- Department of Urology, First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, China
Background: The association between MTHFR gene polymorphisms (C677T and A1298C) and prostate cancer risk remains controversial.
Methods: Two independent researchers searched the PubMed, Embase, Cochrane and Web of Science databases for all papers published up to 12/19/2023 and used various genetic models to evaluate the relationship between MTHFR polymorphisms and prostate cancer risk.
Results: The meta-analysis included 26 case‒control studies with a total of 12,455 cases and 13,900 controls with the C677T polymorphism and 6,396 cases and 8,913 controls with the A1298C polymorphism. Overall, no significant association was found between the MTHFR gene polymorphisms and prostate cancer risk. However, the C677T polymorphism was associated with reduced prostate cancer risk in the Asian population (T allele vs. C allele: OR = 0.759, 95% CI 0.669–0.861, p < 0.001; TT + CT vs. CC: OR = 0.720, 95% CI 0.638–0.812, p < 0.001; TT vs. CC + CT: OR = 0.719, 95% CI 0.617–0.838, p < 0.001; TT vs. CC: OR = 0.620, 95% CI 0.522–0.737, p < 0.001); however, the A1298C polymorphism was associated with an increased risk in the mixed race group from the United States (CC + AC vs. AA: OR = 1.464, 95% CI 1.052–2.037, p = 0.024; AC vs. AA: OR = 1.615, 95% CI 1.037–2.514, p = 0.034).
Conclusion: The meta-analysis suggested that MTHFR gene polymorphisms (C677T and A1298C) may have different effects on prostate cancer risk in specific populations.
1 Introduction
Prostate cancer is a common malignant disease in males that originates from cancer cells within prostate tissue; prostate tumours often grow slowly and remain asymptomatic (Ferlay et al., 2019). According to data from the World Health Organization (WHO), prostate cancer ranks second in incidence among males worldwide, second only to lung cancer (Ferlay et al., 2021). The incidence of prostate cancer is greater among older men, especially those aged >60 years (Siegel et al., 2022). Prostate cancer is also associated with higher socioeconomic status (Chen et al., 2018). In China, the incidence of prostate cancer has been increasing, with an annual incidence rate of 39.09 cases per 100,000 persons and a 5-year survival rate of 79.8% according to the 2018 China Cancer Registry Annual Report (Descotes, 2019).
Prostate cancer is a multifactorial disease associated with risk factors, including age, family history, insulin-like growth factor levels, dietary habits, lifestyle factors, environmental factors, and occupational exposure (Perdana et al., 2016). A descriptive epidemiological study conducted by Pernar et al. emphasized that age, family history, race, and genetics were significant risk factors for prostate cancer (Malik et al., 2018). Hence, genetic polymorphisms play a crucial role in prostate cancer susceptibility. Methylenetetrahydrofolate reductase (MTHFR) is a crucial enzyme involved in DNA methylation. Mutations in the MTHFR gene may lead to changes in enzyme activity, resulting in alterations in DNA methylation levels and an increased risk of prostate cancer. A1298C and C677T are two common mutation sites in the MTHFR gene. The A1298C mutation may cause decreased enzyme activity, affecting the extent of DNA methylation and increasing the risk of prostate cancer. The C677T mutation, on the other hand, can affect enzyme stability and function, leading to disruption of the maintenance of DNA methylation levels and an increased risk of prostate cancer (Choi and Friso, 2006; Dong et al., 2008). Therefore, understanding the relationship between these two mutation sites and prostate cancer may provide new insights for early diagnosis and treatment.
However, the results of previous meta-analyses vary across studies. For example, some studies found no association between the A1298C or C677T locus and prostate cancer, while others showed a clear association (Collin et al., 2009; Li et al., 2012; Li and Xu, 2012; Zhang et al., 2012; Chen et al., 2015). In addition, previous meta-analyses did not perform subgroup analyses for different races, genetic testing methods, or sample types, which may also lead to differences in results. Although some studies have explored the relationship between the A1298C and C677T loci and prostate cancer, additional studies are needed to determine the importance of this association. To address this significant and controversial question, we performed an updated meta-analysis based on current clinical evidence to assess the association between MTHFR gene polymorphisms (C677T and A1298C) and prostate cancer risk.
2 Methods and materials
2.1 Search strategy
Two independent researchers searched the PubMed, Embase, Cochrane and Web of Science databases for all papers published up to 12/19/2023. The following keywords were used: “prostate tumour”, “prostatic neoplasms”, “prostate cancer”, “MTHFR”, “methylenetetrahydrofolate reductase”, “A1298C″, “rs1801131″, “Glu429Ala”, “C677T″, “rs1801133″, and “Ala222Val”. To ensure a comprehensive literature search, the recommended MeSH terms were used for each keyword with similar meanings. In addition, the reference lists of the included studies were manually searched to identify other published articles not indexed in public databases (Supplementary Appendix S1).
2.2 Data extraction
Two researchers extracted and crosschecked the data based on predefined inclusion and exclusion criteria. If the data were insufficient or uncertain, attempts were made to contact the original authors to supplement and verify the accuracy of the data. Incomplete studies were excluded, and only the highest-quality studies were retained among those with duplicate publications, replication, or similar data.
The following information was extracted: the surname of the first author, publication year, country, and participant race. Additionally, the number of cases and controls, matching variables, and the genotype distributions of the cases and controls from the data sources were recorded.
2.3 Quality assessment
On this quality assessment scale, six items were evaluated (Ferlay et al., 2019): appropriateness of the case definition and diagnosis (Ferlay et al., 2021), representativeness of cases (Siegel et al., 2022), selection of controls (Chen et al., 2018), definition of controls (Descotes, 2019), investigation and assessment of exposure, and (Perdana et al., 2016) similarity of investigation methods between cases and controls. The quality score ranged from 0 to 9. Studies scoring less than 4 points were considered low-quality studies, studies scoring 4–6 points were considered moderate-quality studies, and those scoring >6 pints were considered high-quality studies.
2.4 Data analysis
By calculating the combined odds ratios (ORs) and corresponding 95% confidence intervals (95% CIs) of the gene frequencies for each genetic model and considering p < 0.05 to indicate statistical significance, we evaluated the association between the MTHFR C677T and A1298C polymorphisms and the risk of prostate cancer. We compared five genetic models (Ferlay et al., 2019): an allelic model (C677T: T allele vs. C allele; A1298C: C allele vs. A allele) (Ferlay et al., 2021); a dominant model [C677T: (TT + CT) vs. CC; A1298C: (CC + AC) vs. AA] (Siegel et al., 2022); a recessive model (C677T: TT vs. (CC + CT); A1298C: CC vs. (AA + AC); (Chen et al., 2018) an overdominant model (C677T: CT vs. TT; A1298C: AC vs. CC); and (Descotes, 2019) an additive model (C677T: TT vs. CC; A1298C: CC vs. AA). We assessed heterogeneity among the included studies using the chi-square-based Q test and I2 test. A p-value < 0.10 indicated heterogeneity among the studies (Cumpston et al., 2019). If the I2 value was <40%, the heterogeneity was likely unimportant, whereas if the I2 value was >75%, it was believed that there was considerable heterogeneity (Chang et al., 2022). Due to unavoidable differences in heterogeneity, a random effects model (REM) was used for quantitative meta-analysis (Khandagale et al., 2023). Additionally, subgroup analyses were performed to investigate the potential sources of heterogeneity. Subgroup analyses were performed based on race, genotyping method, and sample type, and Hardy-Weinberg equilibrium (HWE) was calculated using the chi-square goodness-of-fit test, with p > 0.05 indicating HWE in the control group; otherwise, Hardy-Weinberg disequilibrium (HWD) was indicated. Sensitivity analysis was conducted by sequentially excluding each study, and if the sensitivity analysis results were not robust within an acceptable range, the study was removed for reanalysis. Begg’s funnel plot and Egger’s test were employed to assess the publication bias risk in the selected studies. All the abovementioned statistical analyses were performed using STATA 15.0.
3 Results
3.1 Study characteristics
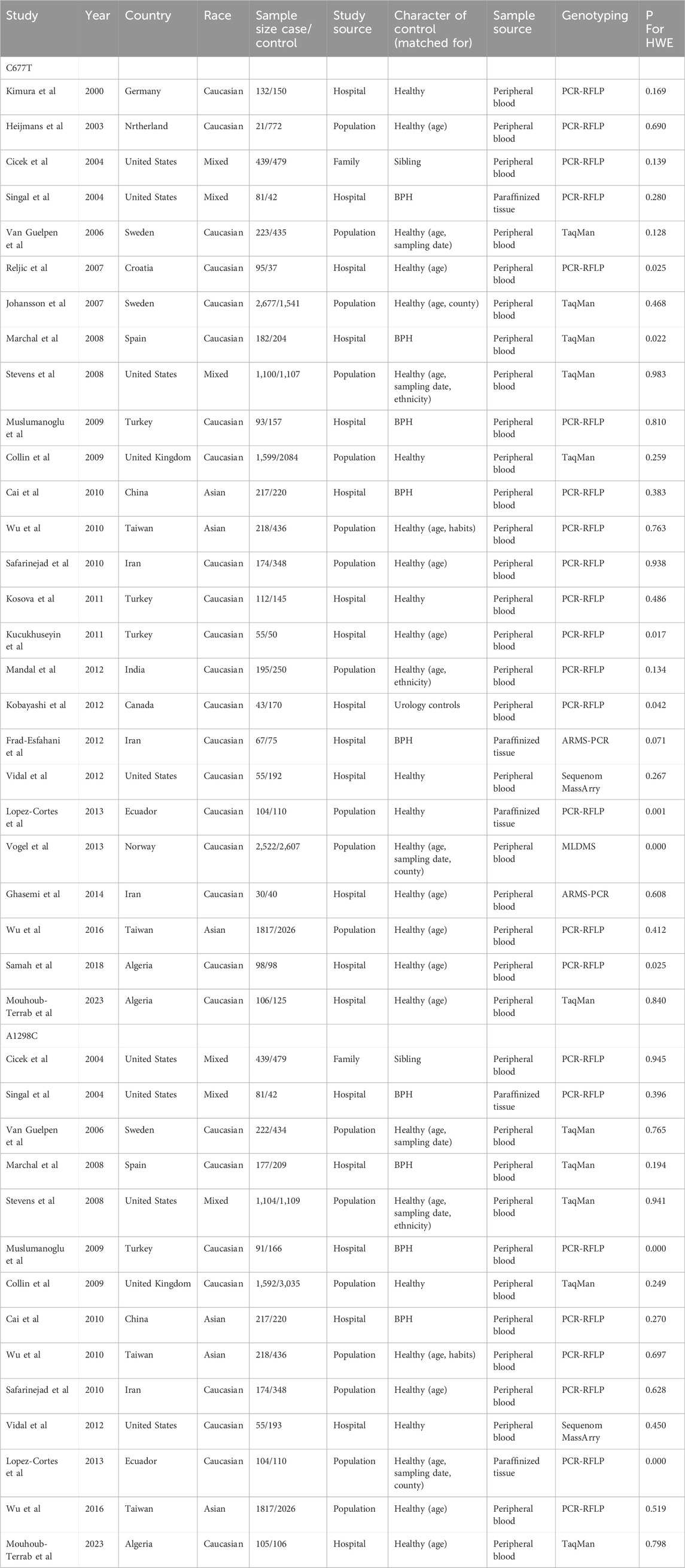
A literature search and screening of the Embase, PubMed, Web of Science, and Cochrane databases resulted in a total of 39 articles from Embase, 26 articles from PubMed, 60 articles from Web of Science, and no articles from the Cochrane database or manual search. The detailed screening process is illustrated in Figure 1. After reading the titles and abstracts, 99 articles were excluded; 35 were duplicates, 40 were irrelevant to the topic, 19 were reviews or meta-analyses, 4 were conference abstracts, and 1 had incomplete data. Overall, our meta-analysis included 26 eligible case‒control studies (Figure 1). Among the 26 studies, all studies investigated C677T, while 14 studies investigated A1298C. The genotype frequencies of C677T and A1298C were separately reported; thus, these reports were considered individual studies in this meta-analysis. Therefore, all 26 studies (Kimura et al., 2000; Heijmans et al., 2003; Cicek et al., 2004; Singal et al., 2004; Van Guelpen et al., 2006; Johansson et al., 2007; Reljic et al., 2007; Marchal et al., 2008; Stevens et al., 2008; Collin et al., 2009; Muslumanoglu et al., 2009; Cai et al., 2010; Safarinejad et al., 2010; Wu et al., 2010; Kosova et al., 2011; Küçükhüseyin et al., 2011; Fard-Esfahani et al., 2012; Kobayashi et al., 2012; Mandal et al., 2012; Vidal et al., 2012; de Vogel et al., 2013; López-Cortés et al., 2013; Ghasemi et al., 2014; Wu et al., 2016; TELLOUCHE-BOUHOUHOU et al., 2018; Mouhoub-Terrab et al., 2023) included 12,455 cases and 13,900 controls with the C677T polymorphism and 6,396 cases and 8,913 controls with the A1298C polymorphism (Table 1).
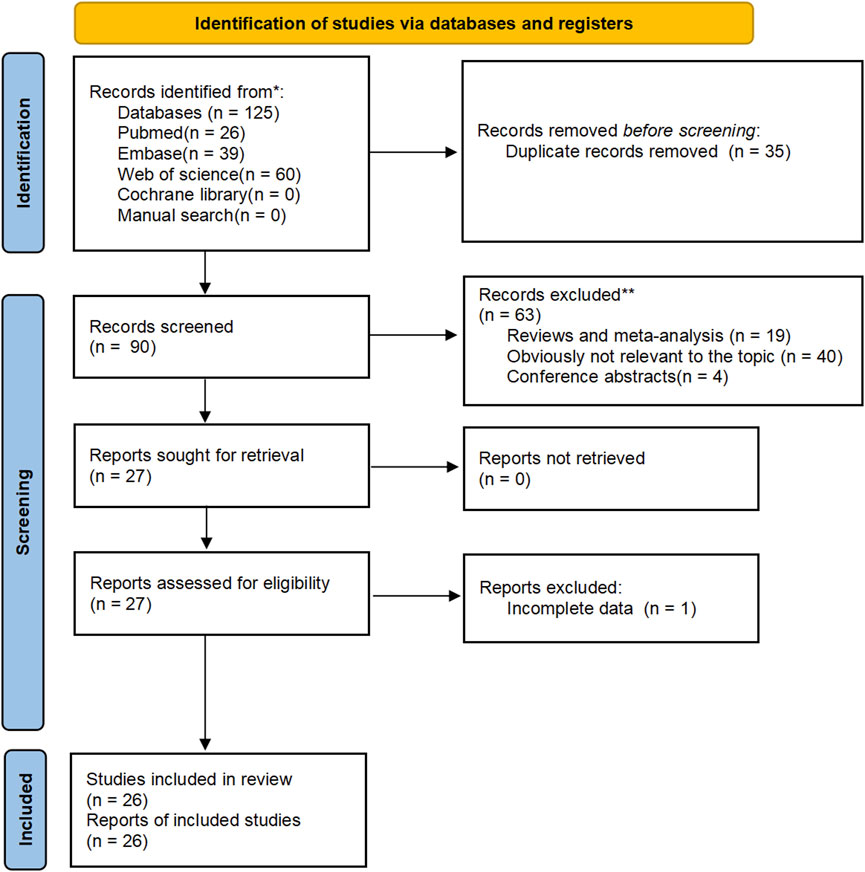
FIGURE 1. Flow chart of meta-analysis to identify associations between MTHFR gene polymorphisms (C677T, A1298C) and prostate cancer risk.
The participants included Caucasians, Asians, and individuals of mixed races. Genotyping of the included SNPs was performed using TaqMan, polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), an amplification refractory mutation system (ARMS), Sequenom MassArray, and matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS). According to the methodological quality assessment, all studies had scores above 4. The genotype distributions of the control groups in all studies were tested for Hardy‒Weinberg equilibrium (HWE) (Table 1), and studies deviating from HWE were excluded from subsequent analyses. Thus, the final analysis included 19 studies for the C677T polymorphism, comprising 9,356 cases and 10,624 controls, and 12 studies for the A1298C polymorphism, comprising 6,201 cases and 8,637 controls.
3.2 Association between the C677T (rs1801133) polymorphism and prostate cancer susceptibility
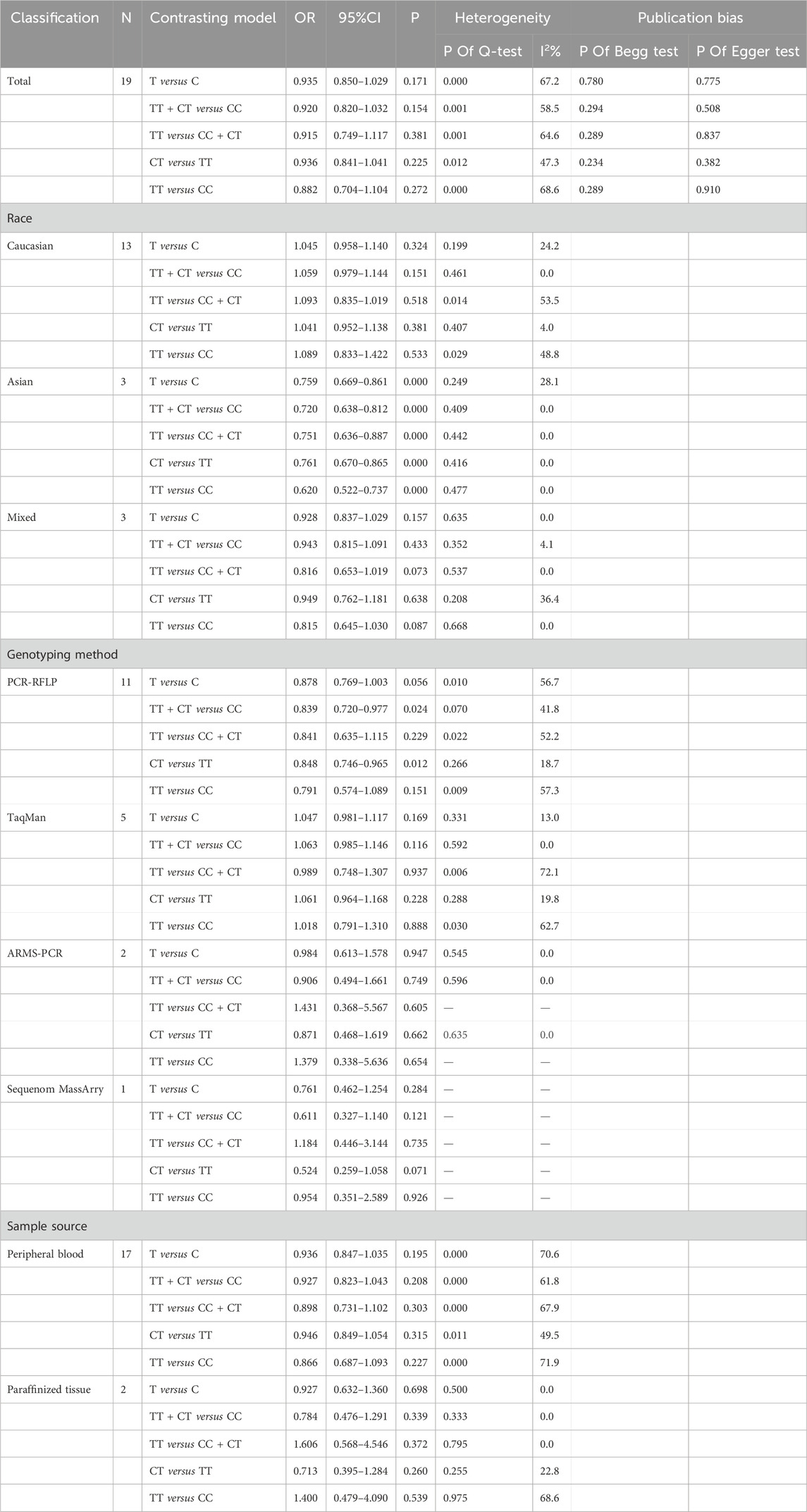
The strength of the association between the C677T (rs1801133) polymorphism and prostate cancer risk is presented in Table 2. As shown in Table 2, no significant associations were detected in any of the genetic models (T allele vs. C allele: OR = 0.935, 95% CI 0.850–1.029, p = 0.171, I2 = 62.7%, pH < 0.001; TT + CT vs. CC: OR = 0.920, 95% CI 0.820–1.032, p = 0.154, I2 = 58.5%, pH = 0.001; TT vs. CC + CT: OR = 0.915, 95% CI 0.749–1.117, p = 0.381, I2 = 64.6%, pH < 0.001; CT vs. CC: OR = 0.936, 95% CI 0.841–1.041, p = 0.225, I2 = 47.3%, pH = 0.012; TT vs. CC: OR = 0.882, 95% CI 0.704–1.104, p = 0.272, I2 = 68.6%, pH < 0.001) (Table 2) (Figures 2A–E).

FIGURE 2. (A) Forest plot of the association between MTHFR gene C677T polymorphism and PCa risk under Allelic model. (B) Forest plot of the association between MTHFR gene C677T polymorphism and PCa risk under Dominant model. (C) Forest plot of the association between MTHFR gene C677T polymorphism and PCa risk under recessive model. (D) Forest plot of the association between MTHFR gene C677T polymorphism and PCa risk under over-dominant model. (E) Forest plot of the association between MTHFR gene C677T polymorphism and PCa risk under additive model.
Subgroup analysis (Supplementary Appendix S2) was performed to evaluate the heterogeneity among the studies based on race, genotyping method, and sample type. According to our subgroup analysis, heterogeneity was significantly reduced among all the race subgroups. In the Asian population, a decreased risk of prostate cancer was associated with the C677T polymorphism (T allele vs. C allele: OR = 0.759, 95% CI 0.669–0.861, p < 0.001, I2 = 28.1%, pH = 0.249; TT + CT vs. CC: OR = 0.720, 95% CI 0.638–0.812, p < 0.001, I2 = 0.0%, pH = 0.409; TT vs. CC + CT: OR = 0.719, 95% CI 0.617–0.838, p < 0.001, I2 = 0.0%, pH = 0.442; CT vs. CC: OR = 0.761, 95% CI 0.670–0.865, p < 0.001, I2 = 0.0%, pH = 0.416; TT vs. CC: OR = 0.620, 95% CI 0.522–0.737, p=<0.001, I2 = 0.0%, pH = 0.477). With respect to the different genotyping methods, heterogeneity in the dominant and allelic models was reduced in the TaqMan and ARMan-ARMS-PCR groups. The overdominant model showed reduced heterogeneity in the PCR-RFLP, TaqMan, and ARMan-ARMS-PCR groups. However, the heterogeneity remained relatively high in the other models. In the PCR-RFLP group, a decreased risk of prostate cancer was observed in the comparison of the dominant and overdominant models (TT + CT vs. CC: OR = 0.839, 95% CI 0.720–0.977, p = 0.024, I2 = 41.8%, pH = 0.070; CT vs. CC: OR = 0.848, 95% CI 0.746–0.965, p = 0.012, I2 = 18.7%, pH = 0.266). In the subgroup analysis based on sample type, even after excluding two studies that extracted genes from paraffin-embedded tissues (Singal and Vidal), heterogeneity remained high. However, in the paraffinized tissue subgroup, heterogeneity significantly decreased in all models except for the additive model. No significant correlation was found in any of the genetic models (Table 2).
3.3 Association between the A1298C (rs1801131) polymorphism and prostate cancer susceptibility
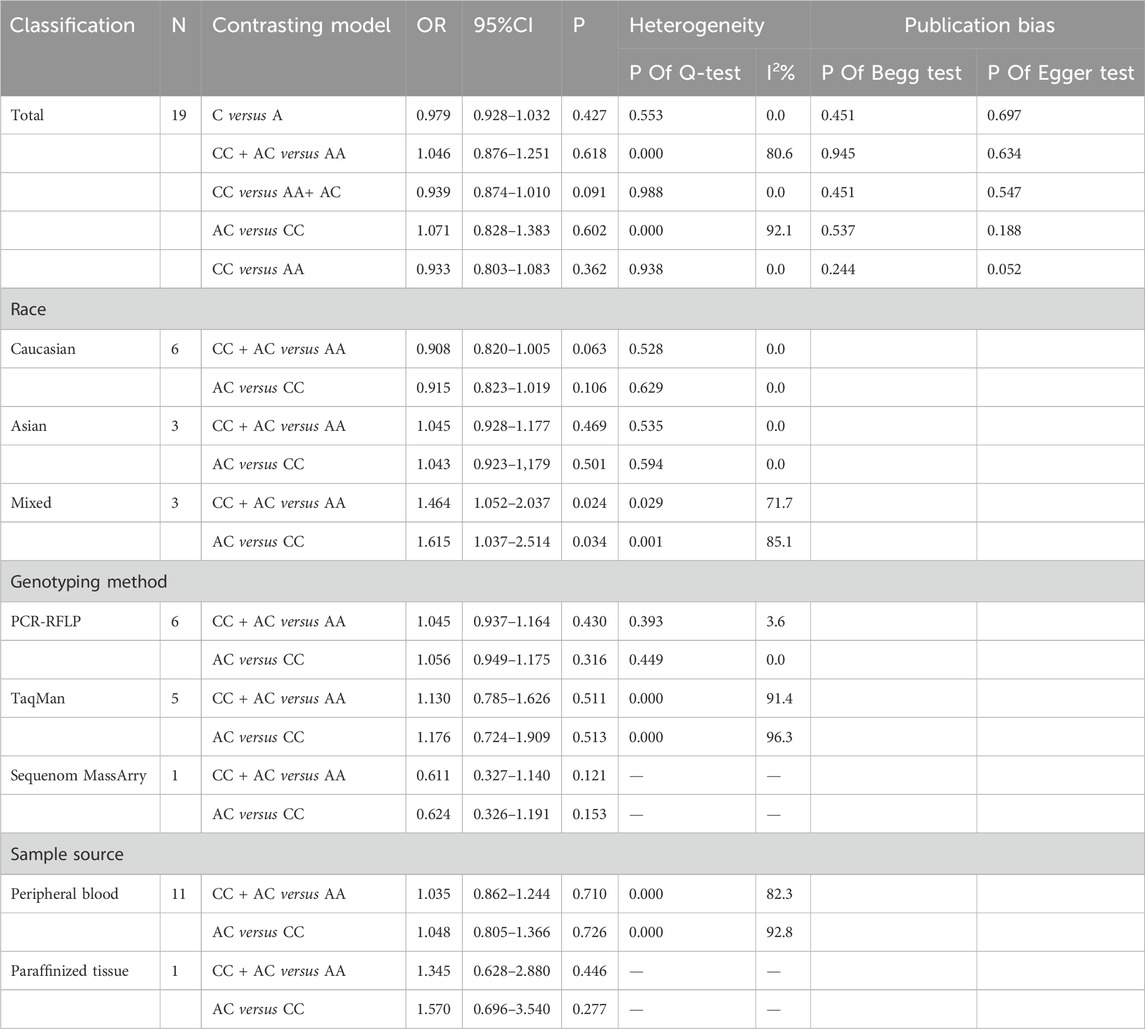
The association between the A1298C (rs1801131) polymorphism and prostate cancer risk was not found to be significant in any of the genetic models (Table 2) (C allele vs. A allele: OR = 0.979, 95% CI 0.928–1.032, pH = 0.553, I2 = 0.0%, p = 0.427; CC + AC vs. AA: OR = 1.046, 95% CI 0.876–1.251, pH < 0.001, I2 = 80.6%, p = 0.618; CC vs. AA + AC: OR = 0.939, 95% CI 0.874–1.010, pH = 0.988, I2 = 0.0%, p = 0.091; AC vs. AA: OR = 1.071, 95% CI 0.828–1.383, pH < 0.001, I2 = 92.1%, p = 0.602; CC vs. AA: OR = 0.933, 95% CI 0.803–1.083, pH = 0.938, I2 = 0.0%, p = 0.362) (Table 3) (Figures 3A–E).
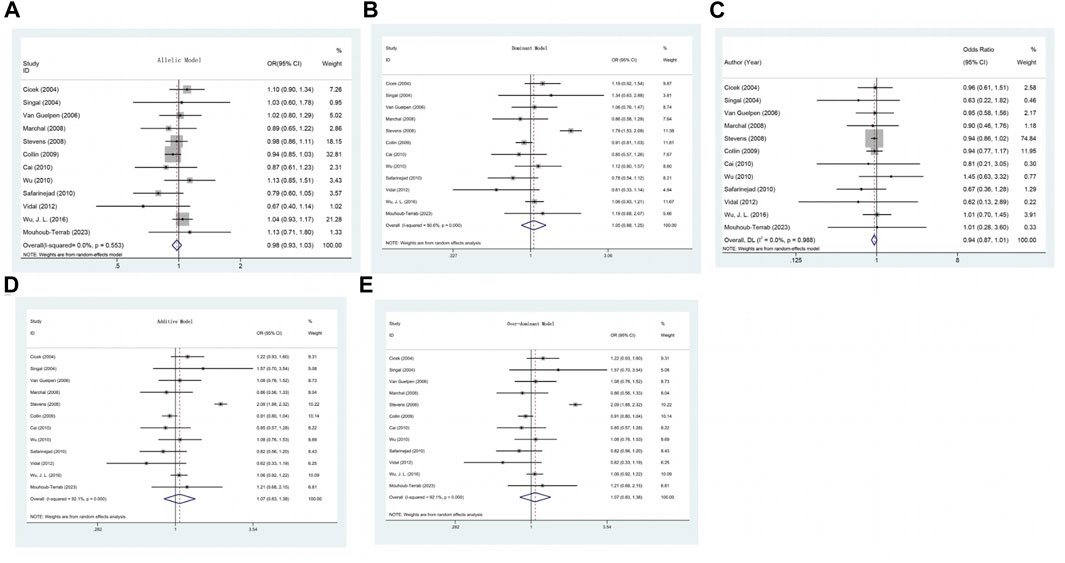
FIGURE 3. (A) Forest plot of the association between MTHFR gene A1298C polymorphism and PCa risk under allelic model. (B) Forest plot of the association between MTHFR gene A1298C polymorphism and PCa risk under dominant model. (C) Forest plot of the association between MTHFR gene A1298C polymorphism and PCa risk under recessive model. (D) Forest plot of the association between MTHFR gene A1298C polymorphism and PCa risk under over-dominant model. (E) Forest plot of the association between MTHFR gene A1298C polymorphism and PCa risk under additive model.
Furthermore, in the models with high heterogeneity, subgroup analysis (Supplementary Appendix S2) based on race, genotyping method, and sample type did not reveal any significant associations. However, it is worth noting that the studies by Cicek, Singal, and Stevens included multiethnic populations in the United States, and the subgroup analysis indicated an increased risk of prostate cancer in the comparison of the dominant and overdominant models (CC + AC vs. AA: OR = 1.464, 95% CI 1.052–2.037, p = 0.024, I2 = 71.7%, pH = 0.029; AC vs. AA: OR = 1.615, 95% CI 1.037–2.514, p = 0.034, I2 = 85.1%, pH = 0.001). For the dominant and overdominant models, a significant reduction in heterogeneity was observed among Caucasians and Asians. In studies using different genotyping methods, heterogeneity was significantly reduced compared with that in those utilizing PCR-RFLP. According to the subgroup analysis based on sample type, heterogeneity remained high even after excluding two studies (Singal) that extracted genes from paraffin-embedded tissues (Table 3).
3.4 Sensitivity analysis and publication bias
After systematically excluding each individual study and conducting a second meta-analysis to pool effect sizes, it was found that the analysis results for all models regarding C677T remained consistent with the primary analysis, demonstrating the robustness of the main findings (Porta et al., 2014). However, during the sensitivity analysis of A1298C, a notable alteration was observed in the homozygous (CC vs. AA) model comparison when the study by Stevens was excluded due to its high sensitivity.
We employed both Begg’s and Egger’s tests to assess publication bias, and all studies demonstrated publication bias within an acceptable range (p-value > 0.05) (Supplementary Appendix S3).
4 Statistical analysis summary
This study investigates the relationship between MTHFR gene polymorphisms (C677T and A1298C) and genetic susceptibility to prostate cancer through a meta-analysis. Independent researchers searched databases like PubMed, Embase, Cochrane, and Web of Science until 19 December 2023, analyzing the association between MTHFR polymorphisms and prostate cancer risk using various genetic models. The meta-analysis included 26 case-control studies, involving 12,455 cases and 13,900 controls for C677T polymorphism, and 6,396 cases and 8,913 controls for A1298C polymorphism. Overall, no significant association was found between MTHFR polymorphisms and prostate cancer risk. However, in Asian populations, C677T polymorphism was associated with a reduced risk of prostate cancer, while in mixed-race groups in the USA, A1298C polymorphism was linked to an increased risk.
5 Discussion
In this meta-analysis, we evaluated the relationship between two SNPs (C677T and A1298C) in the methylenetetrahydrofolate reductase gene and prostate cancer risk based on 26 published studies. Overall, our analysis did not find evidence to suggest an association between the C677T or A1298C polymorphisms and the risk of prostate cancer in any genetic model. However, in subgroup analysis, we found that the C677T polymorphism may be associated with a reduced risk of prostate cancer in Asian populations, while the A1298C polymorphism may be associated with an increased risk in the American population.
The present analysis revealed that the C677T polymorphism is associated with a reduced risk of prostate cancer according to various genetic models, including the allele, dominant, recessive, and additive models. Li et al. (2012) also proposed a protective effect of the C677T allele polymorphism (Li and Xu, 2012). However, Chen et al. (2015) reported that the C677T polymorphism is associated with an increased risk of prostate cancer in East Asian populations, which contradicts previous findings (Chen et al., 2015). Moreover, Colin et al. (2009) and Zhang et al. (2012) did not find any association between the C677T polymorphism and the risk of prostate cancer. On the one hand, the present study incorporated the latest research and included a larger number of cases and controls, which enhances the reliability and statistical significance of the results (Collin et al., 2009; Zhang et al., 2012). On the other hand, the exclusion of studies with controls that deviated from Hardy-Weinberg equilibrium (HWE) might account for the divergent conclusions. HWE is an important principle in genetics that describes a stable state where genotype frequencies remain constant. A deviation of the control group from HWE may affect the accuracy of the study’s conclusions.
Regarding the A1298C polymorphism, previous meta-analyses showed that there is no association between this polymorphism and the risk of prostate cancer, regardless of whether the analysis was performed on the overall data or based on different regions, populations, genotyping methods, or prostate cancer stages (Li et al., 2012; Li and Xu, 2012). In our study, a subgroup analysis revealed increased susceptibility to prostate cancer in the mixed-race group when comparing the dominant and overdominant models. It is worth noting that the mixed-race group comprised individuals from various ethnic backgrounds, all of whom happen to be from the United States. Therefore, the correlation between the A1298C polymorphism and prostate cancer risk may be influenced by certain regional factors, such as dietary habits, climate conditions, and even cultural and socioeconomic factors.
Significant heterogeneity was observed in the association between the C677T and A1298C polymorphisms and prostate cancer risk. Therefore, subgroup analyses were performed based on race, genotyping method, and sample type. The heterogeneity was significantly reduced in different race groups, and a reduced risk of prostate cancer associated with the C677T polymorphism was observed in the Asian population. Furthermore, studies using the PCR-RFLP technique revealed that the C677T polymorphism decreased susceptibility to prostate cancer. These findings emphasize the importance of using consistent genotyping methods for accurate assessment of the association between the C677T polymorphism and prostate cancer risk. According to the subgroup analysis of the association between the A1298C polymorphism and prostate cancer risk, no significant associations were found. It is worth noting that while the studies by Cicek, Singal, and Stevens included a multiethnic population from the United States, which may consist of Caucasian, Asian, and black individuals, the analysis indicated an increased risk of prostate cancer in the comparison of the dominant and overdominant models. This finding suggested that the association between the A1298C polymorphism and prostate cancer susceptibility may be influenced by regional factors such as diet and requires further investigation.
In the race-based subgroup analysis for C667T and A1298C, a notable reduction in heterogeneity was observed, clearly highlighting the role of race as a factor contributing to heterogeneity in various studies. This underscores the importance of performing race-specific subgroup analyses in such research. According to the genotyping method-based subgroup analysis for C667T, the dominant and allelic models showed less heterogeneity in the TaqMan and ARMan-ARMS-PCR groups, while the overdominant model demonstrated reduced heterogeneity across the PCR-RFLP, TaqMan, and ARMan-ARMS-PCR groups. Similarly, for A1289C, studies utilizing the PCR-RFLP method displayed a marked decrease in heterogeneity. These observations indicate that the choice of genotyping method significantly influences heterogeneity, making it a crucial consideration in genetic research. Additionally, recognizing and accounting for methodological differences is vital when comparing or integrating study outcomes. According to our subgroup analysis focused on the sample source for C667T, all the models, except for the additive model, showed significantly less heterogeneity in the paraffin-embedded tissue subgroup. These findings suggest that the choice of sample source can contribute to heterogeneity and imply that paraffin-embedded tissues may be more consistent and reliable as a sample source in genetic studies.
Our meta-analysis is subject to several limitations. First, despite our efforts to incorporate the latest research, the number of included studies remained relatively small. Second, due to insufficient data availability, we were unable to adjust our results for other factors, such as patient age, sex, and environmental variables. Third, this meta-analysis exclusively examined the polymorphisms of two loci within Caucasian and Asian populations, thus precluding the assessment of the relationship between the C667T and A1298C polymorphisms and prostate cancer in other ethnic groups. Moreover, we must acknowledge the potential presence of publication bias since our analysis encompasses only published studies, and statistically nonsignificant results are often less likely to be published. Finally, the observed heterogeneity among the included studies may be attributed to various factors, including geographic distribution, participant demographics, study design, and methodological disparities.
6 Conclusion
In conclusion, our study provides an updated meta-analysis estimating the association between MTHFR gene polymorphisms and prostate cancer risk, incorporating a larger sample size than did previous studies. The C677T polymorphism may be associated with a reduced risk of prostate cancer in Asian populations, while the presence of the A1298C polymorphism may be associated with an increased risk in the U.S. population. Future studies should focus on large-scale, well-designed research incorporating regional factors such as diet and climate to confirm the association between MTHFR polymorphisms and prostate cancer susceptibility.
Author contributions
JY: Conceptualization, Formal Analysis, Investigation, Methodology, Visualization, Writing–original draft, Writing–review and editing. YH: Conceptualization, Formal Analysis, Investigation, Methodology, Visualization, Writing–original draft, Writing–review and editing. XS: Conceptualization, Software, Validation, Writing–review and editing. YC: Validation, Writing–review and editing. XD: Conceptualization, Data curation, Supervision, Validation, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We would like to thank the researchers and study participants for their contributions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1343687/full#supplementary-material
References
Cai, D., Ning, L., Pan, C., Liu, X., Bu, R., Chen, X., et al. (2010). Association of polymorphisms in folate metabolic genes and prostate cancer risk: a case-control study in a Chinese population. J. Genet. 89 (2), 263–267. eng. Epub 2010/09/24. Cited in: Pubmed; PMID 20861582. doi:10.1007/s12041-010-0037-7
Chang, Y., Phillips, M. R., Guymer, R. H., Thabane, L., Bhandari, M., Chaudhary, V., et al. (2022). The 5 min meta-analysis: understanding how to read and interpret a forest plot. Eye (Lond) 36 (4), 673–675. eng. Epub 2022/01/05.Cited in:Pubmed;PMID 34987196. doi:10.1038/s41433-021-01867-6
Chen, P. L., Li, W. T., Wang, J., Jiang, Y. D., Wu, P., Chen, T., et al. (2015). Association between MTHFR gene polymorphisms (C677T, A1298C) and genetic susceptibility to prostate cancer: a meta-analysis. GMR 14 (4), 19191–19202. eng. Epub 2016/01/20. Cited in: Pubmed; PMID 26782572. doi:10.4238/2015.December.29.29
Chen, W. Q., Li, H., Sun, K. X., Zheng, R. S., Zhang, S. W., Zeng, H. M., et al. (2018). Report of cancer incidence and mortality in China, 2014. Zhonghua zhong liu za zhi Chin. J. Oncol. 40 (1), 5–13. chi. Epub 2018/01/25. Cited in: Pubmed; PMID 29365411. doi:10.3760/cma.j.issn.0253-3766.2018.01.002
Choi, S.-W., and Friso, S. (2006). Interaction between folate and methylene-tetrahydrofolate reductase gene in cancer. Nutrient-gene Interact. cancer, 57–74. doi:10.1201/9781420004847-4
Cicek, M. S., Nock, N. L., Li, L., Conti, D. V., Casey, G., and Witte, J. S. (2004). Relationship between methylenetetrahydrofolate reductase C677T and A1298C genotypes and haplotypes and prostate cancer risk and aggressiveness. a Publ. Am. Assoc. Cancer Res. cosponsored by Am. Soc. Prev. Oncol. 13 (8), 1331–1336. eng. Epub 2004/08/10. Cited in: Pubmed; PMID 15298954. doi:10.1158/1055-9965.1331.13.8
Collin, S. M., Metcalfe, C., Zuccolo, L., Lewis, S. J., Chen, L., Cox, A., et al. (2009). Association of folate-pathway gene polymorphisms with the risk of prostate cancer: a population-based nested case-control study, systematic review, and meta-analysis. a Publ. Am. Assoc. Cancer Res. cosponsored by Am. Soc. Prev. Oncol. 18 (9), 2528–2539. eng. Epub 2009/08/27. Cited in: Pubmed; PMID 19706844. doi:10.1158/1055-9965.Epi-09-0223
Cumpston, M., Li, T., Page, M. J., Chandler, J., Welch, V. A., Higgins, J. P., et al. (2019). Updated guidance for trusted systematic reviews: a new edition of the Cochrane Handbook for Systematic Reviews of Interventions. Cochrane Database Syst. Rev. 10 (10), ED000142. ED000142. eng. Epub 2019/09/12.Cited in: Pubmed; PMID 31643080. doi:10.1002/14651858.ED000142
Descotes, J. L. (2019). Diagnosis of prostate cancer. Asian J. urology 6 (2), 129–136. eng. Epub 2019/05/08. Cited in: Pubmed; PMID 31061798. doi:10.1016/j.ajur.2018.11.007
de Vogel, S., Meyer, K., Fredriksen Å, U. A., Ueland, P. M., Nygård, O., Vollset, S. E., et al. (2013). Serum folate and vitamin B12 concentrations in relation to prostate cancer risk–a Norwegian population-based nested case-control study of 3000 cases and 3000 controls within the JANUS cohort. Int. J. Epidemiol. 42 (1), 201–210. eng. Epub 2013/03/20. Cited in: Pubmed; PMID 23508410. doi:10.1093/ije/dys199
Dong, L. M., Potter, J. D., White, E., Ulrich, C. M., Cardon, L. R., and Peters, U. (2008). Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. Jama 299 (20), 2423–2436. eng. Epub 2008/05/29. Cited in: Pubmed; PMID 18505952. doi:10.1001/jama.299.20.2423
Fard-Esfahani, P., Mohammadi Torbati, P., Hashemi, Z., Fayaz, S., and Golkar, M. (2012). Analysis of relation between C677T genotype in MTHFR gene and prostatic cancer in Iranian males. Acta medica Iran. 50 (10), 657–663. eng. Epub 2013/01/01. Cited in: Pubmed; PMID 23275280.
Ferlay, J., Colombet, M., Soerjomataram, I., Mathers, C., Parkin, D. M., Piñeros, M., et al. (2019). Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. cancer 144 (8), 1941–1953. eng. Epub 2018/10/24. Cited in: Pubmed; PMID 30350310. doi:10.1002/ijc.31937
Ferlay, J., Ervik, M., Lam, F., Colombet, M., Mery, L., and Piñeros, M. (2021). Global cancer observatory: cancer today. Lyon, France: International Agency for Research on Cancer.
Ghasemi, S., Tavakoli, A., Moghadam, M., Zargar, M. A., Abbaspour, M., Hatamnejadian, N., et al. (2014). Risk of prostate cancer and thrombosis-related factor polymorphisms. Biomed. Rep. 2 (1), 53–56. eng. Epub 2014/03/22. Cited in: Pubmed; PMID 24649068. doi:10.3892/br.2013.180
Heijmans, B. T., Boer, J. M., Suchiman, H. E., Cornelisse, C. J., Westendorp, R. G., Kromhout, D., et al. (2003). A common variant of the methylenetetrahydrofolate reductase gene (1p36) is associated with an increased risk of cancer. Cancer Res. 63 (6), 1249–1253. eng. Epub 2003/03/22. Cited in: Pubmed; PMID 12649184.
Johansson, M., Van Guelpen, B., Hultdin, J., Wiklund, F., Adami, H. O., Bälter, K., et al. (2007). The MTHFR 677C --> T polymorphism and risk of prostate cancer: results from the CAPS study. CCC 18 (10), 1169–1174. eng. Epub 2007/09/12. Cited in: Pubmed; PMID 17846906. doi:10.1007/s10552-007-9055-z
Khandagale, S. B., Kamble, V. S., Oza, C., Bhor, S., Khadilkar, A. V., and Khare, S. P. (2023). Surrogate markers of metabolic syndrome and insulin resistance in children and young adults with type 1 diabetes: a systematic review and meta-analysis (MetS and IR in T1DM). Int. J. Diabetes Dev. Ctries. doi:10.1007/s13410-023-01284-3
Kimura, F., Franke, K. H., Steinhoff, C., Golka, K., Roemer, H. C., Anastasiadis, A. G., et al. (2000). Methyl group metabolism gene polymorphisms and susceptibility to prostatic carcinoma. Prostate 45 (3), 225–231. eng. Epub 2000/11/14. 3<225::aid-pros4>3.0.co;2-7. Cited in: Pubmed; PMID 11074524. doi:10.1002/1097-0045(20001101)45:3<225::aid-pros4>3.0.co;2-7
Kobayashi, L. C., Limburg, H., Miao, Q., Woolcott, C., Bedard, L. L., Massey, T. E., et al. (2012). Folate intake, alcohol consumption, and the methylenetetrahydrofolate reductase (MTHFR) C677T gene polymorphism: influence on prostate cancer risk and interactions. Front. Oncol. 2, 100. eng. Epub 2012/08/23. Cited in: Pubmed; PMID 22912935. doi:10.3389/fonc.2012.00100
Kosova, B., Ozel, R., Kaymaz, B., and Aktan, C. (2011). Bozok çetintaş V, sen S, çal Ç. Effects of the catechol-O-methyltransferase val108/158Met and methylenetetrahydrofolate reductase C677T gene polymorphisms on prostate cancer susceptibility. Turkiye Klinikleri J. Med. Sci. 08/01 (31), 943–950. doi:10.5336/medsci.2010-17861
Küçükhüseyin, Ö., Kurnaz, Ö., Akadam-Teker, A. B., Narter, F., Yılmaz-Aydoğan, H., and İsbir, T. (2011). Effects of the MTHFR C677T polymorphism on prostate specific antigen and prostate cancer. APJCP 12 (9), 2275–2278. eng. Epub 2012/02/03. Cited in: Pubmed; PMID 22296369.
Li, D., Tian, T., Guo, C., Ren, J., Yan, L., Liu, H., et al. (2012). No association of the MTHFR gene A1298C polymorphism with the risk of prostate cancer: a meta-analysis. Exp. Ther. Med. 3 (3), 493–498. eng. Epub 2012/09/13. Cited in: Pubmed; PMID 22969917. doi:10.3892/etm.2012.445
Li, X. L., and Xu, J. H. (2012). MTHFR polymorphism and the risk of prostate cancer: a meta-analysis of case-control studies. Prostate cancer prostatic Dis. 15 (3), 244–249. eng. Epub 2012/03/01. Cited in: Pubmed; PMID 22370724. doi:10.1038/pcan.2012.5
López-Cortés, A., Jaramillo-Koupermann, G., Muñoz, M. J., Cabrera, A., Echeverría, C., Rosales, F., et al. (2013). Genetic polymorphisms in MTHFR (C677T, A1298C), MTR (A2756G) and MTRR (A66G) genes associated with pathological characteristics of prostate cancer in the Ecuadorian population. Am. J. Med. Sci. 346 (6), 447–454. eng. Epub 2013/03/06. Cited in: Pubmed; PMID 23459165. doi:10.1097/MAJ.0b013e3182882578
Malik, S. S., Batool, R., Masood, N., and Yasmin, A. (2018). Risk factors for prostate cancer: a multifactorial case-control study. Curr. problems cancer 42 (3), 337–343. eng. Epub 2018/02/13. Cited in: Pubmed; PMID 29433825. doi:10.1016/j.currproblcancer.2018.01.014
Mandal, R. K., Nissar, K., and Mittal, R. D. (2012). Genetic variants in metabolizing genes NQO1, NQO2, MTHFR and risk of prostate cancer: a study from North India. Mol. Biol. Rep. 39 (12), 11145–11152. eng. Epub 2012/10/12. Cited in: Pubmed; PMID 23054000. doi:10.1007/s11033-012-2023-z
Marchal, C., Redondo, M., Reyes-Engel, A., Perea-Milla, E., Gaitan, M. J., Machuca, J., et al. (2008). Association between polymorphisms of folate-metabolizing enzymes and risk of prostate cancer. Eur. J. Surg. Oncol. 34 (7), 805–810. eng. Cited in: Pubmed; PMID 17967524. doi:10.1016/j.ejso.2007.09.008
Mouhoub-Terrab, R., Chibane, A. A., and Khelil, M. (2023). No association between MTHFR gene C677T/A1298C polymorphisms, serum folate, vitamin B12, homocysteine levels, and prostate cancer in an Algerian population. Mol. Genet. genomic Med. 11 (9), e2194. eng. Epub 2023/05/14. Cited in: Pubmed; PMID 37182212. doi:10.1002/mgg3.2194
Muslumanoglu, M. H., Tepeli, E., Demir, S., Uludag, A., Uzun, D., Atli, E., et al. (2009). The analysis of the relationship between A1298C and C677T polymorphisms of the MTHFR gene with prostate cancer in Eskisehir population. Genet. Test. Mol. biomarkers 13 (5), 641–645. eng. Epub 2009/10/10. Cited in: Pubmed; PMID 19814618. doi:10.1089/gtmb.2009.0046
Perdana, N. R., Mochtar, C. A., Umbas, R., and Hamid, A. R. (2016). The risk factors of prostate cancer and its prevention: a literature review. Acta medica Indones. 48 (3), 228–238. eng. Epub 2016/11/15. Cited in: Pubmed; PMID 27840359.
Porta, M., Greenland, S., Hernán, M., Silva, I. S., Last, J. M., and Burón, A. (2014). A dictionary of epidemiology. New York: Oxford University Press.
Reljic, A., Simundic, A. M., Topic, E., Nikolac, N., Justinic, D., and Stefanovic, M. (2007). The methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and cancer risk: the Croatian case-control study. Clin. Biochem. 40 (13-14), 981–985. eng. Epub 2007/06/19. Cited in: Pubmed; PMID 17573062. doi:10.1016/j.clinbiochem.2007.05.005
Safarinejad, M. R., Shafiei, N., and Safarinejad, S. (2010). Relationship between three polymorphisms of methylenetetrahydrofolate reductase (MTHFR C677T, A1298C, and G1793A) gene and risk of prostate cancer: a case-control study. Prostate 70 (15), 1645–1657. eng. Epub 2010/06/22. Cited in: Pubmed; PMID 20564317. doi:10.1002/pros.21200
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics. CA a cancer J. Clin. 72 (1), 7–30. eng. Epub 2022/01/13. Cited in: Pubmed; PMID 35020204. doi:10.3322/caac.21442
Singal, R., Ferdinand, L., Das, P. M., Reis, I. M., and Schlesselman, J. J. (2004). Polymorphisms in the methylenetetrahydrofolate reductase gene and prostate cancer risk. Int. J. Oncol. 25 (5), 1465–1471. eng. Epub 2004/10/20. Cited in: Pubmed; PMID 15492840. doi:10.3892/ijo.25.5.1465
Stevens, V. L., Rodriguez, C., Sun, J., Talbot, J. T., Thun, M. J., and Calle, E. E. (2008). No association of single nucleotide polymorphisms in one-carbon metabolism genes with prostate cancer risk. a Publ. Am. Assoc. Cancer Res. cosponsored by Am. Soc. Prev. Oncol. 17 (12), 3612–3614. eng. Epub 2008/12/10. Cited in: Pubmed; PMID 19064578. doi:10.1158/1055-9965.Epi-08-0789
Tellouche-Bouhouhou, S., Chellat-Rezgoune, D., Abadi, N., Satta, D., and Dahdouh, A. (2018). Methylenetetrahydrofolate reductase c677t gene polymorphism and prostate cancer risk. Asian J. Pharm. Clin. Res. 11 (5), 387–389. doi:10.22159/ajpcr.2018.v11i5.24390
Van Guelpen, B. R., Wirén, S. M., Bergh, A. R., Hallmans, G., Stattin, P. E., and Hultdin, J. (2006). Polymorphisms of methylenetetrahydrofolate reductase and the risk of prostate cancer: a nested case-control study. official J. Eur. Cancer Prev. Organ. (ECP) 15 (1), 46–50. eng. Epub 2005/12/24. Cited in: Pubmed; PMID 16374229. doi:10.1097/01.cej.0000186640.19872.4d
Vidal, A. C., Grant, D. J., Williams, C. D., Masko, E., Allott, E. H., Shuler, K., et al. (2012). Associations between intake of folate, methionine, and vitamins B-12, B-6 and prostate cancer risk in American veterans. J. cancer Epidemiol. 2012, 957467. eng. Epub 2012/08/29. Cited in: Pubmed; PMID 22927849. doi:10.1155/2012/957467
Wu, H. C., Chang, C. H., Tsai, R. Y., Lin, C. H., Wang, R. F., Tsai, C. W., et al. (2010). Significant association of methylenetetrahydrofolate reductase single nucleotide polymorphisms with prostate cancer susceptibility in taiwan. Anticancer Res. 30 (9), 3573–3577. eng. Epub 2010/10/15. Cited in: Pubmed; PMID 20944139.
Wu, J. L., Zhou, S. X., Zhao, R., Zhang, X., Chang, K., Gu, C. Y., et al. (2016). MTHFR c.677C>T inhibits cell proliferation and decreases prostate cancer susceptibility in the han Chinese population in shanghai. Sci. Rep. 6, 36290. eng. Epub 2016/11/08. Cited in: Pubmed; PMID 27819322. doi:10.1038/srep36290
Zhang, W. B., Zhang, J. H., Pan, Z. Q., Yang, Q. S., and Liu, B. (2012). The MTHFR C677T polymorphism and prostate cancer risk: new findings from a meta-analysis of 7306 cases and 8062 controls. APJCP 13 (6), 2597–2604. eng. Epub 2012/09/04. Cited in: Pubmed; PMID 22938427. doi:10.7314/apjcp.2012.13.6.2597
Keywords: MTHFR, C677T, A1298C, genetic polymorphism, prostate cancer, rs1801133, rs1801131
Citation: You J, Huang Y, Shen X, Chen Y and Ding X (2024) Associations between MTHFR gene polymorphisms (C677T and A1298C) and genetic susceptibility to prostate cancer: a systematic review and meta-analysis. Front. Genet. 15:1343687. doi: 10.3389/fgene.2024.1343687
Received: 24 November 2023; Accepted: 15 January 2024;
Published: 26 January 2024.
Edited by:
Qing Lin, Johns Hopkins University, United StatesReviewed by:
Jing Liu, Johns Hopkins University, United StatesNihal Inandiklioğlu, Bozok University, Türkiye
Copyright © 2024 You, Huang, Shen, Chen and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Ding, eXV5YW5nNzM3QDEyNi5jb20=
 Jianan You
Jianan You Yuhua Huang
Yuhua Huang Xinyu Shen
Xinyu Shen