- 1ICAR - Central Institute of Fisheries Education, Mumbai, Maharashtra, India
- 2ICAR - Central Institute of Freshwater Aquaculture, Bhubaneswar, Odisha, India
Cyprinid species are the most cultured aquatic species around the world in terms of quantity and total value. They account for 25% of global aquaculture production and significantly contribute to fulfilling the demand for fish food. The aquaculture of these species is facing severe concerns in terms of seed quality, rising feed costs, disease outbreaks, introgression of exotic species, environmental impacts, and anthropogenic activities. Numerous researchers have explored biological issues and potential methods to enhance cyprinid aquaculture. Selective breeding is extensively employed in cyprinid species to enhance specific traits like growth and disease resistance. In this context, we have discussed the efforts made to improve important cyprinid aquaculture practices through genetic and genomic approaches. The recent advances in DNA sequencing technologies and genomic tools have revolutionized the understanding of biological research. The generation of a complete genome and other genomic resources in cyprinid species has significantly strengthened molecular-level investigations into disease resistance, growth, reproduction, and adaptation to changing environments. We conducted a comprehensive review of genomic research in important cyprinid species, encompassing genome, transcriptome, proteome, metagenome, epigenome, etc. This review reveals that considerable data has been generated for cyprinid species. However, the seamless integration of this valuable data into genetic selection programs has yet to be achieved. In the upcoming years, genomic techniques, gene transfer, genome editing tools are expected to bring a paradigm shift in sustainable cyprinid aquaculture production. The comprehensive information presented here will offer insights for the cyprinid aquaculture research community.
1 Introduction
Aquaculture is a rapidly growing fastest food producing sectors and is vital for ensuring sustainable livelihoods, nutrition, and global food security (FAO, 2022). Over the past two decades, this sector has experienced substantial growth in global food output. As the world’s population approaches 9.7 billion by 2050, there is a rising demand for protein-rich food, making aquaculture crucial in meeting this challenge (Cottrell et al., 2018; Searchinger et al., 2018; Wang et al., 2023). The per capita consumption of fish has doubled from 9.0 kg in 1961 to 20.2 kg in 2020, contributing to almost 20% of animal protein intake in human diets (FAO, 2022). Aquaculture made a historic contribution to worldwide aquatic animal production in 2020, hitting 49.2 percent (FAO, 2022). This achievement was driven by approximately 500 cultured species, with 22 species contributing 75% of the total production (FAO, 2022; Naylor et al., 2021).
Global aquaculture production has surged from 17.3 million metric tons (MMT) in 1990 to 122.6 MMT in 2020, with significant contributions from seaweeds (4.2%), carps (27.8%), bivalves (2.8%), shrimps (16.4%), catfish (12.6%), and tilapia (15.9%) (FAO, 2022). Asia dominates this production, accounting for over 92%, led by China, which contributes 90% of aquaculture production in the region (Naylor et al., 2021). Global finfish production has risen from 20.8 MMT in 2000 to 57.5 MMT in 2020, with 13 finfish species representing around 72% of the total production. Carp production in Asia, particularly catla, rohu, grass carp, silver carp, common carp, bighead carp, and black carp, has experienced significant growth, reaching 27.6 MMT in 2020 (Figure 1) (FAO, 2022). Notably, catla (Labeo catla), rohu (Labeo rohita), grass carp (Ctenopharyngodon idellus), silver carp (Hypoththalmichthys molitrix), common carp (Cyprinus carpio), bighead carp (Hypophthalmichthys nobilis), and black carp (Mylopharyngodon piceus) account for roughly 50.6% of total inland aquaculture finfish output (FAO, 2022).
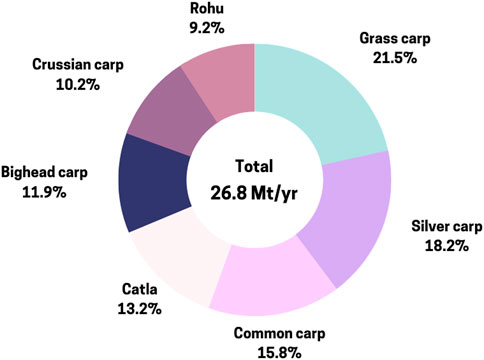
Figure 1. Annual production of cyprinid species with production above 1 Mt/yr (FAO/FishStatJ, 2022).
Cyprinid aquaculture in Asian countries exhibits a high level of diversity concerning the cultured species and production types and has undergone rapid evolution. The contribution of carp species in total production is dominated by grass carp, silver carp, common carp, catla, bighead carp, crucian carps (Carassius spp.), and rohu etc. The feeding and breeding biology of these important carp species is well studied in Asian countries and is being used in aquaculture production through seed production and grow-out culture. In addition to these, various attempts were made to domesticate cyprinid species and genetically improve using selective breeding programs such as rohu in India and Bangladesh (Gjerde et al., 2002; Das Mahapatra et al., 2007) common carp in China, Indonesia, and Vietnam (Ninh et al., 2011) and silver barb (Puntius gonionotus) in Bangladesh and Thailand (Hussain and Mazid, 2002). In China, Ministry of Agriculture of China established a national committee for the examination and approval of original improved varieties of aquatic species and releases new aquatic species every year (Hu et al., 2021).
In order to meet global food demand, selective breeding of cyprinid species focuses on production factors like body growth and immunity. The systematic genetic improvement program in cyprinid species based on pedigreed populations and genetic merit resulted in an increased genetic gain of 12.5% per generation (Gjedrem and Robinson, 2014). The heritability for body weight in cyprinid species was observed to range from 0.2 to 0.4, which is considered good for the genetic selection program (Ninh et al., 2013). Initially, genetic variation in cyprinid species was studied based on morphometric characteristics, and later, new molecular genetic technologies were utilized for parentage assignment and stock variation analysis (Dey et al., 2010). The genetically improved cyprinid species demonstrated a significant improvement in the growth performance as compared to wild or local stock for the same species (Ponzoni et al., 2008; Das Mahapatra et al., 2007).
Molecular genetics and genomics studies in cyprinid species help to gain insight into the genetic basis of production and performance traits for implementing a genetic improvement program (Heras, 2021; Houston et al., 2020). Parentage analysis based on microsatellite markers supports the communal rearing of families of fish in the early stages, which reduces common environmental effects during the selection program (Ninh et al., 2013; Mahapatra et al., 2018). The genetic improvement program in cyprinid species such as rohu relied on phenotypic and pedigree data, which was restricted to a few production traits. During the last decade, advancements in molecular tools and genome sequencing enabled the development of diverse genomic resources for the important cyprinid species. These resources can be used for the enhancement of production efficiency, sustainability, product quality, and profitability of cyprinid culture by integrating them into the existing breeding programs.
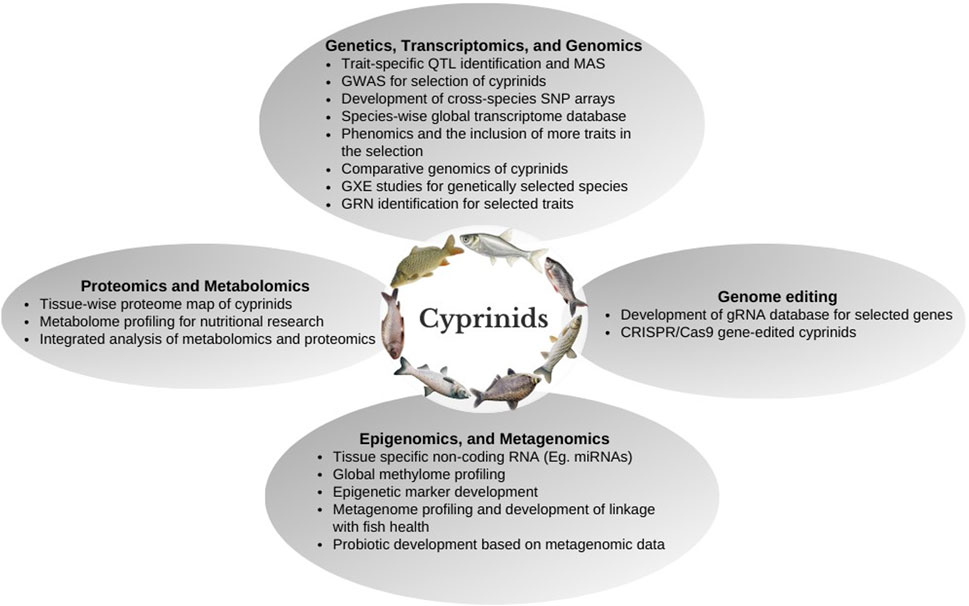
In this paper, the progress made in genetic improvement programs and genomic resources developed at the genome, transcriptome, proteome, metagenome, epigenome, etc., in cyprinid species have been reviewed. The utilization of molecular markers such as microsatellite and single nucleotide polymorphisms (SNPs) developed in cyprinid species for production enhancement and evolutionary studies have been discussed. The complete genome and transcriptome analysis of important cyprinid species is a valuable resource for studying the genetic basis of characteristics and their inter-relationships. Functional annotations, genome-wide marker identification, linkage map construction, and genotyping might all be accomplished using a collection of genomic techniques and resources. Genome-editing tools could be used to elucidate the importance of epigenetic control of genes whose expression affects aquaculture performance. Cyprinid species’ genetic and genomic research is needed to shift towards more functional molecular marker discovery and understanding of gene regulatory networks for economically important traits. In addition, selection for feed conversion efficiency (FCE), reproductive parameters, and resilience to environmental stressors such as low DO, salinity, and temperature must be included in the cyprinid breeding programs. Further, the integration of omics data such as transcriptomics, proteomics, and metabolomics with a system biology approach would help to understand complex biological traits and their functional pathways, which contribute to a significant genetic variation among the stocks for enhancing cyprinid aquaculture production. This paper provides a comprehensive overview of cyprinid species genetic research and outlines the future research needs in their aquaculture, with a particular emphasis on genetics and genomics approaches.
2 Status of selective breeding program in cyprinids
Selective breeding in aquaculture is a long-term process that aims to genetically improve fish species for economically important traits such as growth rate, disease resistance, feed conversion efficiency, and reproductive fitness (Gjedrem and Robinson, 2014; Gjedrem and Baranski, 2010; Lind et al., 2012). This approach involves the meticulous selection of individuals based on their genetic parameters and pedigree information. Selective breeding aims to increase the prevalence of favorable alleles by allowing carriers to produce more offspring, leading to the gradual accumulation of genetic improvements over generations. This process relies on additive genetic variation, the hereditary contribution passed from one generation to the next. The history of selective breeding in aquaculture traces back to the 1920s, with early reports focusing on brook trout and common carp. Over the past 5 decades, selective breeding in aquatic species has shown remarkable success in achieving genetic improvement using family/individual selection, resulting in a significant 12.5% (average) increase in growth rate per generation. According to a report, genetically selected aquatic species contributed to 10%–20% of aquaculture production (FAO, 2022; Olesen et al., 2015).
In particular, cyprinids are an economically important species, and numerous selective breeding efforts has been carried out. The selective breeding programs in cyprinid species have primarily focused on enhancing the growth rate, followed by disease resistance and other traits. These programs have predominantly employed the family selection method for genetic improvement. The documented selective breeding programs for some of the commercially important cyprinid species worldwide are discussed below.
2.1 Common carp
The common carp, originating from Asia and Europe, is the third most globally introduced fish species and has been farmed since Roman times (Welcomme, 1988). Genetic improvement began in Hungary in 1962 under Dr. János Bakos, with the establishment of the common carp gene bank in Szarvas in 1963. Hybridization efforts, including mirror Szarvas 215, scaly Szarvas P. 31, and two-line scaly hybrid Szarvas P. 34, have demonstrated productivity surpassing parental lines by 20%–25% (Bakos and Gorda, 1995). The first instance of selective breeding among carp involved the common carp, aiming to enhance its growth rate by utilizing mass selection (Kirpichnikov and Faktorovich, 1974). Attempts to reduce the number of intermuscular bones of common carp by selection have not been successful in earlier times (Kossmann, 1972; Moav et al., 1975). In another study, Wohlfarth et al. (1975) observed that certain common carp populations from the river Amur showed resistance to dropsy, and crossing between local and Siberian wild carp led to the development of three stocks of Krasnodar common carp. These stocks were further utilized for commercial production through heterotic crossbreeding. The Jian carp were subjected to a six-generation mixed breeding procedure that included family selection, inter-line crossover, and gynogenesis. Individual selection for growth rate in common carp resulted in an increase in body weight over two generations in Vietnam, with estimated heritability ranging from 0.2 to 0.29 in just one of the three selected lines. Notably, after five generations of selection, the selected lines of common carp growth rate improved by 33% compared to the base population (Taniguchi et al., 1996).
It was observed significant growth differences among half-sibling families of common carp, with heritability estimates for growth at 0.47 (Brody et al., 1981). Early age body weight heritability was reported as zero (Vandeputte, 2003) and 0.12 (Nielsen et al., 2010). While a moderate heritability of 0.33 was noted for length, weight, and Fulton’s condition factor (K) at 2 months, and ranged between 0.39 and 0.49 at 4 months. A high heritability of 0.44 was reported for harvest body weight in Hungarian synthetic mirror carp over three generations (Vandeputte et al., 2008). Survival until harvest showed heritability of 0.2 (Nielsen et al., 2010). Significant genetic variation supports the notion that selective breeding can enhance carp growth and survival (Nielsen et al., 2010; Vandeputte et al., 2008; Vandeputte et al., 2004). Broodstock from the river Tisa and a Serbian fish farm displayed heritabilities of 0.34–0.45 in the first year and 0.44–0.49 in the second year for growth-related traits. Genetic correlations varied from 0.54–0.91 in the first year and 0.24–0.74 in the second year, generally remaining high. Close correlations between length and weight (0.80–0.98) suggest that indirect selection for length could facilitate genetic improvement for weight (Vandeputte et al., 2004; Vandeputte et al., 2008; Ninh et al., 2011). In contrast, it was reported relatively low genetic correlations (−0.54 to 0.47) for body weights recorded across different seasons in common carp, attributing the variance to temperature variations (Nielsen et al., 2010). Negative correlations were found between Fulton’s condition factor (K) and length (−0.38) and K and weight (−0.17) (Vandeputte et al., 2004).
Estimates for growth-related features in the Oujiang color common carp population have been reported (Wang et al., 2006) with heritabilities in the range of 0.14–0.30. The USSR developed the Ropsha carp, a fast-growing, cold-tolerant hybrid (Kirpichnikov and Faktorovich, 1974; Babouchkine and Tiews, 1987). Another study revealed a heritability of 0.44 for body weight in common carp based on data spanning three generations (Vandeputte et al., 2008). Researchers obtained a genetic gain of 7% relative to the base population of each generation after studying four generations of genetic selection for harvest body weight in common carp between 2004 and 2014. This study comprised the production, tagging, and rearing of 78 carp families over 20 months (Dong et al., 2015). A study on the heritability and genetic relationships between the weight, length, and height of common carp (C. carpio L.) throughout a three-year growing period was carried out (Spasić et al., 2010). The weight, length, and height of the 50 families of common carp that were born in 2007 were measured at tagging, first autumn, and second autumn. High heritability estimates based on univariate models were discovered for all traits during the second production year (Chen et al., 2022).
The high heritability for final weight (W3) and length (L3) in common carp (0.49, 0.50) and 0.34 ± 0.09 was reported for survival, suggesting successful selective breeding for these traits. Selective breeding programs for disease resistance for dropsy, in Krasnodar carp have been reported. It was estimated moderate heritability for body weight and low heritability for survival in common carp (Dong et al., 2015). A breeding program on Hungarian synthetic mirror carp resulted in heritabilities for body weight at 0.62 and total muscle fat at 0.23 to 0.41 (Prchal et al., 2018). High positive genetic correlations between growth and fillet yield indicated that selection for faster growth would improve fillet yield. In 2017, the base population of Amur mirror carp was established in the Czech Republic, and predictors for genetic improvement of slaughter yields were identified (Prchal et al., 2021). Earlier, low heritability for body weight for common carp at tagging was reported (Nielsen et al., 2010; Ninh et al., 2011). The predictors recorded on market-sized fish showed slightly better heritability (0.41, 0.44), genetic correlations to the slaughter yields (0.78–0.86), and expected genetic gains (1.29%–1.54%) of the slaughter yields than two-year old fish which suggests that the indirect selection for improved slaughter yields could be performed (Prchal et al., 2021). Overall, these studies indicated that significant genetic improvement in production traits such as body weight and processing traits were achieved in common carp through selective breeding.
Groundwater salinization is a global challenge, but it opens opportunities for cultivating saline-tolerant species like common carp, which can tolerate salinity up to 12 ppt. ICAR-CIFE in India has launched a selective breeding program to develop a faster-growing, low saline-tolerant common carp strain for inland saline aquaculture (Lalramnunsanga et al., 2024). The program uses diverse populations from across India, with genetic diversity assessed through truss morphometry and mitochondrial markers. The findings will aid in reducing inbreeding risks and support the genetic management of common carp in the ongoing breeding efforts.
2.2 Rohu
Rohu has become well-known as a promising aquaculture species in countries such as India, Bangladesh, Myanmar, and Pakistan. Due to effective induced breeding and domestication of this species, India and Bangladesh have used crossbreeding and selective breeding methods for genetic improvement. The ICAR-Central Institute of Freshwater Aquaculture (CIFA) in Bhubaneswar, Odisha, India, and the AKVAFORSK (Institute of Aquaculture Research) Norway jointly launched the selective breeding program for rohu in India in 1992 (Rasal and Sundaray, 2020). The process involved establishing a base population using six stocks collected from various rivers, including Ganga, Yamuna, Sutlej, Gomati, and Brahmaputra, along with a farmed stock of CIFA (Gjedrem and Robinson, 2014; Das Mahapatra et al., 2007). Full-sib families were generated using diallele crossing, and the fingerlings were tagged with a passive integrated transponder (PIT) for the evaluation of genetic parameters. Genetic improvement was carried out through a family selection method. The two complete diallele crosses involved five stocks of rohu carp to assess the impact of heterosis on growth and survival (Gjerde et al., 2002). The study reported that for harvest weight and survival, the total heterosis for each of the stock crosses was low or negative.
Over eight generations, an average genetic gain of 18% per generation was achieved in rohu, resulting in the development of a variety called “Jayanti” rohu (Mahapatra et al., 2016; Rasal et al., 2017a). The selective breeding program has continued, with 12 generations completed to date, and each generation consists of 50–60 families in each year class (Mahapatra et al., 2017). Furthermore, a disease resistance trait against the bacterial pathogen Aeromonas hydrophila was incorporated into the rohu selective breeding program, where arrays of single nucleotide polymorphisms (SNPs) were identified using transcriptome data from a study (Mahapatra et al., 2008; Robinson et al., 2014). Estimated heritabilities for challenge test survival obtained from the threshold model in Jayanti rohu is 0.11 ± 0.04 (Mahapatra et al., 2008). Experimental infection with Aeromonas hydrophila (9.55 × 106 cfu g-1 fish) through the intraperitoneal route produced higher survival in the resistant line (73.33%) as compared to the susceptible line (16.67%) (Sahoo et al., 2011). The resistant line showed 58% greater survival in the challenge test than the susceptible line (Sahoo et al., 2004). ICAR-CIFA received a Trade Mark Registration for “AhR Jayanti” rohu (Aeromonas hydrophila-resistant Jayanti rohu) in April 2022.
In Bangladesh, the WorldFish Carp Genetic Improvement Program (WFCGIP) initiated a selective breeding program for rohu focusing on the trait of harvest body weight in 2014 (Keus et al., 2017; Hamilton et al., 2019b). A base population developed utilizing 14 high-ranking full-sib families through family-based (pedigree-based) selection after procuring stocks from the Halda, Jamuna, and Padma rivers. After three generations (G3), significant realized genetic gains of 38.6% and 34.9% were observed for body harvest weight in the Jashore and Natore–Rajshahi populations, respectively (Hamilton et al., 2019b).
The genetic parameters for growth and survival in a rohu breeding program were studied under both mono and polyculture conditions. The estimated heritabilities (and common environmental effect for full-sibs, c2) across both production systems was 0.34 ± 0.10 (0.23 ± 0.04) for body weight at harvest. Genetic correlations between body weight at tagging and at sampling/harvest were of medium magnitude (0.38–0.49), while the correlation between body weight at sampling and harvest was very high (0.98), indicating that selection for increased harvest body weight would result in correlated genetic responses for early growth. The high magnitude of c2 (ranging from 0.66 to 0.78) for body weight at tagging emphasizes the need to standardize the rearing environment (Gjerde et al., 2019). A recent report suggested early selection of G1 individuals by visual identification and non-random sampling to enhance genetic gain in rohu harvest weight (Hamilton et al., 2022).
2.3 Catla
Catla, a highly significant aquaculture species, is predominantly cultivated in South Asia, often in combination with other species (Penman et al., 2005). However, factors including excessive levels of inbreeding, unchecked interspecific hybridization, and negative selection have traditionally had an impact on the quality of the seed produced in hatcheries (Khan et al., 2018). A project financed by the United States Agency for International Development (USAID) was started in 2012 to address these issues. Fertilized spawn from the Halda, Jamuna, and Padma (Ganges) rivers was gathered as part of this initiative to supply Bangladeshi catla hatcheries with genetically varied and non-inbred broodstock (Keus et al., 2017). Fin-clipping the fish and genotyping them with the DArTseq technology led to the discovery of 3048 single nucleotide polymorphisms (SNPs) and 4,726 silicoDArT markers. Intriguingly, the percentage of those without common putative parents was lower in the Jamuna and Padma fish sources, at 18.4% and 8.0%, respectively, compared to the Halda source, with 46.8% (Hamilton et al., 2019a).
The ICAR-Central Institute of Freshwater Aquaculture (ICAR-CIFA) initiated a selective breeding program on Catla (Labeo catla) in 2010 to enhance body weight at harvest, addressing the needs of the fish farming community. For the base population, nine strains/populations of L. catla, including two riverine strains (Ganga and Subernarekha) from different geographical regions (West Bengal, Bihar, Odisha, Andhra Pradesh, and Uttar Pradesh), were collected (Mahapatra et al., 2018). Phenotypic information on growth traits and microsatellite markers was utilized to infer relationships within and between strains. Combined family selection method was employed to select superior animals for each generation based on their breeding value (Mahapatra et al., 2016). A nested mating design, using the dry stripping method, produced full-sib and half-sib families. PIT tagging was conducted on enhanced catla fingerlings (10–150 g), revealing that the most effective size for PIT tagging in catla was 20–30 g (Mahapatra et al., 2022). A mixed linear animal model, with a pond as a fixed effect, was employed to estimate genetic parameters, and heritability for body weight at harvest was estimated to be 0.29 ± 0.03. After two generations of selection, a 15% genetic gain per generation was achieved in genetically improved catla (Mahapatra et al., 2022). A study examining the genotype-by-culture system interaction in catla revealed no evidence of heterosis or differences between their genetic groups concerning harvest weight or survival (Hamilton et al., 2023a).
2.4 Silver carp
Silver carp is the second-most significant species of farmed fish (FAO, 2020). The yearly silver carp production in Bangladesh is around 0.2 MT (DoF, 2019). WorldFish conducted a family-based (i.e., pedigree-based) genetic improvement initiative to enhance the growth rate in polyculture production systems, thereby improving the genetic quality of silver carp (Hamilton et al., 2021). Also, there exists a substantial additive genetic variation in the growth rate of about 43% greater average weight and 11% higher average length observed in Bangladeshi silver carp (Gheyas et al., 2009).
As “candidate founders” for a family-based genetic enhancement initiative in 2015–16, WorldFish obtained silver carp individuals from 21 Bangladeshi hatcheries. For generating the base population, 230 broodfish were used. The mean pedigree-derived additive genetic relationship between actual founders was low (0.0093) based on microsatellite markers, indicating that relationships between actual founders are unlikely to have a significant influence on future parent selection, mating choices, or inbreeding rates (Hamilton et al., 2021). The genetic variation and correlations between harvestage and a set of secondary traits (i.e., gill raker sponginess, gut length, the extent of overlap of pectoral and pelvic fins, presence of Lernaea and prevalence of red spots - sites of inflammation/hemorrhaging) were quantified in silver carp. The result suggested to have no significant correlation between the selected traits (Hamilton et al., 2023b). In silver carp, a higher heritability estimate of 0.67 at 6 months of age was reported by (Gheyas et al., 2009) Gheyas and his colleagues. For other traits, Hamilton and his team found moderate heritabilities of 0.24 for harvest-age weight and 0.22 for pectoral/pelvic fin overlap. In contrast, lower heritabilities were observed for the gill raker score (0.12) and relative gut length (0.09). Notably, there were no genetic correlations between harvest-age weight and secondary traits such as gill raker sponginess, gut length as a ratio of standard length, extent of overlap of pectoral and pelvic fins, presence of Lernaea, and prevalence of red spots (Hamilton et al., 2023b). This suggests that indirect selection or correlated responses cannot be obtained for these two traits.
2.5 Bighead carp
The bighead carpis an important freshwater food cyprinid species predominantly in China, contributing to over 90% of the world’s annual production, which exceeded 3.2 million tons in 2020 (FAO, 2020). It is renowned for its large size, high fecundity, and long maturity age (4–5 Years). Notably, its head makes up approximately 34% of the whole fish, contributing to its higher market value due to its rich nutritional and culinary qualities (Hong et al., 2013). It is a major aquaculture species in China and is selectively bred for faster growth and larger head size (Zhou et al., 2021b). To enhance its economic value, selective breeding for fast growth and larger head size is considered an important goal in bighead carp aquaculture (Zhou et al., 2021b). Genetic analysis of bighead carp populations (eight) in the Yangtze River using 15 microsatellite DNA markers revealed high diversity, weak differentiation among populations (Fst = 0.02, p < 0.01), and individual-level genetic admixture (Zhu et al., 2022). A genetic linkage map for bighead carp was constructed with 905 microsatellites assigned to 24 linkage groups. QTL mapping identified significant and suggestive QTL associated with growth traits on LG9 and LG17, explaining 18.6%–25.5% of phenotypic variance (Liu et al., 2016). A cross-breeding program conducted in Hungary among cyprinids revealed that common carp and bighead carp had good growth rates (BaKos et al., 1978). The reciprocal crosses between silver carp and bighead carp have higher survival, yield, and tameness than both the parents (Issa et al., 1986), and these hybrids are fertile but have low efficiency in controlling microalgal blooms due to the larger spacing of the gill filter apparatus (Marian et al., 1986).
A parentage test was conducted in bighead carp using ten microsatellite markers, resulting in a high success rate of 98.96% and 100% in two designed mating groups. These markers exhibited high polymorphism and heterozygosity (Zhang et al., 2019a). In a study on bighead carp, a high-density genetic map was created with 3,121 SNP markers from 117 individuals in an F1 family. This map spanned 2,341.2 cM with an average marker interval of 0.7 cM. Heritability estimates for body weight (BW) and standard length (SL) in 30-day-old bighead carp were 0.47 and 0.49, respectively. The study detected 37 suggestive QTL for five growth-related traits, explaining phenotypic variance from 15% to 38% (Fu et al., 2016a). The long sexual maturity time of bighead carp (4–5 years or more) poses challenges for selective breeding efforts, prompting the need for efficient breeding strategies. In a selective breeding program involving bighead carp (840 fishes), researchers estimated the heritability of growth-related traits and identified growth-related QTL using resequencing data (Chen et al., 2023). High phenotypic (0.70–0.95) and genetic (0.77–0.97) correlations were found among all growth-related traits. The estimated heritability values were as follows: 0.2 ± 0.11 for body-weight, 0.28 ± 0.14 for body-height, 0.32 ± 0.16 for head-length, 0.14 ± 0.11 for total length, and 0.12 ± 0.09 for body-length in seven-month-old bighead carp. These findings provide valuable insights for improving the efficiency of bighead carp breeding and production.
2.6 Grass carp
Grass carp is a highly productive freshwater fish in China, with local adaptation to diverse environments (Shen et al., 2019). It is primarily cultivated through pond culture, accounting for 71% of freshwater aquaculture production (Wang et al., 2015). Studies have focused on genetic parameters for growth, disease-resistance, and muscle protein and fat content (Zhang et al., 2019c; Fu et al., 2015; Xie et al., 2018). In previous studies, it was found that heritability estimates for growth traits ranged from 0.24 to 0.38, indicating a significant genetic variation (P < 0.01). High genetic and phenotypic correlations were observed among growth traits (0.81–0.99, P < 0.01), suggesting the potential for significant genetic improvement in these traits through selective breeding (Fu et al., 2016b). Similarly, in another study, heritability estimates were determined for early growth traits, and it was observed 0.304 for standard length (SL), 0.307 for body-weight (BW), and 0.150 for condition factor (K) at 40 days post-hatch (dph) (P < 0.05). Significant genetic and phenotypic correlations were observed between SL and BW, with values of 0.83 and 0.95, respectively (Fu et al., 2015).
For the selective breeding of grass carp, researchers identified 15 growth-related quantitative trait loci (QTLs) across 7 linkage-groups. These QTLs were associated with body-weight, body- length, body-height, and body-width based on microsatellite data. They explained varying percentages of phenotypic variance and provided valuable genetic insights for selective breeding (Yu et al., 2020). In the recent study on reducing intramuscular bones (IBs) in C. idella through selective breeding, researchers used high polymorphic microsatellite loci. The heritability of IBs number was also low at 0.137, suggesting that low heritability signifies that the genetic influence on IBs number was modest, making it challenging to substantially alter this trait through selective breeding efforts (Xiong et al., 2023). The genetic correlation among growth traits in grass carp ranged from 0.81 to 0.99 (Fu et al., 2016b; Rothbard and Wohlfarth, 1993). Rothbard and Wohlfarth (1993) reported albinotic and red-colored strains for grass carp (Rothbard and Wohlfarth, 1993). Wang et al. (2022c) demonstrated that disease-resistant grass carp (DR-GC) exhibited a 22.18% higher survival rate and a 16.31% faster growth rate than normal grass carp (GC) (Wang et al., 2022a). They also reported that the input–output ratio of DR-GC to GC is 1.00:1.30, showcasing significant cost-saving, efficiency-increasing, and higher-breeding benefits in DR-GC (Wang et al., 2022b).
Hybrids of grass carp and bighead carp were reported to be triploid (Márián and Krasznai, 1979; Beck et al., 1980), with diploids showing reduced viability. Triploids are preferred when both controlling aquatic vegetation and preventing its naturalization in ecosystems are necessary (Shireman and Smith, 1983).
2.7 Crucian carp
Crucian carp underwent domestication in 12th century in China (Kirpichnikov, 1981). Goldfish, reared during the Tang and Song Dynasties, became symbolic (Chen, 1956; Chen et al., 2020). Initially named Cyprinus auratus, it was later renamed Carassius auratus, closer to Crucian carp (Komiyama et al., 2009). Phylogenetic analysis showed Japanese goldfish originated from the Chinese Crucian carp “Gibelio” (Komiyama et al., 2009). Comet, a Goldfish variant, was produced in 1872 in the United States (Smartt, 2001). Various color and morphology variants developed through crossbreeding and selection (Tave, 1986; Kirpichnikov, 1981; Yamamoto, 1973). Selection of goldfish focused on features like double or triple tails (Komiyama et al., 2009). Pengze crucian carp, bred since the 1980s, underwent over 10 generations of selection (Zhou and Gui, 2002; Zhou et al., 2000). Artificial selection for goldfish traits, such as dorsal fin loss and diverse eye features, occurred independently (Komiyama et al., 2009). Currently, over 180 variants and 70 strains are produced globally (Nasu and Ohuchi, 2016). Some goldfish strains exhibit unique phenotypes (Omori and Kon, 2019). Intergeneric hybrids between common carp and crucian carp show male sterility but accelerated growth. Hybrids between crucian carp and grass carp hybrids are triploid (Kasama and Kobayasi, 1989). Intergeneric hybrids between Carassius auratus and Gnathopogon elongatus elongates resemble C. auratus (Suzuki, 1963).
2.8 Wuchang bream
The blunt snout bream (Megalobrama amblycephala), also known as Wuchang bream, is an endemic species in China. It has been a major economic species in Chinese freshwater aquaculture due to its high larval survival rate, rapid growth, and tender flesh (Wang and Gao, 2018). Selective breeding programs have focused on growth traits and disease resistance (Xun et al., 2023; Zhang et al., 2014; Xiong et al., 2019; Luo et al., 2014b). The study estimated heritabilities for growth-related traits (body-weight, total length, body-length, and body-height) in Wuchang Bream using a microsatellite-based pedigree approach. Heritabilities were found to be high (0.5–0.6), and there were strong genetic correlations among these traits. Selective breeding for growth in this species, particularly focusing on body-length, is considered feasible based on the findings (Luo et al., 2014a). Additionally, the selectively bred F5 population of Wuchang bream exhibited improved tolerance to hypoxic stress (Wu et al., 2020). In previous studies, the heritability of intramuscular bones (IB) in blunt snout bream was assessed for reducing IBs. This work resulted in a wide range of IB counts in the test population based on microsatellite data. The genetic correlation between IB numbers in different sections was high (0.9), while the phenotypic correlation was low (0.2). The heritability of IB numbers was medium (0.2–0.3) for some sections, indicating the potential for selective breeding to reduce IBs in this fish species (Xiong et al., 2019).
2.9 Black carp
The black carp is a large and fast-growing cyprinid, and native to eastern Asia (Feng et al., 2006). It is a preferred species for biological control of molluscans (Pieterse et al., 2017). In order to establish molecular selective breeding in black carp, QTL for four growth-related traits were identified, and a high correlation coefficient of 0.984, 0.97, 0.958, and 0.946 has been observed between the traits such as BL and BWI, BWE, and BL, BWE and BH, BWE and BWI, respectively (Guo et al., 2022). These studies reveal that selection for one of the four traits may also result in the improvement of others. A total of 10%–15% phenotypic variation was observed based on QTL. The QTL regions controlling the growth-related traits were concentrated on certain linkage groups such as LG10, LG17, and LG20, and suggested that LG17 could be exploited for MAS in the selective breeding programs of black carp (Guo et al., 2022).
2.10 Tench
Tench (Tinca L.) is one of the original European cyprinid species and is reared in farm ponds, either in monoculture or polyculture (Svobodova and Kolarova, 2004). The selective breeding and domestication of tench was started in the former Czech, and in addition to the five original strains, new strains from Hungary, France and Romania have also been introduced (Svobodova and Kolarova, 2004). The formation of 6 tench strains using a diallel crossing system among 6 strains from 4 populations has been carried out (Kvasnicka et al., 1998). Various color mutants has been reported in tench (Kvasnicka et al., 1998; Rothbard et al., 2010).
3 Genomics selection and genome-wide association studies (GWAS) in cyprinid species
Genomic selection (GS) is an advanced molecular breeding method in aquaculture that uses markers, like single-nucleotide polymorphisms (SNPs), to enhance the accuracy of breeding values. This method harnesses genomic relationships and markers associated with essential genes, with its application steadily expanding across various aquaculture species. GS demonstrates efficacy in refining critical traits such as growth and disease resistance, driven by the overarching objective of bolstering sustainability and fortifying resilience against the impacts of climate change. Despite its considerable potential, the widespread integration of GS remains somewhat constrained, though the prospect of cost-effective strategies holds promise for broader implementation.
Certain desirable traits (i.e., feed efficiency, disease resistance, fillet/carcass yields, and flesh quality) are challenging to measure directly and require evaluations of siblings. In contrast to classical pedigree-based selection, GS excels at capturing within-family genetic variation, thereby enhancing the overall genetic response. GS also endeavours to reduce the generation interval and mitigate the challenges associated with inbreeding. The comprehensive GS program encompasses the establishment of breeding populations, individual phenotyping, estimation of genomic estimated breeding values (gEBV), trait-based selection, and the development of training populations involving genotyping. The single-step Genomic Best Linear Unbiased Prediction (GBLUP) method optimally utilizes available information for precise predictions.
Thus, genomic selection involves selecting individuals based on estimated breeding values (gEBV) derived from genotypic (genome-wide genetic markers) and phenotypic information (Yáñez et al., 2015). The use of genomic selection in significant aquaculture species has been made feasible with high-quality reference genomes and genome-wide markers. The development of SNP arrays and genotyping methods has enabled the integration of the findings into genomic selection programs for aquaculture species. In particular, for qualitative characteristics like disease resistance and stress tolerance, these techniques have a substantial potential to increase genetic gain and improve the accuracy of anticipated breeding values, as noted in several review articles (Zenger et al., 2019; Houston et al., 2020; Yanez et al., 2023; Cao et al., 2020). Several studies hypothesized that genome-wide association studies (GWAS) may be used to assess the underlying genetic variation of complex traits.
Genomic selection is a recent advancement being utilized in aquaculture species, which have available marker panels/arrays (Yanez et al., 2023). Initially, genotype by sequencing with 12,311 SNP markers was applied in common carp for evaluating the potential of GS for growth improvement by estimating breeding values for 1,425 juvenile carp. The heritability for body length at 120 days was found to be 0.33 (Palaiokostas et al., 2018). The study led to an 18% improvement in the genomic prediction accuracy of estimated breeding values (EBV). To enhance the prediction accuracy of estimated breeding values and genetic gain against Koi Herpesvirus disease (KHVD) in common carp, the genomic selection was conducted on 1,425 juveniles using phenotypic and SNP markers. It was discovered that a QTL on LG 44 accounted for 7% of the genetic variance in KHVD resistance. Comparing the genetic prediction to the pedigree BLUP technique, the accuracy ranged from 8% to 18% higher (Palaiokostas et al., 2019). In yellow river carp, genomic selection using an SNPs array increased growth-related trait prediction by 2% (Wang et al., 2021a). In Amur mirror carp, low-density SNP panels (218 SNPs) showed high heritability (0.42–0.96) for KHVD resistance in selected populations (Prchal et al., 2023). This shows that genomic selection strategies for enhancing KHVD resistance in carps can be successful when using low-density SNP arrays.
The role of genomics in enhancing various traits in aquaculture has been explored through several studies. In crucian carp, a study utilizing 8 K SNP markers with 50 linkage groups (LGs) demonstrated their association with feed conversion efficiency, explaining 14.0%–20.9% of phenotypic variations (Pang et al., 2017). Gibel carp (C. gibelio) showcased genomic markers linked to feed conversion efficiency, indicating potential reductions in fishmeal consumption and improved fish growth (Shi et al., 2022). In common carp, the identification of 28 K SNP markers associated with Feed Conversion Efficiency (FCE) and genotyping with the carp 250 K SNP revealed nine key genes linked with feed efficiency traits (Zhang et al., 2021). Additionally, QTL mapping and GWAS in a Mirror carp family challenged with CyHV-3 highlighted the potential roles of mTOR, herpes simplex infection pathways, and homoeologous expression divergence in immune responses (Chen et al., 2022). Genome-wide studies on common carp families identified significant loci and candidate genes associated with muscle fat content (Zheng et al., 2016). In yellow river carp, GWAS associated with abnormal scale patterns, using the common carp 250 K SNP, identified key mutations in the fibroblast growth factor receptor 1 a1 (fgfr1a1) gene, suggesting potential for selection based on these mutations (Zhou and Gui, 2018). Additional, GWAS studies in common carp focused on muscle fat content and abdominal fat traits, revealing the complex genetic basis of fat metabolism and deposition (Zheng et al., 2016). In grass carp, GWAS with a 21 K array showed positive genetic correlation (0.9) and moderate to high heritability (0.4–0.5) among growth-related traits (Hao et al., 2023). Similarly, in bighead carp, GWAS associated with head-size and head-shape traits identified six significant SNP markers, suggesting their potential use in selection programs (Zhou et al., 2021b). Skin color, a crucial trait in cultured species, was investigated in yellow river carp through GWAS using the common carp 250 K SNP array, identifying eighteen significant SNP markers and shedding light on the genetic basis of abnormal skin coloration (Jiang et al., 2022a).
Despite challenges such as genotyping costs and limitations in the size of the reference population, genomic selection (GS) stands out as a promising approach for optimizing aquaculture breeding programs. The implementation process involves constructing a genomic prediction equation, estimating genomic breeding values (gEBV), and selecting candidates based on these values. Rigorous independent validation ensures the accuracy of predictions, facilitating the identification of the most suitable animals for breeding. However, GS faces constraints related to mating design, the number and size of families, the number of generation intervals, genome size, marker panel density, and selection methods. Parameters like genotype by environment interactions, the application of heterosis in multi-trait selection, realistic LD, modeling, and index selection have been largely overlooked in simulation studies to date. These aspects warrant investigation to efficiently optimize potential GS breeding programs.
4 Genomic resources in cyprinid species
4.1 Whole-genome and mitochondrial genome sequence of important cyprinids
The field of aquaculture and fisheries has experienced a revolution in genomic research owing to advances in sequencing technology. A genome encompasses all the DNA content, both coding and non-coding, within an organism. The year 2011 marked a significant milestone when the first genome of Atlantic cod was successfully sequenced (Star et al., 2011). Since then, the genomes of many significant aquaculture species have been sequenced and made available in public databases (Star et al., 2011; Lu and Luo, 2020). The progress in genome sequencing of important fish species has been extensively reviewed by various authors (Lu and Luo, 2020; Fan et al., 2020). In 2009, the Fish 10 K Genome Project was launched with the ambitious goal of sequencing and assembling genomes of approximately 10,000 vertebrate species, including 4,000 fish species (Bernardi et al., 2012).
Among the cyprinid species, seven important carp genomes have been sequenced, as depicted in Figure 2, and are available for various applications such as growth, development, disease resistance, reproduction, etc. The domesticated gynogenetic C. carpio (strain Songpu) genome was successfully sequenced in 2014, making it the first carp genome based on several NGS technologies and a hybrid assembly method. This genome was analyzed to have a GC content of 37.0%, which was somewhat greater than Danio rerio’s but much lower than the GC contents of other sequenced teleost genomes. In addition, scientists used resequencing on four wild and six domestic strains from Asia and Europe to explore the existing genetic diversity (Xu et al., 2014b).
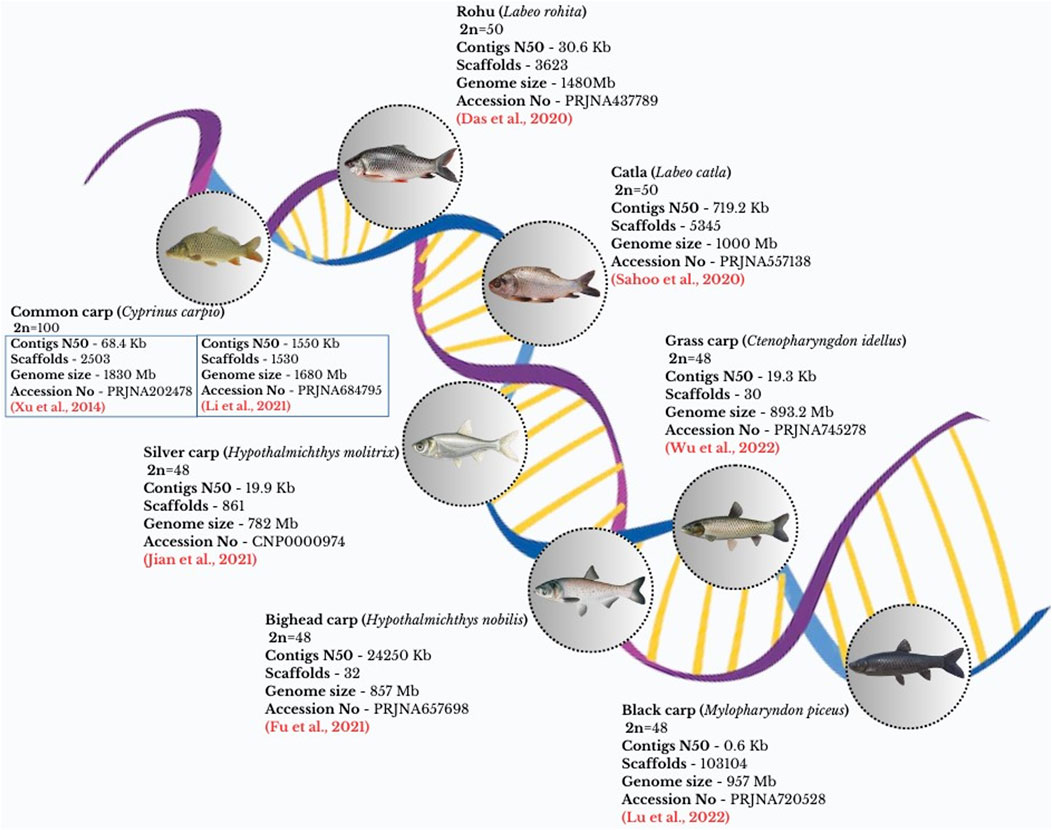
Figure 2. Whole genome sequence of important cyprinid species indicating genome size, Contigs N50, Scaffolds number.
The grass carp’s genome was sequenced and found to be 1.07 Gb for a female and 0.9 Gb for a male. The genome’s N50 was 6.5 Mb, distributed across 114 scaffolds (Wang et al., 2015). Nevertheless, to address the low continuity level and absence of chromosomal-level assembly, resequencing was performed using the PacBio system (Wu et al., 2022a). Sequencing of the double-haploid silver carp and bighead carp’s genomes produced 837 Mb and 845 Mb draft genome assemblies, respectively. According to the results of the phylogenetic study, the silver and bighead carp formed a clade, and their estimated period of divergence was around 3.6 million years ago (Jian et al., 2021).
A total of 1.01 Gb assembled genome size was reported for Labeo catla. This serves as an important resource for research on the comparative biology, evolution, and genomes of cyprinid species (Sahoo et al., 2020). In the case of Rohu, a draft genome of 1.48 Gb, along with five million SNPs, were reported using resequencing of wild populations (Das et al., 2020). Recently, high-quality de novo genome sequencing of rohu was carried out using next-generation sequencing platforms, resulting in 946 Mb-sized final genome and 2,844 unplaced scaffolds (Arick et al., 2023). Black carp’s genome survey assembly was performed by using Illumina sequencing technology. This centered on the evolution of genes or gene families linked to innate immunity as well as developmental distinctions from other carps (Lu et al., 2022). Recently, HiFi genome assembly of four major Asian domestic carps have been generated and available for various purposes including speciation, adaptation and evolutionary dynamic studies (Wang et al., 2024; Chen et al., 2024).
Complete mitochondrial genome sequencing of various carp species such as L. catla of 16,597 bp in length (Kibria et al., 2020) L., rohita of 16,606 bp in length (Das et al., 2020), C. mrigala of 16,594 bp in length (Bej et al., 2013) mud carp, C. molitorella of 16,602 bp in length (Zhang et al., 2015), C. idella of 16,609 bp in length (Wang et al., 2008a), black carp, M. piceus of 16,581 bp in length (Lin et al., 2016), and L. fimbriatus of 16,614 bp in length (Sahoo et al., 2016) has been reported. Mitochondrial DNA (mtDNA) finds extensive applications in conservation genetics and phylogenetic analysis of numerous freshwater fish species. Its high mutation rate makes it a valuable tool for detecting evolution and species boundaries (Das et al., 2013). In population genetics, mtDNA has proven to be useful for assessing genetic diversity, population structure, species identification, evolutionary studies, and estimating introgression from restocking efforts (Wenne, 2023a) as listed in Table 1. The most widely employed molecular method for species identification is DNA barcoding, which involves sequencing, amplifying, and analyzing specific regions such as the Cytochrome C oxidase 1 (COI) gene of the mitochondrial genome (Hallerman, 2021a).
Over the last decade, significant efforts have been devoted to sequencing and re-sequencing genomes of various carp species, employing multiple sequencing platforms and computational tools. Integration of transcriptomics, epigenomics, and other functional genomics techniques with whole-genome sequencing (WGS), researchers have made significant progress in identifying and characterizing genes linked to specific traits (Misra et al., 2019). However, the potential of this genomic information has not been harnessed for genomic selection for production enhancement or fisheries management.
4.2 Transcriptomics studies in cyprinid species
The transcriptome represents the complete set of genes in particular cells, tissues, or organisms at specific physiological conditions, developmental stages, and external environmental stimuli (Wang et al., 2009; Lindberg and Lundeberg, 2010; Heras, 2021; Qian et al., 2014). With the rapid development in the NGS and computational tools, several transcriptome studies in model and non-model fish were performed (Chandhini and Rejish Kumar, 2019). The transcriptomic analysis enables a systematic understanding of the gene profile and their expression patterns in fish (Barange et al., 2018; Chandhini and Rejish Kumar, 2019). RNA-seq approach is immensely used for analyzing dynamic transcriptome in non-model fish including carp species where genomic resources are not available (Qian et al., 2014). Numerous biological processes in carp species, including immunological response, biotic and abiotic stressors, developmental biology, etc., have been uncovered through RNA-seq. Fish-T1K project initiated by Beijing Genomics Institute, China, in November 2013 in collaboration with several research institutes, aimed at generating 1,000 transcriptome sequences of diverse fishes using RNA-seq approach (Sun et al., 2016). Under this initiative, transcriptome data of more than 200 species have been completed and the majority of species belonged to three orders such as Cypriniformes, Perciformes, and Cyprinodontiformes. Studies at the transcriptome level can assist in locating important loci or biomarkers to understand their function in physiological processes and reveal the way they respond to fluctuating environmental factors, including hypoxia, pollution, temperature, and salinity. We have given brief details of transcriptome studies undertaken in carp species in Table 2.
The first transcriptome sequencing in common carp to identify innate immune genes responding to Mycobacterium marinum discovered 39 TIR (Toll/interleukin-1 receptor) domain-containing transcripts/genes (Zhang et al., 2011). Similarly, in rohu, transcriptome sequencing revealed several transcripts associated with the immune response to A. hydrophila challenge, including HSP70, HSP90, HSP30, major histocompatibility (MH), and glycoproteins or serum lectin (Robinson et al., 2012). In the last 5–6 years, there has been significant progress in transcriptome work in carps, particularly focusing on metabolism, growth, disease, biotic or abiotic stress, immunity, and related areas.
In black carp, transcriptome sequencing of liver and brain tissues revealed 13,737 differentially expressed genes (DEGs) in response to fasting (Dai et al., 2021). In grass carp, liver and brain transcriptome analysis under heat stress (36°C) vs. control (28°C) exposure identified 2,534 DEGs in the liver and 1,622 in brain tissue (Zhang et al., 2022). A liver transcriptome analysis in genetically selected Jayanti rohu (L. rohita) revealed 15,232 and 15,360 differently expressed transcripts when fed with a high carbohydrate diet (40% and 60% starch level) as compared to the control diet (20% starch level), demonstrating upregulation of critical transcripts related to glycogen production and de novo lipogenesis (Rasal et al., 2020). “FishGET,” a recent development, offers a comprehensive gene expression and transcriptome database for eight species, including grass carp. It facilitates the discovery of new genes and non-coding RNAs, serving as a valuable reference for transcriptome annotations (Guo et al., 2023). The “FishGET” RNA-seq database contains 1,362 paired-end data from 97 bio-projects across 8 species, providing extensive sequence data for getting valuable insights into various aspects of carp biology, such as growth, reproduction, nutrition, stress response, adaptation, and immunity.
4.3 Epigenetic factors (DNA methylation and miRNAs) in cyprinid species
Epigenetics is the study of changes in the expression of genes and/or phenotypes of organisms that occur without a change in the underlying DNA sequence (Gibney and Nolan, 2010; Liu et al., 2022). Earlier, epigenetic research in aquaculture species was often ignored, but in recent years it has drawn attention to the biological responses of fish to stress, pollution, disease, and food (Metzger and Schulte, 2016; Gavery and Roberts, 2017; Rasal et al., 2023). The epigenetic alterations of DNA methylation, histone modification, and micro RNAs (miRNAs) are significant in a variety of biological processes, including development, reproduction, and growth (Feil and Fraga, 2012; Xu and Xie, 2018). DNA methyltransferases (DNMT) are involved in the addition of a methyl group to the cytosine base during DNA methylation (Luo et al., 2018). Methylation of CpG sites in gene coding regions or promoter sequences can inhibit transcription, while intragenic methylation may stimulate transcription (Jones et al., 2015; Han et al., 2022). The accessibility of fish whole genome sequences enables the identification of DNA methylation sites.
Previous studies suggested that epigenetic factors might be included in selective breeding programs since they operate as a mediator between phenotype and genotype (Anastasiadi et al., 2021; Granada et al., 2018). During the last 3–4 years, epigenetic research has been concentrated on carp species in response to environmental conditions or metabolic stressors, disease, etc. Recently, great efforts have been made to decipher epigenetic regulation in disease control in candidate aquaculture fish, including carp. In grass carp, genome-wide DNA methylation studies depicted that age-dependent epigenetic factors play a crucial role in grass carp reovirus (GCRV) susceptibility (He et al., 2022). Using the whole-genome bisulfite sequencing (WGBS) approach, this study identified 6,214 differentially methylated regions (DMRs) and 4,052 differentially methylated genes in five-month-old (FMO) and resistant three-year-old (TYO) grass carp, respectively. This work suggests the role of epigenetics in gene regulation, which is influenced by the age of fish during viral disease infection, and that can be helpful in the development of breeding programs for disease-resistance traits. In common carp, methylation studies at the global and gene-specific levels in fresh and old spermatozoa reveal that sperm aging impacts the effectiveness and rate of fertilization (Cheng et al., 2021). Similarly, increased H4K12ac expression level was found to be related to aging oocytes in common carp (Waghmare et al., 2021).
Studies on DNA methylation in Crisp grass carp (CGC) and improved varieties of grass carp (GC) have shown that faba bean (Vicia faba L.) diet-induced epigenetic alterations contribute to the development of muscle (Li et al., 2022). This work identified a total of 14 key differentially methylated genes associated with muscle development and provided insights for nutritional programming research. Studies have shown that the growth feature is crucial for aquaculture output and that allotriploid carp heterosis and growth are regulated by DNA methylation (Ren et al., 2022). In Supplementary Table S1, we have compiled a summary of DNA methylation studies conducted in carp species.
MicroRNAs are a type of epigenetic factor, non-coding single-stranded (18–23 nt) small RNA involved in the regulation of gene expression during cell development and other biological events (Rasal et al., 2016). These microRNAs predominantly bind to the 3′untranslated region (UTR) of target genes, leading to the repression or degradation of their target mRNA, thereby facilitating regulatory mechanisms. In teleost, numerous miRNAs have been identified and documented in the miRNA database (http://www.mirbase.org/), with 280 miRNA entries reported for the common carp. Similarly, in carp species, the presence of miRNAs has been reported in response to several biological events and depicted their regulatory role. Earlier studies in carps revealed that environmental or metabolic factors may influence the transcriptional processes via mRNA-miRNAs interactions. Based on the rapid refeeding response after fasting in grass carp, eight miRNAs (miRNA-1a, miRNA-133b, miRNA-133a, miRNA-181a, miRNA-214, miRNA-146, miRNA-206, and miRNA-26a) in fast skeletal muscle were identified, and expression analysis suggested their role during myogenesis (Zhu et al., 2014).
MicroRNA profiling in carp species provides insight into the role of miRNAs in numerous biological functions. Profiling of microRNAs in the liver of common carp identified five major miRNAs (miRNA-137, miRNA-143, miRNA-21, miRNA-146a, and miRNA-125b) to be related to oxidative stress in response to different diets (Yang et al., 2019). Similar to this, prolonged exposure to 1-methyl-3-octylimidazolium bromide (C8mimBr) affected the expression of miRNA-125b, miRNA-155, and miRNA-21 in the spleen of silver carp, which suggested their role during inflammation and oxidative stress (Ma et al., 2019). In Labeo bata, a total of 231 conserved and 445 novel liver-specific microRNAs linked to the carbohydrate metabolic pathway were identified (Rasal et al., 2019). Similarly, in rohu, miRNA studies identified 138 conserved and 161 novel liver-specific miRNAs associated with carbohydrate metabolism. Further expression analysis of five key miRNAs, namely miR-22, miR-122, miR-365, miR-200, and miR-146, suggested their involvement in metabolism when fed with high carbohydrate diets (Rasal et al., 2020). To provide a comprehensive overview of the miRNA studies conducted in carp species, we have compiled the information in the Supplementary Table S1.
MicroRNA studies in carp species are at an early stage and have emerged as a fundamental factor or biomarker for understanding their role in gene regulation. In response to various environmental stresses, diseases, and feeding habits, research on miRNA in a selected carps has considerably increased during the past several years. Their molecular interaction with the target genes must be defined in more detail to serve as a biomarker for the improvement of aquaculture practices.
4.4 Microbial characterization in cyprinid species
The term “metagenomics” refers to the study of the structure and function of entire nucleotide sequences isolated and analyzed from all the organisms (often but not always microbes) in a bulk sample (Thomas et al., 2012). This allows the scientific community to explore the diverse range of microbes that inhabit in mammals, oceans, soils, fishes, and the environment (Frank, 2008; Ashwath et al., 2022). Advances in DNA sequencing and computing methods have achieved paradigm shifts in metagenomics, enabling comprehensive study of the complex microbial system. Genomics analysis has undergone a significant shift towards metagenomics, allowing the study of entire microbial communities without the necessity of isolating and culturing individual species. Given the vast diversity of fishes in terms of species and habitats, it is crucial to conduct ecological assessments that identify microbes and their diversity both in the aquatic environment and within the fish gut. Fish gut microbiota is play an important role in the microbial fermentation of feed which adds to the host’s health (Flint et al., 2008; Round and Mazmanian, 2009; Wu et al., 2012; Ringø et al., 2003).
The diversity of microbes was studied based on the 16sRNA gene sequencing, which is highly diverse among microbes, but it does not provide a functional aspect (Vaishampayan et al., 2010; Johny et al., 2021). The bacterial diversity has been studied in grass carp, rohu, common carp, gibel carp, etc., by harboring 16sRNA sequencing (Mukherjee et al., 2020). In common carp, bacterial diversity was identified based on various aspects such as the effect of pollutants, bacterial or viral challenges, antibiotics, etc. As the skin-mucus bacterial diversity determines the health of the host, skin-mucus bacterial diversity in common carp was assessed and showed the presence of dominant phyla such as Proteobacteria, followed by Actinobacteria, Bacteroidota, Firmicutes, and Cyanobacteria (Papp et al., 2023). In grass carp, the majority of Proteobacteria was replaced by Bacteroidetes during larval feeding as opposed to the egg stage (Wang et al., 2015). Gut microbial diversity in silver carp demonstrated seasonal clustering (Ye et al., 2014). In goldfish Bacteroidetes were observed dominantly in response to pentachlorophenol (PCP) exposure (Kan et al., 2015). Studies suggest that the feeding behavior of various carp species has a notable impact on the composition of their gut microbiota (Li et al., 2017a). Studies carried out on grass carp revealed correlated gut microbiota composition following the regime (Ni et al., 2014). The bacterial diversity in the gut of fish influences fish physiology as well as health. In the crucian carp, opportunistic bacteria like Vibrio, Aeromonas, and Shewanella were found to be more during infection with the red-operculum disease (Li et al., 2017b). Similarly, changes in gut microbiota were seen in rohu in response to argulus infection, revealing the presence of Actinobacteria, Patescibacteria, Stenotrophomonas and Pirellula in infected fish (Mondal et al., 2022).
The chronic toxicity of Norfloxacin (NOR) concentrations on the gut microbiota of juvenile common carp was evaluated using 16sRNA sequencing. The prevalent bacteria in the gut of common carp included Proteobacteria, Bacteroidetes, Fusobacteria, Firmicutes, and Actinobacteria (Li et al., 2023). Analyses of acquired antibiotic-resistance genes (ARGs) in the intestinal microbiome of common carp indicate a possible link between acquired ARGs in domestic and wild animal populations (Libisch et al., 2022). Meta-transcriptomic viral survey of invasive and native common carp across the Murray–Darling Basin did not detect CyHV-3 or any closely related viruses. This suggests that there is little virus transmission from common carp to native fish species (Costa et al., 2021). Herewith, we have summarized the identification of bacterial diversity in carp species using 16 S RNA sequencing approaches in Supplementary Table S2. Overall, 16sRNA sequencing approach in carp species revealed that bacteria in the fish gut microbiome are dominated by the phyla Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, Bacilli, Clostridia, and Verrucomicrobia (Xia et al., 2014; Wu et al., 2012; Roeselers et al., 2011). While Proteobacteria, Firmicutes, and Bacterioidetes contribute around 90% of fish gut bacterial diversity which showed their physiological role during digestion, immunity, and nutrient acquisition (Ingerslev et al., 2014; Johny et al., 2021).
Hologenome concept views the holobiont comprising a host and its microbiomes as the primary unit of selection in evolution (Alberdi et al., 2022; Limborg et al., 2018). Earlier research underscores the critical role that microbiomes play in host fitness and local adaptation. Hologenomics studies, which examine the genetic interactions between hosts and their microbial communities, hold great promise but remain an underexplored field. This approach could significantly improve aquaculture by enhancing growth, health, and sustainability. For instance, studies on invasive carp species in the Mississippi River Basin suggest that hybrid carp gut microbiomes contribute to their successful adaptation and invasion by aiding in food resource digestion (Zhu et al., 2021). While hologenomics shows great potential, especially in aquaculture, it remains in its early stages and is not yet widely applied.
4.5 Proteomics and metabolomics in cyprinid biological research
A proteome is a representation of the whole protein complement of the genome. Proteome composition varies over time between tissues and is studied through proteomics, which is the study of all proteins in a certain cell, tissue, or organism (Rodrigues et al., 2018). Thus, although the genome is constant for organisms, proteome content is dynamic and changes as per the developmental stage or in response to external stimuli. The studies suggest that this field is useful for understanding fish developmental biology and could be useful for the identification of novel biomolecules or proteins. Several researchers attempted to review progress made in proteomics research in fisheries (Nissa et al., 2021; Jaiswal et al., 2023). In carp species, proteomic research is at an infancy level, and recently it gained attention to study proteome of biological systems in response to disease, stress, or physiological stages. In addition to these, this field is also being utilized for examining fish food quality and authentication using protein biomarkers (Mazzeo and Siciliano, 2016). Earlier studies showed that proteomic information could be used for toxicological research in carps, such as in goldfish (Wang et al., 2008b).
Mass spectrometry was used for the first time to analyze the spermatozoa of common carp, identifying 348 proteins, including numerous proteins such as Dynein, tubulin, HSP70, HSP90, HSP60, NKEF-B, adenosylhomocysteinase, brain type creatine kinase, mitochondrial ATP synthase, and valosin (Dietrich et al., 2014).
In cyprinid aquaculture, diseases are the major hurdle in production, and thus understanding fish immunity and host-parasite relationship could be helpful to devise a strategy for disease control. The proteomic study of host tissues linked to pathogenic infection is a powerful approach to identifying signature proteins as shown in common carp, grass carp, and rohu. In rohu, liver proteomic analysis revealed 158 differentially expressed proteins in response to A. hydrophila infection (Nissa et al., 2023). Similarly, in common carp, alteration of proteome was observed in skin mucus due to Ichthyophthirius multifiliis infection, and several key biomarker proteins were identified, such as dermatopontin, lumican, olfactomedin 4, papilin I, and cytoskeletal 18 (Saleh et al., 2018).
Despite these efforts, there is still a need to investigate carp fish proteomes in response to environmental stressors, pollutants, pathogens, etc. The research communities need to have access to a comprehensive proteome of significant carp species. Recently, an open-source database named “Rohu PeptideAtlas” has been released. This resource contains 6,015 proteins, 2.9 million PSMs (Peptide-Spectrum Matches), and approximately 150 thousand peptides sourced from 17 distinct normal tissues, plasma, and embryos of rohu. This valuable repository will play a crucial role in supporting the carp aquaculture communities (Nissa et al., 2022).
Metabolomics employs high-throughput mass spectrometry (MS) techniques to analyze and estimate the content of metabolites, with a molecular weight below 1,500 Da, within an organism (Alfaro and Young, 2018; Kumari et al., 2022). Earlier, those sorts of studies were applied in humans, but recently metabolomics studies have gained consideration in animals, including fishes for environmental, health, and nutritional research (Brennan, 2013; Goldansaz et al., 2017; Roques et al., 2020). In fish nutritional aspects, metabolomic provides insights of metabolic effects in response to different feed and nutrient content as reviewed earlier (Kumari et al., 2022). In metabolomic studies, several compounds such as carbohydrates, fatty acids, amino acids, nucleotides, steroids, organic acids, and carotenoids can be studied. In a holistic approach, this field helps to identify the complexity of fish species in response to nutrition and environmental conditions (Sébédio, 2017; Young and Benton, 2018).
In cyprinid species, metabolomic research is very limited, and recently few studies depicted its use in understanding the impact on fish nutrition and health (Alfaro and Young, 2018). When fed, The metabolic profile of grass carp in response to imbalanced diet with low levels of protein showed a change in lipid content with SFA (Saturated Fatty Acids), MUFA (Mono Unsaturated Fatty Acids), and PUFA (Poly Unsaturated Fatty Acids owing to regulation of choline and end products of glycerol and glycerol-3-phosphate (Jin et al., 2015b). Thus, diet plays a significant role in the metabolic profile of fish. In grass carp, metabolomic studies revealed dietary effects on flesh quality when fed with grass and artificial feeding. While grass-fed animals had greater levels of n-3 unsaturated fatty acids (UFAs) such as eicosapentaenoic acid, alpha-linolenic acid, and gamma-linolenic acid, those fed artificially had higher levels of arachidonic acid, docosapentaenoic acid, and gamma-linolenic acid (Zhao et al., 2018).
It was suggested that environmental conditions, pollutants, and abiotic factors could also affect the health/welfare and metabolism of fish. To understand the physiological mechanism of anoxia in crucian carp, the metabolic profile investigated high levels of succinate in different tissues, including brain, liver, heart, and blood plasma, as compared to normoxic (control) fish (Dahl et al., 2021). Recently, in Qingtian paddy field carp (C. carpio var qingtianensis), metabolite profile in response to hypoxia revealed 131 differentially expressed metabolites as compared to control groups, and a total of 63 metabolites with differential expression were noticed in reoxygenated and the hypoxic fish (Jiang et al., 2022b). The impact of toxicant such as endosulfan sulfate was studied on dead or live common carp using GC–MS-based metabolome profiling and a total of 30 significant metabolites (biomarkers) associated with starch, galactose, glycerolipid, and sucrose metabolism were identified among dead and live carp (Lee et al., 2020). Similarly, metabolomics explored the harmful effects of 17-Ethynylestradiol (EE2) in crucian carp. This led to the identification of 24 metabolites in the gonad and 16 metabolites in the kidney, as well as the disruption of lipid metabolism. A specialized database (reference spectra of the metabolites) is still needed for fish metabolomics investigations, particularly in carp species, to examine inter- and intra-species metabolome profiles (Goldansaz et al., 2017).
5 Molecular maker discovery in cyprinid species
Molecular markers are DNA sequences in the gene or genome that show genetic alterations at a particular location leading to genetic variation. This depicts the differentiation in the form of DNA polymorphism between organisms, species, population genetics studies at population levels that can be used for mapping and identifying individuals. Several authors reviewed the progress made in molecular marker discovery and their applications in the teleost species (Chauhan and Rajiv, 2010; Wenne, 2023a). In carp species, several genetic markers were identified for assessment of genetic diversity, parentage assignments, species detection, population structure analysis, evolutionary study, linkage map construction, and selective breeding. It includes RAPD, RFLP, AFLP, microsatellites (SSRs), ESTs,SNPs, etc. Genetic markers also have been used for the conservation of genetic resources, stock differentiation, finding quantitative trait loci (QTL), and fisheries management. The use of proteins and allozymes as markers arose in the 1960s, but since has declined.
However, it has been observed that most of these phenotypic and biochemical markers exhibit limited polymorphism when studied in the majority of carp strains (Crooijmans et al., 1997), which restricted its application. RAPD and AFLP have limited applicability due to their drawbacks, including dominant nature and irreproducible findings, respectively. On the other hand, genetic markers such as microsatellites, single nucleotide polymorphisms (SNPs), expressed sequence tags (ESTs), and mitochondrial (mtDNA) markers have become prevalent in aquaculture-related research. These new markers offer more reliable and diverse options for studying genetic variations in aquatic species. The use of molecular markers has augmented classical phenotype-based selection. These molecular markers increase accuracy in identification of superior individuals or families. Although, several studies implicated the use of markers in the cyprinid aquaculture and management, recently developed NGS and computational tools have enabled precise DNA marker discovery associated with traits of interest (Sundaray et al., 2016; Rasal et al., 2017b). This section aims to provide an overview of the most recent applications of genetic markers in studying cyprinid species, which consistently rank as the top performers in Indian aquaculture productivity.
Microsatellites have found extensive use in research focusing on fisheries and aquaculture since the early 1990s. The microsatellite also known as simple sequence repeats (SSRs) are highly polymorphic DNA markers, which are neutral, co-dominant, high polymorphism, consisting of short 2-4 bp nucleotide repeats and wide distribution in organisms’ genomes (Sundaray et al., 2016; Wenne, 2023a). In earlier studies, they have been widely used for population genetic structure analysis, QTL mapping, stock characterization, marker-assisted selection, and breeding. The next-generation sequencing (NGS) genome assay identifies multiple microsatellites, of which a few highly variable loci serve adequate for population genetics and aquaculture applications (Mojekwu and Anumudu, 2013).
A total of 17 microsatellite loci showed 86.0%–96.0% genetic identity during the genetic differentiation among hatchery and wild populations of Oujiang common carp, C. carpio var. color (Ma et al., 2011). Similarly, 10 microsatellite loci were used during the analysis of population structure and genetic diversity of 11 carp strains from Germany (Scheuerman and Glinzig mirror carp), France (Forez and Dombez scaly carp), Amur river farmed stock, and one wild stock from the Ebro river (Spain), and it demonstrated 89.6% accuracy bayesian clustering (Ma et al., 2011). To analyze population structure and genetic diversity in silver carp, a total of 159 SSRs (27 polymorphic loci) were identified using transcriptome sequences, and a total of 134 SSRs (65 loci being polymorphic) markers were developed using wild silver carp populations (Feng et al., 2014; Guo et al., 2013).
Recently, SSRs were generated using the genome of silver carp to assess the genetic variation of the four populations, and they found a total of 368,572 SSRs or 0.77% of the genome. The most prevalent SSRs were discovered to be di-nucleotide repeats (55.59%) with AC/GT, and 13 pairs of SSRs were used to illustrate the genetic variation across four groups (Wang et al., 2022c). Similarly, in common carp, analysis of whole genome shotgun (WGS) sequences revealed a total of 79,014 microsatellite loci, and genotyping with 192 individuals showed 963 polymorphic markers that could be used for high-density linkage mapping (Ji et al., 2012). To evaluate genetic purity and genetic diversity of the 13 strains of Hungarian common carp, 12 SSRs markers were used and showed a significant level of genetic differentiation (3.79%) between strains with 93.64% accuracy in bayesian clustering analysis (Tóth et al., 2020).
Initially, 21 SSRs were identified for population structure analysis in rohu (Patel et al., 2008). Subsequently, an extensive investigation of rohu transcriptome data revealed a total of 22,383 EST-SSRs, with 29 of them significantly associated with reproduction-related transcripts (Sahoo et al., 2016). FishMicrosat: a microsatellite database was developed for various fishes, which includes important carp species such as L. rohita, L. catla, C. idella, and H. molitrix (Nagpure et al., 2013). This database serves as an important resource of microsatellite markers for evolutionary, population structure, cross-species loci identification, and genetic relatedness among carp species. Microsatellite markers were used to examine genetic variation among nine catla strains before the start of a selective breeding program. The study found a genetic heterogeneity with 0.4137 FST estimates, and 58.63% of the variation was observed among the individuals (Mahapatra et al., 2018). These markers were successfully used for base population establishment during the catla selective breeding program. Several SSRs in black carp were found, and their application in genetic diversity analysis across wild and farmed populations was demonstrated (Zhou et al., 2020). We have summarized the identification of SSRs markers in carp species with their uses in Supplementary Table S3.
High abundance SNP markers is a popular choice for population genetic studies in aquaculture (Wenne, 2023b; Abdelrahman et al., 2017; Rasal et al., 2017b). Millions of SNPs have been identified in carp species due to advancements in NGS technology, as shown in Supplementary Table S3. These SNPs are useful in generating linkage maps, identifying key phenotypic QTLs, and marker-assisted selection (MAS) in aquaculture (Yáñez et al., 2016). It is possible to decode connections between SNP markers and production or performance attributes using genome-wide association studies (GWAS) (Robledo et al., 2018). For the analysis of 68 individuals from second-generation mirror carp hybrids, C. carpio, 336 SNPs were taken into account (Jin et al., 2012), and further studies implicated that a total of 978 SSRs could be used for genetic map construction, consisting of 50 linkage groups (Jin et al., 2015a). A genomic map of the common carp was created utilizing 8,487 SNPs spanning 3762.88 cM and 50 linkage groups (Laghari et al., 2014; Xu et al., 2014a; Liu et al., 2017).
Carp genome re-sequencing reveals millions of SNPs associated with growth or disease resistance. In rohu, through whole genome sequencing and re-sequencing of 10 wild populations, 4.95 million SNPs (380,991 to 679,963) were identified in each population (Das et al., 2020). A total of 9,157 SNPs were found in rohu breeding populations from Halda, Jamuna, and Padma rivers. Among them, 1985 SNPs exhibited slight genetic divergence between the river populations (Hamilton et al., 2019b). The rohu genome and transcriptome resequencing revealed 39,158 high-quality coding SNPs. Twelve of these SNPs were linked to genes involved in cellular development and proliferation, including myf 6, fgf, gfr-bound protein 10-like, IGF-1-like, and TGF β, among others (Nandanpawar et al., 2023). In grass carp, two SNPs were found to be associated with increased resistance to reovirus and tlr7 expression (Su et al., 2018). Recent resequencing of grass carp genomes identified a total of 9,510,648 SNPs across three populations: Yangtze, Pearl river, and mono-female population, indicating a moderate genetic differentiation between them (Zhang et al., 2023a). In common carp, a 250 k SNP array was developed using re-sequenced genome data, with 84,933 (34.0%) SNPs found to be polymorphic (Xu et al., 2014a). The performance of 24 morphological and growth-related parameters in yellow river carp was evaluated using this SNPs array. The gBLUP method showed medium to high heritabilities of these traits as compared to the traditional BLUP method, with an increase in prediction abilities by 2% (Jiang et al., 2022a).
6 Gene transfer in cyprinid species
Transgenesis, a revolutionary science, involves creating genetically modified organisms by integrating a foreign DNA sequence into its chromosomal DNA (Wang et al., 2021b). Various techniques, such as microinjection, electroporation, transposable elements, and retroviral-mediated transfer, have been employed to produce transgenic individuals. The primary objectives of genetic manipulation in aquaculture include intensifying growth and improving food conservation efficiency, enhancing tolerance to environmental variables such as temperature and salinity, introducing new color variants in ornamental fishes, and developing disease-resistant forms. Microinjection and electroporation have proven to be reliable techniques for gene transfer in fish, with goldfish being the first reported transgenic fish, followed by advancements in species like rainbow trout and common carp (Zhu et al., 1985). Growth hormone (GH) genes, known for their highly conserved sequences, have been a central focus in transgenic studies, targeting over 20 teleost species. Transgenic applications in ornamental fish, particularly those utilizing reporter genes like green fluorescent protein (GFP) or red fluorescent protein (RFP), have led to the commercialization of species like the “GloFish” series, featuring attractive fluorescent colors. While much of the transgenic research in food fish has centered around growth-related traits, other aspects such as disease resistance, tolerance to environmental conditions, and improvements in feed conversion ratio have been explored. The AquAdvantage Salmon, a growth-enhanced transgenic food fish, stands out as the first transgenic fish commercially approved by the US Food and Drug Administration (FDA).
The journey into transgenic fish production in carps began in the mid-1980s when researchers achieved a breakthrough by integrating recombinant human growth hormone (rGH) into red common carp embryos (Wu et al., 2003). These transgenic fish exhibited a remarkable growth rate, surpassing four times that of the control group, showcasing the potential and efficiency of gene transfer in fish breeding. The incorporation of specific promoters, such as the β-actin promoter from carp, facilitated the construction of all-fish transgenic constructs. The “auto transgenesis” approach, utilizing regulatory elements and targeted genes from the same species, proved beneficial in transgenesis. This has led to the development of an “all-fish (CAgcGH, grass carp growth hormone fused with common carp β-actin promoter)” transgenic yellow river carp, where the GH from grass carp was introduced, adhering to biosafety and bioethics standards (Wu et al., 2003). This transgenic common carp not only demonstrated faster growth and enhanced food conversion efficiency but also addressed environmental concerns by producing infertile transgenic triploid fish. These findings were further expanded the scope by introducing the GH of black carp into hybrids of C. auratus red var. and C. carpio, which exhibited rapid growth. Similarly, researchers enhanced the growth of rohu using an “all-fish” transgene construct (GH cDNA from rohu attached to a β-actin promoter from grass carp), resulting in transgenic individuals exhibiting 5–6 times greater growth than their non-transgenic counterparts (Devlin et al., 2009; Rajesh and Majumdar, 2005). The β-actin promoter, showing variable but sometimes ubiquitous expression, was studied in medaka (Cho et al., 2011). The transgenesis using rohu β-actin gene and promoter, demonstrated the potential of endogenous promoters in driving foreign gene expression (Barman et al., 2017). Further studies, mylz2 promoter of rohu carp (1.2 kb) was successfully cloned and direct injection into skeletal muscle showcased the potential for tissue-specific and desired gene expression in transgenic rohu (Barman et al., 2017).
In common carp, other traits such as feed conversion efficiency, appetite, and elevated post-feeding routine oxygen consumption rates are being explored. Transgenic studies extended to the manipulation of fatty acid profiles, as demonstrated by the successful generation of fat1 and fat2 single- and double-transgenic EPC cell lines (Zhang et al., 2019b). These studies resulted in “all-fish” fat1-transgenic common carp populations with significantly increased n-3 PUFA content, emphasizing the potential of transgenic technology to enhance the production of essential fatty acids for human consumption. Moreover, exploring genetically modified fish as bioreactors for producing target proteins is gaining attention. For instance, transgenic zebrafish expressing the human fat1 gene have successfully produced omega-3 long-chain polyunsaturated fatty acids (n-3 LCPUFA), crucial for neural development and human health (Pang et al., 2014). The optimization of Tg (CA: fat1) common carp, achieved through a sperm sample screening method, holds promise as a novel dietary source of omega-3 polyunsaturated fatty acids (n3-PUFAs) (Zhang et al., 2019b). Dietary protein effects on growth and feed utilization were studied in GH transgenic and non-transgenic common carp (Guo et al., 2021). Higher protein improved growth, reduced body lipid, and enhanced metabolic processes in transgenic fish, indicating better overall performance compared to non-transgenic counterparts. Recent studies, GH transgenic common carp demonstrated notable aquaculture benefits, boasting a 12% higher specific growth rate (SGR) and 17% higher feed efficiency (FE) compared to their wild-type counterparts. This investigation explores lipid metabolism in GH transgenic and wild-type common carp, uncovering diminished lipid content across diverse tissues in GH transgenic carp (Wu et al., 2022b). The findings showcase enhanced lipid metabolism and utilization pathways in their liver, providing valuable insights for optimizing lipid efficiency in aquaculture. Interestingly, studies also reported enhancement of food intake GH common carp mediated by upregulating the hypothalamic Agouti-related protein (AgRP) (Zhong et al., 2013).
Transgenic fish with enhanced disease resistance contribute to increased aquaculture production and improved fish welfare (Dunham, 2009). Efforts have been made in this direction by incorporating genes for bactericidal compounds, such as cecropin in channel catfish and human lactoferrin (hLF) in grass carp, to bolster the nonspecific disease resistance of cultured fishes (Wang et al., 2022a). One notable example is the transfer of the hLF gene, creating transgenic grass carp with resistance to Aeromonas hydrophila (Wang et al., 2021b).
The development of transgenic fish has witnessed significant progress in carps, addressing not only growth-related traits but also essential fatty acid composition and tissue-specific gene expression. However, transgenic fish commercialization is limited due to ecological concerns and social issues. To mitigate ecological risks, creating sterile transgenic fish has been explored through ploidy manipulation or hormonal treatment. Confinement strategies, both biological and physical, are proposed to address the potential escape of transgenic fish into the wild. Although several transgenic food fish have been developed, only the AquAdvantage Salmon has been globally commercialized, mainly due to societal and ecological considerations. Biosafety assessments are crucial, and adherence to international guidelines is necessary for the responsible development and deployment of transgenic fish technologies. The challenges include achieving stable transgene integration, mosaicism, and optimizing regulations for safe laboratory practices and biosafety. The integration of traditional selective breeding and transgenic approaches may be essential to meet the growing demands for future food requirements.
7 Genome editing in cyprinid species
Genome editing also known as gene editing is a method that involves making specific changes to the DNA or region of the genome of a cell or organism. Genome editing technologies like transcriptional activator-like effector nucleases (TALENs) and zinc finger nuclease (ZFN) were early methods for targeted genome editing (Cong et al., 2013; Beerli and Barbas, 2002). However, they have been largely replaced by the more efficient and versatile CRISPR/Cas9 system i.e., CRISPR (clustered regulatory interspaced short palindromic repeats)/CRISPR associated nucleases (Cong et al., 2013). CRISPR/Cas9 allows for precise editing of genomes, including gene knockout, activation, inhibition, and even epigenetic modifications, revolutionizing biological research and offering new possibilities for understanding and modifying DNA. Several authors have reviewed the progress of genome editing in aquaculture species and explored new possibilities for enhancing genetic improvements in cultured aquatic species (Hallerman, 2021b; Barman et al., 2017; Gratacap et al., 2019; Gutási et al., 2023; Ferdous et al., 2022). This signifies the potential for cutting-edge genetic technologies to advance aquaculture and improve the quality, growth, and sustainability of farmed aquatic species.
Selective breeding in cultured species is constrained by factors like low heritability of traits, long generation-intervals, the complexity of targeting multiple traits, and limited genetic variation in broodstock. CRISPR/Cas9 offers the potential for rapid genetic improvement in aquaculture by targeting specific traits like sterility, growth, and disease resistance. CRISPR/Cas9 and TALEN genome editing techniques have been effectively used in several aquaculture species, including cyprinids such as common carp, rohu, and grass carp.
In common carp, genome editing tools were used to target genes related to bone development (SP7, SPP1, RUNX2, and OPG) and muscle growth (Myostatin) (Zhong et al., 2016b). TALEN induced mutations in specific target sites, while CRISPR-Cas9 caused severe bone-defects when targeting SP7 genes and increased muscle growth when targeting MSTNba. Moreover, CRISPR-Cas9 enabled the creation of double mutant fish with high efficiency (SP7A; MSTNba). These technologies in common carp represent a significant first step forward in genetic engineering for cyprinid species, with the potential to enhance their qualities and economic value in aquaculture. Recently, CRISPR/Cas9 was used to knockout the MSTN gene in common carp, resulting in fish with increased muscle growth (Shahi et al., 2022). This study was supported by the observed increase in skeletal muscle fiber-density and the upregulation of myogenic regulatory factors.
CRISPR-Cas9 was used to engineer Melanocortin receptor-1 (MC1R) knock-out mutants in Oujiang color common carp (Mandal et al., 2020). These mutants exhibited altered melanophore production, resulting in grayish or albino skin patches. This result suggests that MC1R plays a vital role in melanogenesis in fish. In another study, two Agouti signaling protein (ASIP) genes were targeted by CRISPR/Cas9 technology in Oujiang color common carp and found that they are involved in regulating melanin distribution and aggregation during the formation of black patches on the skin (Chen et al., 2019). Similarly, CRISPR-Cas9 was used to target the Tyrosinase (TYR) gene in white crucian carp (WCC) and its hybrid progeny (WR) (Liu et al., 2019). They successfully reduced TYR protein levels, leading to a reduction in melanin and altered pigmentation in the fish. Additionally, key pigment synthesis genes were downregulated in the mutant fish, highlighting the role of tyr in melanin synthesis.
In gibel carp, CRISPR/Cas9 disrupted foxl2 gene variants and demonstrated differences in their expression, leading to significant effects on ovary development and germ cell depletion (Gan et al., 2021). This work sheds light on gene diversification and evolution in polyploid gibel carp. Recently, CRISPR/Cas9 gene editing targeted two runt-related transcription factor 2b (runx2b homeologs), that play a key role in the development of intermuscular bones (IBs) in gibel carp (Gan et al., 2023). The study demonstrates that simultaneous disruption of both genes led to the complete loss of IBs. In a recent breakthrough, scientists successfully employed genome editing techniques to modify the bmp6 gene in diploid crucian carp (Kuang et al., 2023). This achievement marks a significant milestone as it led to the creation of a specialized strain of crucian carp devoid of intermuscular bones (IMBs). The edited strain exhibited accelerated growth rates when compared to their wild-type counterparts, opening new possibilities for genome editing applications in other cyprinid fish species to eliminate IMBs.
This technique holds promise for creating models to study immune response, and it represents the first evidence of CRISPR/Cas9’s high efficiency in targeted gene-disruption through homologous recombination (HR). The study in rohu carp demonstrates the successful application of the CRISPR/Cas9 system to disrupt the Toll-like receptor 22 (TLR22) gene, marking a notable advance in teleost fish genetics (Chakrapani et al., 2016). In grass carp, CRISPR/Cas9 genome editing was used to target and knockout the gcJAM-A (Junctional Adhesion Molecule-A) gene (Ma et al., 2018). This work reduced grass carp reovirus (GCRV) infection in permissive grass carp kidney cells (CIK), showing promise for controlling the disease in aquaculture.
Therefore, recent studies demonstrate the successful use of genome editing, particularly CRISPR/Cas9, in cyprinid species for trait improvement. The availability of reference genomes, which will allow better design of guide RNAs for the CRISPR-Cas9 vector, resulting in minimized off-target effects. These techniques hold great potential for targeting complex traits like immunity and disease resistance, crucial for aquaculture improvements and disease management.
8 Future perspectives and concluding remarks
Achieving sustainable aquaculture production depends on utilizing a diverse range of genetic and genomic resources. We have comprehensively highlighted future perspectives and way forward in cyprinid species for increasing aquaculture production in Figure 3. The selective breeding program in cyprinid species has successfully improved traits like growth rate and disease resistance. Nevertheless, striking a balance between maintaining high selection intensity and keeping low inbreeding rates remains a long-term challenge. Some breeding programs failed due to a weak base population or limited stocks involved, leading to inbreeding and loss of genetic viability. For successful selective breeding in cyprinid species, it is essential to generate a comprehensive set of heritability values and genetic correlations for important traits. Overlooking genotype-by-environment interactions may result in a range of phenotypes, leading to a shortfall in the desired performance of genetically selected cyprinid species. Therefore, selection strategies should consider GxE interactions during cyprinid breeding programs. Both the Intergovernmental Panel on Climate Change (IPCC) and the Food and Agriculture Organization (FAO) have issued warnings about the severe impact of climate change on global warming and its implications for the aquaculture sector (Cochrane et al., 2009). As a result, it is essential to identify adaptive genes or traits in cyprinid species that can thrive in changing environmental conditions and disease outbreaks.
In cyprinid species aquaculture, the major concern is feed cost, which constitutes 50%–60% of the total production cost. Therefore, enhancing feed use efficiency through selective breeding is essential to reduce input costs in aquaculture. Accurate estimation of FCR traits and feed use efficiency is challenging in fish due to their rearing environment, and incorrect methodologies in many studies, resulting in low heritability estimates. As a solution, an indirect method of selection, such as measuring growth during feeding/re-feeding, could be used to improve these traits in cyprinids. In addition to feed-use efficiency, the selective breeding program for cyprinid species should also consider other important traits like tolerance to temperature, stress, or salinity to achieve genetic improvement. However, implementing and disseminating genetic improvement programs in developing countries face serious constraints, including limited financial resources, infrastructure, and skilled manpower.
The integration of genomic resources into cyprinid species genetic improvement programs is in the early stages. Whole genome sequences and phenotypic data are important for genomic selection. It is crucial to validate markers linked to production and performance traits using larger sample sizes and efficient genotyping methods. Cyprinid species genome sequences are often in draft form, requiring the generation of annotated databases with functional assignment. Combining sequenced reads to create a reference genome for each cyprinid species is essential. Also, various gene transfer techniques were used for genetic improvement and manipulation in aquatic species. Looking at the ecological concerns, sterile transgenic fish need to be produced through ploidy manipulation or hormonal treatment for trait improvement in carps. Investigating the function of predicted genes through expression studies or gene knock-out approaches, like CRISPR/cas9, can further enhance genetic improvement in aquaculture industries. Molecular markers play a crucial role in some fish species’ selective breeding programs, enabling the construction of linkage maps linked to economically important traits. It is imperative to identify genome-wide high-density markers associated with key traits, including disease resistance, feed conversion efficiency, reproductive fitness, and responses to abiotic stressors like temperature, dissolved oxygen, and salinity. Although medium or high-density SNP arrays have been reported for some carp species, the adoption of techniques like restriction-site associated DNA sequencing (RAD-Seq) and genotyping by sequencing (GBS) is essential to generate SNP arrays in other cyprinid species that are relevant to the traits of interest. These advanced genotyping methods will aid in the identification and utilization of valuable genetic variations in cyprinid breeding programs. It is estimated that an SNPs array can increase an average of 25% genetic gain with high selection accuracy in aquaculture species (Allal and Nguyen, 2022).
Polymorphic markers enable high-density linkage maps, while QTL for production traits needs to be identified and validated in cyprinid species. Genomic selection requires genotypic and phenotypic data for accurate GEBV prediction. Reliable phenotypic data is crucial for estimating genetic parameters and improving selection accuracy and genetic gain. GxE relationship data aids in precise genomic selection in cyprinid species. Understanding epigenetic regulation in cyprinid species is limited but important for estimating genetic variation and gene expression modulation. Climate change and environmental factors affect traits involving epigenetic regulation through DNA methylation or miRNAs. Further research on DNA methylation’s mechanism and association with an important traits in cyprinid species is necessary. Epigenome editing can establish causal relationships between epigenetic factors and traits of interest.
To understand complex biological processes and traits in cyprinids, integrating multi-omics data (transcriptome, epigenome, proteome, metabolome, etc.) is crucial. Understanding the gene-regulatory network for growth and disease resistance aids cyprinid aquaculture. Additionally, each cyprinid species should have an integrated database for assessing genomic, proteomic, expression profiles, SNP/microsatellite markers, and linkage mapping data. The tissue-specific proteome data needs to be generated to decipher the key role of expressed proteins in particular situations. This database facilitates comparative genomics and key gene mapping for production traits and aids in population dynamics and fisheries management. The cost of sequencing and GBS has significantly reduced, enabling broader application. However, a downstream analysis may face challenges in dealing with genome complexity as the cost continues to decrease.
Genomic selection has been suggested to yield increased genetic gain with high selection accuracy. However, its implementation in cyprinid species breeding programs is still in its early stages and requires further exploration. Many academic and research institutes have developed genomic resources for important cyprinid species, but greater collaboration between academia and industry is needed to fully harness their potential for the genetic improvement of regional cyprinid species. Looking ahead, growing carp genetics and genomics data can enhance selection accuracy, production, and sustainability in aquaculture. Genomic selection will be pivotal in future cyprinid species breeding programs, surpassing traditional pedigree-based selection methods in terms of accuracy.
In conclusion, this review provides comprehensive information on genetic and genomic progress achieved in cyprinid species. Although selective breeding techniques achieved considerable progress in carp genetic research, further genomic selection holds a promising approach for enhancing the accuracy of breeding values. NGS, TGS (Third-Generation Sequencing) and computational tools have revolutionized the understanding of carps and other species biological research and generated a vast amount of genomic resources. This has further led to molecular-level studies concerning disease resistance, growth, reproduction, and adaptation to changing environments in cyprinids. This review depicts the progress of genomics in important cyprinid species at the level of genome, transcriptome, proteome, metabolome, metagenome, epigenome, etc. Gene transfer and genome editing applications steadily expanding across various aquaculture species, including carps, for genetic improvement and understanding of biological functions. This review serves as a vital resource for carp aquaculture research communities.
Author contributions
KR: Conceptualization, Investigation, Resources, Writing–original draft, Writing–review and editing. PK: Data curation, Visualization, Writing–original draft, Writing–review and editing. SR: Writing–original draft. PA: Writing–original draft. MH: Writing–original draft. AA: Writing–original draft. SS: Writing–original draft. SD: Writing–original draft. AR: Writing–review and editing. AS: Writing–review and editing. MB: Conceptualization, Investigation, Supervision, Writing–original draft, Writing–review and editing. JS: Writing–review and editing. NN: Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors acknowledge the Director, ICAR-Central Institute of Fisheries Education, Mumbai, and the Director, ICAR-Central Institute of Freshwater Aquaculture, Bhubaneswar, Orissa for providing the necessary support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1398084/full#supplementary-material
References
Abdelrahman, H., Elhady, M., Alcivar-Warren, A., Allen, S., Al-Tobasei, R., Bao, L., et al. (2017). Aquaculture genomics, genetics and breeding in the United States: current status, challenges, and priorities for future research. BMC Genomics 18, 191–223. doi:10.1186/s12864-017-3557-1
Alberdi, A., Andersen, S. B., Limborg, M. T., Dunn, R. R., and Gilbert, M. T. P. (2022). Disentangling host–microbiota complexity through hologenomics. Nat. Rev. Genet. 23, 281–297. doi:10.1038/s41576-021-00421-0
Alfaro, A. C., and Young, T. J. (2018). Showcasing metabolomic applications in aquaculture: a review. Rev. Aquac. 10, 135–152. doi:10.1111/raq.12152
Allal, F., and Nguyen, N. H. (2022). “Genomic selection in aquaculture aquaculture species,” in Genomic prediction of complex traits: methods and protocols (Springer).
Anastasiadi, D., Venney, C. J., Bernatchez, L., and Wellenreuther, M. (2021). Epigenetic inheritance and reproductive mode in plants and animals. Trends Ecol. Evol. 36, 1124–1140. doi:10.1016/j.tree.2021.08.006
Arick, M. A., Grover, C. E., Hsu, C.-Y., Magbanua, Z., Pechanova, O., Miller, E. R., et al. (2023). A high-quality chromosome-level genome assembly of rohu carp Labeo rohita, and its utilization in SNP-based exploration of gene flow and sex determination. G3 (Bethesda). 13, jkad009. doi:10.1093/g3journal/jkad009
Ashwath, P., Premanath, R., Dharnappa Sannejal, A., Tammisetti, V. N., Gupta, S. K., and Alla, D. R. (2022). “Metagenomic approaches to identify fish gut microbiome and the effect of prebiotic supplements on gut microbes and health management,” in Biotechnological advances in aquaculture health management (Springer).
Bakos, J., and Gorda, S. (1995). Genetic improvement of common carp strains using intraspecific hybridization. Aquaculture 129, 183–186. doi:10.1016/0044-8486(94)00245-j
Bakos, J., Krasznai, Z., and Marian, T. (1978). Cross-breeding experiments with carp, tench and Asian phytophagous cyprinids. Aquac. Hung. Hung.
Barange, M., Bahri, T., Beveridge, M. C., Cochrane, K. L., Funge-Smith, S., and Poulain, F. (2018). Impacts of climate change on fisheries and aquaculture. Food Agric. Organ. 12, 628–635.
Barman, H. K., Rasal, K. D., Chakrapani, V., Ninawe, A., Vengayil, D. T., Asrafuzzaman, S., et al. (2017). Gene editing tools: state-of-the-art and the road ahead for the model and non-model fishes. J Transgenic Res. 26, 577–589. doi:10.1007/s11248-017-0030-5
Beck, M. L., Biggers, C. J., and Dupree, H. K. (1980). Karyological analysis of Ctenopharyngodon idella, Aristichthys nobilis, and their F1 hybrid. Trans. Am. Fish. Soc. 109, 433–438. doi:10.1577/1548-8659(1980)109<433:kaocia>2.0.co;2
Beerli, R. R., and Barbas, C. F. (2002). Engineering polydactyl zinc-finger transcription factors. J. Nat. Biotechnol. 20, 135–141. doi:10.1038/nbt0202-135
Bej, D., Sahoo, L., Das, S. P., Swain, S., Jayasankar, P., and Das, P. (2013). Complete mitochondrial genome sequence of Cirrhinus mrigala (Hamilton, 1822). Mitochondrial DNA 24, 91–93. doi:10.3109/19401736.2012.722998
Bernardi, G., Wiley, E. O., Mansour, H., Miller, M. R., Orti, G., Haussler, D., et al. (2012). The fishes of Genome 10K. Mar. Genomics 7, 3–6. doi:10.1016/j.margen.2012.02.002
Brennan, L. (2013). Metabolomics in nutrition research: current status and perspectives. Biochem. Soc. Trans. 41, 670–673. doi:10.1042/BST20120350
Brody, T., Wohlfarth, G., Hulata, G., and Moav, R. (1981). Application of electrophoretic genetic markers to fish breeding: IV. Assessment of breeding value of full-sib families. Aquaculture 24, 175–186. doi:10.1016/0044-8486(81)90054-5
Byadgi, O., Chen, Y.-C., Maekawa, S., Wang, P.-C., and Chen, S.-C. (2018). Immune-related functional differential gene expression in koi carp (Cyprinus carpio) after challenge with Aeromonas sobria. Int. J. Mol. Sci. 19, 2107. doi:10.3390/ijms19072107
Cao, L., Liu, H., Mulder, H. A., Henryon, M., Thomasen, J. R., Kargo, M., et al. (2020). Genomic breeding programs realize larger benefits by cooperation in the presence of genotype× environment interaction than conventional breeding programs. Front. Genet. 11, 251. doi:10.3389/fgene.2020.00251
Chakrapani, V., Patra, S. K., Panda, R. P., Rasal, K. D., Jayasankar, P., and Barman, H. K. (2016). Establishing targeted carp TLR22 gene disruption via homologous recombination using CRISPR/Cas9. J. Dev. Comp. Immunol. 61, 242–247. doi:10.1016/j.dci.2016.04.009
Chandhini, S., and Rejish Kumar, V. J. (2019). Transcriptomics in aquaculture: current status and applications. Rev. Aquac. 11, 1379–1397. doi:10.1111/raq.12298
Chauhan, T., and Rajiv, K. (2010). Molecular markers and their applications in fisheries and aquaculture. Adv. Biosci. Biotechnol. 1, 281–291. doi:10.4236/abb.2010.14037
Chen, S. C. (1956). A history of the domestication and the factors of the varietal formation of the common goldfish, Carassius auratus. Sci. Sin. 6, 89.
Chen, D., Zhang, Q., Tang, W., Huang, Z., Wang, G., Wang, Y., et al. (2020). The evolutionary origin and domestication history of goldfish (Carassius auratus). Proc. Natl. Acad. Sci. U. S. A. 117, 29775–29785. doi:10.1073/pnas.2005545117
Chen, G., Liu, H., Yu, X., Luo, W., and Tong, J. (2023). Estimation of heritabilities and quantitative trait loci for growth traits of bighead carp (Hypophthalmichthys nobilis). J. Aquac. 566, 739213. doi:10.1016/j.aquaculture.2022.739213
Cheng, Y., Vechtova, P., Fussy, Z., Sterba, J., Linhartová, Z., Rodina, M., et al. (2021). Changes in phenotypes and DNA methylation of in vitro aging sperm in common carp Cyprinus carpio. Int. J. Mol. Sci. 22, 5925. doi:10.3390/ijms22115925
Chen, H., Wang, J., Du, J., Si, Z., Yang, H., Xu, X., et al. (2019). ASIP disruption via CRISPR/Cas9 system induces black patches dispersion in Oujiang color common carp. J. Aquac. 498, 230–235. doi:10.1016/j.aquaculture.2018.08.057
Chen, L., Li, C., Li, B., Zhou, X., Bai, Y., Zou, X., et al. (2024). Evolutionary divergence of subgenomes in common carp provides insights into speciation and allopolyploid success. Fundam. Res. 4, 589–602. doi:10.1016/j.fmre.2023.06.011
Chen, L., Xu, J., Sun, X., and Xu, P. (2022). Research advances and future perspectives of genomics and genetic improvement in allotetraploid common carp. J Rev. Aquac. 14, 957–978. doi:10.1111/raq.12636
Cho, Y. S., Lee, S. Y., Kim, Y. K., Kim, D. S., and Nam, Y. K. (2011). Functional ability of cytoskeletal β-actin regulator to drive constitutive and ubiquitous expression of a fluorescent reporter throughout the life cycle of transgenic marine medaka Oryzias dancena. Transgenic Res. 20, 1333–1355. doi:10.1007/s11248-011-9501-2
Cochrane, K., De Young, C., Soto, D., and Bahri, T. (Editors) (2009). Climate change implications for fisheries and aquaculture. FAO Fisheries and Aquaculture Technical Paper. No. 530. Rome. FAO, 212.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. J Sci. 339, 819–823. doi:10.1126/science.1231143
Costa, V. A., Mifsud, J. C., Gilligan, D., Williamson, J. E., Holmes, E. C., and Geoghegan, J. L. (2021). Metagenomic sequencing reveals a lack of virus exchange between native and invasive freshwater fish across the Murray-Darling Basin, Australia. Virus Evol. 7, veab034. doi:10.1093/ve/veab034
Cottrell, R. S., Fleming, A., Fulton, E. A., Nash, K. L., Watson, R. A., and Blanchard, J. L. (2018). Considering land–sea interactions and trade-offs for food and biodiversity. Glob. change Biol. 24, 580–596. doi:10.1111/gcb.13873
Crooijmans, R., Poel, J. V. D., Groenen, M., Bierbooms, V., and Komen, J. (1997). Microsatellite markers in common carp (Cyprinus carpio L.). Anim. Genet. 28, 129–134. doi:10.1111/j.1365-2052.1997.00097.x
Dahl, H.-A., Johansen, A., Nilsson, G. E., and Lefevre, S. (2021). The metabolomic response of crucian carp (Carassius carassius) to anoxia and reoxygenation differs between tissues and hints at uncharacterized survival strategies. Metabolites 11, 435. doi:10.3390/metabo11070435
Dai, Y.-F., Shen, Y.-B., Wang, S.-T., Zhang, J.-H., Su, Y.-H., Bao, S.-C., et al. (2021). RNA-Seq transcriptome analysis of the liver and brain of the black carp (Mylopharyngodon piceus) during fasting. Mar. Biotechnol. 23, 389–401. doi:10.1007/s10126-021-10032-9
Das Mahapatra, K., Saha, J., Sarangi, N., Jana, R., Gjerde, B., Nguyen, N. H., et al. (2007). Genetic improvement and dissemination of Rohu (Labeo rohita, Ham.) in India. Assoc. Adv. Animal Breed. Genet.
Das, P., Sahoo, L., Das, S. P., Bit, A., Joshi, C. G., Kushwaha, B., et al. (2020). De novo assembly and genome-wide SNP discovery in rohu carp, Labeo rohita. Front. Genet. 11, 386. doi:10.3389/fgene.2020.00386
Das, T., Sehar, S., and Manefield, M. (2013). The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ. Microbiol. Rep. 5, 778–786. doi:10.1111/1758-2229.12085
Devlin, R. H., Raven, P. A., Sundström, L. F., and Uh, M. (2009). Issues and methodology for development of transgenic fish for aquaculture with a focus on growth enhancement. Mol. Res. Aquac., 217–260. doi:10.1002/9780813807379.ch9
Dey, M. M., Kumar, P., Paraguas, F. J., Li, C. O., Khan, M. A., and Srichantuk, N. (2010). Performance and nature of genetically improved carp strains in Asian countries. Aquac. Econ. and Manag. 14, 3–29. doi:10.1080/13657300903566845
Dietrich, M. A., Arnold, G. J., Fröhlich, T., and Ciereszko, A. (2014). In-depth proteomic analysis of carp (Cyprinus carpio L) spermatozoa. PROTEOMICS 12, 10–15. doi:10.1016/j.cbd.2014.09.003
DOF (2019). Yearbook of fisheries statistics of Bangladesh, 2018–2019. Fisheries resources survey system (FRSS). Dhaka: Department of Fisheries.
Dong, Z., Nguyen, N. H., and Zhu, W. (2015). Genetic evaluation of a selective breeding program for common carp Cyprinus carpio conducted from 2004 to 2014. BMC Genet. 16, 94–99. doi:10.1186/s12863-015-0256-2
Dunham, R. A. (2009). Transgenic fish resistant to infectious diseases, their risk and prevention of escape into the environment and future candidate genes for disease transgene manipulation. Comp. Immunol. Microbiol. Infect. Dis. 32, 139–161. doi:10.1016/j.cimid.2007.11.006
Du, X., Zhang, W., Wu, J., You, C., and Dong, X. (2023). Full-length RNA sequencing provides insights into goldfish evolution under artificial selection. Int. J. Mol. Sci. 24, 2735. doi:10.3390/ijms24032735
Fan, G., Song, Y., Yang, L., Huang, X., Zhang, S., Zhang, M., et al. (2020). Initial data release and announcement of the 10,000 fish genomes project (Fish10K). GigaScience 9, giaa080. doi:10.1093/gigascience/giaa080
FAO (2022). The state of world fisheries and aquaculture 2022 2. Towards Blue Transformation. Rome: FAO.
Feil, R., and Fraga, M. F. (2012). Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13, 97–109. doi:10.1038/nrg3142
Feng, H., Cheng, J., Luo, J., Liu, S. J., and Liu, Y. (2006). Cloning of black carp beta-actin gene and primarily detecting the function of its promoter region. Yi Chuan Xue Bao 33, 133–140. doi:10.1016/S0379-4172(06)60032-2
Feng, X., Yu, X., Fu, B., He, S., and Tong, J. (2014). Development of 159 transcript-associated microsatellite markers in silver carp (Hypophthalmichthys molitrix). Conserv. Genet. Resour. 6, 111–113. doi:10.1007/s12686-013-0017-3
Ferdous, M. A., Islam, S. I., Habib, N., Almehmadi, M., Allahyani, M., Alsaiari, A. A., et al. (2022). CRISPR-cas genome editing technique for fish disease management: current study and future perspective. J. Microorg. 10, 2012. doi:10.3390/microorganisms10102012
Flint, H. J., Bayer, E. A., Rincon, M. T., Lamed, R., and White, B. A. (2008). Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6, 121–131. doi:10.1038/nrmicro1817
Frank, D. N. (2008). XplorSeq: a software environment for integrated management and phylogenetic analysis of metagenomic sequence data. BMC Bioinforma. 9, 420–516. doi:10.1186/1471-2105-9-420
Fu, B., Liu, H., Yu, X., and Tong, J. (2016a). A high-density genetic map and growth related QTL mapping in bighead carp (Hypophthalmichthys nobilis). J. Sci. Rep. 6, 28679. doi:10.1038/srep28679
Fu, B., Zhou, Y., Liu, H., Yu, X., and Tong, J. (2021). Updated genome assembly of bighead carp (Hypophthalmichthys nobilis) and its differences between male and female on genomic, transcriptomic, and methylation level. Front. Genet. 12, 728177. doi:10.3389/fgene.2021.728177
Fu, J., Shen, Y., Xu, X., and Li, J. (2016b). Genetic parameter estimates for growth of grass carp, Ctenopharyngodon idella, at 10 and 18 months of age. J. Aquac. 450, 342–348. doi:10.1016/j.aquaculture.2015.08.018
Fu, J., Shen, Y., Xu, X., Liu, C., and Li, J. (2015). Genetic parameter estimates and genotype by environment interaction analyses for early growth traits in grass carp (Ctenopharyngodon idella). J. Aquac. Int. 23, 1427–1441. doi:10.1007/s10499-015-9894-7
Gan, R.-H., Li, Z., Wang, Z.-W., Li, X.-Y., Wang, Y., Zhang, X.-J., et al. (2023). Creation of intermuscular bone-free mutants in amphitriploid gibel carp by editing two duplicated runx2b homeologs. J. Aquac. 567, 739300. doi:10.1016/j.aquaculture.2023.739300
Gan, R.-H., Wang, Y., Li, Z., Yu, Z.-X., Li, X.-Y., Tong, J.-F., et al. (2021). Functional divergence of multiple duplicated Foxl2 homeologs and alleles in a recurrent polyploid fish. J. Mol. Biol. Evol. 38, 1995–2013. doi:10.1093/molbev/msab002
Gavery, M. R., and Roberts, S. B. (2017). Epigenetic considerations in aquaculture. PeerJ 5, e4147. doi:10.7717/peerj.4147
Ge, G., Long, Y., Shi, L., Ren, J., Yan, J., Li, C., et al. (2020). Transcriptomic profiling revealed key signaling pathways for cold tolerance and acclimation of two carp species. BMC genomics 21, 539–615. doi:10.1186/s12864-020-06946-8
Gheyas, A. A., Woolliams, J. A., Taggart, J. B., Sattar, M. A., Das, T. K., Mcandrew, B. J., et al. (2009). Heritability estimation of silver carp (Hypophthalmichthys molitrix) harvest traits using microsatellite based parentage assignment. Aquaculture 294, 187–193. doi:10.1016/j.aquaculture.2009.06.013
Gibney, E., and Nolan, C. (2010). Epigenetics and gene expression. Hered. (Edinb). 105, 4–13. doi:10.1038/hdy.2010.54
Gjedrem, T., and Baranski, M. (2010). Selective breeding in aquaculture: an introduction. Springer Science and Business Media.
Gjedrem, T., and Robinson, N. (2014). Advances by selective breeding for aquatic species: a review. Agric. Sci. 5, 1152–1158. doi:10.4236/as.2014.512125
Gjerde, B., Mahapatra, K. D., Reddy, P. V. G. K., Saha, J. N., Jana, R. K., Meher, P. K., et al. (2019). Genetic parameters for growth and survival in rohu carp (Labeo rohita). Aquaculture 503, 381–388. doi:10.1016/j.aquaculture.2019.01.029
Gjerde, B., Reddy, P. V., Mahapatra, K. D., Saha, J. N., Jana, R. K., Meher, P. K., et al. (2002). Growth and survival in two complete diallele crosses with five stocks of Rohu carp (Labeo rohita). Aquaculture 209, 103–115. doi:10.1016/s0044-8486(01)00848-1
Goldansaz, S. A., Guo, A. C., Sajed, T., Steele, M. A., Plastow, G. S., and Wishart, D. (2017). Livestock metabolomics and the livestock metabolome: a systematic review. A Syst. Rev. 12, e0177675. doi:10.1371/journal.pone.0177675
Granada, L., Lemos, M. F., Cabral, H. N., Bossier, P., and Novais, S. C. (2018). Epigenetics in aquaculture–the last frontier. Rev. Aquac. 10, 994–1013. doi:10.1111/raq.12219
Gratacap, R. L., Wargelius, A., Edvardsen, R. B., and Houston, R. D. (2019). Potential of genome editing to improve aquaculture breeding and production. J Trends Genet. 35, 672–684. doi:10.1016/j.tig.2019.06.006
Guo, C., Duan, Y., Ye, W., Zhang, W., Cheng, Y., Shi, M., et al. (2023). FishGET: a fish gene expression and transcriptome database with improved accuracy and visualization. iScience 26, 106539. doi:10.1016/j.isci.2023.106539
Guo, J., Wang, A., Mao, S., Xu, X., Li, J., and Shen, Y. (2022). Construction of high-density genetic linkage map and QTL mapping for growth performance in black carp (Mylopharyngodon piceus). Aquaculture 549, 737799. doi:10.1016/j.aquaculture.2021.737799
Guo, W., Fu, L., Wu, Y., Liu, H., Yang, Y., Hu, W., et al. (2021). Effects of dietary protein levels on growth and feed utilization in non-transgenic and growth-hormone-gene transgenic common carp (Cyprinus carpio L.). Aquac. Rep. 21, 100854. doi:10.1016/j.aqrep.2021.100854
Guo, W., Yu, X., and Tong, J. (2013). Development of 134 novel polynucleotide-repeat microsatellite markers in silver carp (Hypophthalmichthys molitrix). Conserv. Genet. Resour. 5, 525–528. doi:10.1007/s12686-012-9843-y
Gutási, A., Hammer, S. E., El-Matbouli, M., and Saleh, M. (2023). Review: recent applications of gene editing in fish species and aquatic medicine. J. Anim. 13, 1250. doi:10.3390/ani13071250
Hallerman, E. (2021a). Applications and limitations of DNA barcoding in environmental biology. J. Environ. Biol. 42, 1–13. doi:10.22438/jeb/42/1/mrn-1710
Hallerman, E. (2021b). Genome editing in cultured fishes. J. CABI Agric. Biosci. 2, 46–19. doi:10.1186/s43170-021-00066-3
Hamilton, M. G., Mekkawy, W., Alam, M. B., Barman, B. K., Karim, M., and Benzie, J. A. (2023a). Genotype-by-culture-system interaction in catla and silver carp: monoculture and biculture. Aquaculture 562, 738846. doi:10.1016/j.aquaculture.2022.738846
Hamilton, M. G., Mekkawy, W., Alam, M. B., and Benzie, J. A. H. (2022). Early selection to enhance genetic gain in a rohu (Labeo rohita) genetic improvement program. Aquaculture 553, 738058. doi:10.1016/j.aquaculture.2022.738058
Hamilton, M. G., Mekkawy, W., Barman, B. K., Alam, M. B., Karim, M., and Benzie, J. A. (2021). Genetic relationships among founders of a silver carp (Hypophthalmichthys molitrix) genetic improvement program in Bangladesh. Aquaculture 540, 736715. doi:10.1016/j.aquaculture.2021.736715
Hamilton, M. G., Mekkawy, W., and Benzie, J. A. (2019a). Sibship assignment to the founders of a Bangladeshi Catla catla breeding population. Genet. Sel. Evol. 51, 17. doi:10.1186/s12711-019-0454-x
Hamilton, M. G., Mekkawy, W., Kilian, A., and Benzie, J. A. (2019b). Single nucleotide polymorphisms (SNPs) reveal sibship among founders of a Bangladeshi rohu (Labeo rohita) breeding population. Front. Genet. 10, 597. doi:10.3389/fgene.2019.00597
Hamilton, M. G., Yeasin, M., Chadag, V. M., Delamare-Deboutteville, J., Debnath, P. P., and Benzie, J. A. (2023b). Genetic correlations between harvest weight and secondary traits in a silver carp (Hypophthalmichthys molitrix) genetic improvement program. Aquac. Int. 31, 333–348. doi:10.1007/s10499-022-00977-6
Han, X., Xiao, C., Yi, S., Li, Y., Chen, M., and Huang, D. (2022). Accurate age estimation from blood samples of Han Chinese individuals using eight high-performance age-related CpG sites. Int. J. Leg. Med. 136, 1655–1665. doi:10.1007/s00414-022-02865-3
Hao, Y., Jia, X., Yuan, L., Liu, Y., Gui, L., Shen, Y., et al. (2023). Genome-wide association study reveals growth-related SNPs and candidate genes in grass carp (Ctenopharyngodon idella). Aquaculture 577, 739979. doi:10.1016/j.aquaculture.2023.739979
He, L., Liang, X., Wang, Q., Yang, C., Li, Y., Liao, L., et al. (2022). Genome-wide DNA methylation reveals potential epigenetic mechanism of age-dependent viral susceptibility in grass carp. Immun. Ageing 19, 28–17. doi:10.1186/s12979-022-002
Heras, J. (2021). Fish transcriptomics: applied to our understanding of aquaculture. Preprints, 2020010332. doi:10.20944/preprints202001.0332.v1
Hong, H., Luo, Y., Zhou, Z., Bao, Y., Lu, H., and Shen, H. (2013). Effects of different freezing treatments on the biogenic amine and quality changes of bighead carp (Aristichthys nobilis) heads during ice storage. Food Chem. 138, 1476–1482. doi:10.1016/j.foodchem.2012.11.031
Houston, R. D., Bean, T. P., Macqueen, D. J., Gundappa, M. K., Jin, Y. H., Jenkins, T. L., et al. (2020). Harnessing genomics to fast-track genetic improvement in aquaculture. Nat. Rev. Genet. 21, 389–409. doi:10.1038/s41576-020-0227-y
Hu, F., Zhong, H., Wu, C., Wang, S., Guo, Z., Tao, M., et al. (2021). Development of fisheries in China. Reproduction Breed. 1, 64–79. doi:10.1016/j.repbre.2021.03.003
Hu, M., Qu, X., Pan, L., Fu, C., Jia, P., Liu, Q., et al. (2017). Effects of toxic Microcystis aeruginosa on the silver carp Hypophthalmichtys molitrix revealed by hepatic RNA-seq and miRNA-seq. Sci. Rep. 7, 10456. doi:10.1038/s41598-017-10335-9
Hussain, M., and Mazid, M. (2002). “Genetic status and improvement strategies for endemic and exotic carps of Bangladesh,” in Proceedings of a workshop on genetic management and improvement strategies for exotic carps in Asia (Dhaka, Bangladesh: Citeseer), 9–27.
Hu, X., Bai, J., Liu, R., and Lv, A. (2023). Comprehensive transcriptomics and proteomics analysis of Carassius auratus gills in response to Aeromonas hydrophila. Fish. Shellfish Immunol. Rep. 4, 100077. doi:10.1016/j.fsirep.2022.100077
Ingerslev, H.-C., Strube, M. L., Von Gersdorff Jørgensen, L., Dalsgaard, I., Boye, M., and Madsen, L. (2014). Diet type dictates the gut microbiota and the immune response against Yersinia ruckeri in rainbow trout (Oncorhynchus mykiss). Fish. Shellfish Immunol. 40, 624–633. doi:10.1016/j.fsi.2014.08.021
Issa, M. A., Horvath, L., Kosba, M. A., and Sharrabi, M. (1986). A note on the survival, growth, feed conversion and some morphological characters of the reciprocal hybrids of silver carp (Hypophthalmichthys molitrix Val.) and bighead carp (Aristichthys nobilis Rich.) raised in polyculture. Aquacult. Hung 5, 7–14.
Jaiswal, S., Rasal, K. D., Chandra, T., Prabha, R., Iquebal, M. A., Rai, A., et al. (2023). Proteomics in fish health and aquaculture productivity management: status and future perspectives. Aquaculture 566, 739159. doi:10.1016/j.aquaculture.2022.739159
Jiang, Y., Li, B., Yu, M., Chang, S., Li, S., Xu, J., et al. (2022a). Genome-wide association study and gene editing reveals the causal gene responsible for abnormal red skin color in Yellow River carp. Aquaculture 560, 738530. doi:10.1016/j.aquaculture.2022.738530
Jiang, Y., Qi, M., Zhang, J., Wen, Y., Sun, J., and Liu, Q. (2022b). Metabolomic profiling analysis of physiological responses to acute hypoxia and reoxygenation in juvenile qingtian paddy field carp Cyprinus carpio var qingtianensis. Front. Physiol. 13, 853850. doi:10.3389/fphys.2022.853850
Jian, J., Yang, L., Gan, X., Wu, B., Gao, L., Zeng, H., et al. (2021). Whole genome sequencing of silver carp (Hypophthalmichthys molitrix) and bighead carp (Hypophthalmichthys nobilis) provide novel insights into their evolution and speciation. Mol. Ecol. Resour. 21, 912–923. doi:10.1111/1755-0998.13297
Jia, Y., Nan, P., Zhang, W., Wang, F., Zhang, R., Liang, T., et al. (2018). Transcriptome analysis of three critical periods of ovarian development in Yellow River carp (Cyprinus carpio). Theriogenology 105, 15–26. doi:10.1016/j.theriogenology.2017.08.027
Jin, S., Zhang, X., Jia, Z., Fu, H., Zheng, X., and Sun, X. (2012). Genetic linkage mapping and genetic analysis of QTL related to eye cross and eye diameter in common carp (Cyprinus carpio L.) using microsatellites and SNPs. Aquaculture 358, 176–182.
Jin, S., Zhang, X., Lu, J., Fu, H., Jia, Z., and Sun, X. (2015a). Genetic analysis of QTL for eye cross and eye diameter in common carp (Cyprinus carpio L.) using microsatellites and SNPs. Genet. Mol. Res. 14, 3557–3569. doi:10.4238/2015.April.17.5
Jin, Y., Tian, L.-X., Xie, S.-W., Guo, D.-Q., Yang, H.-J., Liang, G.-Y., et al. (2015b). Interactions between dietary protein levels, growth performance, feed utilization, gene expression and metabolic products in juvenile grass carp (Ctenopharyngodon idella). Aquaculture 437, 75–83. doi:10.1016/j.aquaculture.2014.11.031
Ji, P., Zhang, Y., Li, C., Zhao, Z., Wang, J., Li, J., et al. (2012). High throughput mining and characterization of microsatellites from common carp genome. Int. J. Mol. Sci. 13, 9798–9807. doi:10.3390/ijms13089798
Johny, T. K., Puthusseri, R. M., and Bhat, S. G. (2021). A primer on metagenomics and next-generation sequencing in fish gut microbiome research. Aquac. Res. 52, 4574–4600. doi:10.1111/are.15373
Jones, M. J., Goodman, S. J., and Kobor, M. S. (2015). DNA methylation and healthy human aging. Aging Cell 14, 924–932. doi:10.1111/acel.12349
Kan, H., Zhao, F., Zhang, X.-X., Ren, H., and Gao, S. (2015). Correlations of gut microbial community shift with hepatic damage and growth inhibition of Carassius auratus induced by pentachlorophenol exposure. Environ. Sci. Technol. 49, 11894–11902. doi:10.1021/acs.est.5b02990
Kasama, M., and Kobayasi, H. (1989). Hybridization experiments in Cyprinida - II. Hybridization experiment between female crucian carp and male grass carp. Nippon. Suisan Gakkaishi Jpn. Ed. 55, 1001–1006. doi:10.2331/suisan.55.1001
Keus, E., Subasinghe, R., Aleem, N., Sarwer, R., Islam, M., Hossain, M., et al. (2017). Aquaculture for income and nutrition: final report. Penang, Malaysia: WorldFish. Program Report.
Khan, M. R. I., Parvez, M. T., Talukder, M. G. S., Hossain, M. A., and Karim, M. S. (2018). Production and economics of carp polyculture in ponds stocked with wild and hatchery produced seeds. J. Fish. 6, 541–548. doi:10.17017/jfish.v6i1.2018.306
Kibria, M., Islam, N., Billah, M., Shawrob, K. S. M., Rumi, M., and Siddiki, A. Z. (2020). Complete mitochondrial genome sequence of Catla catla (Hamilton, 1822) from the Halda river of Bangladesh. Mitochondrial DNA Part B 5, 3215–3217. doi:10.1080/23802359.2020.1809542
Kirpichnikov, V., and Faktorovich, K. (1974). Increase in the resistance of carp to dropsy by means of breeding. Communication II. The course of the selection and evaluation of the breeding groups selected. Sov. Genet. 8, 592–600.
Kolder, I., Van Der Plas-Duivesteijn, S., Tan, G., Wiegertjes, G., Forlenza, M., Guler, A., et al. (2016). A full-body transcriptome and proteome resource for the European common carp. BMC genomics 17, 1–12.
Komiyama, T., Kobayashi, H., Tateno, Y., Inoko, H., Gojobori, T., and Ikeo, K. (2009). An evolutionary origin and selection process of goldfish. Gene 430, 5–11. doi:10.1016/j.gene.2008.10.019
Kossmann, H. (1972). Studies on genetic variability of intermuscular bones in the carp. Theor. Appl. Genet. 42, 130–135. doi:10.1007/BF00583415
Kuang, Y., Zheng, X., Cao, D., Sun, Z., Tong, G., Xu, H., et al. (2023). Generate a new crucian carp (Carassius auratus) strain without intermuscular bones by knocking out bmp6. J. Aquac. 569, 739407. doi:10.1016/j.aquaculture.2023.739407
Kumar, P. V., Rasal, K. D., Acharya, A., Dey, D., Sonwane, A. A., Reang, D., et al. (2023). Muscle transcriptome sequencing revealed thermal stress-responsive regulatory genes in farmed rohu, Labeo rohita (Hamilton, 1822). Mar. Biotechnol. (NY) 25, 1057–1075. doi:10.1007/s10126-023-10259-8
Kumari, R., Gm, S., and Saurabh, S. (2022). “Metabolomic advances in fish nutritional research and health management,” in Biotechnological advances in aquaculture health management (Springer).
Kvasnicka, P., Flajshans, M., Rab, P., and Linhart, O. (1998). Brief communication. Inheritance studies of blue and golden varieties of tench (Pisces: Tinca tinca L.). J. Hered. 89, 553–556. doi:10.1093/jhered/89.6.553
Laghari, M., Lashari, P., Zhang, X., Xu, P., Xin, B., Zhang, Y., et al. (2014). Mapping quantitative trait loci (QTL) for body weight, length and condition factor traits in backcross (BC1) family of Common carp (Cyprinus carpio L.). Mol. Biol. Rep. 41, 721–731. doi:10.1007/s11033-013-2911-x
Lalramnunsanga, , Mishra, A., Singh, A. L., Prakash, S., Salvi, A., Kumar, A. P., et al. (2024). Genetic diversity of common carp Cyprinus carpio in the base population of a selective breeding programme in India. Discov. Anim. 1, 3. doi:10.1007/s44338-024-00004-3
Lee, H.-K., Lee, J., Lee, J., Lee, H., and Kim, J.-H. (2020). Metabolite profiles of live or dead carp (Cyprinus carpio) exposed to endosulfan sulfate using a targeted GC–MS analysis. Appl. Biol. Chem. 63, 11–19. doi:10.1186/s13765-020-00495-0
Li, J. T., Wang, Q., Huang Yang, M. D., Li, Q. S., Cui, M. S., Dong, Z., et al. (2021). Parallel subgenome structure and divergent expression evolution of allo-tetraploid common carp and goldfish. Nat. Genet. 53, 1493–1503. doi:10.1038/s41588-021-00933-9
Libisch, B., Abdulkadir, S., Keresztény, T., Papp, P. P., Olasz, F., Fébel, H., et al. (2022). Detection of acquired antibiotic resistance genes in domestic pig (Sus scrofa) and common carp (Cyprinus carpio) intestinal samples by metagenomics analyses in Hungary. Antibiot. (Basel). 11, 1441. doi:10.3390/antibiotics11101441
Limborg, M. T., Alberdi, A., Kodama, M., Roggenbuck, M., Kristiansen, K., and Gilbert, M. (2018). Applied hologenomics: feasibility and potential in aquaculture. Trends Biotechnol. 36, 252–264. doi:10.1016/j.tibtech.2017.12.006
Lindberg, J., and Lundeberg, J. (2010). The plasticity of the mammalian transcriptome. Genomics 95, 1–6. doi:10.1016/j.ygeno.2009.08.010
Lind, C., Ponzoni, R., Nguyen, N. H., and Khaw, H. (2012). Selective breeding in fish and conservation of genetic resources for aquaculture. Reproduction Domest. animals 47, 255–263. doi:10.1111/j.1439-0531.2012.02084.x
Lin, M., Zou, J., and Wang, C. (2016). Complete mitochondrial genomes of domesticated and wild common carp (Cyprinus carpio L.). Mitochondrial DNA Part A 27, 50–51. doi:10.3109/19401736.2013.869690
Li, P., Chen, C.-Z., Zhao, X.-L., Liu, L., and Li, Z.-H. (2023). Metagenomics analysis reveals the effects of norfloxacin on the gut microbiota of juvenile common carp (Cyprinus carpio). Chemosphere 325, 138389. doi:10.1016/j.chemosphere.2023.138389
Li, T., Long, M., Li, H., Gatesoupe, F.-J., Zhang, X., Zhang, Q., et al. (2017a). Multi-omics analysis reveals a correlation between the host phylogeny, gut microbiota and metabolite profiles in cyprinid fishes. Front. Microbiol. 8, 454. doi:10.3389/fmicb.2017.00454
Liu, H., Fu, B., Pang, M., Feng, X., Wang, X., Yu, X., et al. (2016). QTL fine mapping and identification of candidate genes for growth-related traits in bighead carp (Hypophthalmichehys nobilis). Aquaculture 465, 134–143. doi:10.1016/j.aquaculture.2016.08.039
Liu, H., Fu, B., Pang, M., Feng, X., Yu, X., and Tong, J. (2017). A high-density genetic linkage map and QTL fine mapping for body weight in crucian carp (Carassius auratus) using 2b-RAD sequencing. G3 (Bethesda). 7, 2473–2487. doi:10.1534/g3.117.041376
Liu, Q., Qi, Y., Liang, Q., Song, J., Liu, J., Li, W., et al. (2019). Targeted disruption of tyrosinase causes melanin reduction in Carassius auratus cuvieri and its hybrid progeny. J Sci. China Life Sci. 62, 1194–1202. doi:10.1007/s11427-018-9404-7
Liu, Z., Zhou, T., and Gao, D. (2022). Genetic and epigenetic regulation of growth, reproduction, disease resistance and stress responses in aquaculture. Front. Genet. 13, 994471. doi:10.3389/fgene.2022.994471
Li, X., Zhou, L., Yu, Y., Ni, J., Xu, W., and Yan, Q. (2017b). Composition of gut microbiota in the gibel carp (Carassius auratus gibelio) varies with host development. Microb. Ecol. 74, 239–249. doi:10.1007/s00248-016-0924-4
Li, Y., Fu, B., Zhang, J., Wang, G., Tian, J., Li, H., et al. (2022). Comparative genome-wide methylation analysis reveals epigenetic regulation of muscle development in grass carp (Ctenopharyngodon idellus) fed with whole faba bean. PeerJ 10, e14403. doi:10.7717/peerj.14403
Lu, G., and Luo, M. (2020). Genomes of major fishes in world fisheries and aquaculture: status, application and perspective. Aquac. Fish. 5, 163–173. doi:10.1016/j.aaf.2020.05.004
Luo, C., Hajkova, P., and Ecker, J. R. (2018). Dynamic DNA methylation: in the right place at the right time. Science 361, 1336–1340. doi:10.1126/science.aat6806
Luo, W., Wang, J., Zhou, Y., Pang, M., Yu, X., and Tong, J. (2022). Dynamic mRNA and miRNA expression of the head during early development in bighead carp (Hypophthalmichthys nobilis). BMC genomics 23, 168. doi:10.1186/s12864-022-08387-x
Luo, W., Zeng, C., Deng, W., Robinson, N., Wang, W., and Gao, Z. (2014a). Genetic parameter estimates for growth-related traits of blunt snout bream (Megalobrama amblycephala) using microsatellite-based pedigree. J Aquac. Res. 45, 1881–1888.
Luo, W., Zeng, C., Yi, S., Robinson, N., Wang, W., and Gao, Z. (2014b). Heterosis and combining ability evaluation for growth traits of blunt snout bream (Megalobrama amblycephala) when crossbreeding three strains. J Chin. Sci. Bull. 59, 857–864. doi:10.1007/s11434-014-0115-y
Luo, W., Zhou, Y., Wang, J., Yu, X., and Tong, J. (2021). Identifying candidate genes involved in the regulation of early growth using full-length transcriptome and RNA-Seq analyses of frontal and parietal bones and vertebral bones in bighead carp (Hypophthalmichthys nobilis). Front. Genet. 11, 603454. doi:10.3389/fgene.2020.603454
Lu, X., Chen, H.-M., Qian, X.-Q., and Gui, J.-F. (2020). Transcriptome analysis of grass carp (Ctenopharyngodon idella) between fast-and slow-growing fish. Comp. Biochem. Physiol. Part D. Genomics Proteomics 35, 100688. doi:10.1016/j.cbd.2020.100688
Lu, Y., Xia, H., Zhai, W., Liu, J., Zhou, Y., Xu, X., et al. (2022). Genome survey sequence of black carp provides insights into development-related gene duplications. J. World Aquac. Soc. 53, 1197–1214. doi:10.1111/jwas.12870
Mahapatra, K. D., Gjerde, B., Sahoo, P., Saha, J., Barat, A., Sahoo, M., et al. (2008). Genetic variations in survival of rohu carp (Labeo rohita, Hamilton) after Aeromonas hydrophila infection in challenge tests. Aquaculture 279, 29–34. doi:10.1016/j.aquaculture.2008.03.054
Mahapatra, K. D., Jayasankar, P., Saha, J., Murmu, K., Rasal, A., Nandanpawar, P., et al. (2016). JayantiRohu: glimpses from the journey of first genetically improved fish in India, 60.
Mahapatra, K. D., Rasal, A., Murmu, K., Sundaray, J. K., Das, P., Mohanty, J., et al. (2022). Genetically improved catla: superior strain for sustainable aquaculture production in India. Bhubaneswar, Odisha: ICAR-CIFA.
Mahapatra, K. D., Sahoo, L., Nath Saha, J., Murmu, K., Rasal, A., Nandanpawar, P., et al. (2018). Establishment of base population for selective breeding of catla (Catla catla) depending on phenotypic and microsatellite marker information. J. Genet. 97, 1327–1337. doi:10.1007/s12041-018-1034-5
Mahapatra, K., Saha, J., Murmu, K., Rasal, A., Nandanpawar, P., and Patnaik, M. (2017). “JAYANTI” rohu-a promising fish variety for improving aquaculture production. J. Inl. Fish. Soc. India 49, 03–10.
Ma, J., Chen, X., Xin, G., and Li, X. (2019). Chronic exposure to the ionic liquid [C8mim] Br induces inflammation in silver carp spleen: involvement of oxidative stress-mediated p38MAPK/NF-κB signalling and microRNAs. Fish. Shellfish Immunol. 84, 627–638. doi:10.1016/j.fsi.2018.09.052
Ma, J., Fan, Y., Zhou, Y., Liu, W., Jiang, N., Zhang, J., et al. (2018). Efficient resistance to grass carp reovirus infection in JAM-A knockout cells using CRISPR/Cas9. J. Fish. shellfish Immunol. 76, 206–215. doi:10.1016/j.fsi.2018.02.039
Mandal, B. K., Chen, H., Si, Z., Hou, X., Yang, H., Xu, X., et al. (2020). Shrunk and scattered black spots turn out due to MC1R knockout in a white-black Oujiang color common carp (Cyprinus carpio var. color). J. Aquac. 518, 734822. doi:10.1016/j.aquaculture.2019.734822
Márián, T., and Krasznai, Z. (1979). Comparative karyological studies on Chinese carps. Aquaculture 18, 325–336. doi:10.1016/0044-8486(79)90036-x
Marian, T., Krasznai, Z., and Olah, J. (1986). Characteristic karyological, biochemical and morphological markers of silver carp (Hypophthalmichthys molitrix Val.), bighead carp (Aristichthys nobilis Rich.) and their hybrids. Aquac. Hung. 5, 15–30.
Ma, Y., Wang, C., Wang, J., Yang, X., Bi, X., and Xu, L. (2011). Genetic differentiation of wild and hatchery Oujiang color common carp: potential application to the identification of escapees. Fish. Sci. 77, 591–597. doi:10.1007/s12562-011-0359-2
Mazzeo, M. F., and Siciliano, R. A. (2016). Proteomics for the authentication of fish species. J. Proteomics 147, 119–124. doi:10.1016/j.jprot.2016.03.007
Metzger, D. C., and Schulte, P. M. (2016). Epigenomics in marine fishes. Epigenomics Mar. fishes 30, 43–54. doi:10.1016/j.margen.2016.01.004
Misra, B. B., Langefeld, C., Olivier, M., and Cox, L. A. (2019). Integrated omics: tools, advances and future approaches. J. Mol. Endocrinol. 62, R21–R45. doi:10.1530/jme-18-0055
Moav, R., Hulata, G., and Wohlfarth, G. (1975). Genetic differences between the Chinese and European races of the common carp.I. Analysis of genotype-environment interactions for growth rate. Heredity 34, 323–340. doi:10.1038/hdy.1975.42
Mojekwu, T., and Anumudu, C. (2013). Microsatellite markers in aquaculture: application in fish population genetics.
Mondal, H. K., Maji, U. J., Mohanty, S., Sahoo, P. K., and Maiti, N. K. (2022). Alteration of gut microbiota composition and function of Indian major carp, rohu (Labeo rohita) infected with Argulus siamensis. Microb. Pathog. 164, 105420. doi:10.1016/j.micpath.2022.105420
Mukherjee, A., Rodiles, A., Merrifield, D. L., Chandra, G., and Ghosh, K. (2020). Exploring intestinal microbiome composition in three Indian major carps under polyculture system: a high-throughput sequencing based approach. Aquaculture 524, 735206. doi:10.1016/j.aquaculture.2020.735206
Nagpure, N. S., Rashid, I., Pati, R., Pathak, A. K., Singh, M., Singh, S. P., et al. (2013). FishMicrosat: a microsatellite database of commercially important fishes and shellfishes of the Indian subcontinent. BMC genomics 14, 630–710. doi:10.1186/1471-2164-14-630
Nandanpawar, P., Sahoo, L., Sahoo, B., Murmu, K., Chaudhari, A., Das, P., et al. (2023). Identification of differentially expressed genes and SNPs linked to harvest body weight of genetically improved rohu carp, Labeo rohita. Front. Genet. 14, 1153911. doi:10.3389/fgene.2023.1153911
Naylor, R. L., Hardy, R. W., Buschmann, A. H., Bush, S. R., Cao, L., Klinger, D. H., et al. (2021). A 20-year retrospective review of global aquaculture. Nature 591, 551–563. doi:10.1038/s41586-021-03308-6
Nielsen, H. M., Ødegård, J., Olesen, I., Gjerde, B., Ardo, L., Jeney, G., et al. (2010). Genetic analysis of common carp (Cyprinus carpio) strains: I: genetic parameters and heterosis for growth traits and survival. Aquaculture 304, 14–21.
Ni, J., Yan, Q., Yu, Y., and Zhang, T. (2014). Factors influencing the grass carp gut microbiome and its effect on metabolism. FEMS Microbiol. Ecol. 87, 704–714. doi:10.1111/1574-6941.12256
Ninh, N. H., Ponzoni, R. W., Nguyen, N. H., Woolliams, J. A., Taggart, J. B., Mcandrew, B. J., et al. (2011). A comparison of communal and separate rearing of families in selective breeding of common carp (Cyprinus carpio): estimation of genetic parameters. Aquaculture 322, 39–46. doi:10.1016/j.aquaculture.2011.09.031
Ninh, N. H., Ponzoni, R. W., Nguyen, N. H., Woolliams, J. A., Taggart, J. B., Mcandrew, B. J., et al. (2013). A comparison of communal and separate rearing of families in selective breeding of common carp (Cyprinus carpio): responses to selection. Aquaculture 408, 152–159. doi:10.1016/j.aquaculture.2013.06.005
Nissa, M. U., Pinto, N., Ghosh, B., Singh, U., Goswami, M., and Srivastava, S. J. (2023). Proteomic analysis of liver tissue reveals Aeromonas hydrophila infection mediated modulation of host metabolic pathways in Labeo rohita. J. Proteomics 279, 104870. doi:10.1016/j.jprot.2023.104870
Nissa, M. U., Pinto, N., Parkar, H., Goswami, M., and Srivastava, S. (2021). Proteomics in fisheries and aquaculture: an approach for food security. Food control. 127, 108125. doi:10.1016/j.foodcont.2021.108125
Nissa, M. U., Reddy, P. J., Pinto, N., Sun, Z., Ghosh, B., Moritz, R. L., et al. (2022). The PeptideAtlas of a widely cultivated fish Labeo rohita: a resource for the Aquaculture Community. Sci. Data 9, 171. doi:10.1038/s41597-022-01259-9
Olesen, I., Bentsen, H. B., Phillips, M., and Ponzoni, R. W. (2015). Can the global adoption of genetically improved farmed fish increase beyond 10%, and how? J. Mar. Sci. Eng. 3, 240–266. doi:10.3390/jmse3020240
Omori, Y., and Kon, T. (2019). Goldfish: an old and new model system to study vertebrate development, evolution and human disease. J. Biochem. 165, 209–218. doi:10.1093/jb/mvy076
Palaiokostas, C., Kocour, M., Prchal, M., and Houston, R. D. (2018). Accuracy of genomic evaluations of juvenile growth rate in common carp (Cyprinus carpio) using genotyping by sequencing. Front. Genet. 9, 82. doi:10.3389/fgene.2018.00082
Palaiokostas, C., Vesely, T., Kocour, M., Prchal, M., Pokorova, D., Piackova, V., et al. (2019). Optimizing genomic prediction of host resistance to koi herpesvirus disease in carp. Front. Genet. 10, 543. doi:10.3389/fgene.2019.00543
Pang, S.-C., Wang, H.-P., Li, K.-Y., Zhu, Z.-Y., Kang, J. X., and Sun, Y.-H. (2014). Double transgenesis of humanized fat1 and fat2 genes promotes omega-3 polyunsaturated fatty acids synthesis in a zebrafish model. Mar. Biotechnol. 16, 580–593. doi:10.1007/s10126-014-9577-9
Pang, M., Fu, B., Yu, X., Liu, H., Wang, X., Yin, Z., et al. (2017). Quantitative trait loci mapping for feed conversion efficiency in crucian carp (Carassius auratus). Sci. Rep. 7, 16971. doi:10.1038/s41598-017-17269-2
Pang, M., Luo, W., Fu, B., Yu, X., Zhou, Y., and Tong, J. (2018). Transcriptomic profiles of brain provide insights into molecular mechanism of feed conversion efficiency in Crucian carp (Carassius auratus). Int. J. Mol. Sci. 19, 858. doi:10.3390/ijms19030858
Papp, M., Tóth, A. G., Nagy, S. Á., Erdélyi, K., Maróti, G., Cox, N., et al. (2023). Shotgun metagenomic analysis of the skin mucus bacteriome of the common carp (Cyprinus carpio). bioRxiv 2023.06. 12.544578.
Penman, D. J., Gupta, M. V., and Dey, M. M. (2005). Carp genetic resources for aquaculture in Asia, WorldFish.
Pieterse, A., Malan, A. P., and Ross, J. L. (2017). Nematodes that associate with terrestrial molluscs as definitive hosts, including Phasmarhabditis hermaphrodita (Rhabditida: rhabditidae) and its development as a biological molluscicide. J. Helminthol. 91, 517–527. doi:10.1017/S0022149X16000572
Ponzoni, R. W., Nguyen, N. H., Khaw, H. L., and Ninh, N. H. (2008). Accounting for genotype by environment interaction in economic appraisal of genetic improvement programs in common carp Cyprinus carpio. Aquaculture 285, 47–55. doi:10.1016/j.aquaculture.2008.08.012
Pradhan, P., Verma, D. K., Peruzza, L., Gupta, S., Haq, S. A., Shubin, S. V., et al. (2020). Molecular insights into the mechanisms of susceptibility of Labeo rohita against oomycete Aphanomyces invadans. Sci. Rep. 10, 19531. doi:10.1038/s41598-020-76278-w
Prchal, M., Palaiokostas, C., Gela, D., Piačková, V., Reschová, S., and Kocour, M. J. (2023). Genetic parameters and genomic prediction of resistance to koi herpesvirus disease using a low-density SNP panel on two Amur mirror carp populations. Aquac. Rep. 30, 101582. doi:10.1016/j.aqrep.2023.101582
Prchal, M., Vandeputte, M., Gela, D., Dolezal, M., Blichtova, H., Rodina, M., et al. (2018). Estimation of genetic parameters of fatty acids composition in flesh of market size common carp (cyprinus Carpio L.) and their relation to performance traits revealed that selective breeding can indirectly affect flesh quality. Czech J. Animal Sci. 63, 280–291. doi:10.17221/30/2018-cjas
Prchal, M., Zhao, J., Gela, D., Kašpar, J., Lepič, P., Kašpar, V., et al. (2021). Simplified method for genetic slaughter yields improvement in common carp under European pond conditions. Aquac. Rep. 21, 100832. doi:10.1016/j.aqrep.2021.100832
Qian, X., Ba, Y., Zhuang, Q., and Zhong, G. (2014). RNA-Seq technology and its application in fish transcriptomics. OMICS 18, 98–110. doi:10.1089/omi.2013.0110
Qu, X., Hu, M., Shang, Y., Pan, L., Jia, P., Fu, C., et al. (2018). Liver transcriptome and miRNA analysis of silver carp (Hypophthalmichthys molitrix) intraperitoneally injected with microcystin-LR. Front. Physiology 9, 381. doi:10.3389/fphys.2018.00381
Rajesh, R., and Majumdar, K. C. (2005). Transgene integration - an analysis in autotransgenicLabeo rohita Hamilton (Pisces: cyprinidae). Fish Physiology Biochem. 31, 281–287. doi:10.1007/s10695-006-0037-y
Rasal, K. D., Chakrapani, V., Pandey, A. K., Rasal, A. R., Sundaray, J. K., Ninawe, A., et al. (2017b). Status and future perspectives of single nucleotide polymorphisms (SNPs) markers in farmed fishes: way ahead using next generation sequencing. Gene Rep. 6, 81–86. doi:10.1016/j.genrep.2016.12.004
Rasal, K. D., Iquebal, M. A., Dixit, S., Vasam, M., Raza, M., Sahoo, L., et al. (2020). Revealing alteration in the hepatic glucose metabolism of genetically improved carp, Jayanti rohu Labeo rohita fed a high carbohydrate diet using transcriptome sequencing. Int. J. Mol. Sci. 21, 8180. doi:10.3390/ijms21218180
Rasal, K. D., Iquebal, M. A., Jaiswal, S., Dixit, S., Vasam, M., Nandi, S., et al. (2019). Liver-specific microRNA identification in farmed carp, Labeo bata (Hamilton, 1822), fed with starch diet using high-throughput sequencing. Mar. Biotechnol. 21, 589–595. doi:10.1007/s10126-019-09912-y
Rasal, K. D., Mohapatra, S., Kumar, P. V., K, S. R., Asgolkar, P., Acharya, A., et al. (2023). DNA methylation profiling of ovarian tissue of climbing perch (anabas testudienus) in response to monocrotophos exposure. Mar. Biotechnol. (NY) 25, 1123–1135. doi:10.1007/s10126-023-10264-x
Rasal, K. D., Nandanpawar, P. C., Swain, P., Badhe, M. R., Sundaray, J. K., and Jayasankar, P. (2016). MicroRNA in aquaculture fishes: a way forward with high-throughput sequencing and a computational approach. Rev. Fish Biol. Fish. 26, 199–212. doi:10.1007/s11160-016-9421-6
Rasal, K. D., and Sundaray, J. K. (2020). Status of genetic and genomic approaches for delineating biological information and improving aquaculture production of farmed rohu, Labeo rohita (Ham, 1822). Aquaculture 12, 2466–2480. doi:10.1111/raq.12444
Rasal, A., Patnaik, M., Murmu, K., Nandanpawar, P., Sundaray, J., and Mahapatra, K. (2017a). Genetically improved Jayanti Rohu: a boon to freshwater aquaculture in India. World Aqaculture Soc. Mag. 48, 23–25.
Ren, L., Zhang, H., Luo, M., Gao, X., Cui, J., Zhang, X., et al. (2022). Heterosis of growth trait regulated by DNA methylation and miRNA in allotriploid fish. Epigenetics and Chromatin 15, 19. doi:10.1186/s13072-022-00455-6
Ringø, E., Olsen, R. E., Mayhew, T. M., and Myklebust, R. (2003). Electron microscopy of the intestinal microflora of fish. Aquaculture 227, 395–415. doi:10.1016/j.aquaculture.2003.05.001
Robinson, N., Baranski, M., Mahapatra, K. D., Saha, J. N., Das, S., Mishra, J., et al. (2014). A linkage map of transcribed single nucleotide polymorphisms in rohu (Labeo rohita) and QTL associated with resistance to Aeromonas hydrophila. BMC Genomics 15, 541–623. doi:10.1186/1471-2164-15-541
Robinson, N., Sahoo, P. K., Baranski, M., Mahapatra, K. D., Saha, J. N., Das, S., et al. (2012). Expressed sequences and polymorphisms in rohu carp (Labeo rohita, Hamilton) revealed by mRNA-seq. Mar. Biotechnol. 14, 620–633. doi:10.1007/s10126-012-9433-8
Robledo, D., Matika, O., Hamilton, A., and Houston, R. D. (2018). Genome-wide association and genomic selection for resistance to amoebic gill disease in Atlantic salmon. G3 (Bethesda). 8, 1195–1203. doi:10.1534/g3.118.200075
Rodrigues, P. M., Martin, S. A., Silva, T. S., Boonanuntanasarn, S., Schrama, D., Moreira, M., et al. (2018). Proteomics in fish and aquaculture research, 311–338.
Roeselers, G., Mittge, E. K., Stephens, W. Z., Parichy, D. M., Cavanaugh, C. M., Guillemin, K., et al. (2011). Evidence for a core gut microbiota in the zebrafish. ISME J. 5, 1595–1608. doi:10.1038/ismej.2011.38
Roques, S., Deborde, C., Richard, N., Skiba-Cassy, S., Moing, A., and Fauconneau, B. (2020). Metabolomics and fish nutrition: a review in the context of sustainable feed development. Rev. Aquac. 12, 261–282. doi:10.1111/raq.12316
Rothbard, S., Biton, I., and Kulikovski, Z. (2010). Breeding, production and marketing of golden tench (Tinca tinca (L.)) in Gan shmuel fish breeding center, Israel. Rev. fish Biol. Fish. 20, 367–373. doi:10.1007/s11160-009-9132-3
Rothbard, S., and Wohlfarth, G. W. (1993). Inheritance of albinism in the grass carp, Ctenopharyngodon idella. Aquaculture 115, 13–17. doi:10.1016/0044-8486(93)90354-2
Round, J. L., and Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323. doi:10.1038/nri2515
Sahoo, P. K., Meher, P. K., Mahapatra, K. D., Saha, J. N., Jana, R. K., and Reddy, P. V. (2004). Immune responses in different fullsib families of Indian major carp, Labeo rohita, exhibiting differential resistance to Aeromonas hydrophila infection. Aquaculture 238, 115–125. doi:10.1016/j.aquaculture.2004.06.008
Sahoo, P. K., Rauta, P. R., Mohanty, B. R., Mahapatra, K. D., Saha, J. N., Rye, M., et al. (2011). Selection for improved resistance to Aeromonas hydrophila in Indian major carp Labeo rohita: survival and innate immune responses in first generation of resistant and susceptible lines. Fish and Shellfish Immunol. 31, 432–438. doi:10.1016/j.fsi.2011.06.014
Sahoo, L., Das, P., Sahoo, B., Das, G., Meher, P. K., Udit, U. K., et al. (2020). The draft genome of Labeo catla. BMC Res. Notes 13, 411. doi:10.1186/s13104-020-05240-w
Sahoo, L., Das, S., Bit, A., Patnaik, S., Mohanty, M., Das, G., et al. (2022). De novo assembly, transcriptome characterization and marker discovery in Indian major carp, Labeo rohita through pyrosequencing. Genetica 150, 59–66. doi:10.1007/s10709-021-00141-7
Sahoo, L., Swain, S. K., Bej, D., Das, S. P., Jayasankar, P., and Das, P. (2016). Complete mitochondrial genome sequence of Labeo fimbriatus (Bloch, 1975). Mitochondrial DNA Part A 27, 4019–4020. doi:10.3109/19401736.2014.1003824
Saleh, M., Kumar, G., Abdel-Baki, A.-A., Dkhil, M. A., El-Matbouli, M., and Al-Quraishy, S. (2018). Quantitative shotgun proteomics distinguishes wound-healing biomarker signatures in common carp skin mucus in response to Ichthyophthirius multifiliis. Vet. Res. 49, 37–12. doi:10.1186/s13567-018-0535-9
Searchinger, T., Waite, R., Hanson, C., Ranganathan, J., Dumas, P., and Matthews, E. (2018). Creating a sustainable food future.
Sébédio, J.-L. (2017). Metabolomics, nutrition, and potential biomarkers of food quality, intake, and health status. Adv. Food Nutr. Res. 82, 83–116. doi:10.1016/bs.afnr.2017.01.001
Shahi, N., Mallik, S. K., and Sarma, D. (2022). Muscle growth in targeted knockout common carp (Cyprinus carpio) mstn gene with low off-target effects. J. Aquac. 547, 737423. doi:10.1016/j.aquaculture.2021.737423
Shen, Y., Wang, L., Fu, J., Xu, X., Yue, G. H., and Li, J. (2019). Population structure, demographic history and local adaptation of the grass carp. J. BMC genomics 20, 467–516. doi:10.1186/s12864-019-5872-1
Shi, C., Lou, Q., Fu, B., Jin, J., Huang, J., Lu, Y., et al. (2022). Genomic polymorphisms at the crhr2 locus improve feed conversion efficiency through alleviation of hypothalamus-pituitary-interrenal axis activity in gibel carp (Carassius gibelio). Sci. China Life Sci. 65, 206–214. doi:10.1007/s11427-020-1924-4
Shi, M., Huang, R., Du, F., Pei, Y., Liao, L., Zhu, Z., et al. (2014). RNA-seq profiles from grass carp tissues after reovirus (GCRV) infection based on singular and modular enrichment analyses. Mol. Immunol. 61, 44–53. doi:10.1016/j.molimm.2014.05.004
Shireman, J. V., and Smith, C. R. (1983). Synopsis of biological data on the grass carp, Ctenopharyngodon idella (Cuvier and Valenciennes, 1844). Rome: FAO. Available at: pubs.usgs.gov/publication/1014542.
Song, X., Hu, X., Sun, B., Bo, Y., Wu, K., Xiao, L., et al. (2017). A transcriptome analysis focusing on inflammation-related genes of grass carp intestines following infection with Aeromonas hydrophila. Sci. Rep. 7, 40777. doi:10.1038/srep40777
Spasić, M. M., Poleksić, V., Stanković, M., Dulić, Z., Rašković, B., Živić, I., et al. (2010). Selective breeding programme of common carp (Cyprinus carpio L.) in Serbia: preliminary results. J. Agric. Sci. BGD. 55, 243–251. doi:10.2298/jas1003243s
Star, B., Nederbragt, A. J., Jentoft, S., Grimholt, U., Malmstrøm, M., Gregers, T. F., et al. (2011). The genome sequence of Atlantic cod reveals a unique immune system. Nature 477, 207–210. doi:10.1038/nature10342
Su, J., Shang, X., Wan, Q., and Su, J. (2018). SNP-based susceptibility–resistance association and mRNA expression regulation analyses of tlr7 to grass carp Ctenopharyngodon idella reovirus. J. Fish. Biol. 92, 1505–1525. doi:10.1111/jfb.13607
Sundaray, J. K., Rasal, K. D., Chakrapani, V., Swain, P., Kumar, D., Ninawe, A. S., et al. (2016). Simple sequence repeats (SSRs) markers in fish genomic research and their acceleration via next-generation sequencing and computational approaches. Aquac. Int. 24, 1089–1102. doi:10.1007/s10499-016-9973-4
Sun, Y., Huang, Y., Li, X., Baldwin, C. C., Zhou, Z., Yan, Z., et al. (2016). Fish-T1K (Transcriptomes of 1,000 Fishes) Project: large-scale transcriptome data for fish evolution studies. Gigascience 5, 18. doi:10.1186/s13742-016-0124-7
Suzuki, R. (1963). Hybridization experiments in cyprinid fishes. VI reciprocal crosses between gvathopogon elongatus elongatus and Gnathopogon japonicus. Jpn. J. Ichthyology 10, 39–42.
Svobodova, Z., and Kolarova, J. (2004). A review of the diseases and contaminant related mortalities of tench (Tinca tinca L.). Veterinarni Med. 49, 19–34. doi:10.17221/5671-vetmed
Tadmor-Levi, R., Doron-Faigenboim, A., Marcos-Hadad, E., Petit, J., Hulata, G., Forlenza, M., et al. (2019). Different transcriptional response between susceptible and resistant common carp (Cyprinus carpio) fish hints on the mechanism of CyHV-3 disease resistance. Bmc Genomics 20, 1019–1117. doi:10.1186/s12864-019-6391-9
Taniguchi, N., Yamasaki, M., Takagi, M., and Tsujimura, A. (1996). Genetic and environmental variances of body size and morphological traits in communally reared clonal lines from gynogenetic diploid ayu, Plecoglossus altivelis. Aquaculture 140, 333–341. doi:10.1016/0044-8486(95)01204-4
Tave, D. (1986). Genetics for fish hatchery managers. AVI Publishing Co., Inc. Available at: https://link.springer.com/book/9780442004170.
Thomas, T., Gilbert, J., and Meyer, F. (2012). Metagenomics-a guide from sampling to data analysis. Microb. Inf. Exp. 2, 3–12. doi:10.1186/2042-5783-2-3
Tóth, B., Khosravi, R., Ashrafzadeh, M. R., Bagi, Z., Fehér, M., Bársony, P., et al. (2020). Genetic diversity and structure of common carp (Cyprinus carpio L.) in the Centre of Carpathian Basin: implications for conservation. Genes 11, 1268. doi:10.3390/genes11111268
Vaishampayan, P. A., Kuehl, J. V., Froula, J. L., Morgan, J. L., Ochman, H., and Francino, M. P. (2010). Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol. Evol. 2, 53–66. doi:10.1093/gbe/evp057
Vandeputte, M. (2003). Selective breeding of quantitative traits in the common carp (Cyprinus carpio): a review. Aquat. Living Resour. 16, 399–407. doi:10.1016/s0990-7440(03)00056-1
Vandeputte, M., Kocour, M., Mauger, S., Dupont-Nivet, M., De Guerry, D., Rodina, M., et al. (2004). Heritability estimates for growth-related traits using microsatellite parentage assignment in juvenile common carp (Cyprinus carpio L.). Aquaculture 235, 223–236. doi:10.1016/j.aquaculture.2003.12.019
Vandeputte, M., Kocour, M., Mauger, S., Rodina, M., Launay, A., Gela, D., et al. (2008). Genetic variation for growth at one and two summers of age in the common carp (Cyprinus carpio L.): heritability estimates and response to selection. Aquaculture 277, 7–13. doi:10.1016/j.aquaculture.2008.02.009
Waghmare, S. G., Samarin, A. M., Samarin, A. M., Danielsen, M., Møller, H. S., Policar, T., et al. (2021). Histone acetylation dynamics during in vivo and in vitro oocyte aging in common carp cyprinus carpio. Int. J. Mol. Sci. 22, 6036. doi:10.3390/ijms22116036
Wang, C.-H., Li, S.-F., Xiang, S.-P., Wang, J., Liu, Z.-G., Pang, Z.-Y., et al. (2006). Genetic parameter estimates for growth-related traits in Oujiang color common carp (Cyprinus carpio var. color). Aquaculture 259, 103–107. doi:10.1016/j.aquaculture.2006.05.031
Wang, C., Chen, Q., Lu, G., Xu, J., Yang, Q., and Li, S. (2008a). Complete mitochondrial genome of the grass carp (Ctenopharyngodon idella, Teleostei): insight into its phylogenic position within Cyprinidae. Gene 424, 96–101. doi:10.1016/j.gene.2008.07.011
Wang, C., Yang, L., Lu, Y., Fang, C., Gan, X., Chen, Y., et al. (2024). Genomic features for adaptation and evolutionary dynamics of four major Asian domestic carps. Sci. China Life Sci. 67, 1308–1310. doi:10.1007/s11427-023-2479-2
Wang, J., Chen, G., Yu, X., Zhou, X., Zhang, Y., Wu, Y., et al. (2023). Transcriptome analyses reveal differentially expressed genes associated with development of the palatal organ in bighead carp (Hypophthalmichthys nobilis). Comp. Biochem. Physiol. Part D. Genomics Proteomics 46, 101072. doi:10.1016/j.cbd.2023.101072
Wang, J., Chen, L., Li, B., Xu, J., Feng, J., Dong, C., et al. (2021a). Performance of genome prediction for morphological and growth-related traits in Yellow River carp. Aquaculture 536, 736463. doi:10.1016/j.aquaculture.2021.736463
Wang, J., Gaughan, S., Lamer, J. T., Deng, C., Hu, W., Wachholtz, M., et al. (2020). Resolving the genetic paradox of invasions: preadapted genomes and postintroduction hybridization of bigheaded carps in the Mississippi River Basin. Evol. Appl. 13, 263–277. doi:10.1111/eva.12863
Wang, J., Su, B., and Dunham, R. A. (2022a). Genome-wide identification of catfish antimicrobial peptides: a new perspective to enhance fish disease resistance. Rev. Aquac. 14, 2002–2022. doi:10.1111/raq.12684
Wang, J., Wei, Y., Wang, D., Chan, L. L., and Dai, J. (2008b). Proteomic study of the effects of complex environmental stresses in the livers of goldfish (Carassius auratus) that inhabit Gaobeidian Lake in Beijing, China. Ecotoxicology 17, 213–220. doi:10.1007/s10646-007-0187-4
Wang, W., and Gao, Z. (2018). Recent developments in bream culture: culture systems and genetic improvement. J Aquac. China Success Stories Mod. Trends, 158–173. doi:10.1002/9781119120759.ch2_5
Wang, Y., Hamid, N., Jia, P.-P., and Pei, D.-S. (2021b). A comprehensive review on genetically modified fish: key techniques, applications and future prospects. Rev. Aquac. 13, 1635–1660. doi:10.1111/raq.12538
Wang, Y., Liao, A. M., Tan, H., Li, M., Geng, C., Wang, S., et al. (2022b). The comparative studies on growth rate and disease resistance between improved grass carp and common grass carp. Aquaculture 560, 738476. doi:10.1016/j.aquaculture.2022.738476
Wang, Y., Lu, Y., Zhang, Y., Ning, Z., Li, Y., Zhao, Q., et al. (2015). The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 47, 625–631. doi:10.1038/ng.3280
Wang, Y., Sha, H., Li, X., Zhou, T., Luo, X., Zou, G., et al. (2022c). Microsatellite characteristics of silver carp (Hypophthalmichthys molitrix) genome and genetic diversity analysis in four cultured populations. Genes 13, 1267. doi:10.3390/genes13071267
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi:10.1038/nrg2484
Welcomme, R. L. (1988). International introductions of inland aquatic species. Rome: Food and Agriculture Org, 294.
Wenne, R. (2023a). Microsatellites as molecular markers with applications in exploitation and conservation of aquatic animal populations. Genes 14, 808. doi:10.3390/genes14040808
Wenne, R. (2023b). Single nucleotide polymorphism markers with applications in conservation and exploitation of aquatic natural populations. Animals. 13, 1089. doi:10.3390/ani13061089
Wohlfarth, G., Moav, R., and Hulata, G. (1975). Genetic differences between the Chinese and European races of the common carp. II. Multi-character variation-a response to the diverse methods of fish cultivation in Europe and China. Hered. (Edinb). 34, 341–350. doi:10.1038/hdy.1975.43
Wu, C.-S., Ma, Z.-Y., Zheng, G.-D., Zou, S.-M., Zhang, X.-J., and Zhang, Y.-A. (2022a). Chromosome-level genome assembly of grass carp (Ctenopharyngodon idella) provides insights into its genome evolution. BMC genomics 23, 271. doi:10.1186/s12864-022-08503-x
Wu, C., Huang, X., Chen, Q., Hu, F., Zhou, L., Gong, K., et al. (2020). The formation of a new type of hybrid culter derived from a hybrid lineage of Megalobrama amblycephala (♀) × Culter alburnus (♂). J. Aquac. 525, 735328. doi:10.1016/j.aquaculture.2020.735328
Wu, G., Sun, Y., and Zhu, Z. (2003). Growth hormone gene transfer in common carp. Aquat. Living Resour. 16, 416–420. doi:10.1016/s0990-7440(03)00087-1
Wu, S., Wang, G., Angert, E. R., Wang, W., Li, W., and Zou, H. (2012). Composition, diversity, and origin of the bacterial community in grass carp intestine. PLoS One 7, e30440. doi:10.1371/journal.pone.0030440
Wu, Y., Li, R., Wu, X., Guo, W., Li, Y., Song, Y., et al. (2022b). Improved liver lipid catabolism and utilization in growth hormone transgenic common carp (Cyprinus carpio L.) through enhanced lipolytic and fatty acid β-oxidation pathways. Front. Endocrinol. 13. doi:10.3389/fendo.2022.982488
Xia, J. H., Lin, G., Fu, G. H., Wan, Z. Y., Lee, M., Wang, L., et al. (2014). The intestinal microbiome of fish under starvation. BMC Genomics 15, 266–311. doi:10.1186/1471-2164-15-266
Xie, C., Li, J., Li, D., Shen, Y., Gao, Y., and Zhang, Z. (2018). Grass carp: the fish that feeds half of China. J Aquac. China Success Stories Mod. Trends, 93–115. doi:10.1002/9781119120759.ch2_1
Xiong, X.-M., Huang, W.-J., Dong, Q., Zhang, D.-Y., Wan, S.-M., and Gao, Z.-X. (2023). Genetic parameter estimates for intermuscular bone trait in grass carp (Ctenopharyngodon idella). J. Aquac. 563, 739011. doi:10.1016/j.aquaculture.2022.739011
Xiong, X.-M., Robinson, N. A., Zhou, J.-J., Chen, Y.-L., Wang, W., Wang, X.-B., et al. (2019). Genetic parameter estimates for intermuscular bone in blunt snout bream (Megalobrama amblycephala) based on a microsatellite-based pedigree. J. Aquac. 502, 371–377. doi:10.1016/j.aquaculture.2018.12.072
Xu, J., Zhao, Z., Zhang, X., Zheng, X., Li, J., Jiang, Y., et al. (2014a). Development and evaluation of the first high-throughput SNP array for common carp (Cyprinus carpio). BMC Genomics 15, 307–310. doi:10.1186/1471-2164-15-307
Xun, W., Ji, M., Ma, Z., Deng, T., Yang, W., Hou, G., et al. (2023). Dietary emodin alleviates lipopolysaccharide-induced intestinal mucosal barrier injury by regulating gut microbiota in piglets. J. Anim. Nutr. 14, 152–162. doi:10.1016/j.aninu.2023.05.004
Xu, P., Zhang, X., Wang, X., Li, J., Liu, G., Kuang, Y., et al. (2014b). Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 46, 1212–1219. doi:10.1038/ng.3098
Xu, Q., and Xie, W. (2018). Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol. 28, 237–253. doi:10.1016/j.tcb.2017.10.008
Yamamoto, T.-O. (1973). Inheritance of albinism in the goldfish, Carassius auratus. Jpn. J. Genet. 48, 53–64. doi:10.1266/jjg.48.53
Yanez, J. M., Barria, A., Lopez, M. E., Moen, T., Garcia, B. F., Yoshida, G. M., et al. (2023). Genome-wide association and genomic selection in aquaculture. Rev. Aquac. 15, 645–675. doi:10.1111/raq.12750
Yáñez, J. M., Naswa, S., López, M., Bassini, L., Correa, K., Gilbey, J., et al. (2016). Genomewide single nucleotide polymorphism discovery in Atlantic salmon (Salmo salar): validation in wild and farmed American and European populations. Mol. Ecol. Resour. 16, 1002–1011. doi:10.1111/1755-0998.12503
Yáñez, J. M., Newman, S., and Houston, R. D. (2015). Genomics in aquaculture to better understand species biology and accelerate genetic progress. Front. Genet. 6, 128. doi:10.3389/fgene.2015.00128
Yang, S., Luo, J., Long, Y., Du, J., Xu, G., Zhao, L., et al. (2019). Mixed diets reduce the oxidative stress of common carp (Cyprinus carpio): based on microRNA sequencing. Front. Physiol. 10, 631. doi:10.3389/fphys.2019.00631
Ye, L., Amberg, J., Chapman, D., Gaikowski, M., and Liu, W.-T. (2014). Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME J. 8, 541–551. doi:10.1038/ismej.2013.181
Young, H. A., and Benton, D. (2018). Heart-rate variability: a biomarker to study the influence of nutrition on physiological and psychological health? Behav. Pharmacol. 29, 140–151. doi:10.1097/FBP.0000000000000383
Yu, C., Xu, X., Li, J., and Shen, Y. (2020). Primary mapping of QTL for growth-related traits in grass carp (Ctenopharyngodon idellus). J. Aquac. Int. 28, 2275–2285. doi:10.1007/s10499-020-00594-1
Zenger, K. R., Khatkar, M. S., Jones, D. B., Khalilisamani, N., Jerry, D. R., and Raadsma, H. W. (2019). Genomic selection in aquaculture: application, limitations and opportunities with special reference to marine shrimp and pearl oysters. Front. Genet. 9, 693. doi:10.3389/fgene.2018.00693
Zhang, D.-Y., Liu, X.-M., Huang, W.-J., Wang, Y., Anwarullah, K., Luo, L.-F., et al. (2023a). Whole-genome resequencing reveals genetic diversity and signatures of selection in mono-female grass carp (Ctenopharyngodon idella). Aquaculture 575, 739816. doi:10.1016/j.aquaculture.2023.739816
Zhang, Y.-L., Duan, X.-D., Jiang, W.-D., Feng, L., Wu, P., Liu, Y., et al. (2019c). Soybean glycinin decreased growth performance, impaired intestinal health, and amino acid absorption capacity of juvenile grass carp (Ctenopharyngodon idella). J. Fish. Physiol. Biochem. 45, 1589–1602. doi:10.1007/s10695-019-00648-z
Zhang, Y.-Y., Liu, B., Ge, X.-P., Liu, W.-B., Xie, J., Ren, M.-C., et al. (2014). Effects of dietary emodin supplementation on growth performance, non-specific immune responses, and disease resistance to Aeromonas hydrophila in juvenile Wuchang bream (Megalobrama amblycephala). Israeli J. Aquac. - Bamidgeh 66. doi:10.46989/001c.20738
Zhang, D., Fu, J., Zhang, L., Fang, M., Zhu, W., Wang, L., et al. (2019a). The parentage analysis of bighead carp (Hypophthalmichthys nobilis) based on ten microsatellite markers. Genomics Appl. Biol. 38, 2949–2957.
Zhang, D., Guo, H., Zhu, C., Cui, K., Zhang, N., and Jiang, S. (2015). The complete mitochondrial genome of mud carp Cirrhinus molitorella (Cypriniformes, Cyprinidae). Mitochondrial DNA 26, 149–150. doi:10.3109/19401736.2013.819497
Zhang, J., Shen, Y., Xu, X., Dai, Y., and Li, J. (2020a). Transcriptome analysis of the liver and muscle tissues of black carp (Mylopharyngodon piceus) of different growth rates. Mar. Biotechnol. 22, 706–716. doi:10.1007/s10126-020-09994-z
Zhang, W., Xu, X., Li, J., and Shen, Y. (2022). Transcriptomic analysis of the liver and brain in grass carp (Ctenopharyngodon idella) under heat stress. Mar. Biotechnol. 24, 856–870. doi:10.1007/s10126-022-10148-6
Zhang, X., Luan, P., Cao, D., and Hu, G. (2021). A high-density genetic linkage map and fine mapping of QTL for feed conversion efficiency in common carp (Cyprinus carpio). Front. Genet. 12, 778487. doi:10.3389/fgene.2021.778487
Zhang, X., Pang, S., Liu, C., Wang, H., Ye, D., Zhu, Z., et al. (2019b). A novel dietary source of EPA and DHA: metabolic engineering of an important freshwater species-common carp by fat1-transgenesis. Mar. Biotechnol. (NY) 21, 171–185. doi:10.1007/s10126-018-9868-7
Zhang, X., Shen, Y., Xu, X., Zhang, M., Bai, Y., Miao, Y., et al. (2018). Transcriptome analysis and histopathology of black carp (Mylopharyngodon piceus) spleen infected by Aeromonas hydrophila. Fish. shellfish Immunol. 83, 330–340. doi:10.1016/j.fsi.2018.09.047
Zhang, Y., Lu, Y., Zhang, P., Shang, X., and Li, Y. (2023b). Brain injury induced by mercury in common carp: novel insight from transcriptome analysis. Biol. Trace Elem. Res. 201, 403–411. doi:10.1007/s12011-022-03161-2
Zhang, Y., Stupka, E., Henkel, C. V., Jansen, H. J., Spaink, H. P., and Verbeek, F. J. (2011). Identification of common carp innate immune genes with whole-genome sequencing and RNA-Seq data. J. Integr. Bioinform 8, 169. doi:10.2390/biecoll-jib-2011-169
Zhang, Y., Zhang, P., Yu, P., Shang, X., Lu, Y., and Li, Y. (2020b). Transcriptome analysis reveals the mechanism of common carp brain injury after exposure to lead. Sci. Total Environ. 743, 140796. doi:10.1016/j.scitotenv.2020.140796
Zhang, Y., Zhang, P., Yu, P., Shang, X., Lu, Y., and Li, Y. (2020c). Transcriptome analysis reveals the mechanism of fluorine exposure on memory loss of common carp. Environ. Pollut. 265, 114927. doi:10.1016/j.envpol.2020.114927
Zhao, H., Gao, Z., Wang, L., Wang, J., Wang, S., Fei, B., et al. (2018). Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). Gigascience 7, giy115. doi:10.1093/gigascience/giy115
Zheng, X., Kuang, Y., Lv, W., Cao, D., Sun, Z., and Sun, X. (2016). Genome-wide association study for muscle fat content and abdominal fat traits in common carp (Cyprinus carpio). PLOS ONE 11, e0169127. doi:10.1371/journal.pone.0169127
Zhong, C., Song, Y., Wang, Y., Zhang, T., Duan, M., Li, Y., et al. (2013). Increased food intake in growth hormone-transgenic common carp (Cyprinus carpio L.) may be mediated by upregulating Agouti-related protein (AgRP). Gen. Comp. Endocrinol. 192, 81–88. doi:10.1016/j.ygcen.2013.03.024
Zhong, H., Li, J., Zhou, Y., Li, H., Tang, Y., Yu, J., et al. (2016a). A transcriptome resource for common carp after growth hormone stimulation. Mar. genomics 25, 25–27. doi:10.1016/j.margen.2015.11.004
Zhong, Z., Niu, P., Wang, M., Huang, G., Xu, S., Sun, Y., et al. (2016b). Targeted disruption of sp7 and myostatin with CRISPR-Cas9 results in severe bone defects and more muscular cells in common carp. J. Sci. Rep. 6, 22953. doi:10.1038/srep22953
Zhou, L., and Gui, J. F. (2002). Karyotypic diversity in polyploid gibel carp, Carassius auratus gibelio Bloch. Genetica 115, 223–232. doi:10.1023/a:1020102409270
Zhou, L., and Gui, J. F. (2018). “Applications of genetic breeding biotechnologies in chinese aquaculture,” in Aquaculture in China. Editors Q. Gui, Z. Tang, J. Li, Liu, and S. S. De Silva doi:10.1002/9781119120759.ch6_1
Zhou, L., Wang, Y., and Gui, J. F. (2000). Analysis of genetic heterogeneity among five gynogenetic clones of silver crucian carp, Carassius auratus gibelio Bloch, based on detection of RAPD molecular markers. Cytogenet. Cell Genet. 88, 133–139. doi:10.1159/000015506
Zhou, S., Liu, Y., Dong, J., Yang, Q., Xu, N., Yang, Y., et al. (2021a). Transcriptome analysis of goldfish (Carassius auratus) in response to Gyrodactylus kobayashii infection. Parasitol. Res. 120, 161–171. doi:10.1007/s00436-020-06827-9
Zhou, Y., Fu, B., Yu, X., Chen, G., Wang, J., Luo, W., et al. (2021b). Genome-wide association study reveals genomic regions and candidate genes for head size and shape in bighead carp (Hypophthalmichthys nobilis). Aquaculture 539, 736648. doi:10.1016/j.aquaculture.2021.736648
Zhou, Y., Tong, J., Wang, J., and Yu, X. (2020). Development of microsatellite markers and genetic diversity in wild and cultured populations of black carp (Mylopharyngodon piceus) along the Yangtze River. Aquac. Int. 28, 1867–1882. doi:10.1007/s10499-020-00563-8
Zhou, Y., Yu, W., Zhong, H., Li, J., Li, H., He, F., et al. (2016). Transcriptome analysis reveals that insulin is an immunomodulatory hormone in common carp. Fish. shellfish Immunol. 59, 213–219. doi:10.1016/j.fsi.2016.10.018
Zhu, L., Zhang, Z., Chen, H., Lamer, J. T., Wang, J., Wei, W., et al. (2021). Gut microbiomes of bigheaded carps and hybrids provide insights into invasion: a hologenome perspective. Evol. Appl. 14, 735–745. doi:10.1111/eva.13152
Zhu, W., Fu, J., Luo, M., Wang, L., Wang, P., Liu, Q., et al. (2022). Genetic diversity and population structure of bighead carp (Hypophthalmichthys nobilis) from the middle and lower reaches of the Yangtze River revealed using microsatellite markers. Aquac. Rep. 27, 101377. doi:10.1016/j.aqrep.2022.101377
Zhu, X., Chu, W. Y., Wu, P., Yi, T., Chen, T., and Zhang, J. S. (2014). MicroRNA signature in response to nutrient restriction and re-feeding in fast skeletal muscle of grass carp (Ctenopharyngodon idella). Dongwuxue Yanjiu 35, 404–410. doi:10.13918/j.issn.2095-8137.2014.5.404
Keywords: selective breeding, genetics, genomics, cyprinids, genomic selection
Citation: Rasal KD, Kumar PV, Risha S, Asgolkar P, Harshavarthini M, Acharya A, Shinde S, Dhere S, Rasal A, Sonwane A, Brahmane M, Sundaray JK and Nagpure N (2024) Genetic improvement and genomic resources of important cyprinid species: status and future perspectives for sustainable production. Front. Genet. 15:1398084. doi: 10.3389/fgene.2024.1398084
Received: 11 March 2024; Accepted: 02 September 2024;
Published: 19 September 2024.
Edited by:
Suxu Tan, Qingdao University, ChinaReviewed by:
Tao Zhou, Xiamen University, ChinaPrabhugouda Siriyappagouder, Nord University, Norway
Manu Kumar Gundappa, Wageningen University and Research, Netherlands
Copyright © 2024 Rasal, Kumar, Risha, Asgolkar, Harshavarthini, Acharya, Shinde, Dhere, Rasal, Sonwane, Brahmane, Sundaray and Nagpure. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manoj Brahmane, bWJyYWhtYW5lQGNpZmUuZWR1Lmlu
 Kiran D. Rasal1
Kiran D. Rasal1 Arpit Acharya
Arpit Acharya Avinash Rasal
Avinash Rasal Manoj Brahmane
Manoj Brahmane Jitendra K. Sundaray
Jitendra K. Sundaray