- 1Zhejiang University—University of Edinburgh Institute, Zhejiang University, Haining, China
- 2Faculty of Biomedical Sciences, School of Medicine and Veterinary Medicine, University of Edinburgh Institute, Edinburgh, United Kingdom
- 3Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, United Kingdom
Mapping genetic variations to phenotypic variations poses a significant challenge, as mutations often combine unexpectedly, diverging from assumed additive effects even in the same environment. These interactions are known as epistasis or genetic interactions. Sign epistasis, as a specific type of epistasis, involves a complete reversal of mutation effects within altered genetic backgrounds, presenting a substantial hurdle to phenotype prediction. Despite its importance, there is a limited systematic overview of the mechanistic causes of sign epistasis. This review explores the mechanistic causes, highlighting its occurrence in signalling cascades, peaked fitness landscapes, and physical interactions. Moving beyond theoretical discussions, we delve into the practical applications of sign epistasis in agriculture, evolution, and antibiotic resistance. In conclusion, this review aims to enhance the comprehension of sign epistasis and molecular dynamics, anticipating future endeavours in systematic biology engineering that leverage the knowledge of sign epistasis.
Introduction
The mapping of genotype-to-phenotype has been at the core of genetics, yet the relationship between them remains complex and challenging to predict. Even in controlled environments, mutations often interact unexpectedly, deviating from the conventional assumption of additive mutational effects, a phenomenon referred to as genetic interactions or epistasis (Fisher, 1919; Phillips, 2008; Domingo et al., 2019).
Among various types of epistasis, sign epistasis, as a severe form, poses the greatest challenge to phenotype prediction and thus warrants special attention. Sign epistasis occurs when the effect of one mutation completely switches direction from positive to negative, and vice versa, within altered genetic backgrounds (Weinreich et al., 2005). This phenomenon can manifest within a single gene or between different genes. While not always anticipated, such occurrences are common (de Visser et al., 2011; Poelwijk et al., 2011) and significantly constrain evolutionary paths (Weinreich et al., 2005; Weinreich et al., 2006). For example, sign epistasis plays a crucial role in protein evolution, where negative sign epistasis may lead to evolutionary dead ends, and negative reciprocal sign epistasis is responsible for the divergence and branching of evolutionary pathways (Miton et al., 2021; Buda et al., 2023).
Regardless of whether single mutations have detrimental or beneficial effects individually, their combinations can result in sign epistasis. Here, we categorise the occurrence of sign epistasis into three types based on the effects of the single mutations, as depicted in Figure 1.
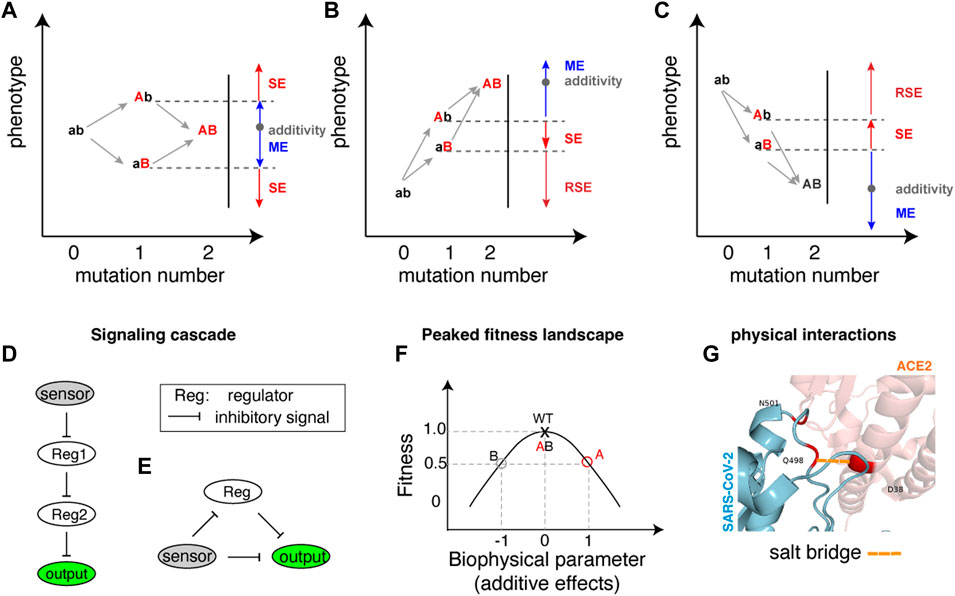
FIGURE 1. Illustration of sign epistasis and mechanisms. (A–C) Sign epistasis in the perspective of double mutations in different mutation combination scenarios where one mutation is detrimental (lower phenotype) while the other mutation is beneficial (higher phenotype) (A), both single mutations are beneficial (B) and both single mutations are detrimental (C). The blue colour represents Magnitude Epistasis (ME), red indicates Sign Epistasis (SE), and dark red signifies Reciprocal Sign Epistasis (RSE). Additivity (shown as a grey dot in each panel) implies that the phenotype resulting from a double mutation equals the sum of phenotypic changes by individual mutations. (D, E) Architectures of gene regulatory networks generating sign epistasis, with D and E showcasing different architectures in E. coli. (F) A peaked fitness landscape can generate sign epistasis. The red and grey circles each represent single mutations A and B with fitness values of 0.5 compared to the wildtype fitness value of 1. The horizontal arrows indicate biophysical parameter changes in two opposite directions. The double mutant AB fitness becomes the same as the wild type due to the combined effect of underlying biochemical parameters. (G) An example of protein residue physical interactions at the binding interface generating sign epistasis. A salt bridge is highlighted in a dashed line coloured in orange shade.
For scenarios where a beneficial (Mutation A) and a detrimental (Mutation B) mutation combine (Figure 1A), sign epistasis occurs if the combined phenotype (AB) surpasses the beneficial effect of the single mutant (Ab). In this case, Mutation B changes its sign from negative to positive in the presence of Mutation A. Conversely, if the double mutant (AB) effect is inferior to the detrimental effect of the single mutant (aB), Mutation A changes its sign while Mutation B retains its negative effect. The other cases of deviation from additivity, termed magnitude epistasis (ME), occur when the double mutants’ phenotype falls within the boundary defined by the two single mutations (Figure 1A).
When two beneficial mutations combine (Figure 1B), sign epistasis arises if the combined effects are worse than the better phenotype of the two single mutations. As illustrated in Figure 1B, if the double mutant (AB) phenotype falls between the two single mutants, the less beneficial mutant (Mutation B) changes its sign from beneficial to detrimental in the presence of the more beneficial mutant (Mutation A). If the double mutant (AB) phenotype is not as good as the less beneficial mutant (Mutation B), it indicates reciprocal sign epistasis (RSE), where both Mutation A and B independently exhibit sign epistasis effects, showcasing opposite effects when the other mutation is present.
Combining two detrimental mutations (Figure 1C) can also lead to both sign and reciprocal sign epistasis, with the combined effect surpassing either one of the single mutational effects.
The exploration of sign epistasis in quantitative genetics unravels a complex tapestry of genetic interactions, molecular dynamics, and broader implications across various biological systems. Despite the unique characteristics of sign epistasis compared to other forms of epistasis, its causes have seldom been systematically discussed separately from general epistasis (Phillips, 2008; de Visser et al., 2011; Domingo et al., 2019).
In this review, we elucidate the mechanistic causes of sign epistasis and discuss its broader implications in the realm of genetics.
Signaling cascade generates sign epistasis between genes
Epistasis between genes is often generated from the upstream-downstream relationship between genes, and recent studies highlight that these relationships also produce sign epistasis.
The occurrence of sign epistasis was demonstrated in a synthetic signalling cascade in bacteria (Figure 1D), consisting of a sensor for induction signal (arabinose for instance), repressors (tetR and lacI as two regulators for instance) and a reporter gene (YFP), (Nghe et al., 2018). Within this gene regulatory network, the two repressors (shown as Reg1 and Reg2 in Figure 1D) coordinate the integration of the induction signal to gene expression hierarchically. Mutations introduced to either upstream or downstream regulatory repressors, or both could switch the direction of mutational effects, resulting in sign epistasis. Biochemical modelling reveals that specific combinations of mutations on transcription factors’ binding affinities to response elements predispose the system to exhibit sign epistasis (Nghe et al., 2018). In essence, sign epistasis is anticipated when certain mutation combinations are introduced to both the upstream and downstream genes in a hierarchical signalling cascade.
Another architecture of synthetic network using the same components (sensing arabinose, regulation via tetR and lacI, controlling reporter gene) in the bacterial system can also lead to the frequent manifestation of sign epistasis(Figure 1E). Unlike the linear upstream-downstream cascade, this synthetic gene regulatory network involves direct inhibitory control and delayed positive regulation (via double inhibitory regulation) of an output (Figure 1E). In this system that leads to a peaked response with increasing inducer concentration, over 50% of the significant epistatic pairs exhibit sign epistasis when two mutations are introduced into two of the three components. Notably, when two mutations enhancing output gene expression combine (as depicted in Figure 1B), the majority of significant epistatic pairs manifest as negative reciprocal sign epistasis pairs (Baier et al., 2023). This observation suggests that combinations of beneficial mutations in two genes can frequently result in detrimental effects in such gene regulatory networks.
Peaked fitness landscapes generate sign epistasis between and within genes
Many biological systems exhibit peaked fitness landscapes that are not monotonically related to gene expression levels or protein activities (Keren et al., 2016), or inducer signals (as the gene regulatory network depicted in Figure 1E). Due to the non-monotonic nature of the system (i.e., a non-monotonic relationship between the protein activity and fitness), combining the two mutations can often produce unexpected results—sign epistasis. Such phenomena have not only been theorized (Gjuvsland et al., 2013) but also observed frequently. Metabolic flux systems, featuring multiple enzymes, commonly generate peaked fitness landscapes (Dykhuizen et al., 1987). Examples include frequent sign epistasis observed between flhA and fghA genes within the GSH-linked metabolic pathway (Chou et al., 2014) and between araA and araB genes in the Arabinose utilization pathway (Chou et al., 2014; Kemble et al., 2020).
Figure 1F illustrates how a peaked fitness landscape generates sign epistasis. As shown in Figure 1F, the wildtype sits on the peak of the fitness landscape, and two detrimental mutations A and B each decrease the fitness to half of the wildtype value (0.5 in the y-axis). Although the fitness effects of the two mutations are the same, their effects on underlying biophysical parameters or protein activity/levels can be opposite (shown in the x-axis), the combined effect pushes the protein activity back to the fitness peak, resulting in seemingly sign epistatic effect (Figure 1F). Such examples can be found in maintaining balanced levels of autophagy, where both insufficient and excessive amounts can lead to detrimental effects (Kang and Avery, 2008).
Even in the context of a single gene system, the expression-fitness landscapes (Keren et al., 2016) and protein stability-fitness relationship (DePristo et al., 2005) can take on a peaked form. Studies on protein folding energy unveil a neutral range, typically within 1 kcal/mol-1 of the wildtype stability (DePristo et al., 2005). Decreased stability beyond this range may lead to reduced protein concentration, while increased stability beyond the range could result in aggregation and, thus, reduced fitness. Consequently, a mutation could have either a positive or negative impact depending on the stability of the genetic background of the protein (DePristo et al., 2005). Although we did not find experimentally demonstrated examples, it is theoretically plausible to expect that mutations within the same gene, each altering expression level, could generate sign epistasis (Li and Lehner, 2020).
Physical interactions generate sign epistasis between and within genes
Physical interactions among atoms within a protein or a protein complex lead to sign epistasis (Figure 1G). Sign epistasis induced by structural contacts can be considered a subcategory of specific epistasis (Domingo et al., 2019), sometimes referred to as contact epistasis (Wonderlick et al., 2023) or idiosyncratic epistasis (Johnson et al., 2023), signifying epistasis resulting from structural interactions between two residues within the protein’s configuration.
Taking the SARS-CoV-2 mutation Q498R as an example, this mutation weakly reduces the binding affinity of the alpha variant to its receptor (ACE2) but enhances the binding affinity with the N501Y mutation (Starr et al., 2022). Molecular dynamic simulations have revealed that the Q498R and N501Y mutations collectively restore individually disrupted salt bridges. Additionally, they establish a new salt bridge with the D38 residue of the ACE2 receptor, augmenting its receptor-bound stability (Starr et al., 2022). The coexistence of both mutations induces a significant alteration in the protein’s structural configuration. A parallel scenario is observed with the HIV-1 protease gene, where a detrimental mutation (L10I) enhances fitness when G48V and L90M mutations are present through structural modification of the encoded protein (Mammano et al., 2000).
Sign epistasis of this kind also frequently occurs between two mutations, each in one of the two interacting molecules. A classic example is evident in the barnase and barstar protein-protein interaction model system, where two individually detrimental mutations (E76R in barstar and R59E in barnase) combine to restore the stable complex through interchanging charges between the interacting positions (Jucovic and Hartley, 1996). Bacterial toxin-antitoxin pairs also exhibit abundant sign epistasis (Aakre et al., 2015) via the same mechanism.
Besides frequently observed in protein coding genes, this type of sign epistasis arising from structural contacts is also observed for RNA phenotypes, such as mutational effects on alternative splicing (Baeza-Centurion et al., 2019) and tRNA function (Domingo et al., 2018).
Application of sign epistasis
In the realm of agriculture, artificial intervention, including artificial selection for favourable traits, is crucial. However, the genetic architecture and the causal effects of genetic interaction on traits of interest are often overlooked, leading to unexpected variance in crop yields (Goldringer et al., 1997; Blanc et al., 2006) and maize flowering (Buckler et al., 2009). The significance of sign epistasis, with its sign-reverting property on different genetic backgrounds, lies in trait selection and optimization. Studies by Vagne and colleagues (Vagne et al., 2015) emphasize the importance of introducing reciprocal sign epistasis into analysis of critical recombination rates to fix optimal genotypes. This shed light on a strategy to better understand the relationship between key parameters and the fixation of fittest traits. Sign epistasis can also contribute to maximizing heterosis, indicating its potential utility for optimizing traits in crops.
Sign epistasis between alleles can even influence the formation of nascent species. Postzygotic reproductive isolation, a key factor in this context, is often attributed to the Bateson-Dobzhansky-Muller (BDM) incompatibilities (Orr, 1995; Orr, 1996; Presgraves, 2010). The genetics underlying these incompatibilities align with reciprocal sign epistasis, where the introduction of alleles from other species underperforms alleles with favoured phenotypes in hybrid individuals, resulting in hybrid infertility or lethality (Ono et al., 2017).
Furthermore, the nature of sign epistasis plays a crucial role in shaping the broad-scale evolutionary phenomenon in terms of the shape and “ruggedness” of the fitness landscape (Poelwijk et al., 2007; de Visser et al., 2011; Poelwijk et al., 2011). Hypothetically, if all possible alleles exert the same effects across all genetic backgrounds, the population’s variance is expected to converge toward a similar genotype of optimal fitness. Conversely, when some alleles are only beneficial or harmful in specific genetic backgrounds, the chronological order of genetic changes may result in a diverse population both genetically and phenotypically. Viral genomes appear to be dominated by sign epistasis (Molla et al., 1996; Cong et al., 2007; Martinez-Picado and Martínez, 2008; Lalić and Elena, 2012), indicating the significant role sign epistasis plays in viral genome evolution.
Sign epistasis is also prevalent in bacteria, offsetting the cost of antibiotic resistance including resistance to nalidixic acid (gyrA mutation), rifampicin resistance (rpoB mutation), and streptomycin resistance (rpsL mutation) (Schrag et al., 1997; Maisnier-Patin et al., 2002; Silva et al., 2011; Wong, 2017). Specifically, mutations conferring resistance to antibiotics become advantageous when coexisting with other drug-resistant mutations or an additional resistance plasmid. Recognizing the power of sign epistasis, these antagonistic interactions highlight the necessity for more effective resistance reversal policies.
Lastly, insights into intra-genic sign epistasis also contribute to advanced protein design strategies, enhancing our understanding of protein evolution and conformation (Domingo et al., 2019; Starr et al., 2022).
Discussion
In this review, we have delved into sign epistasis from a quantitative genetics perspective, offering a mechanistic understanding of the seemingly intricate interplay of effect-switching genetic mutations and their impacts across diverse biological systems. Understanding the mechanisms giving rise to sign epistasis, rooted in signalling cascades, peaked fitness landscapes, and physical Interactions (Table 1), contributes to advancing genotype-phenotype predictions and developing strategies to address challenges in agriculture (Blanc et al., 2006), antibiotic resistance (Wong, 2017), protein design (Miton and Tokuriki, 2016), and design of sophisticated genetic circuits (Baier et al., 2023).
However, there is still a long way to go until we can leverage the knowledge of sign epistasis for the systematic engineering of biology. Besides the inherent challenges of predicting phenotypes posed by sign epistasis, the potential influence of the environment adds further complexity. It has been demonstrated that the occurrence or magnitude of sign epistasis can be altered by the environment, including the presence of inducers or inhibitors (Baier et al., 2023) and antibiotics (Ghenu et al., 2023). Nevertheless, the impact of other environmental factors on sign epistasis remains unclear.
In summary, unveiling the mechanisms of sign epistasis contributes to narrowing the gap between genotypes to phenotypes, providing insights into both the fundamental principles of molecular biology and practical applications across diverse fields.
Author contributions
JZ: Conceptualization, Formal Analysis, Investigation, Visualization, Writing–original draft, Writing–review and editing. FC: Investigation, Writing–original draft, Writing–review and editing. XL: Conceptualization, Funding acquisition, Project administration, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. XL is supported by the Young Scientists Fund of the National Natural Science Foundation of China (NSFC Grant No.32100478).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aakre, C. D., Herrou, J., Phung, T. N., Perchuk, B. S., Crosson, S., and Laub, M. T. (2015). Evolving new protein-protein interaction specificity through promiscuous intermediates. Cell 163, 594–606. doi:10.1016/j.cell.2015.09.055
Baeza-Centurion, P., Miñana, B., Schmiedel, J. M., Valcárcel, J., and Lehner, B. (2019). Combinatorial genetics reveals a scaling law for the effects of mutations on splicing. Cell 176, 549–563. doi:10.1016/j.cell.2018.12.010
Baier, F., Gauye, F., Perez-Carrasco, R., Payne, J. L., and Schaerli, Y. (2023). Environment-dependent epistasis increases phenotypic diversity in gene regulatory networks. Sci. Adv. 9, eadf1773. doi:10.1126/sciadv.adf1773
Blanc, G., Charcosset, A., Mangin, B., Gallais, A., and Moreau, L. (2006). Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor. Appl. Genet. 113, 206–224. doi:10.1007/s00122-006-0287-1
Buckler, E. S., Holland, J. B., Bradbury, P. J., Acharya, C. B., Brown, P. J., Browne, C., et al. (2009). The genetic architecture of maize flowering time. Science 325, 714–718. doi:10.1126/science.1174276
Buda, K., Miton, C. M., Fan, X. C., and Tokuriki, N. (2023). Molecular determinants of protein evolvability. Trends Biochem. Sci. 48, 751–760. doi:10.1016/j.tibs.2023.05.009
Chou, H.-H., Delaney, N. F., Draghi, J. A., and Marx, C. J. (2014). Mapping the fitness landscape of gene expression uncovers the cause of antagonism and sign epistasis between adaptive mutations. PLoS Genet. 10, e1004149. doi:10.1371/journal.pgen.1004149
Cong, M., Heneine, W., and García-Lerma, J. G. (2007). The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J. Virology 81, 3037–3041. doi:10.1128/jvi.02712-06
DePristo, M. A., Weinreich, D. M., and Hartl, D. L. (2005). Missense meanderings in sequence space: a biophysical view of protein evolution. Nat. Rev. Genet. 6, 678–687. doi:10.1038/nrg1672
de Visser, J. A. G. M., Cooper, T. F., and Elena, S. F. (2011). The causes of epistasis. Proc. R. Soc. B 278, 3617–3624. doi:10.1098/rspb.2011.1537
Domingo, J., Baeza-Centurion, P., and Lehner, B. (2019). The causes and consequences of genetic interactions (epistasis). Annu. Rev. Genomics Hum. Genet. 20, 433–460. doi:10.1146/annurev-genom-083118-014857
Domingo, J., Diss, G., and Lehner, B. (2018). Pairwise and higher-order genetic interactions during the evolution of a tRNA. Nature 558, 117–121. doi:10.1038/s41586-018-0170-7
Dykhuizen, D. E., Dean, A. M., and Hartl, D. L. (1987). Metabolic flux and fitness. Genetics 115, 25–31. doi:10.1093/genetics/115.1.25
Fisher, R. A. (1919). XV.—the correlation between relatives on the supposition of mendelian inheritance. Earth Environ. Sci. Trans. R. Soc. Edinb. 52, 399–433. doi:10.1017/S0080456800012163
Ghenu, A.-H., Amado, A., Gordo, I., and Bank, C. (2023). Epistasis decreases with increasing antibiotic pressure but not temperature. Philosophical Trans. R. Soc. B Biol. Sci. 378, 20220058. doi:10.1098/rstb.2022.0058
Gjuvsland, A. B., Wang, Y., Plahte, E., and Omholt, S. W. (2013). Monotonicity is a key feature of genotype-phenotype maps. Front. Genet. 4, 216. doi:10.3389/fgene.2013.00216
Goldringer, I., Brabant, P., and Gallais, A. (1997). Estimation of additive and epistatic genetic variances for agronomic traits in a population of doubled-haploid lines of wheat. Heredity 79, 1–11. doi:10.1038/sj.hdy.6881860
Johnson, M. S., Reddy, G., and Desai, M. M. (2023). Epistasis and evolution: recent advances and an outlook for prediction. BMC Biol. 21, 120. doi:10.1186/s12915-023-01585-3
Jucovic, M., and Hartley, R. W. (1996). Protein-protein interaction: a genetic selection for compensating mutations at the barnase-barstar interface. Proc. Natl. Acad. Sci. U.S.A. 93, 2343–2347. doi:10.1073/pnas.93.6.2343
Kang, C., and Avery, L. (2008). To be or not to be, the level of autophagy is the question: dual roles of autophagy in the survival response to starvation. Autophagy 4, 82–84. doi:10.4161/auto.5154
Kemble, H., Eisenhauer, C., Couce, A., Chapron, A., Magnan, M., Gautier, G., et al. (2020). Flux, toxicity, and expression costs generate complex genetic interactions in a metabolic pathway. Sci. Adv. 6, eabb2236. doi:10.1126/sciadv.abb2236
Keren, L., Hausser, J., Lotan-Pompan, M., Vainberg Slutskin, I., Alisar, H., Kaminski, S., et al. (2016). Massively parallel interrogation of the effects of gene expression levels on fitness. Cell 166, 1282–1294. doi:10.1016/j.cell.2016.07.024
Lalić, J., and Elena, S. F. (2012). Magnitude and sign epistasis among deleterious mutations in a positive-sense plant RNA virus. Heredity 109, 71–77. doi:10.1038/hdy.2012.15
Li, X., and Lehner, B. (2020). Biophysical ambiguities prevent accurate genetic prediction. Nat. Commun. 11, 4923. doi:10.1038/s41467-020-18694-0
Maisnier-Patin, S., Berg, O. G., Liljas, L., and Andersson, D. I. (2002). Compensatory adaptation to the deleterious effect of antibiotic resistance in Salmonella typhimurium. Mol. Microbiol. 46, 355–366. doi:10.1046/j.1365-2958.2002.03173.x
Mammano, F., Trouplin, V., Zennou, V., and Clavel, F. (2000). Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74, 8524–8531. doi:10.1128/JVI.74.18.8524-8531.2000
Martinez-Picado, J., and Martínez, M. A. (2008). HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: a view from the clinic and ex vivo. Virus Res. Retroviral Reverse Transcr. 134, 104–123. doi:10.1016/j.virusres.2007.12.021
Miton, C. M., Buda, K., and Tokuriki, N. (2021). Epistasis and intramolecular networks in protein evolution. Curr. Opin. Struct. Biol. Eng. Des. ● Membr. 69, 160–168. doi:10.1016/j.sbi.2021.04.007
Miton, C. M., and Tokuriki, N. (2016). How mutational epistasis impairs predictability in protein evolution and design. Protein Sci. 25, 1260–1272. doi:10.1002/pro.2876
Molla, A., Korneyeva, M., Gao, Q., Vasavanonda, S., Schipper, P. J., Mo, H.-M., et al. (1996). Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2, 760–766. doi:10.1038/nm0796-760
Nghe, P., Kogenaru, M., and Tans, S. J. (2018). Sign epistasis caused by hierarchy within signalling cascades. Nat. Commun. 9, 1451. doi:10.1038/s41467-018-03644-8
Ono, J., Gerstein, A. C., and Otto, S. P. (2017). Widespread genetic incompatibilities between first-step mutations during parallel adaptation of Saccharomyces cerevisiae to a common environment. PLOS Biol. 15, e1002591. doi:10.1371/journal.pbio.1002591
Orr, H. A. (1995). The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139, 1805–1813. doi:10.1093/genetics/139.4.1805
Orr, H. A. (1996). Dobzhansky, Bateson, and the genetics of speciation. Genetics 144, 1331–1335. doi:10.1093/genetics/144.4.1331
Phillips, P. C. (2008). Epistasis — the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9, 855–867. doi:10.1038/nrg2452
Podgornaia, A. I., and Laub, M. T. (2015). Pervasive degeneracy and epistasis in a protein-protein interface. Science 347, 673–677. doi:10.1126/science.1257360
Poelwijk, F. J., Kiviet, D. J., Weinreich, D. M., and Tans, S. J. (2007). Empirical fitness landscapes reveal accessible evolutionary paths. Nature 445, 383–386. doi:10.1038/nature05451
Poelwijk, F. J., Tănase-Nicola, S., Kiviet, D. J., and Tans, S. J. (2011). Reciprocal sign epistasis is a necessary condition for multi-peaked fitness landscapes. J. Theor. Biol. 272, 141–144. doi:10.1016/j.jtbi.2010.12.015
Presgraves, D. C. (2010). The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11, 175–180. doi:10.1038/nrg2718
Schrag, S. J., Perrot, V., and Levin, B. R. (1997). Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc. R. Soc. Lond. Ser. B Biol. Sci. 264, 1287–1291. doi:10.1098/rspb.1997.0178
Silva, R. F., Mendonça, S. C. M., Carvalho, L. M., Reis, A. M., Gordo, I., Trindade, S., et al. (2011). Pervasive sign epistasis between conjugative plasmids and drug-resistance chromosomal mutations. PLOS Genet. 7, e1002181. doi:10.1371/journal.pgen.1002181
Starr, T. N., Greaney, A. J., Hannon, W. W., Loes, A. N., Hauser, K., Dillen, J. R., et al. (2022). Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution. Science 377, 420–424. doi:10.1126/science.abo7896
Vagne, C., David, J., Tavaud, M., and Fontez, B. (2015). Reciprocal sign epistasis and truncation selection: when is recombination favorable in a pre-breeding program with a selfing species? J. Theor. Biol. 386, 44–54. doi:10.1016/j.jtbi.2015.08.013
Weinreich, D. M., Delaney, N. F., Depristo, M. A., and Hartl, D. L. (2006). Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114. doi:10.1126/science.1123539
Weinreich, D. M., Watson, R. A., and Chao, L. (2005). Perspective:Sign epistasis and genetic constraint on evolutionary trajectories. Evol 59, 1165–1174. doi:10.1554/04-272
Wonderlick, D. R., Widom, J. R., and Harms, M. J. (2023). Disentangling contact and ensemble epistasis in a riboswitch. Biophysical J. 122, 1600–1612. doi:10.1016/j.bpj.2023.01.033
Keywords: epistasis, sign epistasis, genetics, genetic mutations, genotype-phenotype mapping
Citation: Zhang J, Chen F and Li X (2024) Mechanistic causes of sign epistasis and its applications. Front. Genet. 15:1366917. doi: 10.3389/fgene.2024.1366917
Received: 07 January 2024; Accepted: 08 February 2024;
Published: 28 February 2024.
Edited by:
Lukasz Jaroszewski, University of California, Riverside, United StatesReviewed by:
Xinru Qiu, University of California, Riverside, United StatesCopyright © 2024 Zhang, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianghua Li, djF4bGkyMjZAZXhzZWVkLmVkLmFjLnVr
 Jinqiu Zhang
Jinqiu Zhang Feiyu Chen1,2
Feiyu Chen1,2 Xianghua Li
Xianghua Li