
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 05 June 2024
Sec. Genetics of Common and Rare Diseases
Volume 15 - 2024 | https://doi.org/10.3389/fgene.2024.1266210
 Angie C. Jelin1,2*†
Angie C. Jelin1,2*† Nikolai Sopko3*†
Nikolai Sopko3*† Nara Sobreira2,4
Nara Sobreira2,4 Simeon A. Boyadjiev5
Simeon A. Boyadjiev5 Elizabeth Wohler2
Elizabeth Wohler2 Christian Morrill3
Christian Morrill3 P. Dane Witmer2
P. Dane Witmer2 Jason Michaud3
Jason Michaud3 David Valle2
David Valle2 John Gearhart3
John Gearhart3 Heather Dicarlo3
Heather Dicarlo3Introduction/background: Bladder exstrophy epispadias complex (BEEC) is a rare congenital anomaly of unknown etiology, although, genetic and environmental factors have been associated with its development. Variants in several genes expressed in the urogenital pathway have been reported as causative for bladder exstrophy in human and murine models. The expansion of next-generation sequencing and molecular genomics has improved our ability to identify the underlying genetic causes of similarly complex diseases and could thus assist with the investigation of the molecular basis of BEEC.
Objective: The objective was to identify the presence of rare heterozygous variants in genes previously implicated in bladder exstrophy and correlate them with the presence or absence of bladder regeneration in our study population.
Patients and Methods: We present a case series of 12 patients with BEEC who had bladder biopsies performed by pediatric urology during bladder neck reconstruction or bladder augmentation. Cases were classified as “sufficient” or “insufficient” (n = 5 and 7, respectively) based on a bladder volume of greater than or less than 40% of expected bladder size. Control bladder tissue specimens were obtained from patients (n = 6) undergoing biopsies for conditions other than bladder exstrophy. Whole exome sequencing was performed on DNA isolated from the bladder specimens. Based on the hypothesis of de novo mutations, as well as the potential implications of autosomal dominant conditions with incomplete penetrance, each case was evaluated for autosomal dominant variants in a set of genes previously implicated in BEEC.
Results: Our review of the literature identified 44 genes that have been implicated in human models of bladder exstrophy. Our whole exome sequencing data analysis identified rare variants in two of these genes among the cases classified as sufficient, and seven variants in five of these genes among the cases classified as insufficient.
Conclusion: We identified rare variants in seven previously implicated genes in our BEEC specimens. Additional research is needed to further understand the cellular signaling underlying this potentially genetically heterogeneous embryological condition.
BEEC is a complex embryological defect of the abdominal wall that requires staged urological reconstructive surgeries (Stewart et al., 2015). BEEC has been proposed to be polygenic in the Comparative Toxicogenomics Database (CTD)-gene associations data set includes 4,457 genes/proteins associated with urogenital anomalies (Davis et al., 2015). In fact, 44 are implicated as causative for BEEC in either human or murine models, but their role in the disease mechanism is largely unknown (Stevenson, 2021).
Surgical planning and methodology for patients with BEEC is complex. Although bladder regeneration or expansion are critical predictors of outcome, they can only be measured over time (Bazinet et al., 2024). Patients with sufficient bladder size do not require augmentation cystoplasty later in life, as their bladders have grown with age to achieve adequate storage volumes (Khandge et al., 2023). In contrast, bladders of insufficient size demonstrate minimal growth with age and require augmentation cystoplasty later in life to achieve adequate storage volumes (Khandge et al., 2023). Identifying potential genotypic differences between these two clinical populations may aid in understanding factors important to bladder growth biology in the BEEC population and potentially assist in patient counseling of functional outcomes and future surgical requirements.
Whole exome sequencing (WES) has emerged as a cost-effective technique to identify single nucleotide variants in genes that encode proteins (Bekheirnia et al., 2016). This has allowed for significant advances in uncovering the genetic etiology of both single gene and complex diseases (Petrovski et al., 2019). WES can also be used to identify somatic mosaic variants in an affected tissue such as the bladder tissue from patients with bladder exstrophy.
We sought to evaluate bladder tissue from patients with bladder exstrophy, with and without sufficient bladder size, by whole exome sequencing.
Our objective was to identify the presence of rare heterozygous variants in genes previously implicated in BEEC and correlate them with the presence or absence of bladder regeneration in our study population.
Bladder tissue was obtained from patients with BEEC and controls who were undergoing care at a tertiary university-based setting. Patients who presented to pediatric urology for bladder neck reconstruction or bladder augmentation between July 2009 and July 2015 were consented for bladder biopsy. Full thickness bladder biopsies were obtained by a pediatric urologist in the operating room at the time of the procedure. Control samples were obtained from patients undergoing ureteral implantation for vesico-urinary reflux (VUR) with normal size and morphology of the urinary bladder. This study was approved by the Institutional Review Board of the Johns Hopkins University.
We queried medical records to determine the expansion of bladder growth over time in addition to patient demographics. We also obtained the initial bladder size and initial operative procedure. Patients presenting for a biopsy with expected bladder growth greater than 40% were classified as “sufficient” while those presenting with minimal growth, <40%, were classified as “insufficient”. All of our BEEC patients were non-syndromic, without associated anomalies or developmental delays. Additional patients undergoing ureteral implantation for VUR were also consented as controls.
Whole exome sequencing was performed as previously described (Li et al., 2016) using the illumina HiSeq2500 Platform. Variants were analyzed utilizing the PhenoDB (Sobreira et al., 2015) tool to filter for rare, heterozygous variants with a minor allele frequency (MAF) < 0.01 in the 1,000 Genome Project, Exome Variant Server, gnomAD, and our collective in-house dataset (Sobreira et al., 2015). Following completion of the variant filtering pipeline, variants present in both BEEC cases and controls were excluded. Only variants meeting the following criteria were included: (1) filter = pass, (2) quality ≥ 100, (3) depth coverage ≥ 20X, and (4) variant fraction ≥ 20%. We utilized the integrated genomic viewer to ensure the variant read was supported. Variants identified in sufficient or insufficient cases from the curated list of 44 genes associated with BEEC in human models were selected for further analysis.
Patients with non-syndromic BEEC who underwent whole exome sequencing on DNA isolated from bladder biopsies included: five classified as sufficient, seven classified as insufficient, and six controls. Demographics of the three groups are provided in Table 1. The average percentage expected bladder volume of cases with sufficient bladder size was 67.7% and those with insufficient was 25.1%. In the majority of cases the initial procedure was performed in the neonatal period (8/12) (Table 2).
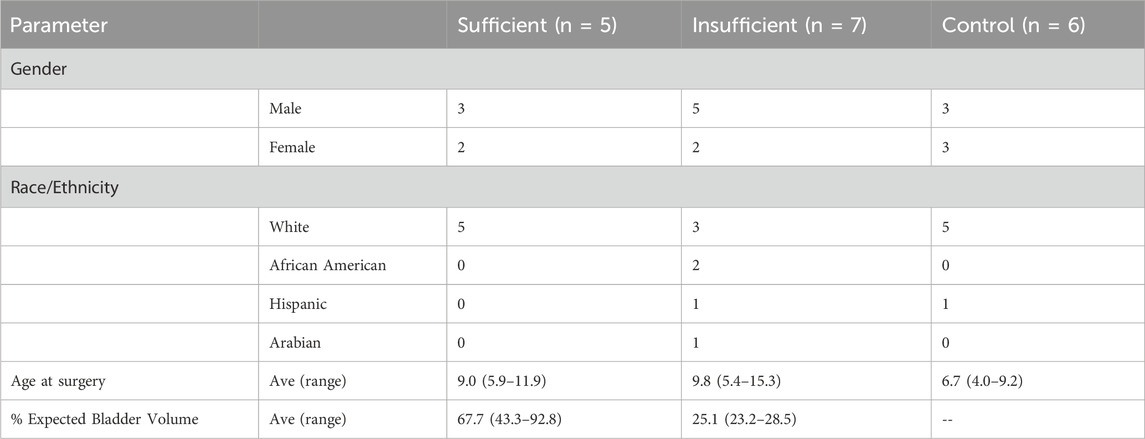
Table 1. Demographics of bladder exstrophy with sufficient or insufficient bladder volumes and controls.
Our review of the literature identified 44 human genes implicated in the etiology of BEEC. We identified rare, candidate causative variants in seven genes on this list (BMP10, WIZ, CYP4F22, CELSR1, CELSR3, TP63 and WNT11) in 2/5 cases classified as sufficient and in 5/7 cases classified as insufficient (Table 3). CELSR3 variants were present in three cases with insufficient bladders; two in Caucasian patients, and one in a Hispanic patient. One of the patients with a variant in CELSR3 also had variants in two other genes identified as associated with BEEC (TP63 and WNT11). A CELSR1 variant was identified in an African-American patient with insufficient bladder capacity. Variants in two cases with sufficient bladder capacity were identified in BMP10 and WIZ, respectively (Table 3).
BEEC is a rare congenital anomaly that requires multi-specialty care and surgical reconstruction. New technologies, including whole exome sequencing, are elucidating the underlying genetic etiologies of this complex condition (Petrovski et al., 2019). We identified rare single nucleotide candidate causative variants in DNA isolated from bladder tissue of patients with BEEC that may be related to the bladder regenerative capacity.
Three of our patients had a rare candidate causative variant in CELSR3, which encodes a cadherin epidermal growth factor-like laminin G-like seven-pass G-type receptor that is involved in polarity signaling (Fenstermaker et al., 2010). These were rare missense variants [c.5875G>A (p.Ala1959Thr), c.9098G>A (p.Arg3033His), c.9566G>A (p.Arg3189Gln)] in three unrelated patients with insufficient bladder regeneration (Table 3). Interestingly, Reutter et al. identified a patient with cloacal exstrophy, a subtype of BEEC, who was a compound heterozygote for two CELSR3 coding variants [c.5470G>A (p.Val1824Met) and c.6950T>C (p.Met2317Tyr)] (Reutter et al., 2016). CELSR3 is involved in the WNT pathway and Celsr3 knockout mice exhibit perinatal lethality (Pitsava et al., 2021). De novo variants in TUBE1 another gene on our BEEC gene list (Table 2) also functions in centriole positioning, thus strengthening their classification as candidate genes (Pitsava et al., 2021).
The WIZ variant identified in patient S1 (sufficient bladder) and the CYP4F22 and TP63 variants identified in patient l1 and l5, respectively (insufficient bladders) are classified as variants of uncertain significance by Varsome (Kopanos et al., 2019). WIZ is perhaps the most interesting candidate to evaluate for causation because homozygosity for null alleles results in embryonic lethality in mouse models (Daxinger et al., 2013).
TP63 has been previously reported to be associated with bladder exstrophy (Qi et al., 2013) and the mouse Tp63 knockout (Yang et al., 1999; Cheng et al., 2006) model presents with classic exstrophy of the bladder (Ching et al., 2010). Furthermore, Ching et al. (2010) documented reproducible dysregulation of variable tissue-specific TP63 isoform expression in 11 of 15 BEEC patients without obvious coding p63 gene variants, suggesting alterations in TP63 expression may be causative (Ching et al., 2010). These observations add support for a pathogenic role for the TP63 variant c.1518G>T (p.Met506Ile)identified in one of our patients.
We performed whole exome sequencing of DNA isolated from bladder tissue in 12 BEEC patients from several different ancestries. We also provide vital information regarding the demographics and phenotypic information of patients with BEEC including bladder filling capacity. Our utilization of DNA isolated from the affected bladder tissue can also capture somatic mutations that could be present only in the bladder and absent in DNA from blood and saliva. Unfortunately, our access was limited to patient’s samples only, so we were not able to determine if a variant was inherited or de novo. For that reason, that we chose to limit analysis to rare heterozygous variants in genes that have been previously implicated in bladder exstrophy.
In conclusion, we present a unique series of BEEC cases that were categorized by bladder size and evaluated by whole exome sequencing. We identified rare variants in genes previously been proposed as BEEC candidates. Future steps will include obtaining parental samples and sequencing blood or saliva DNA samples of cases to assist in determining whether the mutations identified in this study are confined to the bladder (somatic) or germline. Additional investigation into the functional analysis of the identified variants will be necessary to determine causality.
The original contributions presented in the study are publicly available. This data can be found here: ClinVar repository, accession number SUB14445977, https://www.ncbi.nlm.nih.gov/clinvar/?term=SUB14445977.
The studies involving humans were approved by the Johns Hopkins School of Medicine IRB. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
AJ: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Writing–original draft, Writing–review and editing. NiS: Conceptualization, Data curation, Investigation, Methodology, Writing–original draft. NaS: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft. SB: Data curation, Methodology, Writing–review and editing. EW: Data curation, Formal Analysis, Investigation, Writing–original draft. CM: Data curation, Methodology, Writing–review and editing. DW: Data curation, Formal Analysis, Methodology, Writing–original draft. JM: Conceptualization, Methodology, Writing–original draft. DV: Methodology, Supervision, Writing–original draft. JG: Conceptualization, Methodology, Resources, Writing–original draft. HD: Conceptualization, Investigation, Methodology, Resources, Writing–original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. AJ is supported by grant 5K23DK119949-02 from the National Institutes of Health (NIH). The contents of the publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1266210/full#supplementary-material
Bazinet, A., Filfilan, A., Mokhtari, N., Lenfant, L., Elghoneimi, A., and Chartier-Kastler, E. (2024). Adult patients treated for bladder exstrophy at a young age: what are their current demands? Can. Urol. Assoc. J. 18. Epub ahead of print. PMID: 38466862. doi:10.5489/cuaj.8601
Bekheirnia, M. R., Bekheirnia, N., Bainbridge, M. N., Gu, S., Coban Akdemir, Z. H., Gambin, T., et al. (2016). Whole-exome sequencing in the molecular diagnosis of individuals with congenital anomalies of the kidney and urinary tract and identification of a new causative gene. Genet. Med. 19, 412–420. doi:10.1038/gim.2016.131
Cheng, W., Jacobs, W. B., Zhang, J. J. R., Moro, A., Park, J. H., Kushida, M., et al. (2006). DeltaNp63 plays an anti-apoptotic role in ventral bladder development. Development 133, 4783–4792. doi:10.1242/dev.02621
Ching, B. J., Wittler, L., Proske, J., Yagnik, G., Qi, L., Draaken, M., et al. (2010). p63 (TP73L) a key player in embryonic urogenital development with significant dysregulation in human bladder exstrophy tissue. Int. J. Mol. Med. 26 (6), 861–867. PMID: 21042780. doi:10.3892/ijmm_00000535
Davis, A. P., Grondin, C. J., Lennon-Hopkins, K., Saraceni-Richards, C., Sciaky, D., King, B. L., et al. (2015). The Comparative Toxicogenomics Database's 10th year anniversary: update 2015. Nucleic Acids Res. 43, D914–D920. doi:10.1093/nar/gku935
Daxinger, L., Harten, S. K., Oey, H., Epp, T., Isbel, L., Huang, E., et al. (2013). An ENU mutagenesis screen identifies novel and known genes involved in epigenetic processes in the mouse. Genome Biol. 14, R96. doi:10.1186/gb-2013-14-9-r96
Fenstermaker, A. G., Prasad, A. A., Bechara, A., Adolfs, Y., Tissir, F., Goffinet, A., et al. (2010). Wnt/planar cell polarity signaling controls the anterior-posterior organization of monoaminergic axons in the brainstem. J. Neurosci. 30 (47), 16053–16064. PMID: 21106844; PMCID: PMC3073573. doi:10.1523/JNEUROSCI.4508-10.2010
Khandge, P., Morrill, C. C., Wu, W. J., Harris, K. T., Haffar, A., Maruf, M., et al. (2023). Achieving goal capacity for continence surgery: a cumulative event analysis of bladder exstrophy patients. J. Pediatr. Urol. 19 (5), 563.e1–563.e8. Epub 2023 May 6. PMID: 37246118. doi:10.1016/j.jpurol.2023.04.039
Kopanos, C., Tsiolkas, V., Kouris, A., Chapple, C. E., Albarca Aguilera, M., Meyer, R., et al. (2019). VarSome: the human genomic variant search engine. Bioinformatics 35 (11), 1978–1980. doi:10.1093/bioinformatics/bty897
Li, R., Sobreira, N., Witmer, P. D., Pratz, K. W., and Braunstein, E. M. (2016). Two novel germline DDX41 mutations in a family with inherited myelodysplasia/acute myeloid leukemia. Haematologica 101, e228–e231. doi:10.3324/haematol.2015.139790
Petrovski, S., Aggarwal, V., Giordano, J. L., Stosic, M., Wou, K., Bier, L., et al. (2019). Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet 393 (10173), 758–767. Epub 2019 Jan 31. PMID: 30712878. doi:10.1016/S0140-6736(18)32042-7
Pitsava, G., Feldkamp, M. L., Pankratz, N., Lane, J., Kay, D. M., Conway, K. M., et al. (2021). Exome sequencing of child-parent trios with bladder exstrophy: findings in 26 children. Am. J. Med. Genet. A 185 (10), 3028–3041. Epub 2021 Aug 5. PMID: 34355505; PMCID: PMC8446314. doi:10.1002/ajmg.a.62439
Qi, L., Wang, M., Yagnik, G., Mattheisen, M., Gearhart, J. P., Lakshmanan, Y., et al. (2013). Candidate gene association study implicates p63 in the etiology of nonsyndromic bladder-exstrophy-epispadias complex. Birth Defects Res. A Clin. Mol. Teratol. 97 (12), 759–763. Epub 2013 Aug 2. PMID: 23913486. doi:10.1002/bdra.23161
Reutter, H., Keppler-Noreuil, K., E Keegan, C., Thiele, H., Yamada, G., and Ludwig, M. (2016). Genetics of bladder-exstrophy-epispadias complex (BEEC): systematic elucidation of mendelian and multifactorial phenotypes. Curr. Genomics 17 (1), 4–13. PMID: 27013921; PMCID: PMC4780475. doi:10.2174/1389202916666151014221806
Sobreira, N., Schiettecatte, F., Boehm, C., Valle, D., and Hamosh, A. (2015). New tools for mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 36 (4), 425–431. doi:10.1002/humu.22769
Stevenson, R. E. (2021). Common pathogenesis for sirenomelia, OEIS complex, limb-body wall defect, and other malformations of caudal structures. Am. J. Med. Genet. A 185 (5), 1379–1387. doi:10.1002/ajmg.a.62103
Stewart, D., Inouye, B. M., Goldstein, S. D., Shah, B. B., Massanyi, E. Z., DiCarlo, H., et al. (2015). Pediatric surgical complications of major genitourinary reconstruction in the exstrophy-epispadias complex. J. Pediatr. Surg. 50 (1), 167–170. doi:10.1016/j.jpedsurg.2014.10.036
Keywords: bladder exstrophy, genetic variants, Celsr3, exome sequence, bladder growth
Citation: Jelin AC, Sopko N, Sobreira N, Boyadjiev SA, Wohler E, Morrill C, Witmer PD, Michaud J, Valle D, Gearhart J and Dicarlo H (2024) Rare exonic CELSR3 variants identified in Bladder Exstrophy Epispadias Complex. Front. Genet. 15:1266210. doi: 10.3389/fgene.2024.1266210
Received: 28 July 2023; Accepted: 28 March 2024;
Published: 05 June 2024.
Edited by:
Corrado Romano, University of Catania, ItalyReviewed by:
Glenda Maria Beaman, The University of Manchester, United KingdomCopyright © 2024 Jelin, Sopko, Sobreira, Boyadjiev, Wohler, Morrill, Witmer, Michaud, Valle, Gearhart and Dicarlo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angie C. Jelin, YWplbGluMUBqaG1pLmVkdQ==; Nikolai Sopko, bmlrb2xhaS5zb3Brb0BnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.