
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Genet., 06 December 2023
Sec. RNA
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1270983
This article is part of the Research TopicReviews in Non-Coding RNA: 2023View all 7 articles
One of the most frequent epigenetic modifications of RNA in eukaryotes is N6 methyladenosine (m6A), which is mostly present in messenger RNAs. Through the influence of several RNA processing stages, m6A modification is a crucial approach for controlling gene expression, especially in cancer progression. It is universally acknowledged that numerous non-coding RNAs (ncRNAs), such as microRNAs, circular RNAs, long non-coding RNAs, and piRNAs, are also significantly affected by m6A modification, and the complex genetic regulatory relationship between m6A and ncRNAs plays a pivotal role in the development of cancer. The connection between m6A modifications and ncRNAs offers an opportunity to explore the oncogene potential regulatory mechanisms and suggests that m6A modifications and ncRNAs could be vital biomarkers for multiple cancers. In this review, we discuss the mechanisms of interaction between m6A methylation and ncRNAs in cancer, and we also summarize diagnostic and prognostic biomarkers for clinical cancer detection. Furthermore, our article includes some methodologies for identifying m6A sites when assessing biomarker potential.
In messenger RNAs (mRNAs) or non-coding RNAs (ncRNAs), the sixth N atom of adenine A) gets methylated to form the m6A modification (Ping et al., 2014). With an average of around 3-5 m6A sites per mRNA, m6A modifications are the most prevalent and significant mRNA alteration in mammals, accounting for over 50% of all methyl-labeled RNAs (Wei et al., 1975), 0.1%–0.4% of all adenosine among all cellular RNAs, and an average of 3-5 m6A sites per mRNA (Rottman et al., 1974). The m6A modifications are particularly prevalent in the 3′-UTR, the stop codon, and the long internal exon in the mRNAs and frequently occur in the generally observed order RRACH (where R: A or G and H: A, C or U) (Fu et al., 2014). The splicing, export, translation, and degradation of mRNAs are all significantly impacted by these modifications (Wei and He, 2021). By regulating gene expression, m6A-modified RNAs influence a broad range of pathological and physiological processes (Frye et al., 2018). The occurrence and growth of cancer are highly correlated with abnormalities in m6A modification (He et al., 2019).
MicroRNAs (miRNAs), circular RNAs (circRNAs), and long non-coding RNAs (lncRNAs) have all been identified to have specific patterns that include the m6A modification, thanks to advancements in high-throughput genetic sequencing and mathematical techniques (Coker et al., 2019). Over ninety percent of the genome of mammalian cells is thought to be transcribed by ncRNAs, according to one research (Robinson et al., 2020). These ncRNAs operate as regulatory molecules that mediate various kinds of genetics activities, such as signal transduction, transcription, post-transcriptional changes, and chromatin remodeling. Therefore, the m6A modification of ncRNAs with modulatory capabilities is an instrumental element of epigenetic regulation. By altering the structure, biogenesis, and function of ncRNAs, m6A modifications impact diverse dimensions of pathophysiologic events (Huang et al., 2020). Dysregulation of the relationship between m6A modifying complements and ncRNAs has been linked to a variety of studies that have shown how cancer processes, including growth, aggression, and drug susceptibility, are affected (Ma et al., 2017; Li et al., 2019). This reveals a complicated web of prospective relationships connecting m6A and ncRNAs in the formation of cancer, whereby m6A and ncRNAs control target mRNAs by competing, cooperating, or both.
Despite being in its early stages, an abundant amount of dedication has gone into understanding the interaction between m6A regulators and non-coding RNAs. In this review, we summarize recent advances in the oncological field through our knowledge of the interactions between m6A and ncRNAs, concentrating on the potential regulatory mechanisms and biological changes of m6A-modified ncRNAs, together with the influence of ncRNAs on m6A regulators. Finally, we have generalized some diagnostic and prognostic biomarkers as well as methods for identifying the m6A sites, hoping that this will help in the detection and treatment of cancer.
M6A-related proteins, which are also known as “writers,” “erasers,” and “readers,” are composed of the following components: m6A methyltransferases, m6A demethylases, and m6A identification factors (Figure 1). They have the capacity to add, delete or selectively identified m6A locations on non-coding RNAs respectively, thus altering important biological functions and affecting cancer development and progression (Shi et al., 2019).
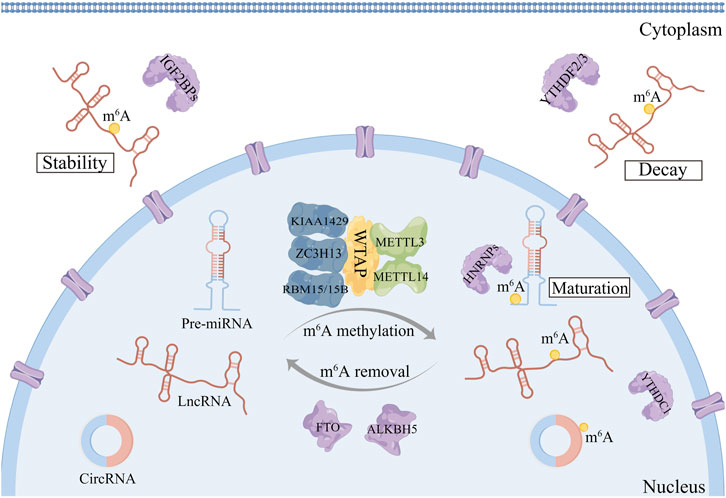
FIGURE 1. The dynamic and reversible process of m6A modifications on ncRNAs. M6A modifications are catalyzed by a writer complex consisting of METTL3-METTL14-WTAP core components as well as their cofactors KIAA1429 and RBM15/15B. The removal of m6A modifications depends on m6A erasers: FTO and ALKBH5. The function of m6A is mediated partly by m6A methylation recognition proteins, also called m6A readers, such as YTHDF1-3, YTHDC1-2, IGF2BP1-3, and HNRNPA2B1.
The methyltransferase complex (MTC), which consists of the proteins methyltransferase-like 3 (METTL3) (Chai et al., 2021), methyltransferase-like 14 (METTL14) (Wang et al., 2014), KIAA1429 (Jiang et al., 2021), Wilms tumor 1 associated protein (WTAP) (Jia et al., 2020), RNA-binding motif protein 15/15B (RBM15/15B) (Patil et al., 2016), and zinc finger CCCH structural domain-containing protein 13 (ZC3H13) (Wen et al., 2018), catalyzes m6A. As the catalytic core, the methyltransferase structural domain of METTL3 has catalytic activity (Wang et al., 2016). In addition to stabilizing MTC, METTL14 also recognizes certain RNA structures as catalytic substrates. In order to catalyze m6A methyltransferase, WTAP ligates METTL3/14 to create a complex that attaches to the nucleus (Schöller et al., 2018). In contrast, RBM15/15B recruits METTL3/14 complexes through WTAP-dependent methylation of U-rich sites (Yue et al., 2018), while KIAA1499 plays a clear role in the methylation deposition of the 3′-UTR (Knuckles et al., 2018). Meanwhile, ZC3H13 drives “writers” to the nucleus (Knuckles et al., 2018). However, it is still obscure how precisely these proteins regulate MTC development. It is also important to note that further research is still needed to determine if these methyltransferases may be employed as possible diagnostic biomarkers for cancer.
M6A methylation is a dynamic and reversible RNA alteration that may be removed by m6A demethylases, which are primarily composed of the proteins fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5) (Jia et al., 2011; Zheng et al., 2013). FTO and ALKBH5 are both members of the family of AlkB dioxygenases that remove m6A modifications in the presence of Fe(III) and 2OG, which is reliant on Fe(II) and 2-oxoglutarate (2OG) (Kurowski et al., 2003; Gerken et al., 2007). Labeled m6A annotations are removed from mRNAs and ncRNAs by the demethylation features included in FTO and ALKBH5. They encourage the demethylation response, which is crucial for the dynamic homeostasis of the m6A regulatory network, and they lower the degree of m6A modification in cellular mRNAs and ncRNAs. Notably, FTO possesses multiple substrates, including tRNA/U RNAs (Wei et al., 2018) and RNAs transcribed from repetitive elements (Wei et al., 2022). The functions of m6A writers and erasers as oncogenes and tumor suppressors in cancer have been established by several investigations. Mutations or dysregulation of m6A erasers are intrinsically linked to human cancer. Significant and valid RNA transcripts may experience aberrant m6A modifications due to instability brought on by the writer alone, the eraser alone, or both. These changes play a crucial part in the formation and advancement of tumors. However, the exact mechanisms regarding m6A erasers in cancer are less studied. Moreover, it may be worthwhile to investigate the possibility of more erasers in the future.
The m6A regulatory system features a unique RNA-binding protein termed the m6A reader, in addition to writers and erasers. The reader is composed of the YT521-B homology (YTH) structural domain family (YTHDF1/2/3) (Shi et al., 2019), YTH structural domain-containing proteins (YTHDC1/2) (Haussmann et al., 2016), the heterogeneous nuclear ribonucleoprotein (HNRNP) protein family and insulin-like growth factor-2 mRNA-binding protein 1/2/3 (IGF2BP1/2/3, also known as IMP1/2/3) (Huang et al., 2018). Readers coordinate m6A-dependent control of RNA processing and performance, whereas writers and erasers collaborate to establish the spatial organization of m6A modifications on RNA. YTHDF1 and YTHDF2 increase the synthesis and degradation of m6A methylated mRNAs in the cytoplasm, respectively. On the contrary, YTHDF3 can help YTHDF2 accelerate the degradation of m6A-modified RNAs or promote m6A-modified RNA translation together with YTHDF1 (Li et al., 2017). YTHDC1 performs a variety of tasks, mostly in the nucleus, including boosting mRNA export, accelerating the degradation of particular transcripts, and coordinating mRNA splicing via the optimal assembly for particular splicing factors (Xiao et al., 2016). On the other hand, YTHDC2 is associated with the maintenance of mRNA and synthesis (Hsu et al., 2017). The most prevalent member of the HNRNP family that identifies m6A modification sites on specific primary miRNAs and comes into contact with drosha and DGCR8 to enhance pri-miRNA processing is HNRNPA2/B1 (Alarcón et al., 2015). To improve the integrity of mRNA and synthesis efficiency, IGF2BPs detect m6A modifications as well (Huang et al., 2018). Readers serve an essential regulatory function in how genes are expressed in the whole genome and significantly influence the formation of tumors or typical physiological processes (Chen et al., 2019). Because m6A needs to be recognized by readers to perform its corresponding function, inhibiting reader production or preventing reader identification of m6A can present a novel approach for cancer therapy.
MicroRNAs (miRNAs) are a category of short endogenous non-coding RNA transcript that ranges in length from 18 to 25 nucleotides (Saliminejad et al., 2019). MiRNAs attach to the 3′-untranslated region (3′UTR) of specific mRNAs, causing the breakdown of mRNA or inhibiting mRNA translation (Guajardo et al., 2020). There is emerging evidence that miRNAs possess oncogenic or antitumor capabilities by repressing tumor suppressors and oncogenes and are engaged in a broad range of biological and pathological events in human cancer (Van Roosbroeck and Calin, 2017). A great deal of researches have additionally concentrated on miRNA modifications in cellular malignant transformation and tumor progression, in which m6A is implicated.
Three phases make up the miRNAs biological genesis. Transcription is the initial step, during which miRNAs are converted into pri-miRNAs in the nucleus. Second, pri-miRNAs are transformed into pre-miRNAs with the aid of DGCR8 and Drosha. Pre-miRNAs are eventually transported to the cytoplasm for subsequent processing. Pre-miRNAs have been cleaved by Dicer to originate mature miRNAs in the cytoplasm (Mirihana Arachchilage et al., 2015). According to research, miRNA RNA maturation requires the m6A modification (Tang et al., 2021). To aid in miRNA maturation, METTL3 collaborates with the microprocessor protein DGCR8. Wen Peng et al. found that over-expression of METTL3 encourages pri-miR-1246 to mature and boosts the amount of miR-1246 in the body. Colorectal cancer cells migrate and invade when the oncogene SPRED2 is expressed less, as a result of miR-1246 (Peng et al., 2019). In pancreatic cancer, a high level of miR-380-3p expression is maintained by m6A modifications mediated by METTL3 and METTL14, which upregulate PTEN degradation and activate the Akt pathway, promoting tumor development (Jiang et al., 2022). Similar to this, METTL3 may boost miR-143-3p splicing in lung cancer (LC) cells to create more mature miRNAs and encourage lung cancer brain metastasis (Wang et al., 2019a). Interestingly, Rucheng Yan et al. found that bee toxin-induced selective METTL3 inhibition led to a reduction of miR-146a-5p, which ultimately activated the NOTCH2 pathway and induced apoptosis in bladder cancer (BC) cells (Yan et al., 2022). Deoxycholic acid, which binds to METTL3, inhibits the METTL3-METTL14-WTAP complex, according to Ruing Lin et al. This results in a reduction in miR92b-3p by interfering with the methylation of pri-miR-92b (Lin et al., 2020). These methylation-associated tumor suppressors provide innovative viewpoints on cancer therapy and may develop into a novel approach to the management of cancers.
M6A readers also facilitate the maturation of miRNAs. According to some reports, HNRNPA2B1 functions as an m6A reader that binds to the primary miRNAs and influences how miRNAs are processed. In lung adenocarcinoma (LUAD), HNRNPA2B1 reads the m6A region on pri-miR-106b to promote the developmental stage of miR-106b-5p, which suppresses SFRP2 and stimulates Wnt-β/catenin, thereby promoting cell stemness and LUAD progression (Rong et al., 2022). The processing of miRNAs in myeloma cells is also mediated by HNRNPA2B1, according to research by Rui Liu et al. The HNRNPA2B1-DGCR8 complex attaches to either pri-miR-92a-2-5p or pri-miR-373-3p, which promotes the maturation of miRNAs and upregulates their expression. Subsequently, both microRNAs are then transported via exosomes to recipient monocytes or mesenchymal stem cells, which activate osteoclastogenesis and inhibit osteoblastogenesis by blocking IRF8 or RUNX2 thereby causing bone damage (Liu et al., 2022a). It has been shown that YTHDC1 works with different pathways to control the expression of miRNAs. By opposing MCPIP1-mediated termination of miRNA biogenesis, YTHDC1 may facilitate the degradation associated with pri-miR-30d, followed by subsequent processing of mature miRNAs in pancreatic ductal adenocarcinoma. MiR-30d regulates the expression of SLC2A1 and HK1 by specifically aiming at RUNX1, which binds to the promoters of SLC2A1 and HK1 genes and thus inhibits aerobic glycolysis (Hou et al., 2021).
Taken together, we can find that the reader selectively recognizes the m6A motifs labeled by the writer and influences the interaction with DGCR8 to increase the output of pri-miRNAs through a microprocessor mechanism. The absence of m6A readers or m6A writers slows down the processing of the pri-miRNAs. It has been known for a long time that miRNAs exert their biological functions through direct binding to the 3′UTR of mRNAs. Studies have shown that there is a large enrichment of m6A peaks at the 3′UTR of mRNAs, and it is conceivable that the proximity of m6A to miRNA binding sites affects the miRNA-mediated mechanism of transcriptional repression (Meyer et al., 2012). The specific distribution of m6A may be a way to regulate the biological function of miRNAs.
Sun et al. discovered that METTL3 boosts pri-miR-17-92 processing by modifying the A879 site via m6A modification. As an oncogenic miRNA cluster, the miR-17–92 cluster activates the AKT/mTOR pathway by inhibiting PTEN and TMEM127 to enhance GC development and metastasis. It is worth noting that this mechanism also increases the susceptibility of GC to treatment with everolimu (Sun et al., 2020a). Most of the studies were instead associated with increased cancer drug resistance.
The m6A eraser can modulate the feedback loop of molecules. Some studies have shown that m6A erasers can function both directly within molecular feedback loops and indirectly outside of molecular feedback loops. ALKBH5 and miR-193a-3p form a positive feedback loop in which miR-193a-3p downregulates ALKBH5 expression, while ALKBH5 can prevent DGCR8 from recognizing precursor miR-193a-3p and the activation of mature miR-193a-3p. As miR-193a-3p increases, its mRNA degradation of ALKBH5 is enhanced, and the inhibitory effect of ALKBH5 on miR-193a-3p is weakened, which ultimately creates a positive feedback loop resulting in the accumulation of miR-193a-3p. A positive feedback loop between miR-193a-3p and ALKBH5 promotes ESCC growth and metastasis (Xue et al., 2021). Comparatively, FTO may encourage the proliferation of glioma cells by upregulating the levels of MYC, miR-155, and miR-23 through the MYC-miR-155/23a cluster-MXI1 positive feedback loop (Xiao et al., 2020). To be specific, as a demethylase, FTO can strengthen the stability of MYC mRNA by decreasing its m6A level. Overexpression of MYC significantly raised the levels of pri-miR-155 and pri-miR-23a, leading to increased miR-155 and miR-23a production. The tumor suppressor MXI1 was supposed to inhibit MYC levels by binding to the promoter of MYC; however, a large amount of miR-155 and miR-23a binds to the 3′UTR of MXI1, causing a decrease in its expression. In this way, MYC continues to accumulate, eventually leading to the proliferation of gliomas.
A family of lengthy transcripts known as long non-coding RNAs (lncRNAs) has in excess of 200 nucleotides and either no or minimal potential to code for proteins. The human genome contains several lncRNAs that are broadly transcribed (Deniz and Erman, 2017). The majority of research have discovered the biological functions of lncRNAs, which include the functionality of proteins, RNA equilibrium, and transcription modulation (Statello et al., 2021). An increasing body of research suggests that lncRNAs are crucial in regulating various cellular functions connected to cancer, including cell growth, invasion, migration, cell death, and stemness (Wang et al., 2020a; Luo et al., 2020; Monteiro et al., 2021).
Multiple research investigations have demonstrated that m6A modifications possess a substantial impact on the long-term stability of ncRNAs, particularly lncRNAs. In head and neck cell carcinoma (HNSCC), METTL3 and METTL14-mediated m6A modifications to enhance the stability of LNCAROD in HNSCC cells (Ban et al., 2020). Not only that, in gastric cancer (GC), KIAA1429 overexpression promotes LINC00958 RNA residue levels and KIAA1429 regulates LINC00958 enrichment, which accelerates aerobic glycolysis in GC cells (Yang et al., 2021a). A substantial m6A modification site was discovered on FOXD2-AS1 by Zhipeng Ren et al. WTAP boosted the methylation modification, which improved the long-term viability of FOXD2-AS1 transcripts (Ren et al., 2022). Notably, the m6A writer not only enhances the stability of lncRNAs but also decreases it. METTL14 was found to downregulate XIST expression, thereby inhibiting CRC proliferation and metastasis (Yang et al., 2020). This suggests that the m6A writer can affect the stability of lncRNAs by altering the methylation level of lncRNAs.
M6A erasers affect the stability of lncRNAs in a variety of ways. Erasers can both improve the stability of ncRNAs, thereby upregulating their expression, and accelerate their degradation, thereby downregulating their expression. This seems to be related to the fact that lncRNAs can bind both different m6A readers and RNA-binding proteins. Research demonstrated that increased FTO in esophageal squamous cell carcinoma (ESCC) lowered the m6A methylation of LINC00022 transcripts, causing LINC000222 degradation to be repressed by YTHDF2. FTO overexpression has been found to promote LINC00022-dependent cell growth and tumor development (Cui et al., 2021). However, in hepatocellular carcinoma, ALKBH5 downregulates the lncRNA LINC02551, which is associated with the function of the demethyltransferase of ALKBH5 (Zhang et al., 2022a).
Numerous investigations have revealed that the effects of m6A methylation on RNA fate and function are mainly controlled through readers, and m6A readers may make the greatest contribution to the regulation of ncRNA stability. Senxu Lu et al. discovered that IMP2 functioned as a reader for m6A-modified ZFAS1 and enhanced the stability of lncRNA ZFAS1. Stable ZFAS1 binds to the OBG-type functional domain of OLA1, exposing the ATP binding site and increasing protein activity, eventually accelerating ATP hydrolysis and the Warburg effect in colorectal cancer (Lu et al., 2021). On the other hand, increasing m6A levels in the lncRNA TSUC7 and YTHDF2’s detection of the m6A peak in TSUCC7 led to the degradation of TSUC7, which made lung cancer more resistant to erlotinib (Li et al., 2021a). Notably, Hongmei Liu et al. discovered that YTHDF1 and YTHDF2 balance function in the transcription or degradation of lncRNA THOR by binding to different sites and thus in lncRNA THOR. Specifically, modifications of THOR sites 2 and 3 promoted LC cell proliferation, whereas modifications of THOR sites 4 and 5 inhibited lung cancer cell proliferation (Liu et al., 2020). Interestingly, YTHDF1 could adhere to m6A modification sites to stimulate lncRNA translation. It has been shown that the Y-linked lncRNA LINC00278 has the ability to encode the neuropeptide YY1BM. Additionally, LINC00278 contains a traditional m6A modification motif adjacent to the YY1BM stop codon, which interacts with YTHDF1 and facilitates the translation of YY1BM. By preventing the transcription of eEF2K, which enhances eEF2 function and causes ESCC apoptosis, YY1BM suppresses the connection between YY1 and AR (Wu et al., 2020).
M6A writers can regulate the endogenous competition mechanism of lncRNAs. According to an increasing number of research projects, lncRNAs may control the expression of miRNAs and hence affect the expression of downstream target mRNAs by acting as molecular sponges for miRNAs. This process is known as the ceRNA mechanism (Qi et al., 2020). The longevity of lncRNAs may be controlled by m6A modifications, which can also have an impact on the ceRNA machinery as previously mentioned (Li Z. et al., 2021). It has been demonstrated that LINC00958 is upregulated by METTL3-mediated N6 methyladenosine modification, which stabilizes its RNA transcripts. Furthermore, LINC00958 can uptake miR3619-5p to increase HDGF expression, which promotes adipogenesis and the progression of hepatocellular carcinoma (HCC) (Zuo et al., 2020). In addition to this, METTL3 maintains the upregulation of LNCAROD by increasing m6A methylation-mediated RNA stability, while LNCAROD increases PKM2 levels by simultaneously enhancing SRSF3-mediated PKM to PKM2 conversion and uptake of miR-145-5p to increase PKM2 levels, ultimately increasing aerobic glycolysis in HCC cells (Jia et al., 2021). In contrast, in cholangiocarcinoma, m6A-modified NKILA attenuates its inhibition of miR-582-3p (Zheng et al., 2022).
In breast cancer (BRC), WTAP binds to the m6A site of DLGAP1-AS1 and promotes its stability, while lncRNA DLGAP1-AS1 in turn increases its adriamycin resistance through the WTAP/DLGAP1-AS1/miR-299-3p feedback loop (Huang et al., 2021). Additionally, the MALAT1-miR-1914-3p-YAP axis is regulated by METTL3-initiated m6A mRNA methylation, which enhances YAP activity and directly stimulates YAP translation in non-small cell lung cancer (NSCLC) (Jin et al., 2021). This results in treatment resistance and metastasis. Previous research has also discovered that lncRNA ANRIL splicing, which is dependent on the m6A methylation of ANRIL, is how SRSF3 gene expression desensitizes tumor cells in the pancreas to the chemotherapy medication gemcitabine (Wang et al., 2022a). The influence of m6A writers on lncRNAs sheds light on the molecular processes behind cancer chemotherapy, defining a sound direction for advancing cancer therapies in the future.
The m6A erasers can also regulate cancer sensitivity to drugs. By controlling the degree of stability of lncRNAs, m6A erasers, like m6A writers, may affect how responsive cancer is to chemotherapy. Chengjie Lin et al. found that ALKBH5-mediated m6A demethylation promoted SH3BP5-AS1 degradation, which inhibited the Wnt signaling pathway and thus reduced chemoresistance to gemcitabine in pancreatic cancer (Lin et al., 2022a). In esophageal squamous cell carcinoma, ALKBH5-mediated m6A removal is linked to regulating the lncRNA CASC8, which enhances cell growth and cisplatin resistance in ESCC (Wu et al., 2022). Although current studies on the regulation of drug sensitivity by m6A erasers are still lacking, immunotherapy related to m6A erasers may offer new opportunities for cancer treatment (Yang et al., 2019).
M6A readers contribute to the creation of molecular negative feedback loops. Unlike m6A erasers that often regulate molecular positive feedback loops, m6A readers are involved in molecular negative feedback loops. Wen Ni et al. discovered that the LncRNA GAS5 binds straight to YAP, boosting its phosphorylation and ubiquitin-mediated degeneration and therefore impeding YAP-mediated production of YTHDF3. Whereas YTHDF3 attaches m6A-methylated GAS5 temporarily and preferentially to cause its decay and establish a negative feedback loop that slows CRC growth (Ni et al., 2019). Furthermore, it has been reported that a stronger relationship between the m6A reader YTHDF3 and DICER1-AS1 leads to DICER1-AS1 degeneration as a consequence of glucose exhaustion, and YTHDF3 has an essential focus of miR-5586-5p through which it can form a negative feedback loop with DICER1-AS1 to promote the process of glycolysis in pancreatic cancer (Hu et al., 2022). The specificity of the feedback loop provides new possibilities for the control of tumor malignant behavior.
Recently, several studies have revealed that m6A regulators can promote the binding of lncRNAs to certain RNA-binding proteins by adjusting their local structures, ultimately affecting tumorigenesis and progression. For example, as a lncRNA highly expressed in the nucleus, MALAT1 modified by m6A can alter its localization and activity in the nucleus and bind to different regulatory proteins through changes in RNA structure. In the clip structure of MALAT1, there are some sites that can be modified by m6A. As the level of m6A modification increases, the U-A pairing effect will diminish, and the hairpin structure will become slack. This augments the binding of the lncRNA MALAT1 to m6A readers. To be specific, methylation of sites A2577 and A2515 in MALAT1 allows the surrounding RNA sequences to bind HNRNPC and HNRNPG more readily. Lowering of METTL3/METTL14 reduces the exposure of MALAT1 to these proteins, hence suppressing the proliferation of tumors (Liu et al., 2017).
The production of circular RNAs (circRNAs), a novel class of endogenous RNA molecules that do not code with a covalent closed-loop structure, is predominantly accomplished by a process known as reverse splicing (Sanger et al., 1976). Circular RNAs (circRNAs) lack a 5′cap and a 3′poly(a) tail, therefore being chemically closed in contrast to normal linear RNAs (Grabowski et al., 1981), which makes them more resistant to nucleic acid exonucleases. CircRNAs are significant in several diseases, especially human cancer, because they are plentiful, durable, evolutionarily preserved, and have spatiotemporal-specific expression patterns (Memczak et al., 2013; Li et al., 2015). CircRNAs control many different processes, including collaborating with RNA-binding proteins, serving as sponges for microRNAs, controlling gene transcription, and modulating the translation of genes into amino acids (Chen, 2020).
In hepatocellular carcinoma, IGF2BP1 may attach to circMDK and enhances the transcriptome stability of circMDK. CircMDK uptake miR-346 and miR-874-3p to upregulated ATG16L1, which activates the PI3K/AKT/mTOR signaling pathway and promotes cell growth, migration as well as invasion (Du et al., 2022). M6A modifications in colorectal cancer are also present on circ3823, and the m6A recognition protein YTHDF3 and the demethylase ALKBH5 regulate the rate of degradation of circ3823. Degradation of circ3823 inhibits proliferation, metastasis, and angiogenesis of CRC cells (Guo et al., 2021). Additionally, by making circCCDC134 more stable via YTHDF2, which encourages cervical cancer (CC) spread, ALKBH5-mediated m6A methylation is in charge of its overexpression (Liang et al., 2022).
CircRNAs may, like lncRNAs, operate as molecular sponges for miRNAs, which may enable them to control the synthesis of miRNAs and have an influence on the expression of downstream target mRNAs. Similarly, m6A regulators can modulate this process. In gastric cancer, silencing of METTL14 decreased m6A levels of circORC5 but increased circORC5 expression. circORC5 could take up miR-30c-2-3p and reverse METTL14-induced miR-30c-2/3p upregulation and AKT1S1 and EIF4B downregulation. This suggests that METTL14-mediated m6A modification of circORC5 inhibits GC progression by regulating the miR-30c2-3p/AKT1S1 axis (Fan et al., 2022). On the contrary, in colorectal cancer, METTL3-mediated m6A modification enhances the binding of circALG1 to miR-342-5p, thereby upregulating placental growth factor (PGF) expression, which accelerates cancer cell metastasis (Lin et al., 2022b).
M6A writers can also increase cancer resistance to radiotherapy. In hypopharyngeal squamous cell cancer (HPSCC), Ping Wu et colleagues. observed that METTL3-mediated m6A modification preserved circCUX1 expression, suppressing caspase 1 production and imparting radiation resistance (Wu et al., 2021a).
Previous studies have shown that, with the help of m6A regulators, some of the circRNAs can exhibit the ability to encode proteins. This is due to the fact that some of the m6A methylation sites in circRNAs can operate as internal ribosomal entry sites (IRESs) to drive protein translation (Tang et al., 2020). Specifically, the writers METTL3 and METTL14 facilitate the process of translation by raising the m6A methylation level of circRNAs; in contrast, the eraser FTO-mediated demethylation process hinders translation. As an m6A reader, YTHDF3 plays an equally pivotal role in this process (Yang et al., 2017). Translation of circRNAs under m6A modification opens new perspectives, which may become a significant breakthrough in research.
In conclusion, m6A regulators play important roles in several cancers by influencing different biological functions in ncRNAs (Table 1).
With the advancement of ncRNAs and m6A regulators studies, more and more occurrences demonstrate that not only can m6A regulators impact ncRNAs, but ncRNAs may additionally influence the generation and activities of m6A regulators (Figure 2). Abnormal levels of ncRNAs can correspondingly lead to changes in m6A levels, which have an important role in cancer development.
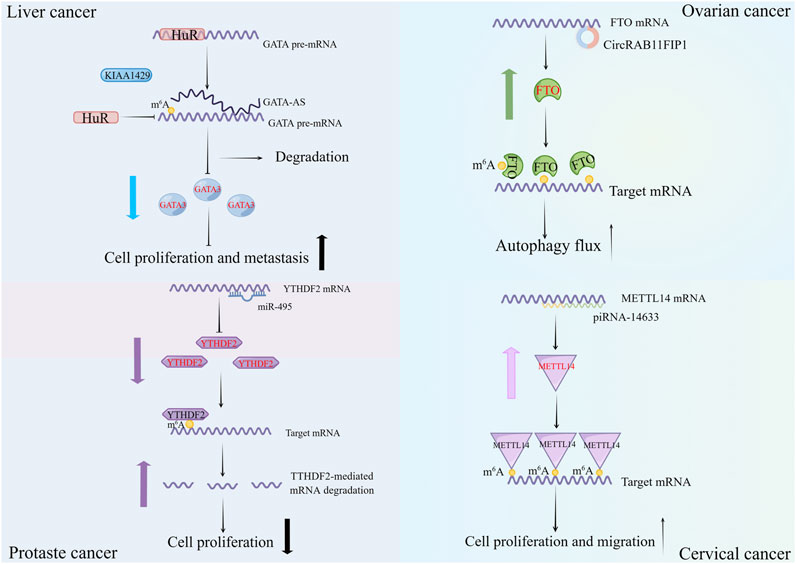
FIGURE 2. Examples of non-coding RNAs (lncRNA, circRNA, miRNA and piRNA) modulating m6A regulators. Different classes of non-coding RNAs can directly modulate the expression of m6A regulators or indirectly guide the function of m6A regulators to influence the malignant behavior of cancers.
MiRNAs downregulate the expression levels of m6A regulators. For m6A writers, it was shown that in NSCLC, miR-4443 modulates cisplatin resistance in vivo by means of METTL3/FSP1 pathway. MiR-4443 overexpression suppresses cisplatin-induced ferroptosis by negatively regulating METTL3-induced FSP1 m6A modification (Song et al., 2021). Interestingly, Liyan Wang et al. discovered that exosomes produced from M1 macrophages transported miR-628-5p to hepatocellular carcinoma cells to restrict METTL14 expression, which lowered the degree of circFUT8 m6A modification, hence reducing HCC growth (Wang et al., 2022b). For m6A readers, YTHDF2 molecule is downregulated by miR-6125, which boosts the degree of durability of GSK3 mRNA. MiR-6125 aims at the 3′-UTR of YTHDF2. In contrast, elevated GSK3 concentrations of protein restricts the production of components connected to the Wnt/-catenin/Cyclin D1 pathway, causing termination in the G0-G1 phase and ultimately suppressing CRC cell proliferation (Li et al., 2021c). Similarly, miR-495 has YTHDF2 as a target gene, which it may use to interfere with the production of YTHDF2 in prostate cancer cells. On the other hand, amplification of YTHDF2 might disrupt with miR-495 suppressive effects on growth, aggression, and movement of prostate cancer cells as well as its triggering of apoptosis (Du et al., 2020). In terms of m6A erasers, Jiazhu Sun et al. found that miR-5581-3p dramatically decreased FTO expression, which subsequently hindered BC cell lines proliferation and mobility (Sun et al., 2022). Notably, miRNAs could, in an indirect manner, upregulate the expression of m6A erasers. It has been shown that miR-96 may inhibit AMPK2, which increases the expression of FTO and, by preventing its m6A modification, upregulates the expression of MYC. The pro-proliferative and anti-apoptotic actions of miR-96 in colorectal cancer cells are also connected to this mechanism (Yue et al., 2020).
MiRNAs have an impact on the activity of m6A regulators. Besides binding to mRNAs to promote their degradation, miRNAs were found to recruit m6A erasers. In glioblastoma, miR-145 recruited FTO and induced the attachment of FTO and CLIP3 mRNA, and increased intracellular demethylase action, while the translation efficiency of m6A-deficient transcripts was significantly increased (Zepecki et al., 2021). Not only that, miRNAs can compete with each other for binding sites with m6A readers, thus inhibiting the translation of related proteins. For example, miR455-3p and METTL3 might contend for the sequence change of the HSF1 mRNA, which would prevent HSF1 protein synthesis. The HSF1 gene expression and pharmaceutical treatment both diminish the development of colorectal cancer in mice (Song et al., 2020). On the one hand, the creation of miRISC as a result of miRNA binding may obstruct interactions with METTL3. On the other hand, once mRNA is formed, the m6A modification modifies its molecular shape and construction, which has an effect on how miRNA and mRNA pair up (Roost et al., 2015). However, how exactly the binding of miRNA to mRNA interferes with the function of METTL3 still needs to be investigated.
LncRNAs have the ability to recruit m6A regulators to control the metabolism of downstream molecules. Further research has revealed that lncRNAs may recruit m6A writers and readers to control the durability of downstream mRNAs. Regarding the regulation of m6A writers, it has been shown that LNC942 straightforwardly attracts METTL14 by carrying particular recognition sequences, thereby maintaining the expression of LNC942 downstream targets in vitro and in vivo through post-transcriptional m6A methylation modifications, which exerted a robust oncogenic effect in promoting breast cancer cell growth and development of colonies along with inhibiting apoptosis potent oncogenic effects (Sun et al., 2020b). The long non-coding RNA GATA3-AS, which is translated from the GATA3 gene’s antisense strand, promotes tumor development and metastasis in hepatocellular carcinoma by interacting with KIAA1429 and GATA3 pre-mRNA and instructing KIAA1422 to preferentially induce m6A modification of GATA3 pre-mRNA (Lan et al., 2019). METTL3 is reportedly brought in by ARHGAP5-AS1 to promote m6A modification of ARHGAP5 mRNA, which stabilizes ARHGAP5 in its cytoplasm as well. Because of this, ARHGAP5 is overexpressed to foster chemoresistance, and this overexpression is associated with a poor prognosis in GC (Zhu et al., 2019). According to Xinyu Wang et al.'s research on the regulation of m6A readers, m6A residues attached to MALAT1 served as a scaffold for drawing YTHDC1 to nuclear patches, largely enhancing the ability of esophageal cancer cells to spread (Wang et al., 2021a). By attaching to the m6A reader IGF2BP1, which is crucial to in vivo stemness and tumorigenesis in BCSCs, LncRNA KB-1980E6.3 enhances the endurance of the c-Myc mRNA in breast cancer (Zhu et al., 2021). Related studies have shown that NEAT1 prevents TRIM25-mediated ubiquitination of RPRD1B and lowers RPRD1B protein decomposition by attracting hnRNPA2B1 (Jia et al., 2022). In contrast, via the c-Jun/c-Fos/SREBP1 axis, RPRD1B boosts fatty acid consumption in gastric cancer and assists the initial tumor colonization in lymphatic system. Similarly, in the GC, lncRNA CBSLR recruits YTHDF2 to establish the CBSLR/YTHDF2/CBS signaling axis, which decreases the longevity of CBS by increasing YTHDF2 adhering to the m6A-modified coding sequence (CDS) of CBS mRNA. Patients with gastric cancer with a high CBSLR/low CBS level had inferior clinical outcomes and treatment responsiveness (Yang et al., 2022a).
LncRNAs enhance the stability of m6A regulators. The effect of lncRNAs on the stability of m6A regulators is mainly achieved by increasing the stability of m6A readers. By inhibiting IGF2BP2’s ubiquitination at K139 and shielding it from the autophagic lysosomal pathway’s (ALP) destruction, LINRIS preserves IGF2BP2’s stability. In CRC cells, MYC-mediated glycolysis is mainly affected by IGF2BP2’s downstream effects, which are attenuated when LINRIS is knocked down (Wang et al., 2019b). Similarly, LCAT1 attaches to IGF2BP2 and stops it from degrading in the lysosome. The cellular production of CDC6 is then upregulated by stabilized IGF2BP2 in a m6A-dependent mechanism. Lung cancer cells may multiply, remain alive, and migrate with greater efficiency when CDC6 accumulates (Yang et al., 2022b). In renal clear cell carcinoma, DMDRMR specifically enhances IGF2BP3 activity in a way associated with m6A modification, stimulating the G1-S transition and thus promoting cell proliferation (Gu et al., 2021). Specifically, Song Zhu et al. found that lncRNAs can also interact with m6A readers through their own translated peptides. The LINC00266-1 encodes a 71-amino acid peptide. Because this peptide primarily cooperates with RNA-binding proteins, such as the m6A reader IGF2BP1, it is known as an “RNA-binding regulatory peptide” (RBRP). RBRP attaches IGF2BP1 and promotes m6A identification of c-myc by IGF2BP1, increasing the lifespan of mRNA and up-regulating c-myc expression, facilitating carcinogenesis (Zhu et al., 2020). A handful of lncRNAs have been discovered to modulate m6A writers as well. The lncRNA UCA1, for example, accelerates the course of acute myeloid leukemia by connecting to METTL14 to elevate m6A quantities and augment CXCR4 and CYP1B1 expression (Li et al., 2022a). Nevertheless, the modulation of m6A erasers associated with lncRNAs has only occasionally been documented, which could offer fresh perspectives on the course of future research and open up opportunities for the treatment of cancer.
CircRNAs have the capacity to control how m6A regulators coordinate with downstream molecules. Numerous studies have shown that circRNAs can also recruit m6A readers to affect the activity of downstream mRNAs. According to research, circRHBDD1 increases the metabolic reprogramming of HCC and reduces the effectiveness of anti-PD-1 treatment by attaching YTHDF1 to PIK3R1 mRNA and accelerating PIK3R1 translation in a way that depends on m6A (Cai et al., 2022). In addition to this, in NONO-TFE3 fused renal cell carcinoma, circMET recruits YTHDF2 by directly binding CDKN2A mRNA, which significantly impairs CDKN2A expression and leads to CDKN2A mRNA attenuation (Yang et al., 2021b). Notably, circRNAs can also regulate mRNA stability by switching m6A readers. For instance, rtcisE2F hampers the binding of E2F6/E2F3 mRNAs with YTHDF2 and enhances their connection with IGF2BP2. E2F6/E2F3 mRNA decay is prevented by IGF2BP2, but E2F6/E3F3 mRNA decay is accelerated by YTHDF2. By converting the m6A reader, rtcisE2F increases the stability of E2F6/E2F3 mRNA, which promotes liver tumorigenesis and metastasis by triggering self-regeneration of liver tumor-initiating cells (Chen et al., 2022). CircRNAs also interfere with the recognition of m6A sites by m6A readers. Fei Xie et al. found that circPTPRA partially interfered with IGF2BP1-mediated recognition of m6A-modified RNAs through interaction with the KH structural domain of IGF2BP1. The interaction of circPTPRA with IGF2BP1 attenuates the regulation of MYC and FSCN1 by IGF2BP1, thereby suppressing the progression of BC (Xie et al., 2021). Similarly, circRNAs can regulate the role of m6A writers. In BC cells, it has been shown that circ0008399 associates with the WTAP protein to foster the development of the WTAP/METTL3/METTL14 compound. By boosting the mRNA stabilization of the intended gene TNFAIP3 in a m6A-dependent manner, Circ0008399/WTAP compound stimulates its expression (Wei et al., 2021). Furthermore, circ-CTNNB1 interacts with the writer RBM15, promoting the generation of HK2, GPI, and PGK1 via m6A modification to accelerate the glycolytic procedure and stimulate osteosarcoma advancement (Yang et al., 2023).
CircRNAs have the ability to influence the synthesis of m6A regulators. CircRNAs have a wide range of effects on the expression levels of m6A regulators, whether they are writers, erasers, or readers, which are regulated by it. According to a publication, circRERE regulates GBX2 expressing themselves in HCC via ZC3H13/m6A, and down-regulating circRERE significantly boosts ZC3H13 production (Lin et al., 2022c). Furthermore, circRAB11FIP1 connects to FTO as well as advocates its translation, which increases autophagy and malignant behavior in ovarian cancer (EOC), according to research by Zhanqin Zhang et al. (Zhang et al., 2021). CircNDUFB2 is a scaffold in NSCLC to strengthen the bond between TRIM25 and IGF2BPs. This ternary combination of TRIM25, circNDUFB2, and IGF2BPs fosters the ubiquitination and breakdown of IGF2BPs, eventually preventing NSCLC progress and dissemination (Li et al., 2021d). CircRNAs, in particular, may serve as miRNA sponges that indirectly modulate the production levels of m6A regulators. For instance, CircVMP1 functions as an absorbent for miR-524-5p and upregulates the levels of METTL3 and SOX2, enhancing the growth of NSCLC and cisplatin resistance (Xie et al., 2022a). According to Lianyong Liu et al., circGPR137B, which shared a location with miR-4739 in the cytoplasm, served as a sponge for miR-4739 and upgraded the level of its target FTO, inhibiting cell proliferation and cancer metastasis in HCC patients. Patients who had high levels of circGPR137B expression also had better survival rates (Liu et al., 2022b). It has been observed that circEZH2 works as a sponge for miR-133b, causing IGF2BP2 to be upregulated in addition to its association with the IGF2BP2 and blocking its ubiquitination-dependent destruction. CircEZH2/IGF2BP2 facilitates the advancement of CRC by boosting CREB1 mRNA stability (Yao et al., 2022).
A tiny non-coding RNA known as piwi-interacting RNA (piRNA) is described as binding selectively to Piwi family proteins in testicular germ cells (Rayford et al., 2021). It regulates mRNA turnover and also directs transposon silencing by regulating DNA methylation (Wang et al., 2021b). Evidence from recent studies has shown that piRNAs are effectively generated in both natural tissue cells and tumor cells (Liu et al., 2019). Additionally, PiRNA has been demonstrated to possess pro- or anti-cancer effects by simply attaching to RNA-binding proteins called PIWI proteins (Zeng et al., 2020). As piRNA was progressively explored, it was found that it could affect a variety of processes in cancer cells by controlling m6A regulators gene expression.
PiRNAs upregulate the expression levels of m6A regulators. It has been shown that piRNA accumulation has concentration-dependent effects on the production of METTL14 in cervical carcinoma. PiRNA-14633 mimics elevated levels of m6A RNA methylation and stabilized METTL14 mRNA, which eventually aided in the formation of cervical cancers (Xie et al., 2022b). Similar to this, piRNA-30473 triggers m6A methylation in diffuse large B-cell lymphoma (DLBCL) via enhancing WTAP production. In particular, by adhering to the 3′UTR of WTAP, piRNA-30473 decreases WTAP mRNA degradation and ultimately improves mRNA balance, thus prompting WTAP to perform an oncogenic function in DLBCL(134). Moreover, piRNA-17560, an exosome generated by senescent neutrophils, increases the replication of FTO in breast tumor cells. Augmentation of FTO facilitates transition from epithelial to mesenchymal and chemotherapy resistance in cancer cells by lowering m6A levels and improving ZEB1 transcript stabilization and activation (Ou et al., 2022). Unfortunately, studies on piRNAs have all been very scarce in recent years, and further investigation of piRNAs is still needed. In addition, whether there are other modalities of regulation of m6A regulators by piRNAs is also a direction that needs to be explored in the future.
Taken together, these discoveries suggest that ncRNAs can impact m6A modifications on mRNAs in terms of their expression and function by targeting m6A writers, erasers, and readers (Table 2), hence offering a basis for future investigation of the role of RNA epigenetic modulation mechanisms in cancers.
Multiple research projects have proven that m6A methylation occurs frequently in tumors, and ncRNAs modified by m6A and m6A-associated proteins have the potential to be applied as biomarkers for the diagnosis and prognosis of cancer. In this part, we summarized some common biomarkers from the respiratory system, digestive system, and reproductive system for clinical diagnosis and prognosis (Figure 3).
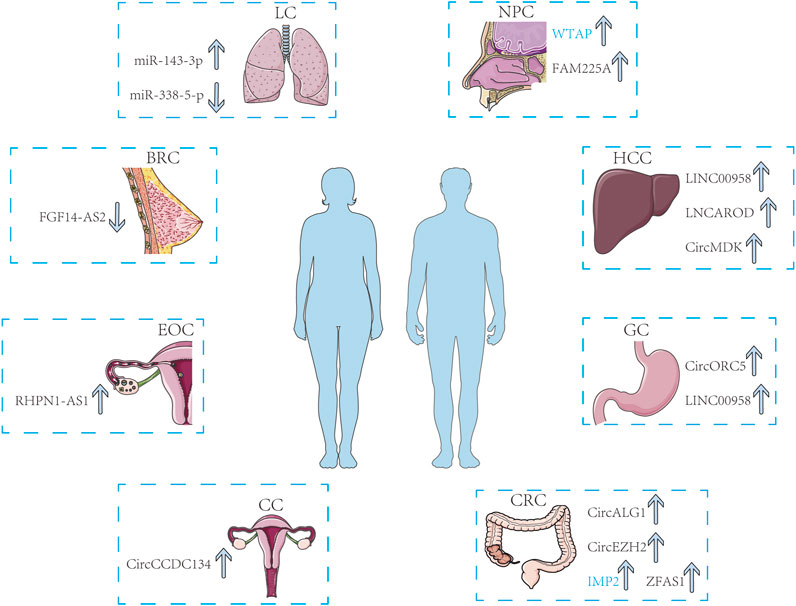
FIGURE 3. M6A-related ncRNAs and m6A regulators can be recognized as biomarkers in multiple cancers including LC, NPC, HCC, GC, CRC, CC, EOC, and BRC. The arrows show the changes in biomarkers as cancer progresses.
According to the latest studies, the stability of ncRNAs modified by m6A and m6A regulators is crucial to the advancement of respiratory cancers. As a result of WTAP m6A-dependent maintenance of lncRNA DIAPH1-AS1 stabilization in nasopharyngeal cancer (NPC), NPC development and metastasis are eventually aided. This research has identified WTAP as a possible biomarker for nasopharyngeal cancer and emphasizes the function of WTAP-mediated m6A modification in nasopharyngeal carcinoma (Li et al., 2022b). Zi-Qi Zheng et al. also discovered that FAM225A was substantially linked to survival in NPC. M6A was enormously concentrated in FAM225A, which boosted the long-term sustainability of its RNA and eventually promoted tumorigenesis and metastasis in NPC. We now have a new prognostic indication thanks to the clinical importance of FAM225 in NPC (Zheng et al., 2019).
In lung cancer, it has been shown that METTL3 can boost progenitor miR-143-3p splicing to support its biosynthesis and that m6A may stimulate progenitor miR-143-3p splicing to facilitate the production of mature miRNAs. While the miR-143-3p can indicate a poorer prognostic element for LC progression along with overall survival in vivo, this provides us with a predictive marker for LC metastasis (Wang et al., 2019a). Interestingly, not only m6A writers can regulate miRNAs, but miRNAs can also regulate m6A writers in turn. According to Hongyu Wu et al., overexpression of METTL3 decreased the blocking effect of miR-338-5p upon LC metastasis. MiR-338-5p was observed to suppress METTL3 production. It indicates that miR-338-5p offers a novel regulatory pathway for lung cancer and may possibly serve as a biomarker for its detection and prognosis (Wu et al., 2021b).
In conclusion, these findings reveal the roles of ncRNAs modified by m6A and m6A regulators in the development of respiratory cancers and herald their potential as predictive markers for nasopharyngeal and lung cancers.
Endogenous competitive mechanisms of circRNAs and lncRNAs are strongly linked to cancer cell growth and migration in malignancies of the digestive system. In colorectal cancer, circALG1 was abundantly expressed in the tumor tissues and peripheral blood of CRC patients, and it was intimately correlated with the spread of cancer cells. Additionally, the ability of circALG1 to attach to miR-342-5p was improved by m6A modification, enhancing the capacity of circALG1 as a ceRNA. This shows that circALG1 could function as a prognostic indicator in CRC (Lin et al., 2022b). Similar to this, both in vitro and in vivo, circEZH2 could foster the growth and motility of CRC cells. A crucial function for circEZH2 in controlling the advancement of CRC has been indicated by the mechanism by which it may serve as a sponge for miR-133b, upregulating IGF2BP2. CircEZH2 could be another useful diagnostic marker for CRC (Yao et al., 2022). In addition, Senxu Lu et al. found significant overexpression of IMP2 and ZFAS1 in CRC cells elevated m6A levels. These clues may be employed independently of other biomarkers to forecast the prognosis of CRC (Lu et al., 2021). There is no denying that m6A modification is fundamental to CRC formation and advancement (Jiang et al., 2023). On top of that, the mutual interaction mechanism between circRNAs and lncRNAs make them promising biomarkers in CRC.
In hepatocellular carcinoma, it was shown that METTL3-mediated N6-methyladenosine modification leads to upregulation of LINC00958, which uptakes miR3619-5p and thus upregulates HDGF expression, which facilitates adipogenesis and advancement of HCC. LINC00958 plays a crucial role and highlights its value as a prognostic predictor of HCC (Zuo et al., 2020). Similar to this, it was shown that HCC patients had considerably higher expression levels of LNCAROD modified by m6A, which advocates HCC cell proliferation, migration, invasion, and chemoresistance. LNCAROD offers a hopeful diagnostic marker for those with HCC (Jia et al., 2021). Similarly, HCC patients’ poor survival was linked to overexpression of circMDK and modification of m6A. CircMDK might function as a malignant biomarker that accelerates the development of HCC (Du et al., 2022).
In gastric cancer, Hui-Ning Fan et al. discovered that circORC5 are able to take up miR-30c-2-3p while the m6A modification of circORC5 suppresses GC advancement by controlling the miR-30c-2-3p, suggesting that circORC5 may provide a guaranteed prognostic factor for GC (Fan et al., 2022). In addition to this, it has been asserted that LINC00958 promotes aerobic glycolysis in GC and that KIAA1429 monitors the enrichment of LINC00958 in a way that link to m6A modification. This shows that LINC00958 is a crucial neoplasms-promoting inducer of GC and it may be exploited as a predictive marker for clinical examination and prognostic assessment (Yang et al., 2021a).
Taken together, these reports illustrate the involvement of ncRNAs modified by m6A in the pathological process of digestive system tumors. Some ncRNAs modified by m6A can be used as diagnostic and predictive factors for digestive system tumors, which can made patients more adequately for timely treatment.
Related evidence suggests that tumor-malignant behaviors in the reproductive system are connected to fluctuations in the production of ncRNAs modified by m6A (Huang et al., 2022). In breast cancer, FGF14-AS2 is downregulated in an m6A-dependent manner through YTHDF2-mediated RNA degradation. Patients who had diminished FGF14-AS2 expression exhibited poorer DMFS, according to clinical data. As a result, it is possible that FGF14-AS2 might be employed as an indicator for bone metastasis in breast cancer. It seems to possess a substantial function in osteolysis metastasis (Zhang et al., 2022b). In contrast, circCCDC134 gets elevated in cervical cancer as a consequence of m6A methylation, which enhances its durability through YTHDF2. CircCCDC134 stimulates growth and dissemination of CC cells, indicating that it may be an innovative possible biomarker. It also offers new regulatory model insights for exploring the oncogenic mechanisms of circCCDC134 in CC(90). Besides, in ovarian cancer, N6-methyladenosine increased the longevity of RHPN1-AS1 methylated transcripts by hampering its deterioration, leading to the elevation of RHPN1-AS1. Furthermore, the increase of RHPN1-AS1 facilitated in the growth and spread of EOC cells. This study provides a promising prognostic indicator for EOC treatment (Wang et al., 2020b).
The above studies help us to understand the pathological process and biological changes of germline tumors from the perspective of m6A-related ncRNAs, and provide the m6A-related references of diagnostic and prognostic markers in reproductive system tumors.
With the in-depth study of tumor genesis and progression, more and more non-coding RNAs associated with m6A modifications have surfaced. Therefore, exploring the specific modification sites of non-coding RNAs is indispensable for the revelation of concrete regulatory mechanisms of m6A, the testing of biomarker utility, and the development of therapeutic approaches. For example, it has been shown that the methylation site (A783) is crucial for lncRNA HOTAIR to actuate the proliferation and aggression of breast cancer cells. Methylated A783 promotes chromatin binding of HOTAIR, breast cancer growth and migration, and gene silencing via interactions with YTHDC1 (Porman et al., 2022). Fortunately, the development of a series of m6A experimental assays in recent years has further advanced m6A-related research (Table 3).
In the early days of research, the methods for detecting m6A sites were almost exclusively antibody-dependent, and this has played an essential role up to now. Methylated RNA immunoprecipitation sequencing (MeRIP), the earliest m6A detection method, is a combination of m6A-methylated mRNA segments and high-throughput sequencing, which can detect the m6A-modified areas of approximately 100 nt–200 nt (Dominissini et al., 2015). The emergence of MeRIP has tremendously boosted the development of research on m6A. However, it is undeniable that the method has low resolution and cannot reflect the precise location of m6A sites.
With the rapid advancement of science and technology, cross-linking and immunoprecipitation (CLIP), another antibody-dependent method for detecting the m6A locus, is becoming well-known and widely used. Through engagement with ultraviolet (UV) irradiation, CLIP establishes covalent bonds between proteins and RNA, which can stabilize protein-RNA interactions. Photo-cross-linking-assisted m6A sequencing strategy (PA-m6A-Seq), m6A individual-nucleotide resolution UV crosslinking and immunoprecipitation (miCLIP), and m6A-level and isoform-characterization sequencing (m6A-LAICseq) are all associated with CLIP. PA-m6A-seq can increase the resolution around the m6A modification site to 30 nt by 4SU-enhanced cross-linking (Chen et al., 2015). Nevertheless, PA-m6A-seq can merely detect m6A-modified sites around the 4SU operator site; moreover, due to the metabolism of 4SU, PA-m6A-seq seems to be only available for use in cells. Overcoming some of the drawbacks of PA-m6A-seq, miCLIP couples immunoprecipitation and high-throughput sequencing to accurately test for m6A remnants with single-base resolution (Linder et al., 2015). However, the quantity of m6A sites recognized by miCLIP was restricted owing to the low cross-linking efficiency. Comparatively, m6A-LAIC-seq measures m6A at the transcriptome scale and is mainly used to differentiate between methylated and unmethylated transcripts (Molinie et al., 2016).
In recent years, antibody-independent methods have gained popularity in the search for more effective m6A site detection methods. For instance, MazF is an m6A-sensitive ribonucleic acid endonuclease that can cut ACA sequences but not m6ACA sequences. MAZTER-seq and m6A- REF-seq were created because of this attribute (Bezrukov et al., 2021). Although m6A-REF-seq can quantize m6A levels with one-base resolution, it only recognizes particular m6A motifs (ACA), and thus its detection efficiency is poor.
Another antibody-free technique called DART-seq uses a fusion constructed from APOBEC1 (cytidine deaminase) and the YTH structural domain to detect the m6A site by inducing C-to-U deamidation at a site adjacent to the m6A residue. DART-seq can separate thousands of m6A sites in whole RNA and characterize the quantitative m6A accumulation in cells (Meyer, 2019). Obviously, the bonding capacity between the integrated APOBEC1-YTH protein and the m6A modification may influence the testing precision of the target.
Besides, m6A-label-seq is a metabolic labeling approach. It detects m6A at a monobase resolution level by metabolically tagging the candidate substrate adenosine during m6A production (Shu et al., 2020). Unfortunately, this method can only be applied to cellular systems.
The FTO-assisted m6A selective chemical labeling method, also known as m6A -SEAL, is also an innovative technique. In the presence of dithiothreitol (DTT)-mediated thiol addition reactions, unstable N6-hydroxymethyladenosine (hm6A) can be converted to ultrastable N6-dithiocarbonylmethyladenosine (dm6A). And m6A-SEAL is associated with the role of hm6A in RNA that can be oxidized by the FTO enzyme of m6A, which can effectively detect m6A modification sites (Wang et al., 2020c). Compared to the previous technique, m6A-SEAL has higher sensitivity and specificity; however, this technique has not yet reached the condition of single-base resolution.
So far, although there are numerous methods for the detection of the m6A sites, they all have their own defective aspects. Therefore, only through the comprehensive utilization of different detection methods and the pursuit of more efficient and accurate m6A site detection approaches can we further boost the progress of m6A-related studies.
In this review, we concluded, on the one hand, the role that m6A regulators play in tumorigenesis and development by modulating non-coding RNAs. On the other hand, we also outlined the ability of non-coding RNAs to influence m6A modifications that promote or attenuate malignant changes in cancer. In addition, we summarized some non-coding RNAs and m6A-related proteins that may be employed as diagnostic and prognostic indicators in the hopes that this information will help clinical practice. Considering the practical application of biomarkers, we also aggregated some useful m6A site detection methods in recent years, which can be helpful for researchers to better choose the experimental methods.
The epigenetic processes of cancer cells have undergone fundamental changes (López et al., 2022). The m6A modification of RNAs, one of the epigenetic research hotspots, is crucial for the regulation of cancer. Multiple researches have proven that m6A-related proteins have the ability to control the production and biological functions of ncRNAs by altering their methylation levels. In cancer, this can manifest as tumor growth, metastasis, and stem cell differentiation, among others. Some progress has been made in the study of the biological effects of m6A on tumors, but there are still some limitations that require further breakthroughs. Currently, the gene knock-out of tissue-specific m6A-related proteins in mice can help reveal the mechanisms of m6A modification in related cancer cells. However, we still need more clinical trials to measure the diagnostic value and therapeutic effects of m6A modification on patients. With the development of more innovative high-end research tools and research methodologies based on m6A, we believe that in the near future we can conduct more basic and clinical studies to fill the gap in current research (Meng et al., 2023).
NcRNAs may control the gene expression level and function of m6A-related proteins as well. This may be related to the influence of ncRNAs on their transcription, translation, and protein capacity of m6A regulators. Correlations between m6A modifications and ncRNAs offer a fresh route for us to explore precise processes underlying the emergence of cancer. Some studies, however, were only able to analyze publicly accessible datasets statistically. The non-coding RNAs’ capacity to predict cancer continues to be constrained due to the comparatively small number of statistical samples (Mo et al., 2021).
With the gradual increase in the importance of m6A modification in the medical community, abnormally expressed m6A-related non-coding RNAs have recently been identified in peripheral bloodstreams of patients who suffer from cancers, which tremendously improves the convenience and utility of m6A-modified non-coding RNAs as biomarkers with good prospects for clinical application (Eun et al., 2023). In addition, cancer has inappropriate expression of m6A-related proteins, which accordingly affects the m6A level of ncRNAs. Taking into account the views expressed above, it is reasonable to assume that m6A-related proteins and m6A-modified non-coding RNAs may be exploited as promising clinical diagnostic and prognostic markers. However, current studies on m6A-related ncRNAs and m6A regulators are still incomplete, and the proposed potential m6A-related diagnostic and prognostic markers still need to be validated by more advanced m6A site detection methods.
SL: Conceptualization, Investigation, Methodology, Writing–original draft. DX: Funding acquisition, Supervision, Validation, Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
m6A, N6-methyladenosine; METTL3, methyltransferase-like 3; METTL14, methyltransferase-like 14; WTAP, Wilms tumor 1-associating protein; KIAA1429, vir-like m6A methyltransferase-associated protein; RBM15/15B, RNA-binding motif protein 15/15B; ZC3H13, zinc finger CCCH structural domain-containing protein 13; FTO, fat mass and obesity-associated protein; ALKBH5, alkylation repair homolog protein 5; YTHDF, YT521-B homology domain family; YTHDC, YT521-B homology domain-containing family; HNRNPC, heterogeneous nuclear ribonucleoprotein C; IGF2BP, insulin-like growth factor 2 binding proteins 2; mRNA, messenger RNA; miRNA, microRNA; lncRNA, long noncoding RNA; circRNA, circular RNA; piRNA, piwi-interacting RNA.
1For Original Research articles, please note that the Material and Methods section can be placed in any of the following ways: before Results, before Discussion or after Discussion
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015). HNRNPA2B1 is a mediator of m(6)a-dependent nuclear RNA processing events. Cell 162 (6), 1299–1308. doi:10.1016/j.cell.2015.08.011
Ban, Y., Tan, P., Cai, J., Li, J., Hu, M., Zhou, Y., et al. (2020). LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol. Oncol. 14 (6), 1282–1296. doi:10.1002/1878-0261.12676
Bezrukov, F., Prados, J., Renzoni, A., and Panasenko, O. O. (2021). MazF toxin causes alterations in Staphylococcus aureus transcriptome, translatome and proteome that underlie bacterial dormancy. Nucleic Acids Res. 49 (4), 2085–2101. doi:10.1093/nar/gkaa1292
Cai, J., Chen, Z., Zhang, Y., Wang, J., Zhang, Z., Wu, J., et al. (2022). CircRHBDD1 augments metabolic rewiring and restricts immunotherapy efficacy via m(6)A modification in hepatocellular carcinoma. Mol. Ther. Oncolytics 24, 755–771. doi:10.1016/j.omto.2022.02.021
Chai, R. C., Chang, Y. Z., Chang, X., Pang, B., An, S. Y., Zhang, K. N., et al. (2021). YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m(6)A modification to activate NF-κB and promote the malignant progression of glioma. J. Hematol. Oncol. 14 (1), 109. doi:10.1186/s13045-021-01124-z
Chen, K., Lu, Z., Wang, X., Fu, Y., Luo, G. Z., Liu, N., et al. (2015). High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. Int. Ed. Engl. 54 (5), 1587–1590. doi:10.1002/anie.201410647
Chen, L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21 (8), 475–490. doi:10.1038/s41580-020-0243-y
Chen, X. Y., Zhang, J., and Zhu, J. S. (2019). The role of m(6)A RNA methylation in human cancer. Mol. Cancer 18 (1), 103. doi:10.1186/s12943-019-1033-z
Chen, Z., Huang, L., Wang, K., Zhang, L., Zhong, X., Yan, Z., et al. (2022). rtcisE2F promotes the self-renewal and metastasis of liver tumor-initiating cells via N(6)-methyladenosine-dependent E2F3/E2F6 mRNA stability. Sci. China Life Sci. 65 (9), 1840–1854. doi:10.1007/s11427-021-2038-5
Coker, H., Wei, G., and Brockdorff, N. (2019). m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim. Biophys. Acta Gene Regul. Mech. 1862 (3), 310–318. doi:10.1016/j.bbagrm.2018.12.002
Cui, Y., Zhang, C., Ma, S., Li, Z., Wang, W., Li, Y., et al. (2021). RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 40 (1), 294. doi:10.1186/s13046-021-02096-1
Deniz, E., and Erman, B. (2017). Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct. Integr. Genomics 17 (2-3), 135–143. doi:10.1007/s10142-016-0524-x
Dominissini, D., Moshitch-Moshkovitz, S., Amariglio, N., and Rechavi, G. (2015). Transcriptome-wide mapping of N⁶-Methyladenosine by m⁶A-seq. Methods Enzymol. 560, 131–147. doi:10.1016/bs.mie.2015.03.001
Du, A., Li, S., Zhou, Y., Disoma, C., Liao, Y., Zhang, Y., et al. (2022). M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol. Cancer 21 (1), 109. doi:10.1186/s12943-022-01575-z
Du, C., Lv, C., Feng, Y., and Yu, S. (2020). Activation of the KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis facilitates prostate cancer progression. J. Exp. Clin. Cancer Res. 39 (1), 223. doi:10.1186/s13046-020-01735-3
Eun, J. W., Cheong, J. Y., Jeong, J. Y., and Kim, H. S. (2023). A new understanding of long non-coding RNA in hepatocellular carcinoma-from m(6)A modification to blood biomarkers. Cells 12 (18), 2272. doi:10.3390/cells12182272
Fan, H. N., Chen, Z. Y., Chen, X. Y., Chen, M., Yi, Y. C., Zhu, J. S., et al. (2022). METTL14-mediated m(6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol. Cancer 21 (1), 51. doi:10.1186/s12943-022-01521-z
Frye, M., Harada, B. T., Behm, M., and He, C. (2018). RNA modifications modulate gene expression during development. Science 361 (6409), 1346–1349. doi:10.1126/science.aau1646
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat. Rev. Genet. 15 (5), 293–306. doi:10.1038/nrg3724
Gerken, T., Girard, C. A., Tung, Y. C., Webby, C. J., Saudek, V., Hewitson, K. S., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318 (5855), 1469–1472. doi:10.1126/science.1151710
Grabowski, P. J., Zaug, A. J., and Cech, T. R. (1981). The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena. Cell 23 (2), 467–476. doi:10.1016/0092-8674(81)90142-2
Gu, Y., Niu, S., Wang, Y., Duan, L., Pan, Y., Tong, Z., et al. (2021). DMDRMR-mediated regulation of m(6)a-modified CDK4 by m(6)A reader IGF2BP3 drives ccRCC progression. Cancer Res. 81 (4), 923–934. doi:10.1158/0008-5472.CAN-20-1619
Guajardo, L., Aguilar, R., Bustos, F. J., Nardocci, G., Gutiérrez, R. A., van Zundert, B., et al. (2020). Downregulation of the polycomb-associated methyltransferase Ezh2 during maturation of hippocampal neurons is mediated by MicroRNAs let-7 and miR-124. Int. J. Mol. Sci. 21 (22), 8472. doi:10.3390/ijms21228472
Guo, Y., Guo, Y., Chen, C., Fan, D., Wu, X., Zhao, L., et al. (2021). Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol. Cancer 20 (1), 93. doi:10.1186/s12943-021-01372-0
Han, H., Fan, G., Song, S., Jiang, Y., Qian, C., Zhang, W., et al. (2021). piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 137 (12), 1603–1614. doi:10.1182/blood.2019003764
Haussmann, I. U., Bodi, Z., Sanchez-Moran, E., Mongan, N. P., Archer, N., Fray, R. G., et al. (2016). m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540 (7632), 301–304. doi:10.1038/nature20577
He, L., Li, H., Wu, A., Peng, Y., Shu, G., and Yin, G. (2019). Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 18 (1), 176. doi:10.1186/s12943-019-1109-9
Hou, Y., Zhang, Q., Pang, W., Hou, L., Liang, Y., Han, X., et al. (2021). YTHDC1-mediated augmentation of miR-30d in repressing pancreatic tumorigenesis via attenuation of RUNX1-induced transcriptional activation of Warburg effect. Cell Death Differ. 28 (11), 3105–3124. doi:10.1038/s41418-021-00804-0
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27 (9), 1115–1127. doi:10.1038/cr.2017.99
Hu, Y., Tang, J., Xu, F., Chen, J., Zeng, Z., Han, S., et al. (2022). A reciprocal feedback between N6-methyladenosine reader YTHDF3 and lncRNA DICER1-AS1 promotes glycolysis of pancreatic cancer through inhibiting maturation of miR-5586-5p. J. Exp. Clin. Cancer Res. 41 (1), 69. doi:10.1186/s13046-022-02285-6
Huang, H., Weng, H., and Chen, J. (2020). m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell 37 (3), 270–288. doi:10.1016/j.ccell.2020.02.004
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Huang, T., Cao, L., Feng, N., Xu, B., Dong, Y., and Wang, M. (2021). N(6)-methyladenosine (m(6)A)-mediated lncRNA DLGAP1-AS1enhances breast canceradriamycin resistance through miR-299-3p/WTAP feedback loop. Bioengineered 12 (2), 10935–10944. doi:10.1080/21655979.2021.2000198
Huang, W., Kong, F., Li, R., Chen, X., and Wang, K. (2022). Emerging roles of m(6)A RNA methylation regulators in gynecological cancer. Front. Oncol. 12, 827956. doi:10.3389/fonc.2022.827956
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Jia, G., Wang, Y., Lin, C., Lai, S., Dai, H., Wang, Z., et al. (2021). LNCAROD enhances hepatocellular carcinoma malignancy by activating glycolysis through induction of pyruvate kinase isoform PKM2. J. Exp. Clin. Cancer Res. 40 (1), 299. doi:10.1186/s13046-021-02090-7
Jia, G. X., Lin, Z., Yan, R. G., Wang, G. W., Zhang, X. N., Li, C., et al. (2020). WTAP function in sertoli cells is essential for sustaining the spermatogonial stem cell niche. Stem Cell Rep. 15 (4), 968–982. doi:10.1016/j.stemcr.2020.09.001
Jia, Y., Yan, Q., Zheng, Y., Li, L., Zhang, B., Chang, Z., et al. (2022). Long non-coding RNA NEAT1 mediated RPRD1B stability facilitates fatty acid metabolism and lymph node metastasis via c-Jun/c-Fos/SREBP1 axis in gastric cancer. J. Exp. Clin. Cancer Res. 41 (1), 287. doi:10.1186/s13046-022-02449-4
Jiang, X., Jin, Z., Yang, Y., Zheng, X., Chen, S., Wang, S., et al. (2023). m6A modification on the fate of colorectal cancer: functions and mechanisms of cell proliferation and tumorigenesis. Front. Oncol. 13, 1162300. doi:10.3389/fonc.2023.1162300
Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z., et al. (2021). The role of m6A modification in the biological functions and diseases. Signal Transduct. Target Ther. 6 (1), 74. doi:10.1038/s41392-020-00450-x
Jiang, Z., Song, X., Wei, Y., Li, Y., Kong, D., and Sun, J. (2022). N(6)-methyladenosine-mediated miR-380-3p maturation and upregulation promotes cancer aggressiveness in pancreatic cancer. Bioengineered 13 (6), 14460–14471. doi:10.1080/21655979.2022.2088497
Jin, D., Guo, J., Wu, Y., Du, J., Yang, L., Wang, X., et al. (2021). m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 14 (1), 32. doi:10.1186/s13045-021-01048-8
Knuckles, P., Lence, T., Haussmann, I. U., Jacob, D., Kreim, N., Carl, S. H., et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 32 (5-6), 415–429. doi:10.1101/gad.309146.117
Kurowski, M. A., Bhagwat, A. S., Papaj, G., and Bujnicki, J. M. (2003). Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics 4 (1), 48. doi:10.1186/1471-2164-4-48
Lan, T., Li, H., Zhang, D., Xu, L., Liu, H., Hao, X., et al. (2019). KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 18 (1), 186. doi:10.1186/s12943-019-1106-z
Li, A., Chen, Y. S., Ping, X. L., Yang, X., Xiao, W., Yang, Y., et al. (2017). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 27 (3), 444–447. doi:10.1038/cr.2017.10
Li, B., Zhu, L., Lu, C., Wang, C., Wang, H., Jin, H., et al. (2021d). circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 12 (1), 295. doi:10.1038/s41467-020-20527-z
Li, H., Zhang, N., Jiao, X., Wang, C., Sun, W., He, Y., et al. (2021c). Downregulation of microRNA-6125 promotes colorectal cancer growth through YTHDF2-dependent recognition of N6-methyladenosine-modified GSK3β. Clin. Transl. Med. 11 (10), e602. doi:10.1002/ctm2.602
Li, J., Li, Z., Bai, X., Chen, X., Wang, M., Wu, Y., et al. (2022a). LncRNA UCA1 promotes the progression of AML by upregulating the expression of CXCR4 and CYP1B1 by affecting the stability of METTL14. J. Oncol. 2022, 2756986. doi:10.1155/2022/2756986
Li, K., Peng, Z. Y., Gao, S., Wang, Q. S., Wang, R., Li, X., et al. (2021a). M6A associated TSUC7 inhibition contributed to Erlotinib resistance in lung adenocarcinoma through a notch signaling activation dependent way. J. Exp. Clin. Cancer Res. 40 (1), 325. doi:10.1186/s13046-021-02137-9
Li, Y., Xiao, J., Bai, J., Tian, Y., Qu, Y., Chen, X., et al. (2019). Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol. Cancer 18 (1), 137. doi:10.1186/s12943-019-1066-3
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 (3), 256–264. doi:10.1038/nsmb.2959
Li, Z., Yang, H. Y., Dai, X. Y., Zhang, X., Huang, Y. Z., Shi, L., et al. (2021b). CircMETTL3, upregulated in a m6A-dependent manner, promotes breast cancer progression. Int. J. Biol. Sci. 17 (5), 1178–1190. doi:10.7150/ijbs.57783
Li, Z. X., Zheng, Z. Q., Yang, P. Y., Lin, L., Zhou, G. Q., Lv, J. W., et al. (2022b). WTAP-mediated m(6)A modification of lncRNA DIAPH1-AS1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death Differ. 29 (6), 1137–1151. doi:10.1038/s41418-021-00905-w
Liang, L., Zhu, Y., Li, J., Zeng, J., and Wu, L. (2022). ALKBH5-mediated m6A modification of circCCDC134 facilitates cervical cancer metastasis by enhancing HIF1A transcription. J. Exp. Clin. Cancer Res. 41 (1), 261. doi:10.1186/s13046-022-02462-7
Lin, C., Ma, M., Zhang, Y., Li, L., Long, F., Xie, C., et al. (2022b). The N(6)-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol. Cancer 21 (1), 80. doi:10.1186/s12943-022-01560-6
Lin, C., Wang, Y., Dong, Y., Lai, S., Wang, L., Weng, S., et al. (2022a). N6-methyladenosine-mediated SH3BP5-AS1 upregulation promotes GEM chemoresistance in pancreatic cancer by activating the Wnt signaling pathway. Biol. Direct 17 (1), 33. doi:10.1186/s13062-022-00347-5
Lin, R., Zhan, M., Yang, L., Wang, H., Shen, H., Huang, S., et al. (2020). Deoxycholic acid modulates the progression of gallbladder cancer through N(6)-methyladenosine-dependent microRNA maturation. Oncogene 39 (26), 4983–5000. doi:10.1038/s41388-020-1349-6
Lin, Y. H., Zhang, B. Y., and Chen, Z. S. (2022c). circRERE regulates the expression of GBX2 through miR-1299 and ZC3H13/N(6)-methyladenosine (m(6)A) to promote growth and invasion of hepatocellular carcinoma cells. J. Biosci. 47, 52. doi:10.1007/s12038-022-00298-z
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12 (8), 767–772. doi:10.1038/nmeth.3453
Liu, H., Xu, Y., Yao, B., Sui, T., Lai, L., and Li, Z. (2020). A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death Dis. 11 (8), 613. doi:10.1038/s41419-020-02833-y
Liu, L., Gu, M., Ma, J., Wang, Y., Li, M., Wang, H., et al. (2022b). CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol. Cancer 21 (1), 149. doi:10.1186/s12943-022-01619-4
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45 (10), 6051–6063. doi:10.1093/nar/gkx141
Liu, R., Zhong, Y., Chen, R., Chu, C., Liu, G., Zhou, Y., et al. (2022a). m(6)A reader hnRNPA2B1 drives multiple myeloma osteolytic bone disease. Theranostics 12 (18), 7760–7774. doi:10.7150/thno.76852
Liu, Y., Dou, M., Song, X., Dong, Y., Liu, S., Liu, H., et al. (2019). The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 18 (1), 123. doi:10.1186/s12943-019-1052-9
López, C., Barnon, M. T., Beacon, T. H., Nardocci, G., and Davie, J. R. (2022). The key role of differential broad H3K4me3 and H3K4ac domains in breast cancer. Gene 826, 146463. doi:10.1016/j.gene.2022.146463
Lu, S., Han, L., Hu, X., Sun, T., Xu, D., Li, Y., et al. (2021). N6-methyladenosine reader IMP2 stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect: implication in colorectal cancer. J. Hematol. Oncol. 14 (1), 188. doi:10.1186/s13045-021-01204-0
Luo, M. L., Li, J., Shen, L., Chu, J., Guo, Q., Liang, G., et al. (2020). The role of APAL/ST8SIA6-AS1 lncRNA in PLK1 activation and mitotic catastrophe of tumor cells. J. Natl. Cancer Inst. 112 (4), 356–368. doi:10.1093/jnci/djz134
Ma, J. Z., Yang, F., Zhou, C. C., Liu, F., Yuan, J. H., Wang, F., et al. (2017). METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 65 (2), 529–543. doi:10.1002/hep.28885
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 (7441), 333–338. doi:10.1038/nature11928
Meng, Q., Schatten, H., Zhou, Q., and Chen, J. (2023). Crosstalk between m6A and coding/non-coding RNA in cancer and detection methods of m6A modification residues. Aging (Albany NY) 15 (13), 6577–6619. doi:10.18632/aging.204836
Meyer, K. D. (2019). DART-seq: an antibody-free method for global m(6)A detection. Nat. Methods 16 (12), 1275–1280. doi:10.1038/s41592-019-0570-0
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Mirihana Arachchilage, G., Dassanayake, A. C., and Basu, S. (2015). A potassium ion-dependent RNA structural switch regulates human pre-miRNA 92b maturation. Chem. Biol. 22 (2), 262–272. doi:10.1016/j.chembiol.2014.12.013
Mo, W. L., Deng, L. J., Cheng, Y., Yu, W. J., Yang, Y. H., and Gu, W. D. (2021). Circular RNA hsa_circ_0072309 promotes tumorigenesis and invasion by regulating the miR-607/FTO axis in non-small cell lung carcinoma. Aging (Albany NY) 13 (8), 11629–11645. doi:10.18632/aging.202856
Molinie, B., Wang, J., Lim, K. S., Hillebrand, R., Lu, Z. X., Van Wittenberghe, N., et al. (2016). m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat. Methods 13 (8), 692–698. doi:10.1038/nmeth.3898
Monteiro, L. J., Peñailillo, R., Sánchez, M., Acuña-Gallardo, S., Mönckeberg, M., Ong, J., et al. (2021). The role of long non-coding RNAs in trophoblast regulation in preeclampsia and intrauterine growth restriction. GenesGenes (Basel) 12 (7), 970. doi:10.3390/genes12070970
Ni, W., Yao, S., Zhou, Y., Liu, Y., Huang, P., Zhou, A., et al. (2019). Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer 18 (1), 143. doi:10.1186/s12943-019-1079-y
Ou, B., Liu, Y., Gao, Z., Xu, J., Yan, Y., Li, Y., et al. (2022). Senescent neutrophils-derived exosomal piRNA-17560 promotes chemoresistance and EMT of breast cancer via FTO-mediated m6A demethylation. Cell Death Dis. 13 (10), 905. doi:10.1038/s41419-022-05317-3
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373. doi:10.1038/nature19342
Peng, W., Li, J., Chen, R., Gu, Q., Yang, P., Qian, W., et al. (2019). Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 38 (1), 393. doi:10.1186/s13046-019-1408-4
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 (2), 177–189. doi:10.1038/cr.2014.3
Porman, A. M., Roberts, J. T., Duncan, E. D., Chrupcala, M. L., Levine, A. A., Kennedy, M. A., et al. (2022). A single N6-methyladenosine site regulates lncRNA HOTAIR function in breast cancer cells. PLoS Biol. 20 (11), e3001885. doi:10.1371/journal.pbio.3001885
Qi, X., Lin, Y., Chen, J., and Shen, B. (2020). Decoding competing endogenous RNA networks for cancer biomarker discovery. Brief. Bioinform 21 (2), 441–457. doi:10.1093/bib/bbz006
Rayford, K. J., Cooley, A., Rumph, J. T., Arun, A., Rachakonda, G., Villalta, F., et al. (2021). piRNAs as modulators of disease pathogenesis. Int. J. Mol. Sci. 22 (5), 2373. doi:10.3390/ijms22052373
Ren, Z., Hu, Y., Sun, J., Kang, Y., Li, G., and Zhao, H. (2022). N(6)-methyladenosine methyltransferase WTAP-stabilized FOXD2-AS1 promotes the osteosarcoma progression through m(6)A/FOXM1 axis. Bioengineered 13 (4), 7963–7973. doi:10.1080/21655979.2021.2008218
Robinson, E. K., Covarrubias, S., and Carpenter, S. (2020). The how and why of lncRNA function: an innate immune perspective. Biochim. Biophys. Acta Gene Regul. Mech. 1863 (4), 194419. doi:10.1016/j.bbagrm.2019.194419
Rong, L., Xu, Y., Zhang, K., Jin, L., and Liu, X. (2022). HNRNPA2B1 inhibited SFRP2 and activated Wnt-β/catenin via m6A-mediated miR-106b-5p processing to aggravate stemness in lung adenocarcinoma. Pathol. Res. Pract. 233, 153794. doi:10.1016/j.prp.2022.153794
Roost, C., Lynch, S. R., Batista, P. J., Qu, K., Chang, H. Y., and Kool, E. T. (2015). Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 137 (5), 2107–2115. doi:10.1021/ja513080v
Rottman, F., Shatkin, A. J., and Perry, R. P. (1974). Sequences containing methylated nucleotides at the 5' termini of messenger RNAs: possible implications for processing. Cell 3 (3), 197–199. doi:10.1016/0092-8674(74)90131-7
Saliminejad, K., Khorram Khorshid, H. R., Soleymani Fard, S., and Ghaffari, S. H. (2019). An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 234 (5), 5451–5465. doi:10.1002/jcp.27486
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 73 (11), 3852–3856. doi:10.1073/pnas.73.11.3852
Schöller, E., Weichmann, F., Treiber, T., Ringle, S., Treiber, N., Flatley, A., et al. (2018). Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. Rna 24 (4), 499–512. doi:10.1261/rna.064063.117
Shi, H., Wei, J., and He, C. (2019). Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74 (4), 640–650. doi:10.1016/j.molcel.2019.04.025
Shu, X., Cao, J., Cheng, M., Xiang, S., Gao, M., Li, T., et al. (2020). A metabolic labeling method detects m(6)A transcriptome-wide at single base resolution. Nat. Chem. Biol. 16 (8), 887–895. doi:10.1038/s41589-020-0526-9
Song, P., Feng, L., Li, J., Dai, D., Zhu, L., Wang, C., et al. (2020). β-catenin represses miR455-3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol. Cancer 19 (1), 129. doi:10.1186/s12943-020-01244-z
Song, Z., Jia, G., Ma, P., and Cang, S. (2021). Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci. 276, 119399. doi:10.1016/j.lfs.2021.119399
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Sun, J., Ma, X., Ying, Y., Wang, W., Shen, H., Wang, S., et al. (2022). SMAD3 and FTO are involved in miR-5581-3p-mediated inhibition of cell migration and proliferation in bladder cancer. Cell Death Discov. 8 (1), 199. doi:10.1038/s41420-022-01010-8
Sun, T., Wu, Z., Wang, X., Wang, Y., Hu, X., Qin, W., et al. (2020b). LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene 39 (31), 5358–5372. doi:10.1038/s41388-020-1338-9
Sun, Y., Li, S., Yu, W., Zhao, Z., Gao, J., Chen, C., et al. (2020a). N(6)-methyladenosine-dependent pri-miR-17-92 maturation suppresses PTEN/TMEM127 and promotes sensitivity to everolimus in gastric cancer. Cell Death Dis. 11 (10), 836. doi:10.1038/s41419-020-03049-w
Tang, C., Xie, Y., Yu, T., Liu, N., Wang, Z., Woolsey, R. J., et al. (2020). m(6)A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 30 (3), 211–228. doi:10.1038/s41422-020-0279-8
Tang, Y., Chen, K., Song, B., Ma, J., Wu, X., Xu, Q., et al. (2021). m6A-Atlas: a comprehensive knowledgebase for unraveling the N6-methyladenosine (m6A) epitranscriptome. Nucleic Acids Res. 49 (D1), D134–D143. doi:10.1093/nar/gkaa692
Van Roosbroeck, K., and Calin, G. A. (2017). Cancer hallmarks and MicroRNAs: the therapeutic connection. Adv. Cancer Res. 135, 119–149. doi:10.1016/bs.acr.2017.06.002
Wang, H., Deng, Q., Lv, Z., Ling, Y., Hou, X., Chen, Z., et al. (2019a). N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol. Cancer 18 (1), 181. doi:10.1186/s12943-019-1108-x
Wang, J., Ding, W., Xu, Y., Tao, E., Mo, M., Xu, W., et al. (2020b). Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany NY) 12 (5), 4558–4572. doi:10.18632/aging.102911
Wang, K., Wang, T., Gao, X. Q., Chen, X. Z., Wang, F., and Zhou, L. Y. (2021b). Emerging functions of piwi-interacting RNAs in diseases. J. Cell Mol. Med. 25 (11), 4893–4901. doi:10.1111/jcmm.16466
Wang, L., Yi, X., Xiao, X., Zheng, Q., Ma, L., and Li, B. (2022b). Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell Mol. Biol. Lett. 27 (1), 106. doi:10.1186/s11658-022-00406-9
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534 (7608), 575–578. doi:10.1038/nature18298
Wang, X., Liu, C., Zhang, S., Yan, H., Zhang, L., Jiang, A., et al. (2021a). N(6)-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev. Cell 56 (5), 702–715.e8. doi:10.1016/j.devcel.2021.01.015
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16 (2), 191–198. doi:10.1038/ncb2902
Wang, Y., Lu, J. H., Wu, Q. N., Jin, Y., Wang, D. S., Chen, Y. X., et al. (2019b). LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol. Cancer 18 (1), 174. doi:10.1186/s12943-019-1105-0
Wang, Y., Wu, S., Zhu, X., Zhang, L., Deng, J., Li, F., et al. (2020a). LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J. Exp. Med. 217 (3), jem.20190950. doi:10.1084/jem.20190950
Wang, Y., Xiao, Y., Dong, S., Yu, Q., and Jia, G. (2020c). Antibody-free enzyme-assisted chemical approach for detection of N(6)-methyladenosine. Nat. Chem. Biol. 16 (8), 896–903. doi:10.1038/s41589-020-0525-x
Wang, Z. W., Pan, J. J., Hu, J. F., Zhang, J. Q., Huang, L., Huang, Y., et al. (2022a). SRSF3-mediated regulation of N6-methyladenosine modification-related lncRNA ANRIL splicing promotes resistance of pancreatic cancer to gemcitabine. Cell Rep. 39 (6), 110813. doi:10.1016/j.celrep.2022.110813
Wei, C. M., Gershowitz, A., and Moss, B. (1975). Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell 4 (4), 379–386. doi:10.1016/0092-8674(75)90158-0
Wei, J., and He, C. (2021). Chromatin and transcriptional regulation by reversible RNA methylation. Curr. Opin. Cell Biol. 70, 109–115. doi:10.1016/j.ceb.2020.11.005
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m(6)A, m(6)A(m), and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71 (6), 973–985. doi:10.1016/j.molcel.2018.08.011
Wei, J., Yu, X., Yang, L., Liu, X., Gao, B., Huang, B., et al. (2022). FTO mediates LINE1 m(6)A demethylation and chromatin regulation in mESCs and mouse development. Science 376 (6596), 968–973. doi:10.1126/science.abe9582
Wei, W., Sun, J., Zhang, H., Xiao, X., Huang, C., Wang, L., et al. (2021). Circ0008399 interaction with WTAP promotes assembly and activity of the m(6)A methyltransferase complex and promotes cisplatin resistance in bladder cancer. Cancer Res. 81 (24), 6142–6156. doi:10.1158/0008-5472.CAN-21-1518
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69 (6), 1028–1038. doi:10.1016/j.molcel.2018.02.015
Wu, H., Li, F., and Zhu, R. (2021b). miR-338-5p inhibits cell growth and migration via inhibition of the METTL3/m6A/c-Myc pathway in lung cancer. Acta Biochim. Biophys. Sin. (Shanghai). 53 (3), 304–316. doi:10.1093/abbs/gmaa170
Wu, P., Fang, X., Liu, Y., Tang, Y., Wang, W., Li, X., et al. (2021a). N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 12 (4), 298. doi:10.1038/s41419-021-03558-2
Wu, Q., Zhang, H., Yang, D., Min, Q., Wang, Y., Zhang, W., et al. (2022). The m6A-induced lncRNA CASC8 promotes proliferation and chemoresistance via upregulation of hnRNPL in esophageal squamous cell carcinoma. Int. J. Biol. Sci. 18 (13), 4824–4836. doi:10.7150/ijbs.71234
Wu, S., Zhang, L., Deng, J., Guo, B., Li, F., Wang, Y., et al. (2020). A novel micropeptide encoded by Y-linked LINC00278 links cigarette smoking and AR signaling in male esophageal squamous cell carcinoma. Cancer Res. 80 (13), 2790–2803. doi:10.1158/0008-5472.CAN-19-3440
Xiao, L., Li, X., Mu, Z., Zhou, J., Zhou, P., Xie, C., et al. (2020). FTO inhibition enhances the antitumor effect of temozolomide by targeting MYC-miR-155/23a cluster-MXI1 feedback circuit in glioma. Cancer Res. 80 (18), 3945–3958. doi:10.1158/0008-5472.CAN-20-0132
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61 (4), 507–519. doi:10.1016/j.molcel.2016.01.012
Xie, F., Huang, C., Liu, F., Zhang, H., Xiao, X., Sun, J., et al. (2021). CircPTPRA blocks the recognition of RNA N(6)-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol. Cancer 20 (1), 68. doi:10.1186/s12943-021-01359-x
Xie, H., Yao, J., Wang, Y., and Ni, B. (2022a). Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 29 (1), 1257–1271. doi:10.1080/10717544.2022.2057617
Xie, Q., Li, Z., Luo, X., Wang, D., Zhou, Y., Zhao, J., et al. (2022b). piRNA-14633 promotes cervical cancer cell malignancy in a METTL14-dependent m6A RNA methylation manner. J. Transl. Med. 20 (1), 51. doi:10.1186/s12967-022-03257-2
Xue, J., Xiao, P., Yu, X., and Zhang, X. (2021). A positive feedback loop between AlkB homolog 5 and miR-193a-3p promotes growth and metastasis in esophageal squamous cell carcinoma. Hum. Cell 34 (2), 502–514. doi:10.1007/s13577-020-00458-z
Yan, R., Dai, W., Wu, R., Huang, H., and Shu, M. (2022). Therapeutic targeting m6A-guided miR-146a-5p signaling contributes to the melittin-induced selective suppression of bladder cancer. Cancer Lett. 534, 215615. doi:10.1016/j.canlet.2022.215615
Yang, D., Chang, S., Li, F., Ma, M., Yang, J., Lv, X., et al. (2021a). m(6) A transferase KIAA1429-stabilized LINC00958 accelerates gastric cancer aerobic glycolysis through targeting GLUT1. IUBMB Life 73 (11), 1325–1333. doi:10.1002/iub.2545
Yang, F., Liu, Y., Xiao, J., Li, B., Chen, Y., Hu, A., et al. (2023). Circ-CTNNB1 drives aerobic glycolysis and osteosarcoma progression via m6A modification through interacting with RBM15. Cell Prolif. 56 (1), e13344. doi:10.1111/cpr.13344
Yang, H., Hu, Y., Weng, M., Liu, X., Wan, P., Hu, Y., et al. (2022a). Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer. J. Adv. Res. 37, 91–106. doi:10.1016/j.jare.2021.10.001
Yang, J., Qian, X., Qiu, Q., Xu, L., Pan, M., Li, J., et al. (2022b). LCAT1 is an oncogenic LncRNA by stabilizing the IGF2BP2-CDC6 axis. Cell Death Dis. 13 (10), 877. doi:10.1038/s41419-022-05316-4
Yang, L., Chen, Y., Liu, N., Shi, Q., Han, X., Gan, W., et al. (2021b). Low expression of TRAF3IP2-AS1 promotes progression of NONO-TFE3 translocation renal cell carcinoma by stimulating N(6)-methyladenosine of PARP1 mRNA and downregulating PTEN. J. Hematol. Oncol. 14 (1), 46. doi:10.1186/s13045-021-01059-5
Yang, S., Wei, J., Cui, Y. H., Park, G., Shah, P., Deng, Y., et al. (2019). m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 10 (1), 2782. doi:10.1038/s41467-019-10669-0
Yang, X., Zhang, S., He, C., Xue, P., Zhang, L., He, Z., et al. (2020). METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer 19 (1), 46. doi:10.1186/s12943-020-1146-4
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 27 (5), 626–641. doi:10.1038/cr.2017.31
Yao, B., Zhang, Q., Yang, Z., An, F., Nie, H., Wang, H., et al. (2022). CircEZH2/miR-133b/IGF2BP2 aggravates colorectal cancer progression via enhancing the stability of m(6)A-modified CREB1 mRNA. Mol. Cancer 21 (1), 140. doi:10.1186/s12943-022-01608-7
Yue, C., Chen, J., Li, Z., Li, L., Chen, J., and Guo, Y. (2020). microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J. Exp. Clin. Cancer Res. 39 (1), 240. doi:10.1186/s13046-020-01731-7
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10. doi:10.1038/s41421-018-0019-0
Zeng, Q., Wan, H., Zhao, S., Xu, H., Tang, T., Oware, K. A., et al. (2020). Role of PIWI-interacting RNAs on cell survival: proliferation, apoptosis, and cycle. IUBMB Life 72 (9), 1870–1878. doi:10.1002/iub.2332
Zepecki, J. P., Karambizi, D., Fajardo, J. E., Snyder, K. M., Guetta-Terrier, C., Tang, O. Y., et al. (2021). miRNA-mediated loss of m6A increases nascent translation in glioblastoma. PLoS Genet. 17 (3), e1009086. doi:10.1371/journal.pgen.1009086
Zhang, H., Liu, Y., Wang, W., Liu, F., Wang, W., Su, C., et al. (2022a). ALKBH5-mediated m(6)A modification of lincRNA LINC02551 enhances the stability of DDX24 to promote hepatocellular carcinoma growth and metastasis. Cell Death Dis. 13 (11), 926. doi:10.1038/s41419-022-05386-4
Zhang, M., Wang, J., Jin, Y., Zheng, Q., Xing, M., Tang, Y., et al. (2022b). YTHDF2-mediated FGF14-AS2 decay promotes osteolytic metastasis of breast cancer by enhancing RUNX2 mRNA translation. Br. J. Cancer 127 (12), 2141–2153. doi:10.1038/s41416-022-02006-y
Zhang, Z., Zhu, H., and Hu, J. (2021). CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis. 12 (2), 219. doi:10.1038/s41419-021-03486-1
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49 (1), 18–29. doi:10.1016/j.molcel.2012.10.015
Zheng, H., Zhu, M., Li, W., Zhou, Z., and Wan, X. (2022). m(5) C and m(6) A modification of long noncoding NKILA accelerates cholangiocarcinoma progression via the miR-582-3p-YAP1 axis. Liver Int. 42 (5), 1144–1157. doi:10.1111/liv.15240
Zheng, Z. Q., Li, Z. X., Zhou, G. Q., Lin, L., Zhang, L. L., Lv, J. W., et al. (2019). Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 79 (18), 4612–4626. doi:10.1158/0008-5472.CAN-19-0799
Zhu, L., Zhu, Y., Han, S., Chen, M., Song, P., Dai, D., et al. (2019). Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 10 (6), 383. doi:10.1038/s41419-019-1585-2
Zhu, P., He, F., Hou, Y., Tu, G., Li, Q., Jin, T., et al. (2021). A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene 40 (9), 1609–1627. doi:10.1038/s41388-020-01638-9
Zhu, S., Wang, J. Z., Chen, D., He, Y. T., Meng, N., Chen, M., et al. (2020). An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat. Commun. 11 (1), 1685. doi:10.1038/s41467-020-15403-9
Keywords: biomarker, cancer, genetic regulation, m6A modification, non-coding RNA
Citation: Liu S and Xiang D (2023) New understandings of the genetic regulatory relationship between non-coding RNAs and m6A modification. Front. Genet. 14:1270983. doi: 10.3389/fgene.2023.1270983
Received: 01 August 2023; Accepted: 20 November 2023;
Published: 06 December 2023.
Edited by:
Sandip Mukherjee, Washington University in St. Louis, United StatesReviewed by:
Jiangbo Wei, The University of Chicago, United StatesCopyright © 2023 Liu and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayong Xiang, eGR5NzVAc211LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.