
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 05 July 2024
Sec. RNA
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1429411
Pest control heavily relies on chemical pesticides has been going on for decades. However, the indiscriminate use of chemical pesticides often results in the development of resistance in pests. Almost all pests have developed some degree of resistance to pesticides. Research showed that the mechanisms of insecticide resistance in insects encompass metabolic resistance, behavioral resistance, penetration resistance and target-site resistance. Research on the these mechanisms has been mainly focused on the cis-regulatory or trans-regulatory for the insecticide resistance-related genes, with less attention paid to non-coding RNAs (ncRNAs), such as microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA). There has been increased studies focus on understanding how these ncRNAs are involved in post-transcriptional regulation of insecticide resistance-related genes. Besides, the formatted endogenous RNA (ceRNA) regulatory networks (lncRNA/circRNA-miRNA-mRNA) has been identified as a key player in governing insect resistance formation. This review delves into the functions and underlying mechanisms of miRNA, lncRNA, and circRNA in regulating insect resistance. ncRNAs orchestrate insect resistance by modulating the expression of detoxification enzyme genes, insecticide target genes, as well as receptor genes, effectively regulating both target-site, metabolic and penetration resistance in insects. It also explores the regulatory mechanisms of ceRNA networks in the development of resistance. By enhancing our understanding of the mechanisms of ncRNAs in insecticide resistance, it will not only provide valuable insights into the new mechanisms of insecticide resistance but also help to enrich new directions in ncRNAs gene regulation research.
Pests globally cause a significant reduction, estimated to be between 10% and 30% in annual food production, resulting in an economic loss of approximately $20 billion (Pimentel et al., 2000; Oerke, 2006). Pesticides are considered the primary and efficient method of directly controlling pests, with an annual usage surpassing one billion kilograms and an upward trend in specific countries (Alavanja, 2009). Nevertheless, the extensive use of insecticides has resulted in varying levels of resistance in over 600 insect species (Connor et al., 2011). Particularly, the diamondback moth (Plutella xylostella, DBM) exhibited the highest level of resistance, being resistant to 101 insecticides with different active ingredients. Additionally, other common pests in vegetables and rice crops, including Bemisia tabaci, Aphis gossypii Glover, and the rice stem bore Chilo suppressalis, demonstrated resistance to 69, 52, and 39 insecticides, respectively (APRD, 2024). The simultaneous application of new and traditional pesticides in agriculture frequently hastens the development of resistance to newer pesticides (Sayyed and Wright, 2006). Insecticide resistance has been a common and intractable problem in the world’s agricultural production. Furthermore, the use of new crop protection chemistries faces challenges due to concerns regarding environmental safety, human health, and the escalating costs associated with their discovery, development, and registration. Therefore, there is an urgent requirement for prudent pesticide utilization and a thorough comprehension of the mechanisms that drive insect resistance to pesticides (R4P Network, 2016).
The research conducted in the last few decades has revealed that insecticide resistance in insects involves various mechanisms, including metabolic resistance, behavioral resistance, penetration resistance, target-site resistance and sequestration resistance (Siddiqui et al., 2022; Pu and Chung, 2024). Indeed, research on insecticide resistance has traditionally focused on protein-coding genes, particularly mutations or changes in the insecticide target genes and increased expression of detoxification enzyme genes (Siddiqui et al., 2022). In other words, research has focused on the cis-regulatory or trans-regulatory for these genes. However, biological processes are regulated by not only protein-coding genes but also non-coding RNAs (ncRNAs). In recent years, there has been an increased focus on studying how ncRNAs involved in posttranscriptional regulation of insecticide resistance-related genes can impact resistance. These studies aim to unravel the mechanisms by which ncRNAs influence the expression and function of genes involved in insecticide resistance and explore their potential contributions to resistance mechanisms. By considering the regulatory role of ncRNAs, researchers hope to gain a more comprehensive understanding of the molecular processes underlying insecticide resistance (Pu and Chung, 2024).
ncRNAs have been shown to target both DNA and RNA substrates, modulating signaling pathways involved in critical biological processes such as development and differentiation (Muthu Lakshmi Bavithra et al., 2023). Insects harbor a large number of ncRNA genes, and among them, microRNA (miRNA) has emerged as a crucial regulator in their life processes, impacting key aspects including metamorphosis, reproduction, immune responses, and the detoxification of plant toxic substances (Zhang et al., 2021b). Moreover, long non-coding RNA (lncRNA) actively participates in insect development, innate immunity, and chromosome dosage compensation processes by coordinating gene expression at epigenetic, transcriptional, and post-transcriptional levels (Li et al., 2019). Circular RNA (circRNA), a relatively novel molecule, also contributes to the regulatory landscape, influencing the development, reproduction, and innate immune responses of insects (Muthu Lakshmi Bavithra et al., 2023). Furthermore, ncRNAs, particularly the lncRNA/circRNA-miRNA-mRNA (competing endogenous RNAs, ceRNAs) regulatory network, has been identified as a key player in governing the formation of insect resistance, offering a potential strategy for pest control (Muthu Lakshmi Bavithra et al., 2023). LncRNAs and circRNAs can indeed function as ceRNA by sequestering miRNAs, thereby relieving the inhibition of detoxification enzyme genes and target genes. This regulatory mechanism plays a crucial role in shaping the metabolic and target-site resistance observed in insects. Additionally, lncRNAs have been discovered to serve as miRNA sponges, regulating the expression of cuticle proteins. This modulation, in turn, modulating insect penetration resistance to insecticides by regulating epidermal thickness (Pu and Chung, 2024).
With the progress of next-generation sequencing technologies and bioinformatics tools, researchers now have the capability to investigate the molecular basis of insecticide resistance. This includes the analysis of diverse regulators and modifiers of gene expression levels, such as miRNA, lncRNA, and circRNA, as well as the networks that form among these regulators and protein-coding genes (Pu and Chung, 2024). The primary focus of this review is to investigate the intricate mechanisms by which miRNA, lncRNA, and circRNA influence the development of insect resistance against various insecticides. Based on the findings, it will highlight the multifaceted roles of lncRNAs and circRNAs serving as ceRNA in orchestrating insect resistance mechanisms and provide insights into potential strategies for managing and combating insecticide resistance.
The ncRNAs can be classified into different categories according to their length and intended function. These categories include ribosomal RNA (rRNA), transfer RNA (tRNA), lncRNAs, small ncRNAs (sncRNAs) such as miRNA, piwi-interacting RNA (piRNA) and small nuclear RNA (snRNA), as well as a newly class of ncRNA known as circRNA (Zhang et al., 2019).
MiRNA, a class of endogenous small RNA molecules ranging from 22 to 25 nucleotides in length, plays a crucial role in the post-transcriptional regulation of genes. It exerting its influence by negatively regulating target genes through mRNA cleavage or inhibiting the translation of target proteins, ultimately leading to gene silencing (Bartel, 2004; Bartel, 2005). The advancement of miRNA sequencing technologies has facilitated the discovery of a multitude of miRNAs in insects. The InsectBase 2.0 database contains an impressive 112,162 mature miRNA sequences from 807 species, highlighting the widespread prevalence and functional diversity of miRNAs in the insect kingdom (Mei et al., 2022).
LncRNAs are a class of non-protein-coding RNAs that are typically longer than 200 nucleotides in length. Unlike protein-coding genes, lncRNA genes lack long open reading frames. However, they undergo a similar processing mechanism as protein-coding genes. Transcribed by RNA polymerase II and subjected to splicing, they acquire a 5′ cap structure and a 3′ polyA tail, culminating in a gene structure resembling mRNA (Guttman et al., 2009). It is now recognized as a widely distributed element in eukaryotes, playing a pivotal role in regulating gene expression (Hung and Chang, 2010; Rinn and Chang, 2012; Batista and Chang, 2013). The InsectBase 2.0 database contains a vast array of 1,293,430 lncRNA sequences from 376 species, attesting to the widespread prevalence of lncRNA in the insect kingdom (Mei et al., 2022).
CircRNAs, a recently discovered class of non-coding RNAs, distinguish themselves from linear counterparts like miRNAs and lncRNAs due to their unique covalently closed-loop structure. This distinctive structure, formed through circularization, splicing, and shearing of parental mRNA precursor (pre-mRNA), results in a closed loop where the 5′ and 3′ ends of the pre-mRNA target sequences are connected. Unlike linear RNA molecules, circRNAs lack the 5′ cap and 3′ poly(A) tail, making them resistant to degradation by RNA exonucleases. As a result, circRNAs exhibit a longer half-life and greater stability (Cocquerelle et al., 1992; Capel et al., 1993; Chen, 2020). CircRNAs can originate from the exons, introns and intergenic sequences of coding genes (Salzman, 2016; Kristensen et al., 2019). Their involvement in various biological processes and signaling pathways is primarily achieved through the regulation of gene transcription (Li et al., 2021e), miRNA adsorption (Xu et al., 2018), binding to RNA binding proteins (RBPs) (Xia et al., 2018) and encoding polypeptides (Chen and Sarnow, 1995).
Non-coding RNAs play a crucial roles in the development of insecticide resistance (Pu and Chung, 2024). In this review, the focus is on three types of ncRNAs: miRNAs, lncRNAs, circRNAs. Each of these ncRNAs has been found to regulate the development of insecticide resistance in insects. Moreover, there is evidence of a regulatory network involving lncRNA/circRNA-miRNA-mRNA that is involved in regulating insect resistance to pesticides.
The mechanisms of insecticide resistance in insects encompass metabolic resistance, behavioral resistance, penetration resistance, target-site resistance (Siddiqui et al., 2022), sequestration resistance (Pu and Chung, 2024), and other types. However, the majority of studies in this field have predominantly focused on the target-site resistance and metabolic resistance.
Mutations and duplications in the acetylcholinesterase gene and mutations in the genes encoding insecticide target receptors, such as voltage-gated sodium channels, chloride channels, nicotinic acetylcholine receptors, ryanodine receptor, and octopamine receptors, can lead to target-site resistance in insects. Mutations in chitin syntheses. Furthermore, mutations in chitin synthesis genes and lipid biosynthesis enzymes are also implicated in target-site resistance. (Siddiqui et al., 2022). Regarding resistance to Bacillus thuringiensis (Bt) Cry proteins, the main mechanism observed is the mutation or down-regulation of the expression of midgut receptor-binding protein genes such as cad, ALP, APN, and ABC transporter proteins (Jurat-Fuentes et al., 2021). This mechanism allows insects to resist the toxic effects of Bt Cry proteins.
Detoxification enzyme systems play a crucial role in insecticide resistance to metabolizinge pesticides into compounds of less toxic or non-toxic compounds (Nauen et al., 2022). In resistant populations, these systems can be enhanced through various mechanisms. Mutations and amplifications of detoxification enzymes, activation of related regulatory factors, and over-expression of detoxification enzymes can all contribute to increased detoxification capabilities. Several enzyme systems have been implicated in insecticide detoxification, including carboxylesterases (CarE), glutathione S-transferases (GSTs), Cytochrome P450 monooxygenases (P450s), ABC and UDP glucosyl transferase (UGT) (Karunaratne and Surendran, 2022). Resistant populations may possess an abundance of these enzymes or enzyme systems with heightened activity, allowing them to efficiently metabolize a broader spectrum of pesticides (Vyas et al., 2023). Specific mutations in detoxification enzymes can also lead to resistance. For example, the F116V mutation in the substrate recognition region SRS1 of the CYP9A186 enzyme in the avermectin-resistant strain of the beet armyworm enables the insect to detoxify and metabolize avermectin, contributing to pest resistance (Zuo et al., 2021). In some cases, resistance can be linked to signaling pathways and gene expression. Activation of the ERK and p38 signals of the serine kinase signaling pathway MAPK in tobacco whitefly phosphorylates the serine 111 of the CREB protein, leading to the activation of the downstream target gene CYP6CM1. This activation results in the over-expression of CYP6CM1, which enhances the metabolism of insecticides and ultimately leads to neonicotinoid resistance in tobacco whitefly (Yang et al., 2020). Furthermore, mutations in the 5′-UTR region of the resistance gene CYP4C64 have been associated with thiamethoxam resistance in insects. These mutations cause the gene to be over-expressed, leading to resistance to the neonicotinoid insecticide thiamethoxam (Yang et al., 2021b).
Absolutely, insects can also develop penetration resistance as a defense mechanism to reduce the entry of insecticides into their bodies. By creating a barrier using their outer epidermis, insects can protect themselves from a wide range of chemical pesticides. This mechanism is associated with the up-regulation of the expression of CYP genes, which in turn alter the composition of cuticular hydrocarbons (CHCs) in the insect epidermis (Pu and Chung, 2024). For instance, in the mosquito species Anopheles gambiae, changes in the CYP4G enzyme can alter the composition of CHCs in the insects’ epidermis. This alteration accelerates the synthesis of CHCs, leading to a thickening of the epidermis. As a result, the amount of insecticides penetrating into the body is reduced, thereby conferring resistance to deltamethrin. This penetration resistance mechanism has also been observed in other insect species such as field whitefly and Blattella germanica (Chen et al., 2020; Liang et al., 2022).
In addition to those well-known modes of resistance, a newly emerged mode called sequestration resistance has been identified. Sequestration resistance occurs when olfactory proteins such as chemosensory proteins (CSPs) and odorant-binding proteins (OBPs), are over-expressed in certain tissues, such as the legs of insects. This over-expression allows the insect to develop resistance by binding the insecticide to these proteins and sequestering it before the insecticide reaches its target protein (Pu and Chung, 2024).
Metabolic resistance involves the enzymatic degradation or sequestration of insecticides by insects, constituting a critical detoxification mechanism that diminishes their toxic effects. miRNAs have been demonstrated to precisely regulate the expression of enzymes implicated in detoxification pathways, thereby affecting the metabolic fate of insecticides (Muthu Lakshmi Bavithra et al., 2023). Given that miRNA can exert negative regulation on target genes, miRNA-mediated regulation of a detoxification metabolic enzyme can lead to reduced expression or inhibited translation. Consequently, the enzyme’s capacity to metabolize pesticides decreases, resulting in heightened sensitivity to these substances. Consequently, the expression levels of miRNAs targeting detoxification metabolic enzymes can be precisely regulated, exhibiting a negative correlation with resistance levels (Zhang et al., 2021b). Our findings emphasize that miRNAs target diverse insect detoxification enzymes, modulate the expression of target genes, and accordingly regulate insect metabolic resistance to different insecticides (Table 1).
P450s constitute a family of enzymes ubiquitously found across all life forms, catalyzing diverse oxidative transformations of both endogenous and exogenous substrates, and playing a pivotal role in detoxification processes (Nelson, 2018). Metabolic resistance development in insects is tightly associated with upregualted expression of P450 genes. However, recent studies have revealed that metabolic resistance in insects can also be linked to decreased expression of P450 genes, exemplified by Varroa destructor coumaphos-resistant populations. These populations evade organophosphorus toxicity by down-regulating the expression of the CYP4EP4 gene, one of the mere 26 p450s in their genome. Consequently, this downregulation diminishes proinsecticide activation, ultimately leading to resistance against coumaphos (Vlogiannitis et al., 2021). Culex pipiens pallens emerges as a prominent pathogenic parasite accountable for malaria transmission. In the prevention and control of this parasite, pyrethroid agents such as deltamethrin play a pivotal role. The regulation of insect resistance to chemical pesticides by miRNA has been extensively studied. In the case of C. pipiens, miRNAs play a crucial role in governing the development of deltamethrin resistance by regulating P450 genes. In the deltamethrin-resistant strain of C. pipiens, there are five low-expressed miRNAs (miR-13664, miR-2, miR-71, miR-4448, and miR-278-3p) that individually target five P450 genes (CYP314A1, CYP9J35, CYP325BG3, CYP4H31, and CYP6AG11) (Hong et al., 2014; Lei et al., 2015; Guo et al., 2017; Sun et al., 2019; Li et al., 2021c). On the other hand, there are five highly expressed miRNAs (miR-92a, miR-285, miR-932, miR-13, and miR-279-3p) that respectively target cytochrome P450 reductase (CPR) 4, CYP6N23, CPR5, CYP9J35, and CYP325BB1, orchestrating the development of resistance to deltamethrin in C. pipiens (Liu et al., 2016; Tian et al., 2016; Ma et al., 2017; Fahmy et al., 2020; Li et al., 2021d). These studies highlight the intricate regulatory role of miRNAs in modulating the expression of specific P450 genes, ultimately influencing the development of resistance to deltamethrin in C. pipiens. In D. melanogaster, the metabolism of dichlorodiphenyltrichloroethane (DDT) is primarily associated with P450 gene. Low resistance to DDT is linked to the CYP6G1 gene (Le Goff and Hilliou, 2017), while medium and high levels of DDT resistance involve multiple genes (Kim et al., 2018). D. melanogaster employs post-transcriptional regulation of target gene expression to detoxify DDT. This regulation includes the downregulation of miR-310-3p, miR-311-3p, miR-312-3p, miR-313-3p, and miR-92a-3p, which orchestrates the expression of CYP6AG1, CYP6AG2, CYP6A8, and CYP4G1 (Seong et al., 2019; Seong et al., 2020). These findings demonstrate the intricate miRNA regulatory mechanisms involved in DDT metabolism and resistance in D. melanogaster.
Three low-expressed miRNAs (miR-8533-3p, miR-8534-5p, and miR-375-5p) in chlorantraniliprole-resistant strains of DBM target LCP-30, CYP6B6, and CYP4G15 respectively, thereby regulating the high expression levels of these genes (Zhu et al., 2017b). Moreover, the downregulation of miR-2b-3p in DBM treated with chlorantraniliprole lead to the inhibition of the detoxification enzyme gene CYP9F2. Subsequent analysis involving both deltamethrin-resistant and -sensitive strains of DBM revealed that miR-2b-3p regulate the sensitivity to both chlorantraniliprole and deltamethrin through the modulation of CYP9F2 gene expression (Etebari et al., 2018). The resistance of fall armyworm (Spodoptera frugiperda, FAW) to chlorantraniliprole is strongly linked to the regulation of CYP6K2 gene expression by miR-190-5p, which binds to the 3′UTR of the CYP6K2 gene. The injection of a miR-190-5p agomir results in a significantly reduction in CYP6K2 gene expression and the sensitivity of FAW to chlorantraniliprole. Conversely, inhibiting miR-190-5p expression significantly increases the expression of CYP6K2, thereby enhancing the tolerance of FAW larvae to chlorantraniliprole (Zhang et al., 2022a). Additionally, miR-278-5p may be involved in resistance to the novel insecticide tetraniliprole (Zhang et al., 2022b). Moreover, miR-278-5p, miR-13b-3p, miR-10485-5p, and miR-10483-5p are potential contributors to resistance against cyantraniliprole and emamectin benzoate in FAW (Yang et al., 2021c). Downregulated novel-miR-4 targets CYP4C1, potentially influencing the sensitivity of Lymantria dispar to cyantraniliprole in cyantraniliprole-treated insects, based on the miRNA regulation patterns observed in DBM and FAW (Zhang et al., 2023). miRNA plays a critical role in regulating resistance, primarily through post-translational modification of CYP genes. For example, miR-316 exhibited differential downregulation in spirotetramat-resistant and -sensitive strains of Sitobion miscanthi. It binds to the 3′ UTR of CYP4CJ6, and inhibition of miR-316 expression leads to an increase in CYP4CJ6 expression. This regulatory mechanism affects the sensitivity of S. miscanthi to spirotetramat (Zhang et al., 2021a). Additionally, a novel miRNA, novel_miR-1517, regulates the expression of CYP6CM1 and influences the resistance of B. tabaci to imidacloprid (Gong et al., 2023). Planthoppers, such as Nilaparvata lugens and Laodelphax striatellus, are significant migratory pests that affect rice crops (Kil and Kim, 2023). They have developed resistance to imidacloprid, chlorpyrifos, deltamethrin, and other agents (Tang et al., 2022; Wen et al., 2022). In N.lugens, the low-expressed miRNA novel_85 can reverse-regulate the expression of CYP6ER1 (Mao et al., 2022). Furthermore, the resistance of L. striatellus to triflumezopyrim involves miRNA PC-5p-30_205949 up-regulating the expression of CYP419A1, and PC-3p-2522_840 up-regulating CYP6FL1 (Yang et al., 2022; Wang et al., 2023a).
CarE-mediated metabolic resistance in insects primarily results from the over-expression of the CarE gene and mutations in the encoded amino acid sequence (Cui et al., 2011). Juvenile hormone esterase (JHE) plays a crucial role in the metamorphosis of insects (Li et al., 2021b) and belongs to the CarE subfamily, one of eight subfamilies that regulate insect metabolic resistance (Ranson et al., 2002). Recent research reveals that miRNAs regulate the formation of resistance to Cry1Ac in DBM by influencing the expression of JHE. PxJHE is differentially expressed in Cry1Ac-resistant and -sensitive strains of DBM. Interfering with PxJHE expression increases the sensitivity of DBM to Cry1Ac. In the resistant strains, the highly expressed miRNAs miR-108 and miR-234 bind to the CDS region of PxJHE. Injection of miR-108 and miR-234 agomir significantly reduces the expression of PxJHE, thereby enhancing sensitivity to Cry1Ac. Conversely, inhibiting the expression of miR-108 or miR-234 increases PxJHE expression, reducing tolerance to Cry1Ac. Additionally, injection of miR-108 and miR-234 agomir results in reduced body length and weight, as well as a significant decrease in pupation rate of DBM (Yang et al., 2023). Furthermore, miRNA novel_191 was downregulated in mediating the resistance to nitenpyram in N. lugens by reverse regulating CarE1 (Mao et al., 2022).
The detoxification enzyme GST utilizes multiple mechanisms to regulate the formation and development of insecticide resistance (Pavlidi et al., 2018). Pyrethroids, abamectin, and DDT can induce oxidative stress, leading to the formation of numerous peroxides upon entering the insect body. GST effectively eliminates these peroxides, thereby reducing their harmful effects on insects (Lumjuan et al., 2005; Liao et al., 2016; Li et al., 2021a). A highly expressed GST gene, PxGSTS1, was identified in field populations of the DBM resistant to chlorantraniliprole. The high expression of this gene was found to mediate the resistance of the DBM to various insecticides. DBM resists the toxicity of insecticides through two distinct mechanisms: direct degradation and counteracting oxidative stress mediated by the regulation of PxGSTS1. Further miRNA target prediction revealed that pxy-miR-8528a targets the 3′UTR of PxGSTS1, inhibiting its expression. In vitro and in vivo experiments further demonstrated the relationship between pxy-miR-8528a and PxGSTS1. Expression profile results showed that pxy-miR-8528a was highly expressed in the field-resistant populations. Field populations injected with agomir-8528a exhibited increased sensitivity to chlorantraniliprole and a significant increase in the insecticide-induced accumulation of various types of reactive oxygen species (Liu et al., 2024). Additionally, GSTs4 was highly over-expressed in the indoxacarb-resistant strain of FAW. Dual-luciferase reporter assays demonstrated the target relationship between miR-317-3p, miR-283-5p and GSTs4. Injection of miR-317-3p and miR-283-5p agomirs reduces the expression level of GSTs4 and increases the susceptibility of FAW to indoxacarb. Conversely, injection of miR-317-3p and miR-283-5p antagomirs increases GSTs4 expression and reduces the larval susceptibility to indoxacarb (Wu et al., 2024).
Tetranychus cinnabarinus is a significant agricultural pest mite with a broad range, feeding on over 100 plant species. Chemical agents and acaricides are currently the primary methods used to control this mite (Jia et al., 2011). Cyflumetofen, a benzoyl acetonitrile acaricide, was first registered in Japan in 2007 and has shown effective control against various mite species (Zhou et al., 2020). The main targets of cyflumetofen include CarE, mixed-functional oxidase (MFO), GSTs, and esterases (Wang et al., 2014; Wei et al., 2016a). In the context of resistance, miR-1-3p was found to be downregulated in cyflumetofen-resistant strains of Tetranychus cinnabarinus. This miRNA specifically targets the detoxification enzyme gene GST (TCGSTM4, a mu-class GST gene) (Zhang et al., 2018). Overexpression of miR-1-3p results in a significant downregulation of the TCGSTM4 gene and decreases the sensitivity of both cyflumetofen-resistant and -sensitive strains of T. cinnabarinus to cyflumetofen. Conversely, reducing the expression of miR-1-3p enhances the sensitivity of both resistant and sensitive strains to cyflumetofen (Zhang et al., 2018).
In conclusion, the research underscores the pivotal role of P450 enzymes, CarE, and GSTs in the development of metabolic resistance in various insect species, highlighting the complexity and diversity of regulatory mechanisms involved. The up/down regulated miRNAs specifically target the detoxification enzyme genes such as P450, CarE, and GSTs. This, in turn, modulates the expression of detoxification enzyme genes, which are involved in the development of metabolic resistance in insects and mites (Figure 1). This comprehensive understanding of the genetic and post-transcriptional regulatory mechanisms underlying insecticide resistance can inform the development of more effective pest management strategies and novel insecticides.
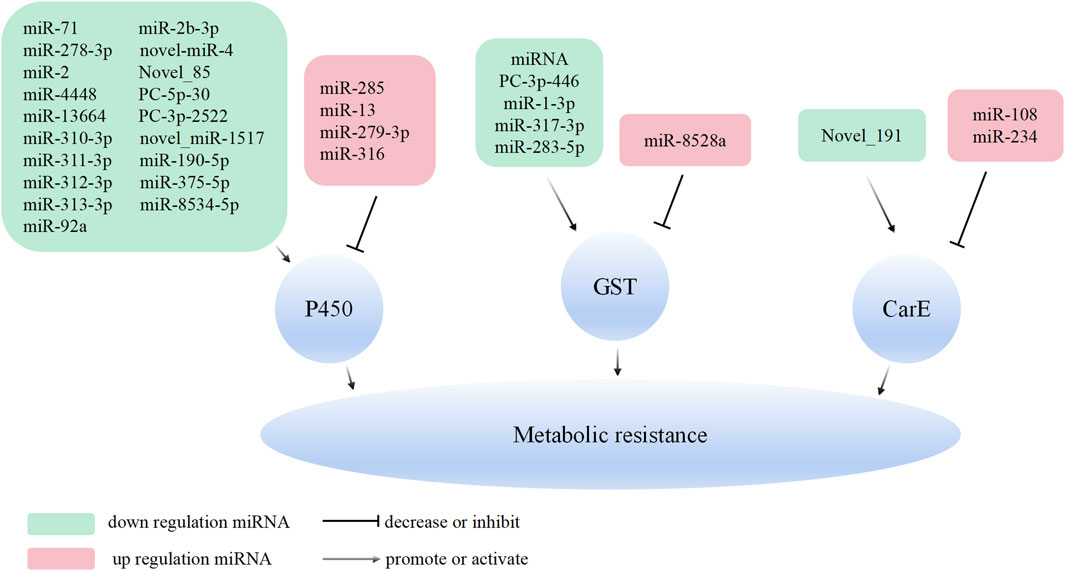
Figure 1. Overview of the molecular mechanisms of miRNAs in metabolic resistance. MiRNA can bind to the 3′UTR or CDS of the target gene (P450, GSTs, and CarE) and inhibit its expression.
miRNAs play a crucial role in regulating target-site resistance by primarily exerting post-transcriptional control over insecticide target genes. These target genes encompass a range of important molecular targets, including acetylcholinesterase (AChE), voltage-gated sodium channel (VGSC), nicotinic acetylcholine receptor (nAChR) and ryanodine receptor (RyR) (Siddiqui et al., 2022). To provide an overview of the regulatory interactions, Table 2 presents a summary of miRNAs that target different insecticide target genes, ultimately contributing to the modulation of insect resistance to various insecticides.
Aedes aegypti is susceptible to pyrethroids and DDT, with voltage-gated sodium channels (VGSC) serving as the primary target (Du et al., 2013). In contrast, miR-33, targets VGSC. Overexpression of miR-33 in both permethrin-resistant and sensitive strains of A. aegypti downregulates the VGSC, gene expression, leading to reduced mortality caused by permethrin (Kubik et al., 2021).
Insecticide target receptor genes, particularly ryanodine receptors (RyRs), are involved in conferring in insect resistance to chlorantraniliprole. Studies conducted on DBM have demonstrated that field-collected resistant lines displayed 2.28–4.14 times greater RyR expression compared to chlorantraniliprole-sensitive lines. Following a 12-hour exposure to chlorantraniliprole at LC50 and LC75 concentrations, there was a 5-fold and 7.2-fold upregulation in RyR, respectively. Subsequent investigations have unveiled that miR-7a and miR-8519 selectively target the 3′UTR region of the RyR gene, leading to the suppression of its expression and exerting an impact on the sensitivity of DBM to chlorantraniliprole (Li et al., 2015).
Fufenozide is a dibenzoylhydrazine-type non-steroidal ecdysone agonists, functions by targeting insect ecdysone receptors (EcR), primarily for the control of Lepidopteran insects (Sun et al., 2010). The 3′UTR of the DBM EcR-B gene is targeted by miR-189942, and its over-expression significantly suppresses EcR-B gene expression. Consequently, the increased expression or knockout of miR-189942 modulates the sensitivity of DBM to fufenozide, resulting in enhanced tolerance of DBM to this insecticide (Li et al., 2020). In A. gossypii, both miR-276 and miR-3016 act together to upregulate the expression of acetyl-CoA carboxylase (ACC), which plays a crucial role in conferring resistance to spirotramat in A. gossypii (Wei et al., 2016b). Furthermore, miR-263b and miR-278 in S. miscanthi are involved in mediating resistance to imidacloprid by suppressing the expression of insecticide receptor genes, specifically nAChRβ1 and nAChRα1A genes, respectively (Zhang et al., 2021a; Zhang et al., 2022c).
The receptor gene encoding ABC transporter proteins of Bt Cry proteins can be classified into eight subfamilies ranging from ABCA to ABCH (Dean and Annilo, 2005). Among these subfamilies, ABCA, ABCB, ABCC, ABCD and ABCG have been implicated d in resistance to various insecticides (Labbe et al., 2011). Analysis of miRNA in Cry1Ac-resistant and sensitive DBM strains has revealed the involvement of miRNA in resistance development by targeting the coding sequence (CDS) region of ABC transporters (Yang et al., 2021a). In particular, PxABCG20 and PxABCG3, which exhibit differential expression between the two strains, are targeted by novel-miR-310 and miR-8510a-3p, respectively. Upregulation of novel-miR-310 and miR-8510a-3p expression suppresses the expression of PxABCG20 and PxABCG3, respectively, thereby enhancing the tolerance to Cry1Ac in the sensitive strain. Conversely, by suppressing novel-miR-310 and miR-8510a-3p, the upregulation of PxABCG20 and PxABCG3 occurs, leading to a reduction in Cry1Ac tolerance (Yang et al., 2022a; Yang et al., 2024). Furthermore, miR-998-3p targets the CDS region of ABCC2, which is conserved across three lepidopteran insects: Helicoverpa armigera, Spodoptera exigua, and DBM. Injection of miR-998-3p agomir results in a reduction of ABCC2 expression, thereby enhancing tolerance to Cry1Ac in all three insects. Conversely, injection of miR-998-3p antagomir increases the expression of ABCC2, reducing the resistance of Cry1Ac-resistant DBM strains to Cry1Ac (Zhu et al., 2020). After treating with sublethal concentrations of Cry1Ac, the PxTrypsin-9 gene exhibits high expression in third instar DBM larvae, and silencing PxTrypsin-9 significantly reduces Cry1Ac toxicity in DBM. Both in vitro and in vivo experiments have demonstrated that miR-2b-3p targets the CDS region of PxTrypsin-9, suppressing its expression and enhancing Cry1Ac tolerance in DBM (Zhang et al., 2024).
p38, xbp-1, and ire-1 have been identified as receptor genes for Cry toxins. Interfering with the expression of these genes, enhances the effect of Cry toxins on the sensitivity of different insects such as C. suppressalis, Aedes aegypti, and Manduca sexta (Cancino-Rodezno et al., 2010; Bedoya-Perez et al., 2013; Qiu et al., 2017). In the case of C. suppressalis, further research have shown that miR-7322-5p is downregulated in insects that were fed Cry1Ca transgenic rice. This miRNA targets 3′UTR of p38, inhibiting its expression and negatively regulating the sensitivity of stem borers to Cry1Ca. Feeding on Cry1Ca enhances the phosphorylation of the Hsp19 protein in stem borers, which is downstream of p38. The downregulation of hsp19 gene expression increases the sensitivity of C. suppressalis to Cry1Ca, suggesting that the activation of hsp19 through the miR-7322-5p/p38/Hsp19 pathway promotes the sequestration of Cry1Ca in C. suppressalis (Wu et al., 2023). While most Cry1 genes are effective against lepidopteran insects, Cry3 protein primarily acts on coleopteran insects (Jouzani et al., 2017). The interaction between Cry3Aa protein and ADAM10 metalloprotease, located on the midgut membrane, is crucial for the toxin’s function in Colorado potato beetle larvae (Ruiz-Arroyo et al., 2017). In Colorado potato beetle larvae, lde-miR-12-1-5p, lde-miR-12-2-5p, lde-miR-252a-5p, lde-miR-2796-5p, lde-miR-316-5p, and lde-miR-998-5p target ADAM10 and may be involved in regulating susceptibility to Cry3Aa (Robles-Fort et al., 2022).
Similar to the miRNA in the regulation of metabolic resistance, miRNAs conferred in the target-site resistance were closely related to miRNA regulation of insecticide target gene expression (Figure 2).
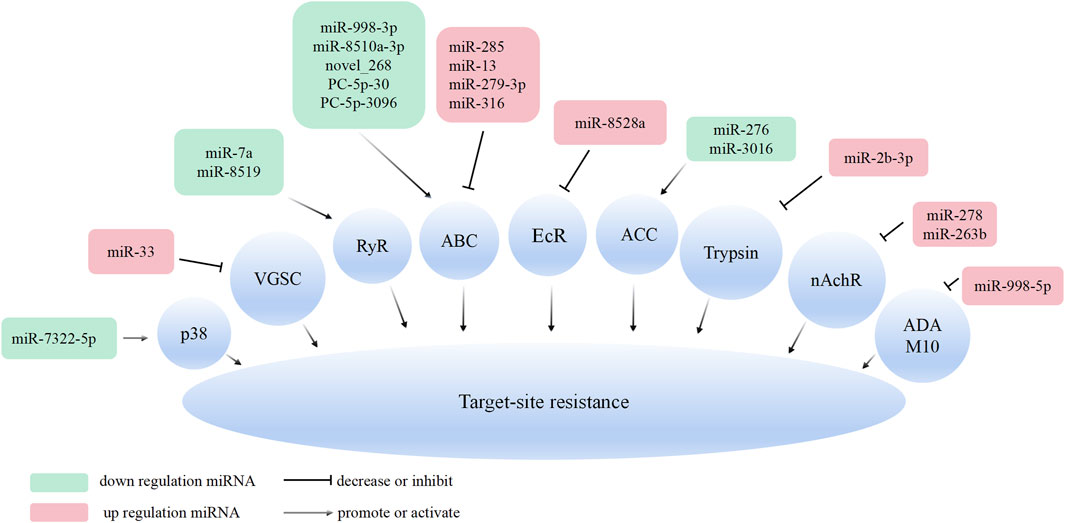
Figure 2. Overview of the molecular mechanisms of miRNAs in insect target-site resistance. MiRNA can bind to the 3′UTR or CDS of the insecticide target gene and inhibit its expression.
The development of insecticides resistance in insect is closely linked to the highly expression of lncRNA genes in resistant strains. Transcriptome analysis of resistant and sensitive strains of DBM exposed to various insecticide, including Bt, chlorpyrifos, fipronil, and deltamethrin, have revealed the association of specific lncRNAs with resistance. For instance, Lawrie et al. (2021) found 358 highly expressed lncRNAs associated with chlorpyrifos resistance, 280 lncRNAs associated with fipronil resistance, and 59 lncRNAs associated with Bt resistance. Similarly, Wang et al. (2023b) observed differential expression of multiple lncRNAs in FAW larvae treated with 23 pesticides. In another study, Wu et al. (2022) demonstrated the differential expression of a significant number of lncRNAs in Tribolium castaneum exposed to terpinene-4-ol. Cotton bollworm resistant to Bt also exhibited the presence of multiple highly expressed lncRNA genes (Rahman et al., 2024). Notably, the lncRNA gene lnc377.4 was found to be highly expressed in malathion-resistant Bactrocera dorsalis strains, and its downregulation increased susceptibility to malathion (Meng et al., 2021). Similarly, highly expressed lncRNAs LNC_004867 and LNC_006576 were closely associated with the development of indoxacarb resistance in Spodoptera litura (Shi et al., 2022). Similar observations were made in chlorantraniliprole-resistant DBM strains, where the high expression of lncRNAs TCONS_00013329 and TCONS_00056155 was associated with the expression of the RyR gene, contributing to resistance development (Zhu et al., 2017b). In chlorantraniliprole-resistant and sensitive strains of C. suppressalis, the identification of 3,470 lncRNAs led to the discovery that the high expression of lncRNA MSTRG.7482.1 contributes to chlorantraniliprole resistance, as demonstrated by RNAi and bioassay experiments (Huang et al., 2022).
In-depth research suggests that lncRNA genes primarily mediate the development of insect resistance through the regulation of detoxification enzyme genes and insecticide target genes (Figure 3). For instance, Li S. et al. (2019a) demonstrated the influence of the lncRNA PgCad1 on the susceptibility of Pectinophora gossypiella to the Bt toxin protein Cry1Ac by regulating the expression of the PgCad1 gene. Similarly, Shen et al. (2021) identified 25 lncRNAs in Phyllotreta striolata that target detoxification enzyme genes. In the spirotetramat-resistant strain of A. gossypii, the highly expressed detoxification enzyme genes CYP4CJ1, CYP6CY7, and CYP6CY21 play a significant role. Down-regulation of these genes increases the mortality of nymphs and adults in the resistant strain. Peng et al. (2022) found that silencing of lncRNAs MSTRG.36649.2/5 and MSTRG.71880.1 alters the expression of CYP6CY21 and CYP380C6, which in turn affects the susceptibility of the resistant A. gossypii strain to spirotetramat. This suggests that these two lncRNAs are involved in the development of spirotetramat resistance in A. gossypii by regulating detoxification enzyme genes. Furthermore, lncRNA ACC contributes to spirotetramat resistance in A. gossypii by regulating ACC transcription (Peng et al., 2021).
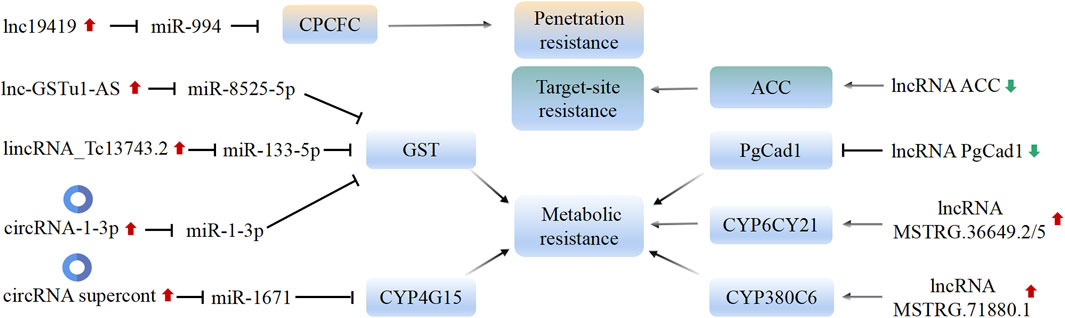
Figure 3. Mechanism of insecticide resistance mediated by lncRNA and circRNA in insects. The common mechanisms by which lncRNA/circRNA affecting insecticide resistance include: 1) acting as a sponge of miRNAs, which affects the expression of target genes; 2) influencing the expression of target or downstream genes. Black arrows represent promotion or activation; black blunt arrows represent decrease or inhibition. The green or red arrows indicate whether the function of lncRNA/circRNA in insects promotes or inhibits the insecticide resistance.
In-depth research on lncRNAs in insects has revealed one mechanism by which insecticide resistance is regulated, namely, the up-regulation or down-regulation of lncRNAs, which in turn leads to the up-regulation or inhibition of target or downstream genes (Figure 3). However, there is an exception with the lncRNA ACC. The insecticidal mechanism of spirotetramat involves the inhibition of ACC expression (Nauen et al., 2008). Interestingly, over-expression and novel mutations in the ACC gene have been found to contribute to high spirotetramat resistance in A. gossypii (Pan et al., 2017). Furthermore, the downregulation of the lncRNA ACC has been observed in spirotetramat-resistant A. gossypii, which in turn affects the expression of genes related to ACC (Peng et al., 2021). Additionally, two transcription factors, C/EBP and C/EBPzeta, have been found to regulate the transcription level of the lncRNA ACC (Peng et al., 2021). In cancer research, the mechanisms of therapy resistance mediated by lncRNAs often involve direct interactions with transcription factors or RNA-binding proteins, thereby affecting the expression of downstream genes and pathways (Singh et al., 2022). The specific mechanisms by which transcription factors bind to and regulate the expression of lncRNAs, as well as the interactions between lncRNAs and proteins, remain to be explored in insects.
The competing endogenous RNA (ceRNA) mechanism was initially discovered in pseudogenes, which possess highly conserved microRNA response elements (MREs) that can bind to miRNA along with protein-coding genes. Pseudogenes act as decoys, attracting miRNA binding and reducing the inhibitory effect of miRNA on mRNA translation (Poliseno et al., 2010). In theory, any RNA molecule containing miRNA binding sites can function as a ceRNA. Therefore, ceRNA can include pseudogenes, protein-coding transcripts, lncRNAs, and circRNAs (Wang et al., 2016). It has been demonstrated that the over-expression of lncRNAs is essential for their “sponge” function and initiation of the ceRNA regulatory pathway. For example, the over-expressed lncRNA-NEAT1 acts as a “sponge” for miR-377-3p, thereby promoting the expression of E2F3 (Sun et al., 2016). CircRNAs are a recent addition to the ceRNA mechanism and their unique characteristics make them well-suited for ceRNA formation. These characteristics include a lack of free ends, predominant cytoplasmic localization, and a predominantly non-coding nature (Wang et al., 2016). While the ceRNA mechanism has been extensively studied in human diseases and cancers, its regulatory networks in insects remain relatively unexplored (Fan et al., 2023; Meng et al., 2023; Ren et al., 2023). In the chlorantraniliprole-resistant DBM strain, both the GSTu1 gene and its anti-sense transcript, lnc-GSTu1-AS, are upregulated. lnc-GSTu1-AS forms a double-stranded duplex with GSTu1, occupying the miR-8525-5p binding site in the GSTu1-3′UTR. This prevents miR-8525-5p-induced degradation of GSTu1, thereby maintaining GSTu1 mRNA stability and contributing to the development of resistance to chlorantraniliprole in DBM (Zhu et al., 2021).
The research conducted by Meng et al. (2023) suggests that in B. dorsalis, the lnc19419-miR-994-CPCFC ceRNA regulatory network plays a role in regulating the penetration resistance to malathion. The injection of miR-994 leads to an increase in the thickness of the epidermis in B. dorsalis, enhancing its tolerance to malathion. This effect is achieved through the targeting of the CPCFC gene by miR-994. Additionally, the lnc19419 molecule is shown to upregulate the expression of the CPCFC gene in the malathion-resistant strain by adsorbing miR-994. Overall, this lnc19419-miR-994-CPCFC ceRNA regulatory network influences the penetration resistance of B. dorsalis to pesticides by modulating the epidermal thickness.
The studies conducted by Lv et al. (2022) and Feng et al. (2020, 2022) provide evidence for the involvement of circular RNAs and long intergenic non-coding RNAs (lincRNAs) in regulating insecticide resistance in different insect species. In the deltamethrin-resistant strains of C. pipiens, the circular RNA supercont3.352:252102|253283 is upregulated. This circRNA acts as a sponge for miR-1671, inhibiting its activity and leading to the down-regulation of the CYP4G15 gene. This regulatory mechanism plays a role in regulating the resistance of C. pipiens to deltamethrin (Lv et al., 2022). Similarly, in cyflumetofen-resistant strains of Tetranychus cinnabarinus, the lincRNA_Tc13743.2 functions as a ceRNA by binding to miR-133-5p. This interaction regulates the expression of the TcGSTm02 gene, which is involved in cyflumetofen resistance (Feng et al., 2020). Furthermore, the circRNA-1-3p in T. cinnabarinus also acts as a ceRNA by sequestering miR-1-3p. This interaction promotes the over-expression of the TcGSTm02 gene, contributing to the resistance of T. cinnabarinus to cyflumetofen (Feng et al., 2022).
The results from the studies mentioned in the previous conversation support the notion that the regulation of insecticide penetration and metabolic resistance in insects involves a cascade of interactions between lncRNAs/circRNAs, miRNAs, and mRNAs. In this regulatory cascade, the abundant lncRNAs/circRNAs act as competitive sponges for miRNAs, preventing their negative interaction with cuticle protein or detoxification enzyme genes and ultimately leading to increased expression of these genes (Figure 3). For cuticle protein, the accumulated protein in the cuticle forms a complex with synthesized chitin, resulting in the thickening of the pronotum cuticle. This thickened cuticle serves as a more effective barrier against insecticide penetration, thus contributing to increased insecticide resistance (Meng et al., 2023). In the case of detoxification enzymes, the over-expression of lncRNAs/circRNAs promotes the expression of these enzymes by sequestering miRNAs. This regulatory mechanism plays a role in the evolution of insecticide resistance in insects. Overall, this regulatory cascade involving lncRNAs/circRNAs, miRNAs, and mRNAs contributes to the modulation of insecticide penetration and metabolic resistance in insects (Figure 3); (Feng et al., 2020; Feng et al., 2022; Lv et al., 2022).
Investigating the role of regulatory factors and ncRNAs in the development of insecticide resistance is indeed crucial. Understanding the molecular mechanisms by which ncRNAs regulate insect resistance provides valuable insights for pest resistance research. In this article, the role of miRNAs, lncRNAs, and the lncRNA/circRNA-miRNA-mRNA (ceRNA) regulatory network in regulating insecticide resistance in insects is reviewed. One primary mechanisms by which ncRNA regulate insect resistance is through the modulation of target genes such as RyR, VGSC, and nAchR. By regulating the expression of these target genes, miRNAs can influence insect susceptibility to chemical pesticides. Another way ncRNAs contribute to insect resistance is by up-regulating the expression of detoxification metabolism enzyme genes, such as P450, GSTs, and CarE, which leads to increased metabolism of chemical pesticides. The ceRNA regulatory network, involving lncRNA/circRNA-miRNA-mRNA interactions, has been identified as a regulator mechanisms in insects resistance. This network influences the expression of detoxification enzyme genes, such as P450 and GSTs, thereby affecting the insect’s metabolic resistance. Additionally, changes in cuticle thickness, regulated by this network, can influence the penetration resistance of insects to insecticides (Figures 1–3).
The majority of protein-coding genes are likely regulated by miRNAs (Ha and Kim, 2014), and miRNA have unique functions. miRNAs have the capacity to regulate the expression of multiple genes within the signaling pathway, both upstream and downstream (Krol et al., 2010). The development of insect resistance to insecticides involves co-regulation of multiple genes. For instance, after feeding on Bt pathogens, JH/20E in the midgut epithelium of DBM binds to the respective Met-Tai and EcR-USP heterodimeric receptor complexes, activating the key gene of the downstream of MAPK signaling pathway. As a result, MAP4K4 orchestrates the significant up-regulation of the transcription factors GATAd and PxmALP genes while inversely regulating the differential expression of the midgut receptor genes ALP and ABCC (ABCC2 and ABCC3). This intricate process ultimately leads to the development of high resistance to Cry1Ac in the DBM without affecting its growth and development (Guo et al., 2015; Guo et al., 2020; Guo et al., 2022). However, the current study only found that miR-998-3p as binding to the CDS region of ABCC2 and regulating the development of resistance to Cry1Ac in DBM (Zhu et al., 2020). It remains unclear whether other miRNAs are involved in the MAPK signaling pathway or if miR-998-3p regulates the expression of other genes within the pathway. Despite the fact that individual miRNAs have been shown to modulate various aspects of insecticide resistance, our understanding of the intricate interactions within these regulatory pathways remains limited. Furthermore, the dysregulation of miRNA expression has been observed in insects under insecticide stress, with most characterized by downregulated expression (Tables 1, 2). The regulatory mechanism underlying this down-regulated expression are yet to be fully elucidated. Additionally, studies on insect lncRNAs and circRNAs are still primarily focused on gene sequence mining, with the poor conservation of lncRNAs and circRNA among different species posing a significant hindrance. This limitation further restricts our understanding of the functions and mechanisms of lncRNAs and circRNAs (Ebbesen et al., 2017; Choudhary et al., 2021). Given the lncRNA/circRNA-miRNA-mRNA (ceRNA) regulatory network has been implicated in the insecticide resistance, integrating multi-omics approaches, such as transcriptomics, proteomics, and metabolomics can provide a systems-level perspective on the regulatory mechanism of ncRNAs in responses to insecticide exposure (Feng et al., 2020; Feng et al., 2022; Lv et al., 2022; Meng et al., 2023). By combining data from multiple platforms, researchers can unravel complex regulatory networks, identify upstream regulators for lncRNAs and circRNAs, and elucidate downstream signaling pathways involved in the development of insecticide resistance (Muthu Lakshmi Bavithra et al., 2023). Future research should prioritize systematic cross-species comparisons to elucidate the evolutionary dynamics of ncRNA-mediated resistance mechanisms. Comparative genomics analyses across diverse insect species can unveil conserved ncRNA regulatory networks and identify novel targets for managing insecticide resistance.
HX: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Supervision, Writing–original draft, Writing–review and editing. CM: Conceptualization, Investigation, Writing–original draft. RP: Formal Analysis, Writing–review and editing. MX: Investigation, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Science Foundation of China (32360669, 31760514) and Jiangxi Provincial Natural Science Foundation of China (20224BAB205016).
Thanks are due to Dr. Jian Pu for assistance with the suggestions in revising the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alavanja, M. C. (2009). Introduction: pesticides use and exposure extensive worldwide. Rev. Environ. Health 24 (4), 303–309. doi:10.1515/reveh.2009.24.4.303
APRD (2024). Arthropod pesticide resistance database. Michigan state university. Available at: https://www.pesticideresistance.org/ (Accessed April 5, 2024).
Bartel, B. (2005). MicroRNAs directing siRNA biogenesis. Nat. Struct. Mol. Biol. 12 (7), 569–571. doi:10.1038/nsmb0705-569
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2), 281–297. doi:10.1016/s0092-8674(04)00045-5
Batista, P. J., and Chang, H. Y. (2013). Long noncoding RNAs: cellular address codes in development and disease. Cell 152 (6), 1298–1307. doi:10.1016/j.cell.2013.02.012
Bedoya-Perez, L. P., Cancino-Rodezno, A., Flores-Escobar, B., Soberon, M., and Bravo, A. (2013). Role of UPR pathway in defense response of Aedes aegypti against Cry11Aa toxin from Bacillus thuringiensis. Int. J. Mol. Sci. 14 (4), 8467–8478. doi:10.3390/ijms14048467
Cancino-Rodezno, A., Alexander, C., Villasenor, R., Pacheco, S., Porta, H., Pauchet, Y., et al. (2010). The mitogen-activated protein kinase p38 is involved in insect defense against Cry toxins from Bacillus thuringiensis. Insect Biochem. Mol. Biol. 40 (1), 58–63. doi:10.1016/j.ibmb.2009.12.010
Capel, B., Swain, A., Nicolis, S., Hacker, A., Walter, M., Koopman, P., et al. (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73 (5), 1019–1030. doi:10.1016/0092-8674(93)90279-y
Chen, C. Y., and Sarnow, P. (1995). Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268 (5209), 415–417. doi:10.1126/science.7536344
Chen, L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21 (8), 475–490. doi:10.1038/s41580-020-0243-y
Chen, N., Pei, X. J., Li, S., Fan, Y. L., and Liu, T. X. (2020). Involvement of integument-rich CYP4G19 in hydrocarbon biosynthesis and cuticular penetration resistance in Blattella germanica (L.). Pest Manag. Sci. 76 (1), 215–226. doi:10.1002/ps.5499
Choudhary, C., Sharma, S., Meghwanshi, K. K., Patel, S., Mehta, P., Shukla, N., et al. (2021). Long non-coding RNAs in insects. Anim. (Basel) 11 (4), 1118. doi:10.3390/ani11041118
Cocquerelle, C., Daubersies, P., Majerus, M. A., Kerckaert, J. P., and Bailleul, B. (1992). Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 11 (3), 1095–1098. doi:10.1002/j.1460-2075.1992.tb05148.x
Connor, D. J., Loomis, R. S., and Cassman, K. G. (2011). Crop ecology: productivity and management in agricultural systems.
Cui, F., Lin, Z., Wang, H., Liu, S., Chang, H., Reeck, G., et al. (2011). Two single mutations commonly cause qualitative change of nonspecific carboxylesterases in insects. Insect biochem. Mol. Biol. 41 (1), 1–8. doi:10.1016/j.ibmb.2010.09.004
Dean, M., and Annilo, T. (2005). Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 6, 123–142. doi:10.1146/annurev.genom.6.080604.162122
Du, Y., Nomura, Y., Satar, G., Hu, Z., Nauen, R., He, S. Y., et al. (2013). Molecular evidence for dual pyrethroid-receptor sites on a mosquito sodium channel. Proc. Natl. Acad. Sci. 110 (29), 11785–11790. doi:10.1073/pnas.1305118110
Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2017). Insights into circular RNA biology. RNA Biol. 14 (8), 1035–1045. doi:10.1080/15476286.2016.1271524
Etebari, K., Afrad, M. H., Tang, B., Silva, R., Furlong, M. J., and Asgari, S. (2018). Involvement of microRNA miR-2b-3p in regulation of metabolic resistance to insecticides in Plutella xylostella. Insect Mol. Biol. 27 (4), 478–491. doi:10.1111/imb.12387
Fahmy, N. T., Osman, A., Badr, M. S., Morcos, N., Diclaro, J. W., and Abd-ElSamie, E. M. (2020). Deciphering pyrethroid resistance in Cx. pipiens (L): implications of cytochrome P450; expression profiling and regulatory microRNA. Mol. Cell Probes. 52, 101579. doi:10.1016/j.mcp.2020.101579
Fan, X., Gao, X., Zang, H., Guo, S., Jing, X., Zhang, Y., et al. (2023). Diverse regulatory manners and potential roles of lncRNAs in the developmental process of asian honey bee (Apis cerana) larval guts. Int. J. Mol. Sci. 24 (20), 15399. doi:10.3390/ijms242015399
Feng, K., Jiang, Z., Liu, P., Liu, J., Wen, X., and He, L. (2022). Circular RNA, circ1-3p, is involved in cyflumetofen resistance by acting as a competitive RNA against miR-1-3p in Tetranychus cinnabarinus. J. Agric. Food Chem. 70 (4), 1068–1078. doi:10.1021/acs.jafc.1c07155
Feng, K., Liu, J., Wei, P., Ou, S., Wen, X., Shen, G., et al. (2020). lincRNA_Tc13743.2-miR-133-5p-TcGSTm02 regulation pathway mediates cyflumetofen resistance in Tetranychus cinnabarinus. Insect biochem. Mol. Biol. 123, 103413. doi:10.1016/j.ibmb.2020.103413
Gong, P. P., Wei, X. G., Liu, S. N., Yang, J., Fu, B. L., Liang, J. J., et al. (2023). Novel_miR-1517 mediates CYP6CM1 to regulate imidacloprid resistance in Bemisia tabaci (Hemiptera: gennadius). Pestic. Biochem. Physiol. 194, 105469. doi:10.1016/j.pestbp.2023.105469
Guo, L., Cheng, Z., Qin, J., Sun, D., Wang, S., Wu, Q., et al. (2022). MAPK-mediated transcription factor GATAd contributes to Cry1Ac resistance in diamondback moth by reducing PxmALP expression. PLoS Genet. 18 (2), e1010037. doi:10.1371/journal.pgen.1010037
Guo, Q., Huang, Y., Zou, F., Liu, B., Tian, M., Ye, W., et al. (2017). The role of miR-2 approximately 13 approximately 71 cluster in resistance to deltamethrin in Culex pipiens pallens. Insect biochem. Mol. Biol. 84, 15–22. doi:10.1016/j.ibmb.2017.03.006
Guo, Z., Kang, S., Chen, D., Wu, Q., Wang, S., Xie, W., et al. (2015). MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS Genet. 11 (4), e1005124. doi:10.1371/journal.pgen.1005124
Guo, Z., Kang, S., Sun, D., Gong, L., Zhou, J., Qin, J., et al. (2020). MAPK-dependent hormonal signaling plasticity contributes to overcoming Bacillus thuringiensis toxin action in an insect host. Nat. Commun. 11 (1), 3003. doi:10.1038/s41467-020-16608-8
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458 (7235), 223–227. doi:10.1038/nature07672
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15 (8), 509–524. doi:10.1038/nrm3838
Hong, S., Guo, Q., Wang, W., Hu, S., Fang, F., Lv, Y., et al. (2014). Identification of differentially expressed microRNAs in Culex pipiens and their potential roles in pyrethroid resistance. Insect biochem. Mol. Biol. 55, 39–50. doi:10.1016/j.ibmb.2014.10.007
Huang, S., Jing, D., Xu, L., Luo, G., Hu, Y., Wu, T., et al. (2022). Genome-wide identification and functional analysis of long non-coding RNAs in Chilo suppressalis reveal their potential roles in chlorantraniliprole resistance. Front. Physiol. 13, 1091232. doi:10.3389/fphys.2022.1091232
Hung, T., and Chang, H. Y. (2010). Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 7 (5), 582–585. doi:10.4161/rna.7.5.13216
Jia, F. L., Chen, Y. J., Chen, J., Wang, D. D., and Dai, G. H. (2011). Biological activity of extracts from 8 species of plants against Tetranychus cinnabarinus. Chin. Agri. Sci. Bull. 27 (24), 286–291.
Jouzani, G. S., Valijanian, E., and Sharafi, R. (2017). Bacillus thuringiensis: a successful insecticide with new environmental features and tidings. Appl. Microbiol. Biotechnol. 101 (7), 2691–2711. doi:10.1007/s00253-017-8175-y
Jurat-Fuentes, J. L., Heckel, D. G., and Ferre, J. (2021). Mechanisms of resistance to insecticidal proteins from Bacillus thuringiensis. Annu. Rev. Entomol. 66, 121–140. doi:10.1146/annurev-ento-052620-073348
Karunaratne, S. H. P. P., and Surendran, S. N. (2022). Mosquito control: a review on the past, present and future strategies. J. Nat. Sci. Found. Sri. 50, 277–292. doi:10.4038/jnsfsr.v50i0.11244
Kil, E. J., and Kim, D. (2023). The small brown planthopper (Laodelphax striatellus) as a vector of the rice stripe virus. Arch. Biochem. 112 (2), e21992. doi:10.1002/arch.21992
Kim, J. H., Moreau, J. A., Zina, J. M., Mazgaeen, L., Yoon, K. S., Pittendrigh, B. R., et al. (2018). Identification and interaction of multiple genes resulting in DDT resistance in the 91-R strain of Drosophila melanogaster by RNAi approaches. Pestic. Biochem. Physiol. 151, 90–99. doi:10.1016/j.pestbp.2018.03.003
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20 (11), 675–691. doi:10.1038/s41576-019-0158-7
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11 (9), 597–610. doi:10.1038/nrg2843
Kubik, T. D., Snell, T. K., Saavedra-Rodriguez, K., Wilusz, J., Anderson, J. R., Lozano-Fuentes, S., et al. (2021). Aedes aegypti miRNA-33 modulates permethrin induced toxicity by regulating VGSC transcripts. Sci. Rep. 11 (1), 7301. doi:10.1038/s41598-021-86665-6
Labbe, R., Caveney, S., and Donly, C. (2011). Genetic analysis of the xenobiotic resistance-associated ABC gene subfamilies of the Lepidoptera. Insect Mol. Biol. 20 (2), 243–256. doi:10.1111/j.1365-2583.2010.01064.x
Lawrie, R. D., Mitchell, R. D., Deguenon, J. M., Ponnusamy, L., Reisig, D., Pozo-Valdivia, A. D., et al. (2021). Characterization of long non-coding RNAs in the bollworm, Helicoverpa zea, and their possible role in Cry1Ac-resistance. Insects 13 (1), 12. doi:10.3390/insects13010012
Le Goff, G., and Hilliou, F. (2017). Resistance evolution in Drosophila: the case of CYP6G1. Pest Manag. Sci. 73 (3), 493–499. doi:10.1002/ps.4470
Lei, Z., Lv, Y., Wang, W., Guo, Q., Zou, F., Hu, S., et al. (2015). MiR-278-3p regulates pyrethroid resistance in Culex pipiens pallens. Parasitol. Res. 114 (2), 699–706. doi:10.1007/s00436-014-4236-7
Li, D., He, C., Xie, L., Kong, F., Wu, Y., Shi, M., et al. (2021a). Functional analysis of SlGSTE12 in pyrethroid and organophosphate resistance in Spodoptera litura. J. Agric. Food. Chem. 69 (21), 5840–5848. doi:10.1021/acs.jafc.1c00723
Li, H., Liu, S., Chen, L., Luo, J., Zeng, D., and Li, X. (2021b). Juvenile hormone and transcriptional changes in honey bee worker larvae when exposed to sublethal concentrations of thiamethoxam. Ecotoxicol. Environ. Saf. 225, 112744. doi:10.1016/j.ecoenv.2021.112744
Li, K., Tian, Y., Yuan, Y., Fan, X., Yang, M., He, Z., et al. (2019a). Insights into the functions of lncRNAs in Drosophila. Int. J. Mol. Sci. 20 (18), 4646. doi:10.3390/ijms20184646
Li, S., Hussain, F., Unnithan, G. C., Dong, S., UlAbdin, Z., Gu, S., et al. (2019b). A long non-coding RNA regulates cadherin transcription and susceptibility to Bt toxin Cry1Ac in pink bollworm, Pectinophora gossypiella. Pestic. Biochem. Physiol. 158, 54–60. doi:10.1016/j.pestbp.2019.04.007
Li, X., Guo, L., Zhou, X., Gao, X., and Liang, P. (2015). miRNAs regulated overexpression of ryanodine receptor is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Sci. Rep. 5, 14095. doi:10.1038/srep14095
Li, X., Hu, S., Yin, H., Zhang, H., Zhou, D., Sun, Y., et al. (2021c). MiR-4448 is involved in deltamethrin resistance by targeting CYP4H31 in Culex pipiens pallens. Parasit. Vectors 14 (1), 159. doi:10.1186/s13071-021-04665-x
Li, X., Hu, S., Zhang, H., Yin, H., Wang, H., Zhou, D., et al. (2021d). MiR-279-3p regulates deltamethrin resistance through CYP325BB1 in Culex pipiens pallens. Parasit. Vectors 14 (1), 528. doi:10.1186/s13071-021-05033-5
Li, X., Ren, X., Liu, Y., Smagghe, G., Liang, P., and Gao, X. (2020). MiR-189942 regulates fufenozide susceptibility by modulating ecdysone receptor isoform B in Plutella xylostella (L.). Pestic. Biochem. Physiol. 163, 235–240. doi:10.1016/j.pestbp.2019.11.021
Li, X., Zhang, J. L., Lei, Y. N., Liu, X. Q., Xue, W., Zhang, Y., et al. (2021e). Linking circular intronic RNA degradation and function in transcription by RNase H1. Sci. China Life Sci. 64 (11), 1795–1809. doi:10.1007/s11427-021-1993-6
Li, Z., Mao, K., Jin, R., Cai, T., Qin, Y., Zhang, Y., et al. (2022). miRNA novel_268 targeting NlABCG3 is involved in nitenpyram and clothianidin resistance in Nilaparvata lugens. Int. J. Biol. Macromol. 217, 615–623. doi:10.1016/j.ijbiomac.2022.07.096
Liang, J. J., Yang, J., Hu, J. Y., Fu, B. L., Gong, P. P., Du, T. H., et al. (2022). Cytpchrome P450 CYP4G68 is associated with imidacloprid and thiamethoxam resistance in field whitefly, Bemisia tabaci (Hemiptera: gennadius). Agriculture 12, 473. doi:10.3390/agriculture12040473
Liao, C. Y., Xia, W. K., Feng, Y. C., Li, G., Liu, H., Dou, W., et al. (2016). Characterization and functional analysis of a novel glutathione S-transferase gene potentially associated with the abamectin resistance in Panonychus citri (McGregor). Pestic. Biochem. Physiol. 132, 72–80. doi:10.1016/j.pestbp.2015.11.002
Liu, B., Tian, M., Guo, Q., Ma, L., Zhou, D., Shen, B., et al. (2016). MiR-932 regulates pyrethroid resistance in Culex pipiens pallens (Diptera: Culicidae). J. Med. Entomol. 53 (5), 1205–1210. doi:10.1093/jme/tjw083
Liu, J., Tian, Z., Li, R., Ni, S., Sun, H., Yin, F., et al. (2024). Key contributions of the overexpressed Plutella xylostella sigma glutathione s-transferase 1 Gene (PxGSTs1) in the resistance evolution to multiple insecticides. J. Agric. Food. Chem. 72 (5), 2560–2572. doi:10.1021/acs.jafc.3c09458
Lumjuan, N., McCarroll, L., Prapanthadara, L. A., Hemingway, J., and Ranson, H. (2005). Elevated activity of an Epsilon class glutathione transferase confers DDT resistance in the dengue vector, Aedes aegypti. Insect biochem. Mol. Biol. 35 (8), 861–871. doi:10.1016/j.ibmb.2005.03.008
Lv, Y., Li, X., Zhang, H., Zou, F., and Shen, B. (2022). CircRNA expression profiles in deltamethrin-susceptible and -resistant Culex pipiens pallens (Diptera: Culicidae). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 261, 110750. doi:10.1016/j.cbpb.2022.110750
Ma, K., Li, X., Hu, H., Zhou, D., Sun, Y., Ma, L., et al. (2017). Pyrethroid-resistance is modulated by miR-92a by targeting CpCPR4 in Culex pipiens pallens. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 203, 20–24. doi:10.1016/j.cbpb.2016.09.002
Mao, K., Jin, R., Ren, Z., Zhang, J., Li, Z., He, S., et al. (2022). miRNAs targeting CYP6ER1 and CarE1 are involved in nitenpyram resistance in Nilaparvata lugens. Insect Sci. 29 (1), 177–187. doi:10.1111/1744-7917.12910
Mei, Y., Jing, D., Tang, S., Chen, X., Chen, H., Duanmu, H., et al. (2022). InsectBase 2.0: a comprehensive gene resource for insects. Nucleic. acids. Res. 50 (D1), D1040–D1045. doi:10.1093/nar/gkab1090
Meng, L. W., Yuan, G. R., Chen, M. L., Dou, W., Jing, T. X., Zheng, L. S., et al. (2021). Genome-wide identification of long non-coding RNAs (lncRNAs) associated with malathion resistance in Bactrocera dorsalis. Pest. Manag. Sci. 77 (5), 2292–2301. doi:10.1002/ps.6256
Meng, L. W., Yuan, G. R., Chen, M. L., Zheng, L. S., Dou, W., Peng, Y., et al. (2023). Cuticular competing endogenous RNAs regulate insecticide penetration and resistance in a major agricultural pest. BMC Biol. 21 (1), 187. doi:10.1186/s12915-023-01694-z
Muthu Lakshmi Bavithra, C., Murugan, M., Pavithran, S., and Naveena, K. (2023). Enthralling genetic regulatory mechanisms meddling insecticide resistance development in insects: role of transcriptional and post-transcriptional events. Front. Mol. Biosci. 10, 1257859. doi:10.3389/fmolb.2023.1257859
Nauen, R., Reckmann, U., Thomzik, J., and Thielert, W. (2008). Biological profile of spirotetramat (Movento®)–a new two-way systemic (ambimobile) insecticide against sucking pest species. Bayer. Crop. J. 61, 245–278.
Nelson, D. R. (2018). Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta. Proteins Proteom. 1866 (1), 141–154. doi:10.1016/j.bbapap.2017.05.003
Pan, Y., Zhu, E., Gao, X., Nauen, R., Xi, J., Peng, T., et al. (2017). Novel mutations and expression changes of acetyl-coenzyme A carboxylase are associated with spirotetramat resistance in Aphis gossypii Glover. Insect. Mol. Biol. 26 (4), 383–391. doi:10.1111/imb.12300
Pavlidi, N., Vontas, J., and Van Leeuwen, T. (2018). The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr. Opin. Insect Sci. 27, 97–102. doi:10.1016/j.cois.2018.04.007
Peng, T., Liu, X., Tian, F., Xu, H., Yang, F., Chen, X., et al. (2022). Functional investigation of lncRNAs and target cytochrome P450 genes related to spirotetramat resistance in Aphis gossypii Glover. Pest Manag. Sci. 78 (5), 1982–1991. doi:10.1002/ps.6818
Peng, T., Pan, Y., Tian, F., Xu, H., Yang, F., Chen, X., et al. (2021). Identification and the potential roles of long non-coding RNAs in regulating acetyl-CoA carboxylase ACC transcription in spirotetramat-resistant Aphis gossypii. Pestic. Biochem. Physiol. 179, 104972. doi:10.1016/j.pestbp.2021.104972
Pimentel, D., Lach, L., Zuniga, R., and Morrison, D. (2000). Environmental and economic costs of nonindigenous species in the United States. Bioscience 50 (1), 53–65. doi:10.1641/0006-3568(2000)050[0053:Eaecon]2.3.Co;2
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W. J., and Pandolfi, P. P. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465 (7301), 1033–1038. doi:10.1038/nature09144
Pu, J., and Chung, H. (2024). New and emerging mechanisms of insecticide resistance. Curr. Opin. Insect Sci. 63, 101184. doi:10.1016/j.cois.2024.101184
Qiu, L., Fan, J., Liu, L., Zhang, B., Wang, X., Lei, C., et al. (2017). Knockdown of the MAPK p38 pathway increases the susceptibility of Chilo suppressalis larvae to Bacillus thuringiensis Cry1Ca toxin. Sci. Rep. 7, 43964. doi:10.1038/srep43964
R4P Network (2016). Trends and challenges in pesticide resistance detection. Trends Plant Sci. 21 (10), 834–853. doi:10.1016/j.tplants.2016.06.006
Rahman, M. M., Omoto, C., and Kim, J. (2024). Genome-Wide exploration of long non-coding RNAs of Helicoverpa armigera in response to pyrethroid insecticide resistance. Insects 15 (3), 146. doi:10.3390/insects15030146
Ranson, H., Claudianos, C., Ortelli, F., Abgrall, C., Hemingway, J., Sharakhova, M. V., et al. (2002). Evolution of supergene families associated with insecticide resistance. Science 298 (5591), 179–181. doi:10.1126/science.1076781
Ren, Y., Chen, J., Wang, Y., Fu, S., Bu, W., and Xue, H. (2023). The lncRNA-mediated ceRNA network of Altica viridicyanea is involved in the regulation of the Toll/Imd signaling pathway under antibiotic treatment. Front. Physiol. 14, 1244190. doi:10.3389/fphys.2023.1244190
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi:10.1146/annurev-biochem-051410-092902
Robles-Fort, A., Pescador-Dionisio, S., Garcia-Robles, I., Sentandreu, V., Martinez-Ramirez, A. C., Real, M. D., et al. (2022). Unveiling gene expression regulation of the Bacillus thuringiensis Cry3Aa toxin receptor ADAM10 by the potato dietary miR171c in Colorado potato beetle. Pest Manag. Sci. 78 (9), 3760–3768. doi:10.1002/ps.6743
Ruiz-Arroyo, V. M., Garcia-Robles, I., Ochoa-Campuzano, C., Goig, G. A., Zaitseva, E., Baaken, G., et al. (2017). Validation of ADAM10 metalloprotease as a Bacillus thuringiensis Cry3Aa toxin functional receptor in Colorado potato beetle (Leptinotarsa decemlineata). Insect Mol. Biol. 26 (2), 204–214. doi:10.1111/imb.12285
Salzman, J. (2016). Circular RNA expression: its potential regulation and function. Trends Genet. 32 (5), 309–316. doi:10.1016/j.tig.2016.03.002
Sayyed, A. H., and Wright, D. J. (2006). Genetics and evidence for an esterase-associated mechanism of resistance to indoxacarb in a field population of diamondback moth (Lepidoptera: plutellidae). Pest Manag. Sci. 62 (11), 1045–1051. doi:10.1002/ps.1270
Seong, K. M., Coates, B. S., and Pittendrigh, B. R. (2019). Impacts of sub-lethal DDT exposures on microRNA and putative target transcript expression in DDT resistant and susceptible Drosophila melanogaster strains. Front. Genet. 10, 45. doi:10.3389/fgene.2019.00045
Seong, K. M., Coates, B. S., and Pittendrigh, B. R. (2020). Post-transcriptional modulation of cytochrome P450s, Cyp6g1 and Cyp6g2, by miR-310s cluster is associated with DDT-resistant Drosophila melanogaster strain 91-R. Sci. Rep. 10 (1), 14394. doi:10.1038/s41598-020-71250-0
Shen, G. M., Ou, S. Y., He, C., Liu, J., and He, L. (2021). Full length sequencing reveals novel transcripts of detoxification genes along with related alternative splicing events and lncRNAs in Phyllotreta striolata. PLoS One 16 (3), e0248749. doi:10.1371/journal.pone.0248749
Shi, L., Li, W. L., Zeng, H. X., Shi, Y., and Liao, X. L. (2022). Systematic identification and functional analysis of long noncoding RNAs involved in indoxacarb resistance in Spodoptera litura. Insect Sci. 29 (6), 1721–1736. doi:10.1111/1744-7917.13015
Siddiqui, J. A., Fan, R., Naz, H., Bamisile, B. S., Hafeez, M., Ghani, M. I., et al. (2022). Insights into insecticide-resistance mechanisms in invasive species: challenges and control strategies. Front. Physiol. 13, 1112278. doi:10.3389/fphys.2022.1112278
Singh, D., Assaraf, Y. G., and Gacche, R. N. (2022). Long non-coding RNA mediated drug resistance in breast cancer. Drug resist. updat. 63, 100851. doi:10.1016/j.drup.2022.100851
Sun, C., Li, S., Zhang, F., Xi, Y., Wang, L., Bi, Y., et al. (2016). Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget 7, 51784–51814. doi:10.18632/oncotarget.10108
Sun, J., Liang, P., and Gao, X. (2010). Inheritance of resistance to a new non-steroidal ecdysone agonist, fufenozide, in the diamondback moth, Plutella xylostella (Lepidoptera: plutellidae). Pest Manag. Sci. 66 (4), 406–411. doi:10.1002/ps.1891
Sun, X. H., Xu, N., Xu, Y., Zhou, D., Sun, Y., Wang, W. J., et al. (2019). A novel miRNA, miR-13664, targets CpCYP314A1 to regulate deltamethrin resistance in Culex pipiens pallens. Parasitology 146 (2), 197–205. doi:10.1017/S0031182018001002
Tang, B., Xu, K., Liu, Y., Zhou, Z., Karthi, S., Yang, H., et al. (2022). A review of physiological resistance to insecticide stress in Nilaparvata lugens. 3 Biotech. 12 (3), 84. doi:10.1007/s13205-022-03137-y
Tian, M., Liu, B., Hu, H., Li, X., Guo, Q., Zou, F., et al. (2016). MiR-285 targets P450 (CYP6N23) to regulate pyrethroid resistance in Culex pipiens pallens. Parasitol. Res. 115 (12), 4511–4517. doi:10.1007/s00436-016-5238-4
Vlogiannitis, S., Mavridis, K., Dermauw, W., Snoeck, S., Katsavou, E., Morou, E., et al. (2021). Reduced proinsecticide activation by cytochrome P450 confers coumaphos resistance in the major bee parasite Varroa destructor. Proc. Natl. Acad. Sci. 118 (6), e2020380118. doi:10.1073/pnas.2020380118
Vyas, T., Singh, V., Kodgire, P., and Joshi, A. (2023). Insights in detection and analysis of organophosphates using organophosphorus acid anhydrolases (OPAA) enzyme-based biosensors. Crit. Rev. Biotechnol. 43 (4), 521–539. doi:10.1080/07388551.2022.2052012
Wang, A., Yang, Y., Zhou, Y., Zhang, Y., Xue, C., Zhao, Y., et al. (2023a). A microRNA, PC-5p-30_205949, regulates triflumezopyrim susceptibility in Laodelphax striatellus (Fallen) by targeting CYP419A1 and ABCG23. Pestic. Biochem. Physiol. 192, 105413. doi:10.1016/j.pestbp.2023.105413
Wang, Q. N., Xiao, W. Q., Yao, Y., Kong, X. D., and Sun, Y. (2023b). Response patterns of lncRNAs of the Spodoptera frugiperda (Lepidoptera: noctuidae) larvae under 23 pesticide treatments. J. Insect Sci. 23 (4), 8. doi:10.1093/jisesa/iead059
Wang, Y., Hou, J., He, D., Sun, M., Zhang, P., Yu, Y., et al. (2016). The emerging function and mechanism of ceRNAs in cancer. Trends Genet. 32 (4), 211–224. doi:10.1016/j.tig.2016.02.001
Wang, Y., Zhao, S., Shi, L., Xu, Z., and He, L. (2014). Resistance selection and biochemical mechanism of resistance against cyflumetofen in Tetranychus cinnabarinus (Boisduval). Pestic. Biochem. Physiol. 111, 24–30. doi:10.1016/j.pestbp.2014.04.004
Wei, P., Shi, L., Shen, G., Xu, Z., Liu, J., Pan, Y., et al. (2016a). Characteristics of carboxylesterase genes and their expression-level between acaricide-susceptible and resistant Tetranychus cinnabarinus (Boisduval). Pestic. Biochem. Physiol. 131, 87–95. doi:10.1016/j.pestbp.2015.12.007
Wei, X., Zheng, C., Peng, T., Pan, Y., Xi, J., Chen, X., et al. (2016b). miR-276 and miR-3016-modulated expression of acetyl-CoA carboxylase accounts for spirotetramat resistance in Aphis gossypii Glover. Insect biochem. Mol. Biol. 79, 57–65. doi:10.1016/j.ibmb.2016.10.011
Wen, S., Liu, C., Wang, X., Wang, Y., Liu, C., Wang, J., et al. (2022). Resistance selection of triflumezopyrim in Laodelphax striatellus (fallen): resistance risk, cross-resistance and metabolic mechanism. Front. Physiol. 13, 1048208. doi:10.3389/fphys.2022.1048208
Wu, H., Yue, S., Huang, Y., Zhao, X., Cao, H., and Liao, M. (2022). Genome-wide identification of the long noncoding RNAs of Tribolium castaneum in response to terpinen-4-ol fumigation. Insects 13 (3), 283. doi:10.3390/insects13030283
Wu, M., Lv, H., Guo, Z., Li, S., Tang, J., Li, J., et al. (2024). miR-317-3p and miR-283-5p play a crucial role in regulating the resistance to indoxacarb in Spodoptera frugiperda by targeting GSTs4. J. Agric. Food Chem. 72 (13), 6889–6899. doi:10.1021/acs.jafc.3c06531
Wu, Y., Weng, Z., Yan, H., Yao, Z., Li, Z., Sun, Y., et al. (2023). The microRNA-7322-5p/p38/Hsp19 axis modulates Chilo suppressalis cell-defences against Cry1Ca: an effective target for a stacked transgenic rice approach. Plant Biotechnol. J. 21 (9), 1827–1838. doi:10.1111/pbi.14095
Xia, P., Wang, S., Ye, B., Du, Y., Li, C., Xiong, Z., et al. (2018). A Circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48 (4), 688–701. doi:10.1016/j.immuni.2018.03.016
Xu, B., Yang, T., Wang, Z., Zhang, Y., Liu, S., and Shen, M. (2018). CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag. Res. 10, 4871–4880. doi:10.2147/CMAR.S178213
Yang, J., Chen, S., Xu, X., Lin, G., Lin, S., Bai, J., et al. (2022a). Novel-miR-310 mediated response mechanism to Cry1Ac protoxin in Plutella xylostella (L.). Int. J. Biol. Macromol. 219, 587–596. doi:10.1016/j.ijbiomac.2022.08.017
Yang, J., Chen, S., Xu, X., Lin, S., Wu, J., Lin, G., et al. (2023). Novel miR-108 and miR-234 target juvenile hormone esterase to regulate the response of Plutella xylostella to Cry1Ac protoxin. Ecotoxicol. Environ. Saf. 254, 114761. doi:10.1016/j.ecoenv.2023.114761
Yang, J., Xu, X., Lin, S., Chen, S., Lin, G., Song, Q., et al. (2021a). Profiling of microRNAs in midguts of Plutella xylostella provides novel insights into the Bacillus thuringiensis resistance. Front. Genet. 12, 739849. doi:10.3389/fgene.2021.739849
Yang, J., Xu, X., Wu, J., Champer, J., and Xie, M. (2024). Involvement of miR-8510a-3p in response to Cry1Ac protoxin by regulating PxABCG3 in Plutella xylostella. Int. J. Biol. Macromol. 263 (Pt 1), 130271. doi:10.1016/j.ijbiomac.2024.130271
Yang, X., Deng, S., Wei, X., Yang, J., Zhao, Q., Yin, C., et al. (2020). MAPK-directed activation of the whitefly transcription factor CREB leads to P450-mediated imidacloprid resistance. Proc. Natl. Acad. Sci. 117 (19), 10246–10253. doi:10.1073/pnas.1913603117
Yang, X., Wei, X., Yang, J., Du, T., Yin, C., Fu, B., et al. (2021b). Epitranscriptomic regulation of insecticide resistance. Sci. Adv. 7 (19), eabe5903. doi:10.1126/sciadv.abe5903
Yang, Y., Wang, A., Zhang, Y., Xue, C., Zhao, M., and Zhang, J. (2022b). Activating pathway of three metabolic detoxification phases via down-regulated endogenous microRNAs, modulates triflumezopyrim tolerance in the small brown planthopper, Laodelphax striatellus (Fallen). Int. J. Biol. Macromol. 222 (Pt B), 2439–2451. doi:10.1016/j.ijbiomac.2022.10.029
Yang, Y., Zhang, Y., Wang, A., Duan, A., Xue, C., Wang, K., et al. (2021c). Four MicroRNAs, miR-13b-3p, miR-278-5p, miR-10483-5p, and miR-10485-5p, Mediate insecticide tolerance in Spodoptera frugiperda. Front. Genet. 12, 820778. doi:10.3389/fgene.2021.820778
Zhang, B. Z., Hu, G. L., Lu, L. Y., Hu, S. F., Li, Y. S., Su, X., et al. (2021a). Identification of differentially expressed microRNAs under imidacloprid exposure in Sitobion miscanthi. Pestic. Biochem. Physiol. 177, 104885. doi:10.1016/j.pestbp.2021.104885
Zhang, B. Z., Zhang, M. Y., Li, Y. S., Hu, G. L., Fan, X. Z., Guo, T. X., et al. (2022c). MicroRNA-263b confers imidacloprid resistance in Sitobion miscanthi (Takahashi) by regulating the expression of the nAChRbeta1 subunit. Pestic. Biochem. Physiol. 187, 105218. doi:10.1016/j.pestbp.2022.105218
Zhang, C., Liu, P., Sun, L., and Cao, C. (2023). Integration of miRNA and mRNA expression profiles in Asian spongy moth Lymantria dispar in response to cyantraniliprole. Pestic. Biochem. Physiol. 191, 105364. doi:10.1016/j.pestbp.2023.105364
Zhang, J., Liu, M., Wen, L., Hua, Y., Zhang, R., Li, S., et al. (2024). MiR-2b-3p downregulated PxTrypsin-9 expression in the larval midgut to decrease Cry1Ac susceptibility of the diamondback moth, Plutella xylostella (L.). J. Agric. Food Chem. 72 (4), 2263–2276. doi:10.1021/acs.jafc.3c07678
Zhang, M. Y., Zhang, P., Su, X., Guo, T. X., Zhou, J. L., Zhang, B. Z., et al. (2022a). MicroRNA-190-5p confers chlorantraniliprole resistance by regulating CYP6K2 in Spodoptera frugiperda (Smith). Pestic. Biochem. Physiol. 184, 105133. doi:10.1016/j.pestbp.2022.105133
Zhang, P. J., Wu, W. Y., Chen, Q., and Chen, M. (2019). Non-coding RNAs and their integrated networks. J. Integr. Bioinform. 16 (3), 20190027. doi:10.1515/jib-2019-0027
Zhang, Q., Dou, W., Taning, C. N. T., Smagghe, G., and Wang, J. J. (2021b). Regulatory roles of microRNAs in insect pests: prospective targets for insect pest control. Curr. Opin. Biotechnol. 70, 158–166. doi:10.1016/j.copbio.2021.05.002
Zhang, Y., Feng, K., Hu, J., Shi, L., Wei, P., Xu, Z., et al. (2018). A microRNA-1 gene, tci-miR-1-3p, is involved in cyflumetofen resistance by targeting a glutathione S-transferase gene, TCGSTM4, in Tetranychus cinnabarinus. Insect Mol. Biol. 27 (3), 352–364. doi:10.1111/imb.12375
Zhang, Y., Wang, A., Yu, L., Yang, Y., Duan, A., Xue, C., et al. (2022b). Systematic identification and characterization of differentially expressed microRNAs under tetraniliprole exposure in the fall armyworm, Spodoptera frugiperda. Spodoptera Frugiperda. Arch. Insect Biochem. Physiol. 110 (1), e21875. doi:10.1002/arch.21875
Zhou, C., Cheng, J., Beadle, R., Earley, F. G., Li, Z., and Maienfisch, P. (2020). Design, synthesis and acaricidal activities of Cyflumetofen analogues based on carbon-silicon isosteric replacement. Bioorg. Med. Chem. 28 (11), 115509. doi:10.1016/j.bmc.2020.115509
Zhu, B., Li, L., Wei, R., Liang, P., and Gao, X. (2021). Regulation of GSTu1-mediated insecticide resistance in Plutella xylostella by miRNA and lncRNA. PLoS Genet. 17 (10), e1009888. doi:10.1371/journal.pgen.1009888
Zhu, B., Li, X., Liu, Y., Gao, X., and Liang, P. (2017a). Global identification of microRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L.). Sci. Rep. 7, 40713. doi:10.1038/srep40713
Zhu, B., Sun, X., Nie, X., Liang, P., and Gao, X. (2020). MicroRNA-998-3p contributes to Cry1Ac-resistance by targeting ABCC2 in lepidopteran insects. Insect biochem. Mol. Biol. 117, 103283. doi:10.1016/j.ibmb.2019.103283
Zhu, B., Xu, M., Shi, H., Gao, X., and Liang, P. (2017b). Genome-wide identification of lncRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L.). BMC Genomics 18 (1), 380. doi:10.1186/s12864-017-3748-9
Keywords: insects, insecticide resistance, ncRNAs, ceRNA network, ncRNA regulation
Citation: Xiao H, Ma C, Peng R and Xie M (2024) Insights into the role of non-coding RNAs in the development of insecticide resistance in insects. Front. Genet. 15:1429411. doi: 10.3389/fgene.2024.1429411
Received: 08 May 2024; Accepted: 10 June 2024;
Published: 05 July 2024.
Edited by:
Yuan Zhou, Peking University, ChinaCopyright © 2024 Xiao, Ma, Peng and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huamei Xiao, eGlhb2h1YW1laTYyNUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.