- The Quzhou Affiliated Hospital of Wenzhou Medical University, Quzhou People’s Hospital, Quzhou, Zhejiang, China
Objective: Recent studies have revealed increasing evidence that the long non-coding RNA bladder cancer associated transcript 1 (LncRNA BLACAT1) plays an essential role in the emergence of different malignancies. This meta-analysis aimed to evaluate the prognostic significance of LncRNA BLACAT1 in various cancers.
Methods: Six electronic databases (PubMed, Embase, Medline, Web of Science, China National Knowledge Infrastructure (CNKI), and the Chinese WanFang database) were comprehensively searched for relevant studies. The analysis of overall survival (OS) and clinicopathological characteristics was conducted.
Results: Nineteen studies with 1,559 patients were eventually eligible to be included in this meta-analysis. High expression level of LncRNA BLACAT1 was identified to be linked with shorter OS (HR: 2.02, 95% CI: 1.66–2.46, p < 0.001) and PFS (HR: 2.424, 95% CI: 1.827–3.020, p < 0.001) in cancer patients as opposed to low expression levels. Subgroup analysis showed that analysis model (multivariate or univariate), cut-off value (mean or median), sample size (more or fewer than 100), and cancer type had little effect on OS in multiple tumors. Moreover, high LncRNA BLACAT1 expression was associated with positive lymph node metastasis (HR: 2.29, 95% CI: 1.66–3.16, p < 0.00001), advanced clinical stage (HR: 2.29, 95% CI: 1.65–3.19, p < 0.00001) and worse differentiation status (HR: 0.58, 95% CI: 0.37–0.92, p = 0.02), compared to low LncRNA BLACAT1 expression.
Conclusion: The findings highlight that high LncRNA BLACAT1 expression might be detrimental and induce a worse prognosis for cancer patients.
Introduction
Cancer is a major threat to human health worldwide and brings a heavy economic burden to the world every year. According to Cancer Statistics, approximately 28.4 million cases will be diagnosed in 2040 (an increase of 47% compared to 2020) (Sung et al., 2021). In the United States, approximately 1,918,030 cancer cases were diagnosed, and 609,360 cancer-related deaths were reported, according to the American Cancer Society (Siegel et al., 2022). Despite tremendous progress in the diagnosis and treatment of cancer in recent years, the overall survival rate of cancer patients has not improved significantly. The lack of effective biomarkers to predict prognosis is considered an important reason for poor prognosis in cancer patients.
Long noncoding RNAs (LncRNA) is a type of non-coding RNAs composed of more than 200 nucleic acids (Chen, 2016), that play a crucial role in regulating gene expression at various points in the transcription/translation process. Numerous studies have found that lncRNA plays is essential for the pathogenesis and prognosis of malignancies as well as several biological processes, including the regulation of the proliferation, invasion, and metastasis of cancer (Schmitz et al., 2016). Thus, lncRNA can be used as biomarkers and therapeutic targets for cancer.
Long noncoding RNA bladder cancer associated transcript 1 (LncRNA BLACAT1), also known as linc-UBC1, which was first reported to be located at human chromosome 1q32.1, is 2616 bp in length (He et al., 2013). Many emerging studies demonstrated that BLACAT1 was aberrantly expressed in different cancers, including gastric cancer, glioma, lung cancer, and cervical cancer (Huang et al., 2019; Liu et al., 2019; Ju et al., 2020; Wang et al., 2020). Although LncRNA BLACAT1 expression was associated with poor overall survival and clinicopathological features (Chen et al., 2018; Cheng et al., 2020), the significance of these associations has not been adequately estimated due to the limitation of sample sizes from individual studies. Therefore, we conducted this meta-analysis to explore the prognostic value of LncRNA BLACAT1 in cancer patients.
Methods
Search strategy
Potentially literature was searched through PubMed, Embase, Web of Science, Cochrane Library, China National Knowledge Infrastructure (CNKI), and WanFang database. The last literature search was conducted on 5 March 2024. The search terms were as follows: (“long noncoding RNA BLACAT1” OR “lncRNA BLACAT1” OR “BLACAT1” OR “bladder cancer associated transcript 1” OR “linc-UBC1”) AND (“cancer” OR “tumor” OR “carcinoma” OR “neoplasm” OR “neoplasia”). The detailed search strategy was described in Supplementary Table S1. The references of included studies will also be checked to avoid potential omissions.
Inclusion and exclusion criteria
The inclusion criteria were as follows: 1) retrospective or prospective cohort studies; 2) LncRNA BLACAT1 expression was identified in cancerous tissues and corresponding non-cancerous tissues through qRT-PCR; 3) patients were divided into high LncRNA BLACAT1 expression group and low LncRNA BLACAT1 expression group; 4) reported sufficient data about the relationship between LncRNA BLACAT1 expression level and overall survival (OS), disease-free survival (DFS), or clinicopathologic parameters; 5) reported sufficient data for the assessment of hazard ratios (HRs) and 95% confidence intervals (CIs). The exclusion criteria were as follows: 1) duplicate articles; 2) reviews, letters, conference reports, and animal studies; 3) articles with insufficient data (e.g., without OS, DFS, or clinicopathologic parameters).
Data extraction and quality assessment
Two researchers searched and evaluated the studies independently according to the inclusion criteria. The following items were extracted: first author, publication year, country or region, cancer type, number of cases, detection method, cut-off value, follow-up months, and analysis method. In addition, clinicopathological parameters including age, gender, tumor size, lymph node metastasis, clinical stage, and distant metastasis were also extracted. The prognostic endpoints included overall survival (OS), progression-free survival (PFS), recurrence-free survival (RFS), and disease-free survival (DFS). HRs and 95%CIs were directly obtained from the univariate or multivariate analysis. If HRs and 95% CIs were not provided, Engauge Digitizer V4.1 software was used to extract HR and 95% CI. The quality of the studies was assessed using the Newcastle–Ottawa Scale (NOS) criteria. Articles with a NOS score ≥6 were considered high-quality; otherwise, they were regarded as low-quality (Stang, 2010). Any disagreements between the two reviewers were resolved by discussion and consensus.
Statistical analysis
All statistical analyses were performed using Review Manager version 5.4 software (RevMan, The Cochrane Collaboration, Oxford, UK) and Stata 16 software (StataCorp, College Station, TX, USA). Pooled HRs and 95% CIs were used to assess the association between LncRNA BLACAT1 expression and the clinical prognosis of the cancer patients. The inter-study heterogeneity was assessed by I2 statistics. If I2 ≤ 50% indicated there is no obvious heterogeneity among included studies, a fixed-effects model was used. Otherwise, the random-effects model was used. Sensitivity analysis was also applied to assess the stability of the combined results and to determine the source of any heterogeneity. Egger’s funnel regression test and Begg’s funnel plot were performed to evaluate the publication bias. A two-tailed p-value < 0.05 indicated statistically significant.
Results
Study search
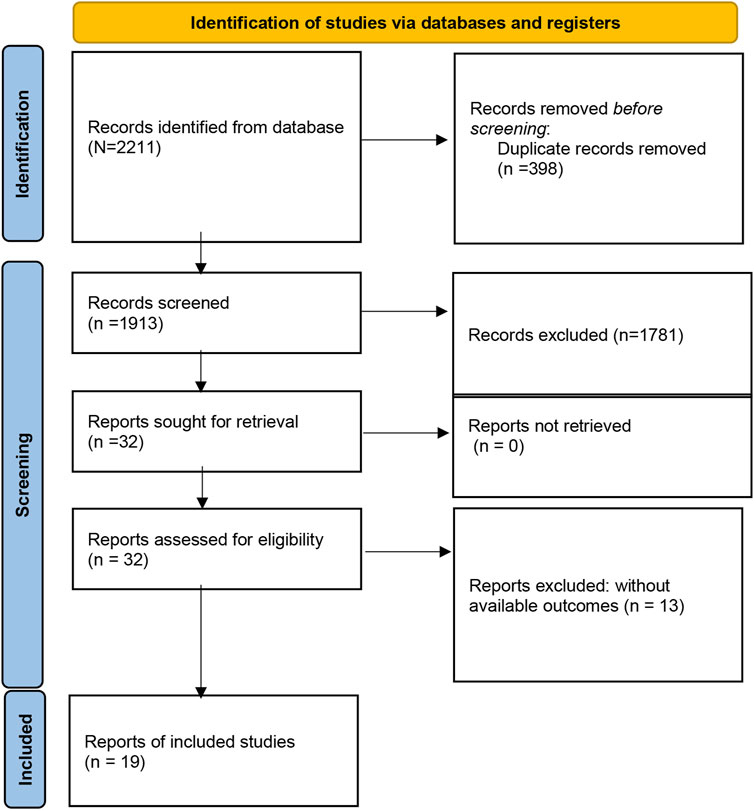
A total of 2211 studies were preliminarily retrieved by searching the PubMed, Embase, Medline, Web of Science, CNKI, and Wanfang databases. 298 articles were excluded for duplicates. Of the remaining 1813 papers, 1981 were directly excluded by scanning the titles or abstracts. Then, 32 potentially relevant articles were selected for full-text review, and 13 articles were removed because of inefficient data to estimate HR for further analysis. Finally, nineteen studies comprising 1,559 patients were eligible to be included in this meta-analysis (Figure 1).
Characteristics of included studies
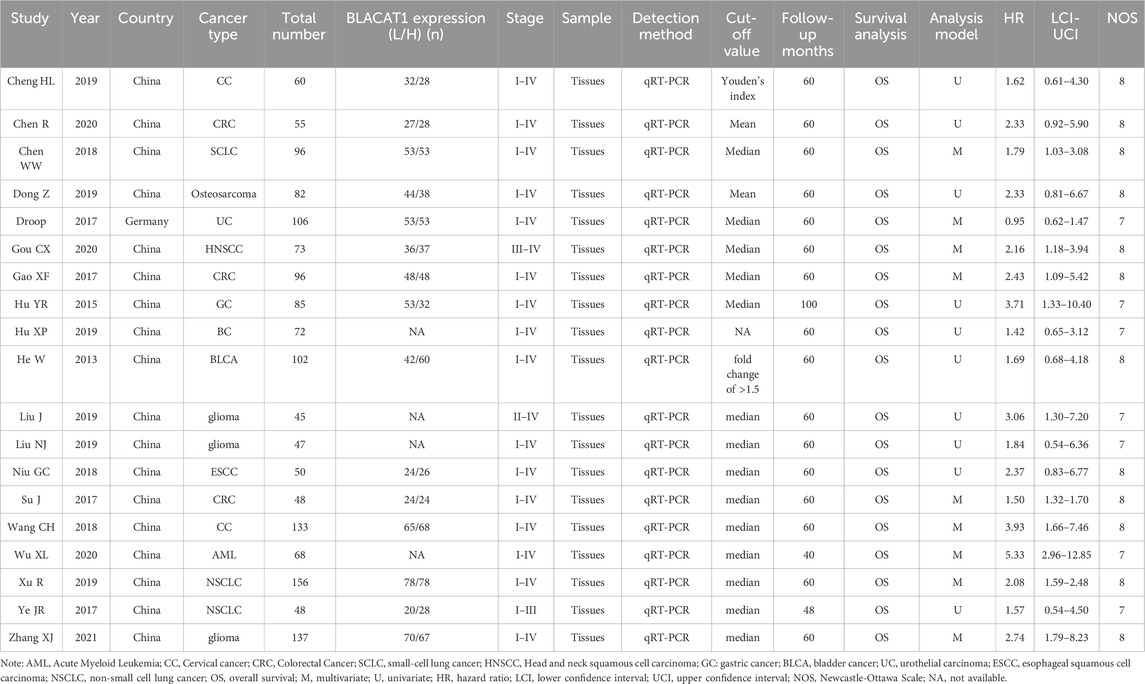
Detailed characteristics of the included studies were listed in Table 1. Eighteen studies were conducted in China, but one in Germany. The included studies had a wide range of sample sizes, ranging from 45 to 156. The following cancers were investigated: acute myeloid leukemia (AML) (Wu et al., 2020), colorectal cancer (CRC) (Gao et al., 2017; Chen et al., 2020), small-cell lung cancer (SCLC) (Chen et al., 2018), head and neck squamous cell carcinoma (HNSCC) (Gou et al., 2020), gastric cancer (GC) (Hu et al., 2015), osteosarcoma (Dong and Wang, 2019), glioma (Liu et al., 2019; Zhang et al., 2021), breast cancer (BC) (Hu et al., 2019), bladder cancer (BLCA) (He et al., 2013), urothelial carcinoma (UC) (Droop et al., 2017), esophageal squamous cell carcinoma (ESCC) (Niu et al., 2018), cervical cancer (CC) (Wang et al., 2018; Cheng et al., 2020) and non-small cell lung cancer (NSCLC) (Ye et al., 2017; Xu et al., 2019). The LncRNA BLACAT1 expression was detected by qRT-PCR in all studies. The cut-off value for calculating LncRNA BLACAT1 expression was the mean, median, fold change, or Youden’s index, and one study gave no details about the cut-off value. Additionally, all included studies got NOS scores of 7 or more, illustrating their high methodological quality.
The relationship between the LncRNA BLACAT1 expression and survival outcomes
Nine studies reported the OS assessed by multivariate analysis and ten studies reported the OS in the form of a survival curve, therefore, nineteen studies were finally included in the meta-analysis of OS. As shown in Figure 2, due to obvious heterogeneity among included studies (I2 = 50.4%, p = 0.006), a random-effects model was adopted. The pooled results showed that high LncRNA BLACAT1 expression was significantly correlated with inferior OS (HR: 2.02, 95% CI: 1.66–2.46, p < 0.0001), which means the cancer patients with low BLACAT1 expression had a better prognosis. Moreover, two studies reported PFS data. As shown in Figure 3, the pooled results indicated that high LncRNA BLACAT1 expression was related to poor PFS (HR: 2.424, 95% CI: 1.827–3.020, p < 0.0001, I2 = 13.9%).
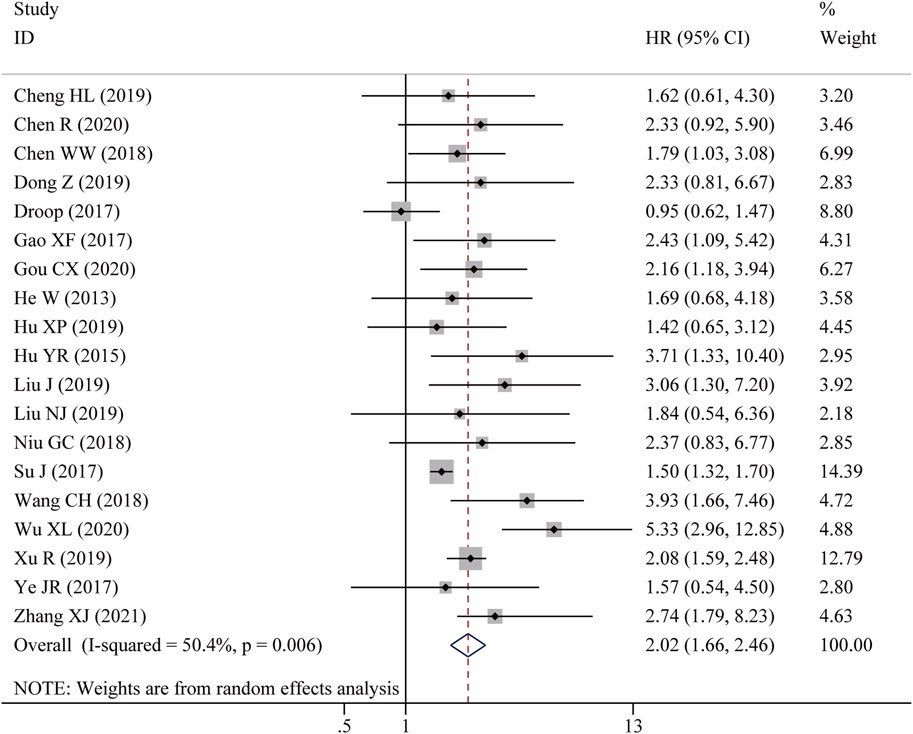
Figure 2. Forest plot shows the association between the LncRNA BLACAT1 expression and overall survival (OS). The rhombus represents the pooled effect, the center of the rhombus represents the pooled effect size, and the width of the rhombus represents the confidence interval. The vertical line represents HR = 1, and any confidence interval crossing this vertical line represents non-statistically significant.

Figure 3. Forest plot shows the association between the LncRNA BLACAT1 expression and progression-free survival (PFS). The rhombus represents the pooled effect, the center of the rhombus represents the pooled effect size, and the width of the rhombus represents the confidence interval. The vertical line represents HR = 1, and any confidence interval crossing this vertical line represents non-statistically significant.
As listed in Table 2, subgroup analyses stratified by survival analysis method, cut-off value, sample size, and cancer type were also performed. For the survival analysis method, the HRs of nine studies were calculated using multivariate analysis, and the subgroup analysis showed that LncRNA BLACAT1 expression was significantly associated with the OS (p < 0.0001), Similarly, high LncRNA BLACAT1 expression was an unfavorable independent prognostic factor for OS based on univariate analysis (p < 0.0001). After stratifying based on cut-off values, the results indicated that no matter how to cut off the expression, there was a distinct association between LncRNA BLACAT1 expression and OS in cancer patients (p < 0.0001). After stratifying based on sample size, LncRNA BLACAT1 was found to be a poor prognostic factor in studies with a sample size less than 100 (HR: 1.55, 95% CI: 1.37–1.73) or more than 100 (HR: 1.80, 95 CI: 0.92–2.68). In addition, subgroup analyses based on cancer type were also carried out, and results revealed that LncRNA BLACAT1 expression was significantly associated with poor OS in patients with specific cancers, including colorectal cancer (HR: 1.51, 95% CI: 1.32–1.70, p < 0.0001) and lung cancer (HR: 2.02, 95% CI: 1.62–2.42, p < 0.0001).
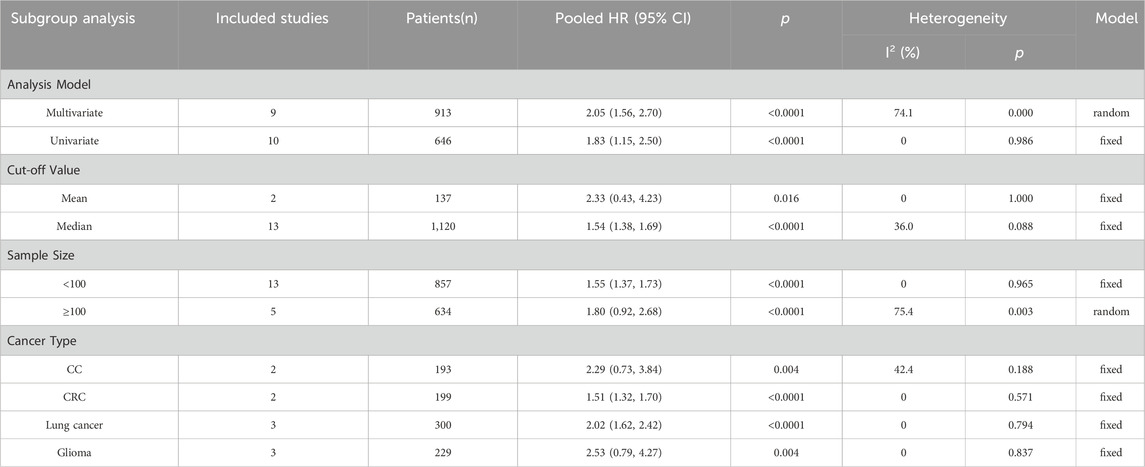
Table 2. Stratified analyses of the pooled HRs of overall survival by analysis model, cut-off value, sample size, and cancer type.
Relationship between LncRNA BLACAT1 overexpression and clinicopathological characteristics
To further elucidate the clinical implications of LncRNA BLACAT1 overexpression, we examined the association between LncRNA BLACAT1 expression and clinicopathological characteristics (Table 3; Figure 4). The results showed that high LncRNA BLACAT1 expression was significantly associated with positive lymph node metastasis (p < 0.00001), advanced clinical stage (p < 0.00001) and poor differentiation status (p = 0.02). However, there was no discrepancy in age (p = 0.30), gender (p = 0.74), smoking (p = 0.37), tumor size (p = 0.41) and distant metastasis (p = 0.07).
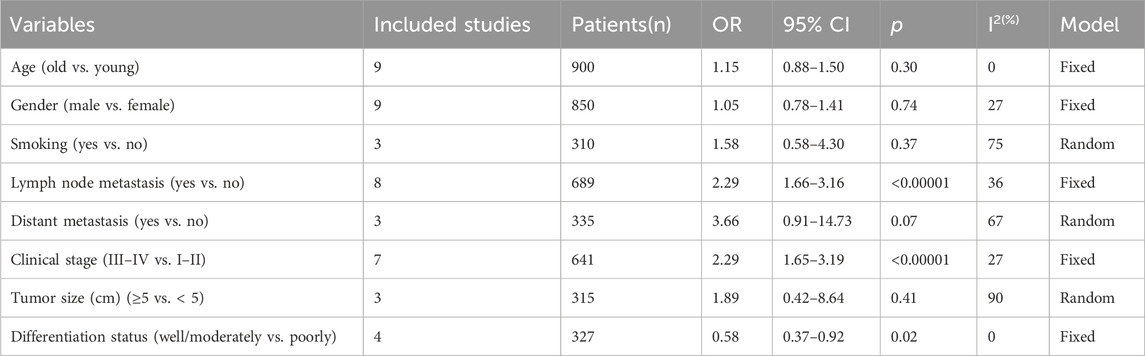
Table 3. Correlation between LncRNA BLACAT1 expression and clinicopathological characteristics of solid tumors.
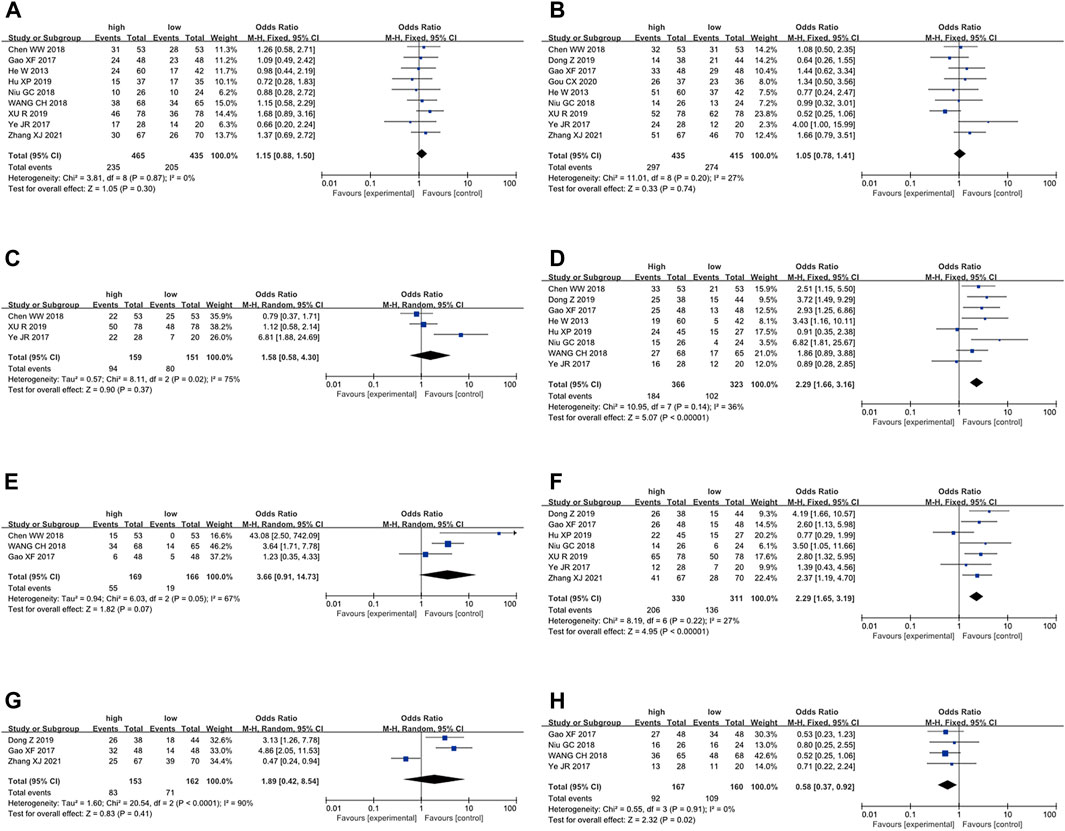
Figure 4. Forest plot shows the association between the LncRNA BLACAT1 expression and clinicopathological characteristics of solid tumors: (A) age; (B) gender; (C) smoking; (D) lymph node metastasis; (E) distant metastasis; (F) clinical stage; (G) tumor size; (H) differentiation status. The rhombus represents the pooled effect, the center of the rhombus represents the pooled effect size, and the width of the rhombus represents the confidence interval. The vertical line represents HR = 1, and any confidence interval crossing this vertical line represents non-statistically significant.
Publication bias
Begg’s funnel plot and Egger’s test were utilized to evaluate publication bias in the current meta-analysis. The result of Begg’s test showed the absence of significant publication bias. The shape of the funnel plot was also symmetrically inverted funnels (Figure 5). Besides, funnel plots demonstrated that there was no obvious publication bias in clinicopathological characteristics (Figure 6).
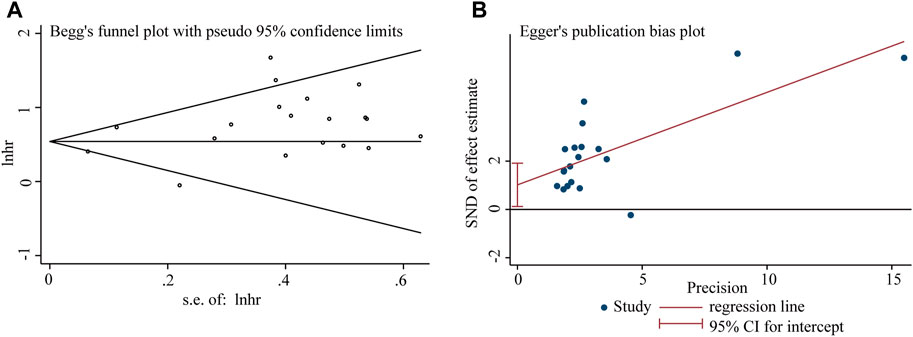
Figure 5. Publication bias analysis of overall survival (OS) data: (A) Begg’s funnel plot of overall survival (OS) in the published studies. (B) Egger’s publication bias plot of OS in the published studies. The X-axis represents the standard error, which is used to measure accuracy. The higher the standard error, the less accurate the test. HR is plotted on the Y-axis. The black dots indicate the impact of LncRNA BLACAT1 on the outcome.
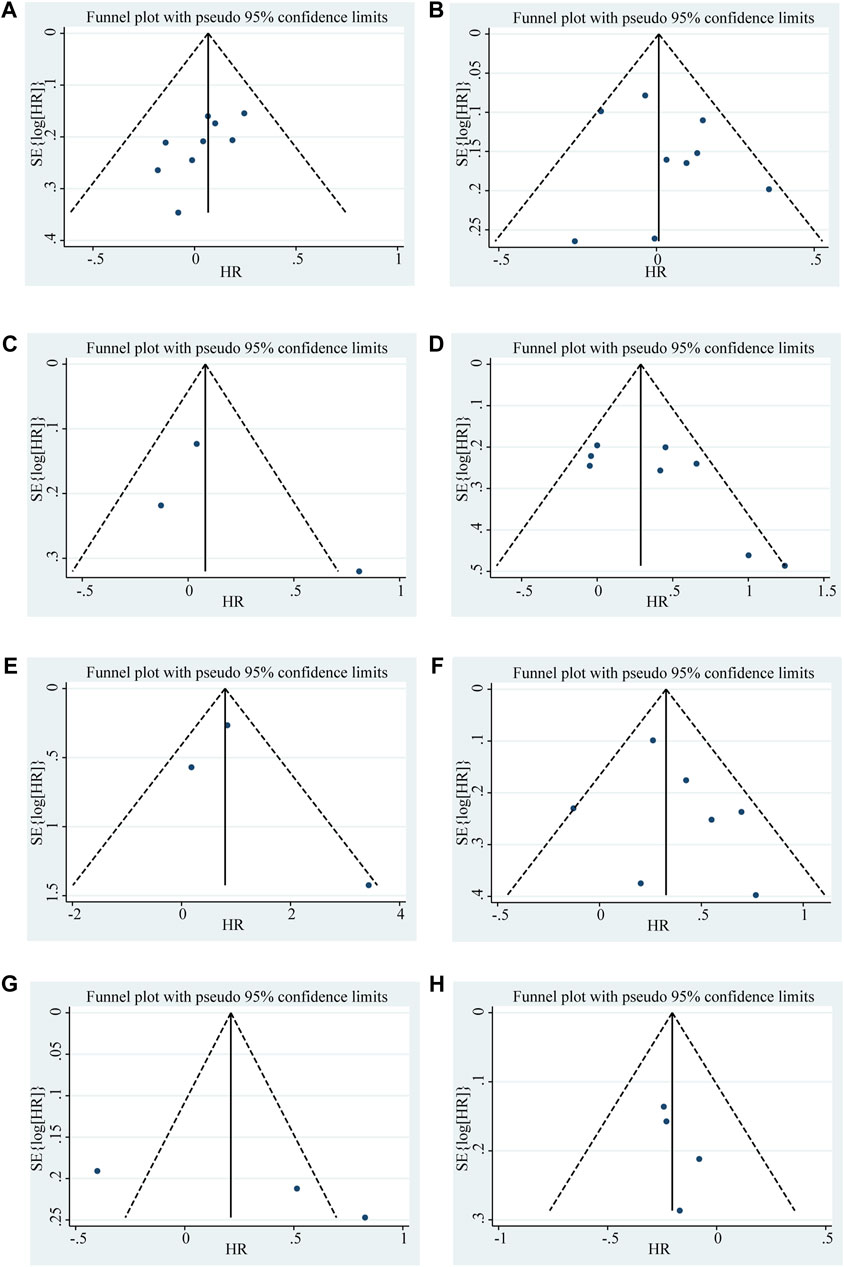
Figure 6. Funnel plots shows the association between the LncRNA BLACAT1 expression and clinicopathological characteristics: (A) age; (B) gender; (C) smoking; (D) lymph node metastasis; (E) distant metastasis; (F) clinical stage; (G) tumor size; (H) differentiation status. The Y-axis represents the standard error, which is used to measure accuracy. The higher the standard error, the less accurate the test. HR is plotted on the X-axis. The black dots indicate the impact of LncRNA BLACAT1 on the outcome.
Sensitivity analysis
Sensitivity analysis was conducted by excluding one study every time which indicated that the pooled HRs were not significantly affected by the removal of any single study (Figure 7).
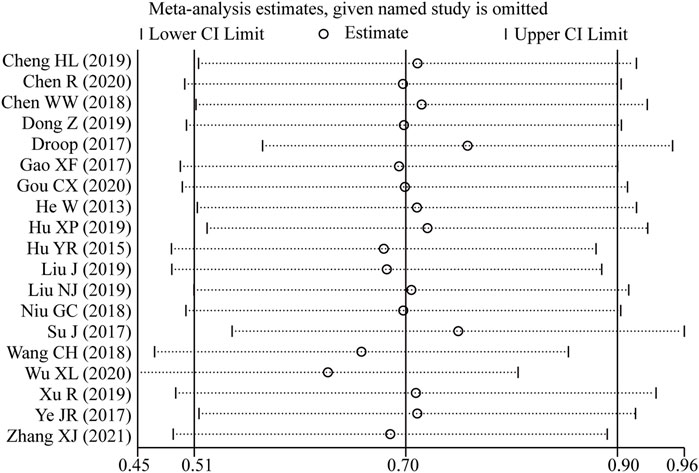
Figure 7. Sensitivity analysis of the pooled HRs of LncRNA BLACAT1 expression and overall survival (OS).
Discussion
Cancer remains a major issue in healthcare and poses significant threats to human health (Siegel et al., 2019). Cancer metastasis, the process involving the dissemination of cancer cells from a primary lesion to distal organs, is the major cause of treatment failure and mortality in individuals with malignant tumors. It is considered as a multi-scale phenomenon, including molecular signaling networks, protein-protein interactions, metabolism, and cell-ECM interactions (Jia et al., 2022). Moreover, cancer lack of effective prognostic, metastasis, and diagnosis biomarkers, is primarily due to tumor heterogeneity (Khatib et al., 2020). Therefore, cancer metastasis has continued to confound researchers due to its complex, and not entirely predictable, pathological trajectory (Dudjak, 1992; Suhail et al., 2019). Currently, the precise metastasis mechanisms of human cancer are still unclear. Increasing evidence suggests that LncRNAs play an essential role in all stages of carcinogenesis and metastasis (Weidle et al., 2017), thus, it is considered as one of the most valuable biological markers (Soudyab et al., 2016; Deng et al., 2017; Aalijahan and Ghorbian, 2019; Galamb et al., 2019). LncRNA BLACAT1 has been demonstrated to be significantly upregulated in various cancers, including gastric cancer, cervical cancer (CC), breast cancer, and bladder cancer, etc. (Wang et al., 2018; Wu et al., 2018; Hu et al., 2019; Chen et al., 2020; Wang et al., 2020). The aberrant expression of LncRNA BLACAT1 resulted in a variety of oncogenic processes, including apoptosis, proliferation, cell cycle arrest, migration, etc. Thus, LncRNA BLACAT1 serves as an oncogene and plays critical roles in various biological and pathological processes. Wang et al. identified that the knock-down of LncRNA BLACAT1 significantly suppressed CC cell proliferation, migration, and invasion, while high LncRNA BLACAT1 expression served as a poor prognostic factor for CC patients (Wang et al., 2018). Another study indicated that LncRNA BLACAT1 overexpression could promote cervical cancer by downregulating miR-424 and miR-143 (Cheng et al., 2020). As for ovarian cancer, LncRNA BLACAT1 knockdown suppressed proliferation, migration, and invasion by suppressing the Wnt/β-catenin signaling in ovarian cancer cells (Yang et al., 2020). In colorectal cancer, high LncRNA BLACAT1 expression contributed to the poor overall survival rate and tumor progression by miR-519d-3p/CREB1 axis (Chen et al., 2020). These studies demonstrated that BLACAT1 plays a crucial role in tumor development and progression, and inspired us to investigate the correlation between LncRNA BLACAT1 expression level and cancer prognosis. In this meta-analysis, we comprehensively evaluate the relationship between BLACAT1 expression and prognosis and clinicopathological characteristics of tumors. The results illustrated that increased LncRNA BLACAT1 expression was significantly associated with unfavorable OS (HR: 2.02, 95% CI: 1.66–2.46) and PFS (HR: 2.424, 95% CI: 1.827–3.020), and similar results were obtained in further subgroup analysis. In addition, our results indicated that elevated level of LncRNA BLACAT1 was significantly related to positive lymph node metastasis, advanced clinical stage (p < 0.00001), and poor differentiation status, while no significant correlations were observed in age, gender, smoking, tumor size and distant metastasis. Furthermore, funnel plot and sensitivity analysis results revealed that our conclusion was reliable and robust. Taken together, these results demonstrated that LncRNA BLACAT1 could be applied as a novel predictor for prognosis in various tumors.
Nevertheless, several limitations in this study should be noted. First, the number of included studies and sample sizes were relatively small, especially for specific types of cancer, which might reduce the stringency of our conclusion. Second, there was no consensus on the cut-off value to define high lncRNA BLACAT1 expression and low lncRNA BLACAT1 expression. The included patients were divided into two groups based on the mean or median of BLACAT1 expression. Third, HRs with 95% CIs were extracted indirectly from survival curves rather than being directly obtained from the original studies, which might inevitably cause possible deviations; Fourth, most of the studies were from China, which limits the generalizability of our results. Thus, our results may only apply to the Chinese population. Despite some limitations mentioned above, our study confirmed the unfavorable role of elevated LncRNA BLACAT1 expression in malignant tumors and inspired researchers to further investigate the underlying mechanisms.
The use of biomarkers to predict therapeutic response is fundamental in precision medicine, thus more high-quality multicenter studies are required to further clarify the significance of LncRNA BLACAT1 in future cancer prognosis. There are several points to consider in this respect. First, the cut-off value should be demonstrated and unified to divide the high/low expression of biomarkers. Second, HRs with 95% CIs should be presented in the form of specific figures to avoid possible deviations. Third, further large-scale studies are needed in different populations. Fourth, the effect of covariates (age, gender, histological type, TNM stage, smoking, and so on) on outcomes should be considered.
In this study, we concluded that patients with high LncRNA BLACAT1 expression had shorter OS, suggesting that high levels of the LncRNA BLACAT1 may be detrimental to cancer patients. Moreover, when compared to patients with low expression, those with high LncRNA BLACAT1 expression tended to have an advanced clinical stage, earlier lymph node metastasis, and poorer differentiation status. All these results suggested that elevated LncRNA BLACAT1 expression may act as an oncogene factor and induce a worse prognosis in various cancers.
Conclusion
In summary, this study suggested that elevated LncRNA BLACAT1 expression was negatively associated with OS, PFS, tumor differentiation, clinical stage, and lymph node metastasis. Therefore, LncRNA BLACAT1 may serve as a promising prognostic biomarker for cancer. Considering the limitations of this study, large-scale and multicenter studies will be needed to confirm these results and validate their clinical significance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
XY: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. NZ: Data curation, Formal Analysis, Investigation, Software, Writing–original draft. GW: Data curation, Formal Analysis, Writing–original draft. JW: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1362420/full#supplementary-material
References
Aalijahan, H., and Ghorbian, S. (2019). Long non-coding RNAs and cervical cancer. Exp. Mol. Pathol. 106, 7–16. doi:10.1016/j.yexmp.2018.11.010
Chen, L. L. (2016). Linking long noncoding RNA localization and function. Trends Biochem. Sci. 41 (9), 761–772. doi:10.1016/j.tibs.2016.07.003
Chen, R., Zhou, S., Chen, J., Lin, S., Ye, F., and Jiang, P. (2020). LncRNA BLACAT1/miR-519d-3p/CREB1 Axis mediates proliferation, apoptosis, migration, invasion, and drug-resistance in colorectal cancer progression. Cancer Manag. Res. 12, 13137–13148. doi:10.2147/CMAR.S274447
Chen, W., Hang, Y., Xu, W., Wu, J., Chen, L., Chen, J., et al. (2018). BLACAT1 predicts poor prognosis and serves as oncogenic lncRNA in small-cell lung cancer. J. Cell Biochem. 120, 2540–2546. doi:10.1002/jcb.27548
Cheng, H., Tian, J., Wang, C., Ren, L., and Wang, N. (2020). LncRNA BLACAT1 is upregulated in cervical squamous cell carcinoma (CSCC) and predicts poor survival. Reprod. Sci. 27 (2), 585–591. doi:10.1007/s43032-019-00058-9
Deng, H., Wang, J. M., Li, M., Tang, R., Tang, K., Su, Y., et al. (2017). Long non-coding RNAs: new biomarkers for prognosis and diagnosis of colon cancer. Tumour Biol. 39 (6), 1010428317706332. doi:10.1177/1010428317706332
Dong, Z., and Wang, Y. (2019). LncRNA BLACAT1 accelerates the proliferation and migration of osteosarcoma cells through regulating STAT3. Pathol. Res. Pract. 215 (3), 571–579. doi:10.1016/j.prp.2019.01.017
Droop, J., Szarvas, T., Schulz, W. A., Niedworok, C., Niegisch, G., Scheckenbach, K., et al. (2017). Diagnostic and prognostic value of long noncoding RNAs as biomarkers in urothelial carcinoma. PLoS One 12 (4), e0176287. doi:10.1371/journal.pone.0176287
Dudjak, L. A. (1992). Cancer metastasis. Semin. Oncol. Nurs. 8 (1), 40–50. doi:10.1016/0749-2081(92)90007-pp
Galamb, O., Barták, B. K., Kalmár, A., Nagy, Z. B., Szigeti, K. A., Tulassay, Z., et al. (2019). Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J. Gastroenterol. 25 (34), 5026–5048. doi:10.3748/wjg.v25.i34.5026
Gao, X., Wen, J., Gao, P., Zhang, G., and Zhang, G. (2017). Overexpression of the long non-coding RNA, linc-UBC1, is associated with poor prognosis and facilitates cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 10, 1017–1026. doi:10.2147/ott.S129343
Gou, C., Han, P., Li, J., Gao, L., Ji, X., Dong, F., et al. (2020). Knockdown of lncRNA BLACAT1 enhances radiosensitivity of head and neck squamous cell carcinoma cells by regulating PSEN1. Br. J. Radiol. 93 (1108), 20190154. doi:10.1259/bjr.20190154
He, W., Cai, Q., Sun, F., Zhong, G., Wang, P., Liu, H., et al. (2013). linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim. Biophys. Acta 1832 (10), 1528–1537. doi:10.1016/j.bbadis.2013.05.010
Hu, X., Liu, Y., Du, Y., Cheng, T., and Xia, W. (2019). Long non-coding RNA BLACAT1 promotes breast cancer cell proliferation and metastasis by miR-150-5p/CCR2. Cell Biosci. 9, 14. doi:10.1186/s13578-019-0274-2
Hu, Y., Pan, J., Wang, Y., Li, L., and Huang, Y. (2015). Long noncoding RNA linc-UBC1 is negative prognostic factor and exhibits tumor pro-oncogenic activity in gastric cancer. Int. J. Clin. Exp. Pathol. 8 (1), 594–600.
Huang, F. X., Chen, H. J., Zheng, F. X., Gao, Z. Y., Sun, P. F., Peng, Q., et al. (2019). LncRNA BLACAT1 is involved in chemoresistance of non-small cell lung cancer cells by regulating autophagy. Int. J. Oncol. 54 (1), 339–347. doi:10.3892/ijo.2018.4614
Jia, Q., Wang, A., Yuan, Y., Zhu, B., and Long, H. (2022). Heterogeneity of the tumor immune microenvironment and its clinical relevance. Exp. Hematol. Oncol. 11 (1), 24. doi:10.1186/s40164-022-00277-y
Ju, Z. S., Sun, B., Bao, D., and Zhang, X. F. (2020). Effect of lncRNA-BLACAT1 on drug resistance of non-small cell lung cancer cells in DDP chemotherapy by regulating cyclin D1 expression. Eur. Rev. Med. Pharmacol. Sci. 24 (18), 9465–9472. doi:10.26355/eurrev_202009_23031
Khatib, S., Pomyen, Y., Dang, H., and Wang, X. W. (2020). Understanding the cause and consequence of tumor heterogeneity. Trends Cancer 6 (4), 267–271. doi:10.1016/j.trecan.2020.01.010
Liu, N., Hu, G., Wang, H., Wang, Y., and Guo, Z. (2019). LncRNA BLACAT1 regulates VASP expression via binding to miR-605-3p and promotes giloma development. J. Cell Physiol. 234 (12), 22144–22152. doi:10.1002/jcp.28778
Niu, G., Zhuang, H., Li, B., and Cao, G. (2018). Long noncoding RNA linc-UBC1 promotes tumor invasion and metastasis by regulating EZH2 and repressing E-cadherin in esophageal squamous cell carcinoma. J. buon 23 (1), 157–162.
Schmitz, S. U., Grote, P., and Herrmann, B. G. (2016). Mechanisms of long noncoding RNA function in development and disease. Cell Mol. Life Sci. 73 (13), 2491–2509. doi:10.1007/s00018-016-2174-5
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72 (1), 7–33. doi:10.3322/caac.21708
Siegel, R. L., Miller, K. D., and Jemal, A. (2019). Cancer statistics, 2019. CA Cancer J. Clin. 69 (1), 7–34. doi:10.3322/caac.21551
Soudyab, M., Iranpour, M., and Ghafouri-Fard, S. (2016). The role of long non-coding RNAs in breast cancer. Arch. Iran. Med. 19 (7), 508–517.
Stang, A. (2010). Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25 (9), 603–605. doi:10.1007/s10654-010-9491-z
Suhail, Y., Cain, M. P., Vanaja, K., Kurywchak, P. A., Levchenko, A., Kalluri, R., et al. (2019). Systems biology of cancer metastasis. Cell Syst. 9 (2), 109–127. doi:10.1016/j.cels.2019.07.003
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Wang, C. H., Li, Y. H., Tian, H. L., Bao, X. X., and Wang, Z. M. (2018). Long non-coding RNA BLACAT1 promotes cell proliferation, migration and invasion in cervical cancer through activation of Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 22 (10), 3002–3009. doi:10.26355/eurrev_201805_15057
Wang, Z., Liu, X., Liu, X., and Niu, D. (2020). Long non-coding RNA BLACAT1 promotes the tumorigenesis of gastric cancer by sponging microRNA-149-5p and targeting KIF2A. Cancer Manag. Res. 12, 6629–6640. doi:10.2147/cmar.S258178
Weidle, U. H., Birzele, F., Kollmorgen, G., and Rüger, R. (2017). Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics 14 (3), 143–160. doi:10.21873/cgp.20027
Wu, X., Wang, J., Li, D., and Xu, Z. (2020). Expression and diagnostic value of lncRNA BLACAT1 in peripheral blood of patients with acute myeloid leukemia. Clin. Lab. 66 (4). doi:10.7754/Clin.Lab.2019.190802
Wu, X., Zheng, Y., Han, B., and Dong, X. (2018). Long noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin resistance of gastric cancer via sponging miR-361. Biomed. Pharmacother. 99, 832–838. doi:10.1016/j.biopha.2018.01.130
Xu, R., Cao, X. R., Zhang, B. Q., Wang, J. L., Wang, L., and Sun, W. Q. (2019). BLACAT1 is negatively associated with prognosis in patients with NSCLC and inhibits cell progression, metastasis and epithelial-mesenchymal transition through down-regulating Wnt/β-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (14), 6217–6225. doi:10.26355/eurrev_201907_18439
Yang, H., Qi, Y., Wang, X. L., Gu, J. J., and Shi, T. M. (2020). Down-regulation of lncRNA BLACAT1 inhibits ovarian cancer progression by suppressing the Wnt/β-catenin signaling pathway via regulating miR-519d-3p. Mol. Cell Biochem. 467 (1-2), 95–105. doi:10.1007/s11010-020-03704-y
Ye, J. R., Liu, L., and Zheng, F. (2017). Long noncoding RNA bladder cancer associated transcript 1 promotes the proliferation, migration, and invasion of nonsmall cell lung cancer through sponging miR-144. DNA Cell Biol. 36 (10), 845–852. doi:10.1089/dna.2017.3854
Keywords: cancer, long non-coding RNA BLACAT1, meta-analysis, prognosis, survival
Citation: Yan X, Zhang N, Wang G and Wang J (2024) The prognostic significance of LncRNA BLACAT1 overexpression in various tumors: a meta-analysis. Front. Genet. 15:1362420. doi: 10.3389/fgene.2024.1362420
Received: 28 December 2023; Accepted: 15 March 2024;
Published: 27 March 2024.
Edited by:
Sandip Mukherjee, Washington University in St. Louis, United StatesReviewed by:
Aishiki Banerjee, University of Florida, United StatesAnjali Garg, Washington University in St. Louis, United States
Copyright © 2024 Yan, Zhang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiaheng Wang, MTg2NTcwMTU2NjhAMTYzLmNvbQ==
 Xuefen Yan
Xuefen Yan Jiaheng Wang
Jiaheng Wang