
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 02 July 2024
Sec. RNA
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1416772
This article is part of the Research TopicThe Evolution in RNA: 2023View all 5 articles
Glioblastoma poses a formidable challenge among primary brain tumors: its tumorigenic stem cells, capable of self-renewal, proliferation, and differentiation, contribute substantially to tumor initiation and therapy resistance. These glioblastoma stem cells (GSCs), resembling conventional stem and progenitor cells, adopt pathways critical for tissue development and repair, promoting uninterrupted tumor expansion. Long non-coding RNAs (lncRNAs), a substantial component of the human transcriptome, have garnered considerable interest for their pivotal roles in normal physiological processes and cancer pathogenesis. They display cell- or tissue-specific expression patterns, and extensive investigations have highlighted their impact on regulating GSC properties and cellular differentiation, thus offering promising avenues for therapeutic interventions. Consequently, lncRNAs, with their ability to exert regulatory control over tumor initiation and progression, have emerged as promising targets for innovative glioblastoma therapies. This review explores notable examples of GSC-associated lncRNAs and elucidates their functional roles in driving glioblastoma progression. Additionally, we delved deeper into utilizing a 3D in vitro model for investigating GSC biology and elucidated four primary methodologies for targeting lncRNAs as potential therapeutics in managing glioblastoma.
Glioblastoma, classified as a grade IV glioma by the World Health Organization, is the epitome of complexity, lethality, and therapeutic resistance among brain tumors (Louis et al., 2007). Constituting 30% of all primary brain tumors and accounting for 80% of malignant cases, glioblastoma is the leading factor contributing to mortality among patients afflicted with primary brain tumors (Ostrom et al., 2023). The current standard of care for glioblastoma is multimodal, starting with maximal safe surgical resection, followed by postoperative radiation therapy and concurrent and adjuvant temozolomide (TMZ) administration (Khosla, 2016). Despite this comprehensive treatment approach, the median survival is a mere 14.6 months, with a 2-year overall survival rate of only 27%, and fewer than 5% of patients surviving beyond 5 years (Stupp et al., 2005; McNamara et al., 2014; Ostrom et al., 2023). The dismal prognosis for glioblastoma patients is primarily due to extensive intra-tumoral cellular heterogeneity, the induction of angiogenesis, uncontrolled cellular proliferation, and the diffuse infiltration of glioblastoma cells into the surrounding brain tissue, all of which contribute to a high recurrence rate (Aparicio-Blanco et al., 2020; Prager et al., 2020; Alves et al., 2021). Many of these characteristics can be attributed to glioblastoma stem cells (GSCs), also referred to as tumor-initiating or tumor-propagating cells. The first experimental evidence supporting the presence of stem cells within human cortical glial tumors was reported in 2002 (Ignatova et al., 2002). Subsequently, numerous studies have corroborated the existence of GSCs in human brain tumors, underscoring their pivotal roles in invasion, angiogenesis, recurrence, and therapeutic resistance (Singh et al., 2003; Singh et al., 2004; Bao et al., 2006a; Bao et al., 2006b; Wakimoto et al., 2009; Cheng et al., 2013). These findings highlight their significance as prime targets for glioblastoma therapy.
Long non-coding RNAs (lncRNAs), typically exceeding 500 nucleotides, are predominantly transcribed by RNA polymerase II and exhibit classical promoter and enhancer elements (Mattick et al., 2023). A significant portion of these lncRNAs undergo processes such as capping, splicing, and polyadenylation, resulting in their characterization as “mRNA-like” (Wang and Chang, 2011; Rinn and Chang, 2012; Mattick et al., 2023). LncRNAs, characterized by cell- or tissue-specific expression, are multifunctional molecules involved in chromatin remodeling, transcriptional regulation, mRNA stabilization, and protein scaffolding, impacting cellular processes like proliferation, differentiation, apoptosis, and metabolism. They also play pivotal roles in developmental processes, immune responses, and the pathogenesis of various diseases, including cancer (Guttman et al., 2010; Guttman et al., 2011; Kung et al., 2013; Long et al., 2017; Yao et al., 2019; Guo et al., 2020; Statello et al., 2021). Recently numerous studies have highlighted their function as molecular sponges, termed competing endogenous RNAs, effectively sequestering microRNAs through competitive interactions with microRNA response elements. This regulatory mechanism modulates gene expression, contributing to the regulation of physiological functions and disease processes (Xiong et al., 2018; Kim et al., 2018; Yang et al., 2022; Zhang et al., 2023a; Wu et al., 2023). In cancer, lncRNAs can act as oncogenes, promoting cancer progression, or conversely, as tumor suppressors, inhibiting tumor growth. Understanding the paradoxical roles that lncRNAs play in cancer biology has paved the way for novel therapeutic strategies, such as silencing oncogenic lncRNAs or restoring the function of tumor-suppressing lncRNAs. Moreover, advances in RNA-based therapeutics, such as antisense oligonucleotides, RNA interference (RNAi), and CRISPR/Cas strategies, offer potential methods for precisely modulating lncRNA activity.
Based on clinical data, as well as in vitro and in vivo evidence, we selected several well-studied examples of lncRNAs involved in the acquired capabilities of GSCs and discussed their regulation in glioblastoma (Figure 1; Table 1).
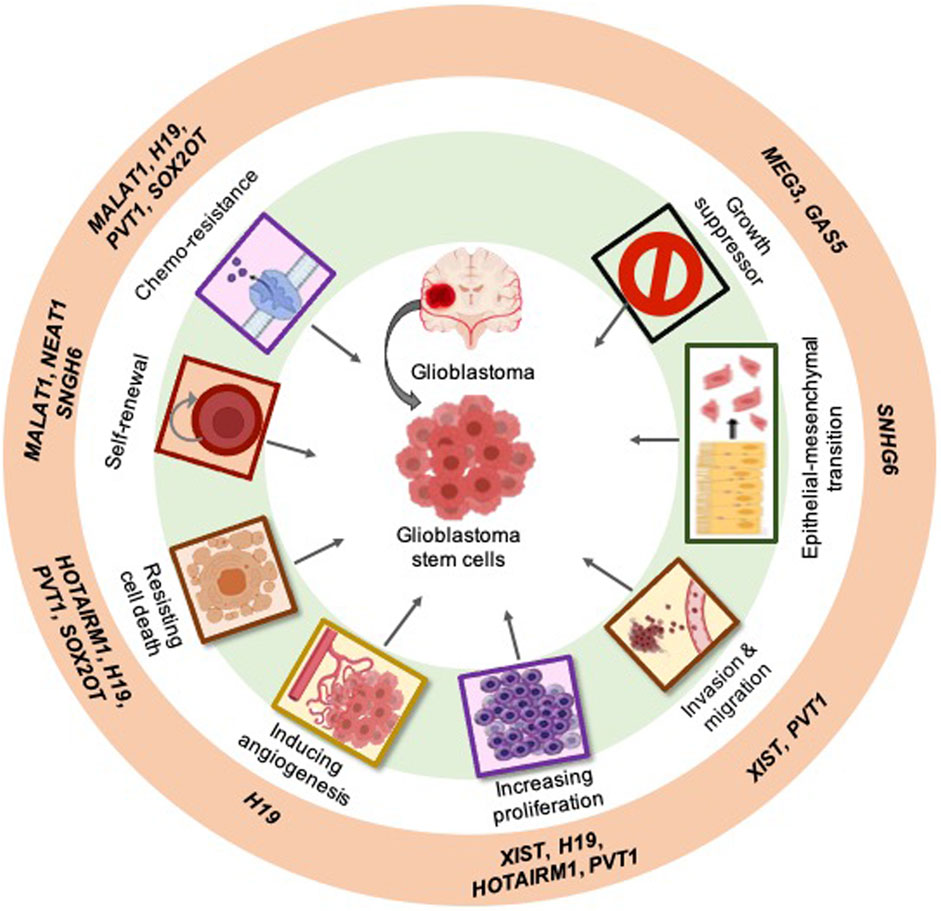
Figure 1. Examples of the most studied long non-coding RNAs (lncRNAs) linked to the hallmark characteristics of cancer within glioblastoma: Examples of the most studied GSC-associated lncRNAs (outer ring). The classical hallmarks of cancer in which lncRNAs play a role (middle ring). lncRNAs can specifically act on GSCs in glioblastoma (center).
X-inactive-specific transcript (XIST) RNA, discovered in the early 1990s, stands as one of the earliest identified lncRNAs, predating the revelation from the Human Genome Project that a significant portion of our genome comprises noncoding sequences (Brown, 1991; Lander et al., 2001). Despite its original roles in X-chromosome dosage compensation, XIST also plays a significant role in regulating cell growth, development, and is implicated in various cancers, including glioblastoma (Yao et al., 2015; Patrat et al., 2020; Yang et al., 2021; Sadagopan et al., 2022). Research has shown that XIST is highly expressed in glioma tissues and GSCs. Knockdown of XIST significantly reduces cell proliferation, migration, and invasion, while also inducing apoptosis (Yao et al., 2015). Furthermore, in vivo studies demonstrate that XIST knockdown suppresses tumor growth and improves survival rates in mice (Yao et al., 2015). Another study has demonstrated the involvement of the XIST/miR-133a/SOX4 axis, wherein XIST interacts with SOX4 and miR-133a, thereby regulating SOX4 expression. This regulatory axis ultimately governs proliferation, migration, metastasis, and the maintenance of glioblastoma stem cells (Luo et al., 2020). In addition, XIST, post-transcriptionally regulated by Steroid Receptor Coactivator 1, influences the stemness of glioblastoma cells by modulating the expression of Kruppel-like factor 4 through the XIST/miR-152 axis (Gong et al., 2021). Therefore, targeting XIST may represent a promising therapeutic approach in glioblastoma.
The imprinted oncofetal lncRNA H19, originating from a paternally imprinted gene, is among the earliest identified upregulated lncRNAs observed across various cancer types, including glioblastoma. This lncRNA is expressed during embryonic development, downregulated at birth, and subsequently reappears in tumors (Bartolomei et al., 1991; Raveh et al., 2015; Jiang et al., 2016). One study demonstrated that knocking down H19 significantly reduces both mRNA and protein levels of GSC markers, such as CD133, NANOG, OCT4, and SOX2 (Li et al., 2016). Additionally, overexpression of H19 in CD133+ GSCs enhances neurosphere formation and tumor growth (Jiang et al., 2016), suggesting its role in GSC maintenance. Furthermore, H19 promotes invasion and angiogenesis in glioblastoma, and its overexpression is associated with poor overall survival and progression-free survival (Jiang et al., 2016; Fawzy et al., 2018). Silencing H19 by TMZ reduces cellular proliferation and increases the apoptosis rate (Li et al., 2016). In addition, H19 regulates High Mobility Group AT-Hook 2 (HMGA2) expression and microRNAs, both of which enhance the mesenchymal transition of glioblastoma and self-renewal of GSCs. Because H19 acts as a competing endogenous RNA for microRNAs like let-7, it modulates their availability and indirectly reduces HMGA2 expression (Jiang et al., 2016; Lecerf et al., 2020). This regulatory mechanism orchestrated by H19 contributes to the maintenance of GSCs and promotes tumor aggressiveness. Hence, H19 may be a valuable therapeutic target in glioblastoma.
The lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has garnered significant attention as a therapeutic target in various cancers, including glioblastoma. In glioblastoma, MALAT1 regulates numerous facets of tumor biology, particularly about GSCs, which are pivotal in tumor initiation and progression (Han et al., 2016; Xiong et al., 2018). Moreover, MALAT1 promotes the self-renewal and maintenance of GSCs by interacting with SOX2 (Williamson, 1989; Xiong et al., 2018; Liao et al., 2019). MALAT1 modulates key signaling pathways, including the Wnt/β-catenin, Notch, and Hedgehog pathways, which are crucial in GSC biology (Fu et al., 2020). Implicated in mediating therapeutic resistance in GSCs, MALAT1 confers resistance to chemotherapy, such as TMZ, and radiotherapy—standard treatments for glioblastoma (Chen et al., 2017; Kim et al., 2018). MALAT1 achieves this by modulating multiple molecular pathways involved in DNA repair, apoptosis, and drug efflux, thus diminishing treatment efficacy and contributing to tumor recurrence and progression (Kim et al., 2018; Baspinar et al., 2020). Strategies aimed at inhibiting MALAT1 expression or function have shown promising results in preclinical studies, demonstrating the potential for MALAT1-targeted therapies to sensitize GSCs to conventional treatments and improve patient outcomes. Further research is necessary to unravel the specific mechanisms by which MALAT1 regulates GSC behavior and to develop effective MALAT1-targeting strategies for treating glioblastoma.
Nuclear enriched abundant transcript 1 (NEAT1) has emerged as a pivotal regulator in GSCs and thus a promising therapeutic target for glioblastoma. Upregulated in GSCs, NEAT1 contributes to various aspects of tumorigenesis and therapy resistance. NEAT1 regulates GSC proliferation, self-renewal, stem cell maintenance, and resistance (Bi et al., 2020; Gao et al., 2021; Ghafouri-Fard et al., 2021). Suppressing NEAT1 impedes the migration and invasion capabilities of glioma cells by modulating the expression of SOX2, targeted by miR-132, underscoring NEAT1’s significance in glioma progression and its potential as a therapeutic target (Zhou et al., 2018). Furthermore, NEAT1 modulates the expression of crucial genes in key signaling pathways, such as Notch, Wnt, and Stat3, which are imperative for GSC maintenance and survival (Zang et al., 2020; Gao et al., 2021; Yuan et al., 2022). By bolstering GSCs and undermining the efficacy of standard treatments, NEAT1 contributes to tumor progression and unfavorable patient outcomes in glioblastoma (Bi et al., 2020; Gao et al., 2021). Targeting NEAT1 in GSCs presents an avenue for sensitizing these cells to conventional therapies and enhancing treatment outcomes in glioblastoma.
HOX antisense intergenic RNA myeloid 1 (HOTAIRM1), a transcript previously implicated as a cis-acting factor, is expressed in the myeloid lineage (Zhang et al., 2009) and induced during neuronal differentiation (Lin et al., 2011). Despite its initial identification in myeloid cells, emerging research has revealed its significance as a cancer-related lncRNA, aberrantly expressed in various tumors, including glioblastoma (Zhang et al., 2014; Diaz-Beya et al., 2015; Wan et al., 2016; Li et al., 2018). It is significantly upregulated in GSCs (Li et al., 2018; Xie et al., 2020). Silencing HOTAIRM1 impairs the proliferation, apoptosis, self-renewal, and tumorigenesis of GSCs, suggesting its crucial role in GSC stemness (Lin et al., 2020; Xie et al., 2020), and thus glioblastoma progression. Additionally, HOTAIRM1 interacts with EZH2, G9a, and DNA methyltransferases, sequestering them away from the transcription start sites of the HOXA1 gene, thereby activating HOXA1 oncogene expression (Li et al., 2018). Therefore, the oncogenic HOTAIRM1/HOXA1 axis in glioblastoma may offer promise as a novel therapeutic target. Another study demonstrated that HOTAIRM1 promotes cell proliferation and invasion in human glioblastoma by upregulating SP1 via sponging miR-137 (Hao et al., 2020). Furthermore, HOTAIRM1 has been associated with shorter survival in glioblastoma patients, independent of IDH mutation and O6-methylguanine-DNA-methyltransferase promoter methylation (Ahmadov et al., 2021), indicating its role as a driver of biological aggressiveness, radioresistance, and poor outcomes in glioblastoma. Hence, targeting HOTAIRM1 may represent a promising therapeutic approach in glioblastoma, as it may function as an oncogene.
The plasmacytoma variant translocation 1 (PVT1) lncRNA, located on human chromosome 8q24, is co-amplified with MYC and has been identified as an oncogene in several cancers (Tseng and Bagchi, 2015). PVT1 is upregulated in glioblastoma cells, tissues, and GSCs (Jin et al., 2020; Lv et al., 2022; Hazra et al., 2023). It is associated with TMZ resistance and poor prognosis in glioblastoma patients (Roh et al., 2023; Huang L. et al., 2024). Recent in vitro and in vivo studies have demonstrated that PVT1 promotes glioblastoma cell proliferation, invasion, and tumor growth in mice while inhibiting apoptosis (Jin et al., 2020; Lv et al., 2022). In addition, PVT1 regulates E74-like factor 4 (ELF4) expression by competitively binding to miR-365 and controls the stemness of GSCs via the miR-365/ELF4/SOX2 axis (Gong et al., 2021) Thus, PVT1 may serve as a potential therapeutic target and prognostic indicator in glioblastoma.
SOX2 overlapping transcript (SOX2OT) is highly expressed in embryonic stem cells and upregulated in various cancer types, including glioblastoma (Wang et al., 2020). SOX2OT is significantly upregulated in GSCs and glioblastoma tissues, and decreased expression levels of SOX2OT impede the malignant biological activities of GSCs by elevating miR-194-5p and miR-122 levels (Su et al., 2017). Another study showed that SOX2OT decreases cell apoptosis, enhances cell proliferation, and confers resistance to TMZ by increasing the expression of SOX2, which in turn activates the Wnt5a/β-catenin signaling pathway. These results suggest that SOX2OT could potentially serve as a new prognostic biomarker and represent a promising therapeutic target for glioblastoma treatment (Liu et al., 2020).
The lncRNA small nucleolar RNA host gene 6 (SNHG6) has been implicated in various cancer types, yet its role in glioblastoma remains relatively unexplored. Two studies have reported frequent upregulation of SNHG6 in glioblastoma tissues and cell lines compared to normal brain tissues (Nie et al., 2021; Rahmani et al., 2023), suggesting its involvement in glioblastoma development. Additionally, the downregulation of SNHG6 in glioblastoma cells induces the expression of the stem cell marker SOX2 and modulates epithelial-mesenchymal transition through interactions with Notch1 and β-catenin (Nie et al., 2021). This transition is critical for tumor metastasis and invasion in glioblastoma. Moreover, high expression levels of SNHG6 in glioblastoma have been associated with advanced tumor stage, poor prognosis, and shorter overall survival of patients (Nie et al., 2021). Overall, the dysregulation of SNHG6 in glioblastoma suggests its potential as a diagnostic biomarker and therapeutic target for the disease. However, further research into the precise mechanisms underlying SNHG6’s involvement in glioblastoma pathogenesis is warranted to better assess its therapeutic promise.
Maternally expressed gene 3 (MEG3), recognized as a tumor suppressor lncRNA, is downregulated across various cancer types, including glioblastoma. In GSCs, this reduced MEG3 expression corresponds with increased tumorigenicity and stemness (Balci et al., 2016). Restoring MEG3 function in vitro impaired the tumorigenic abilities of GSCs, evidenced by inhibited cell growth, migration, and colony formation, alongside reduced in vivo tumor growth, thereby attenuating infiltration. These effects were linked to MEG3’s modulation of genes involved in cell adhesion and epithelial-mesenchymal transition (Yang et al., 2020; Leung et al., 2023). Moreover, epigenetic alterations, particularly hypermethylation within the MEG3-DMR region, predominantly drive the downregulation of genes within the DLK1-DIO3 locus in glioblastoma (Buccarelli et al., 2020). In addition, decreased expression of MEG3 is significantly correlated with shorter survival in GBM patients (Jin et al., 2018). Thus, elucidating the functional impact of MEG3 on GSCs holds significant promise for advancing our understanding of GBM progression and developing targeted therapeutic strategies.
Growth arrest-specific transcript 5 (GAS5) is one of the most highly expressed lncRNAs present in human tissues and is implicated in embryogenesis (Hudson et al., 2014). However, its expression is notably reduced across various cancer types, including glioblastoma, and is inversely correlated with clinicopathological characteristics, such as tumor size, staging, or metastasis (Pickard and Williams, 2015). Diminished expression of GAS5 is observed in high-grade GBM tissues and cells (Zhao et al., 2017), suggesting implications for poorer prognosis. GAS5 exerts its suppressive effect on GSC malignancy by binding to miR-196a-5p, an onco-miRNA known to promote GSC proliferation, migration, and invasion while reducing apoptosis (Zhao et al., 2017). Moreover, GAS5 overexpression inhibits cell viability, migration, and invasion in glioblastoma cells, along with impairing stemness and proliferation in GSCs. Furthermore, GAS5 exhibits anti-oncogenic properties in GBM by modulating the IL-6/STAT3 pathway both in vitro and in vivo, leading to the inhibition of tumor growth in a xenograft model (Wu et al., 2022). These findings underscore the significant role of GAS5 in GBM progression, highlighting its potential as a prognostic biomarker and therapeutic target for glioblastoma.
The primary challenge in lncRNA research lies in the limited conservation observed across many lncRNA species. This lack of conservation manifests at the primary sequence, small sequence block, syntenic, and sometimes, structural levels. While certain cancer-associated lncRNAs, such as MALAT1, NEAT1, and H19, show notable conservation, the majority of human cancer-associated lncRNAs lack sequence conservation across mammalian species. This poses a significant barrier to translating findings from human cancer studies to preclinical mouse models. Therefore, alternative models must be explored for preclinical studies aimed at assessing GSC lncRNAs (Figure 2).
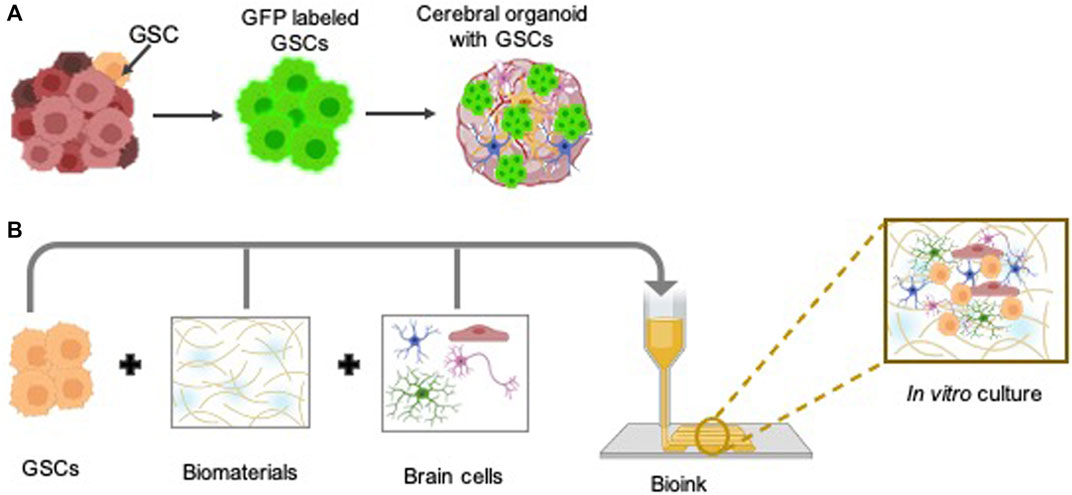
Figure 2. Three-dimensional (3D) preclinical in vitro models to study GSC biology: The intricate biology of GSCs within the human brain environment can be effectively explored through the utilization of advanced 3D in vitro models, such as glioblastoma cerebral organoids (GLICO) (A), in conjunction with bioprinting techniques (B). Some parts of the figures were created using BioRender.com.
Three-dimensional (3D) in vitro models are essential for studying GSC biology owing to their capacity to replicate the intricate tumor microenvironment more accurately compared to conventional 2D cultures. Unlike murine models, 3D in vitro models provide enhanced throughput, cost-effectiveness, and the ability to mimic human-specific responses, rendering them invaluable for investigating glioblastoma pathophysiology and assessing potential therapeutic interventions. While conventional culture methods offer simplicity, affordability, and speed, they fail to replicate crucial interactions between neural stem cells, perivascular endothelial cells, GSCs, and components of the tumor microenvironment, such as microglia, tumor-activated macrophages, and neurons, which regulate fundamental cancer cell processes like metabolism, proliferation and invasiveness (Bikfalvi et al., 2023). Additionally, GSCs cultured in laminin-adhered 2D monolayers or 3D spheroids in suspension fail to replicate tumor invasion characteristics due to the absence of surrounding matrices. Further, long-term TMZ treatment is impeded by cell overgrowth (Ozturk et al., 2020). Presently, glioblastoma cerebral brain organoids and 3D bio-printed models represent preclinical platforms promising for investigating GSC lncRNAs in glioblastoma.
Glioblastoma cerebral brain organoids (GLICO) are innovative approaches for modeling glioblastoma within a physiologically relevant human brain environment. The generation of GLICO entails a multi-step procedure. Firstly, cerebral brain organoids are generated from human embryonic stem cells or fibroblast-derived induced pluripotent stem cells and recapitulate key features of human brain tissue, including its cellular diversity and complex architecture (Lancaster and Knoblich, 2014). Next, GSCs are isolated from freshly obtained patient tumor biopsies (Dundar et al., 2019) or patient-derived xenografts (Hazra et al., 2023) and labeled with a fluorescent marker to facilitate the observation of tumor development within the organoid in real-time. Finally, co-culturing labeled GSCs with cerebral brain organoids (Ogawa et al., 2018; Linkous et al., 2019) or region-specific brain organoids (Fan et al., 2024) presents numerous advantages for comprehensive glioblastoma research. By providing a microenvironment that closely mimics the intricate architecture and cellular interactions of the human brain, cerebral brain organoids offer a physiologically relevant platform to study GSC behavior within a context that mirrors the native tumor niche. Furthermore, these organoids enable researchers to explore GSC responses to various stimuli and therapeutic interventions in a controlled and reproducible manner, thereby facilitating the identification of potential therapeutic targets and the development of personalized treatment strategies for glioblastoma patients. Additionally, by manipulating cerebral brain organoids to replicate the complex tumor microenvironment (TME), characterized by the interplay of immune cells, predominantly microglia, one of the most abundant cellular constituents of the TME, blood vessels, astrocytes, and the extracellular matrix, which exhibits spatial variation across distinct tumor regions, researchers gain valuable insights into the fundamental mechanisms driving glioblastoma initiation and advancement (Bikfalvi et al., 2023). Thus, GLICO models, as advanced preclinical models, hold significant promise for advancing our understanding of glioblastoma pathogenesis, discovering novel therapeutic targets, and evaluating treatment strategies within a biologically relevant context. Furthermore, the GLICO model system provides an unprecedented opportunity to investigate the involvement of GSC lncRNAs by manipulating their expression levels through CRISPR/Cas technology and co-culturing with cerebral organoids, both in the presence and absence of GSC lncRNAs in glioblastoma. However, further research is imperative, given that only three studies have investigated the role of lncRNAs utilizing cerebral brain organoids as model systems in glioblastoma (Liu et al., 2020; Han et al., 2020; Tang et al., 2020). However, one notable disadvantage of cerebral brain organoids is the absence of functional blood vessels, which limits their ability to fully recapitulate the complexity of the tumor microenvironment found in vivo. Without a vascular network, organoids lack the physiological perfusion and nutrient exchange seen in actual brain tissue. This deficiency hinders the accurate modeling of aspects such as tumor angiogenesis, drug delivery, and interactions with circulating immune cells. Consequently, while GLICO models offer valuable insights into glioblastoma biology, their utility may be somewhat constrained when studying certain aspects reliant on vascular interactions.
In pursuit of patient-specific glioblastoma models that faithfully represent the multifaceted complexity of tumors and facilitate accurate therapeutic response assessments, researchers have developed a novel 3D bio-printed glioblastoma model. This innovative model, incorporating patient tumor-derived GSCs, macrophages, astrocytes, neural stem cells, vascular endothelial cells, and decellularized extracellular matrix from brain tissue, accurately recapitulates the structural, biochemical, and biophysical properties of real tumors, encompassing cellular crosstalk, invasion, context-specific functional dependencies, and immunologic interactions within a species-matched neural environment (Yi et al., 2019; Parra-Cantu et al., 2020; Tang et al., 2020). It serves as a dependable platform for exploring drug sensitivity in glioblastoma. Despite significant advancements, traditional 3D bioprinting still faces challenges. However, as this technology continues to evolve, it holds tremendous potential in revolutionizing the combat against brain cancers, offering prospects for enhanced diagnostics, therapies, and ultimately, better patient prognoses. Traditional 3D bioprinting methods are static and may not fully capture the dynamic nature of glioblastoma progression and microenvironmental interactions. Nonetheless, advancements in bioprinting technology, such as 4D, 5D, and even 6D printing (Chadwick et al., 2020; Amiri et al., 2023), present exciting prospects for overcoming these limitations. By introducing temporal and additional spatial dimensions, these advanced models can better mimic the dynamic changes that occur within the tumor microenvironment over time (Yi et al., 2019; Chadwick et al., 2020; Amiri et al., 2023; Yeo et al., 2023). This capability not only improves our understanding of glioblastoma biology but also facilitates the development of more precise and effective therapeutic interventions tailored to individual patient needs.
While 3D bioprinting has been used extensively in various fields of research, including tissue engineering and drug screening, its application to lncRNA study is relatively limited. Given the versatility and precision of 3D bioprinting, researchers could use it to create intricate tissue structures that accurately mimic the in vivo microenvironment, facilitating the study of lncRNA functions within their biologically relevant context. Yet despite its promise, further exploration and development are necessary to fully exploit the capabilities of 3D bioprinting for lncRNA research.
RNA therapeutics offer a promising avenue for mitigating various ailments, including cancer. Recent strides in genome editing, oligonucleotide chemistry, multi-omics methodologies, and RNA manipulation are streamlining the creation of efficient and economical drug discovery pathways tailored to target lncRNAs. As the exploration in this domain expands, the potential for lncRNA-based therapeutics to revolutionize cancer management has become increasingly apparent. Promising therapeutic strategies to target lncRNAs include small interfering RNAs (siRNAs), antisense oligonucleotides (ASOs), CRISPR gene editing, and small molecules that recognize RNA structures (Figure 3).
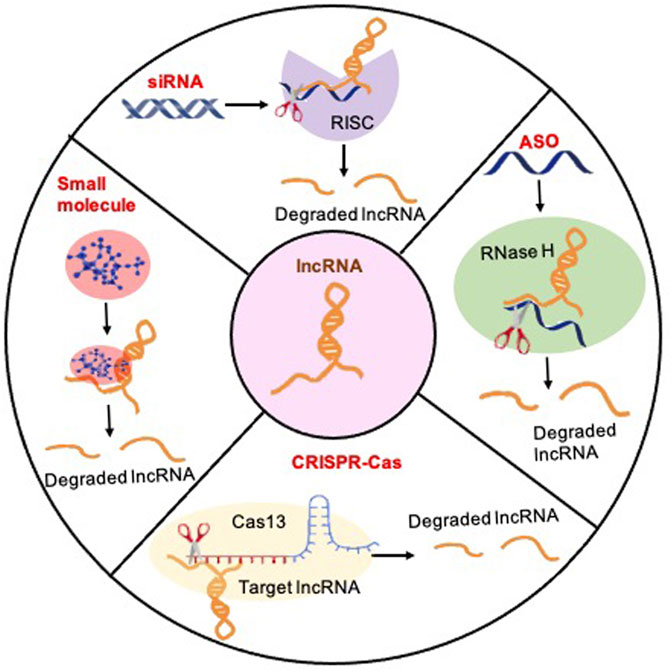
Figure 3. Methods for therapeutically targeting lncRNAs to treat glioblastoma: Four principal therapeutic strategies for lncRNA loss of function: (1) The siRNA-mediated knockdown strategy can be applied to target cytoplasmic lncRNA transcripts by introducing synthetic siRNAs that specifically bind to and degrade the targeted transcripts, thereby effectively suppressing their expression levels. The targeted lncRNA binds to the siRNA-loaded RNA-induced silencing Complex (RISC), which induces the degradation of the lncRNA. (2) The ASO-mediated reduction of nuclear lncRNA transcription levels entails the utilization of antisense oligonucleotides (ASOs) meticulously crafted to bind with nuclear lncRNA transcripts, inducing their degradation or inhibition, consequently diminishing the overall transcriptional activity of the specified lncRNAs within the nucleus. ASOs exploit the cellular enzyme RNase H to catalyze the cleavage of target lncRNAs. (3) CRISPR/Cas strategies represent a potent and accurate approach for targeting lncRNAs. Illustrated here, CRISPR-Cas13 employs a guide RNA to direct the Cas13 protein to designated lncRNA sequences, facilitating the cleavage of the target lncRNA with precision, thereby presenting a promising avenue for precise RNA editing and gene regulation. (4) Small molecules can be directed towards a specific lncRNA to hinder its binding with molecular partners or disrupt proper structure formation.
Ranging from 19 to 30 nucleotides in length, siRNAs are short double-stranded RNAs that initiate gene silencing by engaging the RNA-induced silencing complex to degrade the target gene through posttranscriptional mechanisms (Hannon and Rossi, 2004). The therapeutic potential of siRNAs in human disease spans a wide spectrum, including applications in genetic disorders, viral infections, and neurodegenerative diseases (Gavrilov and Saltzman, 2012). Several therapeutic drugs utilizing siRNAs have been developed and are either available on the market or in clinical trials. Notably, patisiran (ONPATTRO™) harnesses siRNA technology to treat hereditary transthyretin-mediated amyloidosis (Coelho et al., 2013; Hoy, 2018; Urits et al., 2020). By targeting and reducing the production of abnormal transthyretin protein, patisiran effectively mitigates disease progression, showcasing the potential of siRNA-based therapies. While there are currently no FDA-approved siRNA-based therapeutic drugs for cancer, many are in various stages of preclinical and clinical development. For instance, systemic delivery of MALAT1 siRNA via a nano complex that targets GSCs significantly reduced the growth, infiltration, and stemness of GSCs improved the sensitivity of GSCs to TMZ, and increased the survival of animals (Kim et al., 2018). This suggests that MALAT1 could serve as a preclinical model for glioblastoma. Although lncRNA-based siRNA therapeutics hold promise for GBM treatment, there are concerns regarding their clinical applications. One challenge is efficiently delivering siRNA molecules across the blood-brain barrier and into tumor cells within the brain. Additionally, off-target effects and the potential immune stimulation associated with siRNA therapy may pose risks to healthy tissues and compromise the overall safety of the treatment. Lastly, the development of resistance to siRNA-mediated gene silencing could limit the long-term efficacy of this therapeutic approach.
ASOs are synthetic single-stranded DNA or RNA molecules, typically 15–21 nucleotides in length, specifically engineered to bind to complementary RNA sequences through standard Watson-Crick base pairing mechanisms (Crooke et al., 2021). This binding capability allows ASOs to exert their effects by modulating gene expression at the posttranscriptional level. ASOs have emerged as powerful tools for manipulating gene expression, including the knockdown of lncRNAs (Lang et al., 2021; Adewunmi et al., 2023). Various types of ASO modifications, such as phosphorothioate (PS), 2′-methoxyethyl (2′-MOE), 2′-constrained ethyl (2′-cEt), and locked nucleic acid, have been developed to enhance their efficacy and specificity in RNA knockdown (Crooke et al., 2021). A recent study demonstrated the promising preclinical therapeutic target of a high-grade pediatric glioma known as diffuse intrinsic pontine glioma (DIPG) using 2′-MOE ASO (Zhang et al., 2023b). The hallmark genetic alteration of DIPG is a mutation in the genes encoding histone H3.3, which results in a lysine-to-methionine substitution at position 27 (K27M); this alteration is found in approximately 80% of DIPG cases (Wan et al., 2018). The 2′-MOE-PS ASOs efficiently degrade H3-3AK27M mRNA, resulting in reduced levels of H3.3K27M protein in both in vitro and in vivo systems (Zhang et al., 2023b). These findings suggest the potential of ASO therapy as a viable treatment option for DIPG (Zhang et al., 2023b). In glioblastoma, intravenous treatment with ASOs targeting TUG1, in conjunction with a sophisticated drug delivery system, induces GSC differentiation and efficiently inhibits GSC growth in vivo, indicating that TUG1 may be a potent therapeutic approach to eradicate the GSC population (Katsushima et al., 2016). Another preclinical study demonstrated that targeting MALAT1 with modified ASOs promotes the differentiation of tumor cells and decreases tumor growth in a breast cancer mouse model (Arun et al., 2016)—a strategy that may be suitable for other solid cancers, including glioblastoma. These examples highlight the potential of ASOs and lncRNA-targeted therapies for precise and targeted intervention in glioblastoma, offering new avenues for therapeutic development and personalized treatment strategies.
The advent of CRISPR-Cas systems has effectively mitigated the limitations inherent in RNAi for identifying cancer-promoting lncRNAs (Meira et al., 2021). CRISPR technology facilitates the precise perturbation of lncRNA expression in both the nucleus and cytoplasm, enabling reversible or permanent modifications (Li et al., 2020). Furthermore, CRISPR offers scalability for high-throughput assays, both in vitro and in vivo, owing to its compatibility with pooled screening formats (Bock et al., 2022). This scalability obviates the need for costly arrayed libraries or screening robots, as CRISPR-based assays yield direct functional readouts and adaptable targeting libraries, regularly updated to incorporate evolving gene annotations (Bock et al., 2022). Nevertheless, generating loss-of-function alleles for lncRNAs with a conventional CRISPR-Cas9 system is slightly more intricate than for protein-coding genes due to lncRNA’s inherent lack of long open reading frames. To overcome this challenge, a dual guide RNA approach is employed, facilitating the simultaneous creation of two DNA double-strand breaks, potentially resulting in the deletion of the entire lncRNA gene or its promoter region, a technique known as CRISPR deletion (Hazra et al., 2022). CRISPR technology can manipulate lncRNA genes without requiring DNA mutations. Using CRISPR interference (CRISPRi), a recent study identified lncGRS-1 as a glioma-specific therapeutic target. Knockdown of lncGRS-1 effectively impedes the proliferation of primary adult and pediatric glioma cells while preserving the integrity of normal brain cells (Liu et al., 2020). This finding underscores the utility of a swift and precise methodology for identifying promising therapeutic targets within the non-coding genome, thereby advancing the potential of radiation therapy in glioma treatment (Liu et al., 2020). Furthermore, CRISPR can be harnessed to directly target RNA using systems like single-stranded RNA-binding CRISPR-Cas13. This approach exhibits knockdown efficiency comparable to RNAi but with significantly reduced off-target effects (Abudayyeh et al., 2017). Recently, A Cas13d/CasRx-based screening platform was developed for genome-scale interrogation of lncRNAs (Montero et al., 2024), which overcomes the limitations observed in other lncRNA perturbation approaches, such as low targeting efficiency, off-target effects, and inadvertent perturbation of other DNA elements. The system has been further refined to enhance its applicability across various cancer types, enabling the systematic exploration of lncRNA biology, identification of context-specific traits, and discovery of lncRNA vulnerabilities suitable for therapeutic targeting (Montero et al., 2024). This study highlights that CasRx-based lncRNA screening effectively addresses major limitations associated with alternative perturbation methods. For example, RNAi is hindered by low nuclear activity and high off-target rates, whereas CasRx, fused with a nuclear localization signal, demonstrates robust nuclear activity and precise targeting of lncRNAs. Notably, CasRx exhibits high specificity in targeting lncRNAs, with the optimal guide RNA length ranging from 20 to 23 nucleotides, compared to the 2- to 7-nucleotide seed region typical of siRNAs (Konermann et al., 2018; Li et al., 2021). Taking into account all of these advantages, along with the potential for RNA-degrading drugs, CRISPR-CasRx may emerge as the preferred option for targeting lncRNAs.
Targeting lncRNAs with small molecules or natural compounds represents a promising therapeutic approach for glioblastoma. For example, small molecule inhibitors can target the lncRNA HOTAIR, recognized for its role in promoting metastasis through chromatin remodeling and gene expression regulation (Li et al., 2019). In a notable preclinical study, targeting the MALAT1 ENE (Exonuclease Protection Element) triplex with small molecules holds great promise for developing novel anticancer therapies and molecular tools to investigate MALAT1-driven cancers. The MALAT1 ENE triplex, a specific structural motif within the MALAT1 RNA molecule, is known to play a crucial role in cancer progression and metastasis. Disruption of this motif has the potential to inhibit cancer cell growth and metastasis, offering new avenues for cancer treatment. Additionally, these small molecules serve as valuable molecular probes in research settings, allowing for precise study of the mechanisms underlying MALAT1-driven cancers (Abulwerdi et al., 2019). These endeavors underscore the growing significance of small molecule-based approaches in modulating lncRNA function, offering innovative avenues for effective cancer therapies targeting these non-coding transcripts. Understanding higher-order RNA structures is imperative for designing small molecules targeting lncRNAs (Childs-Disney et al., 2022). While the feasibility of targeting RNAs with small molecules has been explored with some success, a deeper comprehension of RNA-ligand interactions, additional RNA targets, and active compounds targeting RNAs is necessary for further advancements.
Embarking on advancements in radiotherapy, surgery, and contemporary medical modalities, the challenge of glioblastoma persists as a significant concern due to its high mortality rates. Exploring the potential synergy between GSC lncRNAs and emerging therapies offers hope for enhancing treatment outcomes, yet demands a profound mechanistic comprehension of their roles in glioblastoma. Studies have unveiled associations between specific lncRNA expression and glioblastoma malignancy grade, histological differentiation, and diagnosis (Zhang et al., 2012). While harnessing lncRNAs for novel diagnostics and therapeutics in glioblastoma holds promise, many aspects of lncRNA biology remain elusive. Therefore, employing lncRNAs for diagnostic and therapeutic purposes without a comprehensive grasp of their multifaceted functions may be premature.
The detailed characterization of each lncRNA and the development of novel therapeutics for glioblastoma present significant challenges. Nevertheless, recognizing lncRNAs as crucial contributors to tumorigenesis provides an opportunity to explore their expression patterns and mechanisms of action for targeted cancer therapies. Moreover, the rapid progress in sequence-based nucleic acid therapeutics offers avenues for exploiting lncRNAs as therapeutic targets in cancer treatment. Therefore, identifying and evaluating lncRNA targets within the glioblastoma context represent vital steps toward translating research insights into clinical applications.
RH: Conceptualization, Supervision, Writing–original draft, Writing–review and editing. RD: Writing–original draft. AT: Visualization, Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., et al. (2017). RNA targeting with CRISPR-Cas13. Nature 550 (7675), 280–284. doi:10.1038/nature24049
Abulwerdi, F. A., Xu, W., Ageeli, A. A., Yonkunas, M. J., Arun, G., Nam, H., et al. (2019). Selective small-molecule targeting of a triple helix encoded by the long noncoding RNA, MALAT1. ACS Chem. Biol. 14 (2), 223–235. doi:10.1021/acschembio.8b00807
Adewunmi, O., Shen, Y., Zhang, X. H. F., and Rosen, J. M. (2023). Targeted inhibition of lncRNA Malat1 alters the tumor immune microenvironment in preclinical syngeneic mouse models of triple-negative breast cancer. Cancer Immunol. Res. 11 (11), 1462–1479. doi:10.1158/2326-6066.CIR-23-0045
Ahmadov, U., Picard, D., Bartl, J., Silginer, M., Trajkovic-Arsic, M., Qin, N., et al. (2021). The long non-coding RNA HOTAIRM1 promotes tumor aggressiveness and radiotherapy resistance in glioblastoma. Cell Death Dis. 12 (10), 885. doi:10.1038/s41419-021-04146-0
Alves, A. L. V., Gomes, I. N. F., Carloni, A. C., Rosa, M. N., da Silva, L. S., Evangelista, A. F., et al. (2021). Role of glioblastoma stem cells in cancer therapeutic resistance: a perspective on antineoplastic agents from natural sources and chemical derivatives. Stem Cell Res. Ther. 12 (1), 206. doi:10.1186/s13287-021-02231-x
Amiri, E., Sanjarnia, P., Sadri, B., Jafarkhani, S., and Khakbiz, M. (2023). Recent advances and future directions of 3D to 6D printing in brain cancer treatment and neural tissue engineering. Biomed. Mater 18 (5), 052005. doi:10.1088/1748-605X/ace9a4
Aparicio-Blanco, J., Sanz-Arriazu, L., Lorenzoni, R., and Blanco-Prieto, M. J. (2020). Glioblastoma chemotherapeutic agents used in the clinical setting and in clinical trials: nanomedicine approaches to improve their efficacy. Int. J. Pharm. 581, 119283. doi:10.1016/j.ijpharm.2020.119283
Arun, G., Diermeier, S., Akerman, M., Chang, K. C., Wilkinson, J. E., Hearn, S., et al. (2016). Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30 (1), 34–51. doi:10.1101/gad.270959.115
Balci, T., Yilmaz Susluer, S., Kayabasi, C., Ozmen Yelken, B., Biray Avci, C., and Gunduz, C. (2016). Analysis of dysregulated long non-coding RNA expressions in glioblastoma cells. Gene 590 (1), 120–122. doi:10.1016/j.gene.2016.06.024
Bao, S., Wu, Q., McLendon, R. E., Hao, Y., Shi, Q., Hjelmeland, A. B., et al. (2006a). Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444 (7120), 756–760. doi:10.1038/nature05236
Bao, S., Wu, Q., Sathornsumetee, S., Hao, Y., Li, Z., Hjelmeland, A. B., et al. (2006b). Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 66 (16), 7843–7848. doi:10.1158/0008-5472.CAN-06-1010
Bartolomei, M. S., Zemel, S., and Tilghman, S. M. (1991). Parental imprinting of the mouse H19 gene. Nature 351 (6322), 153–155. doi:10.1038/351153a0
Baspinar, Y., Elmaci, I., Ozpinar, A., and Altinoz, M. A. (2020). Long non-coding RNA MALAT1 as a key target in pathogenesis of glioblastoma. Janus faces or Achilles' heal? Gene 739, 144518. doi:10.1016/j.gene.2020.144518
Bi, C. L., Liu, J. F., Zhang, M. Y., Lan, S., Yang, Z. Y., and Fang, J. S. (2020). LncRNA NEAT1 promotes malignant phenotypes and TMZ resistance in glioblastoma stem cells by regulating let-7g-5p/MAP3K1 axis. Biosci. Rep. 40 (10). doi:10.1042/BSR20201111
Bikfalvi, A., da Costa, C. A., Avril, T., Barnier, J. V., Bauchet, L., Brisson, L., et al. (2023). Challenges in glioblastoma research: focus on the tumor microenvironment. Trends Cancer 9 (1), 9–27. doi:10.1016/j.trecan.2022.09.005
Bock, C., Datlinger, P., Chardon, F., Coelho, M. A., Dong, M. B., Lawson, K. A., et al. (2022). High-content CRISPR screening. Nat. Rev. Methods Prim. 2 (1), 9. doi:10.1038/s43586-022-00098-7
Brodie, S., Lee, H. K., Jiang, W., Cazacu, S., Xiang, C., Poisson, L. M., et al. (2021). Correction: the novel long non-coding RNA TALNEC2, regulates tumor cell growth and the stemness and radiation response of glioma stem cells. Oncotarget 12 (26), 2546–2547. doi:10.18632/oncotarget.27383
Brown, S. D. (1991). XIST and the mapping of the X chromosome inactivation centre. Bioessays 13 (11), 607–612. doi:10.1002/bies.950131112
Buccarelli, M., Lulli, V., Giuliani, A., Signore, M., Martini, M., D'Alessandris, Q. G., et al. (2020). Deregulated expression of the imprinted DLK1-DIO3 region in glioblastoma stemlike cells: tumor suppressor role of lncRNA MEG3. Neuro Oncol. 22 (12), 1771–1784. doi:10.1093/neuonc/noaa127
Cao, Y., Chai, W., Wang, Y., Tang, D., Shao, D., Song, H., et al. (2021). lncRNA TUG1 inhibits the cancer stem cell-like properties of temozolomide-resistant glioma cells by interacting with EZH2. Mol. Med. Rep. 24 (1), 533. doi:10.3892/mmr.2021.12172
Chadwick, M., Yang, C., Liu, L., Gamboa, C. M., Jara, K., Lee, H., et al. (2020). Rapid processing and drug evaluation in glioblastoma patient-derived organoid models with 4D bioprinted arrays. iScience 23 (8), 101365. doi:10.1016/j.isci.2020.101365
Chen, W., Xu, X. K., Li, J. L., Kong, K. K., Li, H., Chen, C., et al. (2017). MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget 8 (14), 22783–22799. doi:10.18632/oncotarget.15199
Cheng, L., Huang, Z., Zhou, W., Wu, Q., Donnola, S., Liu, J. K., et al. (2013). Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell 153 (1), 139–152. doi:10.1016/j.cell.2013.02.021
Childs-Disney, J. L., Yang, X., Gibaut, Q. M. R., Tong, Y., Batey, R. T., and Disney, M. D. (2022). Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 21 (10), 736–762. doi:10.1038/s41573-022-00521-4
Coelho, T., Maurer, M. S., and Suhr, O. B. (2013). THAOS - the Transthyretin Amyloidosis Outcomes Survey: initial report on clinical manifestations in patients with hereditary and wild-type transthyretin amyloidosis. Curr. Med. Res. Opin. 29 (1), 63–76. doi:10.1185/03007995.2012.754348
Crooke, S. T., Liang, X. H., Baker, B. F., and Crooke, R. M. (2021). Antisense technology: a review. J. Biol. Chem. 296, 100416. doi:10.1016/j.jbc.2021.100416
Diaz-Beya, M., Brunet, S., Nomdedéu, J., Pratcorona, M., Cordeiro, A., Gallardo, D., et al. (2015). The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget 6 (31), 31613–31627. doi:10.18632/oncotarget.5148
Dundar, T. T., Hatiboglu, M. A., Ergul, Z., Seyithanoglu, M. H., Sozen, E., Tuzgen, S., et al. (2019). Glioblastoma stem cells and comparison of isolation methods. J. Clin. Med. Res. 11 (6), 415–421. doi:10.14740/jocmr3781
Fan, Q., Wang, H., Gu, T., Liu, H., Deng, P., Li, B., et al. (2024). Modeling the precise interaction of glioblastoma with human brain region-specific organoids. iScience 27 (3), 109111. doi:10.1016/j.isci.2024.109111
Fawzy, M. S., Ellawindy, A., Hussein, M. H., Khashana, M. S., Darwish, M. K., Abdel-Daim, M. M., et al. (2018). Long noncoding RNA H19, and not microRNA miR-326, is over-expressed and predicts survival in glioblastoma. Biochem. Cell Biol. 96 (6), 832–839. doi:10.1139/bcb-2018-0122
Fu, S., Wang, Y., Li, H., Chen, L., and Liu, Q. (2020). Regulatory networks of LncRNA MALAT-1 in cancer. Cancer Manag. Res. 12, 10181–10198. doi:10.2147/CMAR.S276022
Gandhi, S., Bhushan, A., Shukla, U., Pundir, A., Singh, S., and Srivastava, T. (2023). Downregulation of lncRNA SNHG1 in hypoxia and stem cells is associated with poor disease prognosis in gliomas. Cell Cycle 22 (9), 1135–1153. doi:10.1080/15384101.2023.2191411
Gao, X. Y., Zang, J., Zheng, M. H., Zhang, Y. F., Yue, K. Y., Cao, X. L., et al. (2021). Temozolomide treatment induces HMGB1 to promote the formation of glioma stem cells via the TLR2/NEAT1/wnt pathway in glioblastoma. Front. Cell Dev. Biol. 9, 620883. doi:10.3389/fcell.2021.620883
Gavrilov, K., and Saltzman, W. M. (2012). Therapeutic siRNA: principles, challenges, and strategies. Yale J. Biol. Med. 85 (2), 187–200.
Ghafouri-Fard, S., Agabalazadeh, A., Abak, A., Shoorei, H., Hassanzadeh Taheri, M. M., Taheri, M., et al. (2021). Role of long non-coding RNAs in conferring resistance in tumors of the nervous system. Front. Oncol. 11, 670917. doi:10.3389/fonc.2021.670917
Goenka, A., Song, X., Tiek, D., Iglesia, R. P., Lu, M., Zeng, C., et al. (2023). Oncogenic long noncoding RNA LINC02283 enhances PDGF receptor A-mediated signaling and drives glioblastoma tumorigenesis. Neuro Oncol. 25 (9), 1592–1604. doi:10.1093/neuonc/noad065
Gong, M., Wang, X., Mu, L., Wang, Y., Pan, J., Yuan, X., et al. (2021a). Steroid receptor coactivator-1 enhances the stemness of glioblastoma by activating long noncoding RNA XIST/miR-152/KLF4 pathway. Cancer Sci. 112 (2), 604–618. doi:10.1111/cas.14685
Gong, R., Li, Z. Q., Fu, K., Ma, C., Wang, W., and Chen, J. C. (2021b). Long noncoding RNA PVT1 promotes stemness and temozolomide resistance through miR-365/ELF4/SOX2 Axis in glioma. Exp. Neurobiol. 30 (3), 244–255. doi:10.5607/en20060
Guo, C. J., Xu, G., and Chen, L. L. (2020). Mechanisms of long noncoding RNA nuclear retention. Trends Biochem. Sci. 45 (11), 947–960. doi:10.1016/j.tibs.2020.07.001
Guttman, M., Donaghey, J., Carey, B. W., Garber, M., Grenier, J. K., Munson, G., et al. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477 (7364), 295–300. doi:10.1038/nature10398
Guttman, M., Garber, M., Levin, J. Z., Donaghey, J., Robinson, J., Adiconis, X., et al. (2010). Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 28 (5), 503–510. doi:10.1038/nbt.1633
Han, M., Wang, S., Fritah, S., Wang, X., Zhou, W., Yang, N., et al. (2020). Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/β-catenin signalling. Brain 143 (2), 512–530. doi:10.1093/brain/awz406
Han, Y., Zhou, L., Wu, T., Huang, Y., Cheng, Z., Li, X., et al. (2016). Downregulation of lncRNA-MALAT1 affects proliferation and the expression of stemness markers in glioma stem cell line SHG139S. Cell Mol. Neurobiol. 36 (7), 1097–1107. doi:10.1007/s10571-015-0303-6
Hannon, G. J., and Rossi, J. J. (2004). Unlocking the potential of the human genome with RNA interference. Nature 431 (7006), 371–378. doi:10.1038/nature02870
Hao, Y., Li, X., Chen, H., Huo, H., Liu, Z., and Chai, E. (2020). Over-expression of long noncoding RNA HOTAIRM1 promotes cell proliferation and invasion in human glioblastoma by up-regulating SP1 via sponging miR-137. Neuroreport 31 (2), 109–117. doi:10.1097/WNR.0000000000001380
Hazra, R., Brine, L., Garcia, L., Benz, B., Chirathivat, N., Shen, M. M., et al. (2022). Platr4 is an early embryonic lncRNA that exerts its function downstream on cardiogenic mesodermal lineage commitment. Dev. Cell 57 (21), 2450–2468 e7. doi:10.1016/j.devcel.2022.10.002
Hazra, R., Utama, R., Naik, P., Dobin, A., and Spector, D. L. (2023). Identification of glioblastoma stem cell-associated lncRNAs using single-cell RNA sequencing datasets. Stem Cell Rep. 18 (11), 2056–2070. doi:10.1016/j.stemcr.2023.10.004
Hoy, S. M. (2018). Patisiran: first global approval. Drugs 78 (15), 1625–1631. doi:10.1007/s40265-018-0983-6
Huang, H., Shah, H., Hao, J., Lin, J., Prayson, R. A., Xie, L., et al. (2024b). LncRNA LUCAT1 promotes glioblastoma progression by enhancing HIF1α activity. Neuro Oncol., noae036. doi:10.1093/neuonc/noae036
Huang, L., Wang, Z., Liao, C., Zhao, Z., Gao, H., Huang, R., et al. (2024a). PVT1 promotes proliferation and macrophage immunosuppressive polarization through STAT1 and CX3CL1 regulation in glioblastoma multiforme. CNS Neurosci. Ther. 30 (1), e14566. doi:10.1111/cns.14566
Hudson, W. H., Pickard, M. R., de Vera, I. M. S., Kuiper, E. G., Mourtada-Maarabouni, M., Conn, G. L., et al. (2014). Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate. Nat. Commun. 5, 5395. doi:10.1038/ncomms6395
Ignatova, T. N., Kukekov, V. G., Laywell, E. D., Suslov, O. N., Vrionis, F. D., and Steindler, D. A. (2002). Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 39 (3), 193–206. doi:10.1002/glia.10094
Jiang, W., Finniss, S., Cazacu, S., Xiang, C., Brodie, Z., Mikkelsen, T., et al. (2016b). Repurposing phenformin for the targeting of glioma stem cells and the treatment of glioblastoma. Oncotarget 7 (35), 56456–56470. doi:10.18632/oncotarget.10919
Jiang, X., Yan, Y., Hu, M., Chen, X., Wang, Y., Dai, Y., et al. (2016a). Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J. Neurosurg. 124 (1), 129–136. doi:10.3171/2014.12.JNS1426.test
Jiang, X., Zhang, Y., Yuan, Y., Jin, Z., Zhai, H., Liu, B., et al. (2023). LncRNA GSCAR promotes glioma stem cell maintenance via stabilizing SOX2 expression. Int. J. Biol. Sci. 19 (6), 1681–1697. doi:10.7150/ijbs.80873
Jin, L., Cai, Q., Wang, S., Wang, S., Mondal, T., Wang, J., et al. (2018). Long noncoding RNA MEG3 regulates LATS2 by promoting the ubiquitination of EZH2 and inhibits proliferation and invasion in gallbladder cancer. Cell Death Dis. 9 (10), 1017. doi:10.1038/s41419-018-1064-1
Jin, Z., Piao, L. H., Sun, G. C., Lv, C. X., Jing, Y., and Jin, R. H. (2020). Long non-coding RNA plasmacytoma variant translocation 1 (PVT1) promotes glioblastoma multiforme progression via regulating miR-1301-3p/TMBIM6 axis. Eur. Rev. Med. Pharmacol. Sci. 24 (22), 11658–11665. doi:10.26355/eurrev_202011_23810
Katsushima, K., Natsume, A., Ohka, F., Shinjo, K., Hatanaka, A., Ichimura, N., et al. (2016). Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 7, 13616. doi:10.1038/ncomms13616
Khosla, D. (2016). Concurrent therapy to enhance radiotherapeutic outcomes in glioblastoma. Ann. Transl. Med. 4 (3), 54. doi:10.3978/j.issn.2305-5839.2016.01.25
Kim, S. S., Harford, J. B., Moghe, M., Rait, A., Pirollo, K. F., and Chang, E. H. (2018). Targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide. Nucleic Acids Res. 46 (3), 1424–1440. doi:10.1093/nar/gkx1221
Konermann, S., Lotfy, P., Brideau, N. J., Oki, J., Shokhirev, M. N., and Hsu, P. D. (2018). Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173 (3), 665–676. doi:10.1016/j.cell.2018.02.033
Kung, J. T., Colognori, D., and Lee, J. T. (2013). Long noncoding RNAs: past, present, and future. Genetics 193 (3), 651–669. doi:10.1534/genetics.112.146704
Lancaster, M. A., and Knoblich, J. A. (2014). Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9 (10), 2329–2340. doi:10.1038/nprot.2014.158
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409 (6822), 860–921. doi:10.1038/35057062
Lang, C., Yin, C., Lin, K., Li, Y., Yang, Q., Wu, Z., et al. (2021). m(6) A modification of lncRNA PCAT6 promotes bone metastasis in prostate cancer through IGF2BP2-mediated IGF1R mRNA stabilization. Clin. Transl. Med. 11 (6), e426. doi:10.1002/ctm2.426
Lecerf, C., Peperstraete, E., Le Bourhis, X., and Adriaenssens, E. (2020). Propagation and maintenance of cancer stem cells: a major influence of the long non-coding RNA H19. Cells 9 (12), 2613. doi:10.3390/cells9122613
Leung, D. H. L., Phon, B. W. S., Sivalingam, M., Radhakrishnan, A. K., and Kamarudin, M. N. A. (2023). Regulation of emt markers, extracellular matrix, and associated signalling pathways by long non-coding RNAs in glioblastoma mesenchymal transition: a scoping review. Biol. (Basel) 12 (6), 818. doi:10.3390/biology12060818
Li, H., Yang, Y., Hong, W., Huang, M., Wu, M., and Zhao, X. (2020). Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct. Target Ther. 5 (1), 1. doi:10.1038/s41392-019-0089-y
Li, L., Li, Z., Meng, X., Wang, X., Song, D., Liu, Y., et al. (2023). Histone lactylation-derived LINC01127 promotes the self-renewal of glioblastoma stem cells via the cis-regulating the MAP4K4 to activate JNK pathway. Cancer Lett. 579, 216467. doi:10.1016/j.canlet.2023.216467
Li, Q., Dong, C., Cui, J., Wang, Y., and Hong, X. (2018). Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J. Exp. Clin. Cancer Res. 37 (1), 265. doi:10.1186/s13046-018-0941-x
Li, S., Li, X., Xue, W., Zhang, L., Yang, L. Z., Cao, S. M., et al. (2021). Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat. Methods 18 (1), 51–59. doi:10.1038/s41592-020-01011-4
Li, W., Jiang, P., Sun, X., Xu, S., Ma, X., and Zhan, R. (2016). Suppressing H19 modulates tumorigenicity and stemness in U251 and U87MG glioma cells. Cell Mol. Neurobiol. 36 (8), 1219–1227. doi:10.1007/s10571-015-0320-5
Li, Y., Ren, Y., Wang, Y., Tan, Y., Wang, Q., Cai, J., et al. (2019). A compound AC1Q3QWB selectively disrupts HOTAIR-mediated recruitment of PRC2 and enhances cancer therapy of DZNep. Theranostics 9 (16), 4608–4623. doi:10.7150/thno.35188
Liao, K., Lin, Y., Gao, W., Xiao, Z., Medina, R., Dmitriev, P., et al. (2019). Blocking lncRNA MALAT1/miR-199a/ZHX1 Axis inhibits glioblastoma proliferation and progression. Mol. Ther. Nucleic Acids 18, 388–399. doi:10.1016/j.omtn.2019.09.005
Lin, M., Pedrosa, E., Shah, A., Hrabovsky, A., Maqbool, S., Zheng, D., et al. (2011). RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One 6 (9), e23356. doi:10.1371/journal.pone.0023356
Lin, Y. H., Guo, L., Yan, F., Dou, Z. Q., Yu, Q., and Chen, G. (2020). Long non-coding RNA HOTAIRM1 promotes proliferation and inhibits apoptosis of glioma cells by regulating the miR-873-5p/ZEB2 axis. Chin. Med. J. Engl. 133 (2), 174–182. doi:10.1097/CM9.0000000000000615
Linkous, A., Balamatsias, D., Snuderl, M., Edwards, L., Miyaguchi, K., Milner, T., et al. (2019). Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 26 (12), 3203–3211. doi:10.1016/j.celrep.2019.02.063
Liu, B., Zhou, J., Wang, C., Chi, Y., Wei, Q., Fu, Z., et al. (2020a). LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 11 (5), 384. doi:10.1038/s41419-020-2540-y
Liu, L., Liu, Z., Liu, Q., Wu, W., Lin, P., Liu, X., et al. (2023). LncRNA INHEG promotes glioma stem cell maintenance and tumorigenicity through regulating rRNA 2'-O-methylation. Nat. Commun. 14 (1), 7526. doi:10.1038/s41467-023-43113-5
Liu, S. J., Malatesta, M., Lien, B. V., Saha, P., Thombare, S. S., Hong, S. J., et al. (2020b). CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol. 21 (1), 83. doi:10.1186/s13059-020-01995-4
Long, Y., Wang, X., Youmans, D. T., and Cech, T. R. (2017). How do lncRNAs regulate transcription? Sci. Adv. 3 (9), eaao2110. doi:10.1126/sciadv.aao2110
Louis, D. N., Ohgaki, H., Wiestler, O. D., Cavenee, W. K., Burger, P. C., Jouvet, A., et al. (2007). The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114 (2), 97–109. doi:10.1007/s00401-007-0243-4
Luo, C., Quan, Z., Zhong, B., Zhang, M., Zhou, B., Wang, S., et al. (2020). lncRNA XIST promotes glioma proliferation and metastasis through miR-133a/SOX4. Exp. Ther. Med. 19 (3), 1641–1648. doi:10.3892/etm.2020.8426
Lv, T., Jin, Y., Miao, Y., Xu, T., Jia, F., Feng, H., et al. (2022). LncRNA PVT1 promotes tumorigenesis of glioblastoma by recruiting COPS5 to deubiquitinate and stabilize TRIM24. Mol. Ther. Nucleic Acids 27, 109–121. doi:10.1016/j.omtn.2021.11.012
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24 (6), 430–447. doi:10.1038/s41580-022-00566-8
Mazor, G., Levin, L., Picard, D., Ahmadov, U., Carén, H., Borkhardt, A., et al. (2019). The lncRNA TP73-AS1 is linked to aggressiveness in glioblastoma and promotes temozolomide resistance in glioblastoma cancer stem cells. Cell Death Dis. 10 (3), 246. doi:10.1038/s41419-019-1477-5
McNamara, M. G., Lwin, Z., Jiang, H., Chung, C., Millar, B. A., Sahgal, A., et al. (2014). Conditional probability of survival and post-progression survival in patients with glioblastoma in the temozolomide treatment era. J. Neurooncol 117 (1), 153–160. doi:10.1007/s11060-014-1368-7
Meira, S. Z., Hartford, C. C. R., and Lal, A. (2021). Interrogating lncRNA functions via CRISPR/Cas systems. RNA Biol. 18 (12), 2097–2106. doi:10.1080/15476286.2021.1899500
Montero, J. J., Trozzo, R., Sugden, M., Öllinger, R., Belka, A., Zhigalova, E., et al. (2024). Genome-scale pan-cancer interrogation of lncRNA dependencies using CasRx. Nat. Methods 21, 584–596. doi:10.1038/s41592-024-02190-0
Nie, J., Feng, Y., Wang, H., Lian, X. Y., and Li, Y. F. (2021). Long non-coding RNA SNHG6 supports glioma progression through upregulation of Notch1, Sox2, and EMT. Front. Cell Dev. Biol. 9, 707906. doi:10.3389/fcell.2021.707906
Ogawa, J., Pao, G. M., Shokhirev, M. N., and Verma, I. M. (2018). Glioblastoma model using human cerebral organoids. Cell Rep. 23 (4), 1220–1229. doi:10.1016/j.celrep.2018.03.105
Ostrom, Q. T., Price, M., Neff, C., Cioffi, G., Waite, K. A., Kruchko, C., et al. (2023). CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2016-2020. Neuro Oncol. 25 (12 Suppl. 2), iv1–iv99. doi:10.1093/neuonc/noad149
Ozturk, M. S., Lee, V. K., Zou, H., Friedel, R. H., Intes, X., and Dai, G. (2020). High-resolution tomographic analysis of in vitro 3D glioblastoma tumor model under long-term drug treatment. Sci. Adv. 6 (10), eaay7513. doi:10.1126/sciadv.aay7513
Parra-Cantu, C., Li, W., Quiñones-Hinojosa, A., and Zhang, Y. S. (2020). 3D bioprinting of glioblastoma models. J. 3D Print Med. 4 (2), 113–125. doi:10.2217/3dp-2019-0027
Patrat, C., Ouimette, J. F., and Rougeulle, C. (2020). X chromosome inactivation in human development. Development 147 (1), dev183095. doi:10.1242/dev.183095
Pickard, M. R., and Williams, G. T. (2015). Molecular and cellular mechanisms of action of tumour suppressor GAS5 LncRNA. Genes (Basel) 6 (3), 484–499. doi:10.3390/genes6030484
Prager, B. C., Bhargava, S., Mahadev, V., Hubert, C. G., and Rich, J. N. (2020). Glioblastoma stem cells: driving resilience through chaos. Trends Cancer 6 (3), 223–235. doi:10.1016/j.trecan.2020.01.009
Rahmani, F., Mohammed Al-Asady, A., Hanaie, R., Zandigohar, M., Faridnejad, H., Payazdan, M., et al. (2023). Interplay between lncRNA/miRNA and Wnt/ss-catenin signaling in brain cancer tumorigenesis. EXCLI J. 22, 1211–1222. doi:10.17179/excli2023-6490
Raveh, E., Matouk, I. J., Gilon, M., and Hochberg, A. (2015). The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol. Cancer 14, 184. doi:10.1186/s12943-015-0458-2
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi:10.1146/annurev-biochem-051410-092902
Roh, J., Im, M., Kang, J., Youn, B., and Kim, W. (2023). Long non-coding RNA in glioma: novel genetic players in temozolomide resistance. Anim. Cells Syst. Seoul. 27 (1), 19–28. doi:10.1080/19768354.2023.2175497
Sadagopan, A., Nasim, I. T., Li, J., Achom, M., Zhang, C. Z., and Viswanathan, S. R. (2022). Somatic XIST activation and features of X chromosome inactivation in male human cancers. Cell Syst. 13 (11), 932–944 e5. doi:10.1016/j.cels.2022.10.002
Singh, S. K., Clarke, I. D., Terasaki, M., Bonn, V. E., Hawkins, C., Squire, J., et al. (2003). Identification of a cancer stem cell in human brain tumors. Cancer Res. 63 (18), 5821–5828.
Singh, S. K., Hawkins, C., Clarke, I. D., Squire, J. A., Bayani, J., Hide, T., et al. (2004). Identification of human brain tumour initiating cells. Nature 432 (7015), 396–401. doi:10.1038/nature03128
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Stupp, R., Mason, W. P., van den Bent, M. J., Weller, M., Fisher, B., Taphoorn, M. J. B., et al. (2005). Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352 (10), 987–996. doi:10.1056/NEJMoa043330
Su, R., Cao, S., Ma, J., Liu, Y., Liu, X., Zheng, J., et al. (2017). Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol. Cancer 16 (1), 171. doi:10.1186/s12943-017-0737-1
Tang, M., Xie, Q., Gimple, R. C., Zhong, Z., Tam, T., Tian, J., et al. (2020). Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res. 30 (10), 833–853. doi:10.1038/s41422-020-0338-1
Tseng, Y. Y., and Bagchi, A. (2015). The PVT1-MYC duet in cancer. Mol. Cell Oncol. 2 (2), e974467. doi:10.4161/23723556.2014.974467
Urits, I., Swanson, D., Swett, M. C., Patel, A., Berardino, K., Amgalan, A., et al. (2020). A review of patisiran (ONPATTRO®) for the treatment of polyneuropathy in people with hereditary transthyretin amyloidosis. Neurol. Ther. 9 (2), 301–315. doi:10.1007/s40120-020-00208-1
Wakimoto, H., Kesari, S., Farrell, C. J., Curry, W. T., Zaupa, C., Aghi, M., et al. (2009). Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 69 (8), 3472–3481. doi:10.1158/0008-5472.CAN-08-3886
Wan, L., Kong, J., Tang, J., Wu, Y., Xu, E., Lai, M., et al. (2016). HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J. Cell Mol. Med. 20 (11), 2036–2044. doi:10.1111/jcmm.12892
Wan, Y. C. E., Liu, J., and Chan, K. M. (2018). Histone H3 mutations in cancer. Curr. Pharmacol. Rep. 4 (4), 292–300. doi:10.1007/s40495-018-0141-6
Wang, K. C., and Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43 (6), 904–914. doi:10.1016/j.molcel.2011.08.018
Wang, L., and He, Z. (2019). Functional roles of long non-coding RNAs (LncRNAs) in glioma stem cells. Med. Sci. Monit. 25, 7567–7573. doi:10.12659/MSM.916040
Wang, Y., Cheng, Y., Yang, Q., Kuang, L., and Liu, G. (2022). Overexpression of FOXD2-AS1 enhances proliferation and impairs differentiation of glioma stem cells by activating the NOTCH pathway via TAF-1. J. Cell Mol. Med. 26 (9), 2620–2632. doi:10.1111/jcmm.17268
Wang, Y., Wu, N., Luo, X., Zhang, X., Liao, Q., and Wang, J. (2020). SOX2OT, a novel tumor-related long non-coding RNA. Biomed. Pharmacother. 123, 109725. doi:10.1016/j.biopha.2019.109725
Wei, P., Jiang, J., Xiao, M., Zeng, M., Liu, X., Zhao, B., et al. (2022). The transcript ENST00000444125 of lncRNA LINC01503 promotes cancer stem cell properties of glioblastoma cells via reducing FBXW1 mediated GLI2 degradation. Exp. Cell Res. 412 (1), 113009. doi:10.1016/j.yexcr.2022.113009
Williamson, J. F. (1989). Radiation transport calculations in treatment planning. Comput. Med. Imaging Graph 13 (3), 251–268. doi:10.1016/0895-6111(89)90132-8
Wu, M., Lu, L., Dai, T., Li, A., Yu, Y., Li, Y., et al. (2023). Construction of a lncRNA-mediated ceRNA network and a genomic-clinicopathologic nomogram to predict survival for breast cancer patients. Cancer Biomark. 36 (1), 83–96. doi:10.3233/CBM-210545
Wu, S., Ren, K., Zhao, J., Li, J., Jia, B., Wu, X., et al. (2022). LncRNA GAS5 represses stemness and malignancy of gliomas via elevating the SPACA6-miR-125a/let-7e Axis. Front. Oncol. 12, 803652. doi:10.3389/fonc.2022.803652
Xie, P., Li, X., Chen, R., Liu, Y., Liu, D., Liu, W., et al. (2020). Upregulation of HOTAIRM1 increases migration and invasion by glioblastoma cells. Aging (Albany NY) 13 (2), 2348–2364. doi:10.18632/aging.202263
Xiong, D. D., Li, Z. Y., Liang, L., He, R. Q., Ma, F. C., Luo, D. Z., et al. (2018a). The LncRNA NEAT1 accelerates lung adenocarcinoma deterioration and binds to mir-193a-3p as a competitive endogenous RNA. Cell Physiol. Biochem. 48 (3), 905–918. doi:10.1159/000491958
Xiong, Z., Wang, L., Wang, Q., and Yuan, Y. (2018b). LncRNA MALAT1/miR-129 axis promotes glioma tumorigenesis by targeting SOX2. J. Cell Mol. Med. 22 (8), 3929–3940. doi:10.1111/jcmm.13667
Yang, J., Qi, M., Fei, X., Wang, X., and Wang, K. (2021). Long non-coding RNA XIST: a novel oncogene in multiple cancers. Mol. Med. 27 (1), 159. doi:10.1186/s10020-021-00421-0
Yang, W., Liang, Y., Zheng, Y., Luo, H., Yang, X., and Li, F. (2022). Identification of novel multi-omics expression landscapes and meta-analysis of landscape-based competitive endogenous RNA networks in ALDH+ lung adenocarcinoma stem cells. Biomed. Res. Int. 2022, 9545609. doi:10.1155/2022/9545609
Yang, W., Yu, H., Shen, Y., Liu, Y., Yang, Z., and Sun, T. (2016). MiR-146b-5p overexpression attenuates stemness and radioresistance of glioma stem cells by targeting HuR/lincRNA-p21/β-catenin pathway. Oncotarget 7 (27), 41505–41526. doi:10.18632/oncotarget.9214
Yang, Z., Bian, E., Xu, Y., Ji, X., Tang, F., et al. (2020). Meg3 induces EMT and invasion of glioma cells via autophagy. Onco Targets Ther. 13, 989–1000. doi:10.2147/OTT.S239648
Yao, R. W., Wang, Y., and Chen, L. L. (2019). Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21 (5), 542–551. doi:10.1038/s41556-019-0311-8
Yao, Y., Ma, J., Xue, Y., Wang, P., Li, Z., Liu, J., et al. (2015). Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 359 (1), 75–86. doi:10.1016/j.canlet.2014.12.051
Yeo, M., Sarkar, A., Singh, Y. P., Derman, I. D., Datta, P., and Ozbolat, I. T. (2023). Synergistic coupling between 3D bioprinting and vascularization strategies. Biofabrication 16 (1), 012003. doi:10.1088/1758-5090/ad0b3f
Yi, H. G., Jeong, Y. H., Kim, Y., Choi, Y. J., Moon, H. E., Park, S. H., et al. (2019). A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 3 (7), 509–519. doi:10.1038/s41551-019-0363-x
Yuan, E., Liu, K., Lee, J., Tsung, K., Chow, F., and Attenello, F. J. (2022). Modulating glioblastoma chemotherapy response: evaluating long non-coding RNA effects on DNA damage response, glioma stem cell function, and hypoxic processes. Neurooncol Adv. 4 (1), vdac119. doi:10.1093/noajnl/vdac119
Zang, J., Zheng, M. H., Cao, X. L., Zhang, Y. Z., Zhang, Y. F., Gao, X. Y., et al. (2020). Adenovirus infection promotes the formation of glioma stem cells from glioblastoma cells through the TLR9/NEAT1/STAT3 pathway. Cell Commun. Signal 18 (1), 135. doi:10.1186/s12964-020-00598-7
Zhang, Q., Wang, C., Li, S., Li, Y., Chen, M., and Hu, Y. (2023a). Screening of core genes prognostic for sepsis and construction of a ceRNA regulatory network. BMC Med. Genomics 16 (1), 37. doi:10.1186/s12920-023-01460-8
Zhang, Q., Yang, L., Liu, Y. H., Wilkinson, J. E., and Krainer, A. R. (2023b). Antisense oligonucleotide therapy for H3.3K27M diffuse midline glioma. Sci. Transl. Med. 15 (691), eadd8280. doi:10.1126/scitranslmed.add8280
Zhang, X., Lian, Z., Padden, C., Gerstein, M. B., Rozowsky, J., Snyder, M., et al. (2009). A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113 (11), 2526–2534. doi:10.1182/blood-2008-06-162164
Zhang, X., Sun, S., Pu, J. K. S., Tsang, A. C. O., Lee, D., Man, V. O. Y., et al. (2012). Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 48 (1), 1–8. doi:10.1016/j.nbd.2012.06.004
Zhang, X., Weissman, S. M., and Newburger, P. E. (2014). Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 11 (6), 777–787. doi:10.4161/rna.28828
Zhao, X., Liu, Y., Zheng, J., Liu, X., Chen, J., Liu, L., et al. (2017). GAS5 suppresses malignancy of human glioma stem cells via a miR-196a-5p/FOXO1 feedback loop. Biochim. Biophys. Acta Mol. Cell Res. 1864 (10), 1605–1617. doi:10.1016/j.bbamcr.2017.06.020
Zheng, J., Li, X. d., Wang, P., Liu, X. b., Xue, Y. x., Hu, Y., et al. (2015). CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget 6 (28), 25339–25355. doi:10.18632/oncotarget.4509
Keywords: glioblastoma, cancer stem cells, glioblastoma stem cells, long non-coding RNAs, lncRNAs, therapeutic targets, 3D in vitro model
Citation: Hazra R, Debnath R and Tuppad A (2024) Glioblastoma stem cell long non-coding RNAs: therapeutic perspectives and opportunities. Front. Genet. 15:1416772. doi: 10.3389/fgene.2024.1416772
Received: 13 April 2024; Accepted: 27 May 2024;
Published: 02 July 2024.
Edited by:
Ameneh Jafari, Shahid Beheshti University of Medical Sciences, IranReviewed by:
Mohsen Yazdani, University of Tehran, IranCopyright © 2024 Hazra, Debnath and Tuppad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rasmani Hazra, cmhhenJhQG5ld2hhdmVuLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.