
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 08 January 2025
Sec. Genomics of Plants and the Phytoecosystem
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1492434
This article is part of the Research Topic Genetic Dissection and Improvement of Crop Quality and Stress Adaptation View all 3 articles
 Sameer Pokhrel1
Sameer Pokhrel1 Prasanna Kharel1
Prasanna Kharel1 Swikriti Pandey1
Swikriti Pandey1 Stephanie Botton1
Stephanie Botton1 Gema Takbir Nugraha1
Gema Takbir Nugraha1 Corley Holbrook2
Corley Holbrook2 Peggy Ozias-Akins1*
Peggy Ozias-Akins1*Peanut is a vital source of protein, particularly in the tropical regions of Asian and African countries. About three-quarters of peanut production occurs worldwide in arid and semi-arid regions, making drought an important concern in peanut production. In the US about two-thirds of peanuts are grown in non-irrigated lands, where drought accounts for 50 million USD loss each year. The looming threat of climate change exacerbates this situation by increasing erratic rainfall. Drought not only reduces yield but also degrades product quality. Peanuts under drought stress exhibit higher levels of pre-harvest aflatoxin contamination, a toxic fungal metabolite detrimental to both humans and animals. One way to sustain peanut production in drought-prone regions and address pre-harvest aflatoxin contamination is by developing drought-tolerant peanut cultivars, a process that can be accelerated by understanding the underlying physiological and genetic mechanisms for tolerance to drought stress. Different physiological attributes and genetic regions have been identified in drought-tolerant cultivars that help them cope with drought stress. The advent of precise genetic studies, artificial intelligence, high-throughput phenotyping, bioinformatics, and data science have significantly improved drought studies in peanuts. Yet, breeding peanuts for drought tolerance is often a challenge as it is a complex trait significantly affected by environmental conditions. Besides technological advancements, the success of drought-tolerant cultivar development also relies on the identification of suitable germplasm and the conservation of peanut genetic variation.
Peanut (Arachis hypogaea L.), also known as groundnut, is an important legume and oilseed crop. It is rich in macronutrients with 40%–50% fat, 12%–36% protein, 10%–20% carbohydrate, and micronutrients including calcium, phosphorus, vitamin E, iron, magnesium, and potassium (Rathnakumar et al., 2017). Peanut is a vital protein source in arid and tropical regions of Asian and African countries, where dietary protein predominantly comes from plant sources such as legumes (Singh and Singh, 1992). The top five peanut-producing countries in the world are China, India, Nigeria, the United States, and Senegal with 38%, 13%, 9%, 6%, and 3%, respectively of global peanut production (USDA-FAS, 2024).
About 70 percent of the total peanut production areas include arid and semi-arid regions where drought has a substantial impact (Reddy et al., 2003). Access to water to mitigate drought is limited in most of the peanut-growing parts of the world. As a result, water and heat stress are major limiting factors to peanut growth, pod yield, and seed quality (Aboelill et al., 2012; Hamidou et al., 2012). In fact, drought is the greatest abiotic stress that reduces the yield of peanuts (Boyer, 1982; Araus et al., 2002). With advancing climate change resulting in erratic rainfall and anomalies in global temperature, it is important to understand how plants can adapt to tolerate these changes. Drought, extreme temperatures, salinity, flooding, and altered CO2 concentration are some of the plant abiotic stresses exacerbated in the era of climate change (Onyekachi et al., 2019; Chaudhry and Sidhu, 2022). These environmental aberrations due to climate change influence plant growth and development, their phenology and distribution, and host-pathogen interaction (Coakley et al., 1999; Bertin, 2008; Ghini et al., 2008; Kelly and Goulden, 2008; Raza and Bebber, 2022).
Drought affects plant morphology, physiology, and biochemistry, consequently decreasing plant production (Seleiman et al., 2021). Developing drought-tolerant peanut cultivars can offer a sustainable solution for utilizing scarce water to produce higher yields in drought-prone peanut production areas. It is crucial to dissect the physiological and genetic mechanisms underlying drought tolerance to develop a successful drought-tolerant peanut variety. This review paper discusses the negative effect of drought on peanut production and seed quality and delves into the physiological parameters that enable drought-tolerant peanut cultivars to perform superior to susceptible cultivars in water-stressed conditions. It further explores the genetic basis of drought tolerance, examining the molecular mechanisms by which drought is detected and signaled, and how the interaction of genes, hormones, and several other biochemicals contribute to drought tolerance. The current progress in the utilization of genetic engineering, exploration of the peanut germplasm from different genetic pools, molecular breeding, and technological advancements that successfully developed drought-tolerant cultivars have also been elucidated. Based on the recent achievements, insights have also been drawn on the potential research areas for future peanut genetic improvements.
Drought has a detrimental effect on peanut production resulting in as much as 85% reduction in pod yield (Nageswara Rao et al., 1988). It severely diminishes the uptake of nutrients such as nitrogen, phosphorus, and potassium. Consequently, peanut growth and development are affected, resulting in reduced biomass, pod yield, and quality (Reddy et al., 2003; Junjittakarn et al., 2013; Htoon et al., 2014; Nageswara Rao et al., 1985; Aydinsakir et al., 2016). Along with reduction in pod yield, the seeds produced by water-stressed peanut plants also have poor grades, vigor, and viability (Pallas Jr et al., 1979).
The severity of drought on peanut plants is dependent on the timing, duration, and intensity of drought stress (Hamidou et al., 2012). At different developmental stages, like flowering, peg initiation, and seed maturity, peanut yield was found to vary under drought stress (Nageswara Rao et al., 1988; Hamidou et al., 2013). Reduced irrigation causes the soil to dry up making peg penetration for pod formation difficult. Even when the pegs penetrate the soil, reduced moisture in the root zone may hinder pod formation. In contrast, reduced water supply at the pre-flowering stage followed by adequate watering causes the plants to initiate a flush of flowering which results in increased pod yields by 13%–19% as compared to peanuts with regular irrigation regimes (Nageswara Rao et al., 1985; Nautiyal et al., 1999; Puangbut et al., 2010). However, prolonged drought from flowering initiation to seed filling stage results in up to 50% yield reduction (Stansell and Pallas, 1985). This information could be important in planning water regimes to increase irrigation efficiency and reduce drought stress.
Besides yield, drought stress also negatively impacts the nutritional quality of peanut seeds. The impact is particularly observed in the composition of fatty acids. High oleic acid composition is advantageous not only for enhancing flavor and nutritional value but also for extending shelf-life. The ratio of oleic to linoleic (O/L) in conventional US varieties is 1.5–2.4 (Dwivedi et al., 1996; Davis and Dean, 2016). The impact of drought on the O/L ratio has been conflicting over the years. Some studies report increased O/L ratio at the end-of-season drought in peanut seeds (Dwivedi et al., 1993; Dwivedi et al., 1996; Chaiyadee et al., 2013) while others report decreased O/L ratio (Hashim et al., 1993). The overall oil content can decrease when peanuts encounter mid-season or end-of-season drought (Conkerton et al., 1989; Dwivedi et al., 1996). While the oil content decreases at the end of the season drought, it can result in the increased protein content at the expense of oil content (Dwivedi et al., 1996).
Prolonged periods of drought and heat stress can cause increased aflatoxin contamination in peanut seeds deteriorating their quality (Hamidou et al., 2014). Aflatoxin B1 produced by Aspergillus flavus is highly carcinogenic to both animals and humans, resulting in suppressed immunity, malnutrition, and cancer. As a result, peanuts produced for consumption and commercialization are closely monitored all over the world (Cole et al., 1995; Girdthai et al., 2010; Yu, 2012; Hou et al., 2014; Holbrook et al., 2016). USDA permits no more than 15 ppb (parts per billion) of aflatoxin in any peanut products while the European Union limits it to 4 ppb (Office of the Federal Register, 2004; European Commission, 2024). Elevated temperatures and lowered soil moisture have been linked to an increase in Aspergillus populations in the soil. If the environmental conditions persist, the fungus can infect pods and seeds leaving growers and producers with unusable and potentially aflatoxin-contaminated peanuts. This is more pronounced if there is a moisture deficit during the pod-filling stage (Arunyanark et al., 2009).
Development and utilization of drought tolerant peanut cultivars can help ameliorate aflatoxin contamination. Girdthai et al. (2010) observed that drought-tolerant cultivars had lower Aspergillus colonization and pre-harvest aflatoxin contamination. Holbrook et al. (2000) also reported a similar trend where some genotypes that avoided drought exhibited 92% lower aflatoxin contamination as compared to a check cultivar. Since aflatoxin contamination in peanut genotypes is highly dependent on the genotype by environment interaction, the studies that correlate drought stress and aflatoxin contamination are not always consistent (Hamidou et al., 2014). Drought stress and aflatoxin contamination are closely related and there is a need to dissect genetic mechanisms for drought stress to understand its relation to aflatoxin contamination.
Drought elicits various physiological and Morphological changes in peanuts that constrain its growth and development. The changes are often exhibited as a decrease in water content, cell expansion and division, nutrient absorption, and compromised photosynthetic capability (Hou et al., 2014). In response, peanuts exhibit escape, avoidance, and tolerance strategies to cope with drought stress (Figure 1). Escape refers to strategies such as early flowering, early maturing, and short growth periods where plants complete the life cycle before the onset of stress. Avoidance strategies include structural or physiological adaptations that reduce plants exposure to drought stress, such as thicker waxes or numerous hairs, larger or deeper root systems, efficient use of water for survival, and limited productivity. Tolerance is the ability of plants to endure stress conditions through survival mechanisms such as plant protection gene induction, vascular and anatomical changes, and osmotic adjustment (Bueckert and Clarke, 2013; Dang et al., 2013).
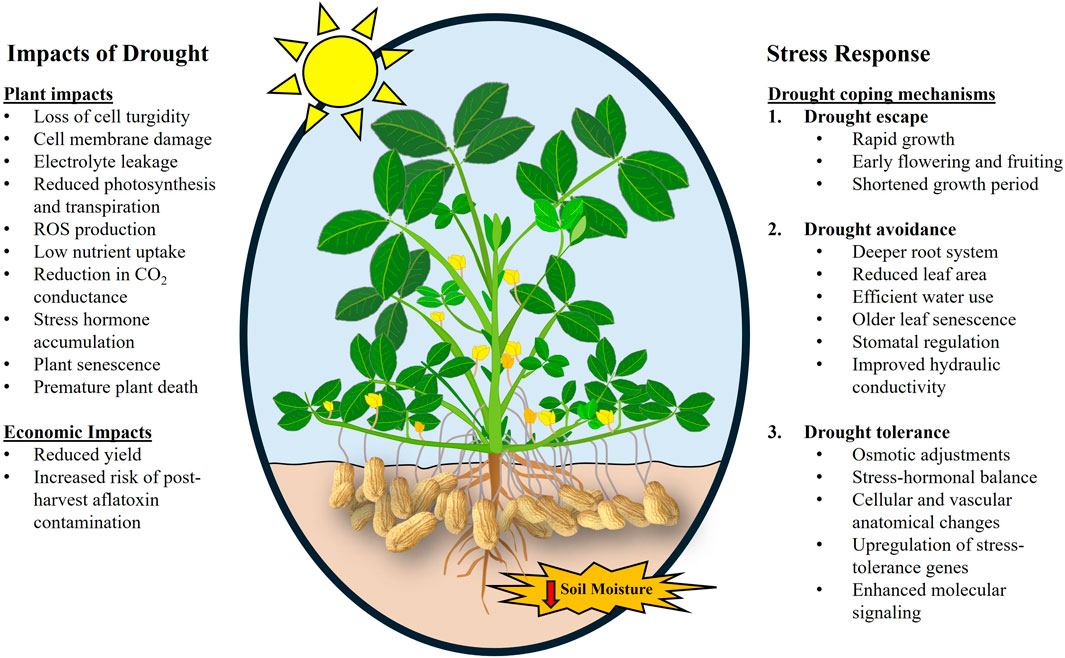
Figure 1. Image showing the impacts of drought in peanuts and tolerance mechanisms of drought-tolerant peanut cultivars. Image modified from https://www.ruralsprout.com/grow-peanuts/.
Root architecture and physiology can play a significant role in coping with drought stress. Peanuts grown under water-deficit conditions have longer and thicker roots, which increases the surface area and enhances their capacity to access moisture in water-stressed conditions (Rowland et al., 2012). Peanuts exhibited a greater root density at 0.4–0.8 m soil depth during water deficit as a strategy to cope with drought (Pandey et al., 1984). Songsri et al. (2008) also observed similar results, where drought-tolerant peanut cultivars had increased root length density (total root length/soil volume) at 40–100 cm deep soil, which helped them avoid drought stress and maintain pod yield. Anatomical dissections of peanut roots under drought conditions showed a decrease in the area of xylem vessels (Thangthong et al., 2018). This trait in drought-tolerant cultivars helps the plant to improve water uptake and reduces the risk of xylem cavitation (bubble formation). As a leguminous crop, peanut roots develop root nodules which help in fixing atmospheric nitrogen into a usable form for plants. Several studies support the fact that roots of drought-tolerant peanut cultivars have a higher nitrogen fixation ability than the susceptible cultivars in a drought-stressed environment (Pimratch et al., 2008; Dinh et al., 2013; Furlan et al., 2017).
Besides roots, leaf morphological and anatomical modifications can also be observed in peanuts tolerant to drought stress. Sankar et al. (2013) reported reduced leaf thickness, epidermis, and number of cells in the palisade and spongy regions when peanuts were subjected to drought stress. Peanut cultivars with greater drought tolerance had greater leaf thickness, higher palisade to spongy tissue ratio, specific leaf weight (dry weight per unit leaf area), and leaf area compared to drought susceptible cultivars (Li et al., 2014).
Drought tolerance in peanuts is also determined by stomatal regulation, where stomatal opening/closing is adjusted to minimize water loss. Stomata are microscopic pores on leaves and stems that allow and regulate the gaseous exchange, influencing photosynthesis and controlling water loss during transpiration (Richardson et al., 2017; Driesen et al., 2020). As plants adjust stomatal aperture to minimize water loss during water stress, such stomatal closure reduces the gaseous exchange and photosynthesis rate, affects plant growth and development, and builds up harmful oxidative molecules (Chaves et al., 2008; Evans, 2013; Gururani et al., 2015; Pirasteh-Anosheh et al., 2016). Soba et al. (2024) observed that the earliest reactions to water stress were reduction in stomatal conductance and photosynthesis. As the drought progressed, a reduction in fluorescence parameters and later changes in biochemical traits impacting carbon fixation were observed. Although stomatal conductance is a common indicator of drought stress in peanuts, the decline in the net photosynthesis is mostly attributed to other metabolic constraints such as disruption of thylakoid processes and limitation in electron transport through photosystem II (Pilon et al., 2018). Drought-tolerant peanut cultivars can cope with water scarcity by closing the stomata faster during water stress (Jiang et al., 2021) and can also maintain photosynthesis efficiency enhancing yield in limited water conditions (Zhen et al., 2022).
Plants accumulate osmolytes which help them in the osmotic adjustments, enable water uptake, maintain cellular homeostasis, and facilitate redox mechanisms by removing excess reactive oxygen species (Ghosh et al., 2021). During drought stress, drought-tolerant peanut cultivars accumulate several osmolytes to significantly higher levels than the drought susceptible cultivars (Celikkol Akcay et al., 2010; Padmavathi and Rao, 2013). Such osmolytes include proline, soluble sugar, free amino acids, and soluble proteins (Zhang et al., 2017; Furlan et al., 2020). Gundaraniya et al. (2020) mentioned that 46 different drought-responsive metabolites including polyamines such as agmatine and cadaverine were present in the roots and leaves of drought-tolerant peanuts. Drought-tolerant peanuts maintain the integrity and stability of cell membranes under drought stress by utilizing antioxidative defenses such as catalase and ascorbate peroxidase along with proline to prevent cellular oxidative damage (Celikkol Akcay et al., 2010). This is important as the cell membrane is one of the primary targets of plant stressors and water stress can damage the cell membrane resulting in the leakage of cellular electrolytes limiting the photosynthesis (Agarie et al., 1995; Lauriano et al., 2000; Bajji et al., 2002).
Physiological traits such as specific leaf area (SLA, ratio of leaf area to dry weight), specific leaf nitrogen (SLN, the ratio of nitrogen content to leaf area), harvest index (HI, the proportion of economic yield in total biomass), water use efficiency (WUE, amount of carbon assimilated as grain/biomass per unit water used), soil plant analysis development (SPAD) chlorophyll meter reading (SCMR, indication of light-transmittance characteristics of the leaf dependent on the leaf chlorophyll content) are indicators of drought resistance/susceptibility in peanuts. Traits such as WUE are difficult to measure on a large scale hence surrogate traits, such as SLA and SCMR, are often used in peanut research. It has been established that SCMR and WUE are correlated, indicating the possibility of using SCMR as a surrogate trait to screen WUE in large populations (Nageswara Rao et al., 2001; Nigam et al., 2005). SCMR was also observed to be strongly correlated to pod yield (Upadhyaya, 2005). Traits such as leaf area index (LAI), SCMR, transpiration efficiency, relative water content (RWC), and canopy temperature (CT) have been used as surrogate traits to study drought tolerance in peanuts (Krishnamurthy et al., 2007; Shaibu et al., 2020). LA is also associated with RWC. Drought-stressed peanut plants have lower RWC (30%) than non-stressed plants (85%–90%) (Reddy et al., 2003; Kambiranda et al., 2011). Nautiyal et al. (2002) observed that drought tolerance was higher in peanut cultivars with low SLA as they maintained higher RWC, suggesting low SLA peanut cultivars are more water-use efficient. Transpiration efficiency (TE) measured using lysimeters in peanuts found that high TE increased pod yield during intermittent drought under dry and hot conditions but was less pronounced under more wet and cool conditions (Vadez and Ratnakumar, 2016). The study demonstrated that TE was not correlated with its commonly used surrogate traits (SCMR and SLA) and hence a direct TE rather than its surrogate evaluation was suggested. The impacts of drought on peanuts have also been illustrated in Figure 1.
The molecular and biochemical basis of drought stress perception and response has not been fully elucidated in peanuts (Bhogireddy et al., 2020; Jiang et al., 2021). Therefore, insights from studies on model plants and other crops need to be applied to understand the mechanism of drought tolerance in peanuts. Broadly, drought tolerance occurs through two main steps: drought stress signal reception and transduction followed by initiation of stress response (Ul-Allah et al., 2021). These responses can occur at physiological, morphological, biochemical, or molecular levels (Mahmood et al., 2019). The physiological and morphological basis for tolerance has been discussed in the previous sections. In this section, the molecular and biochemical basis of stress tolerance in plants is discussed followed by the studies specific to drought tolerance in peanuts. It concludes with the review of biotechnological approaches applied to develop drought-tolerant peanuts.
It is imperative to understand the fundamental mechanisms by which plants sense and respond to drought stress, from initial stress perception to final adaptive responses. This knowledge is crucial for guiding informed efforts to breed drought-tolerant crops, including peanuts (Huang et al., 2012; Shaibu et al., 2020). Significant effort has been invested to elucidate the complex mechanism of drought signaling and response in many plants, yet there are a lot of unknowns to decipher (Takahashi et al., 2020; Yang et al., 2021). Some of the mechanisms for drought sensing and response shown by the plants may be common with other abiotic stresses such as cold, and salt stress (Shinozaki and Yamaguchi-Shinozaki, 2000; Huang et al., 2012). In general, upon stress perception by the cell membrane receptors, many intracellular secondary messengers like Ca+2, and reactive oxygen species (ROS) are released (Shinozaki and Yamaguchi-Shinozaki, 2000; Huang et al., 2012). These secondary messengers, with the help of different protein kinases and protein phosphatases, activate the transcription factors which then induce the expression of certain genes in response to stress (Huang et al., 2012).
The initial signal of water deficiency originates in the roots when the roots perceive decreased water potential in the soil as a dehydration stress (Christmann et al., 2013; Takahashi et al., 2018a). This signal is then transmitted to the leaf tissue via several intercellular messenger signals, including hydraulic pressure, ROS/Ca+2 waves, and peptide signals such as Clavata3/Embryo-Surrounding Region-Related 25 (CLE25) peptide (Takahashi et al., 2018b; Takahashi et al., 2020). Upon recognition of these signals by the receptors in the cell membranes of the leaf cells, intracellular secondary messengers such as ROS and Ca+2 amplify and transmit the signal. This leads to several biochemical, physiological, and molecular changes in the tissue (Lata et al., 2011; Nataraja and Parvathi, 2016; Yang et al., 2021). The most important change is the increase in the level of abscisic acid (ABA) in the cells (Lata et al., 2011; Takahashi et al., 2020; Yang et al., 2021).
Accumulation of ABA is sensed by the stomatal guard cells which induce stomatal closure preventing further loss of water through transpiration (Figure 2). While ABA is the primary hormone regulating stomatal closure, other hormones, including auxin, cytokinin, ethylene, brassinosteroids (BRs), jasmonic acid (JA), and salicylic acid (SA), can act synergistically or antagonistically in this process (Khan et al., 2020; Bandurska, 2022). In addition to their role in stomatal closure, they also affect the morphology and growth patterns of shoots and roots (Chen et al., 2006; Pierik et al., 2007; Zhou et al., 2018; Zhang et al., 2022b). During drought, auxin and cytokinin levels decline in leaves and shoots but accumulate in roots (Havlová et al., 2008; Xu et al., 2016; Abuzeineh and Abualia, 2023). This causes reduction in the shoot growth and an increase in primary root growth by suppressing the lateral root growth. The role ethylene plays in drought response may vary and can be species dependent (Wilkinson and Davies, 2010; Müller, 2021). JA has been shown to act synergistically with ABA to provide drought response while the role of SA and BRs in drought response is still arguable (Zhao et al., 2023; Yoshida and Fernie, 2024). The complex crosstalk between these hormones complicates understanding the role of individual hormones in drought tolerance (Wilkinson et al., 2012).
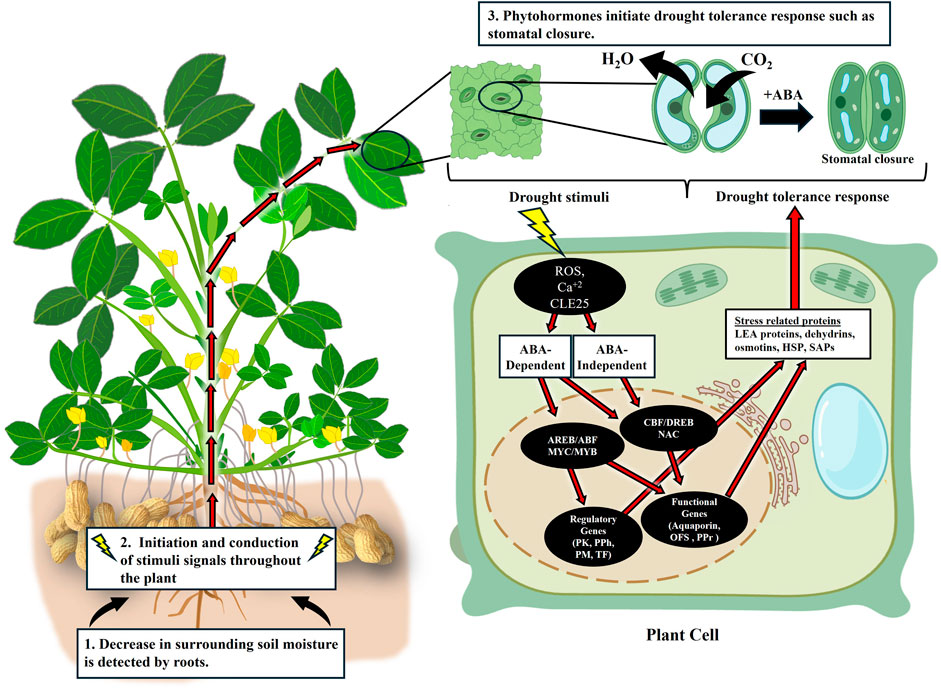
Figure 2. Image showing the molecular mechanism of drought tolerance, from drought perception to activation of response related to drought tolerance. Image modified from Biorender.com and stress related protein listed from Priya et al. (2019). ROS, Reactive Oxygen Species, CLE25, Clavata3/Embryo Surrounding Region-Related 25, AREB/ABF, ABA-Responsive Element-Binding Protein/ABA Binding Factor, MYC/MYB, Myelocytomatosis Oncogene/ Myeloblastosis Oncogene, PK, Protein Kinase genes, PPh, Protein Phosphate genes, PM, Phospholipid Metabolism-related genes, TF, Transcription Factors, OFS, Osmoregulatory Factor Synthase genes, PPr, Protective Protein genes, LEA, Late Embryogenesis-related proteins, HSP, Heat Shock Proteins, SAPs, Stress-Associated Proteins.
Although stomatal closure prevents water loss, it disrupts photosynthesis. This intensifies ROS production in the cells due to photorespiration, electron leakage in the Mehler reaction, and reduction in the Calvin cycle (Helena and Carvalho, 2008). ABA also contributes to ROS accumulation and stimulates the plant’s antioxidant defense system in a positive feedback loop (Jiang and Zhang, 2002; Mittler and Blumwald, 2015). Under normal conditions, the oxidative effect of these ROS is balanced by the enzymatic and non-enzymatic antioxidant defense systems of the plants (Sachdev et al., 2021). The enzymatic defense system includes genes like superoxide dismutase (SOD), catalase (CAT), and ascorbate peroxidase (APX), whereas the non-enzymatic system includes vitamins, carotenoids, and flavonoids (Hasanuzzaman et al., 2020). However, the increased production of ROS caused by drought stress can overwhelm the plants antioxidant capacity leading to oxidative damage of the cells (Miller et al., 2010). To mitigate these effects, expression of multiple drought-responsive genes is crucial.
Upon drought stress signal perception and transduction, plants activate their response to drought through ABA-dependent and ABA-independent pathways (Shinozaki and Yamaguchi-Shinozaki, 2000; Takahashi et al., 2018a) (Figure 2). In the ABA-dependent pathway, ABA directly participates in the expression of the drought stress responsive genes (Soma et al., 2021). The increased level of ABA inside the cells due to drought stress leads to binding of ABA with the ABA receptor, pyrabactin resistance1/pyr1-like/regulatory components of ABA receptor (PYR/PYL/RCAR), forming an ABA-PYR/PYL/RCAR complex. This complex then interacts with protein phosphatase 2C (PP2C). Under normal conditions, PP2C prevents the activation of subclass III sucrose non-fermenting 1-related protein kinases 2 (SnRK2s). Due to the interaction of the ABA-PYR/PYL/RCAR complex and PP2C, SNRK2s are released and become activated either by autophosphorylation or phosphorylation in the presence of other kinases. These activated SNRK2s can phosphorylate transcription factors such as ABA-responsive element-binding protein/ABA binding factor (AREB/ABF) and the myelocytomatosis oncogene/myeloblastosis oncogene (MYC/MYB), which then regulate the expression of drought stress responsive genes (Lata et al., 2011; Soma et al., 2021).
The ABA-independent pathway is activated during the early stages of drought stress before sufficient ABA accumulation occurs to trigger the ABA-dependent pathway (Soma et al., 2021). The osmoreceptors that perceive the drought stress and induce downstream responses are not clear (Soma et al., 2021); however, calcium, JA, and ROS, have been linked to ABA-independent drought sensing and responses in cotton (Mahmood, 2020). This pathway also contributes to the early production of ABA by upregulating the expression of the 9-cis-epoxycarotenoid dioxygenase 3 (NCED3) gene. Unlike the ABA-dependent pathway, which relies on subclass III SnRK2 protein kinases, the ABA-independent pathway employs subclass I SnRK2 protein kinases.
In the initial stages of osmotic stress, B4 Raf-like kinases phosphorylate the subclass I SnRK2s and activate them. In addition, the activity of subclass I SnRK2s is also influenced by stress-induced accumulation of phosphatidic acid (PA), which is regulated by phospholipase Dα (PLDα1) gene. The activated subclass I SnRK2s phosphorylate the mRNA decapping activator VARICOSE (VCS), which degrades mRNA by removing the 5′ cap. This interaction of subclass I SnRK2s and VCS forms the SnRK2-VCS signaling module which positively regulate the expression of stress responsive genes by controlling the post-translational mRNA population (Soma et al., 2021). Even though the ABA-independent pathway is not fully understood, transcription factors like cold-binding factor/dehydration-responsive element binding (CBF/DREB), NAC (NAM, ATAF, and CUC) and ZF-HD (zinc-finger homeodomain) have been associated in the regulation of drought responsive genes in the absence of ABA (Saibo et al., 2009; Lata et al., 2011).
These two pathways do not work mutually exclusive of each other. There exists a crosstalk between ABA-dependent pathways and ABA-independent pathways to coordinate drought stress responses (Figure 2) (Nakashima et al., 2014). For instance, some NAC transcription factors and DREBs have been reported to participate in both ABA-dependent and independent pathways (Lata and Prasad, 2011; Liu S. et al., 2018). This highlights the complex network underlying drought stress signal transduction and expression of drought responsive genes.
The activation of the transcription factors triggers the activation and expression of the stress-responsive genes. The drought stress-responsive genes can be categorized into two categories: functional genes, the products of which are directly engaged in resisting drought stress and providing stress tolerance, and regulatory genes, which indirectly respond to drought stress via signal transduction and regulation of gene expression (Huang et al., 2012; Yang et al., 2021). Functional genes include those encoding aquaporins, osmoregulatory factor synthases, and protective proteins. In contrast, regulatory genes encompass protein kinase genes, protein phosphatase genes, phospholipid metabolism-related genes, and transcription factor genes (Shekoofa and Sinclair, 2018; Mahmood, 2020; Yang et al., 2021).
As mentioned earlier, comprehensive studies that describe the entire process from drought perception by the peanut plant leading up to the responses against the drought stress are still limited. This section details studies conducted on peanuts to identify genes related to drought. Earlier studies on genes associated with drought tolerance in peanuts focused on a few genes that were already discovered in model crops. Later, with advancements in sequencing technologies, several transcriptomic and metabolomic studies have been conducted on peanuts.
One of the earliest studies to identify genes contributing to drought tolerance in peanuts was the characterization of the phospholipase D (PLD) gene (Guo et al., 2006). PLD gene encodes phospholipase D enzyme associated with lipid degradation and drought stress signal transduction (Katagiri et al., 2001; Wang et al., 2014). Guo et al. (2006) associated the role of PLD enzyme in drought sensitivity and tolerance in peanuts which was further confirmed by other studies (Dramé et al., 2007; Zhang et al., 2023). An early increase in PLD accumulation was detected in susceptible peanut cultivars leading to membrane degradation, while a higher accumulation of protective proteins like “late embryogenesis abundant” (LEA) was observed in drought-tolerant cultivars (Guo et al., 2006; Dramé et al., 2007). Two highly expressed genes, serine-rich protein (AhSrp) and leucine-rich protein (AhLrp), were discovered during water stress in drought-tolerant peanut cultivars (Devaiah et al., 2007). These genes encode proteins containing a signal peptide which likely show their participation in drought stress signaling.
An extensive study on the preferential expression of drought-induced genes under gradual water stress in peanuts established the involvement of several upstream signaling components, such as calmodulins, G protein, and receptor kinases, during drought stress (Govind et al., 2009). These signaling components are the core regulatory genes linked with drought signal transduction in peanuts. They also reported induction of several regulatory transcription factors such as NAC, basic helix-loop-helix (bHLH), APETALA 2/ethylene-responsive element binding factor (AP2/ERF), basic leucine zipper (bZIP), CCAAT box, Homeobox, Jumonji, and several zinc finger protein genes under water stress. These transcription factors are components of the ABA-dependent and ABA-independent pathways as mentioned in the earlier section. The presence of ABA-dependent and ABA-independent pathways for drought tolerance in peanuts was further confirmed through a transcriptome analysis on peanut seedlings under water deficit conditions in the presence or absence of ABA pretreatment (Li et al., 2014). Subsequent research showed upregulation in the expression of genes (like AhNCED1, AhZEP, AhBG12, AhBG24, AhAAO2, AhABA3) linked to ABA production, and genes (such as AhABCG22.1 and AhABCG22.2) linked to ABA transport under drought conditions (Long et al., 2019). These studies provide evidence for the presence of ABA-dependent and ABA-independent pathways for drought tolerance in peanuts like other plants as mentioned earlier.
Several studies in peanuts have confirmed the role of transcription factors in regulation of drought responsive genes. The role of AP2/ERF in drought response in peanuts was confirmed through the study involving drought stress and drought release conditions (Dang et al., 2012). Yuan et al. (2020) studied the NAC transcription factor gene family in cultivated and diploid wild peanuts and characterized 164 NAC proteins in cultivated peanuts. During drought stress, 39 of those genes were upregulated, and 13 genes were downregulated. Zhao et al. (2020) evaluated the WRKY gene family in peanuts under drought treatments and reported 158 AhWRKY genes out of which 73 were differentially expressed. These studies demonstrate the involvement of genes and their interactions contributing to the complexity of molecular dissection of drought tolerance in peanuts.
In addition to the transcription factors, genes encoding functional proteins contribute to drought tolerance in peanuts. Genes for heat shock proteins (HSPs), DnaJ-like proteins, aldehyde reductase, proline amino peptidase, choline kinase, defensins, and so on, have been described to play a role in drought tolerance in peanuts (Govind et al., 2009; Pruthvi et al., 2013). Enhanced expression of proline biosynthesis genes (P5CS1, P5CS2a, P5CS2b, P5CR) and relatively lower expression of proline catabolism genes (ProDH1, ProDH2) has been linked to drought tolerance in peanut cultivars under drought stress conditions (Furlan et al., 2020). Hou et al. (2014) also identified functional genes involved in drought response, including those related to stress and hormone signaling, membrane transport, and photosystems.
Protein expression analysis in peanut leaf tissue under drought stress showed five major classes of proteins: signal transduction proteins (e.g., calcium ion binding protein), molecular chaperones (e.g., LEA-1 and HSP protein), photosynthetic proteins (e.g., photosystem I (PSI) proteins), defense proteins (e.g., lectins), and detoxification proteins (e.g., APX-1) (Thangella et al., 2018). This shows that a wide range of drought responsive proteins are necessary to provide drought tolerance in peanuts. A study on the metabolomic profiles of two peanut genotypes with varying drought tolerance, conducted under simulated drought conditions using polyethylene glycol-6000 (PEG-6000), revealed an increase in sugars, organic acids, alcohols, and fatty acids in response to drought stress (Gundaraniya et al., 2020). These compounds include mannose, pentitol, cinnamic acid, caffeic acid, salicylic acid, and fatty acid desaturase enzymes, which provides evidence that a broad range of organic compounds are associated with drought tolerance mechanisms in peanuts. A transcriptome analysis of two peanut cultivars with contrasting drought tolerance traits under fully irrigated and water deficit conditions showed more than 4,500 differentially expressed genes associated with 104 pathways. These pathways were involved in the biosynthesis of plant antibiotics, amino acids, and flavonoids or metabolism of sugars and amino acids (Bhogireddy et al., 2020). The same study revealed that differences in the levels of ABA and expression of sucrose metabolic pathway genes determined their drought tolerance phenotypes. Clearly a large number of genes involved in multiple pathways work in synergy to provide tolerance to drought in peanuts.
In addition to genes and their products, microRNAs (miRNAs), small non-coding RNAs typically 20–24 nucleotides in length, play a role in enhancing drought tolerance in many crops including peanuts (Ferdous et al., 2015). The miRNAs can lead to translation inhibition/mRNA degradation of the target genes resulting in enhanced stress tolerance (Singh et al., 2023). In a bioinformatic evaluation, 33 conserved and 33 novel candidate miRNA families associated with drought and heat stresses were identified in peanuts (Mittal et al., 2023). Ren et al. (2021) identified 73 miRNAs differentially expressed in drought-tolerant peanut variety NH5 under drought stress, of which, novel miR_73 and novel miR_416 were identified as key miRNAs linked to drought adaptation. Functional analysis of miRNAs and their target genes under drought identified miR2111 and miR482 families that targeted genes like lipid transfer protein (LTP) and NAC transcription factor, involved in drought tolerance in peanuts (Zhang et al., 2017). Root tissues were isolated from ‘C76-16’, a drought-tolerant peanut cultivar, to study the mRNA and miRNA expression, which identified 350 differentially expressed miRNAs under drought and revealed that miRNA target genes were mostly linked to redox reactions, modification of cell wall, and transcription regulation during drought stress and drought adaptation (Kumar, 2017).
Above studies indicate the presence of biochemical and molecular pathways involved in drought tolerance in peanuts. While not comprehensive, they have contributed to the partial understanding of molecular basis of drought tolerance in peanuts. Similar to other plants, Ca+2 ions, ROS, and different kinases are involved in drought signal perception and transduction in peanuts (Bhogireddy et al., 2020). Once the drought signal is perceived, the ABA-dependent and ABA-independent pathways are triggered (Li et al., 2014). This activates numerous transcription factors which enhance the peanut’s response to drought by regulating the expression of several functional and regulatory genes. Further studies are required to unravel the complete sequence of events, from initial drought perception to the development of drought tolerance, to fully understand the molecular mechanism of drought tolerance in peanuts.
Most of the drought-related transgenic studies in peanuts employ Agrobacterium-mediated transformation and have utilized genes from many organisms to study their response in peanuts (Krishna et al., 2015; Mallikarjuna et al., 2016). These studies have mostly focused on overexpressing genes. Despite the prospect of CRISPR/Cas9[clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)] for peanut genetic improvement, there are no CRISPR/Cas9 genome editing studies on peanuts published for drought tolerance till date (Puppala et al., 2023).
The first transgenic peanut line for drought tolerance was developed by Bhatnagar-Mathur et al. (2007) when transcription factor DREB1 was introduced from Arabidopsis thaliana under the control of stress-inducible rd29A promoter into a drought sensitive peanut cultivar (Sun et al., 2013). These transgenic peanut lines exhibited increased transpiration efficiency under water deficit conditions indicating that they were more drought tolerant compared to the wild types. Such increased transpiration efficiency and better performance was due to enhanced root growth and their ability to extract water from deeper soil depths in water deficit conditions (Vadez et al., 2007; Vadez et al., 2013). Further evaluation of these lines in field conditions for 4 years confirmed that these transgenic lines consistently yielded better under drought stress compared to wild types (Bhatnagar-Mathur et al., 2014). In a different study, DREB1A transgenic peanuts, produced similarly as Bhatnagar-Mathur et al. (2007), were characterized (Sarkar et al., 2014). These lines were reported to exhibit higher osmotic potential, proline content, leaf chlorophyll content, photosynthesis ability, and shoot biomass compared to wild type, which led to enhanced performance under drought stress. Another study on peanut lines overexpressing DREB1A demonstrated higher transpiration efficiency, increased antioxidant enzymes, and higher proline synthesis, which collectively contributed to better drought tolerance in transgenic lines than wild type lines (Bhalani et al., 2019).
Multiple studies have overexpressed either single or multiple regulatory genes to improve the drought tolerance capacity in transgenic peanut lines. Overexpressing NAC transcription factor (MuNAC4) from horse gram (Macrotyloma uniflorum) in peanut produced transgenic lines with better root system, improved biochemical and phsiological parameters, and increased antioxidant capacity contributing to enhanced drought tolerance (Pandurangaiah et al., 2014). Higher yield under water deficit conditions was also observed in transgenic peanut lines expressing the NAC2 gene from A. thaliana (Bhalani et al., 2019). Transgenic peanuts developed by expressing homeodomain-leucine zipper transcription factor (HDG11) from A. thaliana showed greater yield under drought and salt stress through upregulation of proline, antioxidant enzymes, stress-responsive genes, and enhanced physio-morphological traits (Banavath et al., 2018). Similar findings were noted when WRKY transcription factor from horse gram, MuWRKY3, was overexpressed in peanut (Kiranmai et al., 2018). Introduction of GmMYB3a, a MYB repressor protein from soybean (Glycine max L.) produced transgenic peanuts with efficient photosynthesis, higher WUE and RWC, indicating better adaptation in a water deficit condition (He et al., 2020). Overexpressing peanut bHLH transcription factor (AhHLH112) resulted in transgenic peanut lines with increase in antioxidant enzyme activity, ABA, and ABA-related genes and simultaneous reduction in ROS accumulation, MDA content, and water loss under deficit water conditions (Li et al., 2021).
Patil et al. (2014) overexpressed stress-responsive DNA helicase 45 (PDH45), a transcription activator from pea (Pisum sativum), controlled by 35 S promoter from the Cauliflower Mosaic Virus (CaMV35S) in a peanut genotype with superior water relation traits to pyramid drought tolerance traits. This led to the development of transgenic lines with improved water use efficiency, chlorophyll stability, better growth rate, rooting, and greater productivity compared to the wild type. Expression of eukaryotic translational initiation Factor 4A (eIF4A) gene from pearl millet (Pennisetum glaucum) using rd29A promoter in peanut produced transgenic lines with a better shoot and root growth, augmented ROS scavenging ability, and increased membrane stability in both drought and salt stress (Tiwari et al., 2015; Rao et al., 2017). Overexpression of the abscisic acid stress ripening-1 (ASR-1) gene from a halophyte, Salicornia brachiate, produced transgenic peanut lines with increased concentrations of beneficial metabolites such as sugar, proline, and starch under water stress. These lines also showed a reduction in the accumulation of malondialdehyde (MDA) and ROS, increasing drought tolerance (Tiwari et al., 2015). Overexpression of the KCS1 gene, a critical gene for epicuticular wax biosynthesis, from drought tolerant peanut cultivar into drought sensitive peanut cultivar under the control of CaMV35S promoter increased the deposition of wax on the leaves of transgenic lines preventing water loss (Lokesh et al., 2019). The transgenic lines also displayed increased proline, alcohols, fatty acids, aldehydes, alkanes, and ketones content and reduced oxidative damage and MDA content improving their drought tolerance.
Multiple transgenes have been stacked together to improve drought tolerance in peanuts. Pruthvi et al. (2014) simultaneously expressed three transcription factors, AtDREB2A, AtHB7, and AtABF3 that regulate downstream stress-related genes, from A. thaliana under CaMV35S promoter. These transgenic plants accumulated greater biomass than the wild type along with the production of higher amount of proline, expression of cellular tolerance genes, and their superior ability to scavenge ROS arising from stress. Similarly, co-expression of three regulatory genes related to drought tolerance traits: alfalfa zinc finger 1 (Alfin1), a transcription factor related to root growth under 35 S promoter, P. glaucum heat-shock factor (PgHSF4) under stress-inducible rd29A, and pea DNA helicase (PDH45) under 2 × 35SCaMV promoter produced transgenic peanut lines with better growth, elevated RWC, and enhanced expression of stress responsive genes indicating greater drought tolerance in transgenic lines than the wild-type peanut lines (Ramu et al., 2016). In another multigene transgenic study, three stress responsive transcription factors from horse gram were concurrently expressed: MuWRKY3, related to defense against ROS under CaMV2x35S promoter, MuNAC4, related to drought tolerance under ubiquitin promoter, and MuMYB96, related to cuticular wax synthesis under ribulose-1,5-bisphosphate carboxylase small subunit (rbcS) promoter (Venkatesh et al., 2022). These multigene transgenic plants displayed increased tolerance to drought than wild types through increased wax layer accumulation, better ROS scavenging, and greater shoot and root growth.
Apart from the overexpression of regulatory genes, transgenic approaches have been utilized to enhance drought tolerance through increase in solute concentration in the plant vacuoles. Arabidopsis vacuolar H + -pyrophosphatase gene (AVP1) associated with pumping protons on the vacuolar membrane was overexpressed in peanut under 35 S promoter (Qin et al., 2013). The resulting transgenic peanuts outperformed their wild type counterparts in photosynthetic rate, biomass production, and yield under water deficit conditions arising from drought or salt stress. Overexpressing vacuolar Na+/H+ antiporter gene (AtNHX1) under the control of 35 S promoter in peanut not only improved salt tolerance through build-up of salts and proline in leaf tissue but also the plants performed better upon drought stress (Asif et al., 2011). Overexpression of AhRabG3f gene, associated with lysosomes, under CaMV35s promoter in peanut enhanced their tolerance to drought stress through the expression of multiple stress-responsive genes (Song et al., 2012).
In addition to the plant genes, genes from bacteria have also been introduced in peanuts to enhance their response to drought stress. Qin et al. (2011) introduced isopentenyltransferase (IPT) gene, related to cytokinin synthesis, from Agrobacterium tumefaciens under the control of water-stress inducible senescence-associated receptor protein kinase (SARK) promoter from bean (Phaseolus vulgaris). They reported that transgenic peanut lines were significantly more drought tolerant than the wild types under drought stress in a greenhouse, growth chamber, and in the field conditions as manifested by their larger roots, higher transpiration rate, photosynthetic rate, stomatal conductance, and greater yield. The ability of Escherichia coli to convert fructose to mannitol, an important compound to mitigate abiotic stress, has been utilized by introducing mannitol-1-phosphate dehydrogenase (MtlD) gene into peanut (Bhauso et al., 2014). Transgenic peanuts expressing mtlD gene had better accumulation of compounds like mannitol and proline, higher chlorophyll content, RWC, and osmotic potential and reduced cell membrane leakage under salt and water stress condition. Patel et al. (2017) characterized the transgenic peanut lines developed by Bhauso et al. (2014) under water deficit conditions. They reported that the transgenic lines accumulated higher amounts of mannitol and suffered lower oxidative injuries, showed better photosynthetic performance, and yielded more than wild types. In summary, overexpression of multiple genes in peanuts has developed transgenic peanut lines with improved drought tolerance. However, no transgenic peanut lines have been released (Mallikarjuna et al., 2016). Bio-safety assessment, public perception and acceptance, and governmental policies will determine the release and successful adoption of transgenic and genome edited drought tolerant peanut lines (Singh et al., 2024).
The cultivated peanut is an allotetraploid derived from the natural hybridization of its diploid progenitors, A. duranensis (A-subgenome) and A. ipaensis (B-subgenome). A. hypogaea has a strong linkage disequilibrium (LD) and limited genetic diversity (Bertioli et al., 2020). Following their domestication, cultivated peanuts spread worldwide, where intensive selection over generations led to narrow genetic variation. For instance, the Florunner cultivar, which can be traced to four founder lines, dominated the US market for two decades after its release in 1969 and was extensively used in many breeding programs (Norden et al., 1969). After the appearance of tomato spotted wilt virus (TSWV) in the southeastern US, a plant introduction (PI) line introduced from Brazil, PI 203396, contributed at least 25% of its genetics to most modern peanut cultivars (Clevenger et al., 2017). One way of mitigating this narrow genetic diversity in peanuts is by expanding variation through introgression from wild relatives possessing beneficial alleles.
The availability of germplasm collections, genomic resources, bioinformatics tools, and advanced technologies such as high throughput phenotyping facilitate characterization of diversity and breeding for desirable peanuts. Genome sequences of the cultivated peanuts, the A-genome and B-genome progenitors, and some other wild relatives have significantly contributed to peanut breeding programs (Bertioli et al., 2016; Bertioli et al., 2019; Bertioli et al., 2021). These resources are crucial for peanut breeders worldwide to evaluate breeding populations and to identify beneficial germplasm (Leal-Bertioli et al., 2009; Stalker, 2017; Dutra et al., 2018).
Wild relatives offer a diverse genetic pool that can be explored to improve drought tolerance in cultivated peanuts. Several genetic and transcriptomic studies have reported that the A-genome donor of cultivated peanuts (A. duranensis) exhibits tolerance to drought stress (Guimarães et al., 2012; Brasileiro et al., 2015; Thoppurathu et al., 2022). Leal-Bertioli et al. (2012) investigated key traits related to drought adaptation in the cultivated peanut progenitors, A. duranensis and A. ipaensis, and their derived synthetic allotetraploid. Both morphological (e.g., leaf thickness, trichome density) and physiological (e.g., transpiration profiles under water deficit conditions) traits at both ploidies were evaluated suggesting that morphological and physiological changes may occur upon hybridization and polyploidization. Cason et al. (2020) identified A. dardani as a potential donor for drought tolerance traits and analyzed differentially expressed transcription factors that may enhance the plant’s ability to withstand water deficit conditions.
Another wild peanut species, A. stenosperma, was reported to be a potential donor for drought tolerance, as it possesses a candidate endochitinase-encoding gene (AsECHI), that enhanced post-drought recovery in transgenic Arabidopsis (Mota et al., 2021). However, a comparative transcriptome study between A. stenosperma and A. duranensis revealed that A. duranensis had a better drought tolerance than A. stenosperma (Vinson et al., 2018). The expression profiling of two wild peanut species A. duranensis and A. magna focused on transpiration rates and gene expression levels in roots and leaves under progressive water deficit conditions and identified genes such as Expansin, Nitrilase, NAC, and bZIP transcription factors differentially expressed during the drought stress (Brasileiro et al., 2015). While offering promising alleles for drought tolerance, wild peanuts are mostly diploid and often associated with linkage drag, complicating their introduction into breeding programs.
Germplasm screening for drought tolerance-related traits has been conducted by several researchers to identify accessions that can be used as sources of drought tolerance (Rucker et al., 1995; Upadhyaya, 2005; Bennett et al., 2022). In a series of greenhouse and field trials conducted by Rucker et al. (1995), two genotypes, Tifton 8 and PI 315628 were identified as high-yielding genotypes under water-limited conditions, exhibiting drought avoidance traits such as low visual stress rating, low canopy temperature, and large root systems. Highly drought-tolerant genotypes were also identified based on their drought susceptibility index (DSI), which included ICGV 91114, K 1375, and ICGV 02125 (Pavithradevi et al., 2015). Recently, a multi-parent advanced generation inter-cross (MAGIC) population was created for studying and developing drought tolerance peanuts. Derived from 8 parents, including ICGV 02022, ICG 7190, ICGV 97183, ICG 3053, ICG 14482, ICG 11515, TAG 24 and ICGV 0266, this population generated some drought tolerance advance lines that can serve as valuable resource for peanut breeding programs (Sharma et al., 2024). Sinclair et al. (2018) studied Virginia-type peanut germplasm to identify the genotype that can reduce its transpiration rate earlier when limited water is available in the soil, consequently conserving moisture at an early stage of drought. They found that genotype N12006ol had a high threshold for fraction of transpirable soil water (total available soil water/total transpirable soil water), delayed wilting, and higher yield than the control genotype “Bailey” under a limited water environment due to an early decrease in transpiration rate.
A genotype GP-NC WS 17 was found to be drought-tolerant, which maintained healthy leaves with low epidermal conductance and high SPAD chlorophyll content, SLA, and unsaturated/saturated fatty acid content under severe water deficit. Another genotype C431-1-1 demonstrated drought tolerance with high thermotolerance and maintained overall pigment concentrations under early drought stress (Tallury et al., 2014; Fabreti et al., 2018). Cheek (2024) also confirmed that GP-NC WS 17 and C431-1-1 possess both drought tolerance and reduced aflatoxin contamination. Cultivated peanut germplasms tolerant to drought stress from different countries, including the United States of America, China, and India, are summarized in Puppala et al. (2023). With the advancement in genomic studies, several genomic regions associated with drought tolerance have been identified and molecular markers have been developed to facilitate the selection process.
Drought tolerance in peanuts is a quantitative trait resulting from the cumulative effects of multiple genes and interaction with the environment (Ravi et al., 2011). Advances of genotyping and sequencing technologies have enabled the discovery of QTLs and markers associated with drought tolerance in peanuts leading to marker-assisted selection (MAS). Such markers enable breeders to select drought-tolerance traits at early stages, accelerating the breeding cycle, and increasing the likelihood of developing successful drought-resistant cultivars. This approach has been adopted and has benefited several peanut breeding programs worldwide. A simple sequence repeat (SSR)-based linkage map for cultivated peanuts was first developed in 2009 using a RIL mapping population derived from ICGV 86031 × TAG 24 (Varshney et al., 2009). It was utilized to discover 2-5 QTLs each for surrogate traits for drought tolerance including transpiration, transpiration efficiency, SLA and SCMR, explaining 3.5%–14.1% of phenotypic variation. Axiom_Arachis SNP array version 1 (58 K) and version 2 (48 K) were developed in 2017 and 2018 using different cultivated peanut lines, their progenitors, and wild diploid species (Clevenger et al., 2017; Pandey et al., 2017; Clevenger et al., 2018; Korani et al., 2019). Genome-wide QTL analysis based on the Axiom_Arachis 58 K array identified 16 major main effect QTLs pertaining to drought tolerance (Pandey et al., 2021). Several candidate genes encoding transcription factors like MYB, NAM, MADS-box, bHLH, and other signaling and stress response-related genes were identified in these QTL regions. Fonceka et al. (2012) investigated a synthetic allotetraploid peanut population under well-watered and water-limiting conditions and discovered 13 significant QTLs for stress tolerance indices like total biomass, seed, haulm, and pod weight. Their study revealed that several QTLs were contributed by the amphidiploid parent highlighting the importance of using wild relatives for improving drought tolerance in breeding of cultivated peanuts.
In a genome-wide association study (GWAS) of 300 peanut genotypes, 68 marker-trait associations (MTAs) linked to traits such as leaf area, leaf dry weight, SPAD chlorophyll meter readings (SCMR), harvest index, haulm weight, and seed weight were found under limited water conditions, with phenotypic variation explained (PVE) ranging from 8.24% to 90.09% (Pandey et al., 2014). Shaibu et al. (2020) identified markers associated with traits including LAI, canopy temperature, SCMR, and NDVI, with marker related to SCMR explained the highest phenotypic variation of 20.8% while markers for other traits explained 6%–10% phenotypic variation. Abady et al. (2024) conducted a GWAS study on peanut populations in water-stressed and well-watered conditions. This led to the identification of 48 markers of which 47 were detected under drought and under non-stressed conditions. A single nucleotide polymorphism (SNP) was identified for each trait including leaf area, LAI, SLA, number of primary branches and 43 SNPs for relative water content in leaves. These SNPs were located within or near candidate genes includes Araip.9NG64 (RNA-binding protein), Aradu. PKW10 (CD2 antigen cytoplasmic tail-binding-like protein), Araip57P4D (Chitinase), Araip.4J8RL (Polynucleotide phosphatase/kinase), Araip. SVH5H (ABC transporter protein), Aradu. VIU0I (Zinc finger MYM-type 1-like protein), and Aradu. ML3P3 (P-type ATPase), all of which are implicated in abiotic stress response and drought tolerance (Abady et al., 2024). Additionally, a GWAS conducted on a peanut MAGIC population developed for drought tolerance discovered 37 MTAs, most of which are located on chromosomes 03, 07, 10 and 18 (Sharma et al., 2024). Candidate genes in these regions were found to be associated with leaf senescence, flowering, chlorophyll biosynthesis, stomatal regulation, and yield-related traits.
Ravi et al. (2011) identified 105 and 65 main effect QTLs related to drought tolerance in peanuts by QTL Cartographer and QTLNetwork with PVE ranging between 1.3% and 33.36%, indicating that drought tolerance in peanuts is influenced by multiple small effect QTLs. This study evaluated a wide range of traits related to drought tolerance, including transpiration, transpiration efficiency, SLA, leaf area, SCMR, carbon isotope discrimination ratio, biomass, canopy conductance, total dry matter, dry weight, pod weight, seed weight, and haulm weight. Gautami et al. (2012) used three RIL populations and identified 153 main effect QTL and 25 epistatic QTLs. These QTLs were associated with important drought-related traits, such as transpiration efficiency, transpiration, total dry weight, shoot dry weight, SCMR, HI, and vegetative weight per plant (VegWt/pL) but none of them were major QTLs (Varshney et al., 2009; Ravi et al., 2011). Faye et al. (2015) identified 52 QTLs with low effects, each explaining less than 12% of the phenotypic variance for nine yield-related traits and SPAD chlorophyll meter readings. As multiple studies found small effect QTLs for drought tolerance in peanuts, genomic selection has been proposed as an alternative strategy to marker-assisted backcrossing and marker-assisted recurrent selection (MARS) to breed drought-tolerant peanut varieties (Ravi et al., 2011; Faye et al., 2015).
High-throughput phenotyping (HTP) technologies have become more accessible and started being adopted in peanut breeding over the past few years. Although still under developmental phase, HTP platforms hold potential for accelerating the breeding process by enabling rapid and precise trait assessment (Balota and Oakes, 2017; Sarkar et al., 2021; Balota et al., 2024). Conventional phenotyping methods in peanut breeding involve manual measurement using specialized devices to evaluate plant responses to drought stress. Manual assessment methods commonly used in peanuts include visual ratings, specific leaf area, specific dry weight, total dry matter content, pod yield, chlorophyll fluorescence, SPAD chlorophyll meter reading, normalized difference vegetation index, canopy temperature, relative water content, photosynthesis rate, stomatal conductance, and so on (Upadhyaya, 2005; Sullivan and Holbrook, 2007; Luis et al., 2016; Buezo et al., 2019; Ghosh et al., 2022; Zhang et al., 2022a). These methods are laborious, time inefficient, allow only a limited number of genotypes assessment, and are prone to inconsistency (Baslam et al., 2020; Kim, 2020; Jangra et al., 2021). Automated data collection can reduce labor costs and increase the throughput of phenotypic evaluations, enabling breeders to screen larger populations more efficiently. Puppala et al. (2023) summarized recent HTP technologies that have been adopted for phenotyping drought-associated traits in peanuts, including the utilization of sensors such as RGB (red, green, blue), multispectral, hyperspectral, thermal, LiDAR (light detection and ranging) and GPR (ground penetrating radar). Unmanned aerial vehicles (UAV) with RGB, multispectral, and hyperspectral sensors can estimate various drought-tolerance-related traits in peanuts including leaf wilting rating, leaf area index, and biomass once correlations with ground-truth data are established (Balota and Oakes, 2017; Sarkar et al., 2020; Sarkar et al., 2021). Other platforms including pushcart, minirhizotron, handheld, and UGV (unmanned ground vehicle) with various sensors that can measure pod yield, root architecture, transpiration, and plant height have also been implemented in peanut (Balota and Oakes, 2017; Yuan et al., 2019; Xu et al., 2020; Bidese Puhl et al., 2021; Dobreva et al., 2021). Combining UAV-based hyperspectral imaging and machine learning, a model was created that can predict different traits efficiently under drought stress (Bagherian et al., 2023). Balota et al. (2024) investigated 21 peanut accessions from the US peanut mini-core germplasm collection to identify performance in rainfed and limited water conditions using drone and manual measurements. The correlation between manual and aerial evaluation ranged from 0.02 to 0.94, where traits such as CO2 assimilation, stomatal conductance and transpiration rates had a higher correlation between manual and aerial analysis. High throughput phenotyping holds immense potential for drought evaluation. As technology advances, HTP can be more reliable than conventional phenotyping methods, offering more efficient and precise measurement.
Drought is a polygenic and complex trait. The complexity of studying drought stress is further exacerbated in field-based research due to the differences in soil type, soil moisture, weather conditions, and rainfall. As a result, a variety released as a drought-tolerant variety in one location may not perform well in another due to strong genotype by environment interaction. This strong environmental impact also complicates the genetic dissection of drought tolerance.
Phenotyping drought stress in peanuts presents significant challenges. Various methods have been implemented to evaluate drought-related stress responses, including the measurements of leaf water potential, stomatal conductance, relative water content, chlorophyll content, hydraulic conductance, and investigating root growth parameters. While these traits are vital to understanding the physiological response to drought, they often require significant labor and time to measure.
Tools to measure such physiological traits are anticipated to become more user-friendly, precise, and less labor demanding. The advent of UAV imaging and field sensors has significantly improved the efficiency drought tolerance studies (Bagherian, 2022; Balota et al., 2024). The use of aerial sensing will allow plant breeders to perform more efficient artificial selection from large populations. Soil moisture content and temperature play vital roles in evaluating drought stress. Sensor-based irrigation systems, remote sensing, and geographic information systems (GIS) will be common implements for drought studies in the future.
One of the major challenges in drought studies is to evaluate below-ground root growth. Imaging tools such as minirhizotron system (Vienna Scientific Instruments), ground penetrating radar (GPR) (Liu X. et al., 2018), and magnetic resonance imaging (MRI) (van Dusschoten et al., 2016), combined with artificial intelligence (AI) may offer promising solutions for conducting peanut root studies under drought conditions. Another complexity in peanut root-system study is that the economic yield component (pod) also lies below ground. Peanuts bloom above ground but when flowers are fertilized, a peg is formed, which starts growing underground and forms fruit below the soil surface. In studying peanut yield or root growth using non-destructive methods, it is vital to distinguish between roots and pods and some advancement has already been made (Gimode et al., 2023).
With sequencing technologies becoming more precise and the costs decreasing exponentially, marker-assisted selection is expected to become widely utilized in peanut breeding programs. It will be more common to observe the amalgamation of genetics and data science as sequencing becomes routine work for drought studies. These advancements will facilitate predictive breeding and genomic selection to develop drought-tolerant peanut cultivars. Simulated drought studies, machine learning, AI, and modeling of environmental, genomic and phenomics data will be commonly integrated with drought studies.
The availability of extensive phenotypic data and error-free sequencing technology enables precise discovery of genomic regions conferring drought tolerance in peanuts. Such discoveries could be facilitated by the construction of several pangenomes as reference genomes in peanuts. A pangenome can represent a complete set of genes present in a species and can capture structural variants and genetic diversity better than a single reference approach (Danilevicz et al., 2020).
Although not for drought studies, CRISPR/Cas9 gene editing has been successfully deployed in peanuts (Li et al., 2023; Conner et al., 2024). As we precisely identify genetic loci, CRISPR technology could potentially edit genes associated with drought stress. Advancements in the use of such multi-omics and gene editing technologies will expand the opportunities to exploit variation to improve drought tolerance.
Genetic variation is a key to breeding successful cultivars in any crop. Cultivated peanuts have a very narrow genetic base and a strong linkage disequilibrium which challenges breeding for complex traits like drought tolerance. Exploration and conservation of wild and local landraces of peanuts is critical. Several approaches to broaden the peanut genetic base that include MAGIC, nested association mapping (NAM), mutant line development, and wild introgression will continue to have their space in future peanut genetic improvement.
SaP: Conceptualization, Writing–original draft. PK: Writing–original draft. SwP: Writing–original draft. SB: Writing–original draft. GN: Writing–original draft. CH: Conceptualization, Funding acquisition, Writing–review and editing. PO-A: Conceptualization, Funding acquisition, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. We acknowledge funding support from the Peanut Research Foundation, Georgia Peanut Commission, National Peanut 704 Board, and USDA-NIFA-AFRI awards 2023-67013-39841, 2022-67013-37365, and 2023-78408-39694.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abady, S., Shimelis, H., Janila, P., Wankhade, A., and Chimote, V. P. (2024). Genome-wide association analysis for drought tolerance and component traits in groundnut gene pool. Euphytica 220 (5), 76–29. doi:10.1007/s10681-024-03324-3
Aboelill, A., Mehanna, H., Kassab, O., and Abdallah, E. (2012). The response of peanut crop to foliar spraying with potassium under water stress conditions. Aust. J. Basic Appl. Sci. 6 (8), 626–634.
Abuzeineh, A., and Abualia, R. (2023). Unlocking Auxin’s potential-how plants use auxin transport to beat drought. Biomed. J. Sci. Tech. Res. 51 (3), 42712–42715. doi:10.26717/BJSTR.2023.51.008104
Agarie, S., Hanaoka, N., Kubota, F., Agata, W., and Kaufman, P. B. (1995). Measurement of cell membrane stability evaluated by electrolyte leakage as a drought and heat tolerance test in rice (Oryza sativa L.). J. Fac. Agric. 40, 233–240. Kyushu University. doi:10.5109/24109
Araus, J., Slafer, G., Reynolds, M., and Royo, C. (2002). Plant breeding and drought in C3 cereals: what should we breed for? Ann. Bot. 89 (7), 925–940. doi:10.1093/aob/mcf049
Arunyanark, A., Jogloy, S., Wongkaew, S., Akkasaeng, C., Vorasoot, N., Wright, G., et al. (2009). Association between aflatoxin contamination and drought tolerance traits in peanut. Field Crops Res. 114 (1), 14–22. doi:10.1016/j.fcr.2009.06.018
Asif, M. A., Zafar, Y., Iqbal, J., Iqbal, M. M., Rashid, U., Ali, G. M., et al. (2011). Enhanced expression of AtNHX1, in transgenic groundnut (Arachis hypogaea L.) improves salt and drought tolerence. Mol. Biotechnol. 49 (3), 250–256. doi:10.1007/s12033-011-9399-1
Aydinsakir, K., Dinc, N., Buyuktas, D., Bastug, R., and Toker, R. (2016). Assessment of different irrigation levels on peanut crop yield and quality components under Mediterranean conditions. J. Irrigation Drainage Eng. 142 (9), 1–9. doi:10.1061/(asce)ir.1943-4774.0001062
Bagherian, K. (2022). “Rapid peanut phenotyping and water quality monitoring using remote sensing and machine learning techniques,” in Masters of science (Auburn, Alabama: Auburn University).
Bagherian, K., Bidese-Puhl, R., Bao, Y., Zhang, Q., Sanz-Saez, A., Dang, P. M., et al. (2023). Phenotyping agronomic and physiological traits in peanut under mid-season drought stress using UAV-based hyperspectral imaging and machine learning. Plant Phenome J. 6 (1-20). doi:10.1002/ppj2.20081
Bajji, M., Kinet, J.-M., and Lutts, S. (2002). The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 36, 61–70. doi:10.1023/a:1014732714549
Balota, M., and Oakes, J. (2017). “UAV remote sensing for phenotyping drought tolerance in peanuts,” in Autonomous air and ground sensing systems for agricultural optimization and phenotyping II: spie, 81–87.
Balota, M., Sarkar, S., Bennett, R. S., and Burow, M. D. (2024). Phenotyping peanut drought stress with aerial remote-sensing and crop index data. Agriculture 14 (4), 565–618. doi:10.3390/agriculture14040565
Banavath, J. N., Chakradhar, T., Pandit, V., Konduru, S., Guduru, K. K., Akila, C. S., et al. (2018). Stress inducible overexpression of AtHDG11 leads to improved drought and salt stress tolerance in peanut (Arachis hypogaea L.). Front. Chem. 6, 34. doi:10.3389/fchem.2018.00034
Bandurska, H. (2022). Drought stress responses: coping strategy and resistance. Plants 11 (7), 922–1013. doi:10.3390/plants11070922
Baslam, M., Mitsui, T., Hodges, M., Priesack, E., Herritt, M. T., Aranjuelo, I., et al. (2020). Photosynthesis in a changing global climate: scaling up and scaling down in crops. Front. Plant Sci. 11, 882–929. doi:10.3389/fpls.2020.00882
Bennett, R. S., Burow, M. D., Balota, M., Chagoya, J., Sarkar, S., Sung, C. J., et al. (2022). Response to drought stress in a subset of the U.S. peanut mini-Core evaluated in Oklahoma, Texas, and Virginia. Peanut Sci. 49 (1), 71–87. doi:10.3146/0095-3679-491-ps21-14
Bertin, R. I. (2008). Plant phenology and distribution in relation to recent climate change. J. Torrey Botanical Soc. 135 (1), 126–146. doi:10.3159/07-rp-035r.1
Bertioli, D. J., Abernathy, B., Seijo, G., Clevenger, J., and Cannon, S. B. (2020). Evaluating two different models of peanut's origin. Nat. Genet. 52 (6), 557–559. doi:10.1038/s41588-020-0626-1
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G. D., Farmer, A. D., Cannon, E. K. S., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48 (4), 438–446. doi:10.1038/ng.3517
Bertioli, D. J., Clevenger, J., Godoy, I. J., Stalker, H. T., Wood, S., Santos, J. F., et al. (2021). Legacy genetics of Arachis cardenasii in the peanut crop shows the profound benefits of international seed exchange. Proc. Natl. Acad. Sci. 118 (38), e2104899118–e2104899119. doi:10.1073/pnas.2104899118
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51 (5), 877–884. doi:10.1038/s41588-019-0405-z
Bhalani, H., Thankappan, R., Mishra, G. P., Sarkar, T., Bosamia, T. C., and Dobaria, J. R. (2019). Regulation of antioxidant mechanisms by AtDREB1A improves soil-moisture deficit stress tolerance in transgenic peanut (Arachis hypogaea L.). Plos One 14 (5), e0216706–e0216720. doi:10.1371/journal.pone.0216706
Bhatnagar-Mathur, P., Devi, M. J., Reddy, D. S., Lavanya, M., Vadez, V., Serraj, R., et al. (2007). Stress-inducible expression of at DREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep. 26 (12), 2071–2082. doi:10.1007/s00299-007-0406-8
Bhatnagar-Mathur, P., Rao, J. S., Vadez, V., Dumbala, S. R., Rathore, A., Yamaguchi-Shinozaki, K., et al. (2014). Transgenic peanut overexpressing the DREB1A transcription factor has higher yields under drought stress. Mol. Breed. 33, 327–340. doi:10.1007/s11032-013-9952-7
Bhauso, T. D., Thankappan, R., Kumar, A., Mishra, G. P., Dobaria, J. R., and Venkat Rajam, M. (2014). Over-expression of bacterial'mtlD'gene confers enhanced tolerance to salt-stress and water-deficit stress in transgenic peanut ('Arachis hypogaea') through accumulation of mannitol. Aust. J. Crop Sci. 8 (3), 413–421.
Bhogireddy, S., Xavier, A., Garg, V., Layland, N., Arias, R., Payton, P., et al. (2020). Genome-wide transcriptome and physiological analyses provide new insights into peanut drought response mechanisms. Sci. Rep. 10 (1), 4071. doi:10.1038/s41598-020-60187-z
Bidese Puhl, R., Bao, Y., Sanz-Saez, A., and Chen, C. (2021). “Infield peanut pod counting using deep neural networks for yield estimation,” in 2021 ASABE annual international virtual meeting. (St. Joseph, Michigan: ASABE).
Boyer, J. S. (1982). Plant productivity and environment. Science 218 (4571), 443–448. doi:10.1126/science.218.4571.443
Brasileiro, A. C. M., Morgante, C. V., Araujo, A. C. G., Leal-Bertioli, S. C. M., Silva, A. K., Martins, A. C. Q., et al. (2015). Transcriptome profiling of wild Arachis from water-limited environments uncovers drought tolerance candidate genes. Plant Mol. Biol. Report. 33 (6), 1876–1892. doi:10.1007/s11105-015-0882-x
Bueckert, R. A., and Clarke, J. M. (2013). Review: annual crop adaptation to abiotic stress on the Canadian prairies: six case studies. Can. J. Plant Sci. 93 (3), 375–385. doi:10.4141/cjps2012-184
Buezo, J., Sanz-Saez, A., Moran, J. F., Soba, D., Aranjuelo, I., and Esteban, R. (2019). Drought tolerance response of high-yielding soybean varieties to mild drought: physiological and photochemical adjustments. Physiol. Plant. 166 (1), 88–104. doi:10.1111/ppl.12864
Cason, J. M., Simpson, C. E., Rooney, W. L., and Brady, J. A. (2020). Drought-tolerant transcription factors identified in Arachis dardani and Arachis ipaënsis. Agrosystems Geosciences and Environ. 3 (1), 1–12. doi:10.1002/agg2.20069
Celikkol Akcay, U., Ercan, O., Kavas, M., Yildiz, L., Yilmaz, C., Oktem, H., et al. (2010). Drought-induced oxidative damage and antioxidant responses in peanut (Arachis hypogaea L.) seedlings. Plant Growth Regul. 61, 21–28. doi:10.1007/s10725-010-9445-1
Chaiyadee, S., Jogloy, S., Songsri, P., Singkham, N., Vorasoot, N., Sawatsitang, P., et al. (2013). Soil moisture affects fatty acids and oil quality parameters in peanut. Int. J. Plant Prod. 7 (1), 81–96. doi:10.22069/IJPP.2012.923
Chaudhry, S., and Sidhu, G. P. S. (2022). Climate change regulated abiotic stress mechanisms in plants: a comprehensive review. Plant Cell Rep. 41 (1), 1–31. doi:10.1007/s00299-021-02759-5
Chaves, M. M., Flexas, J., and Pinheiro, C. (2008). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103 (4), 551–560. doi:10.1093/aob/mcn125
Cheek, B. (2024). “Expanding genetic resources for aflatoxin contamination resistance in peanut (Arachis hypogaea L.),” in Masters of science M.S. University of Georgia.
Chen, C.-W., Yang, Y.-W., Lur, H.-S., Tsai, Y.-G., and Chang, M.-C. (2006). A novel function of abscisic acid in the regulation of rice (oryza sativa L.) root growth and development. Plant Cell Physiology 47 (1), 1–13. doi:10.1093/pcp/pci216
Christmann, A., Grill, E., and Huang, J. (2013). Hydraulic signals in long-distance signaling. Curr. Opin. Plant Biol. 16 (3), 293–300. doi:10.1016/j.pbi.2013.02.011
Clevenger, J., Chu, Y., Chavarro, C., Agarwal, G., Bertioli, D. J., Leal-Bertioli, S. C., et al. (2017). Genome-wide SNP genotyping resolves signatures of selection and tetrasomic recombination in peanut. Mol. Plant 10 (2), 309–322. doi:10.1016/j.molp.2016.11.015
Clevenger, J. P., Korani, W., Ozias-Akins, P., and Jackson, S. (2018). Haplotype-based genotyping in polyploids. Front. Plant Sci. 9, 564. doi:10.3389/fpls.2018.00564
Coakley, S. M., Scherm, H., and Chakraborty, S. (1999). Climate change and plant disease management. Annu. Rev. Phytopathology 37 (1), 399–426. doi:10.1146/annurev.phyto.37.1.399
Cole, R. J., Dorner, J. W., and Holbrook, C. C. (1995). “Advances in mycotoxin elimination and resistance,” in Advances in peanut science. Editor H. E. P. A. H. T. Stalker (Stillwater OK: American Peanut Researcha and Education Society), 456–474.
Conkerton, E., Ross, L., Daigle, D., Kvien, C., and McCombs, C. (1989). The effect of drought stress on peanut seed composition II. Oil, protein and minerals. Oléagineux Paris. 44 (12), 593–602.
Conner, J. A., Guimaraes, L. A., Zhang, Z., Marasigan, K., Chu, Y., Korani, W., et al. (2024). Multiplexed silencing of 2S albumin genes in peanut. Plant Biotechnol. J. 22 (9), 2438–2440. doi:10.1111/pbi.14357
Dang, P. M., Chen, C. Y., and Holbrook, C. C. (2012). Identification of drought-induced transcription factors in peanut (Arachis hypogaea L.). J. Mol. Biochem. 1 (3), 196–205.
Dang, P. M., Chen, C. Y., and Holbrook, C. C. (2013). Evaluation of five peanut (Arachis hypogaea) genotypes to identify drought responsive mechanisms utilising candidate-gene approach. Funct. Plant Biol. 40 (12), 1323–1333. doi:10.1071/FP13116
Danilevicz, M. F., Fernandez, C. G. T., Marsh, J. I., Bayer, P. E., and Edwards, D. (2020). Plant pangenomics: approaches, applications and advancements. Curr. Opin. Plant Biol. 54, 18–25. doi:10.1016/j.pbi.2019.12.005
Davis, J. P., and Dean, L. L. (2016). “Peanut composition, flavor and nutrition,” in Peanuts genetics, processing, and utilization. Editors H. T. Stalker, and R. F. Wilson (United States of America: Elsevier Inc.), 289–345.
Devaiah, K., Bali, G., Athmaram, T., and Basha, M. (2007). Identification of two new genes from drought tolerant peanut up-regulated in response to drought. Plant Growth Regul. 52, 249–258. doi:10.1007/s10725-007-9195-x
Dinh, H., Kaewpradit, W., Jogloy, S., Vorasoot, N., and Patanothai, A. (2013). Biological nitrogen fixation of peanut genotypes with different levels of drought tolerance under mid-season drought. SABRAO J. Breed. Genet. 45 (3), 491–503.
Dobreva, I. D., Ruiz-Guzman, H. A., Barrios-Perez, I., Adams, T., Teare, B. L., Payton, P., et al. (2021). Thresholding analysis and feature extraction from 3D ground penetrating radar data for noninvasive assessment of peanut yield. Remote Sens. 13 (10), 1896. doi:10.3390/rs13101896
Dramé, K. N., Clavel, D., Repellin, A., Passaquet, C., and Zuily-Fodil, Y. (2007). Water deficit induces variation in expression of stress-responsive genes in two peanut (Arachis hypogaea L.) cultivars with different tolerance to drought. Plant Physiology Biochem. 45 (3-4), 236–243. doi:10.1016/j.plaphy.2007.02.002
Driesen, E., Van den Ende, W., De Proft, M., and Saeys, W. (2020). Influence of environmental factors light, CO2, temperature, and relative humidity on stomatal opening and development: a review. Agronomy 10 (12), 1975. doi:10.3390/agronomy10121975
Dutra, W. F., Guerra, Y. L., Ramos, J. P. C., Fernandes, P. D., Silva, C. R. C., Bertioli, D. J., et al. (2018). Introgression of wild alleles into the tetraploid peanut crop to improve water use efficiency, earliness and yield. PLoS One 13 (6), 01987766–e198815. doi:10.1371/journal.pone.0198776
Dwivedi, S., Nigam, S., Jambunathan, R., Sahrawat, K., Nagabhushanam, G., and Raghunath, K. (1993). Effect of genotypes and environments on oil content and oil quality parameters and their correlation in peanut (Arachis hypogaea L.). Peanut Sci. 20 (2), 84–89. doi:10.3146/i0095-3679-20-2-5
Dwivedi, S., Nigam, S., Rao, R. N., Singh, U., and Rao, K. (1996). Effect of drought on oil, fatty acids and protein contents of groundnut (Arachis hypogaea L.) seeds. Field Crops Res. 48 (2-3), 125–133. doi:10.1016/s0378-4290(96)01027-1
European Commision, C. R. (2024). Commission Regulation (EU) 2023/915 of 25 April 2023 on maximum levels for certain contaminants in food and repealing Regulation (EC) No 1881/2006. Editor E. Commision (Brussels, Belgium: Official Journal of the European Union. Available at: https://eur-lex.europa.eu/eli/reg/2023/915/2024-04-25 (Accessed July 4, 2024).
Evans, J. R. (2013). Improving photosynthesis. Plant Physiol. 162 (4), 1780–1793. doi:10.1104/pp.113.219006
Fabreti, B. S. P. C., Virk, G. K., Thangthong, N., Kvien, K., Holbrook, C. C., and Ozias-Akins, P. (2018). Drought stres effects on physiological mechanisms of peanut genotypes. Williamsburg, VA: American Peanut Research and Education Society, Inc., 147.
Faye, I., Pandey, M. K., Hamidou, F., Rathore, A., Ndoye, O., Vadez, V., et al. (2015). Identification of quantitative trait loci for yield and yield related traits in groundnut (Arachis hypogaea L.) under different water regimes in Niger and Senegal. Euphytica 206, 631–647. doi:10.1007/s10681-015-1472-6
Ferdous, J., Hussain, S. S., and Shi, B. J. (2015). Role of microRNAs in plant drought tolerance. Plant Biotechnol. J. 13 (3), 293–305. doi:10.1111/pbi.12318
Fonceka, D., Tossim, H.-A., Rivallan, R., Vignes, H., Faye, I., Ndoye, O., et al. (2012). Fostered and left behind alleles in peanut: interspecific QTL mapping reveals footprints of domestication and useful natural variation for breeding. BMC Plant Biol. 12, 26–16. doi:10.1186/1471-2229-12-26
Furlan, A. L., Bianucci, E., Castro, S., and Dietz, K.-J. (2017). Metabolic features involved in drought stress tolerance mechanisms in peanut nodules and their contribution to biological nitrogen fixation. Plant Sci. 263, 12–22. doi:10.1016/j.plantsci.2017.06.009
Furlan, A. L., Bianucci, E., Giordano, W., Castro, S., and Becker, D. F. (2020). Proline metabolic dynamics and implications in drought tolerance of peanut plants. Plant Physiology Biochem. 151, 566–578. doi:10.1016/j.plaphy.2020.04.010
Gautami, B., Pandey, M., Vadez, V., Nigam, S., Ratnakumar, P., Krishnamurthy, L., et al. (2012). Quantitative trait locus analysis and construction of consensus genetic map for drought tolerance traits based on three recombinant inbred line populations in cultivated groundnut (Arachis hypogaea L.). Mol. Breed. 30, 757–772. doi:10.1007/s11032-011-9660-0
Ghini, R., Hamada, E., and Bettiol, W. (2008). Climate change and plant diseases. Sci. Agric. 65, 98–107. doi:10.1590/s0103-90162008000700015
Ghosh, S., Mahadevaiah, S. S., Gowda, S. A., Gangurde, S. S., Jadhav, M. P., Hake, A. A., et al. (2022). Genetic mapping of drought tolerance traits phenotyped under varying drought stress environments in peanut (Arachis hypogaea L.). Euphytica 218 (12), 168. doi:10.1007/s10681-022-03120-x
Ghosh, U. K., Islam, M. N., Siddiqui, M. N., and Khan, M. A. R. (2021). Understanding the roles of osmolytes for acclimatizing plants to changing environment: a review of potential mechanism. Plant Signal. and Behav. 16 (8), 1913306. doi:10.1080/15592324.2021.1913306
Gimode, D., Chu, Y., Holbrook, C. C., Fonceka, D., Porter, W., Dobreva, I., et al. (2023). High-throughput canopy and belowground phenotyping of a set of peanut CSSLs detects lines with increased pod weight and foliar disease tolerance. Agronomy 13 (5), 1223. doi:10.3390/agronomy13051223
Girdthai, T., Jogloy, S., Vorasoot, N., Akkasaeng, C., Wongkaew, S., Holbrook, C., et al. (2010). Associations between physiological traits for drought tolerance and aflatoxin contamination in peanut genotypes under terminal drought. Plant Breed. 129 (6), 693–699. doi:10.1111/j.1439-0523.2009.01738.x
Govind, G., Vokkaliga ThammeGowda, H., Jayaker Kalaiarasi, P., Iyer, D. R., Muthappa, S. K., Nese, S., et al. (2009). Identification and functional validation of a unique set of drought induced genes preferentially expressed in response to gradual water stress in peanut. Mol. Genet. Genomics 281, 591–605. doi:10.1007/s00438-009-0432-z
Guimarães, P. M., Brasileiro, A. C., Morgante, C. V., Martins, A. C., Pappas, G., Silva, O. B., et al. (2012). Global transcriptome analysis of two wild relatives of peanut under drought and fungi infection. BMC Genomics 13, 387–415. doi:10.1186/1471-2164-13-387
Gundaraniya, S. A., Ambalam, P. S., and Tomar, R. S. (2020). Metabolomic profiling of drought-tolerant and susceptible peanut (Arachis hypogaea L.) genotypes in response to drought stress. ACS Omega 5 (48), 31209–31219. doi:10.1021/acsomega.0c04601
Guo, B., Xu, G., Cao, Y., Holbrook, C., and Lynch, R. (2006). Identification and characterization of phospholipase D and its association with drought susceptibilities in peanut (Arachis hypogaea). Planta 223, 512–520. doi:10.1007/s00425-005-0112-0
Gururani, M. A., Venkatesh, J., and Tran, L. S. P. (2015). Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 8 (9), 1304–1320. doi:10.1016/j.molp.2015.05.005
Hamidou, F., Halilou, O., and Vadez, V. (2013). Assessment of groundnut under combined heat and drought stress. J. Agron. Crop Sci. 199 (1), 1–11. doi:10.1111/j.1439-037x.2012.00518.x
Hamidou, F., Rathore, A., Waliyar, F., and Vadez, V. (2014). Although drought intensity increases aflatoxin contamination, drought tolerance does not lead to less aflatoxin contamination. Field Crops Res. 156, 103–110. doi:10.1016/j.fcr.2013.10.019
Hamidou, F., Ratnakumar, P., Halilou, O., Mponda, O., Kapewa, T., Monyo, E., et al. (2012). Selection of intermittent drought tolerant lines across years and locations in the reference collection of groundnut (Arachis hypogaea L.). Field Crops Res. 126, 189–199. doi:10.1016/j.fcr.2011.10.009
Hasanuzzaman, M., Bhuyan, M. H. M. B., Zulfiqar, F., Raza, A., Mohsin, S. M., Al Mahmud, J., et al. (2020). Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants 9 (8), 681. doi:10.3390/antiox9080681
Hashim, I., Koehler, P., Eitenmiller, R., and Kvien, C. (1993). Fatty acid composition and tocopherol content of drought stressed Florunner peanuts. Peanut Sci. 20 (1), 21–24. doi:10.3146/i0095-3679-20-1-6
Havlová, M., Dobrev, P. I., Motyka, V., Storchová, H., Libus, J., Dobrá, J., et al. (2008). The role of cytokinins in responses to water deficit in tobacco plants over-expressing trans-zeatin O-glucosyltransferase gene under 35S or SAG12 promoters. Plant, Cell and Environ. 31 (3), 341–353. doi:10.1111/j.1365-3040.2007.01766.x
He, Y., Mu, S., He, Z., Wang, B., and Li, Y. (2020). Ectopic expression of MYB repressor GmMYB3a improves drought tolerance and productivity of transgenic peanuts (Arachis hypogaea L.) under conditions of water deficit. Transgenic Res. 29 (5), 563–574. doi:10.1007/s11248-020-00220-z
Helena, M., and Carvalho, C. d. (2008). Drought stress and reactive oxygen species: production, scavenging and signaling. Plant Signal. and Behav. 3 (3), 156–165. doi:10.4161/psb.3.3.5536
Holbrook, C. C., Burow, M. D., Chen, C. Y., Pandey, M. K., Liu, L., Chagoya, J. C., et al. (2016). “Recent advances in peanut breeding and genetics,” in Peanuts: genetics, processing, and utilization. Editors H. T. Stalker, and R. F. Wilson (United States of America: Academic Press and AOCS Press), 111–145.
Holbrook, C. C., Kvien, C. K., Rucker, K. S., Wilson, D. M., Hook, J. E., and Matheron, M. E. (2000). Preharvest aflatoxin contamination in drought-tolerant and drought-intolerant peanut genotypes. Peanut Sci. 27 (2), 45–48. doi:10.3146/i0095-3679-27-2-1
Hou, L., Liu, W., Li, Z., Huang, C., Fang, X., Wang, Q., et al. (2014). Identification and expression analysis of genes responsive to drought stress in peanut. Russ. J. Plant Physiology 61, 842–852. doi:10.1134/s1021443714060089
Htoon, W., Jogloy, S., Vorasoot, N., Toomsan, B., Kaewpradit, W., Puppala, N., et al. (2014). Nutrient uptakes and their contributions to yield in peanut genotypes with different levels of terminal drought resistance. Turkish J. Agric. For. 38 (6), 781–791. doi:10.3906/tar-1402-26
Huang, G.-T., Ma, S.-L., Bai, L.-P., Zhang, L., Ma, H., Jia, P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987. doi:10.1007/s11033-011-0823-1
Jangra, S., Chaudhary, V., Yadav, R. C., and Yadav, N. R. (2021). High-throughput phenotyping: a platform to accelerate crop improvement. Phenomics 1 (2), 31–53. doi:10.1007/s43657-020-00007-6
Jiang, C., Li, X., Zou, J., Ren, J., Jin, C., Zhang, H., et al. (2021). Comparative transcriptome analysis of genes involved in the drought stress response of two peanut (Arachis hypogaea L.) varieties. BMC Plant Biol. 21, 64–14. doi:10.1186/s12870-020-02761-1
Jiang, M., and Zhang, J. (2002). Involvement of plasma-membrane NADPH oxidase in abscisic acid-and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 215, 1022–1030. doi:10.1007/s00425-002-0829-y
Junjittakarn, J., Pimratch, S., Jogloy, S., Htoon, W., Singkham, N., Vorasoot, N., et al. (2013). Nutrient uptake of peanut genotypes under different water regimes. Int. J. Plant Prod. 7 (4), 677–692. doi:10.22069/IJPP.2013.1264
Kambiranda, D. M., Vasanthaiah, H. K., Katam, R., Ananga, A., Basha, S. M., and Naik, K. (2011). Impact of drought stress on peanut (Arachis hypogaea L.) productivity and food safety. Plants Environ. 1, 249–272. doi:10.5772/27917
Katagiri, T., Takahashi, S., and Shinozaki, K. (2001). Involvement of a novel Arabidopsis phospholipase D, AtPLDdelta, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 26 (6), 595–605. doi:10.1046/j.1365-313x.2001.01060.x
Kelly, A. E., and Goulden, M. L. (2008). Rapid shifts in plant distribution with recent climate change. Proc. Natl. Acad. Sci. 105 (33), 11823–11826. doi:10.1073/pnas.0802891105
Khan, N., Bano, A., Ali, S., and Babar, M. A. (2020). Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 90 (2), 189–203. doi:10.1007/s10725-020-00571-x
Kim, J. Y. (2020). Roadmap to high throughput phenotyping for plant breeding. J. Biosyst. Eng. 45 (1), 43–55. doi:10.1007/s42853-020-00043-0
Kiranmai, K., Lokanadha Rao, G., Pandurangaiah, M., Nareshkumar, A., Amaranatha Reddy, V., Lokesh, U., et al. (2018). A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum Lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front. Plant Sci. 9, 346. doi:10.3389/fpls.2018.00346
Korani, W., Clevenger, J. P., Chu, Y., and Ozias-Akins, P. (2019). Machine learning as an effective method for identifying true single nucleotide polymorphisms in polyploid plants. Plant Genome 12 (1), 1–10. doi:10.3835/plantgenome2018.05.0023
Krishna, G., Singh, B. K., Kim, E. K., Morya, V. K., and Ramteke, P. W. (2015). Progress in genetic engineering of peanut (Arachis hypogaea L.)-A review. Plant Biotechnol. J. 13 (2), 147–162. doi:10.1111/pbi.12339
Krishnamurthy, L., Vadez, V., Devi, M. J., Serraj, R., Nigam, S., Sheshshayee, M., et al. (2007). Variation in transpiration efficiency and its related traits in a groundnut (Arachis hypogaea L.) mapping population. Field Crops Res. 103 (3), 189–197. doi:10.1016/j.fcr.2007.06.009
Kumar, P. (2017). “MicroRNA regulation of gene expression under drought stress acclimation response in peanut (Arachis hypogaea L.) roots,” in Master of science (Lubbock, TX: Texas Tech University).
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62 (14), 4731–4748. doi:10.1093/jxb/err210
Lata, C., Yadav, A., and Prasad, M. (2011). “Role of plant transcription factors in abiotic stress tolerance,” in Abiotic stress response in plants - physiological, biochemical and genetic perspectives. Editors A. Shanker, and B. Venkateswarlu (Rijeka, Croatia: INTECH Open Access Publishers), 269–296.
Lauriano, J., Lidon, F., Carvalho, C., Campos, P., and do Céu Matos, M. (2000). Drought effects on membrane lipids and photosynthetic activity in different peanut cultivars. Photosynthetica 38, 7–12. doi:10.1023/a:1026775319916
Leal-Bertioli, S. C., Bertioli, D. J., Guimarães, P. M., Pereira, T. D., Galhardo, I., Silva, J. P., et al. (2012). The effect of tetraploidization of wild Arachis on leaf morphology and other drought-related traits. Environ. Exp. Bot. 84, 17–24. doi:10.1016/j.envexpbot.2012.04.005
Leal-Bertioli, S. C. M., José, A. C. V. F., Alves-Freitas, D. M. T., Moretzsohn, M. C., Guimarães, P. M., Nielen, S., et al. (2009). Identification of candidate genome regions controlling disease resistance in Arachis. BMC Plant Biol. 9 (1), 112. doi:10.1186/1471-2229-9-112
Li, A., Zhou, M., Liao, G., Li, X., Wang, A., Xiao, D., et al. (2023). CRISPR/Cas9 gene editing in peanut by Agrobacterium tumefaciens-mediated pollen tube transformation. Plant Cell, Tissue Organ Cult. 155 (3), 883–892. doi:10.1007/s11240-023-02607-2
Li, C., Yan, C., Sun, Q., Wang, J., Yuan, C., Mou, Y., et al. (2021). The bHLH transcription factor AhbHLH112 improves the drought tolerance of peanut. BMC Plant Biol. 21, 540–612. doi:10.1186/s12870-021-03318-6
Li, X., Lu, J., Liu, S., Liu, X., Lin, Y., and Li, L. (2014). Identification of rapidly induced genes in the response of peanut (Arachis hypogaea) to water deficit and abscisic acid. BMC Biotechnol. 14, 58–59. doi:10.1186/1472-6750-14-58
Liu, S., Lv, Z., Liu, Y., Li, L., and Zhang, L. (2018a). Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet. Mol. Biol. 41, 624–637. doi:10.1590/1678-4685-GMB-2017-0229
Liu, X., Dong, X., Xue, Q., Leskovar, D. I., Jifon, J., Butnor, J. R., et al. (2018b). Ground penetrating radar (GPR) detects fine roots of agricultural crops in the field. Plant Soil 423, 517–531. doi:10.1007/s11104-017-3531-3
Lokesh, U., Venkatesh, B., Kiranmai, K., Nareshkumar, A., Amarnathareddy, V., Rao, G. L., et al. (2019). Overexpression of ß-ketoacyl Co-A Synthase1 gene improves tolerance of drought susceptible groundnut (Arachis hypogaea L.) cultivar K-6 by increased leaf epicuticular wax accumulation. Front. Plant Sci. 9, 1869. doi:10.3389/fpls.2018.01869
Long, H., Zheng, Z., Zhang, Y., Xing, P., Wan, X., Zheng, Y., et al. (2019). An abscisic acid (ABA) homeostasis regulated by its production, catabolism and transport in peanut leaves in response to drought stress. PLoS One 14 (6), e0213963. doi:10.1371/journal.pone.0213963
Luis, J. M., Ozias-Akins, P., Holbrook, C. C., Kemerait, J. R. C., Snider, J. L., and Liakos, V. (2016). Phenotyping peanut genotypes for drought tolerance. Peanut Sci. 43 (1), 36–48. doi:10.3146/0095-3679-43.1.36
Mahmood, T., Khalid, S., Abdullah, M., Ahmed, Z., Shah, M. K. N., Ghafoor, A., et al. (2019). Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 9 (1), 105. doi:10.3390/cells9010105
Mahmood, Y. (2020). Drought effects on leaf canopy temperature and leaf senescence in barley. Iraqi J. Agric. Sci. 51 (6), 1684–1693. doi:10.36103/ijas.v51i6.1197
Mallikarjuna, G., Rao, T. S. R. B., and Kirti, P. B. (2016). Genetic engineering for peanut improvement: current status and prospects. Plant Cell Tissue Organ Cult. 125 (3), 399–416. doi:10.1007/s11240-016-0966-9
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33 (4), 453–467. doi:10.1111/j.1365-3040.2009.02041.x
Mittal, M., Dhingra, A., Dawar, P., Payton, P., and Rock, C. D. (2023). The role of microRNAs in responses to drought and heat stress in peanut (Arachis hypogaea). Plant Genome 16 (3), e20350. doi:10.1002/tpg2.20350
Mittler, R., and Blumwald, E. (2015). The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27 (1), 64–70. doi:10.1105/tpc.114.133090
Mota, A. P. Z., Brasileiro, A. C. M., Vidigal, B., Oliveira, T. N., da Cunha Quintana Martins, A., Saraiva, M. A. D. P., et al. (2021). Defining the combined stress response in wild Arachis. Sci. Rep. 11 (1), 11097. doi:10.1038/s41598-021-90607-7
Müller, M. (2021). Foes or friends: ABA and ethylene interaction under abiotic stress. Plants 10 (3), 448. doi:10.3390/plants10030448
Nageswara Rao, R., Singh, S., Sivakumar, M., Srivastava, K., and Williams, J. (1985). Effect of water deficit at different growth phases of peanut. I. Yield responses. Agron. J. 77 (5), 782–786. doi:10.2134/agronj1985.00021962007700050026x
Nageswara Rao, R., Talwar, H., and Wright, G. (2001). Rapid assessment of specific leaf area and leaf nitrogen in peanut (Arachis hypogaea L.) using a chlorophyll meter. J. Agron. Crop Sci. 186 (3), 175–182. doi:10.1046/j.1439-037x.2001.00472.x
Nageswara Rao, R., Williams, J., Sivakumar, M., and Wadia, K. (1988). Effect of water deficit at different growth phases of peanut. II. Response to drought during preflowering phase. Agron. J. 80, 431–438. doi:10.2134/agronj1988.00021962008000030010x
Nataraja, K. N., and Parvathi, M. S. (2016). “Tolerance to drought stress in plants: unravelling the signaling networks,” in Drought stress tolerance in plants. Molecular and genetic perspectives. Editors M. A. Hossain, S. H. Wani, S. Bhattacharjee, D. J. Burritt, and L.-S. P. Tran (Switzerland: Springer International Publishing), 2, 71–90. doi:10.1007/978-3-319-32423-4_3
Nautiyal, P., Rachaputi, N. R., and Joshi, Y. (2002). Moisture-deficit-induced changes in leaf-water content, leaf carbon exchange rate and biomass production in groundnut cultivars differing in specific leaf area. Field Crops Res. 74 (1), 67–79. doi:10.1016/s0378-4290(01)00199-x
Nautiyal, P., Ravindra, V., Zala, P., and Joshi, Y. (1999). Enhancement of yield in groundnut following the imposition of transient soil-moisture-deficit stress during the vegetative phase. Exp. Agric. 35 (3), 371–385. doi:10.1017/s0014479799003075
Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant. Sci. 5, 170. doi:10.3389/fpls.2014.00170
Nigam, S., Chandra, S., Sridevi, K. R., Bhukta, M., Reddy, A., Rachaputi, N. R., et al. (2005). Efficiency of physiological trait-based and empirical selection approaches for drought tolerance in groundnut. Ann. Appl. Biol. 146 (4), 433–439. doi:10.1111/j.1744-7348.2005.040076.x
Norden, A., Lipscomb, R., and Carver, W. (1969). Registration of florunner Peanuts1 (reg. No. 2). Crop Sci. 9, 850. doi:10.2135/cropsci1969.0011183x000900060070x
Office of the Federal Register (2004). 7 CFR 996 - minimum quality and handling standards for domestic and imported peanuts marketed in the United States. Available at: https://www.govinfo.gov/app/details/CFR-2005-title7-vol8/CFR-2005-title7-vol8-part996/summary (Accessed July 4, 2024).
Onyekachi, O. G., Boniface, O. O., Gemlack, N. F., and Nicholas, N. (2019). “The effect of climate change on abiotic plant stress: a review,” in Abiotic and biotic stress in plants. Editor A. B. D. Oliveira (London, United Kingdom: IntechOpen)).
Padmavathi, T. A., and Rao, D. M. (2013). Differential accumulation of osmolytes in 4 cultivars of peanut (Arachis hypogaea L.) under drought stress. J. Crop Sci. Biotechnol. 16, 151–159. doi:10.1007/s12892-012-0102-2
Pallas, J., Stansell, J., and Koske, T. (1979). Effects of drought on Florunner peanuts. Agron. J. 71 (5), 853–858. doi:10.2134/agronj1979.00021962007100050034x
Pandey, M. K., Agarwal, G., Kale, S. M., Clevenger, J., Nayak, S. N., Sriswathi, M., et al. (2017). Development and evaluation of a high density genotyping ‘Axiom_Arachis’ array with 58 K SNPs for accelerating genetics and breeding in groundnut. Sci. Rep. 7 (1), 40577. doi:10.1038/srep40577
Pandey, M. K., Gangurde, S. S., Sharma, V., Pattanashetti, S. K., Naidu, G. K., Faye, I., et al. (2021). Improved genetic map identified major QTLs for drought tolerance- and iron deficiency tolerance-related traits in groundnut. Genes 12 (1), 37. doi:10.3390/genes12010037
Pandey, M. K., Upadhyaya, H. D., Rathore, A., Vadez, V., Sheshshayee, M., Sriswathi, M., et al. (2014). Genomewide association studies for 50 agronomic traits in peanut using the ‘reference set’comprising 300 genotypes from 48 countries of the semi-arid tropics of the world. PLoS one 9 (8), e105228. doi:10.1371/journal.pone.0105228
Pandey, R., Herrera, W., Villegas, A., and Pendleton, J. (1984). Drought response of grain legumes under irrigation gradient: III. Plant growth. Agron. J. 76 (4), 557–560. doi:10.2134/agronj1984.00021962007600040011x
Pandurangaiah, M., Lokanadha Rao, G., Sudhakarbabu, O., Nareshkumar, A., Kiranmai, K., Lokesh, U., et al. (2014). Overexpression of horsegram (Macrotyloma uniflorum Lam.Verdc.) NAC transcriptional factor (MuNAC4) in groundnut confers enhanced drought tolerance. Mol. Biotechnol. 56 (8), 758–769. doi:10.1007/s12033-014-9754-0
Patel, K. G., Thankappan, R., Mishra, G. P., Mandaliya, V. B., Kumar, A., and Dobaria, J. R. (2017). Transgenic peanut (Arachis hypogaea L.) overexpressing mtlD gene showed improved photosynthetic, physio-biochemical, and yield-parameters under soil-moisture deficit stress in lysimeter system. Front. Plant Sci. 8, 1881. doi:10.3389/fpls.2017.01881
Patil, M., Ramu, S. V., Jathish, P., Sreevathsa, R., Reddy, P. C., Prasad, T. G., et al. (2014). Overexpression of AtNAC2 (ANAC092) in groundnut (Arachis hypogaea L.) improves abiotic stress tolerance. Plant Biotechnol. Rep. 8 (2), 161–169. doi:10.1007/s11816-013-0305-0
Pavithradevi, S., N, M., Varman, P., and Kulandaivelu, G. (2015). Evaluation of groundnut genotypes for late season drought tolerance. Legume Res. - Int. J. 38 (6), 763–766. doi:10.18805/lr.v38i6.6721
Pierik, R., Sasidharan, R., and Voesenek, L. A. (2007). Growth control by ethylene: adjusting phenotypes to the environment. J. Plant Growth Regul. 26, 188–200. doi:10.1007/s00344-006-0124-4
Pilon, C., Snider, J. L., Sobolev, V., Chastain, D. R., Sorensen, R. B., Meeks, C. D., et al. (2018). Assessing stomatal and non-stomatal limitations to carbon assimilation under progressive drought in peanut (Arachis hypogaea L.). J. Plant Physiology 231, 124–134. doi:10.1016/j.jplph.2018.09.007
Pimratch, S., Jogloy, S., Vorasoot, N., Toomsan, B., Patanothai, A., and Holbrook, C. (2008). Relationship between biomass production and nitrogen fixation under drought-stress conditions in peanut genotypes with different levels of drought resistance. J. Agron. Crop Sci. 194 (1), 15–25. doi:10.1111/j.1439-037x.2007.00286.x
Pirasteh-Anosheh, H., Saed-Moucheshi, A., Pakniyat, H., and Pessarakli, M. (2016). “Stomatal responses to drought stress,” in Water Stress and crop plants: a sustainable approach. Editor P. Ahmad (John Wiley and Sons, Ltd), 24–40.
Priya, M., Dhanker, O. P., Siddique, K. H., HanumanthaRao, B., Nair, R. M., Pandey, S., et al. (2019). Drought and heat stress-related proteins: an update about their functional relevance in imparting stress tolerance in agricultural crops. Theor. Appl. Genet. 132, 1607–1638. doi:10.1007/s00122-019-03331-2
Pruthvi, V., Narasimhan, R., and Nataraja, K. N. (2014). Simultaneous expression of abiotic stress responsive transcription factors, AtDREB2A, AtHB7 and AtABF3 improves salinity and drought tolerance in peanut (Arachis hypogaea L.). PLoS One 9 (12), e111152. doi:10.1371/journal.pone.0111152
Pruthvi, V., Rama, N., Govind, G., and Nataraja, K. N. (2013). Expression analysis of drought stress specific genes in Peanut (Arachis hypogaea L.). Physiology Mol. Biol. Plants 19 (2), 277–281. doi:10.1007/s12298-012-0156-0
Puangbut, D., Jogloy, S., Toomsan, B., Vorasoot, N., Akkasaeng, C., Kesmala, T., et al. (2010). Drought stress: physiological basis for genotypic variation in tolerance to and recovery from pre-flowering drought in peanut. J. Agron. Crop Sci. 196 (5), 358–367. doi:10.1111/j.1439-037x.2010.00426.x
Puppala, N., Nayak, S. N., Sanz-Saez, A., Chen, C., Devi, M. J., Nivedita, N., et al. (2023). Sustaining yield and nutritional quality of peanuts in harsh environments: physiological and molecular basis of drought and heat stress tolerance. Front. Genet. 14, 1121462–1121524. doi:10.3389/fgene.2023.1121462
Qin, H., Gu, Q., Kuppu, S., Sun, L., Zhu, X., Mishra, N., et al. (2013). Expression of the Arabidopsis vacuolar H+-pyrophosphatase gene AVP1 in peanut to improve drought and salt tolerance. Plant Biotechnol. Rep. 7, 345–355. doi:10.1007/s11816-012-0269-5
Qin, H., Gu, Q., Zhang, J., Sun, L., Kuppu, S., Zhang, Y., et al. (2011). Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions. Plant Cell Physiology 52 (11), 1904–1914. doi:10.1093/pcp/pcr125
Ramu, V. S., Swetha, T. N., Sheela, S. H., Babitha, C. K., Rohini, S., Reddy, M. K., et al. (2016). Simultaneous expression of regulatory genes associated with specific drought-adaptive traits improves drought adaptation in peanut. Plant Biotechnol. J. 14 (3), 1008–1020. doi:10.1111/pbi.12461
Rao, T. S. R. B., Naresh, J. V., Reddy, P. S., Reddy, M. K., and Mallikarjuna, G. (2017). Expression of pennisetum glaucum eukaryotic translational initiation factor 4A (PgeIF4A) confers improved drought, salinity, and oxidative stress tolerance in groundnut. Front. Plant Sci. 8, 1–15. doi:10.3389/fpls.2017.00453
Rathnakumar, A., Bera, S., K, G., and Kumar, N. (2017). “Botany, systematics, characterization and utilization of groundnut germplasm,” in Training manual on hybridization techniques in groundnut. Editor N. K. L. Rathnakumar (Gujarat, India: ICAR-Directorate of Groundnut Research), 6–15.
Ravi, K., Vadez, V., Isobe, S., Mir, R. R., Guo, Y., Nigam, S. N., et al. (2011). Identification of several small main-effect QTLs and a large number of epistatic QTLs for drought tolerance related traits in groundnut (Arachis hypogaea L.). Theor. Appl. Genet. 122 (6), 1119–1132. doi:10.1007/s00122-010-1517-0
Raza, M. M., and Bebber, D. P. (2022). Climate change and plant pathogens. Curr. Opin. Microbiol. 70, 102233. doi:10.1016/j.mib.2022.102233
Reddy, T., Reddy, V., and Anbumozhi, V. (2003). Physiological responses of groundnut (Arachis hypogea L.) to drought stress and its amelioration: a critical review. Plant Growth Regul. 41, 75–88. doi:10.1023/a:1027353430164
Ren, J., Zhang, H., Shi, X., Ai, X., Dong, J., Zhao, X., et al. (2021). Genome-wide identification of key candidate microRNAs and target genes associated with peanut drought tolerance. DNA Cell Biol. 40 (2), 373–383. doi:10.1089/dna.2020.6245
Richardson, F., Brodribb, T. J., and Jordan, G. J. (2017). Amphistomatic leaf surfaces independently regulate gas exchange in response to variations in evaporative demand. Tree Physiol. 37 (7), 869–878. doi:10.1093/treephys/tpx073
Rowland, D. L., Faircloth, W. H., Payton, P., Tissue, D. T., Ferrell, J. A., Sorensen, R. B., et al. (2012). Primed acclimation of cultivated peanut (Arachis hypogaea L.) through the use of deficit irrigation timed to crop developmental periods. Agric. Water Manag. 113, 85–95. doi:10.1016/j.agwat.2012.06.023
Rucker, K. S., Kvien, C. K., Holbrook, C. C., and Hook, J. E. (1995). Identification of peanut genotypes with improved drought avoidance traits. Peanut Sci. 22 (1), 14–18. doi:10.3146/pnut.22.1.0003
Sachdev, S., Ansari, S. A., Ansari, M. I., Fujita, M., and Hasanuzzaman, M. (2021). Abiotic stress and reactive oxygen species: generation, signaling, and defense mechanisms. Antioxidants 10 (2), 277. doi:10.3390/antiox10020277
Saibo, N. J. M., Lourenco, T., and Oliveira, M. M. (2009). Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann. Bot. 103 (4), 609–623. doi:10.1093/aob/mcn227
Sankar, B., Karthishwaran, K., and Somasundaram, R. (2013). Leaf anatomical changes in peanut plants in relation to drought stress with or without paclobutrazol and ABA. J. Phytology 5, 25–29.
Sarkar, S., Cazenave, A.-B., Oakes, J., McCall, D., Thomason, W., Abbot, L., et al. (2020). High-throughput measurement of peanut canopy height using digital surface models. Plant Phenome J. 3 (1), e20003. doi:10.1002/ppj2.20003
Sarkar, S., Cazenave, A. B., Oakes, J., McCall, D., Thomason, W., Abbott, L., et al. (2021). Aerial high-throughput phenotyping of peanut leaf area index and lateral growth. Sci. Rep. 11 (1), 21661. doi:10.1038/s41598-021-00936-w
Sarkar, T., Thankappan, R., Kumar, A., Mishra, G. P., and Dobaria, J. R. (2014). Heterologous expression of the AtDREB1A gene in transgenic peanut-conferred tolerance to drought and salinity stresses. PLoS One 9 (12), e110507. doi:10.1371/journal.pone.0110507
Seleiman, M. F., Al-Suhaibani, N., Ali, N., Akmal, M., Alotaibi, M., Refay, Y., et al. (2021). Drought stress impacts on plants and different approaches to alleviate its adverse effects. Plants 10 (2), 259–325. doi:10.3390/plants10020259
Shaibu, A. S., Sneller, C., Motagi, B. N., Chepkoech, J., Chepngetich, M., Miko, Z. L., et al. (2020). Genome-wide detection of SNP markers associated with four physiological traits in groundnut (Arachis hypogaea L.) mini core collection. Agronomy-Basel 10 (2), 192–214. doi:10.3390/agronomy10020192
Sharma, V., Mahadevaiah, S. S., Latha, P., Gowda, S. A., Manohar, S. S., Jadhav, K., et al. (2024). Dissecting genomic regions and underlying candidate genes in groundnut MAGIC population for drought tolerance. BMC Plant Biol. 24 (1), 1044–1121. doi:10.1186/s12870-024-05749-3
Shekoofa, A., and Sinclair, T. R. (2018). Aquaporin activity to improve crop drought tolerance. Cells 7 (9), 123. doi:10.3390/cells7090123
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3 (3), 217–223. doi:10.1016/s1369-5266(00)80068-0
Sinclair, T. R., Shekoofa, A., Isleib, T. G., Balota, M., and Zhang, H. (2018). Identification of Virginia-type peanut genotypes for water-deficit conditions based on early decrease in transpiration rate with soil drying. Crop Sci. 58 (6), 2607–2612. doi:10.2135/cropsci2018.05.0293
Singh, A., Jain, D., Pandey, J., Yadav, M., Bansal, K. C., and Singh, I. K. (2023). Deciphering the role of miRNA in reprogramming plant responses to drought stress. Crit. Rev. Biotechnol. 43 (4), 613–627. doi:10.1080/07388551.2022.2047880
Singh, S., Sangh, C., Kona, P., and Bera, S. K. (2024). Genome editing in peanuts: advancements, challenges and applications. Nucl. 67 (1), 127–139. doi:10.1007/s13237-024-00482-6
Singh, U., and Singh, B. (1992). Tropical grain legumes as important human foods. Econ. Bot. 46, 310–321. doi:10.1007/bf02866630
Soba, D., Parker, S., Chen, C., Shekoofa, A., and Sanz-Saez, A. (2024). Peanut photosynthesis response to drought can include diffusive and biochemical limitations depending on cultivar. Physiol. Plant. 176 (4), e14489. doi:10.1111/ppl.14489
Soma, F., Takahashi, F., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2021). Cellular phosphorylation signaling and gene expression in drought stress responses: ABA-dependent and ABA-independent regulatory systems. Plants 10 (4), 756. doi:10.3390/plants10040756
Song, L., Li, R., Xiang, X., Wang, J., Qiao, L., Song, X., et al. (2012). Overexpression of stress-inducible small GTP-binding protein AhRab7 (AhRabG3f) in peanut (Arachis hypogaea L.) enhances abiotic stress tolerance. J. Food Agric. and Environ. 10 (3-4), 888–894.
Songsri, P., Jogloy, S., Vorasoot, N., Akkasaeng, C., Patanothai, A., and Holbrook, C. (2008). Root distribution of drought-resistant peanut genotypes in response to drought. J. Agron. Crop Sci. 194 (2), 92–103. doi:10.1111/j.1439-037x.2008.00296.x
Stalker, H. T. (2017). Utilizing wild species for peanut improvement. Crop Sci. 57 (3), 1102–1120. doi:10.2135/cropsci2016.09.0824
Stansell, J., and Pallas, J. (1985). Yield and quality response of Florunner peanut to applied drought at several growth stages. Peanut Sci. 12 (2), 64–70. doi:10.3146/pnut.12.2.0005
Sullivan, D. G., and Holbrook, C. C. (2007). Using ground-based reflectance measurements as selection criteria for drought and aflatoxin-resistant peanut genotypes. Crop Sci. 47 (3), 1040–1050. doi:10.2135/cropsci2006.08.0511
Sun, L., Hu, R., Shen, G., and Zhang, H. (2013). Genetic engineering peanut for higher drought-and salt-tolerance. Food Nutr. Sci. 4 (6A), 1–7. doi:10.4236/fns.2013.46a001
Takahashi, F., Kuromori, T., Sato, H., and Shinozaki, K. (2018a). “Regulatory gene networks in drought stress responses and resistance in plants,” in Survival strategies in extreme cold and desiccation: adaptation mechanisms and their applications. Editors M. Iwaya-Inoue, M. Sakurai, and M. Uemura. (Singapore: Springer Singapore), 189–214.
Takahashi, F., Kuromori, T., Urano, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2020). Drought stress responses and resistance in plants: from cellular responses to long-distance intercellular communication. Front. Plant Sci. 11, 556972. doi:10.3389/fpls.2020.556972
Takahashi, F., Suzuki, T., Osakabe, Y., Betsuyaku, S., Kondo, Y., Dohmae, N., et al. (2018b). A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556 (7700), 235–238. doi:10.1038/s41586-018-0009-2
Tallury, S., Isleib, T., Copeland, S., Rosas-Anderson, P., Balota, M., Singh, D., et al. (2014). Registration of two multiple disease-resistant peanut germplasm lines derived from Arachis cardenasii Krapov. and WC Gregory, GKP 10017. J. Plant Registrations 8 (1), 86–89. doi:10.3198/jpr2013.04.0017crg
Thangella, P. A., Pasumarti, S. N., Pullakhandam, R., Geereddy, B. R., and Daggu, M. R. (2018). Differential expression of leaf proteins in four cultivars of peanut (Arachis hypogaea L.) under water stress. 3 Biotech. 8, 157–221. doi:10.1007/s13205-018-1180-8
Thangthong, N., Jogloy, S., Jongrungklang, N., Kvien, C., Pensuk, V., Kesmala, T., et al. (2018). Root distribution patterns of peanut genotypes with different drought resistance levels under early-season drought stress. J. Agron. Crop Sci. 204 (2), 111–122. doi:10.1111/jac.12249
Thoppurathu, F. J., Ghorbanzadeh, Z., Vala, A. K., Hamid, R., and Joshi, M. (2022). Unravelling the treasure trove of drought-responsive genes in wild-type peanut through transcriptomics and physiological analyses of root. Funct. and Integr. Genomics 22 (2), 215–233. doi:10.1007/s10142-022-00833-z
Tiwari, V., Chaturvedi, A. K., Mishra, A., and Jha, B. (2015). Introgression of the SbASR-1 gene cloned from a halophyte Salicornia brachiate enhances salinity and drought endurance in transgenic groundnut (arachis hypogaea)and acts as a transcription factor [corrected]. Plos One 10 (8), e0131567. doi:10.1371/journal.pone.0131567
Ul-Allah, S., Rehman, A., Hussain, M., and Farooq, M. (2021). Fiber yield and quality in cotton under drought: effects and management. Agric. Water Manag. 255, 106994. doi:10.1016/j.agwat.2021.106994
Upadhyaya, H. D. (2005). Variability for drought resistance related traits in the mini core collection of peanut. Crop Sci. 45 (4), 1432–1440. doi:10.2135/cropsci2004.0389
USDA-FAS (2024). Peanut explorer. USDA-NAS. Available at: https://ipad.fas.usda.gov/cropexplorer/cropview/commodityView.aspx?cropid=2221000 (Accessed February 7, 2024).
Vadez, V., Rao, J. S., Bhatnagar-Mathur, P., and Sharma, K. (2013). DREB1A promotes root development in deep soil layers and increases water extraction under water stress in groundnut. Plant Biol. 15 (1), 45–52. doi:10.1111/j.1438-8677.2012.00588.x
Vadez, V., Rao, S., Sharma, K., Bhatnagar-Mathur, P., and Devi, M. J. (2007). DREB1A allows for more water uptake in groundnut by a large modification in the root/shoot ratio under water deficit. J. SAT Agric. Res. 5 (1), 1–5.
Vadez, V., and Ratnakumar, P. (2016). High transpiration efficiency increases pod yield under intermittent drought in dry and hot atmospheric conditions but less so under wetter and cooler conditions in groundnut (Arachis hypogaea (L.)). Field Crops Res. 193, 16–23. doi:10.1016/j.fcr.2016.03.001
van Dusschoten, D., Metzner, R., Kochs, J., Postma, J. A., Pflugfelder, D., Bühler, J., et al. (2016). Quantitative 3D analysis of plant roots growing in soil using magnetic resonance imaging. Plant Physiol. 170 (3), 1176–1188. doi:10.1104/pp.15.01388
Varshney, R. K., Bertioli, D. J., Moretzsohn, M. C., Vadez, V., Krishnamurthy, L., Aruna, R., et al. (2009). The first SSR-based genetic linkage map for cultivated groundnut (Arachis hypogaea L.). Theor. Appl. Genet. 118 (4), 729–739. doi:10.1007/s00122-008-0933-x
Venkatesh, B., Vennapusa, A. R., Kumar, N. J., Jayamma, N., Reddy, B. M., Johnson, A., et al. (2022). Co-expression of stress-responsive regulatory genes, MuNAC4, MuWRKY3 and MuMYB96 associated with resistant-traits improves drought adaptation in transgenic groundnut (Arachis hypogaea L.) plants. Front. Plant Sci. 13, 1055851. doi:10.3389/fpls.2022.1055851
Vinson, C. C., Mota, A. P. Z., Oliveira, T. N., Guimaraes, L. A., Leal-Bertioli, S. C. M., Williams, T. C. R., et al. (2018). Early responses to dehydration in contrasting wild Arachis species. PLoS One 13 (5), e0198191. doi:10.1371/journal.pone.0198191
Wang, J., Ding, B., Guo, Y., Li, M., Chen, S., Huang, G., et al. (2014). Overexpression of a wheat phospholipase D gene, TaPLDα, enhances tolerance to drought and osmotic stress in Arabidopsis thaliana. Planta 240, 103–115. doi:10.1007/s00425-014-2066-6
Wilkinson, S., and Davies, W. J. (2010). Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant, Cell and Environ. 33 (4), 510–525. doi:10.1111/j.1365-3040.2009.02052.x
Wilkinson, S., Kudoyarova, G. R., Veselov, D. S., Arkhipova, T. N., and Davies, W. J. (2012). Plant hormone interactions: innovative targets for crop breeding and management. J. Exp. Bot. 63 (9), 3499–3509. doi:10.1093/jxb/ers148
Xu, W. H., Yu, G. H., Zare, A., Zurweller, B., Rowland, D., Reyes-Cabrera, J., et al. (2020). Overcoming small minirhizotron datasets using transfer learning. Comput. Electron. Agric. 175, 105466. doi:10.1016/j.compag.2020.105466
Xu, Y., Burgess, P., Zhang, X., and Huang, B. (2016). Enhancing cytokinin synthesis by overexpressing ipt alleviated drought inhibition of root growth through activating ROS-scavenging systems in Agrostis stolonifera. J. Exp. Bot. 67 (6), 1979–1992. doi:10.1093/jxb/erw019
Yang, X., Lu, M., Wang, Y., Wang, Y., Liu, Z., and Chen, S. (2021). Response mechanism of plants to drought stress. Horticulturae 7 (3), 50. doi:10.3390/horticulturae7030050
Yoshida, T., and Fernie, A. R. (2024). Hormonal regulation of plant primary metabolism under drought. J. Exp. Bot. 75 (6), 1714–1725. doi:10.1093/jxb/erad358
Yu, J. (2012). Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 4 (11), 1024–1057. doi:10.3390/toxins4111024
Yuan, C., Li, C., Lu, X., Zhao, X., Yan, C., Wang, J., et al. (2020). Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut. BMC Plant Biol. 20, 454–521. doi:10.1186/s12870-020-02678-9
Yuan, H. B., Bennett, R. S., Wang, N., and Chamberlin, K. D. (2019). Development of a peanut canopy measurement system using a ground-based LiDAR sensor. Front. Plant Sci. 10, 203. doi:10.3389/fpls.2019.00203
Zhang, H., Yu, Y., Wang, S., Yang, J., Ai, X., Zhang, N., et al. (2023). Genome-wide characterization of phospholipase D family genes in allotetraploid peanut and its diploid progenitors revealed their crucial roles in growth and abiotic stress responses. Front. Plant Sci. 14, 1102200. doi:10.3389/fpls.2023.1102200
Zhang, Q., Dang, P., Chen, C., Feng, Y., Batchelor, W., Lamb, M., et al. (2022a). Tolerance to mid-season drought in peanut can be achieved by high water use efficiency or high efficient use of water. Crop Sci. 62 (5), 1948–1966. doi:10.1002/csc2.20806
Zhang, Q., Yuan, W., Wang, Q., Cao, Y., Xu, F., Dodd, I. C., et al. (2022b). ABA regulation of root growth during soil drying and recovery can involve auxin response. Plant, Cell and Environ. 45 (3), 871–883. doi:10.1111/pce.14137
Zhang, T., Hu, S., Yan, C., Li, C., Zhao, X., Wan, S., et al. (2017). Mining, identification and function analysis of microRNAs and target genes in peanut (Arachis hypogaea L.). Plant Physiology Biochem. 111, 85–96. doi:10.1016/j.plaphy.2016.11.018
Zhao, N., He, M., Li, L., Cui, S., Hou, M., Wang, L., et al. (2020). Identification and expression analysis of WRKY gene family under drought stress in peanut (Arachis hypogaea L.). PLoS One 15 (4), e0231396. doi:10.1371/journal.pone.0231396
Zhao, W., Huang, H., Wang, J., Wang, X., Xu, B., Yao, X., et al. (2023). Jasmonic acid enhances osmotic stress responses by MYC2-mediated inhibition of protein phosphatase 2C1 and response regulators 26 transcription factor in tomato. Plant J. 113 (3), 546–561. doi:10.1111/tpj.16067
Zhen, X., Zhang, Q., Sanz-Saez, A., Chen, C. Y., Dang, P. M., and Batchelor, W. D. (2022). Simulating drought tolerance of peanut varieties by maintaining photosynthesis under water deficit. Field Crops Res. 287, 108650. doi:10.1016/j.fcr.2022.108650
Keywords: drought, peanut, physiology, genetics, breeding, tolerance
Citation: Pokhrel S, Kharel P, Pandey S, Botton S, Nugraha GT, Holbrook C and Ozias-Akins P (2025) Understanding the impacts of drought on peanuts (Arachis hypogaea L.): exploring physio-genetic mechanisms to develop drought-resilient peanut cultivars. Front. Genet. 15:1492434. doi: 10.3389/fgene.2024.1492434
Received: 06 September 2024; Accepted: 23 December 2024;
Published: 08 January 2025.
Edited by:
Liwei Zheng, Zhengzhou University, ChinaReviewed by:
Yang Xie, Hebei Normal University of Science and Technology, ChinaCopyright © 2025 Pokhrel, Kharel, Pandey, Botton, Nugraha, Holbrook and Ozias-Akins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peggy Ozias-Akins, cG96aWFzQHVnYS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.