
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 21 June 2024
Sec. Applied Genetic Epidemiology
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1386411
This article is part of the Research Topic Women in Applied Genetic Epidemiology View all 7 articles
 Hongyan Liu1†
Hongyan Liu1† Junxia Li1†
Junxia Li1† Chenchen Guan1,2
Chenchen Guan1,2 Wenjie Gao1
Wenjie Gao1 Yan Li1
Yan Li1 Jianmei Wang1*†
Jianmei Wang1*† Yang Yang1,2*†
Yang Yang1,2*† Yongrui Du1*†
Yongrui Du1*†Background: Endometriosis, characterized by extrauterine endometrial tissue, leads to irregular bleeding and pelvic pain. Menstrual retrograde theory suggests fragments traverse fallopian tubes, causing inflammation and scar tissue. Prevalent among infertile women, risk factors include fewer pregnancies, delayed childbirth, irregular cycles, and familial predisposition. Treatments, medication, and surgery entail side effects. Studies link gut microbiota alterations to endometriosis, necessitating research to establish causation. We used Mendelian randomization to investigate the potential link between endometriosis and gut microbiota through genetic variants.
Methods: Two-sample Mendelian randomization analyzed gut microbiota’s potential causal effects on endometriosis. Instrumental variables, robustly associated with exposures, leveraged GWAS data from MiBioGen for gut microbiota and FinnGen R8 release for endometriosis. SNPs strongly associated with exposures were instrumental variables. Rigorous assessments ensured SNP impact scrutiny on endometriosis.
Results: At the genus level, Anaerotruncus, Desulfovibrio, Haemophilus, and Holdemania showed causal association with endometriosis. Specific gut microbiota exhibited causal effects on different endometriosis stages. Holdemania and Ruminococcaceae UCG002 exerted reversible, stage-specific impacts.
Conclusion: Mendelian randomization provides evidence for the causal link between specific gut microbiotas and endometriosis, emphasizing the pivotal role of gut microbiota dysbiosis. Modulating gut microbiota emerges as a promising strategy for preventing and treating endometriosis.
Endometriosis, defined as the presence of endometrial tissue outside the uterine cavity, typically lines various locations such as the ovaries, fallopian tubes, vagina, or other parts of the uterus (Vercellini et al., 2014), manifests with ectopic endometrial tissue that develops and detaches during the menstrual cycle, leading irregular bleeding. While the prevalence of endometriosis in fertile women ranges from 10% to 15%, the incidence rate significantly rises to 20%–50% in cases of infertility (Tanbo and Fedorcsak, 2017; Zondervan et al., 2020). The widely accepted theory of menstrual retrograde posits, proposed by Sampson (Sampson, 2024), that endometrial fragments retrograde through the fallopian tube, contributing significantly to the etiology of endometriosis. This process can induce inflammation and scar tissue formation, resulting in pelvic pain, sexual discomfort, menstrual irregularities, and even infertility (Tanbo and Fedorcsak, 2017; Peiris et al., 2018). Research has revealed differences in both the quantity and activation status of immune cells within the endometrium between patients with endometriosis and normal females (Agic et al., 2006; Saraswat et al., 2017; Patel et al., 2018; Vallvé-Juanico et al., 2019; Bunis et al., 2022). The aberrant expression of these immune cells may contribute to the development and progression of endometriosis, exerting adverse effects on embryo implantation and reproductive outcomes (Saraswat et al., 2017). Additionally, the ectopic endometrium of endometriosis patients harbors a spectrum of immune cells associated with both innate and adaptive immune systems, collectively creating a conducive environment for the ectopic endometrial growth (Vallvé-Juanico et al., 2019; Bunis et al., 2022).
The symbiotic association between the host and microbiota is characterized by mutual benefits (Shi et al., 2017). The host serves as a vital habitat, supplying essential nutrients to sustain the microbiome, while the gut microbiota, in turn, aid in the development of the metabolic system and facilitate the maturation of the immune system through the provision of advantageous nutrients (Kau et al., 2011). Nevertheless, alterations in the equilibrium of gut microbiota communities can lead to dysbiosis, triggering diseases (Rooks and Garrett, 2016; Shi et al., 2017). Numerous studies have underscored the correlation between alterations in gut microbiota and diverse disorders, emphasizing their impact on systemic inflammation and immune cell function (Karmarkar and Rock, 2013; Costello et al., 2015). Certain studies have specifically explored the correlation between the intestinal flora and the etiology of endometriosis (Chadchan et al., 2021; Salliss et al., 2021). Particularly noteworthy is the study by Ata et al., which identified elevated levels of Gardnerella, Streptococcus, Escherichia, Shigella, and Ureoplasma in women with endometriosis (Ata et al., 2019). Additionally, Acidovorax, Devosia, Methylobacterium, Phascolarctobacterium, and Streptococcus abundance in the peritoneal fluid of endometriosis patients surpassed that in controls (Yuan et al., 2022). The imbalance of gut microbiota can lead to an imbalance of the immune system, resulting in a dysregulation of immune cells and their corresponding pathways. While the microbiota composition in the vaginal, cervical, and intestinal regions exhibits similarities between ASRM (American Society for Reproductive Medicine) stage 3/4 endometriosis patients and controls, variations at the genus level are evident (Author Anonymous, 1997; Ata et al., 2019). They substantiated an association between gut microbiota, serum hormones, and inflammatory factors in endometriosis (Shan et al., 2021). Current primary treatment methods for endometriosis include medication and surgery. Hormonal therapies, such as GnRH agonists and Dienogest, are commonly employed to manage symptoms but come with side effects like mood instability, perimenopausal symptoms, and decreased bone density (Vercellini et al., 2014). Surgery, often utilized to remove endometriosis lesions and scar tissue, is associated with a high recurrence rate (Shakiba et al., 2008; Guo, 2009; Horne and Missmer, 2022). Frequent surgeries not only pose a risk to surrounding organs but also impose substantial economic and psychological burdens on patients. In addition, Zizolfi et al. have provided a comprehensive review of the current research on the interplay between microbiota and endometriosis, and suggested potential therapeutic interventions, including antibiotics, probiotics, and prebiotics, as well as novel approaches such as fecal, vaginal, or uterine microbial transplantation, to restore a dysbiotic state to a more favorable genital microenvironment (Zizolfi et al., 2023).
In this study, we explored the potential relationship between the microbiota and endometriosis and emphasizing this perspective as a focal point in future research to uncover underlying mechanisms, holds promise for paving the way towards the development of new approaches in preventing and treating endometriosis. This would bring hope to patients who bear the burden of endometriosis.
Mendelian randomization (MR), a novel statistical approach utilizing genetic variants as instrumental variables, provides a robust method to explore the causal link between exposure and outcome. This study employed Mendelian randomization analysis to explore the connection between endometriosis and gut microbiota. The analysis considered nine phyla, 16 classes, 20 orders, 32 families, and 119 genera (excluding 3 unknown families and 12 unknown genera). Endometriosis GWAS summary statistics from the FinnGen consortium R8 release data of European ancestry and gut microbiota GWAS summary statistics from the MiBioGen collaboration, predominantly of European ancestry, were collected for this purpose.
Three prerequisites must be met in Mendelian randomization (MR) analysis: 1) instrumental variables should exhibit an association with the exposure; 2) instrumental variables should not be correlated with confounders; and 3) genetic variants must solely influence the outcome through the exposure. The flow of MR analysis is illustrated in Figure 1. Leveraging publicly available GWAS data on relevant gut microbiota, we explored the causal association between endometriosis and gut microbiota. For each taxonomic group, an instrumental variable was selected based on summary statistics. Summary-level data for endometriosis were sourced from FinnGen. The genus was the lowest taxonomic level in the study of Kurilshikov A et al., so we mainly analyzed the causal effects between the genus level of gut microbiota and endometriosis. Endometriosis is classified into four stages by the American Society of Reproductive Medicine (ASRM) based on the disease’s location, extent, and depth within pelvic structures (Author Anonymous, 1997; Zondervan et al., 2020). Stage I involves superficial lesions, Stage II includes both superficial and some deep lesions, Stage III comprises lesions with adhesions between the ovaries and fallopian tubes, and Stage IV is characterized by severe adhesions in the pelvic region and damage to the pouch of Douglas. Subsequently, MR analysis was conducted to estimate the causal effects of the genus on different stages of endometriosis.
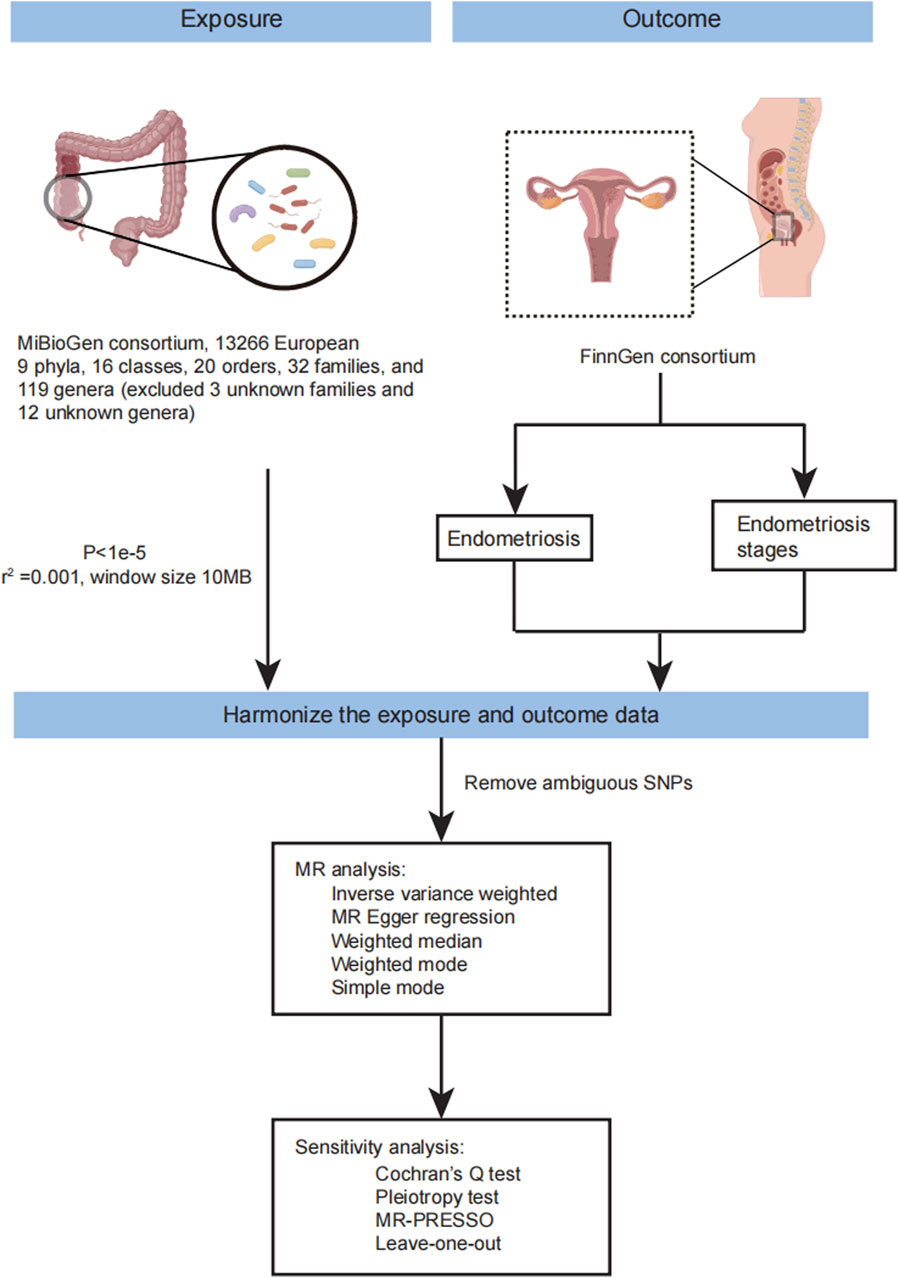
Figure 1. The workflow of MR analysis in this study. The flow chart showing the step of MR analysis in this study.
Genetic variants of gut microbiota were obtained from the large-scale association analyses by MiBioGen consortium (Kurilshikov et al., 2021). The data was collected from 24 cohorts, including 18,340 individuals, most of whom had European ancestry (13,266 individuals). This study included nine phyla, 16 classes, 20 orders, 32 families, and 119 genera for analysis (we excluded the 3 unknown families and 12 unknown genera). In this study, the genus was the main analysis object at the lowest taxonomic level. GWAS summary statistics for endometriosis were obtained from FinnGen consortium R8 release data (Kurki et al., 2023). The GWAS summary statistics data of endometriosis included 13,456 cases. Endometriosis was defined by E80 in ICD-10 and 617 in ICD-9, and 6253 in ICD-8. In addition, we also acquired the GWAS summary statistic data in different stages of endometriosis based on ASRM. FinnGen combined stages 1 and 2 (5122 cases, 185,757 controls) and merged stages 3 and 4 (6751 cases, 184,128 controls).
In this MR analysis, the genus of gut microbiota was divided into 119 taxa (we eliminated the 12 unknown genera). (https://mibiogen.gcc.rug.nl/). We selected genetic variants significantly associated with gut microbiota using a threshold of P < 1e-5, which is consistent with previous studies (Li et al., 2022; Liu et al., 2022; Yu et al., 2023) as instrumental variables for further MR analysis (Supplementary Material S1; Supplementary Table S1). We also considered the linkage disequilibrium (LD) between SNPs. SNPs with high LD may have correlated effects and may inflate the statistical significance of the MR analysis. Then we remove SNPs in strong linkage disequilibrium (LD) which
Harmonizing Single-Nucleotide Polymorphism effects is crucial in MR analyses to ensure that the effect alleles are the same for both exposures and outcomes. Common sources of bias include wrong-effect alleles, palindromic SNPs, and incompatible alleles. Palindromic SNPs pose challenges in identifying the effect allele and should be excluded. This study used various methods, including the random-effects inverse variance weighted method, MR-Egger regression, weighted median (WM), Simple mode, and weighted mode. The primary analysis used the inverse variance weighted method, which meta-analyses the Wald estimates for each SNP to evaluate the association. To acquire a robust result, we also performed MR-PRESSO. Additionally, we conducted MR analyses based on the summary statistics data of different stages of endometriosis from FinnGen.
MR-Egger regression is a method based on the inverse variance weighted method that satisfies the assumptions of instrument strength independent of direct effect (InSIDE) and no measurement error (NOME) (Bowden et al., 2015; Spiller et al., 2019). Compared to the IVW method, the MR-Egger regression method includes an intercept, which helps determine the presence of pleiotropy between instrumental variables and outcome. Therefore, we used MR-Egger regression to detect horizontal pleiotropy. Based on Cochran’s Q test, heterogeneity was evaluated using MR-Egger regression and IVW. MR-PRESSO detected and corrected horizontal pleiotropy by removing outliers in the casual relationship (Verbanck et al., 2018). We used MR-PRESSO global test to detect pleiotropy, and if significant (p < 0.05), outliers identified by the MR-PRESSO outlier test were removed, and the MR analysis was repeated. Finally, we used leave-one-out analysis to assess whether the impact of a single SNP affects the results and has the horizontal pleiotropic effect. We eliminate SNP individually and recalculate the MR analysis to identify which SNP is causing change (Emdin et al., 2017).
To avoid multiple comparison problems, we use the FDR method to adjust p-value in this study. MR-PRESSO global test and outlier test were implemented in R package MR-PRESSO. All analyses were performed by TwoSampleMR (v 0.5.6) package in R software (version 4.0.3) (R Core Team (2020), R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/).
We selected genome-wide significant SNPs association with gut microbiota from MiBioGen consortium. These gut microbiotas were divided into five levels: phylum, class, order, family, and genus. We set the p-value threshold at 1e-5, consistent with previous studies (Li et al., 2022; Liu et al., 2022; Yu et al., 2023). Then, we remove the single nucleotide polymorphisms with a linkage disequilibrium (LD). To keep the direction of effect allele of exposure and outcome are same, ambiguous SNPs with non-concordant alleles and palindromic SNPs with ambiguous strands that cannot be corrected were discarded in the harmonizing step. So, the SNPs we used may be equal to or less than that listed in Supplementary Material S1; Supplementary Table S1. We calculated the F statistic for each SNP and remained the SNPs with the F-statistic greater than 10 for the following analysis. (Supplementary Material S1; Supplementary Table S1).
We analyzed MR to explore the causal association between gut microbiota and endometriosis. The MR estimates used different methods, shown in Supplementary Material S1; Supplementary Tables S2–S7. In order level of gut microbiota, the Burkholderiales (IVW: OR = 1.21, 95%CI: 1.02–1.42, p = 0.027) and Rhodospirillales (IVW: OR = 0.90, 95%CI: 0.82–0.99, p = 0.029; WM: OR = 0.87, 95%CI: 0.76–0.99, p = 0.039) have a causal relationship with endometriosis at the order level of the gut microbiota. (Supplementary Material S1; Supplementary Table S4) We performed the MR-Egger intercept test and Cochran’s Q test; we did not observe evident horizontal pleiotropy and heterogeneity and no potential, influential instrumental variable in the leave-one-out analysis for Burkholderiales and Rhodospirillales. In brief, Burkholderiales is a risk factor in endometriosis progression, it can increase the risk of endometriosis, and Rhodospirillales can decrease the risk of endometriosis; it is a protecting factor in the development of endometriosis.
In family level of gut microbiota, Clostridialesvadin BB60 group decreased the risk of endometriosis (IVW: OR = 0.86, 95%CI: 0.78–0.95, p = 0.003; WM: OR = 0.86, 95%CI: 0.75–0.99, p = 0.041). Oxalobacteraceae can reduce the risk of endometriosis (IVW: OR = 0.91, 95%CI: 0.85–0.98, p = 0.014). Porphyromonadaceae increases the risk of endometriosis (IVW: OR = 1.27, 95%CI: 1.03–1.56, p = 0.027) and lower by Rhodospirillaceae (IVW: OR = 0.91, 95%CI: 0.83–1.00, p = 0.048). (Supplementary Material S1: Supplementary Table S5) Clostridialesvadin BB60 group, Oxalobacteraceae, Porphyromonadaceae, and Rhodospirillaceae had no horizontal pleiotropy and heterogeneity. In a word, Porphyromonadaceae is a risk factor for endometriosis; it can increase the risk or aggravate endometriosis. The rest of these significant gut microbiota are protecting factors in the development of endometriosis.
In genus level of gut microbiota, Anaerotruncus increase the risk of endometriosis (IVW: OR = 1.29, 95%CI: 1.07–1.55, p = 8.31E-3; WM: OR = 1.22, 95%CI: 1.00, p = 0.047). In MR-PRESSO, Anaerotruncus still have a causal association with endometriosis (p = 0.013). (Supplementary Material S1; Supplementary Tables S6–S7) (Figure 2) Desulfovibrio decreases the risk of endometriosis (IVW: OR = 0.88, 95%CI: 0.78–1.00, p = 0.046). Haemophilus decrease the risk of endometriosis (IVW: OR = 0.89, 95%CI: 0.80–0.99, p = 0.039). Holdemania decrease the risk of endometriosis (IVW: OR = 0.88, 95%CI: 0.78–0.98, p = 0.025).
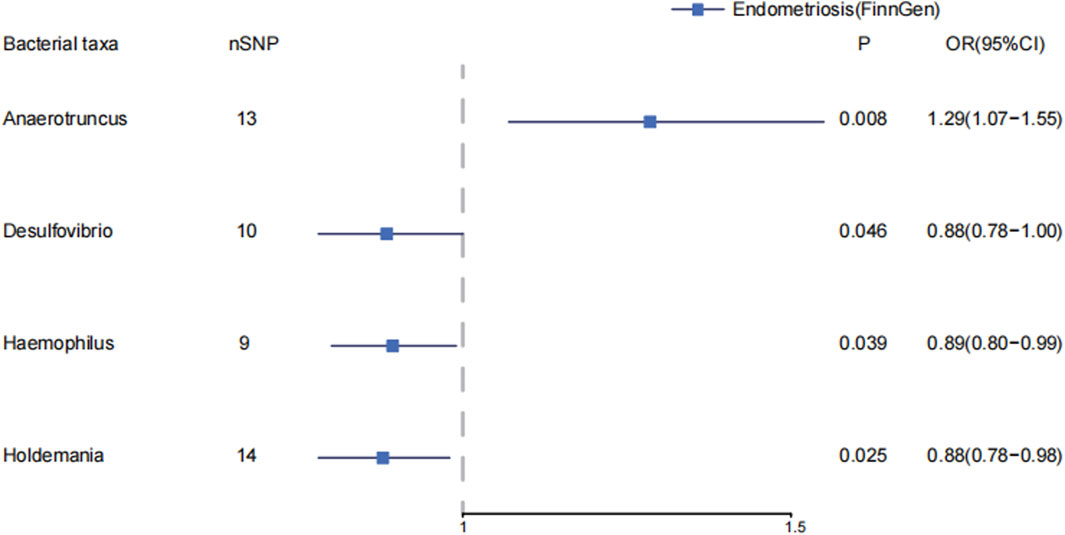
Figure 2. The causal effects of the genus level of gut microbiota on endometriosis in FinnGen by MR analysis. Forest plot depicting the significant causal effects of the genus level of gut microbiota on endometriosis in FinnGen. The odds ratio (OR) was estimated using IVW method. The horizontal bars represent 95% confidence intervals (CI).
Candidatus Soleaferrea, Eubacterium brachy group, Family_XIII AD3011 group, Ruminococcaceae NK4A214 group, Ruminococcaceae UCG002, and Sutterella were associated with the risk of stage 1–2 of endometriosis (Supplementary Material S1; Supplementary Table S8) (Figure 3A). Candidatus Soleaferrea, Ruminococcaceae NK4A214 group, and Sutterella showed significant causal associations with decreased risk of endometriosis in stage 1–2. The OR (95%CI) of Candidatus Soleaferrea in inverse variance weighted was 0.84 (0.72–0.98) (p = 0.029). The inverse variance weighted estimate of the Ruminococcaceae NK4A214 group and Sutterella showed their protective effects on endometriosis in stages 1–2. The ORs (95%CI) were 0.77(0.61–0.97), 0.77(0.60–1.00), respectively. The inverse variance weighted estimate suggests that the Eubacterium brach group (OR = 1.16, 95%CI: 1.03–1.31, p = 0.015), Family_XIII AD3011 group (OR = 1.26, 95%CI: 1.02–1.55, p = 0.032), and Ruminococcaceae UCG002 (OR = 1.21, 95%CI: 1.02–1.43, p = 0.025) were risk factors on endometriosis in stages 1–2. We did not observe significant horizontal pleiotropy and heterogeneity on these gut microbiotas. There is no significant causal association between other gut microbiota and endometriosis in stages 1–2. (Supplementary Material S1; Supplementary Table S8).
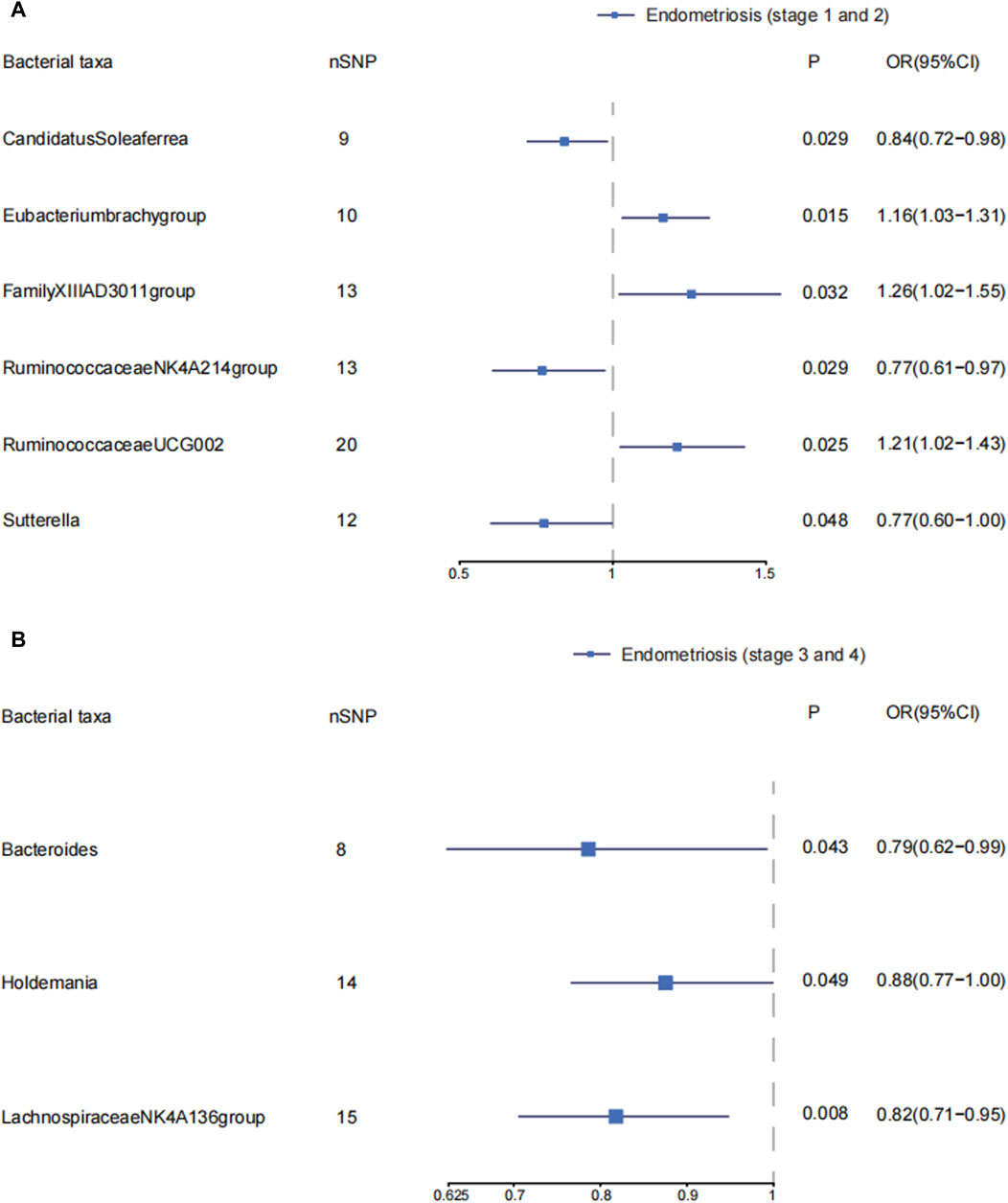
Figure 3. The causal effects of the genus level of gut microbiota on different endometriosis stages. Forest plots depicting causal estimates of the genus level of gut microbiota on different stages of endometriosis in FinnGen. (A) The significant causal association between genus level of gut microbiota and stage 1 and 2 of endometriosis. (B) The significant causal association between genus level of gut microbiota and stage 3 and 4 of endometriosis. The odds ratio (OR) was estimated using IVW method. The horizontal bars represent 95% confidence intervals (CI).
We explored the causal effects of genus level of gut microbiota on endometriosis in ASRM stages 3–4. Bacteroides, Holdemania, and Lachnospiraceae NK4A136 group were protective factors in developing endometriosis in stages 3–4. The ORs (95%CI) were 0.79(0.62–0.99), 0.88(0.77–1.00), and 0.82 (0.71–0.95), respectively. We found no obvious horizontal pleiotropy and heterogeneity. The inverse variance weighted estimates did not support the causal associations of other gut microbiota on endometriosis in stages 3–4. (Supplementary Material S1; Supplementary Table S10) (Figure 3B).
Studies indicated that changes in the composition of gut microbiota may affect the development and progression of endometriosis. Comprehending these links could assist in identifying potential biomarkers for swift diagnosis and in developing personalized treatment approaches for patients with endometriosis. Furthermore, understanding the associations between gut microbiota and endometriosis might lead to the development of innovative treatment methods and improve clinical outcomes for individuals affected by this condition.
Recently, Muraoka et al. found that Fusobacterium infection activates the transforming growth factor-β (TNF-β) signal, promoting the progression of endometriosis. Antibiotic treatment effectively prevented the disease’s progression in a mouse model (Muraoka et al., 2023). Lipopolysaccharide (LPS) derived from gut microbiota can activate Toll-like receptor 4 (TLR4) and induce an inflammatory response, promoting the growth of endometriotic lesions. The production of TNF-α and IL-8, triggered by LPS activating TLR4, is crucial for endometrial tissue adhesion and angiogenesis (Nothnick, 2001). Meanwhile, Iba et al. found that treating endometrial stromal cells (ESCs) obtained from ovarian endometriosis with an NF-kB inhibitor resulted in reduced production of TNF-α and IL-8 and decreased proliferation (Iba et al., 2004). Khan et al. compared peritoneal fluid and menstrual fluid samples in endometriosis patients and controls, showing higher levels of LPS in the menstrual fluid of individuals with endometriosis (Khan et al., 2010). The presence of LPS may contribute to TLR4-mediated growth of endometriosis.
The gut microbiota can impact the structure and function of the intestinal epithelium and has been linked to various diseases, including hypertension, Parkinson’s disease, and autoimmune diseases (Sampson et al., 2016; Li et al., 2017; Janney et al., 2020; Mu et al., 2020; Christovich and Luo, 2022). The precise mechanism by which gut microbiota affects endometriosis remains uncertain. In this study, gut microbiota had causal effects on endometriosis, specifically Holdemania and Ruminococcaceae UCG002, which are associated with different stages of endometriosis.
Certain bacterial families, including Burkholderiales, Oxalobacteraceae, Porphyromonadaceae, and Desulfovibrio, may play a role in the development of endometriosis through the LPS-TLR4 pathway. They can produce LPS which is an essential component of the outer membrane. LPS activates immune cells or immune cell receptors, causing systemic inflammation responses and contributing to the development of endometriosis (Iba et al., 2004; Clemente et al., 2018; Gao et al., 2023).
LPS can activate the PD-L1 pathway, promote overexpression of PD-1 and PD-L1, and suppress T cell activation and proliferation. On the other hand, the Clostridialesvadin BB60 group lead to decreased PD-L1 levels, which can, in turn, suppress the growth of ectopic endometrial tissue in the pelvic cavity (Huang et al., 2023). As a result, the Clostridialesvadin BB60 group may reduce the incidence of endometriosis or alleviate its symptoms. The ectopic endometrial tissue can lead to inflammation and immune dysregulation, which can in turn affect the microbiome composition.
Xue Q et al. found that the abundance of Haemophilus increased significantly in patients with acute exacerbation of chronic obstructive pulmonary disease (COPD) (Xue et al., 2023). AECOPD is characterized by Haemophilus enrichment and a high level of TNF-α. Haemophilus is a respiratory pathogen specific to neutrophils (Sethi and Murphy, 2001). Neutrophils both promote tumor growth and inhibit tumor progression (Gungabeesoon et al., 2023). It exhibits anti-tumorigenic characteristics in the state of acute inflammation (Hirschhorn et al., 2023). Surprisingly, in this study, Haemophilus showed a protective effect in endometriosis. We hypothesize that endometriosis leads to inflammation in the body, and Haemophilus, through neutrophils, restricts the growth of ectopic endometrial tissue.
Holdemania and Ruminococcaceae UCG002 are involved in gut butyrate production (Sampson et al., 2016). Butyrate plays a vital role in maintaining intestinal homeostasis and anti-inflammation. It can regulate the interaction between dendritic cells and DC-T cells and promote Treg T cell differentiation, thereby maintaining immune balance. This is achieved by HDACi suppressing the expression of NF-κB and inducing anti-inflammatory gene transcription to activate dendritic cells (Park et al., 2015).
We further investigated the causal link between the genus of gut microbiota and the different stages of endometriosis, revealing that Holdemania acted as a protective factor against stage 3–4 endometriosis. Conversely, higher levels of Ruminococcaceae UCG002 increased the risk of stage 1–2 endometriosis. These findings were consistent with previous analyses and suggested that Holdemania and Ruminococcaceae UCG002 may play essential roles in developing endometriosis. As mentioned, Holdemania and Ruminococcaceae UCG002 participated in gut butyrate production (Gomez-Arango et al., 2018). These findings further indicated that gut microbiota worked in the occurrence and development of endometriosis. Notably, Ruminococcaceae UCG002 is positively correlated with Treg cells (Zhi et al., 2022), and its abundance is closely related to the levels of pro-inflammatory cytokines. Exploring the detailed mechanisms by which Holdemania and Ruminococcaceae UCG002 influence the progression of endometriosis should be focus on future research. The results are different between endometriosis states 1 and 2 and stages 3 and 4. We thought the lifestyle and treatment methods of patients with different stages of endometriosis may lead to changes in the composition of intestinal flora. Recent research has shown that lifestyle, diet, and other factors can affect gut microbiota composition. A comparison of the Hadza hunter and population in Nepal and California has revealed the impact of these factors (Carter et al., 2023).
In conclusion, these risk factors in gut microbiota mainly affect endometriosis through the LPS-TRL4 pathway. They induce local or systemic inflammation via releasing LPS, the concentrations of inflammatory mediators, cytokines, and chemokines are increased, and TRL4 is activated. This process promotes ectopic endometrial tissue angiogenesis and, thus, colonization. As for protective factors, they can reduce PD-1L levels, prevent angiogenesis, and inhibit the growth of ectopic endometrial tissue. Our study offers a new approach to treating endometriosis, reducing financial burden and relieving pain compared to conventional treatments. This benefits both patients and advances understanding of the condition.
There are several strengths in this study. Firstly, our study examines the relationship between gut microbiota and endometriosis at the genetic level, which reduces confounding bias and reverse causality. This study provides a new perspective for exploring the pathogenesis of endometriosis. Secondly, we have ensured two-sample Mendelian randomization (MR) analyses by selecting separate samples for exposure and outcome data. Thirdly, we have conducted multiple supplementary analyses, including heterogeneity, pleiotropy, leave-one-out sensitivity analyses, and MR-PRESSO, to ensure the robustness of our results.
Of course, several limitations also need to be acknowledged. Firstly, there are no data available at the species level. We cannot explicitly identify which bacteria affect endometriosis. Secondly, the summary statistic of gut microbiota includes multiple ancestries, which may introduce interference from population stratification in our results. Thirdly, the GWAS summary statistic for gut microbiota was not restricted to female participants. Although the exposure data have excluded the sex chromosomes, potential bias due to sex cannot be avoided entirely. In this study, some results were significant for only the IVW analysis. According to the supplementary analysis, there is horizontal pleiotropy or (and) heterogeneity. However, further MR-PRESSO analysis did not find any significant outliers. Therefore, it is necessary to verify our results through further clinical and basic research.
This two-sample MR analysis revealed a causal association between certain gut microbiotas and endometriosis. These microbiotas primarily influence endometriosis through the LPS-TRL4 pathway and inflammatory factors. Moving forward, it is essential to consider the design of randomized controlled trials (RCTs) and fundamental studies to elucidate further the risk and protective factors associated with endometriosis and its underlying mechanisms.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
HL: Data curation, Formal Analysis, Methodology, Writing–original draft. JL: Data curation, Formal Analysis, Methodology, Writing–original draft. CG: Investigation, Resources, Writing–original draft. WG: Writing–original draft. YL: Writing–original draft. JW: Project administration, Supervision, Writing–review and editing. YY: Project administration, Supervision, Writing–review and editing. YD: Project administration, Supervision, Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
We are grateful to the FinnGen and MiBioGen consortia for their kind and generous contribution in sharing their GWAS data. Figure 1 was skillfully crafted with the use of Figdraw (www.figdraw.com).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1386411/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Plots of leave-one-out for relationship between gut microbiota and endometriosis in FinnGen.
SUPPLEMENTARY FIGURE S2 | Scatter plots for the causal effects of gut microbiota on endometriosis in FinnGen cohort.
SUPPLEMENTARY FIGURE S3 | Plots of leave-one-out for relationship between gut microbiota and endometriosis in stage 1 and 2.
SUPPLEMENTARY FIGURE S4 | Scatter plots for the causal effects of gut microbiota on endometriosis in stage 1 and 2.
SUPPLEMENTARY FIGURE S5 | Plots of leave-one-out for relationship between gut microbiota and endometriosis in stage 3 and 4.
SUPPLEMENTARY FIGURE S6 | Scatter plots for the causal effects of gut microbiota on endometriosis in stage 3 and 4.
SUPPLEMENTARY TABLE S1 | Instrumental variables for genus of gut microbiota in this study.
SUPPLEMENTARY TABLE S2 | MR analysis results between phylum of gut microbiota and endometriosis in FinnGen.
SUPPLEMENTARY TABLE S3 | MR analysis results between class of gut microbiota and endometriosis in FinnGen.
SUPPLEMENTARY TABLE S4 | MR analysis results between order of gut microbiota and endometriosis in FinnGen.
SUPPLEMENTARY TABLE S5 | MR analysis results between family of gut microbiota and endometriosis in FinnGen.
SUPPLEMENTARY TABLE S6 | MR analysis results between genus of gut microbiota and endometriosis in FinnGen.
SUPPLEMENTARY TABLE S7 | MR-PRESSO results between genus of gut microbiota and endometriosis in FinnGen.
SUPPLEMENTARY TABLE S8 | The results of MR analysis between gut microbiota and endometriosis in stage 1 and 2.
SUPPLEMENTARY TABLE S9 | MR-PRESSO results between genus of gut microbiota and endometriosis in stage 1 and 2.
SUPPLEMENTARY TABLE S10 | The results of MR analysis between gut microbiota and endometriosis in stage 3 and 4.
SUPPLEMENTARY TABLE S11 | MR-PRESSO results between genus of gut microbiota and endometriosis in stage 3 and 4.
Agic, A., Xu, H., Finas, D., Banz, C., Diedrich, K., and Hornung, D. (2006). Is endometriosis associated with systemic subclinical inflammation? Gynecol. obstetric investigation 62 (3), 139–147. doi:10.1159/000093121
Ata, B., Yildiz, S., Turkgeldi, E., Brocal, V. P., Dinleyici, E. C., Moya, A., et al. (2019). The endobiota study: comparison of vaginal, cervical and gut microbiota between women with stage 3/4 endometriosis and healthy controls. Sci. Rep. 9 (1), 2204. doi:10.1038/s41598-019-39700-6
Author Anonymous (1997). Revised American society for reproductive medicine classification of endometriosis: 1996. Fertil. Steril. 67 (5), 817–821. doi:10.1016/s0015-0282(97)81391-x
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44 (2), 512–525. doi:10.1093/ije/dyv080
Bowden, J., Del Greco, M. F., Minelli, C., Davey Smith, G., Sheehan, N. A., and Thompson, J. R. (2016). Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int. J. Epidemiol. 45 (6), 1961–1974. doi:10.1093/ije/dyw220
Bunis, D. G., Wang, W., Vallvé-Juanico, J., Houshdaran, S., Sen, S., Ben Soltane, I., et al. (2022). Whole-tissue deconvolution and scRNAseq analysis identify altered endometrial cellular compositions and functionality associated with endometriosis. Sec. Autoimmune Autoinflammatory Disord. 12, 788315. doi:10.3389/fimmu.2021.788315
Carter, M. M., Olm, M. R., Merrill, B. D., Dahan, D., Tripathi, S., Spencer, S. P., et al. (2023). Ultra-deep sequencing of Hadza hunter-gatherers recovers vanishing gut microbes. Cell 186, 3111–3124.e13. doi:10.1016/j.cell.2023.05.046
Chadchan, S. B., Popli, P., Ambati, C. R., Tycksen, E., Han, S. J., Bulun, S. E., et al. (2021). Gut microbiota-derived short-chain fatty acids protect against the progression of endometriosis. Life Sci. alliance 4 (12), e202101224. doi:10.26508/lsa.202101224
Christovich, A., and Luo, X. M. (2022). Gut microbiota, leaky gut, and autoimmune diseases. Front. Immunol. 13, 946248. doi:10.3389/fimmu.2022.946248
Clemente, J. C., Manasson, J., and Scher, J. U. (2018). The role of the gut microbiome in systemic inflammatory disease. BMJ Clin. Res. ed 360, j5145. doi:10.1136/bmj.j5145
Costello, M. E., Robinson, P. C., Benham, H., and Brown, M. A. (2015). The intestinal microbiome in human disease and how it relates to arthritis and spondyloarthritis. Best Pract. Res. Clin. rheumatology 29 (2), 202–212. doi:10.1016/j.berh.2015.08.001
Emdin, C. A., Khera, A. V., and Kathiresan, S. (2017). Mendelian randomization. Jama 318 (19), 1925–1926. doi:10.1001/jama.2017.17219
Gao, X., Zhao, X., Liu, M., Zhao, H., and Sun, Y. (2023). Lycopene prevents non-alcoholic fatty liver disease through regulating hepatic NF-κB/NLRP3 inflammasome pathway and intestinal microbiota in mice fed with high-fat and high-fructose diet. Front. Nutr. 10, 1120254. doi:10.3389/fnut.2023.1120254
Gomez-Arango, L. F., Barrett, H. L., Wilkinson, S. A., Callaway, L. K., McIntyre, H. D., Morrison, M., et al. (2018). Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut microbes 9 (3), 189–201. doi:10.1080/19490976.2017.1406584
Gungabeesoon, J., Gort-Freitas, N. A., Kiss, M., Bolli, E., Messemaker, M., Siwicki, M., et al. (2023). A neutrophil response linked to tumor control in immunotherapy. Cell 186 (7), 1448–1464.e20. doi:10.1016/j.cell.2023.02.032
Guo, S. W. (2009). Recurrence of endometriosis and its control. Hum. Reprod. update 15 (4), 441–461. doi:10.1093/humupd/dmp007
Hirschhorn, D., Budhu, S., Kraehenbuehl, L., Gigoux, M., Schröder, D., Chow, A., et al. (2023). T cell immunotherapies engage neutrophils to eliminate tumor antigen escape variants. Cell 186 (7), 1432–1447.e17. doi:10.1016/j.cell.2023.03.007
Horne, A. W., and Missmer, S. A. (2022). Pathophysiology, diagnosis, and management of endometriosis. BMJ Clin. Res. ed 379, e070750. doi:10.1136/bmj-2022-070750
Huang, Y. F., Zhang, W. M., Wei, Z. S., Huang, H., Mo, Q. Y., Shi, D. L., et al. (2023). Causal relationships between gut microbiota and programmed cell death protein 1/programmed cell death-ligand 1: a bidirectional Mendelian randomization study. Front. Immunol. 14, 1136169. doi:10.3389/fimmu.2023.1136169
Iba, Y., Harada, T., Horie, S., Deura, I., Iwabe, T., and Terakawa, N. (2004). Lipopolysaccharide-promoted proliferation of endometriotic stromal cells via induction of tumor necrosis factor alpha and interleukin-8 expression. Fertil. Steril. 82 (Suppl. 3), 1036–1042. doi:10.1016/j.fertnstert.2004.04.038
Janney, A., Powrie, F., and Mann, E. H. (2020). Host-microbiota maladaptation in colorectal cancer. Nature 585 (7826), 509–517. doi:10.1038/s41586-020-2729-3
Karmarkar, D., and Rock, K. L. (2013). Microbiota signalling through MyD88 is necessary for a systemic neutrophilic inflammatory response. Immunology 140 (4), 483–492. doi:10.1111/imm.12159
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L., and Gordon, J. I. (2011). Human nutrition, the gut microbiome and the immune system. Nature 474 (7351), 327–336. doi:10.1038/nature10213
Khan, K. N., Kitajima, M., Hiraki, K., Yamaguchi, N., Katamine, S., Matsuyama, T., et al. (2010). Escherichia coli contamination of menstrual blood and effect of bacterial endotoxin on endometriosis. Fertil. Steril. 94 (7), 2860–2863. doi:10.1016/j.fertnstert.2010.04.053
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53 (2), 156–165. doi:10.1038/s41588-020-00763-1
Kurki, M. I., Karjalainen, J., Palta, P., Sipilä, T. P., Kristiansson, K., Donner, K. M., et al. (2023). FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613 (7944), 508–518. doi:10.1038/s41586-022-05473-8
Li, J., Zhao, F., Wang, Y., Chen, J., Tao, J., Tian, G., et al. (2017). Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 5 (1), 14. doi:10.1186/s40168-016-0222-x
Li, P., Wang, H., Guo, L., Gou, X., Chen, G., Lin, D., et al. (2022). Association between gut microbiota and preeclampsia-eclampsia: a two-sample Mendelian randomization study. BMC Med. 20 (1), 443. doi:10.1186/s12916-022-02657-x
Liu, K., Zou, J., Fan, H., Hu, H., and You, Z. (2022). Causal effects of gut microbiota on diabetic retinopathy: a Mendelian randomization study. Front. Immunol. 13, 930318. doi:10.3389/fimmu.2022.930318
Mu, Q., Edwards, M. R., Swartwout, B. K., Cabana Puig, X., Mao, J., Zhu, J., et al. (2020). Gut microbiota and bacterial DNA suppress autoimmunity by stimulating regulatory B cells in a murine model of lupus. Front. Immunol. 11, 593353. doi:10.3389/fimmu.2020.593353
Muraoka, A., Suzuki, M., Hamaguchi, T., Watanabe, S., Iijima, K., Murofushi, Y., et al. (2023). Fusobacterium infection facilitates the development of endometriosis through the phenotypic transition of endometrial fibroblasts. Sci. Transl. Med. 15 (700), eadd1531. doi:10.1126/scitranslmed.add1531
Nothnick, W. B. (2001). Treating endometriosis as an autoimmune disease. Fertil. Steril. 76 (2), 223–231. doi:10.1016/s0015-0282(01)01878-7
Park, J., Kim, M., Kang, S. G., Jannasch, A. H., Cooper, B., Patterson, J., et al. (2015). Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 8 (1), 80–93. doi:10.1038/mi.2014.44
Patel, B. G., Lenk, E. E., Lebovic, D. I., Shu, Y., Yu, J., and Taylor, R. N. (2018). Pathogenesis of endometriosis: interaction between Endocrine and inflammatory pathways. Best Pract. Res. Clin. obstetrics Gynaecol. 50, 50–60. doi:10.1016/j.bpobgyn.2018.01.006
Peiris, A. N., Chaljub, E., and Medlock, D. (2018). Endometriosis. Jama 320 (24), 2608. doi:10.1001/jama.2018.17953
Rooks, M. G., and Garrett, W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16 (6), 341–352. doi:10.1038/nri.2016.42
Salliss, M. E., Farland, L. V., Mahnert, N. D., and Herbst-Kralovetz, M. M. (2021). The role of gut and genital microbiota and the estrobolome in endometriosis, infertility and chronic pelvic pain. Hum. Reprod. update 28 (1), 92–131. doi:10.1093/humupd/dmab035
Sampson, J. (2024). Gynecology: peritoneal endometriosis due to the menstrual dissemination of endometrial tissue into the peritoneal cavity. Am. J. Obstet. Gynecol. 14, 422–469. doi:10.1016/s0002-9378(15)30003-x
Sampson, T. R., Debelius, J. W., Thron, T., Janssen, S., Shastri, G. G., Ilhan, Z. E., et al. (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell 167 (6), 1469–1480. doi:10.1016/j.cell.2016.11.018
Saraswat, L., Ayansina, D. T., Cooper, K. G., Bhattacharya, S., Miligkos, D., Horne, A. W., et al. (2017). Pregnancy outcomes in women with endometriosis: a national record linkage study. BJOG Int. J. obstetrics Gynaecol. 124 (3), 444–452. doi:10.1111/1471-0528.13920
Sethi, S., and Murphy, T. F. (2001). Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14 (2), 336–363. doi:10.1128/CMR.14.2.336-363.2001
Shakiba, K., Bena, J. F., McGill, K. M., Minger, J., and Falcone, T. (2008). Surgical treatment of endometriosis: a 7-year follow-up on the requirement for further surgery. Obstetrics Gynecol. 111 (6), 1285–1292. doi:10.1097/AOG.0b013e3181758ec6
Shan, J., Ni, Z., Cheng, W., Zhou, L., Zhai, D., Sun, S., et al. (2021). Gut microbiota imbalance and its correlations with hormone and inflammatory factors in patients with stage 3/4 endometriosis. Archives Gynecol. obstetrics 304 (5), 1363–1373. doi:10.1007/s00404-021-06057-z
Shi, N., Li, N., Duan, X., and Niu, H. (2017). Interaction between the gut microbiome and mucosal immune system. Mil. Med. Res. 4, 14. doi:10.1186/s40779-017-0122-9
Spiller, W., Davies, N. M., and Palmer, T. M. (2019). Software application profile: mrrobust—a tool for performing two-sample summary Mendelian randomization analyses. Int. J. Epidemiol. 48 (3), 684–690. doi:10.1093/ije/dyy195
Tanbo, T., and Fedorcsak, P. (2017). Endometriosis-associated infertility: aspects of pathophysiological mechanisms and treatment options. Acta obstetricia Gynecol. Scand. 96 (6), 659–667. doi:10.1111/aogs.13082
Vallvé-Juanico, J., Houshdaran, S., and Giudice, L. C. (2019). The endometrial immune environment of women with endometriosis. Hum. Reprod. update 25 (5), 564–591. doi:10.1093/humupd/dmz018
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50 (5), 693–698. doi:10.1038/s41588-018-0099-7
Vercellini, P., Viganò, P., Somigliana, E., and Fedele, L. (2014). Endometriosis: pathogenesis and treatment. Nat. Rev. Endocrinol. 10 (5), 261–275. doi:10.1038/nrendo.2013.255
Xue, Q., Xie, Y., He, Y., Yu, Y., Fang, G., Yu, W., et al. (2023). Lung microbiome and cytokine profiles in different disease states of COPD: a cohort study. Sci. Rep. 13 (1), 5715. doi:10.1038/s41598-023-32901-0
Yu, H., Wan, X., Yang, M., Xie, J., Xu, K., Wang, J., et al. (2023). A large-scale causal analysis of gut microbiota and delirium: a Mendelian randomization study. J. Affect. Disord. 329, 64–71. doi:10.1016/j.jad.2023.02.078
Yuan, W., Wu, Y., Chai, X., and Wu, X. (2022). The colonized microbiota composition in the peritoneal fluid in women with endometriosis. Archives Gynecol. obstetrics 305 (6), 1573–1580. doi:10.1007/s00404-021-06338-7
Zhi, T., Hong, D., Zhang, Z., Li, S., Xia, J., Wang, C., et al. (2022). Anti-inflammatory and gut microbiota regulatory effects of walnut protein derived peptide LPF in vivo. Food Res. Int. Ott. Ont 152, 110875. doi:10.1016/j.foodres.2021.110875
Zizolfi, B., Foreste, V., Gallo, A., Martone, S., Giampaolino, P., and Di Spiezio Sardo, A. (2023). Endometriosis and dysbiosis: state of art. Front. Endocrinol. 14, 1140774. doi:10.3389/fendo.2023.1140774
Keywords: gut microbiota, endometriosis, GWAS, two-sample Mendelian randomization, FinnGen
Citation: Liu H, Li J, Guan C, Gao W, Li Y, Wang J, Yang Y and Du Y (2024) Endometriosis is a disease of immune dysfunction, which could be linked to microbiota. Front. Genet. 15:1386411. doi: 10.3389/fgene.2024.1386411
Received: 15 February 2024; Accepted: 29 May 2024;
Published: 21 June 2024.
Edited by:
Kristina Allen-Brady, The University of Utah, United StatesReviewed by:
Sagiri Taguchi, Oak Clinic IVF center, JapanCopyright © 2024 Liu, Li, Guan, Gao, Li, Wang, Yang and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianmei Wang, d2FuZ2ppYW5tZWlAdG11LmVkdS5jbg==; Yang Yang, eXlAdG11LmVkdS5jbg==; Yongrui Du, Z2V2aW4xOTcxQHRtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.