
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 22 January 2024
Sec. Cancer Genetics and Oncogenomics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1260352
Observational studies have shown an association between liver dysfunction and hepatocellular carcinoma (HCC), but the causality relationship between them is unclear. We aimed to determine whether there is a bidirectional causal relationship between liver function indicators (alanine aminotransferase, ALT; aspartate aminotransferase, AST; alkaline phosphatase, ALP; γ-glutamyltransferase, GGT) and HCC. Our two-sample Mendelian randomization (MR) study acquired single nucleotide polymorphisms (SNPs) associated with liver function indicators (ALT, n = 134,182; AST, n = 134,154; GGT, n = 118,309; ALP, n = 105,030) and with HCC (n = 197,611) from publicly available genome-wide association studies (GWAS) of East Asian ancestry in Japan (BioBank Japan, BBJ). Univariable MR analyses were performed to identify whether the genetic evidence of exposure was significantly associated with outcome. Multivariable MR analysis was conducted to estimate the independent effects of exposures on outcome. Univariable MR analysis indicated that the level of ALT, AST, and GGT was the risk factor for HCC incidence. Meanwhile, multivariable MR analysis revealed that AST was an independent risk factor for HCC. The hazard ratio (HR) of the probability of HCC was 3.045 [95% confidence interval (95%CI), 1.697–5.463, p = 0.003] for AST. The results of reverse MR analyses showed that gene-predictive HCC incidence could increase the levels of AST (HR = 1.031, 95%CI: 1.009–1.054, p = 2.52 × 10−4) and ALT (HR = 1.040, 95%CI: 1.019–1.063, p = 0.005). Meanwhile, HCC may be negatively correlated with ALP levels (HR = 0.971, 95%CI: 0.947–0.995, p = 0.018). This study provides evidence to support that genetically predicted higher levels of AST are related to increased risk of HCC, with no strong evidence of a causal effect of genetically predicted ALP, ALP, and GGT on HCC. In addition, genetic predisposition to HCC could influence blood concentration of ALT, AST, and ALP. Thus, this may create a vicious cycle.
The burden of hepatocellular carcinoma (HCC) is an important healthcare problem and continues to be the most common histologic type of primary liver cancer (Toh et al., 2023). Japan has one of the highest rates of HCC in the world, with an estimated 34,000 HCC-related deaths in 2019 (Sung et al., 2021). The prevalence of HCC has also increased in recent years. In recent decades, considerable progress has been made in the study of the epidemiology, risk factors, molecular characteristics, and pathogenesis of HCC. Epidemiological and experimental studies have identified several major risk factors associated with hepatocarcinogenesis, including chronic hepatitis B/C, type 2 diabetes mellitus (T2DM), metabolic liver disease (particularly nonalcoholic fatty liver disease), and cirrhosis. Targeting these risk factors, therapeutic measures such as direct antivirals, and the use of metformin, are associated with risk reduction of HCC, and can even delay the postoperative recurrence of HCC (Wu et al., 2016; Tseng, 2018; Zhang et al., 2021). Identifying new risk factors and taking appropriate treatment measures will contribute to improving the prognosis of patients with HCC.
Serum liver enzymes, such as alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), and γ-glutamyltransferase (GGT), are routinely measured clinical markers that represent different dimensions of liver dysfunction (Pratt and Kaplan, 2000). Physicians generally use significant elevations of liver enzyme levels as complementary markers to aid the diagnosis of various diseases. For example, elevations of ALT and AST may indicate the presence of hepatocellular predominant disorders while elevations of ALP and GGT may implicate cholestatic predominant diseases (Giannini et al., 2005). Epidemiological studies have shown the associations between abnormally high liver enzyme levels and risks and mortalities of many diseases, including HCC (Hann et al., 2012; Wu et al., 2022; Reddy et al., 2023). Several studies have shown that high ALT or AST levels are independent risk factors for the development of cirrhosis and HCC (Kawamura et al., 2012; Hernaez et al., 2013). Liver function abnormalities were also an independent prognostic indicator in patients with HCC (Zhang et al., 2019). Moreover, liver dysfunction may also affect the development of HCC in an indirect fashion (De Silva et al., 2019). Observational studies usually show that some liver function indicators, such as ALT, AST, ALP, and GGT, are associated with high risk of cardiovascular disease and type 2 diabetes, which are risk factors for HCC. Growing evidence shows that liver enzyme levels play important roles in HCC pathogenesis, such as tumorigenesis, local tumor progression, and metastasis. Due to the methodological limitations of traditional observational studies, including confounding and measurement error, these associations may be biased. Since the causal associations between liver function indicators and HCC risk have not been thoroughly investigated, identifying host factors predisposing individuals to HCC is urgently needed to improve primary prevention and develop treatment strategies.
Mendelian randomization (MR) is a method of examining the causal effect of a modifiable exposure to disease by using measured variation in genes of known function in observational data. Because the genotype of an individual is determined at conception and cannot be changed, there is no possibility of reverse causation or confounding bias being responsible for an association between genotype and disease (Davey Smith and Hemani, 2014). In recent years, many MR studies have emerged to provide clinical evidence (Chen et al., 2022; Liu et al., 2022; Pan et al., 2022). This proves that MR is a reliable research method to solve some problems, including finding risk factors for diseases.
We have used the largest available data sets to interrogate the potential effect of liver dysfunction, proxied by multiple biomarkers (ALT, AST, ALP, and GGT), on HCC risk. In addition, we have investigated whether HCC affects circulating liver function markers.
We explored the relationship of four liver function markers (plasma concentration of ALT, AST, ALP, and GGT) with HCC. We also used MR to investigate whether predisposition to HCC is likely to have an impact on circulating ALT, AST, ALP, and GGT. The hypotheses, study design, and data sources used are detailed in Figure 1.
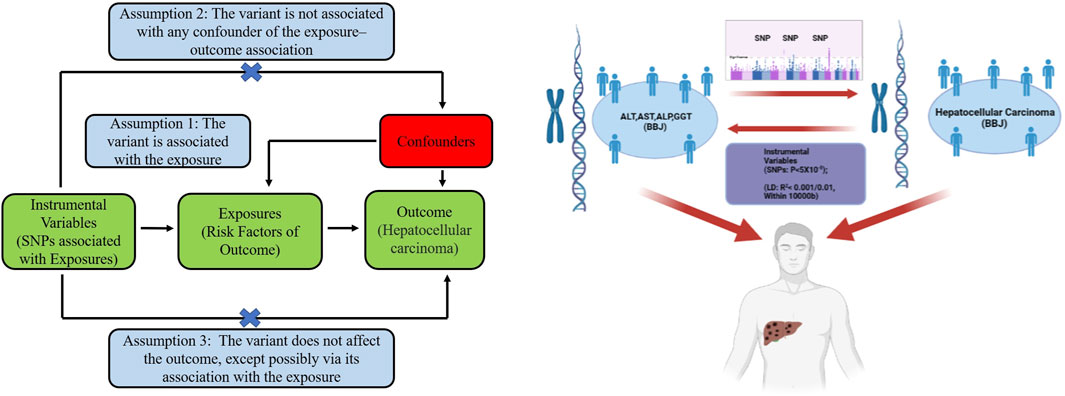
FIGURE 1. The three basic assumptions of Mendelian randomization (left) and the main design of this study (right). ALT: alanine aminotransferase; AST: aspartate aminotransferase; ALP: alkaline phosphatase; GGT: γ-glutamyltransferase. BBJ: Biobank Japan; LD: linkage disequilibrium; SNP: single nucleotide polymorphism.
The exposure-related single nucleotide polymorphisms (SNPs) used in this study were obtained from the Biobank Japan Project (BBJ). BBJ started at the Institute of Medical Science, University of Tokyo, in 2003. To date, the BBJ Project has collected data on approximately 200,000 individuals with 47 different diseases. The genome-wide association studies (GWAS) summary statistics of liver function indicators were extracted from a study conducted by Masahiro Kanai (Kanai et al., 2018), who tested 5,961,600 autosomal variants and 147,353 X-chromosome variants for association with 58 traits in 162,255 Japanese individuals with East-Asian ancestry and identified 1,407 trait-associated loci (p < 5.0 × 10−8), 679 of which were novel. The GWAS summary statistics of liver function indicators in our study included 4 phenotypes: ALT, AST, ALP, and GGT. For ALT GWAS, the participants were 134,182 Japanese individuals. The GWAS summary statistics of AST and GGT comprised 134,154 and 118,309 Japanese individuals. For ALP GWAS, the study included 105,030 Japanese individuals. This study included 126,319 Japanese individuals and 6,108,953 SNPs. They focused on identifying different loci associated with liver function enzymes (Table 1).
Summary-level statistical data for HCC were also obtained from a large GWAS of individuals with East-Asian ancestry in Japan. This study was conducted by Ishigaki et al. (2020) and aimed to address the problem that many participants in current genetic studies are of European ancestry. The study elucidated polygenic disease biology in the East Asian population by conducting a GWAS with 212,453 Japanese individuals across 42 disease traits. In this study, they adjusted for covariates including age, sex, and top five principal components (Table 1).
MR is the use of genetic variants in non-experimental data to make causal inferences about the effect of an exposure on an outcome. In MR, genetic variant(s) are used as instrumental variables (IVs) for assessing the causal effect of the exposure on the outcome. The fundamental conditions for a genetic variant to satisfy to be an IV are as follows: 1) The IVs are associated with the exposures, 2) IVs are not associated with outcomes by means other than exposures, and 3) IVs cannot directly affect outcomes, if only through exposure. We selected the significant genetic variants associated with the exposures from GWAS (significant level p < 5 × 10−8). The minor allele frequency of the SNPs was >0.01. The SNPs used in our study were those that satisfied the linkage disequilibrium in the given genome region and the SNPs with palindromic structure were removed. When evaluating the causal relationship between liver function indicators and HCC, the threshold was r2 < 0.001 and kb > 10,000. When evaluating reverse causality, the threshold was r2 < 0.01 and kb > 10,000. For each variant included in the genetic instruments, variance (R2) represents the variance in exposure explained by the genetic variant and was calculated using the formula R2 = 2 × MAF × (1−MAF) × beta2 (where MAF represents the effect allele frequency and beta represents the effect estimate of the genetic variant in the exposure GWAS) (Palmer et al., 2012). F statistics (F = beta2/se2) were used to evaluate the remaining SNPs’ power. We calculated F statistics for each SNP. SNPs with F statistics <10 were identified to be weak instruments and we excluded them (Figure 1). The SNPs that were included in this analysis are listed in Supplementary Table S1.
Inverse variance weighting (IVW) is a method of weighted average of random variables, where each random variable is weighted by the inverse of its variance. In this study, IVW was the main method adopted in the statistical analysis. Furthermore, the MR-Egger and weighted-median (WM) methods were used as supplements to the IVW method. For univariable MR, IVW, MR-Egger, and WM were used to estimate the effect of exposures on outcomes. For multivariable MR, regression-based IVW was used. The MR-PRESSO global test, outlier test, and distortion test were used to identify and remove SNPs with horizontal pleiotropy. If any outliers existed, we restarted an evaluation of the causal relationship. The intercept test of MR-Egger and Cochran’s Q test in the IVW and MR-Egger models were used to assess pleiotropy and the heterogeneity. In the case of pleiotropy, we preferred to use the MR-Egger. If the p-value in Cochran’s Q test was significant (p < 0.05), the WM model was used to analyze the statistics. Otherwise, a fixed-effects model was performed. Moreover, an online calculator was used to test the statistical power of this study (https://cnsgenomics.shinyapps.io/mRnd/). Genetic variants associated with exposures at genome-wide significance (p < 5 × 10−8) were then LD-pruned using the clump_data command in the “TwoSampleMR” package in R to identify an independent set of variants to serve as a genetic instrument for exposures. The univariable MR analysis was performed by R packages “Two Sample MR” and “Mendelian randomization”. The multivariable MR was performed by R packages “multivariable Mendelian randomization” (“MVMR”) and “Mendelian randomization”. The MR-PRESSO test was conducted using the R package “MRPRESSO”. Data visualization was conducted using R software 4.1.1 (https://www.r-project.org/).
To investigate the causal effects of the liver function indicators on HCC, we constructed a genetic instrument for liver function indicators using 10–49 independent SNPs associated with the above five traits at a genome-wide level of significance (p < 5 × 10−8), which accounted for 1.00–17.35% of the variability in exposures. The mean F-statistic ranged from 34.62 to 168.64, which indicated that no weak instrument bias existed.
In the univariable MR analysis stage, IVW was the main analysis method for MR. Our MR analysis indicated that there was strong evidence to support causality between higher levels of ALT, AST, and GGT with risk of HCC.
The hazard ratios of the probability of HCC were 1.890 (95% confidence interval (CI), 1.209-2.954, p = 0.005) for ALT, 2.909 (95%CI: 1.902-4.451, p = 8.55 × 10−7) for AST, 1.300 (95%CI: 1.048-1.611, p = 0.016) for GGT, and 0.908 (95%CI: 0.821–1.169, p = 0.818) for ALP (Table 2).
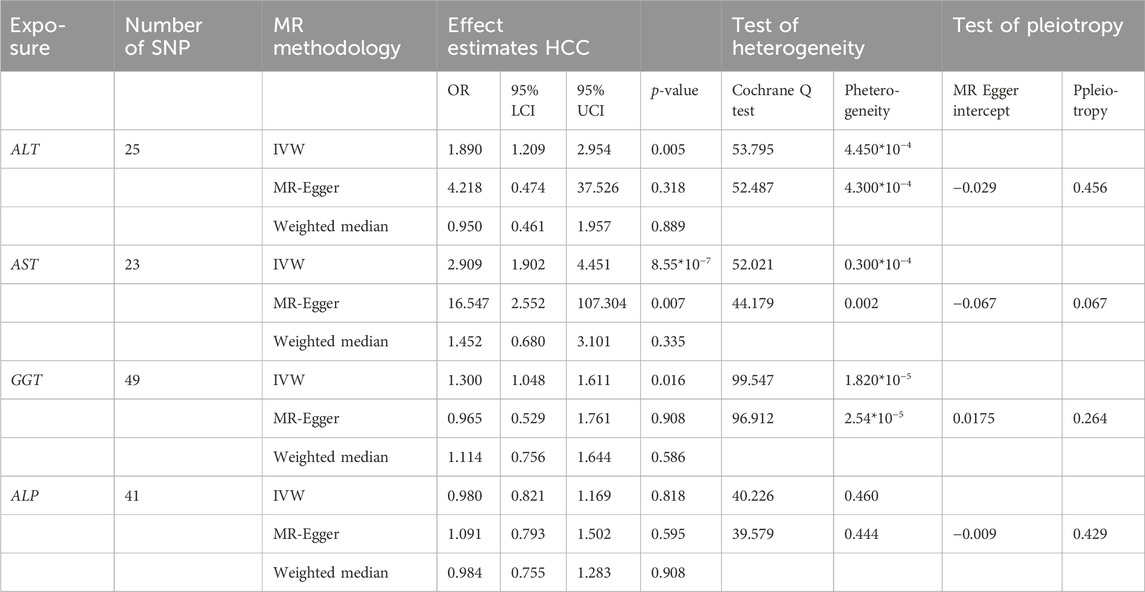
TABLE 2. The effect estimates, test of heterogeneity and test of pleiotropy of liver function on HCC.
Furthermore, the causal relationship between liver function indicators and HCC was explored by conducting multivariable MR analysis. Among the four traits, we had observed that ASP had a causal effect on HCC occurrence when using SNPs-associated exposures. After the adjustment of other traits, GGT and ALT become non-significant. Multivariable MR analysis revealed that the hazard ratios of the probability of HCC were 3.045 (95%CI: 1.697-5.463, p = 2.77 × 10−4) for AST, 1.312 (95%CI: 0.713-2.414, p = 0.385) for ALT, 0.980 (95%CI: 0.0.779-1.232, p = 0.860) for ALP, and 1.296 (95%CI: 0.980-1.714, p = 0.072) for GGT (Table 3).
In order to explore the reverse causality between HCC and liver function indicators, we utilized the data from publicly available large-scale GWAS and deemed that genetically predicted HCC was associated with the levels of ALT, AST, and ALP. Specifically, HCC was associated with higher levels of AST and ALT. In contrast, HCC may have a causal relationship with lower levels of ALP. The MR effects of HCC on liver function indicators were: ALT (OR = 1.031, p = 0.005, 95%CI: 1.009-1.054), AST (OR = 1.040, p = 2.52 × 10−4, CI: 1.019-1.063), ALP (OR = 0.971, p = 0.018, CI: 0.947-0.995), and GGT (OR = 1.014, p = 0.242, CI: 0.991-1.038) (Table 4).
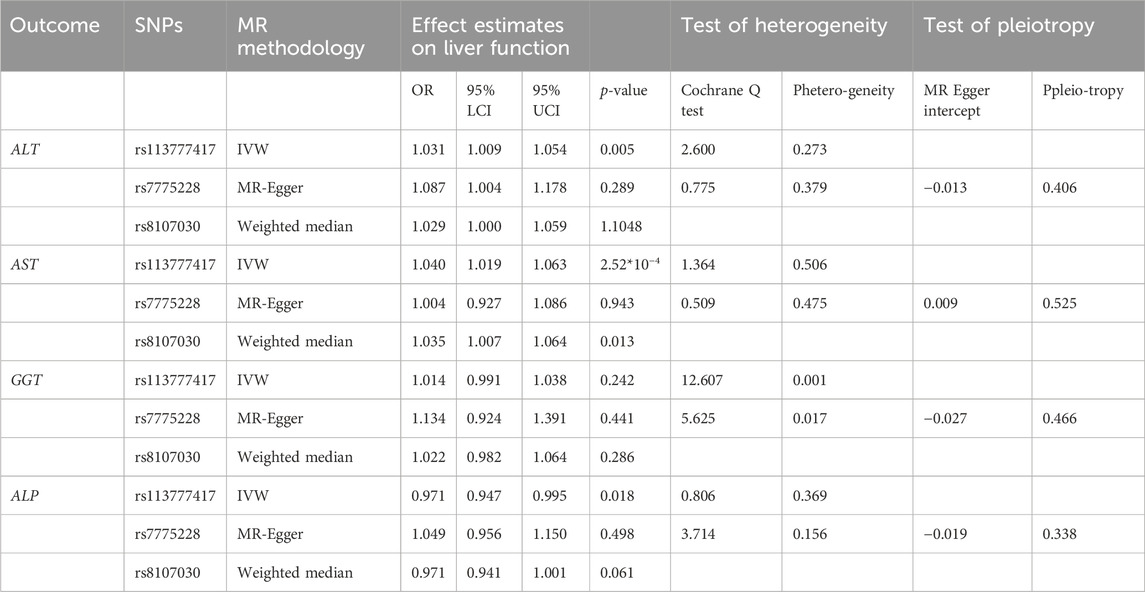
TABLE 4. The effect estimates, test of heterogeneity, and test of pleiotropy of HCC on liver function.
The effects between SNPs-associated exposures and outcomes were visualized using R software.
The pleiotropy of results was not tested in our study. MR-Egger intercept represented the average level of pleiotropy of all SNPs associated exposure. No significant horizontal pleiotropic effects were detected in the MR-Egger test (for the intercept of MR-Egger, all p values were more than 0.05). All the results of these exposures were MR-PRESSO-corrected results if outliers were detected. The statistical power of these exposures was 100%.
HCC causes a heavy disease burden and is the fourth leading cause of cancer-related deaths worldwide (Siegel et al., 2023). Risk factors for the occurrence of HCC are numerous, including HBV and HCV infection, alcohol consumption, aflatoxin B1, and nonalcoholic fatty liver disease (Pan et al., 2022; Liu et al., 2023a; Pan et al., 2023). These conditions are associated with liver dysfunction and can lead to fibrosis, cirrhosis, and eventually HCC (Kotsiliti et al., 2023). Most studies exploring the risk factors for HCC development are based on observational studies and clinical experience. However, the major disadvantage of an observational study is that its validity is threatened by confounding by indication (De Nardi et al., 2022). Furthermore, studies have shown that genetic factors may also independently modulate HCC risk (Shimokawa et al., 2020). Human HCC genome sequencing studies have begun to uncover relationships between risk factors and mutated genes (Sun et al., 2021). MR studies use genetic variants as proxies of non-genetic risk factors to assess whether a risk factor is causally related to a disease. Although MR has already been used successfully in cancer epidemiology to estimate risk factors for overall cancer risk and cancer mortality, it has rarely been applied in the field of HCC study (Yarmolinsky et al., 2022; Wang et al., 2023).
Plasma concentrations of liver enzymes (ALT, AST, ALP, and GGT) are routinely measured clinical markers that represent different dimensions of liver dysfunction. ALT, located in the cytosol, and AST, located in the mitochondria, are released from damaged hepatic cells into the blood after hepatocellular injury or death (Song et al., 2012). ALT and AST are potentially useful surrogates for alcohol-induced liver disease and nonalcoholic fatty liver disease (NAFLD), defined as hepatic steatosis in the absence of excessive alcohol consumption (Kim et al., 2023). ALP is present in the ducts of the liver, and GGT is located on liver cell membranes (Inoue et al., 2023). The combined elevation of ALP and GGT can indicate obstructive or cholestatic liver disease, where bile is not properly transported from the liver because of an obstruction of the bile duct (Takahashi et al., 2023). GGT is also an indicator of alcohol use (De Silva et al., 2019). We conducted this bidirectional MR study to evaluate the potential causal effects between four liver function indicators (ALT, AST, GGT, and ALP) and HCC risk from a genetic perspective and to investigate whether predisposition to HCC might instead lead to liver dysfunction. Our findings from the MR analyses show evidence that genetic predisposition to higher circulating AST is related to higher risk of HCC. There was no strong evidence of a causal effect of genetically predicted ALP, ALP and GGT on HCC. In addition, genetic predisposition to HCC appeared to influence blood concentration of ALT, AST, and ALP. The present bidirectional MR study found that the main indicator of liver dysfunction (AST) increased the risk of HCC, suggesting that liver dysfunction exacerbates hepatocarcinogenesis and HCC could aggravate liver function damage. This may create a vicious cycle.
HCC patients often experience liver dysfunction, thus limiting the application of conventional therapies (Liu et al., 2023b). Therefore, it is particularly important to evaluate liver function in clinical practice. Nevertheless, the molecular mechanisms through which risk factors contribute to hepatocarcinogenesis, for the most part, remain poorly understood. Multiple studies have shown a direct role in liver function abnormalities in hepatic carcinogenesis (Kasprzak and Adamek, 2019). Several studies have shown that high ALT levels are an independent risk factor for the development of cirrhosis and HCC (Ogasawara et al., 2020; Dajti et al., 2021; Tahata et al., 2022). Liver function abnormalities were also an independent prognostic indicator in patients with HCC (Seong et al., 2022; Wong et al., 2023). Moreover, liver dysfunction may also affect the development of HCC in an indirect fashion. Observational studies usually show that some liver function indicators, such as ALT, AST, ALP, and GGT, are associated with a high risk of cardiovascular disease and type 2 diabetes, which are risk factors for HCC (Fard et al., 2022). Consequently, finding effective therapies for liver dysfunction in high-risk populations for HCC is a topic of long-standing interest and importance.
Our bidirectional MR provided comprehensive evidence to interrogate the potential effect of liver dysfunction on HCC risk. However, there are still some limitations in the present study. The limitations of available data hindered our ability to make strong conclusions about the potential association between liver dysfunction and HCC risk. First, because all the included data from GWAS used in this study were primarily focused on participants of East-Asian ancestry, there was bias against other ethnic groups with different lifestyles and cultural backgrounds. Second, all results were derived from genetic levels. There was a lack of prospective multicenter studies to confirm the causal relationship between liver dysfunction and HCC risk. Therefore, more studies are still needed to confirm our conclusions. Finally, although we used large-scale genetic data to obtain instrumental variables for our study, we did not manually check the validity of our instrument. However, we performed sensitivity analyses to assess horizontal pleiotropy and found that our results were robust to potential violations of this assumption.
This study provides a novel finding that individuals with East Asian ancestry who have higher genetic levels of AST are likely at risk of HCC. In addition, genetic predisposition to HCC could influence blood concentration of ALT, AST, and ALP. This may create a vicious cycle. Clinicians should raise awareness of AST in clinical practice.
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
SQ: Writing–original draft. JW: Validation, Writing–review and editing. HY: Conceptualization, Investigation, Writing–original draft. JH: Resources, Visualization, Writing–original draft. SL: Investigation, Conceptualization, Writing–review and editing. YD: Writing–original draft, Validation.
The author(s) declare that no financial support was received for the research in this article.
The authors thank all investigators and participants from Biobank Japan for sharing genetic association estimates for HCC. Furthermore, we would like to thank all investigators contributing to GWAS of liver function indicators.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1260352/full#supplementary-material
Chen, L., Yang, H., Li, H., He, C., Yang, L., and Lv, G. (2022). Insights into modifiable risk factors of cholelithiasis: a Mendelian randomization study. Hepatology 75 (4), 785–796. doi:10.1002/hep.32183
Dajti, E., Marasco, G., Ravaioli, F., Colecchia, L., Ferrarese, A., Festi, D., et al. (2021). Risk of hepatocellular carcinoma after HCV eradication: determining the role of portal hypertension by measuring spleen stiffness. JHEP Rep. 3 (3), 100289. doi:10.1016/j.jhepr.2021.100289
Davey Smith, G., and Hemani, G. (2014). Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23 (R1), R89–R98. doi:10.1093/hmg/ddu328
De Nardi, L., Simeone, R., Torelli, L., Maestro, A., Zanon, D., Barbi, E., et al. (2022). Pediatric males receiving hematopoietic stem cell transplant lose their male disadvantage in disease risk after the procedure: a retrospective observational study. Int. J. Cancer 151 (2), 191–199. doi:10.1002/ijc.33978
De Silva, N. M. G., Borges, M. C., Hingorani, A. D., Engmann, J., Shah, T., Zhang, X., et al. (2019). Liver function and risk of type 2 diabetes: bidirectional mendelian randomization study. Diabetes 68 (8), 1681–1691. doi:10.2337/db18-1048
Fard, M. T., Najafi, F., Rezaeian, S., Kohsari, M., and Moradinazar, M. (2022). Association between serum liver enzymes and hypertension using propensity score matching analysis: evidence from a large Kurdish prospective cohort study. BMC Cardiovasc Disord. 22 (1), 476. doi:10.1186/s12872-022-02884-3
Giannini, E. G., Testa, R., and Savarino, V. (2005). Liver enzyme alteration: a guide for clinicians. Cmaj 172 (3), 367–379. doi:10.1503/cmaj.1040752
Hann, H. W., Wan, S., Myers, R. E., Hann, R. S., Xing, J., Chen, B., et al. (2012). Comprehensive analysis of common serum liver enzymes as prospective predictors of hepatocellular carcinoma in HBV patients. PLoS One 7 (10), e47687. doi:10.1371/journal.pone.0047687
Hernaez, R., Yeh, H. C., Lazo, M., Chung, H. M., Hamilton, J. P., Koteish, A., et al. (2013). Elevated ALT and GGT predict all-cause mortality and hepatocellular carcinoma in Taiwanese male: a case-cohort study. Hepatol. Int. 7 (4), 1040–1049. doi:10.1007/s12072-013-9476-6
Inoue, K., Fujita, R., Nagahara, T., Murakami, S., Nagai, Y., Moriwake, R., et al. (2023). Predictive factors for recovery from alcoholic liver failure. Acta Med. Okayama 77 (2), 169–177. doi:10.18926/AMO/65146
Ishigaki, K., Akiyama, M., Kanai, M., Takahashi, A., Kawakami, E., Sugishita, H., et al. (2020). Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet. 52 (7), 669–679. doi:10.1038/s41588-020-0640-3
Kanai, M., Akiyama, M., Takahashi, A., Matoba, N., Momozawa, Y., Ikeda, M., et al. (2018). Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 50 (3), 390–400. doi:10.1038/s41588-018-0047-6
Kasprzak, A., and Adamek, A. (2019). Mucins: the old, the new and the promising factors in hepatobiliary carcinogenesis. Int. J. Mol. Sci. 20 (6), 1288. doi:10.3390/ijms20061288
Kawamura, Y., Arase, Y., Ikeda, K., Seko, Y., Imai, N., Hosaka, T., et al. (2012). Large-scale long-term follow-up study of Japanese patients with non-alcoholic Fatty liver disease for the onset of hepatocellular carcinoma. Am. J. Gastroenterol. 107 (2), 253–261. doi:10.1038/ajg.2011.327
Kim, B. K., Bergstrom, J., Loomba, R., Tamaki, N., Izumi, N., Nakajima, A., et al. (2023). Magnetic resonance Elastography-Based prediction model for hepatic decompensation in NAFLD; a Multi-Center cohort study. Hepatology 78, 1858–1866. doi:10.1097/HEP.0000000000000470
Kotsiliti, E., Leone, V., Schuehle, S., Govaere, O., Li, H., Wolf, M. J., et al. (2023). Intestinal B-cells license metabolic T-cell activation in NASH microbiota/antigen-independently and contribute to fibrosis by IgA-FcR signalling. J. Hepatol. 79, 296–313. doi:10.1016/j.jhep.2023.04.037
Liu, H., Han, C. L., Tian, B. W., Ding, Z. N., Yang, Y. F., et al. (2023a). Tenofovir versus entecavir on the prognosis of hepatitis B virus-related hepatocellular carcinoma: a systematic review and meta-analysis. Expert Rev. Gastroenterol. Hepatol. 17, 623–633. doi:10.1080/17474124.2023.2212161
Liu, H., Yang, C. C., Ma, Y. L., Yang, Y. F., Yan, L. J., Ding, Z. N., et al. (2023b). Identification of the most effective subgroup of advanced hepatocellular carcinoma from immune checkpoint blocker treatment: a meta-analysis. Immunotherapy 15 (9), 669–678. doi:10.2217/imt-2022-0114
Liu, Y., Xu, H., Zhao, Z., Dong, Y., Wang, X., and Niu, J. (2022). No evidence for a causal link between Helicobacter pylori infection and nonalcoholic fatty liver disease: a bidirectional Mendelian randomization study. Front. Microbiol. 13, 1018322. doi:10.3389/fmicb.2022.1018322
Ogasawara, N., Saitoh, S., Akuta, N., Sezaki, H., Suzuki, F., Fujiyama, S., et al. (2020). Advantage of liver stiffness measurement before and after direct-acting antiviral therapy to predict hepatocellular carcinoma and exacerbation of esophageal varices in chronic hepatitis C. Hepatol. Res. 50 (4), 426–438. doi:10.1111/hepr.13467
Palmer, T. M., Lawlor, D. A., Harbord, R. M., Sheehan, N. A., Tobias, J. H., Timpson, N. J., et al. (2012). Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med. Res. 21 (3), 223–242. doi:10.1177/0962280210394459
Pan, G. Q., Jiao, Y., Meng, G. X., Dong, Z. R., and Li, T. (2023). The relationship between the serum lipid profile and hepatocellular carcinoma in east Asian population: a mendelian randomization study. Heliyon 9 (6), e17126. doi:10.1016/j.heliyon.2023.e17126
Pan, G. Q., Yang, C. C., Shang, X. L., Dong, Z. R., and Li, T. (2022). The causal relationship between white blood cell counts and hepatocellular carcinoma: a Mendelian randomization study. Eur. J. Med. Res. 27 (1), 278. doi:10.1186/s40001-022-00900-y
Pratt, D. S., and Kaplan, M. M. (2000). Evaluation of abnormal liver-enzyme results in asymptomatic patients. N. Engl. J. Med. 342 (17), 1266–1271. doi:10.1056/NEJM200004273421707
Reddy, K. R., McLerran, D., Marsh, T., Parikh, N., Roberts, L. R., Schwartz, M., et al. (2023). Incidence and risk factors for hepatocellular carcinoma in cirrhosis: the multicenter hepatocellular carcinoma early detection strategy (HEDS) study. Gastroenterology 165 (4), 1053–1063.e6. doi:10.1053/j.gastro.2023.06.027
Seong, G., Sinn, D. H., Kang, W., Gwak, G. Y., Choi, M. S., Lee, J. H., et al. (2022). Age and fibrosis index for the prediction of hepatocellular carcinoma risk in patients with high hepatitis B virus DNA but normal alanine aminotransferase. Eur. J. Gastroenterol. Hepatol. 34 (1), 69–75. doi:10.1097/MEG.0000000000001915
Shimokawa, M., Yoshizumi, T., Itoh, S., Iseda, N., Sakata, K., Yugawa, K., et al. (2020). Modulation of Nqo1 activity intercepts anoikis resistance and reduces metastatic potential of hepatocellular carcinoma. Cancer Sci. 111 (4), 1228–1240. doi:10.1111/cas.14320
Siegel, R. L., Miller, K. D., Wagle, N. S., and Jemal, A. (2023). Cancer statistics, 2023. CA Cancer J. Clin. 73 (1), 17–48. doi:10.3322/caac.21763
Song, M., Schuschke, D. A., Zhou, Z., Chen, T., Pierce, W. M., Wang, R., et al. (2012). High fructose feeding induces copper deficiency in Sprague-Dawley rats: a novel mechanism for obesity related fatty liver. J. Hepatol. 56 (2), 433–440. doi:10.1016/j.jhep.2011.05.030
Sun, Y., Wu, L., Zhong, Y., Zhou, K., Hou, Y., Wang, Z., et al. (2021). Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 184 (2), 404–421.e16. doi:10.1016/j.cell.2020.11.041
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Tahata, Y., Sakamori, R., Yamada, R., Kodama, T., Hikita, H., Nozaki, Y., et al. (2022). Risk of hepatocellular carcinoma after sustained virologic response in hepatitis C virus patients without advanced liver fibrosis. Hepatol. Res. 52 (10), 824–832. doi:10.1111/hepr.13806
Takahashi, Y., Seko, Y., Yamaguchi, K., Takeuchi, K., Yano, K., Kataoka, S., et al. (2023). Gamma-glutamyl transferase predicts pemafibrate treatment response in non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 38, 1743–1749. doi:10.1111/jgh.16222
Toh, M. R., Wong, E. Y. T., Wong, S. H., Ng, A. W. T., Loo, L. H., Chow, P. K. H., et al. (2023). Global epidemiology and genetics of hepatocellular carcinoma. Gastroenterology 164 (5), 766–782. doi:10.1053/j.gastro.2023.01.033
Tseng, C. H. (2018). Metformin and risk of hepatocellular carcinoma in patients with type 2 diabetes. Liver Int. 38 (11), 2018–2027. doi:10.1111/liv.13872
Wang, Z., Lu, J., and Hu, J. (2023). Association between antihypertensive drugs and hepatocellular carcinoma: a trans-ancestry and drug-target Mendelian randomization study. Liver Int. 43 (6), 1320–1331. doi:10.1111/liv.15566
Wong, Y. J., Nguyen, V. H., Yang, H. I., Li, J., Le, M. H., Wu, W. J., et al. (2023). Impact of fatty liver on long-term outcomes in chronic hepatitis B: a systematic review and matched analysis of individual patient data meta-analysis. Clin. Mol. Hepatol. 29, 705–720. doi:10.3350/cmh.2023.0004
Wu, C. K., Chang, K. C., Hung, C. H., Tseng, P. L., Lu, S. N., Chen, C. H., et al. (2016). Dynamic α-fetoprotein, platelets and AST-to-platelet ratio index predict hepatocellular carcinoma in chronic hepatitis C patients with sustained virological response after antiviral therapy. J. Antimicrob. Chemother. 71 (7), 1943–1947. doi:10.1093/jac/dkw097
Wu, H. C., Jeng, W. J., Pan, M. H., Hsieh, Y. C., Lu, S. N., Chen, C. J., et al. (2022). Incidence of hepatocellular carcinoma in a community-based Taiwanese population without chronic HBV/HCV infection. JHEP Rep. 4 (2), 100410. doi:10.1016/j.jhepr.2021.100410
Yarmolinsky, J., Díez-Obrero, V., Richardson, T. G., Pigeyre, M., Sjaarda, J., Paré, G., et al. (2022). Genetically proxied therapeutic inhibition of antihypertensive drug targets and risk of common cancers: a mendelian randomization analysis. PLoS Med. 19 (2), e1003897. doi:10.1371/journal.pmed.1003897
Zhang, L. X., Lv, Y., Xu, A. M., and Wang, H. Z. (2019). The prognostic significance of serum gamma-glutamyltransferase levels and AST/ALT in primary hepatic carcinoma. BMC Cancer 19 (1), 841. doi:10.1186/s12885-019-6011-8
Keywords: AST, ALT, GGT, ALP, hepatocellular carcinoma, mendelian randomization
Citation: Qin S, Wang J, Yuan H, He J, Luan S and Deng Y (2024) Liver function indicators and risk of hepatocellular carcinoma: a bidirectional mendelian randomization study. Front. Genet. 14:1260352. doi: 10.3389/fgene.2023.1260352
Received: 20 September 2023; Accepted: 29 December 2023;
Published: 22 January 2024.
Edited by:
Zhifei Cao, Second Affiliated Hospital of Soochow University, ChinaReviewed by:
Aaron Balasingam Koenig, INOVA Health System, United StatesCopyright © 2024 Qin, Wang, Yuan, He, Luan and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiqing Yuan, cm15eWFycm9ubHNqQHdmbWMuZWR1LmNu; Yan Deng, ZGVuZ3lhbjYxNEAxMjYuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.