
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 15 January 2024
Sec. Cancer Genetics and Oncogenomics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1299599
This article is part of the Research Topic Innovative Translational Research to Identify Colorectal Cancer Biomarkers for Personalized Medicine View all 7 articles
 Aleksandra Stanojevic1
Aleksandra Stanojevic1 Jelena Spasic2
Jelena Spasic2 Mladen Marinkovic3,4
Mladen Marinkovic3,4 Suzana Stojanovic-Rundic3,4
Suzana Stojanovic-Rundic3,4 Radmila Jankovic1
Radmila Jankovic1 Ana Djuric1
Ana Djuric1 Jerome Zoidakis5,6
Jerome Zoidakis5,6 Remond J. A. Fijneman7
Remond J. A. Fijneman7 Sergi Castellvi-Bel8
Sergi Castellvi-Bel8 Milena Cavic1*
Milena Cavic1*Background: Methylenetetrahydrofolate reductase (MTHFR) single nucleotide polymorphisms (SNPs) have been suggested as risk, prognostic, and predictive factors for colorectal cancer in various populations, but have not been validated so far. The aim of this study was to examine the association of MTHFR C677T (rs1801133) and A1298C (rs1801131) single nucleotide polymorphisms with the risk of rectal cancer as well as the response to neoadjuvant chemoradiotherapy (nCRT) based on 5-Fluorouracil (5-FU)/leucovorin (LV) in the locally advanced setting.
Patients and methods: This case-control study included 119 healthy controls and 97 patients with locally advanced rectal cancer (LARC). For MTHFR genotyping, restriction fragment length polymorphism analysis (PCR-RFLP) was employed.
Results: In silico analysis highlighted that SNPs C677T and A1298T correlate with MTHFR gene expression, and that gene expression profile correlates with cancer risk and stage. Using dominant and recessive models, it was found that the MTHFR 677CC vs. 677CT+677TT have increased risk of cancer development (odds ratio (OR): 2.27; 95% confidence interval (CI): 1.30–3.95, p = 0.002) as well as 677CC+677CT compared to 677TT (OR: 4.18, 95% CI: 1.16–14.99, p = 0.014). MTHFR 1298AA also shown increased risk for cancer development compared to 1298AC+1298CC (OR:2.0, 95% CI: 1.20–3.59, p = 0.035) Statistical analysis of combined genotypes highlighted the protective role of CT/AC combined genotype (OR: 3.15 95% CI: 1.576–6.279, p = 0.002) while the CC/AA genotype showed an increased risk for rectal cancer development (OR: 2.499, 95% CI: 1.246–5.081, p = 0.016) The carriers of the 677C/1298A haplotype had the highest risk for developing rectal cancer (OR: 1.74; 95% CI: 1.198–2.530, p = 0.002) while the 677T/1298C haplotype seems to provide a protective effect. (OR: 0.44; 95%CI 0.248–0.795, p = 0.003). No significant association with response to chemoradiotherapy was found.
Conclusion: Our data point to MTHFR 667C allele and 1298A alleles as low-penetrance risk factors for rectal cancer in our population. To the best of our knowledge, this is the first study of this type performed on the Slavic population in the Western Balkan, as various population-based factors might also be significant our findings can be used for future meta-analyses and the construction of genetic cancer risk prediction panels.
Colorectal cancer (CRC) is the third most frequently diagnosed cancer with 1.93 million newly diagnosed patients in 2020, and the second leading cause of cancer death worldwide with 935 000 deaths annually (Sung et al., 2021). The situation in Serbia is similar to the global one, CRC is the second most frequent cancer with regards to both incidence and mortality, with around 5000 newly diagnosed CRC cases and around 2400 deaths in 2020 (Serbian Cancer Registry, 2022). It is a multifactorial disease involving both genetic and environmental factors. Approximately 75% of CRCs are sporadic and occur in cases of absence of genetic predisposition or family history of CRC (Yamagishi et al., 2016). Diet plays an important role in the development of colorectal malignancy, as well as smoking history, alcohol consumption, body mass index (BMI), and lifestyle, although the relative significance of each of these factors on its own, or combinations of more than one factor is not clear (Ryan-Harshman and Aldoori, 2007). Rectal and colon cancer are different diseases with regard to diagnosis, sensitivity to treatment, and some risk factors, but few studies addressed risk factors for these two cancers separately.
The role of folic acid deficiency has been investigated in tumorigenesis in general (Kim, 2003). Some of the proposed models of folic depletion influence on cancer development are alterations in DNA methylation, disruption of DNA integrity, and disruption of DNA repair (Choi and Mason, 2000). Interestingly, folate deficiency has been investigated as a factor only in alcohol-related carcinogenesis of rectal cancer, since there is a clear relationship between alcohol consumption and alterations in folate metabolism (Choi and Mason, 2000). Whether dietary intake of folate has a protective effect against selected cancers is not clear, since the results of studies are not consistent. An important protein in the metabolism of folic acid is the methylenetetrahydrofolate reductase (MTHFR). This enzyme converts the 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, the primary form of folate in blood.
Two common functional polymorphisms in the MTHFR gene are C677T (rs1801133; c.788C>T) and A1298C (rs1801131; c.1409A>C). These polymorphisms influence enzyme activity. The C677T polymorphism in exon 4 causes a substitution of C to T nucleotide, leading to the substitution of alanine with valine in codon 222, which in turn affects the active site of the enzyme and thus reduces its activity, with the TT genotype product having a 70% reduced activity in comparison to wild type. A substitution of A to C at nucleotide 1298 (polymorphism A1298C of the MTHFR gene) leads to the substitution of glutamine with alanine at the position 429, also causing reduced enzyme activity (Kennedy et al., 2012). The relationship between these polymorphisms and the risk of developing cancer has been observed and the results are contradictory and inconclusive. Some of the published studies showed differences in connection with ethnicity, as interestingly, there seems to be a higher frequency of the 677, TT genotype in southern Europe than in the north, while in Asia, the frequency is highest in China and lowest in India. Also, African Americans have a lower frequency of the TT genotype than Caucasians. The 1298CC genotype is more frequent in Caucasians (4%–12%) than in China and Japan (1%–4%) (Kennedy et al., 2012).
Preoperative, neoadjuvant chemoradiotherapy (nCRT) based on 5-fluorouracil (5-FU), followed in most cases by operative treatment is the standard of care for LARC (Glynne-Jones et al., 2017). Tumor regression grade (TRG) is an established prognostic factor for local recurrence, disease-free, and overall survival (OS), with significantly better outcomes in patients showing TRG 1–2 (good responders) than those with TRG 3–5 (poor responders) (Vecchio et al., 2005). The cytotoxic activity of 5-FU is exhibited mainly by its active metabolite which forms a complex with thymidylate synthase (TS) and 5,10-methylenetetrahydrofolate (5,10-MTHF) thus causing the inhibition of TS and disrupting normal DNA synthesis (Longley et al., 2003). Elevated intracellular 5,10-MTHF levels are needed for optimal inhibition of TS, and these are controlled by methylenetetrahydrofolate reductase (MTHFR), whose decreased activity results in higher levels of 5,10-MTHF and higher rate of inhibition of TS. There are large inter-individual differences in the efficacy of 5-FU. Considering the described mechanism of action of 5-FU, it is reasonable to assume that certain polymorphisms in genes involved in various points of 5-FU mechanism of action could explain some of these inter-individual differences in clinical response and toxicity to 5-FU (Ulrich et al., 2014). Polymorphisms in the MTHFR gene which cause decreased activity of MTHFR could make these patients more sensitive to 5-FU, therefore there should be a higher efficacy of 5-FU which in turn should lead to better survival. However, most study results do not support this hypothesis. Two polymorphisms of the MTHFR gene, C677T and A1298C, have been investigated in this context in patients with CRC and more extensively in patients with LARC treated with chemo-irradiation.
So far, various research that aimed to profile genetic risk factors of different types of cancer was conducted in Serbia to construct a general predictive risk model (Cavic et al., 2016; Krivokuca et al., 2016; Cavic et al., 2019). Results like these might contribute to the construction of a low-cost and minimally invasive pan-cancer polymorphism screening tool.
Most CRC cases in Serbia are discovered at an advanced stage, when there are few treatment choices and a low chance of survival. In order to improve overall patient care, our group and others have worked to profile the diagnostic, prognostic, and predictive characteristics for colorectal cancer (CRC), rectal and anal cancer (Cavic et al., 2016; Nikolic et al., 2021; Stojanovic- Rundic et al., 2021; Vuletić et al., 2021; Marinkovic et al., 2023; Stanojevic et al., 2023).
The aim of this study was to examine the association of MTHFR C677T and A1298C single nucleotide polymorphisms with the risk of rectal cancer as well as the response to neoadjuvant chemoradiotherapy based on 5-Fluorouracil (5-FU)/leucovorin (LV) in the locally advanced setting, in an effort to provide data from the Western Balkan area which is usually underrepresented in larger meta-analyses.
The interactive online resource for examining cancer transcriptome data from the Cancer Genome Atlas (TCGA-ROAD) (Muzny et al., 2012; National Cancer Institute and the National Human Genome Research Institute, 2021) UALCAN (https://ualcan.path.uab.edu/analysis.html) (UALCAN database, n.d.; Chandrashekar et al., 2017) was employed to analyze MTHFR expression levels in normal and rectal cancer samples. The publicly available database the Human Protein Atlas (HPA) (https://www.proteinatlas.org) (The Human Protein Atlas database V.20.0, 2021; Uhlen et al., 2017) was used to analyze TCGA- ROAD transcriptome data on the expression of MTHFR in relation to its prognostic significance in rectal cancer (Muzny et al., 2012). Kaplan-Meier plots summarize the results of the correlation between MTHFR expression level and patient survival by assigning patients to groups that are either low (under experimental cut-off) or high (above experimental cut-off) groups. Expression cut-off values for the HPA data are presented as the number fragments per kilobase of exon per million reads (FPKM) of MTHFR in the tumor tissue at diagnosis. The transcriptome data of patients with rectal cancer were analyzed using an online tool ROCplotter (www.rocplot.org), to determine the MTHFR expression levels in responders and non-responders (Fekete and Győrffy, 2019). Corresponding images and data were downloaded from the HPA, UALCAN and ROCplotter platforms in the original form. The STRING (https://string-db.org, Szklarczyk et al., 2022) protein network for MTHFR was built based on the highest confidence (0.9) evidence from experimental and biochemical data, co-expression, gene neighborhood, gene co-occurrence, gene fusions, protein homology, manually curated metabolic and signaling pathway databases, and predictive and knowledge text data mining. The network included 5 primary-interaction shell proteins to explore interactions and clustering with other proteins and the effects of these interactions. For the enrichment analysis, the whole genome statistical background was assumed. The analysis was performed using STRING v.11.0 (Szklarczyk et al., 2022), corresponding images and results were exported and statistical significance was considered for p < 0.05. Colonomics web-based tool (https://colonomics.org) was used to access expression quantitative trait loci (eQTL) analysis for MTHFR gene expression depending on SNP C677T and A1298C (Moreno et al., 2018).
Gene Expression Omnibus (GEO) database (Barrett et al., 2012) was used for the search of publicly available datasets for external validation of obtained results. Search criteria included the keyword “rectal cancer” while “genome variation profiling by SNP array” and “SNP genotyping by SNP array” were used as a study type of interest.
A case-control study was performed in a group of 97 patients diagnosed with locally advanced primary rectal adenocarcinoma (age range 29–83 years, median 61; 67 males, 30 females) from several cancer centers in Serbia treated at the Institute for Oncology and Radiology of Serbia from 2018–2019, and 119 healthy control subjects (age range 32–89 years, median 55; 67 males, 52 females) with no previous history of malignancies and no known folate metabolism deficiency, all of Caucasian descent (Table 1). In adherence to the National guidelines for colorectal cancer screening, colonoscopies were conducted on individuals in the control group when there was an indication based on these criteria. Importantly, the results of these colonoscopies were negative for colorectal cancer at the time of examination. All patients were diagnosed with locally advanced rectal cancer, stage II (T3/4N0M0) or III (T1-4N + M0) according to clinical and histological criteria of the 8th edition of the TNM classification of malignant tumors, and ECOG ≤ 2 (Oken et al., 1982). The tumors were located <15cm from the anocutaneous line and were treated with neoadjuvant chemoradiotherapy (5-Fluorouracil 350 mg/m2 i. v. bolus plus Leucovorin 25mg/m2 D1-D5 and D29-D33). Radiotherapy was initiated concurrently with chemotherapy, 50.4 Gy in 28 fractions, conventionally fractioned 1.8Gy/fr, using the technique with 3 or 4 radiation areas (all areas as endorsed by the International Committee of Radiation Units and Measurements (ICRU) 50/62) (Landberg et al., 2016a; Landberg et al., 2016b). Clinical response assessment took place 6–8 weeks after the completion of neoadjuvant therapy, involving pelvic MRI scans, rigid proctoscopy, and digital rectal examinations. Subsequently, patients were referred for surgery. The patohistological assessment of surgical specimens included the determination of histomorphology of the resected tumor (type and grade), tumor invasiveness (ypTNM, R classification) (Wei et al., 2018), pathohistological grading of the tumor regression by the Mandard scale (Siddiqui et al., 2016) with the determination of prognostic categories.
To investigate the predictive role of MTHFR polymorphisms, patients were divided into responders (patients with tumor regression grades (TRG 1 and 2)) and, non-responders (TRG 3–5) based on postoperative specimens. Two out of 97 patients enrolled in the study didn’t have TRG status at the moment of analysis. Patients who achieved a complete clinical response without subsequent operative treatments were excluded from this analysis.
Genomic DNA was isolated from formalin-fixed paraffin-embedded tissue samples (FFPE) obtained by biopsy/resection using the QIAamp® DNA FFPE Tissue isolation kit (Qiagen, United Kingdom). Ethylenediaminetetraacetic acid (EDTA) peripheral blood was drawn from healthy controls by venipuncture and further used for leukocyte isolation using BloodPrep Chemistry for ABI PRISM™ 6100 Nucleic Acid PrepStation (Applied Biosystems, CA, United States). The concordance between germline and somatic DNA in variations of pharmacogenetic genes is around 100%, according to a recent large-scale literature research. Spectrophotometric analysis was used to evaluate the concentration and purity of the extracted DNA samples (Nanodrop, Shimadzu). For MTHFR genotyping, restriction fragment length polymorphism analysis (PCR-RFLP) was employed as previously described (Jakovljevic et al., 2012; Cavic et al., 2014; 2016). The analysis was performed by gel electrophoresis on a chip using the Agilent DNA 1000 Kit on the Agilent 2100 Bioanalyzer. To ensure adequate genotyping, a previously established heterozygote sample was used as a method of control, and genotyping was carried out blind to case-control status. 10% of samples, chosen at random, underwent Sanger sequencing analysis to verify the accuracy of the findings.
The sample data were summarized using descriptive statistical methods (frequencies, percentages, means, medians, standard deviation, SD, and range). The Hardy-Weinberg equilibrium of the analyzed polymorphisms was tested using the Pearson Chi-Square test. Two-sided p values < 0.05 were considered to indicate statistical significance. The associations between the patients’ and healthy controls as well as responders and Non-responders were analyzed by applying Pearson Chi-Square with Yates’ correction. Fisher’s exact test was used to analyze differences between males and females. Combined genotype frequencies were calculated by direct counting while statistical significance in combined genotype distribution between patients and controls was observed using the Chi-Square test with Yates’ correction. In addition, haplotype analysis was used for calculating the interaction between two polymorphic sites of MTHFR. Haplotype frequencies were calculated manually, and data were confirmed using Multiallelic Interallelic Disequilibrium Analysis Software (University of Southampton, Highfield, Southampton, United Kingdom) (Gaunt et al., 2006) and Golden Helix Tree SNP and Variation Suite software (Golden Helix, Bozeman, MT, United States). Statistical significance was obtained using Pearson Chi-Square with Yates’ correction. All statistical analysis was performed using GraphPad Prism 8.0.1(GraphPad Software, Boston, MA, United States) and SNPstat (https://www.snpstats.net/start.html).
In silico analyses using the UALCAN and HPA platforms showed that MTHFR expression was significantly higher in normal compared to rectal cancer tissue (Figure 1A, p = 0.013). Expression of MTHFR in rectal adenocarcinoma by stages 2 and 3 compared to normal tissue showed a statistical significant difference (normal vs. stage 2, p = 0.003; normal vs. stage 3; p = 0.001; normal vs. stage 4, p = 0.001) and low expression correlated with higher cancer stages (Figure 1B; stage 1 vs. stage 3, p = 0.020; stage 1 vs. stage 4, p = 0.050). It was generally not prognostically significant in rectal cancer (p = 0.270), but the 5-year survival rate was found to be 91% for the high expression group and 48% for the low expression group (Figure 1C; expression cut-off 3.14 FPKM, median follow up time 1.75 years).
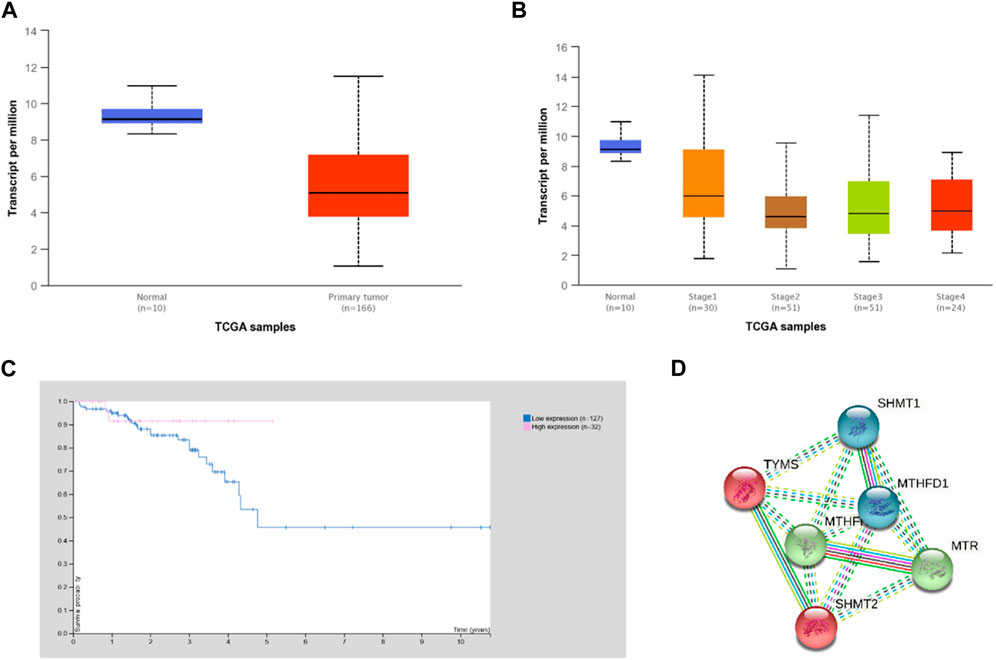
FIGURE 1. Expression of MTHFR in rectal cancer according to the UALCAN and HPA analysis of TCGA data. (A) Expression of MTHFR in rectal adenocarcinoma based on sample type (normal vs. cancer tissue p = 0.013). (B) Expression of MTHFR in rectal adenocarcinoma by stages compared to normal tissue (normal vs. stage 2, p = 0.003; normal vs. stage 3; p = 0.001; normal vs. stage 4, p = 0.001) and between individual cancer stage (stage 1 vs. stage 3, p = 0.020; stage 1 vs. stage 4, p = 0.041). (C) Survival curves of rectal cancer patients in relation to the expression of MTHFR (expression cut-off 3.14 FPKM, p = 0.270). (D) Direct STRING (STRING database, n.d.) network of MTHFR was built based on highest confidence (0.9) evidence from experimental interaction data (pink), co-expression (black), gene neighborhood (green) and co-occurrence (blue) data, curated databases (light blue), predictive and knowledge text mining (light green), protein homology (purple). The network included 5 primary-interaction shell genes to explore their indirect interactions and clustering on all analyzed platforms (PPI enrichment p-value: 0.002). Red nodes–TYMS cluster members; green nodes–MTHFR cluster members; blue nodes–MTHFD1 cluster. Nodes are labeled with HGNC symbols: MTHFD1, methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1 MTHFR - methylenetetrahydrofolate reductase; MTR, methionine synthase; TYMS, thymidylate synthase; SHMT1, serine hydroxymethyltransferase, cytosolic; SHMT2, serine hydroxymethyltransferase, mitochondrial.
ROC analysis included 42 patients in total (19 responders and 23 non-responders) and highlighted that MTHFR expression (Affy ID 7436) is slightly higher in a group of patients who responded poorly to the therapy but without statistical significance. There was weak statistical significance in ROC p-value (p = 0.045) with area under the curve (AUC = 0.648).
The STRING analysis showed that MTHFR has direct and indirect interactions with various proteins that are important for rectal cancerogenesis/homeostasis when gene co-expression, experimental/biochemical data, and text mining were considered at the highest confidence level (0.9) (Figure 1D). Cluster analysis extended to 5 primary-interaction shell genes (MTHFD1 - methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1, MTR - methionine synthase, TYMS - thymidylate synthase, SHMT1 - serine hydroxymethyltransferase, cytosolic and SHMT2 - serine hydroxymethyltransferase, mitochondrial) showed that these proteins form biological clusters across all analyzed platforms. The extended network was found to be enriched in interactions (protein-protein interactions PPI enrichment p-value: 0.002), which indicated that they interact with each other significantly more than is expected for a random set of proteins of similar size and can be thus considered as a biologically interconnected group (Szklarczyk et al., 2012).
Correlation between gene expression and SNPs were evaluated using Colonomics online tool (https://www.colonomics.org/data-browser/dashboard/). eQTL analysis was performed for left colon samples (normal, adjacent and tumor tissue) for both SNPs (C677T and A1298C). As rectal tissue data were not available, left colon was analyzed as anatomically closest to rectal tissue thus expecting similarity in expression profiles. This was confirmed by comparing the MTHFR expression profile of the left colon (Figure 2A) and rectal adenocarcinoma (Figure 1B) MTHFR gene expression of 138 individuals (Normal N = 20; Adjacent N = 59 and Tumor N = 59) confirmed that tumor tissue had overall lower MTHFR expression compared to adjacent and normal tissue, respectively. eQTL analysis highlighted a clear correlation between gene expression and SNPs in both cases. The existence of mutation A>C at 1298 position, positively affected MTHFR expression in every type of tissue (Figure 2B), while C>T at position 677 had an effect of a decrease of MTHFR expression for each type of tissue (Figure 2C).
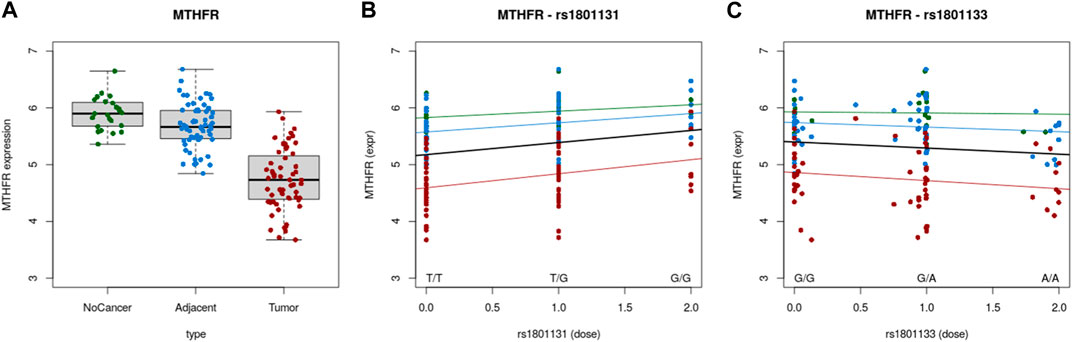
FIGURE 2. (A) Expression of MTHFR in normal, adjacent and tumor tissue. (B) Correlation between MTHFR expression and A1298C polymorphic variants. (C) Correlation between MTHFR expression and C677T polymorphic variants.
Patient characteristics are presented in Table 1. PCR was used to amplify regions of interest using predesigned primers for MTHFR C677T region (forward 5′ TGA AGG AGA AGG TGT CTG CGG GA 3′ and reverse 5′ AGG ACG GTG CGG TGA GAG TG 3′) and for MTHFR A1289C region (forward 5′ CTT TGC CAT GTC CAC AGC ATG 3′ and reverse 5′ AAG GAG GAG CTG CTG AAG ATG 3’). From all patient and control samples, a 198 bp PCR product containing the MTHFR C677T polymorphism site was produced with success (Figure 3A). Digestion was performed using Fast Digest HinfI and Fast Digest MboII enzymes, respectively (Thermo Fisher Scientific, Waltham, Massachusetts, United States). After digestion of the PCR products, homozygotes (CT) formed three bands of 198, 175, and 23 bp, and homozygotes (TT) produced two fragments of 175 and 23 bp. An undigested PCR product (198 bp) showed the presence of a homozygous wild-type genotype (CC). From each patient and control sample, a 163 bp PCR product containing the MTHFR A1298C polymorphism site was produced with success (Figure 3B). Four bands measuring 84, 31, 30, and 18 bp occur when the homozygous wild type A allele is present, while five bands measuring 56, 31, 30, 28, and 18 bp appear when the C allele is present.
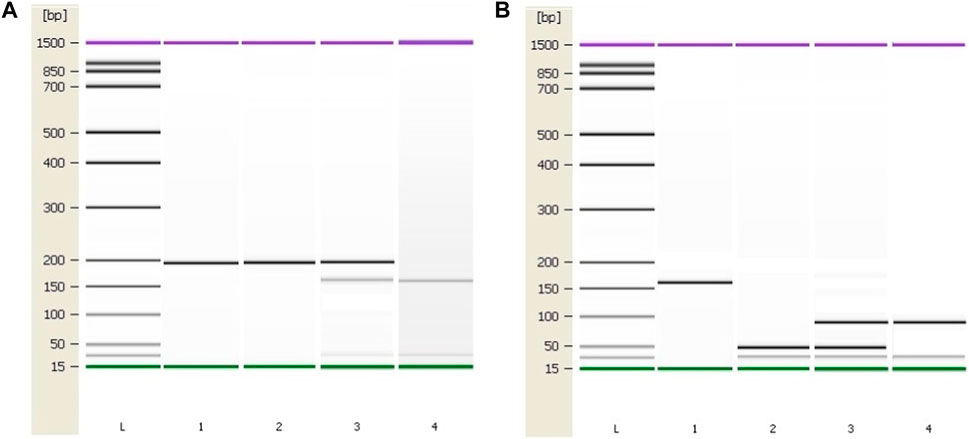
FIGURE 3. Genotyping results of the MTHFR C677T and A1298C polymorphic variants using the Agilent 2100 Bioanalyzer. (A) PCR and RFLP results of the MTHFR C677T polymorphic variant. Column 1: 198 bp PCR product. Column 2: C/C, Column 3: C/T, Column 4: T/T (B) PCR and RFLP genotyping results of the MTHFR A1298C polymorphic variant. Column 1: 163 bp PCR product. Column 2: A/A, Column 3: A/C, Column 4: C/C. L–High-sensitivity DNA ladder (Agilent Technologies). 1500 bp upper and 15 bp lower marker are present in each column.
Distribution of genotypes, MTHFR C677T and MTHFR A1298C distribution in patients and controls did not deviate from the Hardy-Weinberg equilibrium. The allele frequencies of the MTHFR C677T polymorphic variants in patients and controls revealed that patients had a higher frequency of C allele (C = 0.74; T = 0.26) than healthy controls (C = 0.60; T = 0.40) with trend in statistical significance p = 0.050 (Table 2; Figures 4A, B). Analysis of the effects of MTHFR C677T and A1298C polymorphic variants on rectal cancer risk using codominant, dominant, recessive and overdominant models was represented using p value < 0.05; odds ratio (OR) and 95% confidence interval which are shown in Table 3, using a previously published method (Ozretić et al., 2019). The codominant model shown statistical significance between prevalence of 677CC, 677CT and 677TT within patient and control group (p = 0.003). Using dominant and recessive models, it was found that the MTHFR 677CC vs. 677CT+677TT have increased risk of cancer development (OR: 2.27; 95% CI: 1.30–3.95, p = 0.002) as well as 677CC+677CT compared to 677TT (OR: 4.18, 95% CI: 1.16–14.99, p = 0.014). Using codominant (p = 0.013), dominant and recessive models, we found association with cancer risk in group of males. Using recessive models, it was found that 677TT has a protective effect against rectal cancer development (OR: 5.70; 95% CI: 1.20–27.11; p = 0.012). In the female group, using the dominant model it was found that 677TT has a protective effect (OR: 2.57; 95% CI: 1.02–6.50; p = 0.044).
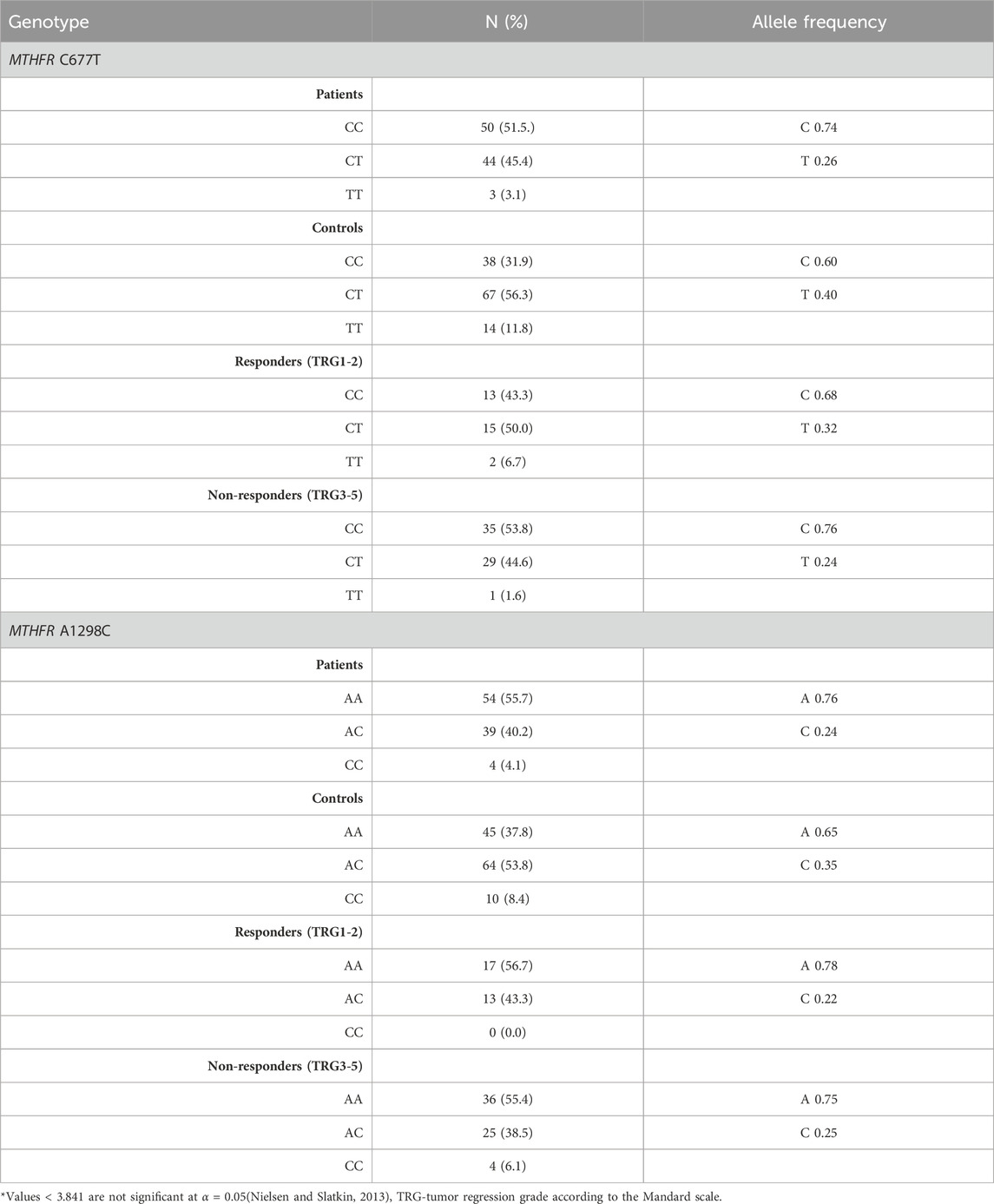
TABLE 2. Genotype and allele frequencies of MTHFR C677T and A1298C polymorphic variants in the patient and healthy control groups.
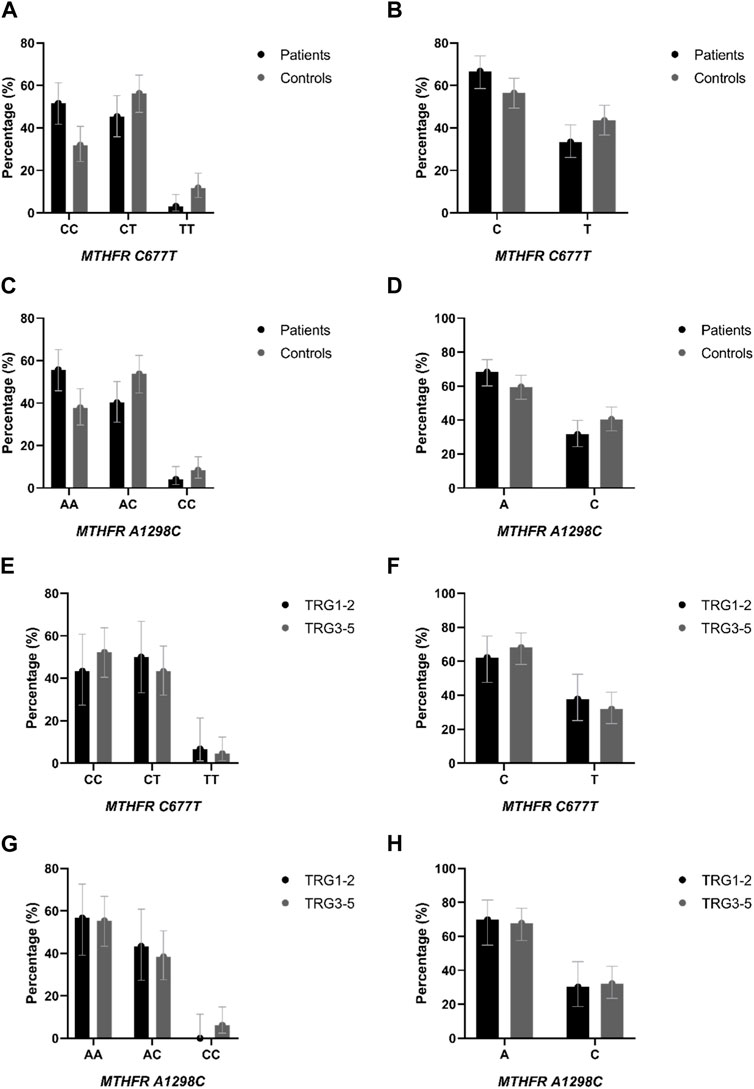
FIGURE 4. (A) Genotype and (B) allele distribution of the MTHFR C677T polymorphic variants in rectal cancer patients and healthy controls. (C) Genotype and (D) allele distribution of the MTHFR A1298C polymorphic variants in rectal cancer patients and healthy controls. (E) Genotype and (F) allele distribution of the MTHFR C677T polymorphic variants in responders and non-responders to neoadjuvant chemoradiotherapy. (G) Genotype and (H) allele distribution of the MTHFR A1298C polymorphic variants in responders and non-responders to neoadjuvant chemoradiotherapy.
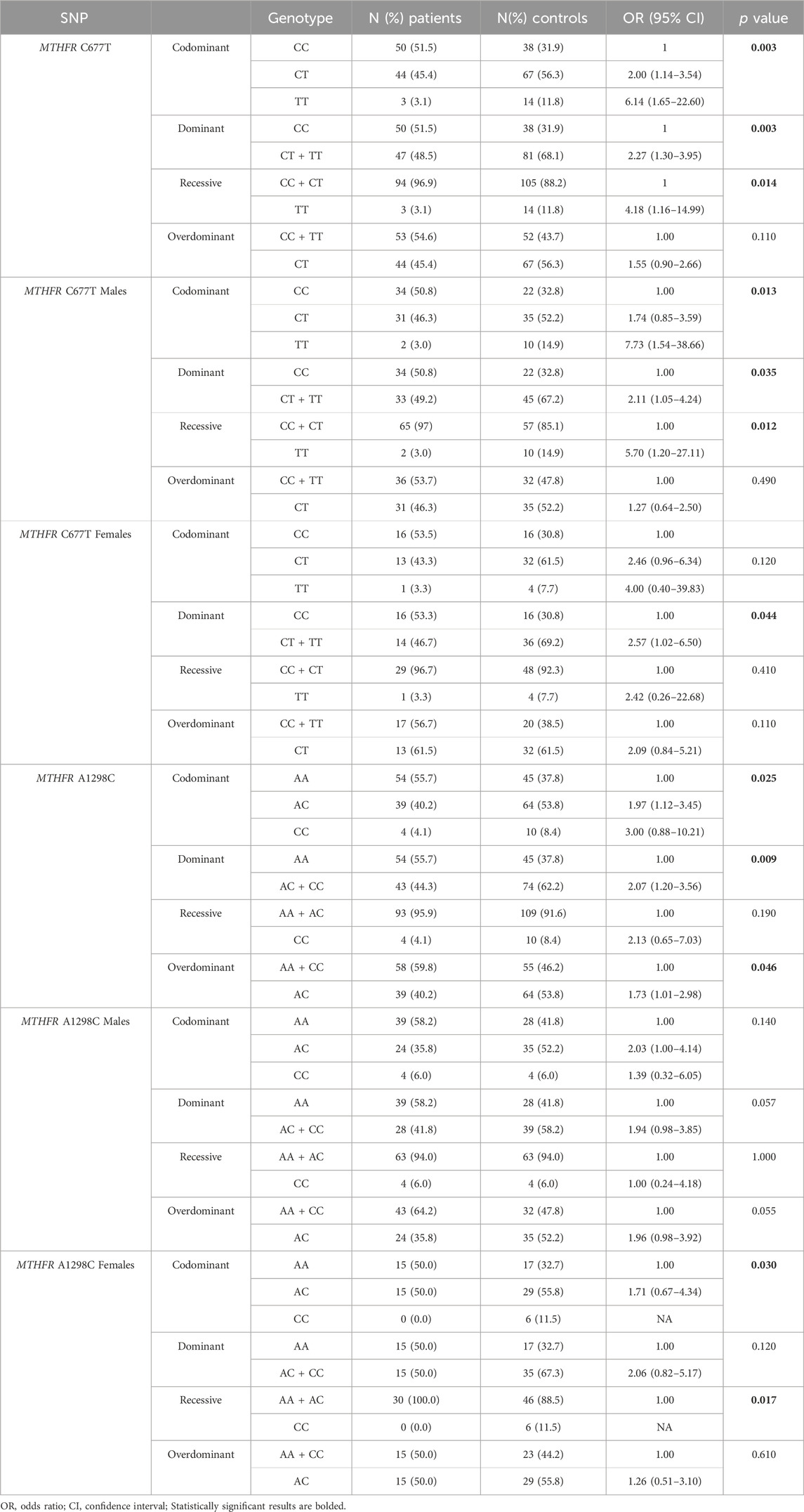
TABLE 3. Analysis of the effects of MTHFR C677T and A1298C polymorphic variants on rectal cancer risk using codominant, dominant, recessive and overdominant models.
The frequency of the A allele was slightly higher in patients (A = 0.76; C = 0.24) than in healthy controls (A = 0.65; C = 0.35) (p = 0.121) (Table 2; Figure 4D). AA homozygosity at the 1298 polymorphic site of the MTFHR gene was associated with a higher risk of developing rectal cancer in the dominant model (Table 3; Figure 4C). MTHFR 1298AA also showed increased risk for cancer development compared to 1298AC+1298CC (OR: 2.07, 95% CI: 1.20–3.56, p = 0.009). It was also found that the overdominant model predicted a protective effect of 1298AC compared to 1298AA and 1298CC (OR: 1.73; 95% CI: 1.01–2.98; p = 0.046). The recessive model in the female population showed that A1298CC had a protective effect for rectal cancer development (p = 0.017). There were no differences in male population with regard to the effect of this polymorphism and risk for rectal cancer.
In groups of patients and controls, the 4 common combined genotypes were CT/AA (29.90% vs. 19.33%), CC/AC (24.74% vs. 15.97%), CC/AA (24.74% vs. 12.61%), and CT/AC (13.40% vs. 32.77%) (Table 4). Statistical analysis of combined genotypes highlighted the protective role of CT/AC combined genotype (p = 0.002) while the CC/AA genotype showed an increased risk for rectal cancer development (p = 0.016) (Table 5).
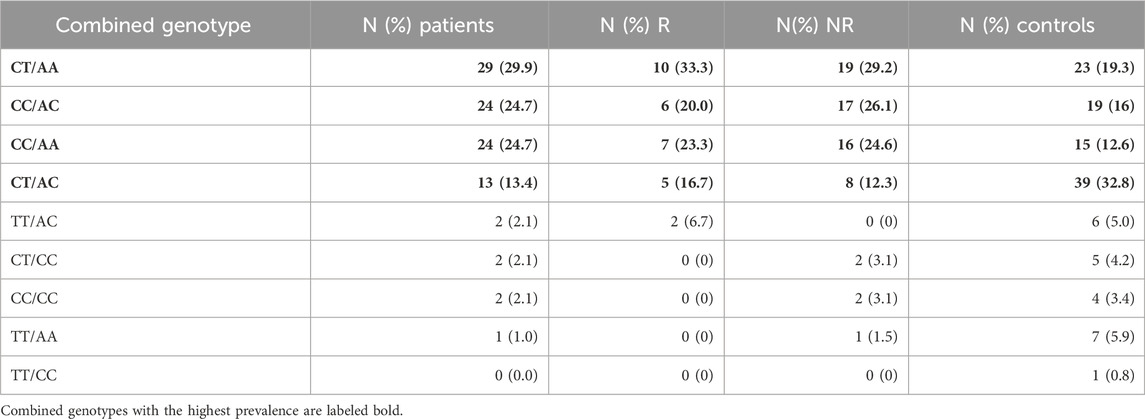
TABLE 4. Combined genotype frequencies of MTHFR C677T and A1298C polymorphic variants within the patients and controls.
Haplotype analysis indicated that the most frequent haplotypes in patients vs. controls were CA (677C-1298A) (46.15% vs. 32.99%) and TA (677T-1298A) (24.10% vs. 26.04%) followed by CC (677C–1298C) (21.03% vs. 23.26%). The rarest haplotype was TC (677T-1298C) (8.72% vs. 17.71%). The carriers of the CA haplotype had the highest risk for developing rectal cancer ((OR): 1.74; 95% (CI) 1.198–2.530, p = 0.002) while the TC haplotype seems to provide a protective effect. (OR: 0.44; 95%CI 0.248–0.795, p = 0.003). These results indicate that the two loci 677 and 1298 share relatively weak linkage disequilibrium in the patient group represented by coefficient of linkage disequilibrium (D′) and correlation coefficient (r2) (D` = 0.27, r2 = 0.00797) (Table 6).
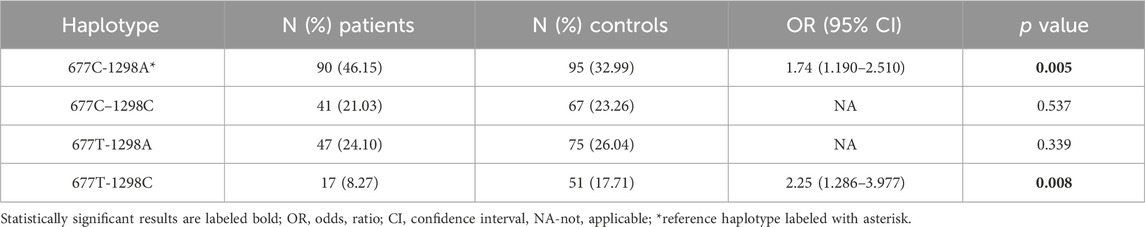
TABLE 6. Estimated haplotype frequencies of MTHFR C677T and A1298C polymorphisms in patients and controls.
Genotypes MTHFR C677T and MTHFR A1298C distribution in responders and non-responders did not deviate from the Hardy-Weinberg equilibrium. The allele frequencies of the MTHFR C677T polymorphic variants in responders and non-responders revealed that responders had a lower frequency of C allele (C = 0.68; T = 0.32) than non-responders (C = 0.76; T = 0.24) (p = 0.270) (Table 2; Figures 4E, F) but no statistical significance was obtained.
The frequency of the A allele in MTHFR A1298C genotype was lower in responders (A = 0.78; C = 0.22) than in non-responders (A = 0.75; C = 0.25) (Table 2; Figures 4G, H) but no statistical significance in predictive potential was obtained (p = 0.739). Analysis of the effects of MTHFR C677T and A1298C polymorphic variants on response to neoadjuvant chemoradiotherapy using codominant, dominant, recessive and overdominant models did not meet statistical significance (Table 7). Within the groups of responders and non-responders, no statistical significance was obtained in terms of predictive potential of combined genotypes as well as haplotype frequencies. Haplotype analysis indicated that the two loci 677 and 1298 show relatively strong linkage disequilibrium in the non-responder group (D` = 0.46, r2 = 0.02163).
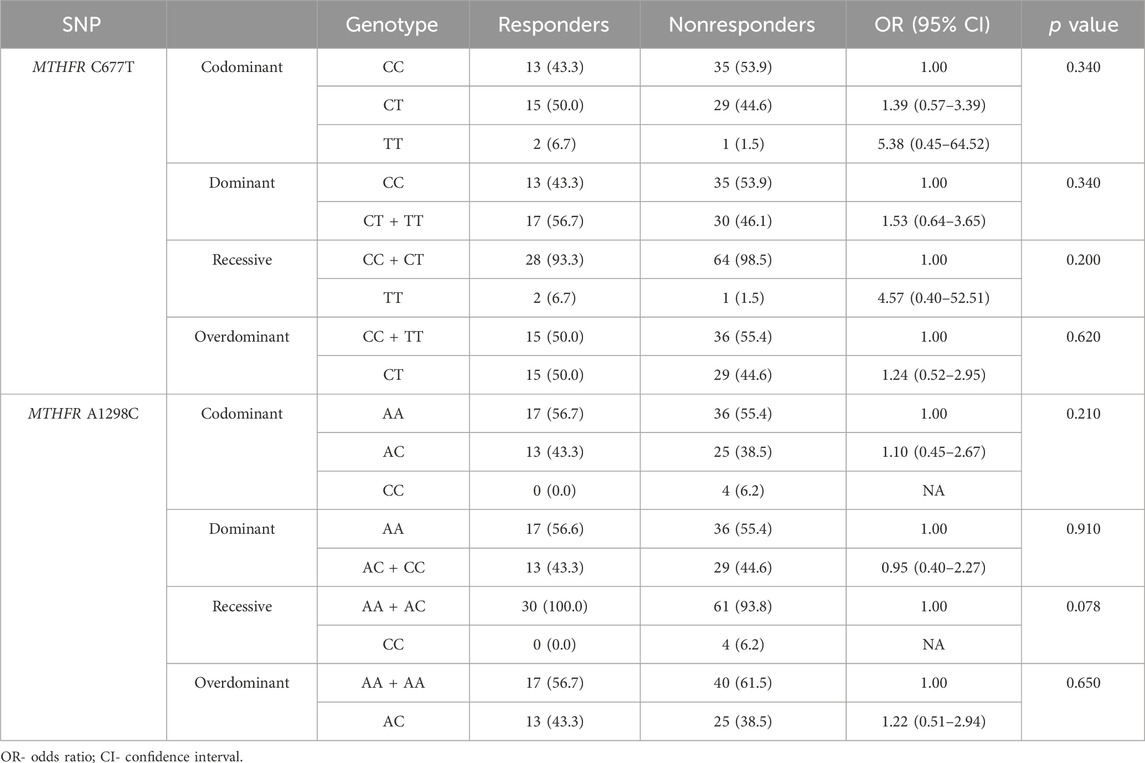
TABLE 7. Analysis of the effects of MTHFR C677T and A1298C polymorphic variants on response to neoadjuvant chemoradiotherapy using codominant, dominant, recessive and overdominant models.
Seven studies were found, but after initial processing one study, GSE35282 met all criteria and included polymorphic variant A1298C data of MTHFR from 43 patients with diagnosed locally advanced rectal cancer (Kim et al., 2013). There was no study available with the C677T polymorphic variant of MTHFR. Patients were divided according to Mandard TRG status into responders (TRG1/2; 41.9%) and non-responders (TR3/4; 58.1%, there were no patients with TRG5). Data obtained in the mentioned study showed high association with our results. Statistical significance was not found within the predictive potential of the A1298C MTHFR polymorphic variant while allele frequencies showed a high association between these two studies (Table 8).
Since colorectal cancer remains one of the most common and deadly malignant diseases worldwide, much effort is directed towards elucidating the etiopathological mechanisms and risk factors. Some risk factors are well known such as diet, alcohol consumption, smoking history and lifestyle but they alone are not enough to explain the development of the disease in all patients.
In silico analysis showed that there was a difference in the expression of normal and tumor tissue. In addition, higher stage of the disease was associated with a lower expression of the MTHFR gene. eQTL analysis indicated that the variant correlated positively (A1298C) and negatively (C677T) with the level of gene expression independent of the tissue type. These results were the basis for examining the predictive potential of MTHFR SNP when it comes to the response to neoadjuvant chemoradiotherapy.
Folate metabolism, more precisely folate deficiency is a very well-known factor of tumorigenesis in general since it plays an important role in maintaining genomic stability by being involved in DNA synthesis, repair, and methylation (Levin and Varga, 2016). 5,10-methylenetetrahydrofolate reductase (MTHFR), one of the key enzymes in the metabolism of folate, is responsible for the irreversible conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which supplies a methyl group for the production of methionine, the lack of which can interfere with DNA synthesis (Ryan and Weir, 2001). Two common functional polymorphisms in the MTHFR gene that have been found to influence the risk of CRC are C677T and A1298C. These SNPs cause reduced activity of the MTHFR enzyme, thus influencing folate metabolism, causing folate depletion and DNA hypomethylation, and disruption of DNA synthesis and repair (Kennedy et al., 2012). DNA hypomethylation is a nearly universal early event in carcinogenesis. It has been suggested that site-specific DNA hypomethylation may be critical, such as the hypomethylation of the coding region (exons 5–8) of the p53 tumor suppressor gene, which is the most frequently mutated in human cancers (Kim et al., 1997; Choi and Mason, 2000).
Data regarding the specific influence of these polymorphisms on CRC risk are varied and inconclusive, and importantly, very few studies investigated the effect of these polymorphisms on the risk of rectal and colon cancer separately, which could be different since rectal and colon cancer are different diseases in their pathogenesis, histology, and sensitivity to treatment. We aimed to investigate the effect of C677T and A1298C polymorphisms on the risk of developing rectal cancer only, in the population of patients in Serbia. The results of this case-control study on 97 patients diagnosed with LARC and 119 healthy volunteers showed a higher frequency of the C677T C allele in patients than in controls (0.74), and the CC homozygosity at the 677 polymorphic sites was more common in patients than in healthy controls. This would suggest a protective effect of the T allele against rectal cancer. These results were confirmed using codominant, dominant, recessive and overdominant model of association. Data from previous studies are inconclusive, with most studies being carried out on single, ethnically homogenous populations, as is ours. Murtaugh et al. in the United States (Murtaugh et al., 2007), Levine et al. in Canada (Levin and Varga, 2016), Sheng et al. (Sheng et al., 2012) and Rai (Rai, 2016) in Asia showed the protective effect of the 677 T allele against CRC and rectal cancer in the American study, as in the situation in our study. In a meta-analysis of 67 studies carried out in 25 countries over the world and comprising all ethnic groups, it was found that the homozygous variant genotype MTHFR 677TT confers a reduced risk of CRC by 12%, but the risk between carriers of 677CT and CC genotypes was similar (Kennedy et al., 2012). On the other hand, a meta-analysis by Teng and colleagues in 2013 done on 71 studies and over 30 thousand patients showed an increased risk of CRC in carriers of the 677TT genotype, but only in Caucasian patients and not Asians (Teng et al., 2013). The same results were found in the Indian and the Hungarian population, with the caveat that this effect was found only on patients with rectal cancer in Hungary, but not colon cancer (Wang et al., 2006; Komlósi et al., 2010). A recent paper by Alanov and colleagues in Azerbaijan showed no effect of this polymorphism on the risk of CRC (Aslanov et al., 2023).
In our group, there was a slightly higher frequency of MTHFR 1298C allele (0.75) than in controls (0.65), and AA homozygosity at the 1298 polymorphic site was more common in patients than in healthy controls. As with the C677T polymorphism, data about the effect of A1298C polymorphism are not consistent. In a study by Jiang and colleagues, carriers of the 1298C allele had a lower risk of rectal cancer (Jiang et al., 2005). Same association was found in the Indian study where carriers of the 1298AC genotype were at a lower risk for colon cancer (OR 0.43, 95% CI 0.22–0.82) and rectal cancer (OR 0.7), and carriers of the homozygous CC genotype had a significantly lower risk for both colon (OR 0.3, 95% CI 0.09–0.80) and rectal cancer (OR 0.43, 95% CI 0.23–0.80) (Wang et al., 2006). This data would suggest a protective effect of the C allele of the A1298C polymorphisms. Again, a 33% reduction in the risk of rectal cancer in individuals carrying the 1298CC genotype was reported (Murtaugh et al., 2007). On the other hand, a Japanese case-control study on 220 patients with rectal cancer and controls found no association between this polymorphism and risk of rectal cancer nor did the meta-analysis of Kennedy and colleagues (Matsuo et al., 2005). However, in this meta-analysis, there was a lower risk of CRC in carriers of the 1298CC genotype in Asian and American studies, but an increased risk of CRC in European countries, which would suggest some geographic differences, possibly related to dietary habits of folate intake or alcohol consumption.
Very few studies stratified results also based on sex, but as before, results are inconclusive and inconsistent. Lightfoot et al. found a reduced risk of CRC in men with the 677CT genotype, and an increased risk in women with the 677TT genotype, while most other studies didn’t find any difference (Lightfoot et al., 2008). Komlosi et al. investigated the Hungarian population and stratified results according to age and sex, finding that the presence of the 677 C allele increased the risk for rectal cancer only in younger (<60) and male patients, but found no influence on colon cancer risk (Komlósi et al., 2010). On the other hand, Murtaugh et al. found a reduced risk of rectal cancer only in female carriers of the 677 T allele, especially in women 60 years or older (OR 0.32, 95% CI) in the United States (Murtaugh et al., 2007). Our analysis highlighted the protective role of the 677TT genotype for rectal cancer development in males (p = 0.030) and females (p = 0.044) as well as 1298CC in females (p = 0.017).
There is not much data on the combined effect of MTHFR C677T and A1298C polymorphisms and the risk of CRC. It has been previously described that the MTHFR C677T and A1298C polymorphisms are in linkage disequilibrium, which means that combinations of 677CT and 1298CC, 677TT and 1298AC, and 677TT and 1298CC are very rare (Kono and Chen, 2005). In this review, it was found that having one variant allele of either 677T or 1298C does not affect the risk of colorectal cancer. In the study by Murtaugh et al. the lowest risk of CRC was found in carriers of 677CC/1298CC. However, in the meta-analysis by Kennedy and the study of Yin et al. carriers of the 677TT/1298AA had the lowest risk of CRC, and carriers of 677CC/1298CC had a non-significantly higher risk of CRC in Yin`s study, again, showing inconsistencies in literature data (Yin et al., 2004). Our results suggest that individuals with haplotypes 677C and 1298A have an increased risk for rectal cancer development compared to any other haplotype while 677T and 1298C haplotypes have a protective role. Combined genotype analysis highlighted that individuals with the CC/AA genotype combination have an increased risk for rectal cancer development while the CT/AC genotype has a protective role.
Although our analysis did not show a statistically significant association between MTHFR C677T and A1298C polymorphisms and response to therapy, our results are somewhat different than most previously reported in the literature. We found a slightly lower frequency of the 677C allele in responders (0.68) than in non-responders (0.76), suggesting that carriers of the C allele are less likely to respond to nCRT. Most other studies (Longley et al., 2003; Vecchio et al., 2005; Balboa et al., 2010; Cecchin et al., 2011; Garcia-Aguilar et al., 2011; Thomas et al., 2011; Ulrich et al., 2014; Zhao et al., 2015; Salnikova and Kolobkov, 2016) found a relationship with the 677C allele and better response to nCRT, which goes further to suggest a very complex and probably multifactorial influence of genetic and other factors on the efficacy of nCRT. The most frequent explanation of the effect of MTHFR polymorphisms on the efficacy of 5FU-based nCRT is that the decreased activity of MTHFR causes elevated levels of 5,10-MTHF, thus reducing TS activity and disrupting DNA and RNA synthesis (Longley et al., 2003; De Mattia et al., 2020).
However, another hypothesis focuses on the higher availability of non-methylated folate substrates in patients with reduced MTHFR activity, which can be used for de novo synthesis of DNA, thus preserving DNA integrity. Also, carriers of the 677TT genotype could be less prone to DNA damage by radiotherapy, thus making the nCRT less effective in these patients (Kawakami et al., 2003; Leopardi, 2006).
All this emphasizes the complexity and multifactorial influences on treatment outcomes, especially in patients treated with combined modalities.
Although this study indicated the importance of MTHFR gene polymorphisms for the risk of developing locally advanced rectal cancer, it has some limitations. The main limitation of the study is the relatively low number of samples, although it is a representative sample of the Serbian LARC population (Flikkema and Toledo-Pereyra, 2012). The single-center and retrospective nature of the study are also limitations, thus validation of our findings and further survival analysis is planned to be performed on a prospective cohort whose collection is in progress at our Institute.
Data obtained in this study points to low-cost, non-invasive, and easily determined factors that might be helpful in identifying specific subgroups of patients that should be monitored more closely, which is especially important in developing countries. The MTHFR 667C allele and 1298A alleles were identified as low-penetrance risk factors for rectal cancer in our population. Our study did not show the influence of these polymorphisms on the efficacy of nCRT, but a further and more detailed analysis using a wider panel of genetic factors is planned. To the best of our knowledge, this is the first study of its kind done on the Slavic population in the Western Balkan region. Knowing that a variety of population-based characteristics may also be significant in this setting, this study could be helpful for future meta-analyses and used for construction of genetic cancer risk prediction panels.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The studies involving humans were approved by the Ethics Board of the Institute for Oncology and Radiology of Serbia and were in accordance with the Helsinki Declaration of 1964 and its later amendments or comparable ethical standards. All patients signed an informed consent. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
AS: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Writing–original draft, Writing–review and editing. JS: Conceptualization, Data curation, Formal Analysis, Investigation, Validation, Writing–original draft, Writing–review and editing. MM: Conceptualization, Data curation, Formal Analysis, Methodology, Software, Writing–original draft, Writing–review and editing. SS-R: Resources, Writing–review and editing, Investigation, Supervision. RJ: Investigation, Project administration, Resources, Supervision, Writing–review and editing. AD: Resources, Writing–review and editing, Project administration, Validation. JZ: Funding acquisition, Project administration, Resources, Supervision, Writing–review and editing. RF: Funding acquisition, Project administration, Resources, Supervision, Writing–review and editing. SC-B: Funding acquisition, Project administration, Resources, Supervision, Writing–review and editing. MC: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing–original draft, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Horizon Europe Twinning Project STEPUPIORS (Agreement No. 101079217) and the Ministry of Education and Science of the Republic of Serbia (Agreement No. 451-03-47/2023-01/200043). This manuscript is available as a preprint (ID MEDRXIV/2023/295916).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aslanov, H., Agaev, R., Bayramov, B., Hadizade, A., Alakbarova, N., Abdulrahimli, S., et al. (2023). Research Article Lack of significant association between MTHFR gene C677T polymorphism and colorectal cancer in the Azerbaijani population. Genet. Mol. Res. 22. doi:10.4238/gmr19100
Balboa, E., Duran, G., Lamas, M. J., Gomez-Caamaño, A., Celeiro-Muñoz, C., Lopez, R., et al. (2010). Pharmacogenetic analysis in neoadjuvant chemoradiation for rectal cancer: high incidence of somatic mutations and their relation with response. Pharmacogenomics 11, 747–761. doi:10.2217/pgs.10.51
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2012). NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41, D991–D995. doi:10.1093/nar/gks1193
Cavic, M., Krivokuca, A., Boljevic, I., Brotto, K., Jovanovic, K., Tanic, M., et al. (2016). Pharmacogenetics in cancer therapy - 8 years of experience at the Institute for Oncology and Radiology of Serbia. J. BUON 21, 1287–1295.
Cavic, M., Krivokuca, A., Spasic, J., Brotto, K., Malisic, E., Radulovic, S., et al. (2014). The influence of methylenetetrahydrofolate reductase and thymidylate synthetase gene polymorphisms on lung adenocarcinoma occurrence. J. BUON 19, 1024–1028.
Cavic, M., Spasic, J., Krivokuca, A., Boljevic, I., Kuburovic, M., Radosavljevic, D., et al. (2019). TP53 and DNA-repair gene polymorphisms genotyping as a low-cost lung adenocarcinoma screening tool. J. Clin. Pathol. 72, 75–80. doi:10.1136/jclinpath-2018-205553
Cecchin, E., Agostini, M., Pucciarelli, S., De Paoli, A., Canzonieri, V., Sigon, R., et al. (2011). Tumor response is predicted by patient genetic profile in rectal cancer patients treated with neo-adjuvant chemo-radiotherapy. Pharmacogenomics J. 11, 214–226. doi:10.1038/tpj.2010.25
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B. V. S. K., et al. (2017). UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658. doi:10.1016/j.neo.2017.05.002
Choi, S.-W., and Mason, J. B. (2000). Folate and carcinogenesis: an integrated scheme. J. Nutr. 130, 129–132. doi:10.1093/jn/130.2.129
De Mattia, E., Roncato, R., Palazzari, E., Toffoli, G., and Cecchin, E. (2020). Germline and somatic pharmacogenomics to refine rectal cancer patients selection for neo-adjuvant chemoradiotherapy. Front. Pharmacol. 11, 897. doi:10.3389/fphar.2020.00897
Fekete, J. T., and Győrffy, B. (2019). ROCplot.org: validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3,104 breast cancer patients. Int. J. Cancer 145, 3140–3151. doi:10.1002/ijc.32369
Flikkema, R. M., and Toledo-Pereyra, L. H. (2012). Sample size determination in medical and surgical research. J. Investig. Surg. Off. J. Acad. Surg. Res. 25, 3–7. doi:10.3109/08941939.2011.648868
Garcia-Aguilar, J., Chen, Z., Smith, D. D., Li, W., Madoff, R. D., Cataldo, P., et al. (2011). Identification of a biomarker profile associated with resistance to neoadjuvant chemoradiation therapy in rectal cancer. Ann. Surg. 254, 486–492. doi:10.1097/SLA.0b013e31822b8cfa
Gaunt, T. R., Rodriguez, S., Zapata, C., and Day, I. N. (2006). MIDAS: software for analysis and visualisation of interallelic disequilibrium between multiallelic markers. BMC Bioinforma. 7, 227. doi:10.1186/1471-2105-7-227
Glynne-Jones, R., Wyrwicz, L., Tiret, E., Brown, G., Rödel, C., Cervantes, A., et al. (2017). Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 28, iv22–iv40. doi:10.1093/annonc/mdx224
Jakovljevic, K., Malisic, E., Cavic, M., Radulovic, S., and Jankovic, R. (2012b). Association between methylenetetrahydrofolate reductase polymorphism C677T and risk of chronic myeloid leukemia in Serbian population. Leuk. Lymphoma 53, 1327–1330. doi:10.3109/10428194.2011.645210
Jiang, Q., Chen, K., Ma, X., Li, Q., Yu, W., Shu, G., et al. (2005). Diets, polymorphisms of methylenetetrahydrofolate reductase, and the susceptibility of colon cancer and rectal cancer. Cancer Detect Prev. 29, 146–154. doi:10.1016/j.cdp.2004.11.004
Kawakami, K., Ruszkiewicz, A., Bennett, G., Moore, J., Watanabe, G., and Iacopetta, B. (2003). The folate pool in colorectal cancers is associated with DNA hypermethylation and with a polymorphism in methylenetetrahydrofolate reductase. Clin. Cancer Res. 9, 5860–5865.
Kennedy, D. A., Stern, S. J., Matok, I., Moretti, M. E., Sarkar, M., Adams-Webber, T., et al. (2012). Folate intake, MTHFR polymorphisms, and the risk of colorectal cancer: a systematic review and meta-analysis. J. Cancer Epidemiol. 2012, 952508–952524. doi:10.1155/2012/952508
Kim, J. C., Ha, Y. J., Roh, S. A., Cho, D. H., Choi, E. Y., Kim, T. W., et al. (2013). Novel single-nucleotide polymorphism markers predictive of pathologic response to preoperative chemoradiation therapy in rectal cancer patients. Int. J. Radiat. Oncol. Biol. Phys. 86, 350–357. doi:10.1016/j.ijrobp.2012.12.018
Kim, Y., Pogribny, I., Basnakian, A., Miller, J., Selhub, J., James, S., et al. (1997). Folate deficiency in rats induces DNA strand breaks and hypomethylation within the p53 tumor suppressor gene. Am. J. Clin. Nutr. 65, 46–52. doi:10.1093/ajcn/65.1.46
Kim, Y.-I. (2003). Role of folate in colon cancer development and progression. J. Nutr. 133, 3731S–3739S. doi:10.1093/jn/133.11.3731S
Komlósi, V., Hitre, E., Pap, É., Adleff, V., Réti, A., Székely, É., et al. (2010). SHMT1 1420 and MTHFR 677 variants are associated with rectal but not colon cancer. BMC Cancer 10, 525. doi:10.1186/1471-2407-10-525
Kono, S., and Chen, K. (2005). Genetic polymorphisms of methylenetetrahydrofolate reductase and colorectal cancer and adenoma. Cancer Sci. 96, 535–542. doi:10.1111/j.1349-7006.2005.00090.x
Krivokuca, A. M., Cavic, M. R., Malisic, E. J., Rakobradovic, J. D., Kolarevic-Ivankovic, D., Tomasevic, Z. I., et al. (2016). Polymorphisms in cancer susceptibility genes XRCC1, RAD51 and TP53 and the risk of breast cancer in Serbian women. Int. J. Biol. Markers 31, e258–e263. doi:10.5301/jbm.5000201
Landberg, T., Chavaudra, J., Dobbs, J., Gerard, J.-P., Hanks, G., Horiot, J.-C., et al. (2016a). Report 62. J. Int. Comm. Radiat. Units Meas. os32. doi:10.1093/jicru/os32.1.Report.62
Landberg, T., Chavaudra, J., Dobbs, J., Hanks, G., Johansson, K.-A., Möller, T., et al. (2016b). Report 50. J. Int. Comm. Radiat. Units Meas. os26. doi:10.1093/jicru/os26.1.Report.50
Leopardi, P., Marcon, F., Caiola, S., Cafolla, A., Siniscalchi, E., Zijno, A., et al. (2006). Effects of folic acid deficiency and MTHFR C677T polymorphism on spontaneous and radiation-induced micronuclei in human lymphocytes. Mutagenesis 21, 327–333. doi:10.1093/mutage/gel031
Levin, B. L., and Varga, E. (2016). MTHFR: addressing genetic counseling dilemmas using evidence-based literature. J. Genet. Couns. 25, 901–911. doi:10.1007/s10897-016-9956-7
Lightfoot, T. J., Barrett, J. H., Bishop, T., Northwood, E. L., Smith, G., Wilkie, M. J. V., et al. (2008). Methylene tetrahydrofolate reductase genotype modifies the chemopreventive effect of folate in colorectal adenoma, but not colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 17, 2421–2430. doi:10.1158/1055-9965.EPI-08-0058
Longley, D. B., Harkin, D. P., and Johnston, P. G. (2003). 5-Fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 3, 330–338. doi:10.1038/nrc1074
Marinkovic, M., Stojanovic-Rundic, S., Stanojevic, A., Ostojic, M., Gavrilovic, D., Jankovic, R., et al. (2023). Exploring novel genetic and hematological predictors of response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Front. Genet. 14, 1245594. doi:10.3389/fgene.2023.1245594
Matsuo, K., Ito, H., Wakai, K., Hirose, K., Saito, T., Suzuki, T., et al. (2005). One-carbon metabolism related gene polymorphisms interact with alcohol drinking to influence the risk of colorectal cancer in Japan. Carcinogenesis 26, 2164–2171. doi:10.1093/carcin/bgi196
Moreno, V., Alonso, M. H., Closa, A., Vallès, X., Díez-Villanueva, A., Valle, L., et al. (2018). Colon-specific eQTL analysis to inform on functional SNPs. Br. J. Cancer 119, 971–977. doi:10.1038/s41416-018-0018-9
Murtaugh, M. A., Curtin, K., Sweeney, C., Wolff, R. K., Holubkov, R., Caan, B. J., et al. (2007). Dietary intake of folate and co-factors in folate metabolism, MTHFR polymorphisms, and reduced rectal cancer. Cancer Causes Control 18, 153–163. doi:10.1007/s10552-006-0099-2
Muzny, D. M., Bainbridge, M. N., Chang, K., Dinh, H., Drummond, J., Fowler, G. R., et al. (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. doi:10.1038/nature11252
National Cancer Institute and the National Human Genome Research Institute (2021). The cancer genome atlas. Available at: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/studied-cancers (Accessed March 3, 2021).
Nielsen, R., and Slatkin, M. (2013). An introduction to population genetics. Oxford: Oxford University Press.
Nikolic, N., Radosavljevic, D., Gavrilovic, D., Nikolic, V., Stanic, N., Spasic, J., et al. (2021). Prognostic factors for post-recurrence survival in stage II and III colorectal carcinoma patients. Med. (B Aires) 57, 1108. doi:10.3390/medicina57101108
Oken, M. M., Creech, R. H., Tormey, D. C., Horton, J., Davis, T. E., McFadden, E. T., et al. (1982). Toxicity and response criteria of the eastern cooperative Oncology group. Am. J. Clin. Oncol. 5, 649–656. doi:10.1097/00000421-198212000-00014
Ozretić, P., Da Silva Filho, M. I., Catalano, C., Sokolović, I., Vukić-Dugac, A., Šutić, M., et al. (2019). Association of NLRP1 coding polymorphism with lung function and serum IL-1β concentration in patients diagnosed with chronic obstructive pulmonary disease (COPD). Genes 10, 783. doi:10.3390/genes10100783
Rai, V. (2016). Evaluation of the MTHFR C677T polymorphism as a risk factor for colorectal cancer in asian populations. Asian Pac. J. Cancer Prev. 16, 8093–8100. doi:10.7314/APJCP.2015.16.18.8093
Ryan, B. M., and Weir, D. G. (2001). Relevance of folate metabolism in the pathogenesis of colorectal cancer. J. Laboratory Clin. Med. 138, 164–176. doi:10.1067/mlc.2001.117161
Ryan-Harshman, M., and Aldoori, W. (2007). Diet and colorectal cancer: review of the evidence. Can. Fam. Physician 53, 1913–1920.
Salnikova, L. E., and Kolobkov, D. S. (2016). Germline and somatic genetic predictors of pathological response in neoadjuvant settings of rectal and esophageal cancers: systematic review and meta-analysis. Pharmacogenomics J. 16, 249–265. doi:10.1038/tpj.2015.46
Serbian Cancer Registry (2022). Malignant tumours in the republic of Serbia. Available at: https://www.batut.org.rs/index.php?content=2096 (Accessed on December 26, 2023).
Sheng, X., Zhang, Y., Zhao, E., Lu, S., Zheng, X., Ge, H., et al. (2012). MTHFR C677T polymorphism contributes to colorectal cancer susceptibility: evidence from 61 case–control studies. Mol. Biol. Rep. 39, 9669–9679. doi:10.1007/s11033-012-1832-4
Siddiqui, M. R. S., Bhoday, J., Battersby, N. J., Chand, M., West, N. P., Abulafi, A.-M., et al. (2016). Defining response to radiotherapy in rectal cancer using magnetic resonance imaging and histopathological scales. World J. Gastroenterol. 22, 8414–8434. doi:10.3748/wjg.v22.i37.8414
Stanojevic, A., Samiotaki, M., Lygirou, V., Marinkovic, M., Nikolic, V., Stojanovic-Rundic, S., et al. (2023). Data-independent acquisition mass spectrometry analysis of FFPE rectal cancer samples offers in-depth proteomics characterization of the response to neoadjuvant chemoradiotherapy. Int. J. Mol. Sci. 24, 15412. doi:10.3390/ijms242015412
Stojanovic-Rundic, S., Marinkovic, M., Cavic, M., Karapandzic, V. P., Gavrilovic, D., Jankovic, R., et al. (2021). The role of haematological parameters in predicting the response to radical chemoradiotherapy in patients with anal squamous cell cancer. Radiol. Oncol. 55, 449–458. doi:10.2478/raon-2021-0039
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., and SoerjomataramJemal, I. A. (2021). Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Szklarczyk, D., Kirsch, R., Koutrouli, M., Nastou, K. C., Mehryary, F., Hachilif, R., et al. (2022). The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. doi:10.1093/nar/gkac1000
Teng, Z., Wang, L., Cai, S., Yu, P., Wang, J., Gong, J., et al. (2013). The 677C.T (rs1801133) polymorphism in the MTHFR gene contributes to colorectal cancer risk: a meta-analysis based on 71 research studies. PLoS One 8, e55332. doi:10.1371/journal.pone.0055332
The Human Protein Atlas database V.20.0 (2021). The human protein atlas database V.20.0. Available at: http://www.proteinatlas.org/pathology (Accessed March 3, 2021).
Thomas, F., Motsinger-Reif, A. A., Hoskins, J. M., Dvorak, A., Roy, S., Alyasiri, A., et al. (2011). Methylenetetrahydrofolate reductase genetic polymorphisms and toxicity to 5-FU-based chemoradiation in rectal cancer. Br. J. Cancer 105, 1654–1662. doi:10.1038/bjc.2011.442
Uhlen, M., Zhang, C., Lee, S., Sjöstedt, E., Fagerberg, L., Bidkhori, G., et al. (2017). A pathology atlas of the human cancer transcriptome. Science 357, eaan2507. doi:10.1126/science.aan2507
Ulrich, C. M., Rankin, C., Toriola, A. T., Makar, K. W., Altug-Teber, Ö., Benedetti, J. K., et al. (2014). Polymorphisms in folate-metabolizing enzymes and response to 5-fluorouracil among patients with stage II or III rectal cancer (INT-0144; SWOG 9304). Cancer 120, 3329–3337. doi:10.1002/cncr.28830
Union for International Cancer Control (2020). The TNM classification of malignant tumours. Available at: https://www.uicc.org/sites/main/files/atoms/files/UICC.TNM.Classification (Accessed March 11, 2021).
Vecchio, F. M., Valentini, V., Minsky, B. D., Padula, G. D. A., Venkatraman, E. S., Balducci, M., et al. (2005). The relationship of pathologic tumor regression grade (TRG) and outcomes after preoperative therapy in rectal cancer. Int. J. Radiat. Oncol. Biol. Phys. 62, 752–760. doi:10.1016/j.ijrobp.2004.11.017
Vuletić, A., Mirjačić Martinović, K., Tišma Miletić, N., Zoidakis, J., Castellvi-Bel, S., and Čavić, M. (2021). Cross-talk between tumor cells undergoing epithelial to mesenchymal transition and natural killer cells in tumor microenvironment in colorectal cancer. Front. Cell Dev. Biol. 9, 750022. doi:10.3389/fcell.2021.750022
Wang, J., Gajalakshmi, V., Jiang, J., Kuriki, K., Suzuki, S., Nagaya, T., et al. (2006). Associations between 5,10-methylenetetrahydrofolate reductase codon 677 and 1298 genetic polymorphisms and environmental factors with reference to susceptibility to colorectal cancer: a case-control study in an Indian population. Int. J. Cancer 118, 991–997. doi:10.1002/ijc.21438
Wei, J., Huang, R., Guo, S., Zhang, X., Xi, S., Wang, Q., et al. (2018). ypTNM category combined with AJCC tumor regression grade for screening patients with the worst prognosis after neoadjuvant chemoradiation therapy for locally advanced rectal cancer. Cancer Manag. Res. 10, 5219–5225. doi:10.2147/CMAR.S179151
Yamagishi, H., Kuroda, H., Imai, Y., and Hiraishi, H. (2016). Molecular pathogenesis of sporadic colorectal cancers. Chin. J. Cancer 35, 4. doi:10.1186/s40880-015-0066-y
Yin, G., Kono, S., Toyomura, K., Hagiwara, T., Nagano, J., Mizoue, T., et al. (2004). Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and colorectal cancer: the fukuoka colorectal cancer study. Cancer Sci. 95, 908–913. doi:10.1111/j.1349-7006.2004.tb02201.x
Keywords: chemoradiotherapy, methylenetetrahydrofolate reductase, MTHFR C677T, MTHFR A1298C, rectal cancer, single nucleotide polymorphism
Citation: Stanojevic A, Spasic J, Marinkovic M, Stojanovic-Rundic S, Jankovic R, Djuric A, Zoidakis J, Fijneman RJA, Castellvi-Bel S and Cavic M (2024) Methylenetetrahydrofolate reductase polymorphic variants C677T and A1298C in rectal cancer in Slavic population: significance for cancer risk and response to chemoradiotherapy. Front. Genet. 14:1299599. doi: 10.3389/fgene.2023.1299599
Received: 22 September 2023; Accepted: 31 December 2023;
Published: 15 January 2024.
Edited by:
Kejian Zhang, Independent Researcher, Beijing, ChinaReviewed by:
Vugar Yagublu, University of Heidelberg, GermanyCopyright © 2024 Stanojevic, Spasic, Marinkovic, Stojanovic-Rundic, Jankovic, Djuric, Zoidakis, Fijneman, Castellvi-Bel and Cavic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Milena Cavic, bWlsZW5hLmNhdmljQG5jcmMuYWMucnM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.