- 1Center for Sickle Cell Disease, Howard University, Washington, DC, United States
- 2The University Centre for Research and Development (UCRD), Chandigarh University, Mohali, Punjab, India
- 3Department of Medicine and Cancer Center, Howard University, Washington, DC, United States
Editorial on the Research Topic
The intricate web of gastrointestinal virome, mycome and archaeome: implications for gastrointestinal diseases
The gastrointestinal (GI) tract is a complex ecosystem containing bacteria, viruses (virome), fungi (mycome), and archaea (archaeome). It plays a significant role in ecological functioning, evolutionary changes, and host-microbiome interactions (Ogilvie and Jones, 2017; Daliri et al., 2020; Piewngam et al., 2020; Vemuri et al., 2020). Recent advances in sequencing technologies and metagenomic approaches have expanded our comprehension of the gastrointestinal-microbiome axis and its roles in maintaining gut homeostasis, health, and disease (Lepage et al., 2013; Gao et al., 2021; Lau and Yu, 2022). While bacteria have been extensively studied, the virome, mycome, and archaeome have recently gained attention. The human gut virome varies between individuals, composition and dynamics (Minot et al., 2011; Foca et al., 2015). They play a crucial role in gut regulation and contributing to disease development through interactions with the microbiota and immune cells (Ogilvie and Jones, 2017; Santiago-Rodriguez and Hollister, 2019; Hou et al., 2022; Tiamani et al., 2022). The gut mycobiome also plays a critical role in human health and disease, with potential implications for infants, obesity, inflammatory bowel disease (IBD) and neurodegenerative diseases (Forbes et al., 2018; Jain et al., 2021). The archaeome, a diverse and potentially important component of human gut host-associated microbiomes, plays an important role in gut physiology and health, in shaping the health and fitness of the animals (Gaci et al., 2014; Lurie-Weinberger and Gophna, 2015; Borrel et al., 2020; Kim et al., 2020). Studies have shown that Archaea play a key role in the pathogenesis of IBD and the risk of pediatric asthma (Barnett et al., 2019; Singh et al., 2023). However, their role in human health remains poorly understood, therefore, understanding these microbial communities offers new therapeutic approaches for GI disorders and enhances strategies for improving GI health as shown in Figure 1.
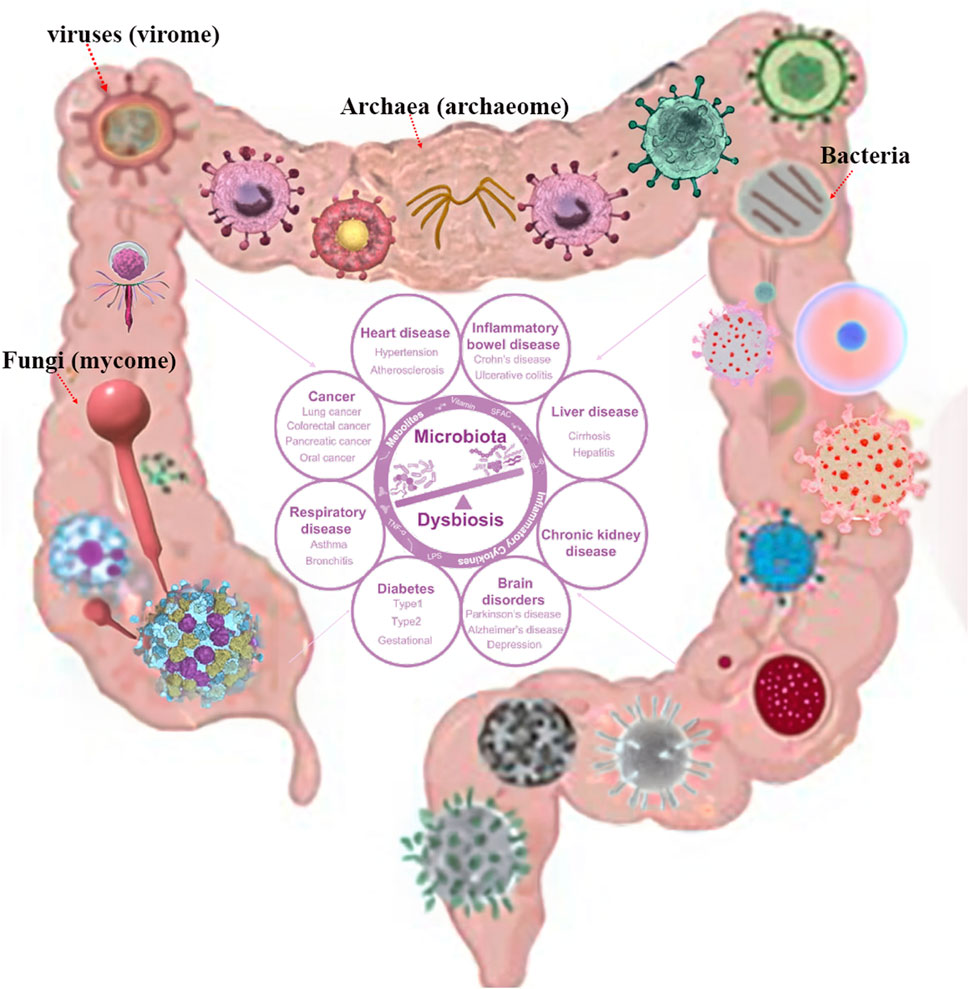
Figure 1. Human microbiota composition with potential consequences. Permission obtained from Hou et al. (2022), Spencer et al. (2022).
The aim of the editorial “The intricate web of gastrointestinal virome, mycobiome, and archaeome: implications for gastrointestinal diseases” is to explore the roles and interactions of the gastrointestinal microbiome in relation to gastrointestinal diseases. The objectives include reviewing current research on these microbial communities, discussing their implications for gastrointestinal health, highlighting potential therapeutic implications, and fostering a deeper understanding among healthcare professionals and researchers and suggest avenues for future research and clinical application in this evolving field. Recent studies have shown that, Postherpetic Neuralgia (PHN) is a chronic neuropathic pain syndrome caused by Herpes Zoster virus (HZV) reactivation, and it is more common in older and immunocompromised individuals, and vaccination is effective in preventing complications (Oxman et al., 2005; Mallick-Searle et al., 2016; Ferreira Sampaio et al., 2023). Additionally, it has been shown that HSV patients with peptic ulcer disease may be at a greater risk for developing PHN due to impaired cellular immunity and low nutritional status (Chen et al., 2013). Zhimin et al. have reported that the causal effect between HZV and PHN and the gut microbiota using bidirectional two-sample Mendelian randomization (MR) analysis. The study revealed the alterations in gut microbiota may influence susceptibility to HSV and the development of PHN. This research highlights the potential for specific gut microbiota to serve as a therapeutic target, offering new avenues for the prevention and treatment of viral infections and associated chronic conditions. This study highlights the importance of considering the gut microbiota not just in isolation but as a critical component in the overall immune response and susceptibility to viral infections. By understanding these causal relationships, new therapeutic strategies could be developed, focusing on modulating the gut microbiota to prevent or mitigate the effects of HSV associated PHN. Additionally, Hanjing et al. carried out the study in China (Jinjiang, Fujian), and reported the interactions between Helicobacter pylori, chronic gastritis, and the gut microbiota. They demonstrate that how Helicobacter. pylori infection and chronic gastritis are intricately linked with changes in gut microbiota composition. These findings emphasize the need for a comprehensive approach in managing Helicobacter. pylori infections and gastritis, considering their impact on the overall gut microbiota health. Moreover, gut microbiome-associated metabolites in serum can accurately distinguish colorectal cancer and adenomas from normal samples (Chen et al., 2022). Also, another study profiled the serum antibody responses of 997 healthy individuals against 244,000 peptide antigens derived from gut microbiota, data indicated that the serum antibody repertoires displayed high diversity and stability against human microbiota and identified several factors influencing the serum antibody response, such as age, gender, body mass index, and geographical location. These findings shed light on the role of the immune system in maintaining homeostasis with the gut microbiota (Vogl et al., 2021). Chen et al. present a differential analysis of serum immunology and gut microbiota in patients suffering from various gastrointestinal diseases. The study identifies distinct immunological and microbiota profiles associated with specific GI conditions, providing valuable insights into the interplay between the immune system and gut microbiota. This research highlights the potential for personalized medicine approaches in GI, tailoring treatments to the unique microbiota like recent report has shown that smart antibiotic (Lolamicin) can target a group of harmful microbes but does not disturb those that live peacefully in the gut (Munoz et al., 2024). Furthermore, Xiao et al. examine the causal relationship between the gut microbiome and liver cirrhosis through a bi-directional two-sample Mendelian randomization analysis. The results highlight significant causal links, suggesting that specific alterations in the gut microbiome may contribute to the development of liver cirrhosis. This study points out the importance of considering gut microbiota health in the prevention and management of liver diseases. Also, other evidence has shown that gut microbiome plays a significant role in liver disorders, potentially contributing to disease process and severity. Prebiotics and probiotics may be effective treatments for complications and liver cirrhosis. Additionally, the gut-liver axis plays a crucial role in liver disease, and progressive changes in the gut microbiome accompany cirrhosis. It became more severe in the setting of decompensation, with the cirrhosis dysbiosis ratio being a useful quantitative index (Bajaj et al., 2014; Giannelli et al., 2014; Qin et al., 2014; Tilg et al., 2016; Albillos et al., 2020; Li et al., 2022). This issue opens a new avenue for preventive strategies and therapeutic interventions aimed at modulating the gut microbiome to improve health and patient quality of life.
Conclusion
The intricate interplay between the gut microbiota, virome, mycome, and archaeome has profound implications for GI diseases and these studies provide a foundation for future research and potential therapeutic innovations. This editorial reveals how these microbial communities influence conditions such as HSV, chronic gastritis, liver cirrhosis and emphasize the potential of targeting gut microbiota for therapeutic interventions. Understanding the intricate web of these microorganisms opens new avenues for personalized medicine, aiming to improve GI health through advanced sequencing technologies and metagenomic analyses (Brim et al., 2017; Liang et al., 2017). We hope this issue inspires further exploration into the intricate web of the GI ecosystem and its impact on health, fostering a deeper understanding that can translate into better clinical practices and holds vast potential for unlocking new paradigms in the treatment and management of gastrointestinal diseases to improve patient outcomes.
Author contributions
AA: Writing–review and editing, Writing–original draft. MA: Formal Analysis, Writing–review and editing. MR: Conceptualization, Data curation, Supervision, Writing–review and editing, Writing–original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albillos, A., de Gottardi, A., and Rescigno, M. (2020). The gut-liver axis in liver disease: pathophysiological basis for therapy. J. Hepatol. 72, 558–577. doi:10.1016/j.jhep.2019.10.003
Bajaj, J. S., Heuman, D. M., Hylemon, P. B., Sanyal, A. J., White, M. B., Monteith, P., et al. (2014). Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 60, 940–947. doi:10.1016/j.jhep.2013.12.019
Barnett, D. J. M., Mommers, M., Penders, J., Arts, I. C. W., and Thijs, C. (2019). Intestinal archaea inversely associated with childhood asthma. J. Allergy Clin. Immunol. 143, 2305–2307. doi:10.1016/j.jaci.2019.02.009
Borrel, G., Brugere, J. F., Gribaldo, S., Schmitz, R. A., and Moissl-Eichinger, C. (2020). The host-associated archaeome. Nat. Rev. Microbiol. 18, 622–636. doi:10.1038/s41579-020-0407-y
Brim, H., Yooseph, S., Lee, E., Sherif, Z. A., Abbas, M., Laiyemo, A. O., et al. (2017). A microbiomic analysis in african Americans with colonic lesions reveals Streptococcus sp.VT162 as a marker of neoplastic transformation. Genes (Basel) 8, 314. doi:10.3390/genes8110314
Chen, F., Dai, X., Zhou, C. C., Li, K. X., Zhang, Y. J., Lou, X. Y., et al. (2022). Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut 71, 1315–1325. doi:10.1136/gutjnl-2020-323476
Chen, J. Y., Chang, C. Y., Lan, K. M., Sheu, M. J., Lu, C. L., and Hu, M. L. (2013). Is peptic ulcer disease a risk factor of postherpetic neuralgia in patients with herpes zoster? Med. Hypotheses 81, 834–838. doi:10.1016/j.mehy.2013.09.007
Daliri, E. B., Ofosu, F. K., Chelliah, R., Lee, B. H., and Oh, D. H. (2020). Health impact and therapeutic manipulation of the gut microbiome. High. Throughput 9, 17. doi:10.3390/ht9030017
Ferreira Sampaio, R., Vinicius da Silva Pereira, M., Nayara Coelho Muniz Cardoso, M., Soares da Costa, D., Saraiva Sales, E., Vinicius Magalhães Guedes, M., et al. (2023). Integrative literature review: prevention of neurological complications of herpes zoster in adults - advances, challenges and perspectives. Health. Soc. 3, 16–30. doi:10.51249/hs.v3i04.1447
Foca, A., Liberto, M. C., Quirino, A., Marascio, N., Zicca, E., and Pavia, G. (2015). Gut inflammation and immunity: what is the role of the human gut virome? Mediat. Inflamm. 2015, 326032. doi:10.1155/2015/326032
Forbes, J. D., Bernstein, C. N., Tremlett, H., Van Domselaar, G., and Knox, N. C. (2018). A fungal world: could the gut mycobiome Be involved in neurological disease? Front. Microbiol. 9, 3249. doi:10.3389/fmicb.2018.03249
Gaci, N., Borrel, G., Tottey, W., O'Toole, P. W., and Brugere, J. F. (2014). Archaea and the human gut: new beginning of an old story. World J. Gastroenterol. 20, 16062–16078. doi:10.3748/wjg.v20.i43.16062
Gao, B., Chi, L., Zhu, Y., Shi, X., Tu, P., Li, B., et al. (2021). An introduction to next generation sequencing bioinformatic analysis in gut microbiome studies. Biomolecules 11, 530. doi:10.3390/biom11040530
Giannelli, V., Di Gregorio, V., Iebba, V., Giusto, M., Schippa, S., Merli, M., et al. (2014). Microbiota and the gut-liver axis: bacterial translocation, inflammation and infection in cirrhosis. World J. Gastroenterol. 20, 16795–16810. doi:10.3748/wjg.v20.i45.16795
Hou, K., Wu, Z. X., Chen, X. Y., Wang, J. Q., Zhang, D., Xiao, C., et al. (2022). Microbiota in health and diseases. Signal Transduct. Target Ther. 7, 135. doi:10.1038/s41392-022-00974-4
Jain, U., Ver Heul, A. M., Xiong, S., Gregory, M. H., Demers, E. G., Kern, J. T., et al. (2021). Debaryomyces is enriched in Crohn's disease intestinal tissue and impairs healing in mice. Science 371, 1154–1159. doi:10.1126/science.abd0919
Kim, J. Y., Whon, T. W., Lim, M. Y., Kim, Y. B., Kim, N., Kwon, M. S., et al. (2020). The human gut archaeome: identification of diverse haloarchaea in Korean subjects. Microbiome 8, 114. doi:10.1186/s40168-020-00894-x
Lau, H. C. H., and Yu, J. (2022). Uncovering novel human gut virome using ultra-deep metagenomic sequencing. Chin. Med. J. Engl. 135, 2395–2397. doi:10.1097/CM9.0000000000002382
Lepage, P., Leclerc, M. C., Joossens, M., Mondot, S., Blottiere, H. M., Raes, J., et al. (2013). A metagenomic insight into our gut's microbiome. Gut 62, 146–158. doi:10.1136/gutjnl-2011-301805
Li, R., Yi, X., Yang, J., Zhu, Z., Wang, Y., Liu, X., et al. (2022). Gut microbiome signatures in the progression of hepatitis B virus-induced liver disease. Front. Microbiol. 13, 916061. doi:10.3389/fmicb.2022.916061
Liang, Q., Chiu, J., Chen, Y., Huang, Y., Higashimori, A., Fang, J., et al. (2017). Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin. Cancer Res. 23, 2061–2070. doi:10.1158/1078-0432.CCR-16-1599
Lurie-Weinberger, M. N., and Gophna, U. (2015). Archaea in and on the human body: health implications and future directions. PLoS Pathog. 11, e1004833. doi:10.1371/journal.ppat.1004833
Mallick-Searle, T., Snodgrass, B., and Brant, J. M. (2016). Postherpetic neuralgia: epidemiology, pathophysiology, and pain management pharmacology. J. Multidiscip. Healthc. 9, 447–454. doi:10.2147/JMDH.S106340
Minot, S., Sinha, R., Chen, J., Li, H., Keilbaugh, S. A., Wu, G. D., et al. (2011). The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625. doi:10.1101/gr.122705.111
Munoz, K. A., Ulrich, R. J., Vasan, A. K., Sinclair, M., Wen, P. C., Holmes, J. R., et al. (2024). A Gram-negative-selective antibiotic that spares the gut microbiome. Nature 630, 429–436. doi:10.1038/s41586-024-07502-0
Ogilvie, L. A., and Jones, B. V. (2017). The human gut virome: form and function. Emerg. Top. Life Sci. 1, 351–362. doi:10.1042/ETLS20170039
Oxman, M. N., Levin, M. J., Johnson, G. R., Schmader, K. E., Straus, S. E., Gelb, L. D., et al. (2005). A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N. Engl. J. Med. 352, 2271–2284. doi:10.1056/NEJMoa051016
Piewngam, P., De Mets, F., and Otto, M. (2020). Intestinal microbiota: the hidden gems in the gut? Asian Pac J. Allergy Immunol. 38, 215–224. doi:10.12932/AP-020720-0897
Qin, N., Yang, F., Li, A., Prifti, E., Chen, Y., Shao, L., et al. (2014). Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64. doi:10.1038/nature13568
Santiago-Rodriguez, T. M., and Hollister, E. B. (2019). Human virome and disease: high-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut. Viruses 11, 656. doi:10.3390/v11070656
Singh, P., Murugesan, S., Kumar, M., Saadaoui, M., Elhag, D. A., Hendaus, M. A., et al. (2023). P915 Uncovering the role of Archaea in the pathogenesis of Inflammatory bowel disease. J. Crohn's Colitis 17, i1026. doi:10.1093/ecco-jcc/jjac190.1045
Spencer, L., Olawuni, B., and Singh, P. (2022). Gut virome: role and distribution in health and gastrointestinal diseases. Front. cell. infect. microbiol. 12, 836706. doi:10.3389/fcimb.2022.836706
Tiamani, K., Luo, S., Schulz, S., Xue, J., Costa, R., Khan Mirzaei, M., et al. (2022). The role of virome in the gastrointestinal tract and beyond. FEMS Microbiol. Rev. 46, fuac027. doi:10.1093/femsre/fuac027
Tilg, H., Cani, P. D., and Mayer, E. A. (2016). Gut microbiome and liver diseases. Gut 65, 2035–2044. doi:10.1136/gutjnl-2016-312729
Vemuri, R., Shankar, E. M., Chieppa, M., Eri, R., and Kavanagh, K. (2020). Beyond just bacteria: functional biomes in the gut ecosystem including virome, mycobiome, archaeome and helminths. Microorganisms 8, 483. doi:10.3390/microorganisms8040483
Keywords: virome, mycome, archaeome, human health and disease, gastroinestinal tract
Citation: Ahmad A, Ashfaq M and Rashid M (2024) Editorial: The intricate web of gastrointestinal virome, mycome and archaeome: implications for gastrointestinal diseases. Front. Genet. 15:1463350. doi: 10.3389/fgene.2024.1463350
Received: 11 July 2024; Accepted: 30 July 2024;
Published: 09 August 2024.
Edited and reviewed by:
Maxim B. Freidin, King’s College London, United KingdomCopyright © 2024 Ahmad, Ashfaq and Rashid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mudasir Rashid, bXVkYXNpci5yYXNoaWRAaG93YXJkLmVkdQ==
 Asrar Ahmad
Asrar Ahmad Mohammad Ashfaq
Mohammad Ashfaq Mudasir Rashid
Mudasir Rashid