
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Genet., 06 March 2025
Sec. Cancer Genetics and Oncogenomics
Volume 16 - 2025 | https://doi.org/10.3389/fgene.2025.1517513
This article is part of the Research TopicComparative Genomics and Functional Genomics Analyses in CancerView all 4 articles
 Gang Jin1*†
Gang Jin1*† Tao Guo1,2,3,4†
Tao Guo1,2,3,4† Jia-Wei Liu1
Jia-Wei Liu1 Han-Yu Yang1,5
Han-Yu Yang1,5 Jian-Guo Xu4,6
Jian-Guo Xu4,6 Yao Pang1
Yao Pang1 Yi Yang1
Yi Yang1 Shao-E. He4,7,8
Shao-E. He4,7,8 Kang Yi2,4,6*
Kang Yi2,4,6*Background: miR-155 is overexpressed in many cancers, highlighting its potential as a biomarker for cancer diagnosis, treatment, and therapeutic evaluation. miR-155 is processed from the miR-155 host gene (MIR155HG). Genetic variations in MIR155HG may influence cancer susceptibility, but existing evidence is inconclusive. This study aimed to evaluate the association of MIR155HG polymorphisms with cancer risk.
Material/Methods: A systematic literature search identified 15 case-control studies on three single nucleotide polymorphisms (SNPs): rs767649 (T > A), rs928883 (G > A), and rs1893650 (T > C). Meta-analysis was performed using RevMan 5.4, with odds ratios (ORs) and 95% confidence intervals (CIs) as effect measures.
Results: No significant association was observed for rs767649 and rs928883 in overall cancer analysis. However, subgroup analysis revealed rs767649 increased susceptibility to respiratory, digestive, and reproductive cancers, while reducing cancer risk after excluding reproductive cancers. rs928883 showed a protective effect for digestive cancers. rs1893650 was not significantly associated with cancer risk.
Conclusion: MIR155HG polymorphisms influence susceptibility to specific cancer subtypes, particularly respiratory and digestive cancers. These findings underscore the importance of genetic and environmental factors in cancer risk and warrant further investigation.
The onset of cancer originates from abnormalities in cell proliferation and apoptosis (Thompson, 1995), which result from dysregulation of intracellular signaling pathways governing these processes (Manzo-Merino et al., 2014). Cancer is a critical public health issue worldwide and remains one of the leading causes of morbidity and mortality globally (Siegel et al., 2023), and poses a significant global health burden. The etiology of cancer is highly complex, with factors such as smoking, alcohol consumption, environmental factors, and viral infections potentially contributing to the development of various types of cancer. Nevertheless, the exact mechanisms underlying cancer development are not yet fully understood. There is increasing evidence of a complex interplay between genetic and environmental factors in the initiation and progression of cancer (Lichtenstein et al., 2000; Pharoah et al., 2004). Among these, single nucleotide polymorphisms (SNPs) have been extensively studied as they may alter cancer susceptibility (Hanahan and Weinberg, 2011), indicating that the detection of genetic risk factors could suggest cancer risk and prognosis, further enhancing cancer diagnostic capabilities.
microRNA (miRNA) refers to small, non-coding single-stranded RNA molecules encoded by endogenous genes, with a length of approximately 20–24 nucleotides. Mature miRNAs bind to the 3′untranslated region (3′UTR) of target gene mRNAs through sequence complementarity. When the miRNA and the target mRNA are not fully complementary, the expression of the target gene can be suppressed at the level of protein translation. In contrast, when the miRNA is fully or nearly fully in a complementary manner to the target mRNA, it leads to the degradation of the target gene (Bartel, 2004), thus regulating the expression of the target gene and mediating processes such as cell proliferation, metabolism, development, and differentiation. In 1993, Lee and colleagues first discovered that lin4 encoded miRNA in C. elegans (Caenorhabditis elegans) (Lee et al., 1993), which binds complementarily to the 3′UTR, controlling the development of C. elegans by inhibiting the expression of target gene mRNAs. microRNA-155 (miR-155) is located in the third exon of the non-coding gene B-cell integration cluster (BIC) on human chromosome 21. It is a non-coding RNA consisting of 23 nucleotides, and its expression is influenced by various steps of BIC transcription and miRNA processing. As a member of the miRNA family, miR-155 exhibits the biological functions typical for miRNAs, not only promote tumorigenesis by inducing mutational phenotypes that lead to a high mutation rate, but also create a gene expression environment that is especially susceptible to malignant transformation (Tili et al., 2011; Witten et al., 2019). miR-155 can directly or indirectly regulate the PI3K signaling pathway, inducing cell proliferation (Wu and Wang, 2020), migration (Fang et al., 2024), invasion, and apoptosis of tumor cells, thereby playing a role in the regulation of cancer development and progression (Kong et al., 2016; Wu et al., 2017) (Figure 1). A large body of research has demonstrated that miR-155 plays a role in various human cancers (Donnem et al., 2011; Sandhu et al., 2012), and circulating plasma levels of miR-155 can serve as a biomarker for predicting cancer risk (Lao and Le, 2019; Wu et al., 2023). Various clinical studies have shown that miR-155 is highly expressed in various malignant tumor tissues, including lung cancer, breast cancer, pancreatic cancer, gallbladder cancer, glioblastoma, and other solid tumors (Bera et al., 2014; Czubak et al., 2015; D'Urso et al., 2012; Erbes et al., 2015; Peng et al., 2015; Poltronieri et al., 2013), as well as in hematological malignancies such as acute myeloid leukemia, multiple myeloma, and B-cell lymphoma (Bi et al., 2015; Khalife et al., 2015; Sandhu et al., 2012). Some scholars have also suggested that miR-155 is under expressed in tumor cells, with low expression observed in esophageal cancer, melanoma, ovarian cancer, and others (Jia et al., 2023; Ling et al., 2013; Qin et al., 2013; Wang et al., 2016). miR-155 is processed from MIR155HG, and genetic variations in MIR155HG can affect the expression of miR-155 (Wu et al., 2017). Currently, most of the research on the polymorphism of MIR155HG focuses on three sites: rs767649 (T > A), located in the promoter region of −1,570 bp upstream of MIR155HG, is the most studied SNP for MIR155HG; rs928883 (G > A) located in 2.3 kb upstream of MIR155HG in the second intron of MIR155HG (Schuetz et al., 2012); rs1893650 (T > C) located in intron 1 of MIR155HG, approximately 1.5 kb upstream of the transcription start site (TSS) of the gene. This positioning plays a crucial role in the regulation biological functions of miR-155, particularly its impact on cancer-related pathways such as the PI3K/Akt signaling pathway (Karajovic et al., 2024; Wu W. et al., 2019). The study reports on the correlation between three SNPs in MIR155HG and cancer (Ji et al., 2016; Schuetz et al., 2012; Wang et al., 2016; Wu H. et al., 2019; Xie et al., 2015; Zou et al., 2020). However, different studies have drawn inconsistent conclusions (Dezfuli et al., 2020; Ji et al., 2016; Xie et al., 2015), and the association between these SNPs and cancer development remains unclear.

Figure 1. Molecular mechanisms of microRNA-155 in cancer progression via the PI3K/AKT/mTOR signaling pathway. microRNA-155 suppresses key negative regulators (SHIP1, PTEN), leading to the activation of the PI3K/AKT/mTOR signaling pathway. This activation drives tumor progression by promoting cell proliferation and survival through enhanced expression of anti-apoptotic molecules (Bcl-2, Mcl-1) and suppression of pro-apoptotic factors (Bax, Bad). It further facilitates invasion and metastasis via epithelial-mesenchymal transition, metabolic reprogramming, immune evasion, and therapy resistance.
The current research has reported the association between three SNPs of MIR155HG and cancer (Dezfuli et al., 2020; Iranparast et al., 2023; Ji et al., 2016; Karajovic et al., 2024; Wu H. et al., 2019; Wu et al., 2017; Xie et al., 2015; Zou et al., 2020), though conclusions from different studies are inconsistent. In addition, the relationship between MIR155HG polymorphisms and overall human cancer risk has not yet been comprehensively analyzed. To further investigate this issue, this study is the first to utilize a systematic review based on meta-analysis to explore the association between MIR155HG polymorphisms and cancer. It also examines the differences among subgroups, including respiratory system-related cancers, digestive system-related cancers, reproductive system-related cancers, as well as cancers in different regions and ethnic groups.
A systematic literature study was conducted on 7 databases including PubMed, Embase, Web of Science, Cochrane Library, China National Knowledge Infrastructure, VIP, Wan Fang to retrieve all relevant articles before 10 October 2024. All retrieval studies are manually searched and selected (Figure 2). The retrieval process is described in the supplementary document (Supplementary Material).
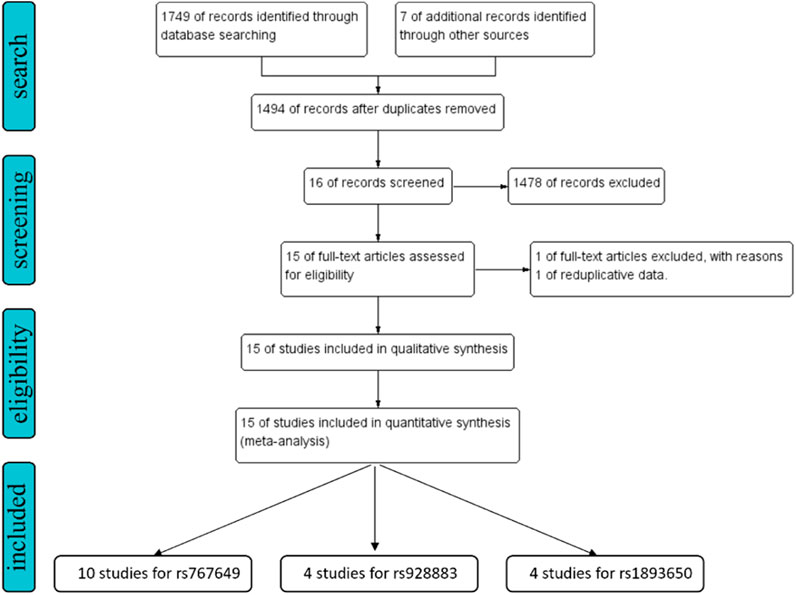
Figure 2. The flow chart of study selection for the present study. We expanded the search scope to “related articles.”
The inclusion criteria for this study were determined before the literature search. The included studies needed to meet the following criteria: (1) association studies between MIR155HG polymorphisms and cancer; (2) case-control studies; (3) detailed genotype data can be obtained by calculated odds ratios (OR) and 95% confidence intervals (CIs); (4) distribution of genotypes in the control group is consistent with Hardy-Weinberg equilibrium (HWE); (5) Include original research written in either English or Chinese.
Exclusion criteria: (1) reviews, letters, comments, expert opinions, case reports, and family-based association studies; (2) repetition of previous publications; (3) animal-based studies or cell line research.
The following data were independently extracted according to inclusion and exclusion criteria: the first author, publication year, country and region of study, genotyping method, type of cancers, source of control population, case and control sample size, genotype frequencies of MIR155HG polymorphisms in case and control, and results of the HWE test (Table 1). The risk of bias in the included literature was referenced to the Newcastle-Ottawa scale scoring standard (Table 3). The scoring system evaluated the included studies from 3 aspects: (1) the selectivity of the case and the control group; (2) the comparability of the case and the control group; (3) the exposure of the risk factors. The highest score achievable is 9 points. It is widely accepted that a research study was considered high-quality when the score was ≥7, and it is considered to be a study with low risk of bias.
All data analysis was performed using RevMan5.4 software. The OR and 95% CIs were calculated among 5 genetic models including allele model, homozygous model, heterozygous model, dominant model, and recessive model. We performed heterogeneity tests on the included studies using the Q test and I2 test. The fixed-effects model was only used for analysis when P > 0.10 and I2 ≤ 50%. Otherwise, the heterogeneity of this study was considered significant, and the random-effects model was used for analysis. We considered the analysis results to be significant when P value <0.05.
The literature search identified 1756 articles, and based on the inclusion and exclusion criteria, 15 studies were included in the final analysis, including 12 English articles (Chao et al., 2020; Dezfuli et al., 2020; Dezfuli et al., 2021; Iranparast et al., 2023; Ji et al., 2016; Jia et al., 2023; Karajovic et al., 2024; Wang et al., 2016; Wu H. et al., 2019; Xie et al., 2015; Zou et al., 2020) and 3 Chinese articles (Supplementary Material). After organizing the data, rs767649 (T > A), rs928883 (G > A), and rs1893650 (T > C) were included in5309 cases and6304 controls, 2176cases and 1957 controls, and 1,554 cases and 1,547 controls, respectively. The characteristics of all selected articles are summarized in Table 1, while Table 2 examines the genotype features of the included studies. Additionally, Table 3 presents the bias risks associated with the results obtained from the 15 included studies.
This analysis aimed to assess the association between the rs767649 (T > A) polymorphism and cancer risk. Ten studies were included, comprising 5,309 cases and 6,304 controls. Due to high heterogeneity, a random-effects model was applied (Table 4). The results showed no statistically significant association across all genetic models: allelic model (T vs. A, OR = 0.91, 95% CI [0.80, 1.04]), homozygous model (TT vs. AA, OR = 0.86, 95% CI [0.65, 1.15]), heterozygous model (TT vs. TA, OR = 0.9, 95% CI [0.75, 1.08]), dominant model (TA + AA vs. TT, OR = 0.88, 95% CI [0.72, 1.08]), and recessive model (TT + TA vs. AA, OR = 0.93, 95% CI [0.79, 1.10]). These results suggested that no significant association was found between the rs767649 polymorphism and cancer risk (P > 0.05) (Figure 3), and further studies are needed to confirm its role in cancer susceptibility.

Table 4. The meta-analysis results of microRNA-155 rs767649 (T > A) polymorphism and its correlation with the risk of cancer and its subgroups.
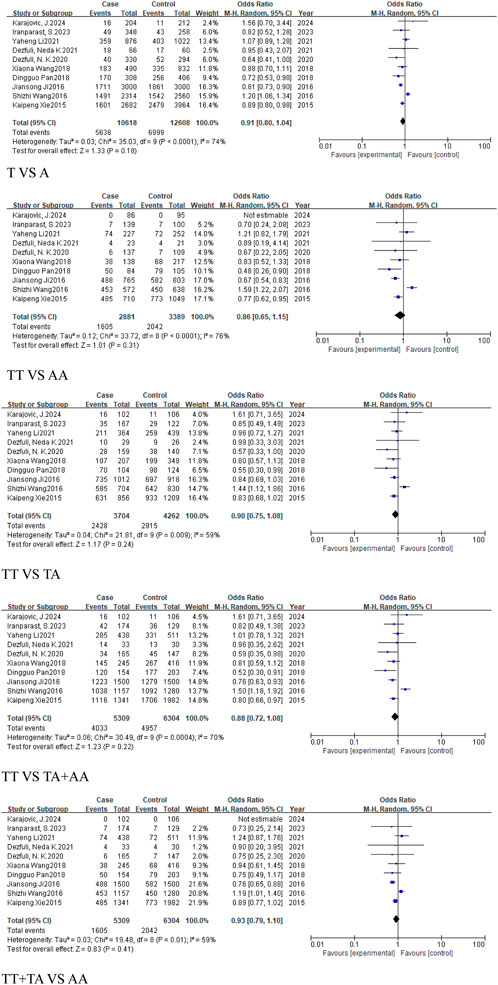
Figure 3. Forest plot of correlation between the microRNA-155 rs767649 (T > A) polymorphism and Cancer.
This analysis examined the association between the rs767649 (T > A) polymorphism and cancer risk in respiratory, digestive, reproductive system-related cancers, and cancers excluding reproductive system cancers.
For the group of respiratory system-related cancers (non-small cell lung cancer), three studies (1,539 cases, 2,159 controls) identified a statistically significant association in the allele model (T vs. A, OR = 0.87, 95%CI [0.79, 0.96], P = 0.007), the homozygous model (TT vs. AA, OR = 0.77, 95%CI [0.63, 0.94], P = 0.01), heterozygous model (TT vs. TA, OR = 0.8, 95%CI [0.66, 0.96], P = 0.02), and the dominant model (TA + AA vs. TT, OR = 0.78, 95%CI [0.65, 0.93], P = 0.005) (Figure 4). No significant association was found in the recessive model.

Figure 4. Forest plot of correlation between the microRNA-155 rs767649 (T > A) polymorphism and respiratory system-related cancers.
Similarly, 2 studies were included in the subgroup of digestive system-related cancers, including 1,654 cases and 1703 controls. The heterogeneity among the five models was low, and a fixed-effect model was used for analysis. Statistically significant associations were identified in some models for digestive system-related cancers: the allele model (T vs. A, OR = 0.8, 95%CI [0.73, 0.88], P < 0.0001), the homozygous model (TT vs. AA, OR = 0.65, 95%CI [0.53, 0.79], P < 0.0001), heterozygous model (TT vs. TA, OR = 0.8, 95%CI [0.66, 0.98], P = 0.03), the dominant model (TA + AA vs. TT, OR = 0.73, 95%CI [0.61, 0.88], P = 0.0008), and the recessive model (TT + TA vs. AA, OR = 0.76, 95%CI [0.66, 0.88], P = 0.0001) (Figure 5), with significant associations in the allele and homozygous models.
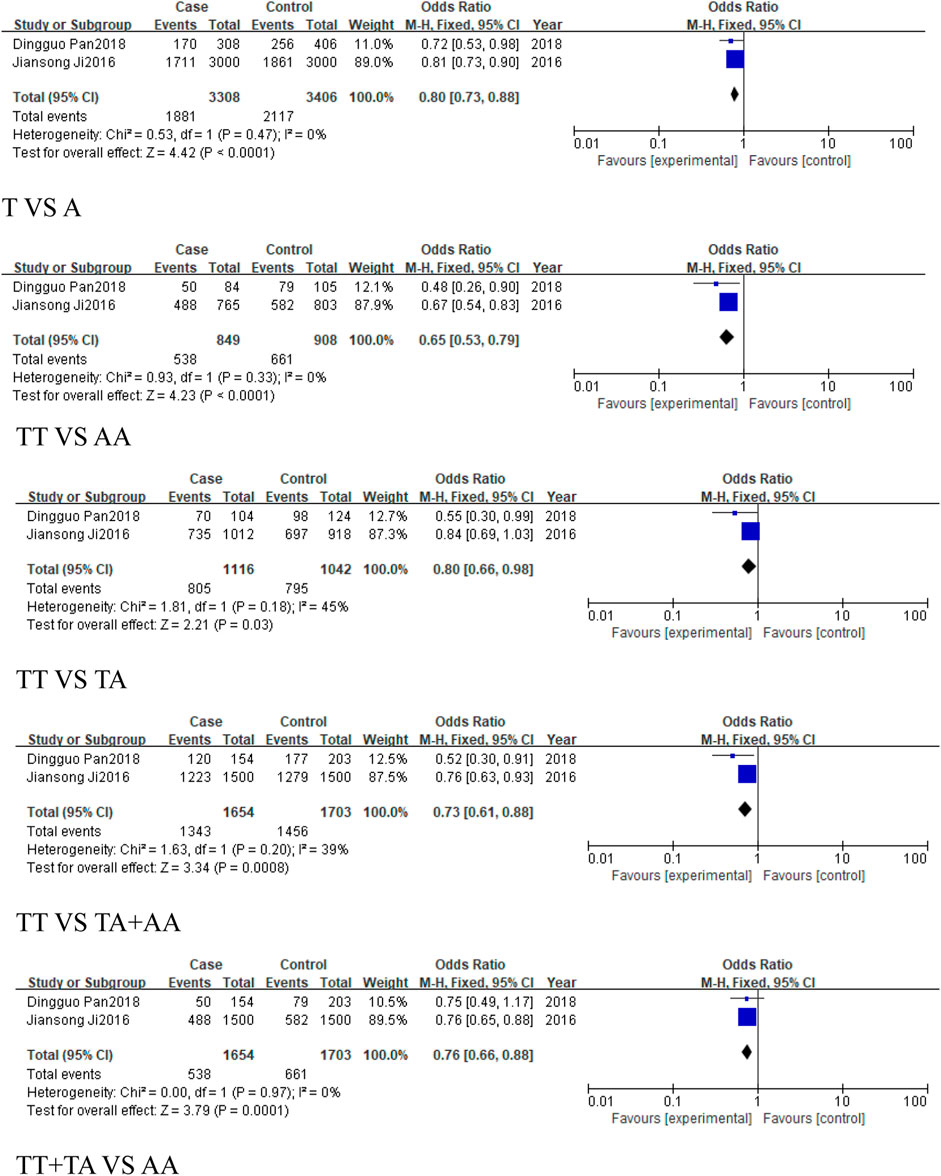
Figure 5. Forest plot of correlation between the microRNA-155 rs767649 (T > A) polymorphism and digestive system-related cancers.
For reproductive system-related cancers subgroup, 4 studies were included, including 2014 cases and 2,336 controls. The results showed that the rs767649 (T > A) polymorphism increased the risk of reproductive system-related cancers in the recessive model (TT + TA vs. AA, OR = 1.16, 95%CI [1.01, 1.33], P = 0.04) (Figure 6), while other models showed no significant association.
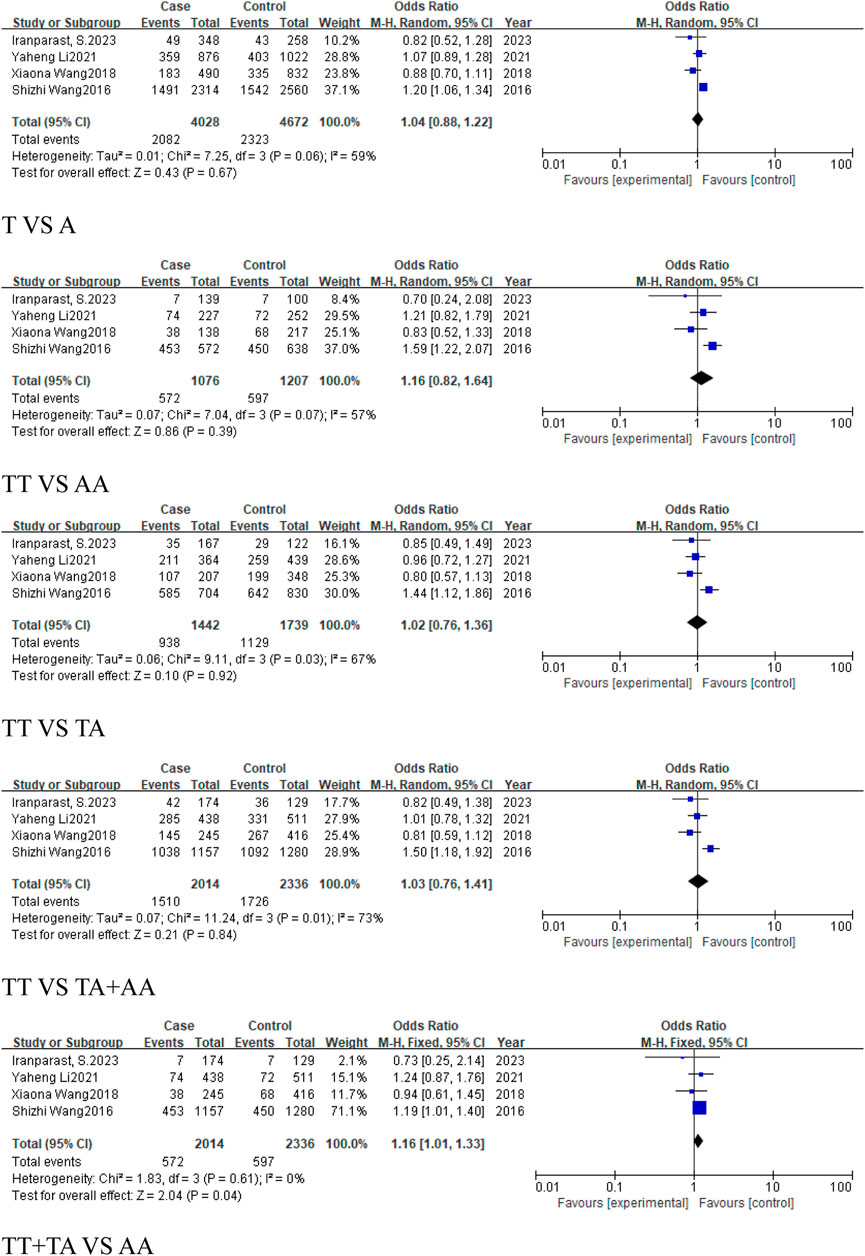
Figure 6. Forest plot of correlation between the microRNA-155 rs767649 (T > A) polymorphism and reproductive system-related cancers.
In the subgroup of cancers, which excludes reproductive system-related cancers, included six studies (3,295 cases, 3,968 controls), all five models showed reduced risk of cancer, including the allele model (T vs. A, OR = 0.84, 95%CI [0.79, 0.90], P < 0.00001), homozygous model (TT vs. AA, OR = 0.7, 95%CI [0.61, 0.81], P < 0.00001), heterozygous model (TT vs. TA, OR = 0.82, 95%CI [0.71, 0.93], P = 0.003), dominant model (TA + AA vs. TT, OR = 0.77, 95%CI [0.68, 0.87], P < 0.0001), and recessive model (TT + TA vs. AA, OR = 0.82, 95%CI [0.74, 0.91], P = 0.0001) (Figure 7).
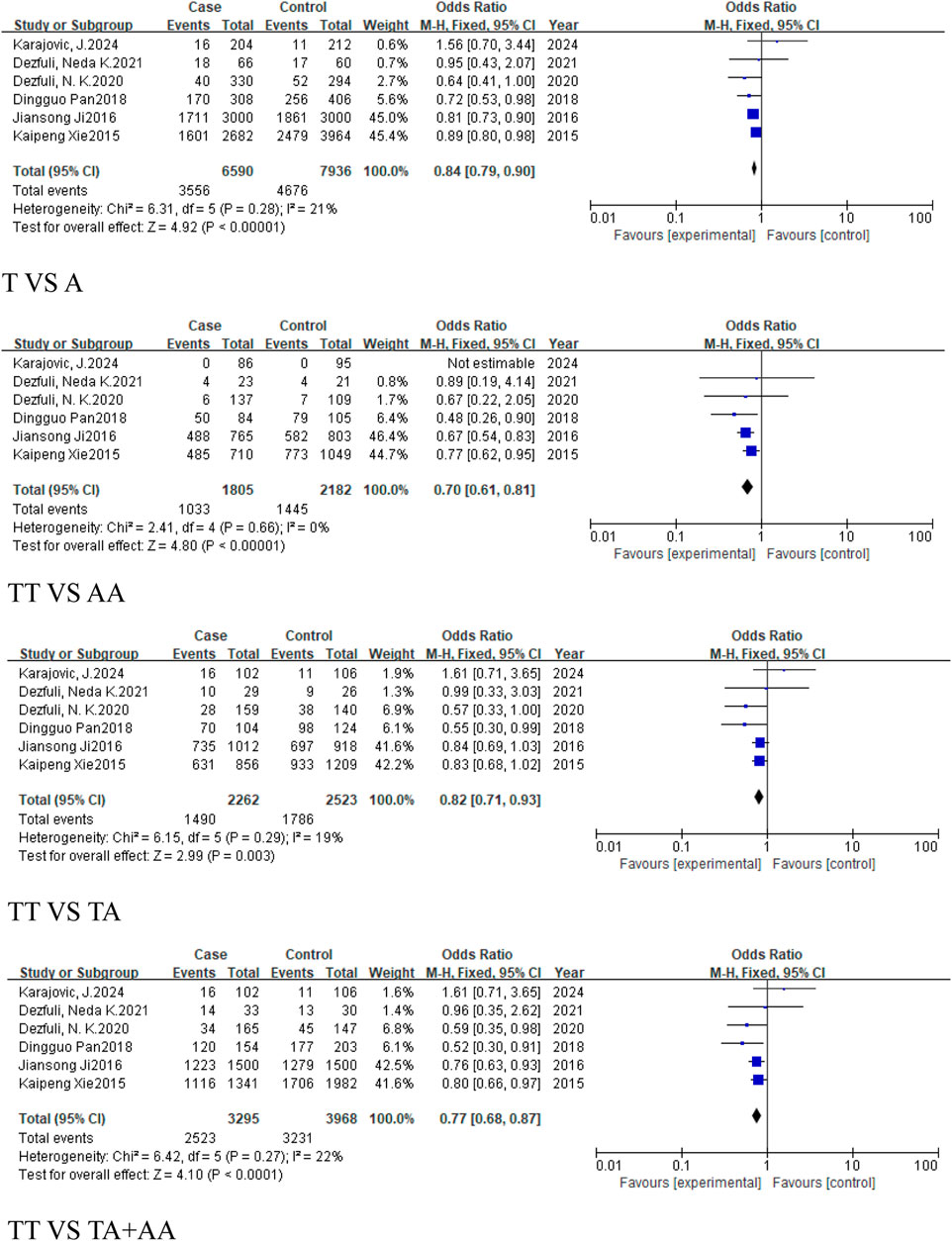
Figure 7. Forest plot of correlation between the microRNA-155 rs767649 (T > A) polymorphism and cancer after exclusion of reproductive system-related cancers.
This analysis aimed to investigate the impact of geographical variation on the association between the rs767649 (T>A) polymorphism and cancer risk. Subgroup analyses were conducted for Asian, European, East Asian, and Caucasian populations.
The meta-analysis of the regional subgroups included a total of 10 studies comprising 5,309 cases and 6,304 controls. Based on geographical differences, the studies were categorized into Asian and European subgroups (Table 2). The Asian subgroup comprised 9 studies, (5,207 cases, 6,198 controls), and no statistically significant associations were observed across any genetic models, including the allele model (T vs. A, OR = 0.9, 95% CI [0.79, 1.03]), homozygous model (TT vs. AA, OR = 0.86, 95% CI [0.65, 1.15]), heterozygous model (TT vs. TA, OR = 0.88, 95% CI [0.73, 1.05]), dominant model (TA + AA vs. TT, OR = 0.86, 95% CI [0.70, 1.05]), and recessive model (TT + TA vs. AA, OR = 0.93, 95%CI [0.79, 1.10]) (Figure 8).
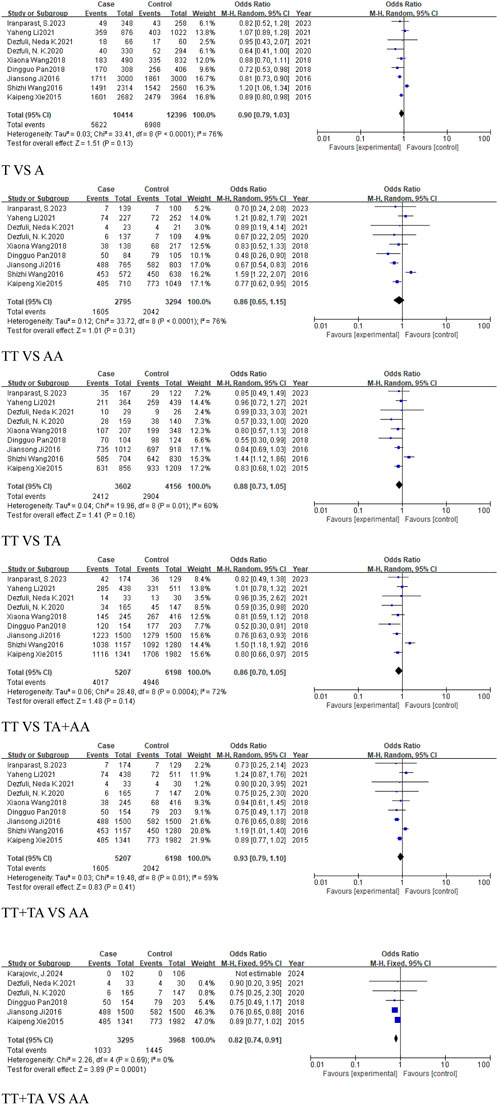
Figure 8. Forest plot of correlation between the microRNA-155 rs767649 (T > A) polymorphism and cancer (Asian subgroup).
The European subgroup consisted of only one study, and no pooled analysis was conducted due to insufficient data (Table 4). The East Asian subgroup included 6 studies (4,835 cases, 5,892 controls), with results showing no statistically significant associations across all genetic models: allele model (T vs. A, OR = 0.93, 95% CI [0.80, 1.08]), homozygous model (TT vs. AA, OR = 0.88, 95% CI [0.64, 1.23]), heterozygous model (TT vs. TA, OR = 0.91, 95% CI [0.73, 1.12]), dominant model (TA + AA vs. TT, OR = 0.89, 95% CI [0.69, 1.13]), and recessive model (TT + TA vs. AA, OR = 0.94, 95%CI [0.79, 1.13]) (Figure 9). For the Caucasian subgroup, which consisted of 4 studies with 474 cases and 412 controls, the rs767649 polymorphism showed no statistically significant associations with cancer risk across all five genetic models: allelic model (T vs. A, OR = 0.82, 95% CI [0.63, 1.08]), homozygous model (TT vs. AA, OR = 0.72, 95% CI [0.36, 1.45]), heterozygous model (TT vs. TA, OR = 0.83, 95% CI [0.59, 1.16]), dominant model (TA + AA vs. TT, OR = 0.82, 95% CI [0.60, 1.12]), and recessive model (TT + TA vs. AA, OR = 0.77, 95% CI [0.39, 1.53]) (Figure 10).
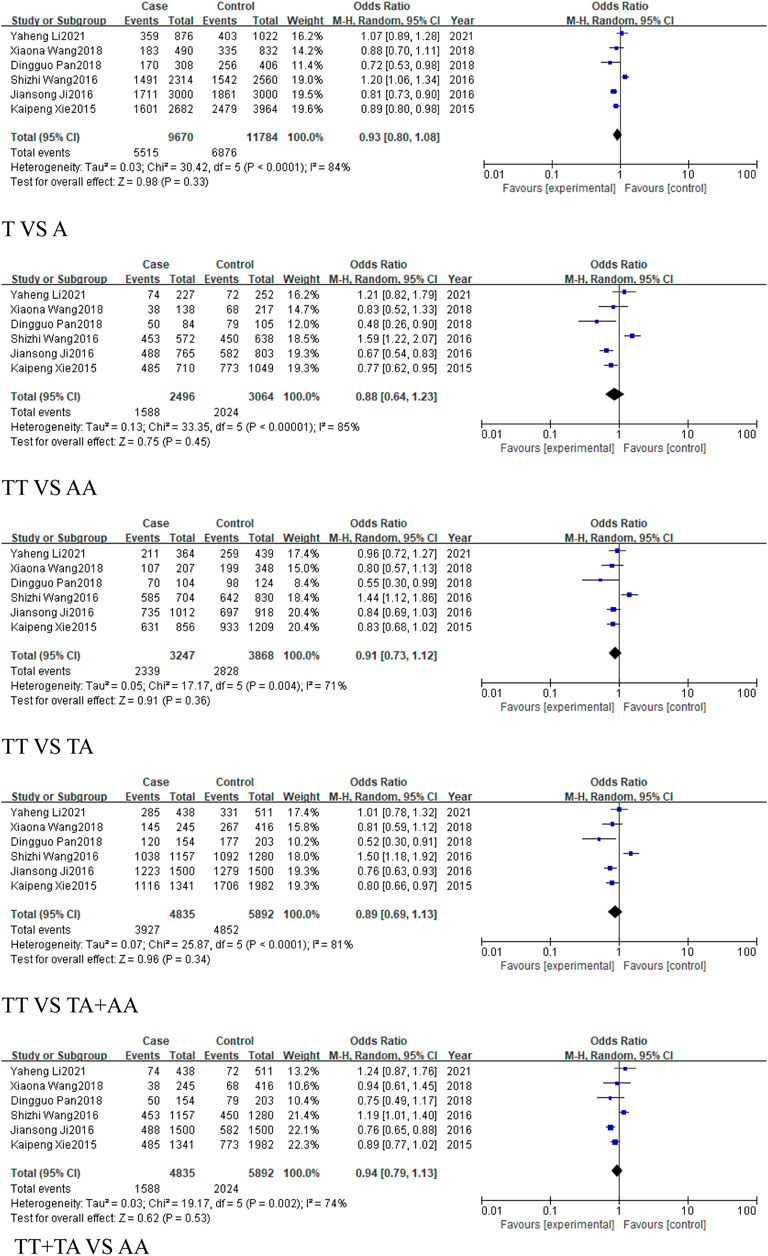
Figure 9. Forest plot of correlation between the microRNA-155 rs767649 (T > A) polymorphism and cancer (East Asian subgroup).
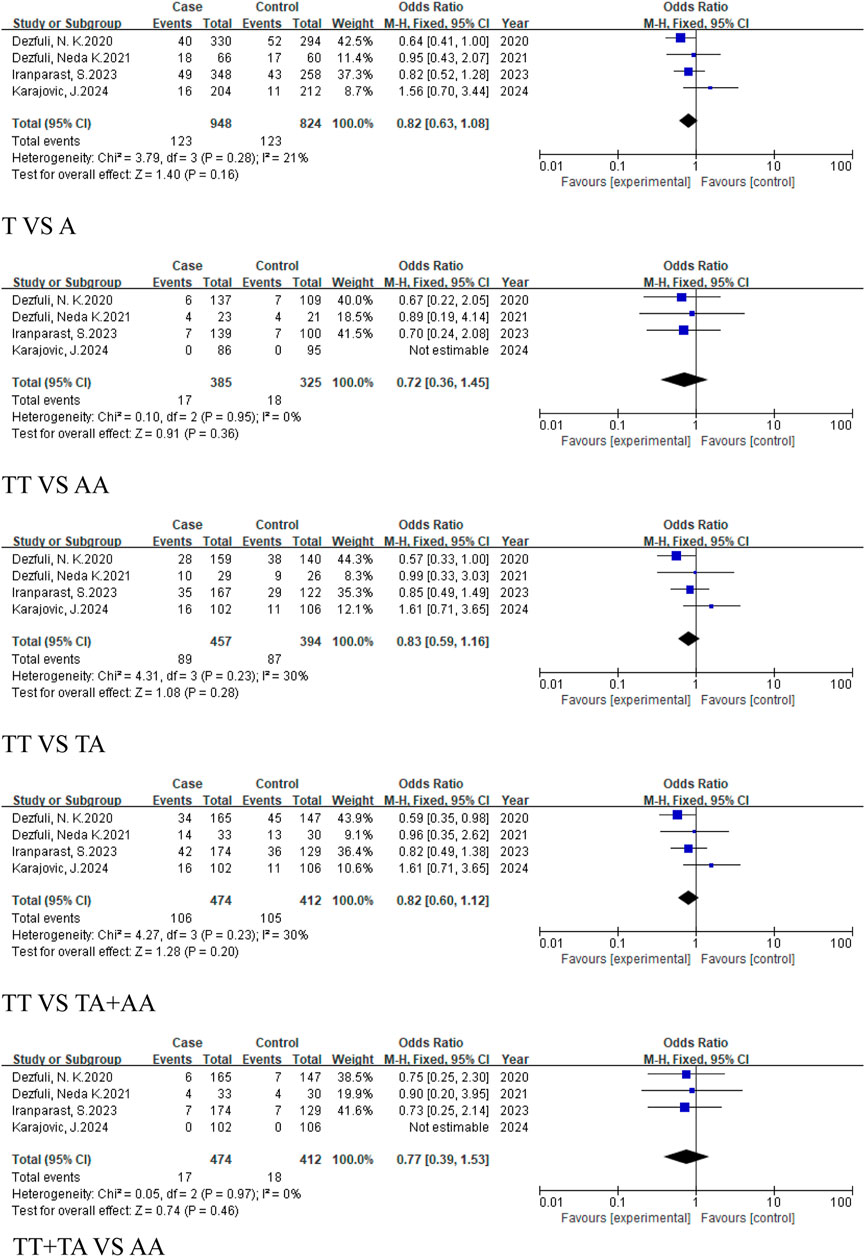
Figure 10. Forest plot of correlation between the microRNA-155 rs767649 (T > A) polymorphism and cancer (Caucasian subgroup).
The meta-analysis of rs928883 (G > A) polymorphism comprised a total of 4 studies, with 2,176 cases and 1957 controls. The heterogeneity test of this study was evaluated using the I2 test, the results indicated that the included studies exhibited high heterogeneity, except for the recessive model. Therefore, a random-effects model was used to combine the research results in four genetic models. The meta-analysis results of the association between rs928883 (G > A) polymorphism and the risk of cancer and its subgroups are shown in the table (Table 5).

Table 5. The meta-analysis results of microRNA-155 rs928883 (G > A) polymorphism and its correlation with the risk of cancer and its subgroups.
Four studies, comprising 2,176 cancer cases and 1,957 controls, were included in the cancer group. The results revealed no statistically significant association across all genetic models: the allele model (G vs. A, OR = 1.01, 95% CI [0.81, 1.27]), homozygous model (GG vs. AA, OR = 1.12, 95% CI [0.63, 2.01]), heterozygous model (GG vs. GA, OR = 1.06, 95% CI [0.77, 1.45]), dominant model (GA + AA vs. GG, OR = 1.04, 95% CI [0.75, 1.45]), and the recessive model (GG + GA vs. AA, OR = 0.96, 95% CI [0.65, 1.43]) (Figure 11). This suggests that there is no significant association between rs928883 (G > A) polymorphism and the risk of cancer in the overall cancer analysis (Table 5).
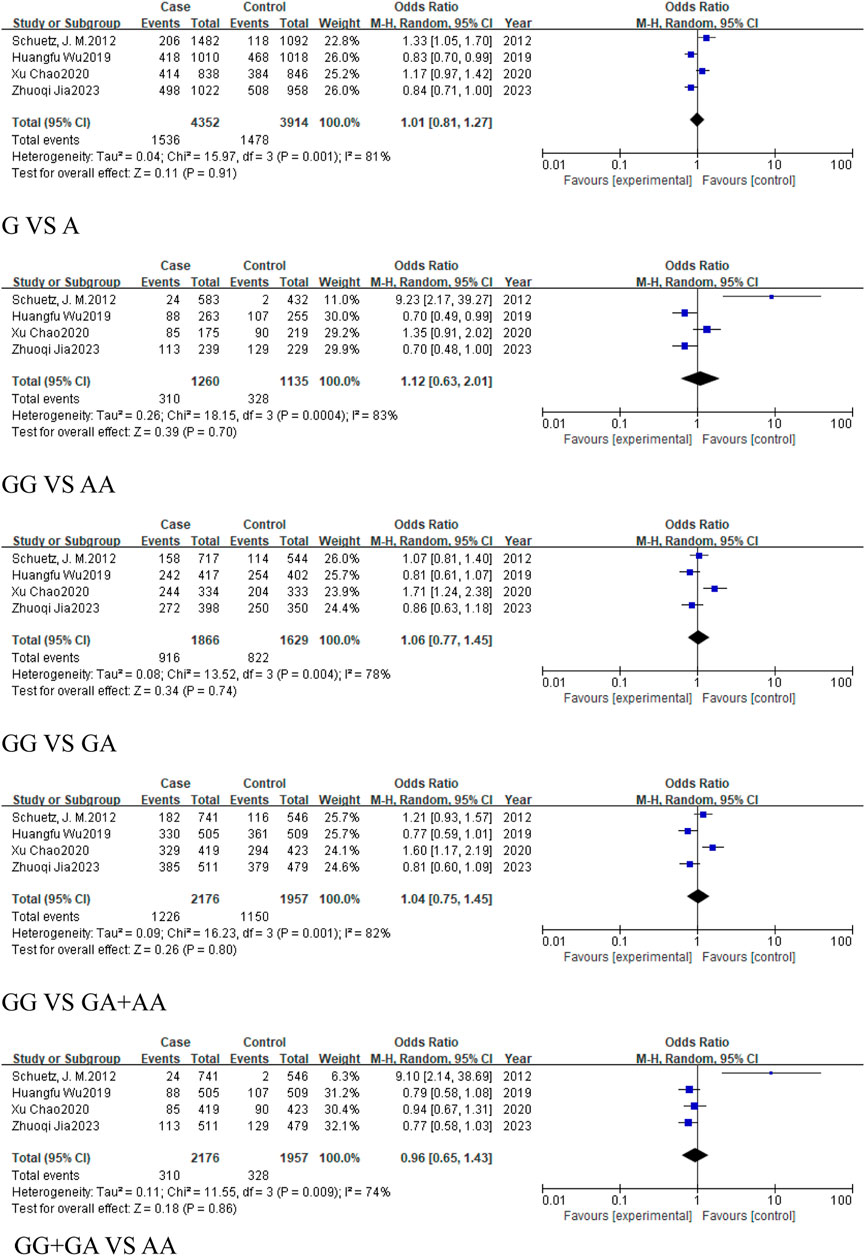
Figure 11. Forest plot of correlation between the microRNA-155 rs928883 (G > A) polymorphism and cancer.
Three studies, comprising 1,435 cases and 1,411 controls, were included in the subgroup of digestive system-related cancers. The results indicated a statistically significant association in the recessive model (GG + GA vs. AA, OR = 0.82, 95%CI [0.69, 0.99], P = 0.03), suggesting that the rs928883 (G > A) polymorphism may play a role in reducing the risk of digestive system-related cancers (Figure 12). Similarly, in the analysis of epithelial-origin malignancies and the subgroup of Asian and East Asian populations, the rs928883 (G > A) polymorphism was found to be associated with cancer only in the recessive model (GG + GA vs. AA, OR = 0.82, 95%CI [0.69, 0.99], P = 0.03), which was statistically significant as a protective factor (Figure 12).
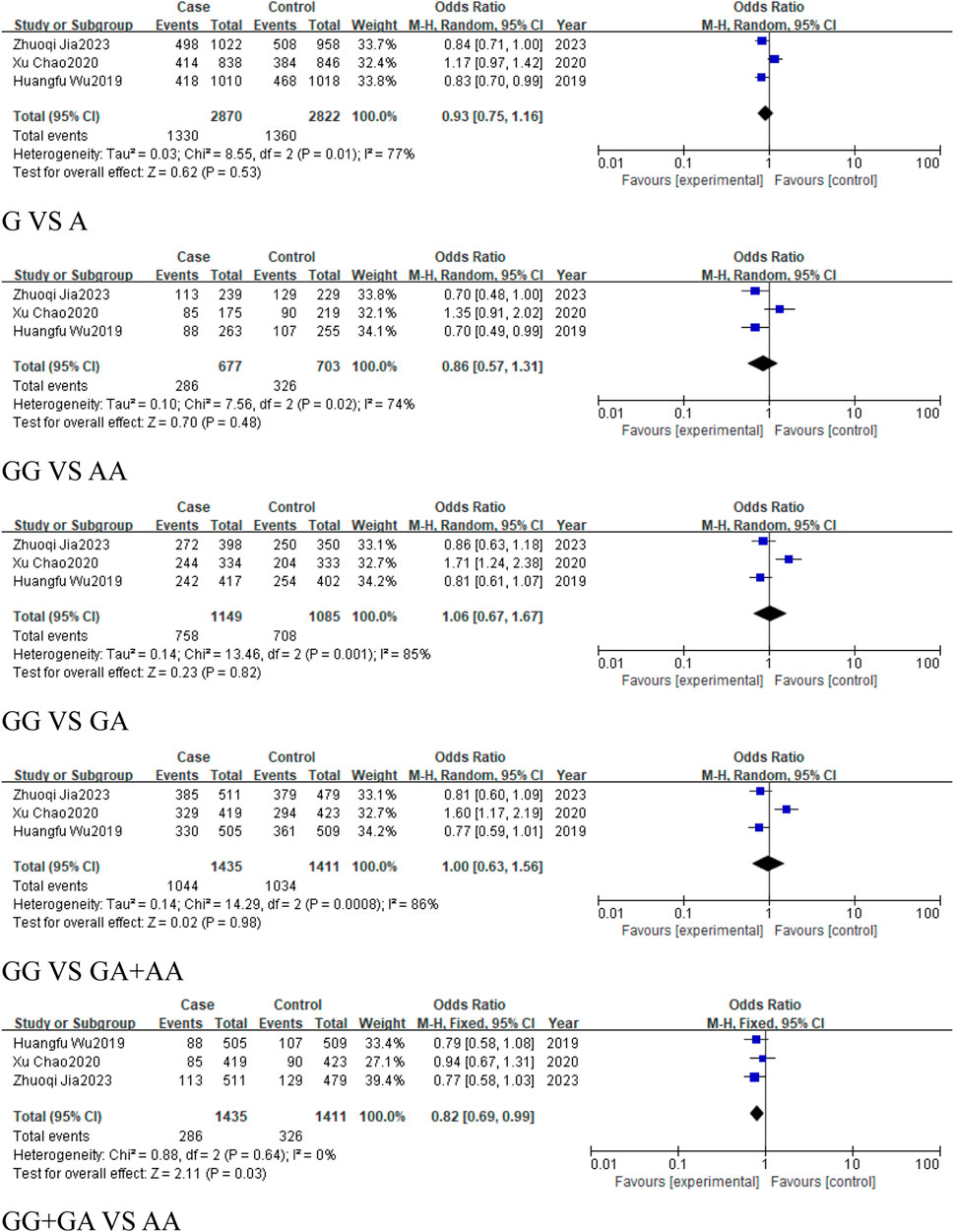
Figure 12. Forest plot of correlation between the microRNA-155 rs928883 (G > A) polymorphism and digestive system-related cancers.
A total of 4 studies were included in the group of cancer-related studies, involving 1,554 cancer cases and 1,547 controls. The included studies, except for the dominant model, exhibited high heterogeneity, and therefore a random-effects model was used for the analysis (Table 6). The meta-analysis results showed no association between rs1893650 (T > C) and cancer in any of the five genetic models (Figure 13). Similarly, in subgroup analyses for digestive system-related cancers, as well as for Asian and East Asian populations, no significant association was found between rs1893650 and cancer risk in any of the five genetic models (Figure 14). These findings suggest that the rs1893650 (T > C) polymorphism is not significantly associated with cancer risk or the risk in its subgroups. Given the limited number of included studies, the meta-analysis results should be interpreted with caution.
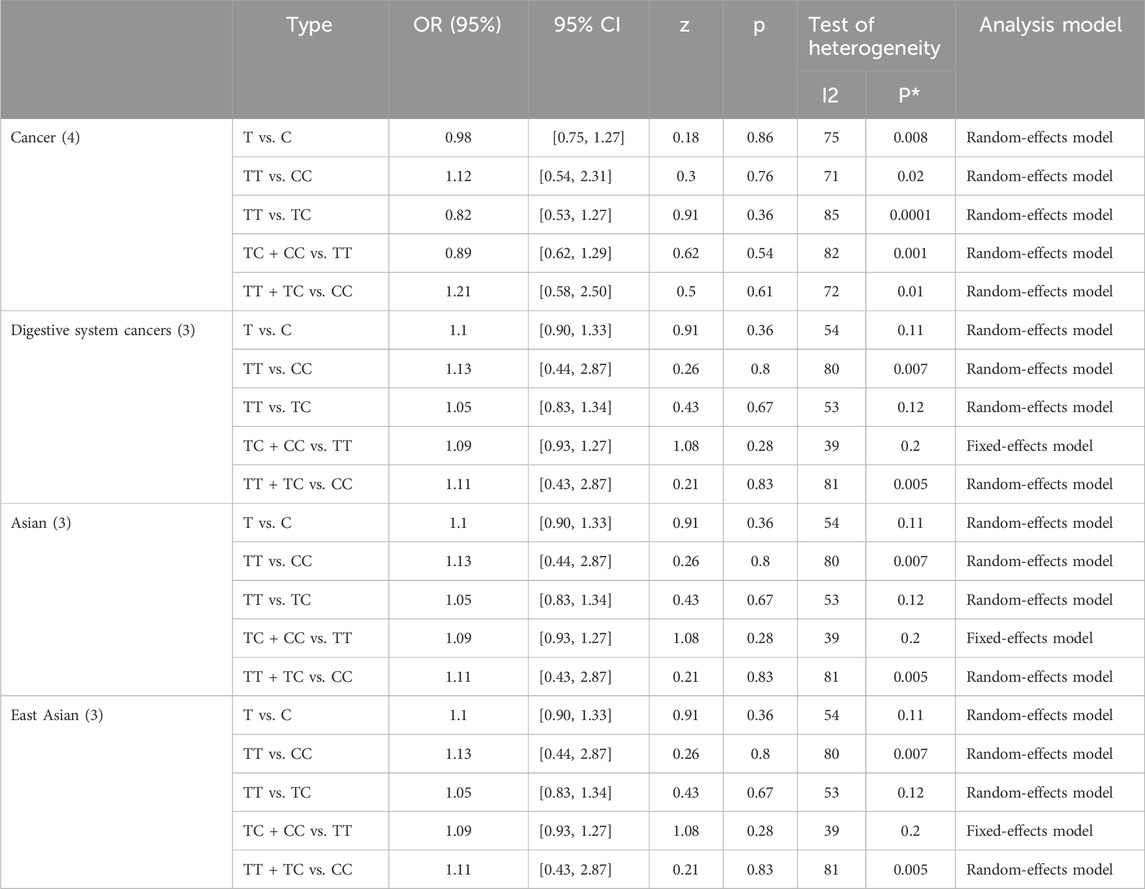
Table 6. The meta-analysis results of microRNA-155 rs1893650 (T > C) polymorphism and its correlation with the risk of cancer and its subgroups.
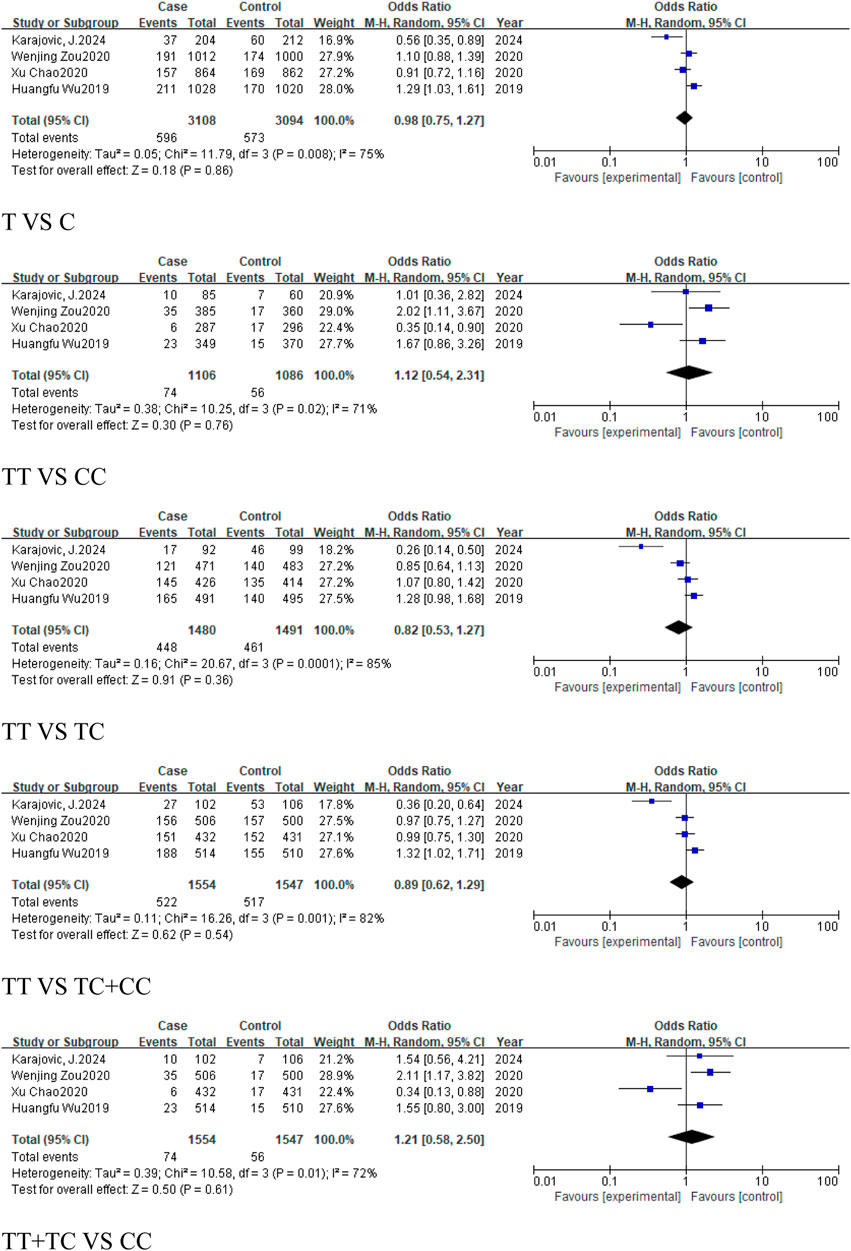
Figure 13. Forest plot of correlation between the microRNA-155 rs1893650 (T > C) polymorphism and cancers.
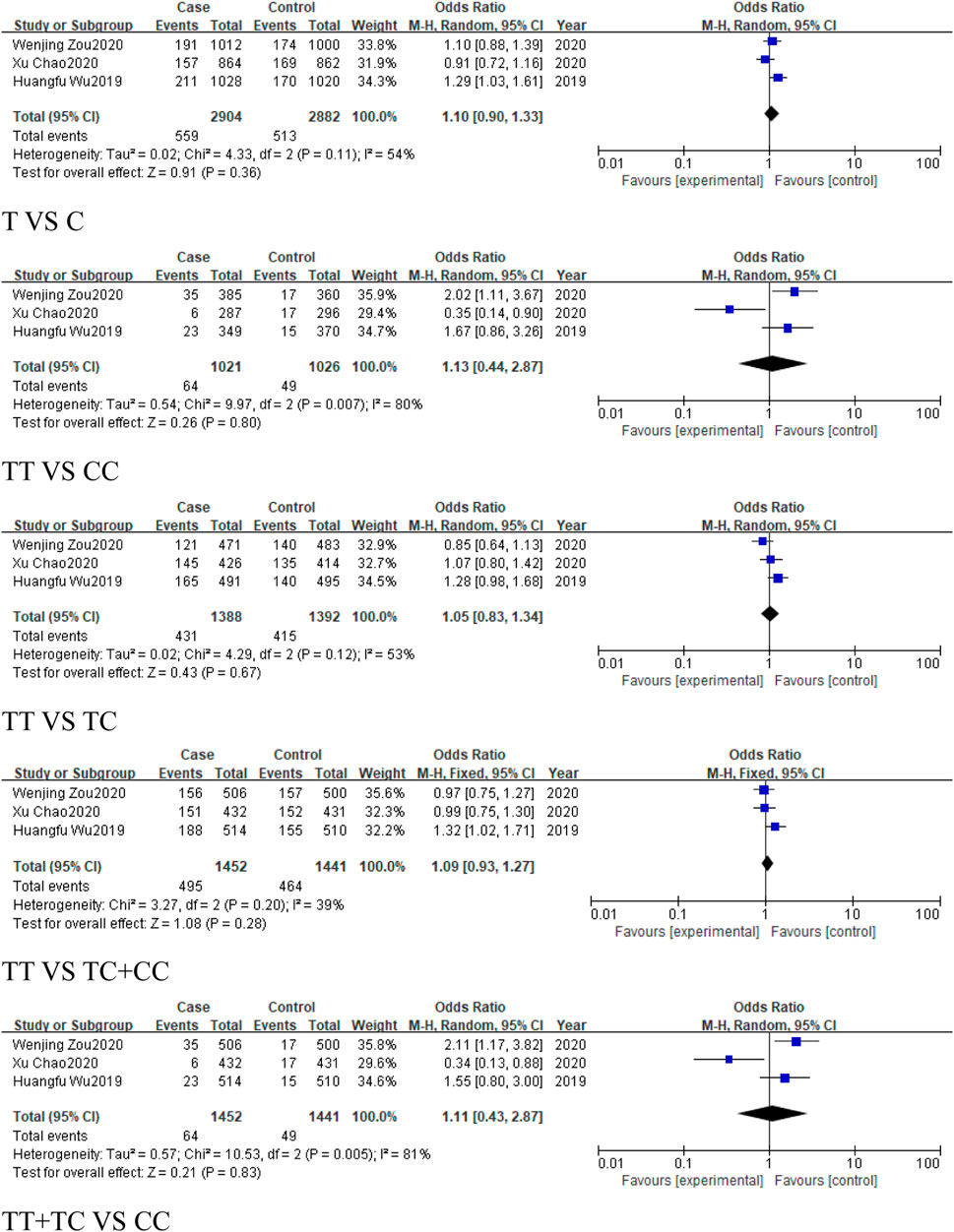
Figure 14. Forest plot of correlation between the microRNA-155 rs1893650 (T > C) polymorphism and digestive system-related cancers.
Due to the limited number of original articles included in this research, as well as the scarcity of studies for rs767649 (T > A), rs928883 (G > A), and rs1893650 (T > C) with no more than 10 studies respectively, we did not evaluate the presence of publication bias (Sterne et al., 2001). Sensitivity analysis was not conducted for studies with low heterogeneity. However, for studies with high heterogeneity, further subgroup analysis is performed based on the existing subtyping to explore potential sources of heterogeneity.
As a significant miRNA, miR-155 was first discovered in 2002 (Lagos-Quintana et al., 2002). Since then, miR-155 has been shown to play a crucial role in various biological processes, including immune regulation, inflammatory responses, and cell proliferation. It regulates the expression of target genes, participating in multiple cellular processes such as proliferation, differentiation, and apoptosis, making it a research focus in fields such as immune diseases and cardiovascular diseases. In recent years, studies have found that miR-155 is an evolutionarily conserved miRNA regulated by the activator protein-1 and nuclear factor-κB complexes. By binding to multiple target genes, miR-155 is involved in the development and progression of cancer (Faraoni et al., 2009; Thai et al., 2007). SNPs may affect gene transcription and/or protein expression, thereby influencing individual susceptibility to diseases. Research suggests that SNPs in the miR-155 gene are closely associated with various malignant tumors (Iranparast et al., 2023; Karajovic et al., 2024; Xie et al., 2015). To the best of our knowledge, rs767649 (T > A), rs928883 (G > A), and rs1893650 (T > C) are the three most studied SNP sites in the MIR155HG. This study represents the first meta-analysis that investigates the correlation between the MIR155HG polymorphism and the susceptibility to cancer and its subgroups. We summarize and analyze the potential impact of mutations at these specific sites on the development of cancer. By systematically reviewing the existing research in this field, this study aims to provide valuable insights and serve as a reference for future clinical investigations in this area.
This meta-analysis, encompassing data from 10 published articles (5,309 cancer cases and 6,304 controls), investigated the association between the rs767649 (T > A) polymorphism and cancer. There is no significant correlation between rs767649 polymorphism and the susceptibility to cancer.
Notably, the aforementioned studies generally exhibit high heterogeneity in genetic models, which may be attributed to differences in the types of diseases chosen by various studies. Different tumor types may have distinct pathogenesis mechanisms, potentially leading to different roles of MIR155HG. To address this possibility, our study further explored the association between the rs767649 (T > A) polymorphism and cancer subgroups, analyzing the correlation of rs767649 with cancers of the respiratory system, digestive system, and reproductive system. When stratified by cancer type, this significant heterogeneity was reduced or disappeared. Three original studies analyzed the association between the rs767649 (T > A) polymorphism and respiratory cancer risk, including 1539 NSCLC cases and 2,159 healthy controls. The analysis showed that the rs767649 polymorphism reduced the risk of NSCLC across four genetic models: allele model, homozygous model, heterozygous model, and dominant model. Compared with the TT genotype, the AA genotype reduced the risk of NSCLC by 23% (P = 0.01), and the TA genotype reduced it by 20% (P = 0.02). The TA + AA genotypes reduced the OR for NSCLC by 22% (P = 0.005), suggesting that the A allele may be a key factor influencing NSCLC risk. Our study also analyzed the association between the rs767649 (T>A) polymorphism and the risk of digestive system-related cancers using data from two reports. Meta-analysis results showed that this SNP was associated with a reduced risk of digestive system cancers across all five genetic models, with statistically significant differences in the allele model, homozygous model, dominant model, and recessive model. These findings suggest that tumor type and sample size play important roles in the study of SNP-cancer associations. Additionally, we examined the association between the rs767649 (T>A) polymorphism and the risk of reproductive system-related cancers using data from four original studies. The results showed that all five genetic models increased the risk of reproductive system cancers, consistent with the findings of Wang et al. (Wang et al., 2016), but this correlation only reached statistical significance in the recessive model (P = 0.04). Given the relatively small sample size in this cancer type, these results should be interpreted with caution. Research has shown that factors such as hormones, lifestyle, and environmental factors lead to differences in cancer incidence between genders (Borg et al., 2024; Fu et al., 2021; Zheng et al., 2017). To minimize the influence of gender on the study results, we excluded original studies related to reproductive system cancers, such as cervical and breast cancers. In the cancer subgroups excluding reproductive system cancers, we found that the rs767649 (T > A) polymorphism reduced cancer risk across all five genetic models (allele model, homozygous model, heterozygous model, dominant model, and recessive model), with statistically significant differences observed in all five models.
The study populations selected in different studies vary, and these populations may possess distinct genetic backgrounds. The role of SNPs at these sites may be influenced by different genetic backgrounds, which could affect the heterogeneity of the genetic models in this study. Research has reported an association between rs767649 (T > A) polymorphism and cancer risk in East Asian populations, while such associations have been less frequently reported in Caucasian populations. A subgroup analysis based on the geographic origin of the samples revealed no statistically significant association between the two in the Asian group. Regarding the heterogeneity in regional groups, we reviewed some details of the included studies, focusing on the racial characteristics of the populations. The populations included both East Asians and Caucasians, which could be a source of heterogeneity. When conducting a subgroup analysis based on race, the heterogeneity in the Caucasian subgroup disappeared, no correlation was found between rs767649 (T>A) polymorphism and cancer risk. This suggests that the relationship between this SNP and cancer risk remains unclear across different regions and ethnicities.
The meta-analysis of this study shows that the rs928883 (G > A) polymorphism is not significantly associated with cancer risk in the overall cancer analysis. When classified according to tumor types, it was found that the recessive model of rs928883 (G > A) reduces the risk of digestive system-related cancers and serves as a protective factor against the occurrence of such cancers. Following this, we conducted analyses by region and ethnic subgroups, finding that in the Asian subgroup and the East Asian ethnic subgroup, the recessive model of rs928883 (G > A) polymorphism is significantly associated with cancer susceptibility. It is noteworthy that the recessive model of this SNP acts as a protective factor in cancer occurrence. The original studies in the European subgroup and Caucasian ethnic subgroup included in this study consisted of only one paper (Schuetz et al., 2012), and we did not perform a meta-analysis on the association between rs928883 polymorphism and cancer risk in this subgroup. However, Schuetz et al. were the first to report the association between rs928883 polymorphism and marginal zone lymphoma.
In existing studies, there is limited original research on the relationship between the rs1893650 (T > C) polymorphism located in the intron region of MIR155HG and cancer risk. Published cancer-related studies indicate an association between rs1893650 (T > C) polymorphism and cancer susceptibility. However, its role varies across different cancer types. Karajovic et al. found that rs1893650 is negatively correlated with thyroid cancer susceptibility, suggesting that rs1893650 could serve as a biomarker for indicating potential risk and prognosis of thyroid cancer (Karajovic et al., 2024). In contrast, Zou et al. reported that the MIR155HG rs1893650 (T > C) polymorphism increases the risk of gastric cancer (Zou et al., 2020). The discrepancy between these findings may be due to the different types of diseases studied, as the mechanisms of different tumor types may vary, leading to different roles for MIR155HG. The results of the meta-analysis in this study do not support the association between rs1893650 polymorphism and cancer or its subtypes. Furthermore, most current studies on the rs1893650 polymorphism focus on its association with the risk of cardiovascular diseases (heart disease, atherosclerosis), metabolic diseases (obesity, diabetes), and autoimmune diseases (rheumatoid arthritis, systemic lupus erythematosus).
Our research has certain limitations. Firstly, the number of studies included in our analysis is limited, which may lead to false positive or false negative results, particularly in terms of sample size for each subgroup. This limitation hinders a comprehensive exploration of the relationship between low-frequency variations and the risk of cancer. Secondly, the control group for the primary studies used in our study is derived from hospitals, which may not accurately represent the general population. It is challenging to completely eliminate selection bias. Finally, we extracted basic information from each study, such as the country of the study subjects, their race, and genetic mutation sites. However, we still cannot avoid the influence of some confounding factors, such as age, familial relationships, smoking, and drug history. These limitations may potentially reduce the accuracy of the final results. Given these limitations, the results of our study should be carefully interpreted and further elaborated upon with a larger sample size in a systematic manner.
This study is the first meta-analysis to systematically examine the associations between MIR155HG polymorphisms and cancer susceptibility. We found that rs767649 (T > A) is associated with reduced risk in NSCLC and digestive system cancers but increased risk in reproductive system cancers, highlighting its tumor-specific effects. Excluding reproductive system cancers further confirmed its protective role in non-reproductive cancers. Similarly, rs928883 (G > A) showed a protective effect in digestive system cancers, particularly in East Asian populations, while no significant associations were observed for rs1893650 (T > C). Our findings provide novel insights into the roles of MIR155HG polymorphisms as potential biomarkers for cancer risk stratification. These results underscore their utility in precision oncology for developing targeted screening and prevention strategies, warranting further validation through large-scale studies.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
GJ: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Software, Supervision, Validation, Writing–original draft, Writing–review and editing. TG: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing–review and editing. J-WL: Data curation, Investigation, Software, Writing–review and editing. H-YY: Data curation, Investigation, Validation, Writing–review and editing. J-GX: Formal Analysis, Investigation, Software, Writing–review and editing. YP: Data curation, Methodology, Writing–review and editing. YY: Data curation, Methodology, Writing–review and editing. SE-H: Data curation, Software, Writing–review and editing. KY: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Software, Supervision, Validation, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Health Industry Research Project of Gansu Province of China (GSWSKY-2019-76); Medicine Research Fund Project of Gansu Provincial Hospital (23GSSYF-30); Natural Science Foundation of Gansu Province (22JR5RA655). The Gansu Provincial Hospital Intramural Scientific Research Fund Project, 24GSSYD-12.
The authors gratefully acknowledge the financial supports by the Health Industry Research Project of Gansu Province of China (GSWSKY-2019-76), The Medicine Research Fund Project of Gansu Provincial Hospital (23GSSYF-30) and the Natural Science Foundation of Gansu Province (22JR5RA655). The Gansu Provincial Hospital Intramural Scientific Research Fund Project, 24GSSYD-12.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1517513/full#supplementary-material
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2), 281–297. doi:10.1016/s0092-8674(04)00045-5
Bera, A., VenkataSubbaRao, K., Manoharan, M. S., Hill, P., and Freeman, J. W. (2014). A miRNA signature of chemoresistant mesenchymal phenotype identifies novel molecular targets associated with advanced pancreatic cancer. PLoS One 9 (9), e106343. doi:10.1371/journal.pone.0106343
Bi, C., Chung, T. H., Huang, G., Zhou, J., Yan, J., Ahmann, G. J., et al. (2015). Genome-wide pharmacologic unmasking identifies tumor suppressive microRNAs in multiple myeloma. Oncotarget 6 (28), 26508–26518. doi:10.18632/oncotarget.4769
Borg, M., Tønnesen, H., Ibsen, R., Hilberg, O., and Løkke, A. (2024). Lung cancer: a nationwide analysis of sex and age incidence trends from 1980 to 2022. Acta Oncol. 63, 526–531. doi:10.2340/1651-226x.2024.34876
Chao, X., Feng, X., Wang, X., Shi, H., Li, H., Wang, Y., et al. (2020). MiRNA155HG polymorphisms influenced the risk of liver cancer among the Han Chinese population. BMC Med. Genet. 21 (1), 134. doi:10.1186/s12881-020-01064-4
Czubak, K., Lewandowska, M. A., Klonowska, K., Roszkowski, K., Kowalewski, J., Figlerowicz, M., et al. (2015). High copy number variation of cancer-related microRNA genes and frequent amplification of DICER1 and DROSHA in lung cancer. Oncotarget 6 (27), 23399–23416. doi:10.18632/oncotarget.4351
Dezfuli, N. K., Adcock, I. M., Alipoor, S. D., Seyfi, S., Salimi, B., Mafi Golchin, M., et al. (2020). The miR-146a SNP Rs2910164 and miR-155 SNP rs767649 are risk factors for non-small cell lung cancer in the Iranian population. Can. Respir. J. 2020, 8179415. doi:10.1155/2020/8179415
Dezfuli, N. K., Alipoor, S. D., Roofchayee, N. D., Seyfi, S., Salimi, B., Adcock, I. M., et al. (2021). Evaluation expression of miR-146a and miR-155 in non-small-cell lung cancer patients. Front. Oncol. 11, 715677. doi:10.3389/fonc.2021.715677
Donnem, T., Eklo, K., Berg, T., Sorbye, S. W., Lonvik, K., Al-Saad, S., et al. (2011). Prognostic impact of MiR-155 in non-small cell lung cancer evaluated by in situ hybridization. J. Transl. Med. 9, 6. doi:10.1186/1479-5876-9-6
D'Urso, P. I., D'Urso, O. F., Storelli, C., Mallardo, M., Gianfreda, C. D., Montinaro, A., et al. (2012). miR-155 is up-regulated in primary and secondary glioblastoma and promotes tumour growth by inhibiting GABA receptors. Int. J. Oncol. 41 (1), 228–234. doi:10.3892/ijo.2012.1420
Erbes, T., Hirschfeld, M., Rücker, G., Jaeger, M., Boas, J., Iborra, S., et al. (2015). Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer 15, 193. doi:10.1186/s12885-015-1190-4
Fang, H., Chi, X., Wang, M., Liu, J., Sun, M., Zhang, J., et al. (2024). M2 macrophage-derived exosomes promote cell proliferation, migration and EMT of non-small cell lung cancer by secreting miR-155-5p. Mol. Cell Biochem. doi:10.1007/s11010-024-05161-3
Faraoni, I., Antonetti, F. R., Cardone, J., and Bonmassar, E. (2009). miR-155 gene: a typical multifunctional microRNA. Biochim. Biophys. Acta 1792 (6), 497–505. doi:10.1016/j.bbadis.2009.02.013
Fu, J., Zhang, G., Xu, P., Guo, R., Li, J., Guan, H., et al. (2021). Seasonal changes of thyroid function parameters in women of reproductive age between 2012 and 2018: a retrospective, observational, single-center study. Front. Endocrinol. (Lausanne) 12, 719225. doi:10.3389/fendo.2021.719225
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144 (5), 646–674. doi:10.1016/j.cell.2011.02.013
Iranparast, S., Tahmasebi-Birgani, M., Motamedfar, A., Amari, A., and Ghafourian, M. (2023). miR-155 rs767649 T > A gene polymorphism is associated with downregulation of miR-155 expression, suppressor of cytokine signaling-1 overexpression, and low probability of metastatic tumor at the time of breast cancer diagnosis. J. Res. Med. Sci. 28, 32. doi:10.4103/jrms.jrms_960_21
Ji, J., Xu, M., Tu, J., Zhao, Z., Gao, J., Chen, M., et al. (2016). MiR-155 and its functional variant rs767649 contribute to the susceptibility and survival of hepatocellular carcinoma. Oncotarget 7 (37), 60303–60309. doi:10.18632/oncotarget.11206
Jia, Z., Zhou, W., Liang, J., Zhang, G., Fu, J., and Sun, X. (2023). The single nucleotide polymorphisms rs4143370 and rs34904192 in MIR155HG were associated with esophageal cancer risk in the Chinese han population. Digestion 104 (3), 222–232. doi:10.1159/000527751
Karajovic, J., Kovacevic, B., Uzelac, B., Stefik, D., Jovanovic, B., Ristic, P., et al. (2024). Association of HOTAIR, MIR155HG, TERC, miR-155, -196a2, and -146a genes polymorphisms with papillary thyroid cancer susceptibility and prognosis. Cancers (Basel) 16 (3), 485. doi:10.3390/cancers16030485
Khalife, J., Radomska, H. S., Santhanam, R., Huang, X., Neviani, P., Saultz, J., et al. (2015). Pharmacological targeting of miR-155 via the NEDD8-activating enzyme inhibitor MLN4924 (Pevonedistat) in FLT3-ITD acute myeloid leukemia. Leukemia 29 (10), 1981–1992. doi:10.1038/leu.2015.106
Kong, X., Liu, F., and Gao, J. (2016). MiR-155 promotes epithelial-mesenchymal transition in hepatocellular carcinoma cells through the activation of PI3K/SGK3/β-catenin signaling pathways. Oncotarget 7 (40), 66051–66060. doi:10.18632/oncotarget.11800
Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12 (9), 735–739. doi:10.1016/s0960-9822(02)00809-6
Lao, T. D., and Le, T. A. H. (2019). Association between LMP-1, LMP-2, and miR-155 expression as potential biomarker in nasopharyngeal carcinoma patients: a case/control study in vietnam. Genet. Test. Mol. Biomarkers 23 (11), 815–822. doi:10.1089/gtmb.2019.0089
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 (5), 843–854. doi:10.1016/0092-8674(93)90529-y
Lichtenstein, P., Holm, N. V., Verkasalo, P. K., Iliadou, A., Kaprio, J., Koskenvuo, M., et al. (2000). Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 343 (2), 78–85. doi:10.1056/nejm200007133430201
Ling, N., Gu, J., Lei, Z., Li, M., Zhao, J., Zhang, H. T., et al. (2013). microRNA-155 regulates cell proliferation and invasion by targeting FOXO3a in glioma. Oncol. Rep. 30 (5), 2111–2118. doi:10.3892/or.2013.2685
Manzo-Merino, J., Contreras-Paredes, A., Vázquez-Ulloa, E., Rocha-Zavaleta, L., Fuentes-Gonzalez, A. M., and Lizano, M. (2014). The role of signaling pathways in cervical cancer and molecular therapeutic targets. Arch. Med. Res. 45 (7), 525–539. doi:10.1016/j.arcmed.2014.10.008
Peng, Y., Dong, W., Lin, T. X., Zhong, G. Z., Liao, B., Wang, B., et al. (2015). MicroRNA-155 promotes bladder cancer growth by repressing the tumor suppressor DMTF1. Oncotarget 6 (18), 16043–16058. doi:10.18632/oncotarget.3755
Pharoah, P. D., Dunning, A. M., Ponder, B. A., and Easton, D. F. (2004). Association studies for finding cancer-susceptibility genetic variants. Nat. Rev. Cancer 4 (11), 850–860. doi:10.1038/nrc1476
Poltronieri, P., D'Urso, P. I., Mezzolla, V., and D'Urso, O. F. (2013). Potential of anti-cancer therapy based on anti-miR-155 oligonucleotides in glioma and brain tumours. Chem. Biol. Drug Des. 81 (1), 79–84. doi:10.1111/cbdd.12002
Qin, W., Ren, Q., Liu, T., Huang, Y., and Wang, J. (2013). MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 587 (9), 1434–1439. doi:10.1016/j.febslet.2013.03.023
Sandhu, S. K., Volinia, S., Costinean, S., Galasso, M., Neinast, R., Santhanam, R., et al. (2012). miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Eμ-miR-155 transgenic mouse model. Proc. Natl. Acad. Sci. U. S. A. 109 (49), 20047–20052. doi:10.1073/pnas.1213764109
Schuetz, J. M., Daley, D., Graham, J., Berry, B. R., Gallagher, R. P., Connors, J. M., et al. (2012). Genetic variation in cell death genes and risk of non-Hodgkin lymphoma. PLoS One 7 (2), e31560. doi:10.1371/journal.pone.0031560
Siegel, R. L., Miller, K. D., Wagle, N. S., and Jemal, A. (2023). Cancer statistics, 2023. CA Cancer J. Clin. 73 (1), 17–48. doi:10.3322/caac.21763
Sterne, J. A., Egger, M., and Smith, G. D. (2001). Systematic reviews in health care: investigating and dealing with publication and other biases in meta-analysis. Bmj 323 (7304), 101–105. doi:10.1136/bmj.323.7304.101
Thai, T. H., Calado, D. P., Casola, S., Ansel, K. M., Xiao, C., Xue, Y., et al. (2007). Regulation of the germinal center response by microRNA-155. Science 316 (5824), 604–608. doi:10.1126/science.1141229
Thompson, C. B. (1995). Apoptosis in the pathogenesis and treatment of disease. Science 267 (5203), 1456–1462. doi:10.1126/science.7878464
Tili, E., Michaille, J. J., Wernicke, D., Alder, H., Costinean, S., Volinia, S., et al. (2011). Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc. Natl. Acad. Sci. U. S. A. 108 (12), 4908–4913. doi:10.1073/pnas.1101795108
Wang, S., Cao, X., Ding, B., Chen, J., Cui, M., Xu, Y., et al. (2016). The rs767649 polymorphism in the promoter of miR-155 contributes to the decreased risk for cervical cancer in a Chinese population. Gene 595 (1), 109–114. doi:10.1016/j.gene.2016.10.002
Witten, L. W., Cheng, C. J., and Slack, F. J. (2019). miR-155 drives oncogenesis by promoting and cooperating with mutations in the c-Kit oncogene. Oncogene 38 (12), 2151–2161. doi:10.1038/s41388-018-0571-y
Wu, D., and Wang, C. (2020). miR-155 regulates the proliferation of glioma cells through PI3K/AKT signaling. Front. Neurol. 11, 297. doi:10.3389/fneur.2020.00297
Wu, H., He, G., Han, H., Xiong, W., Song, T., Chen, H., et al. (2019a). Analysis of MIR155HG variants and colorectal cancer susceptibility in Han Chinese population. Mol. Genet. Genomic Med. 7 (8), e778. doi:10.1002/mgg3.778
Wu, W., Yu, T., Wu, Y., Tian, W., Zhang, J., and Wang, Y. (2019b). The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J. Exp. Clin. Cancer Res. 38 (1), 133. doi:10.1186/s13046-019-1132-0
Wu, X., Wang, Y., Yu, T., Nie, E., Hu, Q., Wu, W., et al. (2017). Blocking MIR155HG/miR-155 axis inhibits mesenchymal transition in glioma. Neuro Oncol. 19 (9), 1195–1205. doi:10.1093/neuonc/nox017
Wu, Y., Hong, Q., Lu, F., Zhang, Z., Li, J., Nie, Z., et al. (2023). The diagnostic and prognostic value of miR-155 in cancers: an updated meta-analysis. Mol. Diagn Ther. 27 (3), 283–301. doi:10.1007/s40291-023-00641-6
Xie, K., Ma, H., Liang, C., Wang, C., Qin, N., Shen, W., et al. (2015). A functional variant in miR-155 regulation region contributes to lung cancer risk and survival. Oncotarget 6 (40), 42781–42792. doi:10.18632/oncotarget.5840
Zheng, B., Zhu, Y. J., Wang, H. Y., and Chen, L. (2017). Gender disparity in hepatocellular carcinoma (HCC): multiple underlying mechanisms. Sci. China Life Sci. 60 (6), 575–584. doi:10.1007/s11427-016-9043-9
Keywords: cancer, MicroRNA-155, MIR155HG, single nucleotide polymorphism, meta-analysis
Citation: Jin G, Guo T, Liu J-W, Yang H-Y, Xu J-G, Pang Y, Yang Y, He S-E and Yi K (2025) The relationship of miR-155 host gene polymorphism in the susceptibility of cancer: a systematic review and meta-analysis. Front. Genet. 16:1517513. doi: 10.3389/fgene.2025.1517513
Received: 26 October 2024; Accepted: 27 January 2025;
Published: 06 March 2025.
Edited by:
Olga V. Anatskaya, Russian Academy of Sciences (RAS), RussiaReviewed by:
Emina Mališić, Institute of Oncology and Radiology of Serbia, SerbiaCopyright © 2025 Jin, Guo, Liu, Yang, Xu, Pang, Yang, He and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Jin, amcxMzg5NDk0NTM0QDE2My5jb20=; Kang Yi, eWlrYW5nMDlAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.