growth factor signaling, and pluripotency pathways associated with HIV infection. These results suggest that gene expression changes driven by HIV set-point viral load associated variants in the chromosome 1 impact multiple genes and, by influencing expression of PRKAB2, may result in altered expression of critical immune signaling processes. These findings advance our understanding of the contribution of host genetics on HIV pathogenesis and identifies new targets for ex vivo functional studies.
The HIV host-pathogen interaction is complex, with hundreds of human genes being implicated as candidate host restriction, inhibitory, proviral, or dependency factors (Hiatt et al., 2022). Recently, a genome-wide association study (GWAS) identified a novel region of chromosome 1 surrounding PRKAB2, FMO5, and CHD1L to be associated with reduced HIV set-point viral load (spVL) and functional follow-up identified CHD1L as a novel HIV inhibitory factor in U2OS and myeloid cells (McLaren et al., 2023). However, the potential of PRKAB2, the β2 subunit of the 5′AMP-activated protein kinase complex (AMPK), located upstream of CHD1L, was not evaluated for its effect on HIV infection.
AMPK is a heterotrimeric protein complex that is comprised of an alpha subunit (PRKAA1/PRKAA2), beta subunit (PRKAB1/PRKAB2), and gamma subunit (PRKAG1/PRKAG2/PRKAG3). This complex acts an essential regulator of cellular metabolism and during viral infection, functions as an inhibitory factor by reducing cholesterol, fatty acid, and protein synthesis of infected cells (Zhang and Wu, 2009; Zhang et al., 2012; Moreira et al., 2016). AMPK has also been shown to repress pro-inflammatory signaling (Jeong et al., 2009; Rutherford et al., 2016), activate anti-inflammatory signaling in macrophages (Sag et al., 2008), and has been shown to influence HIV pathogenesis (Bhutta et al., 2021). Therefore, we hypothesize that PRKAB2 may directly or, through downstream signaling pathways, influence HIV infection.
The advent of large-scale siRNA and CRISPR-Cas9 screens have enabled researchers to screen thousands of genes as HIV factors, but knockdown and knockout screens have not previously implicated CHD1L, PRKAB2, or FMO5 (Brass et al., 2008; Zhou et al., 2008; Yeung et al., 2009; Park et al., 2017). The lack of association suggests that these genes either do not function as HIV factors in the investigated cell types (HeLa, Jurkat T, or CEM-GXRCas9 cells) or may not act directly as HIV factors. In contrast, analysis of closely related genes to PRKAB2, such as PRKAA1, the dominant catalytic AMPKα subunit in myeloid cells (Sag et al., 2008), revealed its function as both an HIV host dependency factor (HDF) (Zhou et al., 2008) and an HIV inhibitory factor (Gélinas et al., 2018). These findings highlight that changes to specific AMPK subunits can affect HIV infection and supports investigating whether genetically regulated changes to PRKAB2 expression are associated with HIV infection.
This study seeks to determine whether functional HIV spVL associated variants identified by the HIV spVL GWAS (McLaren et al., 2023) impact PRKAB2 expression and downstream signaling pathways relevant to HIV infection. Using RNA-sequencing data from PRKAB2−/− immunopluripotent stem cells (iPSC), we assess whether PRKAB2 affects CHD1L expression, downstream signalling pathways, and known HIV factors. The results of this study implicate PRKAB2 as a potential HIV factor by identifying essential immune pathways altered by PRKAB2 loss-of-function, providing additional insight into how the role of how the chromosome 1 region affects HIV infection.
The Immune Variation (ImmVar) project is a collaborative effort to characterize how genetic and environmental factors influence the immune response in healthy individuals (De Jager et al., 2015). Genotype data for 112 African American individuals from the ImmVar project was obtained from the database of Genotypes and Phenotypes (dbGaP; phs000815.v2.p1). Chromosome 1 variants were imputed for individuals from the ImmVar project using the 1000 Genomes reference panel with SHAPEIT (Delaneau et al., 2011) and IMPUTE2 (Howie et al., 2009). Variants with a score of <0.9 and Hardy-Weinberg equilibrium threshold of p < 1 × 10−5 were removed. Genotypes were assessed at rs59784663-A-G, rs72999655-A-G, rs7525622-G-A, and rs7300425-C-T. One individual was removed from subsequent analysis due to an incomplete genotype call at rs59784663-A-G.
Gene expression profiles for individuals from the ImmVar project were evaluated to determine whether HIV spVL associated variants influence gene expression. Gene expression from naïve CD4+ T cells and monocytes, adjusted for batch effects, age, gender, and technical artifacts using principal components (PCs), were obtained from the Gene Expression Omnibus (GEO) (GSE56035). In brief, expression was profiled using Affymetrix GeneChip Human Gene ST 1.0 microarrays with background correction and normalization using the Robust Multichip Average method in Affymetrix Powertools as previously described (Raj et al., 2014). Sample, probe, and expression quality control, as well as the adjustment for non-genetic factors, including the regression of the top 14 PCs in monocytes and top 12 PCs in naïve CD4+ T cells, are detailed in Raj et al. (2014). In monocytes and naïve CD4+ T cells, the residuals are used to compare gene expression following correction for non-genetic factors.
Gene expression, in transcripts per kilo base million, were obtained from Genotype Tissue Expression (GTEx) database v8 in whole blood tissues for all available individuals (N = 756) (GTEx Consortium, 2020). The GTEx database v8 (N = 948) includes individuals of diverse ancestry, with 84.6%, 12.9%, 1.3%, and 1.1% of individuals identifying as white, African American, Asian, or unknown, although the specific ancestries for the 756 individuals were not available for this analysis. In this dataset, whole blood was defined as gathered from the femoral vein, subclavian vein, heart, and other sites (UBERON:0013756).
Raw RNA-seq reads and normalized RNA-seq read counts as transcripts per million were available for wild-type and PRKAB2−/− immunopluripotent stem cells (iPSCs) from the National Center for Biotechnology Information (NCBI) GEO database under the accession (GSE144043). In brief, NCBI generates RNA-seq count data using HISAT2 (Koga et al., 2019) to align sequences to GCA_000001405.15. Read counts were calculated using featureCounts with a 50% alignment threshold.
A summary of cohorts and data types accessed in this meta-analysis are available in Table 1.
Previous studies have suggested that HIV spVL associated variants in the chromosome 1 region regulate CHD1L expression in a cell-type-specific manner (McLaren et al., 2023). Therefore, we sought to investigate genetically regulated PRKAB2 expression across multiple immune cell types. Gene expression data was compared for whole blood samples from GTEx as transcripts per million (TPM) or monocytes and naïve CD4+ T cells from the ImmVar project as residuals following control for non-genetic factors. For each dataset, a two-sided Pearson correlation was used to determine correlation between CHD1L and PRKAB2, with p < 0.05 defined as statistically significant. A linear regression model, along with a 95% confidence interval, was to visualize the magnitude and direction of correlation between PRKAB2 and CHD1L expression. Statistical analysis was performed using R (v4.1.2).
Genomic data from 3,886 individuals of African ancestry were obtained from ten independent GWAS under the International Collaboration for the Genomics of HIV (ICGH) (N = 3,886), as previously described (McLaren et al., 2023). Predictive gene expression models trained on African American whole blood eQTLs from the Genes-environments and Admixture in Latino Americans (GALA II) and the Study of African Americans, Asthma, Genes, and Environments (SAGE) cohorts were applied to the 3,886 individual-level genotypes using PrediXcan (Gamazon et al., 2015). Imputed CHD1L expression was compared amongst individuals with allelic combinations of functional HIV spVL associated variants: rs72999655-A-G, rs7525622-G-A, and rs73004025-C-T from the ICGH cohort. Statistical significance was determined using a Wilcoxon Rank Sum Test at p < 0.05.
Given that GWAS variants typically exert a small effect on gene expression changes, we sought to determine whether PRKAB2 was associated with immune signaling pathways using expression data from PRKAB2 knockout cells to prioritize subsequent association testing. Differential gene expression analyses of PRKAB2 and CHD1L expression were performed using NCBI generated TPM from wild-type iPSCs (N = 4), PRKAB2−/− clone one (C1; N = 4), and PRKAB2−/− clone two (C2; N = 4) from the GEO database (GSE144043). Significant differences determined using a Wilcoxon Rank Sum Exact Test against p < 0.05.
Transcriptome differences between wild-type iPSCs, PRKAB2−/− C1, and PRKAB2−/− C2, were assessed using NCBI generated RNA-seq counts from the GEO database (GSE144043). Lowly expressed genes were filtered based on a minimum count threshold of at least 10 in a given sample. Differential expression was performed using DESeq2 using FDR <0.05, α = 0.05 as statistically significant. Visualization and statistical analysis were performed using R (v4.1.2).
The Database for Annotation, Visualization and Integrated Discovery (DAVID; version v2023q4) was used to perform pathway analysis and functional annotation of DEGs (Huang et al., 2009; Sherman et al., 2022). Functional enrichment was classified for Gene Ontology, Reactome, and KEGG pathway terms using medium classification stringency and an enrichment score of greater than two. Individual annotations were significant with an FDR cut-off threshold of 0.05. HIV interactions were characterized by any overlap of genes with the HIV interaction category.
Regulatory variants can influence the expression of multiple genes which collectively contribute to complex phenotypes (Ying et al., 2023). While previous studies have focused on genetically regulated CHD1L expression (McLaren et al., 2023), we hypothesized that PRKAB2 may be co-regulated due to its proximity to CHD1L. To address this, we analyzed the correlation between PRKAB2 and CHD1L expression in immune cells from the GTEx and ImmVar databases. Gene expression from the GTEx datasets showed a strong positive correlation between PRKAB2 and CHD1L expression from whole blood tissues (r = 0.7, p < 3.1 × 10−110) (Figure 1A). However, we observed no significant correlation in naïve CD4+ T cells (r = 0.17, p < 0.081) (Figure 1B) but a significant negative correlation in monocytes (r = −0.29, p < 0.002) (Figure 1C). These correlations suggest that CHD1L and PRKAB2 likely share regulatory features in immune cells but the magnitude and direction of effect may be cell-type-specific.
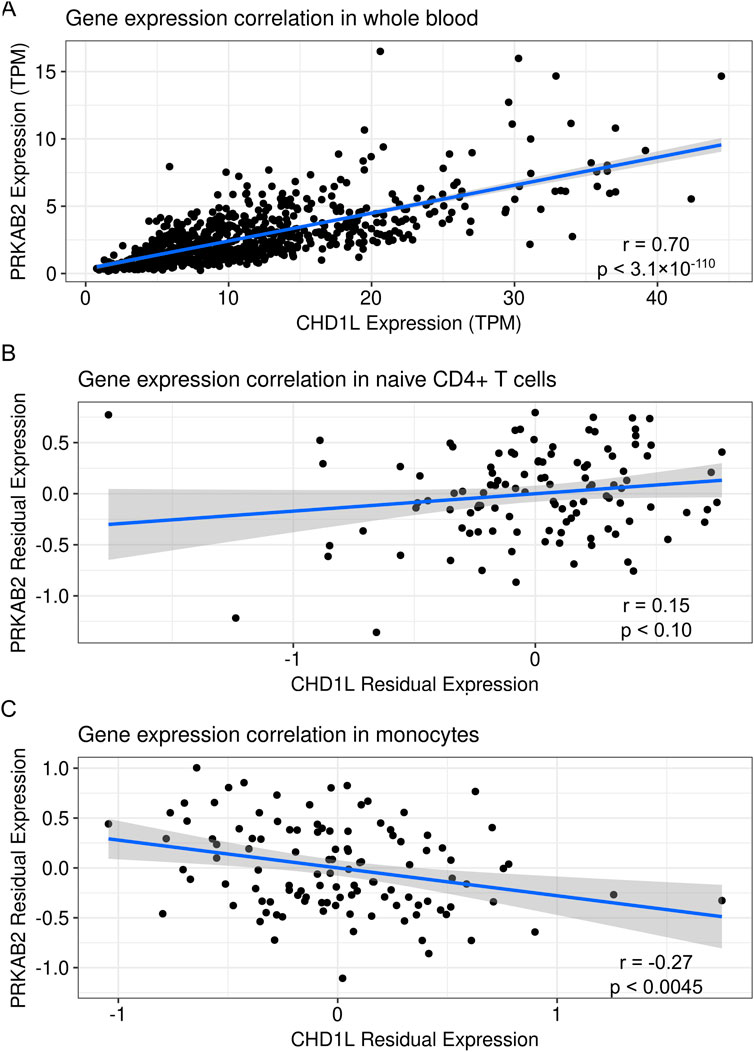
Figure 1. Correlation of CHD1L and PRKAB2 expression in human blood tissues. (A) Transcripts per million for CHD1L and PRKAB2 expression from whole blood in the Genotype-Tissue Expression database (N = 756). (B) Residuals of PRKAB2 and CHD1L expression after controlling for batch, sex, and technical variation in naïve CD4+ T cells. (C) Residuals of PRKAB2 and CHD1L expression after controlling for batch, sex, and technical variation in monocytes. The blue line represents a linear regression of the values surrounded by standard error in grey.
We next assessed the impact of HIV spVL associated variants on PRKAB2 expression to determine whether regulatory features in the chromosome 1 region influence both genes. In monocytes, individuals heterozygous for lead GWAS variant rs59784663-A-G (N = 6), had a significant reduction of PRKAB2 expression compared to homozygous reference individuals (N = 103; p < 0.036) (Figure 2A) but no significant effect in naïve CD4+ T cells (p < 0.56) (Figure 2B).
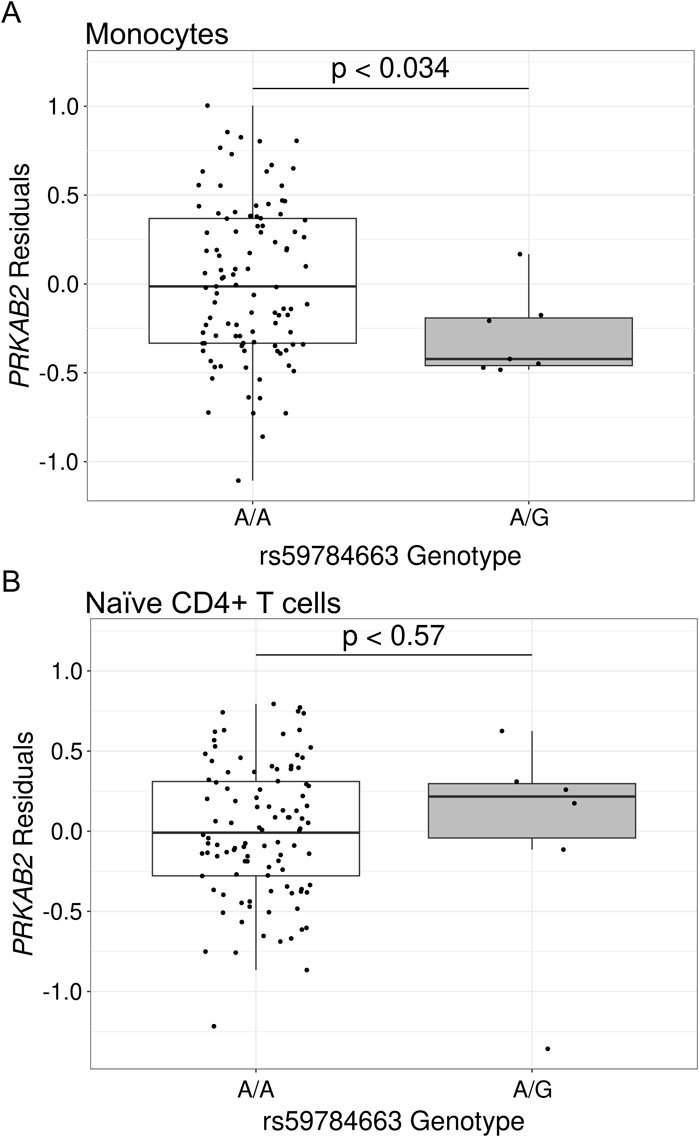
Figure 2. Gene expression of PRKAB2 based on rs59784663-A-G genotype in African populations. Gene expression is the residuals following control for non-genetic factors in African populations for (A) monocytes or (B) naïve CD4+ T cells. Significance was determined using a two-sided Wilcoxon Rank Sum Test.
We then sought to determine the effect of candidate causal variants rs72999655-A-G, rs7525622-G-A, and rs73004025-C-T on PRKAB2 expression, and to have sufficient sample size for analysis, imputed PRKAB2 expression from whole blood models using 3,886 individual-level genotypes from the ICGH cohort (McLaren et al., 2023). Individuals heterozygous for rs72999655-A-G, rs7525622-G-A, and rs73004025-C-T (N = 312) exhibited lower PRKAB2 expression than homozygous reference (N = 3,159; p < 1.65 × 10−26) and homozygous alternate individuals (N = 7) exhibited much lower PRKAB2 expression than homozygous reference (p < 0.00031) (Figure 3). Overall, these results show that HIV spVL associated variants likely influence PRKAB2 expression and highlights the need to investigate potential downstream consequences of HIV infection.
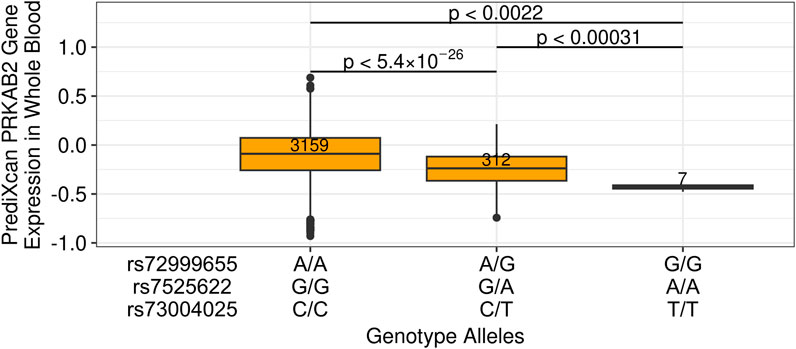
Figure 3. Imputed PRKAB2 expression changes based on alleles of HIV spVL associated genetic variants. PRKAB2 expression was generated using whole blood PrediXcan models from the Genes-Environments and Admixture in Latino Asthmatics and the Study of African Americans, Asthma, Genes, and Environments cohorts. Allelic combinations are shown for individuals homozygous reference, heterozygous, or homozygous alternate at all variants.
Further investigation into the relationship between PRKAB2 and CHD1L was performed using GEO gene expression data from PRKAB2−/− iPSC models (GSE144043) (Ziegler et al., 2020). In PRKAB2−/− iPSCs, PRKAB2 expression was significant reduced compared to wild-type controls (p < 2.2 × 10−5 and p < 9.6 × 10−7 for clones C1 and C2, respectively), while a larger reduction was observed between C2 compared to C1 (p < 8.1 × 10−5) (Figure 4A). However, despite reduced PRKAB2 expression in both clones, CHD1L expression was inconsistent. While CHD1L expression was not significantly different between wild-type and C1 iPSCs (p < 0.068), we observed higher CHD1L expression in C1 clones, but a significant reduction of CHD1L expression observed in C2 compared to wild-type (p < 0.0012) (Figure 4B). The variability between clones may reflect differences in gRNA efficacy or off-target effects during generation of PRKAB2−/− knockouts. While the data is unclear in the potential of AMPKβ2-mediated CHD1L expression, AMPK is known to activate DNA repair pathways (Szewczuk et al., 2020), suggesting that AMPKβ2 may alter expression of CHD1L only under specific physiological conditions.
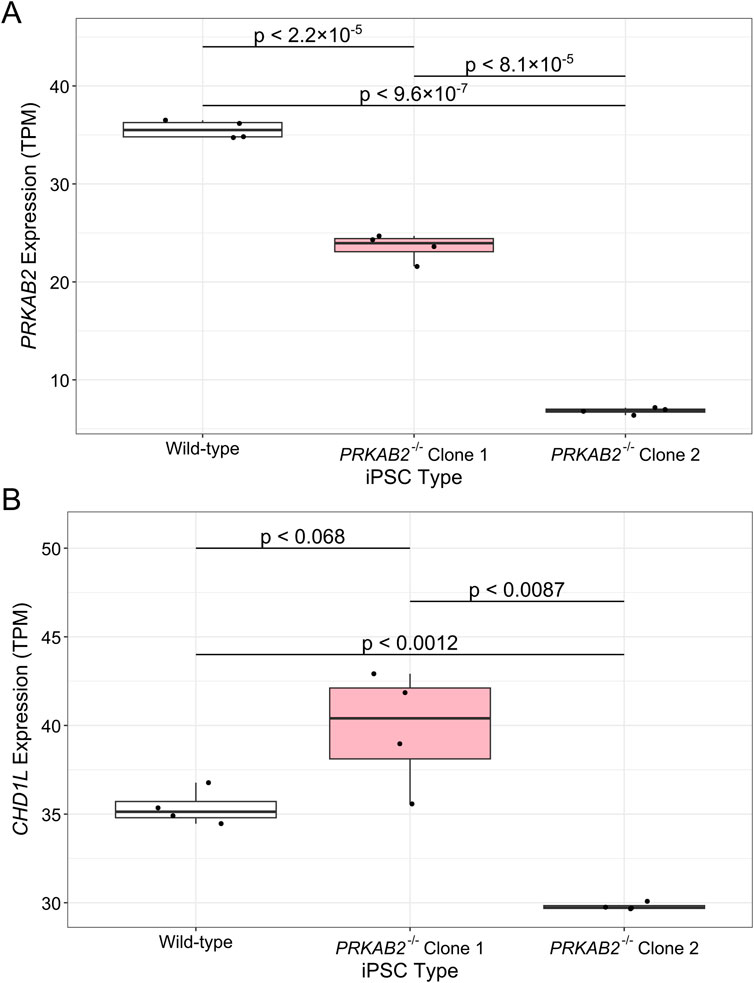
Figure 4. Gene expression differences in PRKAB2−/− loss-of-function iPSCs. Gene expression of (A) PRKAB2 and (B) CHD1L in wild-type iPSCs, PRKAB2 knockout clone 1 iPSCs, and PRKAB2 knockout clone 2 iPSCs. Significance was determined using a Wilcoxon Rank Sum Exact Test.
One of the major challenges in identifying changes to PRKAB2 signaling pathways is that the functional AMPKβ2 complex regulates expression of downstream genes through phosphorylation, which is not detectable through RNA-sequencing analyses. However, AMPK regulates the activation of transcription factors (Cantó and Auwerx, 2010), indicating that signalling pathways downstream of PRKAB2 may be differentially expressed. Therefore, we leveraged gene expression profiles from the wild-type and PRKAB2−/− iPSCs (GSE144043) to determine potential pathways influence by PRKAB2.
We first sought to determine whether PRKAB2−/− C1 and C2 exhibited different gene expression profiles given the differences in PRKAB2 expression observed previously. We observed 58 differentially expressed genes (DEGs) between C1 and C2 suggesting that differences in gRNA efficiency or off-target effects may influence gene expression profiles. Additionally, when comparing DEGs in C2 and wild-type iPSCs, the most significant DEG was PRKAB2 (log2FC = −2.4, FDR < 2.2 × 10−308), but when comparing C1 and wild-type iPSCs, PRKAB2 ranked 44th (log2FC = −0.70, FDR < 7.9 × 10−59). Therefore, we chose to analyze gene expression differences between wild-type and PRKAB2−/− C2, given that C2 exhibited a larger reduction in PRKAB2 expression compared to C1.
There were 170 downregulated and 145 upregulated genes in PRKAB2−/− C2 compared to wild-type iPSCs (FDR < 0.05, |log2 fold change| ≥ 1, Figure 5A). These genes were significantly enriched for functions related to cytokine activity (17 genes, FDR < 9.5 × 10−8) and growth factor activity (10 genes, FDR <0.012), as well as KEGG pathways regulating stem cell pluripotency (9 genes, FDR <0.028; Table 2). Clustering analysis revealed two major groups: one associated with cytokine activity (enrichment score 4.24) and the other with male genitalia development (enrichment score 2.13; Figure 5B). Given the relevance of cytokine activity to HIV infection, we prioritized this group for further analysis. Among the differentially expressed genes in the cytokine activity pathway, TNFSF9 (log2FC = −1.5, FDR < 1.8 × 10−32), TGFB2 (log2FC = −1.0, FDR < 0.000071), and SCG2 (log2FC = 1.003, FDR < 0.0069), stood out as known HIV interactors using DAVID annotation.
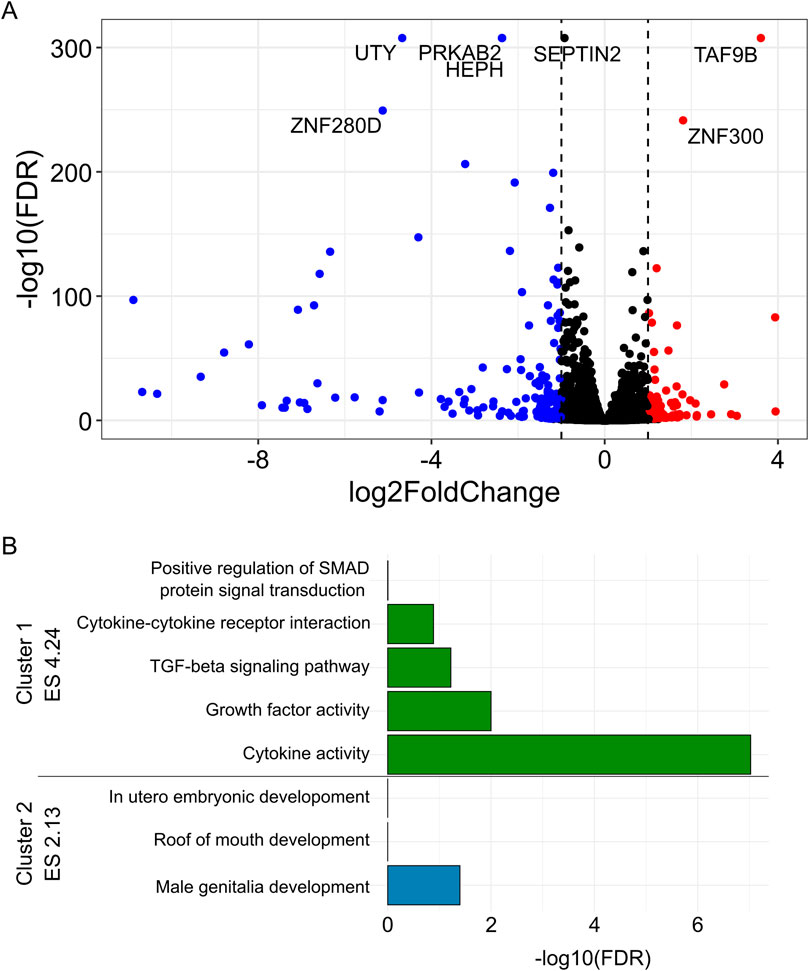
Figure 5. Differentially expressed genes between PRKAB2−/− knockout and wild-type iPSCs. (A) (Blue) Significant downregulated genes (FDR < 0.05, log2 fold change < 1) in PRKAB2 knockout clone two cells. (Red) Significant upregulated genes (FDR < 0.05, log2 fold change > 1) in PRKAB2 knockout clone two cells. (B) Enrichment clustering of differentially expressed genes revealed two distinct clusters with an enrichment score of greater than two.
However, given that PRKAB2−/− C1 and C2 exhibited significant differences in both PRKAB2 and CHD1L expression, we replicated the transcriptomic analysis comparing wild-type iPSCs to both PRKAB2−/− C1 and C2, together. This resulted in 125 downregulated and 160 upregulated genes in PRKAB2−/− knockout iPSCs compared to wild-type iPSCs, but the most significant enriched pathway remained cytokine activity (Supplementary Figure S1). Collectively, these results show that individuals with HIV spVL associated variants exhibit reduced PRKAB2 expression and reduced PRKAB2 expression alters cytokine regulation, suggesting that the chromosome 1 region may regulate HIV spVL through these immune pathways.
During the analysis of a recent GWAS of HIV spVL, CHD1L was identified to be the candidate causal gene and implicated as a novel HIV inhibitory factor (McLaren et al., 2023). However, the question remains as to why CHD1L was not detected in previous screens of HIV inhibitory factors (Gélinas et al., 2018). While cell-type-specific effects may explain some variation, it is also likely that multiple chromosome 1 genes may collectively contribute to control HIV infection through downstream signaling pathways (Broekema et al., 2020). Therefore, we chose to investigate the role of PRKAB2, part of the AMPKβ2 complex, as AMPK has been previously associated with HIV infection (Zhou et al., 2008; Zhang and Wu, 2009).
Our findings indicate that PRKAB2 and CHD1L expression is positively correlated in whole blood but negatively correlated in monocytes, supporting cell-type-specific gene regulation. Previously fine-mapping of a HIV spVL GWAS identified three candidate causal GWAS variants, rs72999655-A-G, rs7525622-G-A, and rs73004025-C-T, and that alleles linked to lower HIV spVL were associated with increased CHD1L expression in monocytes (Tough et al., 2025). In this study, individuals heterozygous for rs72999655-A-G, rs7525622-G-A, and rs73004025-C-T exhibited reduced PRKAB2 expression in monocytes and whole blood, but no differences in naïve CD4+ T cells. While the biological mechanism how these variants drive gene expression changes remains unclear, these results suggest that lower PRKAB2 expression is associated with reduced HIV spVL.
Next, we hypothesized that PRKAB2 may directly influence CHD1L expression, as AMPK activation initiates DNA repair pathways involving CHD1L (Wu et al., 2013; Szewczuk et al., 2020). We observed that PRKAB2 loss-of-function had no effect on CHD1L expression in PRKAB2−/− C1 but resulted in significant downregulation in PRKAB2−/− C2. While both C1 and C2 were identified to have complete protein ablation (Ziegler et al., 2020), it is unclear whether the observed differences were driven by off-target effects, environmental, or physiological conditions. To address this ambiguity, additional studies are necessary to investigate whether AMPKβ2 influences CHD1L expression under conditions of AMPK activation such as DNA damage, low cellular metabolism, or HIV infection.
While our previous experiments showed that individuals with protective HIV spVL associated variants have reduced PRKAB2 expression, the overall impact on downstream signaling pathways is unknown. This is in part because AMPKβ1 and AMPKβ2 exhibit different transcriptome profiles (Ziegler et al., 2020), but specific AMPKβ2 pathways associated with HIV infection are unclear. To address this, we compared the transcriptome profiles from PRKAB2−/− and wild-type iPSCs, which identified the most significant pathway altered by PRKAB2 loss-of-function was regulation of cytokine signaling. Cytokine and chemokine levels, prior to HIV infection, can be strong predictors of disease progression (Huang et al., 2016; Ngcobo et al., 2022) and increased AMPK activity is associated with suppression of pro-inflammatory signaling in macrophages (Sag et al., 2008). Therefore, this provides evidence that HIV spVL associated variants in the chromosome 1 region likely influence HIV spVL through changes to cytokine signaling and immune activation pathways.
Three genes regulating cytokine activity were identified by the DAVID analysis to interact with HIV proteins: TNFSF9, TGFB2, and SCG2. The gene SCG2, upregulated in PRKAB2−/−, plays an important role in regulating the immune response through the assembly and function of the major histocompatibility complex, PI3K-Akt, TGF-β, and JAK-STAT pathways (Wang et al., 2021; Steinfass et al., 2023). The gene TNFSF9, also known as 4-1BBL and downregulated in PRKAB2−/−, produces a pro-inflammatory immune response and a key component of cytotoxic CD8+ T cells, macrophages, and monocytes activation (Ju et al., 2003; Wang et al., 2007; Olofsson et al., 2008). The gene TGFB2, downregulated in PRKAB2−/−, has immunosuppressive functions and regulates activation of dendritic and T cells (Tu et al., 2020). Collectively, these genes implicate additional pathways downstream of AMPK signaling associated with HIV pathogenesis and identifies targets for future research to elucidate mechanisms by which HIV spVL associated variants influence HIV spVL.
However, there are limitations to this study that should be acknowledged. Firstly, the cohort data analyzed here consists of individuals of African American ancestry from the ImmVar and GALA II/SAGE cohorts and requires replication in African cohorts to ensure broader generalizability. Secondly, our analysis of differentially expressed genes utilized unstimulated iPSCs, whereas AMPK is typically activated in response to metabolic stress (Steinberg and Hardie, 2023). While this provides insight into pathways associated with AMPKβ2, future studies should assess the function in primary immune cells and during HIV infection.
Overall, this study highlights that HIV spVL associated variants influence PRKAB2 expression and raises important questions about whether this effect is synergistic, antagonistic, or independent of CHD1L regulation. Furthermore, as AMPK is a regulator of immune function, future studies should investigate whether chromosome 1 genes differentially affect HIV pathogenesis during acute and chronic infection, providing insights into their potential as therapeutic targets.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by University of Manitoba research ethics board. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from were acquired from dbGaP under accession code phs000815.v2.p1 and from each of the cohorts, studies and centers within the ICGH HIV spVL GWAS as described previously. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
RT: Conceptualization, Formal Analysis, Methodology, Visualization, Writing–original draft, Writing–review and editing. PM: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded through the Public Health Agency of Canada (PHAC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1551171/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Differential gene expression profiles between wild-type iPSCs and PRKAB2−/− clones (C1 and C2). (A) (Blue) Significant downregulated genes (FDR <0.05, log2 fold change <1) in PRKAB2 knockout C1 and C2 compared to wild-type iPSCs. (Red) Significant upregulated genes (FDR <0.05, log2 fold change >1) in PRKAB2 knockout C1 and C2 compared to wild-type iPSCs. (B) Enrichment of DAVID terms amongst significant differentially expressed genes.
Bhutta, M. S., Gallo, E. S., and Borenstein, R. (2021). Multifaceted role of AMPK in viral infections. Cells 10, 1118. doi:10.3390/cells10051118
Brass, A. L., Dykxhoorn, D. M., Benita, Y., Yan, N., Engelman, A., Xavier, R. J., et al. (2008). Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926. doi:10.1126/science.1152725
Broekema, R. V., Bakker, O. B., and Jonkers, I. H. (2020). A practical view of fine-mapping and gene prioritization in the post-genome-wide association era. Open Biol. 10, 190221. doi:10.1098/rsob.190221
Cantó, C., and Auwerx, J. (2010). AMP-activated protein kinase and its downstream transcriptional pathways. Cell. Mol. Life Sci. 67, 3407–3423. doi:10.1007/s00018-010-0454-z
De Jager, P. L., Hacohen, N., Mathis, D., Regev, A., Stranger, B. E., and Benoist, C. (2015). ImmVar project: insights and design considerations for future studies of “healthy” immune variation. Semin. Immunol. 27, 51–57. doi:10.1016/j.smim.2015.03.003
Delaneau, O., Marchini, J., and Zagury, J.-F. (2011). A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181. doi:10.1038/nmeth.1785
Gamazon, E. R., Wheeler, H. E., Shah, K. P., Mozaffari, S. V., Aquino-Michaels, K., Carroll, R. J., et al. (2015). A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098. doi:10.1038/ng.3367
Gélinas, J.-F., Gill, D. R., and Hyde, S. C. (2018). Multiple inhibitory factors act in the late phase of HIV-1 replication: a systematic review of the literature. Microbiol. Mol. Biol. Rev. 82. doi:10.1128/MMBR.00051-17
GTEx Consortium (2020). The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330. doi:10.1126/science.aaz1776
Hiatt, J., Hultquist, J. F., McGregor, M. J., Bouhaddou, M., Leenay, R. T., Simons, L. M., et al. (2022). A functional map of HIV-host interactions in primary human T cells. Nat. Commun. 13, 1752. doi:10.1038/s41467-022-29346-w
Howie, B. N., Donnelly, P., and Marchini, J. (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529. doi:10.1371/journal.pgen.1000529
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi:10.1038/nprot.2008.211
Huang, X., Liu, X., Meyers, K., Liu, L., Su, B., Wang, P., et al. (2016). Cytokine cascade and networks among MSM HIV seroconverters: implications for early immunotherapy. Sci. Rep. 6, 36234. doi:10.1038/srep36234
Jeong, H. W., Hsu, K. C., Lee, J.-W., Ham, M., Huh, J. Y., Shin, H. J., et al. (2009). Berberine suppresses proinflammatory responses through AMPK activation in macrophages. Am. J. Physiol. Endocrinol. Metab. 296, E955–E964. doi:10.1152/ajpendo.90599.2008
Ju, S.-W., Ju, S.-G., Wang, F.-M., Gu, Z.-J., Qiu, Y.-H., Yu, G.-H., et al. (2003). A functional anti-human 4-1BB ligand monoclonal antibody that enhances proliferation of monocytes by reverse signaling of 4-1BBL. Hybrid. Hybridomics 22, 333–338. doi:10.1089/153685903322538872
Kachuri, L., Mak, A. C. Y., Hu, D., Eng, C., Huntsman, S., Elhawary, J. R., et al. (2023). Gene expression in African Americans, Puerto Ricans and Mexican Americans reveals ancestry-specific patterns of genetic architecture. Nat. Genet. 55, 952–963. doi:10.1038/s41588-023-01377-z
Koga, Y., Tsurumaki, H., Aoki-Saito, H., Sato, M., Yatomi, M., Takehara, K., et al. (2019). Roles of cyclic AMP response element binding activation in the ERK1/2 and p38 MAPK signalling pathway in central nervous system, cardiovascular system, osteoclast differentiation and mucin and cytokine production. Int. J. Mol. Sci. 20, 1346. doi:10.3390/ijms20061346
McLaren, P. J., Porreca, I., Iaconis, G., Mok, H. P., Mukhopadhyay, S., Karakoc, E., et al. (2023). Africa-specific human genetic variation near CHD1L associates with HIV-1 load. Nature 620, 1025–1030. doi:10.1038/s41586-023-06370-4
Moreira, D., Silvestre, R., Cordeiro-da-Silva, A., Estaquier, J., Foretz, M., and Viollet, B. (2016). AMP-Activated protein kinase as a target for pathogens: friends or foes? Curr. Drug Targets 17, 942–953. doi:10.2174/1389450116666150416120559
Ngcobo, S., Molatlhegi, R. P., Osman, F., Ngcapu, S., Samsunder, N., Garrett, N. J., et al. (2022). Pre-infection plasma cytokines and chemokines as predictors of HIV disease progression. Sci. Rep. 12, 2437. doi:10.1038/s41598-022-06532-w
Olofsson, P. S., Söderström, L. A., Wågsäter, D., Sheikine, Y., Ocaya, P., Lang, F., et al. (2008). CD137 is expressed in human atherosclerosis and promotes development of plaque inflammation in hypercholesterolemic mice. Circulation 117, 1292–1301. doi:10.1161/CIRCULATIONAHA.107.699173
Park, R. J., Wang, T., Koundakjian, D., Hultquist, J. F., Lamothe-Molina, P., Monel, B., et al. (2017). A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nat. Genet. 49, 193–203. doi:10.1038/ng.3741
Raj, T., Rothamel, K., Mostafavi, S., Ye, C., Lee, M. N., Replogle, J. M., et al. (2014). Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523. doi:10.1126/science.1249547
Rutherford, C., Speirs, C., Williams, J. J. L., Ewart, M.-A., Mancini, S. J., Hawley, S. A., et al. (2016). Phosphorylation of Janus kinase 1 (JAK1) by AMP-activated protein kinase (AMPK) links energy sensing to anti-inflammatory signaling. Sci. Signal. 9, ra109. doi:10.1126/scisignal.aaf8566
Sag, D., Carling, D., Stout, R. D., and Suttles, J. (2008). Adenosine 5’-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J. Immunol. 181, 8633–8641. doi:10.4049/jimmunol.181.12.8633
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucl. Acids Res. 50, W216–W221. doi:10.1093/nar/gkac194
Steinberg, G. R., and Hardie, D. G. (2023). New insights into activation and function of the AMPK. Nat. Rev. Mol. Cell Biol. 24, 255–272. doi:10.1038/s41580-022-00547-x
Steinfass, T., Poelchen, J., Sun, Q., Mastrogiulio, G., Novak, D., Vierthaler, M., et al. (2023). Secretogranin II influences the assembly and function of MHC class I in melanoma. Exp. Hematol. Oncol. 12, 29. doi:10.1186/s40164-023-00387-1
Szewczuk, M., Boguszewska, K., Kaźmierczak-Barańska, J., and Karwowski, B. T. (2020). The role of AMPK in metabolism and its influence on DNA damage repair. Mol. Biol. Rep. 47, 9075–9086. doi:10.1007/s11033-020-05900-x
Tough, R. H., and McLaren, P. J.International Collaboration for the Genomics of HIV (2025). Functionally-informed fine-mapping identifies genetic variants linking increased CHD1L expression and HIV restriction in monocytes. Sci. Rep. 15, 2325. doi:10.1038/s41598-024-84817-y
Tu, L., Sun, X., Yang, L., Zhang, T., Zhang, X., Li, X., et al. (2020). TGF-β2 interfering oligonucleotides used as adjuvants for microbial vaccines. J. Leukoc. Biol. 108, 1673–1692. doi:10.1002/JLB.5A0420-491R
Wang, C., Wen, T., Routy, J.-P., Bernard, N. F., Sekaly, R. P., and Watts, T. H. (2007). 4-1BBL induces TNF receptor-associated factor 1-dependent Bim modulation in human T cells and is a critical component in the costimulation-dependent rescue of functionally impaired HIV-specific CD8 T cells. J. Immunol. 179, 8252–8263. doi:10.4049/jimmunol.179.12.8252
Wang, H., Yin, J., Hong, Y., Ren, A., Wang, H., Li, M., et al. (2021). SCG2 is a prognostic biomarker associated with immune infiltration and macrophage polarization in colorectal cancer. Front. Cell Dev. Biol. 9, 795133. doi:10.3389/fcell.2021.795133
Wu, C. L., Qiang, L., Han, W., Ming, M., Viollet, B., and He, Y. Y. (2013). Role of AMPK in UVB-induced DNA damage repair and growth control. Oncogene 32, 2682–2689. doi:10.1038/onc.2012.279
Yeung, M. L., Houzet, L., Yedavalli, V. S. R. K., and Jeang, K. T. (2009). A genome-wide short hairpin RNA screening of Jurkat T-cells for human proteins contributing to productive HIV-1 replication. J. Biol. Chem. 284, 19463–19473. doi:10.1074/jbc.M109.010033
Ying, P., Chen, C., Lu, Z., Chen, S., Zhang, M., Cai, Y., et al. (2023). Genome-wide enhancer-gene regulatory maps link causal variants to target genes underlying human cancer risk. Nat. Commun. 14, 5958. doi:10.1038/s41467-023-41690-z
Zhang, H. S., and Wu, M. R. (2009). SIRT1 regulates Tat-induced HIV-1 transactivation through activating AMP-activated protein kinase. Virus Res. 146, 51–57. doi:10.1016/j.virusres.2009.08.005
Zhang, H.-S., Wu, T.-C., Sang, W.-W., and Ruan, Z. (2012). MiR-217 is involved in Tat-induced HIV-1 long terminal repeat (LTR) transactivation by down-regulation of SIRT1. Biochim. Biophys. Acta 1823, 1017–1023. doi:10.1016/j.bbamcr.2012.02.014
Zhou, H., Xu, M., Huang, Q., Gates, A. T., Zhang, X. D., Castle, J. C., et al. (2008). Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4, 495–504. doi:10.1016/j.chom.2008.10.004
Ziegler, N., Bader, E., Epanchintsev, A., Margerie, D., Kannt, A., and Schmoll, D. (2020). AMPKβ1 and AMPKβ2 define an isoform-specific gene signature in human pluripotent stem cells, differentially mediating cardiac lineage specification. J. Biol. Chem. 295, 17659–17671. doi:10.1074/jbc.RA120.013990
Keywords: genome-wide association studies, bioinformatics, PRKAB2, RNA-sequencing, HIV - human immunodeficiency virus
Citation: Tough RH and McLaren PJ (2025) Chromosome 1 variants associated with decreased HIV set-point viral load correlate with PRKAB2 expression changes. Front. Genet. 16:1551171. doi: 10.3389/fgene.2025.1551171
Received: 24 December 2024; Accepted: 20 February 2025;
Published: 06 March 2025.
Edited by:
Rajkumar S. Kalra, Okinawa Institute of Science and Technology Graduate University, JapanReviewed by:
Aditya Yashwant Sarode, Columbia University, United StatesCopyright © 2025 Tough and McLaren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Riley H. Tough, cmlsZXkudG91Z2hAcGhhYy1hc3BjLmdjLmNh; Paul J. McLaren, cGF1bC5tY2xhcmVuQHBoYWMtYXNwYy5nYy5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.