
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet. , 04 March 2024
Sec. RNA
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1346119
This article is part of the Research Topic Role of Non-coding RNAs (Emphasis on the Emerging Role of Circular RNAs) In Cancer. Application Potential for Molecular Diagnostics and Therapeutics of Cancer. View all 6 articles
Lung cancer remains a global public health concern with significant research focus on developing better diagnosis/prognosis biomarkers and therapeutical targets. Circular RNAs (circRNAs) are a type of single-stranded RNA molecules that covalently closed and have ubiquitous expression. These molecules have been implicated in a variety of disease mechanisms, including lung cancer, as they exhibit oncogenic or tumor suppressor characteristics. Recent research has shown an important role that circRNAs play at different stages of lung cancer, particularly in lung adenocarcinoma. In this review, we summarize the latest research on circRNAs and their roles within lung cancer diagnosis, as well as on disease mechanisms. We also discuss the knowledge gaps on these topics and possible future research directions.
Lung cancer is the leading cause of death among all cancers types, and remains among the top ten causes of death overall (Siegel et al., 2021). Lung cancer is classified into two main group types, non-small cell lung cancer (NSCLC) and small-cell lung cancer (SCLC), with NSCLC making up approximately 85% of all lung cancer cases (Siegel et al., 2021). The NSCLC group is further subdivided into three subtypes: lung adenocarcinoma (LUAD), squamous cell carcinoma (SCC), and large cell carcinoma (LCC), with LUAD making up most of the NSCLC cases (Siegel et al., 2022). While the latest developments in lung cancer diagnosis and treatment methods have improved the 5-year overall survival in lung cancer patients, there is still a need for identification of early diagnosis biomarkers, as well as more accurate prognostic biomarkers and more efficient therapeutic targets (Ettinger et al., 2021).
Circular RNAs (circRNAs) were discovered in the 1976 in the murine respirovirus (formerly known as Sendai virus) (Kolakofsky, 1976). The first discovery in humans, however, took place a decade later (Kos et al., 1986). Since their initial identification, numerous circRNAs have been detected in various organisms, including viruses, archaea, bacteria, and eukaryotic cells (Miao et al., 2021). Their presence has been linked to developmental phases, physiological states, and various diseases and conditions including cancer and cardiometabolic diseases (Giral et al., 2018; Chen et al., 2019; Di et al., 2019; Arthurs et al., 2022; Chen et al., 2022). This has paved the way for a new area of research focused on uncovering how circRNAs are formed, and their roles as fundamental components of gene expression processes. In the following sections, we summarize and discuss the roles that circRNAs have been shown to play in lung cancer initiation, progression, diagnosis, prognosis and response to therapeutics. We also provide a brief summary on the latest reported circRNAs and the potential roles that they can play in tackling lung cancer as oncogenes or tumor suppressors. Finally, we discuss future perspectives for lung cancer biomarker research including circRNAs.
Circular RNAs are single-stranded, covalently closed RNAs that arise from protein-coding genes. During gene transcription, the RNA polymerase II transcribes the pre-mRNA, which then undergoes splicing at splicing sites known as spliceosomes. In this process, the circRNAs are generated when back-splicing takes place in parallel with canonical splicing, and the splice donor joins the splice acceptor to form a circular shaped RNA (Barrett et al., 2015). For a significant period of time, circRNAs were considered to be products of splicing errors (Nigro et al., 1991). The origin of the circRNAs was thought to be either from the lariat or from back-splicing (Zaphiropoulos, 1996; Jeck et al., 2013), however, a recent study proposed a unified model for circRNA biogenesis that includes intron and exon definition and back-splicing (Li L. et al., 2019) (Figure 1).
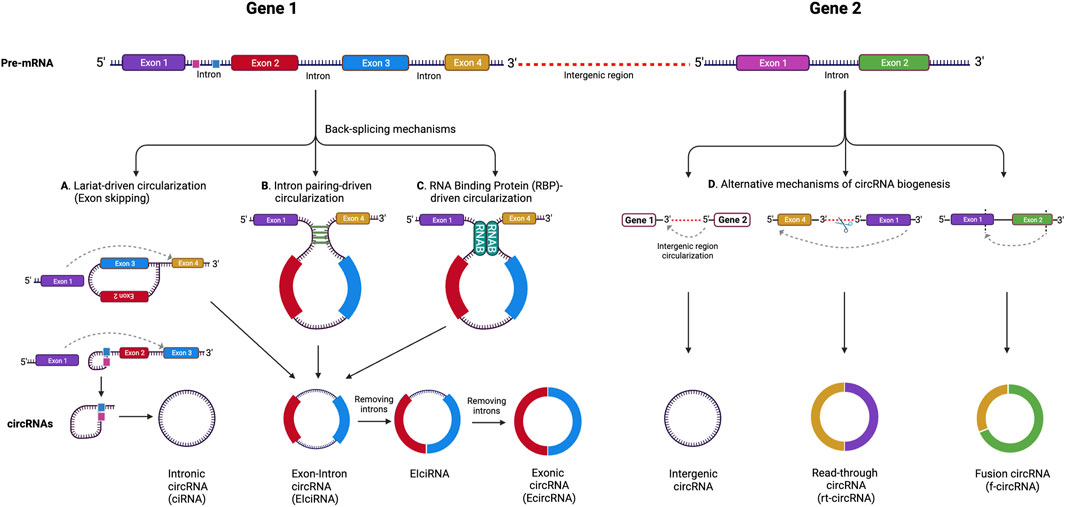
FIGURE 1. CircRNA biogenesis mechanisms. (A) Lariat-driven circularization (exon skipping), (B) intron pairing-driven circularization, (C) RNA binding protein (RBP)-driven circularization. Alternative circRNA biogenesis processes (D) leading to the formation of intergenic circRNA, rt-circRNA and f-circRNA. Adapted from (Pisignano et al., 2023), licensed CC-BY-4.0. Created with BioRender.com.
In addition, while the majority of circRNAs are made of exons from protein-encoding genes, they can also contain introns, intergenic regions, untranslated regions (UTRs), noncoding RNAs (ncRNAs) loci, and antisense locations of known transcripts (Caba et al., 2021) (Figure 1). CircRNA biogenesis can be modulated by RNA-binding proteins (RBPs) through RBP-driven circularization mechanism (Lyu and Huang, 2017; Zang et al., 2020). CircRNA generation can be facilitated via RBP binding to the introns close to the splicing sites (Conn et al., 2015). Some circRNAs can regulate their own expression post-transcriptionally by acting as a RBP sponge. For example, circMbl mediates its expression in a negative feedback loop manner by alternatively splicing its precursor RNA. (Begemann et al., 1997). RBPs are involved in circRNA splicing, folding, processing, localization, and stabilization through interaction with circRNA junctions (Janas et al., 2015). Figure 2 provides a summary of the known biological functions of circRNAs (Pisignano et al., 2023).
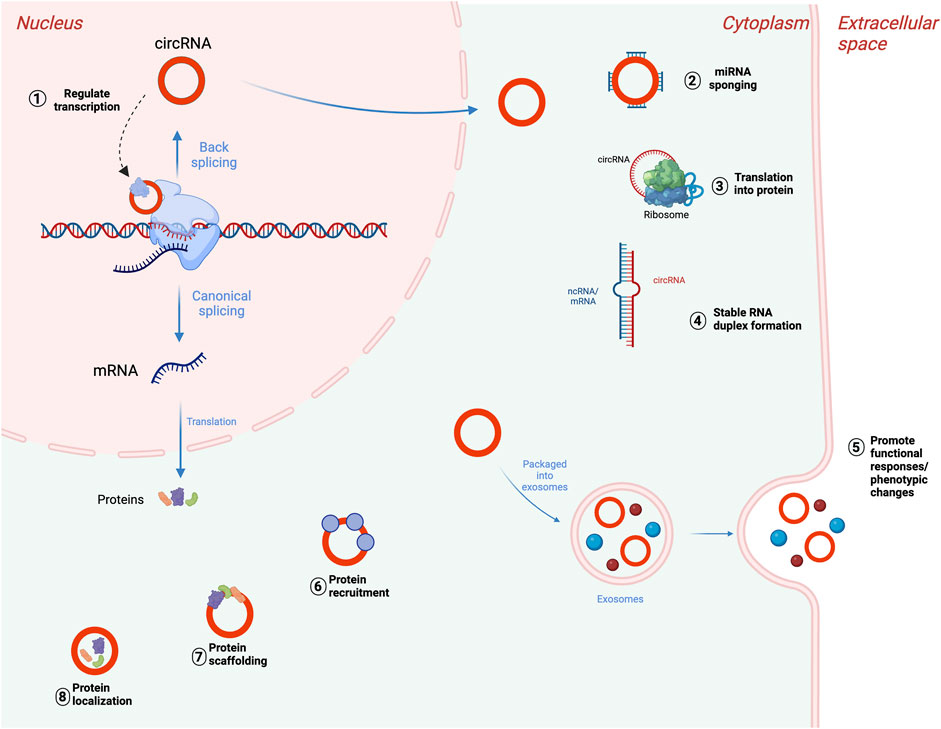
FIGURE 2. Biological functions of circRNAs. (1) Transcriptional regulation of parental genes, (2) posttranscriptional regulation through miRNA sponging, (3) translation of circRNAs into proteins or peptides. CircRNAs can also bind to mRNAs or lncRNAs and impact their stability (4), accumulate in exosomes to mediate cellular response (5), interact with RNA-binding protein (RBPs) (6), participate in protein scaffolding (7), and participating in protein localization pathways (8). Adapted from “DNA vs mRNA Transfection”, by BioRender.com (2022), https://app.biorender.com/biorender-templates, and (Pisignano et al., 2023), licensed CC-BY-4.0. Created with BioRender.com.
CircRNA molecules can be classified into several groups depending on their composition (Figure 1). These include: 1) exonic circRNAs (ecircRNAs), formed by exons exclusively and making up about 85% of all circRNAs, 2) circular intronic RNAs (ciRNAs), made of introns, and 3) exon-intron circRNAs (EIcircRNAs), formed by a combination of exons and introns (Zhou et al., 2018). There are also a few newly described circRNA categories including read-through circRNAs (rt-circRNAs), formed when an acceptor site at 5ʹ end of an exon binds to a donor site at a downstream 3ʹ end of an exon from the adjacent gene (read-through transcription), and fusion circRNAs (f-circRNAs), generated from chromosomal rearrangements such as translocations and deletions (Caba et al., 2021). These different types of circRNAs are found in various cellular compartments, with ciRNAs, EIcirRNAs, and f-circRNAs present in the nucleus, ecircRNAs found in cytoplasm and exosomes, and rt-circRNAs found in cytoplasm (Shang et al., 2019; He et al., 2021).
Even though circRNAs were previously considered non-coding RNAs (ncRNAs), there are several circRNAs that are translated and associated with cellular functions (Chen and Sarnow, 1995; Perriman and Ares, 1998; Pamudurti et al., 2017; Lei et al., 2020). The growing list of circRNA functions include transcriptional regulation (Conn et al., 2017), competing with endogenous RNAs by microRNA sponging (Hansen et al., 2013; Panda, 2018), modulating mRNA stability (Hansen et al., 2011), regulation of translation (Schuller and Green, 2018), translation into proteins (Zhou et al., 2020), and interaction with RNA binding proteins (Du et al., 2016).
A group of circRNAs containing introns are localized in nucleus and participate in transcriptional regulation of parental genes. EIciRNAs and ciRNAs play the cis-regulatory role by modulating parental genes. EIciRNAs perform the mentioned regulation via interaction with U1 small nuclear ribonucleoprotein (snRNP) forming the EIciRNAs-U1 snRNP complex. This complex further combines with polymerase II to modulate host gene transcription via its promoter region (Li et al., 2015; Wang et al., 2020). Another way circRNAs can generate cis-regulatory effects on parental genes is by accumulating at the transcriptional sites.
MiRNAs are significant gene expression regulators at a post-transcriptional level that act by binding to sites within untranslated regions of messenger RNAs (Bartel, 2009). This underscores the importance of the most studied circRNA function, the miRNA sponging. The circRNA have several target sites to which miRNAs can bind, resulting in reduction of their regulatory function. It is also known that some circRNAs can act as a sponge for various miRNAs, acting as tumor suppressors or oncogenes (Chen et al., 2020). For example, the CDR1as, expressed in various human tissues, and lung carcinoma, has more than 60 binding sites and it acts as a competing endogenous RNA (ceRNA) for miR-7. As CDR1as is expressed, it leads to miR-7 activity inhibition with further increase in expression of miR-7 targets (Peng et al., 2015).
Besides interacting with miRNAs, circRNAs have been reported to serve as a binding platform for argonaut proteins (Ago), proteins that are crucial in RNA silencing thorough mRNA cleavage or translation inhibition (Go and Mani, 2012). A study has demonstrated how RBPs are involved in miRNA recruitment and translation by circRNAs. The circBIRC6 was enriched in RBP Ago2 complex and regulated the pluripotency of human embryonic stem cells (hESCs) by combining with miR-34a and miR-145 (Yu et al., 2017). Involvement of Ago2 in the binding between circRNA-ZNF609 and miR-615-5p with further generation of similar effect on vascular endothelial dysfunction is another example of RBP participation in circRNA functional role (Liu et al., 2017). Additionally, with the help of RBPs, circRNAs can mediate mRNA translation or expression through methylation (Yang et al., 2017).
As circRNAs and linear RNAs come from common parental genes, the circRNAs with complete exons can be translatable, but in a cap-independent mechanism with the RBP assistance, different from conventional ribosome scanning mechanism (Du et al., 2017). CircRNAs have also demonstrated other interaction types with RBPs such as regulating RBPs by acting as sponges, competitively binding to RBPs, serving as super transporters, and platforms for RBP assembly (Hentze and Preiss, 2013; Du et al., 2017).
CircRNA can facilitate the interaction between two proteins by acting as a scaffold, but also can dissociate the interaction between proteins by binding to one. Additionally, circRNAs can block protein A from protein B by directly binding to only protein A. For example, CDR1as can block the tumor suppressor protein p53 from MDM2, thus preventing p53 ubiquitination and consequent DNA damage (Lou et al., 2020). MDM2 functions as an oncoprotein by hindering the transcriptional transactivation mediated by the p53 tumor suppressor. It not only guides p53 from the cell nucleus to the cytoplasm but also polyubiquitylates p53. The polyubiquitylated form of p53 undergoes swift degradation in the cytoplasm through the 26 S proteasome (Mendoza et al., 2014).
Given the versatile roles the circRNAs can play within biological processes, recent studies have reported several mechanisms through which circRNAs are involved in lung tumorigenesis. These are summarized in Table 1. For example, a circRNAs named circHIPK3 is formed through circularization of the second exon of the homology domain-interacting protein kinase 3 (HIPK3) gene, and is highly expressed in cytoplasm of human lungs, brains, colon, stomach, heart and other organs (Zhou et al., 2021). This circRNA has been shown to activate the PI3K/AKT signaling pathway by sponging the miR-188-3p in lung cancer patients (Zhao et al., 2022).
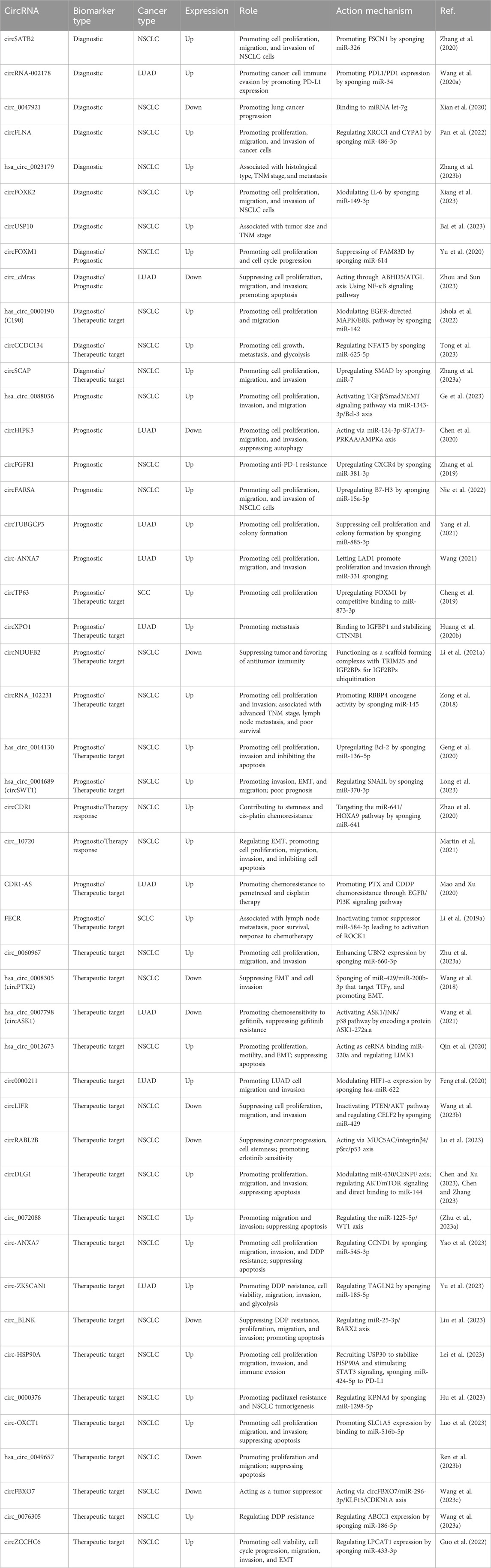
TABLE 1. List of circRNAs as potential biomarkers of diagnosis, prognosis, and therapeutic targets for lung cancer.
Another circRNAs, circTP63, facilitates cell-cycle progression by acting as a competing endogenous RNA (ceRNA) and competitively binding to miR-873-3p (Cheng et al., 2019). Consequently, the binding prevents miR-873-3p from decreasing the levels of FOXM1 (forkhead box protein M1), a proliferation-associated transcription factor, further upregulating CENPA and CENP genes, and thus promoting cell proliferation (Cheng et al., 2019). In a similar fashion, circFOXM acts as a ceRNA, suppressing the FAM83D (Family With Sequence Similarity 83 Member D) gene by sponging miR-614, and consequently promoting cell proliferation and cell cycle progression in NSCLC cells (Yu et al., 2020). In turn, the circFGFR1, which is significantly overexpressed in NSCLC tissues, acts as a tumor promoter by upregulating the C-X-C motif chemokine receptor 4 (CXCR4), a target gene for miR381-3p, and contributing to anti-PD-1 (Programmed cell Death Protein 1) resistance by sponging miR-381-3p (Zhang et al., 2019).
The B7-H3 (also known as CD276), member of the B7 immune checkpoint protein family, is overexpressed in cancer cells (Flem-Karlsen et al., 2018), thus promoting tumorigenesis. Contributing to this mechanism, circFARSA has been found overexpressed in cancer tissue and cell lines, leading to upregulation of B7-H3 via miR-15a-5p sponging (Nie et al., 2022). Similarly, circXPO1, a circRNA formed as a result of back-splicing of the well-established therapeutic agent XPO1 gene (Azizian and Li, 2020), is highly expressed in LUAD tissue. Its expression has been shown to correlate with worse survival in LUAD patients. The associated mechanism involves binding to IGF2BP1 (Insulin like Growth Factor 2 mRNA Binding Protein 1), stabilizing CTNNB1 (Catenin Beta 1), and consequently promoting LUAD progression (Huang et al., 2020).
Contrary to the previously mentioned circRNAs, elevated levels of circNDUFB2 inhibit tumor growth and metastasis in NSCLC cells by acting as a scaffold to enhance the interaction between the TRIM25 (Tripartite Motif Containing 25) and IGFBP2 (Insulin like Growth Factor Binding Protein 2) genes. Additionally, circNDUFB2 is recognized by RIG-I (Retinoic acid-Inducible Gene I) and leads to activation of the RIG-I-MAVS (Mitochondrial Antiviral Signaling protein) signaling cascade, with consequent immune cell recruitment into the tumor microenvironment (TME) (Li et al., 2021). Therefore, circRNAs can also have an inhibitory effect on lung tumorigenesis. In this regard, a circular RNA_ITCH (circ-ITCH) was reported to have significantly low expression in lung cancer tissue, and when expressed ectopically, it elevated levels of its parental cancer-suppressing gene, ITCH (Wan et al., 2016). Further analysis revealed that circ-ITCH enhanced ITCH expression by sponging miR-7 and miR-214, leading to suppression of the Wnt/β-catenin pro-oncogenic signaling pathway.
Another circRNA associated with LUAD is circTUBGCP3. Upregulation of this circRNA, or downregulation of miR-885-3p, was associated with pathological stage and poor survival in LUAD patients (Yang et al., 2021). Mechanistically, sponging of miR-885-3p by circTUBGCP3 prevents the miRNA from suppressing cell proliferation and attenuated the tumor-promoting effects of circTUBGGCP3. Circ-ANXA-7, which is highly expressed in LUAD tissue and cells, also promotes cell proliferation, migration, and invasion of LUAD cells, together with tumor growth (Wang, 2021). On the other hand, circ_0080608, which is under-expressed in tumor tissue, has been associated with suppressing lung cancer progression by regulating the miR-661/ADRA1A (Adrenergic Receptor Alpha 1A) pathway (Ren et al., 2023). Finally, circFOXK2 has been reported to modulate IL-6 via miR-149-3p sponging in NSCLC tissues and cells and thus promoting cell proliferation, migration, and invasion (Xiang et al., 2023).
Overexpression or activation of EGFR has been associated with poor prognosis in NSCLC (Kawai et al., 2005). A circRNA known as has_circ_0000190 (C190) has been found upregulated in both NSCLC clinical samples and cell lines, via a mechanism involving the EGFR pathway (Ishola et al., 2022). The C190 will be discussed more in the following sections having both prognostic biomarker and therapy target potentials.
Stemness pertains to shared molecular mechanisms that enable stem cells to preserve their ability to produce more cells resembling themselves through self-renewal and to generate specialized offspring (Lagunas-Rangel, 2020). In this context, a small subset of cells emerging during cancer development, known as cancer stem cells (CSCs), exhibits both of these characteristics akin to normal stem cells (Yu et al., 2012). CSCs are considered the source of cancer cells, possessing the capacity to support tumor growth, re-establish a tumor upon transplantation into an immunodeficient animal host, instigate resistance to treatment, and contribute to metastatic dissemination (Peiris-Pages et al., 2016). Recent research indicates that the primary sources of these stem cells encompass (1) adult stem cells, (2) tumor cells, (3) differentiated cells, and (4) cell fusion (Feng et al., 2019).
Several investigations have suggested that the malfunction of various signaling pathways, including JAK/STAT, Hedgehog, WNT/β-catenin, Notch, PI3K/PTEN, and NF-κB, can facilitate the self-renewal and differentiation of cancer stem cells (CSCs) (Matsui, 2016). The pivotal aspect lies in the ultimate modification of gene expression patterns through pluripotent transcription factors like OCT4, NANOG, and SOX2, along with collaborating factors such as the KLF family, c-MYC, and Lin28 (Liu et al., 2013). Notably, an increasing body of evidence has showcased the participation of circular RNAs (circRNAs) in regulating signaling pathways associated with CSCs (Feng et al., 2019). These circRNAs function either as oncogenes or tumor suppressors, contingent upon the cell type, implying their potential as therapeutic targets and/or clinical biomarkers for the diagnosis, prognosis, or monitoring of the disease.
The ability of circRNAs to interact and sponge miRNAs and proteins leads to their involvement in CSCs role in cancer progression. For instance, a circ-CPA4 was reported to contribute to the regulation of stem growth, stemness, drug resistance and immune evasion in NSCLC. The regulation was demonstrated to take place via circ-CPA4/let-7 miRNA/PD-L1 axis (Hong et al., 2020). A recent study reported the role of circRNA hsa_circ_0003222, which accelerates stemness and progression of non-small cell lung cancer by sponging miR-527 (Li C. et al., 2021). Contribution to lung cancer stemness was reported for circ_POLA2 via miR-326/GNB1 axis, where the circ_POLA2 was highly expressed in lung cancer tissues and predicted a poor prognostic outcome (Fan et al., 2020).
The current body of evidence indicates that quiescent cancer stem cells (CSCs) play a role in cancer resistance to chemotherapy. Consequently, strategies solely focused on inhibiting CSC stemness may not be adequate to effectively counter post-chemotherapy recurrence. The tumor microenvironment (TME) is pivotal in regulating CSCs, maintaining their characteristics through various signals, and promoting the transition of non-stem cells to stem cell states (de Sousa e Melo et al., 2017). Targeting components of the TME appears to be a potentially more effective approach in overcoming treatment resistance compared to directly inhibiting CSC stemness. However, the complexity of immune cell heterogeneity across cell types poses challenges in precisely understanding CSC-immune cell interactions. Recently, the widespread use of single-cell RNA sequencing has enabled the identification of changing states in both CSCs and immune cells, shedding light on their interactions in different tumor contexts (Manni and Min, 2022). Further investigation is needed to explore the communication among these immune cells within the tumor microenvironment (TME), with the aim of advancing the development of immunotherapies that specifically target cancer stem cells (CSCs) more effectively.
The circRNA circPVT1 is overexpressed in SCC tissue and serum from patients, and it promotes cell proliferation by acting as a ceRNA by sponging miR-30d and miR-30e, mitigating the suppressive effect of miRN-30d/e on cyclin F (CCNF) (Shi et al., 2021). A study that investigated role of circSATB2 in lung cancer reported its overexpression in NSCLC cells and tissue (Zhang et al., 2020). Furthermore, it was found that circSATB2 positively regulated fascin homolog 1 actin-bundling protein 1 (FSCN1) through miR-326 (Zhang et al., 2020). On the other hand, the circular RNA, hsa_circ_100395, was reported to act as a negative regulator of lung cancer development (Chen et al., 2018). Its decreased expression correlated with Tumor, Nodes, and Metastasis (TNM) stage and inversely correlated with poor prognosis. Another circRNA upregulated in LUAD and associated with the advanced TNM stage, lymph node metastasis and poor overall survival is circRNA_102231 (Zong et al., 2018).
One candidate for a prognostic biomarker role in lung cancer is has_circRNA_103809, which promotes cell proliferation in vitro, and its knockdown delays tumor growth in vivo. The mechanism proposed for this circRNA involves acting as a sponge of miR-4302, promoting ZNF121 expression, and consequently enhancing the proto-oncogene MYC protein levels in lung cancer cells (Liu et al., 2018). In NSCLC tissues, the significant upregulation of hsa_circ_0014130 is associated with proliferation and invasion. This circRNA also acts as a sponge of miR-136-5p leading to overexpression of its target gene Bcl-2 (Geng et al., 2020). More recently, the circRNA circFLNA has been found to promote lung cancer progression by acting as a sponge of miR-486-3p and thus regulating the XRCC1 (X-ray Repair Cross Complementing 1) and CYP1A1 (Cytochrome P450 Family 1 Subfamily A Member 1) genes (Pan et al., 2022).
A relationship between several circRNAs, including circ_10720 and epithelial-mesenchymal transition (EMT) process in NSCLC patients was reported in recent years (Martin et al., 2021). Furthermore, the circ_10720 was found to be overexpressed in some cell lines (HCC44, A549), and under-expressed in other (H23, H1299). Circ_10720 was overexpressed in NSCLC tissue, and its high levels were associated with shorter time to relapse (TTR).
Among the circRNAs that could have a prognostic biomarker potential for LUAD is circCRIM1 (hsa_circ_0002346). A study has concluded that this circRNA can act as a prognostic biomarker for LUAD survival as its downregulation in LUAD tissue has been significantly correlated with lymphatic metastasis and TNM stage (Wang et al., 2019). Indeed, circCRIM1 promotes the expression of leukemia inhibitory factor receptor (a tumor suppressor) by sponging miR-93 and miR-182 (Wang et al., 2019). Similarly, a circRNA that is upregulated in LUAD tissue and has prognostic potential is circ-ANXA7. Not only it promotes tumor progression as mentioned in the previous section, but also its high expression predicted poorer outcome for LUAD patients (Wang, 2021).
As liquid biopsies can serve as non-invasive biomarker sources, there is an interest in determining accurate circRNA biomarkers within them. Analysis of circRNA C190 in human blood showed association between high C190 levels with larger tumor size, worse histological type of adenocarcinoma, later cancer stage, more distant metastatic organs, extra-thoracic metastasis, poor survival, and prognosis (Luo et al., 2020). A study that investigated circRNA profile in Tumor-educated platelets (TEPs) has determined 411 circRNAs, out of 4,732 detected, to be significantly (p-value < 0.05) differently expressed in asymptomatic individuals compared to NSCLC patients. The nuclear receptor-interacting protein 1 (NRIP1) circRNA (circNRIP1) was selected as a potential biomarker shown to be significantly downregulated in platelets derived from NSCLC patient (D'Ambrosi et al., 2021). A programmed cell death ligand 1 (PDL1) and programmed death protein 1 (PD1) combine in a PD1/PDL1 pathway to control and maintain immune tolerance within the tumor microenvironment (TME) (Han et al., 2020).
A study focusing on circRNA role in LUAD tissue, found hsa_circRNA_002178 to be upregulated in cancer tissues, and that it enhanced PDL1 expression by sponging miR-34 to induce T-cell exhaustion. Furthermore, in plasma, circRNA-002178 was delivered into CD8+ cells by means of exosomes to promote PD1 expression (Wang J. et al., 2020). A study in Chinese population has shown that circ_0047921, circ_0056285, and circ_0007761 expression in serum exosomes could be used to distinguish early NSCLC cases from healthy controls (Xian et al., 2020). Furthermore, the circ_0047921 could distinguish early NSCLC cases from chronic obstructive pulmonary disease (COPD) controls, whereas combination of circ_0056285 and circ_0007761could distinguish between early NSCLC cases and tuberculosis controls.
A F-circEA, a circRNA generated by EML4-ALK fusion gene, is another potential liquid biopsy biomarker as it specifically exists in the plasma of EML4-ALK-positive NSCLC patients and contributes to tumor development by promoting cell migration and invasion (Tan et al., 2018). An investigation into NSCLC diagnostic potential of a circulating circular RNA hsa_circ_0023179 revealed that its higher expression was connected to histological type, TNM stage, lymph node metastasis, and distal metastasis in NSCLC tissue, serum and cells (Zhang et al., 2023). Moreover, it showed higher sensitivity and specificity when compared to traditional tumor markers.
While numerous circRNAs are introduced with theoretical potential of their diagnostic and prognostic value, an extensive amount of work lays ahead in investigating their reliability, precision and specificity. As of January 2024, the review of active clinical trials on circRNAs yields 12 registered clinical trials with 4 focusing on circRNA role in cancer (ClinicalTrials.gov). Common elements observed in the current clinical studies involve the research phase, specifically the discovery phase, and the necessity for subsequent validation in separate cohorts. Additionally, laboratory examinations are conducted to explore the molecular mechanisms underlying tumor-modifying behavior.
To date, a few circRNAs have been investigated for a potential role of a therapy target. For example, the C190 that could serve as a diagnostic biomarker, also could be considered as a therapy target.
More studies show the important role played by circRNAs in lung cancer therapy response. A study reported that the upregulation of CDR1-AS in LUAD tissue and cell lines was relevant to smoking history, T stage and neoadjuvant chemotherapy with pemetrexed (PTX) and cisplatin (CDDP) in LUAD patients (Mao and Xu, 2020). It was an independent prognostic biomarker therapy response, as it was highly expressed in LUAD tissues and cells (A549/CR) resistant to neoadjuvant chemotherapy with PTX and CDDP. Mechanistic investigation has revealed that CDR1-AS promoted PTX and CDDP chemoresistance through EGFR/PI3K signaling pathway. In another study, the circCDR1 and homeobox protein Hox-A9 (HOXA9) were overexpressed and miR-641 was low-expressed in cisplatin-resistant NSCLC cells (Zhao et al., 2020). The study showed that circCDR1 was regulating HOXA9 in NSCLC cells by acting as a miR-641 sponge, contributing to stemness and DDP chemoresistance. The circCRIM1 mentioned previously for its prognostic potential, was also shown to suppress the invasion and metastasis and in LUAD tissue, which makes it a potential therapeutic target (Wang et al., 2019). Treatment with growth factor receptor tyrosine kinase inhibitors is limited by the acquired resistance in the EGFR-mutant LUAD patients (Kobayashi et al., 2005). The circASK1 (hsa_circ_0007798) was shown to be significantly downregulated in gefitinib-resistant cells. It encodes for protein ASK1-272a.a, which is involved in ASK1/JNK/p38 signaling activation and mediates the chemosensitivity-inducing effect in of circASK1 in LUAD (Wang et al., 2021).
The hsa_circ_0008305 (circPTK2) and the transcriptional intermediary factor γ (TIFγ) have been found significantly downregulated in NSCLC cells undergoing the EMT induced by transforming growth factor β (TGF-β) (Wang et al., 2018). The TGF-β is highly expressed in NSCLCs and it promotes EMT and NSCLC cell invasion, whereas TIFγ acts as a tumor metastasis suppressor by regulating TGF-β/Smad signaling pathway (Dupont et al., 2005; Zhang et al., 2011; Xue et al., 2014). The circPTK2 overexpression was shown to suppress TGF-β-induced EMT as it sponges miR-429/miR-200b-3p, miRNAs that target TIFγ (Wang et al., 2018). These features and role make circPTK2 another potential therapeutical target candidate. On the other hand, circ_0060967, previously reported overexpressed in NSCLC tissue, was shown to promote cell viability, proliferation, migration, and invasion by sponging miR-660-3p and upregulating UBN2 (Zhu Z. et al., 2023). High expression of circ_0060967 implied poor prognosis and it could serve as a potential therapeutic target.
The overexpression of an otherwise under-expressed circLIFR in NSCLC tissue was found to suppress tumor progression and imped cell proliferation, migration, and invasion in vivo (Wang X. et al., 2023). A mechanistic investigation revealed circLIFR suppresses NSCLC progression by regulating CELF2 (CUGBP Elav-Like family Member 2) and inactivating PTEN (Phosphatase and Tensin homolog)/AKT (serine/threonine-protein kinase) signaling pathways by sponging miR-429. Low levels of circRABL2B in plasma exosomes could distinguish early stage lung-cancer patients, and it was revealed that circRABL2B counteracts lung cancer progression via MUC5AC (Mucin 5AC, Oligomeric Mucus/Gel-Forming)/integrin β4/pSrc/p53 axis, making the circRABL2B potential therapeutic target (Lu et al., 2023). An acceleration to cisplatin resistance in NSCLC patients by circ-ANXA7 was also recently reported (Yao et al., 2023). A mechanism behind this was concluded to be the regulation of CCND1 (Cyclin D1) by circ-ANXA7 via miR-545-3p sponging. A comparable finding on the role of circ-ZKSCAN1 on cisplatin resistance reported that circ-ZKSCAN1 promoted LUAD tumorigenesis and DDP resistance by regulating miR-185-5p/TAGLN2 (Transgelin 2) axis (Yu et al., 2023).
In SCLC chemoresistant cells, the circRNA cESRP1 (circular RNA epithelial splicing regulatory protein 1) was found significantly downregulated when compared to the parental chemosensitive cells (Huang et al., 2020). The same study identified cESRP1 as an important player in SCLC chemosensitivity by sponging miR-93-5p and inhibiting the TGF-β pathway (Huang et al., 2020). These findings suggest that cERP1 may serve as a prognostic and a potential therapeutic target in SCLC patients. Similarly, the abnormally upregulated Friend leukemia virus integration 1 (FLI1) is correlated with SCLC malignant phenotype (Li et al., 2017). A study that examined the role of FLI1 exonic circular RNAs (FECR) role as a new SCLC malignant driver, has revealed a positive association between the upregulated FECR1 and FECR2 and lymph node metastasis. It is also worth noting that serum exosomal FECR1 was associated with poor survival and chemotherapy clinical response (Li et al., 2019).
CircRNAs have emerged as pivotal players in the intricate landscape of lung cancer. Our literature review confirmed that circRNA research has promising implications for enhanced diagnosis and novel therapies in the context of lung cancer, given the associations of circRNAs with various physiological processes and cell biology features of tumorigenesis, and their potential roles as biomarkers.
While significant strides have been made in unraveling the role of circRNAs in lung cancer, several knowledge gaps persist. First, the precise mechanisms by which circRNAs exert their regulatory functions in lung cancer remain incompletely understood. Elucidating the specific pathways and interactions involved could deepen our comprehension of their impact on disease progression. Additionally, the clinical utility of circRNAs as reliable biomarkers for lung cancer diagnosis and prognosis requires further validation across diverse patient populations. Standardized methodologies for circRNA detection and quantification are essential for ensuring reproducibility and comparability of results. Furthermore, the dynamic nature of circRNA expression patterns during different stages of lung cancer and their response to therapeutic interventions necessitate longitudinal studies to capture the temporal dynamics accurately. Lastly, translating our current knowledge of circRNAs into effective therapeutic strategies demands a more comprehensive understanding of their interactions with other cellular components and signaling networks.
While circRNAs frequently engage in multiple molecular processes across various tissues and diseases, it is important to note that targeting circRNAs for therapeutic purposes may result in unintended effects on non-cancerous cells and tissues, posing challenges for clinical application. Additionally, the identification of multiple microRNAs sponged by the same circRNAs in different cancer types implies that their impact on a particular cancer phenotype is likely influenced by the specific context. As the computing and artificial intelligence (AI) capabilities grow in hand with body of literature, AI algorithms can be used to analyze vast amounts of genomic and clinical data to identify novel circRNAs associated with lung cancer, aiding in the discovery of potential biomarkers for early diagnosis or prognosis. Machine learning models can also predict the functional roles of circRNAs by integrating diverse data sources, offering insights into their molecular mechanisms in lung cancer progression. Moreover, AI can facilitate the integration of multi-omics data, enabling a more comprehensive understanding of the complex interactions between circRNAs and other molecular components in lung cancer. This holistic approach may uncover previously unnoticed patterns and correlations, guiding researchers towards more targeted investigations. In drug discovery and therapeutic development, AI can expedite the identification of circRNAs that may serve as therapeutic targets. By predicting off-target effects and optimizing treatment strategies, AI can enhance the efficiency and safety of circRNA-targeted therapies.
The collaboration between the fields of data science, artificial intelligence (AI), and cancer research is crucial for unlocking the potential of AI in advancing cancer studies. The National Cancer Institute (NCI) can foster this collaboration by offering suitable funding opportunities and access to data sources. Additionally, facilitating connections between cancer researchers and AI experts, along with supporting the training and growth of a workforce skilled in AI, data science, and cancer, is essential. Drawing inspiration from the NCI–DOE collaboration, a series of workshops can form a community actively involved in pushing the boundaries of existing computational practices in cancer research and developing innovative computational technologies.
Bridging the mentioned gaps in mechanisms and technologies will undoubtedly enhance our ability to harness circRNAs for improved lung cancer diagnosis, prognosis, and treatment. Ongoing research may unveil the dynamic nature of circRNA expression during disease progression and treatment response. Longitudinal studies could provide insights into the temporal changes of circRNA profiles, offering valuable information for monitoring disease evolution and therapeutic efficacy. In essence, the future of circRNA research in lung cancer appears promising, with the potential for transformative contributions to early diagnosis, targeted therapies, and personalized treatment approaches.
MB: Investigation, Methodology, Visualization, Writing–original draft, Writing–review and editing, Investigation, Methodology, Visualization, Writing–original draft, Writing–review and editing, PS: Conceptualization, Data curation, Funding acquisition, Resources, Supervision, Writing–review and editing, Conceptualization, Data curation, Funding acquisition, Resources, Supervision, Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arthurs, A. L., Jankovic-Karasoulos, T., Smith, M. D., and Roberts, C. T. (2022). Circular RNAs in pregnancy and the placenta. Int. J. Mol. Sci. 23 (9), 4551. doi:10.3390/ijms23094551
Azizian, N. G., and Li, Y. (2020). XPO1-dependent nuclear export as a target for cancer therapy. J. Hematol. Oncol. 13 (1), 61. doi:10.1186/s13045-020-00903-4
Bai, H., Jiang, M., Fang, S., Peng, Z., Liang, N., Cai, Y., et al. (2023). Whole blood-derived circUSP10 acts as a diagnostic biomarker in patients with early-stage non-small-cell lung cancer. Cell Transpl. 32, 9636897231193066. doi:10.1177/09636897231193066
Barrett, S. P., Wang, P. L., and Salzman, J. (2015). Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 4, e07540. doi:10.7554/eLife.07540
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 (2), 215–233. doi:10.1016/j.cell.2009.01.002
Begemann, G., Paricio, N., Artero, R., Kiss, I., Perez-Alonso, M., and Mlodzik, M. (1997). muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development 124 (21), 4321–4331. doi:10.1242/dev.124.21.4321
Caba, L., Florea, L., Gug, C., Dimitriu, D. C., and Gorduza, E. V. (2021). Circular RNA-is the circle perfect? Biomolecules 11 (12), 1755. doi:10.3390/biom11121755
Chen, B. J., Huang, S., and Janitz, M. (2019). Changes in circular RNA expression patterns during human foetal brain development. Genomics 111 (4), 753–758. doi:10.1016/j.ygeno.2018.04.015
Chen, C. Y., and Sarnow, P. (1995). Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268 (5209), 415–417. doi:10.1126/science.7536344
Chen, D., Ma, W., Ke, Z., and Xie, F. (2018). CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle 17 (16), 2080–2090. doi:10.1080/15384101.2018.1515553
Chen, L., Huang, C., and Shan, G. (2022). Circular RNAs in physiology and non-immunological diseases. Trends Biochem. Sci. 47 (3), 250–264. doi:10.1016/j.tibs.2021.11.004
Chen, X., Mao, R., Su, W., Yang, X., Geng, Q., Guo, C., et al. (2020). Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy 16 (4), 659–671. doi:10.1080/15548627.2019.1634945
Chen, Y., and Zhang, Y. (2023). CircDLG1 promotes malignant development of non-small cell lung cancer through regulation of the miR-630/CENPF axis. Strahlenther Onkol. 199 (2), 169–181. doi:10.1007/s00066-022-01965-8
Chen, Y. F., and Xu, A. P. (2023). Circular RNA circDLG1 (has_circ_0068706) functions as an oncogene in nonsmall cell lung cancer through regulating AKT/mTOR signaling and direct binding to miR-144. Kaohsiung J. Med. Sci. 39 (5), 446–457. doi:10.1002/kjm2.12662
Cheng, Z., Yu, C., Cui, S., Wang, H., Jin, H., Wang, C., et al. (2019). circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat. Commun. 10 (1), 3200. doi:10.1038/s41467-019-11162-4
Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160 (6), 1125–1134. doi:10.1016/j.cell.2015.02.014
Conn, V. M., Hugouvieux, V., Nayak, A., Conos, S. A., Capovilla, G., Cildir, G., et al. (2017). A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 3, 17053. doi:10.1038/nplants.2017.53
D'Ambrosi, S., Visser, A., Antunes-Ferreira, M., Poutsma, A., Giannoukakos, S., Sol, N., et al. (2021). The analysis of platelet-derived circRNA repertoire as potential diagnostic biomarker for non-small cell lung cancer. Cancers (Basel) 13 (18), 4644. doi:10.3390/cancers13184644
de Sousa e Melo, F., Kurtova, A. V., Harnoss, J. M., Kljavin, N., Hoeck, J. D., Hung, J., et al. (2017). A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature 543 (7647), 676–680. doi:10.1038/nature21713
Di, X., Jin, X., Li, R., Zhao, M., and Wang, K. (2019). CircRNAs and lung cancer: biomarkers and master regulators. Life Sci. 220, 177–185. doi:10.1016/j.lfs.2019.01.055
Du, W. W., Yang, W., Liu, E., Yang, Z., Dhaliwal, P., and Yang, B. B. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44 (6), 2846–2858. doi:10.1093/nar/gkw027
Du, W. W., Zhang, C., Yang, W., Yong, T., Awan, F. M., and Yang, B. B. (2017). Identifying and characterizing circRNA-protein interaction. Theranostics 7 (17), 4183–4191. doi:10.7150/thno.21299
Dupont, S., Zacchigna, L., Cordenonsi, M., Soligo, S., Adorno, M., Rugge, M., et al. (2005). Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell 121 (1), 87–99. doi:10.1016/j.cell.2005.01.033
Ettinger, D. S., Wood, D. E., Aisner, D. L., Akerley, W., Bauman, J. R., Bharat, A., et al. (2021). NCCN guidelines insights: non-small cell lung cancer, version 2.2021. J. Natl. Compr. Canc Netw. 19 (3), 254–266. doi:10.6004/jnccn.2021.0013
Fan, Z., Bai, Y., Zhang, Q., and Qian, P. (2020). CircRNA circ_POLA2 promotes lung cancer cell stemness via regulating the miR-326/GNB1 axis. Environ. Toxicol. 35 (10), 1146–1156. doi:10.1002/tox.22980
Feng, D., Xu, Y., Hu, J., Zhang, S., Li, M., and Xu, L. (2020). A novel circular RNA, hsa-circ-0000211, promotes lung adenocarcinoma migration and invasion through sponging of hsa-miR-622 and modulating HIF1-α expression. Biochem. Biophys. Res. Commun. 521 (2), 395–401. doi:10.1016/j.bbrc.2019.10.134
Feng, Z., Meng, S., Zhou, H., Xu, Z., Tang, Y., Li, P., et al. (2019). Functions and potential applications of circular RNAs in cancer stem cells. Front. Oncol. 9, 500. doi:10.3389/fonc.2019.00500
Flem-Karlsen, K., Fodstad, O., Tan, M., and Nunes-Xavier, C. E. (2018). B7-H3 in cancer - beyond immune regulation. Trends Cancer 4 (6), 401–404. doi:10.1016/j.trecan.2018.03.010
Ge, P., Chen, X., Liu, J., Jing, R., Zhang, X., and Li, H. (2023). Hsa_circ_0088036 promotes nonsmall cell lung cancer progression by regulating miR-1343-3p/Bcl-3 axis through TGFβ/Smad3/EMT signaling. Mol. Carcinog. 62 (7), 1073–1085. doi:10.1002/mc.23547
Geng, Y., Bao, Y., Zhang, W., Deng, L., Su, D., and Zheng, H. (2020). Circular RNA hsa_circ_0014130 inhibits apoptosis in non-small cell lung cancer by sponging miR-136-5p and upregulating BCL2. Mol. Cancer Res. 18 (5), 748–756. doi:10.1158/1541-7786.MCR-19-0998
Giral, H., Landmesser, U., and Kratzer, A. (2018). Into the wild: GWAS exploration of non-coding RNAs. Front. Cardiovasc Med. 5, 181. doi:10.3389/fcvm.2018.00181
Go, G. W., and Mani, A. (2012). Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J. Biol. Med. 85 (1), 19–28. https://www.ncbi.nlm.nih.gov/pubmed/22461740.
Guo, Y., Xue, W., Sun, S., Chen, X., Li, H., and Yan, C. (2022). Circular RNA circZCCHC6 contributes to tumorigenesis by regulating LPCAT1 via miR-433-3p in non-small cell lung cancer. Clin. Exp. Med. 22 (4), 647–659. doi:10.1007/s10238-021-00780-2
Han, Y., Liu, D., and Li, L. (2020). PD-1/PD-L1 pathway: current researches in cancer. Am. J. Cancer Res. 10 (3), 727–742. https://www.ncbi.nlm.nih.gov/pubmed/32266087.
Hansen, T. B., Kjems, J., and Damgaard, C. K. (2013). Circular RNA and miR-7 in cancer. Cancer Res. 73 (18), 5609–5612. doi:10.1158/0008-5472.CAN-13-1568
Hansen, T. B., Wiklund, E. D., Bramsen, J. B., Villadsen, S. B., Statham, A. L., Clark, S. J., et al. (2011). miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30 (21), 4414–4422. doi:10.1038/emboj.2011.359
He, A. T., Liu, J., Li, F., and Yang, B. B. (2021). Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct. Target Ther. 6 (1), 185. doi:10.1038/s41392-021-00569-5
Hentze, M. W., and Preiss, T. (2013). Circular RNAs: splicing's enigma variations. EMBO J. 32 (7), 923–925. doi:10.1038/emboj.2013.53
Hong, W., Xue, M., Jiang, J., Zhang, Y., and Gao, X. (2020). Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J. Exp. Clin. Cancer Res. 39 (1), 149. doi:10.1186/s13046-020-01648-1
Hu, S., Zhang, Q., Sun, J., Xue, J., and Wang, C. (2023). Circular RNA circ_0000376 promotes paclitaxel resistance and tumorigenesis of non-small cell lung cancer via positively modulating KPNA4 by sponging miR-1298-5p. Thorac. Cancer 14 (22), 2116–2126. doi:10.1111/1759-7714.14994
Huang, Q., Guo, H., Wang, S., Ma, Y., Chen, H., Li, H., et al. (2020a). A novel circular RNA, circXPO1, promotes lung adenocarcinoma progression by interacting with IGF2BP1. Cell Death Dis. 11 (12), 1031. doi:10.1038/s41419-020-03237-8
Huang, W., Yang, Y., Wu, J., Niu, Y., Yao, Y., Zhang, J., et al. (2020b). Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-β signalling. Cell Death Differ. 27 (5), 1709–1727. doi:10.1038/s41418-019-0455-x
Ishola, A. A., Chien, C. S., Yang, Y. P., Chien, Y., Yarmishyn, A. A., Tsai, P. H., et al. (2022). Oncogenic circRNA C190 promotes non-small cell lung cancer via modulation of the EGFR/ERK pathway. Cancer Res. 82 (1), 75–89. doi:10.1158/0008-5472.CAN-21-1473
Janas, T., Janas, M. M., Sapon, K., and Janas, T. (2015). Mechanisms of RNA loading into exosomes. FEBS Lett. 589 (13), 1391–1398. doi:10.1016/j.febslet.2015.04.036
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19 (2), 141–157. doi:10.1261/rna.035667.112
Kawai, H., Ishii, A., Washiya, K., Konno, T., Kon, H., Yamaya, C., et al. (2005). Combined overexpression of EGFR and estrogen receptor alpha correlates with a poor outcome in lung cancer. Anticancer Res. 25 (6C), 4693–4698. https://www.ncbi.nlm.nih.gov/pubmed/16334162.
Kobayashi, S., Boggon, T. J., Dayaram, T., Janne, P. A., Kocher, O., Meyerson, M., et al. (2005). EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 352 (8), 786–792. doi:10.1056/NEJMoa044238
Kolakofsky, D. (1976). Isolation and characterization of Sendai virus DI-RNAs. Cell 8 (4), 547–555. doi:10.1016/0092-8674(76)90223-3
Kos, A., Dijkema, R., Arnberg, A. C., van der Meide, P. H., and Schellekens, H. (1986). The hepatitis delta (delta) virus possesses a circular RNA. Nature 323 (6088), 558–560. doi:10.1038/323558a0
Lagunas-Rangel, F. A. (2020). Circular RNAs and their participation in stemness of cancer. Med. Oncol. 37 (5), 42. doi:10.1007/s12032-020-01373-x
Lei, J., Zhu, J., Hui, B., Jia, C., Yan, X., Jiang, T., et al. (2023). Circ-HSP90A expedites cell growth, stemness, and immune evasion in non-small cell lung cancer by regulating STAT3 signaling and PD-1/PD-L1 checkpoint. Cancer Immunol. Immunother. 72 (1), 101–124. doi:10.1007/s00262-022-03235-z
Lei, M., Zheng, G., Ning, Q., Zheng, J., and Dong, D. (2020). Translation and functional roles of circular RNAs in human cancer. Mol. Cancer 19 (1), 30. doi:10.1186/s12943-020-1135-7
Li, B., Zhu, L., Lu, C., Wang, C., Wang, H., Jin, H., et al. (2021a). circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 12 (1), 295. doi:10.1038/s41467-020-20527-z
Li, C., Zhang, J., Yang, X., Hu, C., Chu, T., Zhong, R., et al. (2021b). hsa_circ_0003222 accelerates stemness and progression of non-small cell lung cancer by sponging miR-527. Cell Death Dis. 12 (9), 807. doi:10.1038/s41419-021-04095-8
Li, L., Li, W., Chen, N., Zhao, H., Xu, G., Zhao, Y., et al. (2019a). FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin. Cancer Res. 25 (4), 1302–1317. doi:10.1158/1078-0432.CCR-18-1447
Li, L., Song, W., Yan, X., Li, A., Zhang, X., Li, W., et al. (2017). Friend leukemia virus integration 1 promotes tumorigenesis of small cell lung cancer cells by activating the miR-17-92 pathway. Oncotarget 8 (26), 41975–41987. doi:10.18632/oncotarget.16715
Li, X., Liu, S., Zhang, L., Issaian, A., Hill, R. C., Espinosa, S., et al. (2019b). A unified mechanism for intron and exon definition and back-splicing. Nature 573 (7774), 375–380. doi:10.1038/s41586-019-1523-6
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 (3), 256–264. doi:10.1038/nsmb.2959
Liu, A., Yu, X., and Liu, S. (2013). Pluripotency transcription factors and cancer stem cells: small genes make a big difference. Chin. J. Cancer 32 (9), 483–487. doi:10.5732/cjc.012.10282
Liu, C., Yao, M. D., Li, C. P., Shan, K., Yang, H., Wang, J. J., et al. (2017). Silencing of circular RNA-znf609 ameliorates vascular endothelial dysfunction. Theranostics 7 (11), 2863–2877. doi:10.7150/thno.19353
Liu, W., Ma, W., Yuan, Y., Zhang, Y., and Sun, S. (2018). Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem. Biophys. Res. Commun. 500 (4), 846–851. doi:10.1016/j.bbrc.2018.04.172
Liu, Y., Hu, Y., Zhao, C., and Lu, Q. (2023). CircRNA B cell linker regulates cisplatin sensitivity in nonsmall cell lung cancer via microRNA-25-3p/BarH-like homeobox 2 axis. Anticancer Drugs 34 (5), 640–651. doi:10.1097/CAD.0000000000001349
Long, X., Wang, D. G., Wu, Z. B., Liao, Z. M., and Xu, J. J. (2023). Circular RNA hsa_circ_0004689 (circSWT1) promotes NSCLC progression via the miR-370-3p/SNAIL axis by inducing cell epithelial-mesenchymal transition (EMT). Cancer Med. 12 (7), 8289–8305. doi:10.1002/cam4.5527
Lou, J., Hao, Y., Lin, K., Lyu, Y., Chen, M., Wang, H., et al. (2020). Circular RNA CDR1as disrupts the p53/MDM2 complex to inhibit Gliomagenesis. Mol. Cancer 19 (1), 138. doi:10.1186/s12943-020-01253-y
Lu, L., Zeng, Y., Yu, Z., Chen, S., Xie, J., Rao, B., et al. (2023). EIF4a3-regulated circRABL2B regulates cell stemness and drug sensitivity of lung cancer via YBX1-dependent downregulation of MUC5AC expression. Int. J. Biol. Sci. 19 (9), 2725–2739. doi:10.7150/ijbs.78588
Luo, H., Peng, J., and Yuan, Y. (2023). CircRNA OXCT1 promotes the malignant progression and glutamine metabolism of non-small cell lung cancer by absorbing miR-516b-5p and upregulating SLC1A5. Cell Cycle 22 (10), 1182–1195. doi:10.1080/15384101.2022.2071565
Luo, Y. H., Yang, Y. P., Chien, C. S., Yarmishyn, A. A., Ishola, A. A., Chien, Y., et al. (2020). Plasma level of circular RNA hsa_circ_0000190 correlates with tumor progression and poor treatment response in advanced lung cancers. Cancers (Basel) 12 (7), 1740. doi:10.3390/cancers12071740
Lyu, D., and Huang, S. (2017). The emerging role and clinical implication of human exonic circular RNA. RNA Biol. 14 (8), 1000–1006. doi:10.1080/15476286.2016.1227904
Manni, W., and Min, W. (2022). Signaling pathways in the regulation of cancer stem cells and associated targeted therapy. MedComm 3 (4), e176. doi:10.1002/mco2.176
Mao, Y., and Xu, R. (2020). Circular RNA CDR1-AS contributes to pemetrexed and cisplatin chemoresistance through EGFR/PI3K signaling pathway in lung adenocarcinoma. Biomed. Pharmacother. 123, 109771. doi:10.1016/j.biopha.2019.109771
Martin, J., Castellano, J. J., Marrades, R. M., Canals, J., Vinolas, N., Diaz, T., et al. (2021). Role of the epithelial-mesenchymal transition-related circular RNA, circ-10720, in non-small-cell lung cancer. Transl. Lung Cancer Res. 10 (4), 1804–1818. doi:10.21037/tlcr-20-920
Matsui, W. H. (2016). Cancer stem cell signaling pathways. Med. Baltim. 95 (1 Suppl. 1), S8–S19. doi:10.1097/MD.0000000000004765
Mendoza, M., Mandani, G., and Momand, J. (2014). The MDM2 gene family. Biomol. Concepts 5 (1), 9–19. doi:10.1515/bmc-2013-0027
Miao, Q., Ni, B., and Tang, J. (2021). Coding potential of circRNAs: new discoveries and challenges. PeerJ 9, e10718. doi:10.7717/peerj.10718
Nie, J., Yang, R., Zhou, R., Deng, Y., Li, D., Gou, D., et al. (2022). Circular RNA circFARSA promotes the tumorigenesis of non-small cell lung cancer by elevating B7H3 via sponging miR-15a-5p. Cell Cycle 21 (24), 2575–2589. doi:10.1080/15384101.2022.2105087
Nigro, J. M., Cho, K. R., Fearon, E. R., Kern, S. E., Ruppert, J. M., Oliner, J. D., et al. (1991). Scrambled exons. Cell 64 (3), 607–613. doi:10.1016/0092-8674(91)90244-s
Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., et al. (2017). Translation of CircRNAs. Mol. Cell 66 (1), 9–21. doi:10.1016/j.molcel.2017.02.021
Pan, J., Huang, G., Yin, Z., Cai, X., Gong, E., Li, Y., et al. (2022). Circular RNA FLNA acts as a sponge of miR-486-3p in promoting lung cancer progression via regulating XRCC1 and CYP1A1. Cancer Gene Ther. 29 (1), 101–121. doi:10.1038/s41417-021-00293-w
Panda, A. C. (2018). Circular RNAs act as miRNA sponges. Adv. Exp. Med. Biol. 1087, 67–79. doi:10.1007/978-981-13-1426-1_6
Peiris-Pages, M., Martinez-Outschoorn, U. E., Pestell, R. G., Sotgia, F., and Lisanti, M. P. (2016). Cancer stem cell metabolism. Breast Cancer Res. 18 (1), 55. doi:10.1186/s13058-016-0712-6
Peng, L., Yuan, X. Q., and Li, G. C. (2015). The emerging landscape of circular RNA ciRS-7 in cancer (Review). Oncol. Rep. 33 (6), 2669–2674. doi:10.3892/or.2015.3904
Perriman, R., and Ares, M. (1998). Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA 4 (9), 1047–1054. doi:10.1017/s135583829898061x
Pisignano, G., Michael, D. C., Visal, T. H., Pirlog, R., Ladomery, M., and Calin, G. A. (2023). Going circular: history, present, and future of circRNAs in cancer. Oncogene 42 (38), 2783–2800. doi:10.1038/s41388-023-02780-w
Qin, H., Liu, J., Du, Z. H., Hu, R., Yu, Y. K., and Wang, Q. A. (2020). Circular RNA hsa_circ_0012673 facilitates lung cancer cell proliferation and invasion via miR-320a/LIMK18521 axis. Eur. Rev. Med. Pharmacol. Sci. 24 (4), 1841–1852. doi:10.26355/eurrev_202002_20362
Ren, C., Cui, L., Li, R., Song, X., Li, J., Xi, Q., et al. (2023a). Hsa_circ_0080608 attenuates lung cancer progression by functioning as a competitive endogenous RNA to regulate the miR-661/adra1a pathway. Horm. Metab. Res. 55, 876–884. doi:10.1055/a-2179-0283
Ren, Y., Zhao, Y., Shan, Y., Li, S., Su, N., Cui, Z., et al. (2023b). Circular RNA hsa_circ_0049657 as a potential biomarker in non-small cell lung cancer. Int. J. Mol. Sci. 24 (17), 13237. doi:10.3390/ijms241713237
Schuller, A. P., and Green, R. (2018). Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol. 19 (8), 526–541. doi:10.1038/s41580-018-0011-4
Shang, Q., Yang, Z., Jia, R., and Ge, S. (2019). The novel roles of circRNAs in human cancer. Mol. Cancer 18 (1), 6. doi:10.1186/s12943-018-0934-6
Shi, J., Lv, X., Zeng, L., Li, W., Zhong, Y., Yuan, J., et al. (2021). CircPVT1 promotes proliferation of lung squamous cell carcinoma by binding to miR-30d/e. J. Exp. Clin. Cancer Res. 40 (1), 193. doi:10.1186/s13046-021-01976-w
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2021). Cancer statistics, 2021. CA Cancer J. Clin. 71 (1), 7–33. doi:10.3322/caac.21654
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72 (1), 7–33. doi:10.3322/caac.21708
Tan, S., Gou, Q., Pu, W., Guo, C., Yang, Y., Wu, K., et al. (2018). Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 28 (6), 693–695. doi:10.1038/s41422-018-0033-7
Tong, Z., Wang, Z., Jiang, J., Tong, C., and Wu, L. (2023). A novel molecular mechanism mediated by circCCDC134 regulates non-small cell lung cancer progression. Thorac. Cancer 14 (20), 1958–1968. doi:10.1111/1759-7714.14942
Wan, L., Zhang, L., Fan, K., Cheng, Z. X., Sun, Q. C., and Wang, J. J. (2016). Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the wnt/β-catenin pathway. Biomed. Res. Int. 2016, 1579490. doi:10.1155/2016/1579490
Wang, C., Tan, S., Li, J., Liu, W. R., Peng, Y., and Li, W. (2020a). CircRNAs in lung cancer - biogenesis, function and clinical implication. Cancer Lett. 492, 106–115. doi:10.1016/j.canlet.2020.08.013
Wang, J., Lai, X., and Peng, X. (2023c). CircLIFR inhibits non-small cell lung cancer progression by acting as a miR-429 sponge to enhance CELF2 expression. Biochem. Genet. 61 (2), 725–741. doi:10.1007/s10528-022-10285-6
Wang, J., Zhao, X., Wang, Y., Ren, F., Sun, D., Yan, Y., et al. (2020b). circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 11 (1), 32. doi:10.1038/s41419-020-2230-9
Wang, L., Liang, Y., Mao, Q., Xia, W., Chen, B., Shen, H., et al. (2019). Circular RNA circCRIM1 inhibits invasion and metastasis in lung adenocarcinoma through the microRNA (miR)-182/miR-93-leukemia inhibitory factor receptor pathway. Cancer Sci. 110 (9), 2960–2972. doi:10.1111/cas.14131
Wang, L., Tong, X., Zhou, Z., Wang, S., Lei, Z., Zhang, T., et al. (2018). Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol. Cancer 17 (1), 140. doi:10.1186/s12943-018-0889-7
Wang, T., Liu, Z., She, Y., Deng, J., Zhong, Y., Zhao, M., et al. (2021). A novel protein encoded by circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1-dependent apoptosis. Cancer Lett. 520, 321–331. doi:10.1016/j.canlet.2021.08.007
Wang, X., Wang, H., Jiang, H., Qiao, L., and Guo, C. (2023a). Circ_0011292 enhances paclitaxel resistance in non-small cell lung cancer by regulating miR-379-5p/TRIM65 Axis. Cancer Biother Radiopharm. 38 (5), 84–95. doi:10.1089/cbr.2019.3546
Wang, Y. (2021). circ-ANXA7 facilitates lung adenocarcinoma progression via miR-331/LAD1 axis. Cancer Cell Int. 21 (1), 85. doi:10.1186/s12935-021-01791-5
Wang, Z. H., Ye, L. L., Xiang, X., Wei, X. S., Niu, Y. R., Peng, W. B., et al. (2023b). Circular RNA circFBXO7 attenuates non-small cell lung cancer tumorigenesis by sponging miR-296-3p to facilitate KLF15-mediated transcriptional activation of CDKN1A. Transl. Oncol. 30, 101635. doi:10.1016/j.tranon.2023.101635
Xian, J., Su, W., Liu, L., Rao, B., Lin, M., Feng, Y., et al. (2020). Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of non-small-cell lung cancer in the Chinese population. J. Mol. Diagn 22 (8), 1096–1108. doi:10.1016/j.jmoldx.2020.05.011
Xiang, T., Chen, L., Wang, H., Yu, T., Li, T., Li, J., et al. (2023). The circular RNA circFOXK2 enhances the tumorigenesis of non-small cell lung cancer through the miR-149-3p/IL-6 Axis. Biochem. Genet. doi:10.1007/s10528-023-10394-w
Xue, J., Lin, X., Chiu, W. T., Chen, Y. H., Yu, G., Liu, M., et al. (2014). Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β-dependent cancer metastasis. J. Clin. Invest. 124 (2), 564–579. doi:10.1172/JCI71104
Yang, Y., Fan, X., Nie, Y., Liu, D., Zhu, D., Wu, K., et al. (2021). CircTUBGCP3 facilitates the tumorigenesis of lung adenocarcinoma by sponging miR-885-3p. Cancer Cell Int. 21 (1), 651. doi:10.1186/s12935-021-02356-2
Yang, Z. G., Awan, F. M., Du, W. W., Zeng, Y., Lyu, J., Wu, D., et al. (2017). The circular RNA interacts with STAT3, increasing its nuclear translocation and wound Repair by modulating Dnmt3a and miR-17 function. Mol. Ther. 25 (9), 2062–2074. doi:10.1016/j.ymthe.2017.05.022
Yao, J., Zhang, H. Y., Gu, S., Zou, J. L., Zhang, Q., and Qu, R. C. (2023). Circular RNA-AnnexinA7 accelerates cisplatin resistance in non-small cell lung cancer via modulating microRNA-545-3p to mediate Cyclin D1. Acta Biochim. Pol. 70 (2), 295–304. doi:10.18388/abp.2020_6539
Yu, C., Cheng, Z., Cui, S., Mao, X., Li, B., Fu, Y., et al. (2020). circFOXM1 promotes proliferation of non-small cell lung carcinoma cells by acting as a ceRNA to upregulate FAM83D. J. Exp. Clin. Cancer Res. 39 (1), 55. doi:10.1186/s13046-020-01555-5
Yu, C. Y., Li, T. C., Wu, Y. Y., Yeh, C. H., Chiang, W., Chuang, C. Y., et al. (2017). The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 8 (1), 1149. doi:10.1038/s41467-017-01216-w
Yu, N., Gong, H., Chen, W., and Peng, W. (2023). CircRNA ZKSCAN1 promotes lung adenocarcinoma progression by miR-185-5p/TAGLN2 axis. Thorac. Cancer 14 (16), 1467–1476. doi:10.1111/1759-7714.14889
Yu, Z., Pestell, T. G., Lisanti, M. P., and Pestell, R. G. (2012). Cancer stem cells. Int. J. Biochem. Cell Biol. 44 (12), 2144–2151. doi:10.1016/j.biocel.2012.08.022
Zang, J., Lu, D., and Xu, A. (2020). The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J. Neurosci. Res. 98 (1), 87–97. doi:10.1002/jnr.24356
Zaphiropoulos, P. G. (1996). Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc. Natl. Acad. Sci. U. S. A. 93 (13), 6536–6541. doi:10.1073/pnas.93.13.6536
Zhang, H. J., Wang, H. Y., Zhang, H. T., Su, J. M., Zhu, J., Wang, H. B., et al. (2011). Transforming growth factor-β1 promotes lung adenocarcinoma invasion and metastasis by epithelial-to-mesenchymal transition. Mol. Cell Biochem. 355 (1-2), 309–314. doi:10.1007/s11010-011-0869-3
Zhang, N., Nan, A., Chen, L., Li, X., Jia, Y., Qiu, M., et al. (2020). Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol. Cancer 19 (1), 101. doi:10.1186/s12943-020-01221-6
Zhang, P. F., Pei, X., Li, K. S., Jin, L. N., Wang, F., Wu, J., et al. (2019). Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol. Cancer 18 (1), 179. doi:10.1186/s12943-019-1111-2
Zhang, Q., Qin, S., Peng, C., Liu, Y., Huang, Y., and Ju, S. (2023a). Circulating circular RNA hsa_circ_0023179 acts as a diagnostic biomarker for non-small-cell lung cancer detection. J. Cancer Res. Clin. Oncol. 149 (7), 3649–3660. doi:10.1007/s00432-022-04254-0
Zhang, Y., Qi, W., and Wu, Y. (2023b). EIF4A3-induced circular RNA SCAP facilitates tumorigenesis and progression of non-small-cell lung cancer via miR-7/SMAD2 signaling. Environ. Sci. Pollut. Res. Int. 30 (24), 65237–65249. doi:10.1007/s11356-023-26307-8
Zhao, Y., Zhang, C., Tang, H., Wu, X., and Qi, Q. (2022). Mechanism of RNA circHIPK3 involved in resistance of lung cancer cells to gefitinib. Biomed. Res. Int. 2022, 4541918. doi:10.1155/2022/4541918
Zhao, Y., Zheng, R., Chen, J., and Ning, D. (2020). CircRNA CDR1as/miR-641/HOXA9 pathway regulated stemness contributes to cisplatin resistance in non-small cell lung cancer (NSCLC). Cancer Cell Int. 20, 289. doi:10.1186/s12935-020-01390-w
Zhou, J., Wang, B., Bin, X., Xie, C., Li, B., Liu, O., et al. (2021). CircHIPK3: key player in pathophysiology and potential diagnostic and therapeutic tool. Front. Med. (Lausanne) 8, 615417. doi:10.3389/fmed.2021.615417
Zhou, M. Y., Yang, J. M., and Xiong, X. D. (2018). The emerging landscape of circular RNA in cardiovascular diseases. J. Mol. Cell Cardiol. 122, 134–139. doi:10.1016/j.yjmcc.2018.08.012
Zhou, Q., and Sun, Y. (2023). Circular RNA cMras suppresses the progression of lung adenocarcinoma through ABHD5/ATGL Axis using NF-κB signaling pathway. Cancer Biother Radiopharm. 38 (5), 336–346. doi:10.1089/cbr.2020.3709
Zhou, W. Y., Cai, Z. R., Liu, J., Wang, D. S., Ju, H. Q., and Xu, R. H. (2020). Circular RNA: metabolism, functions and interactions with proteins. Mol. Cancer 19 (1), 172. doi:10.1186/s12943-020-01286-3
Zhu, X., Wan, J., You, X., Yang, W., and Zhao, L. (2023a). Circular non-coding RNA circ_0072088 serves as a ceRNA, targeting the miR-1225-5p/WT1 axis to regulate non-small cell lung cancer cell malignant behavior. Thorac. Cancer 14 (20), 1969–1979. doi:10.1111/1759-7714.14943
Zhu, Z., Zhang, K., Lou, M., Tong, J., Wu, Q., Lu, J., et al. (2023b). Circ_0060967 facilitates proliferation, migration, and invasion of non-small-cell lung cancer cells by sponging miR-660-3p/UBN2. Mol. Cell Biochem. 478 (5), 1129–1140. doi:10.1007/s11010-022-04569-z
Keywords: CircRNAs, circular RNA, lung cancer, lung adenocarcinoma, NSCLC, biomarkers
Citation: Babayev M and Silveyra P (2024) Role of circular RNAs in lung cancer. Front. Genet. 15:1346119. doi: 10.3389/fgene.2024.1346119
Received: 29 November 2023; Accepted: 23 February 2024;
Published: 04 March 2024.
Edited by:
Harikrishnan Radhakrishnan, SRI International, United StatesReviewed by:
Mehak Gupta, Vertex Pharmaceuticals, United StatesCopyright © 2024 Babayev and Silveyra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patricia Silveyra, cHNpbHZleXJAaXUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.