- 1Guizhou Institute of Biotechnology, Guizhou Academy of Agricultural Sciences, Guiyang, China
- 2Ministry of Agriculture and Rural Affairs Key Laboratory of Crop Genetic Resources and Germplasm Innovation in Karst Region, Guiyang, China
- 3Guizhou Key Laboratory of Agriculture Biotechnology, Guiyang, China
The CONSTANS-like (COL) gene plays important roles in plant growth, development, and abiotic stress. A total of 15 COL genes are unevenly distributed on eight chromosomes in the potato genome. The amino acid length of the family members was 347–453 aa, the molecular weight was 38.65–49.92 kD, and the isoelectric point was 5.13–6.09. The StCOL family can be divided into three subfamilies by evolutionary tree analysis, with conserved motifs and similar gene structure positions in each subfamily. The analysis of promoter cis-acting elements showed 17 cis-acting elements related to plant hormones, stress, and light response. Collinearity analysis of COL genes of tomato, potato, and Arabidopsis showed that 13 StCOL genes in the different species may have a common ancestor. A total of 10 conserved motifs and six kinds of post-translational modifications in the 15 StCOL proteins were identified. The 15 StCOL genes exhibit a genomic structure consisting of exons and introns, typically ranging from two to four in number. The results showed that 10 genes displayed significant expression across all potato tissues, while the remaining five genes were down-expressed in potato transcriptome data. The quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis exhibited differential expression of 8 StCOL genes in the potato leaves and tubers at different growth stages, as well as 7 StCOL genes under 2°C treatment conditions. These results suggested that the StCOL gene family may play an important role in regulating potato tuberization and responding to cold stress.
1 Introduction
COL is a zinc finger activator of transcription that contains two conserved elements in the N-terminal B-box domain and the C-terminal CCT domain (Imaizumi et al., 2005; Kikuchi et al., 2012). The CO (CONSTANS)/COLs can be divided into three major groups based on the divergence of conserved domains (Crocco and Botto, 2013). Group I members include two B-box domains and one CCT domain, while group II members possess one B-box domain and one CCT domain. Group III members contain one B-box and one diverged zinc finger structure (Li et al., 2020). COL genes have been reported in many plant species, such as Arabidopsis (Robson et al., 2001), rice (Griffiths et al., 2003), petunia (Khatun et al., 2021), and mango (Guo et al., 2022). COL genes, which are key genes in the photoperiodic pathway, play an important role in plant growth and development, regulating flowering, tuber formation, and abiotic stress (Turck et al., 2008; Kloosterman et al., 2013; Zhang et al., 2023).
Flowering is a vital growth transition period in plant growth and development and is one of the most important agronomic traits for crop yield (Steinbach, 2019). The promotion of flowering in Arabidopsis by CO is influenced by day length and serves as a central integrator in the photoperiodic flowering pathway (Samach et al., 2000). Previous studies have found that CO activates SOC1 through FT to promote flowering in Arabidopsis (Yoo et al., 2005). AtCOL1 and AtCOL2 synergically regulate the circadian rhythm of Arabidopsis (Ledger et al., 2001). AtCOL3 promotes lateral root development and aboveground branching, while AtCOL4 promotes tolerance to abiotic stress (Datta et al., 2006; Min et al., 2015). Overexpression of COL5 affects flowering time and the expression of FT and SOC1 in Arabidopsis (Hassidim et al., 2009). Overexpression of HvCO1 can accelerate the flowering time of wheat and mediate the upregulation of HvFT1 under long-day (LD) conditions (Campoli et al., 2012). The grape gene VvCO plays a role in regulating the seasonal cycle of grape flowering. VvCOL1 is mainly expressed in dormancy, and CO homologs of grapevine are also expressed in this unique organ (Almada et al., 2009). GmCOL1a and GmCOL1b function as flowering repressors in the photoperiodic flowering of soybean under long-day conditions (Cao et al., 2015). The overexpression of mango MiCOL2A and MiCOL2B significantly delayed flowering time in Arabidopsis under LD and SD conditions (Liang et al., 2023).
Cold stress is a major abiotic stress that adversely affects plant growth and crop productivity. Many studies have reported that COL genes play important roles in cold stress response (Mikkelsen and Thomashow, 2009; Jung et al., 2012; Huang et al., 2022). AtCOL1 and AtCOR27 are rapidly induced under cold stress by a CBF-independent pathway in Arabidopsis (Mikkelsen and Thomashow, 2009). HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 1 (HOS1) mediated CO degradation via a ubiquitin/proteasome pathway under cold stress (Jung et al., 2012). The CO transcript levels were increased at night by cooler temperature treatments (Kinmonth-Schultz et al., 2016). Five COL genes (PaCOL2, PaCOL6, PaCOL8, PaCOL10, and PaCOL13) were induced in response to low temperature in petunia (Khatun et al., 2021). A total of 10 CaCOL genes were identified in the pepper genome, five of which were significantly expressed under cold treatment (Huang et al., 2022).
Potato (Solanum tuberosum L.) is the world’s fourth largest food crop and is cultivated worldwide. Many studies have reported that StCO genes play an important role in potato tuber formation and flowering (González-Schain et al., 2012; Kloosterman et al., 2013; Abelenda et al., 2016). Potato StCO affects flowering and stem elongation and the autonomous pathway of potato tuber through different mechanisms (González-Schain and Suárez-López, 2008). Previous studies indicated that StCO was involved in the photoperiodic regulation of tuberization (González-Schain et al., 2012). It was found that StCDF1 represses the transcription of StCO1/2 under LD conditions in potato (Kloosterman et al., 2013). The silence of StCOL1 was strongly associated with the downregulated expression of StSP5G in potato (Abelenda et al., 2016). The overexpression of Nelumbo nucifera COL5 in potato increased the tuber weight and starch content under SD conditions (Cao et al., 2021). StCDF1 is a non-redundant regulator of tuberization and binds to the promoter of StCO1, StCO2, and StCO3 genes in potato (Ramírez and Gonzales, 2021). However, the molecular mechanism of StCOL genes in controlling tuber formation and clod stress response in potato remains unclear.
In this study, the conserved domains, evolutionary relationships, chromosome location, promoter element, and collinearity analysis of StCOL genes were systematically identified. The differential expression patterns of StCOL genes in different tissues and development stages were determined by potato transcriptome data and quantitative reverse transcription polymerase chain reaction (qRT-PCR). The expression of StCOL genes at different treatment times under cold stress was also determined by qRT-PCR assays. The results will provide an important theoretical basis for exploring the molecular mechanism of potato tuberization and cold stress response.
2 Materials and methods
2.1 Identification of COL gene family members in potato
The conserved domain of COL proteins was downloaded from the Pfam database (http://pfam.xfam.org/family/PF06203 and PF00643) and was used as the template region to search StCOL sequences in the potato genome database (http://spuddb.uga.edu/). Hidden Markov model (HMM) software was used to select candidate StCOL genes, which were then submitted to NCBI (https://www.ncbi.nlm.nih.gov/) and UniProt (http://www.uniprot.org) for confirming the final StCOL gene family members.
2.2 Protein sequence and phylogenetic analysis of COLs
The online website cluster (https://pir.georgetown.edu/pirwww/search/multialn.shtml) was used to perform multiple alignments of the predicted protein sequences of the 15 StCOLs in potato, 17 AtCOLs in Arabidopsis, and 13 SlCOLs in tomato (Zobell et al., 2005). The unrooted phylogenetic tree was constructed using the MEGA 7 software with 1000 bootstrap replicates (Hall, 2013). The StCOLs were generated according to the phylogenetic relationships of 17 AtCOL genes with the StCOL gene and a specific gene nomenclature system.
2.3 Sequence analysis of COL genes in potato
The MEME (http://meme.sdsc.edu/meme/cgi-bin/meme.cgi) online software was used to assess the conservative area of the StCOL gene family protein sequence. The maximum ordinal number was set to 10, and the optimum motif width was 6–50 amino acid residues (Bailey et al., 2009). The gene structures of StCOLs were determined to obtain gene exons and containing substructures by Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). In addition, the cis-acting elements of StCOL promoter regions in potato were predicted by PlantCARE (PlantCARE/HTML/http://bioinformatics.psb.ugent.be/webtools/).
2.4 Synteny analysis and chromosomal localization of COLs
Collinearity analysis was conducted through PlantDGD software (http://pdgd.njau.edu.cn:8080/) (Qiao et al., 2019). The Multiple Collinearity Scan toolkit (MCScanX) was used to identify StCOL gene sequence repetition events. BLASTP was used to identify the species, symbiotic homologous pairs of the protein sequence. The protein sequence parameter was set to 1) alignment significance: e-value (default: 1 × 10−5), 2) MATCH_SCORE: final score (default: 50) (Wang et al., 2012). The obtained data were plotted by Circos online software (http://circos.ca/). Then, MapInspect software was used to locate COL genes whose relative positions have been identified in the potato genome database on potato chromosomes.
2.5 StCOL protein characterization
ExPASy online software (http://web.expasy.org/protparam/) was used to predict and analyze the amino acid number, theoretical molecular number, isoelectric point, and other physicochemical properties of the potato COL protein.
2.6 Expression analysis of StCOL genes
The publicly available RNA-seq data in the potato genome (http://spuddb.uga.edu/) were downloaded. The RNA-seq data of the root, tuber pith, tuber cortex, young tuber, mature tuber, shoot apex, tuber sprout, tuber peel, stamen, flower, petiole, leaf, stem, and stolon were selected to analyze the expression of StCOL genes. The “Normalized” function was used to normalize gene expression and construct the heat maps of StCOL gene expression using TBtools (Chen et al., 2020). The expression data are presented in Supplementary Table S1.
2.7 Total RNA extraction and qRT-PCR analysis
The leaves and tuber of the early maturing potato variety “Favorita” at three different growth stages (20, 42, and 65 days) were collected. The low-temperature sensitive variety “Desiree” was subjected to 2°C for 0 h, 1 h, 2 h, 24 h, and 48 h, and the leaves of all treatments were collected. The collected samples were stored in a −80°C refrigerator. The total RNA of all collected samples was isolated by a TaKaRa MiNiBEST Universal RNA Extraction Kit (TaKaRa, Beijing, China). Reverse transcription was performed with StarScript II RT Mix with gDNA Remover (GeneStar, Beijing, China). The PCR amplification was conducted on a CFX96 touch real-time PCR detection system (Bio-Rad, Hercules, CA, United States). β-actin was used as an internal reference gene. The Ct (2−ΔΔCT) method was used to calculate the relative expression of the StCOL genes (Livak and Schmittgen, 2001). The experiment was performed with three biological replicates. The primer sequences for qRT-PCR are listed in Supplementary Table S2. Duncan’s test with p < 0.05 was used to indicate significant differences.
3 Results
3.1 Identification of COL genes and multiple alignment analysis
A total of 15 potato COL genes were identified in the potato genome database. The genes were named StCOL1–StCOL15. The amino acid length of StCOL gene family members ranged from 347 to 453 aa, the molecular weight was 38.65–49.92 kD, and the isoelectric point was 5.13–6.09 (Supplementary Table S3). The isoelectric point of all 15 StCOL proteins was less than 7, the instability index was 6.94–59.96, and the aliphatic index ranged from 57.18 to 71.48. The grand average of hydropathicity ranged from −0.867 to −0.331, all of which were less than 0, indicating that StCOL proteins were hydrophilic.
3.2 Phylogenetic analysis of the COLs
In order to understand the evolutionary relationships among the StCOL gene family, Arabidopsis, and tomato, 15 StCOLs, 17 AtCOLs, and 13 SlCOLs were constructed by the phylogenetic tree (Figure 1). The StCOL protein family can be divided into three subfamilies: Group Ⅰ, Group Ⅱ, and Group Ⅲ. Group Ⅰ contained five StCOLs, six AtCOLs, and four SlCOLs. Group Ⅱ consists of three StCOLs, four AtCOLs, and three SlCOLs. Group III consists of seven StCOLs, seven AtCOLs, and five SlCOLs.
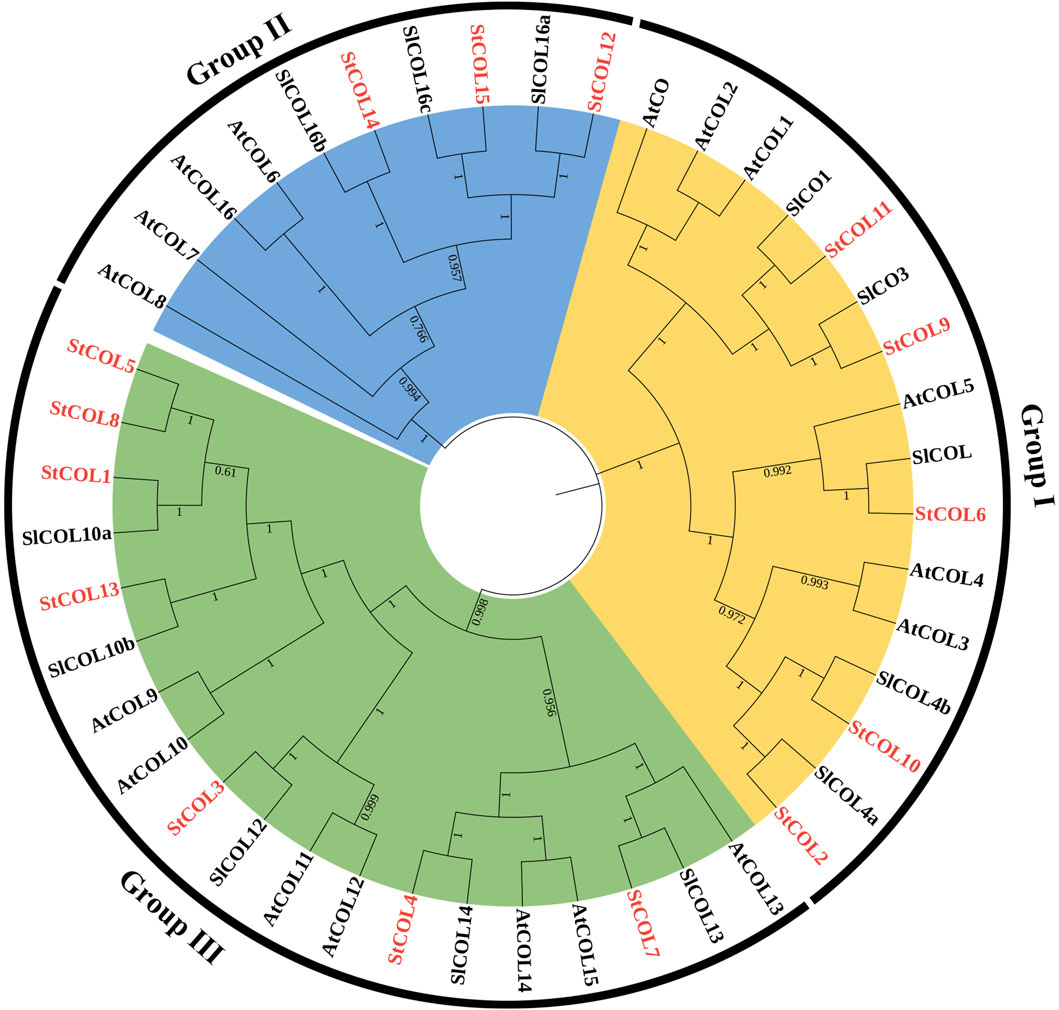
Figure 1. Neighbor-joining phylogenetic tree of COL proteins from S. tuberosum (St), S. lycopersicum (Sl), and A. thaliana (At). The classification of different groups is displayed with different colors.
3.3 Gene structure and conserved motif analysis of StCOLs
A total of 10 conserved motifs were obtained by predicting the StCOL protein conserved motifs through the MEME website (Figure 2A). All 15 StCOL proteins contain motif 1 (CCT domain) and motif 2 (zinc finger B-box domain). The five StCOLs in group I all contain motif 1, motif 2, and motif 3. There is a motif 9 in StCOL9 and StCOL11, but the location is slightly different. In group Ⅱ, StCOL12, StCOL14, and StCOL15 all contain motif 1, motif 2, and motif 7. In group ⅠⅡ, all seven StCOL proteins contained motif 1, motif 2, motif 3, and motif 6, of which four StCOL proteins contained all eight motifs. In order to further understand the structural and conserved characteristics of the StCOL gene family, exons and introns of the StCOL gene family were analyzed. The 15 StCOL genes all contain between two and four exons and introns. The number of introns and exons of genes in group III and group Ⅱ was the same. The number of exons and introns varies in the different groups, which indicates that the functions of the different genes may be specific.
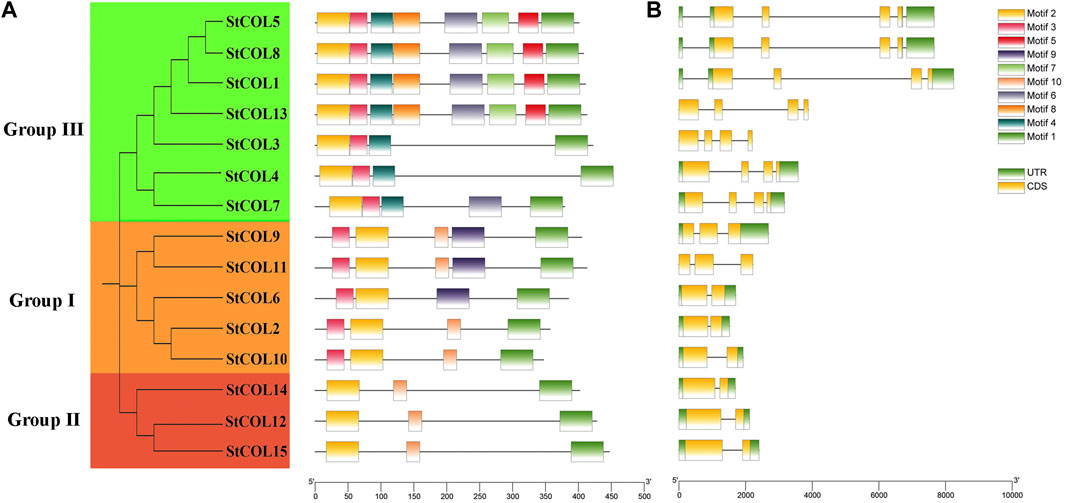
Figure 2. Gene structures and protein motifs of StCOL family genes. (A) The distribution of conserved motifs for StCOL genes. Different motifs and their relative positions are represented by the colored boxes. (B) Exon/intron structures of StCOL genes. Green boxes and yellow boxes represent coding sequence (CDS) and untranslated region (UTR), respectively.
3.4 Promoter sequence analysis
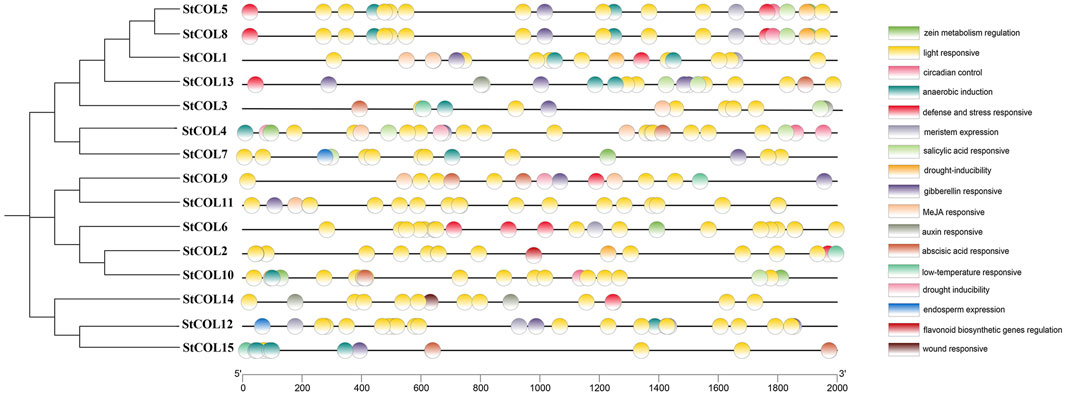
Seventeen cis-acting elements related to stress response, growth and development, and hormone regulation were predicted in the potato COL gene family (Figure 3). Among them were five cis-acting elements related to growth and development, six cis-acting elements related to stress response, and six cis-acting elements related to hormone regulation response. Twelve genes contained response elements necessary for anerobic induction, 10 genes contained abscisic acid response elements, 11 genes contained gibberellin response elements, four genes contained cis-acting elements related to low-temperature stress response, and 15 StCOL contained light response elements. Most of the growth-related response elements are photoregulatory response elements. These results indicate that the COL gene may play an important role in anerobic induction, abscisic acid response, gibberellin response, low-temperature stress response, and light response conditions.
3.5 Prediction of protein sequence features
In order to understand the physicochemical properties of COL proteins in potato, the post-translational modifications were performed at ScanProsite (http://ca.expasy.org/tools/scanprosite/). It mainly includes six kinds of post-translational modifications in the 15 StCOL proteins, including phosphorylation sites protein kinase C (PKC), casein kinase II (CK II), CAMP-CGMP-kinase (cAMP-cGMP), tyrosine kinase (Tyr), N-myristoylation (N-Myr), and N-glycosylation (N-Glyc) (Supplementary Table S4). Each StCOL protein contains at least three and up to six post-translational modifications. The phosphorylation sites protein kinase C, casein kinase II, and N-myristoylation existed in 15 StCOL proteins. A total of eight StCOL proteins had cAMP-cGMP sites, five StCOL proteins with Tyr sites, and 13 StCOL proteins with N-Glyc sites were observed. The results suggested that StCOL proteins may exert their function through phosphorylation sites protein kinase C, casein kinase II, and N-myristoylation.
3.6 Chromosome location and duplication models of StCOL genes
Fifteen COL genes were found to be unequally distributed on eight potato chromosomes through chromosome localization (Figure 4A). Chr5 has the largest distribution with four COL genes. Chr3, Chr4, Chr8, and Chr9 contain only one COL gene. StCOLs were mainly distributed at the top or end of chromosomes. MCScanX was used to analyze the evolution and function of 15 StCOLs in the potato chromosome genome. The results showed that four pairs of the StCOL gene family had gene duplication events (Figure 5). There were no tandem repeats and large fragment replication, including StCOL14 on Chr03, StCOL12 on Chr12, StCOL6 and StCOL1 on Chr07, and StCOL8 gene clusters on Chr12. There were gene duplication events in StCOL5 and StCOL8 on Chr12. Interspecies collinearity analysis of COL genes of tomato, potato, and Arabidopsis identified 17 homologous genes between tomato and potato and 21 homologous genes between potato and Arabidopsis, indicating that there were more direct homologous genes, close relatives, and similar gene functions among these species (Figure 4B). A total of 13 StCOL genes are homologous in tomato and Arabidopsis (Supplementary Table S5), indicating that these StCOL genes may have a common ancestor in different species that evolved from the same ancestor.
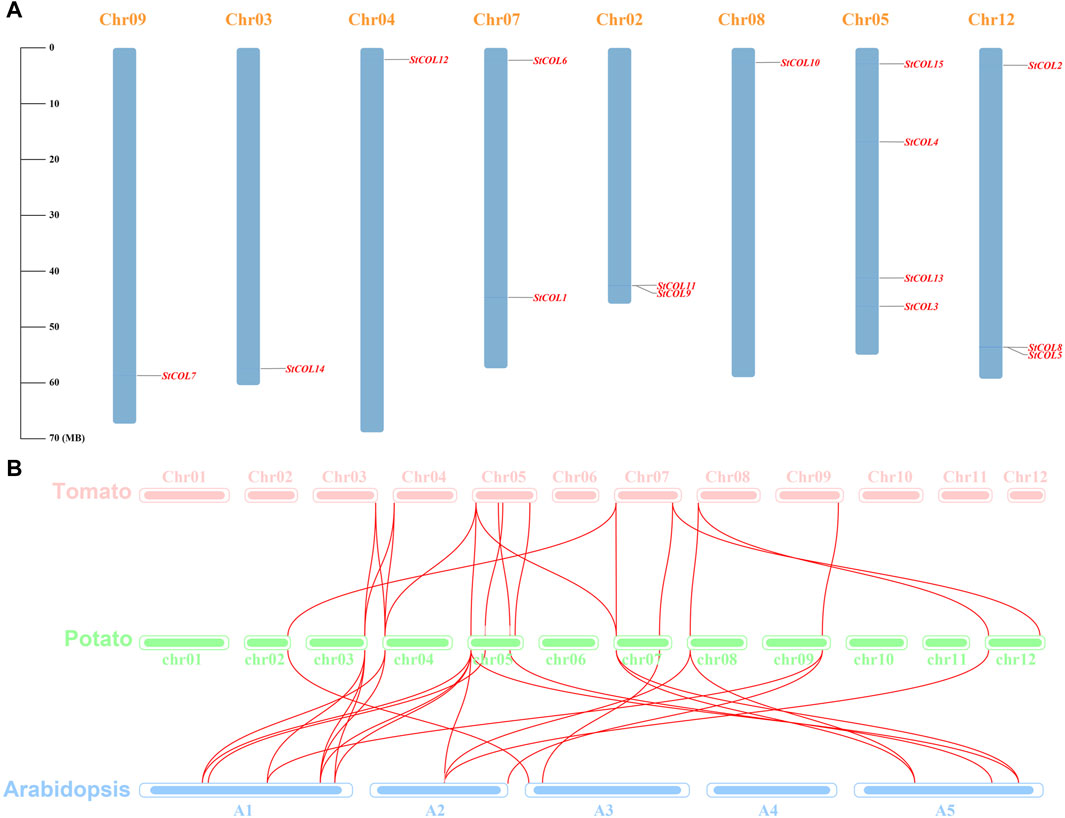
Figure 4. Chromosome location and synteny analysis. (A) Chromosome location of potato StCOL genes on the chromosome. (B) Duplication genes and collinearity orthologs were mapped to each chromosome among potato, tomato, and Arabidopsis.
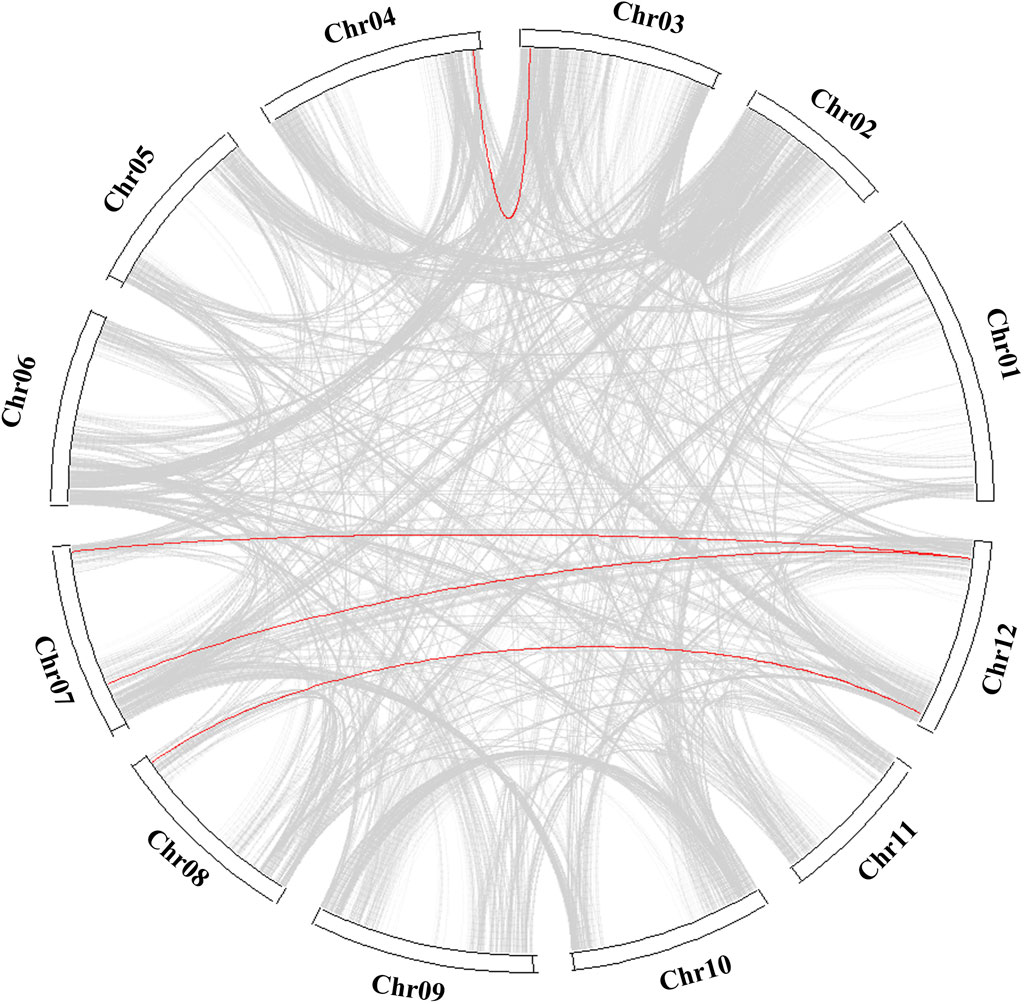
Figure 5. Collinearity analysis of COL genes between tomato, potato, and Arabidopsis. The red line represents collinearity between StCOL genes across species.
3.7 Expression patterns of StCOL genes in different potato tissues
The expression patterns of StCOL family genes in different potato tissues were analyzed using potato transcriptome data. The results showed that 10 genes were up-expressed to a certain extent in all tissues of potato, while the remaining five genes were down-expressed in potato (Figure 6). StCOL2 is highly expressed in root, tuber pith, stem, and other tissues, indicating that these genes play an important role in potato growth and development. High expression levels of StCOL1, StCOL2, StCOL6, StCOL7, and StCOL10 were found in flowers and may play a role in regulating the flowering process in potato. The StCOL2, StCOL4, StCOL7, StCOL10, StCOL11, and StCOL13 were more highly expressed in root, tuber pith, tuber cortex, and young tuber tissues than other genes. It may be that these genes play a major role in the formation of potato tubers.
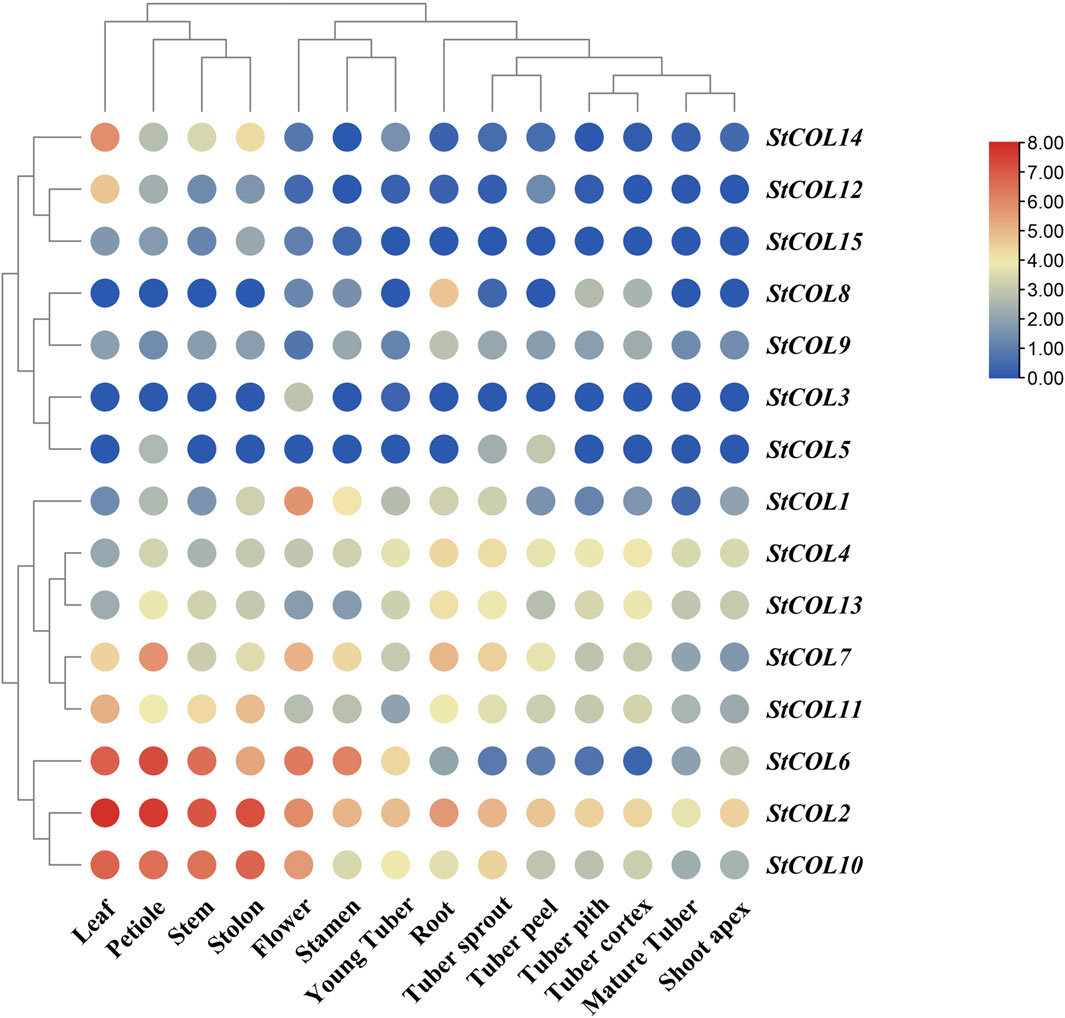
Figure 6. Expression analysis of genes in different tissues by transcriptome data. The horizontal axis represents different tissues in potato, and the vertical axis represents different StCOL genes in potato.
3.8 Expression analysis of the StCOL gene at different maturities of potato leaves and tubers
The relative expression levels of StCOL genes were measured at different maturities of potato leaves and tubers. The qRT-PCR results showed that the StCOL2, StCOL5, StCOL6, StCOL8, and StCOL9 were highly expressed in the leaves of potato growing to day 42 (Figure 7). The expression levels of StCOL4 were significantly higher in potato tubers than in leaves. With increased growth time, the expression level of StCOL7 and StCOL15 decreased in leaves and tubers. The expression levels of StCOL2, StCOL6, and StCOL9 presented opposite patterns in leaves and tuber during the same period. These results indicated that the StCOL genes may play an important role in the formation of potato tubers.
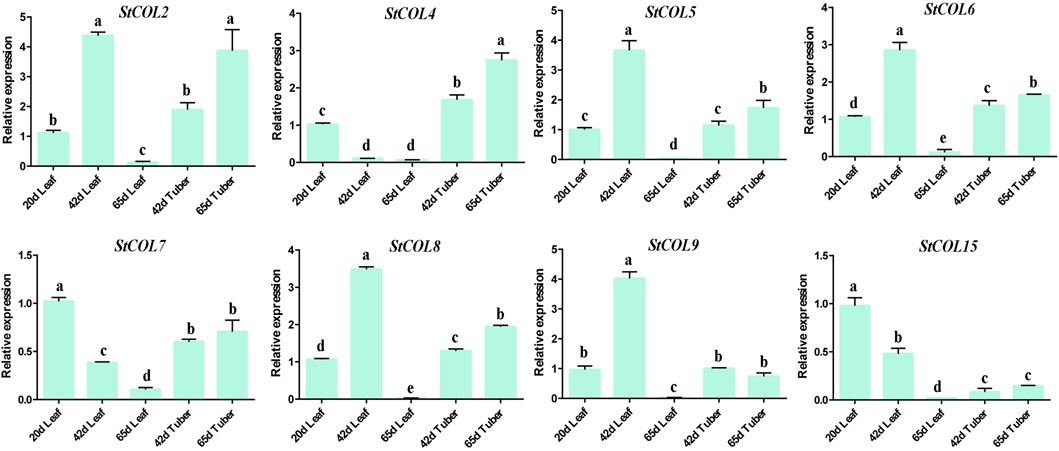
Figure 7. Relative expressions of StCOL genes in leaves (20 days, 42 days, and 65 days) and tuber (42 days and 65 days). Duncan’s test with p < 0.05 was used to indicate significant differences.
3.9 Expression analysis of the StCOL gene after different cold stress treatment times
The relative expression levels of StCOL genes were measured after different cold stress treatment times by qRT-PCR. The results showed that the StCOL1, StCOL6, StCOL12, and StCOL14 were highly expressed after treatment with 2°C for 1 h (Figure 8). Compared with 0 h, the expression levels of StCOL5, StCOL7, and StCOL9 were significantly down-expressed after different lengths of cold stress treatment times. These results indicated that the StCOL genes may play an important role in potato response to cold stress.
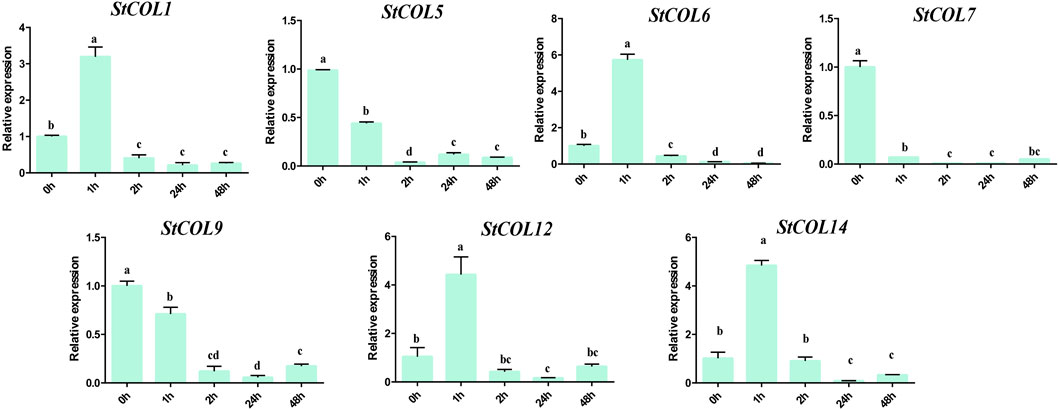
Figure 8. The expression levels of StCOL genes at 0 h, 1 h, 2 h, 24 h, and 48 h after 2°C cold treatment. Duncan’s test with p < 0.05 was used to indicate significant differences.
4 Discussion
Potato is an important food crop worldwide. Potato tubers are rich in nutrients, including starch, protein, and vitamin C. The tuberization of potato varies by geographic region and harvest time. Low-temperature stress seriously affected the growth of potato leaves in the aboveground part and the formation of potato blocks in the underground part. Many studies have indicated that the StCO gene plays an important role in regulating potato formation (González-Schain et al., 2012; Abelenda et al., 2016). Although the genome-wide identification, characterization, and expression profiling of the COL gene family have been reported in potato (Li et al., 2023), the molecular mechanism of the potato COL family involved in the regulation of potato tuberization and response to low-temperature stress is still unclear. In this study, a total of 15 StCOL genes were identified, and the expression patterns of these genes at different maturities of potato leaves and tubers and different cold-stress treatment times were determined.
The CO genes play a vital role in the flowering process of plants. Under long-day conditions, the DELLA protein directly interacts with CO to regulate flowering in Arabidopsis (Xu et al., 2016). Previous studies have shown that Arabidopsis thaliana, as a tolerant long-day (LD) plant, specifically accumulates FT transcripts in LD conditions, and activation of FT transcription is regulated by CCT factor CONSTANS (AtCO) (An et al., 2004). In rice, Hd1 transcripts are similar to AtCO, and the gene activity of Hd1 is mediated by light-activated photopigments because rice se5 mutants with impaired chromophore biosynthesis exhibit a severe early flowering phenotype in LD conditions, and the FT homologous gene Hd3a in rice also regulates flowering (Yano et al., 2000; Hecht et al., 2005). The expression of OsCOL9 enhances rice disease resistance by enhancing the expression of plant hormone biosynthesis genes NPR1, WRKY45, and OsACO1 (Liu et al., 2016). In SD and LD conditions, overexpression of OsCOL15 leads to a delayed flowering phenotype (Wu et al., 2018). The COL gene also plays an important role in leaf growth and tuber formation, and photoperiod signal perception mainly occurs in leaf organs. Previous studies have shown that nine PtCOL genes are widely expressed in various tissues and organs of poplar but are preferentially expressed in leaves (Li et al., 2020). Potato tuber formation requires short-day regulation, and this photoperiod response is related to the activation of the StSP6A gene in leaves. Potato StCOL1 inhibits the formation of storage organs by directly activating the FT-like StSP5G repressor protein (Abelenda et al., 2016). The relative expression of the StCOL gene in leaves and tubers of “Favorita” was detected at different growth stages. The expression levels of StCOL2, StCOL5, StCOL6, StCOL8, and StCOL9 in leaves were significantly higher than those in tubers, suggesting that these genes may play an important role in the development of potato leaves. Moreover, the expression levels of StCOL4 in potato tubers are higher than those in leaves, and the expression levels increase with growth time. Many studies have found that StCDF1 represses the expression of StCO1/2 by binding to the promoter of StCO1, StCO2, and StCO3 genes in potato (Kloosterman et al., 2013; Ramírez Gonzales, 2021). The overexpression of Nelumbo nucifera COL5 in potato led to increased tuber weight and starch content under SD conditions (Cao et al., 2021). The StCOL genes identified in this study provide a data basis for further study of StCOL gene function in tuber formation.
Low-temperature stress seriously affects the production, development, and geographical distribution of plants (Ding et al., 2020). A total of 15 COL genes were identified in petunia, of which six were induced by cold treatment (Khatun et al., 2021). In this study, seven COL genes were differentially expressed under 1 h of cold treatment. Previous studies indicate that CO integrated photoperiodic and cold stress signals into the flowering genetic pathways (Jung et al., 2012). The AtCOL1 and AtCOR27 are rapidly induced in response to low temperature by a CBF-independent pathway (Mikkelsen and Thomashow, 2009). Five CaCOL genes were induced by cold, of which CaCOL02 and CaCOL03 were remarkably upregulated under cold stress and downregulated by heat stress (Huang et al., 2022). These reports indicated that COL genes played an important role in low-temperature stress. The potato SP6A, a homolog of the floral inductor FLOWERING LOCUS T, which controls tuber formation and SP6A expression, is downregulated under high temperatures, preventing tuberization (Lehretz et al., 2019; Park et al., 2022). However, the genes that regulate potato tuberization at low temperatures have not been reported. In this study, the promoter sequences in four StCOL genes (StCOL2, StCOL3, StCOL9, and StCOL15) contained cis-acting elements related to low-temperature stress response. The StCOL9 showed downregulated expression after 1 h of cold treatment by qRT-PCR analysis. These results provide a database for further exploring the molecular mechanism of StCOL genes involved in the regulation of tuberization under cold stress in potato.
5 Conclusion
In this research, a total of 15 COL family genes were identified in the potato genome. These StCOL genes were unevenly distributed on eight chromosomes, and the conserved motifs and gene structure positions of 15 COL genes were identified. Notably, the StCOL genes containing cis-acting elements associated with stress, hormones, and growth and development were identified. Moreover, the expression level of StCOL genes was determined in different development tissues after different low-temperature treatment times in potato. These studies suggested that StCOL family genes might play an important role in growth development, tuber formation, and cold stress response in potato.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Author contributions
WY: investigation, writing–original draft, data curation, methodology, validation. LW: data curation, methodology, validation, writing–original draft, and formal analysis. QS: validation, writing–original draft, and investigation. MC: writing–original draft, formal analysis, and resources. FL: resources, writing–original draft, funding acquisition, investigation, and writing–review and editing. XL: investigation, writing–original draft, writing–review and editing, conceptualization, formal analysis, and visualization.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Guizhou Province Natural Science Foundation key project (Qiankehejichu [2020]1Z015), the Guizhou Academy of Agricultural Sciences Guojihoubuzhu [2021]41, the Innovation Capacity Construction of Breeding Scientific Research platform in Guizhou Province (QianKeHeFuQi [2022]014), the Construction of Biological Breeding Platform for Important Crops in Karst Mountain Areas of Guizhou Province (QianKeHeZhongYinDi; [2023]033), the Guizhou Provincial Science and Technology Plan Project (Qian Kehe Support [2022] key 025 and 026), and the national potato industry technology system at the Guiyang Comprehensive Experimental Station (2023–2024).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1390411/full#supplementary-material
References
Abelenda, J. A., Cruz-Oró, E., Franco-Zorrilla, J. M., and Prat, S. (2016). Potato StCONSTANS-like1 suppresses storage organ formation by directly activating the FT-like StSP5G repressor. Curr. Biol. 26, 872–881. doi:10.1016/j.cub.2016.01.066
Almada, R., Cabrera, N., Casaretto, J. A., Ruiz-Lara, S., and Villanueva, E. G. (2009). VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep. 28, 1193–1203. doi:10.1007/s00299-009-0720-4
An, H., Roussot, C., Sua´rez-Lo´pez, P., Corbesier, L., Vincent, C., Pineiro, M., et al. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615–3626. doi:10.1242/dev.01231
Bailey, T. L., Mikael, B., Buske, F. A., Martin, F., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi:10.1093/nar/gkp335
Campoli, C., Drosse, B., Searle, I., Coupland, G., and Korff, M. V. (2012). Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J. 69, 868–880. doi:10.1111/j.1365-313X.2011.04839.x
Cao, D., Li, Y., Lu, S., Wang, J., Nan, H., Li, X., et al. (2015). GmCOL1a and GmCOL1b function as flowering repressors in soybean under long-day conditions. Plant Cell Physiol. 56, 2409–2422. doi:10.1093/pcp/pcv152
Cao, D., Lin, Z., Huang, L., Damaris, R. N., Li, M., and Yang, P. (2021). A CONSTANS-Like gene of Nelumbo nucifera could promote potato tuberization. Planta 253, 65–11. doi:10.1007/s00425-021-03581-9
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Crocco, C. D., and Botto, J. F. (2013). BBX proteins in green plants: insights into their evolution, structure, feature and functional diversification. Gene 531, 44–52. doi:10.1016/j.gene.2013.08.037
Datta, S., Hettiarachchi, G. H. C. M., Deng, X. W., and Holm, M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18, 70–84. doi:10.1105/tpc.105.038182
Ding, Y. L., Shi, Y. T., and Yang, S. H. (2020). Molecular regulation of plant responses to environmental temperatures. Mol. Plant. 13, 544–564. doi:10.1016/j.molp.2020.02.004
González-Schain, N. D., Díaz-Mendoza, M., Zurczak, M., and Suárez-López, P. (2012). Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. 70, 678–690. doi:10.1111/j.1365-313X.2012.04909.x
González-Schain, N. D., and Suárez-López, P. (2008). CONSTANS delays flowering and affects tuber yield in potato. Biol. Plant 52, 251–258. doi:10.1007/s10535-008-0054-z
Griffiths, S., Dunford, R. P., Coupland, G., and Laurie, D. A. (2003). The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 131, 1855–1867. doi:10.1104/pp.102.016188
Guo, Y. H., Luo, C., Liu, Y., Liang, R. Z., Yu, H. X., Lu, X. X., et al. (2022). Isolation and functional analysis of two CONSTANS-like 1 genes from mango. Plant Physiol. Biochem. 172, 125–135. doi:10.1016/j.plaphy.2022.01.010
Hall, B. G. (2013). Building phylogenetic trees from molecular data with mega. Mol. Biol. Evol. 30, 1229–1235. doi:10.1093/molbev/mst012
Hassidim, M., Harir, Y., Yakir, E., and Green, R. M. (2009). Over-expression of CONSTANS LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 230, 481–491. doi:10.1007/s00425-009-0958-7
Hecht, V., Foucher, F., Ferrandiz, C., Macknight, R., Navarro, C., Morin, J., et al. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137, 1420–1434. doi:10.1104/pp.104.057018
Huang, Z., Bai, X., Duan, W., Chen, B., Chen, G., Xu, B., et al. (2022). Genome-wide identification and expression profiling of CONSTANS-Like genes in Pepper (Capsicum annuum): gaining an insight to their phylogenetic evolution and stress-specific roles. Front. Plant Sci. 13, 828209. doi:10.3389/fpls.2022.828209
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., and Kay, S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi:10.1126/science.1110586
Jung, J., Seo, P. J., and Park, C. (2012). The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J. Biol. Chem. 287, 43277–43287. doi:10.1074/jbc.M112.394338
Khatun, K., Debnath, S., Robin, A. H. K., Wai, A. H., Nath, U. K., Lee, D. J., et al. (2021). Genome-wide identification, genomic organization, and expression profiling of the CONSTANS-like (COL) gene family in petunia under multiple stresses. BMC Genomics 22, 727–817. doi:10.1186/s12864-021-08019-w
Kikuchi, R., Kawahigashi, H., Oshima, M., Ando, T., and Handa, H. (2012). The differential expression of HvCO9, a member of the CONSTANS-like gene family, contributes to the control of flowering under short-day conditions in barley. J. Exp. Bot. 63, 773–784. doi:10.1093/jxb/err299
Kinmonth-Schultz, H. A., Tong, X., Lee, J., Song, Y. H., Ito, S., Kim, S. H., et al. (2016). Cool night-time temperatures induce the expression of CONSTANS and FLOWERING LOCUS T to regulate flowering in Arabidopsis. New Phytol. 211, 208–224. doi:10.1111/nph.13883
Kloosterman, B., Abelenda, J. A., Gomez, M. M., Oortwijn, M., de Boer, J. M., Kowitwanich, K., et al. (2013). Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495, 246–250. doi:10.1038/nature11912
Ledger, S., Strayer, C., Ashton, F., Kay, S. A., and Putterill, J. (2001). Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 26, 15–22. doi:10.1046/j.1365-313x.2001.01003.x
Lehretz, G. G., Sonnewald, S., Hornyik, C., Corral, J. M., and Sonnewald, U. (2019). Post-transcriptional regulation of FLOWERING LOCUS T modulates heat-dependent source-sink development in potato. Curr. Biol. 29, 1614–1624. doi:10.1016/j.cub.2019.04.027
Li, J., Gao, K., Yang, X., Khan, W. U., Guo, B., Guo, T., et al. (2020). Identification and characterization of the CONSTANS-like gene family and its expression profiling under light treatment in Populus. Int. J. Biol. Macromol. 161, 999–1010. doi:10.1016/j.ijbiomac.2020.06.056
Li, R., Li, T., Wu, X., Yao, X., Ai, H., Zhang, Y., et al. (2023). Genome-wide identification, characterization and expression profiling of the CONSTANS-like genes in potato (Solanum tuberosum L.). Genes 14, 1174. doi:10.3390/genes14061174
Liang, R. Z., Luo, C., Liu, Y., Hu, W. L., Guo, Y. H., Yu, H. X., et al. (2023). Overexpression of two CONSTANS-like 2 (MiCOL2) genes from mango delays flowering and enhances tolerance to abiotic stress in transgenic Arabidopsis. Plant Sci. 327, 111541. doi:10.1016/j.plantsci.2022.111541
Liu, H., Dong, S., Sun, D., Liu, W., Gu, F., Liu, Y., et al. (2016). CONSTANS-Like 9 (OsCOL9) interacts with receptor for activated C-Kinase 1(OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. Plos One 11, e0166249. doi:10.1371/journal.pone.0166249
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Mikkelsen, M. D., and Thomashow, M. F. (2009). A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 60, 328–339. doi:10.1111/j.1365-313X.2009.03957.x
Min, J. H., Chung, J. S., Lee, K. H., and Kim, C. S. (2015). The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 57, 313–324. doi:10.1111/jipb.12246
Park, J. S., Park, S. J., Kwon, S. Y., Shin, A. Y., Moon, K. B., Park, J. M., et al. (2022). Temporally distinct regulatory pathways coordinate thermo-responsive storage organ formation in potato. Cell Rep. 38, 110579. doi:10.1016/j.celrep.2022.110579
Qiao, X., Li, Q. H., Yin, H., Qi, K., Li, L., Wang, R., et al. (2019). Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 20, 38. doi:10.1186/s13059-019-1650-2
Ramírez Gonzales, L., Shi, L., Bergonzi, S. B., Oortwijn, M., Franco-Zorrilla, J. M., Solano-Tavira, R., et al. (2021). Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 105, 855–869. doi:10.1111/tpj.15093
Robson, F., Costa, M. M., Hepworth, S. R., Vizir, I., Pin˜eiro, M., Reeves, P. H., et al. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28, 619–631. doi:10.1046/j.1365-313x.2001.01163.x
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. doi:10.1126/science.288.5471.1613
Steinbach, Y. (2019). The Arabidopsis thaliana CONSTANS-LIKE 4 (COL4)-a modulator of flowering time. Front. Plant Sci. 10, 651. doi:10.3389/fpls.2019.00651
Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi:10.1146/annurev.arplant.59.032607.092755
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi:10.1093/nar/gkr1293
Wu, W., Zhang, Y., Zhang, M., Zhan, X., Shen, X., Yu, P., et al. (2018). The rice CONSTANS-like protein OsCOL15 suppresses flowering by promoting Ghd7 and repressing RID1. Biochem. Bioph Res. Co. 495, 1349–1355. doi:10.1016/j.bbrc.2017.11.095
Xu, F., Li, T., Xu, P. B., Li, L., Du, S. S., Lian, H. L., et al. (2016). DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett. 590, 541–549. doi:10.1002/1873-3468.12076
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484. doi:10.1105/tpc.12.12.2473
Yoo, S. K., Chung, K. S., Kim, J., Lee, J. H., Hong, S. M., Yoo, S. J., et al. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through Flowering Locus T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778. doi:10.1104/pp.105.066928
Zhang, B., Feng, M., Zhang, J., and Song, Z. (2023). Involvement of CONSTANS-like proteins in plant flowering and abiotic stress response. Int. J. Mol. Sci. 24, 16585. doi:10.3390/ijms242316585
Keywords: CONSTANS-like, gene family, tuberization, cold stress, qRT-PCR
Citation: Yin W, Wang L, Shu Q, Chen M, Li F and Luo X (2024) Genome-wide identification and expression analysis of the CONSTANS-like family in potato (Solanum tuberosum L.). Front. Genet. 15:1390411. doi: 10.3389/fgene.2024.1390411
Received: 23 February 2024; Accepted: 04 June 2024;
Published: 09 July 2024.
Edited by:
Rongbin Hu, University of California, Riverside, United StatesReviewed by:
Kamran Shah, South China Agricultural University, ChinaWenquan Wang, Hainan University, China
Copyright © 2024 Yin, Wang, Shu, Chen, Li and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Li, Z3psZmVpQHNpbmEuY29t; Xiaobo Luo, eGlhb2x1b2JvcG90YXRvQDE2My5jb20=
†These authors have contributed equally to this work
 Wang Yin1†
Wang Yin1† Xiaobo Luo
Xiaobo Luo