
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 13 September 2024
Sec. Genetics of Common and Rare Diseases
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1447971
 Patrice Djataou1,2
Patrice Djataou1,2 Marceline Djuidje Ngounoue2
Marceline Djuidje Ngounoue2 Carine Nguefeu Nkenfou-Tchinda3
Carine Nguefeu Nkenfou-Tchinda3 Marie Nicole Ngoufack1
Marie Nicole Ngoufack1 Elise Elong1
Elise Elong1 Aline Tiga1
Aline Tiga1 Clifford Muluh4
Clifford Muluh4 Joelle Kadji Kameni1
Joelle Kadji Kameni1 Moussa Djaouda5
Moussa Djaouda5 Alexis Ndjolo1
Alexis Ndjolo1 Celine Nguefeu Nkenfou1,6*
Celine Nguefeu Nkenfou1,6*Background: HIV infection and its progression to AIDS depend on several factors including host genetic factors. The immunological mechanisms of host resistance to HIV infection greatly influence the prevalence of HIV in a given region. Worldwide, Cameroon not exempted, the frequency of AIDS-associated genes varies and may influence this prevalence. The North and Far North Regions of Cameroon have had the lowest HIV prevalence in the country for many years despite risky behaviors associated with their customs and habits. In this work, we seek to explore the contribution of host genes to the HIV low prevalence in these regions.
Methodology: Five genes variants previously described as HIV AIDS related were studied. These genes are: CCR5Δ32, CCR5promoter59029G, CCR2-64I, SDF1-3ʹA and Trim5α(R136Q). A total of 384 consented participants were included in this study. The HIV serological status was confirmed using national algorithm. Genomic DNA was extracted from the buffy coats and used for genotyping. The results obtained were compiled in Excel 2016, Epi Info 7.1 and snpStats software and Chi two tests allowed us to compare the frequencies of the AIDS related alleles in the North with those in other Regions of Cameroon and to measure the impact of these ARGs on protection against HIV.
Results: The frequency of protective alleles CCR5Δ32, CCR5promoter59029G, CCR2-64I, SDF1-3ʹA and Trim5α(R136Q) was the allelic frequencies should be expressed as percentages i.e. 0.52%; 37.56%; 36.46%; 25.19% and 69.33%. These allelic frequencies exhibited a significant difference when compared to those obtained in other regions of Cameroon (p < 0.01). Protective alleles were predominant in the Northern region compared to others and were associated with resistance to HIV [(p < 0.0001); OR = 2.02 CI, 95%].
Conclusion: The higher frequency of HIV-protective alleles in the northern regions may be a contributing factor to the lower prevalence of HIV. Nevertheless, this should be reinforced by other preventive and surveillance methods to guarantee the sustained low prevalence. HIV can develop resistance through the process of mutation, but the host targets themselves are genetically stable. The study of these host genetic restriction factors is of great value in the design of a practical cure for HIV infection or an effective vaccine.
Many years after its discovery, HIV/AIDS remains a major public health problem worldwide. The pandemic has spread rapidly around the world, with a peak in the number of infections recorded in the mid-1990s (Lot, 2008). Today, HIV pandemic is more or less under control, with an overall decline in prevalence and new infections worldwide (UNAIDS, 2024). Preventive measures, awareness raising and antiretroviral treatments are helping to significantly reduce the number of new infections each year. Current treatments aim to limit viral replication and reduce the viral load to an undetectable level (Ahmed et al., 2022; Abrahams et al., 2009); while controlling the emergence of ARV drugs resistance. As a result, the life expectancy of people living with HIV has improved significantly. Research into HIV/AIDS has evolved, and today science already has the means to control the infection with new-generation ARVs. While HIV/AIDS awareness and prevention in communities have produced satisfactory results, research into a definitive vaccine remains a challenge for researchers. In the HIV infection process, chemokine co-receptors (CCR5 or CXCR4) interact with gp120 to fuse with the membrane and integrate the target cell. Several genes have been correlated with the evolution of HIV within populations. Antiretroviral treatment has brought many advances in the fight against HIV, as well as improving the life expectancy of seropositive people (Burgoyne and Tan, 2008). However, with the emergence of resistance to ARVs, the search for new curative technologies remains a challenge. Resistance to ARVs increases the risk of transmission of the virus with anticipated rapid progression of the disease to the AIDS stage because the virus has become resistant to the treatment (Sayan et al., 2016). In fact, studies show a mutation in the selection of viral tropism from R5 to the more virulent R4 (Yandrapally et al., 2021). HIV-associated genes remain a real mine of research because they influence the mechanisms of infection in the primary stages, pathogenesis and response to antiretroviral treatment.
In the history of HIV/AIDS, two cases of remission have been recorded: one in Berlin and the other in London (Gupta et al., 2019; Gupta et al., 2020a). These two patients have received through transplantation HIV-associated resistance gene, CCR5Δ32, which has been associated for many years as host gene for resistance to infection. In host cell infection mechanisms, HIV recruits in addition to the CD4 receptor, a CCR5 coreceptor to fuse with the host membrane. A particular mutation of the CCR5 gene co-receptor to CCR5Δ32 characterized by a deletion of 32 base pairs in its coding region prevent the virus from infecting the cell (Agrawal et al., 2004). After biosynthesis, this mutation produces a truncated receptor that is incapable of being recruited by HIV, so there is no infection. SDF-1 (stromal derived factor-1) is the only known ligand for CXCR4 (Sadri et al., 2022). Binding of SDF1 to CXCR4 leads to internalization of the receptor (Arenzana-Seisdedos, 2015). Thus, the virus with “R4″ tropism cannot bind the receptor and therefore penetrate the target cell. In the homozygous state, SDF1-3′A/A delays the onset of acquired immunodeficiency syndrome (AIDS), according to a genetic association analysis of 2,857 patients enrolled in five AIDS cohort studies (Winkler et al., 1998). The recessive protective effect of SDF1-3′A was increasingly pronounced in individuals infected with HIV-1 for longer periods, was twice as strong as the dominant genetic restriction of AIDS conferred by CCR5 and CCR2 chemokine receptor variants in these populations, and was complementary with these mutations in delaying the onset of AIDS (Winkler et al., 1998). The mechanism of resistance to HIV infection would probably be due to the blockage of R5 to more aggressive R4 HIV-1 receptor switching leading to the AIDS stage (Brambilla et al., 2000). Studies have shown that the SDF1-3′A polymorphism has been associated with high plasma levels of SDF1 (Brambilla et al., 2000; Signoret et al., 1997). SDF1 induces internalisation of CXCR4 receptors, thereby preventing the selection mutation from R5 to R4 tropism (Yandrapally et al., 2021; Soriano et al., 2002). CCR2 is one of the most extensively studied genetic factors confirmed to be linked to host resistance to HIV-1 infection. A CCR2-64I point mutation (rs1799864) characterized by guanine substitution leads to the replacement of the amino acid valine (V) by isoleucine (I) at position 64 in the first transmembrane domain of CCR2. CCR2 can also, under certain conditions, be used as a co-receptor by HIV (Soriano et al., 2002). Numerous studies have reported that the CCR2-64I polymorphism delays the progression of HIV-1 to AIDS by maintaining a stable HIV viral load and a slow decline in CD4 cells (Ammaranond et al., 2011; Juhász et al., 2012). The mechanism of CCR2-64I protection can be explained by a direct modification of the kinetics of infection, or by an indirect effect of the physiological concentration of CCR2 on the availability of CCR5 on the surface of target cells, or by a regulatory binding imbalance in the regulatory or promotion region of this gene (Metodiev, 2012). In addition, studies have shown that the CCR2-64I mutant allele forms heterodimers with CXCR4, and may therefore impair the function of CXCR4 as a co-receptor for X4 viruses (Burton et al., 2005). Thus, individuals in various populations harboring CCR2-64I (Angela Covino et al., 2016), CCR5Δ32 (Gupta and Padh, 2012), and CCR5 promoter A/G (Jang et al., 2008) mutations are less susceptible to HIV-1 infection and progress much more slowly to AIDS. The CCR5 polymorphism results in the absence of cell surface CCR5 expression, whereas the CCR2-64I gene variant confers resistance to AIDS progression, probably due to the ability of this mutant receptor to heterodimerize with the CCR5 and CXCR4 receptors (Mellado et al., 1999).
Trim5α play a critical role in the primate anti-viral defence system. Trim5α in the Rhesus monkey completely blocks HIV-1 infection whereas human Trim5α mediates a low level inhibition to HIV-1 replication (Kim et al., 2019; Chan et al., 2014). The mechanism of HIV-1 restriction is not fully understood. Proposed models include binding of a multimer of Trim5α to the incoming viral capsid that leads to premature uncoating of the capsid or by interaction between Trim5α and the capsid with other undefined cofactors (Sokolskaja and Luban, 2006). Using an in vitro assay system, cells were transduced with different Trim5α missense polymorphisms; R136Q enhanced HIV restriction while H43Y, V112F, and G110E relaxed restriction on HIV replication or had no effect (G249D, H419Y) (Nakajima et al., 2009; Javanbakht et al., 2006; Goldschmidt et al., 2006; Sawyer et al., 2006).
This implies that there is a combination of genes that play a role in the resistance to HIV infection and that these genes must be addressed collectively.
HIV prevalence in Cameroon was estimated at 3.7% in 2018 (DHS cameroon, 2018). However, infection is unevenly distributed across the country. The North and Far North Regions of Cameroon have the lowest seroprevalence rates in Cameroon, at 1.6% and 1.5% respectively (DHS cameroon, 2018) for a national incidence rate of 0.24%. The low prevalence of HIV infection in this region is thought to be due to genetic factors that protect the population against infection. Cameroon is known for its ethnic and genetic diversity (Spínola et al., 2011). AIDS-Related Genes (ARG) genotyping studies have been conducted in the Central and Western Regions of Cameroon, revealing important information about the disease progression. (Nkenfou et al., 2013; Dambaya et al., 2019; Ma et al., 2005). However, these ARG genotyping studies have not yet been carried out in these two northern regions of Cameroon. In order to understand the uneven distribution of HIV seroprevalence in Cameroon, we will compare the distribution of ARGs and susceptibility to HIV infection in the Northern Regions and the Central and Western Regions of Cameroon.
The study was carried out in the Garoua and Maroua Regional Hospitals located respectively in the North and Far North Regions of Cameroon. Samples will be collected between 15 June 2022 and 30 July 2022, following ethical approval and consent from participants.
The epi Info 7 calculation “STATCALC” software was used to calculate the population size (https://www.epivf.fr/calcul_nsn.html accessed July 2024) (Lwanga et al., 1991). We assumed that, given the low prevalence of HIV, the population had a high frequency of allelic ARGs of 50%, with an acceptable margin of error of 5%, 80% of power and a confidence level of 95%.
The population size is given by the following formula:
Where n is the sample size.
N is the estimated size of the total population.
z is the 95% confidence level, p is 50%, the estimated proportion of ARGs in this population.
e the tolerated margin of error 5%.
That’s 384 participants enrolled.
-People aged between 18 and 70 of both sexes who present at the hospital during the study;
-People who have been living in the North and Far North Regions and are native of these regions,
-Volunteers who have signed the informed consent form.
Individuals whose data were incomplete, whose samples could not be analyzed due to insufficient quantity or quality, as well as those whose HIV status was indeterminate, were excluded from the study.
384 participants were enrolled after listening to/reading and signing the consent form to take part in the study and completing a questionnaire. Subsequently, 5 mL of blood in EDTA tubes was collected from each subject, and the buffy coat was separated after centrifuging the blood samples at maximum speed for 1 minute and pipetting the thin whitish layer on top of the red blood cells for the purpose of performing ARG genotyping. HIV serology tests were carried out using the national algorithm (parallel two tests). Reactive Determine HIV ½ tests were repeated using “KHB” test.
The Buffy coats were used as a source of genomic DNA, that was extracted using QiaAmp DNA mini kit (Qiagen S.A. Three Avenue du Canada, LP 809, 91,974 Courtaboeuf Cedex, France), according to the manufacturer’s instructions. DNA concentration was measured by a nanodrop spectrophotometer (Thermo fisher, 3,411 Silverside Road Tatnall Building, Suite 100 Wilmington, DE 19810 U.S.A.).
The CCR5Δ32, CCR5 promoter, CCR2-64I, SDF1-3ʹA and Trim 5α genetic variants in participants were determined by PCR followed by RFLP detection using the specific primers and restriction endonucleases as described previously. Nevertheless, this original protocol was optimized during our study (Nkenfou et al., 2013; Ma et al., 2005; Kristiansen et al., 2001; Manen et al., 2008).
The amplification of CCR5 was done as follows: one cycle for 30 s at 94°C, followed by 40 cycles of 30 s, 30 s, and 1 min at 94°C, 50°C, and 72°C, respectively, followed by a final extension of 10 min at 72°C. CCR5-promoter gene amplification was done as follows: 3 min at 94°C, followed by 40 cycles of 30 s, 30 s, and 45 s at 94°C, 60°C, and 72°C, respectively, and a final extension of 10 min at 72°C. CCR2 gene was amplified in one cycle of 30 s at 94°C, followed by 40 cycles of 30 s, 30 s, and 30 s at 95°C, 63°C, and 72°C, respectively, and a final extension of 10 min at 72°C. To detect SDF-1 gene, the amplification started with a denaturation step of one cycle of 3 min at 94°C, followed by 40 cycles of 30, 30, and 30 s at 94°C, 58°C, and 72°C, respectively, and a final extension of 10 min at 72°C. The amplification of Trim 5α gene fragment was done using the following conditions: one cycle of 3 min at 94°C, followed by 40 cycles of 30 s, 30 s and 1 min at 94°C, 50°C and 72°C, respectively, and a final extension of 10 min at 72°C (Sawyer et al., 2006; DHS cameroon, 2018).
The amplified fragments were run in agarose gel with variable percentage (1%–1.5%) depending on the fragment size, previously stained with ethidium bromide, and visualized under ultraviolet light. The above-mentioned gene fragments size and their specific primers are presented in (Table 1).
Genotyping was carried out with the use of restriction fragment length polymorphism (RFLP) method except for the CCR5-Δ32 deletion based on the respective restriction enzyme sites in the four amplified (PCRs) products of CCR5 promoter, Trim 5α, CCR2 and SDF1 as described previously, (Table 1). The restriction enzymes used in this work were purchased from Thermo Fischer and used according to manufacturer instructions.
Allele and genotype frequencies and the Hardy-Weinberg equilibrium (HWE) were evaluated using SNPstats software (snpStats). All alleles achieved HWE. The differences in the allele frequency of each genetic variant between and within the distinct groups of HIV-1 seronegative and HIV-1 seropositive individuals in Northern and Central, West, and other Cameroon regions were determined by Chi-square or Fisher exact test when indicated. To estimate the association between ARG polymorphisms and participants’ serology, we used several inheritance models (codominant, dominant, recessive, overdominant and additive). For each ARG, odds ratios (OR) and 95% confidence intervals (CI) were calculated using logistic regression analysis.
p-values were calculated, and p < 0.05 was considered statistically significant.
The National Ethics Committee reviewed the protocol for ethical consideration and approval was given under N°2021/06/85/CE/CNERSH/SP. Informed consent or assent/parental consent form was signed when applicable by all the participants before enrolment. As well, this study was conducted in accordance with the Declaration of Helsinki (WMA).
A total of 384 participants were enrolled, regardless of gender, with a mean age of 31.25 years and a standard deviation of 10.2 years. Table 2 presents the sociodemographic characteristics of the study population. The most common age group was between 18 and 35 years. Females comprised n = 203 (52.86%) of the subjects in our study population compared to n = 181 (47.14%) males, a ratio of 1.1 in favor of females. The overall HIV prevalence in this study population was 1.63%. Some participants admitted to having had a sexually transmitted infection such as syphilis (n = 21), gonorrhea (n = 37) and chlamydia (n = 12).
In the overall population, CCR5∆32 double mutation (homozygote mutants) was completely absent in the study population and 1.04% for heterozygous genotype. The double mutation G/G of CCR5 promoter was frequent at 66.93%, with 20.83% for heterozygous genotype.
SDF1-3ʹA double mutation was 2.08%, and the heterozygous genotype was present at 97.92%.
Homozygous wild type for SDF1-3ʹA were absent. The double mutation of Trim5α Q136 was 61.72% (Table 3).
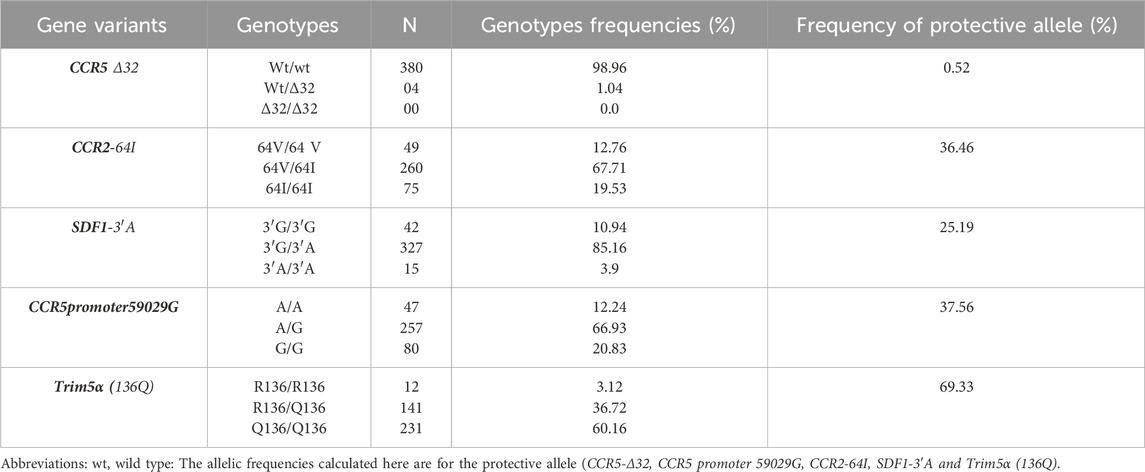
Table 3. Distribution of genotypes and frequencies of ARGs protective allele (CCR5Δ32, CCR2-64I, SDF1-3ʹA, CCR5 Promoter 59029G and Trim 5α(138Q)) in the study population.
The heterozygous genotype for the CCR5-Δ32 mutation was present in our study population although rare or absent in Africa according to several data. This genotype was found in the Moundang (n = 3) and in a Massa (n = 1) tribes in our study population. These two ethnic groups share the same department of origin. It is therefore possible that the homozygous CCR5-Δ32 mutation is present in this locality.
The most represented alleles for CCR5 variant was the wild type (wt), for CCR5p59029 variant it was CCR5p-59029A; for Trim5α, it was Trim5α (136Q); for CCR2 variant the major allele was CCR2-64I and for SDF1 it was SDF1-3′A.
Trim5α(136Q) (OR = 3.3 [CI, 95%; 2.74–3.85]), SDF1-3ʹA (OR = 2.2 [CI, 95%; 1.52–2.87]) and CCR5p-59029A (OR = 2.6 [CI, 95%; 1.98–3.22]) may be strongly associated with resistance to HIV infection. Meanwhile, CCR2-64I (OR = 1.7 [CI 95%; 0.93–2.47]) and CCR5∆32 (OR = 1.3 [ 95% CI; 0.42–2.18]) were less strongly associated with HIV infection (Table 4).
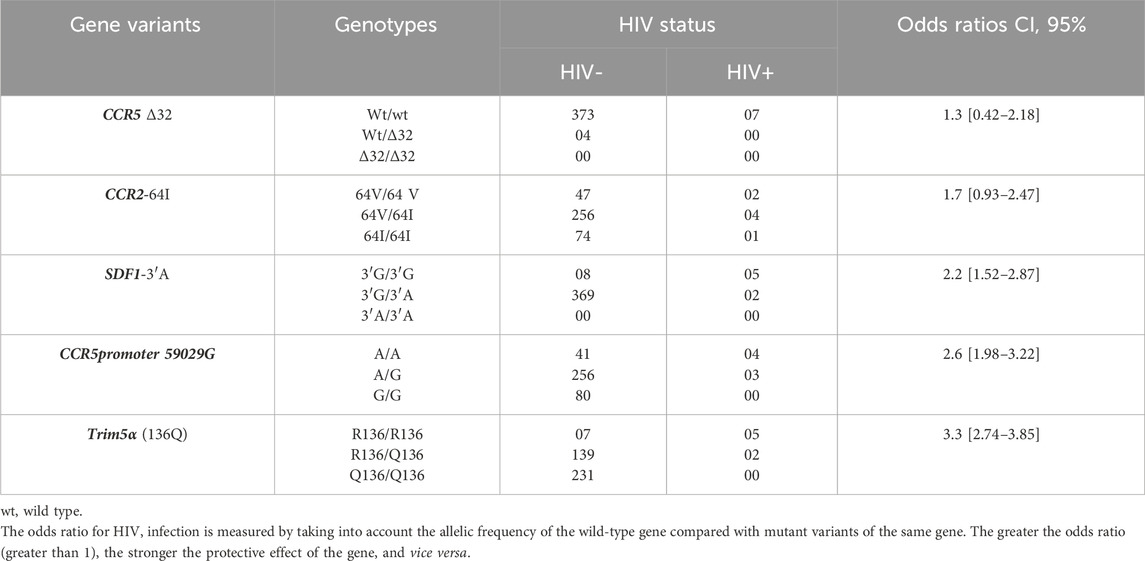
Table 4. Distribution of the genotypes of the 5 AIDS related genes variants in the study population according to HIV status.
The search for a simple association between the presence of an ARG and the onset of HIV using SNPstats online software enabled us to show the protective effect of Trim 5α(136Q) (p < 0.0001) CCR5 Promoter 59029 G (p < 0.001), CCR2-64I (p = 0.022) and SDF1-3ʹA (p < 0.001). In addition, the combination of these ARGs showed a more pronounced protective effect against HIV acquisition (p < 0.00001).
Multitude of studies on ARGs have been conducted in various regions of Cameroon. It was important to compare the allelic frequencies of the ARGs studied to those of other regions, as in Cameroon HIV prevalence va ries according to regions (see Figure 1). The study revealed a significantly higher prevalence of protective gene variants in the study region compared to other regions in Cameroon. Table 5 presents the distribution of ARG frequencies in other parts of the country in comparison to the northern Regions of Cameroon.

Figure 1. Cameroon map showing the distribution of HIV prevalence according to regions in 2020. https://phia.icap.columbia.edu/wp-content/uploads/2017/02/CAMPHIA-Summary-Sheet-EN_ARV-adjusted_Feb2020.pdf.
The overall analysis of genotype data did not show any deviation from the Hardy-Weinberg expected frequency, the χ2 tests showed that all genes’ loci in HIV infected or uninfected are in equilibrium (p = 0.48). The observed genotype frequencies had no significant difference from the frequencies expected in each group, indicating that the five alleles are effectively in genetic equilibrium.
The overall prevalence of HIV infection in this study was 1.82%. This prevalence was not statistically different from that published by the DHS in 2018 (p = 0.88), namely, 1.5% and 1.6% for the North and Far North regions of Cameroon (DHS cameroon, 2018). The northern regions have a high proportion of Muslims, and polygamy is permitted within this religious tradition. In light of the resurgence of other sexually transmitted infections (STIs) in Cameroon (Nkenfou et al., 2018), which are known to be risk factors for HIV, there is a potential for an increase in the transmission of HIV.
This contributes to the elevated prevalence of hepatitis B in these regions, which can be attributed to customary practices of sharing food items, such as eating from the same dish. Furthermore, a comparable study conducted by Mboppi and colleagues in a district of southern Cameroon indicated that muslim religion was associated with a high prevalence of sexually transmitted infections (STIs) and HIV infection (Mbopi-Keou et al., 2014). It is likely that these factors, including polygamy, early marriage, low rates of condom use, and poverty, may contribute to the high prevalence of STIs and HIV in this region. In addition, our study was carried out in hospital settings and not in the community. In addition, these regions have been subjected to armed conflict for an extended period, resulting in an influx of refugees (BANQUE MONDIALE). This could potentially compromise the resilience of the health system and contribute to an increase in HIV cases (Mishra et al., 2021).
The genotypes for the mutant CCR5Δ32 gene were 98.96%, 1.04% and 00% respectively for wild-type, heterozygous and homozygous mutants, with an allelic frequency of the protective gene of 0.52%. This distribution corroborates several studies conducted in Cameroon and Africa where the Δ32 mutation is very rare or even absent (Nkenfou et al., 2013; Dambaya et al., 2019; Ma et al., 2005; Nyambi et al., 2002). However, studies conducted in Cameroon have demonstrated that this mutation is completely absent in the Central, Western, and Eastern Regions. The presence of the mutant and protective allele at a frequency of 0.5% in this region may explain the low HIV prevalence observed. In host cell infection mechanisms, HIV recruits a CCR5 coreceptor in addition to the CD4 receptor to fuse with the host membrane (Kassaye et al., 2009; Weichseldorfer et al., 2022). This specific mutation of the CCR5 co-receptor gene, designated CCR5Δ32, is characterized by a deletion of 32 base pairs in its coding region. This mutation results in the production of a truncated receptor that is incapable of being recruited by HIV, and hence of infection. The CCR5 Δ32 mutation confers strong host resistance to HIV infection (Angela Covino et al., 2016). Indeed, the Berlin patient and London patient have achieved long-term remission of HIV following bone marrow transplants from donors with the CCR5Δ32 mutation (homozygotes) (Gupta et al., 2019; Ding et al., 2021; Prator et al., 2020; Jilg and Li, 2019; Gupta et al., 2020b). Given its role in the pathogenesis of HIV infection, this mutation represents a target for antiretroviral drugs. Additionally, there is hope of a vaccine (Ensoli et al., 2021) or gene therapy (Li et al., 2022; Kaminski et al., 2016). Although this mutation is rare or absent in Africa, it is generally present in populations living in ethnically homogeneous communities, such as Caucasian populations or the Vlachet Gypsies of Hungary (Juhász et al., 2012; Ongadi et al., 2018). This observation may provide an explanation for the presence of this genotype in the Moudang and Massas, who live in ethnically homogeneous communities.
CCR2-64I frequency was 12.76%, 67.71% and 19.53% for wild-type, heterozygous and mutant genotypes respectively with an allelic frequency of the protective mutation of 36.46% (Table 5). The allelic frequency of the CCR2-64I genotype is statistically similar to those studied in 7 ethnic groups in Cameroon, where the proportion ranged from 10.8% to 31.3% (Ma et al., 2005). In a population of children and adolescents living with HIV in the Central Region, the heterozygous CCR2-64I genotype was found in 10.25%, 48.93%, and 80% of rapid progressors (RP), slow progressors (SP), and long-term non-progressors (LTNP), respectively, with a significant difference between rapid progressors and slow progressors (p = 0.0001) and slow progressors and long-term non-progressors LTNP (p = 0.0002), suggesting a protective effect of the mutant allele (DHS cameroon, 2018). The same study showed that the homozygous CCR2-64I protective gene was present only in slow progressors at 2.12% compared with 19.53% in our study (p = 0.003). Moreover, the homozygous CCR2-64I protective gene was present at 17.6% in the study conducted by Nkenfou et al., in 2013 in the West Cameroon Region (Sawyer et al., 2006). Studies around the world have shown the protective effects of this mutation in the acquisition or progression to the AIDS stage certain populations (Köksal et al., 2021; Apostolakis et al., 2005; Farissi et al., 2020). Nevertheless, a study conducted in Korea demonstrated that this mutation is associated with a rapid progression to AIDS or death, contingent on the stage of HIV infection and interactions with other genetic variants (Choi et al., 2007).
The frequencies observed for the SDF1 genotypes were 10.94%, 85.16%, and 3.9%, respectively, for wildtype, heterozygous, and mutant genotypes. The allelic frequency of the SDF1-3′A protective mutation was 25.19%, which was significantly higher than in studies carried out in the Western and Central Regions or in seven ethnic groups in Cameroon (p < 0.001) (Nakajima et al., 2009; Javanbakht et al., 2006; Goldschmidt et al., 2006). SDF1 codes for a ligand of the CXCR4 receptor, and its presence is linked to either delayed progression of AIDS or rapid progression of the disease and death. Additionally, it has been shown to influence the onset of known neurodegenerative diseases (Guerini et al., 2008). SDF1 is a ligand of the CXCR4 receptor that has been demonstrated to influence the progression to the AIDS stage. The SDF-1 gene is distinguished by the substitution of guanine for adenine at position 801 at the 3′untranslated end of the SDF-1 gene (Agrawal et al., 2004). The global, regional, and ethnic frequency distributions of SDF1 genotypes and the SDF1-3′-A allele exhibit considerable variability (Reiche et al., 2006; Modi et al., 2005; Ding et al., 2018). Although this mutation rate is significant, it is within the range found in most studies conducted in Africa and around the world over the years (Su et al., 1999; Tan et al., 2010).
The frequencies of 59029 A/G CCR5 promoter genotypes were 12.24%; 66.93%; 20.83% for wildtype, heterozygous and mutant. The mutated promoter gene, with an allele frequency of 37.56%, was significantly higher than that observed in other studies in Cameroon (p < 0.08). The CCR5-G59029 allele is characterized by a substitution of A-G in the promoter region, resulting in decreased promoter activity and reduced coreceptor expression (Mehlotra et al., 2015). Individuals homozygous for the CCR5-G59029 gene progress to AIDS on average 3.8 years slower than individuals heterozygous for CCR5A59029 (Gornalusse et al., 2015). In fact, some recent data show regulation of CD4+ T lymphocyte apoptosis induced by expression of genes for this co-receptor in patients infected with HIV (Joshi et al., 2017). More specifically, the CCR5-A59029 mutation has been shown to slow down chemokine co-receptor gene expression by inhibiting the recruitment of transcription factors (Jiang et al., 2011) or insensitive to transcriptional activation by HIV (Gornalusse et al., 2015). The presence of this mutation could perhaps explain the low prevalence of HIV in these two regions of Cameroon, in line with other studies conducted worldwide (Mehlotra et al., 2015; Rathore et al., 2009; Coloccini et al., 2014).
The frequency of genotypes of Trim5α gene was 3.12%, 36.72%, and 60.16% for wild-type, heterozygous, and mutant genotypes, respectively, with an allelic frequency of 69.33%. This mutation was significantly higher in the northern population compared to the central population where the allelic frequency was 42.26% (p < 0.001) (Javanbakht et al., 2006). The tripartite interaction motif 5α (Trim5α) is recognized as an inhibitory factor blocking infections by retroviruses such as HIV. It is a factor that can interact with RNA just after decapsidation, forming an addressed proteasome capable of being recognized and digested (Kim et al., 2019; Anderson et al., 2006). This genotype has been identified in several studies as an innate factor restricting HIV infection (Gupta and Padh, 2012; Jang et al., 2008). This mutation could also be associated with the low prevalence of HIV in these two regions of Cameroon.
Overall, protective ARGs interact directly with HIV during the early stages of infection. Their natural antiviral activities are more effective when these genes are present in the majority of individuals in a given population. While homozygous CCR5∆32 individuals are protected against viral infection, heterozygotes progress very slowly towards the AIDS stage. So the more an individual has a variety of protective genes, the more the infection is inhibited and the progression of the disease to AIDS is slowed.
The slow progression of infection to the final stage has implications for the rate of spread and prevalence of HIV. In fact, the rapid progression of HIV to the final stage has been associated with an increase in virulence and pathogenicity (Coloccini et al., 2014; Anderson et al., 2006). In HIV + patients in the AIDS stage, viral tropism has evolved from R5 to R4 or both, meaning that the virus can infect a wide range of immune cells with an increased viral load (Naif, 2013), which favors transmission. HIV transmission depends on donor viral load, viral tropism, and host immunity, among other factors (Anderson et al., 2006; Fraser et al., 2014). Thus, a more virulent transmitted virus will evolve more rapidly in a population that does not have strong immunogenic resistance.
It is important to consider the potential implications of conducting a gene therapy clinical trial in a population that does not have a high frequency of protective genes (ARGs) (Casado et al., 2020). This could result in immunological pressure on the virus, which may lead to viral escape through a change in tropism, from R5 to R4 or R5/R4, or even CCR2 (Islam et al., 2021). Indeed, the potential for viral escape has implications beyond mere therapeutic failure. More significantly, it could result in the selection of pathogenic and virulent mutations (Arif et al., 2017; Johnson et al., 2022). It is therefore suggested to conduct a clinical trial on a population with a high allelic frequency of protective genes. The current HIV treatments are designed to limit viral replication; however, stopping them immediately results in a rebound in viral replication (Bailón et al., 2022; De Scheerder et al., 2019). Gene therapy, as evidenced by the London and Berlin patients, mimicking this using the CRISPR-Cas9 system in a population that is already resistant to HIV infection, could prove to be a viable solution.
The data provided by ARGs are of interest for gene therapy planning because it would be more interesting and effective to conduct clinical trials in a population with a high frequency of protective genes. Modern gene therapies use gene-editing molecular scissors such as the CRISPR-Cas9 system (Bhowmik and Chaubey, 2022; Akram et al., 2023) or ZFN (Didigu et al., 2014; Ishida, 2020). These molecular scissors can be used to induce CCR5∆32 or CCR5proA/G mutations in a population at risk but with a high frequency of other protective genes. However, given the security problems that have prevailed in these regions of northern Cameroon for several years, the influx of refugees (BANQUE MONDIALE) could affect the gene pool by altering the Harding-Weinberg equilibrium of protective ARGs. Three hypotheses may be posited regarding the potential impact of the influx of refugees on the genetic structure of ARGs in these regions. The initial hypothesis is that the refugees will reduce the allelic frequency of the ARGs prior to the commencement of a clinical trial. The consequence would be a high probability of viral escape, therapeutic failure, and, at worst, a selection mutation. The second desirable scenario would be to carry out the same study in refugee populations, to measure the allele frequency of ARG genes before a clinical trial. The third hypothesis is probable that the genetic impact of refugees will be insignificant, given that they exhibited comparable socio-cultural traits to those of the northern Cameroon.
The therapeutic potential of gene therapy will be the subject of several studies over the coming years. The objective is to describe the genetic structure of the entire target population and the associated immuno-genetic mechanisms in several scenarios.
The size of our study population was not large enough. A larger study would yield more conclusive results by incorporating ethnicity as a variable.
HLA-B gene variants, particularly HLA-B27 and HLA-B57 (Loffredo et al., 2009), have been demonstrated to exhibit resistance to HIV in individuals who possess these variants. Further investigation into the role of additional genetic factors, such as HLA genes, as well as defensins gene (Lafferty et al., 2017), KIR (Carr et al., 2007), APOBEC3′A (McDonnell et al., 2021), and others (He and Wu, 2019) (miRNA through epigenetics), in HIV resistance would be of significant interest.
The aim of this study was to understand the low prevalence of HIV in the North and Far North regions of Cameroon. It seems that the five genes associated with HIV infection (CCR5Δ32, CCR2-64I, SDF1-3ʹA, CCR5 promoter 59029 A/G and TRIM 5α138Q) could explain the low prevalence of HIV infection in these two regions. These genes were found at a higher frequency in these two regions compared to the other regions of Cameroon and can explain the low prevalence of HIV in the North of Cameroon. Knowing that HIV infection is the result of a combination of factors, monitoring of the epidemic should be continuous and even intensified, given the frequent displacement of refugees due to political insecurity, which can weaken the health system and jeopardize efforts to eliminate HIV.
In the perspectives of our work where we will be looking for mutation CCR5∆32 in Moudang and Massa ethnic groups.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
The studies involving humans were approved by the Comité National d’Ethique pour la Recherche en Santé Humaine, N°2021/06/85/CE/CNERSH/SP. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
PD: Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. MaD: Formal Analysis, Investigation, Methodology, Writing–review and editing. CN-T: Formal Analysis, Investigation, Methodology, Writing–review and editing. MN: Formal Analysis, Investigation, Methodology, Writing–review and editing. EE: Formal Analysis, Investigation, Methodology, Writing–review and editing. AT: Formal Analysis, Investigation, Methodology, Writing–review and editing. CM: Formal Analysis, Investigation, Methodology, Writing–review and editing. JK: Formal Analysis, Investigation, Methodology, Writing–review and editing. MoD: Formal Analysis, Investigation, Methodology, Writing–review and editing. AN: Formal Analysis, Supervision, Validation, Writing–review and editing. CN: Conceptualization, Formal Analysis, Supervision, Validation, Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ARV, Antiretroviral HIV drugs; ARG, AIDS Related Genes; AIDS, Acquired immunodeficiency syndrome; CCR5 or CD195, C-C chemokine receptor type 5; CRISPR-, Cluster of Regularly Interspaced Short Palindromic Repeats-; Cas9, CRISPR-associated 9; HIV, Human immunodeficiency virus; SDF1, Stromal Derivated Factor-1; Trim 5α, Tripartite motif-containing protein 5alpha; ZFN, Zinc Finger.
1https://phia.icap.columbia.edu/wp-content/uploads/2017/02/CAMPHIA-Summary-Sheet-EN_ARV-adjusted_Feb2020.pdf
Abrahams, M. R., Anderson, J. A., Giorgi, E. E., Seoighe, C., Mlisana, K., Ping, L. H., et al. (2009). Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J. Virol. 83 (8), 3556–3567. doi:10.1128/JVI.02132-08
Agrawal, L., Lu, X., Qingwen, J., VanHorn-Ali, Z., Nicolescu, I. V., McDermott, D. H., et al. (2004). Role for CCR5Delta32 protein in resistance to R5, R5X4, and X4 human immunodeficiency virus type 1 in primary CD4+ cells. J. Virol. 78 (5), 2277–2287. doi:10.1128/jvi.78.5.2277-2287.2004
Ahmed, R. A., Adekoya, K. O., Onwuamah, C. K., Oboh, B. O., Iyer, S. S., Oluwatosin, A. S., et al. (2022). Mechanism of viral suppression among HIV elite controllers and long-term nonprogressors in Nigeria and South Africa. Viruses 14 (6), 1270. doi:10.3390/v14061270
Akram, F., Sahreen, S., Aamir, F., Haq, I. ul, Malik, K., Imtiaz, M., et al. (2023). An insight into modern targeted genome-editing technologies with a special focus on CRISPR/Cas9 and its applications. Mol. Biotechnol. 65, 227–242. doi:10.1007/s12033-022-00501-4
Ammaranond, P., van Bockel, D. J., Petoumenos, K., McMurchie, M., Finlayson, R., Middleton, M. G., et al. (2011). HIV immune escape at an immunodominant epitope in HLA-B*27–Positive individuals predicts viral load outcome. J. Immunol. 186, 479–488. doi:10.4049/jimmunol.0903227
Anderson, J. L., Campbell, E. M., Wu, X., Vandegraaff, N., Engelman, A., and Hope, T. J. (2006). Proteasome inhibition reveals that a functional preintegration complex intermediate can Be generated during restriction by diverse TRIM5 proteins. J. Virol. 80 (19), 9754–9760. doi:10.1128/JVI.01052-06
Angela Covino, D., Sabbatucci, M., and Fantuzzi, L. (2016). The CCL2/CCR2 Axis in the pathogenesis of HIV-1 infection: a new cellular target for therapy? Curr. Drug Targets 17 (1), 76–110. doi:10.2174/138945011701151217110917
Apostolakis, S., Baritaki, S., Krambovitis, E., and Spandidos, D. A. (2005). Distribution of HIV/AIDS protective SDF1, CCR5 and CCR2 gene variants within Cretan population. J. Clin. Virol. 34 (4), 310–314. doi:10.1016/j.jcv.2005.01.010
Arenzana-Seisdedos, F. (2015). SDF-1/CXCL12: a chemokine in the life cycle of HIV. Front. Immunol. 6, 256. doi:10.3389/fimmu.2015.00256
Arif, M. S., Hunter, J., Léda, A. R., Zukurov, J. P. L., Samer, S., Camargo, M., et al. (2017). Pace of coreceptor tropism switch in HIV-1-Infected individuals after recent infection. J. Virol. . 12 Sept. 91 (19). doi:10.1128/jvi.00793-17
Bailón, L., Llano, A., Cedeño, S., Escribà, T., Rosás-Umbert, M., Parera, M., et al. (2022). Safety, immunogenicity and effect on viral rebound of HTI vaccines in early treated HIV-1 infection: a randomized, placebo-controlled phase 1 trial. Nat. Med. 28, 2611–2621. doi:10.1038/s41591-022-02060-2
BANQUE MONDIALE. World Bank. 2023. Cameroun - vue d’ensemble. Available at: https://www.banquemondiale.org/fr/country/cameroon/overview.
Bhowmik, R., and Chaubey, B. (2022). CRISPR/Cas9: a tool to eradicate HIV-1. AIDS Res. Ther. 1 déc 19 (1), 58. doi:10.1186/s12981-022-00483-y
Brambilla, A., Villa, C., Rizzardi, G., Veglia, F., Ghezzi, S., Lazzarin, A., et al. (2000). Shorter survival of SDF1-3′A/3′A homozygotes linked to CD4+ T cell decrease in advanced human immunodeficiency virus type 1 infection. J. Infect. Dis. 182 (1), 311–315. doi:10.1086/315650
Burgoyne, R. W., and Tan, D. H. S. (2008). Prolongation and quality of life for HIV-infected adults treated with highly active antiretroviral therapy (HAART): a balancing act. J. Antimicrob. Chemother. 61 (3), 469–473. doi:10.1093/jac/dkm499
Burton, C. T., Gotch, F. M., and Imami, N. (2005). CCR2/64I mutation detection in a HIV-1-positive patient with slow CD4 T-cell decline and delay in disease progression. Int. J. STD AIDS 16 (5), 392–394. doi:10.1258/0956462053888817
Carr, W. H., Rosen, D. B., Arase, H., Nixon, D. F., Michaelsson, J., and Lanier, L. L. (2007). Cutting edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J. Immunol. 178, 647–651. doi:10.4049/jimmunol.178.2.647
Casado, C., Galvez, C., Pernas, M., Tarancon-Diez, L., Rodriguez, C., Sanchez-Merino, V., et al. (2020). Permanent control of HIV-1 pathogenesis in exceptional elite controllers: a model of spontaneous cure. Sci. Rep. 10 (1), 1902. doi:10.1038/s41598-020-58696-y
Chan, E., Towers, G. J., and Qasim, W. (2014). Gene therapy strategies to exploit TRIM derived restriction factors against HIV-1. Viruses 6 (1), 243–263. doi:10.3390/v6010243
Choi, B. S., Choi, J. H., Kim, S. S., Kee, M. K., and Lee, J. S. (2007). CCR2b-64I allelic polymorphisms in advanced HIV-infected Koreans accelerate disease progression. AIDS Res. Hum. Retroviruses 23 (6), 805–811. doi:10.1089/aid.2006.0133
Coloccini, R. S., Dilernia, D., Ghiglione, Y., Turk, G., Laufer, N., Rubio, A., et al. (2014). Host genetic factors associated with symptomatic primary HIV infection and disease progression among argentinean seroconverters. PLOS ONE 9, e113146. doi:10.1371/journal.pone.0113146
Dambaya, B., Nkenfou, C. N., Mekue, L., Této, G., Ngoufack, N., Ambada, G., et al. (2019). TRIM5α 136Q, CCR5 promoter 59029G and CCR264I alleles impact the progression of HIV in children and adolescents. Appl. Clin. Genet. 12, 203–211. doi:10.2147/TACG.S205335
De Scheerder, M. A., Vrancken, B., Dellicour, S., Schlub, T., Lee, E., Shao, W., et al. (2019). HIV rebound is predominantly fueled by genetically identical viral expansions from diverse reservoirs. Cell Host Microbe 26, 347–358. doi:10.1016/j.chom.2019.08.003
DHS cameroon (2018). Demographic and health survey Cameroon. Available at: https://dhsprogram.com/pubs/pdf/OF44/OF44.HIV.E.pdf.
Didigu, C. A., Wilen, C. B., Wang, J., Duong, J., Secreto, A. J., Danet-Desnoyers, G. A., et al. (2014). Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood 123 (1), 61–69. doi:10.1182/blood-2013-08-521229
Ding, J., Liu, Y., and Lai, Y. (2021). Knowledge from London and Berlin: finding threads to a functional HIV cure. Front. Immunol. 12, 688747. doi:10.3389/fimmu.2021.688747
Ding, J., Zhao, J., Zhou, J., Li, X., Wu, Y., Ge, M., et al. (2018). Association of gene polymorphism of SDF1(CXCR12) with susceptibility to HIV-1 infection and AIDS disease progression: a meta-analysis. PLOS ONE 13, e0191930. doi:10.1371/journal.pone.0191930
Ensoli, B., Moretti, S., Borsetti, A., Maggiorella, M. T., Buttò, S., Picconi, O., et al. (2021). New insights into pathogenesis point to HIV-1 Tat as a key vaccine target. Arch. Virol. 166, 2955–2974. doi:10.1007/s00705-021-05158-z
Farissi, F. Z., El Annaz, H., Melloul, M., El Alaoui, M. A., Tagajdid, M. R., Abi, R., et al. (2020). Analysis of the CCR2-64I (rs1799864) genetic polymorphism distribution and its effect on the risk of HIV-1 infection and immunovirological outcomes in Moroccan ART-treated individuals. Gene Rep. 20, 100715. doi:10.1016/j.genrep.2020.100715
Fraser, C., Lythgoe, K., Leventhal, G. E., Shirreff, G., Hollingsworth, T. D., Alizon, S., et al. (2014). Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 343, 1243727. doi:10.1126/science.1243727
Goldschmidt, V., Bleiber, G., May, M., Martinez, R., Ortiz, M., Telenti, A., et al. (2006). Role of common human TRIM5alpha variants in HIV-1 disease progression. Retrovirology 3, 54. doi:10.1186/1742-4690-3-54
Gornalusse, G. G., Mummidi, S., Gaitan, A. A., Jimenez, F., Ramsuran, V., Picton, A., et al. (2015). Epigenetic mechanisms, T-cell activation, and CCR5 genetics interact to regulate T-cell expression of CCR5, the major HIV-1 coreceptor. Proc. Natl. Acad. Sci. doi:10.1073/pnas.1423228112
Guerini, F. R., Delbue, S., Zanzottera, M., Agliardi, C., Saresella, M., Mancuso, R., et al. (2008). Analysis of CCR5, CCR2, SDF1 and RANTES gene polymorphisms in subjects with HIV-related PML and not determined leukoencephalopathy. Biomed. Pharmacother. 62, 26–30. doi:10.1016/j.biopha.2007.04.005
Gupta, A., and Padh, H. (2012). The global distribution of CCR5 delta 32 polymorphism: role in HIV-1 protection. BMC Infect. Dis. 12, O16. doi:10.1186/1471-2334-12-S1-O16
Gupta, R. K., Abdul-Jawad, S., McCoy, L. E., Mok, H. P., Peppa, D., Salgado, M., et al. (2019). HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature 568 (7751), 244–248. doi:10.1038/s41586-019-1027-4
Gupta, R. K., Peppa, D., Hill, A. L., Gálvez, C., Salgado, M., Pace, M., et al. (2020a). Evidence for HIV-1 cure after CCR5Δ32/Δ32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: a case report. Lancet HIV 7 (5), e340–e347. doi:10.1016/S2352-3018(20)30069-2
Gupta, R. K., Peppa, D., Hill, A. L., Gálvez, C., Salgado, M., Pace, M., et al. (2020b). Evidence for HIV-1 cure after CCR5Δ32/Δ32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: a case report. Lancet HIV 7 (5), e340–e347. doi:10.1016/S2352-3018(20)30069-2
He, S., and Wu, Y. (2019). Relationships between HIV-mediated chemokine coreceptor signaling, cofilin hyperactivation, viral tropism switch and HIV-mediated CD4 depletion. Curr. HIV Res. 1 déc 17 (6), 388–396. doi:10.2174/1570162X17666191106112018
Ishida, T. (2020). Virucidal activities of zinc-finger antiviral proteins and zinc-binding domains for virus entry, DNA/RNA replication and spread. Edelweiss J. Biomed. Res. Rev. doi:10.33805/2690-2613.109
Islam, S., Moni, M. A., Urmi, U. L., Tanaka, A., and Hoshino, H. (2021). C-C Chemokine receptor-like 2 (CCRL2) acts as coreceptor for human immunodeficiency virus-2. Brief. Bioinform 22, bbaa333. doi:10.1093/bib/bbaa333
Jang, D. H., Choi, B. S., and Kim, S. S. (2008). The effects of RANTES/CCR5 promoter polymorphisms on HIV disease progression in HIV-infected Koreans. Int. J. Immunogenet 35, 101–105. doi:10.1111/j.1744-313X.2007.00743.x
Javanbakht, H., An, P., Gold, B., Petersen, D. C., O’Huigin, C., Nelson, G. W., et al. (2006). Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology 354, 15–27. doi:10.1016/j.virol.2006.06.031
Jiang, D., Mummidi, S., Ahuja, S. K., and Jarrett, H. W. (2011). CCR5 promoter haplotype transcription complex characterization. J. Health Care Poor Underserved 22, 73–90. doi:10.1353/hpu.2011.0169
Jilg, N., and Li, J. Z. (2019). On the Road to a HIV cure: moving beyond Berlin and London. Infect. Dis. Clin. 33 (3), 857–868. doi:10.1016/j.idc.2019.04.007
Johnson, M. M., Jones, C. E., and Clark, D. N. (2022). The effect of treatment-associated mutations on HIV replication and transmission cycles. Viruses 15, 107. doi:10.3390/v15010107
Joshi, A., Punke, E. B., Sedano, M., Beauchamp, B., Patel, R., Hossenlopp, C., et al. (2017). CCR5 promoter activity correlates with HIV disease progression by regulating CCR5 cell surface expression and CD4 T cell apoptosis. Sci. Rep. 7, 232. doi:10.1038/s41598-017-00192-x
Juhász, E., Béres, J., Kanizsai, S., and Nagy, K. (2012). The consequence of a founder effect: CCR5-∆32, CCR2-64I and SDF1-3’A polymorphism in vlach gypsy population in Hungary. Pathol. Oncol. Res. 18 (2), 177–182. doi:10.1007/s12253-011-9425-4
Kaminski, R., Bella, R., Yin, C., Otte, J., Ferrante, P., Gendelman, H. E., et al. (2016). Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther. 23 (8), 690–695. doi:10.1038/gt.2016.41
Kassaye, S., Johnston, E., McColgan, B., Kantor, R., Zijenah, L., and Katzenstein, D.(2009). Envelope coreceptor tropism, drug resistance, and viral evolution among subtype C HIV-1-Infected individuals receiving nonsuppressive antiretroviral therapy. JAIDS J. Acquir Immune Defic. Syndr. doi:10.1097/QAI.0b013e31818ffdff
Kim, K., Dauphin, A., Komurlu, S., McCauley, S. M., Yurkovetskiy, L., Carbone, C., et al. (2019). Cyclophilin A protects HIV-1 from restriction by human TRIM5α. Nat. Microbiol. 4 (12), 2044–2051. doi:10.1038/s41564-019-0592-5
Köksal, M. O., Akgül, B., Beka, H., Çiftçi, S., Keskin, F., Eraksoy, H., et al. (2021). Frequency of CCR5-Δ32, CCR2-64I and SDF1-3’A alleles in HIV-infected and uninfected patients in Istanbul, Turkey. J. Infect. Dev. Ctries. 15, 1183–1189. doi:10.3855/jidc.12861
Kristiansen, T. B., Knudsen, T. B., Ohlendorff, S., and Eugen-Olsen, J. (2001). A new multiplex PCR strategy for the simultaneous determination of four genetic polymorphisms affecting HIV-1 disease progression. J. Immunol. Methods 252, 147–151. doi:10.1016/s0022-1759(01)00349-0
Lafferty, M., Sun, L., Christensen-Quick, A., Lu, W., and Garzino-Demo, A. (2017). Human beta defensin 2 selectively inhibits HIV-1 in highly permissive CCR6⁺CD4⁺ T cells. Viruses 9 (5), 111. doi:10.3390/v9050111
Li, S., Holguin, L., and Burnett, J. C. (2022). CRISPR-Cas9-mediated gene disruption of HIV-1 co-receptors confers broad resistance to infection in human T cells and humanized mice. Mol. Ther. - Methods Clin. Dev. 24, 321–331. doi:10.1016/j.omtm.2022.01.012
Loffredo, J. T., Sidney, J., Bean, A. T., Beal, D. R., Bardet, W., Wahl, A., et al. (2009). Two MHC class I molecules associated with elite control of immunodeficiency virus replication, mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J. Immunol. 182, 7763–7775. doi:10.4049/jimmunol.0900111
Lot, F. (2008). Épidémiologie du VIH/sida et des autres infections sexuellement transmissibles chez les femmes. médecine/sciences 24, 7–19. doi:10.1051/medsci/2008242s7
Lwanga, S. K., Lemeshow, S., and Organization, W. H. (1991). Détermination de la taille d’ un échantillon dans les études sanométriques: manuel pratique. Organ. Mond. Santé. Available at: https://apps.who.int/iris/handle/10665/36881.
Ma, L., Marmor, M., Zhong, P., Ewane, L., Su, B., and Nyambi, P. (2005). Distribution of CCR2-64I and SDF1-3′A alleles and HIV status in 7 ethnic populations of Cameroon. JAIDS J. Acquir Immune Defic. Syndr. doi:10.1097/01.qai.0000157008.66584.d6
Manen, D. van, Rits, M. A. N., Beugeling, C., Dort, K. van, Schuitemaker, H., and Kootstra, N. A. (2008). The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLOS Pathog. doi:10.1371/journal.ppat.0040018
Mbopi-Keou, F. X., Nguefack-Tsague, G., Mireille, G. C., Abo’o Abessolo, S., Angwafo, I. I. I. F., and Muna, W. (2014). Facteurs de risque de l’infection par le VIH dans le district de santé de Meyomessala au Cameroun. Pan Afr. Med. J. 18. doi:10.11604/pamj.2014.18.161.3238
McDonnell, M. M., Karvonen, S. C., Gaba, A., Flath, B., Chelico, L., and Emerman, M. (2021). Highly-potent, synthetic APOBEC3s restrict HIV-1 through deamination-independent mechanisms. PLOS Pathog. 17, e1009523. doi:10.1371/journal.ppat.1009523
Mehlotra, R. K., Hall, N. B., Bruse, S. E., John, B., Blood Zikursh, M. J., Stein, C. M., et al. (2015). CCR2, CCR5, and CXCL12 variation and HIV/AIDS in Papua New Guinea. Infect. Genet. Evol. 36, 165–173. doi:10.1016/j.meegid.2015.09.014
Mellado, M., Rodríguez-Frade, J. M., Vila-Coro, A. J., de Ana, A. M., and Martínez, A. C. (1999). Chemokine control of HIV-1 infection. Nature 400, 723–724. doi:10.1038/23382
Metodiev, K. (2012). Immunodeficiency. London, Unite Kingdom: IntechOpen Limited. BoD – Books on Demand, 406.
Mishra, D., O`Laughlin, K., and Spiegel, P. (2021). A systematic review evaluating HIV prevalence among conflict-affected populations, 2005-2020. Aids Rev. 23 (3), 6530. doi:10.24875/aidsrev.200001311
Modi, W. S., Scott, K., Goedert, J. J., Vlahov, D., Buchbinder, S., Detels, R., et al. (2005). Haplotype analysis of the SDF-1 (CXCL12) gene in a longitudinal HIV-1/AIDS cohort study. Genes Immun. 6, 691–698. doi:10.1038/sj.gene.6364258
Naif, H. M. (2013). Pathogenesis of HIV infection. Infect. Dis. Rep. 5, e6. doi:10.4081/idr.2013.s1.e6
Nakajima, T., Nakayama, E. E., Kaur, G., Terunuma, H., Mimaya, J. ich, Ohtani, H., et al. (2009). Impact of novel TRIM5alpha variants, Gly110Arg and G176del, on the anti-HIV-1 activity and the susceptibility to HIV-1 infection. AIDS 23, 2091–2100. doi:10.1097/QAD.0b013e328331567a
Nkenfou, C., Njolle, N. E., Nguefeu, C., Moudourou, S. A., Fainguem, N., Fokam, J., et al. (2018). Syphilis is on the rise among pregnant women in Cameroon. J. Virus Erad. 4, 36–37. doi:10.1016/s2055-6640(20)30432-5
Nkenfou, C. N., Mekue, L. C. M., Nana, C. T., and Kuiate, J. R. (2013). Distribution of CCR5-Delta32, CCR5 promoter 59029 A/G, CCR2-64I and SDF1-3’A genetic polymorphisms in HIV-1 infected and uninfected patients in the West Region of Cameroon. BMC Res. Notes 6 (1), 288. doi:10.1186/1756-0500-6-288
Nkenfou, C. N., Tchakounté, C., Nkenfou-Tchinda, C. N., Ngoufack, M. N., Yatchou, L. G., Elong, E., et al. (2021). Protective effect of the combination of wild-type genotypes (G/G and G/G) of CCR2-64V and SDF1-3A’ genes in serodiscordant couples in yaounde-Cameroon. Curr. HIV Res. 1 juill 19 (4), 342–351. doi:10.2174/1570162X19666210412121143
Nyambi, P., Zekeng, L., Kenfack, H., Tongo, M., Nanfack, A., Nkombe, I., et al. (2002). HIV infection in rural villages of Cameroon. JAIDS J. Acquir Immune Defic. Syndr.
Ongadi, B. A., Obiero, G., Lihana, R. W., and Kiiru, J. N. (2018). Distribution of genetic polymorphism in the CCR5 among caucasians, asians and africans: a systematic review and meta-analysis. Open J. Genet. 08(03):54. doi:10.4236/ojgen.2018.83006
Prator, C. A., Donatelli, J., and Henrich, T. J. (2020). From Berlin to London: HIV-1 reservoir reduction following stem cell transplantation. Curr. HIV/AIDS Rep. 17 (4), 385–393. doi:10.1007/s11904-020-00505-2
Rathore, A., Chatterjee, A., Sivarama, P., Yamamoto, N., Singhal, P. K., and Dhole, T. N. (2009). Association of CCR5-59029 A/G and CCL3L1 copy number polymorphism with HIV type 1 transmission/progression among HIV type 1-seropositive and repeatedly sexually exposed HIV type 1-seronegative North Indians. AIDS Res. Hum. Retroviruses 25 (11), 1149–1156. doi:10.1089/aid.2008.0019
Reiche, E. M. V., Watanabe, M. a. E., Bonametti, A. M., Morimoto, H. K., Morimoto, A. A., Wiechmann, S. L., et al. (2006). Stromal cell-derived factor 1 (SDF1) genetic polymorphism in a sample of healthy individuals, seronegative individuals exposed to human immunodeficiency virus type 1 (HIV-1) and patients infected with HIV-1 from the Brazilian population. Int. J. Immunogenet 33 (2), 127–133. doi:10.1111/j.1744-313X.2006.00583.x
Sadri, F., Rezaei, Z., and Fereidouni, M. (2022). The significance of the SDF-1/CXCR4 signaling pathway in the normal development. Mol. Biol. Rep. 49 (4), 3307–3320. doi:10.1007/s11033-021-07069-3
Sawyer, S. L., Wu, L. I., Akey, J. M., Emerman, M., and Malik, H. S. (2006). High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5alpha in humans. Curr. Biol. Cb. 10 janv 16 (1), 95–100. doi:10.1016/j.cub.2005.11.045
Sayan, M., Sargin, F., Inan, D., Sevgi, D. Y., Celikbas, A. K., Yasar, K., et al. (2016). HIV-1 transmitted drug resistance mutations in newly diagnosed antiretroviral-naive patients in Turkey. AIDS Res. Hum. Retroviruses 32 (1), 26–31. doi:10.1089/AID.2015.0110
Signoret, N., Oldridge, J., Pelchen-Matthews, A., Klasse, P. J., Tran, T., Brass, L. F., et al. (1997). Phorbol esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J. Cell Biol. 139 (3), 651–664. doi:10.1083/jcb.139.3.651
snpStats (2024). Bioconductor. snpStats. Available at: http://bioconductor.org/packages/snpStats/.
Sokolskaja, E., and Luban, J. (2006). Cyclophilin, TRIM5, and innate immunity to HIV-1. Curr. Opin. Microbiol. 9 (4), 404–408. doi:10.1016/j.mib.2006.06.011
Soriano, A., Martínez, C., García, F., Plana, M., Palou, E., Lejeune, M., et al. (2002). Plasma stromal cell–derived factor (SDF)-1 levels, SDF1-3′A genotype, and expression of CXCR4 on T lymphocytes: their impact on resistance to human immunodeficiency virus type 1 infection and its progression. J. Infect. Dis. 186 (7), 922–931. doi:10.1086/343741
Spínola, H., Couto, A. R., Peixoto, M. J., Anagnostou, P., Destro-Bisol, G., Spedini, G., et al. (2011). HLA class-I diversity in Cameroon: evidence for a north-south structure of genetic variation and relationships with african populations. Ann. Hum. Genet. 75 (6), 665–677. doi:10.1111/j.1469-1809.2011.00672.x
Su, B., Jin, L., Hu, F., Xiao, J., Luo, J., Lu, D., et al. (1999). Distribution of two HIV-1–Resistant polymorphisms (SDF1-3′A and CCR2-64I) in east asian and world populations and its implication in AIDS epidemiology. Am. J. Hum. Genet. 65, 1047–1053. doi:10.1086/302568
Tan, X. hua, yu, Z. J., hong, Di C., rong, Hu A., Yang, L., Qu, S., et al. (2010). Distribution of CCR5-Δ32, CCR5m303A, CCR2-64I and SDF1-3′A in HIV-1 infected and uninfected high-risk Uighurs in Xinjiang, China. Infect. Genet. Evol. doi:10.4236/ojgen.2018.83006
UNAIDS (2024). Suivi mondial de la lutte contre le sida 2024 — Indicateurs et questions pour le suivi des progrès de la Déclaration politique sur le VIH et le sida de 2021. Available at: https://www.unaids.org/fr/resources/documents/2024/global-aids-monitoring-guidelines.
Weichseldorfer, M., Tagaya, Y., Reitz, M., DeVico, A. L., and Latinovic, O. S. (2022). Identifying CCR5 coreceptor populations permissive for HIV-1 entry and productive infection: implications for in vivo studies. J. Transl. Med. 20, 39. doi:10.1186/s12967-022-03243-8
Winkler, C., Modi, W., Smith, M. W., Nelson, G. W., Wu, X., Carrington, M., et al. (1998). Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE study, hemophilia growth and development study (HGDS), multicenter AIDS cohort study (MACS), multicenter hemophilia cohort study (MHCS), San Francisco city cohort (SFCC). Science 279 (5349), 389–393. PMID: 9430590. doi:10.1126/science.279.5349.389
WMA The World Medical Association-Déclaration d’Helsinki de l’AMM – Principes éthiques applicables à la recherche médicale impliquant des êtres humains. Available at: https://www.wma.net/fr/policies-post/declaration-dhelsinki-de-lamm-principes-ethiques-applicables-a-la-recherche-medicale-impliquant-des-etres-humains/.
Keywords: AIDS restriction host genes, gene therapy, HIV, Northern Cameroon, risk factors, surveillance
Citation: Djataou P, Djuidje Ngounoue M, Nkenfou-Tchinda CN, Ngoufack MN, Elong E, Tiga A, Muluh C, Kadji Kameni J, Djaouda M, Ndjolo A and Nkenfou CN (2024) Low prevalence of HIV in the northern Cameroon: contribution of some AIDS restriction genes and potential implications for gene therapy. Front. Genet. 15:1447971. doi: 10.3389/fgene.2024.1447971
Received: 12 June 2024; Accepted: 22 August 2024;
Published: 13 September 2024.
Edited by:
Seik-Soon Khor, Nanyang Technological University, SingaporeReviewed by:
Thais Louvain De Souza, Faculdade de Medicina de Campos, BrazilCopyright © 2024 Djataou, Djuidje Ngounoue, Nkenfou-Tchinda, Ngoufack, Elong, Tiga, Muluh, Kadji Kameni, Djaouda, Ndjolo and Nkenfou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Celine Nguefeu Nkenfou, bmtlbmZvdUB5YWhvby5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.