- 1Department of Animal Sciences, Purdue University, West Lafayette, IN, United States
- 2Division of Animal Sciences, College of Agriculture, Food and Natural Resources, University of Missouri, Columbia, MO, United States
- 3Smithfield Premium Genetics, Raleigh, NC, United States
- 4Department of Agriculture, Food, Environment and Forestry, University of Florence, Firenze, Italy
- 5Department of Animal Science, North Carolina State University, Raleigh, NC, United States
Climate change poses a growing threat to the livestock industry, impacting animal productivity, animal welfare, and farm management practices. Thus, enhancing livestock climatic resilience (CR) is becoming a key priority in various breeding programs. CR can be defined as the ability of an animal to be minimally affected or rapidly return to euthermia under thermally stressful conditions. The primary study objectives were to perform genome-wide association studies for 12 CR indicators derived from variability in longitudinal vaginal temperature in lactating sows under heat stress conditions. A total of 31 single nucleotide polymorphisms (SNPs) located on nine chromosomes were considered as significantly associated with nine CR indicators based on different thresholds. Among them, only two SNPs were simultaneously identified for different CR indicators, SSC6:16,449,770 bp and SSC7:39,254,889 bp. These results highlighted the polygenic nature of CR indicators with small effects distributed across different chromosomes. Furthermore, we identified 434 positional genes associated with CR. Key candidate genes include SLC3A2, STX5, POLR2G, and GANAB, which were previously related to heat stress responses, protein folding, and cholesterol metabolism. Furthermore, the enriched KEGG pathways and Gene Ontology (GO) terms associated with these candidate genes are linked to stress responses, immune and inflammatory responses, neural system, and DNA damage and repair. The most enriched quantitative trait loci are related to “Meat and Carcass”, followed by “Production”, “Reproduction”, “Health”, and “Exterior (conformation and appearance)” traits. Multiple genomic regions were identified associated with different CR indicators, which reveals that CR is a highly polygenic trait with small effect sizes distributed across the genome. Many heat tolerance or HS related genes in our study, such as HSP90AB1, DMGDH, and HOMER1, have been identified. The complexity of CR encompasses a range of adaptive responses, from behavioral to cellular. These results highlight the possibility of selecting more heat-tolerant individuals based on the identified SNP for CR indicators.
1 Introduction
Climate change poses an increasing risk to productivity, welfare, and requires farm management practices change without compromising effectiveness in the livestock industry (Rhoads et al., 2009; Yang et al., 2021). Moreover, economically important traits are significantly influenced by genotype-by-environment interactions, and the animals’ performance can deteriorate with rising global temperatures (Haile-Mariam et al., 2008; Braz et al., 2021). Over the past few decades, intensive breeding for greater productivity, such as increased milk yield, growth rate, little size, and body weight in livestock, led to higher metabolic heat production (Cabezón et al., 2017), potentially reducing the ability of livestock (i.e., pigs) to thrive in harsh environments. Consequently, this has caused farm management to become more challenging with declining profitability as animals face heightened risks from the severe effects of global warming. Thus, enhancing CR has become a primary objective in livestock breeding.
Climate resilience refers to the animal’s ability to maintain or quickly return to euthermia under thermally stressful conditions (Colditz and Hine, 2016; Wen et al., 2024). Many studies have investigated CR and proposed novel indicators, such as genetic variance of the slope of reaction norm models (Shi et al., 2021; Waters et al., 2022; Freitas et al., 2024). However, most direct resilience phenotypes are difficult or expensive to measure, leading to less frequent measurements and more issues, such as low phenotypic variability and low to moderate heritability estimates (Guy et al., 2012; Gorssen et al., 2021). In previous studies, various phenotypes related to heat stress (HS) in lactating sows—including vaginal temperature, respiration rate, skin surface temperature, hair density, and body condition score—were measured under HS conditions and considered as useful indicators of HS (Scheffer et al., 2018; Gorssen et al., 2021; Johnson et al., 2023). These phenotypes exhibited low to moderate heritability estimates. However, identifying CR of animals based on these measures alone is challenging. Individuals who exhibit greater consistency in their phenotypes over time are likely to have higher resilience (Scheffer et al., 2018; Berghof et al., 2019). This is because they are expected to deviate less from their optimal production or physiological levels when faced with disruptions, leading to increased survival and reduced disease incidence (Scheffer et al., 2018; Berghof et al., 2019).
Increased availability of longitudinal data from various methods, such as automatic thermometers, feeding stations, and computer vision systems, makes it possible to derive more effective resilience indicators (Chen et al., 2023; Pedrosa et al., 2023). For instance, methods for deriving several new resilience indicators in dairy cattle based on deviations from observed and expected performance, including variance, lag-1 autocorrelation, and skewness of deviations, have been proposed (Poppe et al., 2020). These methods have been applied in resilience studies across various species, including cattle (Poppe et al., 2020; Chen et al., 2023), pigs (Mancin et al., 2024), and dairy goats (Sánchez-Molano et al., 2019).
Fifteen novel CR indicators, such as variance, lag-1 autocorrelation, and skewness of deviations, as well as HS duration, using longitudinal automatically-recorded vaginal temperature were developed in our previous study (Wen et al., 2024). Most of these indicators were moderately heritable and had low to high genetic correlations with each other. Current understanding of the biological mechanisms and genetic factors influencing CR in lactating sows is rather limited. In this context, genome-wide association studies (GWAS) enable the detection of single nucleotide polymorphisms (SNP) associated with traits of interest (Visscher et al., 2012). Many GWAS studies focusing on resilience have been conducted in different species, such as chicken (Doekes et al., 2023), pigs (Putz et al., 2019), sheep (Tsartsianidou et al., 2021), and cattle (Alonso-Hearn et al., 2022; Chen et al., 2024). However, CR is expected to be a polygenic trait influenced by numerous biological mechanisms, which could lead to the identification of many putative quantitative trait loci (QTL), some of them with small effect and located on different chromosomes. GWAS can contribute to a better understanding of the genetic basis underlying phenotypic variability in CR. By undertaking GWAS on different CR metrics, we can delve deeper into the genetic basis of this complex trait, potentially uncovering valuable insights that will not only advance our scientific knowledge but also inform breeding strategies aimed at enhancing CR in sows. Thus, the primary study objectives were to 1) detect SNPs and genomic regions significantly associated with twelve CR indicators derived from automatically-recorded vaginal temperature measured in lactating sows under HS conditions; and 2) identify the underlying biological functions and metabolic pathways these regions are involved in based on functional genomic analyses.
2 Materials and methods
2.1 Datasets
All live animal data collection procedures were approved by the Purdue University Animal Care and Use Committee (Protocol #1912001990). All data collection procedures, physiological data, genotype information, and quality control processes have been previously described in our previous studies (Johnson et al., 2023; Wen et al., 2023; Wen et al., 2024). In brief, 1,639 lactating sows (parities 2–7; Landrace × Large White) were genotyped using the PorcineSNP50K Bead Chip (Illumina, San Diego, CA, United States). The vaginal temperature (TV) of 1,381 sows within the studied population was automatically measured every 10 min from June 5th to July 30th, 2021, using a vaginally implanted thermochron data recorder (Johnson et al., 2023). Ambient temperature and humidity of each barn was automatically collected every 5 minutes (Johnson et al., 2023). The phenotypic and genomic quality control procedures performed can be accessed on our research (Johnson et al., 2023; Wen et al., 2023). Twelve CR indicators were derived based on variability in automatically measured vaginal temperature (Wen et al., 2024). Log-transformed variance [LnVar(Ave) and LnVar(Med)], Lag-1 autocorrelation (Autocor (Ave) and Autocor(Med)], and skewness [Skew(Ave) and Skew(Med)] of the deviations between observed and the average (Ave) or median (Med) values from moving windows consisting of six consecutive observations with a 10-minute interval were calculated for each animal (Poppe et al., 2020). The HS thresholds for individuals under distinct ventilation conditions (mechanical ventilation at 39.76°C and natural ventilation at 39.78°C) were reported (Johnson et al., 2023). Additional traits were derived including the daily maximum vaginal temperature (MaxTv) per individual and the HS duration (HSD), which quantifies the duration of the time interval in which an individual’s vaginal temperature consistently exceeded the HS threshold each day. Two CR indicators were normalized median (
2.2 Genome-wide association studies and functional genomic analyses
Genome-wide association studies between the CR indicators and the SNPs were conducted using the linear mixed animal model in the GCTA software (Yang et al., 2011), with the option of leaving one chromosome out (MLMA-LOCO). The effects included in the GWAS models are the same as those reported previously (Wen et al., 2024). After performing the GWAS, the genomic inflation factor (λ) was calculated to evaluate potential bias in the results, e.g., from unaccounted population stratification. The λ value was calculated as the ratio of the median of the observed distribution of the statistic to the expected median, for which a 95% confidence interval of value was further derived (Devlin and Roeder, 1999). The Bonferroni correction was used for multiple testing corrections (Armstrong, 2014). The genome-wide significance and suggestive significance threshold were set as
The GALLO R package (Fonseca et al., 2020) was used to detect genes located within 500 Kb up and downstream of significant SNP and QTL regions previously cited in the pig QTLdb (Hu et al., 2019) based on the latest genome reference Sscrofa 11.1 assembly (http://useast.ensembl.org/Sus_scrofa/Info/Index). Gene Ontology (GO) (Ashburner et al., 2000) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000) enrichment analyses for candidate genes were carried out using the DAVID platform (Huang et al., 2009).
3 Results and discussion
3.1 GWAS results summary
We first conducted GWAS studies for all traits to investigate the genetic basis and biological mechanisms associated with heritable CR indicators [heritability estimates ranging from 0.084 ± 0.037 to 0.291 ± 0.047 (Wen et al., 2024)]. The traits evaluated were LnVar(Ave), Autocor(Ave), Skew(Ave), LnVar(Med), Autocor(Med), Skew(Med), MaxTv, HSD, Nor_avevar, Nor_medvar, HSUA, and HSUB. The genomic inflation factors ranged from 0.95 to 1.2 for all indicators, showing small inflation of P-values for the estimated SNP effects (Price et al., 2010). Lambda values and Q-Q plots for each CR trait are shown in Figure 1.
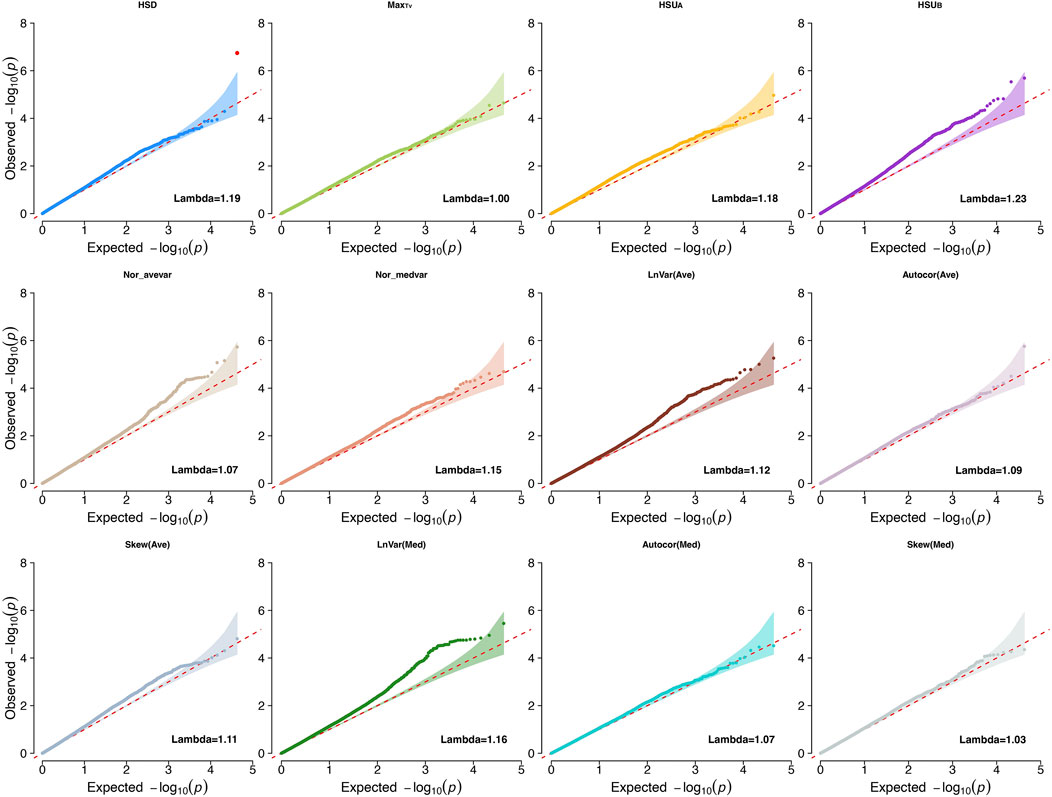
Figure 1. Quantile-quantile plots (QQ-plot) and lambda values for the climatic resilience indicators evaluated1. 1Indicators: LnVar(Ave), log-transformed variance of the deviations between each observation and the average values from moving windows that contains six continuous observations with 10-mins interval in between; Autocor (Ave): Lag-1 autocorrelation of the deviations between the average values from moving windows that contains six continuous observations with 10-mins interval in between; Skew (Ave): skewness of the deviations between each observation and the average values from moving windows that contains six continuous observations with 10-mins interval in between; LnVar(Med): log-transformed variance of the deviations between the median values from moving windows that contains six continuous observations with 10-mins interval in between; Autocor (Med): Lag-1 autocorrelation of the deviations between the median values from moving windows that contains six continuous observations with 10-mins interval in between; Skew (Med): skewness of the deviations between each observation and the median values from moving windows that contains six continuous observations with 10-mins interval in between; Nor_avevar: normalized average TV multiplies the normalized TV variance; Nor_medvar: normalized median TV multiplies the normalized TV variance; HSUA: sum of TV values above the HS threshold during the whole data collection period; HSUB: sum of TV values below the HS threshold during the whole data collection period; HSD: The length of time during which the body temperature remained above the HS threshold value for each collection day; MaxTv: The highest TV of each measurement day.
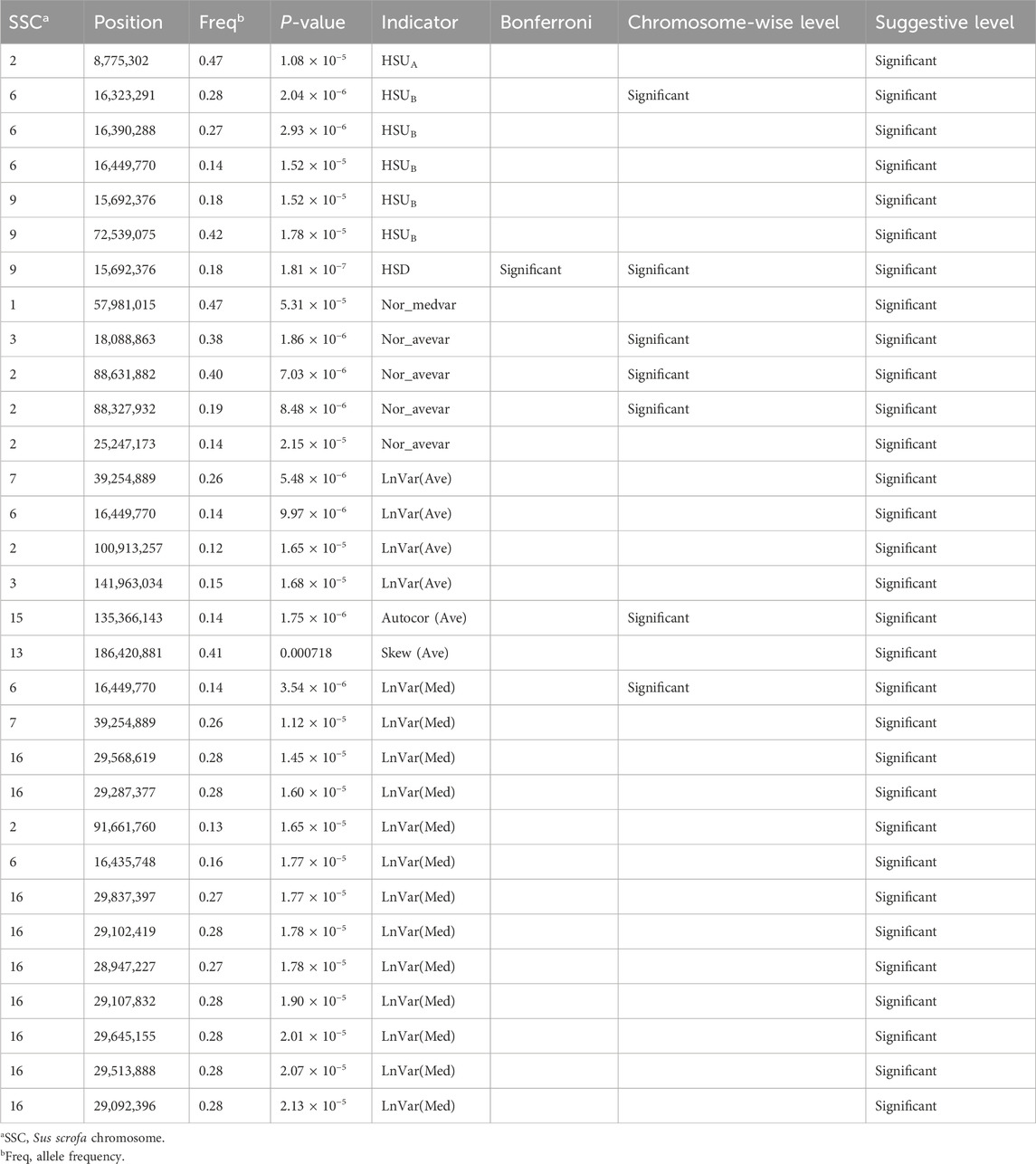
Thirty-one SNPs located on nine Sus scrofa chromosomes (SSC) that reached at least the suggestive significance level were detected for nine CR indicators and presented in Table 1. Four, one, one, 13, one, four, one, five, one SNP at the suggestive threshold was detected for LnVar(Ave), Autocor (Ave), Skew (Ave), LnVar (Med), Nor_medvar, Nor_avevar, HSUA, HSUB, and HSD, respectively. Among these SNPs, one SNP located on SSC15:135,366,143 bp, one SNP on SSC6:16,449,770 bp, one SNP on SSC6:16,323,291 bp, and three SNPs on SSC2:88,327,932 bp and 88,631,882 bp and SSC3:18,088,863 bp, were identified as significant at the chromosome-wise level threshold for four CR indicators: Autocor (Ave), LnVar(Med), HSUB, and Nor_avevar, respectively. Notably, only one SNP (SSC9: 15,692,376 bp) for HSD met the most stringent significance thresholds (Table 1). No significant associations were found for MaxTv, Autocor (Med), and Skew (Med), and this may be lack of power. The small number of suggestive SNPs for each indicator in our study indicates that the evaluated CR indicators are highly polygenic, with many genomic regions of small effects located throughout the different chromosomes. Most HS or heat tolerance related traits in livestock are polygenic (Macciotta et al., 2017; Tiezzi et al., 2020; Cheruiyot et al., 2021), with few major genes identified. Larger sample sizes could be beneficial for identifying these QTLs with smaller effects. These findings are in agreement with our previous research (Freitas et al., 2023; Wen et al., 2023).
Four common SNPs were identified to be associated with more than one CR indicator. The SNPs are located on SSC6:16,449,770 bp and SSC6:16,435,748 bp [HSUB, LnVar(Ave), and LnVar(Med)], SSC7:39,254,889 bp [LnVar(Ave) and LnVar(Med)], and SSC9:15,692,376 (HSUB and HSD). The CR indicators created based on similar metrics are highly correlated at the genetic level, such as LnVar(Ave) and LnVar(Med), or HSUA and HSUB (Wen et al., 2024). Interestingly, the SNPs identified for these correlated indicators are not distributed at similar genomic regions. There are several reasons for that: first, as previously mentioned, these indicators are highly polygenic, and we did not find any major gene control in the CR indicators; Second, the candidate SNPs identified by each indicator are in linkage disequilibrium with their causal variants, explaining why overlapping SNPs are still observed across different indicators.
3.2 Candidate genes and functional genomic analyses
A total of 442 positional genes harboring or adjacent to the significant SNPs were mapped, including 212 protein-coding genes, 225 non-coding RNAs, and 5 pseudogenes (Supplementary Additional File S1: Supplementary Table S1). Specifically, 22, three, one, 30, one, 91, 45, 18, and one protein-coding genes were identified to be associated with LnVar(Ave), Autocor(Ave), Skew(Ave), LnVar(Med), Nor_medvar, Nor_avevar, HSUA, HSUB, and HSD, respectively. For LnVar(Ave), the genomic regions around four significant SNPs (SSC2: 100,913,257 bp; SSC3: 141,963,034 bp; SSC6: 16,449,770 bp; SSC7: 39,254,889 bp) harbors Heat Shock Protein 90 Alpha Family Class B Member 1 (HSP90AB1), Hyperpolarization Activated Cyclic Nucleotide Gated Potassium Channel 1 (HCN1), SPT3 Homolog SAGA And STAGA Complex Component (SUPT3H), and Transmembrane Protein 63B (TMEM63B) genes. HSP90AB1 functions as a chaperone and plays a role in protein transport and degradation. Its expression level decreased significantly in response to HS in pigs (Seibert et al., 2019). Besides, one SNP (SNP g.4338T > C) within HSP90AB1 was found to be significantly related to heat tolerance in Thai indigenous cattle (Charoensook et al., 2012). HCN1, SUPT3H, and TMEM63B were found to be associated with cellular and oxidative stress response in salmon (Beemelmanns et al., 2021), milk production in Holstein cattle (Liu et al., 2021), and residual feed intake in purebred French Large White pigs (Messad et al., 2021), respectively.
One SNP, located at 135,366,143 bp on SSC15, was associated with Autocor (Ave) at chromosome-wise and suggestive significance level. The genomic region around this SNP contains SH3 Domain Binding Protein 4 (SH3BP4) and ArfGAP with GTPase Domain, Ankyrin Repeat and PH Domain 1 (AGAP1). SH3BP4 was identified in a region with copy number variation in South African Nguni cattle, which are recognized for their ability to sustain harsh environmental conditions and resistance to parasites and disease (Wang et al., 2015). There were a few peaks with significant SNPs for LnVar(Med) on SSC2: 91,661,760 bp, SSC6: 16.435–16.449 Mb, SSC7: 39,254,889 bp, and between 28.947 and 29.837 Mb on SSC16. The up and downstream of the significant SNPs covered 15 candidate genes that were enriched for LnVar(Ave) before due to the overlapping SNPs. Strong associations were found on SSC2 and SSC3, with Low-Density Lipoprotein Receptor Class A Domain Containing 3 (LDLRAD3), solute carrier family 1 member 2 (SLC1A2), Dimethylglycine Dehydrogenase (DMGDH), Betaine-Homocysteine S-Methyltransferase 2 (BHMT2), and Homer Scaffold Protein 1 (HOMER1), harboring the most significant SNPs for Nor_avevar. LDLRAD3, known to encode a low-density lipoprotein (LDL) receptor and associated with decreased levels of very low-density lipoprotein receptor, has been linked to HS in chickens (Jastrebski et al., 2017; Wang et al., 2020). Further research in mice has revealed that the very low-density lipoprotein receptor plays a crucial role in regulating thermogenesis in brown adipocytes, suggesting its importance in body temperature regulation (Shin et al., 2022). It has been demonstrated that genes encoding the very low-density lipoprotein receptor are crucial for both lipid metabolism and the response to temperature stress (Álvarez et al., 2020). SLC1A2 was significantly downregulated in the mouse pituitary gland under hot conditions and was related to stress response (Memon et al., 2016). DMGDH was considered a candidate gene for heat tolerance, defined as the rate of decline (slope) in milk, fat, and protein yield in swamp buffalos. Furthermore, DMGDH may be involved in alleviating oxidative stress in heat-stressed cattle (Cheruiyot et al., 2021). BHMT2 is involved in regulating homocysteine metabolism with beneficial effects in heat-stressed animals through its activity against osmotic stress and protection of protein denaturation (Cottrell et al., 2015; Del Vesco et al., 2015). Besides, BHMT2 has been identified as a positively selected candidate gene affecting thermotolerance in African indigenous cattle (Ankole, Ogaden, N'Dama, Boran, and Kenana cattle), using XP-CLR and XP-EHH population statistics (Taye et al., 2017). HOMER1 plays an important role in behavior, particularly concerning adaptation to stress and fear responses (Kamprath et al., 2009). For Skew(Ave) and Nor_medvar, only one protein-coding gene was identified for each trait–ENSSSCG00000042482 and ENSSSCG00000052428, respectively. However, no information regarding their functions was found for these two genes.
The genes Solute Carrier Family 3 Member 2 (SLC3A2), Syntaxin 5 (STX5), RNA Polymerase II Subunit G (POLR2G), and Glucosidase II Alpha Subunit (GANAB) were significantly enriched for HSUA. Previous research observed downregulated SLC3A2 gene expression in bovine mammary epithelial cells under HS conditions (Ma et al., 2018), and this might be an adaptive response to meet increased amino acid requirements during HS (Rhoads et al., 2011). A frameshift mutation in STX5 has been considered a potential causal mutation for cattle’s heat tolerance, and it also significantly impacts milk production (Cheruiyot et al., 2021). Besides, STX5 was linked to tick resistance in Belmont Red cattle (Tabor et al., 2017) and Tunisian indigenous sheep (Ahbara et al., 2022). Tick burdens might correlate with thermal comfort (Rocha et al., 2019), as traits such as skin thickness, hair density, and skin secretions influence both tick resistance and heat regulation (Shyma et al., 2015). GANAB was found to be downregulated in jejunum mucosa of German Holstein cows under HS conditions, and this is related to responses to incorrect protein folding and stabilization processes (Koch et al., 2021). For HSUB, Cytochrome P450 Family 51 Subfamily A Member 1 (CYP51A1), and Cyclin Dependent Kinase 6 (CDK6) were detected in the SSC9. CYP51A1, a gene involved in cholesterol and sterol metabolism, was observed to be upregulated in the plasma of laying hens in response to HS (Zhu et al., 2019). CDK6 was significantly downregulated by HS in duck granulosa cells (Yang et al., 2021).
Only one protein-coding gene (ENSSSCG00000034240) was annotated for the only significant SNP of HSD. The limited overlap between candidate genes identified for various CR indicators in this study and candidate genes from GWAS of resilience for HS is not surprising. First, the traits we used to define resilience, such as HSD and LnVar(Med), differ from those in many other studies (Cheruiyot et al., 2021; Tsartsianidou et al., 2021). Additionally, automatically measured TV enables us to get more accurate CR indicator values. Given the complexity of CR that spans a broad spectrum of adaptive responses, from behavioral to physiological to cellular, it is likely that varying QTLs are captured based on the indicators employed in GWAS studies.
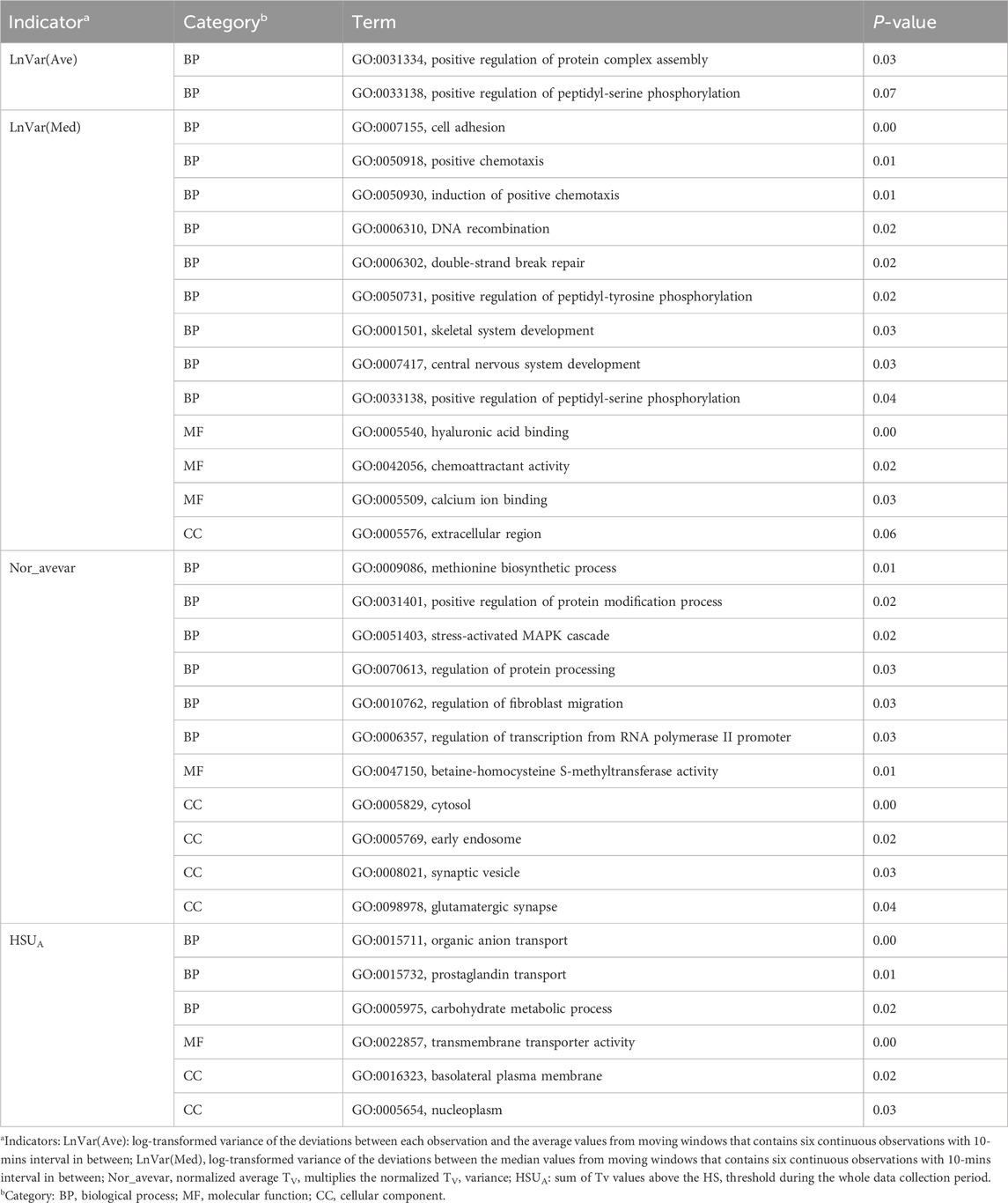
To investigate the biological functions of these candidate genes further, we performed GO and KEGG analysis using DAVID, as shown in Tables 2, 3. Two, one, and one significant KEGG pathways were observed for Lnvar(Med), Lnvar(Ave), and Nor_avevar, respectively. These pathways are related to stress response [e.g., chemical carcinogenesis - receptor activation (Trush and Kensler, 1991)], immune and inflammatory responses [e.g., Th17 cell differentiation (Guo et al., 2018)], cell survival, proliferation, and migration [e.g., PI3K/Akt signaling pathway (Zhang et al., 2016; Yiming et al., 2021)], and nervous system (e.g., Glutamatergic synapse) (Niciu et al., 2012). Heat stress also has been documented to cause a change in animals’ adaptive immune function, transitioning from the typical cell-mediated to humoral immunity (Niciu et al., 2012). This shift can subsequently result in a weakened immune system, making the animal more susceptible to numerous pathogens (Calapre et al., 2013a). Heat stress has been shown to activate heat shock proteins (HSPs), which can promote cell proliferation and survival (Srikanth et al., 2017). Research found that HSPs are overexpressed in various cancers and have been implicated in carcinogenesis (Ciocca and Calderwood, 2005). Heat stress has also been documented to cause a change in animals’ adaptive immune function, transitioning from the typical cell-mediated to humoral immunity (Sophia et al., 2016). This shift can subsequently result in a weakened immune system, making the animal more susceptible to numerous pathogens (Vandana et al., 2019). Additionally, HS can lead to oxidative stress in various livestock, such as dairy cattle (Bernabucci et al., 2002), pigs (Liu et al., 2016), sheep (Chauhan et al., 2016), and poultry (Shakeri et al., 2019). This heightens their vulnerability to numerous pathogens and production-related illnesses.
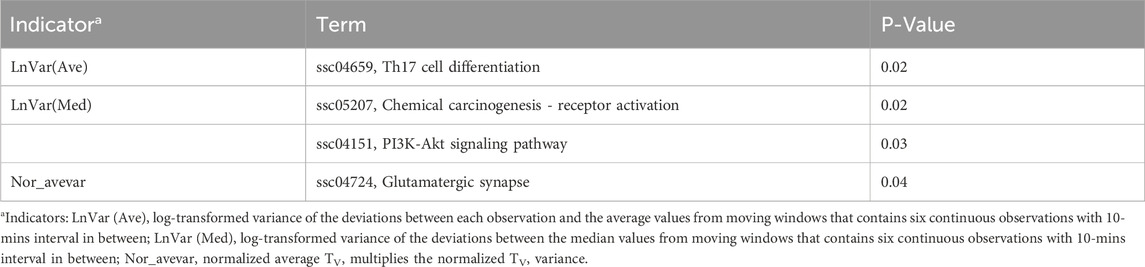
Table 3. Significantly enriched (P < 0.05) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways identified for CR indicators.
In addition, studies have shown that the immune responses in organisms are extremely sensitive to DNA damage that is caused by stressors (Nakad and Schumacher, 2016). The PI3K/AKT signaling pathway is involved in intracellular responses by reactive oxygen species (ROS) and inflammation caused by DNA fragmentation (Datta et al., 2023). Heat stress-induced testicular damage could be alleviated with melatonin, a potent antioxidant, in dairy goats by inhibiting the PI3K/AKT signaling pathway (Liu et al., 2022). Previous research has indicated that thermal stress leads to a reduction in glutamatergic synapse transmission (Popoli et al., 2012). Besides, glutamatergic synapses have been demonstrated to play roles in HSPs synthesis. This synthesis aids in repairing stress-induced synaptic protein damage and bolsters neuroprotective mechanisms (Kiang and Tsokos, 1998). Heat tolerance, defined as the rate of decline in milk production (slope traits) in response to a rising temperature–humidity index, is significantly associated with the enrichment of the glutamatergic synapse pathway in Holstein cows (Cheruiyot et al., 2021).
A total of 13, two, 17, eight, and one significant GO terms were enriched for Lnvar(Med), Lnvar(Ave), Nor_avevar, HSUA, and HSUB, respectively. The functions of enriched GO terms are similar to those of KEGG pathways. These GO terms are related to DNA damage and repair (e.g., DNA recombination, double-strand break repair), stress responses (e.g., stress-activated MAPK cascade), protein modifications (e.g., positive regulation of peptidyl-serine phosphorylation, positive regulation of peptidyl-tyrosine phosphorylation, and positive regulation of protein complex assembly), nervous system (e.g., central nervous system development, glutamatergic synapse, and synaptic vesicle), and cell structure and mechanics. Various types of DNA damage, including the induction of double-strand breaks in DNA (Ning et al., 2021; Habibi et al., 2022), are directly affected by HS. DNA recombination is one mechanism cells use to repair certain types of DNA damage. Previous research showed that the upregulated genes are mainly involved in DNA or protein damage/recombination, cell cycle processes, biogenesis, and stress and immune responses using transcriptome analysis in heat-stressed finishing pigs (Ma et al., 2019).
A substantial number of phosphorylation changes are induced by severe heat stress and occur with kinetics similar to the inhibition of protein synthesis (Duncan and Hershey, 1989). This has been evidenced by the detection of phosphorylation-related GO terms and functions in various species under HS conditions, including buffalos (Muthukumar et al., 2014), broilers (Kim et al., 2022), and swine (Yu et al., 2010; Cross et al., 2018). Notably, phosphorylation is crucial for the transcriptional activity of the heat shock transcription factor 1 and for triggering the heat shock response (Holmberg et al., 2001). Moreover, HS has been shown to activate MAPK phosphorylation in different cell types, such as intestinal cells, lung fibroblasts, and chondrocytes (Liu et al., 2019), and it has also been associated with cell and tissue injury (Banerjee Mustafi et al., 2009).
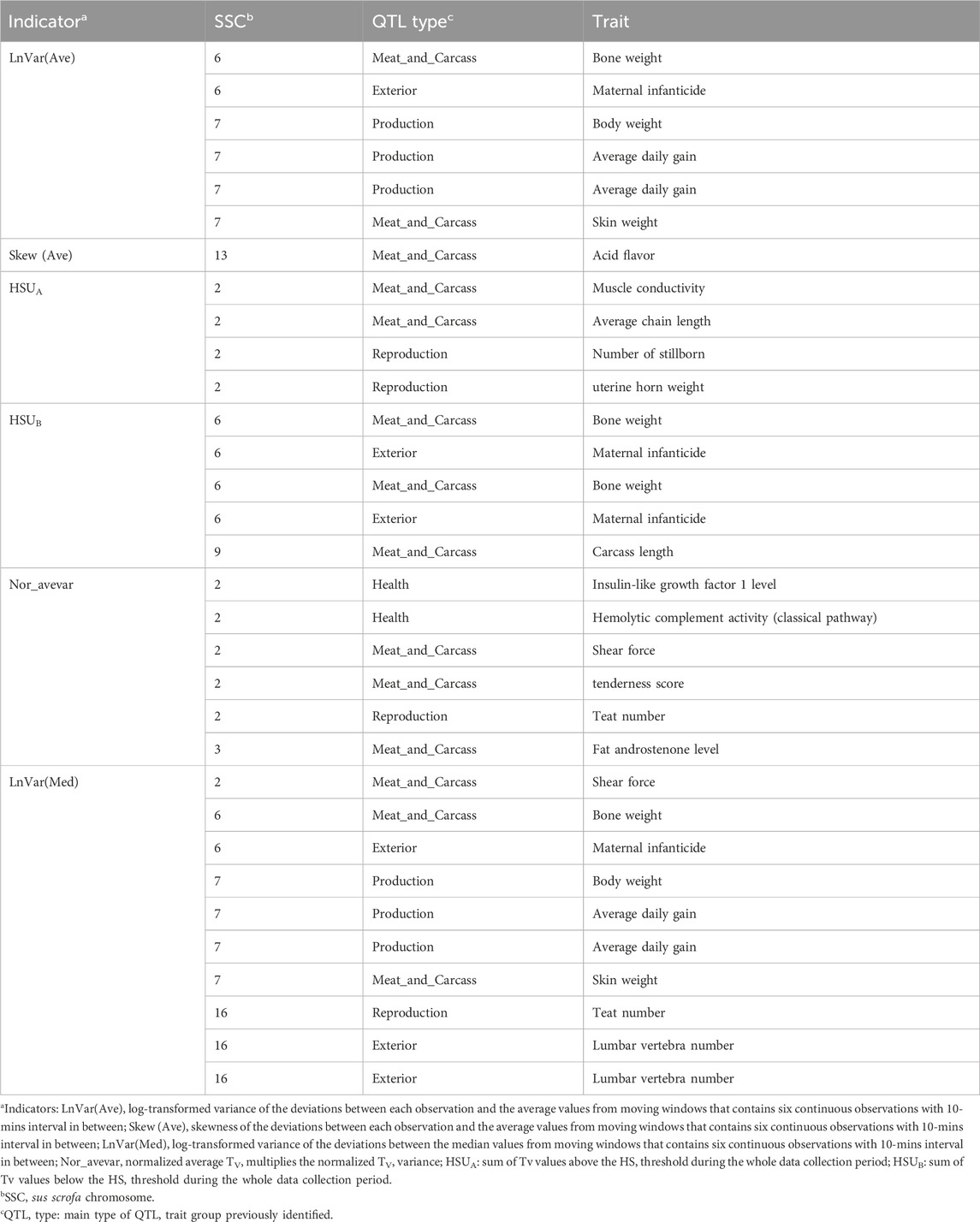
The genomic regions around candidate SNPs are shown to be linked with QTL regions associated with different traits. The major fraction of QTL annotated in this study belonged to the “Meat and Carcass” type, which accounts for 43.75% of the total QTL, and average daily gain and bone weight were the most traits that we enriched for Meat and Carcass QTL. Meanwhile, the genomic regions identified overlapped with several QTLs previously related to production, reproduction, health, and exterior traits (conformation and appearance), as shown in Table 4.
This is the first GWAS for CR indicators derived from automatically measured TV. These significant SNPs hold great potential for enhancing genomic predictions for CR in pigs, by incorporating more SNPs located in the regions of these significant SNPs into existing commercial SNP panels to improve the prediction accuracy. Small sample size may limited the power of analysis, this analysis should be conducted in a larger population. Besides, different weights for these important SNPs or genes could be given through biology-driven genomic prediction methods [e.g., different subsets of SNPs were used for genomic predictions (Li et al., 2018)]. However, even though using different significance thresholds for GWAS, identifying the causal mutations for these CR indicators remains challenging due to the linkage disequilibrium. It would be better to use whole genome sequencing data to fully capture LD patterns, thereby achieving higher GWAS power compared to array-based GWAS (Pengelly et al., 2015). Thus, future research efforts should prioritize additional biological validations. In our study, we used a crossbred population. Similar analyses should be conducted in other populations with different genetic backgrounds to determine if the CR indicators are universally applicable.
4 Conclusion
The study focuses on sows and explores various CR indicators to better understand the genetic factors and biological mechanisms behind climatic resilience. We identified multiple genetic regions associated with different CR indicators, revealing that CR is a highly polygenic trait with small effect sizes distributed across the genome. Furthermore, many heat tolerance or HS related genes in our study, such as HSP90AB1, DMGDH, and HOMER1, have been identified. Additionally, the functional analyses showed the complexity of CR, involving various adaptive responses, from behavioral to cellular. These findings highlight the possibility of selecting more heat-tolerant individuals based on the identified SNP for CR indicators.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: The phenotypic and genomic data used in this study are a property of the industry partner that contributed to the study and therefore are not readily available due to its commercially sensitivity. Requests to access these datasets should be directed to the corresponding author (LB, YnJpdG9sQHB1cmR1ZS5lZHU=).
Ethics statement
The animal study was approved by Purdue University Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HW: Conceptualization, Formal Analysis, Writing–original draft, Writing–review and editing. JJ: Writing–review and editing, Data curation. HM: Supervision, Writing–review and editing. AA: Supervision, Writing–review and editing. FD: Writing–review and editing. AR: Writing–review and editing. YH: Data curation, Resources, Writing–review and editing. FT: Writing–review and editing. CM: Writing–review and editing. AS: Writing–review and editing. LB: Conceptualization, Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Agriculture and Food Research Initiative Competitive Grant number 2022-67021-37022 from the USDA National Institute of Food and Agriculture.
Acknowledgments
The authors acknowledge Caitlin Wager, Sharlene O. Hartman, MaryKate Byrd, Jason R. Graham, Guadalupe Ceja, and Alexis Smith (Purdue University, West Lafayette, IN, United States), Nihya Alston and Dana Cinao (North Carolina State University, Raleigh, NC, United States), John Tyer, Jeremy Howard, Ashley E. DeDecker, Youping Gu, and Laurie Weston (Smithfield Foods, Warsaw, NC, United States) for their contributions to the study planning and data collection. We are also grateful to all the help provided by the Maple Hill farm employees during the data collection period and for the genomic datasets provided by Smithfield Premium Genetics (Raleigh, NC, United States).
Conflict of interest
YH was employed by Smithfield Foods. The remaining authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest. The authors declare that they have no competing interests.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1498380/full#supplementary-material
References
Ahbara, A. M., Khbou, M. K., Rhomdhane, R., Sassi, L., Gharbi, M., Haile, A., et al. (2022). Genome variation in tick infestation and cryptic divergence in Tunisian indigenous sheep. BMC Genomics 23, 167. doi:10.1186/s12864-022-08321-1
Alonso-Hearn, M., Badia-Bringué, G., and Canive, M. (2022). Genome-wide association studies for the identification of cattle susceptible and resilient to paratuberculosis. Front. Vet. Sci. 9, 935133. doi:10.3389/fvets.2022.935133
Álvarez, I., Fernández, I., Traoré, A., Pérez-Pardal, L., Menéndez, N. A., and Goyache, F. (2020). Genomic scan of selective sweeps in Djallonké (West African Dwarf) sheep shed light on adaptation to harsh environments. Sci. Rep. 10, 2824. doi:10.1038/s41598-020-59839-x
Armstrong, R. A. (2014). When to use the Bonferroni correction. Ophthalmic Physiol. Opt. 34, 502–508. doi:10.1111/opo.12131
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi:10.1038/75556
Banerjee Mustafi, S., Chakraborty, P. K., Dey, R. S., and Raha, S. (2009). Heat stress upregulates chaperone heat shock protein 70 and antioxidant manganese superoxide dismutase through reactive oxygen species (ROS), p38MAPK, and Akt. Cell. Stress Chaperones 14, 579–589. doi:10.1007/s12192-009-0109-x
Beemelmanns, A., Zanuzzo, F. S., Xue, X., Sandrelli, R. M., Rise, M. L., and Gamperl, A. K. (2021). The transcriptomic responses of Atlantic salmon (Salmo salar) to high temperature stress alone, and in combination with moderate hypoxia. BMC Genomics 22, 261. doi:10.1186/s12864-021-07464-x
Berghof, T. V. L., Poppe, M., and Mulder, H. A. (2019). Opportunities to improve resilience in animal breeding programs. Front. Genet. 9, 692. doi:10.3389/fgene.2018.00692
Bernabucci, U., Ronchi, B., Lacetera, N., and Nardone, A. (2002). Markers of oxidative status in plasma and erythrocytes of transition dairy cows during hot season. J. Dairy Sci. 85, 2173–2179. doi:10.3168/jds.S0022-0302(02)74296-3
Braz, C. U., Rowan, T. N., Schnabel, R. D., and Decker, J. E. (2021). Genome-wide association analyses identify genotype-by-environment interactions of growth traits in Simmental cattle. Sci. Rep. 11, 13335. doi:10.1038/s41598-021-92455-x
Cabezón, F. A., Schinckel, A. P., Richert, B. T., Peralta, W. A., and Gandarillas, M. (2017). Technical Note: application of models to estimate daily heat production of lactating sows. Prof. Animal Sci. 33, 357–362. doi:10.15232/pas.2016-01583
Calapre, L., Gray, E. S., and Ziman, M. (2013a). Heat stress: a risk factor for skin carcinogenesis. Cancer Lett. 337, 35–40. doi:10.1016/j.canlet.2013.05.039
Charoensook, R., Gatphayak, K., Sharifi, A. R., Chaisongkram, C., Brenig, B., and Knorr, C. (2012). Polymorphisms in the bovine HSP90AB1 gene are associated with heat tolerance in Thai indigenous cattle. Trop. Anim. Health Prod. 44, 921–928. doi:10.1007/s11250-011-9989-8
Chauhan, S. S., Ponnampalam, E. N., Celi, P., Hopkins, D. L., Leury, B. J., and Dunshea, F. R. (2016). High dietary vitamin E and selenium improves feed intake and weight gain of finisher lambs and maintains redox homeostasis under hot conditions. Small Ruminant Res. 137, 17–23. doi:10.1016/j.smallrumres.2016.02.011
Chen, S.-Y., Boerman, J. P., Gloria, L. S., Pedrosa, V. B., Doucette, J., and Brito, L. F. (2023). Genomic-based genetic parameters for resilience across lactations in North American Holstein cattle based on variability in daily milk yield records. J. Dairy Sci. 106, 4133–4146. doi:10.3168/jds.2022-22754
Chen, S.-Y., Gloria, L. S., Pedrosa, V. B., Doucette, J., Boerman, J. P., and Brito, L. F. (2024). Unraveling the genomic background of resilience based on variability in milk yield and milk production levels in North American Holstein cattle through genome-wide association study and Mendelian randomization analyses. J. Dairy Sci. 107, 1035–1053. doi:10.3168/jds.2023-23650
Cheruiyot, E. K., Haile-Mariam, M., Cocks, B. G., MacLeod, I. M., Xiang, R., and Pryce, J. E. (2021). New loci and neuronal pathways for resilience to heat stress in cattle. Sci. Rep. 11, 16619. doi:10.1038/s41598-021-95816-8
Ciocca, D. R., and Calderwood, S. K. (2005). Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell. Stress Chaper 10, 86–103. doi:10.1379/CSC-99r.1
Colditz, I. G., and Hine, B. C. (2016). Resilience in farm animals: biology, management, breeding and implications for animal welfare. Anim. Prod. Sci. 56, 1961. doi:10.1071/AN15297
Cottrell, J. J., Liu, F., Hung, A. T., DiGiacomo, K., Chauhan, S. S., Leury, B. J., et al. (2015). Nutritional strategies to alleviate heat stress in pigs. Anim. Prod. Sci. 55, 1391. doi:10.1071/AN15255
Cross, A. J., Keel, B. N., Brown-Brandl, T. M., Cassady, J. P., and Rohrer, G. A. (2018). Genome-wide association of changes in swine feeding behaviour due to heat stress. Genet. Sel. Evol. 50, 11. doi:10.1186/s12711-018-0382-1
Datta, S., Cano, M., Satyanarayana, G., Liu, T., Wang, L., Wang, J., et al. (2023). Mitophagy initiates retrograde mitochondrial-nuclear signaling to guide retinal pigment cell heterogeneity. Autophagy 19, 966–983. doi:10.1080/15548627.2022.2109286
Del Vesco, A. P., Gasparino, E., Grieser, D. D. O., Zancanela, V., Soares, M. A. M., and De Oliveira Neto, A. R. (2015). Effects of methionine supplementation on the expression of oxidative stress-related genes in acute heat stress-exposed broilers. Br. J. Nutr. 113, 549–559. doi:10.1017/S0007114514003535
Devlin, B., and Roeder, K. (1999). Genomic control for association studies. Biometrics 55, 997–1004. doi:10.1111/j.0006-341X.1999.00997.x
Doekes, H. P., Bovenhuis, H., Berghof, T. V. L., Peeters, K., Visscher, J., and Mulder, H. A. (2023). Research Note: genome-wide association study for natural antibodies and resilience in a purebred layer chicken line. Poult. Sci. 102, 102312. doi:10.1016/j.psj.2022.102312
Duncan, R. F., and Hershey, J. W. (1989). Protein synthesis and protein phosphorylation during heat stress, recovery, and adaptation. J. Cell. Biol. 109, 1467–1481. doi:10.1083/jcb.109.4.1467
Fonseca, P. A. S., Suárez-Vega, A., Marras, G., and Cánovas, Á. (2020). GALLO: an R package for genomic annotation and integration of multiple data sources in livestock for positional candidate loci. GigaScience 9, giaa149. doi:10.1093/gigascience/giaa149
Freitas, P. H. F., Johnson, J. S., Tiezzi, F., Huang, Y., Schinckel, A. P., and Brito, L. F. (2024). Genomic predictions and GWAS for heat tolerance in pigs based on reaction norm models with performance records and data from public weather stations considering alternative temperature thresholds. J. Anim. Breed. Genet. 141, 257–277. doi:10.1111/jbg.12838
Freitas, P. H. F., Johnson, J. S., Wen, H., Maskal, J. M., Tiezzi, F., Maltecca, C., et al. (2023). Genetic parameters for automatically-measured vaginal temperature, respiration efficiency, and other thermotolerance indicators measured on lactating sows under heat stress conditions. Genet. Sel. Evol. 55, 65. doi:10.1186/s12711-023-00842-x
Gorssen, W., Maes, D., Meyermans, R., Depuydt, J., Janssens, S., and Buys, N. (2021). High heritabilities for antibiotic usage show potential to breed for disease resistance in finishing pigs. Antibiotics 10, 829. doi:10.3390/antibiotics10070829
Guo, D., Chen, Y., Wang, S., Yu, L., Shen, Y., Zhong, H., et al. (2018). Exosomes from heat-stressed tumour cells inhibit tumour growth by converting regulatory T cells to Th17 cells via IL-6. Immunology 154, 132–143. doi:10.1111/imm.12874
Guy, S. Z. Y., Thomson, P. C., and Hermesch, S. (2012). Selection of pigs for improved coping with health and environmental challenges: breeding for resistance or tolerance? Front. Gene. 3, 281. doi:10.3389/fgene.2012.00281
Habibi, P., Ostad, S. N., Heydari, A., Aliebrahimi, S., Montazeri, V., Foroushani, A. R., et al. (2022). Effect of heat stress on DNA damage: a systematic literature review. Int. J. Biometeorol. 66, 2147–2158. doi:10.1007/s00484-022-02351-w
Haile-Mariam, M., Carrick, M. J., and Goddard, M. E. (2008). Genotype by environment interaction for fertility, survival, and milk production traits in Australian dairy cattle. J. Dairy Sci. 91, 4840–4853. doi:10.3168/jds.2008-1084
Hall, S. J. G. (2016). Effective population sizes in cattle, sheep, horses, pigs and goats estimated from census and herdbook data. Animal 10, 1778–1785. doi:10.1017/S1751731116000914
Holmberg, C. I., Hietakangas, V., Mikhailov, A., Rantanen, J. O., Kallio, M., Meinander, A., et al. (2001). Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 20, 3800–3810. doi:10.1093/emboj/20.14.3800
Hu, Z.-L., Park, C. A., and Reecy, J. M. (2019). Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 47, D701–D710. doi:10.1093/nar/gky1084
Huang, D. W., Sherman, B. T., Zheng, X., Yang, J., Imamichi, T., Stephens, R., et al. (2009). Extracting biological meaning from Large gene lists with DAVID. CP Bioinforma. 27, Unit 13.11. doi:10.1002/0471250953.bi1311s27
Jastrebski, S. F., Lamont, S. J., and Schmidt, C. J. (2017). Chicken hepatic response to chronic heat stress using integrated transcriptome and metabolome analysis. PLoS ONE 12, e0181900. doi:10.1371/journal.pone.0181900
Johnson, J. S., Wen, H., Freitas, P. H. F., Maskal, J. M., Hartman, S. O., Byrd, M. K., et al. (2023). Evaluating phenotypes associated with heat tolerance and identifying moderate and severe heat stress thresholds in lactating sows housed in mechanically or naturally ventilated barns during the summer under commercial conditions. J. Animal Sci. 101, skad129. doi:10.1093/jas/skad129
Kamprath, K., Plendl, W., Marsicano, G., Deussing, J. M., Wurst, W., Lutz, B., et al. (2009). Endocannabinoids mediate acute fear adaptation via glutamatergic neurons independently of corticotropin-releasing hormone signaling. Genes. Brain Behav. 8, 203–211. doi:10.1111/j.1601-183X.2008.00463.x
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic acids Res. 28, 27–30. doi:10.1093/nar/28.1.27
Kiang, J., and Tsokos, G. C. (1998). Heat shock protein 70 kDa molecular biology, biochemistry, and physiology. Pharmacol. and Ther. 80, 183–201. doi:10.1016/S0163-7258(98)00028-X
Kim, D. Y., Lim, B., Kim, J.-M., and Kil, D. Y. (2022). Integrated transcriptome analysis for the hepatic and jejunal mucosa tissues of broiler chickens raised under heat stress conditions. J. Anim. Sci. Biotechnol. 13, 79. doi:10.1186/s40104-022-00734-y
Koch, F., Albrecht, D., Görs, S., and Kuhla, B. (2021). Jejunal mucosa proteomics unravel metabolic adaptive processes to mild chronic heat stress in dairy cows. Sci. Rep. 11, 12484. doi:10.1038/s41598-021-92053-x
Li, B., Zhang, N., Wang, Y.-G., George, A. W., Reverter, A., and Li, Y. (2018). Genomic prediction of breeding values using a subset of SNPs identified by three machine learning methods. Front. Genet. 9, 237. doi:10.3389/fgene.2018.00237
Li, X., Buitenhuis, A. J., Lund, M. S., Li, C., Sun, D., Zhang, Q., et al. (2015). Joint genome-wide association study for milk fatty acid traits in Chinese and Danish Holstein populations. J. Dairy Sci. 98, 8152–8163. doi:10.3168/jds.2015-9383
Liu, D., Chen, Z., Zhao, W., Guo, L., Sun, H., Zhu, K., et al. (2021). Genome-wide selection signatures detection in Shanghai Holstein cattle population identified genes related to adaption, health and reproduction traits. BMC Genomics 22, 747. doi:10.1186/s12864-021-08042-x
Liu, F., Cottrell, J. J., Furness, J. B., Rivera, L. R., Kelly, F. W., Wijesiriwardana, U., et al. (2016). Selenium and vitamin E together improve intestinal epithelial barrier function and alleviate oxidative stress in heat-stressed pigs: antioxidants protect intestinal barrier integrity in heat-stressed pigs. Exp. Physiol. 101, 801–810. doi:10.1113/EP085746
Liu, Y., Cai, H., Guo, X., Aierken, A., Hua, J., Ma, B., et al. (2022). Melatonin alleviates heat stress-induced testicular damage in dairy goats by inhibiting the PI3K/AKT signaling pathway. Stress Biol. 2, 47. doi:10.1007/s44154-022-00068-9
Liu, Z., Ji, J., Zheng, D., Su, L., and Peng, T. (2019). Calpain-2 protects against heat stress-induced cardiomyocyte apoptosis and heart dysfunction by blocking p38 mitogen-activated protein kinase activation. J. Cell. Physiology 234, 10761–10770. doi:10.1002/jcp.27750
Ma, B., He, X., Lu, Z., Zhang, L., Li, J., Jiang, Y., et al. (2018). Chronic heat stress affects muscle hypertrophy, muscle protein synthesis and uptake of amino acid in broilers via insulin like growth factor-mammalian target of rapamycin signal pathway. Poult. Sci. 97, 4150–4158. doi:10.3382/ps/pey291
Ma, X., Wang, L., Shi, Z., Chen, W., Yang, X., Hu, Y., et al. (2019). Mechanism of continuous high temperature affecting growth performance, meat quality, and muscle biochemical properties of finishing pigs. Genes. Nutr. 14, 23. doi:10.1186/s12263-019-0643-9
Macciotta, N. P. P., Biffani, S., Bernabucci, U., Lacetera, N., Vitali, A., Ajmone-Marsan, P., et al. (2017). Derivation and genome-wide association study of a principal component-based measure of heat tolerance in dairy cattle. J. Dairy Sci. 100, 4683–4697. doi:10.3168/jds.2016-12249
Mancin, E., Maltecca, C., Huang, Y. J., Mantovani, R., and Tiezzi, F. (2024). A first characterization of the microbiota-resilience link in swine. Microbiome 12, 53. doi:10.1186/s40168-024-01771-7
Memon, S. B., Lian, L., Gadahi, J. A., and Genlin, W. (2016). Proteomic response of mouse pituitary gland under heat stress revealed active regulation of stress responsive proteins. J. Therm. Biol. 61, 82–90. doi:10.1016/j.jtherbio.2016.08.010
Messad, F., Louveau, I., Renaudeau, D., Gilbert, H., and Gondret, F. (2021). Analysis of merged whole blood transcriptomic datasets to identify circulating molecular biomarkers of feed efficiency in growing pigs. BMC Genomics 22, 501. doi:10.1186/s12864-021-07843-4
Muthukumar, S., Rajkumar, R., Karthikeyan, K., Liao, C.-C., Singh, D., Akbarsha, M. A., et al. (2014). Buffalo cervico-vaginal fluid proteomics with special reference to estrous cycle: heat shock protein (Hsp)-70 appears to Be an estrus indicator. Biol. Reprod. 90, 97. doi:10.1095/biolreprod.113.113852
Nakad, R., and Schumacher, B. (2016). DNA damage response and immune defense: links and mechanisms. Front. Genet. 7, 147. doi:10.3389/fgene.2016.00147
Niciu, M. J., Kelmendi, B., and Sanacora, G. (2012). Overview of glutamatergic neurotransmission in the nervous system. Pharmacol. Biochem. Behav. 100, 656–664. doi:10.1016/j.pbb.2011.08.008
Ning, Y., Liu, Q., Wang, C., Qin, E., Wu, Z., Wang, M., et al. (2021). Heat stress interferes with formation of double-strand breaks and homolog synapsis. Plant Physiol. 185, 1783–1797. doi:10.1093/plphys/kiab012
Pedrosa, V. B., Boerman, J. P., Gloria, L. S., Chen, S.-Y., Montes, M. E., Doucette, J. S., et al. (2023). Genomic-based genetic parameters for milkability traits derived from automatic milking systems in North American Holstein cattle. J. Dairy Sci. 106, 2613–2629. doi:10.3168/jds.2022-22515
Pengelly, R. J., Tapper, W., Gibson, J., Knut, M., Tearle, R., Collins, A., et al. (2015). Whole genome sequences are required to fully resolve the linkage disequilibrium structure of human populations. BMC Genomics 16, 666. doi:10.1186/s12864-015-1854-0
Popoli, M., Yan, Z., McEwen, B. S., and Sanacora, G. (2012). The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nat. Rev. Neurosci. 13, 22–37. doi:10.1038/nrn3138
Poppe, M., Veerkamp, R. F., Van Pelt, M. L., and Mulder, H. A. (2020). Exploration of variance, autocorrelation, and skewness of deviations from lactation curves as resilience indicators for breeding. J. Dairy Sci. 103, 1667–1684. doi:10.3168/jds.2019-17290
Price, A. L., Zaitlen, N. A., Reich, D., and Patterson, N. (2010). New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11, 459–463. doi:10.1038/nrg2813
Putz, A. M., Harding, J. C. S., Dyck, M. K., Fortin, F., Plastow, G. S., Dekkers, J. C. M., et al. (2019). Novel resilience phenotypes using feed intake data from a natural disease challenge model in wean-to-finish pigs. Front. Genet. 9, 660. doi:10.3389/fgene.2018.00660
Rhoads, M. L., Rhoads, R. P., VanBaale, M. J., Collier, R. J., Sanders, S. R., Weber, W. J., et al. (2009). Effects of heat stress and plane of nutrition on lactating Holstein cows: I. Production, metabolism, and aspects of circulating somatotropin. J. Dairy Sci. 92, 1986–1997. doi:10.3168/jds.2008-1641
Rhoads, R. P., La Noce, A. J., Wheelock, J. B., and Baumgard, L. H. (2011). Short communication: alterations in expression of gluconeogenic genes during heat stress and exogenous bovine somatotropin administration. J. Dairy Sci. 94, 1917–1921. doi:10.3168/jds.2010-3722
Rocha, J. F., Martínez, R., López-Villalobos, N., and Morris, S. T. (2019). Tick burden in Bos taurus cattle and its relationship with heat stress in three agroecological zones in the tropics of Colombia. Parasites Vectors 12, 73. doi:10.1186/s13071-019-3319-9
Sánchez-Molano, E., Kapsona, V. V., Ilska, J. J., Desire, S., Conington, J., Mucha, S., et al. (2019). Genetic analysis of novel phenotypes for farm animal resilience to weather variability. BMC Genet. 20, 84. doi:10.1186/s12863-019-0787-z
Scheffer, M., Bolhuis, J. E., Borsboom, D., Buchman, T. G., Gijzel, S. M. W., Goulson, D., et al. (2018). Quantifying resilience of humans and other animals. Proc. Natl. Acad. Sci. U.S.A. 115, 11883–11890. doi:10.1073/pnas.1810630115
Seibert, J. T., Adur, M. K., Schultz, R. B., Thomas, P. Q., Kiefer, Z. E., Keating, A. F., et al. (2019). Differentiating between the effects of heat stress and lipopolysaccharide on the porcine ovarian heat shock protein response1. J. Animal Sci. 97, 4965–4973. doi:10.1093/jas/skz343
Shakeri, M., Cottrell, J. J., Wilkinson, S., Zhao, W., Le, H. H., McQuade, R., et al. (2019). Dietary betaine improves intestinal barrier function and ameliorates the impact of heat stress in multiple vital organs as measured by evans blue dye in broiler chickens. Animals 10, 38. doi:10.3390/ani10010038
Shi, R., Brito, L. F., Liu, A., Luo, H., Chen, Z., Liu, L., et al. (2021). Genotype-by-environment interaction in Holstein heifer fertility traits using single-step genomic reaction norm models. BMC Genomics 22, 193. doi:10.1186/s12864-021-07496-3
Shin, K. C., Huh, J. Y., Ji, Y., Han, J. S., Han, S. M., Park, J., et al. (2022). VLDL-VLDLR axis facilitates brown fat thermogenesis through replenishment of lipid fuels and PPARβ/δ activation. Cell. Rep. 41, 111806. doi:10.1016/j.celrep.2022.111806
Shyma, K. P., Gupta, J. P., and Singh, V. (2015). Breeding strategies for tick resistance in tropical cattle: a sustainable approach for tick control. J. Parasit. Dis. 39, 1–6. doi:10.1007/s12639-013-0294-5
Sophia, I., Sejian, V., Bagath, M., and Bhatta, R. (2016). Quantitative expression of hepatic toll-like receptors 1–10 mRNA in Osmanabadi goats during different climatic stresses. Small Ruminant Res. 141, 11–16. doi:10.1016/j.smallrumres.2016.06.005
Srikanth, K., Kwon, A., Lee, E., and Chung, H. (2017). Characterization of genes and pathways that respond to heat stress in Holstein calves through transcriptome analysis. Cell. Stress Chaperones 22, 29–42. doi:10.1007/s12192-016-0739-8
Tabor, A. E., Ali, A., Rehman, G., Rocha Garcia, G., Zangirolamo, A. F., Malardo, T., et al. (2017). Cattle tick rhipicephalus microplus-host interface: a review of resistant and susceptible host responses. Front. Cell. Infect. Microbiol. 7, 506. doi:10.3389/fcimb.2017.00506
Taye, M., Lee, W., Caetano-Anolles, K., Dessie, T., Hanotte, O., Mwai, O. A., et al. (2017). Whole genome detection of signature of positive selection in African cattle reveals selection for thermotolerance. Anim. Sci. J. 88, 1889–1901. doi:10.1111/asj.12851
Tiezzi, F., Brito, L. F., Howard, J., Huang, Y. J., Gray, K., Schwab, C., et al. (2020). Genomics of heat tolerance in reproductive performance investigated in four independent maternal lines of pigs. Front. Genet. 11, 629. doi:10.3389/fgene.2020.00629
Trush, M. A., and Kensler, T. W. (1991). An overview of the relationship between oxidative stress and chemical carcinogenesis. Free Radic. Biol. Med. 10, 201–209. doi:10.1016/0891-5849(91)90077-G
Tsartsianidou, V., Sánchez-Molano, E., Kapsona, V. V., Basdagianni, Z., Chatziplis, D., Arsenos, G., et al. (2021). A comprehensive genome-wide scan detects genomic regions related to local adaptation and climate resilience in Mediterranean domestic sheep. Genet. Sel. Evol. 53, 90. doi:10.1186/s12711-021-00682-7
Vandana, G. D., Bagath, M., Sejian, V., Krishnan, G., Beena, V., and Bhatta, R. (2019). Summer season induced heat stress impact on the expression patterns of different toll-like receptor genes in Malabari goats. Biol. Rhythm Res. 50, 466–482. doi:10.1080/09291016.2018.1464638
Visscher, P. M., Brown, M. A., McCarthy, M. I., and Yang, J. (2012). Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24. doi:10.1016/j.ajhg.2011.11.029
Wang, M. D., Dzama, K., Hefer, C. A., and Muchadeyi, F. C. (2015). Genomic population structure and prevalence of copy number variations in South African Nguni cattle. BMC Genomics 16, 894. doi:10.1186/s12864-015-2122-z
Wang, Y., Saelao, P., Kern, C., Jin, S., Gallardo, R. A., Kelly, T., et al. (2020). Liver transcriptome responses to heat stress and newcastle disease virus infection in genetically distinct chicken inbred lines. Genes. 11, 1067. doi:10.3390/genes11091067
Wang, Z., Shen, B., Jiang, J., Li, J., and Ma, L. (2016). Effect of sex, age and genetics on crossover interference in cattle. Sci. Rep. 6, 37698. doi:10.1038/srep37698
Waters, D. L., Clark, S. A., Moghaddar, N., and Van Der Werf, J. H. (2022). Genomic analysis of the slope of the reaction norm for body weight in Australian sheep. Genet. Sel. Evol. 54, 40. doi:10.1186/s12711-022-00734-6
Wen, H., Johnson, J. S., Freitas, P. H. F., Maskal, J. M., Gloria, L. S., Araujo, A. C., et al. (2023). Longitudinal genomic analyses of automatically-recorded vaginal temperature in lactating sows under heat stress conditions based on random regression models. Genet. Sel. Evol. 55, 95. doi:10.1186/s12711-023-00868-1
Wen, H., Johnson, J. S., Gloria, L. S., Araujo, A. C., Maskal, J. M., Hartman, S. O., et al. (2024). Genetic parameters for novel climatic resilience indicators derived from automatically-recorded vaginal temperature in lactating sows under heat stress conditions. Genet. Sel. Evol. 56, 44. doi:10.1186/s12711-024-00908-4
Yang, C., Huang, X., Chen, S., Li, X., Fu, X., Xu, D., et al. (2021). The effect of heat stress on proliferation, synthesis of steroids, and gene expression of duck granulosa cells. Animal Sci. J. 92, e13617. doi:10.1111/asj.13617
Yang, J., Lee, S. H., Goddard, M. E., and Visscher, P. M. (2011). GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. doi:10.1016/j.ajhg.2010.11.011
Yiming, L., Yanfei, H., Hang, Y., Yimei, C., Guangliang, S., and Shu, L. (2021). Cadmium induces apoptosis of pig lymph nodes by regulating the PI3K/AKT/HIF-1α pathway. Toxicology 451, 152694. doi:10.1016/j.tox.2021.152694
Yin, L., Zhang, H., Tang, Z., Xu, J., Yin, D., Zhang, Z., et al. (2021). rMVP: a memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genomics, Proteomics and Bioinforma. 19, 619–628. doi:10.1016/j.gpb.2020.10.007
Yu, J., Yin, P., Liu, F., Cheng, G., Guo, K., Lu, A., et al. (2010). Effect of heat stress on the porcine small intestine: a morphological and gene expression study. Comp. Biochem. Physiology Part A Mol. and Integr. Physiology 156, 119–128. doi:10.1016/j.cbpa.2010.01.008
Zhang, M., Li, N., Qu, X. B., Luo, S., and Drummen, G. P. C. (2016). Total velvet-antler polypeptide extract from Cervus nippon Temminck induces cell proliferation and activation of the PI3K–Akt signalling pathway in human peripheral blood lymphocytes. Anim. Prod. Sci. 56, 1008. doi:10.1071/AN15103
Keywords: climate resilience, heat stress, genome-wide association studies, livestock breeding, genomic regions
Citation: Wen H, Johnson JS, Mulim HA, Araujo AC, De Carvalho FE, Rocha AO, Huang Y, Tiezzi F, Maltecca C, Schinckel AP and Brito LF (2024) Genomic regions and biological mechanisms underlying climatic resilience traits derived from automatically-recorded vaginal temperature in lactating sows under heat stress conditions. Front. Genet. 15:1498380. doi: 10.3389/fgene.2024.1498380
Received: 18 September 2024; Accepted: 28 October 2024;
Published: 07 November 2024.
Edited by:
Maslyn Greene, Clemson University, United StatesReviewed by:
Giustino Gaspa, University of Turin, ItalyBharat Bhushan, Indian Veterinary Research Institute (IVRI), India
Copyright © 2024 Wen, Johnson, Mulim, Araujo, De Carvalho, Rocha, Huang, Tiezzi, Maltecca, Schinckel and Brito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luiz F. Brito, YnJpdG9sQHB1cmR1ZS5lZHU=
 Hui Wen
Hui Wen Jay S. Johnson
Jay S. Johnson Henrique A. Mulim
Henrique A. Mulim Andre C. Araujo
Andre C. Araujo Felipe E. De Carvalho
Felipe E. De Carvalho Artur O. Rocha
Artur O. Rocha Yijian Huang
Yijian Huang Francesco Tiezzi
Francesco Tiezzi Christian Maltecca
Christian Maltecca Allan P. Schinckel
Allan P. Schinckel Luiz F. Brito
Luiz F. Brito