
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 09 April 2025
Sec. Genetics of Common and Rare Diseases
Volume 16 - 2025 | https://doi.org/10.3389/fgene.2025.1485874
 Lin Zhou†
Lin Zhou† Farrukhjon Boboev†
Farrukhjon Boboev† Hui Chen
Hui Chen Fanwen Jiang
Fanwen Jiang Chun Zhang
Chun Zhang Jing Xiao
Jing Xiao Hui Jiang
Hui Jiang Yongchuan Liao*
Yongchuan Liao* Zhuping Xu*
Zhuping Xu*Introduction: Our previous research identified pathogenic variants in RetNet genes in 23.4% of individuals with early-onset high myopia. This study aims to analyze the genetic defects in patients with high myopia complicated by rhegmatogenous retinal detachment.
Method: Whole-exome sequencing was performed on 40 patients with high myopia accompanied by retinal detachment. Variants were filtered from 281 RetNet genes, 178 genes related to syndromic high myopia, 23 non-syndromic high myopia-associated genes, and 29 rhegmatogenous retinal detachment-related genes using a multistep bioinformatics approach. Clinical data were collected for genotype-phenotype correlation analysis.
Results: Pathogenic variants were detected in 47.5% (19/40) in patients with high myopia accompanied by retinal detachment, specifically in RetNet genes (18/40), rhegmatogenous retinal detachment-related genes (11/40), and syndromic high myopia associated genes (10/40). No variants were found in non-syndromic genes. The most prevalent pathogenic genes for high myopia with retinal detachment were Stickler-related genes, including COL2A1 (10.0%, 4/40) and COL11A1 (5.0%, 2/40). Patients with Stickler-related gene variants presented the youngest average age of retinal detachment onset (35.17 ± 18.03 years) and shortest axial length (27.63 ± 1.01 mm).
Conclusion: RetNet genes are the predominant causative genes (18/40, 45.0%) in patients with high myopia and retinal detachment. The findings affirm that Stickler syndrome (15%) is a significant etiological factor for high myopia accompanied by retinal detachment. We recommend enhanced comprehensive systemic and ophthalmic examinations for patients with high myopia to enable early detection and prevention of retinal detachment.
High myopia is defined as a refractive error of at least −6.0 diopters accompanied by an axial length exceeding 26.0 mm (Ohno-Matsui et al., 2021). A significant increase in prevalence of myopia has been reported over the past 2 decades, with 30%–50% of adults affected. Moreover, approximately 20% of myopic individuals could progress to high myopia by 2050 (Morris et al., 2021).
The progression of myopia is influenced by a combination of environmental and genetic factors. Early-onset high myopia, defined as myopia of −6.0 diopters or more in early childhood, is often regarded as a monogenic disease (Wang et al., 2022). To date, hundreds of genes and 26 chromosomal loci have been identified as contributors to high myopia through various methods like next-generation sequencing, genome-wide association studies, and twin studies (Govers et al., 2023). Specifically, 23 genes have been identified as associated with nonsyndromic high myopia. Our previous research indicated that 23.4% early onset high myopia cases were associated with RetNet gene (Zhou et al., 2018).
High myopia is strongly associated with an increased risk of ocular complications due to axial elongation. The most common ocular complications of high myopia are open-angle glaucoma, posterior subcapsular cataract, and rhegmatogenous retinal detachment (RRD) (Johnston et al., 2016). RRD, the most common and vision-affecting complication of high myopia, involves the separation of the retinal neuroepithelium from the pigment epithelium (Kwok et al., 2020; Schick et al., 2020). In a large United Kingdom Biobank study, each additional six diopters of myopia increased the risk of retinal detachment by 7.2 times (Han et al., 2020). High myopia was found in 17.0% of eyes with RRD, a prevalence comparable to that in European populations but lower than that in Asian populations, where it reaches 34.0% (Mitry et al., 2011; Li and Beijing, 2003). The annual incidence of RRD is highest among Chinese residents, at 11.6 cases per 100,000 people (Mowatt et al., 2003). Furthermore, 12%–13% of RRD patients exhibit a family history of retinal detachment in their first-degree relatives (Chandra et al., 2015). Evidence of population differences, possibly related to ethnicity, as well as familial clustering, suggests a genetic predisposition to the occurrence of RRD (Wong et al., 1999). To date, 29 genes have been associated with monogenic disorders involving RRD (Govers et al., 2023).
However, the genetic basis of high myopia accompanied by RRD remains largely unexplored. To elucidate the molecular genetics and the phenotype of high myopia with RRD, we recruited 40 patients. Pathogenetic variants of the RetNet genes, genes related to non-syndromic and syndromic high myopia and genes responsible for RRD were filtered using whole exome sequencing. Genotype-phenotype correlation was analyzed based on clinical presentations.
Patients with high myopia accompanied by RRD were recruited in our study. High myopia was characterized by a refractive error of less than −6.0 diopters and an axial length greater than 26.0 mm. The probands with high myopia complicated by RRD, along with available family members, were collected. Tractional, exudative, and traumatic retinal detachment, as well as individuals with other ocular or systemic diseases, were excluded. Participants were recruited from the clinic of West China Hospital at Sichuan University, Chengdu, China. The study adhered to the principles of the Declaration of Helsinki and received approval from the Ethics Committee of West China Hospital, Sichuan University. Informed consent was obtained from every participant. This study collected peripheral blood samples along with clinical data, including family history, past medical history, refractive error, best corrected visual acuity (BCVA), and axial length.
Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp Blood Midi Kit. Sequencing was performed by DNBSEQ-T7 platform (MGI, Shenzhen, China) with 150 bp paired-end reads. Sanger sequencing was conducted by ABI3730xl sequencer (Applied Biosystems, United States). Post-sequencing, the raw data in FASTQ format underwent quality control to eliminate low-quality reads. Clean reads were then assembled and spliced using the MyGenostics second-generation sequencing analysis platform. Coverage and sequencing quality of the target region were assessed. Pathogenic variants in 281 RetNet genes (https://web.sph.uth.edu/RetNet/), 178 genes related to syndromic high myopia, 23 genes associated with non-syndromic high myopia and 29 genes responsible for RRD were filtered through multi-step bioinformatics analyses as previously reported (Govers et al., 2023; Yi et al., 2022; Jiang et al., 2024) (Supplementary Table S1; Supplementary Figure S1). Sanger sequencing was used to confirm candidate pathogenic variants and conduct co-segregation analysis. The pathogenicity was evaluated according to the American College of Medical Genetics and Genomics (ACMG) genetic variation classification criteria and guidelines (Richards et al., 2015).
Probands with high myopia complicated by RRD underwent ocular examinations by experienced ophthalmologists. A detailed family and ophthalmic history, including duration of low vision and history of refractive or intraocular surgery, was collected. The examinations included assessment of BCVA, refractive error, and axial length. The axial length was measured by an Optical biometer (IOL Master V5.0, Carl Zeiss Meditec AG, Germany).
A genotype-phenotype analysis was conducted on patients harboring different genetic variations. The study focused on three distinct patient groups: (1) individuals carrying known and unknown gene variants, (2) individuals with pathogenic variants in RetNet-associated genes, RRD-related genes, and syndromic high myopia associated genes, (3) individuals with pathogenic variants in Stickler-related genes and other diseases. Comparative analyses were performed on the age of onset, axial length, refractive error, and BCVA among these groups.
The genotypes of patients with high myopia complicated by RRD were compared with those from previous studies involving early-onset and late-onset high myopia using the Chi-square test (χ2). Differences in continuous variables, such as BCVA, refractive error, and axial length, were analyzed using independent sample t-tests for comparisons between two groups and one-way ANOVA for comparisons among more than two groups. All statistical analyses were performed using IBM SPSS Statistics 27 (SPSS Inc., Chicago, IL, United States). A p-value of less than 0.05 was considered statistically significant, with all tests being two-sided.
A total of 40 patients suffered high myopia complicated by RRD was recruited in our study. Whole exome sequencing was performed for all of probands. 281 RetNet genes (https://web.sph.uth.edu/RetNet/), 178 genes related to syndromic high myopia, 23 genes associated with non-syndromic high myopia and 29 genes responsible for RRD were analyzed by multistep bioinformatics analyses. A total of 19 probands (47.5%, 19/40) were detected pathogenic variant in these genes (Table 1). Based on multi-step bioinformatics analysis, including nucleotide and protein changes, frequencies from different databases, pathogenicity prediction, and classification according to ACMG guidelines, we identified 19 pathogenic variants. According to ACMG classification, 16 out of 19 variants were classified as likely pathogenic, and 3 out of 19 were classified as pathogenic. Among these, 16 were missense mutations and three were splice site variants. Of the 19 variants, 5 were previously reported, and 14 were novel.
The pathogenic variant frequencies in RetNet genes and RRD-related genes and genes associated with syndromic myopia was 45.0% (18/40), 27.5% (11/40) and 25.0% (10/40), respectively. The identified pathogenic variants included 14 genes such as: COL2A1, COL11A1, VCAN, CAPN5, EFEMP1, FBN1, FZD4, IMPDH1, LRP5, NR2E3, PDE6B, RDH12, RP1 and SNRNP200. Notably, 13 of these 14 pathogenic genes were RetNet genes, with the exception of FBN1. COL11A1, COL2A1, VCAN, FZD4, LRP5, FBN1 are associated with genes with RRD. COL11A1, COL2A1, VCAN, FBN1, RP1 are linked to syndromic high myopic genes. No pathogenic variants were detected in genes specifically associated with non-syndromic high myopia. The most common pathogenic genes responsible for high myopia with RRD were COL2A1 (10.0%, 4/40) and COL11A1 (5.0%, 2/40) and VCAN (5.0%, 2/40). COL2A1 and COL11A1 were responsible for Stickler syndrome (15%, 6/40) and VCAN was associated with Wagner syndrome (5.0%, 2/40) (Table 1; Figure 1).

Figure 1. Genotype distribution of high myopia with RRD and comparison with early-onset and late-onset high myopia without RRD in previous studies. (A) Distribution of genotypes in high myopia with RRD across different gene categories (including 281 RetNet genes, 178 genes associated with syndromic high myopia, 23 genes linked to non-syndromic high myopia, and 29 genes responsible for rhegmatogenous retinal detachment). The highest proportions of pathogenic genes were found in COL2A1 (4/40, 10.0%), COL11A1 (2/40, 5.0%), and VCAN (2/40, 5.0%). (B) Venn diagram showing the distribution of pathogenic genes in high myopia with RD across different groups. (C) Comparison of the proportion of high myopia with RD versus early-onset high myopia without RD and late-onset high myopia without RD.
The pathogenic variants in RetNet genes identified in our study were compared with those in patients with early-onset and late-onset high myopia from our previous studies.
Although the RetNet website now lists 281 genes, an increase from the previous 234, the pathogenic genes identified in our study remain within the original 234 and are not among the newly added genes. Therefore, they are comparable to the previously catalogued genes. More importantly, patients in the previous studies of early-onset and late-onset high myopia did not experience RRD, making the comparison between these two groups feasible. Therefore, a comparison between these two groups is also feasible. The frequency of the variants in RetNet genes in patient with high myopia associated with RRD was significantly higher than early-onset high myopia without RRD (18/40 VS 76/325, 3.18E-3) and late-onset high myopia without RRD (18/40 VS 14/195, 2.11E-10). The frequency of the stickler syndrome related genes in patient with high myopia associated with RRD was significantly higher than late-onset high myopia without RRD (6/40 VS 0/195, 11.70E-5). The frequency in high myopia with RRD was also higher than early-onset high myopia without RRD, although no significant difference was observed (6/40 VS 21/325, 6.00E-2).
The average age at the onset of RRD was 47.23 ± 16.28 years old, the mean refractive error was −15.30 ± 4.95 diopters, and the average axial length was 29.26 ± 1.87 mm. The best corrected visual acuity of after the surgery of RRD is 3.01 ± 1.28. The onset age of retinal detachment of the patients with pathogenic known variants (45.47 ± 14.02 years old) is lower than patients with pathogenic unknown variants (49.59 ± 17.46 years old), though no significantly difference is detected (Figure 2).
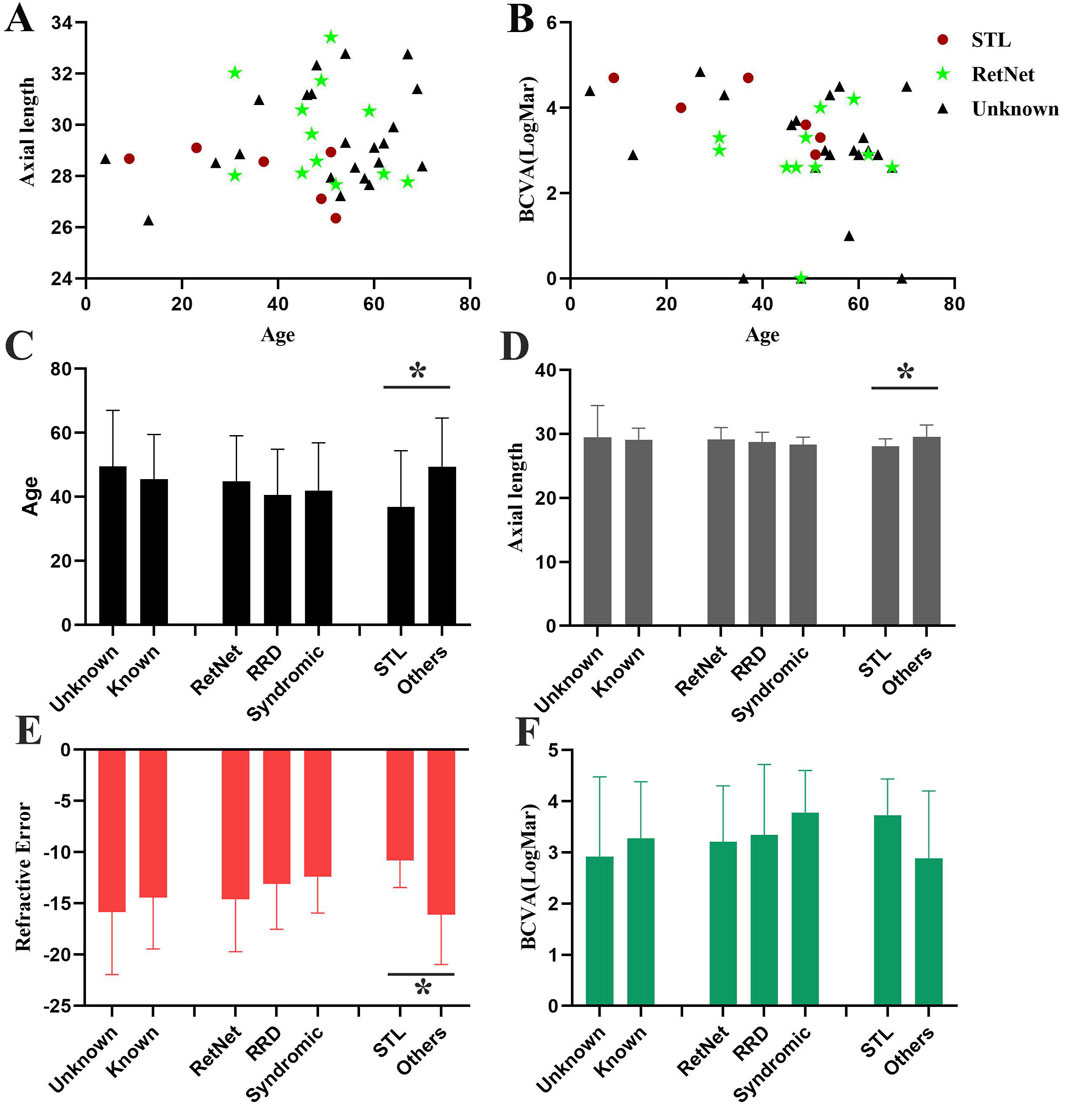
Figure 2. Comparison of phenotypes in patients with high myopia accompanied by RRD. (A) Scatter plot showing the distribution of age and axial length for patients with high myopia accompanied by RRD. (B) Scatter plot displaying the distribution of age and BCVA for patients with high myopia accompanied by RRD. Red circles indicate patients with Stickler syndrome (STL), green stars represent patients carrying RetNet genes other than STL, and black triangles represent patients with no identified pathogenic genes. (C–F). Comparison of age (C), axial length (D), refractive error (E), and BCVA (F) among different patient groups. Three distinct groups are compared: (1) individuals carrying known and unknown gene variants, (2) individuals with pathogenic variants in RetNet-associated genes, RRD-related genes, and syndromic high myopia associated genes, (3) individuals with pathogenic variants in Stickler-related genes and other diseases.
No significant differences were observed in refractive error, axial length, or BCVA after retinal detachment surgery between patients with known and unknown pathogenic variants, or among patients with variants in RetNet genes, RRD-related genes, and syndromic high myopia genes. However, significant difference was found between patients with pathogenic variants in Stickler syndrome-associated genes and other genes. The onset age of the RRD in patients with stickler syndrome (35.17 ± 18.03 years old) was younger than others (49.35 ± 15.26 years old) (p = 4.77E-2). Moreover, the refractive error of the patients with Stickler syndrome is milder (−10.83 ± 2.62 VS −16.09 ± 4.87, p = 1.46E-2) and the axial length is shorter (27.63 ± 1.01 VS 29.54 ± 1.86, p = 1.92E-2) compared to other patients with high myopia and RRD (Figure 2).
Our study enrolled 40 patients with high myopia and concurrent RRD. Pathogenic variants were detected in 47.5% (19/40) of cases across 281 RetNet genes, 178 genes associated with syndromic high myopia, 23 genes linked to non-syndromic high myopia, and 29 genes responsible for RRD.
Variants in RetNet gene were the most common cause of RRD in patients with high myopia, accounting for 45.0% (18/40) of cases. Comparing with our previous study, pathogenic variants in RetNet genes were found in 23.4% of non-syndromic early-onset high myopia patients and 7.2% of non-syndromic late-onset high myopia patients (Zhou et al., 2018). The frequency of pathogenic variants in RetNet genes was significantly higher in patients with high myopia complicated by RRD compared to those with early-onset (18/40 VS 76/325, p = 3.18E-3) or non-syndromic late-onset high myopia (18/40 VS 14/195, p = 2.11E-10) without RRD. Although the RetNet database now includes 281 genes, an increase from the previous 234, the pathogenic variants identified in our study still correspond to the original 234 genes, rather than the newly added ones. Notably, the probands in the previous studies on early-onset and late-onset high myopia did not experience retinal detachment, which supports the validity of comparing these groups. Therefore, these results are directly comparable.
Genes associated with retinal detachment could account for 27.5% (11/40) of patients with high myopia complicated by RRD. This finding is consistent with the well-established association between myopia and an increased risk of RRD. Myopic individuals are more prone to posterior vitreous detachment, retinal tears, lattice degeneration, and retinal thinning (Mitry et al., 2011). These characters may reflect an inherent fragility of the retina, predisposing them to retinal detachment. Therefore, RRD is strongly correlated with high myopia.
Genes associated with syndromic high myopia accounted for 25.0% (10/40) of patients with high myopia complicated by RRD. No variants were detected in genes associated with non-syndromic high myopia. Syndromic high myopia includes high myopia accompanied by either ocular or systemic abnormalities, such as retinitis pigmentosa, congenital night blindness, Stickler syndrome, Marfan syndrome, Weill–Marchesani syndrome, Knobloch syndrome, Cohen syndrome, and Wagner syndrome. Notably, the most common pathogenic genes in high myopia cases with retinal detachment were those related to Stickler syndrome, particularly COL2A1 (10.0%, 4/40) and COL11A1 (5.0%, 2/40). VCAN related to Wagner syndrome accounted for 5.0% (2/40), while RP1 associated with retinitis pigmentosa were found in 2.5% (1/40) of cases.
Genes associated with RRD accounted for 27.5% (11/40) of patients with high myopia complicated by RRD. To date, at least 29 genes have been implicated in retinal detachment (Govers et al., 2023). These include genes related to conditions such as Stickler syndrome, Kniest dysplasia, Marshall syndrome, Knobloch syndrome, Marfan syndrome, among others. The most extensively research has been conducted on genes associated with Stickler Syndrome (Moledina et al., 2022). Stickler Syndrome is a hereditary multisystem connective tissue disorder characterized by a spectrum of phenotypes, all associated with pathogenic variations in genes guiding the production of collagen proteins, specifically type II, XI, and IX. Pathogenic variations affecting these collagen proteins typically result in abnormalities in the eyes, facial features, hearing, and skeletal structure. Stickler Syndrome, primarily caused by variants in COL2A1 and COL11A1, represents the most common form, accounting for approximately 80%–90% of cases (Hoornaert et al., 2010; Soh et al., 2022). Other pathogenic genes include COL9A1, COL9A2, COL9A3, and LOXL3 (Nixon et al., 2022).
Approximately 60% of Stickler syndrome patients eventually develop retinal detachment, often resulting in severe visual impairment and long-term morbidity. Patients with retinal detachment typically require multiple surgical interventions, with a high recurrence rate, leading to an overall poor visual prognosis (Coussa et al., 2019; Ang et al., 2008). In our study, compared to patients with unknown or other pathogenic gene variants, those with Stickler-related gene variants had the youngest average onset age of retinal detachment (35.17 ± 18.03 years old) and shorter axial length (27.63 ± 1.01 mm). Given these severe outcomes, some experts advocate for prophylactic retinal laser treatment in Stickler syndrome patients to reduce or prevent future RRD, although conclusive evidence supporting the absolute benefits of such treatments remains limited (Naravane et al., 2022; Fincham et al., 2014).
In conclusion, our findings indicate that Stickler syndrome, accounting for 15.0% of cases, is a predominant cause of high myopia accompanied by RRD. RetNet genes were implicated in 45.0% of cases in this cohort. As ophthalmologists, we advocate for increased attention to comprehensive systemic and ophthalmic examinations in high myopia patients to facilitate early intervention and reduce the risk of retinal detachment.
The raw sequence data reported in this study have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) within the National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA010850). These data are publicly accessible at the following link: https://ngdc.cncb.ac.cn/gsa-human.
The studies involving humans were approved by the Ethics Committee of West China Hospital, Sichuan University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. This study was conducted in accordance with the principles outlined in the Declaration of Helsinki. The study adhered to the principles of the Declaration of Helsinki and received approval from the Ethics Committee of West China Hospital, Sichuan University. Informed consent was obtained from every participant. All patients received oral and written information and gave their written informed consent prior to collecting the data.
LZ: Writing–review and editing, Conceptualization, Methodology, Project administration; FB: Writing–review and editing, Conceptualization, Methodology; HC: Writing–original draft, Methodology; FJ: Writing–review and editing, Formal Analysis; CZ: Writing–review and editing; JX: Writing–review and editing; HJ: Writing–review and editing; YL: Writing–review and editing, Supervision; ZX: Writing–review and editing, Supervision.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from National Natural Science Foundation of China (82401304). This study was supported by the Sichuan Provincial Science and Technology Department Key R&D Projects (No. 2023YFQ0103) and the Aier Ophthalmology-Sichuan University Scientific Research Fund Project (No. 23JZH039). Clinical Research Incubation Project, West China Hospital, Sichuan University (2021HXFH026).
We used AI-assisted tools for language refinement and editing. The content, analysis, and conclusions presented in this paper remain the sole responsibility of the authors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1485874/full#supplementary-material
Ang, A., Poulson, A. V., Goodburn, S. F., Richards, A. J., Scott, J. D., and Snead, M. P. (2008). Retinal detachment and prophylaxis in type 1 Stickler syndrome. Ophthalmology 115, 164–168. doi:10.1016/j.ophtha.2007.03.059
Chandra, A., Banerjee, P., Davis, D., and Charteris, D. (2015). Ethnic variation in rhegmatogenous retinal detachments. Eye (Lond) 29, 803–807. doi:10.1038/eye.2015.43
Coussa, R. G., Sears, J., and Traboulsi, E. I. (2019). Stickler syndrome: exploring prophylaxis for retinal detachment. Curr. Opin. Ophthalmol. 30, 306–313. doi:10.1097/ICU.0000000000000599
Fincham, G. S., Pasea, L., Carroll, C., McNinch, A. M., Poulson, A. V., Richards, A. J., et al. (2014). Prevention of retinal detachment in Stickler syndrome: the Cambridge prophylactic cryotherapy protocol. Ophthalmology 121, 1588–1597. doi:10.1016/j.ophtha.2014.02.022
Govers, B. M., van Huet, R. A. C., Roosing, S., Keijser, S., Los, L. I., den Hollander, A. I., et al. (2023). The genetics and disease mechanisms of rhegmatogenous retinal detachment. Prog. Retin Eye Res. 97, 101158. doi:10.1016/j.preteyeres.2022.101158
Han, X., Ong, J. S., An, J., Craig, J. E., Gharahkhani, P., Hewitt, A. W., et al. (2020). Association of myopia and intraocular pressure with retinal detachment in European descent participants of the UK Biobank cohort: a mendelian randomization study. JAMA Ophthalmol. 138, 671–678. doi:10.1001/jamaophthalmol.2020.1231
Hoornaert, K. P., Vereecke, I., Dewinter, C., Rosenberg, T., Beemer, F. A., Leroy, J. G., et al. (2010). Stickler syndrome caused by COL2A1 mutations: genotype-phenotype correlation in a series of 100 patients. Eur. J. Hum. Genet. 18, 872–880. doi:10.1038/ejhg.2010.23
Jiang, Y., Yi, Z., Zheng, Y., Ouyang, J., Guo, D., Li, S., et al. (2024). The systemic genotype-phenotype characterization of PAX6-related eye disease in 164 Chinese families. Invest Ophthalmol. Vis. Sci. 65, 46. doi:10.1167/iovs.65.10.46
Johnston, T., Chandra, A., and Hewitt, A. W. (2016). Current understanding of the genetic architecture of rhegmatogenous retinal detachment. Ophthalmic Genet. 37, 121–129. doi:10.3109/13816810.2015.1033557
Kwok, J. M., Yu, C. W., and Christakis, P. G. (2020). Retinal detachment. CMAJ 192, E312. doi:10.1503/cmaj.191337
Li, X., and Beijing, G. (2003). Incidence and epidemiological characteristics of rhegmatogenous retinal detachment in Beijing, China. Ophthalmology 110, 2413–2417. doi:10.1016/s0161-6420(03)00867-4
Mitry, D., Singh, J., Yorston, D., Siddiqui, M. A. R., Wright, A., Fleck, B. W., et al. (2011). The predisposing pathology and clinical characteristics in the Scottish retinal detachment study. Ophthalmology 118, 1429–1434. doi:10.1016/j.ophtha.2010.11.031
Moledina, M., Charteris, D. G., and Chandra, A. (2022). The genetic architecture of non-syndromic rhegmatogenous retinal detachment. Genes (Basel) 13, 1675. doi:10.3390/genes13091675
Morris, R. E., Parma, E. S., Robin, N. H., Sapp, M. R., Oltmanns, M. H., West, M. R., et al. (2021). Stickler syndrome (SS): laser prophylaxis for retinal detachment (modified ora secunda cerclage, OSC/SS). Clin. Ophthalmol. 15, 19–29. doi:10.2147/OPTH.S284441
Mowatt, L., Shun-Shin, G., and Price, N. (2003). Ethnic differences in the demand incidence of retinal detachments in two districts in the West Midlands. Eye (Lond) 17, 63–70. doi:10.1038/sj.eye.6700245
Naravane, A. V., Belin, P. J., Pierce, B., and Quiram, P. A. (2022). Risk and prevention of retinal detachments in patients with stickler syndrome. Ophthalmic Surg. Lasers Imaging Retina 53, 7–11. doi:10.3928/23258160-20211213-02
Nixon, T. R. W., Richards, A. J., Martin, H., Alexander, P., and Snead, M. P. (2022). Autosomal recessive stickler syndrome. Genes (Basel) 13, 1135. doi:10.3390/genes13071135
Ohno-Matsui, K., Wu, P. C., Yamashiro, K., Vutipongsatorn, K., Fang, Y., Cheung, C. M. G., et al. (2021). IMI pathologic myopia. Invest Ophthalmol. Vis. Sci. 62, 5. doi:10.1167/iovs.62.5.5
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and Genomics and the association for molecular pathology. Genet. Med. 17, 405–424. doi:10.1038/gim.2015.30
Schick, T., Heimann, H., and Schaub, F. (2020). Retinal detachment Part 1 - epidemiology, risk factors, clinical characteristics, diagnostic approach. Klin. Monbl Augenheilkd 237, 1479–1491. doi:10.1055/a-1243-1363
Soh, Z., Richards, A. J., McNinch, A., Alexander, P., Martin, H., and Snead, M. P. (2022). Dominant stickler syndrome. Genes (Basel) 13, 1089. doi:10.3390/genes13061089
Wang, Y. M., Lu, S. Y., Zhang, X. J., Chen, L. J., Pang, C. P., and Yam, J. C. (2022). Myopia genetics and heredity. Child. (Basel) 9, 382. doi:10.3390/children9030382
Wong, T. Y., Tielsch, J. M., and Schein, O. D. (1999). Racial difference in the incidence of retinal detachment in Singapore. Arch. Ophthalmol. 117, 379–383. doi:10.1001/archopht.117.3.379
Yi, Z., Li, S., Wang, S., Xiao, X., Sun, W., and Zhang, Q. (2022). Clinical features and genetic spectrum of NMNAT1-associated retinal degeneration. Eye (Lond) 36, 2279–2285. doi:10.1038/s41433-021-01853-y
Keywords: high myopia, rhegmatogenous retinal detachment, RetNet, stickler syndrome, gene ontology, genotype, phenotype
Citation: Zhou L, Boboev F, Chen H, Jiang F, Zhang C, Xiao J, Jiang H, Liao Y and Xu Z (2025) Genetic and clinical profile of high myopia patients with rhegmatogenous retinal detachment. Front. Genet. 16:1485874. doi: 10.3389/fgene.2025.1485874
Received: 25 August 2024; Accepted: 26 February 2025;
Published: 09 April 2025.
Edited by:
Jing Chen, Cincinnati Children’s Hospital Medical Center, United StatesReviewed by:
Li Huang, Sun Yat-sen University, ChinaCopyright © 2025 Zhou, Boboev, Chen, Jiang, Zhang, Xiao, Jiang, Liao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongchuan Liao, eW9uZ2NodWFuMjAwNUAxNjMuY29t; Zhuping Xu, eHV6aHVwaW5nQHNjdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.