- College of Health, Medicine and Life Sciences, Department of Life Sciences, Brunel University London, London, United Kingdom
Histone variants, which generally differ in few amino acid residues, can replace core histones (H1, H2A, H2B, and H3) to confer specific structural and functional features to regulate cellular functions. In addition to their role in DNA packaging, histones modulate key processes such as gene expression regulation and chromosome segregation, which are frequently dysregulated in cancer cells. During the years, histones variants have gained significant attention as gatekeepers of chromosome stability, raising interest in understanding how structural and functional alterations can contribute to tumourigenesis. Beside the well-established role of the histone H3 variant CENP-A in centromere specification and maintenance, a growing body of literature has described mutations, aberrant expression patterns and post-translational modifications of a variety of histone variants in several cancers, also coining the term “oncohistones.” At the molecular level, mechanistic studies have been dissecting the biological mechanisms behind histones and missegregation events, with the potential to uncover novel clinically-relevant targets. In this review, we focus on the current understanding and highlight knowledge gaps of the contribution of histone variants to aneuploidy, and we have compiled a database (HistoPloidyDB) of histone gene alterations linked to aneuploidy in cancers of the The Cancer Genome Atlas project.
1 Structural and functional features of histones and histone variants
Histones are fundamental protein components of the nucleosome, which constitutes the basic unit of chromatin in eukaryotes. Nucleosomes are built as octamers of core histones, namely, H2A, H2B, H3, and H4, and are wrapped by approximately 146 bp of DNA (Figure 1A). Histone H1, known as linker histone, is responsible for modulation of chromatin structure between nucleosomes (Luger et al., 1997). Core histones H2A, H2B, and H3, as well as linker histone H1, have paralogues that often differ in few amino acid residues, known as histone variants. Recently, a larger repertoire of histone genes and variants has been uncovered, including an H4 variant H4.G and several pseudogenes with putative regulatory functions (Long et al., 2019; Seal et al., 2022). As pictured by Maze et al. (Maze et al., 2014), “every amino acid matters” for histone variants, as minor changes in amino acid composition can produce profound changes in functional properties. Histone variants are incorporated into the nucleosome to confer specific structural features to accomplish biological outcomes. In fact, histones and their variants regulate a plethora of functions, including regulation of DNA replication and repair, chromatin organisation, transcription, and chromosome segregation (Bernstein and Hake, 2006; Martire and Banaszynski, 2020). The mechanisms behind the “decision” of which histones to incorporate within the correct spatio-temporal window is still an active research question, which is dependent on a tight regulation of histone availability and the coordination with chaperones and histone modifying enzymes, promoting or disrupting the deposition of certain variants over others (Mendiratta et al., 2019).
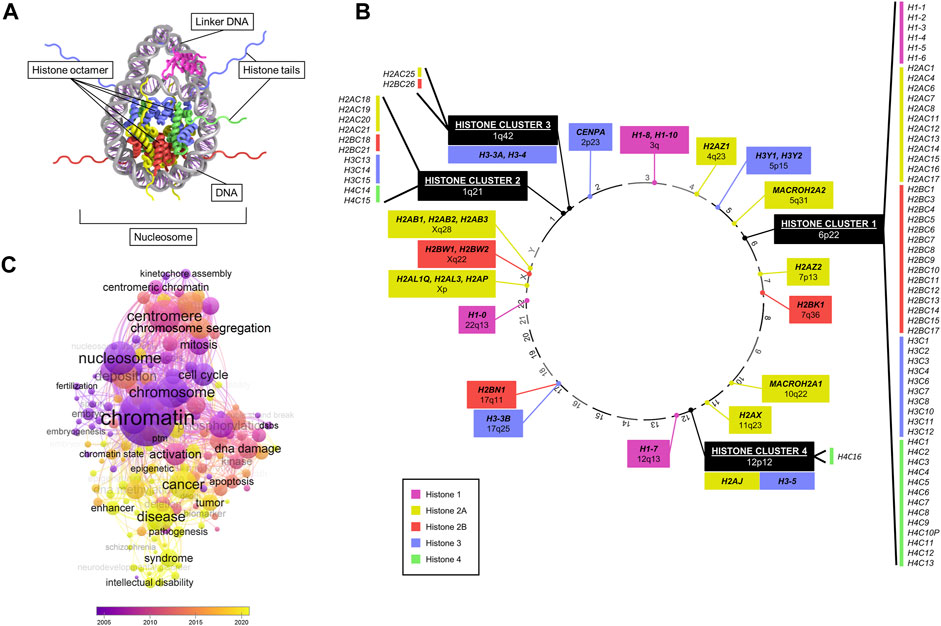
FIGURE 1. Overview of canonical and variant histone genes. (A) Crystal structure of a nucleosome, consisting of histone proteins (forming an octamer) wrapped by DNA. Data adapted from HistoneDB 2.0 (Draizen et al., 2016). (B) Chromosomal location (numbered outside of circle) of all human histone genes. Colours represent different core histones H1, H2A, H2B, H3, and H4. Histone variants are marked in bold text. (C) Bubble plot of numbers of enriched keywords co-occurring in publications searched by “histone variant” in PubMed by year. Generated with VOSviewer.
In humans, histone genes are transcribed from distinct genomic loci (Draizen et al., 2016; Seal et al., 2022) (Figure 1B). The majority of canonical histones are found at a specific locus at 6p22 known as “histone cluster 1”. Three other clusters are located at 1q21, 1q42, and 12p12, harbouring the remainder of canonical histones and a few variants (H3.3A, H3.4, H3.5, and H2A.J). All other histone variants map at various cytobands (Figure 1B). A key difference between canonical and variant histones is the difference in expression during the cell cycle, as canonical histones are replication-dependent, while most of the variants are not (Mei et al., 2017). In addition, almost all canonical histone transcripts do not contain introns or poly(A) tails unlike variants (Maze et al., 2014).
Structurally, core histone proteins consist of a conserved histone-fold domain and a protruding N-terminal tail domain that is the main, yet not exclusive, site of post-translational modifications (PTM). In addition to the variability in nucleosome composition conferred by the incorporation of canonical and variant histones, PTMs also add a further layer of diversity to physical properties and thus functional variability (Bernstein and Hake, 2006; Cavalieri, 2021). Histone variants can possess unique structural features compared to their canonical counterpart, although the majority of variants only present single amino acid substitutions (Maze et al., 2014; Martire and Banaszynski, 2020). The families of H1 and H2A variants show the highest diversity in structural composition compared to H2B and H3 (Maze et al., 2014; El Kennani et al., 2018; Prendergast and Reinberg, 2021).
Due to their role in regulating key cellular processes, the potential of histone variants in contributing to tumourigenic processes has been of great interest (Buschbeck and Hake, 2017). Analysis of PubMed publications featuring “histone variants” revealed an association with a wide range of cellular functions (e.g., cell cycle, mitosis, chromosome segregation, aneuploidy, DNA damage), and a growing association with diseases, notably cancer, in recent years (Figure 1C). In particular, research interest has accrued since the discovery of specific H3 histone driver mutations in glioma disrupting a key methylation site, which coined the term “oncohistone” (Schwartzentruber et al., 2012; Sturm et al., 2012; Wu et al., 2012; Pathania et al., 2017; Nacev et al., 2019). Since then, a large body of literature has grown on histone mutations implicated in several malignancies, including lymphoma (Lohr et al., 2012), chrondroblastoma (Behjati et al., 2013), ovarian cancer (Zhao et al., 2016), and head and neck tumours (Papillon-Cavanagh et al., 2017).
The contribution of histone deregulation to carcinogenesis is multi-faceted and can include effects on gene expression regulation, cell identity, and genomic stability via chromosome integrity and DNA damage response (Buschbeck and Hake, 2017; Ferrand et al., 2020). Genomic instability is tightly correlated, and often precedes, aneuploidy–a hallmark of cancer, broadly defined as numerical and structural aberrations of chromosomes (Sheltzer, 2013; Ben-David and Amon, 2020). Several mechanisms can produce aneuploidy, depending on the type, including defective mitotic machinery, unresolved DNA damage, and centromeric and telomeric dysfunctions (Orr et al., 2015); however, the exact aetiology and contribution of aneuploidy to cancer initiation and progression remains unclear (Molina et al., 2021). The centromeric histone H3 variant CENP-A has been extensively studied as a gatekeeper of centromere maintenance (Renaud-Pageot et al., 2022), however a wider range of variants of other histone families are gaining similar interest and appear to contribute to chromosome stability (Ferrand et al., 2020; Prendergast and Reinberg, 2021; Sales-Gil et al., 2021; Oberdoerffer and Miller, 2023). In particular, histone variants CENP-A, H3.3, H2A.Z, and macroH2A play a crucial role in defining functional chromatin regions (i.e., centromeres, pericentromeric regions, telomeres, inactive X chromosomes), which affect the ability of the cell to faithfully segregate chromosomes by appropriately assembling kinetochore machinery or maintaining telomere length (Ferrand et al., 2020).
In this review, we focus on the contribution and interplay between core histones and histone variants to aneuploidy, by a comprehensive overview of histone variants alterations in cancer. By analysing publicly available cancer data from the Cancer Genome Atlas (TCGA) project, we compiled HistoPloidyDB–a database of correlations between histone gene alterations and aneuploidy (available at https://vagnarelli-lab.github.io/HistoPloidyDB/).
2 Dysregulated expression of histone genes affecting chromosomal stability
Bioinformatics analyses have identified histone gene deregulation as a common core regulatory component at pan-cancer level, together with chromatin remodeling, cell cycle, and immune response (Knaack et al., 2014). Figure 2 illustrates the expression levels of canonical and variant histones in tumour samples of TCGA and healthy tissues from the Genotype-Tissue Expression (GTEx) database, highlighting distinct patterns of expression between canonical and variant genes, as well as by site. Generally, the patterns of expression are distinct in tumours compared to healthy samples, with tissue-specific variability, for both canonical and variant histones. It is known that the regulation of canonical histone gene expression from the major histone clusters is complex, and that the expression of few canonical genes accounts for the majority of transcripts for each core histone (Mei et al., 2017; Flaus et al., 2021). Germline-specific variant histones display unique expression profiles (Doenecke et al., 1997), which is particularly evident in non-cancerous testes samples (Figure 2).
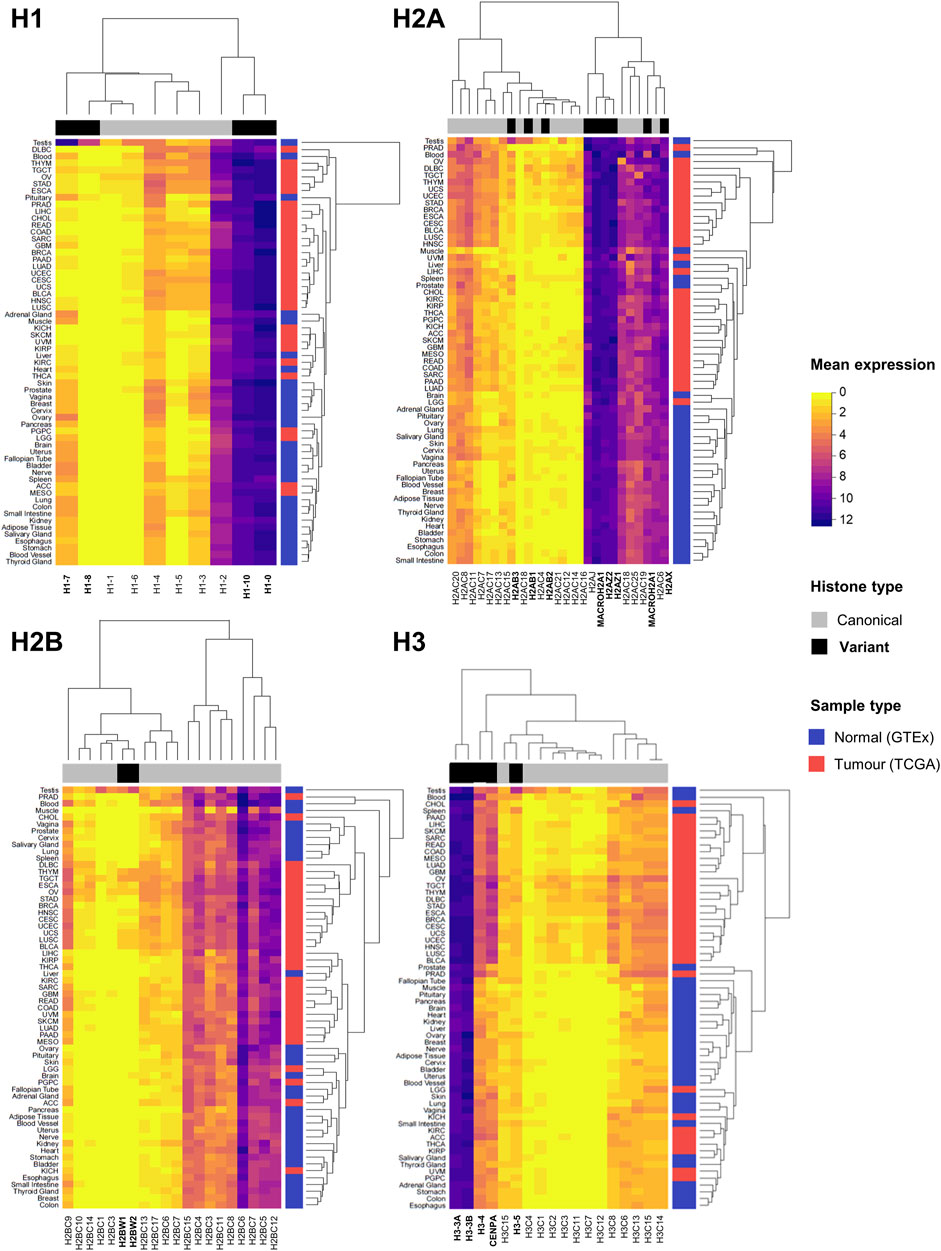
FIGURE 2. Expression overview of histone genes in normal (GTEx) and tumour (TCGA) sites. Heatmaps showing the mean mRNA expression of histone genes in units of log2(norm_count+1) by histone families H1, H2A, H2B, and H3. Hierarchical clustering of Euclidean distances defines similarities and differences in expression patterns, both by canonical and variant genes (indicated on the horizontal side bar in grey and black, respectively), and by site (normal tissue in blue and tumour tissue in red on vertical side bar). Normalised expression data by TOIL pipeline was obtained from University of California Santa Cruz (UCSC) Xena public repository (Goldman et al., 2020).
In cancer, histone genes have a two-fold likelihood of overexpression compared to other genes, however their underexpression is uncommon (Shah et al., 2014; Ferrand et al., 2020; Ghiraldini et al., 2021). Expression data from the TCGA database shows a widespread overexpression of both canonical and variant genes between tumour tissue and matched surrounding healthy tissues, as depicted in Figure 3A by statistically significant fold changes in expression. The upregulation of replication-dependent histones may simply indicate a requirement for higher amounts of histones in proliferating cells undergoing high rates of replication, like cancer cells; however, an imbalanced transcriptional activity of histones can also cause or contribute to oncogenic programmes, as several lines of evidence have highlighted how overexpression of histones can adversely affect gene expression regulation and genomic stability that fuels cancer cells (Miles et al., 2019; Bruhn et al., 2022).
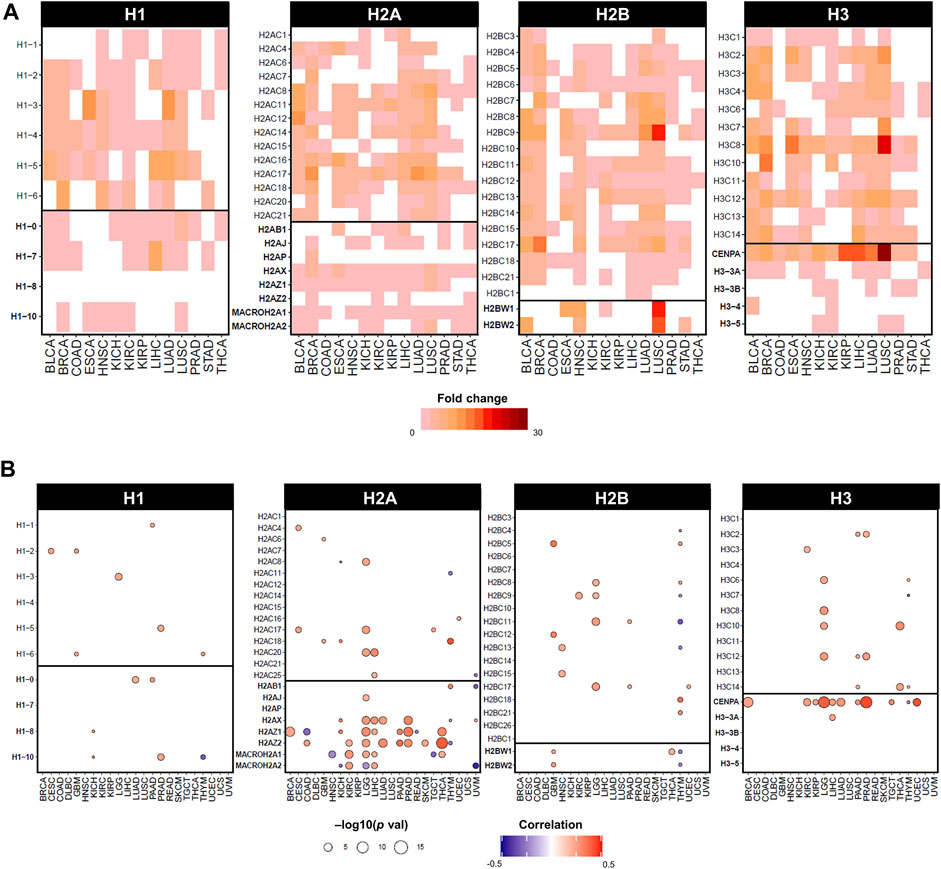
FIGURE 3. Dysregulated expression of histone genes in cancers of TCGA cohort. (A) Statistically significant fold changes in mRNA expression between tumour tissue and matched normal tissue from TCGA. Fold change values were calculated on normalised and batch corrected RSEM values in tumour sites with more than 10 matched tumor and normal samples. The fold change was computed by GSCA (Liu et al., 2023) and calculated as mean(tumour)/mean(normal), statistical significance determined by p-value (<0.05) by t-test. (B) Statistically significant correlations by Pearson correlation coefficients and statistical significance by p-value computed on mRNA expression in RSEM and Aneuploidy Score (i.e., number of altered chromosome arms), obtained from cBioPortal on TCGA cancers from PanCancerAtlas (Cerami et al., 2012; Gao et al., 2013).
In fact, the expression of specific histone genes correlates with aneuploidy levels in several cancers of the TCGA cohort, notably H2A and H3 variants, but also several canonical histones (Figure 3B). Although observational, dysregulation of histone genes occurs in cancers with evidence of genomic/chromosomal instability. For instance, canonical linker histone H1.1 is downregulated and differentially methylated in childhood acute lymphoblastic leukaemia (ALL) with hyperdiploid karyotypes (Davidsson et al., 2009). The canonical histones H1.2, H2AC18, H2BC6, and H2BC21 are upregulated in multiple myeloma, which has an aneuploidy rate of 90% of cases (Zhan et al., 2002). Cancer cells with higher levels of chromosomal instability showed lower levels of expression of canonical H3 and histone variants H1.10, H2A.X, and H2A.Z (Roschke and Kirsch, 2010). Moreover, the entire 6q22 histone locus is overexpressed in aggressive subtypes of liposarcoma that displays high rates of copy number abnormalities (Yoo et al., 2021).
In support of these observations, a large body of literature has described phenotypes of mitotic defects linked to alterations in histone levels. Loss of histone variant H3.3 in murine fibroblasts resulted in defects in chromosome alignment in mitosis and production of multi-lobed nuclei and micronuclei, as well as decreased proliferation and increase in cell death (Ors et al., 2017). Other studies also demonstrated that loss of H3.3 in mouse embryonic stem cells and mouse embryonic fibroblasts produces severe mitotic defects in the form of lagging chromosomes and anaphase bridges (Jang et al., 2015), and that H3.3 is necessary to ensure mitotic fidelity in early mouse development to prevent aneuploidy (Bush et al., 2013; Lin et al., 2013). In p53 deficient backgrounds, these chromosomal defects are accentuated, with higher rates of numerical and structural aneuploidies (Jang et al., 2015). H3.3 was also linked to chromosome instability by numerical aneuploidy by overexpression of Aurora B, which causes an increase in mitotic phosphorylation of H3.3 (Ota et al., 2002). Loss of H1.1 in Xenopus was shown to cause anaphase segregation defects, due to abnormally elongated chromosomes (Maresca et al., 2005). Overall, these studies lay the ground for exploring the dysregulation of these variants and their contribution to genomic instability of tumours.
2.1 Histone excess compromises the balance of histone availability and incorporation
A number of mechanisms have been proposed to account for the excessive supply of histones that can compromise chromosomal stability (Singh et al., 2010; Miles et al., 2019). An accumulation of histones, regardless of canonical or variant forms, can disrupt the balance between nucleosome-bound and free histones, and outcompete the incorporation of the required histones. In S. cerevisae, excessive canonical histone levels result in increased rates of chromosome loss (Meeks-Wagner and Hartwell, 1986; Gunjan and Verreault, 2003; Singh et al., 2010). Also in yeast, prolonged exposure to high levels of H2A and H2B histones produces mitotic defects that can results in whole genome duplication (WGD) via endomitosis, concomitant with a reduced incorporation of the H2A.Z variant into chromatin due to an outcompetition mechanism (Miles et al., 2018). Similarly, excessive levels of canonical H3 leads to chromosome loss by interfering with the normal deposition of the centromere-specifying variant CENP-A (Castillo et al., 2007; Au et al., 2008).
In addition, owing to their basic composition, histones are prone to engage in unwanted interactions with acidic cellular components and negatively charged macromolecules (Singh et al., 2009; Mendiratta et al., 2019). Therefore, an excessive reservoir of histones can induce deleterious effects by inappropriate interactions with histone chaperones, modifying enzymes, or nucleic acids (Gunjan and Verreault, 2003; Singh et al., 2010). In fact, in yeast cells, overexpression of histone chaperones can counteract the cytotoxic effects of histone excess, which include slower proliferation, DNA damage sensitivity, and genomic instability via chromosome loss (Gunjan and Verreault, 2003). Histone chaperones are tightly linked to histone function, as their role in deposition and histone level control ensures an appropriate balance of incorporation and exchange (Hammond et al., 2017), which can be compromised during carcinogenesis. Like histone genes, histone chaperone genes are also dysregulated in cancer, including the H3-H4-cargoing chaperones ASF1, HIRA, CAF-1, DAXX, and DEK, the H3-H4 and H2A-H2B chaperone FACT, and the CENP-A-H4 chaperone HJURP (Ray-Gallet and Almouzni, 2022; Zhao et al., 2022). In terms of contribution to aneuploidy, several studies have shown the importance of histone chaperones in maintaining chromosomal stability, including increased levels of aneuploidy upon loss of FACT (Prendergast et al., 2020), ASF1 (Horard et al., 2018) and HJURP (Filipescu et al., 2017), mitotic defects and micronuclei following overexpression of DEK (Matrka et al., 2015), and expansion of ribosomal DNA (rDNA) repeats in absence of ASF1 (Houseley and Tollervey, 2011), which can be a source of chromosomal aberrations if inappropriately resolved (Daniloski et al., 2019). Clinically, the expression of CAF-1 was found to be correlated with ploidy status in breast cancer, as well as other clinical features of disease progression (Polo et al., 2004). In cancer cell lines, the induction of metastatic programmes resulted in an increased occupancy of the H3.3 variant, with a concomitant reduction of other canonical histones. However, this effect was dependent on CAF-1, as demonstrated by the induction of the metastatic phenotype upon depletion of the chaperone, but not by H3.3 overexpression alone (Gomes et al., 2019). This synergism between chaperones and histones is also supported by a co-operative effect of overexpression of H3.3A and ASF1 inducing proliferation of pancreatic beta cells (Paul et al., 2016). Therefore, the deleterious chromosomal and oncogenic effects resulting from a dysregulated availability of histones are also interlinked with their chaperones.
2.2 Environment-driven changes in histone expression levels
Exposure to carcinogens, including benzene (McHale et al., 2009) and Human T-Lymphotropic Virus Type-1 (HTLV-1) (Bogenberger and Laybourn, 2008), as well as treatments with DNA damage-inducing agents such as ionising radiation, Adriamycin, and cisplatin (Su et al., 2004), have also been linked to changes in canonical histone levels and induction of genomic instability.
One mechanism that has been proposed for the link between environmental exposure and oncogenic histone activity involves aberrant polyadenylation of histone H3.1 transcripts (Brocato et al., 2015; Chen et al., 2020). Replication-dependent histones are not polyadenylated at the 3′ to ensure degradation that is required to maintain normal replication-dependency of canonical histones. Instead, canonical histones possess a stem-loop structure at the 3′ responsible for post-translational processing. When exposed to arsenic, H3.1 mRNA acquires a poly(A) tail, which increases its stability and consequent protein abundance (Brocato et al., 2015; Chen et al., 2020). The excessive availability of H3.1 outcompetes the H3.3 variant from chromatin regulatory regions, resulting in changes in gene expression, cell cycle dysfunctions, and aneuploidy (Chen et al., 2020). The authors highlighted the displacement of H3.3 as a core component of transformation by arsenic, also supported by genetic manipulation of the H3.3 variant alone; in fact, transformation was achieved by knock-down of H3.3 and overexpression of H3.3 could rescue arsenic-dependent transformation (Chen et al., 2020). Exposure to nickel also increases the polyadenylation of H3.1 transcripts, creating a “histone excess” problem that leads to inappropriate incorporation of free histones onto chromatin, inducing genomic instability and transformation (Jordan et al., 2017). The aberrant polyadenylation of canonical histones can also results from decreased levels of the stem-loop binding protein (SLBP), which normally binds to the 3′ stem-loop structure to correctly coordinate histone degradation (Brocato et al., 2015; Jordan et al., 2017). In Drosophila, dysfunctional SLBP leads to chromosomal instability, including chromosomal breaks and numerical aneuploidy, via the accumulation of histones with poly(A) tails (Salzler et al., 2009).
The observed reduction in histone levels (both at transcriptional and protein levels) has also been linked to the response to the DNA damage caused by the carcinogenic agents, which triggers DNA damage response mechanisms and a block of DNA synthesis leading to genomic instability (Su et al., 2004). While this may contradict the effects of excess histone genes, these mechanisms have been attributed to DNA hypomethylation, in a similar fashion to loss of imprinting, which are also causative of deleterious genomic effects, including aneuploidy (Eden et al., 2003; Bogenberger and Laybourn, 2008). In the study on HTLV-1, no effects were detected in ploidy in the infected cells (Bogenberger and Laybourn, 2008), however the appearance of chromosomal anomalies may be a subsequent event in response to an increasingly instable genome. For example, mutations of H3 variants in giant cell tumour of the bone were shown to predate the development of karyotype defects (Fittall et al., 2020). Downregulation of canonical histones (H2AC8, H2AC16, H2BC9, H2BC13, H3C6, H3C7, and H4C6) has been reported to increase genomic instability in giant cell tumour of bone stromal cells (Lau et al., 2017). Therefore, the role of histone deregulation is likely to be dynamic and follow multiple mechanisms.
Another example of environmental influence on histone activity is the modulation of karyotype by C. albicans as a resistance mechanism to antifungal drugs. The induction of numerical and structural aneuploidy is a common mechanism of stress response in the pathogen (Selmecki et al., 2009). C. albicans possesses two H2A homologues, of which one (H2A.1) has evolutionarily lost a key Bub1 phosphorylation site for maintenance of faithful chromosome segregation. Indeed, H2A.1 promotes the occurrence of aneuploidy through antimycotic treatment, which also results in depletion of CENP-A at centromeres (Brimacombe et al., 2018). This is also in agreement with increased chromosomal instability in yeast H2A mutants at C-terminal sites of Bub1 phosphorylation (Kawashima et al., 2010). Karyotype evolution is an important phenomenon in the context of cancer, with consequences on disease progression and resistance to treatment. In particular, the process of polyploidisation can be induced during differentiation or in response to stressors, including chemotherapeutic drugs, ionizing radiation, hypoxia, or reactive oxygen species (Was et al., 2022). Upregulation of both canonical (H1, H2A, H2B, and H3) and variant histones (H1.0, H2A.X and H3.3B) was identified in a mouse erythroleukaemia cell line with a tetraploid phenotype acquired through resistance to differentiation (Fernández-Calleja et al., 2017). Changes in histone composition have also been reported in murine trophoblast giant cells (TGC), which are placental cells with large polyploid nuclei generated by endoreplication. Compared to the undifferentiated (non-polyploid) counterparts, TGCs exhibited a reduction in expression of canonical histones but maintenance of high levels of histone variants including H2A.X, H2A.Z, and H3.3 (Hayakawa et al., 2018). Similarly, several canonical histones (H2AC8, H2AC16, H2BC9, H2BC13, H3C7, H4C6) were found to be upregulated in polyploid giant cancer cells derived from MCF7 breast cancer lines (Buehler et al., 2022), and rat hepatocytes treated with the polyploidy-inducing carcinogen thioacetamide showed a hypermethylation and downregulation of canonical H2AC1 (Mizukami et al., 2017). Similarly, the expression of histone variant H1.0 increases from untransformed to highly transformed astrocytes, with a direct correlation with aneuploidy status (Caradonna et al., 2020), further suggesting a link between environmental stresses and histone deregulation in the context of transformation.
2.3 Overexpression of histone variants causing centromeric dysfunction
The centromeric H3 variant CENP-A is overexpressed in a plethora of human malignancies [recently reviewed by (Renaud-Pageot et al., 2022)], while its under-expression has not been reported. Physiologically, CENP-A participates in centromere maintenance by structurally and functionally defining centromeric regions (Stirpe and Heun, 2023). In virtue of their pivotal role as docking stations for the kinetochore, centromeres and chromosome segregation are tightly linked, therefore an association between CENP-A with aneuploidy is not surprising. It has been demonstrated that overexpression of CENP-A is responsible for its aberrant centromeric localisation, chromosomal instability and aneuploidy (Giunta et al., 2021; Shrestha et al., 2021). CENP-A was found to be overexpressed in colorectal cancer patient samples showing a mislocalisation to non-centromeric regions (Tomonaga et al., 2003), indicating that dysregulation at the transcriptional level can lead to an aberrant centromere specification and subsequently erroneous assembly of mitotic machinery and missegregation.
Functional centromeres also rely on histone variants other than CENP-A, principally the H2A.Z variant. Both CENP-A and H2A.Z occupy specific regions of centromeres and contribute to their structure and function (Greaves et al., 2007; Boyarchuk et al., 2014). The association of H2A.Z with mitotic fidelity, and particularly with the co-ordination of kinetochore attachment, has been well documented (Krogan et al., 2004; Rangasamy et al., 2004; Greaves et al., 2007; Hou et al., 2010; Kawashima et al., 2010; Sharma et al., 2013; Boyarchuk et al., 2014; Sales-Gil et al., 2021). In Schizosaccharomyces pombe, chromosome stability is ensured via co-ordination of CENP-A deposition and requires the presence of H2A.Z-homologue pht1 (Ahmed et al., 2007). Earlier studies in S. pombe reported that deletions in pht1 resulted in reduced chromosome segregation fidelity and chromosome loss (Carr et al., 1994), which was also observed in later studies where depletion of H2A.Z caused genomic instability and chromosome segregation defects including lagging chromosomes, chromatin bridges (Rangasamy et al., 2004), and chromosome losses (Krogan et al., 2004; Hou et al., 2010). Mislocalisation or overexpression of H2A.Z has also been shown to cause defective centromeres and increased aneuploidy in yeast with defective INO80 chromatin remodeling complex (Chambers et al., 2012). Similarly, deletion of H2A.Z resulted in chromosome losses and aneuploidy, together with centromeric defects involving CENP-A (Ling and Yuen, 2019). In addition to mitotic defects, the H2A.Z-deleted fission yeast strains described by Carr et al. (Carr et al., 1994) also exhibited lower proliferative rates, abnormal colony morphology, and improved heat shock resistance, which agrees with H2A.Z’s functions beyond centromeric regulation, including transcriptional regulation that can produce more “global” phenotypic effects.
The H2A variant family comprises of several functionally distinct members involved in a variety of cellular processes (Oberdoerffer and Miller, 2023). Interestingly, expression of H2A variants shows the strongest correlations with aneuploidy second to CENP-A (Figure 3B), and H2AZ.2 and H2A.X have been identified as top 70 genes in signatures defining functional aneuploidy and chromosomal instability at pan-cancer level (Carter et al., 2006). Although the H2A.Z variant has been investigated as a single entity, recent studies have uncovered that its two paralogues H2AZ.1 and H2AZ.2 accomplish distinct functions (Lamaa et al., 2020; Sales-Gil et al., 2021). H2A.Z.1 is mainly involved in transcriptional and cell cycle regulation, whereas H2A.Z.2 is required for faithful chromosome segregation (Sales-Gil et al., 2021). These differences are also reflected in the involvement of specific variants in the pathogenesis of a number of malignancies (Vardabasso et al., 2015; Tang et al., 2020; Han et al., 2022; Zheng et al., 2022). Gastroesophageal adenocarcinoma cells lines showed a positive correlation between high expression of H2A.X and features of chromosomal instability and WGD events (Sahgal et al., 2023). Clinically, H2A.X transcriptional levels had prognostic value in patient cohorts and could predict drug sensitivity to compounds targeting DNA damage response in cell lines (Sahgal et al., 2023). Furthermore, also belonging to the H2A variant family, macroH2A.1 and macroH2A.2 are two peculiar variants characterised by the presence of a macro domain that functions as a binding site for macromolecule docking. They both function in orchestrating gene expression regulation and genome integrity by managing DNA repair and replication stress (Kim et al., 2018). In mice, loss of the splice variant macroH2A.1.2 caused an increase in micronuclei and aneuploidy mainly of the X chromosome, but also on other autosomes albeit at a lower rate, caused by inefficient repair of under-replicated DNA (Sebastian et al., 2020).
2.4 Histone expression dysregulation in ageing-related aneuploidy
Some studies also indicate a possible role of histone alterations in ageing-related aneuploidy. Ageing, in the form of chronological age as well as senescence phenotypes, is associated with chromosomal abnormalities that can promote the development of malignancy (Tanaka et al., 2018). Reduction of canonical histone levels and accumulation of histone variants has been associated with ageing [reviewed by Yi and Kim (Yi and Kim, 2020)]. In particular, histone variants belonging to the H3 and H2A families have been reported at increased levels in aged or senescent cells, namely, H3.3, H2A.Z, H2A.J, and macroH2A (Rogakou and Sekeri–Pataryas, 1999; Kreiling et al., 2011; Maze et al., 2015; Contrepois et al., 2017; Tvardovskiy et al., 2017; Stefanelli et al., 2018). Histone variant H3.3 has been identified as a candidate gene for ageing-related aneuploidy by differential gene expression between young and aged fibroblasts (Geigl et al., 2004). Moreover, high levels of histone variant H2A.J are associated with senescence and a promotion of inflammatory signatures (Contrepois et al., 2017; Isermann et al., 2020), which is a similar phenotype observed through depletion of H2A.Z.1 leading to senescence (Sales-Gil et al., 2021). In yeast, ageing produced a marked reduction in nucleosome occupancy, resulting in a widespread transcriptional activation, genomic instability, and an increased frequency of chromosomal translocations (Hu et al., 2014). Dysfunctional H1.4 histone activity is linked to a premature senescent phenotype, accompanied by high degree aneuploidy (Flex et al., 2019). Also belonging to the linker histone family, histone variant H1.8 functions in ensuring meiotic maturation in oocytes (Furuya et al., 2007) and its expression levels are downregulated in aged oocytes in comparison with young ones (Jiao et al., 2012). An increased expression of the canonical histone H2AC1 has also been documented in older oocytes (Ntostis et al., 2022), while canonical histone H2BC3 is upregulated in aneuploid in vitro fertilisation (IVF) embryos (Huang et al., 2021; Yaacobi-Artzi et al., 2022). Interestingly, aneuploid oocytes have been shown to express a lower level of the histone-like protein HILS1 (Fragouli et al., 2010). In the context of ageing, an inadequate supply of histones can result in altered epigenetic marks, with consequences on the maintenance of transcriptional and genomic control; in fact, ageing is associated with dysregulated epigenetic patterns (Wang et al., 2018), including aberrant acetylation patterns in ageing-related aneuploidy in oocytes specifically (Akiyama et al., 2006). In addition to a dysregulation in expression of H2A and H3 variants, aged fibroblast also present differential expression of genes involved in histone modifications (Geigl et al., 2004).
3 Functional consequences impacting ploidy by mutations of histone genes—PTMs and beyond
While histone genes are often overexpressed in cancer, mutations are less frequent (Shah et al., 2014; Ferrand et al., 2020; Ghiraldini et al., 2021). The most prominent example of cancer-associated histone mutations are H3 missense mutations in gliomas (Schwartzentruber et al., 2012; Sturm et al., 2012; Wu et al., 2012; Pathania et al., 2017; Nacev et al., 2019), with analogous mutations in chondroblastomas (Fang et al., 2016), sarcomas (Lu et al., 2016), and head and neck tumours (Papillon-Cavanagh et al., 2017). However, thanks to the availability of large patient databases with high-throughput sequencing data, the identification of mutations in histone variants has grown considerably (Shah et al., 2014; Bennett et al., 2019; Nacev et al., 2019). Leveraging data from the TCGA cancer cohorts, the mutation frequency of canonical and variant histone genes is generally low, with few cancer-specific exceptions of high percentages in diffuse large B cell lymphomas (DLBL) and uterine carcinoma (UCEC) (Figure 4A). Regardless of canonical or variant, the majority of mutations are missense, followed by truncating mutations (Figure 4B). Among the most frequent mutated genes are canonical H1 histones (Figure 4C). An association between mutation status (regardless of mutation type) and increased aneuploidy was found prevalently for canonical histones of the H2A, H2B and H3 families, and only a few significant associations with variants (i.e., H2AZ.1, H3.3B, and H3.5) (Figure 4C). Certain histone genes were only associated with high levels of aneuploidy (i.e., where at least 50% of chromosome arms were altered), notably the H3.3B variant (Figure 4C). For a comprehensive list of aneuploidy-associated mutations, all mutations associated with high aneuploidy scores (>10) are available on the HistoPloidyDB website.
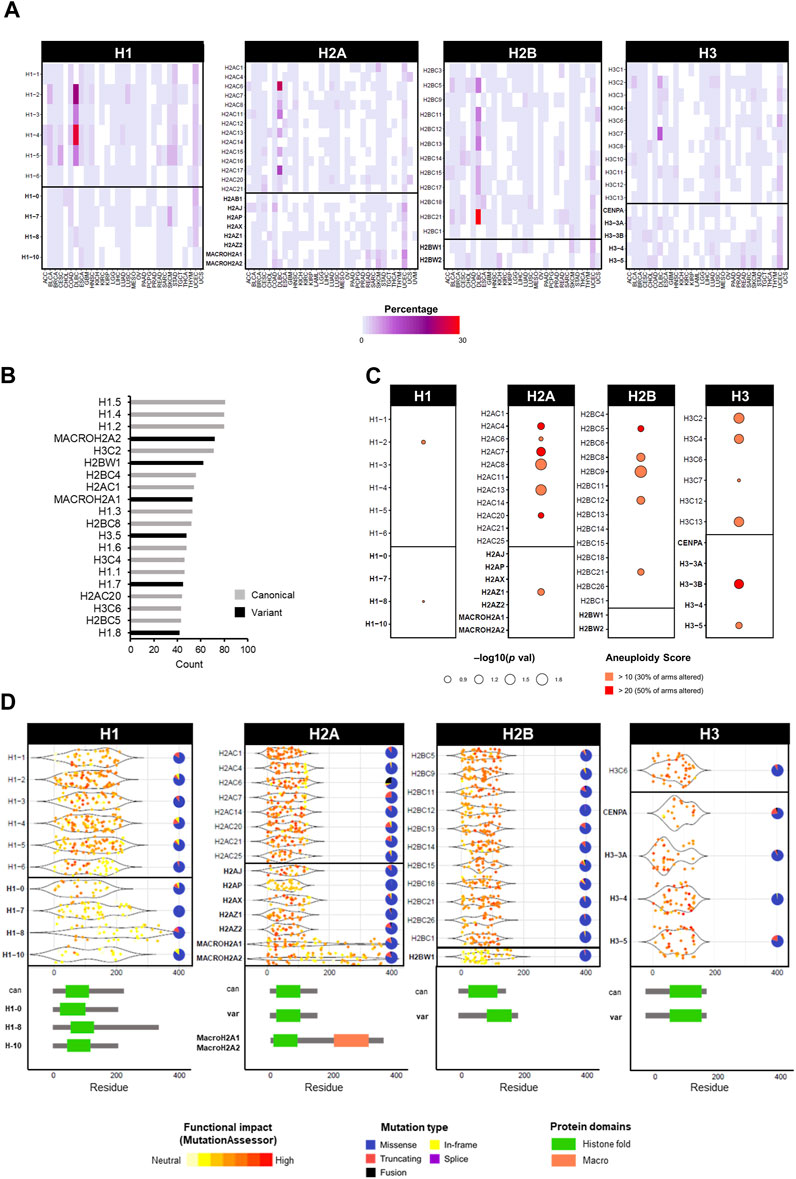
FIGURE 4. Overview of mutations in histone genes in cancers of the TCGA database. (A) Frequency of mutations as single nucleotide variations in TCGA cancers in percentage. Mutation data was obtained from cBioPortal in TCGA cancers from PanCancerAtlas (Cerami et al., 2012; Gao et al., 2013). (B) Top 20 mutated histone genes by number of cases across all cancers, distinguishable by canonical (grey) and variant (black). (C) Correlation between high aneuploidy levels and high expression levels of histone genes calculated by Chi-squared test. The gene expression was defined as “high” or “low” based on the absolute expression value relative to the mean average. (D) Frequency of mutations in histone genes by location on amino acid residue of the protein. Underneath, a schematic representation of protein structure with domains of canonical and specific variants. Each dot represents one mutation and the colour indicates the degree of functional impact predicted by MutationAssessor. Pie charts show the proportion of mutation types for each gene across all cancers.
While the initial oncohistone alterations were first described within the tail region of histones, affecting their PTMs, mutations with functional consequences have also been reported within different histone domains (Bennett et al., 2019; Nacev et al., 2019). Figure 4D shows that the location of mutations in histone genes in the TCGA cancer cohort can occur throughout the gene body, however their predicted functional impact (as predicted by MutationAssessor) varies by domain. Gene-specific differences are notable, however the most impactful mutations map within tail or central fold domains (Figure 4D). In fact, the most prominent glioma-associated H3.3 mutations produce a change from lysine to methionine (K27M and K36M) in either canonical H3 or non-canonical H3.3, causing the inability to undergo crucial methylation modifications, thus perturbing the correct epigenetic marks genome-wide (Lewis et al., 2013). Interestingly, these effects are specific to methionine and not achievable by any other amino acid, as demonstrated by a reduction of H3K27 tri-methylation only with the methionine substitution in HEK293T cells (Castel et al., 2015), highlighting the importance of individual residues for the function of histones. In fact, the substitution to methionine produces structural changes that affect the ability to interact with other epigenetic factors, notably members of the epigenetic writer polycomb repressive complex 2 (PRC2), with major epigenetic reprogramming and repercussions on gene expression control (Lewis et al., 2013; Fang et al., 2016; Harutyunyan et al., 2019). A third class of notable H3 mutations are G34R/V and G34R/W/L, which are associated with high-grade gliomas and bone tumours, respectively, and only occur in the H3.3 variant gene (Behjati et al., 2013; Koelsche et al., 2017; Chen et al., 2020). Despite not being a PTM site per se, the G34 mutations affect the methylation status of the adjacent K36 residue, but only producing a local effect, as opposed to a genome-wide methylation effect by the K27M and K36M counterparts (Lewis et al., 2013; Zhang et al., 2017; Shi et al., 2018). A comprehensive analysis of the glioma mutations of canonical H3 K27M, and H3.3 variant mutations K27M and G34R revealed distinct interactomes and dysregulation of transcription and chromatin dynamics, further strengthening the array of potential oncogenic effects by disruption of single amino acid residues of histone genes (Siddaway et al., 2022). Interestingly, these mutations also exhibit precise clinical manifestations in anatomical location, age presentation, and cell of origin (Kallappagoudar et al., 2015; Bano et al., 2017). In the case of H3.3 G34R/V mutations in gliomas, this specificity has been attributed to a developmental permissive chromatin context that allows the transformation of specific ontogeny, highlighting the potential variability in tumourigenic mechanisms by histone mutations, not only by the amino acids alone but also by cellular context (Chen et al., 2020).
The relationship between altered epigenetic landscapes and aneuploidy may be linked to the onset of abnormal transcriptional programmes affecting chromosome segregation, or aberrant chromatin specification of crucial genomic regions such as centromeres and telomeres, resulting in chromosomal instability (Herrera et al., 2008; Decombe et al., 2021). For example, H3G34R mutations display telomeric alterations potentially linked to altered methylation status of telomeric regions, where H3.3 is normally deposited (Lewis et al., 2010; Schwartzentruber et al., 2012; Sturm et al., 2012). The H3.3-H4 chaperone DAXX has also been implicated in epigenetic dysregulation and chromosomal stability in cancer (Tang et al., 2015; Mahmud and Liao, 2019). DAXX participates in the deposition of H3.3 at pericentromeric regions, and its loss has been linked to features of genomic instability (i.e., micronuclei, nuclear blebbing, and multinucleation) (Ishov et al., 2004; Morozov et al., 2017). DAXX mutations are prevalent in non-functional pancreatic neuroendocrine tumours (NF-PanNETs), where they correlate with disease severity and the presence of copy number variations (Hong et al., 2020) and are associated with altered global DNA methylation patterns (Pipinikas et al., 2015). Tumours with DAXX mutations also exhibit telomeric defects (Heaphy et al., 2011; Tang et al., 2015). Additional insights into epigenetics and aneuploidy come from observations of high aneuploidy rates in miscarriages during embryonic windows where critical epigenetic reprogramming (primarily DNA methylation) occurs (Nikitina et al., 2016; Tolmacheva et al., 2020), also supported by reports describing a deregulation of genes involved in cell cycle and histone modifications in placental tissue undergone pregnancy losses (Zhu et al., 2017). Moreover, DNA methylation patterns have been associated with aneuploidy phenotypes in cancer (Herrera et al., 2008; Ehrlich and Lacey, 2012). Similarly, it has been demonstrated that a disturbed acetylation status of histone H3 at K56 produced whole chromosome duplications and missegregation events in yeast, which was dependent on the activity of the histone chaperone ASF1 (Chan and Kolodner, 2012).
On the other hand, histone mutations within the histone fold domain, which are not primarily targeted by PTMs, may affect nucleosome stability and interactions with chromatin. In turn, these changes in chromatin accessibility can perturb gene expression regulation with oncogenic consequences (Arimura et al., 2018; Bennett et al., 2019), highlighting the importance of investigating histone mutations beyond PTM sites. Comparison of individual mutations against MutationAssessor scores and Aneuploidy Scores identified a wide range of gene alterations with an association with an aneuploid phenotype that also correlates with a functional defect, highlighting the H2A and H3 histone families with the most genes with both properties (Figure 5).

FIGURE 5. Functional and phenotypic properties of mutations in histone genes in the TCGA cohort. Dot plots represent individual mutations by gene and their associated Aneuploidy Score and predicted functional impact by MutationAssessor. Vertical intersect marks Aneuploidy Scores larger than 10 affected chromosome arms, while horizontal dashed intersect marks a medium functional impact score (>2) on MutationAssessor. Mutations falling within the top right quadrant indicate high aneuploidy and potential protein disruption, with some mutation details outlined in text. Colours indicate the histone family (H1, H2A, H2B, or H3). Data was obtained from cBioPortal in TCGA cancers from PanCancerAtlas (Cerami et al., 2012; Gao et al., 2013).
In terms of their impact on genomic stability, it was reported that diffuse gliomas without H3K27M mutations showed a lower degree of chromosomal aberrations compared to mutated patients (Dufour et al., 2020), and that both H3.1 and H3.3 mutations display highly unstable genomes by copy number alterations (Buczkowicz et al., 2014). The target of H3.3 mutations flank a crucial Ser residue that is phosphorylated in early mitosis to ensure faithful chromosome segregation (Hinchcliffe et al., 2016). In vitro, mutants of H3.3 (K27M) presented mitotic aberrations with increased rates of aneuploidy, both numerical (monosomies, tetrasomies, and WGD) and structural abnormalities (Mackay et al., 2017; Bočkaj et al., 2021). H3.3A mutations are associated with p53 alterations, chromosomal gains and losses (specifically loss of 17p), and a complex karyotype, when compared to H3.1-mutated cohorts (Castel et al., 2015). Clinical differences also exist between H3.1 (canonical) and H3.3A (variant) mutations, with the latter being associated with disease aggressiveness and dismal prognosis (Castel et al., 2015). H3.3A and H3.3B mutations are also found in giant cell tumour of the bone where they are reported to behave as initiating events that can lead to the transformation of benign tumours into malignant forms. Malignant GCTBs are characterised by higher mutational rates and aneuploidy, which arise following H3.3 mutations, suggesting a driver role in compromised genomic stability (Fittall et al., 2020). Indeed, in embryonic fibroblasts, mutations of H3.3B were found to be associated with increased aneuploidy (Bush et al., 2013).
Mutations in canonical genes of the linker histone H1 have been reported in multiple myeloma, which has been associated with impacting on chromatin organisation and genomic stability (Hoang et al., 2018; Walker et al., 2018; Maura et al., 2019). Missense mutations in the conserved histone-fold domain in H1.5, H1.3, H1.4, and H2BC12 were identified as drivers, due to the mutations occurring at key sites of nucleosome binding (Pawlyn et al., 2016; Maura et al., 2019). Interestingly, analogous mutations within the fold domain were also described in follicular lymphoma (Li et al., 2014; Krysiak et al., 2017) and chronic lymphocytic leukaemia (Landau et al., 2015). In particular, the lymphoma-associated Ser102Phe H1.2 mutation showed a reduced ability to bind chromatin in mouse embryonic stem cells, indicating an improper function in the modulation of chromatin compaction (Okosun et al., 2014). In multiple myeloma, H1.4 and H1.2 mutations were identified as clonal, which suggests their involvement in initiating and/or maintaining the oncogenic status by affecting genomic stability, also supported by a high rate of structural and numerical aneuploidies and complex cytogenetic rearrangements (Pawlyn et al., 2016; Aksenova et al., 2021).
More mechanistic insights on the effects of H1.4 mutations come from the homonymous HIST1H1E (H1.4) syndrome, characterised by frameshift mutations in the C-terminal tail of H1.4 causative of intellectual disability and multi-organ and skeletal abnormalities (Burkardt et al., 2019). Functionally, these mutations affect a critical portion of Ser/Thr residues that are phosphorylated to modulate chromatin compaction, particularly during mitosis (Hergeth and Schneider, 2015). In vitro, H1.4 mutations led to more uncompact chromatin, replicative senescence, and decreased proliferation with increased susceptibility to DNA damage. Mutated fibroblasts also displayed nucleolar instability and a higher degree of aneuploidy compared to controls, indicating drastic changes in genomic stability through chromatin modelling by dysfunctional histone H1 (Flex et al., 2019). As described earlier, depletion of H1 in Xenopus is associated with aberrant chromosome segregation (Maresca et al., 2005).
While the H2A family is frequently upregulated and correlated with aneuploidy in terms of expression, a lower degree of mutations is observed in cancers (Figure 4A). This is particularly evident for variants of H2A, possibly due to their diverse and non-redundant roles, which could be incompatible with major functional defects. The only H2A member with a significant association with aneuploidy is H2AZ.1 (Figure 4A), in agreement with the role of the H2A.Z variant in chromosomal stability (discussed above). In yeast, H2A.Z was shown to be required for sister chromatid cohesion for accurate chromosome segregation that is dependent on acetylation (Sharma et al., 2013), which could be compromised by mutations at PTMs sites. In particular, acetylation has been shown to be a crucial PTM for H2A.Z (Millar et al., 2006; Halley, 2009; Ishibashi et al., 2009). H2AZ.2-depleted (but not H2AZ.1) HeLa showed genomic instability in the form of micronuclei, chromatin bridges and lagging chromosomes; this role of H2AZ.2 in chromosomal stability was further shown to be dependent on PTMs, as mutated acetylation sites were unable to rescue the defects (Sales-Gil et al., 2021). Moreover, some evidence of H2A mutations and chromosomal instability may come from reports on H2A.X, which is a crucial player in the DNA damage response cascade and a classical marker of double stranded DNA breaks (DSB) (Kuo and Yang, 2008). It has been proposed that H2A.X acts as a dosage-dependent “protector” of tumour-associated chromosomal aberrations (Bassing et al., 2003; Celeste et al., 2003; Franco et al., 2006). In fact, the 11q23 locus where H2A.X is located is deleted in a large number of malignancies (Srivastava et al., 2009) and genetic association analyses revealed that mutations in H2A.X predispose to a susceptibility to Non-Hodgkin lymphoma that is characterised by recurrent chromosomal translocations (Novik et al., 2007). Even a single allele deletion of H2A.X increases genomic instability and promotes tumourigenesis in p53 null mice. Homozygous deletions of H2A.X greatly increased the development of lymphomas and the occurrence of structural aberrations including chromosomal translocations and amplifications (Celeste et al., 2003). Structural aneuploidies (mostly translocations) in the context of H2A.X loss are the result of improper repair of DSBs in both intentional V(D)J-related breaks and generalised DNA repairs (Bassing et al., 2003).
Mutations of the H2B family have also been described in a wide range of malignancies [reviewed by Wan and Chan (Wan and Chan, 2021)], with delirious effects on nucleosome stability and chromatin accessibility. A notable example are E76K and E76Q mutations in the canonical H2B gene, which are located at regions of contact with other histones within the nucleosome, hence affecting the formation and stability of the nucleosomal octamer (Nacev et al., 2019). In turn, destabilised nucleosomes can lead to aberrant histone exchanges and altered chromatin accessibility with consequences on transcription of pro-oncogenic programmes (Arimura et al., 2018; Bennett et al., 2019; Bagert et al., 2021). In terms of H2B mutations affecting chromosome stability, H2BC21 has been identified as a driver event in copy number alteration-based oncogenic events at pan-cancer level (Belikov et al., 2022). Among mutations that affect chromosomal stability, the G53D mutation in canonical H2B genes has been reported in different tumours (Bennett et al., 2019; Nacev et al., 2019), which occurs within the inner fold region of H2B, a site of ubiquitination that affects centromere stability and chromosome segregation in yeast (Maruyama et al., 2006). Similarly, it was shown that aberrant ubiquitination of H2B at Lys120 (via USP22 depletion) leads to mitotic defects, including micronuclei, nuclear abnormalities, and increase in chromosome gains and losses (Jeusset et al., 2021). Mutations at the acetylation site at Lys24 in H2BC3 and H2BC4 have been reported in the neoplastic Extramammary Paget’s disease (EMPD) of the skin (Takeichi et al., 2020). While patient numbers were too limited to draw statistically significant conclusions, both mutated cases showed particularly invasive disease (Takeichi et al., 2020), which is often associated with aneuploidy (Lloyd and Flanagan, 2000). H2BC11 and H2BC8 were also found to be mutated in ovarian carcinosarcoma patients with evidence of high rates of copy number variations (Zhao et al., 2016). Canonical H2B genes are also mutated at a high rate in follicular lymphoma, however the biological significance remains unclear (Krysiak et al., 2017).
4 Discussion
Histones and their variants coordinate crucial cellular processes, including chromosome segregation and the maintenance of genomic stability, which are of particular relevance in cancer. Cancer is characterised and fuelled by defective genomic health, which precedes and maintains states of large genomics rearrangements and aneuploidy. In recent years, histones have gained increasing interest in the development of malignancies. The regulation of quantity and functionality of histones in the cell is fulfilled at several levels, including transcriptional, post-transcriptional, translational, and post-translational levels. Therefore, the alterations of histones that can produce oncogenic effects can be multi-faceted. Here, we have reviewed the literature on the association between alterations in histones and aneuploidy, with the aid of publicly available TCGA cancer database. We gathered all correlations between expression, mutation, and copy number variations in histone genes with aneuploidy in the HistoPloidyDB database.
The majority of studies on histones and aneuploidy have focused on dysregulated expression and an imbalance between the incorporation of canonical and variants. Despite apparently contradictory reports on positive or negative correlation between expression levels and aneuploidy status (also evident from aneuploidy correlations in Figure 3B), it is likely that the overall effect is dependent on the net abundance of canonical and variant histones with respect to their baseline levels, as well as the availability and function of histone chaperones. Considering the differential effects on environmental cues and tissue-specificity, different tumours will display specific combinations of canonical/variant ratios, which can lead to genomic instability and aneuploidy. In summary, the dysregulated expression of histones may affect the balance between canonical and variant forms required to maintain chromosomal that may initiate or maintain tumourigenesis.
While less frequent than changes in gene expression, mutations in histones genes are also found in cancers and can be associated with aneuploidy. Interestingly, some histones only associated with high aneuploidy rates (Figure 4C), which may hint at different mechanisms producing chromosomal instability. This is of relevance functionally, as different scales of aneuploidy can have distinct effects on cellular functions (Weaver and Cleveland, 2009; Silk et al., 2013). In fact, aneuploidy is a double-edged sword in cancer, as it can be a deleterious or advantageous feature to cancer fitness and drug resistance; the exact role of aneuploidy in cancer is also still a topic of debate, with numerous contextual variables determining whether it promotes or suppresses tumourigenesis (Ben-David and Amon, 2020; Lukow and Sheltzer, 2022). Nevertheless, the heterogeneity of histone mutations, which are also not clustered within a particular protein domain (Figure 4D), does not allow for an accurate correlation with cellular phenotypes, but it can provide a platform to further characterise aneuploidy-associated mutations. As with the role of aneuploidy itself, the exact role of histones in tumourigenesis is not yet defined and may depend on the type and level of alteration to produce a specific outcome. Most histone mutations have been categorised as passenger (Bennett et al., 2019; Nacev et al., 2019), however further characterisation of functional effects of these alterations can uncover cooperating mechanisms.
Nevertheless, it appears that aberrations in histones is a yet undefined component of cancer, whether is a cause or a consequence. This is also supported by numerous reports of amplifications of the histone clusters loci in cancers, suggesting a functional advantage; for instance, the histone clusters loci are frequently subjected to copy number variation (mainly amplifications) in gastric cancer (Gorringe et al., 2005), in breast and basal cell carcinomas (Morelle et al., 2014), leukaemia (Holmfeldt et al., 2013; Simonetti et al., 2019), ovarian cancer (Zhao et al., 2016), and osteosarcoma (Sadikovic et al., 2009; Pires et al., 2022).
Overall, this work lays the ground for research into the role of histones and their variants in the disruption of chromosomal stability in cancer. For instance, comprehensive analyses of mutations in chromosome segregation genes have shed light on their contribution to genomic stability in health and disease states (Islam et al., 2022), highlighting the importance of investing functional effects of such alterations. Beside the fundamental and well-characterised contribution of the H3 variant CENP-A to the specification and maintenance of the centromere, the data collected altogether may indicate that other histone variants could also contribute to the specify and/or maintain a correct centromeric/kinetochore function. Future work will determine the exact role in oncogenesis, whether predisposing, promoting, causing, or maintaining the pathogenesis of the disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
DR: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing–original draft, Writing–review and editing. PV: Conceptualization, Funding acquisition, Resources, Supervision, Writing–review and editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a BBSRC grant BB/V013920/1 to PV.
Acknowledgments
The authors acknowledge the help from Professor Viji Draviam and Trupti Gore (Queen Mary University of London) for their help in setting HistoPloidyDB interface.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1290903/full#supplementary-material
References
Ahmed, S., Dul, B., Qiu, X., and Walworth, N. C. (2007). Msc1 acts through histone H2A. Z to promote chromosome stability in Schizosaccharomyces pombe. Genetics 177 (3), 1487–1497. doi:10.1534/genetics.107.078691
Akiyama, T., Nagata, M., and Aoki, F. (2006). Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc. Natl. Acad. Sci. 103 (19), 7339–7344. doi:10.1073/pnas.0510946103
Aksenova, A. Y., Zhuk, A. S., Lada, A. G., Zotova, I. V., Stepchenkova, E. I., Kostroma, , et al. (2021). Genome instability in multiple myeloma: facts and factors. Cancers 13 (23), 5949. doi:10.3390/cancers13235949
Arimura, Y., Ikura, M., Fujita, R., Noda, M., Kobayashi, W., Horikoshi, N., et al. (2018). Cancer-associated mutations of histones H2B, H3. 1 and H2A. Z. 1 affect the structure and stability of the nucleosome. Nucleic Acids Res. 46 (19), 10007–10018. doi:10.1093/nar/gky661
Au, W., Crisp, M. J., DeLuca, S. Z., Rando, O. J., and Basrai, M. A. (2008). Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179 (1), 263–275. doi:10.1534/genetics.108.088518
Bagert, J. D., Mitchener, M. M., Patriotis, A. L., Dul, B. E., Wojcik, F., Nacev, B. A., et al. (2021). Oncohistone mutations enhance chromatin remodeling and alter cell fates. Nat. Chem. Biol. 17 (4), 403–411. doi:10.1038/s41589-021-00738-1
Bano, D., Piazzesi, A., Salomoni, P., and Nicotera, P. (2017). The histone variant H3. 3 claims its place in the crowded scene of epigenetics. Aging (Albany NY) 9 (3), 602–614. doi:10.18632/aging.101194
Bassing, C. H., Suh, H., Ferguson, D. O., Chua, K. F., Manis, J., Eckersdorff, M., et al. (2003). Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 114 (3), 359–370. doi:10.1016/s0092-8674(03)00566-x
Behjati, S., Tarpey, P. S., Presneau, N., Scheipl, S., Pillay, N., Van Loo, P., et al. (2013). Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45 (12), 1479–1482. doi:10.1038/ng.2814
Belikov, A. V., Vyatkin, A. D., and Leonov, S. V. (2022). Novel Driver Strength Index highlights important cancer genes in TCGA PanCanAtlas patients. PeerJ 10, e13860. doi:10.7717/peerj.13860
Ben-David, U., and Amon, A. (2020). Context is everything: aneuploidy in cancer. Nat. Rev. Genet. 21 (1), 44–62. doi:10.1038/s41576-019-0171-x
Bennett, R. L., Bele, A., Small, E. C., Will, C. M., Nabet, B., Oyer, J. A., et al. (2019). A mutation in histone H2B represents a new class of oncogenic driver. Cancer Discov. 9 (10), 1438–1451. doi:10.1158/2159-8290.CD-19-0393
Bernstein, E., and Hake, S. B. (2006). The nucleosome: a little variation goes a long way. Biochem. Cell. Biol. 84 (4), 505–517. doi:10.1139/o06-085
Bočkaj, I., Martini, T. E., de Camargo Magalhães, , Eduardo, S., Bakker, P. L., Meeuwsen-de Boer, T. G., et al. (2021). The H3. 3K27M oncohistone affects replication stress outcome and provokes genomic instability in pediatric glioma. PLoS Genet. 17 (11), e1009868. doi:10.1371/journal.pgen.1009868
Bogenberger, J. M., and Laybourn, P. J. (2008). Human T lymphotropic virus type 1 protein tax reduces histone levels. Retrovirology 5 (1), 9–14. doi:10.1186/1742-4690-5-9
Boyarchuk, E., Filipescu, D., Vassias, I., Cantaloube, S., and Almouzni, G. (2014). The histone variant composition of centromeres is controlled by the pericentric heterochromatin state during the cell cycle. J. Cell. Sci. 127 (15), 3347–3359. doi:10.1242/jcs.148189
Brimacombe, C. A., Burke, J. E., Parsa, J., Witchley, J. N., Burrack, L. S., Madhani, H. D., et al. (2018). Chromatin rewiring mediates programmed evolvability via aneuploidy. bioRxiv, 407841.
Brocato, J., Chen, D., Liu, J., Fang, L., Jin, C., and Costa, M. (2015). A potential new mechanism of arsenic carcinogenesis: depletion of stem-loop binding protein and increase in polyadenylated canonical histone H3. 1 mRNA. Biol. Trace Elem. Res. 166, 72–81. doi:10.1007/s12011-015-0296-5
Bruhn, C., Bastianello, G., and Foiani, M. (2022). Cancer cell histone density links global histone acetylation, mitochondrial proteome and histone acetylase inhibitor sensitivity. Commun. Biol. 5 (1), 882. doi:10.1038/s42003-022-03846-3
Buczkowicz, P., Hoeman, C., Rakopoulos, P., Pajovic, S., Letourneau, L., Dzamba, M., et al. (2014). Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat. Genet. 46 (5), 451–456. doi:10.1038/ng.2936
Buehler, A., Krueger, R., Monavari, M., Fuentes-Chandia, M., Palmisano, R., Schoedel, J., et al. (2022). When mechanical stress matters: generation of polyploid giant cancer cells in tumor-like microcapsules. bioRxiv.
Burkardt, D. D., Zachariou, A., Loveday, C., Allen, C. L., Amor, D. J., Ardissone, A., et al. (2019). HIST1H1E heterozygous protein-truncating variants cause a recognizable syndrome with intellectual disability and distinctive facial gestalt: a study to clarify the HIST1H1E syndrome phenotype in 30 individuals. Am. J. Med. Genet. Part A 179 (10), 2049–2055. doi:10.1002/ajmg.a.61321
Buschbeck, M., and Hake, S. B. (2017). Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell. Biol. 18 (5), 299–314. doi:10.1038/nrm.2016.166
Bush, K. M., Yuen, B. T., Barrilleaux, B. L., Riggs, J. W., O’Geen, H., Cotterman, R. F., et al. (2013). Endogenous mammalian histone H3. 3 exhibits chromatin-related functions during development. Epigenetics chromatin 6, 7–16. doi:10.1186/1756-8935-6-7
Caradonna, F., Schiera, G., Di Liegro, C. M., Vitale, V., Cruciata, I., Ferrara, T., et al. (2020). Establishment and preliminary characterization of three astrocytic cells lines obtained from primary rat astrocytes by sub-cloning. Genes. 11 (12), 1502. doi:10.3390/genes11121502
Carr, A. M., Dorrington, S. M., Hindley, J., Phear, G. A., Aves, S. J., and Nurse, P. (1994). Analysis of a histone H2A variant from fission yeast: evidence for a role in chromosome stability. Mol. General Genet. MGG 245, 628–635. doi:10.1007/BF00282226
Carter, S. L., Eklund, A. C., Kohane, I. S., Harris, L. N., and Szallasi, Z. (2006). A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38 (9), 1043–1048. doi:10.1038/ng1861
Castel, D., Philippe, C., Calmon, R., Le Dret, L., Truffaux, N., Boddaert, N., et al. (2015). Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 130, 815–827. doi:10.1007/s00401-015-1478-0
Castillo, A. G., Mellone, B. G., Partridge, J. F., Richardson, W., Hamilton, G. L., Allshire, R. C., et al. (2007). Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet. 3 (7), e121. doi:10.1371/journal.pgen.0030121
Cavalieri, V. (2021). The expanding constellation of histone post-translational modifications in the epigenetic landscape. Genes. 12 (10), 1596. doi:10.3390/genes12101596
Celeste, A., Difilippantonio, S., Difilippantonio, M. J., Fernandez-Capetillo, O., Pilch, D. R., Sedelnikova, O. A., et al. (2003). H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 114 (3), 371–383. doi:10.1016/s0092-8674(03)00567-1
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2 (5), 401–404. doi:10.1158/2159-8290.CD-12-0095
Chambers, A. L., Ormerod, G., Durley, S. C., Sing, T. L., Brown, G. W., Kent, N. A., et al. (2012). The INO80 chromatin remodeling complex prevents polyploidy and maintains normal chromatin structure at centromeres. Genes. Dev. 26 (23), 2590–2603. doi:10.1101/gad.199976.112
Chan, J. E., and Kolodner, R. D. (2012). Rapid analysis of Saccharomyces cerevisiae genome rearrangements by multiplex ligation–dependent probe amplification. PLoS Genet. 8 (3), e1002539. doi:10.1371/journal.pgen.1002539
Chen, C. C., Deshmukh, S., Jessa, S., Hadjadj, D., Lisi, V., Andrade, A. F., et al. (2020b). Histone H3. 3G34-mutant interneuron progenitors co-opt PDGFRA for gliomagenesis. Cell. 183 (6), 1617–1633. doi:10.1016/j.cell.2020.11.012
Chen, D., Chen, Q. Y., Wang, Z., Zhu, Y., Kluz, T., Tan, W., et al. (2020a). Polyadenylation of histone H3. 1 mRNA promotes cell transformation by displacing H3. 3 from gene regulatory elements. Iscience 23 (9), 101518. doi:10.1016/j.isci.2020.101518
Contrepois, K., Coudereau, C., Benayoun, B. A., Schuler, N., Roux, P., Bischof, O., et al. (2017). Histone variant H2A. J accumulates in senescent cells and promotes inflammatory gene expression. Nat. Commun. 8 (1), 14995. doi:10.1038/ncomms14995
Daniloski, Z., Bisht, K. K., McStay, B., and Smith, S. (2019). Resolution of human ribosomal DNA occurs in anaphase, dependent on tankyrase 1, condensin II, and topoisomerase IIα. Genes. Dev. 33 (5-6), 276–281. doi:10.1101/gad.321836.118
Davidsson, J., Lilljebjörn, H., Andersson, A., Veerla, S., Heldrup, J., Behrendtz, M., et al. (2009). The DNA methylome of pediatric acute lymphoblastic leukemia. Hum. Mol. Genet. 18 (21), 4054–4065. doi:10.1093/hmg/ddp354
Decombe, S., Loll, F., Caccianini, L., Affannoukoué, K., Izeddin, I., Mozziconacci, J., et al. (2021). Epigenetic rewriting at centromeric DNA repeats leads to increased chromatin accessibility and chromosomal instability. Epigenetics Chromatin 14 (1), 35–17. doi:10.1186/s13072-021-00410-x
Doenecke, D., Albig, W., Bode, C., Drabent, B., Franke, K., Gavenis, K., et al. (1997). Histones: genetic diversity and tissue-specific gene expression. Histochem Cell. Biol. 107 (1), 1–10. doi:10.1007/s004180050083
Draizen, E. J., Shaytan, A. K., Mariño-Ramírez, L., Talbert, P. B., Landsman, D., and Panchenko, A. R. (2016). HistoneDB 2.0: a histone database with variants—an integrated resource to explore histones and their variants. Database 2016, baw014. doi:10.1093/database/baw014
Dufour, C., Perbet, R., Leblond, P., Vasseur, R., Stechly, L., Pierache, A., et al. (2020). Identification of prognostic markers in diffuse midline gliomas H3K27M-mutant. Brain Pathol. 30 (1), 179–190. doi:10.1111/bpa.12768
Eden, A., Gaudet, F., Waghmare, A., and Jaenisch, R. (2003). Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300 (5618), 455. doi:10.1126/science.1083557
Ehrlich, M., and Lacey, M. (2012). DNA hypomethylation and hemimethylation in cancer. Epigenetic alterations Oncog. 754, 31–56. doi:10.1007/978-1-4419-9967-2_2
El Kennani, S., Adrait, A., Permiakova, O., Hesse, A., Ialy-Radio, C., Ferro, M., et al. (2018). Systematic quantitative analysis of H2A and H2B variants by targeted proteomics. Epigenetics Chromatin 11, 2–18. doi:10.1186/s13072-017-0172-y
Fang, D., Gan, H., Lee, J., Han, J., Wang, Z., Riester, S. M., et al. (2016). The histone H3. 3K36M mutation reprograms the epigenome of chondroblastomas. Science 352 (6291), 1344–1348. doi:10.1126/science.aae0065
Fernández-Calleja, V., Hernández, P., Schvartzman, J. B., de Lacoba, M. G., and Krimer, D. B. (2017). Differential gene expression analysis by RNA-seq reveals the importance of actin cytoskeletal proteins in erythroleukemia cells. PeerJ 5, e3432. doi:10.7717/peerj.3432
Ferrand, J., Rondinelli, B., and Polo, S. E. (2020). Histone variants: guardians of genome integrity. Cells 9 (11), 2424. doi:10.3390/cells9112424
Filipescu, D., Naughtin, M., Podsypanina, K., Lejour, V., Wilson, L., Gurard-Levin, Z. A., et al. (2017). Essential role for centromeric factors following p53 loss and oncogenic transformation. Genes. Dev. 31 (5), 463–480. doi:10.1101/gad.290924.116
Fittall, M. W., Lyskjær, I., Ellery, P., Lombard, P., Ijaz, J., Strobl, A., et al. (2020). Drivers underpinning the malignant transformation of giant cell tumour of bone. J. Pathol. 252 (4), 433–440. doi:10.1002/path.5537
Flaus, A., Downs, J. A., and Owen-Hughes, T. (2021). Histone isoforms and the oncohistone code. Curr. Opin. Genet. Dev. 67, 61–66. doi:10.1016/j.gde.2020.11.003
Flex, E., Martinelli, S., Van Dijck, A., Ciolfi, A., Cecchetti, S., Coluzzi, E., et al. (2019). Aberrant function of the C-terminal tail of HIST1H1E accelerates cellular senescence and causes premature aging. Am. J. Hum. Genet. 105 (3), 493–508. doi:10.1016/j.ajhg.2019.07.007
Fragouli, E., Bianchi, V., Patrizio, P., Obradors, A., Huang, Z., Borini, A., et al. (2010). Transcriptomic profiling of human oocytes: association of meiotic aneuploidy and altered oocyte gene expression. MHR Basic Sci. reproductive Med. 16 (8), 570–582. doi:10.1093/molehr/gaq033
Franco, S., Gostissa, M., Zha, S., Lombard, D. B., Murphy, M. M., Zarrin, A. A., et al. (2006). H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 21 (2), 201–214. doi:10.1016/j.molcel.2006.01.005
Furuya, M., Tanaka, M., Teranishi, T., Matsumoto, K., Hosoi, Y., Saeki, K., et al. (2007). H1foo is indispensable for meiotic maturation of the mouse oocyte. J. Reproduction Dev. 53 (4), 895–902. doi:10.1262/jrd.19008
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6 (269), pl1. doi:10.1126/scisignal.2004088
Geigl, J. B., Langer, S., Barwisch, S., Pfleghaar, K., Lederer, G., and Speicher, M. R. (2004). Analysis of gene expression patterns and chromosomal changes associated with aging. Cancer Res. 64 (23), 8550–8557. doi:10.1158/0008-5472.CAN-04-2151
Ghiraldini, F. G., Filipescu, D., and Bernstein, E. (2021). Solid tumours hijack the histone variant network. Nat. Rev. Cancer 21 (4), 257–275. doi:10.1038/s41568-020-00330-0
Giunta, S., Hervé, S., White, R. R., Wilhelm, T., Dumont, M., Scelfo, A., et al. (2021). CENP-A chromatin prevents replication stress at centromeres to avoid structural aneuploidy. Proc. Natl. Acad. Sci. 118 (10), e2015634118. doi:10.1073/pnas.2015634118
Goldman, M. J., Craft, B., Hastie, M., Repečka, K., McDade, F., Kamath, A., et al. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38 (6), 675–678. doi:10.1038/s41587-020-0546-8
Gomes, A. P., Ilter, D., Low, V., Rosenzweig, A., Shen, Z., Schild, T., et al. (2019). Dynamic incorporation of histone H3 variants into chromatin is essential for acquisition of aggressive traits and metastatic colonization. Cancer Cell. 36 (4), 402–417. doi:10.1016/j.ccell.2019.08.006
Gorringe, K. L., Boussioutas, A., and Bowtell, D. D.Melbourne Gastric Cancer Group, Peter Mac Micro Array Facility (2005). Novel regions of chromosomal amplification at 6p21, 5p13, and 12q14 in gastric cancer identified by array comparative genomic hybridization. Genes., Chromosomes Cancer 42 (3), 247–259. doi:10.1002/gcc.20136
Greaves, I. K., Rangasamy, D., Ridgway, P., and Tremethick, D. J. (2007). H2A. Z contributes to the unique 3D structure of the centromere. Proc. Natl. Acad. Sci. 104 (2), 525–530. doi:10.1073/pnas.0607870104
Gunjan, A., and Verreault, A. (2003). A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 115 (5), 537–549. doi:10.1016/s0092-8674(03)00896-1
Halley, J. E. (2009). The role of H2A. Z and H2A. Z acetylation in gene regulation and gene expression in Saccharomyces cerevisiae. Berkeley: University of California.
Hammond, C. M., Strømme, C. B., Huang, H., Patel, D. J., and Groth, A. (2017). Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell. Biol. 18 (3), 141–158. doi:10.1038/nrm.2016.159
Han, S., Cao, C., Liu, R., Yuan, Y., Pan, L., Xu, M., et al. (2022). GAS41 mediates proliferation and GEM chemoresistance via H2A. Z. 2 and Notch1 in pancreatic cancer. Cell. Oncol. 45 (3), 429–446. doi:10.1007/s13402-022-00675-8
Harutyunyan, A. S., Krug, B., Chen, H., Papillon-Cavanagh, S., Zeinieh, M., De Jay, N., et al. (2019). H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat. Commun. 10 (1), 1262. doi:10.1038/s41467-019-09140-x
Hayakawa, K., Terada, K., Takahashi, T., Oana, H., Washizu, M., and Tanaka, S. (2018). Nucleosomes of polyploid trophoblast giant cells mostly consist of histone variants and form a loose chromatin structure. Sci. Rep. 8 (1), 5811–5815. doi:10.1038/s41598-018-23832-2
Heaphy, C. M., De Wilde, R. F., Jiao, Y., Klein, A. P., Edil, B. H., Shi, C., et al. (2011). Altered telomeres in tumors with ATRX and DAXX mutations. Science 333 (6041), 425. doi:10.1126/science.1207313
Hergeth, S. P., and Schneider, R. (2015). The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 16 (11), 1439–1453. doi:10.15252/embr.201540749
Herrera, L. A., Prada, D., Andonegui, M. A., and Dueñas-González, A. (2008). The epigenetic origin of aneuploidy. Curr. Genomics 9 (1), 43–50. doi:10.2174/138920208783884883
Hinchcliffe, E. H., Day, C. A., Karanjeet, K. B., Fadness, S., Langfald, A., Vaughan, K. T., et al. (2016). Chromosome missegregation during anaphase triggers p53 cell cycle arrest through histone H3. 3 Ser31 phosphorylation. Nat. Cell. Biol. 18 (6), 668–675. doi:10.1038/ncb3348
Hoang, P. H., Dobbins, S. E., Cornish, A. J., Chubb, D., Law, P. J., Kaiser, M., et al. (2018). Whole-genome sequencing of multiple myeloma reveals oncogenic pathways are targeted somatically through multiple mechanisms. Leukemia 32 (11), 2459–2470. doi:10.1038/s41375-018-0103-3
Holmfeldt, L., Wei, L., Diaz-Flores, E., Walsh, M., Zhang, J., Ding, L., et al. (2013). The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 45 (3), 242–252. doi:10.1038/ng.2532
Hong, X., Qiao, S., Li, F., Wang, W., Jiang, R., Wu, H., et al. (2020). Whole-genome sequencing reveals distinct genetic bases for insulinomas and non-functional pancreatic neuroendocrine tumours: leading to a new classification system. Gut 69 (5), 877–887. doi:10.1136/gutjnl-2018-317233
Horard, B., Sapey-Triomphe, L., Bonnefoy, E., and Loppin, B. (2018). ASF1 is required to load histones on the HIRA complex in preparation of paternal chromatin assembly at fertilization. Epigenetics Chromatin 11 (1), 19–16. doi:10.1186/s13072-018-0189-x
Hou, H., Wang, Y., Kallgren, S. P., Thompson, J., Yates, J. R., and Jia, S. (2010). Histone variant H2A. Z regulates centromere silencing and chromosome segregation in fission yeast. J. Biol. Chem. 285 (3), 1909–1918. doi:10.1074/jbc.M109.058487
Houseley, J., and Tollervey, D. (2011). Repeat expansion in the budding yeast ribosomal DNA can occur independently of the canonical homologous recombination machinery. Nucleic Acids Res. 39 (20), 8778–8791. doi:10.1093/nar/gkr589
Hu, Z., Chen, K., Xia, Z., Chavez, M., Pal, S., Seol, J., et al. (2014). Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes. Dev. 28 (4), 396–408. doi:10.1101/gad.233221.113
Huang, Y., Li, Z., Lin, E., He, P., and Ru, G. (2021). Oxidative damage-induced hyperactive ribosome biogenesis participates in tumorigenesis of offspring by cross-interacting with the Wnt and TGF-β1 pathways in IVF embryos. Exp. Mol. Med. 53 (11), 1792–1806. doi:10.1038/s12276-021-00700-0
Isermann, A., Mann, C., and Rübe, C. E. (2020). Histone variant H2A. J marks persistent DNA damage and triggers the secretory phenotype in radiation-induced senescence. Int. J. Mol. Sci. 21 (23), 9130. doi:10.3390/ijms21239130
Ishibashi, T., Dryhurst, D., Rose, K. L., Shabanowitz, J., Hunt, D. F., and Ausió, J. (2009). Acetylation of vertebrate H2A. Z and its effect on the structure of the nucleosome. Biochem. (N Y ) 48 (22), 5007–5017. doi:10.1021/bi900196c
Ishov, A. M., Vladimirova, O. V., and Maul, G. G. (2004). Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J. Cell. Sci. 117 (17), 3807–3820. doi:10.1242/jcs.01230
Islam, A., Manjarrez-González, J.C., Gore, T., Song, X, and Draviam, V.M. (2022). Human genetic variations reveal Chromosomal Instability aiding Variants (CIVa) in kinetochore-microtubule associated proteins. bioRxiv, 2022-01. doi:10.1101/2022.01.22.477339
Jang, C., Shibata, Y., Starmer, J., Yee, D., and Magnuson, T. (2015). Histone H3. 3 maintains genome integrity during mammalian development. Genes. Dev. 29 (13), 1377–1392. doi:10.1101/gad.264150.115
Jeusset, L. M., Guppy, B. J., Lichtensztejn, Z., McDonald, D., and McManus, K. J. (2021). Reduced USP22 expression impairs mitotic removal of h2b monoubiquitination, alters chromatin compaction and induces chromosome instability that may promote oncogenesis. Cancers 13 (5), 1043. doi:10.3390/cancers13051043
Jiao, Z., Xu, M., and Woodruff, T. K. (2012). Age-associated alteration of oocyte-specific gene expression in polar bodies: potential markers of oocyte competence. Fertil. Steril. 98 (2), 480–486. doi:10.1016/j.fertnstert.2012.04.035
Jordan, A., Zhang, X., Li, J., Laulicht-Glick, F., Sun, H., and Costa, M. (2017). Nickel and cadmium-induced SLBP depletion: a potential pathway to metal mediated cellular transformation. PloS one 12 (3), e0173624. doi:10.1371/journal.pone.0173624
Kallappagoudar, S., Yadav, R. K., Lowe, B. R., and Partridge, J. F. (2015). Histone H3 mutations—a special role for H3. 3 in tumorigenesis? Chromosoma 124, 177–189. doi:10.1007/s00412-015-0510-4
Kawashima, S. A., Yamagishi, Y., Honda, T., Ishiguro, K., and Watanabe, Y. (2010). Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327 (5962), 172–177. doi:10.1126/science.1180189
Kim, J., Oberdoerffer, P., and Khurana, S. (2018). The histone variant macroH2A1 is a splicing-modulated caretaker of genome integrity and tumor growth. Mol. Cell. Oncol. 5 (3), e1441629. doi:10.1080/23723556.2018.1441629
Knaack, S. A., Siahpirani, A. F., and Roy, S. (2014). A pan-cancer modular regulatory network analysis to identify common and cancer-specific network components. Cancer Inf. 13, 69–84. doi:10.4137/CIN.S14058
Koelsche, C., Schrimpf, D., Tharun, L., Roth, E., Sturm, D., Jones, D. T., et al. (2017). Histone 3.3 hotspot mutations in conventional osteosarcomas: a comprehensive clinical and molecular characterization of six H3F3A mutated cases. Clin. sarcoma Res. 7 (1), 9–11. doi:10.1186/s13569-017-0075-5
Kreiling, J. A., Tamamori-Adachi, M., Sexton, A. N., Jeyapalan, J. C., Munoz-Najar, U., Peterson, A. L., et al. (2011). Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell. 10 (2), 292–304. doi:10.1111/j.1474-9726.2010.00666.x
Krogan, N. J., Baetz, K., Keogh, M., Datta, N., Sawa, C., Kwok, T. C., et al. (2004). Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. 101 (37), 13513–13518. doi:10.1073/pnas.0405753101
Krysiak, K., Gomez, F., White, B. S., Matlock, M., Miller, C. A., Trani, L., et al. (2017). Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. J. Am. Soc. Hematol. 129 (4), 473–483. doi:10.1182/blood-2016-07-729954
Kuo, L. J., and Yang, L. (2008). Gamma-H2AX - a novel biomarker for DNA double-strand breaks. Vivo 22 (3), 305–309.
Lamaa, A., Humbert, J., Aguirrebengoa, M., Cheng, X., Nicolas, E., Côté, J., et al. (2020). Integrated analysis of H2A. Z isoforms function reveals a complex interplay in gene regulation. Elife 9, e53375. doi:10.7554/eLife.53375
Landau, D. A., Tausch, E., Taylor-Weiner, A. N., Stewart, C., Reiter, J. G., Bahlo, J., et al. (2015). Mutations driving CLL and their evolution in progression and relapse. Nature 526 (7574), 525–530. doi:10.1038/nature15395
Lau, C. P., Kwok, J. S., Tsui, J. C., Huang, L., Yang, K. Y., Tsui, S. K., et al. (2017). Genome-wide transcriptome profiling of the neoplastic giant cell tumor of bone stromal cells by RNA sequencing. J. Cell. Biochem. 118 (6), 1349–1360. doi:10.1002/jcb.25792
Lewis, P. W., Elsaesser, S. J., Noh, K., Stadler, S. C., and Allis, C. D. (2010). Daxx is an H3. 3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. 107 (32), 14075–14080. doi:10.1073/pnas.1008850107
Lewis, P. W., Müller, M. M., Koletsky, M. S., Cordero, F., Lin, S., Banaszynski, L. A., et al. (2013). Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340 (6134), 857–861. doi:10.1126/science.1232245
Li, H., Kaminski, M. S., Li, Y., Yildiz, M., Ouillette, P., Jones, S., et al. (2014). Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. J. Am. Soc. Hematol. 123 (10), 1487–1498. doi:10.1182/blood-2013-05-500264
Lin, C., Conti, M., and Ramalho-Santos, M. (2013). Histone variant H3. 3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 140 (17), 3624–3634. doi:10.1242/dev.095513
Ling, Y. H., and Yuen, K. W. Y. (2019). Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc. Natl. Acad. Sci. 116 (13), 6270–6279. doi:10.1073/pnas.1821384116
Liu, C., Hu, F., Xie, G., Miao, Y., Li, X., Zeng, Y., et al. (2023). GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Briefings Bioinforma. 24 (1), bbac558. doi:10.1093/bib/bbac558
Lloyd, J., and Flanagan, A. M. (2000). Mammary and extramammary Paget's disease. J. Clin. Pathol. 53 (10), 742–749. doi:10.1136/jcp.53.10.742
Lohr, J. G., Stojanov, P., Lawrence, M. S., Auclair, D., Chapuy, B., Sougnez, C., et al. (2012). Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. 109 (10), 3879–3884. doi:10.1073/pnas.1121343109
Long, M., Sun, X., Shi, W., Yanru, A., Leung, S. T., Ding, D., et al. (2019). A novel histone H4 variant H4G regulates rDNA transcription in breast cancer. Nucleic Acids Res. 47 (16), 8399–8409. doi:10.1093/nar/gkz547
Lu, C., Jain, S. U., Hoelper, D., Bechet, D., Molden, R. C., Ran, L., et al. (2016). Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352 (6287), 844–849. doi:10.1126/science.aac7272
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389 (6648), 251–260. doi:10.1038/38444
Lukow, D. A., and Sheltzer, J. M. (2022). Chromosomal instability and aneuploidy as causes of cancer drug resistance. Trends Cancer 8 (1), 43–53. doi:10.1016/j.trecan.2021.09.002
Mackay, A., Burford, A., Carvalho, D., Izquierdo, E., Fazal-Salom, J., Taylor, K. R., et al. (2017). Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 32 (4), 520–537. doi:10.1016/j.ccell.2017.08.017
Mahmud, I., and Liao, D. (2019). DAXX in cancer: phenomena, processes, mechanisms and regulation. Nucleic Acids Res. 47 (15), 7734–7752. doi:10.1093/nar/gkz634
Maresca, T. J., Freedman, B. S., and Heald, R. (2005). Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J. Cell. Biol. 169 (6), 859–869. doi:10.1083/jcb.200503031
Martire, S., and Banaszynski, L. A. (2020). The roles of histone variants in fine-tuning chromatin organization and function. Nat. Rev. Mol. Cell. Biol. 21 (9), 522–541. doi:10.1038/s41580-020-0262-8
Maruyama, T., Nakamura, T., Hayashi, T., and Yanagida, M. (2006). Histone H2B mutations in inner region affect ubiquitination, centromere function, silencing and chromosome segregation. EMBO J. 25 (11), 2420–2431. doi:10.1038/sj.emboj.7601110
Matrka, M. C., Hennigan, R. F., Kappes, F., DeLay, M. L., Lambert, P. F., Aronow, B. J., et al. (2015). DEK over-expression promotes mitotic defects and micronucleus formation. Cell. Cycle 14 (24), 3939–3953. doi:10.1080/15384101.2015.1044177
Maura, F., Bolli, N., Angelopoulos, N., Dawson, K. J., Leongamornlert, D., Martincorena, I., et al. (2019). Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat. Commun. 10 (1), 3835. doi:10.1038/s41467-019-11680-1
Maze, I., Noh, K., Soshnev, A. A., and Allis, C. D. (2014). Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 15 (4), 259–271. doi:10.1038/nrg3673
Maze, I., Wenderski, W., Noh, K., Bagot, R. C., Tzavaras, N., Purushothaman, I., et al. (2015). Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87 (1), 77–94. doi:10.1016/j.neuron.2015.06.014
McHale, C. M., Zhang, L., Lan, Q., Li, G., Hubbard, A. E., Forrest, M. S., et al. (2009). Changes in the peripheral blood transcriptome associated with occupational benzene exposure identified by cross-comparison on two microarray platforms. Genomics 93 (4), 343–349. doi:10.1016/j.ygeno.2008.12.006
Meeks-Wagner, D., and Hartwell, L. H. (1986). Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 44 (1), 43–52. doi:10.1016/0092-8674(86)90483-6
Mei, Q., Huang, J., Chen, W., Tang, J., Xu, C., Yu, Q., et al. (2017). Regulation of DNA replication-coupled histone gene expression. Oncotarget 8 (55), 95005–95022. doi:10.18632/oncotarget.21887
Mendiratta, S., Gatto, A., and Almouzni, G. (2019). Histone supply: multitiered regulation ensures chromatin dynamics throughout the cell cycle. J. Cell. Biol. 218 (1), 39–54. doi:10.1083/jcb.201807179
Miles, D. M., Desdouets, C., and Géli, V. (2019). Histone stress: an unexplored source of chromosomal instability in cancer? Curr. Genet. 65 (5), 1081–1088. doi:10.1007/s00294-019-00967-x
Miles, D. M., Peñate, X., Olmo, T. S., Jourquin, F., Centeno, M. C. M., Mendoza, M., et al. (2018). High levels of histones promote whole-genome-duplications and trigger a Swe1 WEE1-dependent phosphorylation of Cdc28 CDK1. eLife 7, e35337. doi:10.7554/eLife.35337
Millar, C. B., Xu, F., Zhang, K., and Grunstein, M. (2006). Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes. Dev. 20 (6), 711–722. doi:10.1101/gad.1395506
Mizukami, S., Yafune, A., Watanabe, Y., Nakajima, K., Jin, M., Yoshida, T., et al. (2017). Identification of epigenetically downregulated Tmem70 and Ube2e2 in rat liver after 28-day treatment with hepatocarcinogenic thioacetamide showing gene product downregulation in hepatocellular preneoplastic and neoplastic lesions produced by tumor promotion. Toxicol. Lett. 266, 13–22. doi:10.1016/j.toxlet.2016.11.022
Molina, O., Abad, M. A., Solé, F., and Menéndez, P. (2021). Aneuploidy in cancer: lessons from acute lymphoblastic leukemia. Trends Cancer 7 (1), 37–47. doi:10.1016/j.trecan.2020.08.008
Morelle, A., Cericatto, R., Krepischi, A. C. V., and Ruiz, I. R. G. (2014). Clinical and genetic characterization of basal cell carcinoma and breast cancer in a single patient. Springerplus 3 (1), 454–457. doi:10.1186/2193-1801-3-454
Morozov, V. M., Giovinazzi, S., and Ishov, A. M. (2017). CENP-B protects centromere chromatin integrity by facilitating histone deposition via the H3. 3-specific chaperone Daxx. Epigenetics chromatin 10 (1), 63–18. doi:10.1186/s13072-017-0164-y
Nacev, B. A., Feng, L., Bagert, J. D., Lemiesz, A. E., Gao, J., Soshnev, A. A., et al. (2019). The expanding landscape of ‘oncohistone’mutations in human cancers. Nature 567 (7749), 473–478. doi:10.1038/s41586-019-1038-1
Nikitina, T. V., Sazhenova, E. A., Tolmacheva, E. N., Sukhanova, N. N., Kashevarova, A. A., Skryabin, N. A., et al. (2016). Comparative cytogenetic analysis of spontaneous abortions in recurrent and sporadic pregnancy losses. Biomed. Hub. 1 (1), 1–11. doi:10.1159/000446099
Novik, K. L., Spinelli, J. J., MacArthur, A. C., Shumansky, K., Sipahimalani, P., Leach, S., et al. (2007). Genetic variation in H2AFX contributes to risk of non–hodgkin lymphoma. Cancer Epidemiol. Biomarkers Prev. 16 (6), 1098–1106. doi:10.1158/1055-9965.EPI-06-0639
Ntostis, P., Iles, D., Kokkali, G., Vaxevanoglou, T., Kanavakis, E., Pantou, A., et al. (2022). The impact of maternal age on gene expression during the GV to MII transition in euploid human oocytes. Hum. Reprod. 37 (1), 80–92. doi:10.1093/humrep/deab226
Oberdoerffer, P., and Miller, K. M. (2023). Histone H2A variants: diversifying chromatin to ensure genome integrity. Seminars Cell. & Dev. Biol. 135, 59–72. doi:10.1016/j.semcdb.2022.03.011
Okosun, J., Bödör, C., Wang, J., Araf, S., Yang, C., Pan, C., et al. (2014). Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46 (2), 176–181. doi:10.1038/ng.2856
Orr, B., Godek, K. M., and Compton, D. (2015). Aneuploidy. Curr. Biol. 25 (13), R538–R542. doi:10.1016/j.cub.2015.05.010
Ors, A., Papin, C., Favier, B., Roulland, Y., Dalkara, D., Ozturk, M., et al. (2017). Histone H3. 3 regulates mitotic progression in mouse embryonic fibroblasts. Biochem. Cell. Biol. 95 (4), 491–499. doi:10.1139/bcb-2016-0190
Ota, T., Suto, S., Katayama, H., Han, Z., Suzuki, F., Maeda, M., et al. (2002). Increased mitotic phosphorylation of histone H3 attributable to AIM-1/Aurora-B overexpression contributes to chromosome number instability. Cancer Res. 62 (18), 5168–5177.
Papillon-Cavanagh, S., Lu, C., Gayden, T., Mikael, L. G., Bechet, D., Karamboulas, C., et al. (2017). Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet. 49 (2), 180–185. doi:10.1038/ng.3757
Pathania, M., De Jay, N., Maestro, N., Harutyunyan, A. S., Nitarska, J., Pahlavan, P., et al. (2017). H3. 3K27M cooperates with Trp53 loss and PDGFRA gain in mouse embryonic neural progenitor cells to induce invasive high-grade gliomas. Cancer Cell. 32 (5), 684–700. doi:10.1016/j.ccell.2017.09.014
Paul, P. K., Rabaglia, M. E., Wang, C., Stapleton, D. S., Leng, N., Kendziorski, C., et al. (2016). Histone chaperone ASF1B promotes human β-cell proliferation via recruitment of histone H3. 3. Cell. Cycle 15 (23), 3191–3202. doi:10.1080/15384101.2016.1241914
Pawlyn, C., Kaiser, M. F., Heuck, C., Melchor, L., Wardell, C. P., Murison, A., et al. (2016). The spectrum and clinical impact of epigenetic modifier mutations in myeloma. Clin. Cancer Res. 22 (23), 5783–5794. doi:10.1158/1078-0432.CCR-15-1790
Pipinikas, C. P., Dibra, H., Karpathakis, A., Feber, A., Novelli, M., Oukrif, D., et al. (2015). Epigenetic dysregulation and poorer prognosis in DAXX-deficient pancreatic neuroendocrine tumours. Endocr. Relat. Cancer 22 (3), L13–L18. doi:10.1530/ERC-15-0108
Pires, S. F., de Barros, J. S., Costa, S. S., Scliar, M. O., Lengert, A. H., Boldrini, É., et al. (2022). DNA methylation patterns suggest the involvement of DNMT3B and TET1 in osteosarcoma development. Mol. Genet. Genomics 298, 721–733. doi:10.1007/s00438-023-02010-8
Polo, S. E., Theocharis, S. E., Klijanienko, J., Savignoni, A., Asselain, B., Vielh, P., et al. (2004). Chromatin assembly factor-1, a marker of clinical value to distinguish quiescent from proliferating cells. Cancer Res. 64 (7), 2371–2381. doi:10.1158/0008-5472.can-03-2893
Prendergast, L., Hong, E., Safina, A., Poe, D., and Gurova, K. (2020). Histone chaperone FACT is essential to overcome replication stress in mammalian cells. Oncogene 39 (28), 5124–5137. doi:10.1038/s41388-020-1346-9
Prendergast, L., and Reinberg, D. (2021). The missing linker: emerging trends for H1 variant-specific functions. Genes. Dev. 35 (1-2), 40–58. doi:10.1101/gad.344531.120
Rangasamy, D., Greaves, I., and Tremethick, D. J. (2004). RNA interference demonstrates a novel role for H2A. Z in chromosome segregation. Nat. Struct. Mol. Biol. 11 (7), 650–655. doi:10.1038/nsmb786
Ray-Gallet, D., and Almouzni, G. (2022). H3–H4 histone chaperones and cancer. Curr. Opin. Genet. Dev. 73, 101900. doi:10.1016/j.gde.2022.101900
Renaud-Pageot, C., Quivy, J., Lochhead, M., and Almouzni, G. (2022). CENP-A regulation and cancer. Front. Cell. Dev. Biol. 10, 1165. doi:10.3389/fcell.2022.907120
Rogakou, E. P., and Sekeri–Pataryas, K. E. (1999). Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp. Gerontol. 34 (6), 741–754. doi:10.1016/s0531-5565(99)00046-7
Roschke, A. V., and Kirsch, I. R. (2010). Targeting karyotypic complexity and chromosomal instability of cancer cells. Curr. Drug Targets 11, 1341–1350. doi:10.2174/1389450111007011341
Sadikovic, B., Yoshimoto, M., Chilton-MacNeill, S., Thorner, P., Squire, J. A., and Zielenska, M. (2009). Identification of interactive networks of gene expression associated with osteosarcoma oncogenesis by integrated molecular profiling. Hum. Mol. Genet. 18 (11), 1962–1975. doi:10.1093/hmg/ddp117
Sahgal, P., Patil, D. T., Sztupinszki, Z., Tisza, V., Spisak, S., Huffman, B., et al. (2023). Replicative stress in gastroesophageal adenocarcinoma is associated with chromosomal instability and sensitivity to DNA damage response inhibitors. bioRxiv, 534412.
Sales-Gil, R., Kommer, D. C., de Castro, I. J., Amin, H. A., Vinciotti, V., Sisu, C., et al. (2021). Non-redundant functions of H2A. Z. 1 and H2A. Z. 2 in chromosome segregation and cell cycle progression. EMBO Rep. 22 (11), e52061. doi:10.15252/embr.202052061
Salzler, H. R., Davidson, J. M., Montgomery, N. D., and Duronio, R. J. (2009). Loss of the histone pre-mRNA processing factor stem-loop binding protein in Drosophila causes genomic instability and impaired cellular proliferation. PLoS One 4 (12), e8168. doi:10.1371/journal.pone.0008168
Schwartzentruber, J., Korshunov, A., Liu, X., Jones, D. T., Pfaff, E., Jacob, K., et al. (2012). Driver mutations in histone H3. 3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482 (7384), 226–231. doi:10.1038/nature10833
Seal, R. L., Denny, P., Bruford, E. A., Gribkova, A. K., Landsman, D., Marzluff, W. F., et al. (2022). A standardized nomenclature for mammalian histone genes. Epigenetics Chromatin 15 (1), 34. doi:10.1186/s13072-022-00467-2
Sebastian, R., Hosogane, E. K., Sun, E. G., Tran, A. D., Reinhold, W. C., Burkett, S., et al. (2020). Epigenetic regulation of DNA repair pathway choice by MacroH2A1 splice variants ensures genome stability. Mol. Cell. 79 (5), 836–845. doi:10.1016/j.molcel.2020.06.028
Selmecki, A. M., Dulmage, K., Cowen, L. E., Anderson, J. B., and Berman, J. (2009). Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5 (10), e1000705. doi:10.1371/journal.pgen.1000705
Shah, M. A., Denton, E. L., Arrowsmith, C. H., Lupien, M., and Schapira, M. (2014). A global assessment of cancer genomic alterations in epigenetic mechanisms. Epigenetics chromatin 7 (1), 29–15. doi:10.1186/1756-8935-7-29
Sharma, U., Stefanova, D., and Holmes, S. G. (2013). Histone variant H2A. Z functions in sister chromatid cohesion in Saccharomyces cerevisiae. Mol. Cell. Biol. 33 (17), 3473–3481. doi:10.1128/MCB.00162-12
Sheltzer, J. M. (2013). A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res. 73 (21), 6401–6412. doi:10.1158/0008-5472.CAN-13-0749
Shi, L., Shi, J., Shi, X., Li, W., and Wen, H. (2018). Histone H3. 3 G34 mutations alter histone H3K36 and H3K27 methylation in cis. J. Mol. Biol. 430 (11), 1562–1565. doi:10.1016/j.jmb.2018.04.014
Shrestha, R. L., Rossi, A., Wangsa, D., Hogan, A. K., Zaldana, K. S., Suva, E., et al. (2021). CENP-A overexpression promotes aneuploidy with karyotypic heterogeneity. J. Cell. Biol. 220 (4), e202007195. doi:10.1083/jcb.202007195
Siddaway, R., Canty, L., Pajovic, S., Milos, S., Coyaud, E., Sbergio, S., et al. (2022). Oncohistone interactome profiling uncovers contrasting oncogenic mechanisms and identifies potential therapeutic targets in high grade glioma. Acta Neuropathol. 144 (5), 1027–1048. doi:10.1007/s00401-022-02489-2
Silk, A. D., Zasadil, L. M., Holland, A. J., Vitre, B., Cleveland, D. W., and Weaver, B. A. (2013). Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl. Acad. Sci. 110 (44), E4134–E4141. doi:10.1073/pnas.1317042110
Simonetti, G., Padella, A., do Valle, I. F., Fontana, M. C., Fonzi, E., Bruno, S., et al. (2019). Aneuploid acute myeloid leukemia exhibits a signature of genomic alterations in the cell cycle and protein degradation machinery. Cancer 125 (5), 712–725. doi:10.1002/cncr.31837
Singh, R. K., Liang, D., Gajjalaiahvari, U. R., Kabbaj, M. M., Paik, J., and Gunjan, A. (2010). Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell. Cycle 9 (20), 4236–4244. doi:10.4161/cc.9.20.13636
Singh, R. K., Paik, J., and Gunjan, A. (2009). Generation and management of excess histones during the cell cycle. Front. Bioscience-Landmark 14 (8), 3145–3158. doi:10.2741/3441
Srivastava, N., Gochhait, S., de Boer, P., and Bamezai, R. N. (2009). Role of H2AX in DNA damage response and human cancers. Mutat. Research/Reviews Mutat. Res. 681 (2-3), 180–188. doi:10.1016/j.mrrev.2008.08.003
Stefanelli, G., Azam, A. B., Walters, B. J., Brimble, M. A., Gettens, C. P., Bouchard-Cannon, P., et al. (2018). Learning and age-related changes in genome-wide H2A. Z binding in the mouse hippocampus. Cell. Rep. 22 (5), 1124–1131. doi:10.1016/j.celrep.2018.01.020
Stirpe, A., and Heun, P. (2023). The ins and outs of CENP-A: chromatin dynamics of the centromere-specific histone. Seminars Cell. & Dev. Biol. 135, 24–34. doi:10.1016/j.semcdb.2022.04.003
Sturm, D., Witt, H., Hovestadt, V., Khuong-Quang, D., Jones, D. T., Konermann, C., et al. (2012). Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 22 (4), 425–437. doi:10.1016/j.ccr.2012.08.024
Su, C., Gao, G., Schneider, S., Helt, C., Weiss, C., O'Reilly, M. A., et al. (2004). DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 23 (5), 1133–1143. doi:10.1038/sj.emboj.7600120
Takeichi, T., Okuno, Y., Matsumoto, T., Tsunoda, N., Suzuki, K., Tanahashi, K., et al. (2020). Frequent FOXA1-activating mutations in extramammary Paget’s disease. Cancers 12 (4), 820. doi:10.3390/cancers12040820
Tanaka, K., Goto, H., Nishimura, Y., Kasahara, K., Mizoguchi, A., and Inagaki, M. (2018). Tetraploidy in cancer and its possible link to aging. Cancer Sci. 109 (9), 2632–2640. doi:10.1111/cas.13717
Tang, M., Li, Y., Zhang, Y., Chen, Y., Huang, W., Wang, D., et al. (2015). Disease mutant analysis identifies a new function of DAXX in telomerase regulation and telomere maintenance. J. Cell. Sci. 128 (2), 331–341. doi:10.1242/jcs.159467
Tang, S., Huang, X., Wang, X., Zhou, X., Huang, H., Qin, L., et al. (2020). Vital and distinct roles of H2A. Z isoforms in hepatocellular carcinoma. OncoTargets Ther. 13, 4319–4337. doi:10.2147/OTT.S243823
Tolmacheva, E. N., Vasilyev, S. A., and Lebedev, I. N. (2020). Aneuploidy and DNA methylation as mirrored features of early human embryo development. Genes. 11 (9), 1084. doi:10.3390/genes11091084
Tomonaga, T., Matsushita, K., Yamaguchi, S., Oohashi, T., Shimada, H., Ochiai, T., et al. (2003). Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 63 (13), 3511–3516.
Tvardovskiy, A., Schwämmle, V., Kempf, S. J., Rogowska-Wrzesinska, A., and Jensen, O. N. (2017). Accumulation of histone variant H3. 3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Res. 45 (16), 9272–9289. doi:10.1093/nar/gkx696
Vardabasso, C., Gaspar-Maia, A., Hasson, D., Pünzeler, S., Valle-Garcia, D., Straub, T., et al. (2015). Histone variant H2A. Z. 2 mediates proliferation and drug sensitivity of malignant melanoma. Mol. Cell. 59 (1), 75–88. doi:10.1016/j.molcel.2015.05.009
Walker, B. A., Mavrommatis, K., Wardell, C. P., Ashby, T. C., Bauer, M., Davies, F. E., et al. (2018). Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. J. Am. Soc. Hematol. 132 (6), 587–597. doi:10.1182/blood-2018-03-840132
Wan, Y. C. E., and Chan, K. M. (2021). Histone H2B mutations in cancer. Biomedicines 9 (6), 694. doi:10.3390/biomedicines9060694
Wang, Y., Yuan, Q., and Xie, L. (2018). Histone modifications in aging: the underlying mechanisms and implications. Curr. stem Cell. Res. Ther. 13 (2), 125–135. doi:10.2174/1574888X12666170817141921
Was, H., Borkowska, A., Olszewska, A., Klemba, A., Marciniak, M., Synowiec, A., et al. (2022). Polyploidy formation in cancer cells: how a Trojan horse is born. Seminars Cancer Biol. 81, 24–36. doi:10.1016/j.semcancer.2021.03.003
Weaver, B. A., and Cleveland, D. W. (2009). The role of aneuploidy in promoting and suppressing tumors. J. Cell. Biol. 185 (6), 935–937. doi:10.1083/jcb.200905098
Wu, G., Broniscer, A., McEachron, T. A., Lu, C., Paugh, B. S., Becksfort, J., et al. (2012). Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44 (3), 251–253. doi:10.1038/ng.1102
Yaacobi-Artzi, S., Kalo, D., and Roth, Z. (2022). Association between the morphokinetics of in-vitro-derived bovine embryos and the transcriptomic profile of the derived blastocysts. Plos one 17 (10), e0276642. doi:10.1371/journal.pone.0276642
Yi, S., and Kim, K. (2020). New insights into the role of histone changes in aging. Int. J. Mol. Sci. 21 (21), 8241. doi:10.3390/ijms21218241
Yoo, Y., Park, S., Jo, E. B., Choi, M., Lee, K. W., Hong, D., et al. (2021). Overexpression of replication-dependent histone signifies a subset of dedifferentiated liposarcoma with increased aggressiveness. Cancers 13 (13), 3122. doi:10.3390/cancers13133122
Zhan, F., Hardin, J., Kordsmeier, B., Bumm, K., Zheng, M., Tian, E., et al. (2002). Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. J. Am. Soc. Hematol. 99 (5), 1745–1757. doi:10.1182/blood.v99.5.1745
Zhang, Y., Shan, C., Wang, J., Bao, K., Tong, L., and Jia, S. (2017). Molecular basis for the role of oncogenic histone mutations in modulating H3K36 methylation. Sci. Rep. 7 (1), 43906. doi:10.1038/srep43906
Zhao, S., Bellone, S., Lopez, S., Thakral, D., Schwab, C., English, D. P., et al. (2016). Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial–mesenchymal transition. Proc. Natl. Acad. Sci. 113 (43), 12238–12243. doi:10.1073/pnas.1614120113
Zhao, Z., Cai, Z., Jiang, T., Han, J., and Zhang, B. (2022). Histone chaperones and digestive cancer: a review of the literature. Cancers 14 (22), 5584. doi:10.3390/cancers14225584
Zheng, Y., Han, X., and Wang, T. (2022). Role of H2A. Z. 1 in epithelial-mesenchymal transition and radiation resistance of lung adenocarcinoma in vitro. Biochem. Biophys. Res. Commun. 611, 118–125. doi:10.1016/j.bbrc.2022.03.141
Keywords: histone, variant, aneuploidy, chromosome, cancer, database
Citation: Ragusa D and Vagnarelli P (2023) Contribution of histone variants to aneuploidy: a cancer perspective. Front. Genet. 14:1290903. doi: 10.3389/fgene.2023.1290903
Received: 08 September 2023; Accepted: 27 October 2023;
Published: 23 November 2023.
Edited by:
Oscar Molina, Josep Carreras Leukaemia Research Institute (IJC), SpainReviewed by:
Zhiming Li, Columbia University, United StatesIwona Rzeszutek, University of Rzeszow, Poland
Copyright © 2023 Ragusa and Vagnarelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Denise Ragusa, denise.ragusa@brunel.ac.uk; Paola Vagnarelli, paola.vagnarelli@brunel.ac.uk
 Denise Ragusa
Denise Ragusa Paola Vagnarelli
Paola Vagnarelli