
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 25 June 2024
Sec. Immunogenetics
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1414487
Lung cancer is one of the most prevalent malignancies worldwide, contributing to over a million cancer-related deaths annually. Despite extensive research investigating the genetic factors associated with lung cancer susceptibility and prognosis, few studies have explored genetic predispositions regarding the immune system. This review discusses the most recent genomic findings related to the susceptibility to or protection against lung cancer, patient survival, and therapeutic responses. The results demonstrated the effect of immunogenetic variations in immune system-related genes associated with innate and adaptive immune responses, cytokine, and chemokine secretions, and signaling pathways. These genetic diversities may affect the crosstalk between tumor and immune cells within the tumor microenvironment, influencing cancer progression, invasion, and prognosis. Given the considerable variability in the individual immunegenomics profiles, future studies should prioritize large-scale analyses to identify potential genetic variations associated with lung cancer using highthroughput technologies across different populations. This approach will provide further information for predicting response to targeted therapy and promotes the development of new measures for individualized cancer treatment.
Lung cancer is the primary cause of cancer-related deaths worldwide, accounting for 1.7 million deaths and 2.2 million new cases in 2020, with projections indicating an increase to 3.5 million cases by 2040 (International Agency for Research on Cancer, 2020). It is categorized into two main subgroups, small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), accounting for 10%–15% and 80% of all cases, respectively. SCLC can be histologically subdivided into small cell carcinoma, mixed small cell/large cell cancer, and combined small cell lung cancer. On th other hand, NSCLC is classified into lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large-cell carcinoma (LCC), representing the most common subtypes (Wu et al., 2023). Smoking is widely recognized as the leading cause of lung cancer, with approximately 90% of cases attributed to tobacco consumption (Alberg and Samet, 2003). Moreover, several factors such as age (typically over 65 years), sex, and environmental and genetic factors influence the development of lung cancer (Cheng et al., 2021; Huang et al., 2021). For instance, genetic alterations in oncogenic drivers, such as the Kirsten rat sarcoma (KRAS), v-raf murine sarcoma viral oncogene homolog B (BRAF), and epidermal growth factor receptor (EGFR) genes, are commonly found in patients with NSCLC and can affect their response to treatments (Chevallier et al., 2021). Owing to the absence of typical early symptoms and effective early detection strategies, approximately 70% of lung cancer patients are diagnosed at an advanced-stage or with distant metastasis, leading to a 5-year survival rate of only about 4% (Ren et al., 2021; Wu et al., 2023). Three common treatment options for lung cancer include surgery, chemotherapy, and radiotherapy. Recently, immune checkpoint inhibitors, such as antibodies against programmed death-1 (PD-1), programmed death-ligand 1 (PD-L1), and cytotoxic T lymphocyte-associated antigen 4 (CTLA-4), have greatly improved cancer treatment. However, the response to these therapies is still relatively low (Lahiri et al., 2023).
The immune system plays a significant role in monitoring, recognizing, and destroying tumor cells under homeostatic conditions. Therefore, any dysregulation in immune-related responses and genomic-level mutations should be scrutinized as they may be related to lung cancer pathogenesis (Smok-Kalwat et al., 2023). The lung tumor microenvironment (TME), characterized by a heterogeneous cell population and intricate interactions, plays a significant role in the progression of lung cancer and the efficacy of treatment (Zhang et al., 2021). In recent years, the development of novel techniques, such as single-cell RNA sequencing (scRNA-seq), has enabled scientists to identify genetic and epigenetic changes that may increase lung cancer risk, as well as different aspects of the disease, such as its TME, at the genomic and transcriptomic levels. Furthermore, these advancements have improved personalized immunotherapies and the identification of individuals who can benefit more from specific treatments based on their genetic backgrounds. For instance, investigating single-cell RNA-Seq data from patients with LUAD and cell lines revealed heterogeneous expression of major histocompatibility complex (MHC) class II genes and significant downregulation of interferon (IFN)-γ signaling genes. This downregulation was found to be associated with immunotherapy resistance (Ma et al., 2019). Despite previous investigations, there is a growing need to identify immune response-related genes that may be associated with the susceptibility, treatment response, and prognosis of lung cancer.
The primary cause of death in patients with lung cancer is recurrence after chemotherapy or surgical intervention (Uramoto and Tanaka, 2014). Therefore, identifying novel biomarkers for early detection and developing therapeutic approaches based on immunogenic markers to reduce metastasis and enhance overall survival in patients with lung cancer is crucial. This review aimed to investigate the potential association between genetic variations in innate and adaptive immune cells, cytokines, and chemokines and susceptibility, prognosis, and response to immunotherapy in patients with lung cancer.
The TMB is characterized by the number of somatic coding mutations per megabase in the tumor genome and has emerged as a potential predictor of immunotherapy response in various tumor types (McNamara et al., 2020). The correlation between TMB and tumor immunology is complex and requires further investigation. Tumors with increased TMB often show higher expression of cancer-specific antigens, known as neoantigens. These somatic, non-synonymous mutations can lead to the expression of immunogenic epitopes that are specifically found in cancer cells (Chae et al., 2018; Wang P. et al., 2021). Lung cancers are among the malignancies with high TMB (Chalmers et al., 2017). An analysis of data from patients with LUAD in the Cancer Genome Atlas (TCGA) database revealed that elevated TMB is associated with increased infiltration of various immune cell populations, favoring an anti-tumor immune response. Tumors harboring mutations in DNA repair-associated genes, including mismatch repair genes, homologous recombination genes, and polymerase epsilon (POLE) exhibit significantly higher mutational counts, neoantigen quantity, and T cell infiltration (Chae et al., 2018). Research has also demonstrated an association between high mutational load and the overexpression of immune-related genes, such as interleukin-12 receptor subunit beta-2 (IL12RB2) and IFN-γ, which are related to Th1 anti-tumor reponse and IL-21, IL-23A, and interleukin 17 receptor A (IL17RA), which play roles in Th17-related pathways (Ye et al., 2013; Chae et al., 2018). Th17 may have both pro- and anti-tumorigenic roles in cancer. In contrast, cytokine genes associated with Th2 response such as IL-33 and thymic stromal lymphopoietin (TSLP) have shown an inverse correlation with neoantigen burden. High neoantigen load was also correlated with the overexpression of genes related to the cytotoxic function of CD8+ T cells, including granzyme B (GZMB) and FAS ligand (FASLG), as well as TAP2 which facilitates antigen processing in the MHC I antigen presentation pathway. Additionally, higher mutational load has been positively correlated with the overexpression of the chemokines tumor necrosis factor receptor superfamily member 25 (TNFRSF25), C-C chemokine receptor type 1 (CCR1), and lymphotoxin beta receptor (LTBR) in patients with LUAD. Neoantigens likely promote M1 macrophage polarization, enhancing anti-tumor immunity by increasing interactions between Th1/Th17 cell responses and other innate immune cells. In this regard, a higher mutational load was correlated with both higher and lower expression levels of several M1 and M2 macrophage-associated genes, respectively (Chae et al., 2018). In patients with NSCLC treated with immune checkpoint inhibitors, responders with the highest TMB were found to have mutations in genes associated with DNA repair and replication, including the polymerase delta 1 catalytic subunit (POLD1), POLE, and MutS homolog 2 (MSH2) (Chae et al., 2018).
Several studies have reported a higher TMB in patients with a history of smoking (Xiao et al., 2017; Chae et al., 2018). Moreover, patients of East Asian ancestry with LUAD have less genomic complexity, characterized by fewer mutations and copy number alterations compared to those of European ancestry (Chen et al., 2020).
In patients with NSCLC undergoing treatment with immune checkpoint inhibitors, the number of neoantigens per tumor is correlated with the mutation burden. Tumors from patients who experience sustained clinical benefits showed a remarkably higher neoantigen burden compared to those from individuals without durable benefit. Moreover, an increased neoantigen burden is associated with improved progression-free survival (Rizvi et al., 2015). For patients with NSCLC receiving treatment with immune checkpoint inhibitors, either as monotherapy or in combination, higher levels of TMB have been associated with positive outcomes, such as improved objective response, durable clinical benefit, and prolonged progression-free and overall survival (Rizvi et al., 2015; Hellmann et al., 2018; Rizvi et al., 2018).
In summary, analyzing TMB represents progress in the search for biomarkers of lung cancer. Whole-exome sequencing (WES) with next-generation sequencing (NGS) stands as the gold-standard method for evaluating tissue TMB. However, several limitations such as high cost, prolonged turnaround time, large input DNA requirements, and technological complexity make it impractical for routine clinical practice (Berland et al., 2019). Consequently, targeted gene panels have emerged as viable alternatives in clinical settings, demonstrating a strong correlation with the WES platform (Hellmann et al., 2018; Rizvi et al., 2018). Yet, the diversity in methods and criteria used to assessTMB necessitates the standardization of key aspects of panel-based TMB estimation. Therefore, it is crucial to conduct future clinical trials to confirm TMB as a reliable biomarker and improve its efficacy in guiding clinical decisions.
Chronic obstructive pulmonary disease (COPD) is characterized by chronic inflammation affecting the lung parenchyma and airways, resulting in irreversible and progressive restriction of airflow. There is a strong association between COPD and the development of lung cancer, as individuals diagnosed with COPD are more likely to develop lung cancer (Young and Hopkins, 2018). Neutrophilic inflammation has been identified as a key factor in the progression of COPD (Aloe et al., 2021). Studies have been conducted to identify neutrophil biomarkers that can assist in predicting the prognosis of NSCLC patients and their response to immunotherapies. Integrated data from scRNA-seq and bulk RNA-seq have suggested a prognostic risk model that includes six neutrophil differentiation-related genes based on the rate of four reference genes (ACTB, GAPDH, TFRC, TUBB). The identified genes are MS4A7, CXCR2, CSRNP1, RETN, CD177, and LUCAT1, all of which are primarily associated with immune-related pathways (Pang et al., 2022). Additionally, depletion of Toll-like Receptor (TLR) 2 and TLR4 resulted in a significant decrease in the proportion of neutrophils and their related cytokines in tumor tissues and bronchoalveolar lavage fluid (BALF), respectively (Jungnickel et al., 2017). In the case of cancer, precancerous cells can be activated through endogenous TLR ligands secreted from inflammatory cells, leading to the overexpression of angiogenic factors, cytokines, and growth factors. Therefore, TLRs can create a microenvironment favorable for tumor progression (He et al., 2007; Szajnik et al., 2009; Wang et al., 2010). A significant correlation has been observed between the TLR4 rs4986791 (+1196C/T) gene polymorphism and the risk of lung cancer. This correlation is likely due to the effect of functional polymorphisms on modulating inflammation mechanisms (Kurt et al., 2016). Moreover, the TLR4 rs7869402C > T polymorphism was found to be associated with a higher risk of NSCLC. This SNP is located in the 3′untranslated region of TLR4, which affects its binding to miRNA and subsequently impacts mRNA expression (Wu et al., 2020).
Studies using lung tumor IL-17:KrasG12D murine models have suggested the tumor-promoting role of the IL-17-neutrophil axis, as IL-17A increased the infiltration of neutrophils to the tumor site. These mice were also found to be resistant to treatment with PD-1 blockade (Akbay et al., 2017). Studies have also indicated an association between various polymorphisms in the IL-17A gene and an elevated risk of lung cancer (Ma et al., 2015; He et al., 2017). A higher frequency of IL-17A rs8193037GA and AA genotypes has been linked to a predisposition to NSCLC, as having the rs8193037A allele was associated with higher production of IL-17 (Cheng et al., 2015a). Moreover, patients carrying the T-allele of IL-17A rs8193036(C > T) had a higher risk of developing advanced stages of LUAD than those having the CC genotype of rs8193036, probably through inducing the overexpression of IL-17A (Lee et al., 2021). These studies suggest that polymorphisms that cause IL-17 overexpression are associated with increased susceptibility and enhanced lung tumor progression. However evaluating the expression level of IL-17A and checking the polymorphisms in different populations are required to validate the findings.
Under steady-state conditions, the lung environment must be carefully regulated to tolerate environmental antigens and commensal bacteria (Culley, 2009). Studies have demonstrated that lung NK cells are more mature (CD11brigh CD27low), express lower levels of activating co-stimulatory molecules (such as CD1d, CD86, and B220), and express higher levels of inhibitory receptors (CD94-NKG2A) than activating receptors (NKG2D) compared with those in the bone marrow or spleen, which helps maintain pulmonary homeostasis (Wang et al., 2012). The final activity of NK cells is determined by the signals from various inhibitory or activating receptors (Djaoud and Parham, 2020). Killer-cell immunoglobulin-like receptors (KIR) are germ-line encoded receptors that bind to classical human leukocyte antigen (HLA) class I molecules as their ligands, a process named “licensing,” which plays an important role in NK cell development. KIR and HLA loci show considerable variation among populations (Pyo et al., 2010). A recent study in southern Iran revealed an association between specific KIR gene clusters and HLA ligands, with susceptibility to lung cancer. Higher frequencies of KIR2DL2 and KIR2DS2 have been reported in patients with lung cancer, individually and in combination with their ligand (HLA-C1), compared with healthy controls. This susceptibility could be the result of impaired missing self-recognition in carriers of KIR2DL2/HLA-C1 with lung cancer tumors sustaining HLA-I expression or from hypo-responsiveness owing to KIR2DS2/HLA-C1 interactions. In contrast, higher frequencies of KIR2DS1 and KIR3DS1 genes probably conferred protection against this cancer, as these NK cells have a lower activation threshold, allowing for easier activation in the absence of their respective ligands. Patients with lung cancer also demonstrated higher levels of inhibitory KIR genes than activating genes. The presence of an increased number of inhibitory KIR genes can attenuate NK cell function in lung tumor cells, facilitating tumor evasion within the suppressive TME of lung cancer (Hematian et al., 2022). Further studies have also reported the association of rs1639113 and rs9265821 polymorphisms in the 3’ region of the HLA-C and HLA-B genes with the risk of lung cancer, respectively. These results were based on a genome-wide association study (GWAS) analysis in a Croatian population consisting of 203 patients with lung cancer and 3,60,938 controls (Baranasic et al., 2023). The HLA genotype affects which neoantigen, arising from tumor cell mutations, will be represented to the immune system and stimulate the subsequent elimination of tumor cells. Therefore, mutations that give rise to neoantigens with poor representation by HLA molecules or the downregulation of neoantigen-presenting HLA-I molecules by the tumors, such as NSCLC, can help them evade anti-tumor responses (Perea et al., 2017; Saab et al., 2020). Reduced expression of HLA-I molecules can result from HLA haplotype loss or the downregulation of HLA genes, such as β2-microglobulin (B2M) or antigen processing machinery genes such as TAP1/2, Tapasin, and LMP2/7 (Perea et al., 2017). Further investigations have shown that HLA loss of heterozygosity (LOH) is an immune escape mechanism in approximately 40% of early-stage NSCLC and is associated with an increase in the frequency of sub clonal mutations and neoantigen burden. Tumors with high HLA LOH tend to have more neoantigens that are predicted to bind to the lost HLA molecules, making these neoantigens invisible to the immune system. Additionally, tumors with high HLA LOH showed increased PD-L1 expression in immune cells and RNA signatures that suggest immune activation. Overall, the consideration of HLA LOH allows for the prediction of a set of neoantigens that can potentially stimulate T cell responses more effectively. This understanding may be beneficial for the development of future neoantigen-based immunotherapies (McGranahan et al., 2017). In this context, in silico immunogenomic tools are being developed to predict the HLA class I-binding affinity for each tumor-specific set of peptides, including neoantigens. The stability of these interactions strongly correlates with T cell immunogenicity (Liu and Mardis, 2017). In the case of other NK cell ligands, an in silico study by Kucuk et al. demonstrated decreased expression of several members of the NKG2D ligands in the LUAD group compared with the normal group. This downregulation is considered an anti-tumor escape mechanism, with MHC class I polypeptide-related sequence A (MICA) showing the most significant decrease. Lower expression of MICA transcripts may also be regarded as a prognostic factor for LUAD. Furthermore, MICB transcript expression correlates with immune cell infiltration (Kucuk and Cacan, 2022). These findings suggest that genes encoding NK cell receptors and their ligands can be valuable targets for predicting susceptibility to lung cancer, offering prognostic value and potential therapeutic targets in future immunotherapies.
In addition to NK cell receptors and their ligands, mutations in several other genes can affect NK cell function in lung cancer. For instance, higher expression of erythropoietin-producing hepatocellular receptor A5 (EPHA5), a member of the Eph family of tyrosine kinase receptors, is associated with lymph node metastasis, TNM stage, and LUAD differentiation (Liu et al., 2022). In a recent study by Li et al., the overexpression of wild-type EPHA5 suppressed tumor invasion and migration, in contrast to the effects observed with mutant EPHA5. Further analysis demonstrated that overexpression of wild-type EPHA5 significantly promoted NK cell proliferation and cytotoxicity against NSCLC cells and was associated with decreased cell apoptosis. However, mutant EPHA5 impairs NK cell activity and enhances the migration and invasion of NSCLC cells (Zhang et al., 2020). Fatty acid-binding protein (FABP) 5 regulates lipid metabolism and is overexpressed in various cancers, including lung cancer, with high expression associated with poor prognosis (George Warren et al., 2023). To elucidate the potential association between lipid homeostasis and lung cancer metastasis, the levels of IFN-γ and granzyme B produced by lung NK cells in tumor-bearing Fabp5−/− mice were significantly decreased compared with their wild-type counterparts, suggesting a role for FABP5 in regulating the cytotoxicity of NK cells. Moreover, the maturation of lung-derived NK cells decreased in tumor-bearing Fabp5−/− mice, likely due to the reduced expression of T-bet and Eomes transcription factors. These events lead to enhanced lung cancer metastasis (Yang et al., 2021). Recent studies have also indicated that apolipoprotein E (ApoE), which is involved in cholesterol metabolism, is overexpressed in human lung cancer tissues compared with adjacent non-cancerous tissues (Trost et al., 2008). This overexpression is associated with a poor prognosis due to the promotion of tumor progression and invasion in patients with LUAD (Su et al., 2011). Further investigations have demonstrated that inhibition of ApoE enhances NK cell cytotoxicity and increases NK cell infiltration by upregulating triggering receptors expressed on myeloid cells (TREM)-1 and T-bet (Lee et al., 2019a). Higher expression of APOE has been reported in macrophages within tumor tissues than in normal tissues (He et al., 2021). Serine/threonine kinase 11 (STK11) is a tumor suppressor gene, and loss-of-function mutations in this gene have been associated with enhanced tumor escape, invasion, and metastasis (Mazzaschi et al., 2021). These mutations can affect the function of immune cells and change the composition of the TME. For example, STK11 deletion significantly reduces the cytotoxicity of NK cells and their infiltration into the TME, ultimately promoting the proliferation of LUAD cells (Huang et al., 2022). Consistent with previous research, the overexpression of AXL, a member of the TAM (Tyro3, Axl, and Mer) receptor tyrosine kinase family, in mesenchymal carcinoma cells likely plays a role in the cell-intrinsic immune escape mechanism in NSCLC patients by suppressing the killing effects of NK cells and cytotoxic T lymphocytes (CTLs). Consequently, targeting AXL is a potential strategy for sensitizing mesenchymal NSCLC clones to lymphocyte-mediated cytotoxicity. AXL targeting induces NK-kB activation, resulting in enhanced expression of intercellular adhesion molecule 1 (ICAM1) and UL16 binding protein 1 (ULBP1) along with MAPK inhibition, finally improving the survival of patients with NSCLC. ULBP1 serves as a ligand for NKG2D, an activating receptor on NK cells, and its overexpression facilitates the recognition of stressed cells by the immune system. Therefore, one of the immune escape mechanisms involving AXL in carcinoma cells might be by decreasing the expression of ULBP1 (Terry et al., 2019). Further studies have also revealed the overexpression of the pancreatic progenitor cell differentiation and proliferation factor (PPDPF) in NSCLC tissue and cell lines, which is associated with lower patient survival and increased resistance of lung tumor cells to radiotherapy (Yun et al., 2022). Zheng et al. reported that the PPDPF-induced activation of STAT3 plays a significant role in decreasing NK cell activation and infiltration, creating an immunosuppressive microenvironment in LUAD (Zheng et al., 2022).
Recent investigations using advanced techniques such as scRNA-seq have led to the development of novel gene signatures derived from marker genes of tumor-infiltrating NK cells to predict the prognosis and response to immunotherapy in patients with lung cancer. For instance, Song et al. identified a novel seven-gene signature, including GCSAML, ACTG1, ACOT7, SELENOK, PEBP1, BIRC3, and ACAP1. These genes are primarily involved in the modulation of cell proliferation, migration, regulation of the cell cycle and NK cell functions in patients with LUAD (Song et al., 2022). A recent in silico study also suggested an antigen-presenting cell (APC)/T/NK cell-related gene signature consisting of 16 genes to estimate overall survival in patients with LUAD, which were mostly associated with immune-related pathways and responses. Three genes (NCR3, RAET1E, and SHC1) were closely linked to NK cell cytotoxicity pathways, while another set of three genes (MAP2K1, NRAS, and PTPN6) showed a significant association with T cell and NK cell-related pathways (Huang L. et al., 2023).
Investigation of microRNA editing levels has been suggested as a potential biomarker of LUAD (Maemura et al., 2018). MiRNAs affect the function, survival, and cytotoxicity of NK cells by regulating cytokine production (Shirshev et al., 2017). For instance, miR-218-5p impairs IL-2-induced NK cell cytotoxic capacity against LUAD cells by regulating the expression of serine hydroxymethyl transferase 1 (SHMT1), both in vitro and in vivo (Yang et al., 2019). Zhou et al. demonstrated elevated STAT3 mRNA levels and downregulated miR-130a levels in primary NK cells derived from patients with NSCLC compared with healthy controls. However, miR-130a overexpression in the IL-2-induced NK-92 cell line markedly increased cytokine production and the killing effect of NK cells on A549 cells by affecting STAT3 expression (Zhou et al., 2020). Consistent with previous research on NSCLC, the overexpression of hsa-miR-301a-3p can reduce the cytotoxicity of NK cells and decrease the secretion of IFN-γ and TNF by targeting Runt-related transcription factor 3 (RUNX3). This mechanism has also been shown to promote tumor progression in in vivo models (Zhang et al., 2023). A study conducted by Gao et al. also found decreased expression of miR-30c in primary NK cells from lung cancer patients compared to healthy controls. Overexpression of miR-30c increased NK cell cytotoxicity against A549 cells, while downregulation of miR-30c reduced the secretion of IFN-γ and TNF by these cells. The regulation of NK cell cytotoxicity by miR-30c occurs through targeting N-acetylgalactosaminyltransferase 7 (GALNT7) (Gao et al., 2023), a glycosylation enzyme that has oncogenic effects by binding to various miRNAs in different types of cancers (Nie et al., 2016; Wang JB. et al., 2020; Cao et al., 2020). Moreover, as GALNT7 mediates the phosphorylation of PI3K/AKT signaling, and its inhibition ultimately leads to the inactivation of this pathway, enhancing the killing effects of NK cells (Gao et al., 2023). These studies suggest that miRNAs play a role in regulating NK cell function and lung cancer immunopathogenesis, which could be considered in future NK cell-based anti-tumor immunotherapies. Long non-coding RNAs (lncRNAs) have also been implicated as regulators of immune cell differentiation and infiltration into tumors. In silico studies have suggested that several lncRNAs could be associated with immune-related pathways, specifically cytokines and cytokine receptors, as well as their valuable roles in classifying various cancer subtypes based on molecular and immunological features (Li Y. et al., 2020). A recent study using a computational approach has also revealed the significance of lncRNAs in predicting the response to immune checkpoint inhibitor immunotherapy in patients with NSCLC (Sun et al., 2020). Other studies have also investigated the anti-tumor functions of TME-derived lncRNAs in LUAD tumors. The AC008750.1 non-coding transcript shows high expression in NK cells and CD8+ T cells and is positively correlated with NK cell granule protein 7 (NKG7) expression under stimulatory conditions, a marker associated with the granulation of NK cells and CD8+ T cells. To validate this, AC008750.1 knockdown using siRNA significantly decreased the cytotoxicity of NK cells against LUAD cells (Sage et al., 2020). However, further research is necessary to enhance our comprehension of the roles of lncRNAs in modulating immune-related pathways, thereby improving patient responsiveness to immunotherapies.
Research findings indicate that lung tumor-infiltrating DCs express higher levels of CD11b and the inhibitory molecule PD-L1 than peritumoral lung DCs (Pyfferoen et al., 2017). Additionally, lung tumor-derived cDC1s express lower levels of T cell immunoglobulin mucin-4 (TIM4), leading to decreased tumor-associated antigen uptake and lower activation of CD8+ cells in advanced stages of murine lung tumors (Caronni et al., 2021). NSCLC-derived DCs demonstrate increased secretion of IL-10 and higher expression of B7-H3 T cell co-inhibitory molecules compared with their counterparts in normal lungs, creating an immunosuppressive TME where anti-tumor T cells fail to become effectively activated (Schneider et al., 2011). An immunosuppressive TME may be associated with a higher frequency of plasmacytoid DCs (pDCs) derived from NSCLC tissues, which are characterized by elevated expression levels of CD33 and PD-L1. Tumor-derived pDCs also reduce the production of type I IFNs (Pedroza-Gonzalez et al., 2015; Sorrentino et al., 2015). Previous investigations have indicated that increased secretion of transforming growth factor-beta (TGF-β) by DCs in patients with NSCLC is associated with higher levels of Tregs (Dumitriu et al., 2009), which can further contribute to the development of an immunosuppressive TME in NSCLC (Zhong et al., 2021). Dysfunction of tumor-induced DC in NSCLC may be caused by the inhibition of nuclear factor-kappa B (NF-kB) and signal transducer and activator of transcription 3 (STAT3) signaling pathways, leading to downregulation of downstream genes associated with cytokine and chemokine production, as well as antigen presentation (Li et al., 2017). Overall, DCs derived from lung cancer showed increased expression of genes incolved in creating an immunosuppressive TME, which promotes tumor progression.
Exploring the role of miRNAs in regulating the function of DCs adds another layer of complexity. In silico studies have revealed a potential association between the overexpression of miR-582, reduced expression of CD1B, and lower overall survival in advanced stages of lung cancer. This miRNA regulates CD1B, a marker of resting and activated DCs engaged in the presentation of lipids and glycolipids to T cells via the MHC complex. Identifying potential miRNA biomarkers implicated in regulating immune cell genes, particularly their target immune genes, holds promise for identifying novel immunotherapies to impede tumor progression in patients with lung cancer. Manipulation of the expression of these miRNAs is achievable through interventions, such as small interfering RNA, miRNA mimics, and small molecule inhibitors of miRNAs. However, experiments with larger sample sizes, evaluation of expression levels in tumor tissues and their adjacent counterparts, and further experimental methodologies are required to confirm these findings (Guo et al., 2020).
These findings suggest that changes in the phenotype and function of DCs in lung tumors contribute to an immunosuppressive microenvironment, hindering effective T cell activation. These alterations may be considered potential immune evasion mechanisms in lung cancer cells. Understanding the molecular mechanisms underlying DC dysfunction in lung tumors will enable us to target and manipulate these inhibitory mechanisms, ultimately improving the overall survival of patients with lung cancer.
Tumor-associated macrophages (TAMs) exhibit distinct phenotypes known as inflammatory or classically activated (M1) and anti-inflammatory or alternatively activated (M2) macrophages. The M1 phenotype is associated with both pro- and anti-tumorigenic roles, whereas the M2 phenotype is characterized by tumor-promoting properties. This duality plays a significant role in the cancer immune microenvironment, influencing the growth, progression, and metastasis of lung tumors (Sedighzadeh et al., 2021). M2-like TAMs play a crucial pro-tumor role in lung cancer. The observed decrease in M2-like TAM population within the TME impedes lung cancer growth and metastasis (Kawaguchi et al., 2023). Tumor cells play an important role in M2 phenotype polarization in the TME by releasing cytokines. For instance, IL-37 was found to induce the expression of a scavenger receptor, known as the macrophage receptor with a collagenous structure (MARCO), on TAMs. MARCO+ TAMs are anti-inflammatory macrophages with pro-tumoral effects in the TME (Georgoudaki et al., 2016). Depletion of the IL-37 gene in two distinct lung cancer cell lines (H460 and A549) suppressed MARCO expression and diminished IL-10 production in co-cultured macrophages (La Fleur et al., 2021). Moreover, analysis of RNA-seq data from patients with NSCLC revealed a positive correlation between the gene expression of MARCO and the expression of various genes associated with immunosuppressive TAMs, including CD68, CD163, macrophage scavenger receptor 1 (MSR1), IL4R, TGFB1, and IL10 (La Fleur et al., 2018). IL-17D is another cytokine that facilitates the infiltration of TAM in lung cancer cells. The induction of IL-17D expression in LLC1 cells, which typically lack this gene, resulted in a significant increase in F4/80+ CD11b+ TAMs in a subcutaneous tumor model. M2 macrophages (CD206+ F4/80+ CD11b+ TAMs) within total TAMs were prominently elevated in these tumors. Induction of IL-17D expression in A549 cells, which also do not endogenously express IL-17D, led to elevated expression of macrophage-related genes such as CCL3, CCL4, and CSF1. Further investigations using an IL-17D knockdown lung cancer cell line (H1155) revealed that IL-17D upregulates the genes related to macrophage-recruitment and polarization through the p38 MAPK signaling pathway (Lin et al., 2022).
Another recognized gene linked to M2 macrophage polarization is the one encoding Mincle (Li C. et al., 2020). Mincle or macrophage-inducible C-type lectin (Clec4e) is a transmembrane pattern recognition receptor expressed in myeloid cells (Huang X. et al., 2023). The majority of CD163+ cells in NSCLC are Mincle+ TAMs. Silencing Mincle in bone marrow-derived macrophages (BMDM) significantly increased the expression of M1 markers (iNOS, MCP-1, and TNF) upon stimulation with LLC conditioned medium. Analysis of scRNA-seq data from patients with NSCLC revealed a significant association between Mincle expression in the stroma and tumor, and unfavorable disease-specific survival (Li C. et al., 2020). Clustering of the immune components of LUAD samples in The TCGA database identified the gene encoding nucleotide-binding oligomerization domain-containing protein 2 (NOD2) as an important gene with tumor-suppressing effects. Lower levels of NOD2 expression are associated with unfavorable characteristics, such as larger tumor size, metastatic tumors, and advanced stages. NOD2 knockdown in a human monocytic cell line (THP-1) resulted in decreased expression of M1 markers under lipopolysaccharide stimulation. However, M2 markers increased upon stimulation with IL-4. Therefore, NOD2 deficiency was identified as a driving factor in the induction of protumorigenic macrophages in LUAD (Wang Y. et al., 2021).
The gene encoding estrogen receptor α (ERα) is another identified gene associated with macrophage infiltration and polarization in lung cancer. The expression of ERα has been linked to a poor prognosis in NSCLC (Castellanos et al., 2023). Inducing the overexpression of ERα in NSCLC cells resulted in increased recruitment of macrophages and induced M2 polarization. Conversely, downregulation of ERα using ERα-shRNAs significantly reduced macrophage infiltration and induced M1 polarization. Further investigations revealed that ERα activates the CCL2/CCR2 axis, facilitating macrophage infiltration, M2 polarization, and MMP9 production, thereby enhancing the invasiveness of the NSCLC cells (He et al., 2020). Analysis of mRNA-seq data obtained from the TCGA database indicated that metastasis-associated gene 1 (MTA1), an oncogene involved in NSCLC metastasis, is associated with macrophage infiltration and the malignant phenotype of lung cancer. Moreover, the positive association of MTA1 with CD206 in NSCLC and LUSC suggests the involvement of this gene in the infiltration of the protumorigenic phenotype of macrophages into the TME (Ma et al., 2022).
Studies have identified transcription factors involved in macrophage polarization in lung cancer. One such transcription factor is Krüppel-like factor 4 (KLF4), which plays a role in macrophage infiltration and polarization in NSCLC. KLF4 is expressed at higher levels in M2 macrophages compared to M0 macrophages (Arora et al., 2021). Furthermore, research in c-Maf knockout mice has confirmed the role of this transcription factor in controling M2-related genes. TAMs in the LLC mouse model show elevated levels of c-Maf expression. When c-Maf was silenced in TAMs, there was a decrease in the expression of Il10, Arg1, indoleamine 2,3-dioxygenase (Ido), and Vegfa, along with an increase in the levels of Il12 and Tnf. Moreover, c-Maf knockdown TAMs significantly enhanced the production of IFN-γ from CD4+ and CD8+ T cells. Moreover, inducing LCC in mice with myeloid-specific genetic ablation of c-Maf resulted in a significantly lower tumor burden and smaller tumor size. Notably, c-Maf chromatin immunoprecipitation sequencing indicated that c-Maf directly regulates the colony-stimulating factor 1 receptor (CSF1R) locus in M2 BMDMs (Liu et al., 2020). CSF1, also known as macrophage colony-stimulating factor (M-CSF), and its receptor (CSF1R) play an important role in the polarization of M2 macrophage (Valero et al., 2021). CSF-1R+ macrophages are related to immunosuppression and tumor progression, and the expression of CSF-1R in TAMs is associated with an unfavorable prognosis and resistance to tumor immunotherapy (Koh et al., 2014; Candido et al., 2018; Gyori et al., 2018).
Regarding the implicated signaling pathways, the β–catenin signaling has emerged as a critical pathway in the transition of macrophages from M1 to M2 in lung cancer. The induction of lung cancer in mice with a specific knockout of β-catenin in macrophages led to a significant decrease in tumor growth compared to their wild-type counterparts. TAM analysis of these transgenic mice revealed an increase in the expression of M1 markers and a concomitant reduction in the expression of M2 markers (Sarode et al., 2020a). Additionally, inhibition of Wnt/β-catenin signaling with the molecular inhibitor of Wnt secretion, LGK-974, increased M1 markers and decreased M2 markers in NSCLC cells (A549 and H1299) (Tang et al., 2021). Moreover, the TGF-β1/β-catenin pathway in tumor cells and TAMs was observed to be involved in SCLC pathogenesis (Ahirwar et al., 2023). In gene silencing/upregulating studies, different genes have been discovered downstream of the β-catenin pathway. BMDMs isolated from β-catenin knockout mice showed significant downregulation of FOS-like antigen 2 (FOSL2) and upregulation of the AT-rich interaction domain 5A (ARID5A). FOSL2 and ARID5A have been identified as downstream transcription factors that are crucial for orchestrating the M1 to M2 transition (Sarode et al., 2020a). Furthermore, it was discovered that slit guidance ligand 2 (SLIT2) and its receptor, roundabout guidance receptor 1 (ROBO1), are downstream of the TGF-β1/β-catenin pathway. SLIT2 has been identified as a tumor suppressor gene in SCLC, with its expression significantly reduced in this type of tumor, while ROBO1 expression is significantly higher in SCLC cells compared to adjacent normal cells (Peifer et al., 2012; Ahirwar et al., 2023). Overexpression of SLIT2 and deletion of ROBO1 in SCLC cells have shown the anti-tumor and pro-tumor effects of SLIT2 and ROBO1, respectively, through macrophage polarization (Ahirwar et al., 2023).
M2 macrophages play a crucial role in promoting NSCLC metastasis. Guo et al. identified αB-crystallin (CRYAB) as a key factor in this process (Guo et al., 2019). CRYAB belongs to the small heat shock protein family and is actively involved in various signaling pathways, including apoptosis, inflammation, and oxidative stress (Zhang et al., 2019). When CRYAB-knockout NSCLC cell lines were co-cultured with M2 macrophages, a significant decrease in cell invasion and the epithelial-to-mesenchymal transition was observed. This effect was attributed to the CRYAB-mediated activation of the ERK1/2/Fra-1/Slug pathway (Guo et al., 2019).
Several lncRNAs are involved in macrophage polarization in lung cancer. These lncRNAs, known for their widespread expression, play pivotal roles in the regulation of gene expression. LOC100270746 is a lncRNA located on chromosome 6p22.2. The expression of LOC100270746 was discovered to be lower in LUAD tissues than in normal lung tissues. Low levels of LOC100270746 were associated with unfavorable features, such as larger tumor size, local invasion, advanced TNM stages, and poor overall survival. Silencing LOC100270746 with shRNA in a LUAD cell line (HCC827) revealed the anti-tumor effects of this lncRNA, such as decreased viability, proliferation, migration, and invasion, as well as increased apoptosis of LUAD cells. In vivo studies also demonstrated that A549 cells overexpressing LOC100270746 induced smaller and fewer lung metastatic nodules in mice, with significantly lower macrophage infiltration in the tumors. Furthermore, LOC100270746 inhibited M2 macrophage polarization in LUAD. CSF1 is a downstream target of LOC100270746, with this lncRNA exerting its effects by suppressing CSF1 expression (Li Y. et al., 2022). In contrast, GNAS-AS1, another lncRNA, showed higher expression in M2 macrophages and NSCLC-related TAMs than in M0 and M1 macrophages. The expression of GNAS-AS1 was significantly higher in tumor tissues from patients with NSCLC than in adjacent normal tissues. Moreover, high levels of GNAS-AS1 were associated with unfavorable clinicopathological characteristics, including lymph node involvement, shorter overall survival, and metastasis-free survival. Inducing the overexpression of GNAS-AS1 in THP-1-differentiated macrophages resulted in greater polarization to M2 macrophages upon stimulation with IL-4, which in turn promoted NSCLC migration and invasion. Additionally, the knockdown of GNAS-AS1 using shRNA significantly reduced the A549 and H1299 cells proliferation, invasion, and migration (Li Z. et al., 2020). LINC00313 is another lncRNA involved in M2 macrophage polarization. This lncRNA was detected in exosomes from H1299 cells, and its depletion abrogated M2 macrophage polarization. LINC00313 was found to act as an miR-135a-3p sponge, leading to the upregulation of STAT6 expression (Kong et al., 2022).
In addition to M1 macrophages, exosomes have been found to exert anti-tumor effects, by decreasing the viability, invasiveness, and migration of lung cancer cells (Peng et al., 2023). MiRNAs are one of the functional components of exosomes (Dexheimer and Cochella, 2020) and are involved in various aspects of cancer immunity (Li C. et al., 2022). For example, miR-let-7b-5p was detected at higher levels in the exosomes derived from M1 macrophages compared to M0-derived ones. Investigations have shown that exosomes can transfer miR-let-7b-5p to lung tumor cells, inhibiting tumor cell proliferation and promoting tumor cell apoptosis by regulating GNG5 protein levels (Peng et al., 2023). In another study, miR-181a-5p was identified as a key molecule in M1-derived exosomes that inhibits cell proliferation and induces apoptosis by targeting ETS1 and serine/threonine kinase 16 (STK16). STK16 is highly expressed in LUAD tissues, and its expression is associated with an unfavorable prognosis (Wang X. et al., 2022).
Evidence suggests that macrophage polarization plays a role in shaping the response to lung cancer treatment. For instance, anti-silencing factor 1 (Asf1) is a highly conserved chaperone of histones H3/H4. silencing Asf1a in KRAS-mutant LUAD cells using CRISPR-Cas9 increased tumors sensitivity to anti-PD-1 treatment in orthotopic lung cancer models, without affecting tumor cell proliferation. This enhanced treatment response was attributed to the elevation of M1 macrophages and subsequent T cell activation through granulocyte-macrophage colony-stimulating factor (GM-CSF) upregulation (Li F. et al., 2020). Furthermore, high plasma IL-6 levels in lung cancer patients have been associated with immunotherapy resistance (Kuo et al., 2021). Macrophage-derived IL-6 promotes STAT3-dependent PD-1 expression in CD8+ T cells, involving Rab37, a member of the Rab-GTPase family implicated in membrane trafficking. Dysregulation of Rab37 has been observed in lung cancer cells (Tzeng et al., 2017). Rab37 knockout mice induced with LLC demonstrated the pivotal role of this molecule in PD-1 expression in CD8+ T cells, these mice also showed reduced Tregs and CD206+ M2 macrophages (Kuo et al., 2021). Therefore, numerous various genes regulate macrophage polarization to either promote or inhibit lung tumor progression.
Innate lymphoid cells (ILCs) are a subset of the innate immune system that can be activated without antigen-specific receptors in response to alterations in homeostasis and stimulatory signals, particularly at mucosal sites. Previous studies have demonstrated the important role of murine lung ILCs in preserving the integrity of the epithelial barrier and maintaining tissue homeostasis in the lungs following influenza virus infection (Monticelli et al., 2011). Three distinct subsets of ILCs (CD45+ Lin− CD127+) have been identified in the human lung (De Grove et al., 2016). In the case of NSCLC, a higher frequency of NCR+ ILC3s has been reported in tumor tissues compared to adjacent normal lung tissues. In contrast, the frequency of ILC2s is significantly lower. These tumor infiltrating ILC3s express low levels of KIRs, Perforin, and CD94. They also play an important role in the TME by producing proinflammatory cytokines such as IL-22 and TNF and promoting leukocyte recruitment through the secretion of IL-2 and IL-8. The frequency of NCR+ ILC3 decreases as the tumor progresses, which is potentially associated with poor prognosis (Carrega et al., 2015). Investigations have also revealed the accumulation of ILCs in the central region of human lung tumors, characterized by lower expression levels of immune checkpoint receptors, such as T cell immunoglobulin and mucin-domain containing-3 (TIM-3), T cell immunoreceptor with Ig and ITIM domains (TIGIT), PD-1, and CD39. Decreased expression of CD49a, in contrast to increased expression of CD103, was observed in ILCs of the tumor center compared with those located in tumor-free distal sites (Brownlie et al., 2023). To understand the specific factors regulating ILC plasticity within the TME of NSCLC, IL-23 producing SqCCs (one of the two types of NSCLCs) were found to induce the conversion of ILC1 to ILC3. IL-17 secreted by ILC3 promotes tumor progression, leading to poor prognosis in patients with SqCCs. This suggests a potential therapeutic target in the IL-23/ILC3/IL-17 axis for IL-23-producing lung tumors (Koh et al., 2019). While several studies have investigated the characteristics of ILCs within the TME of lung cancer and their possible association with prognosis, limited research has explored the immunogenomic landscape of ILCs in various lung tumors. Despite the immunoregulatory functions of ILCs, they may not be ideal candidates for immune checkpoint therapies due to their low expression of immune checkpoint receptors.
Human NK T (NKT) cells, identified as CD3+ CD56+ or CD8+CD56+, represent a subpopulation of CD1d-restricted T cells, demonstrating characteristics of both T and NK cells. These cells promote anti-tumor immunity by producing interferon β or participating in the elimination of tumor cells (Yin et al., 2021). NKT1, NKT2, and NKT17 cell subsets have been found in murine lungs, with NKT1 being the most prevalent (Crosby and Kronenberg, 2018). However, the difference in the percentage of NKT cells (CD45+CD3+CD56+) and T cells (CD45+CD3+CD56−) was insignificant between patients with lung cancer and healthy controls (Yin et al., 2021). The immunoregulatory functions of NKT and CD8+ T cells in NSCLC are in agreement with the remarkable expression of PD-1 and lymphocyte activation gene 3 (LAG-3) in these cells, as demonstrated by single-cell CyTOF analysis (Datar et al., 2019). CD3+CD56+CD16+ NKT-like cells are potential biomarkers for predicting the response and toxicity of immune checkpoint inhibitors in advanced-stage NSCLC. Further studies with larger cohorts are required to validate these observations (Lin et al., 2023).
Gamma-delta (γδ) T cells are a subset of T cells that can directly eliminate target cells in an HLA-independent manner, making them efficient players in the front lines of anti-tumor immunity. In contrast, in the context of cancer, a distinctive subset known as Vδ2 Tregs, which are specific types of γδ T cells with regulatory activities and Foxp3 expression, can be induced in the presence of antigen stimulation (IPP/IL-2) and TGF-β1 and IL-15 cytokines in vitro (Casetti et al., 2009). γδT17 cells may increase the accumulation and expansion of polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs), potentially inducing a tumor-suppressive microenvironment that promotes colorectal cancer progression (Wu et al., 2014). Therefore, additional investigations are required to further elucidate the specific roles of γδ T cells in various cancer types. Recent findings indicate that in contrast to most peripheral blood γδ T cells expressing Vδ2, the majority of γδ T cells in lung tissues and tumors express Vδ1. These tissue-resident γδ T cells express higher levels of T cell receptor delta constant (TRDC) compared to other tissue sites, with a higher expression of T cell receptor delta variable 1 (TRDV1) than TRDV2, suggesting that γδ T cells in non-malignant lung tissues are in a steady state. Additionally, CD103 expressing Vδ1 γδ T cells are more frequent in tumors than in non-tumor lung tissues, enhancing their tumor-homing capabilities (Davey et al., 2017; Wu et al., 2022). Based on the expression of CD45RA and CD27, γδ T cells can be divided into two groups: effector and memory. Vδ1 T cells with a CD45RA−CD27− effector memory T cell (TEM) phenotype are more abundant in tumor tissues than in non-tumor tissues, indicating cytolytic and IFN-γ production capabilities. Vδ1 T cells show potential for NSCLC immunotherapy; however, they have limited responsiveness to activation-induced cell death, which restricts their suitability for adoptive cell therapy (Davey et al., 2017; Wu et al., 2022). Studies have also shown that DNA methyltransferase inhibitors can enhance the antitumor effects of MHC-unrestricted γδ T cell therapy by increasing the expression of adhesion molecules such as intercellular adhesion molecule 1 (ICAM-1) or facilitating immune cytoskeleton reorganization and immune synapse formation, finally improving the tumor-killing efficacy of γδ T cells. Therefore, epigenetic modifications could be used to develop novel therapeutic strategies for lung cancer and promote cell-based immunotherapy (Weng et al., 2021). Allogeneic Vγ9Vδ2 T cells, which are the most frequent subset of γδ T cells in humans, have been implicated in adoptive immunotherapy. However, further clinical investigations are needed in this field (Xu et al., 2021). Figure 1 shows the recent immunogenetic factors associated with lung cancer progression in innate immunity.
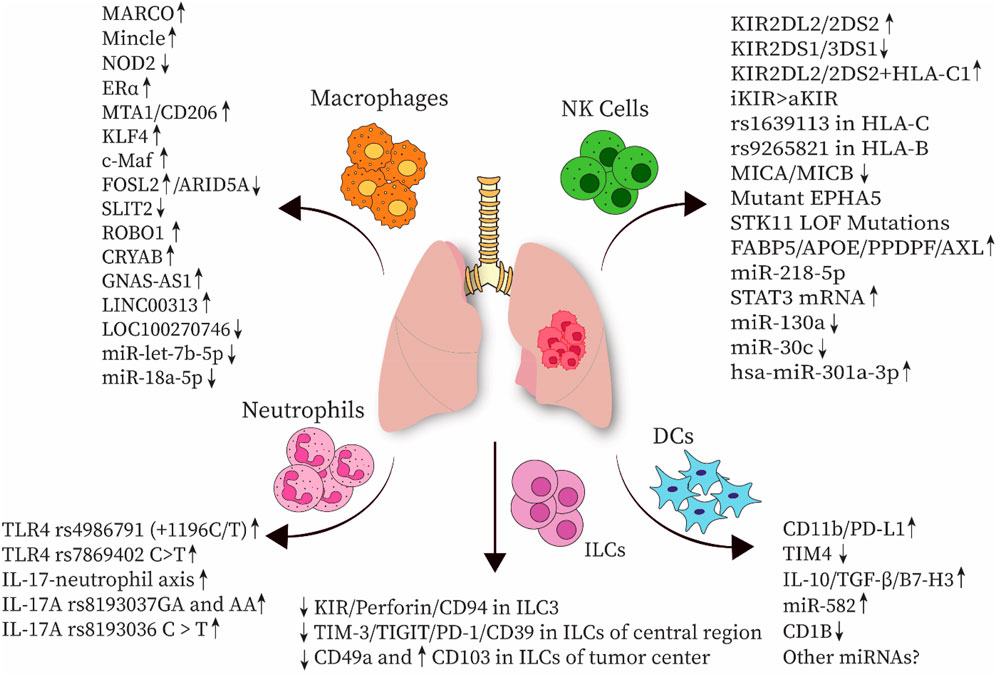
Figure 1. Several immunogenetic factors associated with innate immune cells have been implicated in the progression of lung cancer. NK cell, natural killer cell; DC, dendritic cell; ILC, innate lymphoid cell; MARCO, macrophage receptor with collagenous structure; NOD2, nucleotide-binding oligomerization domain-containing protein 2; Erα, estrogen receptor α; MTA1, metastasis-associated gene 1; KLF4, Krüppel-like factor 4; FOSL2, FOS-like antigen 2; ARID5A, AT-rich interaction domain 5A; SLIT2, slit guidance ligand 2; ROBO1, roundabout guidance receptor 1; CRYAB, αB-crystallin; TLR, Toll-like receptor; KIR, Killer-cell immunoglobulin-like receptor; TIM-3, T-cell immunoglobulin and mucin-domain containing-3; TIGIT, T cell immunoreceptor with Ig and ITIM domains; PD-1, Programmed cell death protein 1; TGF-β, Transforming growth factor-β; TIM4, T-cell immunoglobulin mucin-4; HLA, human leukocyte antigen; MICA, MHC class I polypeptide-related sequence A; EPHA5, erythropoietin-producing hepatocellular receptor A5; STK11, Serine/threonine kinase 11; LOF, loss-of-function mutations; FABP5, Fatty acid-binding protein 5; ApoE, apolipoprotein E; PPDPF, pancreatic progenitor cell differentiation and proliferation factor.
Crosstalk between tumor cells and tumor-infiltrating lymphocytes (TILs) in the TME is mediated by low-molecular-weight secreted proteins, such as cytokines and chemokines. Given their substantial role in creating an inflammatory microenvironment, these TIL-producing proteins can significantly enhance tumor progression, metastasis, and resistance to therapy, thereby emerging as cancer prognostic markers. IL-6, IL-10, IL-2, IL-22, IL-8, IL-32, IL-37, and TNF are the most common cytokines involved in the therapeutic management of lung cancer (Sarode et al., 2020b). Interest in investigating genetic alterations within cytokine and chemokine genes is growing, as these changes can be associated with tumor progression or suppression and may serve as potential prognostic markers. In this regard, the overexpression of tumor necrosis factor superfamily 15 (TNFSF15), a member of TNF superfamily, is linked to suppressed tumor progression in different cancers, primarily through the inhibition of angiogenesis (Hou et al., 2005; Liang et al., 2011). A case-control study on Han Chinese population demonstrated that TNFSF15–638A > G and −358 T > C polymorphisms probably increased susceptibility to SCLC, as opposed to NSCLC (Gao et al., 2019). These polymorphisms affect the expression levels of TNFSF15 and its downstream signaling pathways, ultimately modulating the immune response. For instance, the interaction between death receptor 3 (DR3) and vascular endothelial growth inhibitor (VEGI)-251 splice variant, encoded by TNFSF15, stimulates signaling pathways by inducing NF-kB and Caspase cascade, initiating T cell responses and cell apoptosis, respectively (Migone et al., 2002; Bittner et al., 2016). Additionally, the TNFSF15−358 T > C polymorphism increases the risk of SCLC among nonsmokers rather than smokers, indicating the need for further analysis. Moreover, the TNFSF15–638GG genotype is associated with an increased risk of SCLC in men and individuals over 60 years of age (Gao et al., 2019). Higher mRNA and protein expression of TNFα-induced protein 2 (TNFAIP2), a gene regulated by TNF, has been reported in NSCLC tissues compared to adjacent normal tissues, which is likely epigenetically controlled by miR-145-5p. TNFAIP2 plays a role in promoting tumor proliferation, migration, and metastasis in NSCLC. Therefore, interventions such as silencing TNFAIP2 or overexpressing miR-145-5p may be potential therapeutic approaches for NSCLC (Li J. et al., 2020).
Upregulation of IL-32, a pro-inflammatory cytokine produced by NK cells and functional T cells, has been reported in most LUADs as well as in some large-cell carcinomas and SCLCs, but not in SCCs. TILs and tumor cells expressing IL-32 are associated with lymph node metastasis and poor outcomes (Sorrentino and Di Carlo, 2009). Wang et al. demonstrated an association between the CC homozygote of the rs12934561 IL-32 polymorphism and a higher risk of lung cancer, whereas the TT genotype was associated with poor survival in patients with squamous carcinoma. The T allele of the rs28372698 IL-32 polymorphism is associated with poor prognosis in patients with moderately and well-differentiated lung cancer. Moreover, the expression of IL-32 was significantly inhibited in peripheral blood leukocytes of patients with lung cancer (Wang et al., 2017). Conversely, IL-32γ may suppress lung tumor progression by increasing tissue inhibitor of metalloproteinase 3 (TIMP-3) expression, which acts as a tumor suppressor gene (Yun et al., 2018), or by downregulating integrin alpha V (ITGAV)-mediated STAT5 signaling pathways, leading to the suppression of the growth of CD133+ lung cancer stem cells (Lee et al., 2019b). A recent study conducted in a Brazilian Amazon population has revealed an association between the Ins/Ins genotype of IL-1A (rs3783553) polymorphism and a higher risk of NSCLC (Pereira et al., 2023). In hepatocellular carcinoma, this particular polymorphism has been found to increase the expression of IL-1A, potentially by impairing the binding sites of miRNA-122 and 378 to the 3’ UTR region (Gao et al., 2009). A similar mechanism may occur in other cancer types. Given that IL-1A is a pro-inflammatory cytokine mainly secreted by monocytes and macrophages and plays crucial roles in angiogenesis, proliferation, and tumor migration, its upregulation is associated with a higher risk of cancer (Ma and Zhou, 2016). Similarly, IL-1B, mostly secreted from tumor-associated macrophages as a pro-inflammatory cytokine, enhances the expression of ELF3 in LUADs with EGFR mutations, leading to the activation of the PI3K/Akt/NF-κB pathway and consequently contributes to tumor progression (He et al., 2021). A case-control study conducted on a Spanish population revealed an association between the IL1B rs1143634-TT genotype and a reduced risk of NSCLC, whereas individuals carrying the C allele had an increased risk of developing NSCLC. Several factors, such as the inclusion of different types of lung cancer besides NSCLC, different age groups, and populations from diverse ethnic backgrounds, contributed to the variations observed in the outcomes of this study when compared to previous research. Therefore, further investigations with larger cohorts from different populations are necessary to validate these findings (Pérez-Ramírez et al., 2017). Additionally, high expression of IL-35 was found in the BALF from the tumor site in patients with NSCLC. IL-35 inhibits the function of Th1 and Th17 cells while increasing the regulatory function of CD4+ T cells and reducing the cytolytic activity of CD8+ T cells, highlighting its role in the induction of T cell exhaustion and dysfunction, favoring tumor progression (Wang et al., 2018). Studies on the Han population have suggested an association between an increased frequency of monocyte chemoattractant protein 1 (MCP-1) AA genotype and a decreased frequency of the GG genotype with a higher risk of NSCLC. While they indicateds a role for MCP-1 polymorphisms in the risk of NSCLC, no association was found between CCR2 polymorphisms and NSCLC (Yang et al., 2015). However, a significant association was observed between carrying the −64I allele of CCR2-64I gene polymorphism and higher susceptibility to NSCLC in a Tunisian population, with a higher expression of CCR2 in patients (Rafrafi et al., 2015). An in silico study also indicated lower expression of CCR2 in patients with LUAD, while higher expression was associated with better survival in these patients (Wan et al., 2021). Given these controversies, further research in larger samples is required to clarify the role of CCR2 polymorphisms in lung cancer.
A GWAS conducted in a Croatian population consisting of 203 patients with lung cancer and 360, 938 controls demonstrated a significant association between specific SNPs and increased susceptibility to lung cancer. These SNPs include rs6806802 in CCR9, rs9766026 in F13A1, rs2386841 in IL2RA, rs2495366 in TNRSF14, rs1148471 in TNRSF8, rs1682802 in GNA15, and rs1148038 in IL31RA. These genes and associated SNPs were mainly involved in the activation of NF-κB transcription factor, leading to an inflammatory condition. Given that the higher number of controls compared with cases in this GWAS may increase the possibility of false-positive/negative findings; therefore, further investigation is required to confirm these findings (Baranasic et al., 2023). Table 1 summarizes the other cytokine polymorphisms associated with lung cancer susceptibility and protection.
Various miRNAs have been shown to regulate the expression of cytokine or chemokine genes, influencing tumor progression and ultimately patient survival. The expression of miR-3648 was significantly elevated in LUAD cell lines (PC9 and A549) and LUAD tissues compared to normal lung epithelial cells (BeAS-2B) and noncancerous tissues, respectively. miR-3648 promotes tumor cell progression by suppressing the expression of cytokine signaling 2 (SOCS2), a member of the SOCS family that negatively regulates cytokine receptor signaling. Therefore, miR-3648 could be considered a potential target for LUAD immunotherapy (Tu and Mei, 2022). Similarly, miR-875 was overexpressed in NSCLC tumor tissues compared to normal adjacent tissues. It regulates tumor proliferation and apoptosis by targeting SOCS2, indicating its potential as a therapeutic target (Tong et al., 2019). Studies have also shown higher expression of miR-20a and lower expression of miR-145 in NSCLC serum compared to controls, suggesting them as potential biomarkers. Furthermore, a weak to moderate association between the concentrations of TGF-β and VEGF, and miR-20a and miR-223 has been reported, indicating them as potential targets for lung cancer treatment (Chaniad et al., 2020). In patients with SCC, miR-9-3p negatively regulates the expression of IL-17A. IL-17 is a cytokine with both antitumor and protumor effects; in the case of SCC, miR-9-3p modulates the expression of IL-17A as an antagonist of IL-17A mRNA, which is crucial in SCC development (Migdalska-Sęk et al., 2020). Lower expression of miR-195-5p has been reported in NSCLC tissues and cell lines and is associated with poor survival in patients with NSCLC, suggesting its role as a prognostic marker (Zheng et al., 2019). Previous studies have shown an association between miR-200 family members and decreased overall survival in NSCLC patients, particularly in those with LUAD. Further investigation revealed that miR-200a-3p increased the expression of CX3CR1, leading to the induction of epithelial-to-mesenchymal transition (EMT) and distant metastasis. This finding suggests that the miR-chemokine receptor axis could be considered a therapeutic target in patients with LUAD. The expression of CX3CR1 and CXCR1 chemokine receptors is correlated with the infiltration of M2 macrophages and NKT cells, indicating their potential role in regulating immune responses in the TME of LUAD (Sharma et al., 2023). TGF-β contributes to tumor progression by inducing EMT. LINC00273, a long noncoding RNA, liberates Zinc finger E-box binding homeobox 1 (ZEB1) from miR-200a-3p, promoting TGF-β-induced lung tumor metastasis, suggesting LINC00273 as a target for preventing lung tumor metastasis (Sarkar et al., 2020). IL-13 has been implicated in promoting lung cancer progression by increasing the expression of the Ying Yang 1 (YY1) transcription factor through the activation of the PI3K/AKT pathway in A549 cells, leading to enhanced migration and proliferation of these cells. The pro-tumorigenic effects of IL-13 are inhibited by miR-29a by targeting YY1. However, further in vitro and in vivo studies are required to validate and better understand the therapeutic potential of miR-29a in lung cancer (Zhang et al., 2018).
Environmental factors play a significant role in modulating cytokine secretion, thus supporting the progression of cancer. For instance, prolonged exposure to particulate matter 2.5 (PM2.5), a crucial component of air pollution, results in the upregulation of transmembrane serine protease 2 (TMPRSS2) in lung cancer cells. This, in turn, fascilitates inflammasome formation and increases the secretion of IL-18, a cytokine associated with increased cell proliferation, pro-inflammatory responses, and cancer development (Wang et al., 2023).
In conclusion, the type of lung cancer, the ethnicity of the study population, and the sample size may have contributed to the diverse findings related to cytokine polymorphisms. However, analyzing high-throughput data using GWAS can result in more reliable outcomes, enabling the identification of polymorphisms that can be used as prognostic markers or potential targets for lung cancer treatment. Despite the need for further analysis to better understand the association between different miRNAs and cytokine or chemokine expression, more efforts should be directed towards identifying specific sets of miRNAs that can be used as predictive indicators or potential targets in lung cancer therapy.
In a study by Li et al., a gene signature consisting of pairs of genes related to B cells was constructed by analyzing scRNA-seq data from patients with NSCLC. This gene signature has proven to be a strong prognostic factor for patients with NSCLC, showing a significant correlation with immune scores, tumor purity, clinicopathological characteristics, and various tumor-infiltrating immune cells. Furthermore, this signature has shown promising results as a potential biomarker for predicting the efficacy of immunotherapy in patients with NSCLC (Li X. et al., 2022). Moreover, the samples of 11 patients with NSCLC were categorized into B-cell rich and B-cell poor groups using RNA-seq data. B-cell rich samples exhibited elevated levels of the CXCL13 transcripts, a recognized B-cell chemoattractant, and PD1+ CD8 T cells (Raju Paul et al., 2023). CXCL13 and PD-1+ CD8 T cells play key roles in the formation of tertiary lymphoid structures, which have been identified in several cancers and are typically associated with favorable outcomes (Sautès-Fridman et al., 2019; Workel et al., 2019). B cells, which contribute to and regulate these structures, were significantly correlated with tertiary lymphoid structures in NSCLC samples. Moreover, RNA-seq data of patients with LUAD who underwent surgery, chemotherapy, or radiation revealed that pre-treatment percentage of intratumoral B cells correlated with improved overall survival in all stages of LUAD (Raju Paul et al., 2023). Transcriptomic analysis has also revealed that the presence of B and plasma cells in the TME plays a crucial role in determining the overall survival of patients with NSCLC treated with atezolizumab. B cells and plasma cells are also associated with the presence of tertiary lymphoid structures (Patil et al., 2022). Another study using RNA-seq data extracted from the TCGA database showed that patients with high B cell infiltration in LUAD had longer survival compared to those with low infiltration. However, the presence of PD-L1 may impede the survival advantage among patients with high B cell infiltration (Ho et al., 2018). Future research could focus on exploring the association between B cell receptor (BCR) genetics and lung cancer, as no investigations have been conducted in this area so far.
CD4+ T cells are the most common T cell population in adenocarcinoma and SCC, followed by CD8+ cells. A small portion of TILs consists of double negative T cells, with the CD45RA−CD45RO+ memory/effector phenotype being the most prevalent among T cells (Stankovic et al., 2018). Genetic alterations can significantly modify T cell immune responses, affecting the clinical outcomes of lung cancer patients. Effective immune responses to TILs depend on T cell receptor (TCR)-mediated recognition of mutation-derived neoantigens. Therefore, exploring the TCR repertoire has the potential to predict responses to immune checkpoint blockade and survival in cancer patients. A cold and heterogeneous TCR repertoire is associated with poor prognosis in patients with localized NSCLC. To investigate this correlation, a study conducted TCR sequencing of 45 tumor regions from 11 patients with localized NSCLC and discovered extensive intratumor heterogeneity (ITH) in the TCR repertoire in terms of T cell density and clonality, which are factors related to T cell response and expansion. The observed spatial differences in the TCR repertoire were attributed to the distinct neoantigens in different tumor regions (Reuben et al., 2017; Reuben et al., 2020). High molecular genomic ITH, defined as the presence of various cancer cells and stromal cells with diverse molecular and phenotypic characteristics within the same tumor, has been associated with a higher risk of relapse, lower response to treatment, and increased tumor aggressiveness (Zhang et al., 2014; McGranahan et al., 2016; Stewart et al., 2020). Higher ITH in T cell clonality has also been linked to an increased risk of postsurgical relapse. These mutations, occurring in different regions of a tumor, can result in the differential expression of neoantigens, leading to variations in immunogenicity and the ability of different tumor regions to induce an anti-tumor T cell response. Previous studies have indicated that tumors with high clonal neoantigen burden and low neoantigen ITH are associated with a significantly longer progression-free survival (McGranahan et al., 2016). These findings emphasize the significance of taking into account the quantity and heterogeneity of neoantigens when predicting the prognosis and clinical outcomes of patients with lung cancer. T cell heterogeneity may result from heterogeneity in the architecture of tumors, as well as their vasculature and lymphatics, which in turn can affect the ability of T cells to infiltrate different regions of a tumor. Therefore, analyzing the textural features of patients’ tumors, possibly through noninvasive methods, to detect ITH in the mutational landscape or T cell repertoire could provide valuable insights for predicting how patients will respond to therapies (Reuben et al., 2017). Higher TMB has also been associated with increased T cell clonality, a lower CD4:CD8 ratio, higher levels of granzyme B, higher immunogenicity, and the induction of T cell responses through the creation of neoantigens. Moreover, higher homology in the T cell repertoire between the tumor and uninvolved tumor-adjacent lung in patients with NSCLC is linked to lower survival, likely due to the decreased tumor-focused T cell responses. When performing T cell expansion for therapeutic purposes, it is crucial to consider the overlap of potentially reactive T cells between uninvolved tumor-adjacent lungs and tumor tissues in patients with NSCLC. This approach is essential to avoid non-specific anti-tumor responses and minimize the risk of immune-related adverse events associated with the expansion of T cell subsets unrelated to tumors. It should also be taken into account when developing and implementing TIL-based therapies (Reuben et al., 2020). In case of SCLC, whole-exome sequencing of the TCR was conducted on 50 tumor samples from 19 resected limited-stage SCLC tumors (LS-SCLCs). The results revealed a high variation in TMB and copy number aberration (CNA) burden between patients, while it was similar between different regions within the same tumor. SCLC tumors also demonstrate lower TCR metrics, including diversity, density, and clonality, than tumor-adjacent lung tissues, indicating reduced infiltration, proliferation, and diversity of T cells in tumor tissues. A high CNA burden is negatively associated with the quality and quantity of T cells, probably contributing to a cold TCR repertoire in SCLC and the cold TME. Collectively, higher levels of TCR ITH have been identified in SCLC tumors than in NSCLC, potentially compromising the efficacy of anti-tumor immune responses (Chen et al., 2021). Additionally, copy-number loss of IFN-γ pathway genes has been reported in SCLC samples, positively associated with CNA burden This suggests a potential mechanism underlying immune evasion in SCLC tumors. This phenomenon may be attributed to dual inactivation of TP53 and RB1, chromosome instability, and ultimately the loss of essential immune related genes such as IFN-γ pathway genes This allows SCLC tumors to evade the anti-tumor immune system. Similar to NSCLC, a higher TMB was associated with longer overall survival, while a higher CNA burden was linked to lower overall survival. Furthermore, patients with a more heterogeneous TCR showed a significantly shorter overall survival. In summary, a cold and heterogeneous TCR repertoire may be one of the factors leading to a lower response to immunotherapy in patients with SCLC. Therefore, overcoming the cold and heterogeneous intratumor T cell repertoire may help enhance the efficacy of immunotherapies in patients with SCLC (Chen et al., 2021).
Further research has also been conducted to clarify the correlation between immune system-related genes and T cell response against lung cancer. A study of 2450 SNPs in T cell cancer immune response-related genes in 941 patients with early-stage NSCLC revealed that rs1964986 and rs1573618 SNPs in the TCR beta chain (TRB) were associated with early-stage NSCLC recurrence. The rs10108662 SNP in indoleamine-2,3-dioxygenase 1 (IDO1) was associated with a higher risk of death, potentially affecting NSCLC patient outcomes by modulating the expression of IDO and T cell cytotoxicity. Additionally, rs959260 and rs4789182 SNPs in the growth factor receptor-bound protein 2 (GRB2) gene were associated with survival, likely due to their role in regulating immune cell development and T cell functions. The rs8080546 SNP in the proteasome 26S subunit, non-ATPase 3 (PSMD3), was associated with a higher risk of death in patients who only underwent surgery, possibly because of increased MHC-1 peptide presentation, leading to T cell exhaustion and impaired T cell cytotoxicity (Wang Q. et al., 2020).
RAS guanyl-releasing protein 2 (RASGRP2), one of the guanine nucleotide exchange factors (GEFs), has been implicated in chemokine and antigen-receptor/cytokine-cytokine receptor signaling pathways, T cell activation, and lymphocyte differentiation in LUAD. RASGRP2 expression has been lower in LUAD tissues than in non-tumoral tissues, and lower levels of RASGRP2 correlated with a poor prognosis Studies have also indicated a positive association between the expression of RASGRP2 and the infiltration of immune cells, including cytotoxic T cells, Th1 cells, and B cells, and a negative association with the infiltration of Th2 cells. Positive correlations have also been reported between RASGRP2 expression and certain immunoinhibitors (such as CTLA-4), immunostimulators (such as TNFRSF13B), chemokines (such as CCL14/17/19), and chemokine receptors (such as CCR6/7 and CXCR5). Immunofluorescence experiments have suggested a potential association between higher levels of RASGRP2 and PD-L1 expression. This implyies that patients with increased RASGRP2 expression may benefit more from immunotherapy. The effects of RASGRP2 may occur through the JAK3-STAT5 signaling pathway, although comprehensive animal experiments and clinical trials are necessary for confirmation (Liu et al., 2023). A recent study found that Foxp3 expression is higher in NSCLC tissues compared to normal tissues. Immunohistochemical (IHC) analysis showed that Foxp3 is mainly expressed by Tregs rather than in cancer tissues. The increased expression of Foxp3 is associated with worse overall survival, indicating that Tregs could be a possible target for therapy in patients with NSCLC (Zhu et al., 2022).
Moreover, polymorphisms observed at several T cell immune checkpoints, such as B- and T-lymphocyte attenuator (BTLA), CTLA-4, and PD-1, which are known for their immunosuppressive roles, have been implicated in modulating susceptibility to lung cancer (Wang J. et al., 2021; Andrzejczak et al., 2022). Table 2 illustrates the identified polymorphisms in T cell immune checkpoints and their association with an increased or decreased risk of lung cancer.
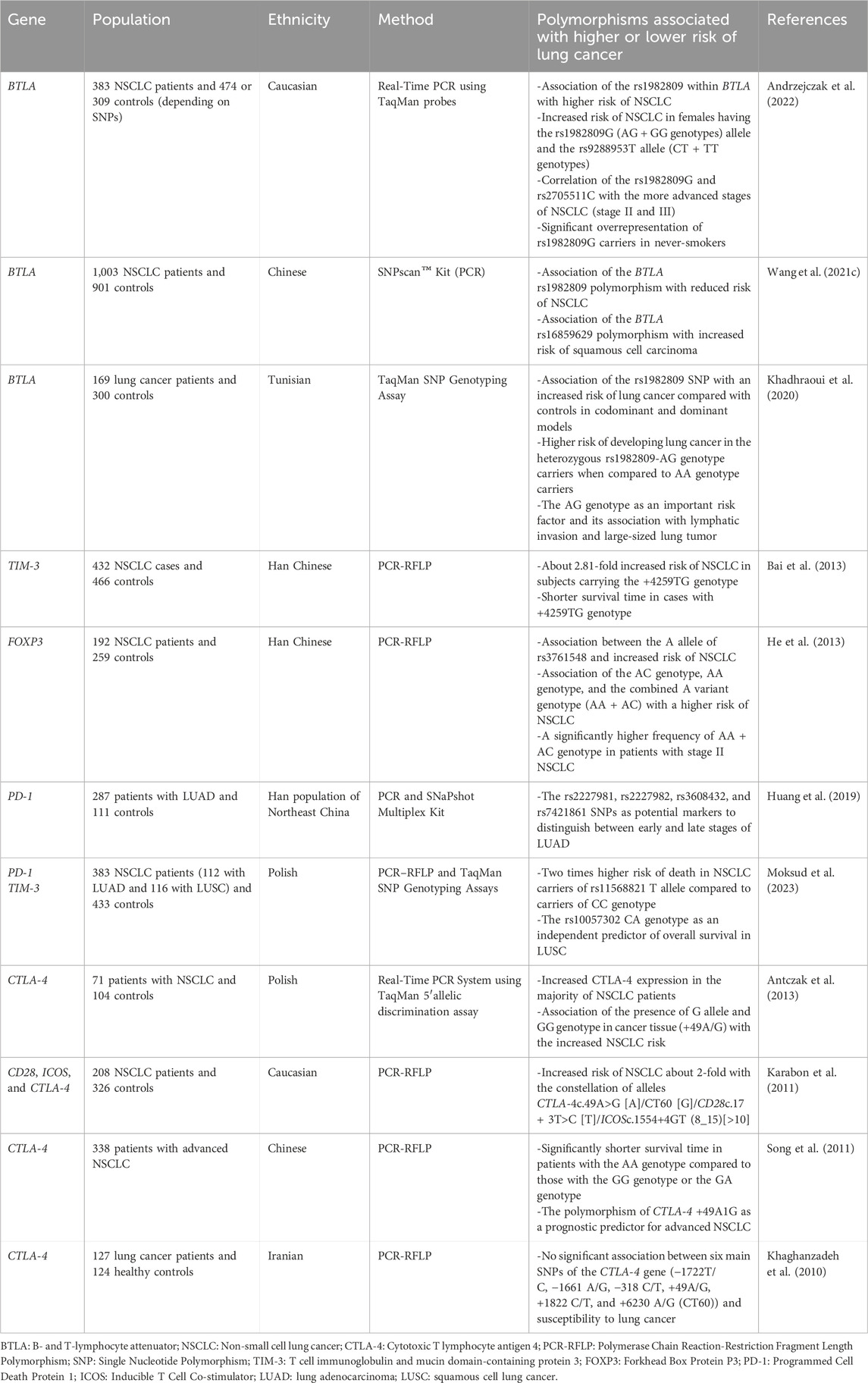
Table 2. Identified polymorphisms in immune checkpoints and FOXP3 associated with lung cancer development or suppression.
PD-L1 polymorphisms have also been associated with the susceptibility to lung cancer. A study of 288 patients with NSCLC and 300 controls found a higher frequency of the PD-L1 8923A/C polymorphism in NSCLC patients compared to controls. These patients also had elevated levels of soluble PD-L1, which were higher in LUAD patients than in those with SCC (Cheng et al., 2015b). Moreover, the CC genotype of rs4143815 and the GG genotype of rs4742098 SNPs in PD-L1 were associated with a nearly two-fold higher risk of squamous cell lung cancer (LUSC). This polymorphism likely affects the interaction between hsa-mir-570 and CD274 mRNA (Wang et al., 2013; Moksud et al., 2023). Another study of PD-L1 polymorphisms was conducted on 126 Egyptian patients with lung carcinoma and 117 healthy controls. The results showed an association between rs2297136 and rs4143815 variants with a higher and lower risk of lung cancer, respectively, suggesting they could be potential biomarkers for lung cancer (Sakran et al., 2023).
Nivolumab, an FDA-approved IgG4 anti-PD-1 monoclonal antibody for NSCLC, is used after platinum-based chemotherapy for patients with high or low PD-L1 expression. Pembrolizumab, another IgG4 anti-PD-1 monoclonal antibody, is used to treat metastatic NSCLC with over 50% PD-L1 expression, but without EGFR or ALK mutations. If PD-L1 expression is less than 50%, it should be combined with other treatments. Atezolizumab, an IgG1 antibody against PD-L1, is suitable for patients with metastatic NSCLC expressing EGFR or ALK mutations, administered during or after platinum-based chemotherapy (Lindeman et al., 2013; Ramos-Esquivel et al., 2017; Siddiqui and Siddiqui, 2023). Nivolumab (IgG4 anti-PD-1) in combination with ipilimumab (fully humanized IgG1 anti-CTLA4) and minimal chemotherapy (only two cycles of platinum-based) has shown efficacy comparable to full-dose chemotherapy in patients with metastatic NSCLC (Shalata et al., 2023). Immunotherapies using anti-PD-1, anti-PD-L1, and anti-CTLA-4 monoclonal antibodies have revolutionized lung cancer treatment; however, only a few advanced-stage patients respond. The high degree of heterogeneity among tumor types and genomic alterations makes it difficult to predict which patients will benefit from immunotherapy (Wang et al., 2022b). Several factors, including mismatch repair deficiency (dMMR)/microsatellite instability (MSI), TMB, tumor PD-L1 expression, and host-specific immunity characteristics, such as the microbiome or germline genetics of the host, can affect the response to immune checkpoint inhibitor-based therapy by determining the immunogenicity of tumors (Le et al., 2015; Ott et al., 2019; Baruch et al., 2021; Doroshow et al., 2021; Sayaman et al., 2021). Additionally, treatment with anti-PD1 blocking antibodies may result in toxicities, including dermatitis, hypothyroidism, colitis, and pneumonitis. Studies aimed at identifying patients more susceptible to such adverse effects could help in selecting personalized therapies. In this regard, Bins et al. investigated whether patients with SNPs in PD-1 and PD-L1-related genes experience severe toxicity with anti-PD1 blocking antibodies. They concluded that these SNPs are unlikely to have clinical implications in predicting nivolumab toxicity in patients with NSCLC (Bins et al., 2018).
Several studies have investigated the mechanisms underlying resistance to immune checkpoint inhibitors. For example, the KIR3DS1 genetic variant of KIR is significantly associated with resistance to PD-1 blockade and progression-free survival in patients with NSCLC, suggesting a potential role for NK cells in responding to PD-1 immunotherapy (Trefny et al., 2019). Lower serum levels of granzyme B and homozygous and heterozygous variants of GZMB rs8192917 are associated with poor prognosis and worse clinical outcomes with PD-1 blockade in patients with NSCLC, highlighting the importance of considering T cell response-related mutations in predicting NSCLC prognosis and response to immunotherapy (Hurkmans et al., 2020). TMB, which is associated with the number of tumor-derived neoantigens, is a significant determinant of a favorable response to immune checkpoint inhibitors. These neoantigens are subsequently presented by MHC molecules, activating T cell anti-tumor responses (McGranahan et al., 2016; Samstein et al., 2019). In addition to TMB, heterozygosity in the repertoire of patient-specific antigen-presenting HLA-I molecules also affects the response to immune checkpoint blockade (ICB) by influencing the selection and clonal expansion of T cells reactive against tumor-derived neoantigens after ICB treatment. Heterozygous HLA-I genotypes facilitate the presentation of a diverse range of tumor-derived antigens to T cells. In this regard, homozygosity at one HLA-I locus (A, B, or C) and low TMB were associated with a significant reduction in overall survival compared to patients with heterozygosity at each HLA-I locus and high TMB. These factors should be considered in future clinical trials (Chowell et al., 2018). Recent studies have also demonstrated that patients with high HLA-I evolutionary divergence (HED), as measured by the sequence divergence between alleles of the HLA-I genotype, are likely to be more responsive to ICB therapies. This factor can be determined by DNA sequencing of normal tissues, as opposed to TMB, which is challenging to evaluate because of tumor purity and clonal fraction. All of these factors, such as TMB, HED, and TCR repertoire, can predict T cell-mediated tumor control and should be considered in future clinical investigations (Chowell et al., 2019). Considering different aspects of the interaction between the tumor and TME is important for predicting the response to immunotherapy. A study involving different tumor regions from 88 patients with early-stage untreated NSCLC demonstrated that the immune system of these patients imposes strong selection pressure during tumor evolution, contributing to immune evasion and poor disease-free survival (Rosenthal et al., 2019). Immunoediting mechanisms may occur by decreasing the expression of neoantigens and HLA LOH, enabling cancer cells to escape immune recognition by deleting HLA alleles, thereby suppressing neoantigen presentation (Pyke et al., 2022) or promoter hypermethylation of genes encoding neoantigens. Therapeutic interventions should aim to mitigate or inhibit immune evasion mechanisms (Rosenthal et al., 2019).
Studies have suggested that patients with NSCLC lacking driver oncogenes may demonstrate a more favorable response to immune checkpoint inhibitor therapy than patients with oncogene-addicted NSCLC, particularly those with EGFR/ALK variations (Lisberg et al., 2018; Addeo et al., 2021). However, whether the immunogenicity and tumor immune microenvironment (TIME) of HER2-amplified LUAD are related to its response to ICI therapy remains unclear. HER2 amplification and mutations occur in approximately 2%–10% of NSCLCs, and effective treatment for this subset of LUAD has not been thoroughly investigated. RNA sequencing analysis indicated significantly higher PD-L1 expression at the mRNA and protein levels in patients with HER2-amplified LUAD than in those with breast invasive carcinoma (BRCA) and stomach adenocarcinoma (STAD). Patients with HER2-amplified LUAD also exhibited higher levels of tissue/blood TMB than the other two groups (Wang et al., 2022b). Moreover, the HER2-amplified LUAD group displayed an inflamed TIME characterized by higher levels of genes expressing classical MHC class I/II antigens, TAP1 and B2M (antigen processing machinery proteins), CD3, co-stimulatory molecules (such as CD28 and ICOS), cytotoxic effect-related genes (CD8A, IFNG, and GZMA), increased infiltration of CD4+ and CD8+ T cells, and M1 MQ, while showing lower infiltration of Tregs. IHC analysis further confirmed increased CD8+ TIL density and decreased FOXP3+ TIL densities in HER2-amplified LUAD specimens. In summary, patients with HER2-amplified LUAD exhibited higher immunogenicity, suggesting that they may benefit from ICI therapy. These patients also demonstrated a more favorable TIME, potentially making them better recognized by the immune system, thereby improving therapeutic efficacy in cancer immunotherapy. However, these studies were limited by the low frequency of HER2 amplification and mutations in patients with NSCLC; therefore, more comprehensive research is required to validate these observations (Wang et al., 2022b).
Some studies have suggested that the effectiveness of immunotherapy may be associated with the expression level of PD-1. In silico studies have categorized patients with lung squamous cell carcinoma into high and low PD-1 expression groups. Significant differences were observed in immune regulation pathways, particularly those associated with T cell activity. In the PD-1 high expression group, the most upregulated pathways, such as the MHC protein complex, were related to the immune system. TMB was high in both high and low PD-1 expression groups; however, no significant difference was observed between them. Owing to the lack of sequencing data for patients undergoing immunotherapy, determining whether these differences were related to the effectiveness of immunotherapy was impossible. However, differential expression of these genes may be a potential predictor of immunotherapy efficacy (Hu et al., 2020).
In summary, investigating the immunogenetic features of lung cancer may help identify potential candidates who can receive the maximum benefit from immunotherapy. Table 3 summarizes the PD-1, PD-L1, and CTLA-4 polymorphisms associated with response to immunotherapy in patients with lung cancer.
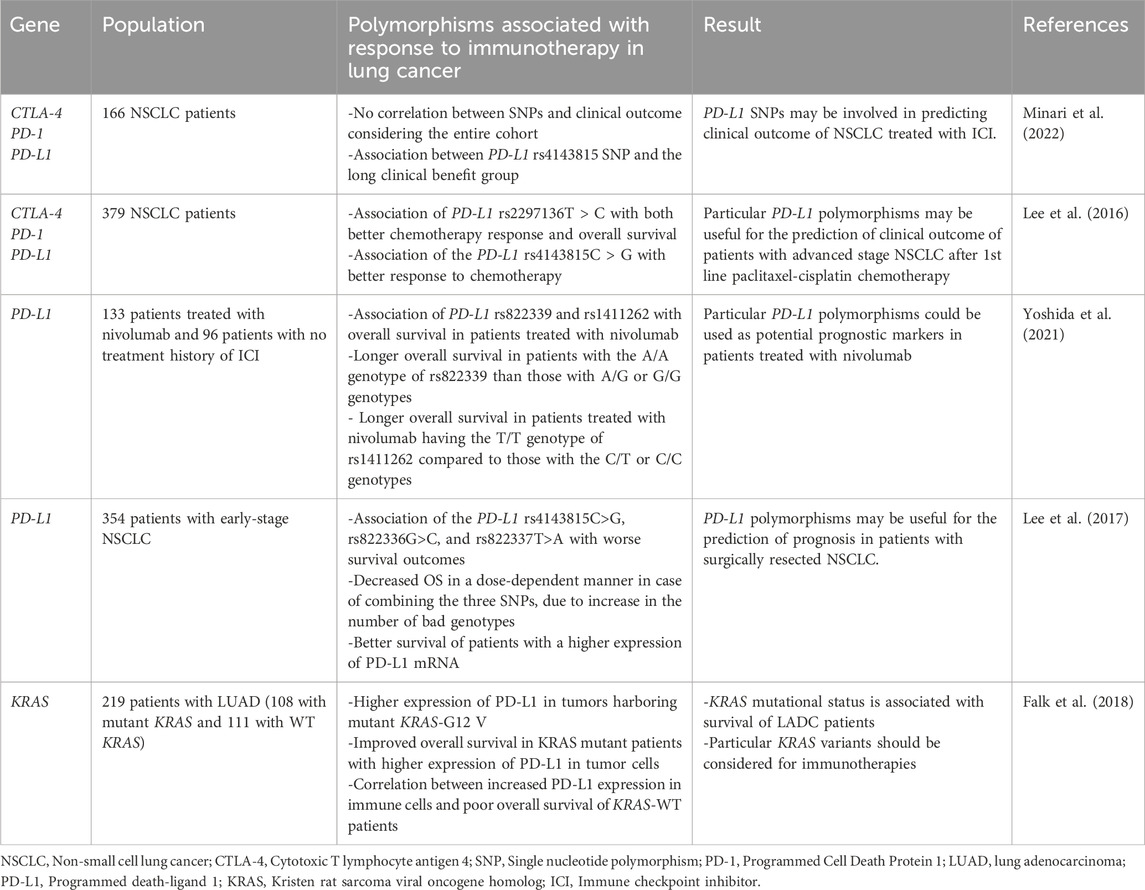
Table 3. Identified polymorphisms in PD-1, PD-L1, and CTLA-4 genes associated with clinical outcome of lung cancer patients.
In conclusion, investigating the cellular and molecular backgrounds of immune cells infiltrating the lung TME can help predict the development and progression of lung cancer. Numerous efforts have been directed towards recognizing the genomic landscape to identify patients who are most responsive to specific immunotherapies. This holds the potential for early intervention and reducing the morbidity and mortality rates. scRNA-seq has emerged as a powerful technique that enables a deeper understanding of immune cell heterogeneity through transcriptomic analysis rather than surface markers, which are widely used in routine flow cytometry methods [9]. Future studies should prioritize large-scale genetic polymorphism analyses of different immune system compartments to determine their association with lung cancer progression, which could facilitate the development of individualized therapies. Given the significant variability in genotype frequencies and allele distributions among populations, more studies are required to identify population-specific and general variants associated with risk, clinical outcomes, and response to lung cancer treatment.
MR-Z: Data curation, Writing–original draft. AK: Resources, Writing–review and editing. KR: Writing–review and editing. AG: Writing–review and editing, Resources, Supervision. RR: Supervision, Writing–review and editing, Conceptualization, Methodology.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Addeo, A., Passaro, A., Malapelle, U., Banna, G. L., Subbiah, V., and Friedlaender, A. (2021). Immunotherapy in non-small cell lung cancer harbouring driver mutations. Cancer Treat. Rev. 96, 102179. doi:10.1016/j.ctrv.2021.102179
Ahirwar, D. K., Peng, B., Charan, M., Misri, S., Mishra, S., Kaul, K., et al. (2023). Slit2/Robo1 signaling inhibits small-cell lung cancer by targeting β-catenin signaling in tumor cells and macrophages. Mol. Oncol. 17 (5), 839–856. doi:10.1002/1878-0261.13289
Akbay, E. A., Koyama, S., Liu, Y., Dries, R., Bufe, L. E., Silkes, M., et al. (2017). Interleukin-17A promotes lung tumor progression through neutrophil attraction to tumor sites and mediating resistance to PD-1 blockade. J. Thorac. Oncol. 12 (8), 1268–1279. doi:10.1016/j.jtho.2017.04.017
Alberg, A. J., and Samet, J. M. (2003). Epidemiology of lung cancer. Chest 123 (1 Suppl. l), 21S–49s. doi:10.1378/chest.123.1_suppl.21s
Aloe, C., Wang, H., Vlahos, R., Irving, L., Steinfort, D., and Bozinovski, S. (2021). Emerging and multifaceted role of neutrophils in lung cancer. Transl. Lung Cancer Res. 10 (6), 2806–2818. doi:10.21037/tlcr-20-760
Andrzejczak, A., Partyka, A., Wiśniewski, A., Porębska, I., Pawełczyk, K., Ptaszkowski, K., et al. (2022). The association of BTLA gene polymorphisms with non-small lung cancer risk in smokers and never-smokers. Front. Immunol. 13, 1006639. doi:10.3389/fimmu.2022.1006639
Antczak, A., Pastuszak-Lewandoska, D., Górski, P., Domańska, D., Migdalska-Sęk, M., Czarnecka, K., et al. (2013). Ctla-4 expression and polymorphisms in lung tissue of patients with diagnosed non-small-cell lung cancer. Biomed. Res. Int. 2013, 576486. doi:10.1155/2013/576486
Arora, S., Singh, P., Ahmad, S., Ahmad, T., Dohare, R., Almatroodi, S. A., et al. (2021). Comprehensive integrative analysis reveals the association of KLF4 with macrophage infiltration and polarization in lung cancer microenvironment. Cells 10 (8), 2091. doi:10.3390/cells10082091
Bai, J., Li, X., Tong, D., Shi, W., Song, H., and Li, Q. (2013). T-cell immunoglobulin- and mucin-domain-containing molecule 3 gene polymorphisms and prognosis of non-small-cell lung cancer. Tumour Biol. 34 (2), 805–809. doi:10.1007/s13277-012-0610-1
Baranasic, J., Niazi, Y., Chattopadhyay, S., Rumora, L., Ćorak, L., Dugac, A. V., et al. (2023). Germline variants of the genes involved in NF-kB activation are associated with the risk of COPD and lung cancer development. Acta Pharm. 73 (2), 243–256. doi:10.2478/acph-2023-0019
Baruch, E. N., Youngster, I., Ben-Betzalel, G., Ortenberg, R., Lahat, A., Katz, L., et al. (2021). Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 371 (6529), 602–609. doi:10.1126/science.abb5920
Berland, L., Heeke, S., Humbert, O., Macocco, A., Long-Mira, E., Lassalle, S., et al. (2019). Current views on tumor mutational burden in patients with non-small cell lung cancer treated by immune checkpoint inhibitors. J. Thorac. Dis. 11 (Suppl. 1), S71–s80. doi:10.21037/jtd.2018.11.102
Bins, S., Basak, E. A., El Bouazzaoui, S., Koolen, S. L. W., Oomen-de Hoop, E., van der Leest, C. H., et al. (2018). Association between single-nucleotide polymorphisms and adverse events in nivolumab-treated non-small cell lung cancer patients. Br. J. Cancer 118 (10), 1296–1301. doi:10.1038/s41416-018-0074-1
Bittner, S., Knoll, G., Füllsack, S., Kurz, M., Wajant, H., and Ehrenschwender, M. (2016). Soluble TL1A is sufficient for activation of death receptor 3. Febs J. 283 (2), 323–336. doi:10.1111/febs.13576
Brownlie, D., von Kries, A., Valenzano, G., Wild, N., Yilmaz, E., Säfholm, J., et al. (2023). Accumulation of tissue-resident natural killer cells, innate lymphoid cells, and CD8(+) T cells towards the center of human lung tumors. Oncoimmunology 12 (1), 2233402. doi:10.1080/2162402X.2023.2233402
Candido, J. B., Morton, J. P., Bailey, P., Campbell, A. D., Karim, S. A., Jamieson, T., et al. (2018). CSF1R(+) macrophages sustain pancreatic tumor growth through T cell suppression and maintenance of key gene programs that define the squamous subtype. Cell Rep. 23 (5), 1448–1460. doi:10.1016/j.celrep.2018.03.131
Cao, Q., Wang, N., Ren, L., Tian, J., Yang, S., and Cheng, H. (2020). miR-125a-5p post-transcriptionally suppresses GALNT7 to inhibit proliferation and invasion in cervical cancer cells via the EGFR/PI3K/AKT pathway. Cancer Cell Int. 20, 117. doi:10.1186/s12935-020-01209-8
Caronni, N., Piperno, G. M., Simoncello, F., Romano, O., Vodret, S., Yanagihashi, Y., et al. (2021). TIM4 expression by dendritic cells mediates uptake of tumor-associated antigens and anti-tumor responses. Nat. Commun. 12 (1), 2237. doi:10.1038/s41467-021-22535-z
Carrega, P., Loiacono, F., Di Carlo, E., Scaramuccia, A., Mora, M., Conte, R., et al. (2015). NCR(+)ILC3 concentrate in human lung cancer and associate with intratumoral lymphoid structures. Nat. Commun. 6, 8280. doi:10.1038/ncomms9280
Casetti, R., Agrati, C., Wallace, M., Sacchi, A., Martini, F., Martino, A., et al. (2009). Cutting edge: TGF-beta1 and IL-15 Induce FOXP3+ gammadelta regulatory T cells in the presence of antigen stimulation. J. Immunol. 183 (6), 3574–3577. doi:10.4049/jimmunol.0901334
Castellanos, M. R., Fanous, E., Thaker, R., Flory, M. J., Seetharamu, N., Dhar, M., et al. (2023). Expression patterns and clinical significance of estrogen receptor in non-small cell lung cancer. Pathol. Res. Pract. 241, 154298. doi:10.1016/j.prp.2022.154298
Chae, Y. K., Anker, J. F., Bais, P., Namburi, S., Giles, F. J., and Chuang, J. H. (2018). Mutations in DNA repair genes are associated with increased neo-antigen load and activated T cell infiltration in lung adenocarcinoma. Oncotarget 9 (8), 7949–7960. doi:10.18632/oncotarget.23742
Chalmers, Z. R., Connelly, C. F., Fabrizio, D., Gay, L., Ali, S. M., Ennis, R., et al. (2017). Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 9 (1), 34. doi:10.1186/s13073-017-0424-2
Chaniad, P., Trakunran, K., Geater, S. L., Keeratichananont, W., Thongsuksai, P., and Raungrut, P. (2020). Serum miRNAs associated with tumor-promoting cytokines in non-small cell lung cancer. PLoS One 15 (10), e0241593. doi:10.1371/journal.pone.0241593
Chen, J., Yang, H., Teo, A. S. M., Amer, L. B., Sherbaf, F. G., Tan, C. Q., et al. (2020). Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 52 (2), 177–186. doi:10.1038/s41588-019-0569-6
Chen, M., Chen, R., Jin, Y., Li, J., Hu, X., Zhang, J., et al. (2021). Cold and heterogeneous T cell repertoire is associated with copy number aberrations and loss of immune genes in small-cell lung cancer. Nat. Commun. 12 (1), 6655. doi:10.1038/s41467-021-26821-8
Cheng, E. S., Weber, M., Steinberg, J., and Yu, X. Q. (2021). Lung cancer risk in never-smokers: an overview of environmental and genetic factors. Chin. J. Cancer Res. 33 (5), 548–562. doi:10.21147/j.issn.1000-9604.2021.05.02
Cheng, S., Shao, Z., Liu, X., Guo, L., Zhang, X., Na, Q., et al. (2015a). Interleukin 17A polymorphism elevates gene expression and is associated with increased risk of nonsmall cell lung cancer. DNA Cell Biol. 34 (1), 63–68. doi:10.1089/dna.2014.2628
Cheng, S., Zheng, J., Zhu, J., Xie, C., Zhang, X., Han, X., et al. (2015b). PD-L1 gene polymorphism and high level of plasma soluble PD-L1 protein may be associated with non-small cell lung cancer. Int. J. Biol. Markers 30 (4), e364–e368. doi:10.5301/jbm.5000170
Chevallier, M., Borgeaud, M., Addeo, A., and Friedlaender, A. (2021). Oncogenic driver mutations in non-small cell lung cancer: past, present and future. World J. Clin. Oncol. 12 (4), 217–237. doi:10.5306/wjco.v12.i4.217
Chowell, D., Krishna, C., Pierini, F., Makarov, V., Rizvi, N. A., Kuo, F., et al. (2019). Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat. Med. 25 (11), 1715–1720. doi:10.1038/s41591-019-0639-4
Chowell, D., Morris, L. G. T., Grigg, C. M., Weber, J. K., Samstein, R. M., Makarov, V., et al. (2018). Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359 (6375), 582–587. doi:10.1126/science.aao4572
Crosby, C. M., and Kronenberg, M. (2018). Tissue-specific functions of invariant natural killer T cells. Nat. Rev. Immunol. 18 (9), 559–574. doi:10.1038/s41577-018-0034-2
Culley, F. J. (2009). Natural killer cells in infection and inflammation of the lung. Immunology 128 (2), 151–163. doi:10.1111/j.1365-2567.2009.03167.x
Datar, I., Sanmamed, M. F., Wang, J., Henick, B. S., Choi, J., Badri, T., et al. (2019). Expression analysis and significance of PD-1, LAG-3, and TIM-3 in human non-small cell lung cancer using spatially resolved and multiparametric single-cell analysis. Clin. Cancer Res. 25 (15), 4663–4673. doi:10.1158/1078-0432.CCR-18-4142
Davey, M. S., Willcox, C. R., Joyce, S. P., Ladell, K., Kasatskaya, S. A., McLaren, J. E., et al. (2017). Clonal selection in the human Vδ1 T cell repertoire indicates γδ TCR-dependent adaptive immune surveillance. Nat. Commun. 8, 14760. doi:10.1038/ncomms14760
De Grove, K. C., Provoost, S., Verhamme, F. M., Bracke, K. R., Joos, G. F., Maes, T., et al. (2016). Characterization and quantification of innate lymphoid cell subsets in human lung. PLoS One 11 (1), e0145961. doi:10.1371/journal.pone.0145961
Dexheimer, P. J., and Cochella, L. (2020). MicroRNAs: from mechanism to organism. Front. Cell Dev. Biol. 8, 409. doi:10.3389/fcell.2020.00409
Djaoud, Z., and Parham, P. (2020). HLAs, TCRs, and KIRs, a triumvirate of human cell-mediated immunity. Annu. Rev. Biochem. 89, 717–739. doi:10.1146/annurev-biochem-011520-102754
Doroshow, D. B., Bhalla, S., Beasley, M. B., Sholl, L. M., Kerr, K. M., Gnjatic, S., et al. (2021). PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 18 (6), 345–362. doi:10.1038/s41571-021-00473-5
Dumitriu, I. E., Dunbar, D. R., Howie, S. E., Sethi, T., and Gregory, C. D. (2009). Human dendritic cells produce TGF-beta 1 under the influence of lung carcinoma cells and prime the differentiation of CD4+CD25+Foxp3+ regulatory T cells. J. Immunol. 182 (5), 2795–2807. doi:10.4049/jimmunol.0712671
Falk, A. T., Yazbeck, N., Guibert, N., Chamorey, E., Paquet, A., Ribeyre, L., et al. (2018). Effect of mutant variants of the KRAS gene on PD-L1 expression and on the immune microenvironment and association with clinical outcome in lung adenocarcinoma patients. Lung Cancer 121, 70–75. doi:10.1016/j.lungcan.2018.05.009
Gao, F., Han, J., Jia, L., He, J., Wang, Y., Chen, M., et al. (2023). MiR-30c facilitates natural killer cell cytotoxicity to lung cancer through targeting GALNT7. Genes Genomics 45 (2), 247–260. doi:10.1007/s13258-022-01306-0
Gao, H., Niu, Z., Zhang, Z., Wu, H., Xie, Y., Yang, Z., et al. (2019). TNFSF15 promoter polymorphisms increase the susceptibility to small cell lung cancer: a case-control study. BMC Med. Genet. 20 (1), 29. doi:10.1186/s12881-019-0762-6
Gao, Y., He, Y., Ding, J., Wu, K., Hu, B., Liu, Y., et al. (2009). An insertion/deletion polymorphism at miRNA-122-binding site in the interleukin-1alpha 3’ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis 30 (12), 2064–2069. doi:10.1093/carcin/bgp283
George Warren, W., Osborn, M., Yates, A., Wright, K., and O’Sullivan, S. E. (2023). The emerging role of fatty acid binding protein 5 (FABP5) in cancers. Drug Discov. Today 28 (7), 103628. doi:10.1016/j.drudis.2023.103628
Georgoudaki, A. M., Prokopec, K. E., Boura, V. F., Hellqvist, E., Sohn, S., Östling, J., et al. (2016). Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep. 15 (9), 2000–2011. doi:10.1016/j.celrep.2016.04.084
Guo, J., Jin, H., Xi, Y., Guo, J., Jin, Y., and Jiang, D. (2020). The miR-582/CD1B Axis is involved in regulation of dendritic cells and is associated with clinical outcomes in advanced lung adenocarcinoma. Biomed. Res. Int. 2020, 4360930. doi:10.1155/2020/4360930
Guo, Z., Song, J., Hao, J., Zhao, H., Du, X., Li, E., et al. (2019). M2 macrophages promote NSCLC metastasis by upregulating CRYAB. Cell Death Dis. 10 (6), 377. doi:10.1038/s41419-019-1618-x
Gyori, D., Lim, E. L., Grant, F. M., Spensberger, D., Roychoudhuri, R., Shuttleworth, S. J., et al. (2018). Compensation between CSF1R+ macrophages and Foxp3+ Treg cells drives resistance to tumor immunotherapy. JCI Insight. 3 (11), e120631. doi:10.1172/jci.insight.120631
He, D., Wang, D., Lu, P., Yang, N., Xue, Z., Zhu, X., et al. (2021). Single-cell RNA sequencing reveals heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations. Oncogene 40 (2), 355–368. doi:10.1038/s41388-020-01528-0
He, M., Yu, W., Chang, C., Miyamoto, H., Liu, X., Jiang, K., et al. (2020). Estrogen receptor α promotes lung cancer cell invasion via increase of and cross-talk with infiltrated macrophages through the CCL2/CCR2/MMP9 and CXCL12/CXCR4 signaling pathways. Mol. Oncol. 14 (8), 1779–1799. doi:10.1002/1878-0261.12701
He, W., Liu, Q., Wang, L., Chen, W., Li, N., and Cao, X. (2007). TLR4 signaling promotes immune escape of human lung cancer cells by inducing immunosuppressive cytokines and apoptosis resistance. Mol. Immunol. 44 (11), 2850–2859. doi:10.1016/j.molimm.2007.01.022
He, Y., Du, Y., Wei, S., Shi, J., Mei, Z., Qian, L., et al. (2017). IL-17A and IL-17F single nucleotide polymorphisms associated with lung cancer in Chinese population. Clin. Respir. J. 11 (2), 230–242. doi:10.1111/crj.12330
He, Y. Q., Bo, Q., Yong, W., Qiu, Z. X., Li, Y. L., and Li, W. M. (2013). FoxP3 genetic variants and risk of non-small cell lung cancer in the Chinese Han population. Gene 531 (2), 422–425. doi:10.1016/j.gene.2013.08.066
Hellmann, M. D., Callahan, M. K., Awad, M. M., Calvo, E., Ascierto, P. A., Atmaca, A., et al. (2018). Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell 33 (5), 853–861.e4. doi:10.1016/j.ccell.2018.04.001
Hematian, L. M., Ashouri, E., Barani, S., Ghayumi, S. M. A., Ghaderi, A., and Rajalingam, R. (2022). KIR-HLA gene diversities and susceptibility to lung cancer. Sci. Rep. 12 (1), 17237. doi:10.1038/s41598-022-21062-1
Ho, K. H., Chang, C. J., Huang, T. W., Shih, C. M., Liu, A. J., Chen, P. H., et al. (2018). Gene landscape and correlation between B-cell infiltration and programmed death ligand 1 expression in lung adenocarcinoma patients from the Cancer Genome Atlas data set. PLoS One 13 (12), e0208459. doi:10.1371/journal.pone.0208459
Hou, W., Medynski, D., Wu, S., Lin, X., and Li, L. Y. (2005). VEGI-192, a new isoform of TNFSF15, specifically eliminates tumor vascular endothelial cells and suppresses tumor growth. Clin. Cancer Res. 11 (15), 5595–5602. doi:10.1158/1078-0432.CCR-05-0384
Hu, Z., Bi, G., Sui, Q., Bian, Y., Du, Y., Liang, J., et al. (2020). Analyses of multi-omics differences between patients with high and low PD1/PDL1 expression in lung squamous cell carcinoma. Int. Immunopharmacol. 88, 106910. doi:10.1016/j.intimp.2020.106910
Huang, K., Hu, E., Li, W., Lv, J., He, Y., Deng, G., et al. (2019). Association of PD-1 polymorphisms with the risk and prognosis of lung adenocarcinoma in the northeastern Chinese Han population. BMC Med. Genet. 20 (1), 177. doi:10.1186/s12881-019-0914-8
Huang, L., Lou, N., Xie, T., Tang, L., Han, X., and Shi, Y. (2023a). Identification of an antigen-presenting cells/T/NK cells-related gene signature to predict prognosis and CTSL to predict immunotherapeutic response for lung adenocarcinoma: an integrated analysis of bulk and single-cell RNA sequencing. Cancer Immunol. Immunother. 72 (10), 3259–3277. doi:10.1007/s00262-023-03485-5
Huang, X., Yu, Q., Zhang, L., and Jiang, Z. (2023b). Research progress on Mincle as a multifunctional receptor. Int. Immunopharmacol. 114, 109467. doi:10.1016/j.intimp.2022.109467
Huang, Y., Zhang, H., Feng, J., and Tang, B. (2022). STK11 mutation affects the killing effect of NK cells to promote the progression of lung adenocarcinoma. Apmis 130 (11), 647–656. doi:10.1111/apm.13271
Huang, Y., Zhu, M., Ji, M., Fan, J., Xie, J., Wei, X., et al. (2021). Air pollution, genetic factors, and the risk of lung cancer: a prospective study in the UK biobank. Am. J. Respir. Crit. Care Med. 204 (7), 817–825. doi:10.1164/rccm.202011-4063OC
Hurkmans, D. P., Basak, E. A., Schepers, N., Oomen-De Hoop, E., Van der Leest, C. H., El Bouazzaoui, S., et al. (2020). Granzyme B is correlated with clinical outcome after PD-1 blockade in patients with stage IV non-small-cell lung cancer. J. Immunother. Cancer 8 (1), e000586. doi:10.1136/jitc-2020-000586
International Agency for Research on Cancer (2020). Global cancer observatory: cancer today. France: World Health Organization. Available at: https://gco.iarc.fr/today.
Jungnickel, C., Schmidt, L. H., Bittigkoffer, L., Wolf, L., Wolf, A., Ritzmann, F., et al. (2017). IL-17C mediates the recruitment of tumor-associated neutrophils and lung tumor growth. Oncogene 36 (29), 4182–4190. doi:10.1038/onc.2017.28
Kaanane, H., Senhaji, N., Berradi, H., Benchakroun, N., Benider, A., Karkouri, M., et al. (2022). The influence of Interleukin-6, Interleukin-8, Interleukin-10, Interleukin-17, TNF-A, MIF, STAT3 on lung cancer risk in Moroccan population. Cytokine 151, 155806. doi:10.1016/j.cyto.2022.155806
Karabon, L., Pawlak, E., Tomkiewicz, A., Jedynak, A., Passowicz-Muszynska, E., Zajda, K., et al. (2011). CTLA-4, CD28, and ICOS gene polymorphism associations with non-small-cell lung cancer. Hum. Immunol. 72 (10), 947–954. doi:10.1016/j.humimm.2011.05.010
Kawaguchi, Y., Ohshio, Y., Watanabe, A., Shiratori, T., Okamoto, K., Ueda, K., et al. (2023). Depletion of tumor-associated macrophages inhibits lung cancer growth and enhances the antitumor effect of cisplatin. Cancer Sci. 114 (3), 750–763. doi:10.1111/cas.15671
Khadhraoui, C., Kaabachi, W., Tritar, F., Daghfous, H., Hamzaoui, K., and Hamzaoui, A. (2020). Association of BTLA rs1982809 polymorphism with lung cancer risk in Tunisian population. Int. J. Immunogenet 47 (6), 554–562. doi:10.1111/iji.12491
Khaghanzadeh, N., Erfani, N., Ghayumi, M. A., and Ghaderi, A. (2010). CTLA4 gene variations and haplotypes in patients with lung cancer. Cancer Genet. Cytogenet 196 (2), 171–174. doi:10.1016/j.cancergencyto.2009.09.001
Koh, J., Kim, H. Y., Lee, Y., Park, I. K., Kang, C. H., Kim, Y. T., et al. (2019). IL23-Producing human lung cancer cells promote tumor growth via conversion of innate lymphoid cell 1 (ILC1) into ILC3. Clin. Cancer Res. 25 (13), 4026–4037. doi:10.1158/1078-0432.CCR-18-3458
Koh, Y. W., Park, C., Yoon, D. H., Suh, C., and Huh, J. (2014). CSF-1R expression in tumor-associated macrophages is associated with worse prognosis in classical Hodgkin lymphoma. Am. J. Clin. Pathol. 141 (4), 573–583. doi:10.1309/AJCPR92TDDFARISU
Kong, W., Zhang, L., Chen, Y., Yu, Z., and Zhao, Z. (2022). Cancer cell-derived exosomal LINC00313 induces M2 macrophage differentiation in non-small cell lung cancer. Clin. Transl. Oncol. 24 (12), 2395–2408. doi:10.1007/s12094-022-02907-7
Kucuk, B., and Cacan, E. (2022). Expressional regulation of NKG2DLs is associated with the tumor development and shortened overall survival in lung adenocarcinoma. Immunobiology. 227 (4), 152239. doi:10.1016/j.imbio.2022.152239
Kuo, I. Y., Yang, Y. E., Yang, P. S., Tsai, Y. J., Tzeng, H. T., Cheng, H. C., et al. (2021). Converged Rab37/IL-6 trafficking and STAT3/PD-1 transcription axes elicit an immunosuppressive lung tumor microenvironment. Theranostics 11 (14), 7029–7044. doi:10.7150/thno.60040
Kurt, H., Ozbayer, C., Bayramoglu, A., Gunes, H. V., Degirmenci, İ., Oner, K. S., et al. (2016). Determination of the relationship between rs4986790 and rs4986791 variants of TLR4 gene and lung cancer. Inflammation 39, 166–171. doi:10.1007/s10753-015-0235-9
La Fleur, L., Botling, J., He, F., Pelicano, C., Zhou, C., He, C., et al. (2021). Targeting MARCO and IL37R on immunosuppressive macrophages in lung cancer blocks regulatory T cells and supports cytotoxic lymphocyte function. Cancer Res. 81 (4), 956–967. doi:10.1158/0008-5472.CAN-20-1885
La Fleur, L., Boura, V. F., Alexeyenko, A., Berglund, A., Pontén, V., Mattsson, J. S. M., et al. (2018). Expression of scavenger receptor MARCO defines a targetable tumor-associated macrophage subset in non-small cell lung cancer. Int. J. Cancer 143 (7), 1741–1752. doi:10.1002/ijc.31545
Lahiri, A., Maji, A., Potdar, P. D., Singh, N., Parikh, P., Bisht, B., et al. (2023). Lung cancer immunotherapy: progress, pitfalls, and promises. Mol. Cancer 22 (1), 40. doi:10.1186/s12943-023-01740-y
Le, D. T., Uram, J. N., Wang, H., Bartlett, B. R., Kemberling, H., Eyring, A. D., et al. (2015). PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372 (26), 2509–2520. doi:10.1056/NEJMoa1500596
Lee, K.-L., Lai, T.-C., Wang, Y.-C., Shih, P.-C., Yang, Y.-C., Tsao, T. C.-Y., et al. (2021). Potential impacts of interleukin-17a promoter polymorphisms on the egfr mutation status and progression of non-small cell lung cancer in Taiwan. Genes 12 (3), 427. doi:10.3390/genes12030427
Lee, S. Y., Jung, D. K., Choi, J. E., Jin, C. C., Hong, M. J., Do, S. K., et al. (2016). PD-L1 polymorphism can predict clinical outcomes of non-small cell lung cancer patients treated with first-line paclitaxel-cisplatin chemotherapy. Sci. Rep. 6, 25952. doi:10.1038/srep25952
Lee, S. Y., Jung, D. K., Choi, J. E., Jin, C. C., Hong, M. J., Do, S. K., et al. (2017). Functional polymorphisms in PD-L1 gene are associated with the prognosis of patients with early stage non-small cell lung cancer. Gene 599, 28–35. doi:10.1016/j.gene.2016.11.007
Lee, Y. S., Kim, K. C., Mongre, R. K., Kim, J. Y., Kim, Y. R., Choi, D. Y., et al. (2019b). IL-32γ suppresses lung cancer stem cell growth via inhibition of ITGAV-mediated STAT5 pathway. Cell Death Dis. 10 (7), 506. doi:10.1038/s41419-019-1737-4
Lee, Y. S., Yeo, I. J., Kim, K. C., Han, S. B., and Hong, J. T. (2019a). Inhibition of lung tumor development in ApoE knockout mice via enhancement of TREM-1 dependent NK cell cytotoxicity. Front. Immunol. 10, 1379. doi:10.3389/fimmu.2019.01379
Li, C., Xue, V. W., Wang, Q. M., Lian, G. Y., Huang, X. R., Lee, T. L., et al. (2020b). The mincle/syk/NF-κB signaling circuit is essential for maintaining the protumoral activities of tumor-associated macrophages. Cancer Immunol. Res. 8 (8), 1004–1017. doi:10.1158/2326-6066.CIR-19-0782
Li, C., Zhou, T., Chen, J., Li, R., Chen, H., Luo, S., et al. (2022b). The role of Exosomal miRNAs in cancer. J. Transl. Med. 20 (1), 6. doi:10.1186/s12967-021-03215-4
Li, C. H., Yang, Y. C., Hsia, T. C., Shen, T. C., Shen, Y. C., Chang, W. S., et al. (2022c). Association of interleukin-8 promoter genotypes with taiwan lung cancer risk. Anticancer Res. 42 (3), 1229–1236. doi:10.21873/anticanres.15590
Li, F., Huang, Q., Luster, T. A., Hu, H., Zhang, H., Ng, W. L., et al. (2020d). In vivo epigenetic CRISPR screen identifies Asf1a as an immunotherapeutic target in kras-mutant lung adenocarcinoma. Cancer Discov. 10 (2), 270–287. doi:10.1158/2159-8290.CD-19-0780
Li, J., Song, Y., Yu, B., and Yu, Y. (2020e). TNFAIP2 promotes non-small cell lung cancer cells and targeted by miR-145-5p. DNA Cell Biol. 39 (7), 1256–1263. doi:10.1089/dna.2020.5415
Li, R., Fang, F., Jiang, M., Wang, C., Ma, J., Kang, W., et al. (2017). STAT3 and NF-κB are simultaneously suppressed in dendritic cells in lung cancer. Sci. Rep. 7, 45395. doi:10.1038/srep45395
Li, X., Wang, R., Wang, S., Wang, L., and Yu, J. (2022d). Construction of a B cell-related gene pairs signature for predicting prognosis and immunotherapeutic response in non-small cell lung cancer. Front. Immunol. 13, 989968. doi:10.3389/fimmu.2022.989968
Li, Y., Chen, C., Liu, H. L., Zhang, Z. F., and Wang, C. L. (2022a). LARRPM restricts lung adenocarcinoma progression and M2 macrophage polarization through epigenetically regulating LINC00240 and CSF1. Cell Mol. Biol. Lett. 27 (1), 91. doi:10.1186/s11658-022-00376-y
Li, Y., Jiang, T., Zhou, W., Li, J., Li, X., Wang, Q., et al. (2020a). Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat. Commun. 11 (1), 1000. doi:10.1038/s41467-020-14802-2
Li, Z., Feng, C., Guo, J., Hu, X., and Xie, D. (2020c). GNAS-AS1/miR-4319/NECAB3 axis promotes migration and invasion of non-small cell lung cancer cells by altering macrophage polarization. Funct. Integr. Genomics 20 (1), 17–28. doi:10.1007/s10142-019-00696-x
Liang, P. H., Tian, F., Lu, Y., Duan, B., Stolz, D. B., and Li, L. Y. (2011). Vascular endothelial growth inhibitor (VEGI; TNFSF15) inhibits bone marrow-derived endothelial progenitor cell incorporation into Lewis lung carcinoma tumors. Angiogenesis 14 (1), 61–68. doi:10.1007/s10456-010-9195-8
Lin, X., Chen, X., Long, X., Zeng, C., Zhang, Z., Fang, W., et al. (2023). New biomarkers exploration and nomogram construction of prognostic and immune-related adverse events of advanced non-small cell lung cancer patients receiving immune checkpoint inhibitors. Respir. Res. 24 (1), 64. doi:10.1186/s12931-023-02370-0
Lin, Z., Huang, Q., Liu, J., Wang, H., Zhang, X., Zhu, Z., et al. (2022). Interleukin-17D promotes lung cancer progression by inducing tumor-associated macrophage infiltration via the p38 MAPK signaling pathway. Aging (Albany NY) 14 (15), 6149–6168. doi:10.18632/aging.204208
Lindeman, N. I., Cagle, P. T., Beasley, M. B., Chitale, D. A., Dacic, S., Giaccone, G., et al. (2013). Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the college of American pathologists, international association for the study of lung cancer, and association for molecular pathology. J. Mol. Diagn 15 (4), 415–453. doi:10.1016/j.jmoldx.2013.03.001
Lisberg, A., Cummings, A., Goldman, J. W., Bornazyan, K., Reese, N., Wang, T., et al. (2018). A phase II study of pembrolizumab in EGFR-mutant, PD-l1+, tyrosine kinase inhibitor naïve patients with advanced NSCLC. J. Thorac. Oncol. 13 (8), 1138–1145. doi:10.1016/j.jtho.2018.03.035
Liu, C., He, Y., Feng, X., Li, J., and Wang, J. (2022). Expression of EPHA5 in lung adenocarcinoma is associated with lymph node metastasis and EGFR mutation. Apmis 130 (6), 338–345. doi:10.1111/apm.13222
Liu, M., Tong, Z., Ding, C., Luo, F., Wu, S., Wu, C., et al. (2020). Transcription factor c-Maf is a checkpoint that programs macrophages in lung cancer. J. Clin. Invest. 130 (4), 2081–2096. doi:10.1172/JCI131335
Liu, X. S., and Mardis, E. R. (2017). Applications of immunogenomics to cancer. Cell 168 (4), 600–612. doi:10.1016/j.cell.2017.01.014
Liu, Y., Ouyang, Y., Feng, Z., Jiang, Z., Ma, J., Zhou, X., et al. (2023). RASGRP2 is a potential immune-related biomarker and regulates mitochondrial-dependent apoptosis in lung adenocarcinoma. Front. Immunol. 14, 1100231. doi:10.3389/fimmu.2023.1100231
Ma, K., Li, Y. X., and Guo, Q. Q. (2022). Correlation between metastasis-associated gene 1 expression and tumor-associated macrophages in non-small cell lung cancer. Neoplasma 69 (5), 1092–1100. doi:10.4149/neo_2022_220615N637
Ma, K. Y., Schonnesen, A. A., Brock, A., Van Den Berg, C., Eckhardt, S. G., Liu, Z., et al. (2019). Single-cell RNA sequencing of lung adenocarcinoma reveals heterogeneity of immune response-related genes. JCI Insight 4 (4), e121387. doi:10.1172/jci.insight.121387
Ma, L., and Zhou, N. (2016). Association between an insertion/deletion polymorphism in IL-1A gene and cancer risk: a meta-analysis. Onco Targets Ther. 9, 1–6. doi:10.2147/OTT.S95887
Ma, Q. Y., Chen, J., Wang, S. H., Wu, N., Hao, Z. H., and Chen, X. F. (2015). Interleukin 17A genetic variations and susceptibility to non-small cell lung cancer. APMIS 123 (3), 194–198. doi:10.1111/apm.12341
Maemura, K., Watanabe, K., Ando, T., Hiyama, N., Sakatani, T., Amano, Y., et al. (2018). Altered editing level of microRNAs is a potential biomarker in lung adenocarcinoma. Cancer Sci. 109 (10), 3326–3335. doi:10.1111/cas.13742
Mazzaschi, G., Leonetti, A., Minari, R., Gnetti, L., Quaini, F., Tiseo, M., et al. (2021). Modulating tumor microenvironment: a review on STK11 immune properties and predictive vs prognostic role for non-small-cell lung cancer immunotherapy. Curr. Treat. Options Oncol. 22 (11), 96. doi:10.1007/s11864-021-00891-8
McGranahan, N., Furness, A. J., Rosenthal, R., Ramskov, S., Lyngaa, R., Saini, S. K., et al. (2016). Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351 (6280), 1463–1469. doi:10.1126/science.aaf1490
McGranahan, N., Rosenthal, R., Hiley, C. T., Rowan, A. J., Watkins, T. B. K., Wilson, G. A., et al. (2017). Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171 (6), 1259–1271.e11. doi:10.1016/j.cell.2017.10.001
McNamara, M. G., Jacobs, T., Lamarca, A., Hubner, R. A., Valle, J. W., and Amir, E. (2020). Impact of high tumor mutational burden in solid tumors and challenges for biomarker application. Cancer Treat. Rev. 89, 102084. doi:10.1016/j.ctrv.2020.102084
Migdalska-Sęk, M., Góralska, K., Jabłoński, S., Kordiak, J., Nawrot, E., Kiszałkiewicz, J. M., et al. (2020). Evaluation of the relationship between the IL-17A gene expression level and regulatory miRNA-9 in relation to tumor progression in patients with non-small cell lung cancer: a pilot study. Mol. Biol. Rep. 47 (1), 583–592. doi:10.1007/s11033-019-05164-0
Migone, T. S., Zhang, J., Luo, X., Zhuang, L., Chen, C., Hu, B., et al. (2002). TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity 16 (3), 479–492. doi:10.1016/s1074-7613(02)00283-2
Minari, R., Bonatti, F., Mazzaschi, G., Dodi, A., Facchinetti, F., Gelsomino, F., et al. (2022). PD-L1 SNPs as biomarkers to define benefit in patients with advanced NSCLC treated with immune checkpoint inhibitors. Tumori 108 (1), 47–55. doi:10.1177/03008916211014954
Moksud, N., Wagner, M., Pawełczyk, K., Porębska, I., Muszczyńska-Bernhard, B., Kowal, A., et al. (2023). Common inherited variants of PDCD1, CD274 and HAVCR2 genes differentially modulate the risk and prognosis of adenocarcinoma and squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 149 (9), 6381–6390. doi:10.1007/s00432-023-04602-8
Monticelli, L. A., Sonnenberg, G. F., Abt, M. C., Alenghat, T., Ziegler, C. G., Doering, T. A., et al. (2011). Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12 (11), 1045–1054. doi:10.1031/ni.2131
Nie, G. H., Luo, L., Duan, H. F., Li, X. Q., Yin, M. J., Li, Z., et al. (2016). GALNT7, a target of miR-494, participates in the oncogenesis of nasopharyngeal carcinoma. Tumour Biol. 37 (4), 4559–4567. doi:10.1007/s13277-015-4281-6
Ott, P. A., Bang, Y. J., Piha-Paul, S. A., Razak, A. R. A., Bennouna, J., Soria, J. C., et al. (2019). T-Cell-Inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J. Clin. Oncol. 37 (4), 318–327. doi:10.1200/JCO.2018.78.2276
Pang, J., Yu, Q., Chen, Y., Yuan, H., Sheng, M., and Tang, W. (2022). Integrating Single-cell RNA-seq to construct a Neutrophil prognostic model for predicting immune responses in non-small cell lung cancer. J. Transl. Med. 20 (1), 531. doi:10.1186/s12967-022-03723-x
Patil, N. S., Nabet, B. Y., Müller, S., Koeppen, H., Zou, W., Giltnane, J., et al. (2022). Intratumoral plasma cells predict outcomes to PD-L1 blockade in non-small cell lung cancer. Cancer Cell 40 (3), 289–300.e4. doi:10.1016/j.ccell.2022.02.002
Pedroza-Gonzalez, A., Zhou, G., Vargas-Mendez, E., Boor, P. P., Mancham, S., Verhoef, C., et al. (2015). Tumor-infiltrating plasmacytoid dendritic cells promote immunosuppression by Tr1 cells in human liver tumors. Oncoimmunology 4 (6), e1008355. doi:10.1080/2162402X.2015.1008355
Peifer, M., Fernández-Cuesta, L., Sos, M. L., George, J., Seidel, D., Kasper, L. H., et al. (2012). Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 44 (10), 1104–1110. doi:10.1038/ng.2396
Peng, J., Li, S., Li, B., Hu, W., and Ding, C. (2023). Exosomes derived from M1 macrophages inhibit the proliferation of the A549 and H1299 lung cancer cell lines via the miRNA-let-7b-5p-GNG5 axis. PeerJ 11, e14608. doi:10.7717/peerj.14608
Perea, F., Bernal, M., Sánchez-Palencia, A., Carretero, J., Torres, C., Bayarri, C., et al. (2017). The absence of HLA class I expression in non-small cell lung cancer correlates with the tumor tissue structure and the pattern of T cell infiltration. Int. J. Cancer 140 (4), 888–899. doi:10.1002/ijc.30489
Pereira, E. E. B., Modesto, A. A. C., Fernandes, B. M., Burbano, R. M. R., Assumpção, P. P., Fernandes, M. R., et al. (2023). Association between polymorphism of genes IL-1A, NFKB1, PAR1, TP53, and UCP2 and susceptibility to non-small cell lung cancer in the Brazilian Amazon. Genes (Basel) 14 (2), 461. doi:10.3390/genes14020461
Pérez-Ramírez, C., Alnatsha, A., Cañadas-Garre, M., Villar, E., Valdivia-Bautista, J., Faus-Dáder, M. J., et al. (2017). Cytokine single-nucleotide polymorphisms and risk of non-small-cell lung cancer. Pharmacogenet Genomics. 27 (12), 438–444. doi:10.1097/FPC.0000000000000307
Pyfferoen, L., Brabants, E., Everaert, C., De Cabooter, N., Heyns, K., Deswarte, K., et al. (2017). The transcriptome of lung tumor-infiltrating dendritic cells reveals a tumor-supporting phenotype and a microRNA signature with negative impact on clinical outcome. Oncoimmunology 6 (1), e1253655. doi:10.1080/2162402X.2016.1253655
Pyke, R. M., Mellacheruvu, D., Dea, S., Abbott, C. W., McDaniel, L., Bhave, D. P., et al. (2022). A machine learning algorithm with subclonal sensitivity reveals widespread pan-cancer human leukocyte antigen loss of heterozygosity. Nat. Commun. 13 (1), 1925. doi:10.1038/s41467-022-29203-w
Pyo, C. W., Guethlein, L. A., Vu, Q., Wang, R., Abi-Rached, L., Norman, P. J., et al. (2010). Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS One 5 (12), e15115. doi:10.1371/journal.pone.0015115
Rafrafi, A., Kaabachi, S., Kaabachi, W., Chahed, B., Amor, A. B., Mbarik, M., et al. (2015). CCR2-64I polymorphism is associated with non-small cell lung cancer in Tunisian patients. Hum. Immunol. 76 (5), 348–354. doi:10.1016/j.humimm.2015.03.003
Raju Paul, S., Valiev, I., Korek, S. E., Zyrin, V., Shamsutdinova, D., Gancharova, O., et al. (2023). B cell-dependent subtypes and treatment-based immune correlates to survival in stage 3 and 4 lung adenocarcinomas. FASEB Bioadv 5 (4), 156–170. doi:10.1096/fba.2023-00009
Ramos-Esquivel, A., van der Laat, A., Rojas-Vigott, R., Juárez, M., and Corrales-Rodríguez, L. (2017). Anti-PD-1/anti-PD-L1 immunotherapy versus docetaxel for previously treated advanced non-small cell lung cancer: a systematic review and meta-analysis of randomised clinical trials. ESMO Open 2 (3), e000236. doi:10.1136/esmoopen-2017-000236
Ren, S., Xiong, X., You, H., Shen, J., and Zhou, P. (2021). The combination of immune checkpoint blockade and angiogenesis inhibitors in the treatment of advanced non-small cell lung cancer. Front. Immunol. 12, 689132. doi:10.3389/fimmu.2021.689132
Reuben, A., Gittelman, R., Gao, J., Zhang, J., Yusko, E. C., Wu, C. J., et al. (2017). TCR repertoire intratumor heterogeneity in localized lung adenocarcinomas: an association with predicted neoantigen heterogeneity and postsurgical recurrence. Cancer Discov. 7 (10), 1088–1097. doi:10.1158/2159-8290.CD-17-0256
Reuben, A., Zhang, J., Chiou, S. H., Gittelman, R. M., Li, J., Lee, W. C., et al. (2020). Comprehensive T cell repertoire characterization of non-small cell lung cancer. Nat. Commun. 11 (1), 603. doi:10.1038/s41467-019-14273-0
Rizvi, H., Sanchez-Vega, F., La, K., Chatila, W., Jonsson, P., Halpenny, D., et al. (2018). Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol. 36 (7), 633–641. doi:10.1200/JCO.2017.75.3384
Rizvi, N. A., Hellmann, M. D., Snyder, A., Kvistborg, P., Makarov, V., Havel, J. J., et al. (2015). Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348 (6230), 124–128. doi:10.1126/science.aaa1348
Rosenthal, R., Cadieux, E. L., Salgado, R., Bakir, M. A., Moore, D. A., Hiley, C. T., et al. (2019). Neoantigen-directed immune escape in lung cancer evolution. Nature 567 (7749), 479–485. doi:10.1038/s41586-019-1032-7
Saab, S., Zalzale, H., Rahal, Z., Khalifeh, Y., Sinjab, A., and Kadara, H. (2020). Insights into lung cancer immune-based biology, prevention, and treatment. Front. Immunol. 11, 159. doi:10.3389/fimmu.2020.00159
Sage, A. P., Ng, K. W., Marshall, E. A., Stewart, G. L., Minatel, B. C., Enfield, K. S. S., et al. (2020). Assessment of long non-coding RNA expression reveals novel mediators of the lung tumour immune response. Sci. Rep. 10 (1), 16945. doi:10.1038/s41598-020-73787-6
Sakran, M. I., Alalawy, A. I., Alharbi, A. A., El-Hefnawy, M. E., Alzahrani, S. M., Alfuraydi, A., et al. (2023). The blockage signal for PD-L1/CD274 gene variants and their potential impact on lung carcinoma susceptibility. Int. Immunopharmacol. 125 (Pt A), 111180. doi:10.1016/j.intimp.2023.111180
Samstein, R. M., Lee, C. H., Shoushtari, A. N., Hellmann, M. D., Shen, R., Janjigian, Y. Y., et al. (2019). Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51 (2), 202–206. doi:10.1038/s41588-018-0312-8
Sarkar, A., Rahaman, A., Biswas, I., Mukherjee, G., Chatterjee, S., Bhattacharjee, S., et al. (2020). TGFβ mediated LINC00273 upregulation sponges mir200a-3p and promotes invasion and metastasis by activating ZEB1. J. Cell Physiol. 235 (10), 7159–7172. doi:10.1002/jcp.29614
Sarode, P., Schaefer, M. B., Grimminger, F., Seeger, W., and Savai, R. (2020b). Macrophage and tumor cell cross-talk is fundamental for lung tumor progression: we need to talk. Front. Oncol. 10, 324. doi:10.3389/fonc.2020.00324
Sarode, P., Zheng, X., Giotopoulou, G. A., Weigert, A., Kuenne, C., Günther, S., et al. (2020a). Reprogramming of tumor-associated macrophages by targeting β-catenin/FOSL2/ARID5A signaling: a potential treatment of lung cancer. Sci. Adv. 6 (23), eaaz6105. doi:10.1126/sciadv.aaz6105
Sautès-Fridman, C., Petitprez, F., Calderaro, J., and Fridman, W. H. (2019). Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer 19 (6), 307–325. doi:10.1038/s41568-019-0144-6
Sayaman, R. W., Saad, M., Thorsson, V., Hu, D., Hendrickx, W., Roelands, J., et al. (2021). Germline genetic contribution to the immune landscape of cancer. Immunity 54 (2), 367–386.e8. doi:10.1016/j.immuni.2021.01.011
Schneider, T., Hoffmann, H., Dienemann, H., Schnabel, P. A., Enk, A. H., Ring, S., et al. (2011). Non-small cell lung cancer induces an immunosuppressive phenotype of dendritic cells in tumor microenvironment by upregulating B7-H3. J. Thorac. Oncol. 6 (7), 1162–1168. doi:10.1097/JTO.0b013e31821c421d
Sedighzadeh, S. S., Khoshbin, A. P., Razi, S., Keshavarz-Fathi, M., and Rezaei, N. (2021). A narrative review of tumor-associated macrophages in lung cancer: regulation of macrophage polarization and therapeutic implications. Transl. Lung Cancer Res. 10 (4), 1889–1916. doi:10.21037/tlcr-20-1241
Shalata, W., Yakobson, A., Dudnik, Y., Swaid, F., Ahmad, M. S., Abu Jama, A., et al. (2023). Multi-center real-world outcomes of nivolumab plus ipilimumab and chemotherapy in patients with metastatic non-small-cell lung cancer. Biomedicines 11 (9), 2438. doi:10.3390/biomedicines11092438
Sharma, A., Singh, P., Jha, R., Almatroodi, S. A., Alrumaihi, F., Rahmani, A. H., et al. (2023). Exploring the role of miR-200 family in regulating CX3CR1 and CXCR1 in lung adenocarcinoma tumor microenvironment: implications for therapeutic intervention. Sci. Rep. 13 (1), 16333. doi:10.1038/s41598-023-43484-1
Shirshev, S. V., Nekrasova, I. V., Gorbunova, O. L., Orlova, E. G., and Maslennikova, I. L. (2017). MicroRNA in hormonal mechanisms of regulation of NK cell function. Dokl. Biochem. Biophys. 474 (1), 168–172. doi:10.1134/S160767291703005X
Siddiqui, F. V. S., and Siddiqui, A. H. (2023). Lung cancer in StatPearls (Treasure Island (FL): StatPearls Publishing). [Updated 2023 May 8].
Smok-Kalwat, J., Mertowska, P., Mertowski, S., Smolak, K., Kozińska, A., Koszałka, F., et al. (2023). The importance of the immune system and molecular cell signaling pathways in the pathogenesis and progression of lung cancer. Int. J. Mol. Sci. 24 (2), 1506. doi:10.3390/ijms24021506
Song, B., Liu, Y., Liu, J., Song, X., Wang, Z., Wang, M., et al. (2011). CTLA-4 +49A>G polymorphism is associated with advanced non-small cell lung cancer prognosis. Respiration 82 (5), 439–444. doi:10.1159/000329345
Song, P., Li, W., Guo, L., Ying, J., Gao, S., and He, J. (2022). Identification and validation of a novel signature based on NK cell marker genes to predict prognosis and immunotherapy response in lung adenocarcinoma by integrated analysis of single-cell and bulk RNA-sequencing. Front. Immunol. 13, 850745. doi:10.3389/fimmu.2022.850745
Sorrentino, C., and Di Carlo, E. (2009). Expression of IL-32 in human lung cancer is related to the histotype and metastatic phenotype. Am. J. Respir. Crit. Care Med. 180 (8), 769–779. doi:10.1164/rccm.200903-0400OC
Sorrentino, R., Terlizzi, M., Di Crescenzo, V. G., Popolo, A., Pecoraro, M., Perillo, G., et al. (2015). Human lung cancer-derived immunosuppressive plasmacytoid dendritic cells release IL-1α in an AIM2 inflammasome-dependent manner. Am. J. Pathol. 185 (11), 3115–3124. doi:10.1016/j.ajpath.2015.07.009
Stankovic, B., Bjørhovde, H. A. K., Skarshaug, R., Aamodt, H., Frafjord, A., Müller, E., et al. (2018). Immune cell composition in human non-small cell lung cancer. Front. Immunol. 9, 3101. doi:10.3389/fimmu.2018.03101
Stewart, C. A., Gay, C. M., Xi, Y., Sivajothi, S., Sivakamasundari, V., Fujimoto, J., et al. (2020). Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat. cancer 1 (4), 423–436. doi:10.1038/s43018-019-0020-z
Su, W. P., Chen, Y. T., Lai, W. W., Lin, C. C., Yan, J. J., and Su, W. C. (2011). Apolipoprotein E expression promotes lung adenocarcinoma proliferation and migration and as a potential survival marker in lung cancer. Lung Cancer 71 (1), 28–33. doi:10.1016/j.lungcan.2010.04.009
Sun, J., Zhang, Z., Bao, S., Yan, C., Hou, P., Wu, N., et al. (2020). Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J. Immunother. Cancer 8 (1), e000110. doi:10.1136/jitc-2019-000110
Szajnik, M., Szczepanski, M. J., Czystowska, M., Elishaev, E., Mandapathil, M., Nowak-Markwitz, E., et al. (2009). TLR4 signaling induced by lipopolysaccharide or paclitaxel regulates tumor survival and chemoresistance in ovarian cancer. Oncogene 28 (49), 4353–4363. doi:10.1038/onc.2009.289
Tang, Y., Jiang, M., Chen, A., Qu, W., Han, X., Zuo, J., et al. (2021). Porcupine inhibitor LGK‑974 inhibits Wnt/β‑catenin signaling and modifies tumor‑associated macrophages resulting in inhibition of the malignant behaviors of non‑small cell lung cancer cells. Mol. Med. Rep. 24 (2), 550. doi:10.3892/mmr.2021.12189
Terry, S., Abdou, A., Engelsen, A. S. T., Buart, S., Dessen, P., Corgnac, S., et al. (2019). AXL targeting overcomes human lung cancer cell resistance to NK- and CTL-mediated cytotoxicity. Cancer Immunol. Res. 7 (11), 1789–1802. doi:10.1158/2326-6066.CIR-18-0903
Tong, J. L., Wang, L. L., Ling, X. F., Wang, M. X., Cao, W., and Liu, Y. Y. (2019). MiR-875 can regulate the proliferation and apoptosis of non-small cell lung cancer cells via targeting SOCS2. Eur. Rev. Med. Pharmacol. Sci. 23 (12), 5235–5241. doi:10.26355/eurrev_201906_18189
Trefny, M. P., Rothschild, S. I., Uhlenbrock, F., Rieder, D., Kasenda, B., Stanczak, M. A., et al. (2019). A variant of a killer cell immunoglobulin-like receptor is associated with resistance to PD-1 blockade in lung cancer. Clin. Cancer Res. 25 (10), 3026–3034. doi:10.1158/1078-0432.CCR-18-3041
Trost, Z., Marc, J., Sok, M., and Cerne, D. (2008). Increased apolipoprotein E gene expression and protein concentration in lung cancer tissue do not contribute to the clinical assessment of non-small cell lung cancer patients. Arch. Med. Res. 39 (7), 663–667. doi:10.1016/j.arcmed.2008.06.009
Tu, Y., and Mei, F. (2022). miR-3648 promotes lung adenocarcinoma-genesis by inhibiting SOCS2 (suppressor of cytokine signaling 2). Bioengineered 13 (2), 3044–3056. doi:10.1080/21655979.2021.2017577
Tzeng, H. T., Tsai, C. H., Yen, Y. T., Cheng, H. C., Chen, Y. C., Pu, S. W., et al. (2017). Dysregulation of rab37-mediated cross-talk between cancer cells and endothelial cells via thrombospondin-1 promotes tumor neovasculature and metastasis. Clin. Cancer Res. 23 (9), 2335–2345. doi:10.1158/1078-0432.CCR-16-1520
Uramoto, H., and Tanaka, F. (2014). Recurrence after surgery in patients with NSCLC. Transl. Lung Cancer Res. 3 (4), 242–249. doi:10.3978/j.issn.2218-6751.2013.12.05
Valero, J. G., Matas-Céspedes, A., Arenas, F., Rodriguez, V., Carreras, J., Serrat, N., et al. (2021). The receptor of the colony-stimulating factor-1 (CSF-1R) is a novel prognostic factor and therapeutic target in follicular lymphoma. Leukemia 35 (9), 2635–2649. doi:10.1038/s41375-021-01201-9
Wan, Y., Wang, X., Liu, T., Fan, T., Zhang, Z., Wang, B., et al. (2021). Prognostic value of CCR2 as an immune indicator in lung adenocarcinoma: a study based on tumor-infiltrating immune cell analysis. Cancer Med. 10 (12), 4150–4163. doi:10.1002/cam4.3931
Wang, C., Zhang, C., Xu, J., Li, Y., Wang, J., Liu, H., et al. (2019). Association between IL-1R2 polymorphisms and lung cancer risk in the Chinese Han population: a case-control study. Mol. Genet. Genomic Med. 7 (5), e644. doi:10.1002/mgg3.644
Wang, E., Qian, Z.-R., Nakasono, M., Tanahashi, T., Yoshimoto, K., Bando, Y., et al. (2010). High expression of Toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. Br. J. cancer 102 (5), 908–915. doi:10.1038/sj.bjc.6605558
Wang, H. M., Zhang, X. H., Feng, M. M., Qiao, Y. J., Ye, L. Q., Chen, J., et al. (2018). Interleukin-35 suppresses the antitumor activity of T cells in patients with non-small cell lung cancer. Cell Physiol. Biochem. 47 (6), 2407–2419. doi:10.1159/000491615
Wang, J., Chen, Z., Cao, R., Zhang, Q., Chen, T., You, C., et al. (2021c). Association of BTLA polymorphisms with susceptibility to non-small-cell lung cancer in the Chinese population. Biomed. Res. Int. 2021, 9121824. doi:10.1155/2021/9121824
Wang, J., Li, F., Zheng, M., Sun, R., Wei, H., and Tian, Z. (2012). Lung natural killer cells in mice: phenotype and response to respiratory infection. Immunology 137 (1), 37–47. doi:10.1111/j.1365-2567.2012.03607.x
Wang, J. B., Chen, X. L., Han, Z. B., Wang, H. W., Wang, Z. H., Li, N. N., et al. (2020a). Long non-coding RNA TP73-AS1 contributes to glioma tumorigenesis by sponging the miR-103a/GALNT7 pathway. Brain Res. 1741, 146886. doi:10.1016/j.brainres.2020.146886
Wang, P., Chen, Y., and Wang, C. (2021a). Beyond tumor mutation burden: tumor neoantigen burden as a biomarker for immunotherapy and other types of therapy. Front. Oncol. 11, 672677. doi:10.3389/fonc.2021.672677
Wang, Q., Gu, J., Wang, L., Chang, D. W., Ye, Y., Huang, M., et al. (2020b). Genetic associations of T cell cancer immune response-related genes with T cell phenotypes and clinical outcomes of early-stage lung cancer. J. Immunother. Cancer 8 (2), e000336. doi:10.1136/jitc-2019-000336
Wang, Q., Mao, Z., Li, W., Wang, S., Wang, L., Chen, L., et al. (2022b). Characteristics of the immunogenicity and tumor immune microenvironment in HER2-amplified lung adenocarcinoma. Front. Immunol. 13, 1042072. doi:10.3389/fimmu.2022.1042072
Wang, T. H., Huang, K. Y., Chen, C. C., Chang, Y. H., Chen, H. Y., Hsueh, C., et al. (2023). PM2.5 promotes lung cancer progression through activation of the AhR-TMPRSS2-IL18 pathway. EMBO Mol. Med. 15 (6), e17014. doi:10.15252/emmm.202217014
Wang, W., Li, F., Mao, Y., Zhou, H., Sun, J., Li, R., et al. (2013). A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum. Genet. 132 (6), 641–648. doi:10.1007/s00439-013-1275-6
Wang, X., Huang, R., Lu, Z., Wang, Z., Chen, X., and Huang, D. (2022a). Exosomes from M1-polarized macrophages promote apoptosis in lung adenocarcinoma via the miR-181a-5p/ETS1/STK16 axis. Cancer Sci. 113 (3), 986–1001. doi:10.1111/cas.15268
Wang, Y., Miao, Z., Qin, X., Li, B., and Han, Y. (2021b). NOD2 deficiency confers a pro-tumorigenic macrophage phenotype to promote lung adenocarcinoma progression. J. Cell Mol. Med. 25 (15), 7545–7558. doi:10.1111/jcmm.16790
Wang, Y., Yang, Y., Zhu, Y., Li, L., Chen, F., and Zhang, L. (2017). Polymorphisms and expression of IL-32: impact on genetic susceptibility and clinical outcome of lung cancer. Biomarkers 22 (2), 165–170. doi:10.1080/1354750X.2016.1252956
Weng, R. R., Lu, H. H., Lin, C. T., Fan, C. C., Lin, R. S., Huang, T. C., et al. (2021). Epigenetic modulation of immune synaptic-cytoskeletal networks potentiates γδ T cell-mediated cytotoxicity in lung cancer. Nat. Commun. 12 (1), 2163. doi:10.1038/s41467-021-22433-4
Workel, H. H., Lubbers, J. M., Arnold, R., Prins, T. M., van der Vlies, P., de Lange, K., et al. (2019). A transcriptionally distinct CXCL13(+)CD103(+)CD8(+) T-cell population is associated with B-cell recruitment and neoantigen load in human cancer. Cancer Immunol. Res. 7 (5), 784–796. doi:10.1158/2326-6066.CIR-18-0517
Wu, H., Gao, H., Li, A., Xie, Y., Jia, Z., Yang, Z., et al. (2020). Impact of genetic variation in TLR4 3′ UTR on NSCLC genetic susceptibility. J. Oncol. 2020, 7593143. doi:10.1155/2020/7593143
Wu, M. F., Wang, Y. C., Li, H. T., Chen, W. C., Liao, C. H., Shih, T. C., et al. (2018). The contribution of interleukin-12 genetic variations to Taiwanese lung cancer. Anticancer Res. 38 (11), 6321–6327. doi:10.21873/anticanres.12989
Wu, P., Wu, D., Ni, C., Ye, J., Chen, W., Hu, G., et al. (2014). γδT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity 40 (5), 785–800. doi:10.1016/j.immuni.2014.03.013
Wu, Y., Biswas, D., Usaite, I., Angelova, M., Boeing, S., Karasaki, T., et al. (2022). A local human Vδ1 T cell population is associated with survival in nonsmall-cell lung cancer. Nat. Cancer 3 (6), 696–709. doi:10.1038/s43018-022-00376-z
Wu, Y., Yuan, M., Wang, C., Chen, Y., Zhang, Y., and Zhang, J. (2023). T lymphocyte cell: a pivotal player in lung cancer. Front. Immunol. 14, 1102778. doi:10.3389/fimmu.2023.1102778
Xiao, D., Li, F., Pan, H., Liang, H., Wu, K., and He, J. (2017). Integrative analysis of genomic sequencing data reveals higher prevalence of LRP1B mutations in lung adenocarcinoma patients with COPD. Sci. Rep. 7 (1), 2121. doi:10.1038/s41598-017-02405-9
Xu, Y., Xiang, Z., Alnaggar, M., Kouakanou, L., Li, J., He, J., et al. (2021). Allogeneic Vγ9Vδ2 T-cell immunotherapy exhibits promising clinical safety and prolongs the survival of patients with late-stage lung or liver cancer. Cell Mol. Immunol. 18 (2), 427–439. doi:10.1038/s41423-020-0515-7
Yang, L., Wang, J., Li, F., Han, M., Chang, X., and Wang, Z. (2015). Relationship between genetic polymorphisms in MCP-1, CCR-2, and non-small-cell lung cancer in the Han nationality of Northern China. Genet. Mol. Res. 14 (2), 3745–3752. doi:10.4238/2015.April.22.2
Yang, Q., Li, J., Hu, Y., Tang, X., Yu, L., Dong, L., et al. (2019). MiR-218-5p suppresses the killing effect of natural killer cell to lung adenocarcinoma by targeting SHMT1. Yonsei Med. J. 60 (6), 500–508. doi:10.3349/ymj.2019.60.6.500
Yang, S., Kobayashi, S., Sekino, K., Kagawa, Y., Miyazaki, H., Kumar, S. S., et al. (2021). Fatty acid-binding protein 5 controls lung tumor metastasis by regulating the maturation of natural killer cells in the lung. FEBS Lett. 595 (13), 1797–1805. doi:10.1002/1873-3468.14106
Yang, Y., Li, L., Chen, F., Zhang, L., and Bu, H. (2018). The role of interleukin-31 polymorphisms in non-small cell lung cancer genetic susceptibility and clinical outcome. Genet. Test. Mol. Biomarkers 22 (5), 314–319. doi:10.1089/gtmb.2017.0256
Ye, J., Livergood, R. S., and Peng, G. (2013). The role and regulation of human Th17 cells in tumor immunity. Am. J. pathology 182 (1), 10–20. doi:10.1016/j.ajpath.2012.08.041
Yin, J., Wang, C., Vogel, U., Ma, Y., Zhang, Y., Wang, H., et al. (2023). Common variants of pro-inflammatory gene IL1B and interactions with PPP1R13L and POLR1G in relation to lung cancer among Northeast Chinese. Sci. Rep. 13 (1), 7352. doi:10.1038/s41598-023-34069-z
Yin, W., Lv, J., Yao, Y., Zhao, Y., He, Z., Wang, Q., et al. (2021). Elevations of monocyte and neutrophils, and higher levels of granulocyte colony-stimulating factor in peripheral blood in lung cancer patients. Thorac. Cancer 12 (20), 2680–2690. doi:10.1111/1759-7714.14103
Yoshida, H., Nomizo, T., Ozasa, H., Tsuji, T., Funazo, T., Yasuda, Y., et al. (2021). PD-L1 polymorphisms predict survival outcomes in advanced non-small-cell lung cancer patients treated with PD-1 blockade. Eur. J. Cancer 144, 317–325. doi:10.1016/j.ejca.2020.11.035
Young, R. P., and Hopkins, R. J. (2018). Chronic obstructive pulmonary disease (COPD) and lung cancer screening. Transl. Lung Cancer Res. 7 (3), 347–360. doi:10.21037/tlcr.2018.05.04
Yun, J., Park, M. H., Son, D. J., Nam, K. T., Moon, D. B., Ju, J. H., et al. (2018). IL-32 gamma reduces lung tumor development through upregulation of TIMP-3 overexpression and hypomethylation. Cell Death Dis. 9 (3), 306. doi:10.1038/s41419-018-0375-6
Yun, M., Yingzi, L., Jie, G., Guanxin, L., Zimei, Z., Zhen, C., et al. (2022). PPDPF promotes the progression and acts as an antiapoptotic protein in non-small cell lung cancer. Int. J. Biol. Sci. 18 (1), 214–228. doi:10.7150/ijbs.65654
Zhang, J., Fujimoto, J., Zhang, J., Wedge, D. C., Song, X., Zhang, J., et al. (2014). Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 346 (6206), 256–259. doi:10.1126/science.1256930
Zhang, J., Liu, J., Wu, J., Li, W., Chen, Z., and Yang, L. (2019). Progression of the role of CRYAB in signaling pathways and cancers. Onco Targets Ther. 12, 4129–4139. doi:10.2147/OTT.S201799
Zhang, J., Song, C., Tian, Y., and Yang, X. (2021). Single-cell RNA sequencing in lung cancer: revealing phenotype shaping of stromal cells in the microenvironment. Front. Immunol. 12, 802080. doi:10.3389/fimmu.2021.802080
Zhang, J., Yang, Y., Wei, Y., Li, L., Wang, X., and Ye, Z. (2023). Hsa-miR-301a-3p inhibited the killing effect of natural killer cells on non-small cell lung cancer cells by regulating RUNX3. Cancer Biomark. 37 (4), 249–259. doi:10.3233/CBM-220469
Zhang, J., Zhang, Z., Song, W., and Liu, J. (2020). EPHA5 mutation impairs natural killer cell-mediated cytotoxicity against non-small lung cancer cells and promotes cancer cell migration and invasion. Mol. Cell Probes 52, 101566. doi:10.1016/j.mcp.2020.101566
Zhang, Y., He, S., Mei, R., Kang, Y., Duan, J., Wei, R., et al. (2018). miR-29a suppresses IL-13-induced cell invasion by inhibiting YY1 in the AKT pathway in lung adenocarcinoma A549 cells. Oncol. Rep. 39 (6), 2613–2623. doi:10.3892/or.2018.6352
Zheng, J., Xu, T., Chen, F., and Zhang, Y. (2019). MiRNA-195-5p functions as a tumor suppressor and a predictive of poor prognosis in non-small cell lung cancer by directly targeting CIAPIN1. Pathol. Oncol. Res. 25 (3), 1181–1190. doi:10.1007/s12253-018-0552-z
Zheng, Q. W., Ni, Q. Z., Zhu, B., Liang, X., Ma, N., Wang, Y. K., et al. (2022). PPDPF promotes lung adenocarcinoma progression via inhibiting apoptosis and NK cell-mediated cytotoxicity through STAT3. Oncogene 41 (36), 4244–4256. doi:10.1038/s41388-022-02418-3
Zhong, Q., Lu, Y., Xu, W., Rong, Z., Chang, X., Qin, L., et al. (2021). The differentiation of new human CD303(+) Plasmacytoid dendritic cell subpopulations expressing CD205 and/or CD103 regulated by Non-Small-Cell lung cancer cells. Int. Immunopharmacol. 99, 107983. doi:10.1016/j.intimp.2021.107983
Zhou, X., Liu, S., Liu, J., Zhang, Z., Mao, X., and Zhou, H. (2020). MicroRNA-130a enhances the killing ability of natural killer cells against non-small cell lung cancer cells by targeting signal transducers and activators of transcription 3. Biochem. Biophys. Res. Commun. 523 (2), 481–486. doi:10.1016/j.bbrc.2019.11.099
Keywords: lung neoplasms, immunogenetics, immunotherapy, genetic susceptibility, genetic variation
Citation: Roshan-Zamir M, Khademolhosseini A, Rajalingam K, Ghaderi A and Rajalingam R (2024) The genomic landscape of the immune system in lung cancer: present insights and continuing investigations. Front. Genet. 15:1414487. doi: 10.3389/fgene.2024.1414487
Received: 09 April 2024; Accepted: 07 June 2024;
Published: 25 June 2024.
Edited by:
Antony Cougnoux, Karolinska Institutet (KI), SwedenReviewed by:
Velizar Stefanov Shivarov, Sofia University, BulgariaCopyright © 2024 Roshan-Zamir, Khademolhosseini, Rajalingam, Ghaderi and Rajalingam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abbas Ghaderi, Z2hhZGVyaWFAc3Vtcy5hYy5pcg==; Raja Rajalingam, cmFqYWxpbmdhbS5yYWphQHVjc2YuZWR1
†ORCID: Mina Roshan-Zamir, orcid.org/0000-0003-3878-5870; Aida Khademolhosseini, orcid.org/0009-0002-5785-1490; Kavi Rajalingam, orcid.org/0009-0006-8654-7956; Abbas Ghaderi, orcid.org/0000-0003-0849-3375; Raja Rajalingam, orcid.org/0000-0001-8821-7877
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.