- 1South Texas Diabetes and Obesity Institute, and Division of Human Genetics, Department of Primary and Community Care, School of Medicine, University of Texas Rio Grande Valley, Brownsville, TX, United States
- 2Haplogenics Corporation, Brownsville, TX, United States
- 3Immunogenetics and Transplantation Laboratory, Department of Surgery, School of Medicine, University of California at San Francisco, San Francisco, CA, United States
- 4CSL Innovation GmbH, Marburg, Germany
- 5Division of Hematology and Oncology, Department of Internal Medicine, School of Medicine, University of California at Davis, Davis, CA, United States
- 6Global Medical Affairs, BioMarin, Novato, CA, United States
- 7CSL Limited Research, Bio21 Institute, Melbourne, VIC, Australia
- 8Cellular Technology Ltd, Shaker Heights, OH, United States
- 9Departments of Pathology and Neurology, Case Western Reserve University School of Medicine, Cleveland, OH, United States
- 10Fine Consultancy, Smyrna, GA, United States
- 11Division of Hematology and Oncology, Department of Medicine, University of Texas Health Science Center, and Gulf States Hemophilia and Thrombophilia Center, Houston, TX, United States
- 12Department of Pathology and Laboratory Medicine, VA-Valley Coastal Bend Healthcare System, Harlingen, TX, United States
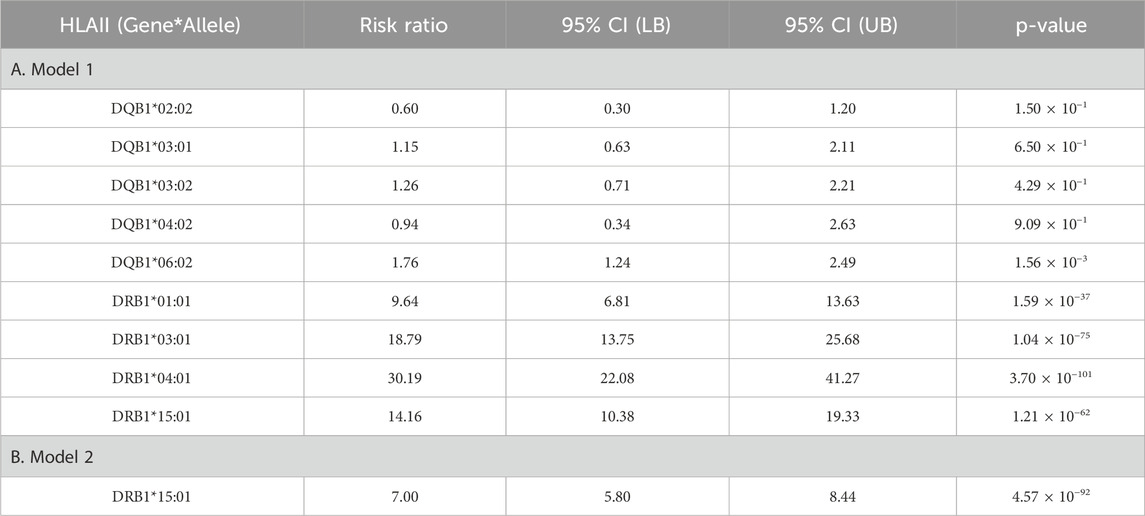
Introduction: Hemophilia A (HA) patients (HAPs) with the human leukocyte antigen (HLA)-class-II (HLAII) haplotype DRB1*15:01/DQB1*06:02, and thus antigen presenting cells which express HLAII β-polypeptide chains that form heterodimers of DR15- and DQ6-serotypes, respectively, have an increased risk of developing factor (F)VIII inhibitors (FEIs)—neutralizing antibodies against the therapeutic-FVIII-proteins (tFVIIIs) infused to prevent/arrest bleeding. As DRB1*15:01 and DQB1*06:02 exist in strong linkage disequilibrium, association analysis cannot determine which is the actual risk allele.
Methods: To establish the true risk allele of this haplotype, we analyzed the tFVIII-derived peptides (tFVIII-dPs) bound to either the DR or DQ molecules that comprise the individual HLAII repertoires expressed by monocyte-derived dendritic cells obtained from 25 normal blood donors and six HAPs, four without and two with FEIs. We performed log-linear mixed model analyses, where the dependent variable is the log of the measured peptide count. Under Model 1, we analyzed an HLAII allele predictor consisting of ten levels (four DRB1 and six DQB1 alleles) in the fixed effects and variables in the random effects to account for non-independence. Model 2—where the HLAII allele variable consisted of only DRB1*15:01 and DQB1*06:02—compares the HLAII alleles.
Results: Relative to the Model 1 reference, DRB1*15:01 and DQB1*06:02 significantly increased tFVIII-derived peptide counts, and DRB1*15:01 contributed significantly more than DQB1*06:02. Reported as risk ratios (RRs) and their 95% confidence interval (CI) lower- (LB) and upper-bound (UB), we found a RR (95% CI-LB, -UB) of 14.16 (10.38, 19.33) and 1.76 (1.24, 2.50) for DRB1*15:01 and DQB1*06:02, respectively. Under Model 2, we found an RR for DRB1*15:01 against DQB1*06:02 of 7.00 (5.80, 8.44).
Discussion/conclusion: Our results suggest that DRB1*15:01 is the offending HLAII allele and that DR15 allotypes underlie the increased FEI risk in HAPs.
Introduction
Hemophilia A (HA) is the X-linked bleeding disorder that results from highly heterogeneous loss-of-function factor (F) VIII (FVIII) gene (F8) mutations and variably deficient-to-absent plasma FVIII coagulant activity (FVIII:C). Life-long regularly repeated infusions of plasma-derived (pd) or recombinant (r) therapeutic FVIII replacement proteins (tFVIIIs) are the standard of care for preventing and arresting bleeding in HA patients (HAPs), but ∼25% will develop neutralizing anti-tFVIII-antibodies, called “FVIII inhibitors (FEIs),” which impair or eliminate their efficacy (Lai and Lillicrap, 2017; Cormier et al., 2020). In addition to well-known environmental variables, FEI risk is influenced by genetic variables, which include the highly heterogeneous set of causative F8 mutations; the functionally distinct single-nucleotide variations (SNVs) in or near loci critical for immune responses which are frequently called immune-mediated disease (IMD) genes; haplotypes of nonsynonymous (ns) SNVs in genes that encode (i) the HLA-class-II (HLAII) molecules used by dendritic cells (DCs), B-cells, and macrophages to present peptide antigens to T cells (i.e., DPA1, DPB1, DQA1, DQB1, DRB1, DRB3, DRB4, and DRB5), and (ii) all or part of FVIII (i.e., F8, F8I22I, and F8B) (Howard et al., 2011; Margaglione and Intrieri, 2018).
In this study, we focused on the HLAII system, particularly on the Chr6 haplotype DRB1*15:01/DQB1*06:02, which contains the *15:01 and *06:02 alleles of DRB1 and DQB1 that exist in strong linkage disequilibrium (LD). This haplotype was found in early studies of HAPs to be positively associated with the risk of developing FEIs (Pavlova et al., 2009). While more recent studies have provided additional support for DRB1*15:01, none have reported that DQB1*06:02 influences FEI risk (Diego et al., 2020; Lessard et al., 2022). Moreover, DRB1*15:01 has also been implicated in the risk for developing autoimmune disorders, such as multiple sclerosis and type 1 diabetes, as well as drug-induced immune-mediated liver disease (Hollenbach and Oksenberg, 2015; Hu et al., 2015; Mangalam et al., 2013; Singer et al., 2010). It cannot be definitively concluded that DRB1*15:01 is the sole risk allele based on genetic association studies alone, however, as it exists in LD with DQB1*06:02. This is especially so in light of findings from a study involving a mouse model of multiple sclerosis in which humanized mice expressing only DQB1*06:02 develop a multiple sclerosis-like neurodegenerative disease mediated by autoimmune targeting of myelin-oligodendrocyte-basic-protein (Kaushansky and Ben-Nun, 2014).
To answer this question and better understand T-cell epitope generation and presentation, we used DC-protein processing and presentation assays (PPPAs), followed by mass spectrometric and peptide proteomic analyses called “MHC-associated peptide proteomics (MAPPs)” to identify and quantify the HLAII-bound/tFVIII-derived peptides (dPs) (Diego et al., 2020; van Haren et al., 2011; V et al., 2013; Sorvillo et al., 2016; Peyron et al., 2018; Jankowski et al., 2019). The DC-PPPA data generated are the HLAII-bound/tFVIII-dPs, with the total number of peptides derived from a specific tFVIII by the DCs of a given HAP, being directly proportional to its immunogenic potential (IP) in that subject (Diego et al., 2020; Jankowski et al., 2019). Because DC-PPPAs and MAPPs analyses yield HLAII-isomer-specific peptide counts—the number of HLAII-bound/tFVIII-dPs being determined for the DP-, DQ-, and DR-isomers separately—we used a generalized linear mixed model to analyze the data (after organization into multiway contingency tables) and draw inferences regarding the relative importance of the DQ- and DR-allotypes to IP. We analyzed HLAII-bound tFVIII-derived peptidomic profiling data generated from DC-PPPAs performed previously in three independent experiments on both the HLA-DQ and -DR isomers (Peyron et al., 2018; Jankowski et al., 2019; Diego et al., 2020). The tFVIII in all three experiments was a full-length (FL)-r-tFVIII, designated “FL-rFVIII” herein.
Methods
Subjects and samples
As described by Peyron et al. (2018), Jankowski et al. (2019), and Diego et al. (2020), peripheral blood mononuclear cells (PBMCs) were isolated from six HAPs—including four without and two with FEIs designated FEI− and FEI+, respectively—and 25 normal donors (NDs) for use in three independent DC-PPPAs (see DC-PPPAs and MAPPs). Information concerning the required Institutional Review Board (IRB) approvals and informed consent have been detailed previously (Peyron et al., 2018; Jankowsky et al., 2019; Diego et al., 2020). Briefly, IRB approval was obtained at either: (1) the University of North Carolina Chapel Hill (USA), which is where PBMCs were isolated from component blood samples obtained from the six HAPs by cytopheresis after each provided informed consent (Jankowski et al., 2019; Diego et al., 2020); (2) Sanquin Research Laboratories (Amsterdam, NLD), where PBMCs were isolated from whole blood (WB) samples obtained by phlebotomy from the nine NDs (Peyron et al., 2018); or (3) Addenbrooke's Hospital (Cambridge, GBR), where PBMCs were isolated from WB samples obtained by phlebotomy from the 16 NDs (Jankowski et al., 2019; Diego et al., 2020). All six HAPs were Caucasian, as detailed in Jankowski et al. (2019), which additionally provides the relevant characteristics of these subjects necessary for clinical/pathologic correlation. Though the racial ancestry of the nine NDs studied by Peyron et al. (2018) and the 16 NDs studied by Jankowski et al. (2019) and Diego et al. (2020) is unknown—as the Ethics Committee at the Sanquin Research Laboratories and Addenbrooke's Hospital, respectively, did not allow this information to be given to the investigators or, for the latter, to ProImmune (www.ProImmune.com) (Oxford, GBR), the fee-for-service company which performed the DC-PPPAs reported in Jankowski et al. (2019) and Diego et al. (2020), i.e., after receiving their WB samples, isolating PBMCs then monocytes, and differentiating their monocytes into immature DCs—most were likely to be Caucasian based on the demographics of US and European blood donors, which, as reported by Wang et al. (2004), Garratty et al. (2004), Glynn et al. (2006), and Benjamin et al. (2008), are typically greater than 80% white. Important details concerning the collection, processing, shipping, and DNA extraction of/from PBMCs have also been described previously by Peyron et al. (2018), Jankowski et al. (2019), and Diego et al. (2020).
DC-PPPAs and MAPPs
The DC-PPPAs used for the research described herein—and in prior investigations referred to collectively as the “FED (FVIII Epitope Determination) Study”—was selected because it directly identifies and relatively quantifies the HLAII-bound/tFVIII-dPs presented by DCs after their uptake and processing of tFVIIIs, doing so separately for the three isomeric forms of HLAII molecules, including DP, DQ, and DR (Diego et al., 2020; Peyron et al., 2018; Jankowski et al., 2019). Note that for the purpose of maximizing our ability to disentangle the allele-specific effects of the DRB1*15:01/DQB1*06:02 haplotype by combining our data with those of Peyron et al. (2018), we were not able to address any DP isomer effects as this other study only isolated/identified the DR- and DQ-bound tFVIII-dPs. The two independent DC-PPPAs and MAPPs analyses reported in Jankowski et al. (2019) and Diego et al. (2020) were conducted under identical experimental conditions at ProImmune using (i) their ProPresent Antigen Presentation Assay (Ventura et al., 2012; Xue et al., 2016; Gouw et al., 2018), (ii) a FL-r-tFVIII which originated as Advate® (Takeda) that we designated “FL-rFVIII”, and (iii) monocyte-derived DCs that were obtained from two different cohorts—one with 12 NDs and the other with four NDs and six HAPs—which are referred to as “S1” and “S2” in Diego et al. (2020). Briefly, immature DCs, which were differentiated from the monocytes isolated from each subject’s PBMCs, were cultured for 7 days with 146 nM of FL-rFVIII in both DC-PPPA-S1 and DC-PPPA-S2 (Diego et al., 2020; Jankowski et al., 2019). After the DCs were induced to maturity with lipopolysaccharide (LPS) overnight, harvested, washed, and then detergent-lysed, their HLAII molecules were affinity-purified as two separate isomer fractions with proprietary anti-DQ and anti-DR monoclonal antibodies whose binding properties were established by ProImmune to be equivalent to the mouse monoclonal antibodies used previously for DC-PPPAs and MAPPs analyses (van Haren et al., 2011; Van Haren et al., 2013; Sorvillo et al., 2016; Peyron et al., 2018; Ventura et al., 2012; Xue et al., 2016; Gouw et al., 2018; van Haren et al., 2012), which include anti-DQ (SPV-L3) (Novus Biologicals) and anti-DR (L243) (Abcam). Peptides eluted from the DQ or DR molecules were analyzed by high-resolution sequencing mass spectrometry (MS), designated herein as liquid chromatography (LC)-tandem MS (LC-MS/MS). The set of tFVIII-derived individual peptide sequences (IPSs) were identified by MAPPs analyses using software to compare the resulting MS data against the UniProt SWISSProt Reference Human Proteome Database supplemented with the amino acid sequences of the commercially available r-tFVIIIs and the known F8 nonsynonymous (ns)-SNPs (Diego et al., 2020).
The DC-PPPAs and MAPPs analyses performed in Peyron et al. (2018)–which were conducted in the laboratories of Drs. Jan Voorberg and Sander Meijer at Sanquin (Amsterdam, NLD) utilizing the same FL-r-tFVIII that originated as Advate® and thus is also designated “FL-rFVIII herein”–used monocyte-derived DCs generated from nine NDs. After pulsing with 100 nM of FL-rFVIII, these DCs were then induced to maturity overnight with LPS, which stabilizes the expression of HLAII-peptide complexes on their cell surface. Analogous to that described above for Diego et al. (2020), the cells were subsequently lysed and their HLA-DR or -DQ molecules were purified using Sepharose beads conjugated with the monoclonal antibodies L243 (DR) or SPV-L3 (DQ). Samples were then analyzed using LC-MS/MS.
As detailed previously in Jankowski et al. (2019), quality control (QC) of the DC-PPPA/MAPPs experiments performed involved confirming that the immature and mature DCs expressed DC markers CD86, DC-SIGN and HLA-DR using immunocytochemistry and flow cytometry (data not shown) and identifying (in the separate DQ- and DR-peptidomes analyzed) HLAII-bound IPSs derived from endogenous proteins known to reside in the endoplasmic reticulum, Golgi, and/or endolysosomal compartment. Specifically, for each of the two isomer groups of HLAII molecules in an experiment—for example, the DR-peptidome of a subject’s DCs cultured with FL-rFVIII—to pass QC within the collection of IPSs identified, some had to have arisen from a minimum of three of the following six endogenous proteins: invariant chain, lysosome-associated membrane proteins-1/-3, transferrin receptor, FCER2/FCGR2, integrin-αM, and apo-lipoprotein B (Diego et al., 2020; Peyron et al., 2018; Jankowski et al., 2019). The likelihood of an LC-MS/MS-identified-peptide being a real identity is described by its expected value and the false discovery rate (Xue et al., 2016; Jeong et al., 2012). In Jankowski et al. (2019), we described how the 1) scoring algorithms and statistical significance determinations were used, 2) residues in tFVIII-derived-IPSs were numbered, and 3) FVIII-derived-IPSs were counted if they contained a minor allele(s) at a variable residue(s). To replicate the DC-PPPAs/MAPPs analyses, a request may be submitted to ProImmune to perform ProPresent assays using the same tFVIIIs and experimental conditions, and similar cellular samples from comparable NDs and HAPs. A similar QC procedure for the DC-PPPA/MAPPs experiments performed at Sanquin are detailed in Peyron et al. (2018)
Statistical analysis
We performed log-linear mixed model analyses under two models, where the dependent variable in both models is the logarithm of the expected peptide count (Knudson et al., 2021; Woodward, 2013; Fienberg and Rinaldo, 2012). Under Model 1, we analyzed in the fixed-effect component of the model a single HLAII-allele predictor variable consisting of ten levels represented by distinct DRB1 and DQB1 alleles (four and six levels, respectively). Under this model, we analyzed in the random effects component of the model variables for individuals (eight and nine levels for Peyron et al. (2018) and Diego et al. (2020), respectively), experiments (two levels for Peyron et al. (2018) and Diego et al. (2020)), and HLAII isomers (two levels for DQ and DR) (Peyron et al., 2018; Jankowski et al., 2019; Diego et al., 2020). The random effects component of the model serves to reduce the error (or “noise”) in our estimation of the predictive effect of HLAII alleles for the peptide counts by accounting for the clustering due to individuals, experiments, and isomers. This approach can therefore be understood as maximizing the signal-to-noise ratio. Model 2 was more focused in that the single HLAII allele predictor variable in the fixed-effect component consisted of only DRB1*15:01 and DQB1*06:02. Model 2 was also simpler in that we only accounted for two random effect variables: for individuals (in this case two and six levels for Peyron et al. (2018) and Diego et al. (2020), respectively) and experiments (Peyron et al., 2018; Jankowski et al., 2019; Diego et al., 2020). Thus, Model 2 is a direct head-to-head comparison of the two main HLAII alleles of interest.
Using data from HAPs, we performed a tetrachoric correlation analysis of dichotomous HA status (affected versus unaffected) (N = 71) and FEI status (affected (i.e., FEI+) versus unaffected (i.e., FEI−)) (N = 64) each time with DRB1*15:01 (presence versus absence) in the Psych package in R. Tetrachoric correlation analysis assumes that the dichotomous variables are each underlain by a normally distributed, latent variable, termed the “liability”, along which there is a threshold, where individuals with liabilities greater than the threshold are affected. The 95% confidence intervals (CIs) and p-values were obtained by bootstrap on 1,000 bootstrap samples.
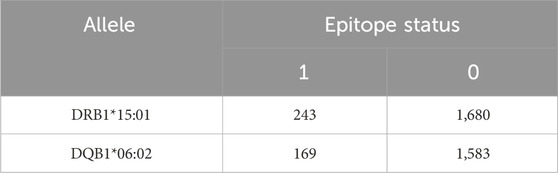
To address allele-specific association with FVIII immunodominant epitopes reported in the literature, we performed a Fisher’s Exact test of a 2×2 contingency table in which the exposure row variable is a dichotomous DRB1*15:01 or DQB1*06:02 status (that is, where an amino acid residue is derived from a peptide bound to the HLAII allele or, more accurately, to the HLAII allotype containing the β-chain encoded by the allele), and the outcome column variable is a dichotomous epitope status scored 1 if a particular tFVIII-dP sequence corresponds to a reported epitope, and 0 otherwise. The immunodominant epitopes considered were found to correspond to sequences from the A2, A3, C1, and C2 domains (Hu et al., 2004; Reding et al., 2003; Reding et al., 2004; Jones et al., 2005; James et al., 2007; Ettinger et al., 2010; Ettinger et al., 2016). Thus, with respect to the dichotomous epitope variable, amino acid residues falling within the published immunodominant epitope sequences were scored 1, and those falling outside those sequences were scored 0.
Results and discussion
In Figure 1A, we plotted the peptide count (per amino acid residue along the tFVIII primary sequence). In Figure 1B, we compared the empirical cumulative distribution functions (eCDFs) of the DQB1 and DRB1 counts. The two are significantly different by a two-sided Kolmogorov–Smirnov test (p < 0.0001) of the difference of eCDFs.
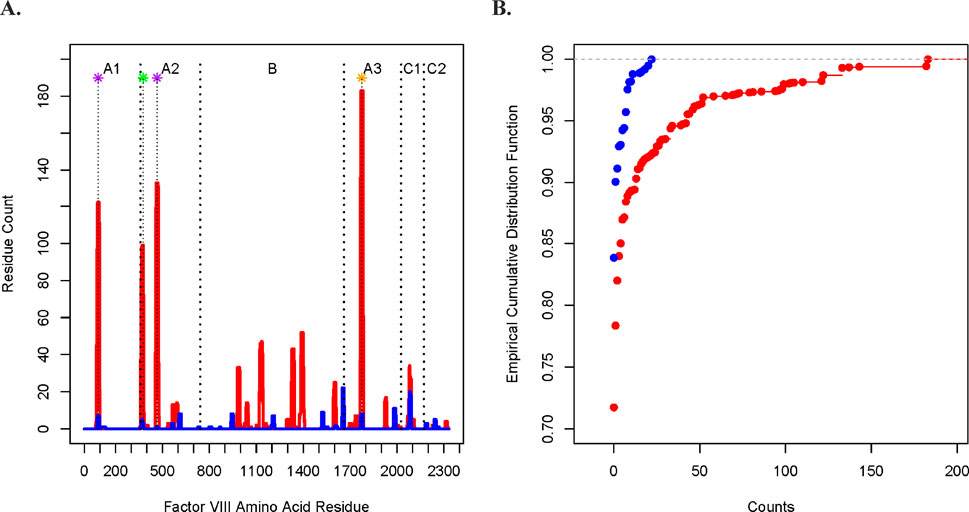
Figure 1. Comparison of the DQ- and DR-derived peptide counts. (A) Peptide counts of the DQB1 (blue) and DRB1 (red) fractions at each amino acid position along the FVIII protein, where the A1, A2, B, A3, C1, and C2 domains are indicated at the top of the figure and where the vertical dashed lines mark contiguous domains. The asterisks at the top indicate the midpoint of ranges from the N-termini to the C-termini of the tFVIII-derived IPSs (relative to the amino acid sequence of the FL-rFVIII parent protein) reported in other studies of potentially immunogenic peptides as well as the corresponding locations from the current study. The purple asterisks indicate peptides that were reported in van Haren et al. with residue midpoints—after rounding to the nearest integer—of 89 and 466 (van Haren et al., 2011; van Haren et al., 2013). The green asterisk located at residue 376 represents a peptide in van Haren et al. (2011), van Haren et al. (2013), Sorvillo et al. (2016), and Hu et al. (2004). The study by Hu et al. (2004) is of interest because they demonstrated in CD4 T-cell stimulation assays that their reported peptide, which spanned residues 371 to 400, consistently had the highest immunogenicity index in NDs as well as in HAPs both those with (FEI+) and without (FEI−) FEIs. The orange asterisk located at midpoint residue 1775 represents a peptide reported in van Haren et al. (2011), van Haren et al. (2013), Sorvillo et al. (2016), and Reding et al. (2004).
The Model 1 results are reported in Figure 2A and Table 1A, where relative to the baseline reference allele provided by DQB1*02:01, both DRB1*15:01 and DQB1*06:02 are at significantly increased risk of contributing to the overall peptide count. Results are reported as risk ratios (RRs) and their associated 95% CI lower and upper bounds (95% CI LB and 95% CI UB). For DRB1*15:01 and DQB1*06:02 respectively, we found an RR (95% CI LB, 95% CI UB) of 14.16 (10.38, 19.33) and 1.76 (1.24, 2.50). Because they are compared to the same baseline, the two RRs may also be directly compared, thus showing that DRB1*15:01 contributes significantly more to the overall peptide count than DQB1*06:02. Under Model 2 (the head-to-head comparison), the RR for the DRB1*15:01 allele against the baseline DQB1*06:02 allele is 7.00 (5.80, 8.44) as reported in Figure 2B and Table 1B.
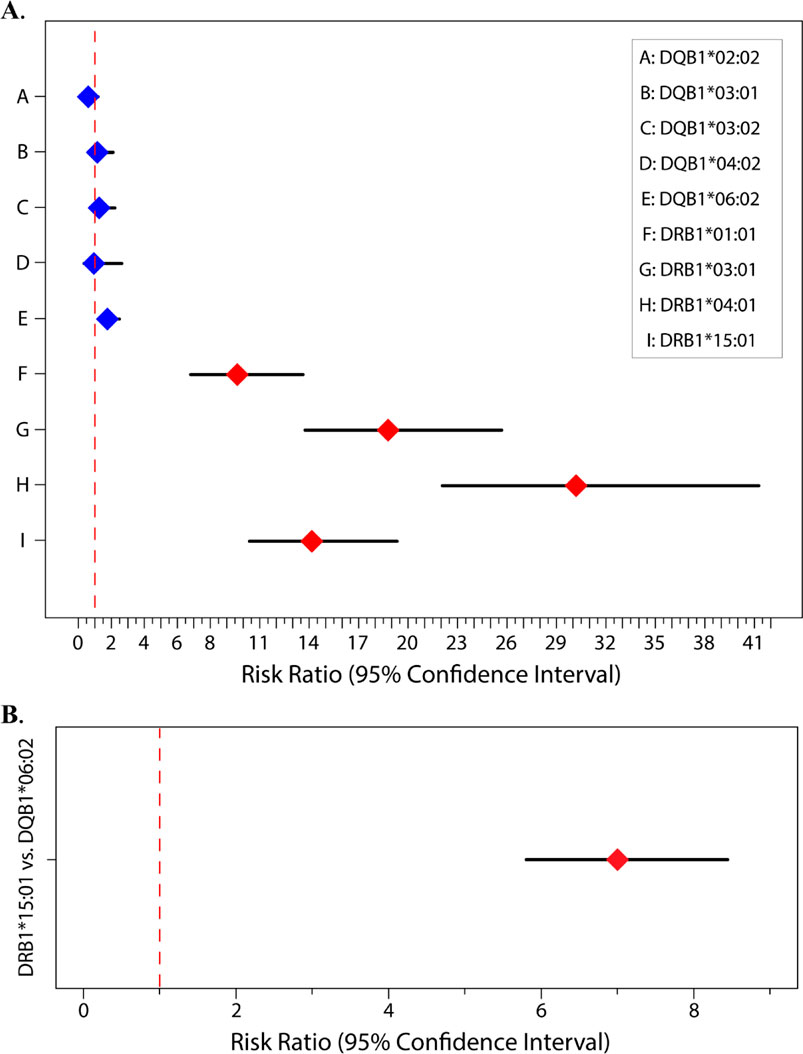
Figure 2. Risk ratios for (A) Model 1. DRB1 alleles (red) and DQB1 alleles (blue) with DQB1*02:01 being the baseline (i.e., reference) allele; (B) Model 2. Head-to-head comparison of DRB1*15:01 (red) to DQB1*06:02 with DQB1*06:02 being the baseline/reference.
We now report the tetrachoric correlation analysis results in terms of the estimate and the LB and UB of its 95% CI, followed by its p-value, in parentheses. We found tetrachoric correlations of 0.91 (0.77, 0.97; 3.7 × 10−4) for HA status and DRB1*15:01 (Figure 3A) and 0.52 (0.23, 0.67; 2.9 × 10−5) for FEI status and DRB1*15:01 (Figure 3B).
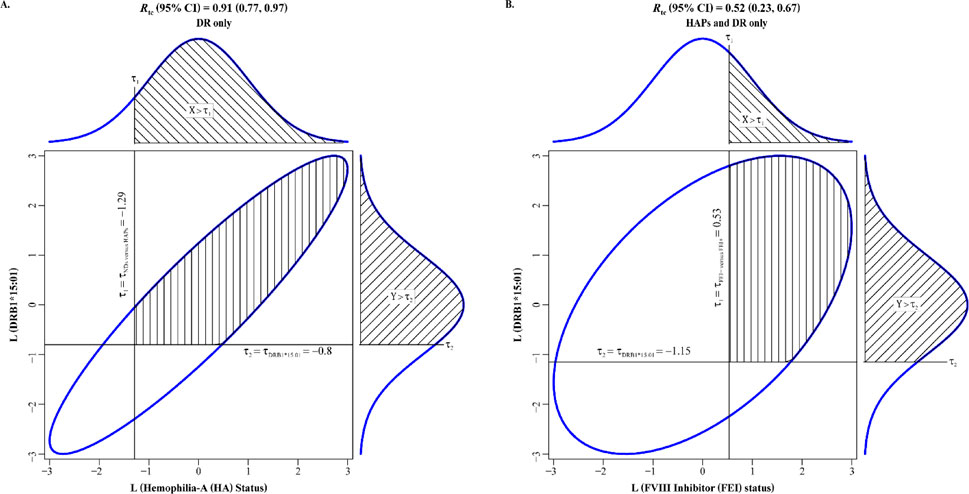
Figure 3. Tetrachoric correlations between DRB1*15:01 (presence versus absence) with (A) hemophilia-A (HA) status (affected versus unaffected) and (B) FVIII inhibitor (FEI) status (yes versus no). Liabilities to HA status and FEI status are on the horizontal axes in (A) and (B), espectively, whereas liability to DRB1*15:01 is on the vertical axis in (A) and (B). The ellipses are the 95% confidence intervals (CIs) for the bivariate normal of the two latent, normally distributed liabilities in (A) and (B).
Results from the statistical analysis of the differential association of DRB1*15:01 and DQB1*06:02 alleles with known immunodominant epitopes in tFVIIIs are shown in Table 2. We found an odds ratio of 1.35 with a 95% CI of 1.10–1.68 (p = 0.005). The inference is that the DRB1*15:01 allele is significantly more associated with epitopes than the DQB1*06:02 allele.
Conclusion
The following two limitations should first be mentioned: (1) These results are from data obtained from DC-PPPAs deriving from Caucasian subjects (or predominantly Caucasian subjects) and so are not generalizable to all groups of people; and (2) Functional T-cell studies are needed to confirm the causality of the DRB1*15:01 allele.
It appears that the strong immunogenicity of the DRB1*15:01/DQB1*06:02 haplotype generally and the DRB1*15:01 allele specifically is underlain at the molecular and cellular levels by a differentially high (i) affinity of DR15 molecules for tFVIII-dPs and (ii) level of DC (and B cell) presentation of DR15-bound/tFVIII-derived peptide complexes to naïve tFVIII-specific CD4 T cells (and to tFVIII-specific helper T cells). The development of FEIs, and thus FEI risk, is complicated as it involves the humoral component of the adaptive immune system and its interactions with the DCs of the innate immune system, which we have hypothesized is regulated by the individually distinct repertoires of HLAII molecules expressed by HAPs via an immunologically relevant gatekeeper role (Diego et al., 2020). Our findings help further characterize the molecular determinants of FVIII immunogenicity as well as the role of HLAII genetics (alleles, genotypes, haplotypes, and allotypes) in the pathogenesis of FEIs, and also provide data to guide the development of precision medicine approaches to improve the management of HAPs. Further investigation is needed to determine the role of these HLAII alleles, genotypes, and haplotypes, as well as HLAII allotypes and isomers, in other diseases, such as the development of multiple sclerosis, narcolepsy, and drug-induced-hypersensitivity with liver injury.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The research involving humans reported here was approved initially by either the: (1) University of North Carolina Chapel Hill (United States) or Addenbrooke’s Hospital (Cambridge, Great Britan) for this and prior investigations referred to collectively as the “FED (Factor VIII Epitope Determination) Study”, which has been described by Jankowski et al. (2019) and Diego et al. (2020); or (2) Sanquin Research Laboratories (Amsterdam, the Netherlands) for the previous investigation described by Peyron et al., 2018. The subjects in these studies, which were conducted in accordance with the local legislation and institutional requirements, provided written informed consent prior to their participation.
Author contributions
VD: conceptualization, data curation, formal analysis, funding acquisition, methodology, resources, software, supervision, validation, visualization, writing–original draft, and writing–review and editing. BL: data curation, funding acquisition, investigation, methodology, project administration, supervision, validation, visualization, writing–review and editing, formal analysis, and software. MA: data curation, formal analysis, software, visualization, writing–review and editing. RR: conceptualization, methodology, software, validation, and writing–review and editing. MH: conceptualization, data curation, investigation, methodology, resources, validation, and writing–review and editing. JG: writing–review and editing. ErM: writing–review and editing. JeP: conceptualization, funding acquisition, and writing–review and editing. LD: investigation, resources, and writing–review and editing. HM: funding acquisition and writing–review and editing. HH: investigation, resources, and writing–review and editing. AV: investigation, resources, and writing–review and editing. JuP: formal analysis, visualization, and writing–review and editing. PL: methodology, resources, and writing–review and editing. SK: methodology and writing–review and editing. EF: writing–review and editing. JC: methodology and writing–review and editing. HG: writing–review and editing. ME: Funding acquisition, Writing–review and editing. SW-B: funding acquisition and writing–review and editing. EuM: conceptualization, data curation, funding acquisition, methodology, project administration, and writing–review and editing. JB: formal analysis, funding acquisition, methodology, software, and writing–review and editing. TH: conceptualization, data curation, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing–original draft, and writing–review and editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by National Institutes of Health (NIH) grants R01 HL169763, RC2 HL101851, R01 HL71130, K08 HL72533, P01 HL045522, R01 MH078143, R01 MH078111, R01 MH083824, R01 MH059490, R01 MD012564, and U54 HG013247, as well as by an investigator-initiated grant from CSL Behring awarded to Haplogenics. Portions of the research were conducted in facilities constructed with support from NIH grants C06 RR013556, C06 RR017515, and C06 RR020547 and leveraged state-of-the-art instrumentation and capabilities established as part of the Valley Baptist Legacy Foundation-supported Project THRIVE and U54 HG013247.
Acknowledgments
We would like to thank Jan Voorberg and Sander Meijer for consenting to our use of their data in Peyron et al. (2018) as well as for generously sharing their expertise on the molecular and cellular processes involved in the uptake, processing, and HLAII presentation of exogenous proteins by monocyte-derived DCs in relation to the PPPAs, and on the technical aspects of peptide sequencing and identification by LC-MS/MS and peptide proteomics. We would also like to thank Laureen Kelley, Martha Hopewell, and Jodi Weeks of Save One Life for their generous gift of the Advate® (FL-rFVIII) used in this study. Finally, we would like to thank Roberta Kellerman, Yara Park, Nigel Key, and Zuben Sauna who carried out the human-subjects research components of this study—which enabled the performance, for the first time, in Jankowsky et al. (2019) and Diego et al. (2020), of DC-PPPAs and MAPPs analyses involving HAPs, both with and without FEIs—that included the acquisition of IRB approval at the UNC Chapel Hill, the recruitment, consent, and enrollment of patients, as well as collecting, processing, and shipping the PBMC samples.
Conflict of interest
These authors have the following positions at Haplogenics Corporation: Director, Technical Operations (BL); Director, Drug Discovery (LD); Chief Medical Officer (JeP); Consultant (HM); and Chief Scientific Officer (TH). Author MH is employed by CSL Innovation GmbH and authors HH, AV, and EM are employed by CSL Limited Research. Author PL is employed by Cellular Technology Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Benjamin, R. J., Busch, M. P., Fang, C. T., Notari, E. P., Puren, A., Schoub, B. D., et al. (2008). Human immunodeficiency virus-1 infection correlates strongly with herpes simplex virus-2 (genital herpes) seropositivity in South African and United States blood donations. Transfusion 48, 295–303. doi:10.1111/j.1537-2995.2007.01523.x
Cormier, M., Batty, P., Tarrant, J., and Lillicrap, D. (2020). Advances in knowledge of inhibitor formation in severe haemophilia A. Br. J. Haematol. 189 (1), 39–53. doi:10.1111/bjh.16377
Diego, V. P., Luu, B. W., Hofmann, M., Dinh, L. V., Almeida, M., Powell, J. S., et al. (2020). Quantitative HLA-class-II/factor VIII (FVIII) peptidomic variation in dendritic cells correlates with the immunogenic potential of therapeutic FVIII proteins in hemophilia A. J. Thrombosis Haemostasis 18 (1), 201–216. doi:10.1111/jth.14647
Ettinger, R. A., James, E. A., Kwok, W. W., Thompson, A. R., and Pratt, K. P. (2010). HLA-DR-restricted T-cell responses to factor VIII epitopes in a mild haemophilia A family with missense substitution A2201P. Haemophilia 16 (102), 44–55. doi:10.1111/j.1365-2516.2008.01905.x
Ettinger, R. A., Paz, P., James, E. A., Gunasekera, D., Aswad, F., Thompson, A. R., et al. (2016). T cells from hemophilia A subjects recognize the same HLA-restricted FVIII epitope with a narrow TCR repertoire. Blood, J. Am. Soc. Hematol. 128 (16), 2043–2054. doi:10.1182/blood-2015-11-682468
Fienberg, S. E., and Rinaldo, A. (2012). Maximum likelihood estimation in log-linear models. Ann. Statistics 40, 996–1023. doi:10.1214/12-aos986
Garratty, G., Glynn, S. A., and McEntire, R.Retrovirus Epidemiology Donor Study (2004). ABO and Rh(D) phenotype frequencies of different racial/ethnic groups in the United States. Transfusion 44, 703–706. doi:10.1111/j.1537-2995.2004.03338.x
Glynn, S. A., Schreiber, G. B., Murphy, E. L., Kessler, D., Higgins, M., Wright, D. J., et al. (2006). Factors influencing the decision to donate: racial and ethnic comparisons. Transfusion 46, 980–990. doi:10.1111/j.1537-2995.2006.00831.x
Gouw, J. W., Jo, J., Meulenbroek, L. A., Heijjer, T. S., Kremer, E., Sandalova, E., et al. (2018). Identification of peptides with tolerogenic potential in a hydrolysed whey-based infant formula. Clin. and Exp. Allergy 48 (10), 1345–1353. doi:10.1111/cea.13223
Hollenbach, J. A., and Oksenberg, J. R. (2015). The immunogenetics of multiple sclerosis: a comprehensive review. J. Autoimmun. 64, 13–25. doi:10.1016/j.jaut.2015.06.010
Howard, T. E., Yanover, C., Mahlangu, J., Krause, A., Viel, K. R., Kasper, C. K., et al. (2011). Haemophilia management: time to get personal? Haemophilia 17 (5), 721–728. doi:10.1111/j.1365-2516.2011.02517.x
Hu, G. L., Okita, D. K., and Conti-Fine, B. M. (2004). T cell recognition of the A2 domain of coagulation factor VIII in hemophilia patients and healthy subjects. J. Thrombosis Haemostasis 2 (11), 1908–1917. doi:10.1111/j.1538-7836.2004.00918.x
Hu, X., Deutsch, A. J., Lenz, T. L., Onengut-Gumuscu, S., Han, B., Chen, W. M., et al. (2015). Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 47 (8), 898–905. doi:10.1038/ng.3353
James, E. A., Kwok, W. W., Ettinger, R. A., Thompson, A. R., and Pratt, K. P. (2007). T-cell responses over time in a mild hemophilia A inhibitor subject: epitope identification and transient immunogenicity of the corresponding self-peptide. J. Thrombosis Haemostasis 5 (12), 2399–2407. doi:10.1111/j.1538-7836.2007.02762.x
Jankowski, W., Park, Y., McGill, J., Maraskovsky, E., Hofmann, M., Diego, V. P., et al. (2019). Peptides identified on monocyte-derived dendritic cells: a marker for clinical immunogenicity to FVIII products. Blood Adv. 3 (9), 1429–1440. doi:10.1182/bloodadvances.2018030452
Jeong, K., Kim, S., and Bandeira, N. (2012). False discovery rates in spectral identification. BMC Bioinforma. 13, S2–S15. doi:10.1186/1471-2105-13-S16-S2
Jones, T. D., Phillips, W. J., Smith, B. J., Bamford, C. A., Nayee, P. D., Baglin, T. P., et al. (2005). Identification and removal of a promiscuous CD4+ T cell epitope from the C1 domain of factor VIII. J. Thrombosis Haemostasis 3 (5), 991–1000. doi:10.1111/j.1538-7836.2005.01309.x
Kaushansky, N., and Ben-Nun, A. (2014). DQB1*06:02-associated pathogenic anti-myelin autoimmunity in multiple sclerosis-like disease: potential function of DQB1*06:02 as a disease-predisposing allele. Front. Oncol. 4, 280. doi:10.3389/fonc.2014.00280
Knudson, C., Benson, S., Geyer, C., and Jones, G. (2021). Likelihood-based inference for generalized linear mixed models: inference with the R package glmm. Stat 10 (1), e339. doi:10.1002/sta4.339
Lai, J., and Lillicrap, D. (2017). Factor VIII inhibitors: advances in basic and translational science. Int. J. Laboratory Hematol. 39, 6–13. doi:10.1111/ijlh.12659
Lessard, S., He, C., Rajpal, D. K., Klinger, K., Loh, C., Harris, T., et al. (2022). Genome-wide association study and gene-based analysis of participants with hemophilia a and inhibitors in the my life, our future research repository. Front. Med. 9, 903838. doi:10.3389/fmed.2022.903838
Mangalam, A. K., Taneja, V., and David, C. S. (2013). HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J. Immunol. 190 (2), 513–518. doi:10.4049/jimmunol.1201891
Margaglione, M., and Intrieri, M. (2018) “Genetic risk factors and inhibitor development in hemophilia: what is known and searching for the unknown,” in Seminars in thrombosis and hemostasis. Thieme Medical Publishers, 509–516.
Pavlova, A., Delev, D., Lacroix-Desmazes, S., Schwaab, R., Mende, M., Fimmers, R., et al. (2009). Impact of polymorphisms of the major histocompatibility complex class II, interleukin-10, tumor necrosis factor-alpha and cytotoxic T-lymphocyte antigen-4 genes on inhibitor development in severe hemophilia A. J. Thrombosis Haemostasis 7 (12), 2006–2015. doi:10.1111/j.1538-7836.2009.03636.x
Peyron, I., Hartholt, R. B., Pedró-Cos, L., van Alphen, F., Brinke, A. T., Lardy, N., et al. (2018). Comparative profiling of HLA-DR and HLA-DQ associated factor VIII peptides presented by monocyte-derived dendritic cells. Haematologica 103 (1), 172–178. doi:10.3324/haematol.2017.175083
Reding, M. T., Okita, D. K., Diethelm-Okita, B. M., Anderson, T. A., and Conti-Fine, B. M. (2003). Human CD4+ T-cell epitope repertoire on the C2 domain of coagulation factor VIII. J. Thrombosis Haemostasis 1 (8), 1777–1784. doi:10.1046/j.1538-7836.2003.00251.x
Reding, M. T., Okita, D. K., Diethelm-Okita, B. M., Anderson, T. A., and Conti-Fine, B. M. (2004). Epitope repertoire of human CD4+ T cells on the A3 domain of coagulation factor VIII. J. Thrombosis Haemostasis 2 (8), 1385–1394. doi:10.1111/j.1538-7836.2004.00850.x
Singer, J. B., Lewitzky, S., Leroy, E., Yang, F., Zhao, X., Klickstein, L., et al. (2010). A genome-wide study identifies HLA alleles associated with lumiracoxib-related liver injury. Nat. Genet. 42 (8), 711–714. doi:10.1038/ng.632
Sorvillo, N., Hartholt, R. B., Bloem, E., Sedek, M., ten Brinke, A., van der Zwaan, C., et al. (2016). von Willebrand factor binds to the surface of dendritic cells and modulates peptide presentation of factor VIII. Haematologica 101 (3), 309–318. doi:10.3324/haematol.2015.137067
van Haren, S. D., Herczenik, E., ten Brinke, A., Mertens, K., Voorberg, J., and Meijer, A. B. (2011). HLA-DR-presented peptide repertoires derived from human monocyte-derived dendritic cells pulsed with blood coagulation factor VIII. Mol. and Cell. Proteomics 10 (6), 002246. doi:10.1074/mcp.M110.002246
van Haren, S. D., Wroblewska, A., Fischer, K., Voorberg, J., and Herczenik, E. (2012). Requirements for immune recognition and processing of factor VIII by antigen-presenting cells. Blood Rev. 26 (1), 43–49. doi:10.1016/j.blre.2011.10.001
Van Haren, S. D., Wroblewska, A., Herczenik, E., Kaijen, P. H., Ruminska, A., ten Brinke, A., et al. (2013). Limited promiscuity of HLA-DRB1 presented peptides derived of blood coagulation factor VIII. PLoS One 8 (11), e80239. doi:10.1371/journal.pone.0080239
Ventura, C., Bisceglia, H., Girerd-Chambaz, Y., Burdin, N., and Chaux, P. (2012). HLA-DR and HLA-DP restricted epitopes from human cytomegalovirus glycoprotein B recognized by CD4+ T-cell clones from chronically infected individuals. J. Clin. Immunol. 32, 1305–1316. doi:10.1007/s10875-012-9732-x
Wang, B., Higgins, M. J., Kleinman, S., Schreiber, G. B., Murphy, E. L., Glynn, S. A., et al. (2004). Comparison of demographic and donation profiles and transfusion-transmissible disease markers and risk rates in previously transfused and non-transfused blood donors. Transfusion 44, 1243–1251. doi:10.1111/j.1537-2995.2004.04034.x
Xue, L., Hickling, T., Song, R., Nowak, J., and Rup, B. (2016). Contribution of enhanced engagement of antigen presentation machinery to the clinical immunogenicity of a human interleukin (IL)-21 receptor-blocking therapeutic antibody. Clin. and Exp. Immunol. 183 (1), 102–113. doi:10.1111/cei.12711
Glossary
CI Confidence interval
DCs Dendritic cells
eCDFs Empirical cumulative distribution functions
F Factor
FVIII Factor VIII
FVIII:C FVIII coagulant activity
FED Study FVIII Epitope Determination Study
FEIs FVIII inhibitors
FEI− Negative for FEIs
FEI+ Positive for FEIs
F8 FVIII gene
FL Full-length
HA Hemophilia A
HAPs HA patients
HLA Human leukocyte antigen
HLAII HLA-class-II
IMD Immune-mediated disease
IP Immunogenic potential
IPSs Individual peptide sequences
LD Linkage disequilibrium
LC Liquid chromatography
LB Lower bound
MHC Major histocompatibility complex
MAPPs MHC-associated peptide proteomics
MS Mass spectrometry
MS/MS Tandem MS
− Negative
NDs Normal donors
ns Nonsynonymous
ORs Odds ratios
PBMCs Peripheral blood mononuclear cells
pd Plasma-derived
PPPAs Protein processing and presentation assays
QC Quality control
RRs Risk ratios
+ Positive
r Recombinant
SNVs Single-nucleotide variations
tFVIII Therapeutic FVIII protein
tFVIII-dPs tFVIII-derived peptides
UB Upper bound
Keywords: Hemophilia A, therapeutic FVIII proteins, FVIII inhibitors, linkage disequilibrium and HLAII haplotype DRB1*15:01/DQB1*06:02, dendritic cell protein processing and presentation assays, therapeutic FVIII derived peptides, MHC-associated peptide proteomics, immunogenic potential
Citation: Diego VP, Luu BW, Almeida MA, Rajalingam R, Hofmann M, Galan JA, Manusov EG, Powell JS, Dinh LV, Mead H, Huynh H, Verhagen AM, Peralta JM, Lehmann PV, Kumar S, Fine EJ, Curran JE, Goring HH, Escobar MA, Williams-Blangero S, Maraskovsky E, Blangero J and Howard TE (2025) Disentangling effects of the DR and DQ isomers encoded by the HLA class II haplotype DRB1*15:01/DQB1*06:02 to help establish the true risk allele for FVIII inhibitor development in Hemophilia A. Front. Genet. 16:1506862. doi: 10.3389/fgene.2025.1506862
Received: 06 October 2024; Accepted: 06 January 2025;
Published: 09 April 2025.
Edited by:
Xi Zhang, Third Affiliated Hospital of Sun Yat-sen University, ChinaReviewed by:
Bing Melody Zhang, Stanford University, United StatesJeane Eliete Laguila, State University of Maringá, Brazil
Copyright © 2025 Diego, Luu, Almeida, Rajalingam, Hofmann, Galan, Manusov, Powell, Dinh, Mead, Huynh, Verhagen, Peralta, Lehmann, Kumar, Fine, Curran, Goring, Escobar, Williams-Blangero, Maraskovsky, Blangero and Howard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vincent P. Diego, dmluY2VudC5kaWVnb0B1dHJndi5lZHU=; Tom E. Howard, dG9tLmhvd2FyZEB1dHJndi5lZHU=
†These authors have contributed equally to this work
 Vincent P. Diego
Vincent P. Diego Bernadette W. Luu2†
Bernadette W. Luu2† Marcio A. Almeida
Marcio A. Almeida Raja Rajalingam
Raja Rajalingam Marco Hofmann
Marco Hofmann Jacob A. Galan
Jacob A. Galan Eron G. Manusov
Eron G. Manusov Juan M. Peralta
Juan M. Peralta Paul V. Lehmann
Paul V. Lehmann Satish Kumar
Satish Kumar Joanne E. Curran
Joanne E. Curran Harald H. Goring
Harald H. Goring Sarah Williams-Blangero
Sarah Williams-Blangero Eugene Maraskovsky
Eugene Maraskovsky John Blangero
John Blangero Tom E. Howard
Tom E. Howard