- The First Affiliated Hospital of Anhui University of Chinese Medicine, Hefei, China
Objective: Autoimmune diseases (ADs) result from an aberrant immune response, in which the body mistakenly targets its own tissues. The association between TGF-β1 gene polymorphisms and risk of developing autoimmune diseases remains to be established. This meta-analysis aimed to reassess the relationship between TGF-β1 T869C gene polymorphisms and susceptibility to autoimmune diseases.
Methods: We conducted a comprehensive search of seven electronic databases for case-control studies investigating the TGF-β1 T869C polymorphism in relation to autoimmune diseases, including rheumatoid arthritis, systemic lupus erythematosus, systemic sclerosis, Sjögren’s syndrome, and juvenile idiopathic arthritis. The search encompassed publications published up to June 2024. Studies were categorized by ethnicity into three groups: Asian, Caucasian, and mixed-ethnicity groups. Five different genetic models were assessed, and the quality of the included studies was evaluated using the Newcastle-Ottawa Scale (NOS). Statistical analyses were performed using Stata 14.0, by calculating the odds ratio (OR) and 95% confidence interval (CI).
Results: A total of 32 case-control studies (31 articles), comprising 4,304 cases and 4,664 controls, were included in this meta-analysis. The overall analysis indicated no significant association between TGF-β1 T869C gene polymorphism and susceptibility to autoimmune diseases. However, subgroup analyses based on race and disease status revealed significant associations. Ethnic subgroup analysis showed that the TGF-β1 T869C allele model (T vs C: OR = 1.422, 95% CI = 1.109–1.824, P = 0.006), homozygous model (TT vs CC: OR = 1.923, 95% CI = 1.232–3.004, P = 0.004), and dominant model (TT + TC vs CC: OR = 1.599, 95% CI = 1.164–2.196, P = 0.004) were associated with autoimmune disease susceptibility in Asians. In the disease subgroup analysis, the results showed that the TGF-β1 T869C allele model (T vs C: OR = 1.468, 95% CI = 1.210–1.781, P = 0.000), recessive model (TT vs TC + CC: OR = 1.418, 95% CI = 1.097–1.832, P = 0.008), dominant model (TT + TC vs CC: OR = 1.747, 95% CI = 1.330–2.295, P = 0.000), homozygous model (TT vs CC: OR = 1.937, 95% CI = 1.373–2.734, P = 0.000), and heterozygous model (TC vs CC: OR = 1.555, 95% CI = 1.199–2.016, P = 0.001) were associated with rheumatoid arthritis susceptibility.
Conclusion: The findings of this meta-analysis suggest that carrying the T allele of the TGF-β1 T869C polymorphism increases the risk of autoimmune diseases in Asian populations. Moreover, individuals carrying the T allele are at higher risk of developing rheumatoid arthritis.
1 Introduction
Autoimmune diseases (ADs) are a class of conditions characterized by immune system dysfunction leading to tissue and organ damage (Rose, 2016). These diseases are broadly categorized into organ-specific autoimmune diseases and systemic autoimmune diseases. Systemic autoimmune diseases such as rheumatoid arthritis, systemic lupus erythematosus, Sjögren’s syndrome, and systemic sclerosis are widespread and pose significant health risks (Vargas-Uricoechea, 2023). Autoimmune diseases affect approximately 10% of the global population and their prevalence is increasing (Cao et al., 2023).
The etiology of ADs is primarily attributed to a combination of genetic predispositions and environmental factors. Among the various factors involved in the pathogenesis of autoimmune diseases, cytokines have garnered considerable attention. Transforming growth factor β1 (TGF-β1) is a key cytokine predominantly expressed by immune cells such as lymphocytes, macrophages, and dendritic cells (Aoki et al., 2005). It has been reported that the TGF-β1 T869C (rs1982073) gene polymorphism is a potential risk factor for various autoimmune diseases, including rheumatoid arthritis, systemic lupus erythematosus, and systemic sclerosis (Hussein et al., 2014; Gómez-Bernal et al., 2022; Xie et al., 2022; Zhang et al., 2020). However, due to discrepancies in experimental data, some studies have found no significant association between TGF-β1 and ADs.
Before 2023, numerous meta-analyses investigated the association between the TGF-β1 promoter T869C polymorphism and the risk of autoimmune diseases. However, these studies frequently faced limitations, including small sample sizes, extended time gaps between analyses, and narrow focus on one or two autoimmune conditions. To address these challenges, the current study sought to include a broader range of autoimmune diseases and incorporate the latest literature, offering a more comprehensive evaluation of the potential connection between the TGF-β1 promoter T869C polymorphism and susceptibility to autoimmune disorders.
2 Materials and methods
2.1 Literature inclusion criteria
The studies included in this analysis were selected based on the following criteria: (i) Study Design: Only case-control studies were considered; (ii) Focus: Studies that evaluated the association between the TGF-β1 T869C polymorphism and susceptibility to autoimmune diseases, specifically rheumatoid arthritis, systemic lupus erythematosus, or systemic sclerosis; (iii) Data Requirements: Studies must provide the genotype frequency distribution for both case and control groups; (iv) Quality Control: The genotype distribution in the control group must adhere to Hardy-Weinberg Equilibrium (HWE).
2.2 Literature exclusion criteria
The following publications were excluded: case reports, review papers, conference abstracts, and similar non-original research articles.
2.3 Literature retrieval method
The meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. A comprehensive search was conducted to identify case-control studies that examined the association between TGF-β1 and autoimmune diseases, including rheumatoid arthritis, systemic lupus erythematosus, systemic sclerosis, Sjögren’s syndrome, and juvenile idiopathic arthritis. The databases searched included PubMed, CNKI, VIP Database, Embase, Web of Science, Wanfang Database, and SCOPUS, covering publications from the inception of each database through June 2024.
For example, the search strategy in PubMed used the following terms: ((((((((Transforming Growth Factor β1 [MeSH Terms]) OR (TGF β1 [Title/Abstract])) OR (TGF-β1 [Title/Abstract])) OR (transforming growth factor beta1 [Title/Abstract])))) AND (((((((((systemic sclerosis [MeSH Terms]) OR (scleroderma [Title/Abstract])) OR (SSC [Title/Abstract]))) OR (((rheumatoid arthritis [MeSH Terms]) OR (RA [Title/Abstract])))) OR (((systemic lupus erythematosus [MeSH Terms]) OR (SLE [Title/Abstract])))) OR (((Juvenile idiopathic arthritis [MeSH Terms]) OR (JIA [Title/Abstract])))) OR (((Sjögren syndrome [MeSH Terms]) OR (SS [Title/Abstract])))))) AND ((((((((single nucleotide [Title/Abstract])) OR (SNP [Title/Abstract])) OR (polymorphism [Title/Abstract])) OR (mutation [Title/Abstract])) OR (variation [Title/Abstract])) OR (variant [Title/Abstract])) OR (polymorphisms [Title/Abstract]))).
2.4 Literature extraction and quality assessment
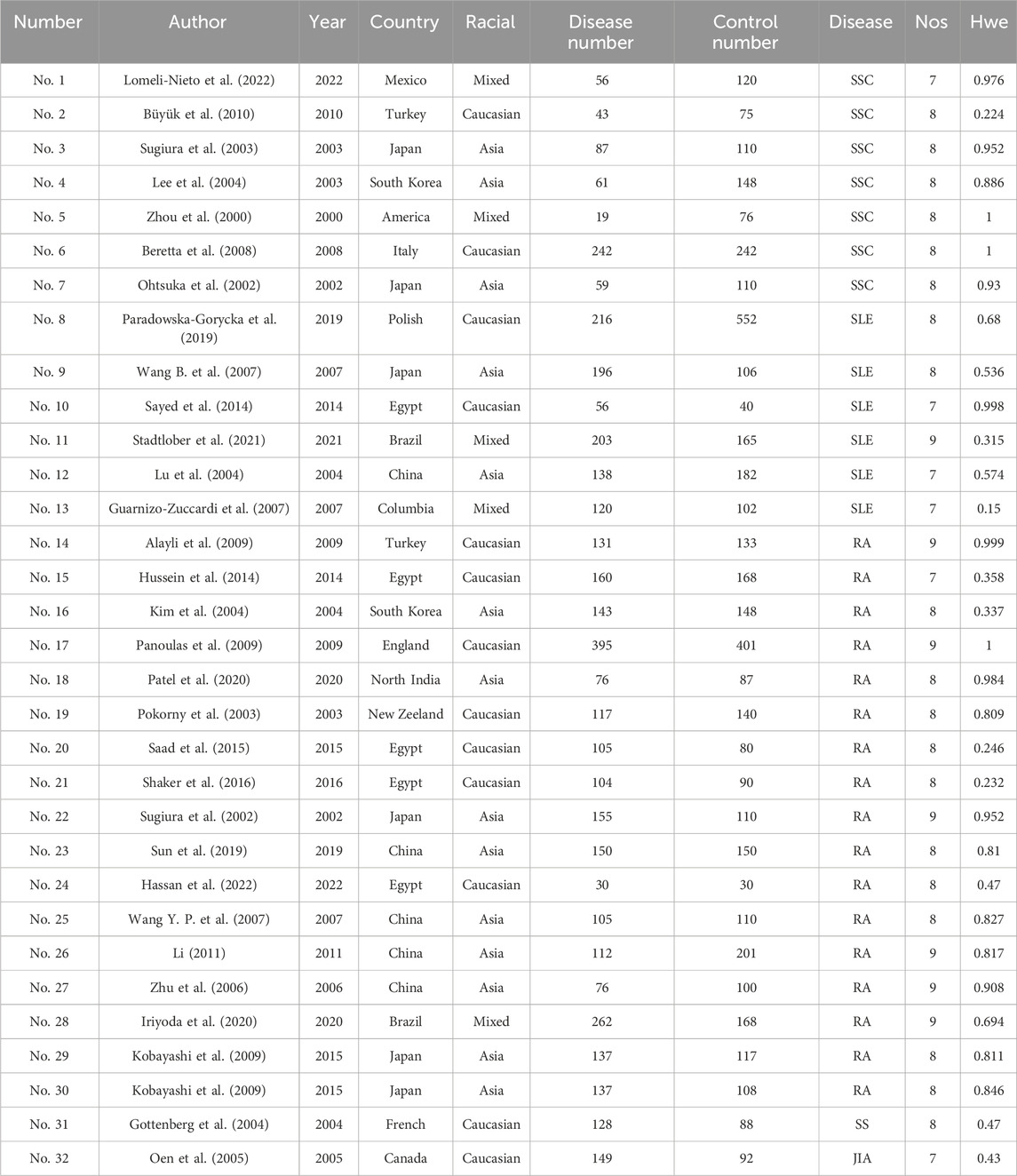
Two researchers, Zhu Yawen and Cheng Yuanyuan, independently extracted relevant data from the articles based on the inclusion and exclusion criteria. The extracted information included the first author, year of publication, type of disease, ethnicity, and number of participants in both the case and control groups. In cases of disagreement between the two researchers, a third researcher, Qian Ai, was consulted to make a final decision. The quality of the included studies was assessed using the Newcastle-Ottawa Scale (NOS), which scores studies from 0 to 9 points. Studies with a score greater than six were considered of high quality (Mirzakhani et al., 2020).
2.5 Statistical analysis
Statistical analyses were performed using STATA 14.0 to assess the association between the TGF-β1 T869C gene polymorphism and autoimmune diseases. The effect size was expressed as odds ratios (OR) with 95% confidence intervals (CI). The analysis focused on five autoimmune diseases: rheumatoid arthritis, systemic lupus erythematosus, systemic sclerosis, Sjögren’s syndrome, and juvenile idiopathic arthritis.
The chi-square test was used to evaluate Hardy-Weinberg Equilibrium (HWE) in each control group. If heterogeneity among studies was indicated by a p-value of less than 0.05, a random-effects model was applied; otherwise, a fixed-effects model was used. The analysis included various genetic models: recessive (TT vs. TC + CC), dominant (TT + TC vs. CC), allele (T vs. C), homozygous (TT vs. CC), and heterozygous (TC vs. CC). Subgroup analysis was conducted when the number of studies was allowed. Publication bias was assessed using the Begg’s and Egger’s tests.
3 Research results
3.1 Literature retrieval
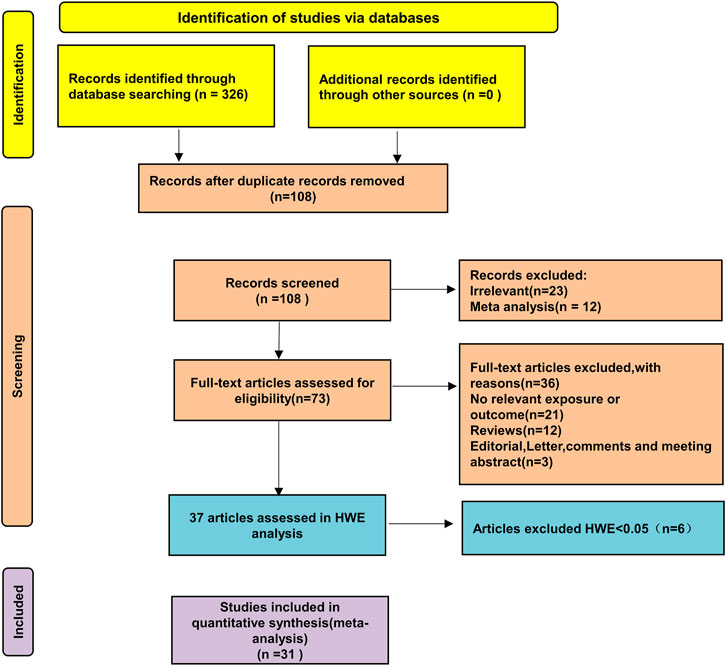
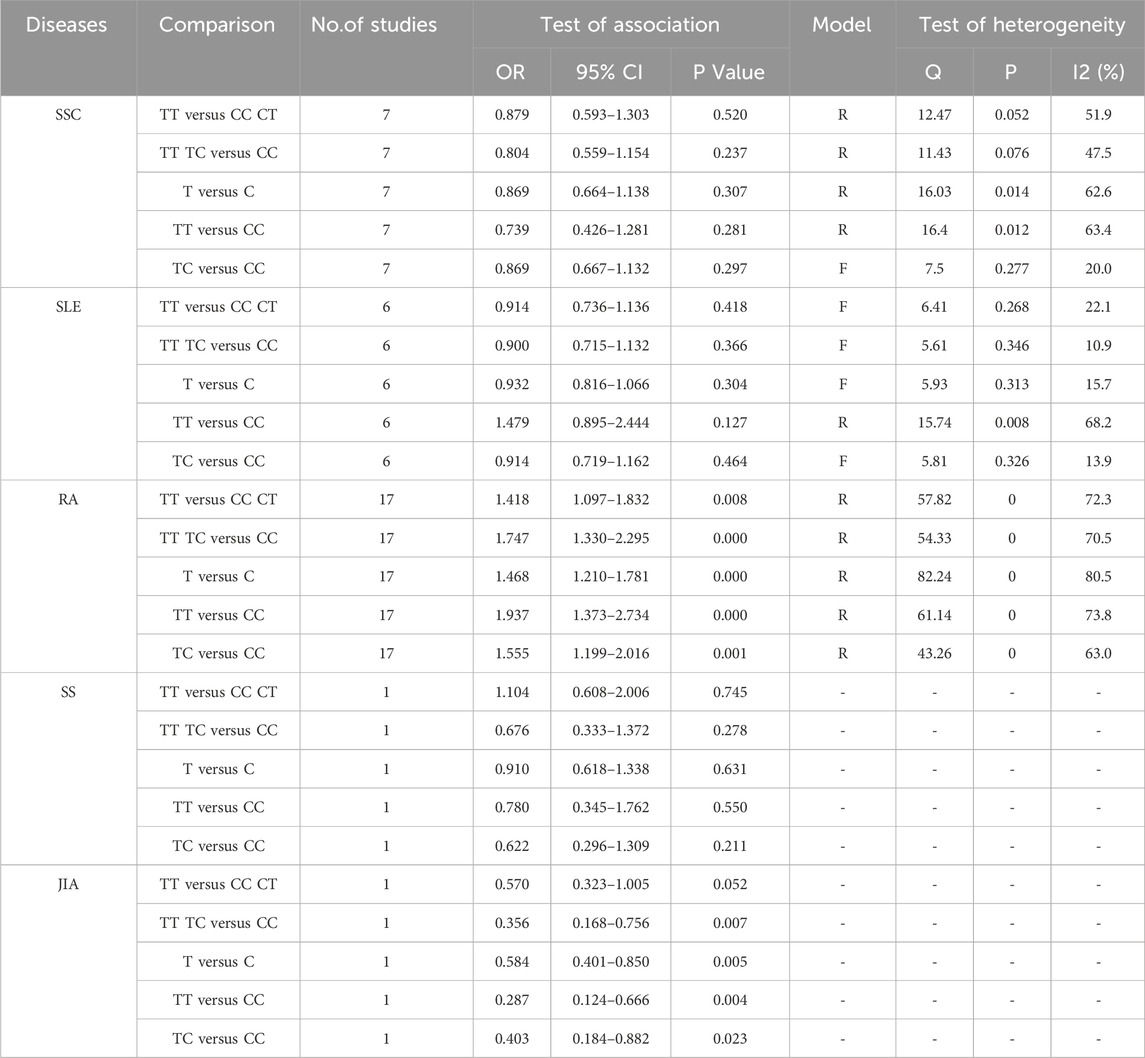
A preliminary search identified 326 published papers (Figure 1). After removing duplicates, 108 papers remained for the next screening phase. Based on a review of the titles and abstracts, 37 articles met the inclusion criteria and were selected for further analysis. After conducting Hardy-Weinberg Equilibrium (HWE) analysis, 31 articles (32 studies) were included in this meta-analysis, comprising 4,304 patients with autoimmune diseases and 4,664 healthy controls (Table 1). The studies included 14 focused on Asian populations, 13 on Caucasian populations, and 5 on mixed ethnic groups. The disease-specific studies included 7 studies on systemic sclerosis, 6 on systemic lupus erythematosus, 17 on rheumatoid arthritis, and one each on Sjögren’s syndrome and juvenile idiopathic arthritis.
3.2 Meta-analysis of TGF-β1 T869C gene polymorphism and susceptibility to autoimmune diseases
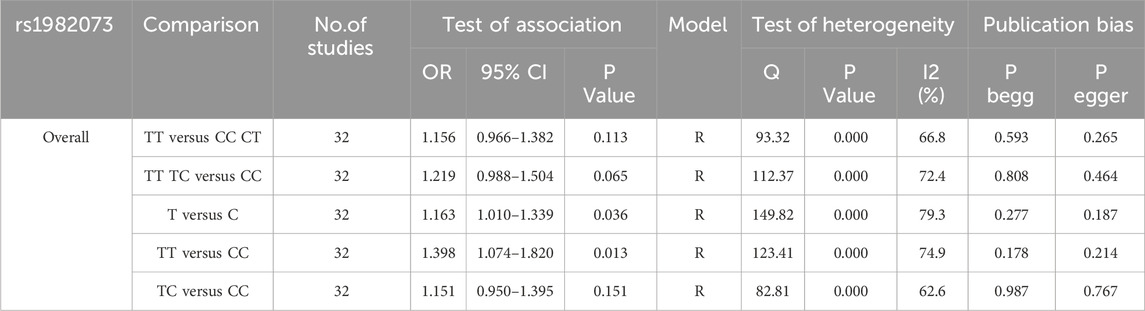
Table 2 presents a summary of the meta-analysis findings regarding the potential association between TGF-β1 T869C gene polymorphism and susceptibility to autoimmune diseases. Because of the heterogeneity observed across all five genetic models, a random effects model was used for the analysis. The findings show no significant association between the TGF-β1 T869C gene polymorphism and susceptibility to autoimmune diseases (T vs. C: OR = 1.163, 95% CI = 1.010-1.339, P = 0.036; TT vs. CC: OR = 1.398, 95% CI = 1.074-1.820, P = 0.013; TT vs. TC + CC: OR = 1.156, 95% CI = 0.966-1.382, P = 0.113; TT + TC vs. CC: OR = 1.219, 95% CI = 0.998-1.504, P = 0.065; TC vs CC: OR = 1.151, 95% CI = 0.950-1.395, P = 0.151).
3.3 Racial asian meta-analysis of TGF-β1 T869C gene polymorphism
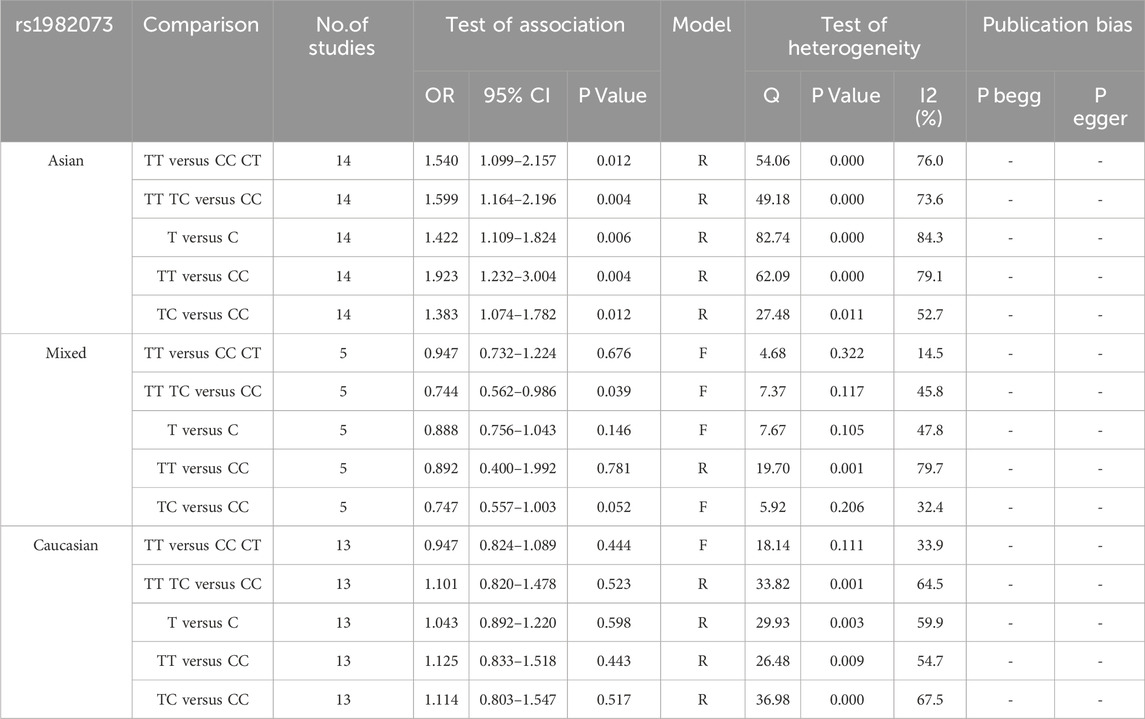
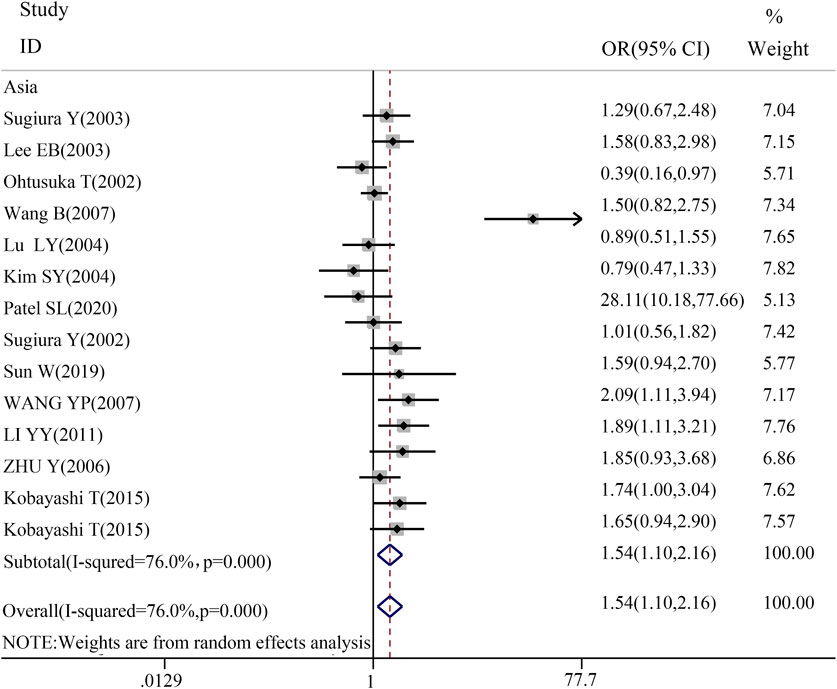
To gain a deeper understanding of the relationship between TGF-β1 T869C gene polymorphism and susceptibility to autoimmune diseases, an ethnic subgroup analysis was conducted. Quantitative analysis of 32 eligible studies identified three distinct ethnic groups: Asians (14 studies), Caucasians (13 studies), and mixed ethnic groups (5 studies). Racial analysis indicated a significant association between TGF-β1 T869C gene polymorphism and susceptibility to autoimmune diseases in the Asian population (Table 3). The results for the Asian subgroup are as follows: TT vs. TC + CC: OR = 1.540, 95% CI = 1.099–2.157, P = 0.012 < 0.01; TT + TC vs. CC: OR = 1.599, 95% CI = 1.164–2.196, P = 0.004 < 0.01; T vs. C: OR = 1.422, 95% CI = 1.109–1.824, P = 0.006 < 0.01; TT vs. CC: OR = 1.923, 95% CI = 1.232–3.004, P = 0.004 < 0.01; TC vs. CC: OR = 1.383, 95% CI = 1.074–1.782, P = 0.012 < 0.01. These findings highlight a significant association between the TGF-β1 T869C polymorphism and susceptibility to autoimmune disease in the Asian population. However, no significant association was observed between Caucasian and mixed-race populations. A forest plot for the Asian group, using the recessive genetic model as an example, is illustrated in Figure 2.
3.4 Disease-specific subgroup analysis of TGF-β1 T869C gene polymorphism
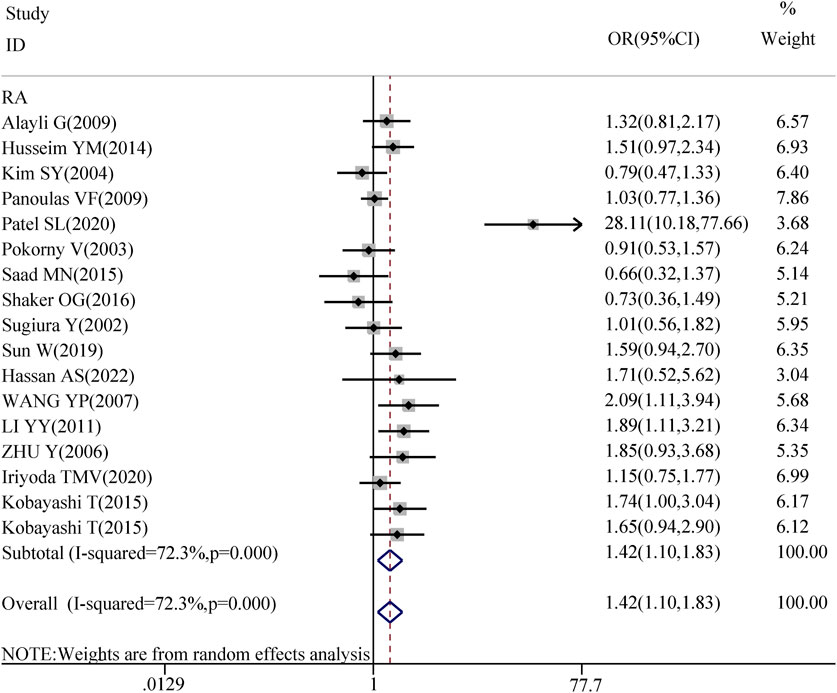
The association between TGF-β1 T869C gene polymorphism and specific autoimmune diseases, including rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and systemic sclerosis (SSC), was assessed using disease-specific subgroup analysis. The analysis included 17 studies on RA, 7 on SLE, and 6 on SSC, as shown in Table 4. In the case of rheumatoid arthritis, all five genetic models revealed a significant correlation: TT vs. TC + CC: OR = 1.418, 95% CI = 1.097–1.832, P = 0.008 < 0.01; TT + TC vs. CC: OR = 1.747, 95% CI = 1.330–2.295, P = 0.000 < 0.01; T vs. C: OR = 1.468, 95% CI = 1.210–1.781, P = 0.001 < 0.01; TT vs. CC: OR = 1.937, 95% CI = 1.373–2.734, P = 0.001 < 0.01; TC vs. CC: OR = 1.555, 95% CI = 1.199–2.016, P = 0.001 < 0.01. However, no significant association was observed between TGF-β1 T869C gene polymorphism and susceptibility to SLE or SSC. A forest plot for the RA subgroup, illustrating the recessive genetic model, is shown in Figure 3.
3.5 Sensitivity analysis and publication bias assessment
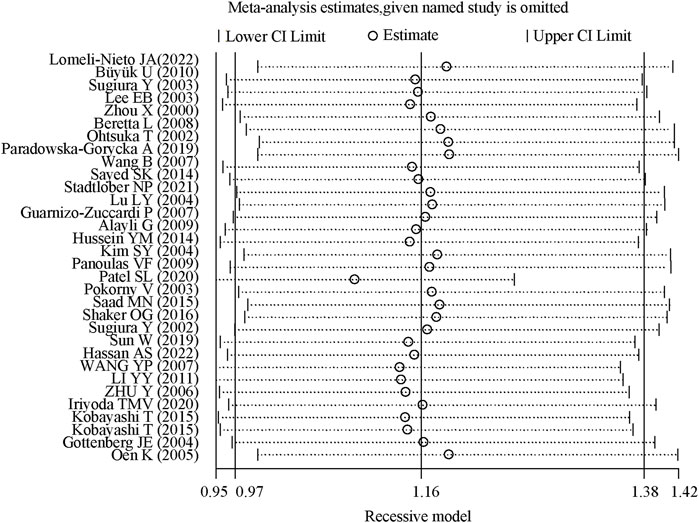
The sensitivity analysis demonstrated that the odds ratios (OR) and 95% confidence intervals (CI) for the comparisons TT + TC vs. CC, TT vs. TC + CC, T vs. C, TT vs. CC, and TC vs. CC remained consistent. This indicates that the results were statistically robust, as illustrated in Figure 4.
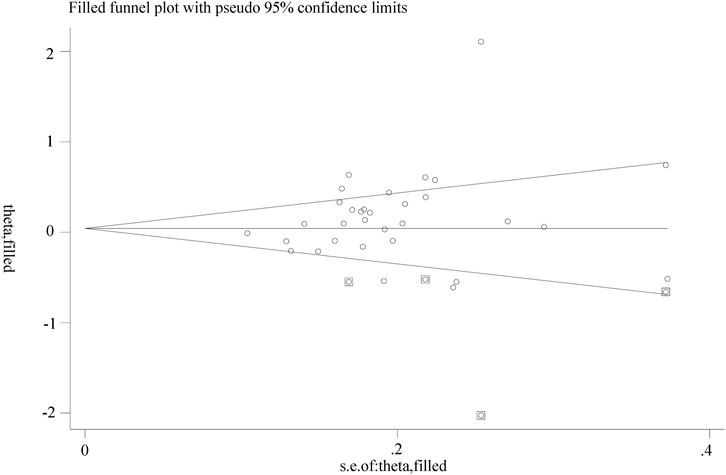
A funnel plot that showed no apparent asymmetry was constructed to assess publication bias. Additionally, both Begg’s test and Egger’s test were performed, and neither test provided evidence of publication bias (T vs. C: Begg’s test, P = 0.277; Egger’s test, P = 0.187), as shown in Figure 5.
4 Discussions
Autoimmune diseases are characterized by the immune system erroneously recognizing normal tissues and cells as foreign entities, leading to an aggressive immune response. Although the precise mechanisms underlying these conditions remain elusive, they are thought to result from a complex interplay between genetic predisposition, environmental influences, gender differences, and epigenetic modifications (Li et al., 2020; Kwiatkowska and Maślińska et al., 2020). Cytokines, a diverse group of small proteins, are central in driving autoimmune responses. Cytokines play a vital role in regulating and mediating immune system functions, and their signaling is essential for a range of biological processes, including cell development, tissue repair, aging, and immune responses (de Gruijter et al., 2022; Lau et al., 2021).
Transforming Growth Factor Beta (TGF-β) is a multifunctional cytokine belonging to a family of growth factors present in the extracellular matrix. Its primary function is to suppress immune cell activity and prevent excessive immune response (Li et al., 2006). The TGF-β family consists of several subtypes including TGF-β1, TGF-β2, and TGF-β3. Among these, TGF-β1 plays a crucial role in the development of autoimmune diseases by inhibiting the activity of T cells, B cells, macrophages, and other immune cells (Prud’homme and Piccirillo et al., 2000). Polymorphisms in TGF-β1 may theoretically affect its expression, potentially increasing the risk of autoimmune diseases. Research has indicated that TGF-β1 produced by regulatory T cells (Tregs) is essential for modulating allergic and autoimmune responses. However, a gradual reduction in TGF-β1 transcription within Tregs leads to a pronounced immune dysregulation (Turner et al., 2020). Additionally, various studies suggest a significant association between elevated TGF-β gene expression and autoimmune disease activity, underscoring its involvement in disease progression (Zaninoni et al., 2023). Currently, four primary genetic polymorphisms of TGF-β1 have been identified: 800G > A, −509C > T, +869T > C, and +915G/C. Among these, the TGF-β1 T869C polymorphism is the most extensively studied polymorphism in the TGF-β1 gene. Numerous studies have demonstrated an association between the T869C polymorphism and susceptibility to various autoimmune diseases such as rheumatoid arthritis, systemic lupus erythematosus, and systemic sclerosis. This polymorphism, located in the first exon of TGF-β1, involves an amino acid substitution from leucine (Leu) to proline (Pro), which may affect the function of the TGF-β1 signal peptide and its immune-modulating effects (Sugiura et al., 2002). The functional significance of this polymorphism in the regulation of TGF-β1 activity has been a central focus of research. Epidemiological evidence further indicates a strong correlation between the T869C polymorphism and the incidence of autoimmune diseases in Asian populations, underscoring the relevance of examining this polymorphism within this demographic. The objective of this study was to undertake a more comprehensive evaluation of the link between TGF-β1 T869C polymorphism and susceptibility to autoimmune diseases by encompassing a wider spectrum of autoimmune conditions and incorporating the latest literature. Investigating the T869C polymorphism not only facilitates comparison and analysis with previous research findings but may also uncover novel therapeutic targets and deepen our understanding of disease mechanisms. Consequently, based on the extensive research in the literature, biological importance, genetic significance, epidemiological data, consistency in study design, clinical relevance, innovation, the necessity of the study, and statistical power, this meta-analysis selected the T869C polymorphism as the focus of this research to explore the role of the TGF-β1 T869C genetic polymorphism in the development of autoimmune diseases, providing a basis for the development of diagnostics and treatments for autoimmune diseases.
Research findings suggest that in Asian populations, carriage of the T allele may confer a risk factor for autoimmune diseases, with individuals possessing this allele being more likely to develop rheumatoid arthritis (RA). Following the initial exploration by Alayli et al. on the association between the TGF-β1 T869C polymorphism and RA in a Turkish population, this polymorphism has garnered increasing attention in the context of autoimmune diseases, providing early evidence for a link between the TGF-β1 T869C polymorphism and an increased risk of RA, which laid the groundwork for subsequent studies (Alayli et al., 2009). Mattey et al. demonstrated that the TGF-β1 T869C polymorphism correlates with disease outcomes and mortality rates among patients with RA, underscoring the potential impact of the T allele on RA severity (Mattey et al., 2005). In the 2010s, Zhang L and colleagues published a meta-analysis encompassing 21 studies, identifying a potential association between the TGF-β1 T869C promoter polymorphism and RA, particularly within Asian populations (OR = 0.81, P = 0.003) (Zhang et al., 2013). This analysis provides compelling evidence supporting the connection between the TGF-β1 T869C polymorphism and increased autoimmune disease risk in Asians, corroborating previous findings and elucidating TGF-β1’s pivotal role in immune response regulation and inflammation promotion. However, the study was not without its limitations, including the inclusion of studies that failed to exclude cases that did not conform to the Hardy-Weinberg equilibrium (HWE), indicating a need for further investigation. Building on this, Hussein et al. highlighted the possible link between the TGF-β1 T869C polymorphism and RA disease progression, whereas Saad et al. presented genetic evidence of associations between multiple polymorphisms, including TGF-β1 and RA. Shaker et al. reaffirmed the link between the TGF-β1 T869C polymorphism and RA susceptibility, with findings supported across a broad spectrum of Asian populations. Recently, Zhu et al. investigated the TGF-β1 mechanism in RA and revealed that TGF-β1 facilitates the migration and invasion of fibroblast-like synoviocytes via the TGF-β1/Smad signaling pathway (Zhu et al. 2019). Their results align with those of this meta-analysis, indicating that the T allele is a risk factor for RA in Asian populations. Furthermore, the T allele of TGF-β1 may be correlated with inflammatory activity, nodular disease, and adverse prognosis in patients with RA. Hassan et al. observed a link between the TGF-β1 T869C polymorphism and disease activity in Egyptian patients with RA, reinforcing the role of the T allele in RA pathogenesis. Collectively, these studies suggest that the TGF-β1 T869C polymorphism could serve as a predictive biomarker for RA and other autoimmune diseases, which is crucial for the early diagnosis and personalized treatment strategy development. While existing research has offered valuable insights, further investigation is essential to explore the interactions between the TGF-β1 T869C polymorphism and additional genetic and environmental factors as well as their impact on autoimmune disease development and progression.
This meta-analysis included 32 case–control studies, including 4,304 cases and 4,664 controls, as documented in 31 articles. The objective of this study was to evaluate the correlation between the TGF-β1 T869C polymorphism and the propensity for autoimmune diseases. Both allele and homozygous models demonstrated significant associations with susceptibility to autoimmune diseases. An ethnic subgroup analysis revealed that across all genetic models, the TGF-β1 T869C polymorphism was markedly associated with autoimmune disease susceptibility in Asian populations, a finding not observed in Caucasian or mixed-race populations. To ascertain the reliability of the study, the researchers evaluated publication bias using both Egger’s and Begg’s tests in addition to performing a sensitivity analysis.
This meta-analysis had several limitations. It included only six studies on systemic lupus erythematosus, seven on systemic sclerosis, one on Sjögren’s syndrome, and a single study on juvenile idiopathic arthritis. The limited number of studies and their inherent heterogeneity necessitate further verification to substantiate the correlation between TGF-β1 T869C polymorphism and autoimmune disease susceptibility. Autoimmune diseases are influenced by a multitude of factors, including genetics, sex, and environmental factors. Additionally, the absence of access to the original data in this study restricts the capacity for a more comprehensive analysis of the interactions between the TGF-β1 T869C polymorphism and environmental influences, lifestyle, and clinical manifestations. No predictive models have been established to account for potential confounding factors, such as sex, age, and environmental conditions. Future research should focus on gene-gene and gene-environment interactions to more effectively assess the relationship between the TGF-β1 T869C polymorphism and autoimmune disease susceptibility. Moreover, the study did not incorporate TGF-β1 levels in its data analysis, which precludes the elucidation of the relationship between TGF-β1 level variations and autoimmune diseases. While this study does not directly evaluate TGF-β1 levels, the existing literature indicates that fluctuations in TGF-β1 levels are associated with disease activity in rheumatoid arthritis (RA). TGF-β1, which plays a dual role in regulating immune responses and suppressing inflammation, may be associated with RA severity and progression. Asian populations, with their distinct genetic backgrounds and environmental exposures, may exhibit a unique relationship between TGF-β1 gene polymorphisms and RA susceptibility. Environmental factors, including infections, lifestyle, and dietary habits, may interact with genetic factors to collectively influence the expression and function of TGF-β1. This study underscores the importance of measuring TGF-β1 levels in future research, particularly in Asian patients with RA. Gaining insight into the relationship between TGF-β1 levels and the T allele could aid in uncovering the molecular mechanisms of RA and inform the development of novel treatment strategies. However, due to the constraints of this study’s design and the data available, direct measurement data of TGF-β1 levels were not provided. This study primarily relies on genetic data, offering insights into the relationship between TGF-β1 gene polymorphisms and RA susceptibility.
In summary, despite certain limitations, the discovery of a significant association between the T allele of the TGF-β1 gene and susceptibility to disease in Asian patients with rheumatoid arthritis (RA) is a noteworthy finding that merits further investigation. This meta-analysis, examining the link between the TGF-β1 T869C polymorphism and autoimmune disease susceptibility across 32 studies (31 articles), substantially expands the scope of previous meta-analyses, thereby bolstering the statistical strength of the collective analysis. Within this dataset, 17 studies specifically addressed rheumatoid arthritis, offering more robust evidence to affirm the connection between the TGF-β1 T869C polymorphism and RA susceptibility. Consequently, the meta-analysis concluded that individuals carrying the T allele might be at an elevated risk of developing autoimmune diseases, particularly RA, in Asian populations. The heightened risk of RA development among patients with the T allele underscores the need for additional research on this genetic correlation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
YZ: Conceptualization, Investigation, Project administration, Software, Writing–original draft, Writing–review and editing. AQ: Investigation, Software, Writing–original draft. YC: Conceptualization, Investigation, Writing–original draft. ML: Data curation, Methodology, Writing–review and editing. CH: Formal Analysis, Methodology, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China [Project No. 82104782] and Collaborative Innovation Project of Universities in Anhui Province, China [Project No. GXXT-2021-085]; Special Project for the Institute of Modernisation of Xin’an Medicine and Traditional Chinese Medicine, Great Health Research Institute [Project No. 2023CXMMTCM004]; Anhui Province Clinical Medical Research Translation Special Project [Project No. 202304295107020114]. This work was supported by the First Affiliated Hospital of Anhui University of Chinese Medicine.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1502921/full#supplementary-material
References
Alayli, G., Kara, N., Tander, B., Canturk, F., Gunes, S., and Bagci, H. (2009). Association of transforming growth factor beta1 gene polymorphism with rheumatoid arthritis in a Turkish population. Jt. bone spine 76 (1), 20–23. doi:10.1016/j.jbspin.2008.02.012
Aoki, C. A., Borchers, A. T., Li, M., Flavell, R. A., Bowlus, C. L., Ansari, A. A., et al. (2005). Transforming growth factor beta (TGF-beta) and autoimmunity. Autoimmun. Rev. 4 (7), 450–459. doi:10.1016/j.autrev.2005.03.006
Beretta, L., Cappiello, F., Moore, J. H., Barili, M., Greene, C. S., and Scorza, R. (2008). Ability of epistatic interactions of cytokine single-nucleotide polymorphisms to predict susceptibility to disease subsets in systemic sclerosis patients. Arthritis rheumatism 59 (7), 974–983. doi:10.1002/art.23836
Büyük, U., Ates, O., Dalyan, L., Müsellim, B., Ongen, G., and Topal-Sarikaya, A. (2010). Analysis of transforming growth factor beta 1 (TGF-beta1) gene polymorphisms in Turkish patients with scleroderma. Cell Biochem. Funct. 28 (4), 274–277. doi:10.1002/cbf.1649
Cao, F., Liu, Y. C., Ni, Q. Y., Chen, Y., Wan, C. H., Liu, S. Y., et al. (2023). Temporal trends in the prevalence of autoimmune diseases from 1990 to 2019. Autoimmun. Rev. 22 (8), 103359. doi:10.1016/j.autrev.2023.103359
de Gruijter, N. M., Jebson, B., and Rosser, E. C. (2022). Cytokine production by human B cells: role in health and autoimmune disease. Clin. Exp. Immunol. 210 (3), 253–262. doi:10.1093/cei/uxac090
Gómez-Bernal, F., Quevedo-Abeledo, J. C., García-González, M., Fernández-Cladera, Y., González-Rivero, A. F., de Vera-González, A., et al. (2022). Serum levels of transforming growth factor beta 1 in systemic lupus erythematosus patients. Biomolecules 13 (1), 73. doi:10.3390/biom13010073
Gottenberg, J. E., Busson, M., Loiseau, P., Dourche, M., Cohen-Solal, J., Lepage, V., et al. (2004). Association of transforming growth factor beta1 and tumor necrosis factor alpha polymorphisms with anti-SSB/La antibody secretion in patients with primary Sjögren's syndrome. Arthritis rheumatism 50 (2), 570–580. doi:10.1002/art.20060
Guarnizo-Zuccardi, P., Lopez, Y., Giraldo, M., Garcia, N., Rodriguez, L., Ramirez, L., et al. (2007). Cytokine gene polymorphisms in Colombian patients with systemic lupus erythematosus. Tissue antigens 70 (5), 376–382. doi:10.1111/j.1399-0039.2007.00917.x
Hassan, A. S., Abdul-Mohymen, A. M., Ahmed, L. A., Seliem, N., Hassan, M. M. R., Ibrahim, H. F., et al. (2022). Study of transforming growth factor beta 1 gene +869T/C polymorphism in Egyptian rheumatoid arthritis patients and its relation to disease activity. Egypt. J. Immunol. 29 (1), 19–28. doi:10.55133/eji.290103
Hussein, Y. M., Mohamed, R. H., El-Shahawy, E. E., and Alzahrani, S. S. (2014). Interaction between TGF-β1 (869C/T) polymorphism and biochemical risk factor for prediction of disease progression in rheumatoid arthritis. Gene 536 (2), 393–397. doi:10.1016/j.gene.2013.11.042
Iriyoda, T. M. V., Flauzino, T., Costa, N. T., Lozovoy, M. A. B., Reiche, E. M. V., and Simão, A. N. C. (2022). TGFB1 (rs1800470 and rs1800469) variants are independently associated with disease activity and autoantibodies in rheumatoid arthritis patients. Clin. Exp. Med. 22 (1), 37–45. doi:10.1007/s10238-021-00725-9
Kim, S. Y., Han, S. W., Kim, G. W., Lee, J. M., and Kang, Y. M. (2004). TGF-beta1 polymorphism determines the progression of joint damage in rheumatoid arthritis. Scand. J. rheumatology 33 (6), 389–394. doi:10.1080/03009740410010344
Kobayashi, T., Murasawa, A., Ito, S., Yamamoto, K., Komatsu, Y., Abe, A., et al. (2009). Cytokine gene polymorphisms associated with rheumatoid arthritis and periodontitis in Japanese adults. J. periodontology 80 (5), 792–799. doi:10.1902/jop.2009.080573
Kwiatkowska, B., and Maślińska, M. (2020). The place of omega-3 and omega-6 acids in supplementary treatment of inflammatory joint diseases. Reumatologia 58 (1), 34–41. doi:10.5114/reum.2020.93511
Lau, S. F., Fu, A. K. Y., and Ip, N. Y. (2021). Cytokine signaling convergence regulates the microglial state transition in Alzheimer's disease. Cell. Mol. life Sci. CMLS 78 (10), 4703–4712. doi:10.1007/s00018-021-03810-0
Lee, E. B., Kim, J. Y., Lee, Y. J., Abdallah, A., Lympany, P., and Song, Y. W. (2004). Transforming growth factor-beta1 polymorphisms in Korean patients with systemic sclerosis. Tissue antigens 63 (5), 491–495. doi:10.1111/j.1399-0039.2004.00185.x
Li, G., Li, Y., Liu, H., Shi, Y., Guan, W., Zhang, T., et al. (2020). Genetic heterogeneity of pediatric systemic lupus erythematosus with lymphoproliferation. Medicine 99 (20), e20232. doi:10.1097/MD.0000000000020232
Li, M. O., Wan, Y. Y., Sanjabi, S., Robertson, A. K., and Flavell, R. A. (2006). Transforming growth factor-beta regulation of immune responses. Annu. Rev. Immunol. 24, 99–146. doi:10.1146/annurev.immunol.24.021605.090737
Li, Y. Y. (2011). Study on the association between transforming growth factor-β1 gene polymorphisms and rheumatoid arthritis. Zhengzhou, China: Zhengzhou University. Master’s thesis.
Lomeli-Nieto, J. A., Muñoz-Valle, J. F., Navarro-Zarza, J. E., Baños-Hernández, C. J., García-Arellano, S., Alvarado-Navarro, A., et al. (2023). TGFB1 mRNA expression and frequency of the + 869T>C and + 915G>C genetic variants: impact on risk for systemic sclerosis. Clin. Exp. Med. 23 (4), 1349–1357. doi:10.1007/s10238-022-00966-2
Lu, L. Y., Cheng, H. H., Sung, P. K., Yeh, J. J., Shiue, Y. L., and Chen, A. (2004). Single-nucleotide polymorphisms of transforming growth factor-beta1 gene in Taiwanese patients with systemic lupus erythematosus. J. Microbiol. Immunol. Infect. = Wei mian yu gan ran za zhi 37 (3), 145–152.
Mattey, D. L., Nixon, N., Dawes, P. T., and Kerr, J. (2005). Association of polymorphism in the transforming growth factor {beta}1 gene with disease outcome and mortality in rheumatoid arthritis. Ann. rheumatic Dis. 64 (8), 1190–1194. doi:10.1136/ard.2004.031674
Mirzakhani, M., Shahbazi, M., Akbari, R., Dedinská, I., Nemati, E., and Mohammadnia-Afrouzi, M. (2020). Soluble CD30, the immune response, and acute rejection in human kidney transplantation: a systematic review and meta-analysis. Front. Immunol. 11, 295. doi:10.3389/fimmu.2020.00295
Oen, K., Malleson, P. N., Cabral, D. A., Rosenberg, A. M., Petty, R. E., Nickerson, P., et al. (2005). Cytokine genotypes correlate with pain and radiologically defined joint damage in patients with juvenile rheumatoid arthritis. Rheumatol. Oxf. Engl. 44 (9), 1115–1121. doi:10.1093/rheumatology/keh689
Ohtsuka, T., Yamakage, A., and Yamazaki, S. (2002). The polymorphism of transforming growth factor-beta1 gene in Japanese patients with systemic sclerosis. Br. J. dermatology 147 (3), 458–463. doi:10.1046/j.1365-2133.2002.04947.x
Panoulas, V. F., Douglas, K. M., Smith, J. P., Stavropoulos-Kalinoglou, A., Metsios, G. S., Nightingale, P., et al. (2009). Transforming growth factor-beta1 869T/C, but not interleukin-6 -174G/C, polymorphism associates with hypertension in rheumatoid arthritis. Rheumatol. Oxf. Engl. 48 (2), 113–118. doi:10.1093/rheumatology/ken443
Paradowska-Gorycka, A., Roszak, M., Stypinska, B., Lutkowska, A., Walczyk, M., Olesinska, M., et al. (2019). IL-6 and TGF-β gene polymorphisms, their serum levels, as well as HLA profile, in patients with systemic lupus erythematosus. Clin. Exp. rheumatology 37 (6), 963–975.
Patel, S. L., Prakash, J., and Gupta, V. (2020). TGF-β1 +869C/T polymorphism increases susceptibility to rheumatoid arthritis in North Indian population. Clin. Rheumatol. 39 (10), 2881–2888. doi:10.1007/s10067-020-05064-w
Pokorny, V., Chau, J., Wu, L., Yeoman, S., Black, P., McQueen, F., et al. (2003). Transforming growth factor beta 1 gene (HSTGFB1) nucleotide T869C (codon 10) polymorphism is not associated with prevalence or severity of rheumatoid arthritis in a Caucasian population. Ann. rheumatic Dis. 62 (9), 907–908. doi:10.1136/ard.62.9.907
Prud'homme, G. J., and Piccirillo, C. A. (2000). The inhibitory effects of transforming growth factor-beta-1 (TGF-beta1) in autoimmune diseases. J. Autoimmun. 14 (1), 23–42. doi:10.1006/jaut.1999.0339
Rose, N. R. (2016). Prediction and prevention of autoimmune disease in the 21st century: a review and preview. Am. J. Epidemiol. 183 (5), 403–406. doi:10.1093/aje/kwv292
Saad, M. N., Mabrouk, M. S., Eldeib, A. M., and Shaker, O. G. (2015). Genetic case-control study for eight polymorphisms associated with rheumatoid arthritis. PloS one 10 (7), e0131960. doi:10.1371/journal.pone.0131960
Sayed, S. K., Galal, S. H., Herdan, O. M., and Mahran, A. M. (2014). Single nucleotide polymorphism T869C of transforming growth factor-beta 1 gene and systemic lupus erythematosus: association with disease susceptibility and lupus nephritis. Egypt. J. Immunol. 21 (2), 9–21.
Shaker, O. G., Alnoury, A. M., Hegazy, G. A., El Haddad, H. E., Sayed, S., and Hamdy, A. (2016). Methylene tetrahydrofolate reductase, transforming growth factor-β1 and lymphotoxin-α genes polymorphisms and susceptibility to rheumatoid arthritis. Rev. Bras. Reumatol. 56 (5), 414–420. doi:10.1016/j.rbre.2016.04.002
Stadtlober, N. P., Flauzino, T., Santos, L. F. D. R. F., Iriyoda, T. M. V., Costa, N. T., Lozovoy, M. A. B., et al. (2021). TGFB1 +869 T > C (rs1800470) variant is independently associated with susceptibility, laboratory activity, and TGF-β1 in patients with systemic lupus erythematosus. Autoimmunity 54 (8), 569–575. doi:10.1080/08916934.2021.1975680
Sugiura, Y., Banno, S., Matsumoto, Y., Niimi, T., Yoshinouchi, T., Hayami, Y., et al. (2003). Transforming growth factor beta1 gene polymorphism in patients with systemic sclerosis. J. rheumatology 30 (7), 1520–1523.
Sugiura, Y., Niimi, T., Sato, S., Yoshinouchi, T., Banno, S., Naniwa, T., et al. (2002). Transforming growth factor beta1 gene polymorphism in rheumatoid arthritis. Ann. rheumatic Dis. 61 (9), 826–828. doi:10.1136/ard.61.9.826
Sun, W., Yi, M., Bai, Y., Wu, L., Chen, J., Ren, Y., et al. (2019). Correlations between the polymorphism of +869T/C in TGF-β1 and rheumatoid arthritis. J. Musculoskelet. and neuronal Interact. 19 (1), 127–132.
Turner, J. A., Stephen-Victor, E., Wang, S., Rivas, M. N., Abdel-Gadir, A., Harb, H., et al. (2020). Regulatory T cell-derived TGF-β1 controls multiple checkpoints governing allergy and autoimmunity. Immunity 53 (6), 1202–1214.e6. doi:10.1016/j.immuni.2020.10.002
Vargas-Uricoechea, H. (2023). Molecular mechanisms in autoimmune thyroid disease. Cells 12 (6), 918. doi:10.3390/cells12060918
Wang, B., Morinobu, A., Kanagawa, S., Nakamura, T., Kawano, S., Koshiba, M., et al. (2007). Transforming growth factor beta 1 gene polymorphism in Japanese patients with systemic lupus erythematosus. Kobe J. Med. Sci. 53 (1-2), 15–23.
Wang, Y. P., Xiao, L. S., Zhou, Y., Fei, Y., and Li, J. P. (2007). Polymorphisms of transforming growth factor-β1 gene and rheumatoid arthritis. Chin. J. Public Health 23 (11), 1405–1406. doi:10.11847/zgggws2007-23-11-68
Xie, X., Gan, H., Tian, J., Li, F., Chen, J., Wang, J., et al. (2022). Iguratimod inhibits skin fibrosis by regulating TGF-β1/Smad signalling pathway in systemic sclerosis. Eur. J. Clin. investigation 52 (8), e13791. doi:10.1111/eci.13791
Zaninoni, A., Fattizzo, B., Pettine, L., Vercellati, C., Marcello, A. P., and Barcellini, W. (2023). Cytokine polymorphisms in patients with autoimmune hemolytic anemia. Front. Immunol. 14, 1221582. doi:10.3389/fimmu.2023.1221582
Zhang, L., Yan, J. W., Wang, Y. X., Wan, Y. N., Li, J. P., Liu, P., et al. (2013). Association of TGF-β1 +869C/T promoter polymorphism with susceptibility to autoimmune diseases: a meta-analysis. Mol. Biol. Rep. 40 (8), 4811–4817. doi:10.1007/s11033-013-2577-4
Zhang, X., Yun, J. S., Han, D., Yook, J. I., Kim, H. S., and Cho, E. S. (2020). TGF-Β pathway in salivary gland fibrosis. Int. J. Mol. Sci. 21 (23), 9138. doi:10.3390/ijms21239138
Zhou, X., Tan, F. K., Stivers, D. N., and Arnett, F. C. (2000). Microsatellites and intragenic polymorphisms of transforming growth factor beta and platelet-derived growth factor and their receptor genes in Native Americans with systemic sclerosis (scleroderma): a preliminary analysis showing no genetic association. Arthritis rheumatism 43 (5), 1068–1073. doi:10.1002/1529-0131(200005)43:5<1068::AID-ANR17>3.0.CO;2-G
Zhu, D., Zhao, J., Lou, A., Huang, Q., OuYang, Q., Zhu, J., et al. (2019). Transforming growth factor β1 promotes fibroblast-like synoviocytes migration and invasion via TGF-β1/Smad signaling in rheumatoid arthritis. Mol. Cell. Biochem. 459 (1-2), 141–150. doi:10.1007/s11010-019-03557-0
Keywords: autoimmune disease, meta-analysis, polymorphism, transforming growth factor-β1, susceptibility
Citation: Zhu Y, Qian A, Cheng Y, Li M and Huang C (2025) Comprehensive systematic review and meta-analysis of the TGF-β1 T869C gene polymorphism and autoimmune disease susceptibility. Front. Genet. 16:1502921. doi: 10.3389/fgene.2025.1502921
Received: 27 September 2024; Accepted: 07 January 2025;
Published: 20 February 2025.
Edited by:
Jordi Pérez-Tur, Spanish National Research Council (CSIC), SpainReviewed by:
Sailu Yellaboina, CR Rao Advanced Institute of Mathematics, Statistics and Computer Science, IndiaAshwin Kotnis, All India Institute of Medical Sciences, Bhopal, India
Copyright © 2025 Zhu, Qian, Cheng, Li and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanbing Huang, Y2h1YW5iaW5oQDE2My5jb20=
 Yawen Zhu
Yawen Zhu Ai Qian
Ai Qian Chuanbing Huang
Chuanbing Huang