- 1College of Chemistry, Chemical Engineering and Resource Utilization, Northeast Forestry University, Harbin, China
- 2Biological Science and Technology Department, Heilongjiang Vocational College for Nationalities, Harbin, China
- 3College of Life Science, Kim Il Sung University, Pyongyang, Democratic People’s Republic of Korea
- 4Botany and Microbiology Department, Faculty of Science, Menoufia University, Shebeen El-Kom, Egypt
Acanthopanax senticosus (Rupr. et Maxim.) Harms is a perennial shrub of the Acanthopanax genus in the Araliaceae family and has a high medicinal value. The application of zinc fertilizer can improve the yield and quality of medicinal materials. However, there are limited reports on approaches to increase the content of medicinal components in A. senticosus, hindering the improvement of its medicinal quality. In this study, A. senticosus was treated with 0.1% (LZn) and 0.4% (HZn) zinc sprayed on the leaf surface. The effects of zinc treatment on the medicinal components in the roots of A. senticosus were analyzed by comprehensive metabolomics and transcriptomics analyses. A total of 316 metabolites were detected, with a prevailing occurrence of terpenoids and phenylpropanoids. We identified metabolites related to the medicinal components that were upregulated after Zn treatment, including 43 terpenoids, 19 phenylpropanoids, eight phenols, and three flavonoids. Combining differential gene expression and K-means analysis, we found 95, 65, and 25 upregulated genes related to phenylpropanoid biosynthesis, terpenoid biosynthesis, and flavonoid biosynthesis, respectively. Under different concentrations of Zn treatment, the upregulated metabolite biosynthesis-related genes and differentially expressed transcription factors varied. Pearson correlation network analysis revealed significant correlations among terpenoids, phenylpropanoids, flavonoids biosynthetic genes, and several transcription factors (ERFs, WRKYs, bHLHs, NACs, and MYBs). This study lays the foundation for understanding the metabolic processes in response to varying levels of zinc foliar spray and provides a theoretical basis for enhancing the efficiency of zinc fertilizer utilization in A. senticosus.
1 Introduction
Acanthopanax senticosus, a perennial shrub of the Acanthopanax genus in the Araliaceae family, is renowned for its high medicinal value (Sithisarn and Jarikasem, 2009; Huang et al., 2011). Its chemical constituents primarily include phenylpropanoids, flavonoids, terpenoids, lignans, sterols, caffeoylquinic acid derivatives, and fatty acids (Shi et al., 2019; Li et al., 2021). A. senticosus has demonstrated various pharmacological effects such as cardioprotection, hypoglycemic activity, hepatoprotection, immunomodulation, neuroprotection, anti-fatigue activity, anti-cancer activity, anti-inflammatory activity, and antioxidant activity (Liang et al., 2010; Bai et al., 2011; Huang et al., 2011; Jiang and Wang, 2015; Kim et al., 2015; Zhang et al., 2015; Zhang et al., 2021).
Zinc (Zn) is a vital trace element for plant growth and development, and it is essential for high yield and quality (Bukhari et al., 2021; Semida et al., 2021). Zn deficiency results in stunted growth as the main symptom in plants (Mattiello et al., 2015). Foliar Zn application can significantly enhance crop quality and yield (Joy et al., 2015; Ahmed et al., 2021). In most studies of medicinal plant fertilization, appropriate fertilization can increase both the yield and quality of medicinal materials and enhance secondary metabolite content. Conversely, inappropriate fertilization can create a stressful environment that negatively impacts medicinal plant growth and quality (Ma et al., 2018; Sun et al., 2021). Foliar fertilization, a widely used agricultural cultivation measure, involves the active absorption of nutrients into leaf interiors (Haslett, 2001). This method is more environmentally friendly than soil application of trace elements, which may produce toxicity (Fernandez and Brown, 2013). Foliar fertilization advantages include low dosage, rapid absorption, minimal pollution, and evident effects (Razzaq et al., 2013; Davarpanah et al., 2016). Zn also maintains the structure of certain protein types, such as transcription factors (TFs) (Xie et al., 2020). Additionally, Zn plays an important role in various physiological functions in plants, such as hormone regulation and signal transduction through mitogen-activated protein kinases (Bhantana et al., 2021; Kaur and Garg, 2021; Ahmad et al., 2022).
Foliar nutrient sprays may vary in their effectiveness depending on their translocation to other plant organs. After foliar application, Zn is taken up by the leaf epidermis, remobilized, and transported to other organs via the phloem (Fernandez and Eichert, 2009; White and Broadley, 2011). However, the remobilization of foliar Zn, from either soil or foliar sources, is influenced by factors such as plant species and genotypes, phenological stage, application method, and environmental conditions (Kutman et al., 2010; Impa et al., 2013). Zn is classified as intermediate or conditionally mobile, but it can still translocate to other organs in many plants following foliar fertilization. This translocation depends on factors such as plant nutritional status, species and variety, or plant phenological state (Wu et al., 2010; Erenoglu et al., 2011; Hegelund et al., 2012).
Currently, transcriptomic and metabolomic analyses have been jointly applied to various medicinal plants, including Bletilla striata (Zou et al., 2021), Dendrobium officinale (Zhang et al., 2021), and Ganoderma lucidum (Meng et al., 2022). Huang et al. (2022) employed a combined transcriptomic and metabolomic analysis to examine changes in flavonoid content and the expression of related biosynthetic pathway genes in Fagopyrum cymosum rhizomes. They uncovered a catechin-related network involving interactions among four metabolites and 14 genes. Liu et al. (2022) discovered that Astragalus membranaceus adapted to saline–alkali stress by upregulating flavonoid biosynthesis through a combined transcriptomic and metabolomic analysis, thereby enhancing its medicinal value. However, insufficient reports exist on the metabolome and transcriptome analyses of A. senticosus.
In this study, we integrated metabolomic and transcriptomic analyses to investigate changes in medicinal component content and differentially expressed genes (DEGs) in A. senticosus roots following foliar application of Zn fertilizer at varying concentrations. By conducting correlation analysis to identify relationships among major differential metabolites, biosynthesis-related genes, and TFs, we provide a reference for enhancing A. senticosus’ medicinal quality through Zn fertilization.
2 Materials and methods
2.1 Plant materials
The test material consisted of 2-year-old A. senticosus plants grown at a cultivation base in Hulan District, Harbin City, Heilongjiang Province, China (N45°52′, E126°36′). The field experiment included a control group with no fertilization and only water spray (CK), a second group with foliar application of a low concentration of 0.1% ZnSO4·7H2O (LZn), and a third group sprayed with a high concentration of 0.4% ZnSO4·7H2O (HZn). Fertilization was applied on June 7th, June 19th, and July 1st between 4 p.m. and 5 p.m. Each seedling received approximately 25 mL of fertilizer spray, covering both leaf surfaces. On October 25th, roots from plants in different treatments were collected and rinsed with clean water followed by distilled water before being frozen in liquid nitrogen and stored at −80°C for future use. For the metabolome analysis, six biological replicates were performed for each treatment, while the transcriptome analysis had three biological replicates.
2.2 Metabolite extraction
The freeze–dried samples were crushed using a mixer mill for 30 s at a frequency of 60 Hz. Then, 100 mg of the sample was added to a solution containing extracted material dissolved in 80% methanol and contained an internal standard of 10 μg/mL. After vortexing for 30 s, the samples were homogenized at a frequency of 45 Hz for 4 minutes and sonicated for 1 hour in an ice-water bath. After being placed in −40°C for 1 hour, the samples were centrifuged at a speed of 12,000 rpm (RCF = 13,800 (×g), R = 8.6 cm) for 15 minutes at 4°C. The supernatant was carefully filtered through a microporous membrane with a pore size of 0.22 μm, and then, 10 μL was taken from each sample and pooled as quality control samples. The samples were stored at −80°C until ultra-high-performance liquid chromatography–mass spectrometry (UHPLC–MS) analysis.
2.3 Liquid chromatography–tandem mass spectrometry conditions
An ultra-high-performance liquid chromatography system (Vanquish, Thermo Fisher Scientific) equipped with a Waters UPLC BEH C18 column (1.7 μm 2.1*100 mm) was used for liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis. The sample injection volume was 5 μL, and the flow rate was 0.5 mL/min. The mobile phase consisted of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The multistep linear elution gradient program was as follows: 85% A for 0–11 min, 25% A for 11–12 min, 2% A for 12–14 min, 2% A for 14–14.1 min, 85% A for 14.1–15 min, and 85% A for 15–16 min.
LC–MS/MS data acquisition was based on the information-dependent acquisition (IDA) mode, and Xcalibur software coupled with a Q Exactive Focus mass spectrometer was used. During each acquisition cycle, the mass range was from 100 to 1,500, and the top three of every cycle were screened, and the corresponding MS/MS data were further acquired. The sheath gas flow rate was set at 45 Arb, the auxiliary gas flow rate was set at 15 Arb, and the capillary temperature was adjusted at 400°C. The full MS resolution was set at 70,000, and the MS/MS resolution was set at 17,500. Collision energy was set at 15/30/45 in the normalized collision energy (NCE) mode, and spray voltage was attuned at +4.0 kV (positive) or −4.0 kV (negative).
2.4 Identification and exploration of differentially accumulated metabolites
The raw mass spectrometry data were imported using XCMS software and underwent retention time correction, peak identification, extraction, integration, and alignment. The peaks containing MS/MS data were identified using a secondary mass spectrometry database built by Shanghai Baiqu Biomedical Technology Co., Ltd., along with corresponding fragmentation matching methods. Variable importance in projection (VIP) values were extracted from the orthogonal partial least squares discriminant analysis (OPLS-DA) results to determine the differentially accumulated metabolites (DAMs) between groups with VIP ≥1 and absolute p-value ≤0.05.
2.5 RNA-seq
The TRIzol reagent (Invitrogen) was used to extract total RNA from tissue samples, and DNase I (TaKara) was used to remove genomic DNA. To monitor RNA degradation and contamination, 1% agarose gels were run. RNA purity was checked using a NanoPhotometer® spectrophotometer (IMPLEN, CA, United States), and RNA concentration was measured with a Qubit® RNA Assay Kit in a Qubit®2.0 Flurometer (Life Technologies, CA, United States). RNA integrity was assessed with an RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, United States).
RNA sample preparation was performed using 1 µg RNA per sample. The NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States) was used to generate sequencing libraries according to the manufacturer’s recommendations. Index codes were added to allocate sequences to each sample. Poly-T oligo-attached magnetic beads were used to purify mRNA from total RNA. Fragmentation was performed using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using a random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends using exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with a hairpin loop structure was ligated to prepare for hybridization. The library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, United States) to select cDNA fragments of 250–300 bp in length. Size-selected, adaptor-ligated cDNA was treated with 3 µL USER Enzyme (NEB, United States) at 37°C for 15 min followed by 5 min at 95°C before PCR. Finally, PCR products were purified (AMPure XP system), and library quality was assessed on the Agilent Bioanalyzer 2100 system.
The original data were filtered using fastp, mainly to eliminate reads containing adapters. Paired reads were removed if the N content in any sequencing read exceeded 10% of the base number of the reads. Paired reads were also removed if the number of low-quality (Q < = 20) bases contained in any sequencing read exceeded 50% of the bases of the read. All subsequent analyses were based on clean reads. Trinity was used to perform transcriptome assembly. Relevant transcripts were regrouped into gene clusters using Corset (https://github.com/trinityrnaseq/trinityrnaseq). TransDecoder (https://github.com/TransDecoder/TransDecoder/wiki) was used to identify candidate coding regions within transcript sequences generated by de novo RNA-Seq transcript assembly using Trinity. Gene function was annotated based on the following databases using diamond or HMMER: Nr, Swiss-Prot, Trembl, KEGG, GO, KOG/COG, and Pfam. Gene expression levels were estimated by RSEM, and FPKM of each gene was calculated based on the gene length.
2.6 Differential expression, gene mining, and enrichment analysis
DESeq2 was used to analyze differential expression between the two groups. A p-value less than 0.05 and |log2foldchange|≥1 were used as the threshold for significant differential expression. We used Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) to perform GO functional enrichment and KEGG pathway analysis for the DEGs to understand their functions. GO terms and metabolic pathways with a Bonferroni-corrected p-value less than 0.05 were considered as significantly enriched by DEGs.
2.7 Correlation network analysis
Principal component analysis (PCA) and Pearson correlation analysis were performed using R’s built-in functions. Subsequently, significant correlation networks (p < 0.05) between biosynthesis genes and metabolites and between biosynthesis genes and TFs were visualized using Cytoscape 3.9 (Cline et al., 2007).
3 Results
3.1 Metabolomic analysis
A total of 316 metabolites were identified in the samples; among them, terpenoids were the most abundant, accounting for 21.8% of the total, followed by phenylpropanoids at 14.6% (Supplementary Table S1). Phenols, alkaloids, flavonoids, amino acid derivatives, fatty acyls, aromatic compounds, organic acids and their derivatives, fatty acids, and other compounds accounted for 6.0%, 5.4%, 4.7%, 2.8%, 1.9%, 1.9%, 1.6%, 1.6%, and 37.7%, respectively (Figure 1A). To compare the metabolic differences among HZn, LZn, and CK, PCA was performed on the metabolomics dataset. In the PCA score plot, the contribution rates of PC1 and PC2 were 39.71% and 28.01%, respectively. Each sample group was clustered together, indicating good repeatability of the samples. The samples between different groups were clearly alienated, indicating significant differences between the metabolomes (Figure 1B). Hierarchical clustering heatmap analysis showed that the replicates of the samples were assembled together, further indicating good data repeatability. In addition, the color differences of the 316 metabolites between different samples were obvious, demonstrating significant differences in metabolites between different samples (Figure 1C).
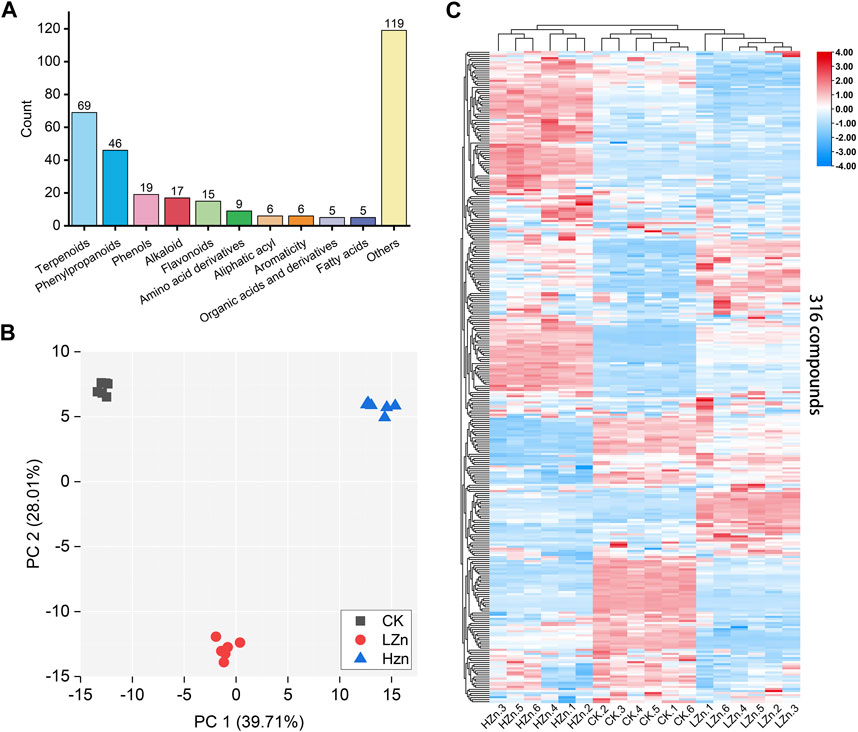
FIGURE 1. Metabolomics analysis based on LC–MS/MS measurement. (A) Types and quantities of metabolites, (B) PCA score plot, and (C) hierarchical clustering heatmap of 316 metabolites in three sample groups.
DAMs were screened using the criteria of VIP >1 and p-value <0.05. A total of 191 DAMs were identified in LZn vs. CK, including 88 upregulated metabolites and 103 downregulated metabolites (Figure 2, Supplementary Table S2). These mainly comprised 53 terpenoids, 25 phenylpropanoids, 12 alkaloids, 10 phenols, seven flavonoids, and 84 other metabolites. Compared with CK, these upregulated compounds in LZn treatment increased by at least 5.86%. Among them, the highest fold changes were observed for isoimperatorin (phenylpropanoid), fraxetin (phenylpropanoid), and flavanone base + 3O, 1Prenyl (flavonoid), which increased by more than 29.98 times compared with CK. In HZn vs. CK, a total of 197 DAMs were identified, including 47 terpenoids, 25 phenylpropanoids, 13 alkaloids, nine phenols, six flavonoids, and 89 other compounds. These upregulated compounds in HZn treatment increased by at least 11.71% compared with CK. The highest fold changes were observed for flavanone base + 3O, 1Prenyl (flavonoid), fraxetin (phenylpropanoid), and cortodoxone (terpenoid), which increased by more than 24.47 times compared with CK. Venn diagram analysis showed that among the upregulated metabolites, HZn vs. CK and LZn vs. CK had 59 and 31 upregulated metabolites, respectively, with a total of 57 shared between them. Among the downregulated metabolites, HZn vs. CK and LZn vs. CK had 28 and 50 downregulated metabolites, respectively, with a total of 53 shared between them. KEGG enrichment analysis revealed that the DAMs of LZn vs. CK were mainly significantly enriched in metabolic pathways such as biosynthesis of secondary metabolites, biosynthesis of cofactors, phenylalanine, tyrosine, and tryptophan biosynthesis, phenylalanine metabolism, and biosynthesis of amino acids. On the contrary, the focal differential metabolites of HZn vs. CK were considerably enriched in metabolic pathways such as biosynthesis of secondary metabolites, biosynthesis of cofactors, and phenylalanine metabolism.
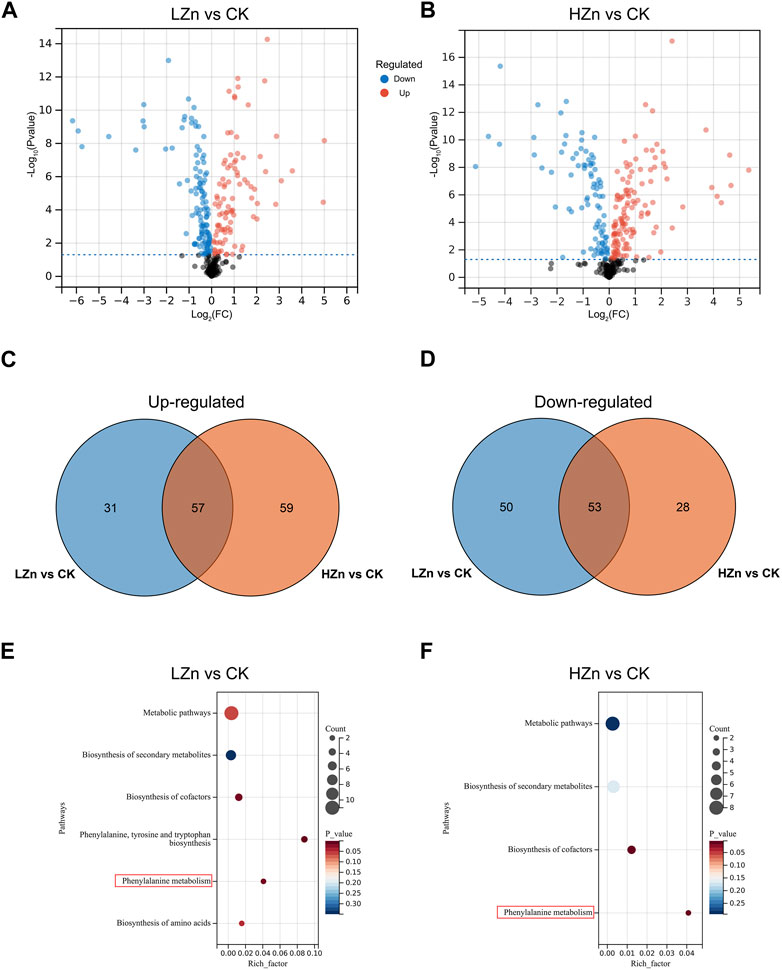
FIGURE 2. Identification and enrichment analysis of DAMs. Identification of DAMs in LZn vs. CK (A) and HZn vs. CK (B) comparisons. Venn diagram of the upregulated (C) and downregulated (D) DAMs between LZn vs. CK and HZn vs. CK. KEGG enrichment analysis in LZn vs. CK (E) and HZn vs. CK (F) comparisons.
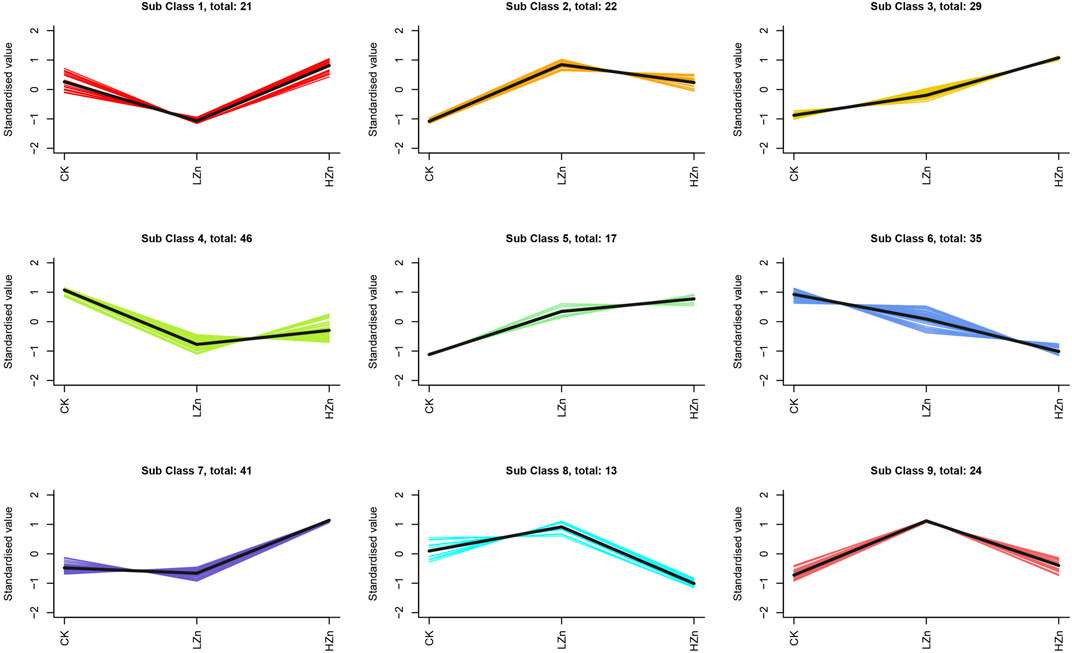
K-means clustering analysis proposed that 228 differential metabolites could be divided into nine classes (Figure 3), numbered as 1–9, and comprised 21, 22, 29, 46, 17, 35, 41, 13, and 24 metabolites, respectively (Supplementary Table S2). Notably, the metabolite content was increased in classes 2, 3, 5, 7, 8, and 9 after Zn treatment. Of these, classes 2 and 5 had a significant metabolite increment in both LZn and HZn treatment groups. Based on the differential metabolite analysis and K-means analysis, we identified medicinally related components of A. senticosus that amplified after Zn treatment, including 43 terpenoids, 19 phenylpropanoids, eight phenols, and three flavonoids (Figure 4). The analysis of the 43 terpenoid compounds revealed that eight compounds belong to class 2, eight to class 3, five to class 5, 13 to class 7, three to class 8, and the remaining six to class 9. Similarly, among the 19 phenylpropanoids studied, class distribution was as follows: three in class 3, two in class 2, six in class 7, two in class 8, and the remaining six in class 9. In the case of the eight phenols analyzed, two were found to belong to class 2, one to class 5, one to class 7, and the remaining four to class 9. Last, of the three flavonoids examined, two were classified into class 5 and one into class 7.
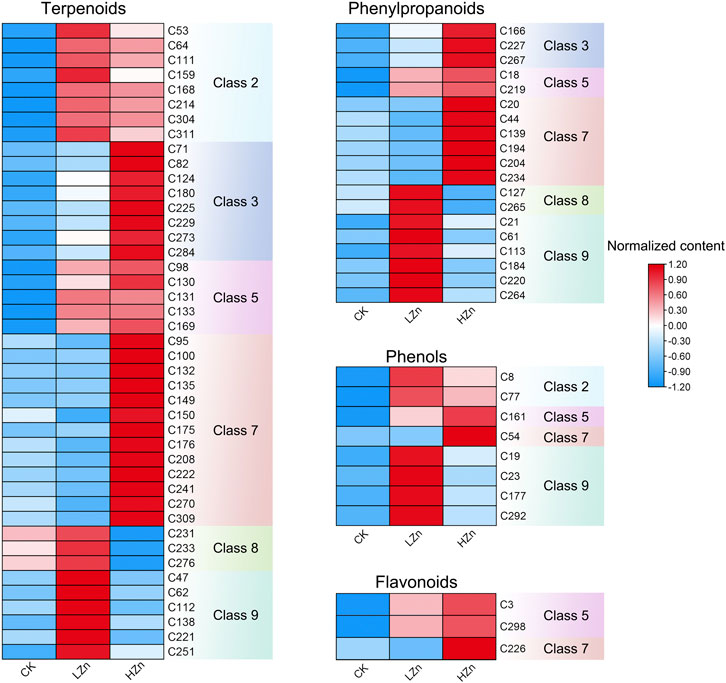
FIGURE 4. Terpenoids, phenylpropanoids, phennols, and flavonoids with upregulated content in the A. senticosus root after foliar application of Zn fertilizer. The classes are derived from the results of the K-means analysis.
3.2 Transcriptome analysis
RNA-seq sequencing yielded 53.95 GB of raw data (Supplementary Table S3). After quality control filtering, 49.62 GB of valid data were obtained (Supplementary Table S4). In the valid data, the Q20 value exceeded 97%, and the Q30 value exceeded 92% (Supplementary Table S4). The GC content ranged from 43.27% to 49.11% (Supplementary Table S4). Gene annotation results showed that a total of 110,322 genes were successfully annotated (Supplementary Table S5). Of these, 54,526; 37,961; 45,013; 45,947; 59,707; and 56,103 genes were annotated in the GO, KEGG, Pfam, Swiss-Prot, and NR databases (Supplementary Table S6). As shown in the volcano plot in Figure 5, red color represents significantly upregulated genes, blue represents significantly downregulated genes, and gray represents genes with no significant difference. In LZn vs. CK, a total of 6,696 differentially expressed genes were obtained, including 3,958 upregulated and 2738 downregulated (Figure 5A). In HZn vs. CK, a total of 6,172 differentially expressed genes were obtained, including 3,440 upregulated and 2,732 downregulated (Figure 5B). Venn diagram analysis showed that among the upregulated differentially expressed genes, HZn vs. CK and LZn vs. CK had 1831 and 2,349 genes, respectively, with a total of 1,609 upregulated genes shared between them (Figure 5C). Among the downregulated differentially expressed genes, HZn vs. CK and LZn vs. CK had 2,707 and 3,933 genes, respectively, with a total of 25 downregulated genes shared between them (Figure 5D).
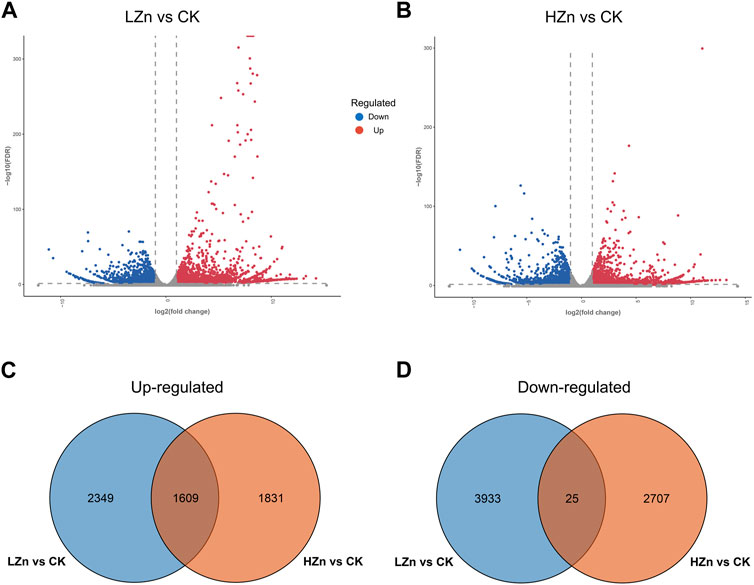
FIGURE 5. Identification of DEGs. (A) DEGs between LZn and CK. (B) DEGs between HZn and CK. Venn diagram of the upregulated (C) and downregulated (D) DEGs between LZn vs. CK and HZn vs. CK.
GO functional enrichment analysis was performed on the differentially expressed genes. Figure 6 shows that in LZn vs. CK, the top five significantly enriched pathways for differentially expressed genes were chloroplast stroma, thylakoid, chloroplast thylakoid membrane, response to cold, and chloroplast (Figure 6A). In HZn vs. CK, the top five significantly enriched pathways for differentially expressed genes were plasmodesma, extracellular region, apoplast, peroxidase activity, and cell wall. According to the KEGG pathway enrichment analysis, the top 20 most significantly enriched metabolic pathways indicating that genes related to the metabolic pathways of medicinal components changed significantly after Zn treatment (Figure 6B). In LZn vs. CK, the significantly enriched pathways related to medicinal metabolites were phenylpropanoid biosynthesis and flavonoid biosynthesis (Figure 6C). In case of HZn vs. CK, phenylpropanoid biosynthesis, flavonoid biosynthesis, and sesquiterpenoid and triterpenoid biosynthesis were spotted as the substantial enriched pathways related to medicinal metabolites (Figure 6D). We also identified differentially expressed TFs. In LZn vs. CK, a total of 192 differentially expressed TFs were identified, including 110 upregulated and 82 downregulated (Supplementary Figure S1, Supplementary Table S7). In HZn vs. CK, a total of 164 differentially expressed TFs were recognized, including 97 upregulated and 67 downregulated (Supplementary Figure S2, Supplementary Table S7). Furthermore, a distinct pattern was observed in the upregulation of genes under different zinc treatments. Specifically, in the DEGs of LZn vs. CK comparison, but not in the DEGs of HZn vs. CK comparison, four flavonoid biosynthesis-related genes, five phenylpropanoid biosynthesis-related genes, and 10 terpenoid biosynthesis-related genes were upregulated. Conversely, in the DEGs of HZn vs. CK comparison, but not in the DEGs of LZn vs. CK, two flavonoid biosynthesis-related genes, 22 phenylpropanoid biosynthesis-related genes, and 12 terpenoid biosynthesis-related genes were upregulated. These findings suggest that varying concentrations of zinc treatment can differentially influence the expression of biosynthetic genes.
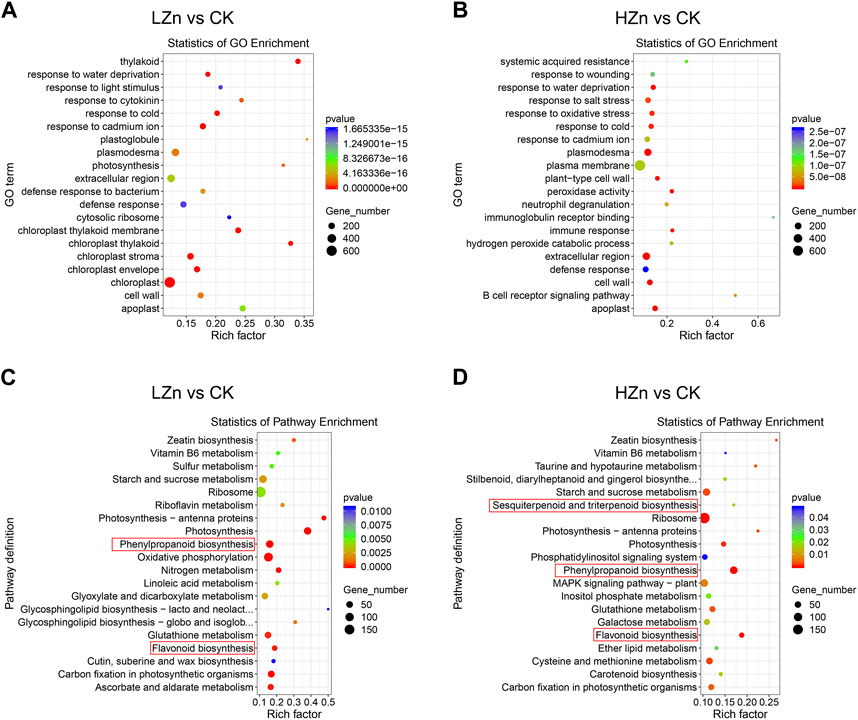
FIGURE 6. GO and KEGG enrichment analysis of differentially expressed genes. GO enrichment analysis in LZn vs. CK (A) and HZn vs. CK (B) comparisons. KEGG enrichment analysis in LZn vs. CK (C) and HZn vs. CK (D) comparisons.
The results of the k-means fuzzy clustering analysis suggested that all differentially expressed genes can be categorized into four classes (Supplementary Figure S3). Class 1 contains 6,102 genes, which exhibited low expression in both CK and LZn and high expression in HZn. Class 2 contains 9,387 genes, whose expression levels decreased after LZn and HZn treatment. Class 3 contains 2596 genes, whose expression levels increased after LZn and HZn treatment. Class 4 contains 9,913 genes, which showed high expression in both CK and HZn treatment but feeble expression after LZn treatment. To reveal the reasons for the upregulation of phenylpropanoid, terpenoid, and flavonoid compounds by zinc treatment, we identified 95, 65, and 25 differentially expressed genes related to phenylpropanoid biosynthesis, terpenoid biosynthesis, and flavonoid biosynthesis in classes 1 and 3, respectively (Figure 7, Supplementary Table S8). Consistent with existing reports, multiple transcription factor families such as ERF, bHLH, MYB, NAC, WRKY, and bZIP have transcriptional regulatory effects on these metabolites. In classes 1 and 3, there are a total of 12 bHLHs, 16 bZIPs, 34 ERFs, 13 MYBs, 14 MYB_related, 13 NACs, and 25 WRKY genes, suggesting that they have potential transcriptional regulatory effects on the biosynthesis of related metabolites (Supplementary Table S9).
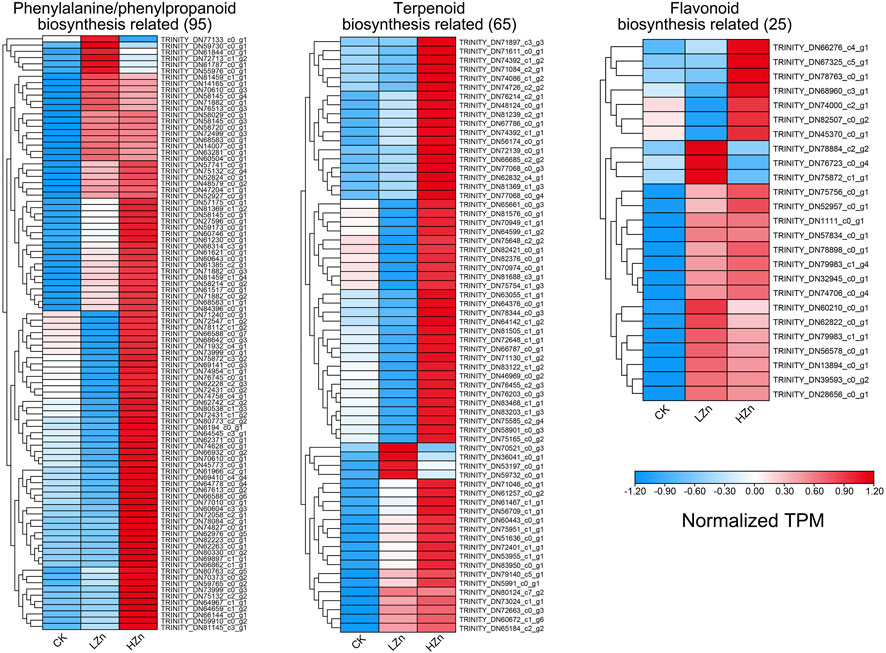
FIGURE 7. Upregulated genes related to the biosynthesis of important medicinal components in A. senticosus after foliar application of Zn fertilizer.
3.3 Integrated analysis of metabolomics and transcriptomics
To better understand the metabolic regulatory network of A. senticosus after treatment with different concentrations of zinc, we performed a Pearson correlation analysis on the differentially expressed genes related to metabolite biosynthesis and TFs (Figure 8). A significant correlation was established between TFs and structural genes when the p-value was less than 0.05 and the correlation coefficient |r| exceeded 0.9.
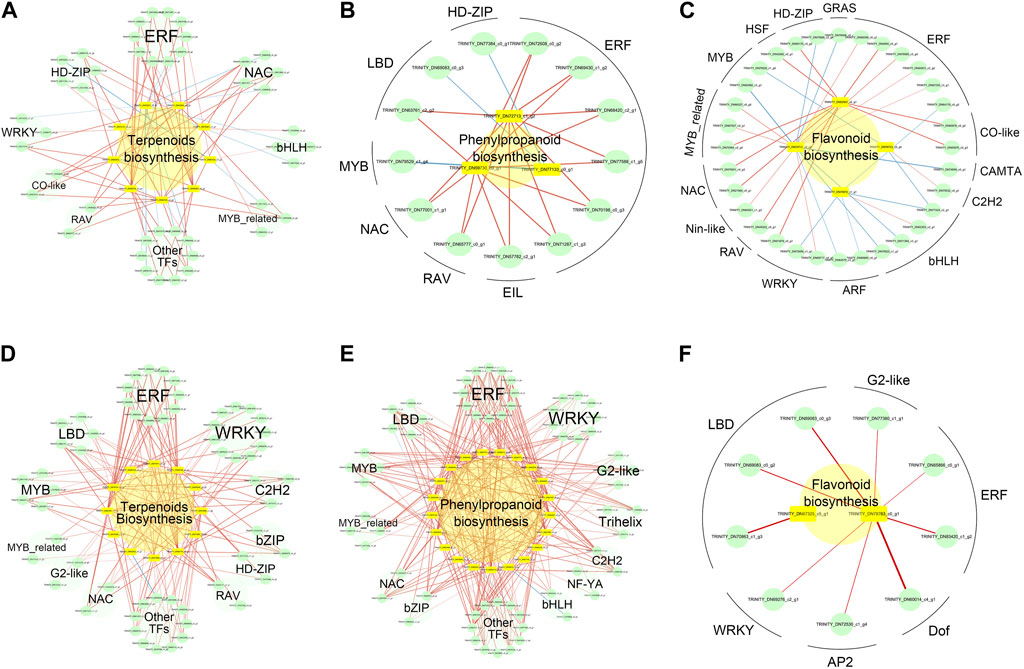
FIGURE 8. Significant correlation network between key metabolite biosynthesis genes and TFs in A. senticosus after foliar application of Zn fertilizer. In the upregulated metabolite biosynthesis, genes between LZn and CK, but not between HZn and CK, correlation networks between terpenoids (A), phenylpropanoids (B), and flavonoids (C) biosynthesis key genes and TFs. In the upregulated metabolite biosynthesis, genes between HZn and CK, but not between LZn and CK, correlation networks between terpenoids (D), phenylpropanoids (E), and flavonoids (F) biosynthesis key genes and TFs.
In the upregulated metabolite biosynthesis genes between LZn and CK, but not between HZn and CK, 44 transcription factors were identified to have a significant correlation with eight terpenoid biosynthesis-related genes (Figure 8A, Supplementary Table S10). These transcription factors included 10 ERFs, five NACs, five HD-ZIPs, four bHLHs, three WRKYs, three MYB_related, two CO-like, two RAVs, and 10 other types of transcription factors. Among them, 30 transcription factors showed a significant positive correlation with eight terpenoid biosynthesis-related genes, while 14 transcription factors had a significant negative correlation with two terpenoid biosynthesis-related genes. Thirteen transcription factors exhibited a significant positive correlation with three phenylpropanoid biosynthesis-related genes, including six ERFs, one EIL, one RAV, one NAC, one MYB, two LBDs, and one HD-ZIP (Figure 8B). Additionally, 32 transcription factors were significantly correlated with four flavonoid biosynthesis-related genes (Figure 8C). These comprised six ERFs, two CO-like, four bHLHs, two ARFs, three WRKYs, two NACs, three MYB-related, two MYBs, two HD-ZIPs, and six other types of transcription factors.
In the upregulated metabolite biosynthesis genes between HZn and CK, but not between LZn and CK, significant correlations were observed between the expression of metabolite biosynthesis-related genes and various transcription factors (Supplementary Table S11). Specifically, 58 transcription factors were significantly correlated with 12 terpenoid biosynthesis-related genes (Figure 8D). These included 11 ERFs, 11 WRKYs, three C2H2s, two bZIPs, two HD-ZIPs, two RAVs, five LBDs, five MYBs, two MYB_relateds, two G2-likes, two NACs, and 11 other types of transcription factors. Among these transcription factors, only one showed a significant negative correlation with a structural gene, while the rest showed a significant positive correlation. A total of 63 transcription factors were significantly correlated with 12 phenylpropanoid biosynthesis-related genes (Figure 8E). These comprised 12 ERFs, nine WRKYs, four G2-likes, three trihelixes, two C2H2s, two NF-YAs, two bHLHs, two bZIPs, two NACs, four MYBs, four MYB_relateds, five LBDs, and 12 other types of transcription factors. Among these transcription factors, only three showed a significant negative correlation with three structural genes. Nine transcription factors demonstrated a significant positive correlation with two flavonoid biosynthesis-related genes (Figure 8F). These included three LBDs, two ERFs, one G2-like, one WRKY, one AP2, and one Dof.
4 Discussion
In this study, we investigated the effect of foliar application of different Zn concentrations on the roots of A. senticosus. Our results revealed that foliar application of a certain Zn concentration can significantly increase the content of medicinal components in the roots of A. senticosus. Compared with the control, foliar application of different Zn levels markedly enhanced the content of many medicinal metabolites in the roots of A. senticosus, including 43 terpenoids, 19 phenylpropanoids, eight phenols, and three flavonoids. In our study, DEG analysis and K-means clustering identified 95 upregulated phenylpropanoid biosynthesis-related genes, 65 terpenoid biosynthesis-related genes, and 25 flavonoid biosynthesis-related genes after Zn treatment. We also observed significant correlations between the expression of metabolite biosynthesis-related genes and various transcription factors in the upregulated metabolite biosynthesis genes between LZn and CK, but not between HZn and CK, or between HZn and CK, but not between LZn and CK. These findings suggest that in A. senticosus, foliar application of Zn fertilizer may regulate the expression of various medicinal compound biosynthesis genes by affecting the expression of related transcription factors, thereby increasing the content of medicinal components in the roots.
Our results are consistent with many previous studies. First, foliar application of zinc fertilizer can increase the content of medicinal components in various plants. For example, in Stevia rebaudiana, foliar application of zinc sulfate significantly increased its stevioside content (Singh and Dwivedi, 2019). In Mentha pulegium, zinc treatment significantly increased the composition of pulegone, cis-isopulegone, a-pinene, sabinene, 1,8-cineole, and borneol in mint essential oil (Lajayer et al., 2016). In Lavandula stoechas, foliar application of zinc fertilizer had a substantial effect on its flavonoid and volatile oil contents (Vojodi Mehrabani et al., 2017). Furthermore, in Melissa officinalis, foliar application of zinc fertilizer significantly increased the plant contents of flavonoids and phenolic compounds (Yadegari, 2017). In Panax ginseng, foliar application of zinc fertilizer considerably improved the root yield and increased its contents of ginsenoside monomer along with other nine saponins (Zhang et al., 2013). Furthermore, trigonelline content was augmented in fenugreek (Trigonella foenum-graecum L.) when zinc fertilizer was applied (Tariverdizadeh et al., 2021). These studies all indicate that foliar application of Zn fertilizer can indeed enhance the quality of medicinal plants. Therefore, foliar application of Zn fertilizer can be widely used as a fertilization technique to improve the quality of medicinal components in A. senticosus roots.
Additionally, preliminary studies indicated that Zn fertilizer treatments could alter the expression of genes encoding enzymes involved in metabolite synthesis, thereby affecting changes in metabolite content. For example, under zinc stress, the activities and gene expression levels of ascorbate peroxidase, catalase, superoxide dismutase, and peroxidase were upregulated in tea plants (Mukhopadhyay et al., 2013). Furthermore, Zn fertilizer treatment considerably increased the antioxidant potentials (ascorbic acid, reduced glutathione, total phenols, and total flavonoids) in wheat flag leaves and enhanced the expression of two antioxidant enzyme genes, four ascorbate-glutathione cycle genes, and two flavonoid biosynthesis-related genes (Sun et al., 2020). Dong et al. (2021) also found that Zn fertilizer treatment could affect the expression of stress-related resistance genes in different barley cultivars. In our study, DEG analysis and K-means clustering emphasized 95 upregulated phenylpropanoid biosynthesis-related genes, 65 terpenoid biosynthesis-related genes, and 25 flavonoid biosynthesis-related genes after Zn treatment. However, the molecular mechanisms underlying the effects of foliar Zn application on the expression of these medicinal metabolite synthesis enzyme genes are still not very clear. Some studies have shown that plant responses to Zn deficiency are regulated by multiple levels, including transcriptional regulation mediated by F-group bZIP proteins, epigenetic regulation at the level of chromatin, and post-transcriptional regulation mediated by small RNAs and alternative splicing (Zeng et al., 2021). These need to be gradually clarified in future studies.
TFs regulate gene expression, allowing plants to respond to stress and modulate developmental processes (Mitsuda and Ohme-Takagi, 2009). Plants have approximately 58 TF families, and six major TF families (AP2/ERF, bHLH, MYB, NAC, WRKY, and bZIP) are involved in biotic and abiotic stress responses (Ng et al., 2018). The MYB family is the largest TF family in plants, and its members play crucial roles in plant development, stress responses, and secondary metabolite biosynthesis (Wang et al., 2021). Members of the ERF (Huang et al., 2021), NAC (Nuruzzaman et al., 2013; Borchers and Pieler, 2022), WRKY (Chi et al., 2013), bZIP (Zhao et al., 2021), and bHLH (Mao et al., 2017) gene families also contribute essentially in plant stress responses and metabolite biosynthesis. In this study, we identified 34 ERFs, 12 bHLHs, 13 NACs, 14 MYB_relateds, 13 MYBs, 16 bZIPs, and 25 WRKY genes with significantly altered expression levels after Zn treatment (Supplementary Table S9). These genes were also significantly correlated with multiple medicinal metabolite biosynthesis-related genes. Proposing that, these TFs may respond to Zn treatment stimuli and regulate the expression of multiple metabolite biosynthesis genes, thereby participating in the production of related metabolites. Additionally, different Zn concentrations resulted in different upregulated terpenoid, phenylpropanoid, and flavonoid biosynthesis genes and differentially expressed transcription factors, indicating that different Zn concentrations could regulate the biosynthesis of metabolites by affecting specific transcription factors that control the expression of metabolite biosynthesis-related genes. Further research on the transcriptional regulatory mechanisms of these TFs would lay the foundation for elucidating the molecular mechanisms underlying the enhancement of medicinal component accumulation in A. senticosus roots by Zn treatment.
5 Conclusion
Our research used metabolomics and transcriptomics to examine change patterns in metabolites and genes in A. senticosus roots following foliar application of varying levels of zinc. A total of 316 metabolites were detected, including 69 terpenoids, 46 phenylpropanoids, 19 phenols, 17 alkaloids, 15 flavonoids, nine amino acid derivatives, six aliphatic acyls, six aromatics, five organic acids and derivatives, five fatty acids, and 119 other compounds. Differential metabolite analysis and K-means clustering identified metabolites related to the medicinal components of A. senticosus that were upregulated after Zn treatment, including 43 terpenoids, 19 phenylpropanoids, eight phenols, and three flavonoids. Transcriptome analysis showed significant changes in the phenylpropanoid and flavonoid biosynthesis pathways after Zn treatment. Differential gene expression analysis and K-means clustering marked a total of 95 phenylpropanoid biosynthesis-related genes, 65 terpenoid biosynthesis-related genes, and 25 flavonoid biosynthesis-related genes. Correlation network analysis revealed significant and positive parallels between three flavonoids and 16 flavonoid biosynthesis genes, 19 phenylpropanoids and 95 phenylpropanoid biosynthesis-related genes, and 43 terpenoids and 66 terpenoid biosynthesis-related genes. Additionally, significant correlations were observed between multiple medicinal component biosynthesis-related genes and some key TFs (ERFs, WRKYs, bHLHs, NACs, and MYBs). This study provides a valuable reference for increasing the medicinal components content in A. senticosus roots through Zn fertilizer application.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA972895.
Author contributions
TS: conceptualization, data curation, project administration, writing–original draft, and writing–review and editing. JS: data curation, formal analysis, investigation, and writing–original draft. YL: formal analysis, investigation, methodology, and writing–review and editing. YR: data curation, formal analysis, software, and writing–review and editing. YL: conceptualization, data curation, investigation, and writing–review and editing. CS: formal analysis, investigation, methodology, and writing–review and editing. AN: formal analysis, investigation, methodology, and writing–review and editing. ZT: funding acquisition, resources, supervision, visualization, and writing–review and editing. AA: supervision, validation, visualization, and writing–review and editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research received funding from Research and Demonstration on Complex Management and Deep Processing Technology of Special Fruit and Medicine Forest in Heilongjiang Province (grant no. JD22A008).
Acknowledgments
The authors are grateful to Shanghai BIOTREE Biological Technology Co., Ltd., for metabolomics and transcriptomics analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1259674/full#supplementary-material
References
Ahmad, S., Mfarrej, M. F. B., El-Esawi, M. A., Waseem, M., Alatawi, A., Nafees, M., et al. (2022). Chromium-resistant staphylococcus aureus alleviates chromium toxicity by developing synergistic relationships with zinc oxide nanoparticles in wheat. Ecotoxicol. Environ. Saf. 230, 113142. doi:10.1016/j.ecoenv.2021.113142
Ahmed, B., Rizvi, A., Ali, K., Lee, J., Zaidi, A., Khan, M. S., et al. (2021). Nanoparticles in the soil–plant system: a review. Chem. Lett. 19, 1545–1609. doi:10.1007/s10311-020-01138-y
Bai, Y., Tohda, C., Zhu, S., Hattori, M., and Komatsu, K. (2011). Active components from Siberian ginseng (Eleutherococcus senticosus) for protection of amyloid beta(25-35)-induced neuritic atrophy in cultured rat cortical neurons. J. Nat. Med. 65, 417–423. doi:10.1007/s11418-011-0509-y
Bhantana, P., Rana, M. S., Sun, X.-C., Moussa, M. G., Saleem, M. H., Syaifudin, M., et al. (2021). Arbuscular mycorrhizal fungi and its major role in plant growth, zinc nutrition, phosphorous regulation and phytoremediation. Symbiosis 84 (1), 19–37. doi:10.1007/s13199-021-00756-6
Borchers, A., Pieler, T., Grierson, D., and Fu, D. Q. (2022). NAC transcription factor family regulation of fruit ripening and quality: a review. Cells 11, 525. doi:10.3390/cells11030525
Bukhari, S. A., Farah, N., Mahmood, S., Altaf, J., Mustafa, G., and Yasin, N. A. (2021). Effects of seed priming with zinc sulfate on nutritional enrichment and biochemical fingerprints of Momordica charantia. J. Food Qual. 2021, 1–13. doi:10.1155/2021/5553278
Chi, Y., Yang, Y., Zhou, Y., Zhou, J., Fan, B., Yu, J. Q., et al. (2013). Protein-protein interactions in the regulation of WRKY transcription factors. Mol. Plant. 6, 287–300. doi:10.1093/mp/sst026
Cline, M. S., Smoot, M., Cerami, E., Kuchinsky, A., Landys, N., Workman, C., et al. (2007). Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382. doi:10.1038/nprot.2007.324
Davarpanah, S., Tehranifar, A., Davarynejad, G., Abadía, J., and Khorasani, R. (2016). Effects of foliar applications of zinc and boron nano-fertilizers on pomegranate (Punica granatum cv. Ardestani) fruit yield and quality. Sci. Hortic. 210, 57–64. doi:10.1016/j.scienta.2016.07.003
Dong, M., Sun, R., Yang, Q., Zhang, L., Yong, Y., Fang, Y., et al. (2021). Phenotype, physiology, and gene expression of barley seedlings in response to nano zinc oxide stress. Phyton 90, 1589–1598. doi:10.32604/phyton.2021.017465
Erenoglu, E. B., Kutman, U. B., Ceylan, Y., Yildiz, B., and Cakmak, I. (2011). Improved nitrogen nutrition enhances root uptake, root-to-shoot translocation and remobilization of zinc (65Zn) in wheat. New Phytol. 189, 438–448. doi:10.1111/j.1469-8137.2010.03488.x
Fernandez, V., and Brown, P. H. (2013). From plant surface to plant metabolism: the uncertain fate of foliar-applied nutrients. Front. Plant. Sci. 4, 289. doi:10.3389/fpls.2013.00289
Fernandez, V., and Eichert, T. (2009). Uptake of hydrophilic solutes through plant leaves: current state of knowledge and perspectives of foliar fertilization. CRC Crit. Rev. Plant Sci. 28, 36–68. doi:10.1080/07352680902743069
Haslett, B. (2001). Zinc mobility in wheat: uptake and distribution of zinc applied to leaves or roots. Ann. Bot. 87, 379–386. doi:10.1006/anbo.2000.1349
Hegelund, J. N., Pedas, P., Husted, S., Schiller, M., and Schjoerring, J. K. (2012). Zinc fluxes into developing barley grains: use of stable Zn isotopes to separate root uptake from remobilization in plants with contrasting Zn status. Plant Soil 361, 241–250. doi:10.1007/s11104-012-1272-x
Huang, B. K., Zhao, H. F., Huang, B. K., Zheng, C. J., Peng, W., and Qin, L. P. (2011a). Acanthopanax senticosus: review of botany, chemistry and pharmacology. Pharmazie 66, 83–97. doi:10.1691/ph.2011.0744
Huang, J., Wang, L., Tang, B., Ren, R., Shi, T., Zhu, L., et al. (2022). Integrated transcriptomics and widely targeted metabolomics analyses provide insights into flavonoid biosynthesis in the rhizomes of golden buckwheat (Fagopyrum cymosum). Front. Plant. Sci. 13, 803472. doi:10.3389/fpls.2022.803472
Huang, J., Zhao, X., Burger, M., Wang, Y., and Chory, J. (2021). Two interacting ethylene response factors regulate heat stress response. Plant Cell 33, 338–357. doi:10.1093/plcell/koaa026
Huang, L. Z., Huang, B. K., Ye, Q., and Qin, L. P. (2011b). Bioactivity-guided fractionation for anti-fatigue property of Acanthopanax senticosus. J. Ethnopharmacol. 133, 213–219. doi:10.1016/j.jep.2010.09.032
Impa, S. M., Morete, M. J., Ismail, A. M., Schulin, R., and Johnson-Beebout, S. E. (2013). Zn uptake, translocation and grain Zn loading in rice (Oryza sativa L.) genotypes selected for Zn deficiency tolerance and high grain Zn. J. Exp. Bot. 64, 2739–2751. doi:10.1093/jxb/ert118
Jiang, Y., and Wang, M. H. (2015). Different solvent fractions of Acanthopanax senticosus harms exert antioxidant and anti-inflammatory activities and inhibit the human Kv1.3 channel. J. Med. Food. 18, 468–475. doi:10.1089/jmf.2014.3182
Joy, E. J. M., Stein, A. J., Young, S. D., Ander, E. L., Watts, M. J., and Broadley, M. R. (2015). Zinc-enriched fertilisers as a potential public health intervention in Africa. Plant Soil 389, 1–24. doi:10.1007/s11104-015-2430-8
Kaur, H., and Garg, N. (2021). Zinc toxicity in plants: a review. Planta 253 (6), 129. doi:10.1007/s00425-021-03642-z
Kim, Y. H., Cho, M. L., Kim, D. B., Shin, G. H., Lee, J. H., Lee, J. S., et al. (2015). The antioxidant activity and their major antioxidant compounds from Acanthopanax senticosus and. A. Koreanum. Mol. 20, 13281–13295. doi:10.3390/molecules200713281
Kutman, U. B., Yildiz, B., Ozturk, L., and Cakmak, I. (2010). Biofortification of durum wheat with zinc through soil and foliar applications of nitrogen. Cereal Chem. 87, 1–9. doi:10.1094/CCHEM-87-1-0001
Lajayer, H. A., Savaghebi, G., Hadian, J., Hatami, M., and Pezhmanmehr, M. (2016). Comparison of copper and zinc effects on growth, micro- and macronutrients status and essential oil constituents in pennyroyal (Mentha pulegium L.). Braz. J. Bot. 40, 379–388. doi:10.1007/s40415-016-0353-0
Li, T., Ferns, K., Yan, Z. Q., Yin, S. Y., Kou, J. J., Li, D., et al. (2021). Acanthopanax senticosus: photochemistry and anticancer potential. Am. J. Chin. Med. 44, 1543–1558. doi:10.1142/S0192415X16500865
Liang, Q., Yu, X., Qu, S., Xu, H., and Sui, D. (2010). Acanthopanax senticosides B ameliorates oxidative damage induced by hydrogen peroxide in cultured neonatal rat cardiomyocytes. Eur. J. Pharmacol. 627, 209–215. doi:10.1016/j.ejphar.2009.10.055
Liu, J., Zhang, X., and Sheng, J. (2022). Integrative analysis of the transcriptome and metabolome reveals the mechanism of saline–alkali stress tolerance in Astragalus membranaceus (Fisch) Bge. var. mongholicus (Bge.) Hsiao. Food Qual. Saf. 6. doi:10.1093/fqsafe/fyac001
Ma, S., Zhu, G., Yu, F., Zhu, G., Wang, D., Wang, W., et al. (2018). Effects of manganese on accumulation of Glycyrrhizic acid based on material ingredients distribution of Glycyrrhiza uralensis. Industrial Crops Prod. 112, 151–159. doi:10.1016/j.indcrop.2017.09.035
Mao, K., Dong, Q., Li, C., Liu, C., and Ma, F. (2017). Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant. Sci. 8, 480. doi:10.3389/fpls.2017.00480
Mattiello, E. M., Ruiz, H. A., Neves, J. C., Ventrella, M. C., and Araujo, W. L. (2015). Zinc deficiency affects physiological and anatomical characteristics in maize leaves. J. Plant. Physiol. 183, 138–143. doi:10.1016/j.jplph.2015.05.014
Meng, L., Zhou, R., Lin, J., Zang, X., Wang, Q., Wang, P., et al. (2022). Transcriptome and metabolome analyses reveal transcription factors regulating ganoderic acid biosynthesis in Ganoderma lucidum development. Front. Microbiol. 13, 956421. doi:10.3389/fmicb.2022.956421
Mitsuda, N., and Ohme-Takagi, M. (2009). Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 50, 1232–1248. doi:10.1093/pcp/pcp075
Mukhopadhyay, M., Das, A., Subba, P., Bantawa, P., Sarkar, B., Ghosh, P., et al. (2013). Structural, physiological, and biochemical profiling of tea plants under zinc stress. Biol. Plant. 57, 474–480. doi:10.1007/s10535-012-0300-2
Ng, D. W., Abeysinghe, J. K., and Kamali, M. (2018). Regulating the Regulators: the control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 19, 3737. doi:10.3390/ijms19123737
Nuruzzaman, M., Sharoni, A. M., and Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 248. doi:10.3389/fmicb.2013.00248
Razzaq, K., Khan, A. S., Malik, A. U., Shahid, M., and Ullah, S. (2013). Foliar application of zinc influences the leaf mineral status, vegetative and reproductive growth, yield and fruit quality of ‘Kinnow’ Mandarin. J. Plant Nutr. 36, 1479–1495. doi:10.1080/01904167.2013.785567
Semida, W. M., Abdelkhalik, A., Mohamed, G. F., Abd El-Mageed, T. A., Abd El-Mageed, S. A., Rady, M. M., et al. (2021). Foliar application of zinc oxide nanoparticles promotes drought stress tolerance in eggplant (Solanum melongena L.). Solanum melongena L. Plants 10, 421. doi:10.3390/plants10020421
Shi, X., Yang, Y., Ren, H., Sun, S., Mu, L. T., Chen, X., et al. (2019). Identification of multiple components in deep eutectic solvent extract of Acanthopanax senticosus root by ultra-high-performance liquid chromatography with quadrupole orbitrap mass spectrometry. Phytochem. Lett. 35, 175–185. doi:10.1016/j.phytol.2019.11.017
Singh, P., and Dwivedi, P. (2019). Micronutrients zinc and boron enhance stevioside content in Stevia rebaudiana plants while maintaining genetic fidelity. Industrial Crops Prod. 140, 111646. doi:10.1016/j.indcrop.2019.111646
Sithisarn, P., and Jarikasem, S. (2009). Antioxidant activity of Acanthopanax trifoliatus. Med. Princ. Pract. 18, 393–398. doi:10.1159/000226294
Sun, J., Li, W., Zhang, Y., Guo, Y., Duan, Z., Tang, Z., et al. (2021). Metabolomics analysis reveals potential mechanisms in bupleurum L. (Apiaceae) induced by three levels of nitrogen fertilization. Agronomy 11, 2291. doi:10.3390/agronomy11112291
Sun, Y., Song, K., Sun, L., Qin, Q., Jiang, T., Jiang, Q., et al. (2020). Morpho-physiological and transcriptome analysis provide insights into the effects of zinc application on nitrogen accumulation and metabolism in wheat (Triticum aestivum L.). Plant Physiol. Biochem. 149, 111–120. doi:10.1016/j.plaphy.2020.01.038
Tariverdizadeh, N., Mohebodini, M., Chamani, E., and Ebadi, A. (2021). Iron and zinc oxide nanoparticles: an efficient elicitor to enhance trigonelline alkaloid production in hairy roots of fenugreek. Industrial Crops Prod. 162, 113240. doi:10.1016/j.indcrop.2021.113240
Vojodi Mehrabani, L., Kamran, R. V., Hassanpouraghdam, M. B., and Pessarakli, M. (2017). Zinc sulfate foliar application effects on some physiological characteristics and phenolic and essential oil contents of lavandula stoechas L. Under sodium chloride (NaCl) salinity conditions. Commun. Soil Sci. Plant Analysis 48, 1860–1867. doi:10.1080/00103624.2017.1406105
Wang, X., Niu, Y., and Zheng, Y. (2021). Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 22, 6125. doi:10.3390/ijms22116125
White, P. J., and Broadley, M. R. (2011). Physiological limits to zinc biofortification of edible crops. Front. Plant. Sci. 2, 80–11. doi:10.3389/fpls.2011.00080
Wu, C. Y., Lu, L. L., Yang, X. E., Feng, Y., Wei, Y. Y., Hao, H. L., et al. (2010). Uptake, translocation, and remobilization of zinc absorbed at different growth stages by rice genotypes of different Zn densities. J. Agric. Food Chem. 58, 6767–6773. doi:10.1021/jf100017e
Xie, R., Zhao, J., Lu, L., Brown, P., Guo, J., and Tian, S. (2020). Penetration of foliar-applied Zn and its impact on apple plant nutrition status: in vivo evaluation by synchrotron-based X-ray fluorescence microscopy. Hortic. Res. 7, 147. doi:10.1038/s41438-020-00369-y
Yadegari, M. (2017). Effects of Zn, Fe, Mn and Cu foliar application on essential oils and morpho-physiological traits of lemon balm (Melissa Officinalis L.). J. Essent. Oil Bear. Plants 20, 485–495. doi:10.1080/0972060x.2017.1325010
Zeng, H., Wu, H., Yan, F., Yi, K., and Zhu, Y. (2021). Molecular regulation of zinc deficiency responses in plants. J. Plant Physiol. 261, 153419. doi:10.1016/j.jplph.2021.153419
Zhang, H., Yang, H., Wang, Y., Gao, Y., and Zhang, L. (2013). The response of ginseng grown on farmland to foliar-applied iron, zinc, manganese and copper. Industrial Crops Prod. 45, 388–394. doi:10.1016/j.indcrop.2012.12.047
Zhang, S. N., Li, X. Z., Liu, S. M., and Lu, F. (2015). Metabonomic study of the effects of Acanthopanax senticosus on peripheral system of rats. Planta. Med. 81, 722–732. doi:10.1055/s-0035-1545915
Zhang, M., Yu, Z., Zeng, D., Si, C., Zhao, C., Wang, H., et al. (2021b). Transcriptome and metabolome reveal salt-stress responses of leaf tissues from Dendrobium officinale. Biomolecules 11, 736. doi:10.3390/biom11050736
Zhang, Y., Zhang, Y., and Liu, Z. (2021a). Effects of Acanthopanax senticosus supplementation on innate immunity and changes of related immune factors in healthy mice. Innate Immun. 27, 461–469. doi:10.1177/1753425920955200
Zhao, K., Chen, S., Yao, W., Cheng, Z., Zhou, B., and Jiang, T. (2021). Genome-wide analysis and expression profile of the bZIP gene family in poplar. Bmc. Plant. Biol. 21, 122. doi:10.1186/s12870-021-02879-w
Keywords: Acanthopanax senticosus, zinc (Zn), foliar fertilization, metabolome, transcriptome
Citation: Sun T, Sun J, Liu Y, Ren Y, Li Y, Shi C, Nasr A, Tang Z and Abozeid A (2023) Metabolome and transcriptome analyses provide new insights into the mechanisms underlying the enhancement of medicinal component content in the roots of Acanthopanax senticosus (Rupr. et Maxim.) Harms through foliar application of zinc fertilizer. Front. Genet. 14:1259674. doi: 10.3389/fgene.2023.1259674
Received: 16 July 2023; Accepted: 31 October 2023;
Published: 17 November 2023.
Edited by:
Muhammad Ali Raza, Islamia University of Bahawalpur, PakistanReviewed by:
Samuel Seaver, Argonne National Laboratory (DOE), United StatesMuhammad Hayder Bin Khalid, Islamia University of Bahawalpur, Pakistan
Copyright © 2023 Sun, Sun, Liu, Ren, Li, Shi, Nasr, Tang and Abozeid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonghua Tang, dGFuZ3poQG5lZnUuZWR1LmNu; Ann Abozeid, YW5uYWJvemVpZEB5YWhvby5jb20=
 Tongze Sun
Tongze Sun Jialin Sun2
Jialin Sun2 Zhonghua Tang
Zhonghua Tang Ann Abozeid
Ann Abozeid