- 1College of Animal Science and Technology, Northwest A&F University, Yangling, Shaanxi, China
- 2Bright Dairy & Food Co., Ltd., Shanghai, China
- 3Shanghai Collaborative Innovation Center of Agri-Seeds, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 4Department of Animal Science, College of Animal Sciences, Zhejiang University, Hangzhou, China
- 5National Beef Cattle Improvement Center, Northwest A&F University, Yangling, Shaanxi, China
Introduction: Chinese Holstein in South China suffer heat stress for a long period, which leads to evolutionary differences from Chinese Holstein in North China. The aim of this study was to estimate the genetic parameters of fertility traits for Chinese Holstein in South China.
Methods: A total of 167,840 Chinese Holstein heifers and cows from Guangming Animal Husbandry Co., LTD farms were used in this study. The fertility traits analyzed were calving interval (CI), days open (DO), age of first service (AFS), age of first calving (AFC), calving to first insemination (CTFS), first insemination to conception (FSTC), gestation length (GL), non-return rate to 56 days (NRR), and number of services (NS).
Results: The descriptive statistics revealed that the same trait in heifers performed better than in cows, which was consistent with the other studies. The heritabilities of fertility traits in this study ranged from close to 0 (for NS of cows) to 0.2474 (for AFC of heifers). The genetic correlation of NRR between heifers and cows was 0.9993, which indicates that the NRR for heifers and cows could be treated as one trait in this population.
Conclusion: The heritabilities of fertility traits in Chinese Holstein in south China were quite different from the heritabilities of fertility traits in North China. NRR56, NS, AFC, and CI were suggested to be included into the selection index to improve fertility performance of Chinsese Holstein of south China. The results of this study could provide genetic parameters for the animal breeding program of Chinese Holstein in the south of China.
1 Introduction
Reproduction traits are the key to postpartum lactation and herd expansion, and directly affect the productive life of dairy cows. Therefore, the reproductive traits should be considered as among the most economically important traits. Most studies showed the genetic correlation between reproductive traits and milk yield traits was negative (Pryce et al., 2004; Walsh et al., 2011). The high-intensity selection of milk yield traits leads to the gradual decline of the reproductive traits of dairy cows. Therefore, it is crucial to improve the selection of reproductive traits in dairy cows. Investigating the genetic parameters is the basic procedure for the selection of reproductive traits. The genetic parameters are population-specific, therefore, it is necessary to estimate the genetic parameters in different populations. The genetic parameters of reproductive traits in Chinese Holstein cattle have been estimated using the Beijing population (Guo et al., 2014; Liu et al., 2017), the Ningxia population (Chen et al., 2021), and so on.
Guo et al. (Guo et al., 2014) evaluated the genetic parameters of five reproductive traits of dairy cows from Sanyuan Lvhe Dairy Cattle Center, Beijing, China. The traits were age at first service (AFS), number of services (NS), days from calving to first service (CTFS), days open (DO), and calving interval (CI). The range of heritabilities for these traits was from 0.034 to 0.100. The heifer and cow records were mixed together for the estimation of genetic parameters in the study of Guo et al. (Guo et al., 2014). Nevertheless, the physiology of cows is significantly different from that of heifers, so the genetic parameters should be estimated separately. Furthermore, Liu et al. (Liu et al., 2017) re-estimated the genetic parameters of interval from calving to first insemination, interval from first to last insemination, days open, conception rate at first insemination, number of inseminations per conception, and non-return rates within 56 days after first insemination for heifers and cows separately using a larger population in 2017 (Liu et al., 2017). The study of Liu et al. (Liu et al., 2017) was also based on the population of the Sanyuan Lvhe Dairy Cattle Center in Beijing.
In southern China, the useful life of dairy cows is shorter than in northern China due to heat stress, and one of the important reasons for culling cows is the passive culling of reproductive traits (Liu et al., 2021). Previous studies have also indicated that reproductive traits in dairy cattle are strongly negatively affected by tropical climate conditions (heat stress). Multiple parameters including large follicles, corpora lutea, ovulation rate, fertilized ova, and transferable embryos were influenced at a temperature–humidity index (THI) of 72 but significantly affected after a THI of 77 (p < 0.05) (Ratchamak et al., 2021).
In our previous study, we found that due to the wide geographical area in China, there was an apparent genetic and environmental interaction between the northern and southern populations for the most economic traits. Therefore, it is necessary to estimate the genetic parameters of reproductive traits in Southern Chinese Holstein cattle populations.
2 Materials and methods
The population used in this study was collected from Guangming Animal Husbandry Co., LTD. The farms were located in South China including Shanghai City, Jiangsu province and Zhejiang province. The data were edited to remove the feasible errors in the records.
Firstly, we merged the reproductive trait records uploaded from 32 farms from 2007 to 2020, and removed duplicate records based on the cow identification number and parity, finally obtaining a sum of 141,933 reproductive records. Secondly, due to the different formats of tables for recording reproductive traits data in each farm, the types of reproductive traits recorded were also different. Therefore, the total table was split into multiple sub-tables for recording different reproductive traits. After removing duplicate records, we got 56,624 records for NS, 91,301 records for CI, 83,727 records for gestation length (GL), and 178,121 records for age of first calving (AFC, part of DHI data). Other reproductive traits can be calculated from the records in the sub-table, such as CTFS, which can be obtained by subtracting the calving time from the first insemination time in the NS file. However, due to the fact that our pedigree file matches milk production traits and does not fully match reproductive traits, we were required to remove individuals from reproductive trait records not included in the pedigree file based on existing pedigree information. At the same time, we also found some mistakes in the record file including the same cattle number corresponding to multiple cows, which was an error in the birth record of cows with abnormal birth times as the cow identification number was consistent while other information records were inconsistent. The records larger than four times the standard deviation were also removed to control the outliers in the dataset. After eliminating the above mistakes, our reproductive trait records included 41,884 records for NS, 49,634 records for CI, 74,180 records for GL, 168,170 records for AFC, 8,501 records for AFS, 41,821 records for first insemination to conception (FSTC), and 49,634 records for CTFS. The non-return rate to 56 days (NRR) was coded as 0 or 1. The animal return to estrus within 56 days after calving was 0, otherwise it was 1. The pedigree was also provided by Guangming Animal Husbandry Co., LTD. The individuals with this phenotype were traced back to as many generations as possible. Finally, the best linear unbiased prediction (BLUP) model was used in this study. DMU software (Madsen et al., 2014) was used to implement the restricted maximum likelihood (REML).
In which
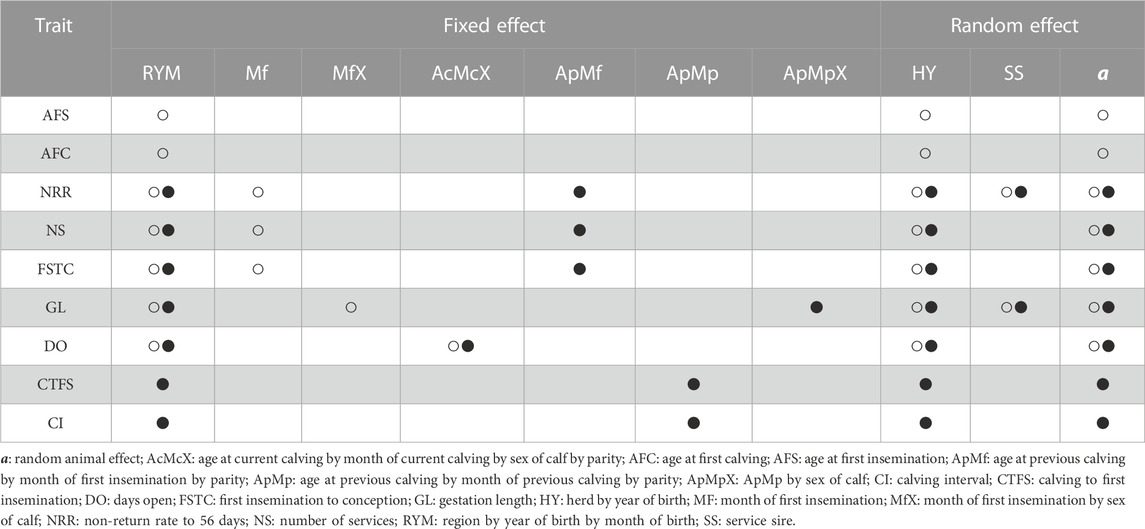
TABLE 1. Fixed and random effects included in the genetic model for genetic parameter estimation for heifer (○)and cow (●) traits.
The two-trait model was used to estimate the genetic correlations of the heifer and cow populations.
In the above formula,
3 Results
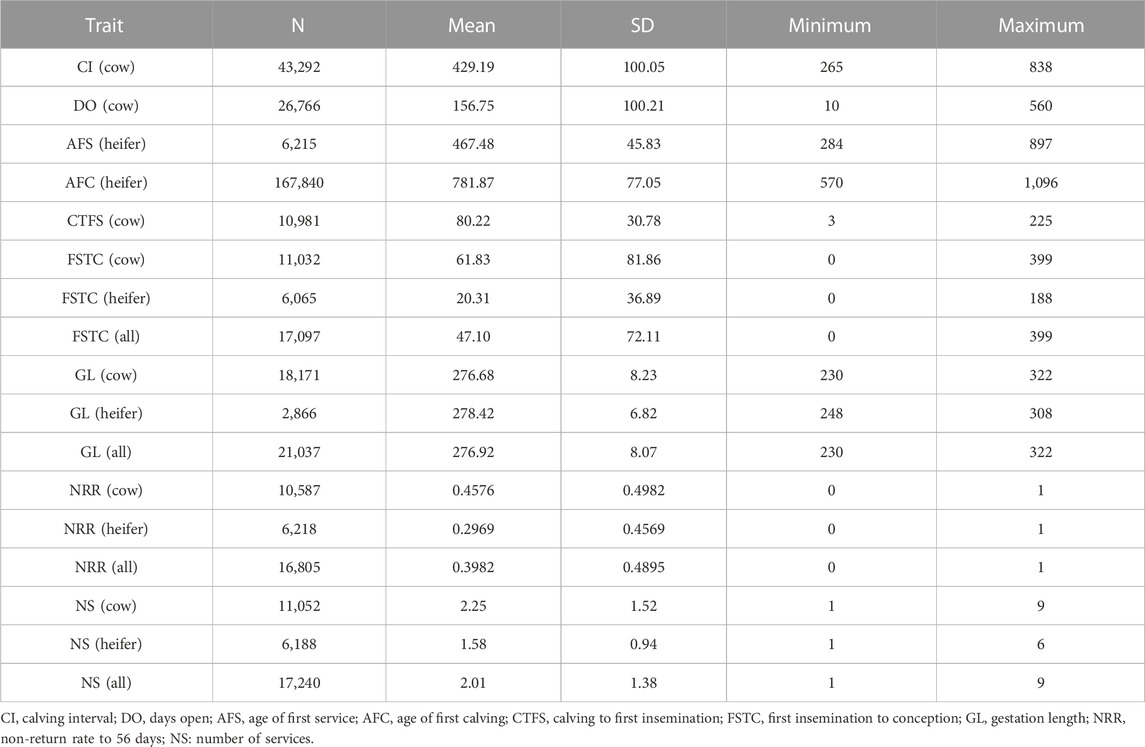
The descriptive statistics are shown in Table 2. The pedigree data contained 529,011 individuals, which indicated that most heifer traits performed better than cows. The NS in heifers was 1.58 on average while it was 2.01 for cows. The larger NS meant that more services were needed for conception. The NRR was 0.2969 on average for heifers and 0.4576 for cows, which suggested that cows were difficult to induce to conceive. The FSTC was 20.31 and 61.83 for heifer and cow respectively, which indicated that cows would need more days from first service to conception.
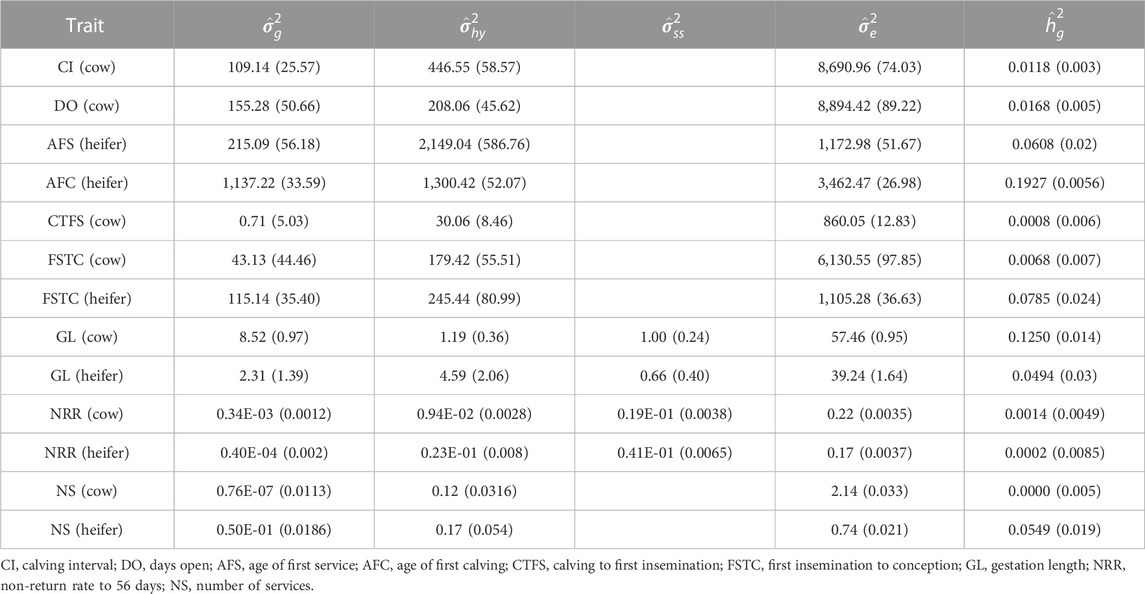
The variance components and heritability of each trait calculated using the animal model BLUP are shown in Table 3. It can be seen from the results that the heritability of each reproductive trait of Holstein cattle in southern China is relatively low. The lowest one was close to 0, which was from the NS of cows. The highest one was 0.2474, which was from the AFC of heifers. AFS and AFC were only recorded for heifers while CI and DO were only recorded for cows. The heritabilities were 0.0608 and 0.2474 for AFS and AFC, and the heritabilities for CI and DO were 0.0118 and 0.0168. Generally, it was difficult to find whether the heritability for heifers was higher or whether that for cows was higher. For example, the heritability for GL in heifers was 0.0494 which was lower than for cows (0.1250). However, the heritability of NS for heifers was 0.0549, which was higher than for cows.
A two-trait model was used to estimate the genetic correlation for each trait between heifers and cows. Unfortunately, only the two-trait model for NRR converged. The data for heritability and genetic correlation of NRR are shown in Table 4. Heritability using the two-trait model was improved for both heifers (0.0031 vs. 0.0002) and cows (0.0022 vs. 0.0014). The genetic correlation was 0.9993 while the phenotypic correlation was 0.1475.
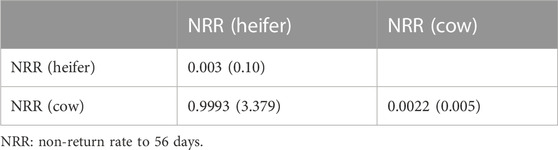
TABLE 4. The heritability (diagonal) and the genetic correlation (lower triangle) of NRR using a two-trait model.
4 Discussion
This study investigated the genetic parameters of heifers and cows in southern China. Animal models were used and genetic correlations between heifer and cow populations were estimated. The heritabilities of reproductive traits in this study were similar to the other Holstein populations. The genetic correlation of NRR between heifers and cows was 0.9993, which indicated that this trait in heifers and cows could be treated as one trait.
Compared with the results from the other studies, the performance of all the traits in the present study was in the middle (Sun et al., 2010; Guo et al., 2014; Buaban et al., 2016; Liu et al., 2017; Haile and Makasha, 2018; Muuttoranta et al., 2019; Alves et al., 2020; Sharko et al., 2022). The average of most traits in this study was superior to that of the Chinese Holstein populations in north China, Thailand, Russia, and Ethiopia which were reported from 2011 to 2022 (Buaban et al., 2016; Liu et al., 2017; Haile and Makasha, 2018; Sharko et al., 2022). The FSTC for heifers and cows was 32.1 and 79.5 in north China respectively, and the FSTC for cows from Thailand and Russia was 55.34 and 41.685 respectively. In this study, the average FSTC for heifers and cows was 20.31 and 61.83, respectively. The difference was more than 10 days compared with cows from north China. However, the FSTC of cows was longer than cows from Thailand and Russia. The DO was 156.75 in this study while it was 161.6 in the population in north China, 156.3 in Thailand, and 140.18 in Russia. The NS for heifers and cows in this study was 1.58 and 2.25, respectively, while they were 1.742 and 2.777 in the north China Holstein population. The CTFS in this study compared to the other study was 80.22, as opposed to 82.1. Both were shorter than that in the Nordic Holstein population (Muuttoranta et al., 2019), the Netherlands population (de Haer et al., 2013), and the American population (Norman et al., 2009). We found that the minimum CTFS was 3 and the maximum was 225 from the descriptive statistics, which showed unreasonable records. Therefore, it is hard to compare phenotypic levels in different countries since the data selection and editing were diverse in these studies. Furthermore, the results did not indicate that dairy farms in China paid more attention to management compared with the other countries. The comparison between heifers and cows was consistent with other studies (Guo et al., 2014; Liu et al., 2017; Muuttoranta et al., 2019). The traits of heifers were superior to those of cows. The reasons could be that the physiological effects of cows altered after calving and the high yield resulted in a negative energy balance (Wiltbank et al., 2006).
The results of this study demonstrated that the heritability of various reproductive traits of Holstein cattle in southern China was smaller than 0.1 except for the AFC of heifers and GL of cows, which was consistent with the previous studies (Jamrozik et al., 2005; Veerkamp and Beerda, 2007; Liu et al., 2008). According to the results from a review study (Pryce and Veerkamp, 2001), the heritabilities of NRR, NS (cow), CTFS, and FSTC were smaller than the average, and/or also smaller than the results from northern China. The heritabilities of NS(cow), CTFS and FSTC were also smaller than the results from Thailand and Russia (Buaban et al., 2016; Liu et al., 2017; Sharko et al., 2022). The reasons might be due to irregular individual management and the greater influence of environmental factors. In southern China, the environmental temperature is too high for raising Holstein cattle, which usually leads to heat stress in summer for most individuals, resulting in a high incidence of individual mastitis and other diseases, and also leads to a short service life (Liu et al., 2021), which might indirectly affect the heritability of reproductive traits. We investigated four traits for both heifers and cows. The heritabilities of NS and FSTC for heifers were higher than those for cows, while the heritabilities of GL and NRR for heifers were smaller than those for cows. These results were consistent with the results from a German Holstein population (Liu et al., 2008) but conflicted with results from a northern China population (Liu et al., 2017). Moreover, in previous studies, the genetic correlation between the NRR of heifers and the NRR of cows was almost 0.65 (Liu et al., 2017), which indicates that these traits should be treated as different traits, corresponding to genotype and environment interaction theories (Mulder, 2007). In the present study, the genetic correlation was 0.9993. One of the possible reasons for this result could be that the number of individuals used here was small for heifers. The other reason could be that the different populations’ genetic parameters were not consistent due to the procedure of selection, management, the climate of the location, and other residual errors. In a word, genetic parameter re-estimation was indeed necessary for different populations.
The heritabilities of the categorical traits (NRR, NS) were smaller than the interval traits, which was mainly caused by the linear model used for the variance components estimation for these traits. Though various non-linear models, such as the threshold model and generalized linear model, have been tested in the variance component estimation for NRR, ICF, etc., the previous results illustrated that the non-linear model might improve the heritability but decrease the reliability and increase computing time (Kadarmideen et al., 2000; Andersen-Ranberg et al., 2005; Sun and Su, 2010). The difference between the linear model and non-linear model could be negligible for multiple category phenotypes such as NS or non-extreme binary traits such as NRR in this study (Meijering and Gianola, 1985).
The benefits of a two-trait model for a certain trait depend on the heritability of this trait and the correlated trait and the genetic correlation between them (Schaeffer, 1984; Falconer and Mackay, 1996; Olasege et al., 2019). The genetic correlation of NRR for cows and heifers was 0.9993 and the heritability of NRR (cow) was relatively higher than the heritability of NRR (heifer). Therefore, the predictive ability for NRR (heifer) was expected to improve using the two-trait model in the prediction of estimated breeding values. When these two traits were merged into one trait, the heritability was 0.0026
It is well-known that a temperature–humidity index can be employed to assess and evaluate heat stress levels in livestock (Fathoni et al., 2022). Therefore, reproduction traits can be fitted in a reaction norm model to measure the impact of THI on reproduction traits. However, as the objective of this study was to estimate the genetic parameters of reproduction traits themselves, the THI was not considered.
CTFS and NRR56 have been suggested to be included in the selection index of the Chinese Holstein population according to the study from Liu et al. (Liu et al., 2017). However, as the heritabilities for these two traits were quite low in this study, it was hard to get genetic improvement when selecting these two traits. These differences could be because these two populations had different genetic backgrounds, and could be also because the size of records was small in this study. Since only a few individuals were overlapped for different traits, the two-trait model was not converged in this study. Therefore, genetic correlations were not obtained between traits. CI included the steps of CTFS, FSTC, and GL. Compared to the heritabilities of these traits, CI had higher heritability than CTFS and FSTC, which indicated that this complex trait in this population was more accurately recorded and had a high heritability. Considering the whether the heritability is high and whether the trait could be optional in a certain period, we suggest including NRR56, NS, AFC, and CI in the selection index in the current stage. Further study is necessary to collect more data to get genetic correlations and to update the selection index.
Data availability statement
The datasets presented in this article are not readily available because the data sets used in this study are the property of Shanghai BrightDairy & Food Co., Ltd. The data are available upon request with permission of the concerned institution through the corresponding author. Requests to access the datasets should be directed to PM, cGVpcGVpLm1hQHNqdHUuZWR1LmNu.
Author contributions
KZ: Conceptualization, Formal Analysis, Writing–original draft, Writing–review and editing. TL: Formal Analysis, Writing–original draft, Writing–review and editing. DL: Data curation, Formal Analysis, Writing–review and editing. SW: Formal Analysis, Writing–review and editing. SW: Formal Analysis, Writing–review and editing. QW: Formal Analysis, Writing–review and editing. YP: Formal Analysis, Writing–review and editing. LZ: Conceptualization, Supervision, Writing–original draft, Writing–review and editing. PM: Conceptualization, Formal Analysis, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was sponsored by Natural Science Foundation of Shanghai (22ZR1431500).
Conflict of interest
Author KZ was employed by Bright Dairy & Food Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alves, K., Brito, L. F., Baes, C. F., Sargolzaei, M., Robinson, J. A. B., and Schenkel, F. S. (2020). Estimation of additive and non-additive genetic effects for fertility and reproduction traits in North American Holstein cattle using genomic information. J. Anim. Breed. Genet. 137, 316–330. doi:10.1111/jbg.12466
Andersen-Ranberg, I. M., Heringstad, B., Gianola, D., Chang, Y. M., and Klemetsdal, G. (2005). Comparison between bivariate models for 56-day nonreturn and interval from calving to first insemination in Norwegian Red. J. Dairy Sci. 88, 2190–2198. doi:10.3168/jds.S0022-0302(05)72894-0
Buaban, S., Duangjinda, M., Suzuki, M., Masuda, Y., Sanpote, J., and Kuchida, K. (2016). Genetic relationships of fertility traits with test-day milk yield and fat-to-protein ratio in tropical smallholder dairy farms. Anim. Sci. J. 87, 627–637. doi:10.1111/asj.12472
Chen, Z., Shi, R., Luo, H., Tian, J., Wei, C., Zhang, W., et al. (2021). Estimation of genetic parameters of reproductive traits of Holstein heifers in Ningxia. Acta Veterinaria Zootechnica Sinica 52, 8. doi:10.11843/j.issn.0366-6964.2021.02.007
de Haer, L. C. M., de Jong, G., and Vessies, P. J. A. (2013). Estimate of genetic parameters of fertility traits, for virgin heifers in The Netherlands. Interbull Bull. 47, 143–146.
Falconer, D. S., and Mackay, T. F. S. (1996). Introduction to quantitative genetics. England: Pearson Education Limited.
Fathoni, A., Boonkum, W., Chankitisakul, V., and Duangjinda, M. (2022). An appropriate genetic approach for improving reproductive traits in crossbred Thai-Holstein cattle under heat stress conditions. Vet. Sci. 9, 163. doi:10.3390/vetsci9040163
Guo, G., Guo, X., Wang, Y., Zhang, X., Zhang, S., Li, X., et al. (2014). Estimation of genetic parameters of fertility traits in Chinese Holstein cattle. Can. J. Animal Sci. 94, 281–285. doi:10.4141/cjas2013-113
Haile, B., and Makasha, Y. (2018). Reproductive performance of Holstein Friesian dairy cows at Alage dairy farm, Ethiopia. J. Dairy Vet. Sci. 7, 555713. doi:10.19080/jdvs.2018.07.555713
Jamrozik, J., Fatehi, J., Kistemaker, G. J., and Schaeffer, L. R. (2005). Estimates of genetic parameters for Canadian Holstein female reproduction traits. J. Dairy Sci. 88, 2199–2208. doi:10.3168/jds.S0022-0302(05)72895-2
Kadarmideen, H., Thompson, R., and Simm, G. (2000). Linear and threshold model genetic parameters for disease, fertility and milk production in dairy cattle. Animal Sci. 71, 411–419. doi:10.1017/s1357729800055338
Liu, A., Lund, M. S., Wang, Y., Guo, G., Dong, G., Madsen, P., et al. (2017). Variance components and correlations of female fertility traits in Chinese Holstein population. J. animal Sci. Biotechnol. 8, 56. doi:10.1186/s40104-017-0189-x
Liu, D., Chen, Z., Zhao, W., Guo, L., Sun, H., Zhu, K., et al. (2021). Genome-wide selection signatures detection in Shanghai Holstein cattle population identified genes related to adaption, health and reproduction traits. BMC Genomics 22, 747. doi:10.1186/s12864-021-08042-x
Liu, Z., Jaitner, J., Reinhardt, F., Pasman, E., Rensing, S., and Reents, R. (2008). Genetic evaluation of fertility traits of dairy cattle using a multiple-trait animal model. J. Dairy Sci. 91, 4333–4343. doi:10.3168/jds.2008-1029
Madsen, P., Milkevych, V., Gao, H., Christensen, O. F., and Jensen, J. (2014). Dmu - a package for analyzing multivariate mixed models in quantitative genetics and Genomics.
Meijering, A., and Gianola, D. (1985). Linear versus nonlinear methods of sire evaluation for categorical traits: a simulation study. Genet. Sel. Evol. 17, 115–132. doi:10.1186/1297-9686-17-1-115
Mulder, H. A. (2007). Methods to optimize livestock breeding programs with genotype by environment interaction and genetic heterogeneity of environmental variance. The Netherland: Wageningen University.
Muuttoranta, K., Tyrisevä, A. M., Mäntysaari, E. A., Pösö, J., Aamand, G. P., and Lidauer, M. H. (2019). Genetic parameters for female fertility in Nordic Holstein and Red Cattle dairy breeds. J. Dairy Sci. 102, 8184–8196. doi:10.3168/jds.2018-15858
Norman, H. D., Wright, J. R., Hubbard, S. M., Miller, R. H., and Hutchison, J. L. (2009). Reproductive status of Holstein and Jersey cows in the United States. J. Dairy Sci. 92, 3517–3528. doi:10.3168/jds.2008-1768
Olasege, B. S., Zhang, S., Zhao, Q., Liu, D., Sun, H., Wang, Q., et al. (2019). Genetic parameter estimates for body conformation traits using composite index, principal component, and factor analysis. J. Dairy Sci. 102, 5219–5229. doi:10.3168/jds.2018-15561
Pryce, J. E., Royal, M. D., Garnsworthy, P. C., and Mao, I. L. (2004). Fertility in the high-producing dairy cow. Livest. Prod. Sci. 86, 125–135. doi:10.1016/s0301-6226(03)00145-3
Pryce, J. E., and Veerkamp, R. F. (2001). The incorporation of fertility indices in genetic improvement programmes. BSAP Occas. Publ. 26, 237–249. doi:10.1017/s0263967x00033711
Ratchamak, R., Ratsiri, T., Chumchai, R., Boonkum, W., and Chankitisakul, V. (2021). Relationship of the temperature-humidity index (THI) with ovarian responses and embryo production in superovulated Thai-Holstein crossbreds under tropical climate conditions. Vet. Sci. 8, 270. doi:10.3390/vetsci8110270
Schaeffer, L. R. (1984). Sire and cow evaluation under multiple trait models. J. Dairy Sci. 67, 1567–1580. doi:10.3168/jds.s0022-0302(84)81479-4
Sharko, F. S., Khatib, A., and Prokhortchouk, E. B. (2022). Genomic estimated breeding value of milk performance and fertility traits in the Russian black-and-white cattle population. Acta Naturae 14, 109–122. doi:10.32607/actanaturae.11648
Sun, C., Madsen, P., Lund, M. S., Zhang, Y., Nielsen, U. S., and Su, G. (2010). Improvement in genetic evaluation of female fertility in dairy cattle using multiple-trait models including milk production traits. J. Animal Sci. 88, 871–878. doi:10.2527/jas.2009-1912
Sun, C., and Su, G. (2010). Comparison on models for genetic evaluation of non-return rate and success in first insemination of the Danish Holstein cows. Livest. Sci. 127, 205–210. doi:10.1016/j.livsci.2009.09.015
Veerkamp, R. F., and Beerda, B. (2007). Genetics and genomics to improve fertility in high producing dairy cows. Theriogenology 68, S266–S273. doi:10.1016/j.theriogenology.2007.04.034
Walsh, S. W., Williams, E. J., and Evans, A. C. (2011). A review of the causes of poor fertility in high milk producing dairy cows. Anim. Reprod. Sci. 123, 127–138. doi:10.1016/j.anireprosci.2010.12.001
Keywords: Chinese Holstein, South China, fertility traits, genetic parameters, heritability
Citation: Zhu K, Li T, Liu D, Wang S, Wang S, Wang Q, Pan Y, Zan L and Ma P (2024) Estimation of genetic parameters for fertility traits in Chinese Holstein of south China. Front. Genet. 14:1288375. doi: 10.3389/fgene.2023.1288375
Received: 04 September 2023; Accepted: 28 November 2023;
Published: 03 January 2024.
Edited by:
Li Ma, University of Maryland, College Park, United StatesReviewed by:
Wuttigrai Boonkum, Khon Kaen University, ThailandJicai Jiang, North Carolina State University, United States
Copyright © 2024 Zhu, Li, Liu, Wang, Wang, Wang, Pan, Zan and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linsen Zan, emFubGluc2VuQDE2My5jb20=; Peipei Ma, cGVpcGVpLm1hQHNqdHUuZWR1LmNu
†These authors have contributed equally to this work
 Kai Zhu
Kai Zhu Tuowu Li
Tuowu Li Dengying Liu
Dengying Liu Shiyi Wang3
Shiyi Wang3 Sihu Wang
Sihu Wang Qishan Wang
Qishan Wang Yuchun Pan
Yuchun Pan Linsen Zan
Linsen Zan Peipei Ma
Peipei Ma