- 1College of Grassland Science, Xinjiang Agricultural University, Urumqi, Xinjiang, China
- 2Institute of Hybrid Wheat, Beijing Academy of Agriculture and Forestry Sciences (BAAFS), Beijing, China
- 3Laboratory of Plant Breeding and Genetics, Department of Agricultural and Environmental Biology, The University of Tokyo, Tokyo, Japan
Phosphorus is indispensable to plant growth and development. Soil phosphorus deficiency poses a substantial constraint on crop yield. SPXs play pivotal roles in phosphate transport and absorption in plants. Yet, the functions of SPXs of oat (Avena sativa L.) under abiotic stresses remain unclear. In this study, we conducted a genome-wide analysis of 169 SPXs from hexaploid oat and five closely related plant species. All homologous AsSPXs were found to arise from duplication events and depict a strong purifying selection. Subcellular localization prediction revealed that AsSPXs were mainly located on the plasma membrane. Seventeen cis-acting elements, predominantly comprising light-, low temperature-, abscisic acid-, and drought-responsive elements, were dispersed in the promoter regions of AsSPXs. Analysis of cis-regulatory elements, protein-protein interaction networks, and qRT-PCR showed that AsSPXs are not solely involved in phosphorus starvation response but also in various stress responses. Notably, AsSPX18-5D (AVESA.00001b.r3.5Dg0002895) exerted pivotal roles in conferring resistance against low phosphorus, salt, and ABA treatments. Our study aimed to explore important stress-resistant genes in oat. Our results could provide a basis for future studies on the evolution and functions of the AsSPX gene family and a crucial foundation for comprehending how oat responds to environmental stresses.
1 Introduction
Global climate changes, such as soil salinization, drought, extreme temperatures, etc., have worsened over the years (Borrelli et al., 2017; He et al., 2023), leading to a decline in the yield and quality of crops. Thus, finding new ways to improve the yield of forage crops under adverse environmental conditions has become a primary concern worldwide. Oat (Avena sativa L.) is an annual herbaceous plant belonging to the Poaceae family. It exhibits remarkable adaptability, thriving in various adverse conditions, such as impoverished soil, saline-alkali, drought, and severe cold stresses. Therefore, oat has become an important genetic resource for studying plant adaptations to harsh environments (Dangi, 2020). It also serves as an ecological forage grass, playing an important role in soil salinization and desertification control, supporting the advancement of the grassland livestock industry (Dangi, 2020). Understanding the response mechanisms of plants to different stresses can help develop practical interventions to improve crop yield.
The SPX proteins, phosphorus receptors, play a crucial regulatory role in plant growth and development. An SPX protein derives its name from the initials of three proteins: Suppressor of yeast gpa 1 (SYG1), phosphate-regulated cyclin-dependent kinase inhibitor 81 (Pho81), and xenotropic and polytropic retrovirus receptor 1 (XPR1) (Wang et al., 2004). SPXs are characterized by the presence of a conserved SPX domain, which exhibits hydrophobic properties, is situated at the N-terminal, and is approximately 165 amino acids (aa) long (Chiou and Lin, 2011; Secco, et al., 2012b). The SPX protein family is divided into four distinct subfamilies: SPX proteins, which contain only the SPX domain and primarily are regulators of plant phosphate starvation (Zhang et al., 2016); SPX-EXS proteins, which contain both the SPX and EXS (deriving its name from the initials of endoplasmic reticulum retention defective 1 (ERD1), XPR1, and SYG1) domains and play an important role in phosphate acquisition, translocation, and assignment (Wang et al., 2004; Zhao et al., 2019); SPX-MFS proteins, which contain the SPX domain bound to the major facilitator superfamily (MFS) domain and are a newly discovered class of vacuolar Pi transporters in plants (Wang C. et al., 2012); and the SPX-RING proteins, which contain the SPX and really interesting new gene (RING)-type zinc finger domains and are involved in phosphate homeostasis and inorganic phosphate (Pi) reaction (Yang et al., 2017; Yue et al., 2017).
Numerous studies have shown that SPX proteins play an important role in the absorption, transport, storage, and signal transduction of Pi in plants. For instance, in Arabidopsis thaliana, AtSPX1/3 (AtSPX1/3), AtSPX2, and AtSPX4 have been shown to be strongly induced, weakly induced, and inhibited, respectively, under low-phosphorus stress conditions (Duan et al., 2008; Secco, et al., 2012a). In rice (Oryza sativa), OsSPX1/2/3/5/6 are induced by phosphorus deficiency in the aerial parts of the plant and the roots. When these genes are simultaneously overexpressed with rice phosphate starvation response 2 (OsPHR2), the phosphorus accumulation phenotype can be restored to wild-type levels (Lv et al., 2014; Shi et al., 2014; Wang et al., 2009; Wang et al., 2014; Zhong et al., 2018). OsSPX1 and OsSPX2 interact with OsPHR2 via their SPX domains and regulate the binding of OsPHR2 to the PHR1-binding sequence (P1BS; GNATATNC). The functional redundancy of this process is phosphorus-dependent, and this process inhibits the phosphate starvation response (Wang et al., 2014). Similarly, in wheat (Triticum aestivum), TaSPX1/5 have been found to interact with TaPHR3 (Sun R. et al., 2023). In addition, nitrogen signaling also plays an important role in regulating the accumulation of SPX proteins. A previous study reported that under high nitrogen conditions, nitrate transporter 1.1 (NRT1.1), the nitrate sensor in rice and Arabidopsis, binds to SPX4 and recruits NBIP1, an E3 ubiquitin ligase, to degrade it. Furthermore, high nitrogen induces nitrate-inducible GARP-type transcriptional repressor 1 (NIGT1), and the NIGT1 protein, in turn, inhibits the expression of SPXs and promotes phosphorus absorption, mediating nitrogen-phosphorus balance in plants (Hu and Chu, 2020; 2019; Kiba et al., 2018; Ueda et al., 2020).
In a saline environment, there can be synergy or antagonism between the salt and nutrient levels in plant roots, resulting in imbalances of several nutrients, such as phosphorus and nitrogen, affecting plant growth and development and accelerating plant aging (Ahmad et al., 2013; Wang et al., 2021). In cotton, Cot-AD50458 encodes SPX1-like protein, and under salt stress, this gene is significantly downregulated, inhibiting phosphorus absorption and accelerating plant aging (Wang et al., 2021). Several phosphate starvation-inducing genes are expressed in senescent leaves of A. thaliana. For instance, AtSPX1 overexpression accelerates leaf senescence, suppresses Pi accumulation, promotes salicylic acid (SA) production, and enhances H2O2 levels in A. thaliana leaves (Yan et al., 2019). Oat exhibits a strong ability to adapt to harsh environments. A previous study showed that drought-resistant oat varieties exhibited elevated abscisic acid (ABA) levels and reduced stomatal conductance, resulting in lower transpiration rates and greater drought resistance (Canales et al., 2021). Another study showed that the overexpression of salt-induced CBF3 in oats can delay leaf aging, reduce the loss of yield, and enhance tolerance to abiotic stresses (Oraby and Ahmad, 2012). OsSPX1 is an important link between the signaling pathways related to low temperature and Pi starvation stresses. OsSPX1 overexpression has been shown to enhance cold tolerance and reduce the total phosphorus content in the leaves of tobacco and A. thaliana (Zhao et al., 2009). Conversely, OsSPX1 downregulation increases the sensitivity of rice seedlings to both cold and oxidative stresses (Wang et al., 2013).
Members of the SPXs have been found across several plant species, including A. thaliana (Duan et al., 2008), rice (Wang et al., 2009), wheat (Kumar et al., 2019), corn (Xiao et al., 2021), tomato (Li et al., 2021), Brassica napus (Du H et al., 2017), giant duckweed (Yang et al., 2022), moso bamboo, and tea oil camellia (Chen et al., 2023; Luo et al., 2023). As core regulatory proteins in phosphorus signaling pathways, SPX proteins have been extensively studied in nitrogen-phosphorus signaling (Hu et al., 2019). The release of high-quality reference genomes for heterohexaploid oat variety “Sanfensan” (A. sativa ssp. nuda, AACCDD, 2n = 6x = 42) and oat “sang” (A. sativa L., AACCDD, 2n = 6x = 42), and their diploid (A. longiglumis, AA, 2n = 14) and tetraploid ancestors (A. insularis, CCDD, 2n = 4x = 28) has accelerated the process of genome-assisted oat breeding (Kamal et al., 2022). However, to the best of our knowledge, genome-wide identification and characterization of members of the SPX gene family in oats have not yet been reported. This study aimed to perform a comprehensive identification and functional analysis of the SPX gene family in oats.
2 Materials and methods
2.1 Identification of SPX genes
The EnsemblePlants database (http://plants.ensembl.org/index.html) was accessed to obtain the genomic data of a dicot (Arabidopsis) and several monocots (oats, barley, rice, wheat, and maize). Firstly, we conducted BLASTP of 20 AtSPXs and 15 OsSPXs. The Hidden Markov Model (HMM) SPX domain (PF03105) in the Pfam database (http://pfam.xfam.org/) were used to search for AsSPXs genes. Then, all the obtained sequences were further verified using the Pfam and SMART (http://smart.embl-heidelberg.de) to determine if they contained SPX domain with the threshold of E < 1e-5. All the gene loci were numbered and mapped onto their corresponding chromosomes. For the polyploid oats, the gene loci were numbered based on the order of A/C/D subgroups.
2.2 Physiochemical analysis and subcellular localization prediction
The ProtParam (http://web.expasy.org/protparam/) program of Expert Protein Analysis System (ExPASy) was used to predict the molecular weights (MWs) and isoelectric points (PIs) of all AsSPX proteins (Garg et al., 2016). The subcellular localization prediction of the oat SPX proteins was predicted using the Bologna Unified Subcellular Component Annotator (BUSCA; http://busca.biocomp.unibo.it) (Savojardo et al., 2018).
2.3 Evolutionary analysis
To determine the evolutionary relationship among the SPX proteins, they were assessed using the Molecular Evolutionary Genetics Analysis 7 (MEGA7) software to construct phylogenetic trees using the neighbor-joining method with 1,000 bootstrap replications (Kumar et al., 2018). Multiple Collinearity Scan toolkit X (MCScan X) was used for collinearity analysis among wheat, Arabidopsis, rice, and oats to identify gene duplication events (Wang Y. et al., 2012). The syntenic relationships were determined using TBtools. The Ka/Ks ratios of collinear gene pairs were calculated using the coding sequences (CDS) and protein sequences. The divergence time was calculated using the following formula: T = Ks/2λ × 10–6 Mya (λ = 6.5 × 10−9) (Gaut et al., 1996).
2.4 Protein-protein interaction (PPI) network analysis
The conserved motifs and domains in the SPX proteins were identified using the Multiple Em for Motif Elicitation (MEME) (https://meme-suite.org/) and conserved domain (CD) searches (https://www.ncbi.nlm.nih.gov/). The potential cis-acting elements were analyzed via the PlantCARE (https://bioinformatics.psb.ugent.be/web--tools/plantcare/html/) database using the 2 kb region upstream of the start codon in all the AsSPXs (Lescot et al., 2002). Finally, the integrated results were visualized using the Gene Structure View function of TBtools. The PPI network of 42 AsSPX protein sequences was constructed using the STRING database. Due to the lack of a protein interaction database of oats, we analyzed the homologous SPX proteins in wheat to predict the interactions among AsSPXs. The Cytoscape software was used to visualize the PPI networks.
2.5 Plant materials and growth conditions
Hexaploid oat plants were used in this study, courtesy of Professor Zhao Guiqin from Gansu Agricultural University. The oat seeds were disinfected using 75% alcohol for 5 min and germinated for 5 days in the growth chamber. The germination conditions included 50% relative humidity and 14-h day/10-h night cycles with temperatures of 24°C/18°C, respectively. The germinated seeds were then cultured in a modified Hogland solution (NS10205-100 mL, Coolaber) for 15 days. Finally, the seedlings were transferred to the following six Hoagland solutions for 24 h for Pi, salt, drought, ABA, cold and heat stress: 0 mol/L Pi (NSP1020-P-500L, Coolaber), 0.2 mol/L sodium chloride (NaCl, Hushi), 20% polyethylene glycol 6,000 (PEG6000, Solarbio), 30 μmol/L abscisic acid (ABA, Sigam), 4°C and 40°C. Hoagland’s solution without any treatment was used as a control (CK). The shoots/leaves and roots of the different groups of seedlings were collected at 0, 1, 3, 6, 9, 12, and 24 h, and immediately frozen in liquid nitrogen and stored in a −80°C freezer.
2.6 RNA isolation, cDNA preparation, and quantitative real-time polymerase chain reaction (qPCR)
Total RNA was extracted from plant extracts using a FastPure® Universal Plant Total RNA Isolation Kit (Vazyme, Nanjing, China) as per the manufacturer’s instructions. First-strand cDNA synthesis was performed using a HiScript® III 1st Strand cDNA Synthesis Kit (+gDNA wiper) with gDNA Eraser (Vazyme, Nanjing, China). qRT-PCR was conducted using a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, United States) with HiScript® III All-in-one RT SuperMix Perfect for qRT-PCR (Vazyme, Nanjing, China). Primers were designed using the Primer3plus online tools (http://www.primer3plus.com/) (Supplementary Table S1). Expression levels of target genes were normalized against oat 18S RNA. The relative expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
3 Results
3.1 Identification and analysis of AsSPXs
We identified a total of 42 AsSPXs via conserved domain analysis and sequence alignment. According to the chromosome where each gene is located on the chromosome, numbered AsSPX1-2A ∼ AsSPX42-7D (Figures 1, 2; Supplementary Table S2). The lengths and molecular weights (MWs) of the identified AsSPXs were in the range of 131–854 aa and 14,709.15∼97,379.45, with the longest and bulkiest variant being AsSPX40-7D, and the shortest and least bulky being AsSPX25-6A. The isoelectric points (PIs) of the identified AsSPXs ranged from 5.27 to 9.27, with an average PI of 7.52, with acidic, basic, and neutral aa accounting for 42.86%, 40.48%, and 16.67%, respectively. The substantial variations in the lengths and properties of AsSPXs proteins indicated that their acidic, alkalinity properties and non conservatism. Subcellular localization prediction revealed that most AsSPXs are mainly located on the plasma membrane, followed by the cytoplasm and nucleus, and a few were located in the mitochondria. These findings suggested that AsSPX family members exhibit multiple biological functions and redundancy mechanisms.
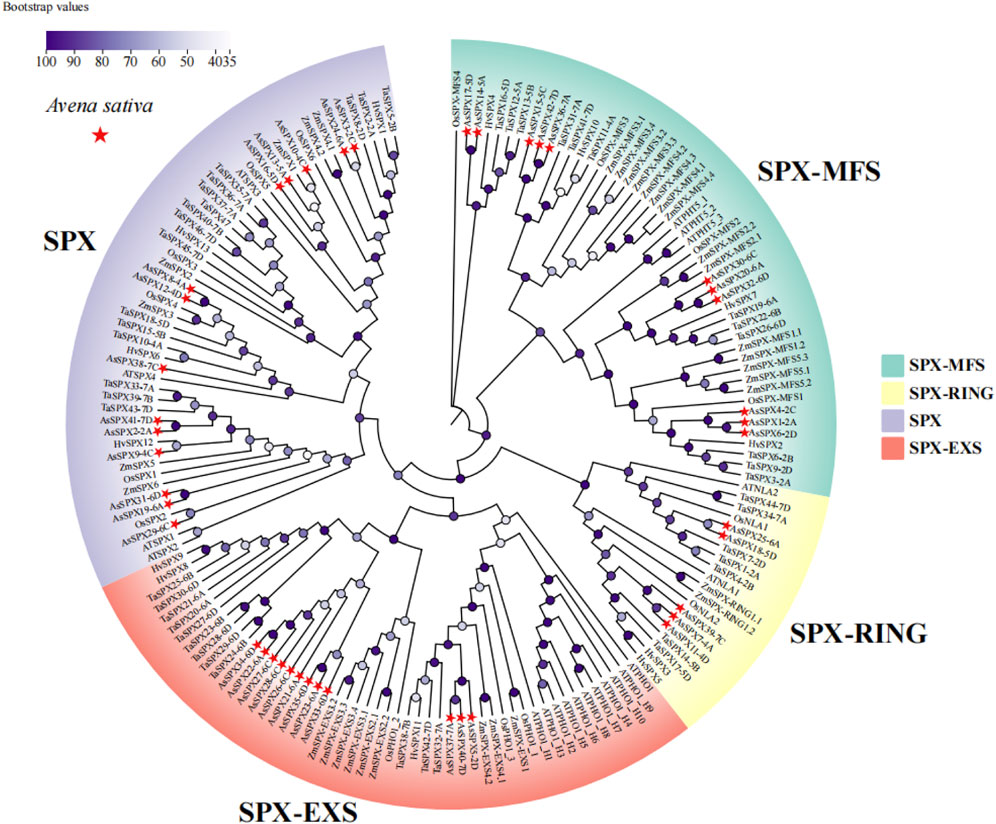
Figure 1. Phylogeny of neighborhood junctions of 169 SPXs from six plant species, Avena sativa, Arabidopsis thaliana, Hordeum vulgare, Oryza sativa, Triticum aestivum, Zea mays (n = 42, 20, 13, 15, 46, and 33 SPXs, respectively). AsSPXs were divided into four subfamilies based on their domains: SPX, SPX-EXS, SPX-MFS, and SPX-RING (comprising 14, 12, 11, and five AsSPXs, respectively). AsSPXs are represented by a red triangle.
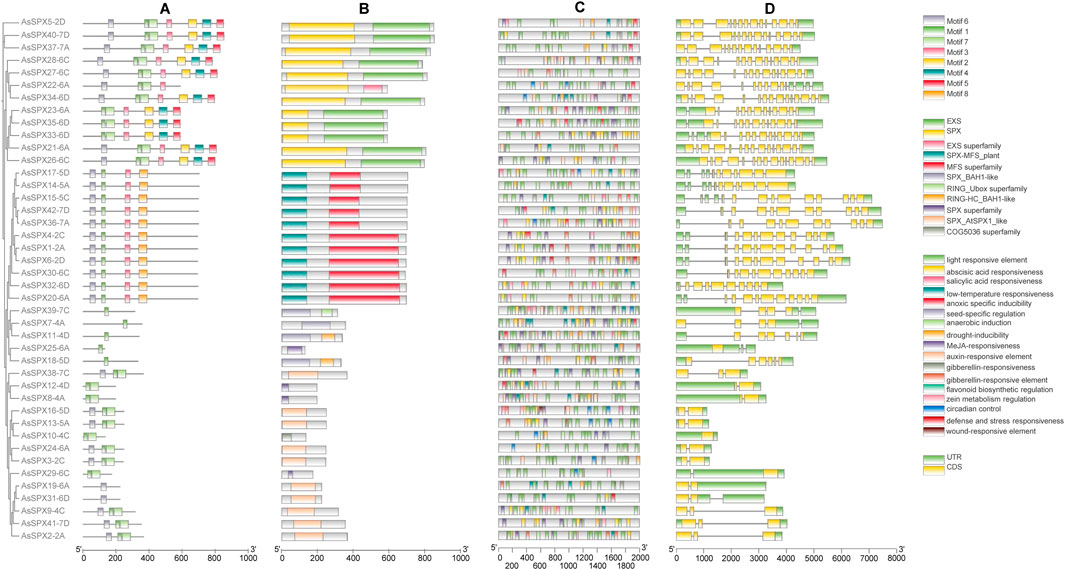
Figure 2. Analysis of phylogeny, motifs, cis-elements in promoters, and gene structures of all 42 AsSPXs. (A) Protein motifs, (B) Protein domains, (C) Cis-elements in the promoters, (D) Gene structures.
3.2 Phylogenetic analysis of the SPXs from oat and other species
To elucidate the evolutionary relationships among the SPXs across different plant species, we assessed a total of 169 SPXs from A. sativa, A. thaliana, barley (Hordeum vulgare), rice, wheat, and maize (Zea mays) (n = 42 AsSPXs, 20 AtSPXs, 13 HvSPXs, 15 OsSPXs, 46 TaSPXs, and 33 ZmSPXs, respectively). Using the neighbor-joining method, we constructed a phylogenetic tree of all SPXs (Figure 1). AsSPXs were divided into four groups based on their domain sequences, SPX, SPX-EXS, SPX-MFS, and SPX-RING subfamilies, comprising 14, 12, 11, and five members, respectively (Figures 1, 2). Similar to oats, most TaSPXs, HvSPXs, and OsSPXs belonged to the SPX group, followed by SPX-EXS, SPX-MFS, and SPX-RING subfamilies. Therefore, the SPXs of these plants harbored conserved SPX domains, thus functioning as regulators of plant phosphate starvation. In contrast, fifteen, nine, seven, and two ZmSPXs were categorized into the SPX-MFS, SPX, SPX-EXS, and SPX-RING subfamilies, respectively. Furthermore, most AtSPXs belonged to the SPX-EXS group (55%), and the fewest belonged to the SPX-RING subfamily (10%). Thus, for all species, the SPX regions were relatively scattered, while the SPX-EXS and SPX-MFS regions were relatively clustered. Since oat, wheat, and barley belong to the Poaceae family, oat is evolutionarily closer to wheat and barley, indicating that their SPX gene families are highly conserved and can be used to assess the evolution and biological functions of the SPX genes.
3.3 Gene structure and conserved motif analysis of AsSPXs
To identify commonly occurring motifs in SPXs, we predicted the composition of the 42 AsSPXs using the MEME tool and identified eight distinct motifs (Figure 2; Supplementary Table S3). Notably, motif 1 was present in all SPXs except for AsSPX19-6A and AsSPX31-6D, implying its potential role in their shared functionality. The SPX-RING group only possessed motif 1. The SPX-MFS subfamily contained motifs 1, 3, 6, and 8; however, motif 8 was solely found in the SPX-MFS subfamily. Motifs 2 to 5 were exclusively present in the SPX-EXS subfamily, with the group members harboring one to seven motifs. These findings indicated that proteins within the same subfamily exhibited similar distribution patterns of motifs. In addition, specific regulatory motifs in the members of each group might contribute to their distinct functions observed across the subfamilies. To comprehensively understand the evolutionary relationships and functional diversity among AsSPXs, we analyzed the intron-exon boundaries within their sequences. We observed that all the genes harbored introns except for the gene encoding AsSPX10-4C. Additionally, 10 genes, such as AsSPX37-7A, AsSPX27-6C, and AsSPX22-6A, lack 5′ introns. This absence is hypothesized to be the result of selective pressures during evolution, where efficient expression for specific functions and increased protein production may have led to the loss of 5′ introns. Furthermore, the SPX-EXS, SPX-MFS, SPX-RING, and SPX group members contained 12–14, 9–13, 2–6, and two introns, respectively. The same subfamily performs similar functions and therefore has similar structures, such as the AsSPX2-2A and AsSPX9-4C genes, which belong to the same SPX subfamily. These results indicated the conservation of gene structure within each SPX subfamily along with the presence of complex structures, implying other potential functionalities associated with the SPX genes.
3.4 Analysis of the cis-acting elements of AsSPXs
Cis-regulatory elements play a crucial role in gene expression regulation. Analysis of the sequences 2000 bp upstream of each AsSPX gene revealed a total of 17 predicted cis-acting elements, providing insights into the regulatory elements of the AsSPX gene family. (Figure 2; Supplementary Table S4). They were categorized into three main groups based on whether they were related to hormone response (n = 5; responsive to ABA, SA, methyl jasmonate (MeJA), auxin, and gibberellin), abiotic stress response (n = 6; responsive to low-temperature, anoxic conditions, anaerobic, drought, defense reactions, and wound), and plant development (n = 4; responsive to seed development, flavonoid biosynthesis, zein metabolism, and circadian control). Similar to the members of the TaPHR (Kumar et al., 2019), ZmSPX (Xiao et al., 2021), and SlPEBP (PEBP gene family of tomato (Solanum lycopersicon)) (Sun Y. et al., 2023) gene families, light-responsive elements were significantly enriched in the promoters of all AsSPXs suggesting their potential involvement in the same signaling pathway. Furthermore, low-temperature-responsive elements were identified in 22 genes, suggesting the crucial role of SPXs as a connecting link between cryogenic stress and Pi starvation signaling pathways (Wang et al., 2013; Zhao et al., 2009). ABA-, MeJA-, and drought-responsive elements were present in 39, 33, and 29 genes, respectively. Though several SPXs are known to be induced by Pi starvation, the presence of P1BS (a conserved Pi stress-responsive element) in SPXs of oat or apple (Malus pumila) has not yet been reported (Kural et al., 2024).
3.5 Collinearity analysis of the gene duplication events in AsSPXs
Genomic collinearity segments within the oat genome were comprehensively analyzed to infer gene replication events during the evolution of the AsSPX protein family. All AsSPXs are mainly derived from genome-wide replication (whole genome duplication; WGD) or fragment replication, followed by tandem duplication (Supplementary Table S5). The 42 identified AsSPXs were distributed across 14 oat chromosomes, with each chromosome harboring a varying number of AsSPXs (Figure 3). Among them, the sixth homologous group harbored the most genes (n = 17). The first and third homologous groups exhibited an uneven gene distribution; however, the second, fourth, fifth, and seventh homologous groups exhibited a relatively stable distribution with 6-7 genes in each group. We detected a total of 35 collinear gene pairs in the oat genome. Furthermore, AsSPXs located on different oat chromosomes exhibited collinearity. This finding suggested that most AsSPXs might be alleles or paralogous genes on homologous chromosomes and have undergone genetic expansion during their evolution. For example, AsSPX1-2A exhibited collinear relationships with AsSPX4-2C, AsSPX6-2D, AsSPX20-6A, AsSPX30-6C, and AsSPX32-6D. Additionally, we observed collinearity between 4C (AsSPX9-4C) and 2A (AsSPX2-2A). These findings suggested that AsSPXs might have evolved from gene replication processes during evolution, with the encoded proteins influenced by varying degrees of truncated duplication events.
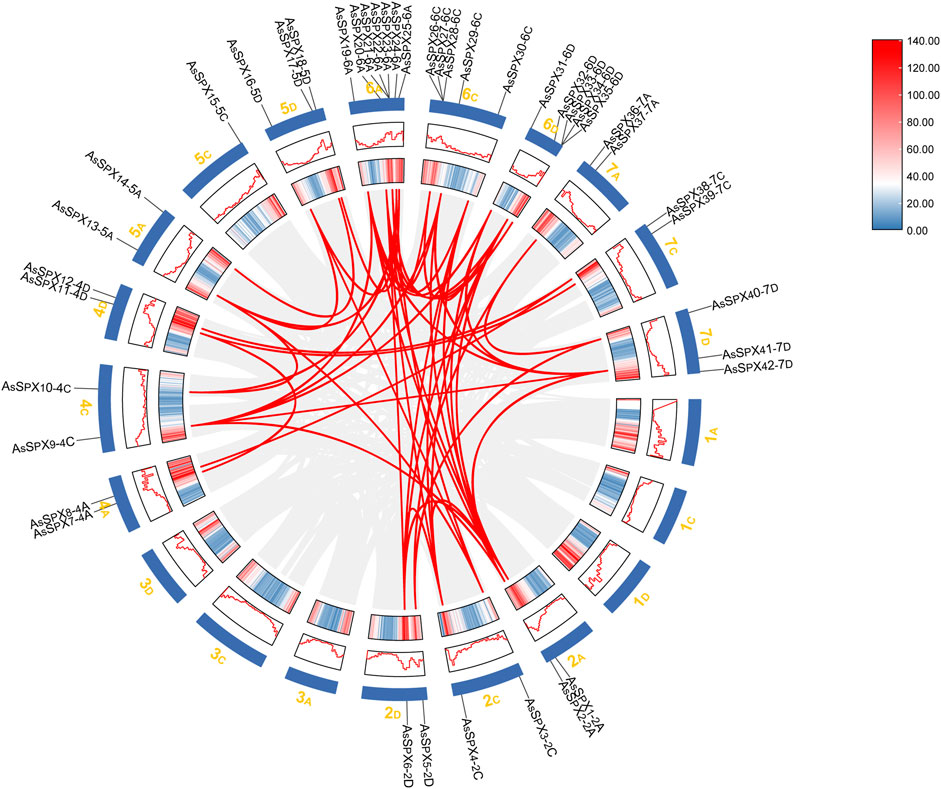
Figure 3. Collinearity analysis of the SPX gene family in oat. The chromosomal distribution of 42 AsSPXs is depicted. Chromosome size and gene density plots are shown in the Circos plot, represented by different shades as plots and heat maps. The collinear relationships among the genes were scanned with MCScanX and connected with red lines.
The inter-specific collinearity analysis revealed the presence of 41 and 79 pairs of homologous genes between hexaploid oat and diploid A. strigosa and hexaploid oat and tetraploid A. insularis, respectively. If a gene in hexaploid oats lacks colinearity with its diploid counterpart, it similarly will not be collinear with the corresponding gene in tetraploid oats (Figure 4A; Supplementary Table S6). There are 93 homologous gene pairs between oat and wheat (Figure 4B). More specifically, orthologous gene pairs were found between the sixth and second homologous groups in oat and the second, sixth, and seventh homologous groups in wheat. Additionally, 15, nine, and eight orthologous gene pairs were detected between the fifth, seventh, and fourth homologous groups in oat and wheat. Notably, no collinearity was observed between the first and third homologous groups in oat and wheat. Furthermore, 44 homologous gene pairs were identified between oats and rice. Similar to the previous studies, we found no collinear relationships between AtSPXs and TaSPXs (Supplementary Figure S1) (Sun R. et al., 2023). Oat is are monocots, while A. thaliana is dicots, and they differ significantly in morphology, structure, and developmental mode, which may be the reason for the lack of collinearity between them. To further evaluate selective pressure on genes associated with these duplication events, the Ka/Ks ratio for homologous SPX pairs was calculated (Supplementary Table S7). The divergence time for AsSPX gene pairs was estimated at ∼35.58 million years ago (MYA). Additionally, all gene pairs had a Ka/Ks < 1, with an average value of 0.25, suggesting strong purification selection upon AsSPX gene pairs.
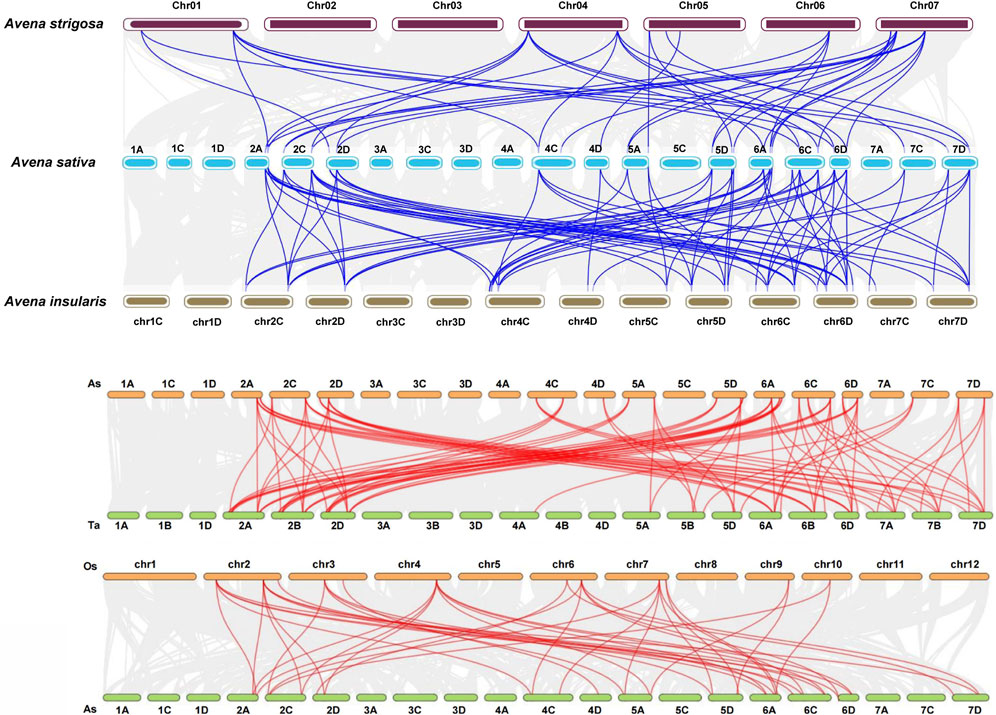
Figure 4. Collinearity analysis between oat and other plant species. (A) There were 41 and 79 pairs of homologous genes between hexaploid oat and Avena strigosa and hexaploid oat and Avena insularis, respectively. (B) There were 93 and 44 pairs of homologous genes between hexaploid oat and wheat and hexaploid oat and rice, respectively.
3.6 Protein-protein interaction (PPI) networks of AsSPXs
Previous studies have demonstrated that SPX4 degradation E3 ligase 1 (SDEL1) and SDEL2 act as ring-fingered ubiquitin E3 ligases that regulate OsSPX4 degradation for phosphate homeostasis and signal transduction (Ruan et al., 2019). Protein ubiquitination has also been reported within the PPI network of ZmSPX (Xiao et al., 2021). Furthermore, AtSPX3 regulates the initiation of embryonic stem apical meristem (SAM) development by interacting with AtYABBY3, suggesting that SPX3 is involved in cell cycle regulation (Stahle et al., 2009). OsSPX1 overexpression can enhance plant cold tolerance (Wang et al., 2013; Zhao et al., 2009). In the present study, PPI networks were used to characterize the cellular function of AsSPX. In our PPI network, seven core interacting proteins were identified: AsSPX5-2D, AsSPX10-4C, AsSPX13-5A, AsSPX18-5D, AsSPX31-6D, AsSPX35-6D, and AsSPX39-7C (Figure 5). These proteins are primarily involved in crucial biological processes, including the regulation of the phosphorus signaling pathway, protein ubiquitination, histone H3-K4 trimethylation and nucleic acid binding, response to external stimuli such as low-temperature reactions, energy and substance metabolisms such as benzoic acid and salicylic acid metabolism, and cell cycle regulation. These findings indicated that, in addition to phosphorus starvation response, these seven AsSPXs are also engaged in various stress responses.
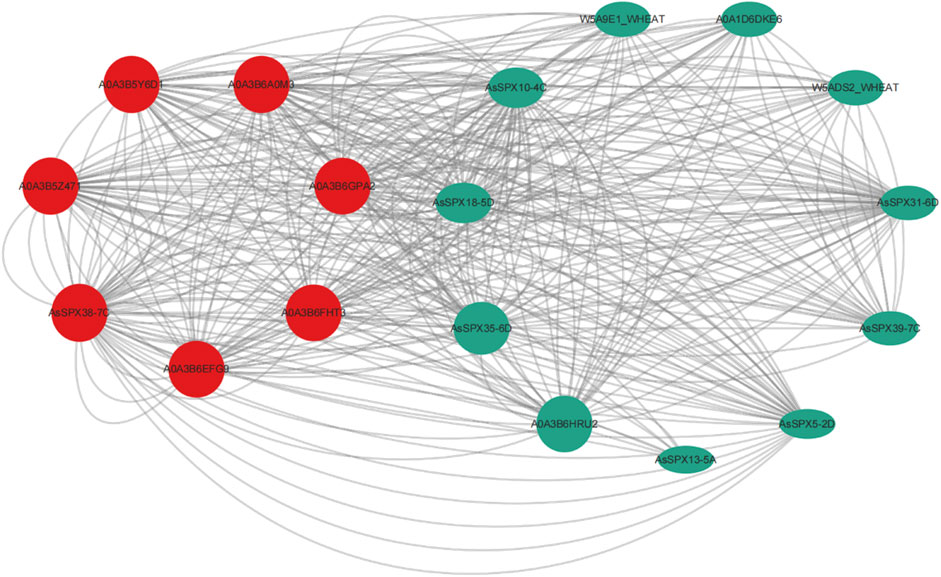
Figure 5. Predicted protein-protein interaction network of AsSPXs. AsSPX5-2D, AsSPX10-4C, AsSPX13-5A, AsSPX18-5D, AsSPX31-6D, AsSPX35-6D, and AsSPX39-7C reference proteins were named based on BLAST results.
3.7 Structure prediction of AsSPXs
Through bioinformatics analysis and protein structure modeling software (Figure 6), we observed that the AsSPXs typically included a series of alpha helices and beta folds, which might play essential roles in protein function. The amphiphilic characteristics of alpha-helices and the diverse structures of beta-sheets endow proteins with a rich array of forms, enabling these four genes to adapt to a variety of environmental stresses (Sun et al., 2004). Concurrently, the complexity of alpha-helices and beta-sheets within these genes correlates with the number of exons they possess (Figure 2). A detailed analysis of the alpha helices revealed specific folding patterns that are potentially associated with the phosphate-sensing ability and signal transduction of AsSPXs. Furthermore, we performed a comparative analysis with homologous SPXs in wheat (Kumar et al., 2019), revealing a certain degree of similarity between the sequences of TaSPXs and AsSPXs. These findings indicated the potential involvement of TaSPXs in phosphate metabolism and signal transduction pathways. These findings are crucial to further elucidating the mechanism of action of SPXs and their role in crop growth and development.
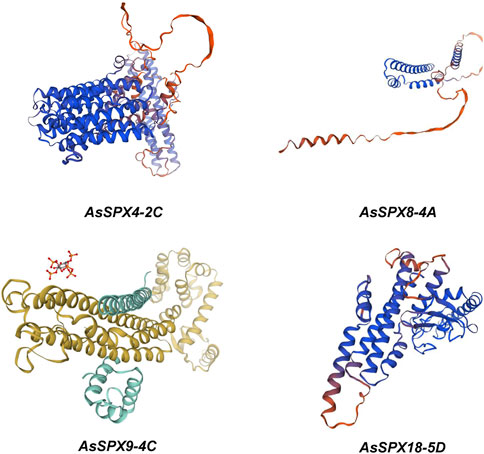
Figure 6. Homology modeling of phosphate starvation response (PHR) proteins encoded by four oat genes, AsSPX4-2C, AsSPX8-4A, AsSPX9-4C, and AsSPX18-5D. The program is generated through ExPaSy (https://swissmodel.expasy.org/interactive).
3.8 Expression of AsSPXs under abiotic stresses
Oat demonstrates a strong ability to adapt to challenging environments. As a gene encoding Pi-responsive protein, the SPX gene plays a crucial role in regulating Pi stress responses. Previously, OsSPX1 has been shown to be involved in low-temperature stress and phosphate deficiency, and OsSPX1 overexpression enhances plant cold tolerance (Wang et al., 2013; Zhao et al., 2009). In Arabidopsis, AtSPX1 acts as a key regulator of the pathways related to leaf senescence, phosphate starvation, and salicylic acid (SA) signaling (Yan et al., 2019). Through homologous evolution analysis, we identified functionally characterized counterparts of SPX genes and examined their response abilities under stress conditions, uncovering potential stress-resistance-related functional genes.
In the present study, we used qRT-PCR to analyze the expression of four AsSPXs under six distinct stress conditions. Compared with the control group (CK), AsSPX4-2C was significantly downregulated in the leaves under salt, drought, cold, and ABA stresses. However, it was induced under heat stress. In the root tissue, AsSPX4-2C was downregulated under drought and heat stresses. At 6 h, it was significantly suppressed in the root under salt and cold stresses.
In leaves and roots, AsSPX8-4A was substantially induced under salt and ABA stresses but was repressed under drought and heat stresses. Furthermore, under cold stress, the gene was generally upregulated except for the decreases at 3 h in the leaves and at 3 and 6 h in the roots.
AsSPX9-4C expression was generally low, but this gene was significantly upregulated in the root tissue under drought stress at 12 h. AsSPX18-5D was considerably upregulated in the leaves and roots at 1, 3, and 9 h under ABA stress and mildly upregulated at 9 h under salt stress. Under heat stress, AsSPX4-2C was upregulated in the leaf tissue, while the other three genes were markedly downregulated in both leaf and root tissues. AsSPX9-4C and AsSPX18-5D were also downregulated under cold stress (Figure 7).
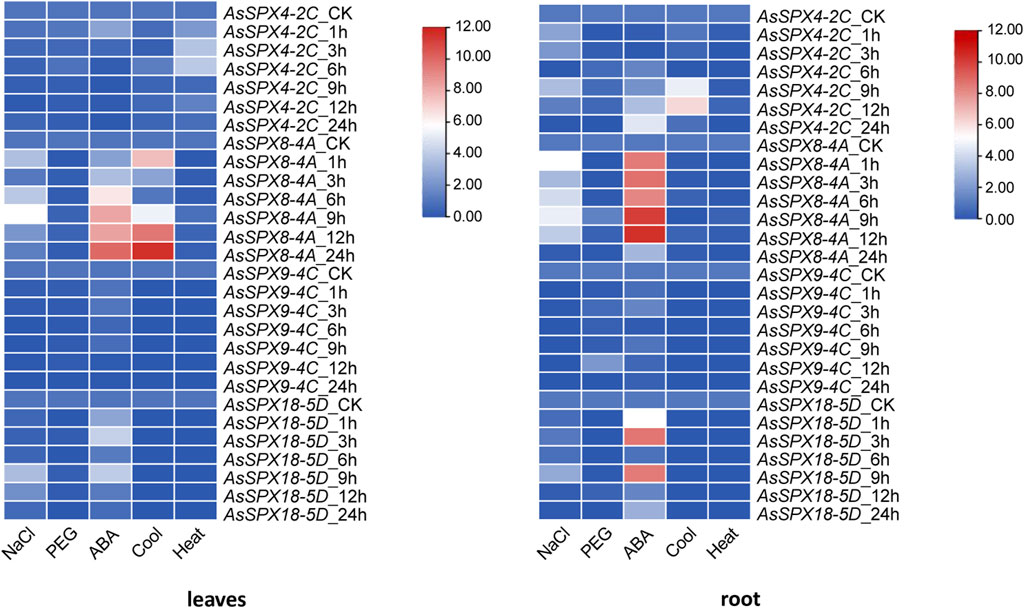
Figure 7. Expression heat map derived from the leaves and roots of oat plants subjected to five types of stresses. The expressions of four AsSPXs were analyzed after treatment with 0.2 mol/L NaCl, 20% polyethylene glycol 600 (PEG6000), 30 μmol/L abscisic acid (ABA), 4°C, and 40°C. Shoots/leaves and roots of the plants were collected at 0, 1, 3, 6, 9, 12, and 24 h post-treatment. In the heat map, the intensity of the red color positively correlated with the gene expression.
In the leaf tissues subjected to Pi stress, AsSPX4-2C was upregulated, with the expression increasing with time. In addition, AsSPX8-4A was strongly induced, especially at 12 h, while AsSPX9-4C and AsSPX18-5D were inhibited. Notably, AsSPX9-4C was also significantly induced at 12 h, suggesting potential functional similarities between AsSPX9-4C and AsSPX8-4A. Furthermore, since both these genes are located on homologous chromosomes, they might be involved in similar pathways.
In the root tissues subjected to Pi stress, the expressions of all four genes were generally high. Notably, AsSPX9-4C expression levels positively correlated with time. The remaining three genes were significantly upregulated at both 6 and 24 h, but their expressions reduced to normal levels at 12 h. This observation suggested the potential involvement of these three genes in similar pathways within the roots (Figure 8). Overall, these findings indicated that SPX plays a crucial role in response to various stresses, including Pi, drought, salt, heat, and cold stresses.
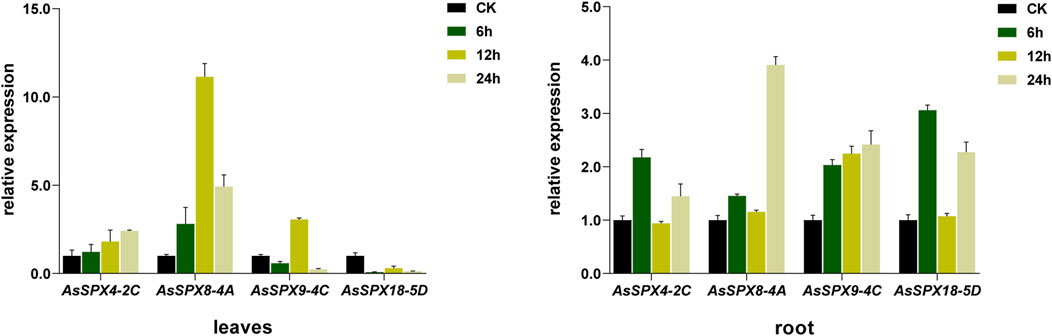
Figure 8. Quantitative real-time PCR of four AsSPXs isolated from plants subjected to inorganic phosphate (Pi) stress. The shoots/leaves and roots of the plants were collected at 0, 6, 12, and 24 h post-treatment. Different colors represent different sampling times.
4 Discussion
4.1 Comparison of the characteristics of the SPX genes from oats and other species
As a high-quality grain and feed crop, oat exhibits resistance to extreme environments, such as salinity, drought, and cold stresses. In the past few decades, effective stress-resistance genes have been continuously excavated from different germplasm resources. Oat is also a high-quality forage and crop germplasm resource, playing an important role in improving the environment (Dangi, 2020; Paudel et al., 2021; Varma et al., 2016). In Northwest China, stresses such as drought, extreme heat, and low phosphorus severely impact plant growth and development (Borrelli et al., 2017; He et al., 2023). Phosphorus promotes seed germination and root growth, regulates nutrient uptake by plants (Ueda et al., 2020), and enhances their ability to cope with environmental stresses (Wang et al., 2013; Zhao et al., 2009). Under phosphorus-deficient conditions, plant growth is slow, root growth is stunted, flowering and fruiting are delayed, and seed setting rate is low. Molecular and genomic studies have shown that many SPX gene family members are involved in Pi signaling and homeostasis (Lv et al., 2014; Shi et al., 2014; Sun R. et al., 2023; Wang et al., 2009; 2014; Zhong et al., 2018). In the current study, the structural analysis of the 42 AsSPXs revealed the presence of eight different motifs, 11 protein compositions, and intron and exon distributions across all AsSPX subfamilies (Figure 2). We found that proteins in the same subfamily exhibited similar distribution patterns, suggesting that they might be conserved and perform similar functions. On the contrary, the proteins in different subfamilies exhibited varying structures, suggesting different functions. These variations might be attributed to the evolution of the SPX proteins, resulting in a high degree of conservation and diversity. Subcellular localization prediction (Supplementary Table S2) showed that AsSPXs are mainly located on the plasma membrane, followed by cytoplasm, nucleus, and mitochondria, suggesting that these proteins exhibit varying functions. Similar distribution of SPXs has been reported in wheat (Kumar et al., 2019), soybean (Nezamivand-Chegini et al., 2021), A. thaliana, and rice (Duan et al., 2008; Duan et al., 2008; Wang et al., 2009). However, the SPXs of maize (Xiao et al., 2021) exhibit a varied distribution on the nucleus and chloroplast. These findings suggested that the members of the SPX protein family also exhibit varying functions across different species. Phylogenetic tree analysis (Figures 1, 2) showed that oat is evolutionarily closer to wheat, followed by barley. Similar to oat, most of the SPXs of wheat, barley, and rice belonged to the SPX group, followed by the SPX-EXS, SPX-MFS, and SPX-RING subfamilies. These results indicated that SPXs primarily function as regulators of phosphate starvation in plants (Zhang et al., 2016). However, in maize, most ZmSPXs primarily belonged to the SPX-MFS subfamily. Previous studies have shown that ZmSPXs mainly act as vacuolar Pi transporters, and cytoplasmic Pi concentration is maintained via Pi transport into vacuoles under normal phosphorus conditions (Wang et al., 2012a). The SPX-EXS subfamily proteins play an important role in phosphate acquisition, translocation, and partitioning (Wang et al., 2004; Zhao et al., 2019). The SPX-RING subfamily proteins maintain phosphate homeostasis and are involved in the Pi response (Yang et al., 2017; Yue et al., 2017). Our findings could be used as a basis for the study of the evolution and function of the SPX protein family.
4.2 SPXs are vital to coping with abiotic stresses
Oat is considered a highly beneficial crop. It is known to regulate blood sugar and lipid levels, exhibit anti-cancer effects, and delay aging (Paudel et al., 2021; Varma et al., 2016). Oat plants are also known for their resilience to extreme environments (Canales et al., 2021; Dangi, 2020; Oraby and Ahmad, 2012). Therefore, it is important to analyze the coping mechanisms of oat to abiotic stresses to improve the ecological environment, promote the development of agriculture and animal husbandry, and improve human health. As a phosphorus-responsive protein, SPX plays an important role under phosphorus-deficient conditions. Previous studies have shown that SPX regulates nitrogen and phosphorus balances (Ueda et al., 2020). However, only a few studies have explored the role of SPXs in enhancing cold resistance (Wang et al., 2013; Zhao et al., 2009) and salinity-alkali tolerance (Oraby and Ahmad, 2012) in plants. In the present study, we analyzed the structural composition, PPIs, and phenotypic characteristics of AsSPXs (Supplementary Figures S2, S3; Supplementary Table S8). Furthermore, qRT-PCR showed that some AsSPXs were also induced under cold and salt stresses, providing a better understanding of the varied roles of AsSPXs.
Phylogenetic tree analysis (Figures 1, 2) and subcellular localization prediction (Supplementary Table S2) showed that AsSPX4-2C belonged to the SPX-MFS subfamily and was located on the plasma membrane, indicating that the SPX-MFS subfamily members primarily function as vacuolar Pi transporters. Furthermore, AsSPX8-4A and AsSPX9-4C belonged to the SPX subfamily, and their genes were located in the fourth chromosome. These findings suggested that they performed similar functions, acting as regulators of phosphate starvation. Finally, AsSPX18-5D belonged to the SPX-RING subfamily, suggesting its potential involvement in the Pi reaction and phosphate homeostasis in the nucleus and extracellularly.
5 Conclusion
In this study, a total of 169 SPX genes were identified across five closely related oat species, distribution uneven across four subfamilies: SPX, SPX-MFS, SPX-EXS, and SPX-RING. The AsSPXs within each subfamily exhibited remarkable similarity in conserved motifs, gene structures, and gene duplications. Furthermore, the promoter regions of AsSPXs were enriched with 17 cis-acting elements associated with responses to light, low temperature, ABA, and drought. Expression analysis revealed the pivotal role of SPX genes in phosphorus sensing and responses to salt, drought, cold, and ABA stresses, underscoring the multifaceted biological functions and redundancy mechanisms within the AsSPX family. Our results provided valuable insights for future investigations on the evolutionary and functional aspects of the AsSPX gene family.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
YD: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. JG: Formal Analysis, Investigation, Methodology, Writing–review and editing. ZD: Validation, Visualization, Writing–review and editing. WZ: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing–review and editing, Writing–original draft. RS: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. SG: Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. “Potential Assessment for Livestock Carrying Capacity and Eco-Livestock Industry Base Construction in Important Grasslands of Xinjiang” (2022xjkk0404), National Natural Science Foundation of China “Study on the root-soil interaction mechanism of nitrogen use efficiency and quality improvement under intercropping between leguminous forage grass and Korla fragrant pear” (32060402), National Natural Science Foundation of autonomous region “R&D, integration, demonstration and promotion of ecological restoration technologies for pastoral grasslands and mining areas” (2022B03030-2), Beijing Nova Program (20220484204) and Creation of transgenic new germplasm for important crops (KJCX20230203).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1469704/full#supplementary-material
References
Ahmad, P., Azooz, M. M., and Prasad, M. N. V. (2013). Ecophysiology and responses of plants under salt stress: biology. Environ. Sci. doi:10.1007/978-1-4614-4747-4
Borrelli, P., Robinson, D. A., Fleischer, L. R., Lugato, E., Ballabio, C., Alewell, C., et al. (2017). An assessment of the global impact of 21st century land use change on soil erosion. Nat. Commun. 8 (1), 2013. doi:10.1038/s41467-017-02142-7
Canales, F. J., Rispail, N., García-Tejera, O., Arbona, V., Pérez-de-Luque, A., and Prats, E. (2021). Drought resistance in oat involves aba-mediated modulation of transpiration and root hydraulic conductivity. Environ. Exp. Bot. 182, 104333. doi:10.1016/j.envexpbot.2020.104333
Chen, J., Han, X., Liu, L., Yang, B., Zhuo, R., and Yao, X. (2023). Genome-wide detection of spx family and profiling of cospx-mfs3 in regulating low-phosphate stress in tea-oil camellia. Int. J. Mol. Sci. 24 (14), 11552. doi:10.3390/ijms241411552
Chiou, T. J., and Lin, S. I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. doi:10.1146/annurev-arplant-042110-103849
Dangi, S. (2020). Oat as green fodder and its intercropping benefits: a review. Agric. Rev. (Karnal, India)(Of). doi:10.18805/ag.R-168
Du, H., Yang, C., Ding, G., Shi, L., and Xu, F. (2017). Genome-wide identification and characterization of spx domain-containing members and their responses to phosphate deficiency in brassica napus. Front. Plant Sci. 8, 35. doi:10.3389/fpls.2017.00035
Duan, K., Yi, K., Dang, L., Huang, H., Wu, W., and Wu, P. (2008). Characterization of a sub-family of arabidopsis genes with the spx domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant. J. 54 (6), 965–975. doi:10.1111/j.1365-313X.2008.03460.x
Garg, V. K., Avashthi, H., Tiwari, A., Jain, P. A., Ramkete, P. W., Kayastha, A. M., et al. (2016). Mfppi - multi fasta protparam interface. Bioinformation 12 (2), 74–77. doi:10.6026/97320630012074
Gaut, B. S., Morton, B. R., McCaig, B. C., and Clegg, M. T. (1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene adh parallel rate differences at the plastid gene rbcl. Proc. Natl. Acad. Sci. U. S. A. 93 (19), 10274–10279. doi:10.1073/pnas.93.19.10274
He, J., Yu, T., and Li, Y. (2023). Biodegradable thermoset poly(lactic acid) resin containing phosphorus: flame retardancy, mechanical properties and its soil degradation behavior. Int. J. Biol. Macromol. 235, 123737. doi:10.1016/j.ijbiomac.2023.123737
Hu, B., and Chu, C. (2020). Nitrogen-phosphorus interplay: old story with molecular tale. New Phytol. 225 (4), 1455–1460. doi:10.1111/nph.16102
Hu, B., Jiang, Z., Wang, W., Qiu, Y., Zhang, Z., Liu, Y., et al. (2019). Nitrate-nrt1.1b-spx4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5 (4), 401–413. doi:10.1038/s41477-019-0384-1
Kamal, N., Tsardakas, R. N., Bentzer, J., Gundlach, H., Haberer, G., Juhasz, A., et al. (2022). The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 606 (7912), 113–119. doi:10.1038/s41586-022-04732-y
Kiba, T., Inaba, J., Kudo, T., Ueda, N., Konishi, M., Mitsuda, N., et al. (2018). Repression of nitrogen starvation responses by members of the arabidopsis garp-type transcription factor nigt1/hrs1 subfamily. Plant. Cell. 30 (4), 925–945. doi:10.1105/tpc.17.00810
Kumar, A., Sharma, M., Gahlaut, V., Nagaraju, M., Chaudhary, S., Kumar, A., et al. (2019). Genome-wide identification, characterization, and expression profiling of spx gene family in wheat. Int. J. Biol. Macromol. 140, 17–32. doi:10.1016/j.ijbiomac.2019.08.105
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega x: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Kural, M., Kiyak, A., Uluisik, S., and Atay, E. (2024). Genome-wide identification and expression analysis of spx domain-containing gene subfamily in response to phosphorus-solubilizing bacteria in apple (malus domestica). Plant Mol. Biol. Rep. 42, 567–584. doi:10.1007/s11105-024-01443-8
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic. acids. Res. 30 (1), 325–327. doi:10.1093/nar/30.1.325
Li, C., You, Q., and Zhao, P. (2021). Genome-wide identification and characterization of spx-domain-containing protein gene family in solanum lycopersicum. PeerJ 9, e12689. doi:10.7717/peerj.12689
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Luo, J., Liu, Z., Yan, J., Shi, W., and Ying, Y. (2023). Genome-wide identification of spx family genes and functional characterization of pespx6 and pespx-mfs2 in response to low phosphorus in phyllostachys edulis. Plants 12 (7), 1496. doi:10.3390/plants12071496
Lv, Q., Zhong, Y., Wang, Y., Wang, Z., Zhang, L., Shi, J., et al. (2014). Spx4 negatively regulates phosphate signaling and homeostasis through its interaction with phr2 in rice. Plant. Cell. 26 (4), 1586–1597. doi:10.1105/tpc.114.123208
Nezamivand-Chegini, M., Ebrahimie, E., Tahmasebi, A., Moghadam, A., Eshghi, S., Mohammadi-Dehchesmeh, M., et al. (2021). New insights into the evolution of spx gene family from algae to legumes; A focus on soybean. BMC Genomics 22 (1), 915. doi:10.1186/s12864-021-08242-5
Oraby, H., and Ahmad, R. (2012). Physiological and biochemical changes of cbf3 transgenic oat in response to salinity stress. Plant Sci. 185-186, 331–339. doi:10.1016/j.plantsci.2012.01.003
Paudel, D., Dhungana, B., Caffe, M., and Krishnan, P. (2021). A review of health-beneficial properties of oats. Foods 10 (11), 2591. doi:10.3390/foods10112591
Ruan, W., Guo, M., Wang, X., Guo, Z., Xu, Z., Xu, L., et al. (2019). Two ring-finger ubiquitin e3 ligases regulate the degradation of spx4, an internal phosphate sensor, for phosphate homeostasis and signaling in rice. Mol. Plant. 12 (8), 1060–1074. doi:10.1016/j.molp.2019.04.003
Savojardo, C., Martelli, P. L., Fariselli, P., Profiti, G., and Casadio, R. (2018). Busca: an integrative web server to predict subcellular localization of proteins. Nucleic. acids. Res. 46 (W1), W459-W466–W466. doi:10.1093/nar/gky320
Secco, D., Wang, C., Arpat, B. A., Wang, Z., Poirier, Y., Tyerman, S. D., et al. (2012a). The emerging importance of the spx domain-containing proteins in phosphate homeostasis. New Phytol. 193 (4), 842–851. doi:10.1111/j.1469-8137.2011.04002.x
Secco, D., Wang, C., Shou, H., and Whelan, J. (2012b). Phosphate homeostasis in the yeast saccharomyces cerevisiae, the key role of the spx domain-containing proteins. FEBS Lett. 586 (4), 289–295. doi:10.1016/j.febslet.2012.01.036
Shi, J., Hu, H., Zhang, K., Zhang, W., Yu, Y., Wu, Z., et al. (2014). The paralogous spx3 and spx5 genes redundantly modulate pi homeostasis in rice. J. Exp. Bot. 65 (3), 859–870. doi:10.1093/jxb/ert424
Stahle, M. I., Kuehlich, J., Staron, L., von Arnim, A. G., and Golz, J. F. (2009). Yabbys and the transcriptional corepressors leunig and leunig_homolog maintain leaf polarity and meristem activity in arabidopsis. Plant. Cell. 21 (10), 3105–3118. doi:10.1105/tpc.109.070458
Sun, P. D., Foster, C. E., and Boyington, J. C. (2004). Overview of protein structural and functional folds. Curr. Protoc. Protein Sci. 35 (1), Unit 17.1–171189. doi:10.1002/0471140864.ps1701s35
Sun, R., Gong, J., Du, Y., Zhang, S., Zheng, W., Ma, J., et al. (2023a). Characterization and evolutionary analysis of phosphate starvation response genes in wheat and other major gramineous plants. Int. J. Biol. Macromol. 225, 63–78. doi:10.1016/j.ijbiomac.2022.11.298
Sun, Y., Jia, X., Yang, Z., Fu, Q., Yang, H., and Xu, X. (2023b). Genome-wide identification of pebp gene family in solanum lycopersicum. Int. J. Mol. Sci. 24 (11), 9185. doi:10.3390/ijms24119185
Ueda, Y., Kiba, T., and Yanagisawa, S. (2020). Nitrate-inducible nigt1 proteins modulate phosphate uptake and starvation signalling via transcriptional regulation of spx genes. Plant. J. 102 (3), 448–466. doi:10.1111/tpj.14637
Varma, P., Bhankharia, H., and Bhatia, S. (2016). Oats: a multi-functional grain. J. Clin. Prev. Cardiol. 5 (1), 9. doi:10.4103/2250-3528.183984
Wang, C., Huang, W., Ying, Y., Li, S., Secco, D., Tyerman, S., et al. (2012a). Functional characterization of the rice spx-mfs family reveals a key role of osspx-mfs1 in controlling phosphate homeostasis in leaves. New Phytol. 196 (1), 139–148. doi:10.1111/j.1469-8137.2012.04227.x
Wang, C., Wei, Q., Zhang, K., Wang, L., Liu, F., Zhao, L., et al. (2013). Down-regulation of osspx1 causes high sensitivity to cold and oxidative stresses in rice seedlings. PLoS One 8 (12), e81849. doi:10.1371/journal.pone.0081849
Wang, Q., Lu, X., Chen, X., Malik, W. A., Wang, D., Zhao, L., et al. (2021). Transcriptome analysis of upland cotton revealed novel pathways to scavenge reactive oxygen species (ros) responding to na(2)so(4) tolerance. Sci. Rep. 11 (1), 8670. doi:10.1038/s41598-021-87999-x
Wang, Y., Ribot, C., Rezzonico, E., and Poirier, Y. (2004). Structure and expression profile of the arabidopsis pho1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135 (1), 400–411. doi:10.1104/pp.103.037945
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012b). Mcscanx: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic. acids. Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Wang, Z., Hu, H., Huang, H., Duan, K., Wu, Z., and Wu, P. (2009). Regulation of osspx1 and osspx3 on expression of osspx domain genes and pi-starvation signaling in rice. J. Integr. Plant Biol. 51 (7), 663–674. doi:10.1111/j.1744-7909.2009.00834.x
Wang, Z., Ruan, W., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014). Rice spx1 and spx2 inhibit phosphate starvation responses through interacting with phr2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. - PNAS 111 (41), 14953–14958. doi:10.1073/pnas.1404680111
Xiao, J., Xie, X., Li, C., Xing, G., Cheng, K., Li, H., et al. (2021). Identification of spx family genes in the maize genome and their expression under different phosphate regimes. Plant Physiol. biochem. 168, 211–220. doi:10.1016/j.plaphy.2021.09.045
Yan, H., Sheng, M., Wang, C., Liu, Y., Yang, J., Liu, F., et al. (2019). Atspx1-mediated transcriptional regulation during leaf senescence in arabidopsis thaliana. Plant Sci. 283, 238–246. doi:10.1016/j.plantsci.2019.03.005
Yang, J., Wang, L., Mao, C., and Lin, H. (2017). Characterization of the rice nla family reveals a key role for osnla1 in phosphate homeostasis. Rice 10 (1), 52. doi:10.1186/s12284-017-0193-y
Yang, J., Zhao, X., Chen, Y., Li, G., Li, X., Xia, M., et al. (2022). Identification, structural, and expression analyses of spx genes in giant duckweed (spirodela polyrhiza) reveals its role in response to low phosphorus and nitrogen stresses. Cells 11 (7), 1167. doi:10.3390/cells11071167
Yue, W., Ying, Y., Wang, C., Zhao, Y., Dong, C., Whelan, J., et al. (2017). Osnla1, a ring-type ubiquitin ligase, maintains phosphate homeostasis in oryza sativa via degradation of phosphate transporters. Plant. J. 90 (6), 1040–1051. doi:10.1111/tpj.13516
Zhang, J., Zhou, X., Xu, Y., Yao, M., Xie, F., Gai, J., et al. (2016). Soybean spx1 is an important component of the response to phosphate deficiency for phosphorus homeostasis. Plant Sci. 248, 82–91. doi:10.1016/j.plantsci.2016.04.010
Zhao, L., Liu, F., Xu, W., Di, C., Zhou, S., Xue, Y., et al. (2009). Increased expression of osspx1 enhances cold/subfreezing tolerance in tobacco and arabidopsis thaliana. Plant Biotechnol. J. 7 (6), 550–561. doi:10.1111/j.1467-7652.2009.00423.x
Zhao, P., You, Q., and Lei, M. (2019). A crispr/cas9 deletion into the phosphate transporter slpho1;1 reveals its role in phosphate nutrition of tomato seedlings. Physiol. Plant. 167 (4), 556–563. doi:10.1111/ppl.12897
Keywords: phosphorus, salt, ABA treatments, AsSPX gene family, Avena sativa L
Citation: Du Y, Gong J, Dou Z, Zheng W, Sun R and Gao S (2024) Genome-wide identification and expression analysis of phosphate-sensing SPX proteins in oats. Front. Genet. 15:1469704. doi: 10.3389/fgene.2024.1469704
Received: 24 July 2024; Accepted: 05 November 2024;
Published: 20 November 2024.
Edited by:
Shailender Kumar Verma, University of Delhi, IndiaReviewed by:
Muhammad Sajjad, Zhejiang Agriculture and Forestry University, ChinaChanderkant Chaudhary, Louisiana State University, United States
Copyright © 2024 Du, Gong, Dou, Zheng, Sun and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zheng, encwNjVAMTYzLmNvbQ==; Renwei Sun, c3J3MDcxOUAxNjMuY29t; Shiqing Gao, Z3NoaXFAMTI2LmNvbQ==
 Yinke Du
Yinke Du Jie Gong
Jie Gong Ziyi Dou1
Ziyi Dou1 Wei Zheng
Wei Zheng Renwei Sun
Renwei Sun Shiqing Gao
Shiqing Gao