
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 05 March 2025
Sec. Livestock Genomics
Volume 16 - 2025 | https://doi.org/10.3389/fgene.2025.1540305
This article is part of the Research TopicEnhancing Livestock Breeding through Advanced Genetic Tools and Phenotyping SystemsView all 6 articles
 Qiaozhen Ke1,2†
Qiaozhen Ke1,2† Yin Li2†
Yin Li2† Huasong Weng3
Huasong Weng3 Baohua Chen2
Baohua Chen2 Jiaying Wang2
Jiaying Wang2 Ji Zhao1,2
Ji Zhao1,2 Pengxin Jiang2
Pengxin Jiang2 Peng Xu2
Peng Xu2 Tao Zhou2*
Tao Zhou2*Large yellow croaker is an economically important carnivorous marine aquaculture fish in China with high protein requirements. Current fish meal - based feeds face issues like high cost and resource depletion, while plant protein sources have potential but also controversies. To explore this, a 120 - day feeding trial was conducted with a standard commercial feed (CF) and a modified feed (PF) where 70% of fish meal was replaced by plant protein. Results showed no significant growth performance differences between the two groups. Transcriptome analysis identified 557 and 308 differentially expressed genes in the liver and intestine respectively. GO and KEGG enrichment analyses indicated their association with immune response, lipid metabolism, and signal transduction. Five key genes related to metabolism and immune regulation were also found. These findings underscore the potential of integrating plant protein into fish diets, which could significantly enhance sustainable practices in global aquaculture while reducing reliance on fish meal. Emphasizing this transition is crucial for fostering environmental sustainability and supporting the future of aquaculture.
Large yellow croaker (Larimichthys crocea) is an important marine aquaculture species in China, with annual production of more than 280,997 tons in 2022 (China Fishery Statistical Yearbooks). As a carnivorous fish, the large yellow croaker has a relatively high demand for protein (He et al., 2017; Gu et al., 2019). Currently, chopped or minced trash fish is the major diet for large yellow croaker (Hardy, 2010; Zhang et al., 2016). However, issues such as its low feed utilization, unsafe quality, pollution of water quality and disease have resulted in a waste of resources and posed a great threat to the water environment and food safety (Shi et al., 2017; Kumar, 2019; Liu and Peng, 2021). The emergence of fish meal in feed formulations has effectively addressed these issues (Liu and Peng, 2021). However, its high cost has become the primary limiting factor for its use. The main ingredients of the complementary feed, such as fish meal and fish oil, heavily rely on imports. Nevertheless, global marine oil and fish meal resources are depleting gradually (Han et al., 2018). Additionally, the aquaculture industry is experiencing exponential growth to provide affordable protein for the world’s ever-increasing population, resulting in heightened feed demand. Large-scale overfishing for fish meal production has led to a decline in marine species (Zhou et al., 2015), and severely disrupted ecological balance. Fish oil (FO) and fish meal (FM) may further exacerbate the pressure on dwindling marine fish resources (Aladetohun, 2013; Oliva-Teles et al., 2022), and the shortage of fish meal has caused persistent price increases. In Asia alone, the consumption of fish meal for Nile tilapia rose from 800,000 tons to 1.7 million tons, while the production of fish feed increased from 40% in 2000 to 60% in 2008 (Tacon and Metian, 2008). According to the Food and Agriculture Organization of the United Nations, reducing the inclusion of fish meal and fish oil in feeds represents a significant advancement in alleviating the strain on global marine resources (Subasinghe, 2001). Therefore, with the expansion of aquaculture, major factors such as increased demand, uncertain supply, and high prices of fishmeal make it necessary to find alternative sources of fish meal (Mu et al., 2017).
Compared with animal protein sources, plant protein sources have the advantages of being widely available and inexpensive. Moreover, plant protein sources can protect marine fishery resources and promote sustainable development of marine fisheries (Shahin et al., 2023). In some feeds for practical applications for carnivorous fish, a reasonable mixture of plant protein ingredients can achieve a complete replacement of fishmeal without affecting their growth performance, such as Sparus aurata (Gómez-Requeni et al., 2004), Rachycentron canadum (Salze et al., 2010), Oncorhynchus mykiss (Lee et al., 2010) and Dicentrarchus labrax (Kaushik et al., 2004). However, compared to animal protein sources, the lack of certain essential amino acids (EAA) in plant protein components and the low utilization and palatability of it, have caused the controversy on the effectiveness of plant protein substitution (Dani, 2018), while the use of certain plant protein components or a high level substitution in fishmeal may slow down the growth of fish (Li et al., 2010; Espe et al., 2012). Therefore, the use of plant proteins in fishmeal should be more rigorous. When applying plant protein to large yellow croaker compound feeds, it is necessary to study the tolerance of large yellow croaker to plant protein, and experimental fish fed with different proportions of plant protein feeds are evaluated and molecular studies are performed.
Transcriptome analysis enables high-throughput screening of thousands of expressed genes in specific tissues (Damon et al., 2013). Through the identification of key genes and pathways, transcriptional regulatory mechanisms can be resolved, providing a theoretical reference and basis for the rational design of efficient large yellow croaker feed formulations (Li et al., 2007). Currently, numerous reports have explored the molecular mechanisms underlying the replacement of fishmeal with plant protein in various aquaculture species using transcriptome analysis. For example, In Salmo salar, Król et al. found that anti-nutritional factors (ANFs) present in plant proteins induce inflammatory responses in the intestines (Król et al., 2016). Caballero-Solares et al. reported that plant a protein-based diet leads to metabolic and immune changes in the liver (Caballero-Solares et al., 2018). In Trachinotus ovatus, Fan et al. observed that a high protein-based diet significantly affects immune and metabolic molecular changes in the liver (Fan et al., 2021). In O. mykiss, Panserat et al. demonstrated that a 100% plant-based diet induces significant metabolic changes in the liver (Panserat et al., 2009). Moreover, Cao et al. integrated transcriptomic and metabolomic analyses to show that plant protein feeding inhibits immune, metabolic, and protein functions (Cao et al., 2022). In large yellow croaker, research studies have shown that a diet with low plant protein replacing fishmeal does not significantly affect growth, immunity, or physiology (Li et al., 2010; Zhang et al., 2008; Wang et al., 2017). As a result, current commercial feed for large yellow croaker typically includes a low amount of plant protein. However, there have been no reports on the effects of high plant protein replacement for fishmeal or on transcriptome analysis.
Transcriptomic research allows us to understand the subtle effects of plant protein substitution for fish meal at the molecular level, enabling the selection of appropriate plant-based feed formulations to promote international sustainable development goals and maintain the healthy growth of the aquaculture industry. Therefore, in this study, we conducted a 120 days feeding experiment by using diet with standard commercial feed and a modified version, comparing the growth performance of large yellow croaker between different feed groups. We investigated differences in intestinal and liver transcript expression in experimental fish between feed groups to gain insight into how feed-induced differences in intestinal and liver transcript levels affect the overall physiology of the fish. This research aims to investigate the molecular-level changes in large yellow croaker resulting from plant protein substitution in the diet, focusing on its impact on metabolism and immunity, and providing a foundation for developing suitable aquaculture feed formulations.
We utilized four primary protein feed ingredients from standard commercial feed: fish meal, vital wheat gluten, dehulled soybean meal, and wheat flour, to adjust the protein composition and design the modified feed formula. Dehulled soybean meal and vital wheat gluten are inexpensive, readily available, and has a high protein content (Zlaugotne et al., 2023), while vital wheat gluten is a high-protein ingredient with an intriguing amino acid profile, particularly high in glutamine, which can improve gut health and modulate immunity (Apper-Bossard et al., 2013). The combination of these two ingredients effectively supplements the necessary amino acids and proteins.
The groups were designated as CF for the commercial feed group, which replaces 30% of the old formulation with plant protein. This formulation has been widely used in feeding large yellow croaker. The PF group, on the other hand, replaces 70% of the old formulation with plant protein, leading to a 37% substitution of plant protein compared to the CF group. The ingredient composition and main nutritional component for the CF and PF groups are shown in Table 1, with fish meal contents of 48% (CF) and 30% (PF). Fish meal, dehulled soybean meal, and vital wheat gluten were used as the main protein sources for the experimental feeds, while fish oil and soy lecithin were used as lipid sources. In the PF group, fish meal was reduced by 70% and replaced with plant protein. The ratio of dehulled soybean meal to vital wheat glutende was 1:3. To maintain equivalent protein and fat content in both feed groups, adjustments were made to the quantities of wheat flour and fish oil. The all experimental feeds were produced in Fujian Gaolong Co., Ltd., and made into puffed pellet feeds with a particle size of 4 mm.
The experimental large yellow croakers were obtained from the mariculture base of Ningde Fufa Fisheries Co., Ltd., (Ningde, Fujian, China), and the whole culture experiment was conducted in the culture sea area of Sandu’ao Dawan, Ningde. Fish from the same breeding batch, Similar sized large yellow croakers were temporarily reared in a large cage (8 m × 8 m × 5 m) to adapt to the experimental conditions, and were fed commercial feed for 2 weeks. Before the start of the different feed feeding culture experiments, the experimental fish were not fed for 24 h. These fish were randomly assigned to two experimental culture cages (8 m × 4 m × 5 m), with 2,000 fish per cage, and all the experimental cages were set up in the same marine environment. The cages were labeled as FC and PC groups in turn and fed with the corresponding labeled feeds. Each feed was fed once a day (17:00) until apparent satiation for 120 days. During the entire feeding experiment period (24 June 2019 to 22 October 2019), the seawater surface temperature was between 24.8°C and 28.6°C. The experimental fish were maintained under natural light conditions, meaning they were exposed solely to the available natural sunlight in the environment, without any artificial lighting. Consequently, the lighting conditions fluctuated in accordance with the natural day-night cycle.
Before the start of the experiment, 60 experimental fish were randomly picked and weighed after being anesthetized with eugenol (1:10,000) (Sinopharm Chemical Reagent Co., Ltd., SCR, Shanghai, China). After 120 days of feeding, 60 large yellow croakers were randomly selected from each group and weighed to determine the final body weight of the two groups of experimental fish to evaluate the growth performance of large yellow croaker. The parametric t-test was used to analyze the significant differences between groups in the initial body weight and final body weight of the experimental fish.
Compared to other organs, the intestine and liver are crucial for fish metabolism and nutrient absorption, and plant protein diets have a direct impact on their functions (Hussein et al., 2023; Dawood, 2021). Therefore, gene expression analysis of the intestine and liver is a primary focus of our study. Six fish (three for liver sampling and three for intestinal sampling) from each feed group were anesthetized with eugenol, then euthanized and sampled to minimize stress and suffering during the trial. During sampling, the abdominal cavity was cut open, and liver samples were immediately snap-frozen in liquid nitrogen and stored in an ultra-low temperature refrigerator at −80°C for subsequent RNA extraction. The intestinal tract was excised and the intestinal contents were removed. The intestinal segment under study was taken from the hindgut, which is defined as the region from the increase in intestinal diameter and presence of visible folds to the rectum, was subsequently cleaned of mesenteric and adipose tissue. Intestine samples were immediately snap-frozen in liquid nitrogen and stored in an ultra-low temperature refrigerator at −80°C for subsequent RNA extraction.
Total RNA of the intestine and liver tissues was extracted using TransZol Up Plus RNA Kit following the manufacturer’s instructions. RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, United States). RNA quantity and integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, United States). The RNA samples with OD260/280 values ranging from 1.67 to 2.16, OD260/230 values ranging from 1.23 to 2.80 and RNA Integrity Number (RIN) ≥ 6.5 were used for the subsequent library construction. A total of 18 RNA-Seq libraries were constructed using the TruSeq RNA Sample Preparation Kit (Illumina) and then sequenced on an Illumina HiSeq X Ten platform, which generates 150 bp pair-end reads.
After sequencing, the adaptor sequences and low-quality reads (quality score ≤ 20) were eliminated to obtain high quality clean reads using fastp (Chen et al., 2018). These clean reads were then aligned to a reference genome of L. crocea (NCBI accession: GCA_003845795.1) using HISAT2 (Kim et al., 2015). Cufflinks was used to assemble genes and identify differential expressed genes (DEGs) between control and treated group (Trapnell et al., 2013). In order to eliminate the influence of different gene lengths and sequencing discrepancies, we measured the gene expression level by the fragments per kilobase of exon model per million mapped reads (FPKM) based on the number of uniquely mapped reads. DESeq2 (Love et al., 2014) was used to analyze differential gene expression, and genes with statistically significant expression differences (p-value < 0.05, |log2(foldchange)| > 2) were considered to be DEGs.
All DEGs were functionally annotated based on the reference genome of L. crocea, which were annotated against the following four databases: NCBI non-redundant protein database (NR), Swiss-Prot database, Gene Ontology (GO) database and Kyoto Encyclopedia of Genes and Genomes (KEGG). In order to understand the function of these DEGs, GO functional enrichment analysis and KEGG pathway analysis were performed using the OmicShare tools (https://www.omicshare.com/tools/). The threshold for significant enrichment of gene sets was set at p < 0.05. Meanwhile, GO and KEGG enrichment analyses were performed on DEGs to explore their corresponding biological functions and associated pathways. The protein-protein interaction (PPI) network was constructed in STRING (https://string-db.org), a search tool for retrieving interacting proteins.
Before feeding the experimental feed 60 experimental fish were randomly picked and the initial average body weight was 150.8 ± 25.7 g. After feeding with the two experimental diets for 120 days, the final body weight of large yellow croaker in each experimental group were collected. The final weight of experimental fish in FC and PC were 372.4 ± 77.2 g and 367.0 ± 65.4 g, respectively (Figure 1). The results showed that there was no significant difference between the two groups (p > 0.05).
Sequencing of 12 RNA samples from liver and intestine tissues yielded a total of 306,592,074 raw reads. After filtering, the number of clean reads of each sample ranged from 21,326,592 to 33,813,448. The clean reads rate of all samples was above 95% (Table 2). All clean reads were mapped to the large yellow croaker genome, and the average mapping rate of the 12 samples was 91.86%.
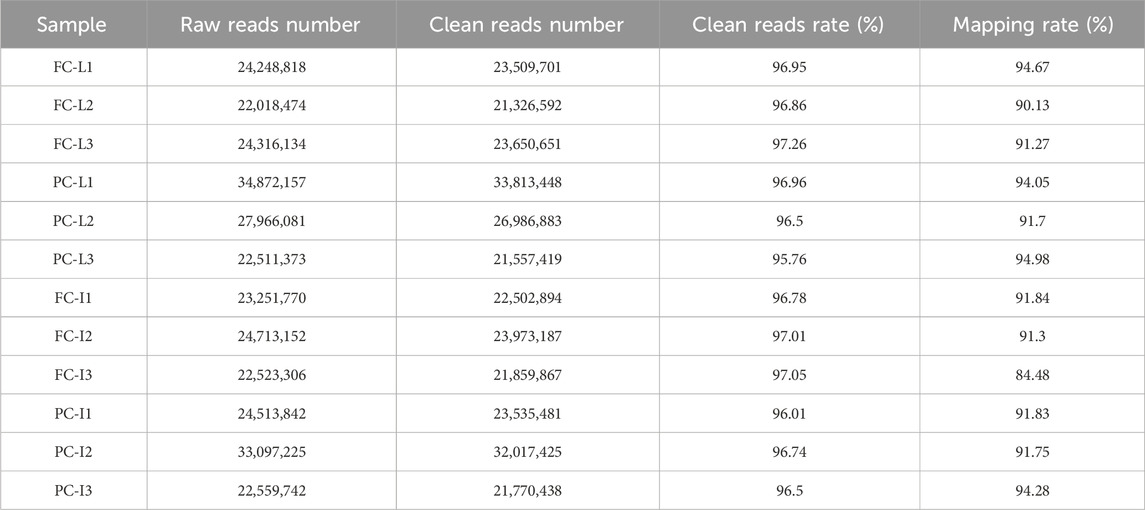
Table 2. Summary of RNA sequencing output and Clean reads mapping rate for liver and intestine samples.
By comparing the gene expression between the FC and PC groups, a total of 865 DEGs were identified. As shown in Figure 2, in the intestine tissue, the number of DEGs was 308, and in the liver tissue, it was 557.
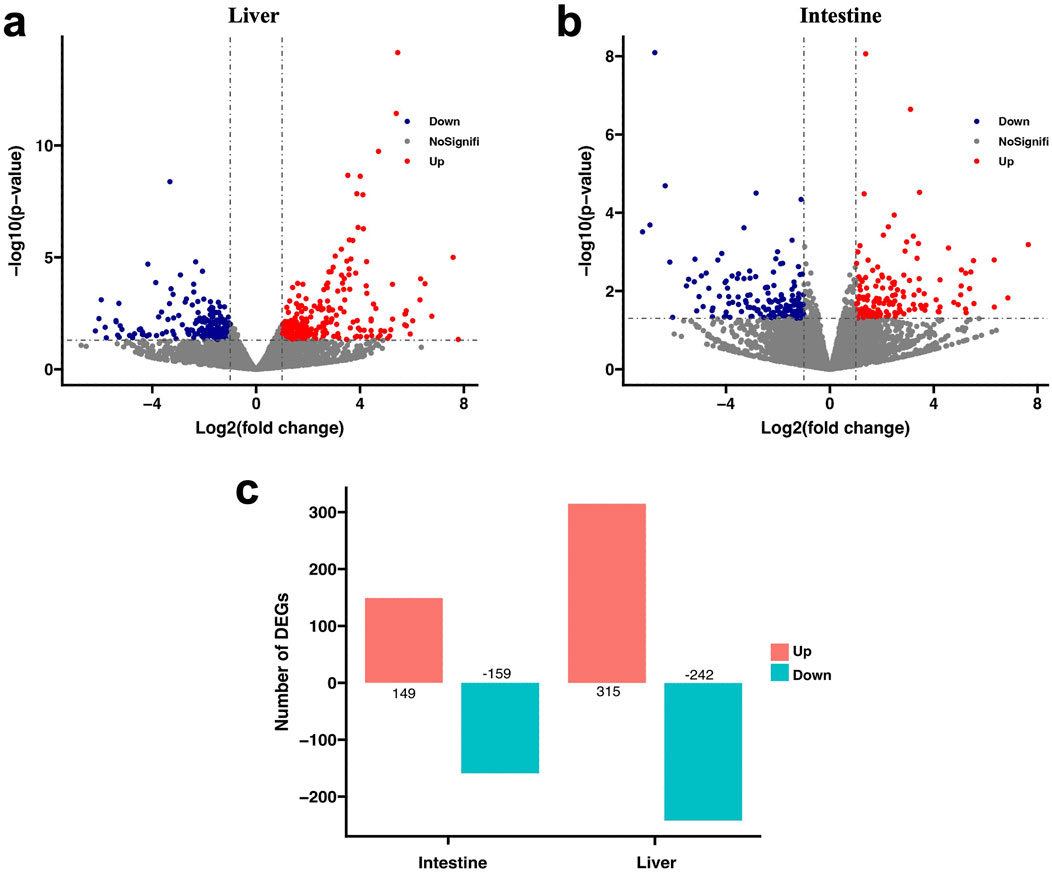
Figure 2. The volcano plot reveals the distribution of DEGs under different plant protein diets. (A) Volcano plot of DEGs in the liver. (B) Volcano plot of DEGs in the intestine. (C) Statistics of number of DEGs. Red spot: log2(fold change) ≥ 1, and p < 0.05; Blue spot: log2(fold change) ≤ −1, and p < 0.05; Black spot: no difference in expression.
GO analysis showed that 557 DEGs in the liver tissue were classified into 671 GO terms (p < 0.05), including 538 in biological process category, 90 in molecular function categories, and 43 cellular component categories. GO terms related to immune and digestive absorption were significantly enriched, such as acid secretion, positive regulation of antigen receptor-mediated signaling pathway, and lymphocyte migration (Figure 3A; Supplementary Table 1). In the intestine tissue, 308 DEGs were classified into 378 GO terms (p < 0.05), including 292 in biological process category, 62 in molecular function category, and 24 cellular component categories. GO terms related to metabolism and intercellular interactions were significantly enriched, such as sterol metabolic process, fatty acid metabolic process, laminin binding, and collagen binding (Figure 3C; Supplementary Table 2).
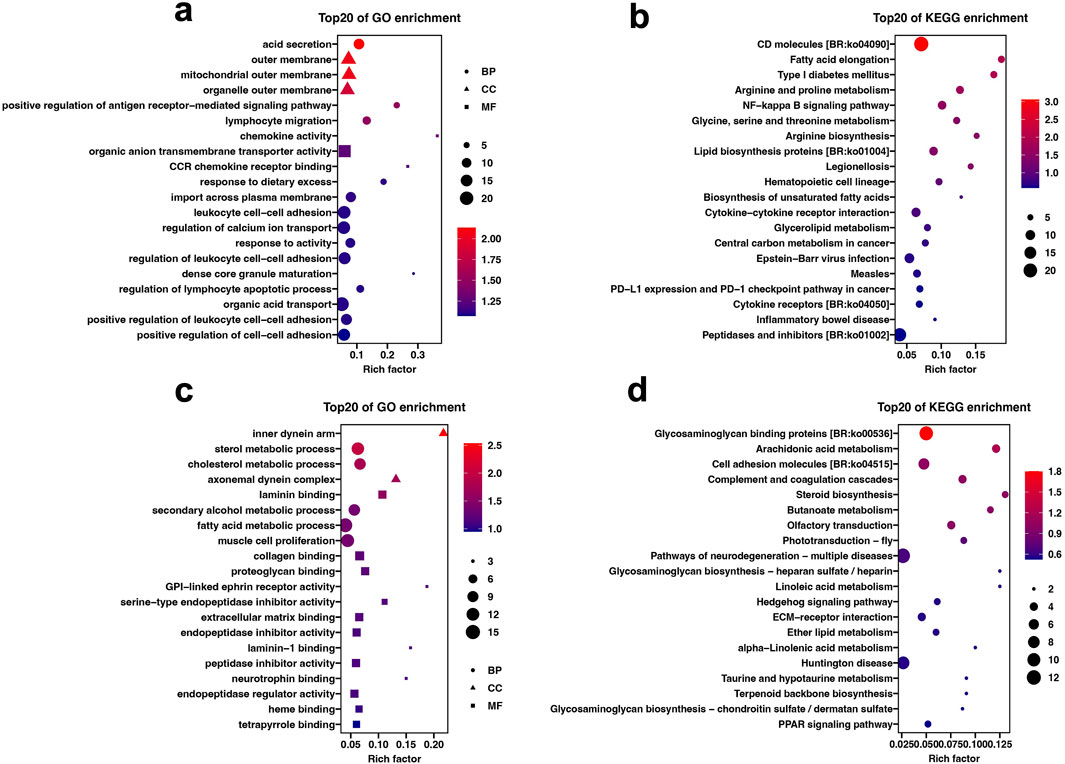
Figure 3. GO and KEGG enrichment analysis results of DEGs under different plant protein diets. (A) GO enrichment of DEGs in the Liver. (B) KEGG enrichment of DEGs in the Liver. (C) GO enrichment of DEGs in the intestine. (D) KEGG enrichment of DEGs in the intestine.
KEGG analysis showed that 32 and 26 KEGG pathways were significantly enriched in the liver and intestine tissue (p < 0.05), respectively. In the liver tissue, pathways related to immune-related processes, such as CD molecules, NF-kappa B signaling pathway, and Cytokine-cytokine receptor interaction were significant enriched. In addition, metabolic pathways such as Fatty acid elongation, Glycine, serine and threonine metabolism, and Biosynthesis of unsaturated fatty acids were also significantly enriched (Figure 3B; Supplementary Table 1). In the intestine tissue, pathways related to cell signaling transduction, such as Glycosaminoglycan binding proteins, ECM-receptor interaction and Glycosaminoglycan biosynthesis-heparan sulfate/heparin were significantly enriched. Additionally, immune and metabolic pathways, including Arachidonic acid metabolism, Steroid biosynthesis and Butanoate metabolism, were also markedly enriched (Figure 3D; Supplementary Table 2).
To further elucidate the impact of high plant protein diet on large yellow croaker, we conducted additional statistical analysis on the KEGG results. In immune-related pathways, we identified a total of 41 genes. Expression analysis revealed a downregulation trend in immune-related genes after high plant protein feeding. In the liver group, 17 genes showed upregulation, while 23 genes exhibited downregulation. CCL25 and Il12b showed the most significant downregulation, while VCAN and IL1R2 showing upregulation (Figure 4A; Supplementary Table 3). In the intestine group, 9 genes showed upregulation, and 31 genes showed downregulation, with DUOX2 gene downregulation being the most pronounced.

Figure 4. Heat map of DEGs expression (log2FC) in different pathways. (A) Immune regulation related gene expression. (B) Lipid metabolism related gene expression. (C) Cell singnaling transduction related gene expression.
In lipid metabolism-related pathways, we identified a total of 18 DEGs. Expression analysis revealed that these genes were primarily upregulated in the liver. For instance, genes such as AACS, LPIN1, GPAM, and DGKI showed upregulation, with only a few genes like GGT5 showing downregulation (Figure 4B; Supplementary Table 4). In contrast, in the intestine, most genes such as AACS, FABP1, and SCD showed significant downregulation, with only a small number of genes like CYP2U1 and GGT5 showing upregulation.
In addition to the significant enrichment of pathways related to immune response and lipid metabolism, our enrichment analysis also revealed a considerable enrichment of genes associated with cell signaling transduction. We identified a total of 15 DEGs, and expression analysis showed that in the liver, 8 genes were upregulated while 7 genes were downregulated. Among them, the upregulation of the VCAN gene was most pronounced, while ll12b showed significant downregulation (Figure 4C; Supplementary Table 5). In the intestine, only 6 genes were upregulated, with SLC8A3 and ITIH2 showing the most notable upregulation, while among the downregulated genes, SPAM1 and VCAN exhibited the most significant downregulation.
Furthermore, we performed a protein-protein interaction (PPI) network analysis on the DEGs related to immune response, lipid metabolism, and cell signaling transduction. As shown in the Figure 5 and Supplementary Table 6, numerous genes exhibit protein-protein interactions. Among them, five protein-coding genes—HADHB, HMGCS1, AACS, HSPA8, and HSPA5—had the highest levels of connectivity, with interaction scores greater than 0.7.
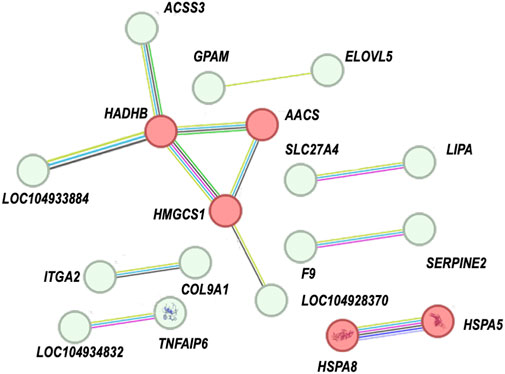
Figure 5. PPI network analysis of DEGs. The thickness of the line indicates the strength of the interaction between any two proteins, red colours indicate interaction scores greater than 0.7.
We compared the effects of standard commercial feed and a modified version, where 70% of the fish meal was replaced with plant protein, on large yellow croaker. We found that under the high plant protein feeding, the fish reached a growth rate similar to that of commercial feed feeding, indicating the feasibility of the high plant protein substitution strategy employed in this research. To deepen our understanding of the effects of plant protein substitution for fishmeal on fish physiology, we conducted an RNA-seq study to investigate the transcriptomic response of large yellow croaker under different plant protein replacement conditions. Extensive DEGs were observed through comparisons among different fishmeal replacement groups. Functional enrichment analysis of these DEGs revealed their association with various pathways, including immune regulation, lipid metabolism, and cell signaling transduction pathways, among others. These biological regulatory functions and processes may enhance our understanding of the response of fish to the substitution of fish meal with plant protein.
Both GO and KEGG analyses revealed that many DEGs are involved in immune regulation pathways and functions, such as the positive regulation of antigen receptor-mediated signaling pathways, CD molecules, and the NF-kappa B signaling pathway. Notably, these pathways exhibited particularly pronounced enrichment in the liver, that may be consistent with the liver’s role as a central hub in immune regulation (Hussein et al., 2023). We observed that many genes were significantly upregulated in the liver, particularly VCAN and IL1R2. VCAN, an extracellular matrix protein, plays a critical role in tissue repair and inflammatory responses (Xu et al., 2021). Its chondroitin sulfate domain can directly bind to CD44, amplifying antigen presentation signals and promoting the production of inflammatory cytokines (Wight et al., 2014). IL1R2, as a key regulatory factor in the IL-1 signaling pathway, may inhibit the excessive activation of the downstream NF-κB pathway by competitively binding to the IL-1β ligand, especially when its expression level is elevated (Zhou et al., 2020; Zhang et al., 2020). Together, they are involved in inflammatory pathways such as CD molecules. The changes in the expression of these genes indicate that a plant-based protein diet has induced an inflammatory response in fish. This is also consistent with previous studies. For instance, Geay et al. found that plant protein diet significantly changed the immune levels in the liver of Dicentrarchus labra through immune indicators (Geay et al., 2011). Novriadi et al. reported that plant protein diet promoted inflammation levels in the liver of Trachinotus carolinus based on serum biochemistry analysis (Novriadi et al., 2018). In transcriptomic studies, changes in the expression of immune genes were also observed in the liver tissues of O. mykiss (Panserat et al., 2009) and S. salar (Caballero-Solares et al., 2018) following a plant protein diet. However, further analysis combining immune, biochemical, and morphological indicators is necessary to determine whether a plant protein diet would cause immune damage in large yellow croaker.
There are not many significantly enriched immune pathways observed in the intestine, and the expression of several key genes, such as DUOX2 and NPTN, is highly downregulated. DUOX2, as a core enzyme for the production of reactive oxygen species, when downregulated, may directly lead to a decline in the intestinal antibacterial capacity (Dobrzyn and Ntambi, 2005; Grasberger et al., 2015). NPTN, as a synaptic regulatory protein, potentially affecting the signaling between the enteric nervous system and immune cells, leading to an imbalance in immune tolerance (Beesley et al., 2014). The significant downregulation of DUOX2 and NPTN suggests that the mucosal barrier function may be compromised. The intestine not only serves as a site for nutrient absorption but also plays a crucial role in immune function. Intestinal cells closely interact with immune cells such as macrophages, participating in antigen presentation and the regulation of T cell responses (Hershberg and Mayer, 2000). The downregulation of the aforementioned genes may indicate an immune imbalance in the intestine caused by a plant protein diet. Further observations will be conducted in conjunction with immune and immunological indicators. Understanding these mechanisms could provide insights into practical dietary formulations that optimize fish health and immune function amidst the growing reliance on plant-based feeds.
Differences in fatty acid composition between plant proteins and fish meal will inevitably affect the efficiency of lipid metabolism when fed over the long term (Dani, 2018). In this study, we identified DEGs related to fatty acid synthesis, transport, metabolism, and lipid synthesis and regulation, with most of them showing downregulation in the intestine. In the intestine, there are significantly more pathways and genes related to lipid metabolism compared to immune regulation. This may be aligned with the intestine’s primary function of nutrient absorption (Dawood, 2021). For example, the FABP1 gene, which is a key carrier involved in intracellular lipid transport, shows significant downregulation that may impair the absorption and intracellular distribution of long-chain fatty acids (Assaily et al., 2011). Additionally, the suppression of SCD expression will directly lead to a reduction in the synthesis of monounsaturated fatty acids (Li et al., 2019). The Acetoacetyl-CoA synthase (AACS) gene is also involved, and defects in AACS function may hinder lipid storage by inhibiting the conversion of ketone bodies to acetyl-CoA (Ohgami et al., 2003). Given these roles, we hypothesize that replacing fish meal with plant protein may result in inadequate absorption of the materials necessary for lipid synthesis in the intestine. Previous transcriptomic studies on other carnivorous fish have yielded similar results. For example, studies on S. salar (Król et al., 2016), Seriola lalandi (Dam et al., 2020) and O. mykiss (Lazzarotto et al., 2018) have shown comparable findings. Estruch et al. also found through proteomic analysis that the intestinal mucosal proteome of S. aurata, L. was disrupted following the feeding of plant protein fishmea (Estruch et al., 2020). Evidently, it can still cause damage to the intestine at a microscopic level.
It is noteworthy that the AACS gene is significantly upregulated in the liver. Fatty acids serve as another crucial precursor in cholesterol synthesis. Consequently, Acetyl-CoA participates in cholesterol synthesis through a series of biochemical reactions (Duan et al., 2022). Cholesterol is exclusively sourced from animal organisms, when plant components replace a significant portion of fishmeal, the dietary cholesterol levels in the feed will decrease substantially, which may lead to alterations in cholesterol regulation (Deng et al., 2009). For instance, in rainbow trout, coping mechanisms against changes in nutritional characteristics and the lack of dietary cholesterol supply involve increasing cholesterol synthesis and limiting cholesterol efflux (Zhu et al., 2018). The liver is the primary center for synthesizing and regulating cholesterol (van der Wulp et al., 2013). Therefore, we speculate that the elevation of cholesterol synthesis-related genes in the liver may be due to the lack of cholesterol supply from plant protein, prompting large yellow croakers to initiate their own biosynthetic mechanisms to produce cholesterol to meet their nutritional needs. This also demonstrates a critical interplay between these organs in maintaining metabolic homeostasis.
In our study, we identified numerous pathways and functional genes associated with cell signaling transduction, specifically the Glycosaminoglycan (GAG) binding proteins pathway, ECM-receptor interaction, and the PPAR signaling pathway. Glycosaminoglycans (GAGs) are integral components of the extracellular matrix (ECM) that significantly influence cell adhesion and signal transduction, functioning in the regulation of cell growth and immune modulation (Gandhi and Mancera, 2008; Zhang and Zhang, 2010). The ECM not only maintains tissue structure but also plays a critical role in lipid metabolism by creating a conducive microenvironment for lipid synthesis. Prior research, particularly in chickens, has demonstrated that ECM components can substantially impact lipid metabolism (Mariman and Wang, 2010), underscoring the interconnectedness between the ECM and lipid regulatory mechanisms. The PPAR signaling pathway is particularly notable for its role in coordinating lipid metabolism and inflammatory responses (Zhang and Young, 2002). PPARs (Peroxisome Proliferator-Activated Receptors) are nuclear receptors that regulate genes involved in fatty acid uptake, storage, and β-oxidation, linking lipid homeostasis with inflammation modulation. The interplay among these pathways is crucial. For example, GAGs may alter ECM properties, thereby influencing PPAR activation. Hence, changes in GAGs or ECM components could directly impact PPAR signaling, affecting both lipid metabolism and inflammatory processes. Although there are no previous studies addressing these pathways, their intrinsic functional relationships suggest that further investigation is warranted.
Moreover, we observed that the DEGs involved in signal transduction were predominantly downregulated, particularly in the intestine. For instance, VCAN, previously linked to immune pathways, is essential for tissue repair and inflammatory responses (Xu et al., 2021). ITGA2, an integrin family member, is critical to the innate immune response and engages in various cellular processes, including cell development and metabolism (Zhu et al., 2013; Takada et al., 2007). Evidence suggests its involvement in cold stress responses in Oreochromis niloticus (El-Sayed et al., 2023) and immune cell migration in S. aurata (Espinosa-Ruíz and Esteban, 2023). The downregulation of the VCAN and ITGA2 genes may be closely related to the expression of immune genes in the intestine, as these genes are likely involved in regulating the expression of immune-related genes. In contrast, the upregulation of these genes in the liver suggests a tissue-specific adaptation to fulfill the metabolic and functional demands of this organ. This stark contrast highlights that while the intestine experiences disruptions in immune and metabolic pathways due to the downregulation of signal transduction genes, the liver compensates by enhancing the expression of genes associated with protective and reparative roles. Understanding the relationships among these genes is critical as we explore the potential of plant protein sources to replace fish meal in aquaculture diets. By optimizing the processes influenced by these genes, we can enhance the nutritional profiles of plant-based diets, thereby supporting fish growth, immunity, and resilience.
Protein-protein interaction (PPI) networks are complex network structures formed by protein interactions. These networks reflect the interactions between proteins in an organism and help us better understand the complexity and regulatory mechanisms of biological systems. In this study, we identified PPI among some genes, and these genes are involved in the immune, metabolic, and cell signaling pathways we mentioned earlier. This indicates that high plant protein feeding leads to a series of integrated responses.
Among them, HADHB, HMGCS1, AACS, HSPA8, and HSPA5 exhibit the highest level of connectivity. HADHB, HMGCS1, and AACS are involved in regulating fatty acid metabolism (Ohgami et al., 2003; Xie et al., 2023; Gu et al., 2024). HSPA8, HSPA5, and others may play a role in immune responses (van der Wulp et al., 2013; Tian et al., 2023). Further research is needed to explore the relationships between their interactions. In conclusion, the discovery of these genes and their related functions further enhances the understanding of the mechanism behind high plant protein substitution for fish meal in feeding large yellow croaker. The interconnectedness of these genes highlights a complex regulatory network that mediates both metabolic and immune functions, illustrating how these processes are vital for adapting to high levels of plant protein substitution in diets. Further research is needed to explore the relationships between their interactions to fully understand how they collectively influence fish physiology. In conclusion, the discovery of these genes and their associated functions significantly enriches our understanding of the mechanisms underpinning the effective substitution of fish meal with plant protein in large yellow croaker diets.
For carnivorous fish such as the large yellow croaker, with the expansion of aquaculture production, feeding fresh mixed fish and high-fishmeal pellet feed not only leads to environmental pollution but also results in a shortage of fishmeal supply and high prices (Mu et al., 2017). However, achieving complete substitution with plant protein diet still poses challenges. It is necessary to have a deeper understanding of the molecular changes caused by plant protein diet. Research on plant protein diets has been conducted in various carnivorous fish species. For instance, in S. salar, replacing 45% of fishmeal with soy protein concentrate does not lead to significant differences in growth or intestinal inflammation. Similarly, in S. lalandi, replacing 50% of fishmeal with soy protein concentrate also does not result in notable differences in growth or intestinal inflammation (Król et al., 2016; Dam et al., 2020). In a high-plant protein fishmeal diet, replacing 60% of fishmeal with defatted cottonseed meal (CSM) and corn protein concentrate in T. ovatus, as well as substituting 80% of fishmeal with soy protein concentrate in S. salar, has been shown to promote inflammation (Caballero-Solares et al., 2018; Fan et al., 2021). Furthermore, high-plant protein diets have been associated with reduced growth rates. Similarly, in O. mykiss, a study by Panserat et al. that replaced 100% of fishmeal with corn protein concentrate and soybean meal over 9.5-week trial found a decrease in growth rates (Panserat et al., 2009). Additionally, Cao et al. conducted a continuous 84-day experiment using 80% soybean meal as a fishmeal substitute and also observed a decline in growth performance (Cao et al., 2022). In our study, we further modified the commercial feed to achieve a higher plant protein content. We found that the body weight changes in large yellow croaker caused by high plant protein diet are comparable to those with low plant protein feeding. This differs from the results observed in O. mykiss and S. salar, suggesting potential species-specific responses. Additionally, the large yellow croaker is currently commonly fed diets with 30% plant protein fishmeal, which may accelerate the adaptation to plant protein diets. However, from the perspective of transcriptome analysis, we also observed changes in immune and metabolic pathways influenced by plant protein. This suggests that a high plant protein diet may have potential effects, such as promoting inflammation, and the long-term effects still require further investigation.
Dehulled soybean meal and vital wheat gluten contain various anti-nutritional factors (ANFs), such as phytic acid, tannins, protease inhibitors, and oligosaccharides, which can affect growth, digestion, absorption, and antioxidant capacity, as well as cause damage to the intestines of aquatic animals (Lin and Cheng, 2017). In this study, we simultaneously analyzed molecular changes in both the liver and intestine and found that although the impact of a high plant protein diet on growth was minimal, there were observable effects at the molecular level on the immune function of the liver and metabolic processes in the intestine of large yellow croaker. We speculate that plant proteins may directly disrupt intestinal absorption and affect the immune function of the liver. While no differences were noted in the short-term feeding trials, the potential for long-term effects cannot be ruled out. Of course, our findings are specific to carnivorous fish, as herbivorous and omnivorous fish have a lower reliance on fishmeal compared to carnivorous species (Dhar et al., 2024). These findings offer significant insights into feed formulation, suggesting that it is crucial to acknowledge the potential trade-offs associated with high plant protein diets. To address the ANFs present in plant proteins, we recommend several actionable solutions. a) processing techniques such as heat treatment, fermentation, and enzymatic hydrolysis can be employed to reduce the levels of these factors, thereby improving nutrient availability (Handa et al., 2017; Mugwanya et al., 2023). b) Additionally, incorporating functional additives like inosine (Hossain et al., 2018) and taurine (Ye et al., 2019) into plant protein-based feeds can significantly enhance the availability of essential amino acids, improving health and disease resistance while mitigating the overall antioxidant effects of plant proteins. c) we suggest that aquaculture enterprises adopt a phased implementation framework, starting with small-scale trials at farming sites to evaluate effectiveness. Based on the results, the application can be gradually expanded, with regular monitoring of fish immune function and metabolic health to make timely adjustments to feed formulations. Continuous monitoring is essential to ensure the health and performance of the fish.
Our future research will focus on investigating the specific impacts of high plant protein diets on the immune system and metabolism of large yellow croaker. We will also explore ways to integrate immune-boosting ingredients, into feed formulations. These efforts aim to optimize feed while addressing concerns related to ANFs. Ultimately, these strategies not only promote improved fish health and growth but also contribute to more sustainable aquaculture practices.
In this study, the response of transcriptome to high levels of fishmeal replacement was examined in large yellow croaker. Based on our research findings, a high plant protein diet for large yellow croaker achieved the same weight gain as a low plant protein diet. Transcriptome analysis results show that 557 and 308 DEGs in the intestine and liver were identified, respectively. GO and KEGG analyses indicated significant enrichment in metabolism-related pathways such as lipid metabolism, immune regulation, and cell signaling transduction pathways. Expression analysis revealed tissue-specific expression of these genes, and the PPI network further revealed their interactions, collectively participating in the molecular regulation of high plant protein diets. These insights are vital for developing optimized aquafeed that enhances sustainability in aquaculture. Future steps should include testing specific feed additives to mitigate immune suppression and extending this analysis to other fish species, particularly carnivorous fish, to enhance the adaptability of various species to high plant protein diets. This work contributes to the advancement of sustainable practices in the aquaculture industry.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1119058.
The animal studies were approved by Animal Care and Use committee at College of Ocean and Earth Sciences, Xiamen University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
QK: Writing–original draft, Writing–review and editing. YL: Writing–original draft, Writing–review and editing. HW: Funding acquisition, Resources, Supervision, Writing–review and editing. BC: Investigation, Software, Writing–review and editing. JW: Formal Analysis, Investigation, Software, Writing–review and editing. JZ: Data curation, Formal Analysis, Investigation, Software, Writing–review and editing. PJ: Investigation, Methodology, Software, Writing–review and editing. PX: Conceptualization, Formal Analysis, Funding acquisition, Resources, Supervision, Visualization, Writing–review and editing. TZ: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Resources, Supervision, Visualization, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the China Postdoctoral Science Foundation (2023M732954), Industry-University-Research Cooperation Project of Ningde (2023C003), National Science Fund for Distinguished Young Scholars (32225049), China Agriculture Research System (CARS-47) and “Science and Technology Innovation 2025” Major Special Project of Ningbo City (No. 2021Z002).
Thanks for all the participants and contributors.
Author HW was employed by Ningde Fufa Fisheries Company Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1540305/full#supplementary-material
SUPPLEMENTARY TABLE 1 | Enrichment results of differentially expressed genes in the liver.
SUPPLEMENTARY TABLE 2 | Enrichment results of differentially expressed genes in the intestine.
SUPPLEMENTARY TABLE 3 | Statistics of immune regulation related gene expression.
SUPPLEMENTARY TABLE 4 | Statistics of lipid metabolism related gene expression.
SUPPLEMENTARY TABLE 5 | Statistics of cell signaling transduction related gene expression.
SUPPLEMENTARY TABLE 6 | Statistical results of the protein-protein interaction network.
Aladetohun, N. (2013). Utilization of blood meal as a protein ingredient from animal waste product in the diet of Oreochromis niloticus. Available at: https://academicjournals.org/journal/IJFA/article-authors/F53855710365 (Accessed January 20, 2025).
Apper-Bossard, E., Feneuil, A., Wagner, A., and Respondek, F. (2013). Use of vital wheat gluten in aquaculture feeds. Aquat. Biosyst. 9, 21. doi:10.1186/2046-9063-9-21
Assaily, W., Rubinger, D. A., Wheaton, K., Lin, Y., Ma, W., Xuan, W., et al. (2011). ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol. Cell 44, 491–501. doi:10.1016/j.molcel.2011.08.038
Beesley, P. W., Herrera-Molina, R., Smalla, K.-H., and Seidenbecher, C. (2014). The Neuroplastin adhesion molecules: key regulators of neuronal plasticity and synaptic function. J. Neurochem. 131, 268–283. doi:10.1111/jnc.12816
Caballero-Solares, A., Xue, X., Parrish, C. C., Foroutani, M. B., Taylor, R. G., and Rise, M. L. (2018). Changes in the liver transcriptome of farmed Atlantic salmon (Salmo salar) fed experimental diets based on terrestrial alternatives to fish meal and fish oil. BMC Genomics 19, 796. doi:10.1186/s12864-018-5188-6
Cao, Y., Gao, Q., Li, X., Zhou, Y., Dong, S., Wang, Y., et al. (2022). Integrated analysis of metabolomics and transcriptomics for assessing effects of fish meal and fish oil replacement on the metabolism of rainbow trout (Oncorhynchus mykiss). Front. Mar. Sci. 9. doi:10.3389/fmars.2022.843637
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi:10.1093/bioinformatics/bty560
Dam, C. T. M., Ventura, T., Booth, M., Pirozzi, I., Salini, M., Smullen, R., et al. (2020). Intestinal transcriptome analysis highlights key differentially expressed genes involved in nutrient metabolism and digestion in yellowtail kingfish (Seriola lalandi) fed terrestrial animal and plant proteins. Genes 11, 621. doi:10.3390/genes11060621
Damon, M., Denieul, K., Vincent, A., Bonhomme, N., Wyszynska-Koko, J., and Lebret, B. (2013). Associations between muscle gene expression pattern and technological and sensory meat traits highlight new biomarkers for pork quality assessment. Meat Sci. 95, 744–754. doi:10.1016/j.meatsci.2013.01.016
Dani, D. (2018). A review on replacing fish meal in aqua feeds using plant protein sources. Int. J. Fish. Aquatic Stud. 6, 164–179.
Dawood, M. A. O. (2021). Nutritional immunity of fish intestines: important insights for sustainable aquaculture. Rev. Aquac. 13, 642–663. doi:10.1111/raq.12492
Deng, J., Mai, K., Ai, Q., Zhang, W., Wang, X., Tan, B., et al. (2009). Interactive effects of dietary cholesterol and protein sources on growth performance and cholesterol metabolism of Japanese flounder (Paralichthys olivaceus): interactive effects of dietary cholesterol and protein sources. Aquacult Nutr. 16, 419–429. doi:10.1111/j.1365-2095.2009.00681.x
Dhar, V., Singh, S. K., Narsale, S. A., Debbarma, S., Saikia, P., and Yirang, Y. (2024). Fishmeal substitutions and their implications for aquatic animal immune and gut function: a review. Comp. Immunol. Rep. 7, 200171. doi:10.1016/j.cirep.2024.200171
Dobrzyn, A., and Ntambi, J. M. (2005). The role of stearoyl-CoA desaturase in the control of metabolism. Prostagl. Leukot. Essent. Fat. Acids 73, 35–41. doi:10.1016/j.plefa.2005.04.011
Duan, Y., Gong, K., Xu, S., Zhang, F., Meng, X., and Han, J. (2022). Regulation of cholesterol homeostasis in health and diseases: from mechanisms to targeted therapeutics. Signal Transduct. Target. Ther. 7, 265. doi:10.1038/s41392-022-01125-5
El-Sayed, A.-F. M., Khaled, A. A., Hamdan, A. M., Makled, S. O., Hafez, E. E., and Saleh, A. A. (2023). The role of antifreeze genes in the tolerance of cold stress in the Nile tilapia (Oreochromis niloticus). BMC Genomics 24, 476. doi:10.1186/s12864-023-09569-x
Espe, M., El-Mowafi, A., Ruohonen, K., Espe, M., El-Mowafi, A., and Ruohonen, K. (2012). Replacement of fishmeal with plant protein ingredients in diets to atlantic salmon (Salmo salar) – effects on weight gain and accretion. Aquac. IntechOpen. doi:10.5772/29975
Espinosa-Ruíz, C., and Esteban, M. Á. (2023). Modulation of cell migration and cell tracking of the gilthead seabream (Sparus aurata) SAF-1 cells by probiotics. Fish. Shellfish Immunol. 142, 109149. doi:10.1016/j.fsi.2023.109149
Estruch, G., Martínez-Llorens, S., Tomás-Vidal, A., Monge-Ortiz, R., Jover-Cerdá, M., Brown, P. B., et al. (2020). Impact of high dietary plant protein with or without marine ingredients in gut mucosa proteome of gilthead seabream (Sparus aurata, L.). J. Proteomics 216, 103672. doi:10.1016/j.jprot.2020.103672
Fan, J.-Q., Lu, K.-C., Chen, G.-L., Li, B.-B., Song, F., and Chen, Y.-H. (2021). Transcriptome analysis of the influence of high plant protein based diet on Trachinotus ovatus liver. Fish. Shellfish Immunol. 119, 339–346. doi:10.1016/j.fsi.2021.10.013
Gandhi, N. S., and Mancera, R. L. (2008). The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des. 72, 455–482. doi:10.1111/j.1747-0285.2008.00741.x
Geay, F., Ferraresso, S., Zambonino-Infante, J. L., Bargelloni, L., Quentel, C., Vandeputte, M., et al. (2011). Effects of the total replacement of fish-based diet with plant-based diet on the hepatic transcriptome of two European sea bass (Dicentrarchus labrax) half-sibfamilies showing different growth rates with the plant-based diet. BMC Genomics 12, 522–618. doi:10.1186/1471-2164-12-522
Gómez-Requeni, P., Mingarro, M., Calduch-Giner, J. A., Médale, F., Martin, S. A. M., Houlihan, D. F., et al. (2004). Protein growth performance, amino acid utilisation and somatotropic axis responsiveness to fish meal replacement by plant protein sources in gilthead sea bream (Sparus aurata). Aquaculture 232, 493–510. doi:10.1016/S0044-8486(03)00532-5
Grasberger, H., Gao, J., Nagao-Kitamoto, H., Kitamoto, S., Zhang, M., Kamada, N., et al. (2015). Increased expression of DUOX2 is an epithelial response to mucosal dysbiosis required for immune homeostasis in mouse intestine. Gastroenterology 149, 1849–1859. doi:10.1053/j.gastro.2015.07.062
Gu, D., Ye, M., Zhu, G., Bai, J., Chen, J., Yan, L., et al. (2024). Hypoxia upregulating ACSS2 enhances lipid metabolism reprogramming through HMGCS1 mediated PI3K/AKT/mTOR pathway to promote the progression of pancreatic neuroendocrine neoplasms. J. Transl. Med. 22, 93. doi:10.1186/s12967-024-04870-z
Gu, Z., Mu, H., Shen, H., Deng, K., Liu, D., Yang, M., et al. (2019). High level of dietary soybean oil affects the glucose and lipid metabolism in large yellow croaker Larimichthys crocea through the insulin-mediated PI3K/AKT signaling pathway. Comparative Biochemistry and Physiology Part B. Biochem. and Mol. Biol. 231, 34–41. doi:10.1016/j.cbpb.2018.12.003
Han, D., Shan, X., Zhang, W., Chen, Y., Wang, Q., Li, Z., et al. (2018). A revisit to fishmeal usage and associated consequences in Chinese aquaculture. Rev. Aquacult 10, 493–507. doi:10.1111/raq.12183
Handa, V., Kumar, V., Panghal, A., Suri, S., and Kaur, J. (2017). Effect of soaking and germination on physicochemical and functional attributes of horsegram flour. J. Food Sci. Technol. 54, 4229–4239. doi:10.1007/s13197-017-2892-1
Hardy, R. W. (2010). Utilization of plant proteins in fish diets: effects of global demand and supplies of fishmeal. Aquac. Res. 41, 770–776. doi:10.1111/j.1365-2109.2009.02349.x
He, J., Wang, P., and Lou, Y. (2017). Effects of replacing fish meal with corn gluten meal on the growth, serum biochemical indices and liver histology of large yellow croaker Larimichthys crocea. Acta Hydrobiol. Sin. 41, 506–515. doi:10.7541/2017.65
Hershberg, R. M., and Mayer, L. F. (2000). Antigen processing and presentation by intestinal epithelial cells – polarity and complexity. Immunol. Today 21, 123–128. doi:10.1016/S0167-5699(99)01575-3
Hossain, S., Koshio, S., Ishikawa, M., Yokoyama, S., Sony, N. M., Islam, J., et al. (2018). Substitution of dietary fishmeal by soybean meal with inosine administration influences growth, digestibility, immunity, stress resistance and gut morphology of juvenile amberjack Seriola dumerili. Aquaculture 488, 174–188. doi:10.1016/j.aquaculture.2018.01.037
Hussein, M. M., Sayed, R. K. A., and Mokhtar, D. M. (2023). Structural and immunohistochemical analysis of the cellular compositions of the liver of molly fish (Poecilia sphenops), focusing on its immune role. Zool. Lett. 9, 1. doi:10.1186/s40851-022-00200-7
Kaushik, S. J., Covès, D., Dutto, G., and Blanc, D. (2004). Almost total replacement of fish meal by plant protein sources in the diet of a marine teleost, the European seabass, Dicentrarchus labrax. Aquaculture 230, 391–404. doi:10.1016/S0044-8486(03)00422-8
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi:10.1038/nmeth.3317
Król, E., Douglas, A., Tocher, D. R., Crampton, V. O., Speakman, J. R., Secombes, C. J., et al. (2016). Differential responses of the gut transcriptome to plant protein diets in farmed Atlantic salmon. BMC Genomics 17, 156. doi:10.1186/s12864-016-2473-0
Kumar, A. (2019). Understanding trash fish value chain: a research on of fish oil and fish meal (fofm) plant in namakkal. Int. J. Innovative Technol. Explor. Eng. 8, 2778–2781. doi:10.35940/ijitee.I8998.078919
Lazzarotto, V., Médale, F., Larroquet, L., and Corraze, G. (2018). Long-term dietary replacement of fishmeal and fish oil in diets for rainbow trout (Oncorhynchus mykiss): effects on growth, whole body fatty acids and intestinal and hepatic gene expression. PLoS ONE 13, e0190730. doi:10.1371/journal.pone.0190730
Lee, K.-J., Powell, M. S., Barrows, F. T., Smiley, S., Bechtel, P., and Hardy, R. W. (2010). Evaluation of supplemental fish bone meal made from Alaska seafood processing byproducts and dicalcium phosphate in plant protein based diets for rainbow trout (Oncorhynchus mykiss). Aquaculture 302, 248–255. doi:10.1016/j.aquaculture.2010.02.034
Li, F., Li, D., Zhang, M., Sun, J., Li, W., Jiang, R., et al. (2019). miRNA-223 targets the GPAM gene and regulates the differentiation of intramuscular adipocytes. Gene 685, 106–113. doi:10.1016/j.gene.2018.10.054
Li, H. T., Mai, K. S., Ai, Q. H., Zhang, L., Zhang, C., Zhang, W., et al. (2007). Apparent digestibility of selected protein ingredients for large yellow croaker. Acta Hydrobiol. Sin. 31, 370–376. doi:10.3724/issn1000-3207-2007-3-370-g
Li, J., Zhang, L., Mai, K., Ai, Q., Zhang, C., Li, H., et al. (2010). Potential of several protein sources as fish meal substitutes in diets for large yellow croaker, Pseudosciaena crocea R. J. World Aquac. Soc. 41, 278–283. doi:10.1111/j.1749-7345.2010.00368.x
Lin, Y.-H., and Cheng, M.-Y. (2017). Effects of dietary organic acid supplementation on the growth, nutrient digestibility and intestinal histology of the giant grouper Epinephelus lanceolatus fed a diet with soybean meal. Aquaculture 469, 106–111. doi:10.1016/j.aquaculture.2016.11.032
Liu, L., and Peng, K. (2021). Research progress on substitution of iced trash fish with aquatic compound feed. Feed Res. 44, 141–144. doi:10.13557/j.cnki.issn1002-2813.2021.21.031
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi:10.1186/s13059-014-0550-8
Mariman, E. C. M., and Wang, P. (2010). Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell Mol. Life Sci. 67, 1277–1292. doi:10.1007/s00018-010-0263-4
Mu, H., Wei, Z., Yi, L., Liang, H., Zhao, L., Zhang, W., et al. (2017). Dietary fishmeal levels affect the volatile compounds in cooked muscle of farmed large yellow croaker Larimichthys crocea. Aquac. Res. 48, 5821–5834. doi:10.1111/are.13405
Mugwanya, M., Dawood, M. A. O., Kimera, F., and Sewilam, H. (2023). Replacement of fish meal with fermented plant proteins in the aquafeed industry: a systematic review and meta-analysis. Rev. Aquac. 15, 62–88. doi:10.1111/raq.12701
Novriadi, R., Rhodes, M., Powell, M., Hanson, T., and Davis, D. A. (2018). Effects of soybean meal replacement with fermented soybean meal on growth, serum biochemistry and morphological condition of liver and distal intestine of Florida pompanoTrachinotus carolinus. Aquacult Nutr. 24, 1066–1075. doi:10.1111/anu.12645
Ohgami, M., Takahashi, N., Yamasaki, M., and Fukui, T. (2003). Expression of acetoacetyl-CoA synthetase, a novel cytosolic ketone body-utilizing enzyme, in human brain. Biochem. Pharmacol. 65, 989–994. doi:10.1016/s0006-2952(02)01656-8
Oliva-Teles, A., Enes, P., Couto, A., and Peres, H. (2022). “8 - replacing fish meal and fish oil in industrial fish feeds,” in Feed and feeding practices in aquaculture. Editor D. A. Davis Second Edition (Oxford: Woodhead Publishing), 231.
Panserat, S., Hortopan, G. A., Plagnes-Juan, E., Kolditz, C., Lansard, M., Skiba-Cassy, S., et al. (2009). Differential gene expression after total replacement of dietary fish meal and fish oil by plant products in rainbow trout (Oncorhynchus mykiss) liver. Aquaculture 294, 123–131. doi:10.1016/j.aquaculture.2009.05.013
Salze, G., McLean, E., Battle, P. R., Schwarz, M. H., and Craig, S. R. (2010). Use of soy protein concentrate and novel ingredients in the total elimination of fish meal and fish oil in diets for juvenile cobia, Rachycentron canadum. Aquaculture 298, 294–299. doi:10.1016/j.aquaculture.2009.11.003
Shahin, S., Okomoda, V., Ma, H., and Abdullah, M. (2023). Sustainable alternative feed for aquaculture: state of the art and future perspective. Planet. Sustain. 1, 62–96. doi:10.46754/ps.2023.07.005
Shi, H., Xie, J., Wang, W., He, J., and Xu, W. (2017). Preliminary study of a new virus pathogen that caused the white-gill disease in cultured Larimichthys crocea. J. Fish. China 41, 1455–1463. doi:10.11964/jfc.20160610430
Subasinghe, R. P., Bueno, P. B., Phillips, M. J., Hough, C., McGladdery, S. E., and Arthur, J. R. (2001). Aquaculture in the third millennium: technical proceedings of the conference on aquaculture in the third millennium. Bangkok: Network of Aquaculture Centres in Asia-Pacific (NACA) and Food and Agriculture Organization of the UN (FAO).
Tacon, A. G. J., and Metian, M. (2008). Global overview on the use of fish meal and fish oil in industrially compounded aquafeeds: trends and future prospects. Aquaculture 285, 146–158. doi:10.1016/j.aquaculture.2008.08.015
Takada, Y., Ye, X., and Simon, S. (2007). The integrins. Genome Biol. 8, 215. doi:10.1186/gb-2007-8-5-215
Tian, H., Ding, M., Guo, Y., Zhu, Z., Yu, Y., Tian, Y., et al. (2023). Effect of HSPA8 gene on the proliferation, apoptosis and immune function of HD11 cells. Dev. Comp. Immunol. 142, 104666. doi:10.1016/j.dci.2023.104666
Trapnell, C., Hendrickson, D. G., Sauvageau, M., Goff, L., Rinn, J. L., and Pachter, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53. doi:10.1038/nbt.2450
van der Wulp, M. Y. M., Verkade, H. J., and Groen, A. K. (2013). Regulation of cholesterol homeostasis. Mol. Cell Endocrinol. 368, 1–16. doi:10.1016/j.mce.2012.06.007
Wang, P., Zhu, J., Feng, J., He, J., Lou, Y., and Zhou, Q. (2017). Effects of dietary soy protein concentrate meal on growth, immunity, enzyme activity and protein metabolism in relation to gene expression in large yellow croaker Larimichthys crocea. Aquaculture 477, 15–22. doi:10.1016/j.aquaculture.2017.04.030
Wight, T. N., Kang, I., and Merrilees, M. J. (2014). Versican and the control of inflammation. Matrix Biol. 35, 152–161. doi:10.1016/j.matbio.2014.01.015
Xie, Y., Zhang, C., Qin, Q., Li, X., Guo, J., Dai, D., et al. (2023). Proteomics analysis of meat to identify goat intramuscular fat deposits potential biomarkers. Food Anal. Methods 16, 1191–1202. doi:10.1007/s12161-023-02483-8
Xu, Q., Li, B., Wang, Y., Wang, C., Feng, S., Xue, L., et al. (2021). Identification of VCAN as hub gene for diabetic kidney disease immune injury using integrated bioinformatics analysis. Front. Physiol. 12, 651690. doi:10.3389/fphys.2021.651690
Ye, H., Xu, M., Liu, Q., Sun, Z., Zou, C., Chen, L., et al. (2019). Effects of replacing fish meal with soybean meal on growth performance, feed utilization and physiological status of juvenile obscure puffer, Takifugu obscurus. Comp. Biochem. Physiology Part C Toxicol. and Pharmacol. 216, 75–81. doi:10.1016/j.cbpc.2018.11.006
Zhang, H., Yi, L., Sun, R., Zhou, H., Xu, W., Zhang, W., et al. (2016). Effects of dietary citric acid on growth performance, mineral status and intestinal digestive enzyme activities of large yellow croaker Larimichthys crocea (Richardson, 1846) fed high plant protein diets. Aquaculture 453, 147–153. doi:10.1016/j.aquaculture.2015.11.032
Zhang, L. (2010). “Glycosaminoglycan (GAG) biosynthesis and GAG-binding proteins,” in Progress in molecular biology and translational science. Glycosaminoglycans in development, health and disease. Editor L. Zhang (Academic Press), 1–17.
Zhang, L., Chu, Q., Liu, X., and Xu, T. (2020). microRNA-21 negatively regulates NF-κB signaling pathway via targeting IL1R1 in miiuy croaker. Dev. Comp. Immunol. 105, 103578. doi:10.1016/j.dci.2019.103578
Zhang, L., Mai, K., Ai, Q., Duan, Q., Zhang, C., Li, H., et al. (2008). Use of a compound protein source as a replacement for fish meal in diets of large yellow croaker, Pseudosciaena crocea R. J. World Aquac. Soc. 39, 83–90. doi:10.1111/j.1749-7345.2007.00134.x
Zhang, X., and Young, H. A. (2002). PPAR and immune system—what do we know? Int. Immunopharmacol. 2, 1029–1044. doi:10.1016/S1567-5769(02)00057-7
Zhou, N., Chen, L.-L., Chen, J., and Guo, Z.-P. (2020). Molecular characterization and expression analysis of IL-1β and two types of IL-1 receptor in barbel steed (Hemibarbus labeo). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 241, 110393. doi:10.1016/j.cbpb.2019.110393
Zhou, S., Smith, A. D., and Knudsen, E. E. (2015). Ending overfishing while catching more fish. Fish Fish. 16, 716–722. doi:10.1111/faf.12077
Zhu, L., Nie, L., Zhu, G., Xiang, L., and Shao, J. (2013). Advances in research of fish immune-relevant genes: a comparative overview of innate and adaptive immunity in teleosts. Dev. Comp. Immunol. 39, 39–62. doi:10.1016/j.dci.2012.04.001
Zhu, T., Corraze, G., Plagnes-Juan, E., Quillet, E., Dupont-Nivet, M., and Skiba-Cassy, S. (2018). Regulation of genes related to cholesterol metabolism in rainbow trout (Oncorhynchus mykiss) fed a plant-based diet. Am. J. Physiology-Regulatory, Integr. Comp. Physiology 314, R58–R70. doi:10.1152/ajpregu.00179.2017
Keywords: Larimichthys crocea, carnivorous fish, high plant protein, transcriptome, nutrition metabolism
Citation: Ke Q, Li Y, Weng H, Chen B, Wang J, Zhao J, Jiang P, Xu P and Zhou T (2025) Differential responses of the intestine and liver transcriptome to high levels of plant proteins in diets for large yellow croaker (Larimichthys crocea). Front. Genet. 16:1540305. doi: 10.3389/fgene.2025.1540305
Received: 05 December 2024; Accepted: 14 February 2025;
Published: 05 March 2025.
Edited by:
Vikash Kumar, Central Inland Fisheries Research Institute (ICAR), IndiaReviewed by:
Sofia Priyadarsani Das, National Taiwan Ocean University, TaiwanCopyright © 2025 Ke, Li, Weng, Chen, Wang, Zhao, Jiang, Xu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Zhou, enRAeG11LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.