
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 11 July 2024
Sec. Genomics of Plants and the Phytoecosystem
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1396375
NHX proteins are transmembrane antiporters belonging to the cation/proton antiporter gene family, with a conserved Na+ (K+)/H+ exchange (PF00999) protein domain. NHXs play a prominent role in plant growth, development, and defense. However, the role of NHX gene family in potato (Solanum tuberosum L.) is yet to be known. In this study, we conducted a genome-wide analysis of the potato NHX gene family. A total of 25 StNHX family members were identified to be unevenly distributed on 10 chromosomes. The proteins ranged in length from 252 to 1,153 amino acids, with molecular masses ranging from 27516.32 to 127860.87 kD, and isoelectric points (pI) ranging from 4.96 to 9.3. Analyses of gene structures and conserved motifs indicated that StNHX genes in the same phylogenetic cluster are conserved. Phylogenetic analysis divided the StNHX genes into three subfamilies (Classes I, II, and III). Synteny analysis indicated that StNHX gene family Class III of NHX and all Arabidopsis thaliana NHXs shared a close evolutionary relationship. Analysis of cis-acting elements in the upstream 1,500 bp promoter region of potato NHX genes showed that these genes could be regulated by light, stress, and hormones such as abscisic acid and gibberellic acid. Protein-protein interaction network analysis indicated that StNHX proteins may participate in the regulation of potato growth and stress response. Besides, To determine a potential role of these genes in tissue development and drought response, we analyzed the RNA-seq data of different DM potato tissues. The results showed that NHX genes exhibited distinct tissue-specific expression patterns. We further examined the expression patterns of StNHX in different tissues (leaves, roots, shoots, tubers, stolons, and flowers) during the flowering stage in ‘Jizhangshu NO.8.’ potato. The qRT-PCR results further confirmed the importance of StNHX genes in potato plant growth and development. We further analyzed the RNA-seq data (DM potato) under different abiotic stresses (salt, drought, and heat), and found that the expression of StNHX genes was induced under abiotic stress. qRT-PCR analysis of shoots and roots of ‘Jizhangshu NO.8.’ potato treated for 0, 6, 12, and 24 h with 15% PEG6000 confirmed that the 25 StNHX genes are involved in the response to drought stress in potato. The results of this study may be useful for selecting appropriate candidate genes for the breeding of new drought-tolerant potato varieties. Furthermore, this study lays a foundation for prospective analysis of StNHX gene functions.
In plants, transmembrane NHX antiporters, which belong to the cation/proton antiporter-1 gene family, contain a conserved Na+ (K+)/H+ exchange (PF00999) protein domain (Ayadi et al., 2020). In order to avoid cellular damage and maintain normal physiological functioning, According to the subcellular localization of its family members, NHXs can be divided into three classes: vacuolar membrane-localized (Vac-class), endosomal membrane-localized (Endo-class), and plasma membrane-localized (PM-class) (Orlowski and Grinstein, 2007). The model plant A. thaliana contains 8 NHX gene family members (AtNHX1-AtNHX8), including six intracellular NHXs (Vac-class: AtNHX1-AtNHX4; Endo-class: AtNHX5 and AtNHX6) and two plasma membrane NHXs (AtNHX7 and AtNHX8) (Yamaguchi et al., 2003). In saline or low water environments, the Vac-class NHXs (AtNHX1-AtNHX4) have the ability to both provide energy through the transmembrane proton gradient formed by vacuolar membrane-bound H+-ATPase or H+-PPase, and mediate Na+ regionalization to minimize cellular damage and stabilize osmotic pressure (Rausch et al., 1996; Apse et al., 1999; Gaxiola et al., 1999; Blumwald, 2000; Blumwald et al., 2000; Yokoi et al., 2002; Li et al., 2009; Liu et al., 2010). The End-class NHXs (AtNHX5 and AtNHX6) are primarily localized to the endosomal compartments, including Golgi bodies and prevacuolar compartments, where they primarily regulate Na+ (K+), pH balance, and potassium nutrition (Gaxiola et al., 1999; Bassil et al., 2011; Bassil et al., 2011; Wu et al., 2016). The plasma membrane NHXs (AtNHX7 and AtNHX8) participate in cytoplasmic Na+ efflux metabolism, which is the first barrier against salt ion toxicity in plants (Yamaguchi et al., 2003; An et al., 2007). NHX antiporters can expel Na+ (K+) out of the cell or partition Na+ (K+) into vacuoles to maintain a low cellular Na+ (K+) concentration. NHX antiporters have the ability to regulate cell turgor, maintain cell Na+ (K+) concentration, prompt cells to absorb water to maintain osmotic balance, regulate intracellular pH, and improve drought and salt tolerance (Elias et al., 2012; Pardo et al., 2006; Wang et al., 2014; Wang et al., 2015; Yamaguchi et al., 2001; Apse et al., 1999). The NHX genes have been identified and characterized in several plant species, including Arabidopsis thaliana (Gaxiola et al., 1999), rice (Atsunori et al., 2004), maize (Zorb et al., 2005), Brassica napus (Wang et al., 2003), apple (Li et al., 2010), Gossypium hirsutum (Li et al., 2010), banana (Xu et al., 2021), cotton (Akram et al.,. 2020), Tomato (Cavusoglu E et al., 2023), wheat (Sharma et al., 2023) among others. To date, many gene sequences encoding salt tolerance proteins in plants have been developed (Mehta et al., 2021; Cavusoglu et al., 2023). The functions of NHX family genes have also been widely studied. For example, applications of 200 mmol L−1 NaCl, 10% PEG, and 100 mmol L−1 ABA were found to improve the transcription level of PbNHX1 in the leaves of Pyrus betulaefolia (Liu et al., 2018). The overexpression of OsNHX1 can improve the salt tolerance of transgenic rice (Zorb et al., 2005). Genome-wide screening of O. sativa Indica subspecies revealed sixteen NHX orthologous, and these were confirmed to be Na+/H+ exchangers, under drought stress, the expression of OsNHX7 and OsNHX16 was significantly higher in transgenic rice leaves than in those of wild type plants (Khare et al., 2021). Guo Huimin et al. (Guo et al., 2012) showed that overexpression of NnNHX1 improved the salt tolerance of transgenic tobacco, with higher expression resulting in greater salt tolerance. Transgenic corn strains overexpressing SsNHX1 exhibited improved seed germination rate and decreased physiological damage when exposed to a 1% (w/v) NaCl solution (Huang et al., 2018). A. thaliana overexpressing AtNHX1 could sustain growth and development in 200 mmol/L NaCl solution (Apse et al., 1999). After utilizing PCR to clone NHX1 from wheat, functional analysis indicated that wheat NHX1 could influence cold tolerance by regulating the expression of reverse transporters (Brini et al., 2005). Overexpression of ZxNHX and ZxVP1-1 can enhance the ion regionalization ability of transgenic beet, thus improving drought tolerance (Wu et al., 2017). Furthermore, silencing ZxNHX leads to the downregulation of ZxAKT1 and ZxSKOR, the overaccumulation of K+ in the tissues of ZxNHX-RNAi plants, and decreased drought tolerance (Yuan et al., 2015). Compared with the wild type, the activity of antioxidant enzymes in transgenic plants was enhanced, the degree of membrane lipid peroxidation was reduced, and the expression of abiotic stress-related genes was significantly upregulated after mannitol treatment (Pardo et al., 2021). The overexpression of NnNHX1 from lotus increased salt tolerance in tobacco (Guo et al., 2012). The GhNHX1 gene plays an important role in salt stress (Wu et al., 2004). Finally, Wang et al. (2016) found that the expression of IbNHX2 was positively correlated with the drought tolerance of transgenic sweet potato (Ipomoea batatas) plants (Wang et al., 2016). These studies demonstrate that overexpression of NHX could improve the drought tolerance of transgenic plants.
Potato (Solanum tuberosum L.; Solanaceae), originating from the Andean regions of Peru and Bolivia (Spooner et al., 2005), is an annual crop cultivated worldwide (Xu et al., 2011), and is the fourth most important food crop after wheat, rice, and maize (Visser et al., 2009; Zhang et al., 2017). Drought is one of the primary environmental factors limiting the productivity and growth of potato plants. Potato is very sensitive to drought, and yield reductions occur even at moderate soil moisture deficits (Gervais et al., 2021; Hill et al., 2021). In this study, we applied bioinformatics to identify 25 NHX gene family members across the whole potato genome. We also studied the chromosomal locations, gene duplication events, evolutionary history, and promoter elements associated with each identified NHX gene. Furthermore, the expression levels of NHX genes in various tissues/organs and under abiotic stress cinditions was also analyzed using publicly available potato RNA-sequencing datasets in double monoploid (DM) potato. Quantitative real-time PCR (qPCR) was used to further analyze the StNHX expression patterns, after which the key gene related to tissue-specific expression and drought tolerance were identified. Overall, these findings should inform the further characterization of StNHXs and provide valuable information for functional elucidation of these genes in potato.
The genome sequences, protein sequences, and coding sequences (CDS) of potato (Stuberosum_448_v4.03) were obtained from the Potato Genome Sequencing Consortium (PGSC; http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml). In the Pfam database (http://pfam.xfam.org/), the Hidden Markov Model (HMM) profile for the NHX conserved domain (PF00999) was downloaded as the search model (Finn et al., 2016). HMMER 3.1 (http://hmmer.org/download.html), with default parameters, was used to obtain candidate genes containing the conserved domain (Finn et al., 2011). SMART (http://smart.embl-heidelberg.de) was used to confirm the existence of the domain, and redundant sequences without NHX family protein domains were removed (Letunic et al., 2015).
The characteristics of StNHX proteins, including the number of amino acids, molecular weight (MW), and theoretical isoelectric point (pI), were estimated using Expasy (https://web.expasy.org/compute_pi/) (Bjellqvist et al., 1993; Gasteiger et al., 2005). Subcellular localization of proteins was predicted using CELLO (http://cello.life.nctu.edu.tw/) (Yu et al., 2006). StNHX protein motifs were analyzed using MEME (https://meme-suite.org/meme/), with a maximum of 20 motifs (Bailey et al., 2009). The exon-intron structures of StNHX genes were examined using the Gene Structure Display Server (GSDS; http://gsds.gao-lab.org/index.php) (Hu et al., 2014) via alignment of the CDSs with their corresponding genomic DNA sequences. The GSDS was also used to graphically display the structure of StNHX genes.
MapChart software was used to create the chromosomal position map and examine the relative distance of StNHX genes (Voorrips, 2002). Gene duplication events were analyzed with MCScanX (Yu et al., 2012). Tandem repeats were determined according to two conditions: 1) the length of the short sequence covers more than 70% of the length of the long sequence; 2) the similarity of the two aligned sequences is greater than 70% (Yang et al., 2008; Zheng et al., 2020). Specifically, tandem repeats refer to two genes located in the same chromosomal segment, where the distance is less than 100 kb and the number of genes between them is less than or equal to five (Wang et al., 2010).
AtNHX protein sequences were obtained from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org). MEGA7.0 was used to construct a root-free phylogenetic tree using the maximum likelihood (ML) method and 1,000 iteration bootstraps (Kumar et al., 2016).
The StNHX gene structure was analyzed, and the gene structure map was drawn based on the gene location information from the whole genome GFF annotation file. The 1,500 bp upstream promoter sequences were extracted in batches by perl script. The StNHX gene promoter sequences (1,500 bp upstream of the ATG translation start codon) were used to predict cis-acting elements using PLANTCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
Network analysis of protein-protein interactions (PPIs) among all identified StNHX genes was performed using STRING v11.5 (https://string-db.org/), with a combined score of ≥0.700 (high confidence) (Szklarczyk et al., 2017). The interaction network was visualized by Cytoscape v3.9.1 (Shannon et al., 2003). Repeated events in potato genes were analyzed by MCScanX (Yupeng et al., 2012).
To study the expression of StNHX genes in different tissues (leaves, roots, shoots, callus, tubers, sepals, stamens, stolons, flowers, petioles, petals, carpels, and fruit) and under different stress conditions (mannitol, salt, and high temperature), publicly-available transcriptomic data of DM potato were downloaded from the PGSC (accession no. SRA030516) (Xu et al., 2011). Gene expression was visualized using TBtools (Chen et al., 2018).
Sterile tissue cultured seedlings of ‘Jizhangshu NO.8.’ potato were grown for 25 days at 22°C and with 16 h light per day. To simulate drought stress, experimental seedlings were treated for 0, 6, 12, and 24 h with 15% PEG6000. Each treatment consisted of three biological replicates. Subsequently, shoot and root tissues were collected and frozen in liquid nitrogen, and stored at −80°C until futher use.
Potatoes (‘Jizhangshu NO.8.’) were field-grown at Hebei North University, China. During the flowering stage, tissue samples (flowers, roots, stems, leaves, tubers, and stolons) were collected and frozen in liquid nitrogen, and stored at −80°C until further use.
To study stress-induced and tissue-specific gene expression, total RNA was extracted using a Plant RNA Reagent Kit (SENO, China) and reverse transcribed into cDNA using a First-Strand cDNA Synthesis Kit (SENO, China). Primers for quantitative fluorescence expression were designed using Primer Premier 6.0 (Supplementary Table S1), with elongation factor 1-alpha (EF-1α) (Tang et al., 2017) used as a reference primer. The real-time PCR system (20 μL) included 1 μL of upstream and downstream primers, 7 μL of ddH2O, 10 μL of SYBR Premix, and 1 μL of cDNA. The reaction procedure was as follows: 30 s at 95°C, 40 cycles of 5 s at 95°C and 30 s at 60°C, and 65°C–95°C melting curve detection. Gene expression was analyzed using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Three replicates were performed for each sample. The results were displayed by means ± standard deviation (SD).
A total of 29 candidate NHX genes were identified by Hidden Markov Model. Using SMART (http://smart.embl-heidelberg.de), a total of 25 NHX family members were found to contain the complete NHX conserved domain. The 25 StNHX genes were divided into three subgroups based on phylogenetic analysis, with 17 genes clustered in subgroup I, two genes clustered in subgroup II, and six genes clustered in subgroup III (Figure 1).
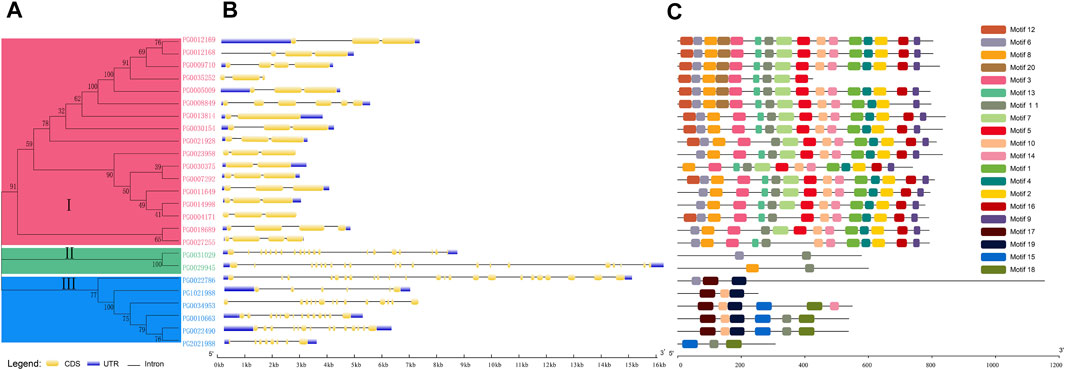
Figure 1. Phylogenetic relationships, gene structures, and conserved motifs of StNHX genes. (A): Multiple alignments of 25 full-length amino acid sequences of StNHX genes. The phylogenetic tree was constructed using the neighbor-joining method, with 1000 bootstrap replicates. Pink: Class I. Green: Class II. Blue: Class III. (B): Exon/intron structure of StNHX genes. Exons are represented by yellow boxes. Introns are represented by black lines. The UTR is marked in blue. (C): StNHX protein motifs. Each motif is represented by a different colored box.
Gene structure was analyzed by GSDS (Figure 1B). Significant differences were observed in the number of introns of StNHX gene family members, with a large gap between the introns in Class I, Class II, and Class III. Class I contained between one and five introns, Class II contained 19 introns, and Class III contained more than five introns. Furthermore, the exon-intron structures were relatively conserved within the same class, indicating that intron loss and gain occurred in the NHX gene family during evolution. This may explain the functional diversity of closely related NHX genes.
MEME was used to analyze the amino acid sequences and predict the full-length protein sequences of StNHX members. The 20 identified motifs were named Motif 1-Motif 20 (Figure 1C), and motifs were found to be conserved within each subgroup. Motifs 1, 2, 3, 5, 8, 12, 13, and 20 all contained 41 conserved amino acids; motifs 4, 6, 10, 11, and 14 all contained 29 amino acids; motif 9 contained at least 21 amino acids; motif 16 contained 33 amino acids; motif 19 contained 47 amino acids; and motifs 7, 15, 17, and 18 contained a maximum of 50 amino acids (Figure 1D). Motif 12 dominated the n-terminal of the first subgroup, and motif 9 dominated the C-terminal in class I, which contained between 9 and 16 motifs, the largest number of any subgroup. The second subgroup contained only two motifs, and the third subgroup contained between three and six motifs. The conserved domain contained motifs 12, 6, 8, 20, 3, 13, 11, 7, 5, 17, 19, 20, 15, and 18. Motifs 3, 5, 7, 12, and 13 were found only in Class I. Motifs 10, 15, 17, 18, and 19 were found only in Class III. Other motifs, including 1, 4, 2, 16, and 9, were found only in Class I. The motif diversity among the NHX classes implies that the functions of these proteins may have changed during evolution. Motifs 6, 8, and 11 were found in all classes. Conserved motifs have a high degree of similarity and genes containing the same motif are likely to be functionally similar, indicating that these conserved features may play crucial functional roles. In general, the motif composition of each class was relatively conserved and the gene structure was similar, which demonstrates the reliability of the evolutionary analysis.
A: Multiple alignments of 25 full-length amino acid sequences of StNHX genes. The phylogenetic tree was constructed using the neighbor-joining method, with 1,000 bootstrap replicates. Pink: Class I. Green: Class II. Blue: Class III.
B: Exon/intron structure of StNHX genes. Exons are represented by yellow boxes. Introns are represented by black lines. The UTR is marked in blue.
C: StNHX protein motifs. Each motif is represented by a different colored box.
Pink: Class Ⅰ. Green: Class Ⅱ. Blue: Class Ⅲ. The scale located on the left panel indicates chromosome size in bases. The chromosome number is indicated at the left of each chromosome Blue box: tandem repeating gene pair.
The chromosomal distribution of candidate StNHX family genes was predicted using GSDS (Figure 2). A total of 25 NHX genes were unevenly distributed across 10 chromosomes, with chromosome 6 containing the most genes (6), followed by chromosomes 4, 5, and 10 (1), and chromosomes 7 and 11 (0). Two Class II genes were located at the top and bottom of chromosomes 3 and 8; three Class III genes were located on chromosome 1; and two Class III genes were located on the distal ends of chromosomes 6 and 10. Members of Class I were mainly concentrated on chromosomes 6 and 8, the rest of the chromosomes contained between one and two genes.
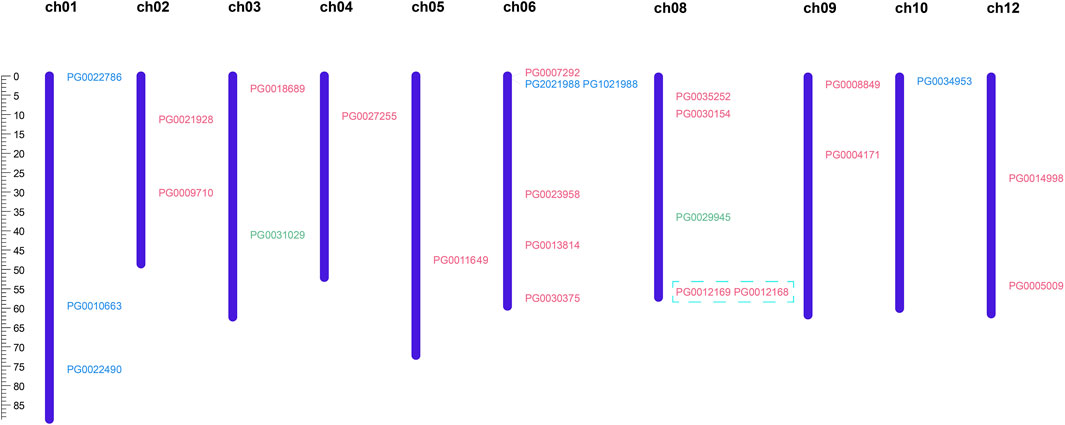
Figure 2. Analysis of chromosomal distribution and duplication events of StNHX genes. Pink: Class I. Green: Class II. Blue: Class III. The scale located on the left panel indicates chromosome size in bases. The chromosome number is indicated at the left of each chromosome Blue box: tandem repeating gene pair.
In order to clarify the evolutionary history of the StNHX gene family, we used MCScanX to study duplication events. In plant genomic evolution, tandem duplication and fragment duplication help to expand both the number of members and functions of a gene family (Niu et al., 2021). While no large fragment replication events were identified, tandem repetition was detected in potato. One tandem repeating gene pair was identified: PG0012168:PG0012169 (Figure 2).
In order to further study the potato NHX gene phylogenetic lineage and functional characteristics, the NHX protein sequences from A. thaliana were analyzed using the neighbor-joining (bootstrap = 1,000) method. The StNHX gene Class III was closely related to A. thaliana in one cluster, and Classes I and II were closely related in one cluster. Cluster analysis indicated that the Class III genes of A. thaliana and potato shared high homology, which suggests that these homologous genes might have similar functions (Figure 3).
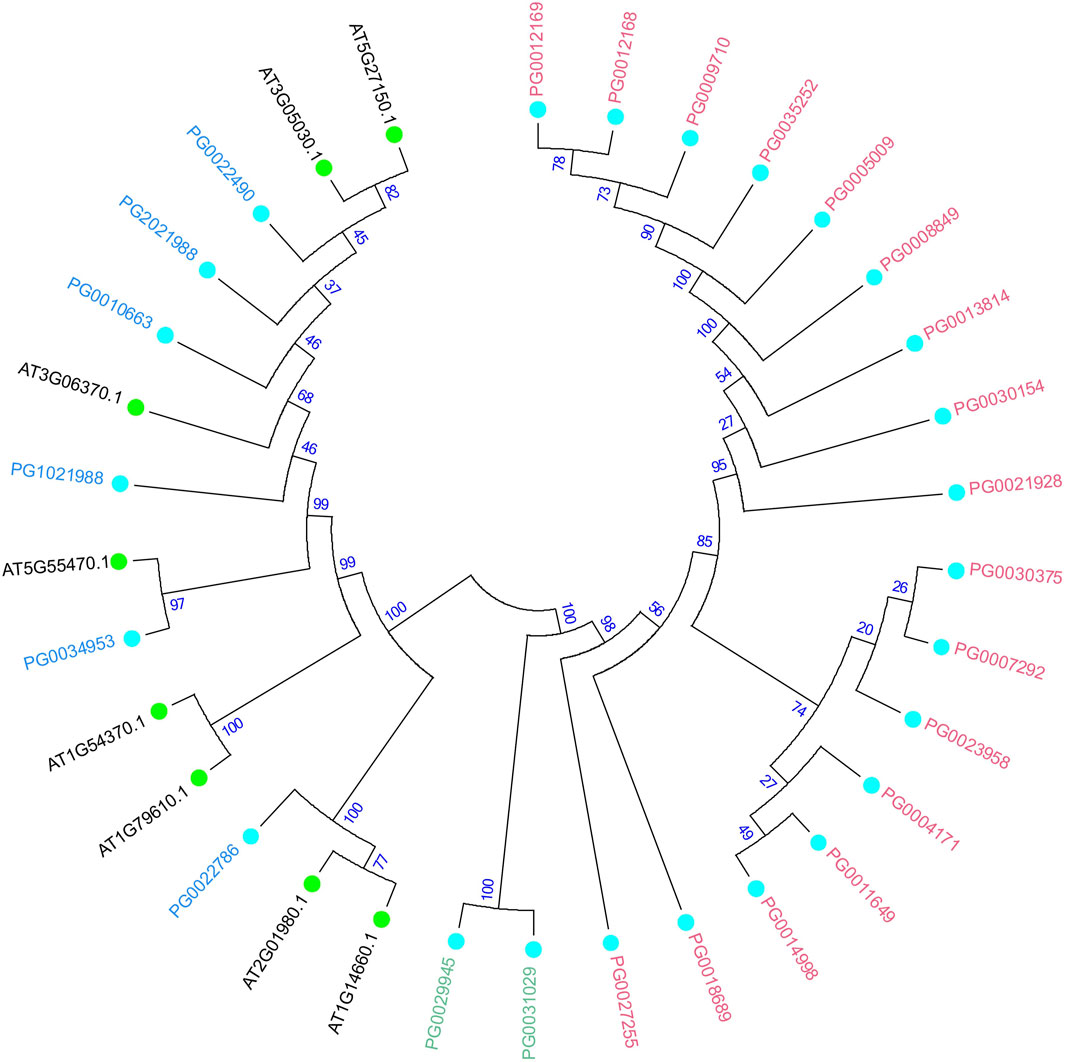
Figure 3. Phylogenetic analysis of NHX proteins in Arabidopsis thaliana and potato. The number at the branch of the evolutionary tree indicates the confidence of the branch, with higher values indicating higher reliability. A. thaliana and potato are shown the different colors, with light blue circles representing StNHX genes and green circles representing AtNHX genes. Pink: Class Ⅰ. Green: Class Ⅱ. Blue: Class Ⅲ.
The StNHX proteins ranged in length between 252 (PG1021988) and 1,153 (PG0022786) amino acids. The molecular weights ranged from 27516.32 (PG1021988) to 127860.87 (PG0022786) kD. The isoelectric point (pI) values ranged from 4.96 (PG1021988) to 9.30 (PG0022786). The isoelectric points of eight StNHX proteins (PG1021988, PG0018689, PG0004171, PG0011649, PG0021928, PG0030154, PG0013814, and PG0022786) are in the acid range (<7). The isoelectric points of 17 StNHX proteins were in the basic range (>7). CELLO v.2.5 subcellular localization analyses indicated that all 25 family members were located in the plasma membrane (Supplementary Table S2).
In plants, cis-regulatory elements control the expression of target genes by interacting with transcription factors (Priest et al., 2009). Therefore, identification of potential cis-regulatory elements in the promoter regions of NHX genes can provide information regarding their transcriptional regulation. The 1,500 bp upstream promoter sequences of NHX genes were selected and analyzed using PLANTCARE. The results showed that besides the two conventional promoter elements (TATA-box and CAAT-box), the StNHX family genes possessed 58 cis-acting elements. These cis-acting elements were divided into six categories: biosynthesis, circadian rhythm regulation, light response, plant hormone regulation, stress response regulation, and growth and development (Figure 4). One cis-acting element, MBSI, is a MYB-binding site involved in the regulation of genes for flavonoid biosynthesis. Another cis-acting element, Circadian, is involved in circadian control in plants. Growth and development-related regulatory elements include O2-site, GCN4_motif, Box 4, RY-element, and CAT-box. Notably, all the selected NHX genes contained the MYB and MYC motif except PG0025352, which are cis-acting regulatory elements involved in involved in environmental adaptation. Twenty-five light response elements were identified, and 13 stress and hormone response elements. The promoters of the NHX gene family were found to all contain these three related elements, suggesting the functional diversity of this gene family.
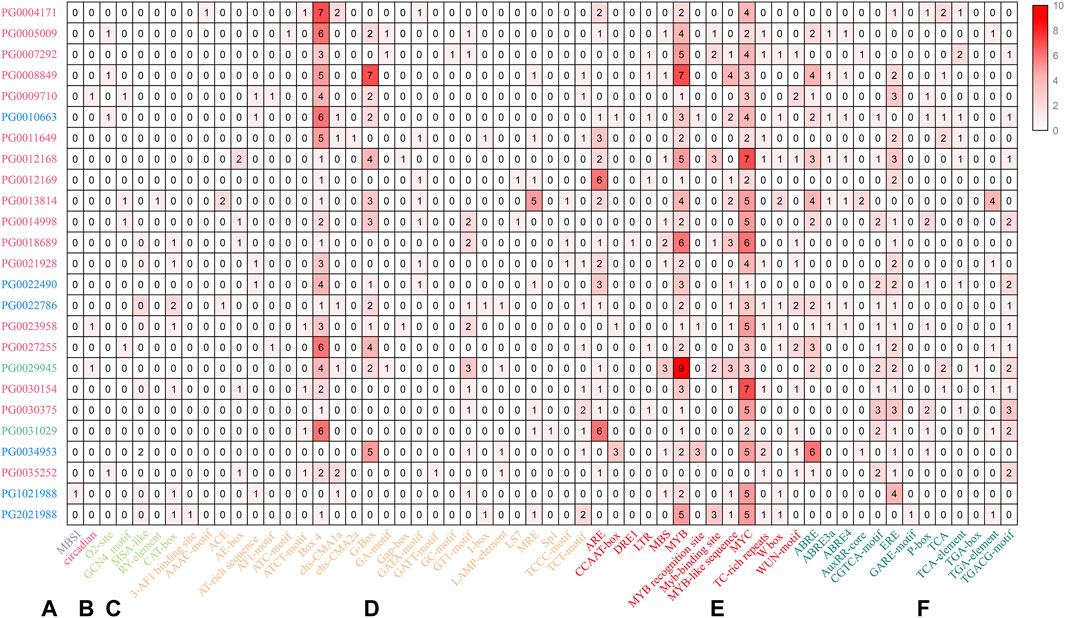
Figure 4. The promoter regions (1,500 bp upstream of the ATG translation start codon) of NHX genes were analyzed. Numbers of cis-acting elements are indicated in red, with darker red shading representing higher numbers. Pink: Class I. Green: Class Ⅱ. Blue: Class Ⅲ. (A) biosynthesis regulation. (B) circadian rhythm regulation. (C) growth and development regulation. (D) light response regulation. (E) plant hormone regulation. (F) stress response regulation.
Both stress response and growth and development elements were selected for further analysis (Figure 5). The results showed that stress-related elements were widely distributed, while growth-related elements only appeared in some StNHX family member promoters. These results suggest that the StNHX gene family plays an important role in stress response, and a few of these genes may play a regulatory role in growth and development.
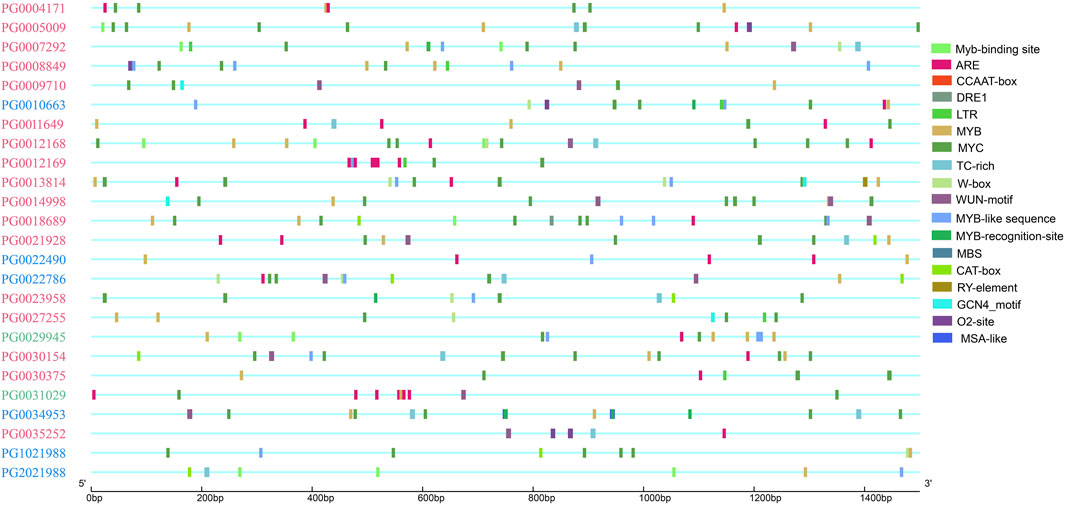
Figure 5. Distribution of cis-elements related to stress response and growth and development in the promoters of NHX genes. Pink: Class Ⅰ. Green: Class Ⅱ. Blue: Class Ⅲ.
In order to identify the function and the mechanism of action of NHX genes, the protein interaction networks (PPIs) of NHX proteins were built using STRING. The results showed that the PPI networks contain 15 StNHXs and can be divided into two groups. For example, PG0005009 was predicted to interact with PGSC0003DMT400024409, PGSC0003DMT400032161, and PGSC0003DMT400082322. PG0013814, PG0021928, PG0022490, PG0030154, PG1021988, PG2021988, PG0029945, PG0012169, PG0035252, PG0034953, PG0010663, PG0022786, PG0012168, and PG0009710 exhibited strong or weak interaction relationships. As shown in Figure 6, some of these proteins interacted strongly, with darker lines indicating higher core PPI values. There are 27 nodes and 29 groups of interaction relationships in the interaction network. PG0022786 was predicted to interact with five proteins, of which three belonged to plasma membrane ATPase (PGSC0003DMT400065168, PGSC0003DMT400010497, and PGSC0003DMT400083041) and one belonged to the protein kinase superfamily (PGSC0003DMT400050430), which play important roles in plant development and stress response. PG0010663 was also predicted to interact with five proteins, of which two belong to the serine/threonine-protein kinase family (PGSC0003DMT400050430 and PGSC0003DMT400052805), indicating that StNHXs may be involved in abiotic stress response. In addition, we found that PGSC0003DMT400027579, which is a sodium/hydrogen exchanger 8-like and salt overly sensitive gene, was predicted to interact with seven StNHXs, indicating that these StNHXs may participate in salt stress response. PG0005009 was predicted to interact with PGSC0003DMT400024409 and PGSC0003DMT400032161, which are heavy metal ATPases and likely participate in copper transport. PGSC0003DMT400082322 belongs to the cytochrome P450 CYP82 gene family, which is involved in growth, metabolism, and stress response regulation. Interestingly, PGSC0003DMT400077052 was located in the center of the whole protein interaction network, and may represent the core of potato StNHX protein family interaction.
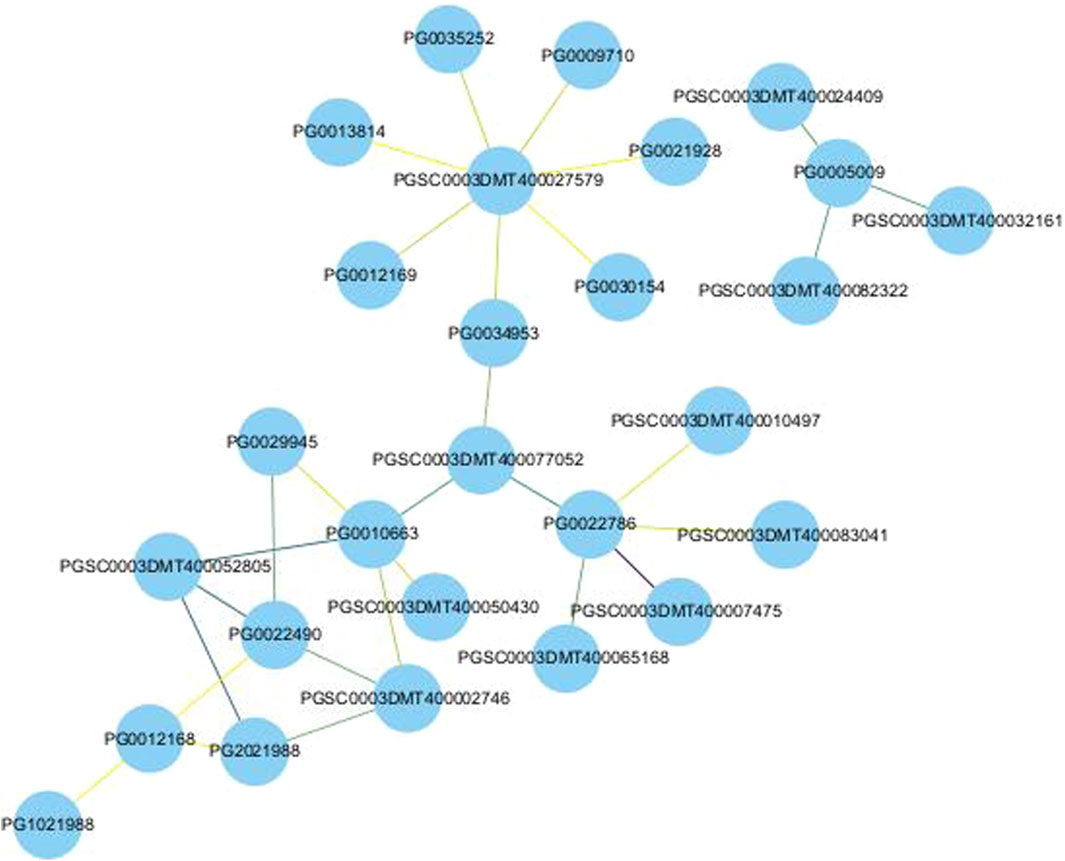
Figure 6. Schematic representation of StNHX protein-protein interaction (PPI) network. Different colored nodes indicate different proteins. Darker colors indicate higher core PPI values. The edge color is shown from yellow to purple in accordance with the combined score (>0.700).
We further constructed a synteny analysis between NHX genes in potato and NHX genes in many plants, including A. thaliana (L.), Brassica oleracea, Oryza sativa、Zea mays (Figure 7). The StNHX genes were homologous to genes in other plants, and syntenic conservation was observed among Solanum lycopersicum (22 orthologous gene pairs dispersed on all chromosomes except chromosome 7 and chromosome 11), B. oleracea (2 orthologous gene pairs dispersed chromosome 6 and chromosome 12), A. thaliana (L.) (2 orthologous gene pairs dispersed chromosome 3 and chromosome 6), Oryza sativa (1 orthologous gene pairs dispersed chromosome 1).
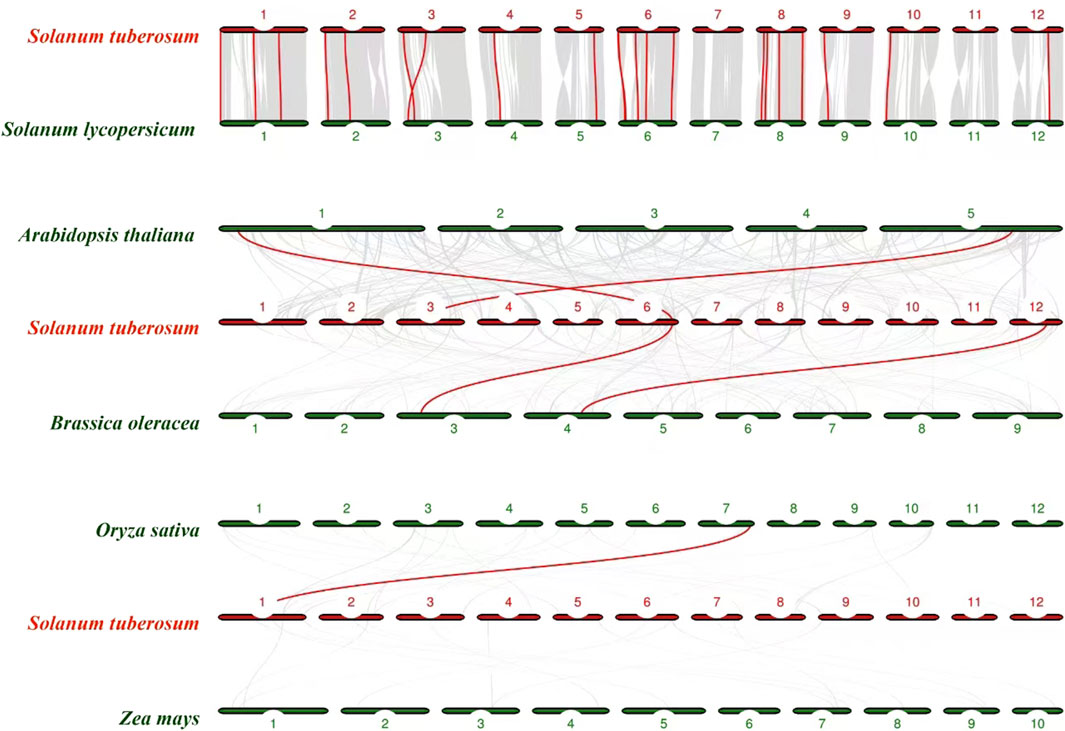
Figure 7. The collinear relationship between potato and other crops. Red lines indicate the collinear gene pairs of potato and other crops.
We also studied the tissue-specific (leaves, roots, shoots, tubers, stolons, and flowers) expression patterns of StNHX genes during the flowing stage in field-grown potatoes. Overall, the expression levels of StNHX genes were different across tissues, and there were also great differences among genes (Figure 9). The expression patterns of PG0023958 and PG0007292 were similar in roots and stolons, with PG0023958 exhibiting the highest expression and PG0007292 exhibiting the lowest. PG0034953 exhibited high expression in flowers and stolons, but low expression in other tissues. In flowers, PG0034953 exhibited the highest expression, followed by PG0005009 with relative expression levels greater than 1, while other genes were weakly expressed. The expression level of PG0030154 was relatively high in stems, but was weakly expressed in other tissues. PG0005009 and PG0022490 were only highly expressed in flowers and stolons, respectively, but were weakly expressed in other tissues. PG0009710 exhibited the highest expression in tubers. The expression level of PG0008849 was higher than other genes, and PG0022490 exhibited the lowest expression in leaves.
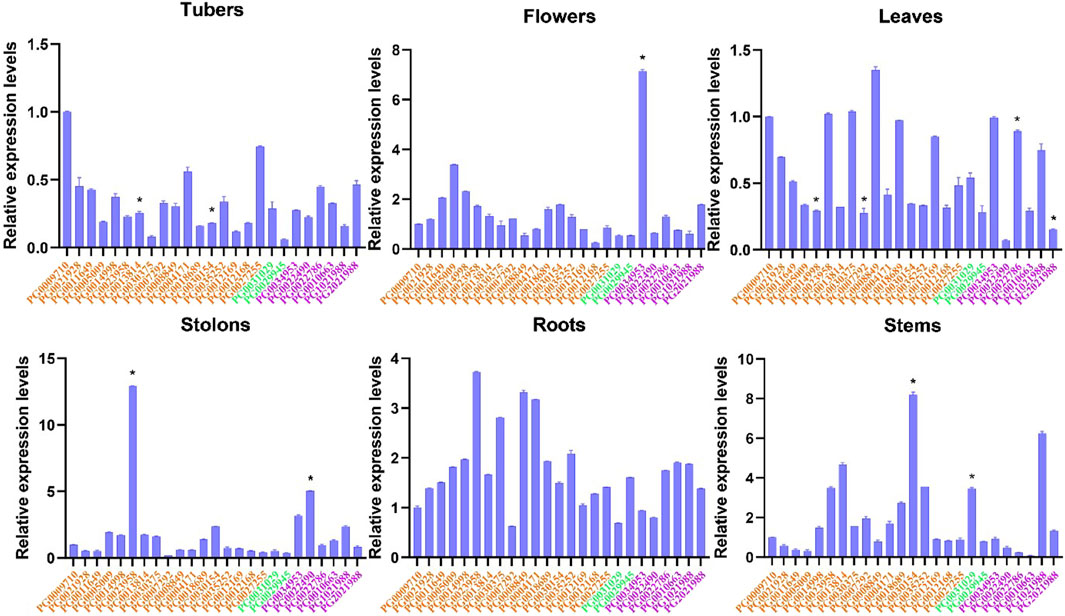
Figure 8. Expression analysis of StNHX gene family in various tissues of ‘Jizhangshu NO.8.’ potatoes at the flowering stage. Expression levels of StNHX genes in various tissues (leaves, roots, shoots, tubers, stolons, and flowers). The x-axis indicates the different genes. The y-axis indicates the relative expression levels. Relative expression values are presented as means ± standard errors based on three biological replicates with three technical replicates. Significant differential expression is indicated by an asterisk (*, p < 0.05).Yellow: Class I. Green: Class II. Purple: Class III.
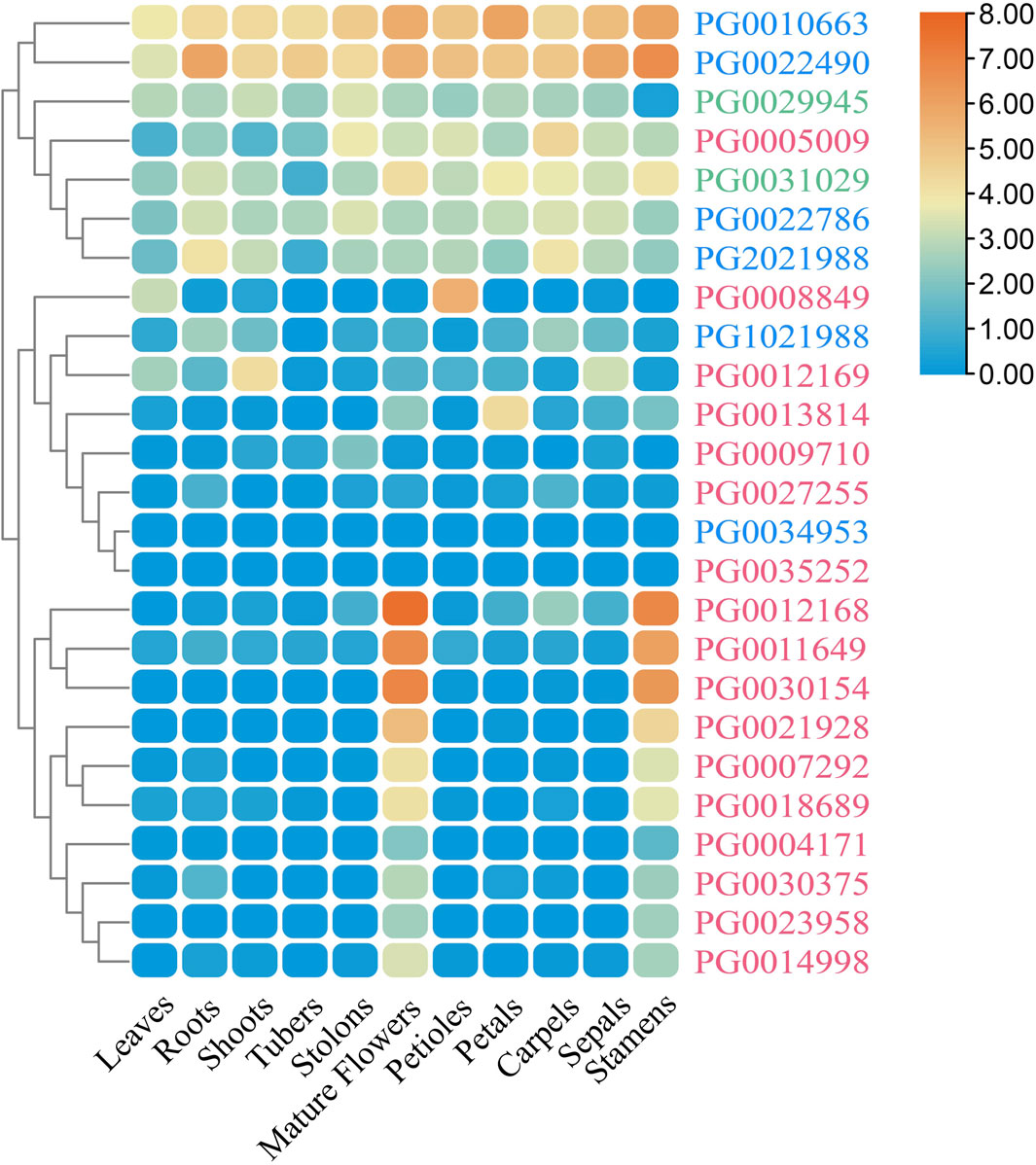
Figure 9. The expression profiles of StNHX genes in various tissues. The relative expression levels of 25 StNHX genes are taken as the logarithm with base 2 for standardization. Different colored patches indicate the relative expression levels of genes in different tissues (DM potato). Red represents high expression and blue represents low expression. Pink: Class I. Green: Class II. Blue: Class III.
To study the roles of StNHX genes in potato, the expression levels of StNHX genes in different tissues were analyzed using publicly-available RNA-Seq data. Of the 25 identified StNHX genes, 23 were found to be expressed in at least one tissue (FPKM > 1). The 25 identified StNHX genes were divided into two major groups based on their expression characteristics. The first group was composed of seven genes (PG0010663, PG0022490, PG0022786, PG2021988, PG0029945, PG0005009, and PG0031029) with relatively high expression in the majority of tissues. The remaining 18 StNHX genes formed the second group, which contained ten genes (PG0004171, PG0023958, PG0030375, PG0014998, PG0007292, PG0018689, PG0021928, PG0011649, PG0030154, and PG0012168) with high expression only in stamens and flowers, and eight genes (PG0034953, PG1021988, PG0035252, PG0009710, PG0008849, PG0027255, PG0012169, and PG0013814) with low expression in most tissues. In addition, some of the StNHX genes exhibited tissue-specific expression. For instance, PG0011649, PG0030154, and PG0012168 exhibited high expression in stamens and flowers (FPKM > 20). PG0004171, PG0023958, PG0030375, PG0014998, PG0007292, PG0018689, and PG0021928 were only expressed in stamens and flowers (FPKM > 1). Both PG0012169 and PG0013814 were highly expressed in shoots and petals (FPKM > 15), respectively, with low or no expression in other tissues (FPKM < 10). PG0034953 and PG0035252 were not expressed in any tissues. These results suggest that StNHX genes are differentially expressed in various tissues. Furthermore, relatively high expression within the same tissue indicates functional conservation, showing the importance of these genes in potato plant growth and development.
We next analyzed the transcriptomic data of salt-, drought-, and heat-stressed DM potato (accession no. SRA030516) to identify potential stress-responsive StNHX genes. According to the log2(FC) value, it was found that StNHX gene expression was induced by salt, drought, and heat, although the expression patterns were different under different stressors. A total of 11, 11, and 8 genes were differentially expressed under NaCl (150 mmol L−1), mannitol (260 μM), and heat (35°C) stress, respectively, with |log2(FC)| > 1 and FPKM >1, of which only 6, 4, and 7 genes were not expressed. The 25 identified StNHX genes were divided into two major groups (Figure 10). The first group was composed of 21 genes. The second group was composed of PG0022490, PG1021988, PG0012169, and PG2021988, which exhibited relatively high expression under abiotic stress.
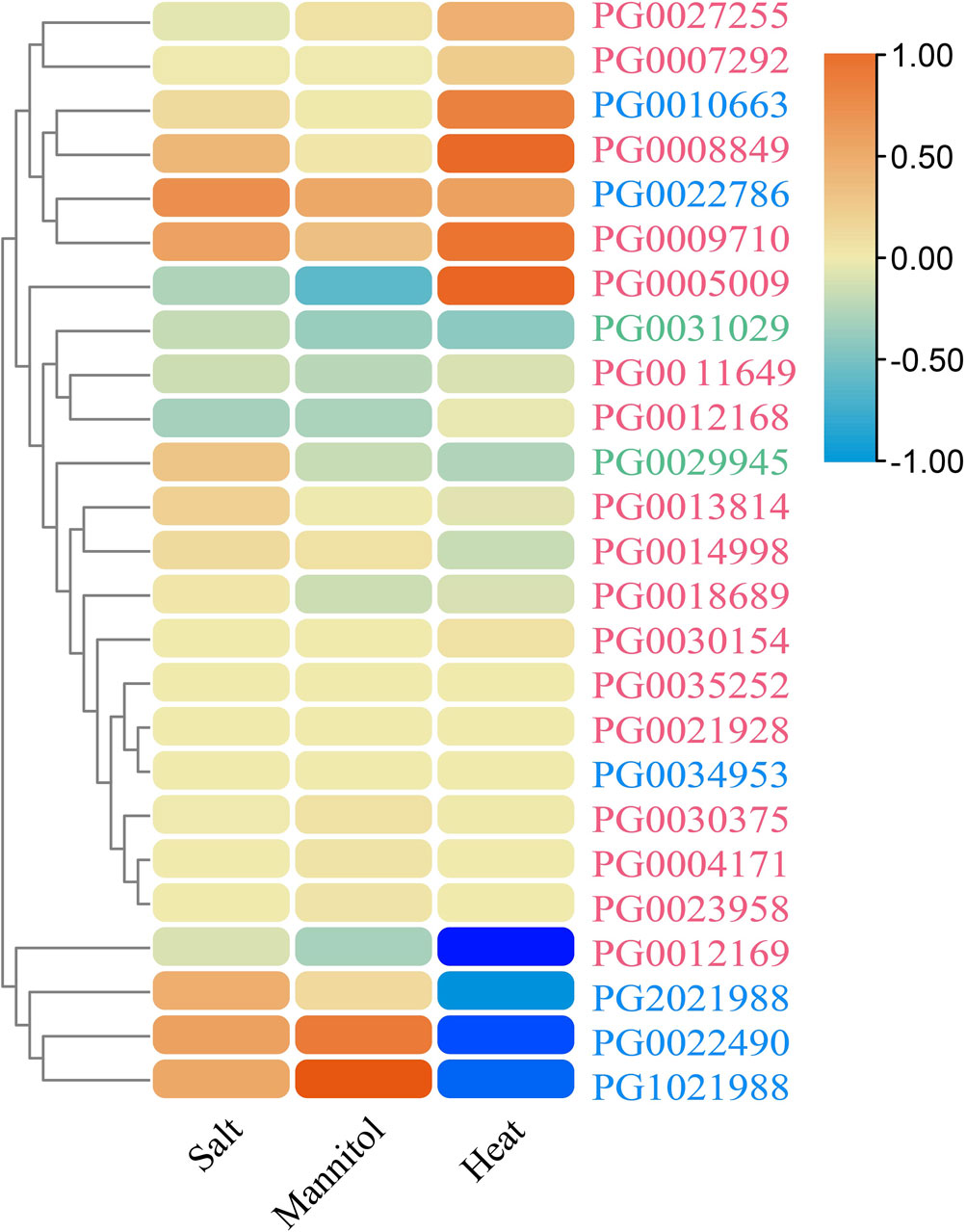
Figure 10. The expression profiles of StNHX genes under salt, drought, and heat stress based on transcriptomic data. The relative expression profiles of StNHX genes under NaCl, mannitol, and heat stress in DM potato. The log2 mean of each gene FPKM+1 was used to draw a color scale. Pink: Class I. Green: Class II. Blue: Class III.
Environmental stress can cause osmotic stress in plants, which can negatively impact both plant growth and yield. In order to further investigate the expression patterns of these NHX family genes in shoots and roots under drought stress, RT-qPCR analysis was performed (Figure 11). Several StNHX genes were found to be regulated by drought stress, exhibiting increased differential expression in response to drought treatment. Several StNHX genes (PG0018689, PG0031029, PG0009710) were significantly downregulated by short-term drought treatment, compared to control. Others were upregulated and peaked at 24 h, including PG000417, PG0022490, PG0007292, PG0014998, and PG0012169, exhibiting a delated response to drought stress. Certain StNHX genes (PG0030375, PG0005009, and PG2021988) were simultaneously upregulated after 6 h of drought stress. Others were induced after 12 h or 24 h, including PG1021988, PG0011649, PG0029945, and PG0021928, suggesting that some genes were actively responsive to drought stress and might be more sensitive to drought. PG0022786 and PG0010663 exhibited similar expression patterns in shoots and roots, with relatively high expression levels only at 6 h. Some genes (PG000971, PG0031029, and PG0018689) were inhibited under drought stress and were downregulated or not expressed. Some StNHX genes (PG0035252 and PG0027255) were upregulated in root tissue, while other genes (PG0008849 and PG0034953) were highly upregulated in leaves. The expression of PG0021928 and PG0029945 gradually increased from 2 h to 24 h in roots, exhibiting a late response to drought stress. These results demonstrate that StNHX genes play crucial roles in drought stress and exhibit different responses in various tissue, and may be involved in the early basal resistance of potato under drought stress.
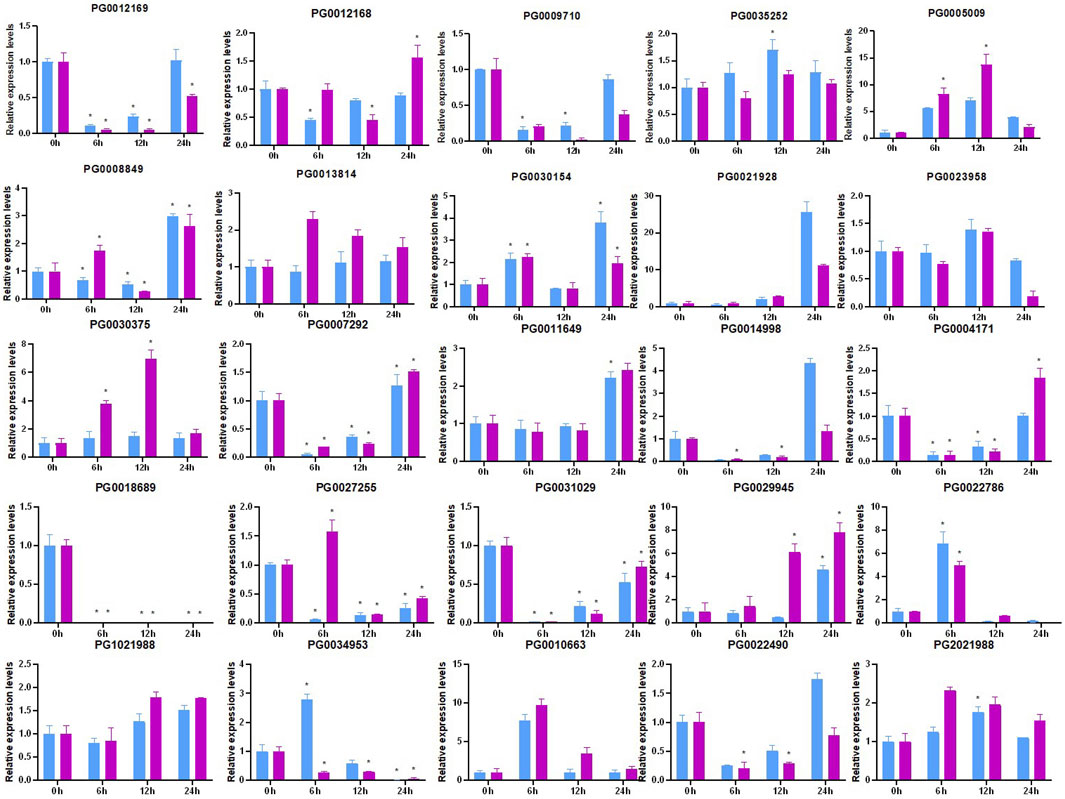
Figure 11. Expression analysis of StNHX genes in drought-stressed‘Jizhangshu NO.8.‘potatoes.Blue column: shoots. Purple column: roots. Expression levels of StNHX genes under dehydration treatment (15% PEG6000) for 0, 6, 12, and 24 h. The x-axis indicates the examined time points after the drought treatment. The y-axis indicates the relative expression levels. Relative expression values are presented as means ± standard errors based on three biological replicates with three technical replicates. Significant differential expression is indicated by an asterisk (*, p < 0.05).
In this study, 25 potato NHX genes were identified, which is a considerably larger number than has been found in other plant species. For example, six members have been in spinach (Li et al., 2021) and sweet orange (Yang et al., 2022), seven in peach (Li et al., 2021) and pepper (Luo et al., 2021), eight in grape (Lu et al., 2019) and A. thaliana (Gaxiola et al., 1999), 11 in banana (Xu et al., 2021) and 16 in P. betulaefolia (Wang et al., 2019). Thus, the potato NHX gene family appears to have expanded substantially during evolution.
The physical and chemical properties, gene structure, conserved motif composition, chromosomal localization, and promoter cis-acting elements of potato NHX gene members were further analyzed. The isoelectric points of StNHX proteins ranged from 4.96 to 9.30, and most proteins were alkaline, which was consistent with the results of previous studies in pepper (Luo et al., 2021) and banana (Xu et al., 2021). It is speculated that StNHXs mainly play a role in alkaline subcellular environments.
The 25 StNHX genes were found to have different number of introns, with the number of introns ranging from 1 to 22 and the number of amino acids ranging from 252 to 1,153. The structural differences were relatively large, which may determine the functional diversity of the NHX genes (Skolnick and Fetrow, 2000). The cluster analysis indicated that the gene structure of closely related members was similar. By analyzing the phylogenetic relationship between A. thaliana and potato, it was found that there were two clusters in total. Class III was in the same clade as the members from A. thaliana, with high homology. It is speculated that the NHX genes in potato and A. thaliana have similar functions. In addition, StNHX genes with similar motif composition tended to cluster together, suggesting that NHX genes with similar evolutionary patterns have similar protein structure and function.
Gene duplication is a common phenomenon in plants (Wendel, 2000; Kondrashov et al., 2003; Conant and Wolfe, 2008). The evolution of duplicate genes and their subsequent differentiation provide the original genetic resources for adaptive evolution, while also contributing to the development of new genes (Ramsey and Schemske, 1998; Flagel and Wendel, 2009). We found that there were no large fragment duplication events in the StNHX gene family, although tandem repetition was detected. A total of one tandem repeating gene pair was identified (PG0012168:PG0012169), which likely contributed to the expansion of this family.
The StNHX promoter regions contained a variety of stress response elements. However, the number and types of elements were different among different genes, indicating that the types and degrees of StNHX response to stress are different, and thus the function of the StNHX genes is different. The identified cis-acting elements present in the potato NHX gene family may play roles in light response, hormone induction, growth and development, and stress response, among other roles. The functions of some of these elements have been confirmed in other species (Yokoi et al., 2002; Adler et al., 2010; Almeida et al., 2017), but the role these elements play in potato remains unknown. Some StNHX genes were found to contain multiple hormone response elements, demonstrating that StNHX proteins may act as intermediate transcription factors to respond to hormonal signals and regulate plant growth under stress. Previous analyses of the cis-acting elements of NHX gene promoter sequences in chili pepper and sweet orange (Li et al., 2021; Luo et al., 2021) were similar to the results of this study. In this study, PPI network analysis also indicated that NHX proteins exhibited has strong or weak interaction, indicating that StNHXs may interact with these proteins and participate in the regulation of potato growth and stress.
Analysis of StNHX gene expression in different tissues indicated that the expression of this gene family was different in different tissues. We speculate that StNHX genes may play regulatory roles in the development of potato flowers, leaves, and fruits, although the specific functional mechanism needs to be verified in subsequent experiments. Tissue-specific gene expression patterns usually reflect their corresponding biological functions. Previous studies postulated that the NHX genes play an important role in plant development and stress response (Wu et al., 2019). Based on the publicly-available RNA-Seq data, StNHX genes exhibited differential expression in the different tissues, suggesting that they may participate in growth and development. In this study, we found that StNHX gene expression was tissue-specific. Therefore, we speculate that StNHX genes may regulate potato development and play a role in the growth and development of different potato tissues.
The StNHX genes play essential roles in the regulation of gene expression to cope with environmental stress (Kang et al., 2022). We found that the StNHX genes were differentially expressed under NaCl, mannitol, and heat stress. Several StNHX genes appeared to take part in the response to only one stress, suggesting that there are different signaling pathways related to the response to abiotic stress. In addition, some genes exhibited opposite expression profiles under different stresses, implying the presence of a complex abiotic stress signal transduction pathway (Li et al., 2020). Previous studies confirmed that the expression of NHX genes could be induced by drought treatments, and that these genes have key functions related to the drought tolerance of various plant species. For example, under 10% PEG stress, the expression of NHX genes in grape leaves was significantly increased to five times that of control (Lu et al., 2019). The expression levels of maize NHX genes increased in leaves and roots under drought and salt stress. Meanwhile, functional identification showed that maize NHX genes play an important role in drought and salt tolerance (Dai et al., 2003). In sweet potato, the expression level of NHX2 was positively correlated with the drought tolerance of transgenic plants (Wang et al., 2016).
In order to further investigate the expression patterns of these NHX family genes in potato shoots and roots under drought treatment, RT-qPCR analysis was performed. We found that several StNHX genes are regulated by drought stress. Most of the studied StNHX genes responding to PEG treatment, with many of these upregulated, although the degree of induction was different. These results indicate that all StNHX genes are involved in the response to drought stress in potato, but different treatment times may have different effects on the expression of these genes. Similarly, shoot- and root-level gene expression was also significantly induced by PEG. However, the regulation pattern was different between the leaves and roots, with some genes upregulated in leaves but downregulated in roots. Such differential expression was attributed to the different ways roots and leaves respond to drought stress. Hence, the specific mechanism should be studied further. Similarly, we observed that the expression of some genes, such as PG0004171, was unaffected in the early stage of drought stress, suggesting these genes may not contribute substantially to the physiological responses associated with drought tolerance.
In summary, a total of 25 NHX genes were identified in potato in this study. The NHX genes were analyzed in depth, including molecular characterization, chromosomal distributions, phylogenetic classification, gene structures, protein domain compositions, conserved motifs, as well as cis-regulatory elements. The StNHX genes exhibited diverse exon/intron structures, motif distributions, and phylogenetic relationships, all of which potentially contributed to their diverse functions. Protein-protein interactions network analysis indicated that these NHX genes had strong or weak interactions, indicating that the StNHXs may interact to be involved in stress responses and potato development. Based on RNA-seq data, NHX genes were found to exhibit distinct tissue specific expression patterns. Additionally, the StNHX genes responded differently to various abiotic stresses, including drought treatment. The qRT-PCR results further confirmed that the StNHX genes are involved in the response to drought stress and showed their importance in potato plant growth and development. These studies and analyses provided an overview information regarding the NHX genes in potato and determined the cadidate NHX genes that regulting drought potato for further investigation.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
JY: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. LZ: Data curation, Writing–review and editing. LC: Investigation, Writing–review and editing. SZ: Investigation, Writing–review and editing. ZN: Investigation, Writing–review and editing. SM: Investigation, Writing–review and editing. LY: Funding acquisition, Investigation, Writing–original draft, Writing–review and editing. WL: Funding acquisition, Investigation, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Hebei North University Project (No. XJ2023020), the Hebei Science and Education Department Project (No. QN2021011), China Agriculture Research System (No. CARS-09-P05), and the Hebei Province Potato Industry Collaborative Innovation Center platform (No. [2016]5).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1396375/full#supplementary-material
Adler, G., Blumwald, E., and Bar-Zvi, D. (2010). The sugar beet gene encoding the sodium/proton exchanger 1 (BvNHX1) is regulated by a MYB transcription factor. Planta 232 (1), 187–195. doi:10.1007/s00425-010-1160-7
Akram, U., Song, Y., Liang, C., Abid, M. A., Askari, M., Myat, A. A., et al. (2020). Genome-wide characterization and expression analysis of NHX gene family under salinity stress in Gossypium barbadense and its comparison with Gossypium hirsutum. Genes (Basel) 11 (7), 803. doi:10.3390/genes11070803
Almeida, D. M., Margarida, O. M., and Saibo, N. (2017). Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 40 (1), 326–345. doi:10.1590/1678-4685-GMB-2016-0106
An, R., Chen, Q. J., Chai, M. F., Lu, P. L., Su, Z., Qin, Z. X., et al. (2007). AtNHX8, a member of the monovalent cation: proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li+/H+ antiporter. Plant J. 49 (4), 718–728. doi:10.1111/j.1365-313X.2006.02990.x
Apse, M. P., Aharon, G. S., Snedden, W. A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 (5431), 1256–1258. doi:10.1126/science.285.5431.1256
Atsunori, F., Atsuko, N., Akemi, T., Hiroshi, T., Akio, M., Hirohiko, H., et al. (2004). Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiology 45 (2), 146–159. doi:10.1093/pcp/pch014
Ayadi, M., Martins, V., Ayed, R. B., Jbir, R., Feki, M., Mzid, R., et al. (2020). Genome wide identification, molecular characterization, and gene expression analyses of grapevine NHX antiporters suggest their involvement in growth, ripening, seed dormancy, and stress response. Biochem. Genet. (1), 58. doi:10.1007/s10528-019-09930-4
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 (Web Server), W202–W208. doi:10.1093/nar/gkp335
Bassil, E., Ohto, M. A., Esumi, T., Tajima, H., Zhu, Z., Cagnac, O., et al. (2011). The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant. Cell 23 (1), 224–239. doi:10.1105/tpc.110.079426
Bjellqvist, B., Hughes, G. J., Pasquali, C., Paquet, N., Ravier, F., Sanchez, J. C., et al. (1993). The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14, 1023–1031. doi:10.1002/elps.11501401163
Blumwald, E. (2000). Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 12, 431–434. doi:10.1016/s0955-0674(00)00112-5
Blumwald, E., Aharon, G. S., and Apse, M. P. (2000). Sodium transport in plant cells. Biochim. Biophys. Acta, Biomembr. 1465, 140–151. doi:10.1016/s0005-2736(00)00135-8
Brini, F., Gaxiola, R. A., Berkowitz, G. A., and Masmoudi, K. (2005). Cloning and characterization of a wheat vacuolar cation/proton antiporter and pyrophosphatase proton pump. Plant Physiol. Biochem. 43 (4), 347–354. doi:10.1016/j.plaphy.2005.02.010
Cavusoglu, E., Sari, U., and Tiryaki, I. (2023). Genome-wide identification and expression analysis of Na+/H+antiporter (NHX) genes in tomato under salt stress. Plant Direct 7 (11), e543. doi:10.1002/pld3.543
Chen, C., Xia, R., Chen, H., and He, Y. (2018). TBtools, a toolkit for biologists integrating various biological HTS-data handling tools with a user-friendly interface. BioRxiv 6, 289660. doi:10.1101/289660
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job:How duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi:10.1038/nrg2482
Dai, Y., Lu, Y., Chen, Y., and Gao, Y. (2003). Cloning and expression analysis of maize MYB family transcription factor genes. J. Yangzhou Univ. Life Sicence Ed. 34 (1), 60–63. doi:10.16872/j.cnki.1671-4652.2013.01.014
Elias, B., Coku, A., and Blumwald, P. (2012). Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 63 (16), 5727–5740. doi:10.1093/jxb/ers250
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi:10.1093/nar/gkr367
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 (D1), D279–D285. doi:10.1093/nar/gkv1344
Flagel, L. E., and Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi:10.1111/j.1469-8137.2009.02923.x
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). Protein identification and analysis tools on the ExPASy server. Proteomics Protoc. Handb. 53, 571–607. doi:10.1385/1-59259-890-0:571
Gaxiola, R. A., Rao, R., Sherman, A., Grisafi, P., Alper, S. L., and Fink, G. R. (1999). The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. U. S. A. 96 (4), 1480–1485. doi:10.1073/pnas.96.4.1480
Gervais, T., Creelman, A., Li, X. Q., Bizimungu, B., De Koeyer, D., and Dahal, K. (2021). Potato response to drought stress: physiological and growth basis. Front. Plant Sci. 12, 698060. doi:10.3389/fpls.2021.698060
Guo, H. M., Gu, C. S., Chen, F. D., Xu, Y. C., and Liu, Z. L. (2012). Salt tolerance verification of Lotus NnNHX1 in transformed tobacco. Acta Hortic. Sin. 39 (2), 323–332. doi:10.16420/j.issn.0513-353x.2012.02.005
Hill, D., Nelson, D., Hammond, J., and Bell, L. (2021). Morphophysiology of potato (Solanum tuberosum) in response to drought stress: paving the way forward. Front. Plant Sci. 11, 597554. doi:10.3389/fpls.2020.597554
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., and Gao, G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi:10.1093/bioinformatics/btu817
Huang, Y., Zhang, X. X., Li, Y. H., Ding, J. Z., Du, H. M., Zhao, Z., et al. (2018). Overexpression of the Suaeda salsa SsNHX1 gene confers enhanced salt and drought tolerance to transgenic Zea mays. J. Integr. Agric. 17 (12), 2612–2623. doi:10.1016/s2095-3119(18)61998-7
Kang, H. X., Wu, G. Q., Wei, M., and Li, S. J. (2022). The role of Na+/H+ antiporter in response of plant to abiotic stress. Plant Physiology Journal 58 (O3), 511–523. doi:10.13592/j.cnki.ppj.2021.0224
Khare, T., Joshi, S., Kaur, K., Srivastav, A., and Kumar, V. (2021). Genome-wide in silico identification and characterization of sodium-proton (Na+/H+) antiporters in Indica rice. Plant Gene 26, 100280. doi:10.1016/j.plgene.2021.100280
Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I., and Koonin, E. V. (2003). Selection in the evolution of gene duplications. Genome Biol. 3, RESEARCH0008. doi:10.1186/gb-2002-3-2-research0008
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Peer, Y. V. D., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Rese 30, 325–327. doi:10.1093/nar/30.1.325
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in. Nucleic Acids Res. 43 (D1), D257–D260. doi:10.1093/nar/gku949
Li, H. T., Liu, H., Gao, X. S., and Zhang, H. (2009). Knock-out of Arabidopsis AtNHX4 gene enhances tolerance to salt stress. Biochem. Biophysical Res. Commun. 382 (3), 637–641. doi:10.1016/j.bbrc.2009.03.091
Li, M., Li, G. X., Liu, W., Gao, X. L., Wang, X. Y., and Zhang, A. N. (2021). Genome-wide identification and expression analysis of NHX gene family in peach. Shandong Agric. Sci. 53 (12), 8–16. doi:10.14083/j.issn.1001-4942.2021.12.002
Li, R., Gu, Y. Y., Li, M., Chen, C. J., Liang, X. Y., and Li, J. L. (2021). Identification of NHX family genes and its expression analysis of response to salt stress in spinach. Shandong Agric. Sci. 53 (9), 1–7. doi:10.14083/j.issn.1001-4942.2021.09.001
Li, Y., Zhang, Y., Feng, F., Dong, L., Cheng, L., Ma, F., et al. (2010). Overexpression of a Malus vacuolar Na+/H+ antiporter gene (MdNHX1) in apple rootstock M.26 and its influence on salt tolerance. Plant Cell Tissue and Organ Cult. 102 (3), 337–345. doi:10.1117/1.601420
Li, Y. M., Wang, K. L., Liu, Z., Allan, A. C., Qin, S. H., Zhang, J. L., et al. (2020). Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 148, 817–832. doi:10.1016/j.ijbiomac.2020.01.167
Liu, H., Tang, R. J., Zhang, Y., Wang, C., Qundan, L. V., Gao, X., et al. (2010). AtNHX3 is a vacuolar K+/H+ antiporter required for low-potassium tolerance in Arabidopsis thaliana. Plant Cell Environ. 33 (11), 1989–1999. doi:10.1111/j.1365-3040.2010.02200.x
Liu, W., Li, H., Lin, J., Yang, Q. S., and Chang, Y. H. (2018). Cloning, expression and functional analysis of PbNHX1 gene in Pyrus betulaefolia. J. Fruit Sci. 35 (2), 137–146. doi:10.13925/j.cnki.gsxb.20170302
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Lu, S. X., Xu, C. M., He, H. H., Liang, G. P., Wang, P., and Chen, B. H. (2019). Identification and expression analysis of NHX genes family in grape. J. Fruit Sci. 36 (3), 266–276. doi:10.13925/j.cnki.gsxb.20180361
Luo, J., Xu, C. M., Zhang, G. B., and Yu, J. H. (2021). Bioinformation analysis and expression analysis of NHX genes family in pepper. Acta Agric. Boreali-Sinica 36 (3), 15–240.
Mehta, G., Muthusamy, S. K., Singh, G. P., and Sharma, P. (2021). Identification and development of novel salt-responsive candidate gene based SSRs (cg-SSRs) and MIR gene based SSRs (mir-SSRs) in bread wheat (Triticum aestivum). Sci. Rep. 11, 2210. doi:10.1038/s41598-021-81698-3
Niu, N., Liu, Z., Huang, P. X., Zhu, J. Y., Li, Z. T., Ma, W. J., et al. (2021). Genome-wide identification and expression analysis of potato GAUT gene family. ACTA AGRON. SIN. 47 (12), 2348–2361. doi:10.3724/SP.J.1006.2021.04268
Orlowski, J., and Grinstein, S. (2007). Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Opin. Cell Biol. 19 (4), 483–492. doi:10.1016/j.ceb.2007.06.001
Pardo, E. M., Toum, L., Pérez-Borroto, L. S., Fleitas, L., Gallino, J. P., Machi, S., et al. (2021). Ectopic expression of GmNHX3 and GmNHX1, encoding two Glycine max Na+/H+ vacuolar antiporters, improves water deficit tolerance in Arabidopsis thaliana. Biol. Plant. 65, 157–166. doi:10.32615/BP.2021.003
Pardo, J. M., Cubero, B., Leidi, E. O., and Quintero, F. J. (2006). Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 57 (5), 1181–1199. doi:10.1093/jxb/erj114
Priest, H. D., Filichkin, S. A., and Mockler, T. C. (2009). cis-Regulatory elements in plant cell signaling. Curr. Opin. Plant Biol. 12 (5), 643–649. doi:10.1016/j.pbi.2009.07.016
Ramsey, J., and Schemske, D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501. doi:10.1146/annurev.ecolsys.29.1.467
Rausch, T., Kirsch, M., Löw, R., Lehr, A., Viereck, R., and Zhigang, A. (1996). Salt stress responses of higher plants: the role of proton pumps and Na+/H+ antiporters. Plant. Physiol. 148, 425–433. doi:10.1016/S0176-1617(96)80275-6
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Sharma, P., Mishra, S., Pandey, B., and Singh, G. (2023). Genome-wide identification and expression analysis of the NHX gene family under salt stress in wheat (Triticum aestivum L). Front. Plant Sci. 14, 1266699. doi:10.3389/fpls.2023.1266699
Skolnick, J., and Fetrow, J. S. (2000). From genes to protein structure and function: novel applications of computational approaches in the genomicera. Trends Biotechnol. 18 (1), 34–39. doi:10.1016/S0167-7799(99)01398-0
Spooner, D. M., Karen, M. L., Gavin, R., Robbie, W., and Bryan, G. J. (2005). A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc. Natl. Acad. Sci. U. S. A. 102 (41), 14694–14699. doi:10.1073/pnas.0507400102
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi:10.1093/nar/gkw937
Tang, X., Zhang, N., Si, H., and Calderónn-Urrea, A. (2017). Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods 13 (1), 85. doi:10.1186/s13007-017-0238-7
Visser, R. G. F., Bachem, C. W. B., Boer, J. M., Bryan, G. J., Chakrabati, S. K., Feingold, S. R., et al. (2009). Sequencing the potato genome: outline and first results to come from the elucidation of the sequence of the world’s third most important food crop. Am. J. Potato Res. 86, 417–429. doi:10.1007/s12230-009-9097-8
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi:10.1093/jhered/93.1.77
Wang, B., Zhai, H., He, S. Z., Zhang, H., Ren, Z., Zhang, D., et al. (2016). A vacuolar Na+/H+ antiporter gene, IbNHX2, enhances salt and drought tolerance in transgenic sweetpotato. Sci. Hortic. 201, 153–166. doi:10.1016/j.scienta.2016.01.027
Wang, J., Zuo, K., Wu, W., Song, J., Sun, X., Lin, J., et al. (2003). Molecular cloning and characterization of a new Na+/H+ antiporter gene from Brassica napus. DNA Seq. 14 (5), 351–358. doi:10.1080/10855660310001596211
Wang, L., Guo, K., Li, Y., Tu, Y., Peng, L., Wang, B., et al. (2010). Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10, 282. doi:10.1186/1471-2229-10-282
Wang, L. G., Feng, X. Y., Zhao, H., Wang, L., An, L., and Qiu, Q. (2014). Functional analysis of the Na+, K+/H+ antiporter PeNHX3 from the tree halophyte Populus euphratica in yeast by modelguided mutagenesis. PLoS ONE 9 (8), e104147. doi:10.1371/journal.pone.0104147
Wang, L. G., Wu, X. X., Liu, Y. F., Qiu, Q. S., and Van, V. (2015). AtNHX5 and AtNHX6 control cellular K+ and pH homeostasis in Arabidopsis: three conserved acidic residues are essential for K + transport. PLoS ONE 10 (12), e0144716. doi:10.1371/journal.pone.0144716
Wang, Y., Li, H., Lin, J., Yang, Q. S., Zhang, S. L., and Chang, Y. H. (2019). Identification of NHX gene family in Pyrus betulaefolia and its expression under abiotic, stress. J. Fruit Sci. 36 (7), 825–836. doi:10.13925/j.cnki.gsxb.20180347
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Wendel, J. F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42 (1), 225–249. doi:10.1023/A:1006392424384
Wu, C. A., Yang, G. D., Meng, Q. W., and Zhang, C. C. (2004). The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiology 45 (5), 600–607. doi:10.1093/pcp/pch071
Wu, G. Q., Feng, R. J., Wei, J. K., Bao, A. K., Wei, L., and Yuan, H. J. (2017). Co-expression of Zygophyllum xanthoxylum ZxNHX and ZxVP1-1 enhances osmotic stress tolerance in transgenic Beta vulgaris. Plant Physiology J. 53 (6), 1007–1014. doi:10.13592/j.cnki.ppj.2016.0234
Wu, G. Q., Wang, J. L., and Li, S. J. (2019). Genome-wide identification of Na+/H+ antiporter (NHX) genes in sugar beet (Beta vulgaris L.) and their regulated expression under salt stress. Genes 10 (5), 401. doi:10.3390/genes10050401
Wu, X. X., Ebine, K., Ueda, T., Qiu, Q. S., and Li, J. X. (2016). AtNHX5 and AtNHX6 are required for the subcellular localization of the SNARE complex that mediates the trafficking of seed storage proteins in Arabidopsis. PLos one 11 (3), e0151658. doi:10.1371/journal.pone.0151658
Xu, X., Pan, S., Cheng, S., Zhang, B., Mu, D., Ni, P., et al. (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi:10.1038/nature10158
Xu, Y., Teng, M. X., He, Y. D., Qiao, F., and Li, X. (2021). Identification and expression analysis of NHX genes family in banana. Plant Physiology J. 57 (3), 681–691. doi:10.13592/j.cnki.ppj.2020.0442
Yamaguchi, T., Apse, M. P., Shi, H., and Blumwald, E. (2003). Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc. Natl. Acad. Sci. 100 (21), 12510–12515. doi:10.1073/pnas.2034966100
Yamaguchi, T., Fukada-Tanaka, S., Inagaki, Y., Saito, N., Yonekura-Sakakibara, K., Tanaka, Y., et al. (2001). Genes encoding the vacuolar Na+/H+ exchanger and flower coloration. Plant. Cell Physiol. 42, 451–461. doi:10.1093/pcp/pce080
Yang, J., Chen, R., Hu, W. J., Wu, Q. L., Tong, X. N., Li, X. T., et al. (2022). Identification and functional analysis of NHX gene family in sweet orange. Jiangsu Agric. Sci. 50 (7), 35–42. doi:10.15889/j.issn.1002-1302.2022.07.005
Yang, S., Zhang, X., Yue, J. X., Tian, D., and Chen, J. Q. (2008). Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genomics 280, 187–198. doi:10.1007/s00438-008-0355-0
Yokoi, S., Quintero, F. J., Cubero, B., Ruiz, M. T., Pardo, J. M., Hasegawa, P. M., et al. (2002). Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 30 (5), 529–539. doi:10.1046/j.1365-313X.2002.01309.x
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinforma. 64 (3), 643–651. doi:10.1002/prot.21018
Yuan, H. J., Ma, Q., Wu, G. Q., Wang, P., Hu, J., and Wang, S. M. (2015). ZxNHX controls Na+ and K+ homeostasis at the whole-plant level in Zygophyllum xanthoxylum through feedback regulation of the expression of genes involved in their transport. Ann. Bot. 115 (3), 495–507doi. doi:10.1093/aob/mcu177
Yupeng, W., Haibao, T., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi:10.1093/nar/gkr1293
Zhang, H., Fen, X. U., Yu, W. U., Hong-Hai, H. U., and Dai, X. F. (2017). Progress of potato staple food research and industry development in China. J. Integr. Agric. 12, 2924–2932. doi:10.1016/s2095-3119(17)61736-2
Zheng, L. G., Andre, C., Feng, C. C., Peter, B., and Wen, H. L. (2020). Extent of gene duplication in the genomes of drosophila, nematode, and yeast. Mol. Biol. Evol. 19, 256–262. doi:10.1093/oxfordjournals.molbev.a004079
Keywords: potato, NHX gene family, bioinformatics analysis, expression profile, drought stress
Citation: Yihong J, Zhen L, Chang L, Ziying S, Ning Z, Meiqing S, Yuhui L and Lei W (2024) Genome-wide identification and drought stress-induced expression analysis of the NHX gene family in potato. Front. Genet. 15:1396375. doi: 10.3389/fgene.2024.1396375
Received: 07 March 2024; Accepted: 10 June 2024;
Published: 11 July 2024.
Edited by:
Ertugrul Filiz, Duzce University, TürkiyeReviewed by:
Pradeep Sharma, Indian Institute of Wheat and Barley Research (ICAR), IndiaCopyright © 2024 Yihong, Zhen, Chang, Ziying, Ning, Meiqing, Yuhui and Lei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liu Yuhui, bHlodWlAZ3NhdS5lZHUuY24=; Wang Lei, d2FuZ2xlaUBoZWJlaW51LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.