
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Genet. , 24 October 2023
Sec. Genetics of Common and Rare Diseases
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1254833
 Mohammed Dashti1
Mohammed Dashti1 Rasheeba Nizam1
Rasheeba Nizam1 Sumi Elsa John1
Sumi Elsa John1 Motasem Melhem2
Motasem Melhem2 Arshad Channanath1
Arshad Channanath1 Hessa Alkandari3,4
Hessa Alkandari3,4 Thangavel Alphonse Thanaraj1*
Thangavel Alphonse Thanaraj1* Fahd Al-Mulla1*
Fahd Al-Mulla1*ONECUT1 gene, encoding hepatocyte nuclear factor 6, is involved in pancreas and liver development. ONECUT1 mutations impair the function of pancreatic β-cells and control a transcriptional/epigenetic machinery regulating endocrine development. Homozygous nonsense and missense mutations at ONECUT1_p.E231 and a homozygous frameshift mutation at ONECUT1_p.M289 were reported in neonatal diabetes individuals of French, Turkish, and Indian ethnicity, respectively. Additionally, heterozygous variants were observed in Northern European T2D patients, and Italian patients with neonatal diabetes and early-/late-onset T2D. Examining diverse populations, such as Arabs known for consanguinity, can generalize the ONECUT1 involvement in diabetes. Upon screening the cohorts of Kuwaiti T1D and MODY families, and of Kuwaiti and Qatari T2D individuals, we observed two homozygous variants—the deleterious missense rs202151356_p.H33Q in one MODY, one T1D, and two T2D individuals, and the synonymous rs61735385_p.P94P in two T2D individuals. Heterozygous variants were also observed. Examination of GTEx, NephQTL, mQTLdb and HaploReg highlighted the rs61735385_p.P94P variant as eQTL influencing the tissue-specific expression of ONECUT1, as mQTL influencing methylation at CpG sites in and around ONECUT1 with the nearest site at 677-bases 3′ to rs61735385_p.P94P; as overlapping predicted binding sites for NF-kappaB and EBF on ONECUT1. DNA methylation profiles of peripheral blood from 19 MODY-X patients versus eight healthy individuals revealed significant hypomethylation at two CpG sites—one located 617-bases 3′ to the p.P94P variant and 8,102 bases away from transcription start; and the other located 14,999 bases away from transcription start. Our study generalizes the association of ONECUT1 with clinical diversity in diabetes.
One Cut Homeobox 1 (ONECUT1), also known as hepatocyte nuclear factor 6 (HNF6), is a pancreatic transcription factor that influences various cellular processes, including glucose metabolism. It belongs to a family of genes that encode key proteins involved in pancreas and liver development. ONECUT1 plays a crucial role in regulating the fate of pancreatic progenitor cells (Heller et al., 2021). Recent research suggests that genetic variations in the ONECUT1 gene contribute to various forms of diabetes by impairing the function of insulin-producing pancreatic β-cells; Philippi et al. (2021) presented an exemplary study on this subject, wherein they observed rare variants within the ONECUT1 gene in individuals with monogenic diabetes (recessive and dominant) and multifactorial type 2 diabetes (T2D). The authors observed the following ONECUT1 variants: i) a homozygous protein-truncating variant, p.E231X, found in a French boy (from a consanguineous family) with severe neonatal syndromic diabetes; ii) a homozygous missense variant, p.E231D, found in a Turkish boy (from a consanguineous family) diagnosed with insulin-requiring diabetes at the age of 14 months; and iii) a set of six heterozygous missense variants in 13 T2D patients, with an early disease onset than noncarriers, of predominantly Northern European ancestry from the Ulm Diabetes Cohort (UDC-T2D cohort); these six variants included p.H33Q, p.G30S, p.G62C, p.P75A, and p.V242A showing association with increased risk of T2D, and p.G96D showing a protective effect but having a modifier effect from p.P75A. A study by Prudente et al. (2022), subsequent to that of Philippi et al. (2021), investigated the role of ONECUT1 variants in Italian patients with various forms of nonautoimmune diabetes mellitus, including permanent neonatal diabetes mellitus (PNDM), familial diabetes, and early- and late-onset T2D. The researchers identified 13 ONECUT1 heterozygous variants in patients with different subtypes of diabetes. A more recent report by Russ-Silsby et al. (2023), identified a homozygous frameshift variant in ONECUT1 as the cause of neonatal diabetes in one individual referred from India to Exeter Monogenic Diabetes Cohort; they further found, by way using a rare variant burden test in the UK Biobank European cohort, a significant association between heterozygous ONECUT1 null variants and type 2 diabetes. The findings from the above-mentioned three studies confirm the critical role of ONECUT1 in human beta-cell function.
Further to examining genetic variants from ONECUT1, Philippi et al. (2021) dissected the functional consequences of defective protein encoded by ONECUT1 in pancreatic development, by way of using genome-edited human embryonic stem cells and patient-specific induced pluripotent stem cells. Their study revealed that loss of ONECUT1 altered transcription factor binding and enhancer activity and NKX2.2/NKX6.1 expression in pancreatic progenitor cells resulting in impaired pancreatic progenitor formation and an impaired subsequent endocrine program. Collectively, they demonstrated that ONECUT1 controls a transcriptional and epigenetic machinery regulating endocrine development, involved in a spectrum of diabetes, Studies involving additional populations can help to further confirm the contribution of ONECUT1 variants to different diabetes types. The Arab population is known for the common practice of consanguineous marriages and inbreeding (Tadmouri et al., 2009; Al-Gazali and Hamamy, 2014). Arab countries, such as Kuwait, have high prevalence of diabetes and metabolic disorders (Channanath et al., 2013; Shaltout et al., 2017; Abu-Farha et al., 2021; Al-Kandari et al., 2021; Elashi et al., 2022). Consanguineous marriages among Arabs often involve cousins or relatives within large extended families or the same tribe, with consanguinity rates reaching as high as 54.3% (Al-Awadi et al., 1985). This practice of consanguinity has led to increased endogamy, homozygosity, and accumulation of deleterious recessive alleles in the gene pool, making the population susceptible to various recessive genetic disorders. Consanguinity and inbreeding may also play a major role in the aetiology of complex disorders, such as diabetes (Rudan et al., 2006), with autosomal recessive alleles potentially playing a larger role in the pathogenesis of complex traits in this population (Bittles and Black, 2010). These deliberations on the population history of Arabs motivated us to explore the broader contribution of ONECUT1 in this indigenous population. In this study, we examined the coding variants from ONECUT1, by way of screening our in-house exome datasets comprising diabetic individuals of Arab ethnicity. These datasets included i) exome data from Kuwaiti families with one or more members diagnosed for maturity-onset diabetes of the young (MODY), ii) exome data from Kuwaiti families with one or more members diagnosed for type 1 diabetes (T1D), and iii) exome data from individuals diagnosed for T2D. We also examined publicly available exome datasets of T2D individuals from Qatar. Publicly available HaploReg databases were used to examine whether the ONECUT1 variants are part of transcription factor binding sites. The GTEx and NephQTL databases were examined to assess whether the ONECUT1 variants have genetic influence on tissue-specific gene expression. Further, to examine the epigenetic involvement of the ONECUT1 variants in diabetes, we used mQTLdb to examine whether the observed ONECUT1 variants are QTLs for methylation sites in and around the gene region. Additionally, we examined the DNA differential methylation profiles of peripheral blood from MODY patients.
The study protocol was approved by the Ethical Review Committee of Dasman Diabetes Institute in accordance with the guidelines of the Declaration of Helsinki and the United States Federal Policy for the Protection of Human Subjects. The cohorts corresponding to the exome datasets examined in this study were recruited as part of earlier published studies, and no participant recruitment took place as part of the current study. At the time of recruitment, written informed consent was obtained from the individuals; in the case of minor individuals, consent was obtained from the parent(s) and assent was obtained from the minor individuals.
We analyzed the ONECUT1 variants in individuals of Arab ethnicity, by way of using our in-house exome datasets of diabetes patients from Kuwait and publicly available exome datasets of diabetes individuals from Qatar. The datasets consisted i) exomes of 47 MODY patients and 32 healthy individuals from 32 Kuwaiti families with one or more members diagnosed with MODY (Al-Kandari et al., 2021); ii) exomes of 63 T1D patients and 62 healthy individuals from 35 Kuwaiti families with one or more members diagnosed with T1D; iii) exomes of 122 T2D patients and 169 healthy individuals from a T2D cohort in Kuwait (John et al., 2018); and iv) publicly available exomes from 503 T2D patients and 261 healthy individuals from Qatar (O’Beirne et al., 2018). Additionally, we performed targeted sequencing of ONECUT1 on an additional sample set of 88 T2D individuals from Kuwait. In order to confirm the genotype calls at examined variants from ONECUT1 in the exome datasets, we performed targeted genotyping on samples displaying genotypes that are heterozygous or homozygous for minor allele at selected variants.
Transcription factor binding motifs, from Haploreg (Ward and Kellis, 2012), overlapping the position of the ONECUT1 variants were identified using the T1D Knowledge Portal (Kudtarkar et al., 2023). Position weight matrices used in the tool are as derived by Kheradpour and Kellis (Kheradpour and Kellis, 2014) using regulatory motifs in ENCODE transcription factor binding experiments. Possible roles of variants in disease aetiology were investigated by analysing data on tissue-specific expression of the quantitative trait locus (eQTL) from resources such as the NEPTUNE patient characteristics for subjects in expression quantitative trait loci (NephQTL) (Gillies et al., 2018) and genotype-tissue expression (GTEx) (GTEx Consortium, 2017) databases. The mQTLdb (Gaunt et al., 2016) (http://www.mqtldb.org/), which is a public resource on lead mQTL loci influencing methylation variation, in children at birth, childhood, adolescence and their mothers during pregnancy and middle age, was used to examine the genetic influence of the ONECUT1 variants on differential methylation at CpG sites at different life stages.
The methodologies for methylation sequencing and DNAm analysis are described in detail in our previous publication (Dashti et al., 2022). A brief summary of the adopted methods is as given below. Methylome libraries were prepared using TruSeq Methyl Capture EPIC Library Prep Kit reagents (Illumina Inc., United States) using the protocol provided by the manufacturer. Indexed libraries were clustered on a cBOT system using TruSeq paired cluster kit V3 (Illumina Inc., United States), and sequencing of paired-end reads was performed on an Illumina HiSeq 2500 sequencing platform. High-quality paired-end trimmed reads, generated using Trim Galore! Version 0.3.3 (Babraham Bioinformatics, United Kingdom), were mapped to the in silico UCSC human assembly HG19 reference genome using Bismark (version 0.22.3) (Krueger and Andrews, 2011) and Bowtie2 (version 2.3.5.1) (Langmead and Salzberg, 2012). Single CpG site analysis was performed on the resulting sequence alignment/map (SAM) files using the methylKit package version 1.2.4 (Akalin et al., 2012). Differentially methylated single CpG sites were detected if the methylKit q-value is ≤0.01 and the difference in the methylation levels between the compared groups was ideally ≥25%; and additional sites exhibiting less differences in methylation levels were considered, as long as the q-value criteria is satisfied.
Upon examining the study exome datasets of individuals with MODY, T1D and T2D for variants from ONECUT1, we observed two homozygous variants, namely, the deleterious missense rs202151356_ p.H33Q, and the synonymous rs61735385_p.P94P (Table 1). The first one was observed in one Kuwaiti individual with MODY, one Kuwaiti individual with T1D, and in two Qatari individuals with T2D. The second homozygous variant was observed in two Qatari individuals with T2D. The minor allele frequency of the rs202151356_ p.H33Q in the five continental populations are particularly very low with a value of ≤0.3% while that of rs61735385_p.P94P is >5% in Europeans and South Asians, and <1% in East Asians. These two variants were also observed in heterozygous form in additional patients from Kuwait and Qatar (see Table 1). Further, we observed two heterozygous variants, namely, rs866368632_5′ UTR (in one Kuwaiti individual with MODY) and rs201286990_p.M161L (in one Kuwaiti individual with T2D) (see Table 1; Figure 1). Both these two heterozygous variants were not seen in continental populations but seen in Middle East (rs866368632_5′ UTR with an MAF of 0.3%) or in Ashkenazi Jews (rs201286990_p.M161L with an MAF of 0.7%). In addition to the variants mentioned above, Qatari T2D patients exhibited the following heterozygous variants: rs2075613_p.G287G (2 individuals), rs147745937_p.G230E (1 individual), rs151292910_p.G188D (1 individual), rs1483480013_p.G186S (1 individual), rs142641519_p.G81D (1 individual), and rs760541486_p.V16L (2 individuals). None of these variants was observed in the datasets of the Kuwaiti cohort, except that the rs2075613_p.G287G was seen in two healthy individuals from Kuwait.
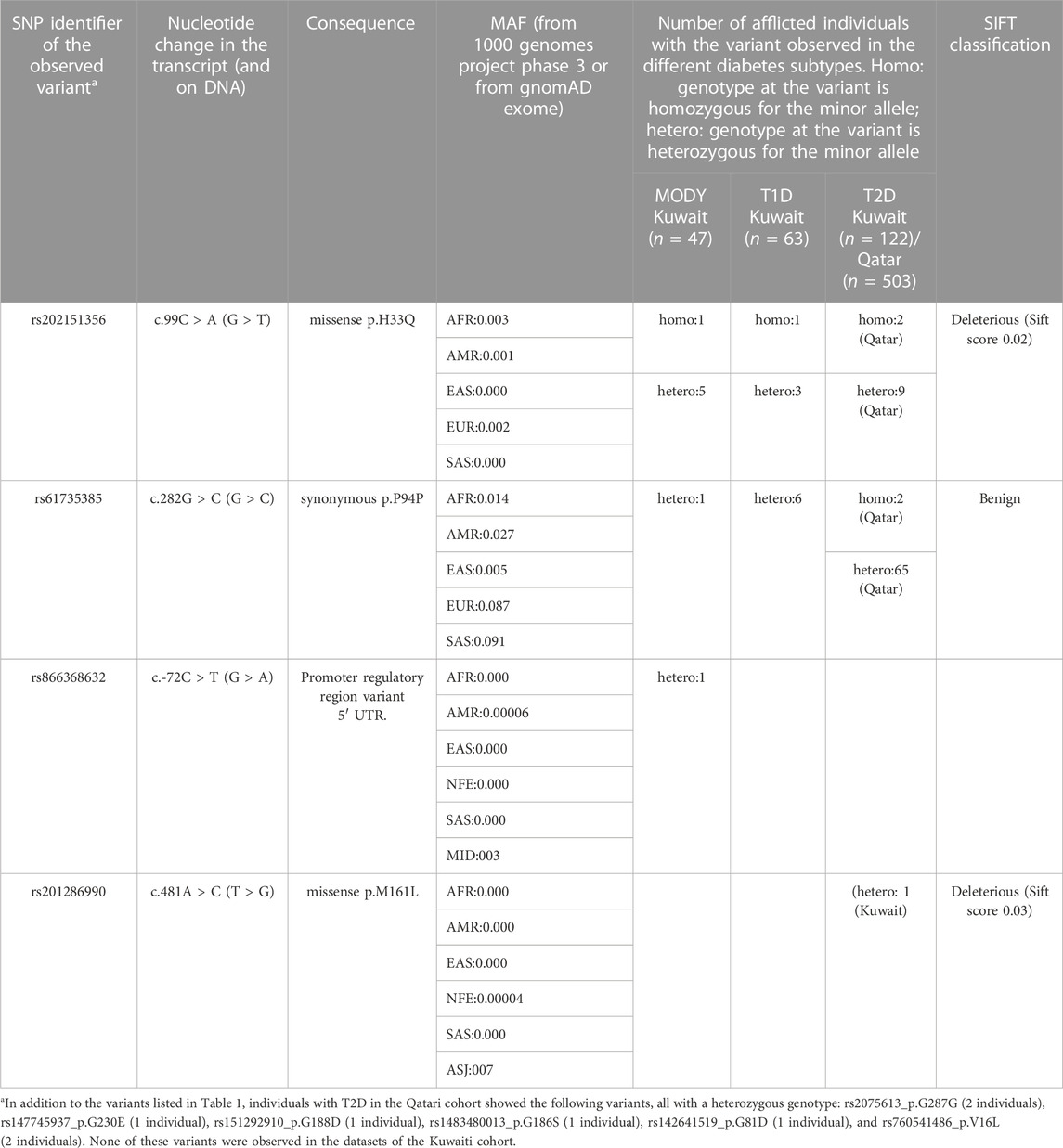
TABLE 1. Variants identified in the coding region of ONECUT1 gene in patients with different subtypes of diabetes mellitus. The corresponding transcript identifier is NM_004498.4.
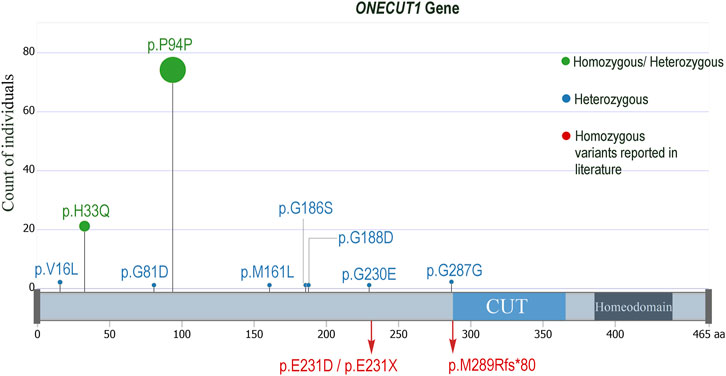
FIGURE 1. Lollypop mutation plot displaying the ONECUT1 variants seen in the examined diabetes datasets from Kuwait and Qatar. The p.H33Q and the p.P94P were seen in homozygous as well as in heterozygous forms, while all the others were seen only in heterozygous forms. The homozygous mutations observed by Philippi et al. (2021), Russ-Silsby et al. (2023) are also indicated.
Familial aggregation of diabetes in Kuwaiti families is amply demonstrated in the pedigrees considered in this study–for example, family history of the MODY patient exhibiting the rs202151356_p.H33Q variant indicates familial aggregation of diabetes in both the maternal and paternal first-degree relatives (Supplementary Figure S1A). The rs202151356_p.H33Q variant was observed (either as homozygous or heterozygous) in four patients with diabetes from three unrelated MODY families (Table 2). Two of these four patients with MODY were siblings. The grandparent of another patient with MODY had been diagnosed with T1D, while the entire family has a high prevalence of T2D. These four patients did not carry mutations and/or structural variants in known MODY genes, as confirmed by using whole-exome sequencing and Multiplex ligation-dependent probe amplification assay, respectively. Moreover, two of these four individuals tested negative for glutamic acid decarboxylase (GAD) and islet antigen 2 (IA2) autoantibodies, while the other two tested positive for GAD and IA2 autoantibodies. Furthermore, the individual who exhibited the homozygous TT genotype had an onset of diabetes at an age in the range of 0–3 years, while the three individuals with the heterozygous GT genotype had onset age in the range of 8–14 years.
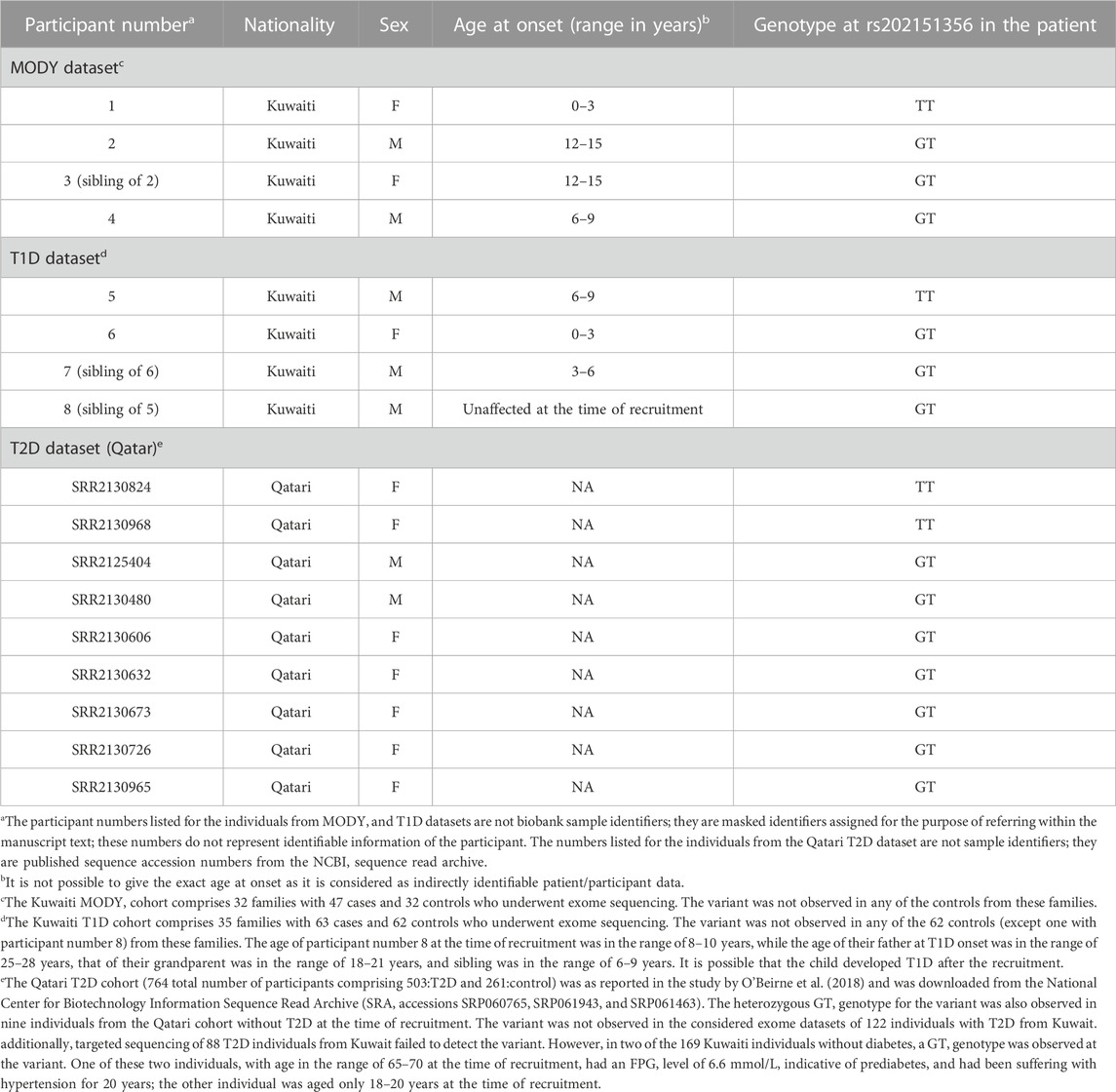
TABLE 2. ONECUT1: rs202151356_p.H33Q) variant observed in datasets of Arab individuals with diabetes.
In the T1D dataset, the rs202151356_p.H33Q variant was observed in three individuals with T1D from two unrelated T1D families in Kuwait. Among these three individuals with T1D, the homozygous TT genotype was observed in one person, with an age at onset in the range of 6–9 years; the other two individuals exhibiting heterozygous GT genotype were siblings with age at onset in the range of 0–3 years and 3–6 years. Additionally, a fourth individual (participant number 8 in Table 2), unaffected at the time of recruitment, also displayed the GT genotype at the variant (see Supplementary Figure S1B for the pedigree).
Upon examining the transcription factor binding motifs from HaploReg (see Methods), it was found that both the homozygous variants overlapped the transcription factor binding motifs (known or predicted using motifs in ENCODE transcription factor binding experiments). The mutations at these sites altered the score (information content) of the motifs by up to >11 bits (Supplementary Table S1). Alternate allele at the homozygous rs202151356_p.H33Q variant reduced the score of the TF motifs Myc_disc9 and Myc_known8; alternate allele at the other homozygous rs61735385_p.P94P variant reduced the scores of TF motifs NF-kappaB_disc2 and EBF_known3. Of the two heterozygous variants seen in Kuwaiti diabetes individuals (see Table 1), the rs201286990_p.M161L overlapped with the TF motif of Mef2_known6 and the alternate allele reduced the score by up to 12 bits.
Examination of GTEx to determine whether the observed variants exhibit genotype-based differential expression of genes revealed that the rs61735385_p.P94P significantly downregulates the expression levels of ONECUT1 (p-value = 5.2E−07) and RP11-209K10.2 (p-value = 1.1E−06) genes in testis (Supplementary Table S2). Examination of the NephQTL database to determine which of the lead variants act as eQTL in kidney revealed that the rs61735385_p.P94P exhibited genotype-based differential expression (downregulation) of MYO5A, MYO5C, ONECUT1, FAM214A, and ARPP19 genes in either glomerular or tubular tissues; however, the p-values for the t-statistics were not significant (Supplementary Table S2).
Examination of the mQTLdb revealed that the rs61735385_p.P94P has genetic influence on DNA methylation at various CpG sites, in or around the ONECUT1 gene region, during different life stages (Supplementary Table S3). The nearest CpG site, namely, cg02061705, is located 677 bases away within the gene from the rs61735385_p.P94P variant; and is differentially methylated in adolescent stage of the life cycle. The farthest CpG is located 19,748 bases downstream to the position of the variant.
We examined data from our ongoing methylation study to determine whether differential methylation events occur in response to diabetes in the genome region harbouring the ONECUT1 gene. We focused on patients with MODY-X non-autoimmune diabetes, who were clinically suspected as having MODY, as per the International Society for Pediatric and Adolescent Diabetes (ISPAD) criteria, and responded well to diabetes treatment, but did not exhibit variants of significance in any known monogenic diabetes-associated genes. By comparing DNA methylation (DNAm) profiling of peripheral blood from 19 individuals with MODY-X (including three patients with the rs202151356_p.H33Q variants) with DNAm profiling of eight control individuals, we found two significantly differentially methylated CpG sites at Chr15:53058083 (q-value = 3.75E−40), and Chr15: 53081183 (q-value = 0.008) (Table 3). Both the sites are hypomethylated in MODY cases, the first one with 35% difference in methylation and the second one with close to 10% difference. This second site, located at Chr15:53081183 is particularly interesting as it is mere 800 bases away from the rs202151356_p.H33Q variant (Chr15:53081983) and 617 bases away from the rs61735385_p.P94P variant (Chr15:53081800). The first site, located at Chr15:53058083 is located 23,900 bases and 24,083 bases away from the rs202151356_p.H33Q and the rs61735385_p.P94P variants, respectively. The first site is located 14,999 bases away from the transcription site and the second site is located at 8,102 bases away from the transcription site.

TABLE 3. CpG sites differentially methylated in ONECUT1 gene between individuals with MODY-X and healthy individuals at a q-value ≤ 0.01 for differences in methylation levels.
Our study observed two homozygous ONECUT1 variants in Arab patients of diabetes—the deleterious missense rs202151356_p.H33Q was observed in 1 individual of MODY, 1 individual of T1D and two individuals of T2D; and the synonymous rs61735385_p.P94P was observed in two T2D individuals. These variants were observed with heterozygous genotypes as well in additional patients. None of the individuals without diabetes (healthy controls) from the T1D or MODY families carried these variants, except for one individual (participant number 8, as mentioned in Table 2, carrying a GT heterozygous genotype at rs202151356_p.H33Q), and is a sibling of a T1D proband with homozygous genotype) who was not diagnosed with T1D at the time of recruitment (see Supplementary Figure S1B). The age at the time of recruitment of this unaffected participant was in the range of 8–10 years; considering that the age at onset of T1D in the grandparent was in the range of 18–21 years, in the father was in the range of 25–28 years, and in the sibling was in the range of 6–9 years, it is possible that the child developed T1D after recruitment.
Prudente et al. (2022) previously reported these two variants in heterozygous forms in Italian patients: the rs202151356_p.H33Q in individuals with early-onset T2D individuals; and the rs61735385_p.P94P in individuals with familial diabetes of adulthood, early-onset T2D, and late-onset T2D. Philippi et al. (2021) also observed the rs202151356_p.H33Q variant in heterozygous form in three T2D individuals of Northern European ancestry from the UDC-T2D cohort. For the first time, we report the presence of these two variants homozygous for the minor allele in individuals with MODY, T1D, or T2D. The rs202151356_p.H33Q is a very rare variant in all the five continental populations, while rs61735385_p.P94P is a common variant in both the European and South Asians while it is rare in East Asians. The SIFT tool predicted the rs202151356_p.H33Q to be deleterious.
Apart from the two homozygous variants, the Kuwaiti datasets revealed two heterozygous variants, namely, the rs866368632_5′UTR in a MODY individual, and the rs201286990_p.M161L in a T2D individual. The Qatari T2D dataset revealed the following 6 heterozygous variants, namely, the rs2075613_p.G287G rs147745937_p.G230E, rs151292910_p.G188D, rs1483480013_p.G186S, rs142641519_p.G81D, and rs760541486_p.V16L. Prudente et al. (2022) had observed two of these heterozygous variants in their study: the rs1483480013_p.G81D variant in an individual with permanent neonatal diabetes and the rs2075613_p.G287G in individuals with permanent neonatal diabetes, familial diabetes of adulthood, and late-onset T2D.
While the rs202151356_p.H33Q is a deleterious missense variant, the rs61735385_p.P94P seems to have implications on transcription-related aspects as indicated by the following findings of our study: i) Both the homozygous variants overlapped the predicted transcription factor (TF) binding motifs (known or predicted using motifs in ENCODE TF binding experiments) on ONECUT1. Alternate allele at the homozygous rs202151356_p.H33Q variant reduced the score of motifs for the binding of TF Myc; alternate allele at the other homozygous rs61735385_p.P94P variant reduced the scores of TF motifs for the binding of transcription factors NF-κB and EBF. It is known that Myc has a potential physiological role on the regulation of the β-cell function (Rosselot et al., 2021). NF-κB activation is a key event involved in diabetes pathogenesis (Patel and Santani, 2009). ii) The genotype-tissue specific expression databases indicated that the rs61735385_p.P94P significantly downregulates the tissue-specific expression levels of ONECUT1 and RP11-209K10.2 genes. The study of Philippi et al. (2021) demonstrated that one of the endocrine regulatory elements affected by T2D-associated variants near ONECUT1 is the above-mentioned lncRNA RP11-209K10.2, and that the lncRNA exhibits similar tissue-specific expression as ONECUT1. iii) The rs61735385_p.P94P is a mQTL regulating the DNA methylation at CpG sites, in or around the ONECUT1 gene region, during different stages of life. The nearest CpG site, is located a mere 677 bases away within the gene from the rs61735385_p P94P variant; and is differentially methylated in adolescent stage of the life cycle.
Further, our data from comparing the DNA methylation profiles of MODY-X nonautoimmune diabetes patients and healthy individuals suggested an epigenetic involvement of ONECUT1 gene via differential DNA methylation in patients with MODY-X. The ONECUT1 emerged as one of the top hypomethylated genes in MODY-X. Of the two observed statistically significant hypomethylation CpG sites in ONECUT1 in MODY-X, one was located 617 bases away from the rs61735385_p.P94P variant. The other CpG is located 14,999 bases away from the transcription start; and is located in an intron within the gene body. The ONECUT gene region is known to exhibit an altered DNA methylation pattern in response to stimuli. For instance, in mice that were fed a high-fat diet, Zhang et al. (2017) observed a significant suppression of ONECUT1 expression in the RNAseq data and differential methylation of CpG sites at multiple sites within 10,000 bp of the transcription start region. Since ONECUT1 is a pancreatic transcription factor, differential methylation events may directly impact the cellular mechanisms that regulate the disease model (Thompson et al., 2015). The work by Heller et al. (2021) reported that ONECUT1 plays a crucial role in pancreatic and endocrine development, and its transcriptional activity is intricately linked to its methylation status. The study by Rimoldi et al. (2022) underscored the interconnections between DNA methylation profiles and transcription factor binding. Our findings further support this association, particularly in the context of diabetes.
There is considerable familial aggregation of diabetes in Kuwaiti population (see Supplementary Figure S1A for an illustration). Our study illustrates familial aggregation of these risk variants: two of the four MODY probands with the homozygous or heterozygous rs202151356_p.H33Q variants are siblings, and two of the three T1D probands with the homozygous or heterozygous rs202151356_p.H33Q variants are siblings (see Table 2). Performing studies such as ours addressing aspects of molecular diagnosis for patients represent a very important task for families.
Over the past few decades, diabetes has evolved to a complex multifactorial disease, characterized by a wide range of clinical and genetic heterogeneity. The dichotomous classification of diabetes as T1D and T2D has been overruled by several distinct subtypes that differ in terms of clinical presentation, genetic variations, penetrance, and disease loci. While T1D and T2D are the most prevalent forms of diabetes in the Arabian Peninsula, the region also accounts for a vast number of MODY cases. Mutations in 14 different genes have been associated with the aetiology of MODY, defining its specific subtypes. Mutations in GCK, HNF1A, HNF1B, and HNF4A account for approximately 80% of the MODY cases (Juszczak et al., 2016). Despite this, a large proportion of suspected MODY cases, referred to as MODY-X, remain unsolved, with no variants of significance detected in known MODY genes. Interestingly, our study highlights ONECUT1 methylation as a potential marker for MODY-X, representing either a unique subtype of MODY or a form of diabetes that is yet to be classified. In this context, it is important to point out that our previous study (Dashti et al., 2022) did not find ONECUT1 as one of the observed differentially methylated and expressed genes in familial T1D. Thus, the observed ONECUT1 methylation in the MODY patients in this study can help to clarify when suspected MODY individuals are misdiagnosed as T1D patients.
In summary, we highlight the significance of ONECUT1 as a recurrent marker for diabetes. Our data from Arab individuals with diabetes support the involvement of ONECUT1 variants, particularly the rs202151356_p.H33Q, in various forms of diabetes, especially MODY, T1D, and T2D. It is suggested that the same monogenic diabetes gene can contribute to different forms of diabetes with early or late onset, depending on the functional impact of the variant. Likewise, the same pathogenic variant can result in several diabetes phenotypes, even within the same family. It is also possible that patients initially diagnosed with T2D may have monogenic diabetes (Bonnefond et al., 2023). Therefore, there may be a common genetic basis for the development of various forms of diabetes, including T2D. For example, certain T2D susceptibility loci, such as the PPARG Pro12Ala variant, MTNR1B, HNF1A, GLIS3, 6q22.32, and novel loci near the MHC, which harbour HLA class II genes, are associated with approximately half of the risk for developing T1D (Eftychi et al., 2004; Raj et al., 2009; Andersen et al., 2014; Mahajan et al., 2014). The TCF7L2 variants (such as rs7903146 and rs12255372) are associated with the pathophysiology of gestational diabetes mellitus (GDM), T1D, latent autoimmune diabetes of adults (LADA), and T2D (Del Bosque-Plata et al., 2022). The findings of Philippi et al. (2021), Prudente et al. (2022), Russ-Silsby et al. (2023), and our study suggest that ONECUT1 is potentially associated with clinical diversity in diabetes, such as neonatal syndromic diabetes, insulin-requiring diabetes, insulin-requiring diabetes, MODY, T1D, and early- and late-onset T2D. By way of including the two homozygous mutations (p.Glu231Asp and p.E231X) observed with neonatal diabetes in the works of Philippi et al. (2021), the one homozygous frameshift mutation (p.Met289Argfs*80) observed with neonatal diabetes in the works of Russ-Silsby et al. (2023), and the two homozygous mutations p.H33Q and p.P94P that we observed with MODY, T1D and T2D, a total of 5 homozygous mutations have been reported so far in ONECUT1 gene as relating to the clinical diversity in diabetes (Supplementary Figure S2).
The present study has certain limitations. Firstly, our study lacked cohorts of patients diagnosed with other types of diabetes such as neonatal diabetes, childhood T2D diabetes, and atypical diabetes. Secondly, the peripheral blood sample is not the ideal biological sample as a) it contains a diverse mixture of cells and DNAm is cell-specific, and b) it may harbour DNA methylation events due to pathological differences relating to autoimmune processes that we could not consider for adjusting the results of differential methylation analysis. However, blood as biological sample has been used in other DNA methylation studies on diabetes (Disanto et al., 2013; Agardh et al., 2015; Johnson et al., 2020; Dashti et al., 2022).
The data from Arab individuals with diabetes further support the recurrent involvement of ONECUT1 variants, particularly the homozygous variants of rs202151356_p.H33Q and rs61735385_p.P94P, in various forms of diabetes. Additionally, our findings demonstrate the involvement of the ONECUT1 variants in regulating the transcriptional and epigenetic machinery relating to diabetes.
The data analyzed in this study is subject to the following licenses/restrictions: The study uses three data sets from previous studies. The first two data sets correspond to minor children diagnosed with MODY and type 1 diabetes—as the participants are minor, their data cannot be made public. However, requests to access these two datasets can be directed to either FA-M (ZmFoZC5hbG11bGxhQGRhc21hbmluc3RpdHV0ZS5vcmc=) or TT (YWxwaG9uc2UudGhhbmdhdmVsQGRhc21hbmluc3RpdHV0ZS5vcmc=). The third data set corresponds to adult individuals from Qatar diagnosed with type 2 diabetes; the raw data pertaining to this cohort is available publicly and can be downloaded from the National Center for Biotechnology Information Sequence Read Archive (SRA accessions SRP060765, SRP061943, and SRP061463). Requests to access these datasets should be directed to Requests for the first two data sets: FA-M (ZmFoZC5hbG11bGxhQGRhc21hbmluc3RpdHV0ZS5vcmc=) or TT (YWxwaG9uc2UudGhhbmdhdmVsQGRhc21hbmluc3RpdHV0ZS5vcmc=); Requests for the third data set on Qatari T2D individuals: from the National Center for Biotechnology Information Sequence Read Archive (SRA accessions SRP060765, SRP061943, and SRP061463).
The studies involving humans were approved by the Ethical Review Committee at Dasman Diabetes Institute. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
MD: Conceptualization, Formal Analysis, Writing–original draft, Writing–review and editing. RN: Formal Analysis, Writing–original draft. SJ: Formal Analysis, Writing–original draft. MM: Data curation, Formal Analysis, Software, Writing–original draft. AC: Data curation, Formal Analysis, Software, Writing–original draft. HA: Data curation, Resources, Writing–review and editing. TT: Conceptualization, Investigation, Writing–original draft, Writing–review and editing. FA-M: Conceptualization, Writing–review and editing, Investigation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by institutional funding from the Kuwait Foundation for the Advancement of Sciences.
We thank all referring physicians and patients for their interest and willingness to contribute to the study cohorts that provided us with exome datasets that were used in this study. We would like to thank Dr. Osama Alsmadi and Dr. Azza Shaltout for their initial support with the clinical recruitment of study participants. We would also like to thank Ms. Lubaina Koti for editing the manuscript for language and structure.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1254833/full#supplementary-material
Abu-Farha, M., Tuomilehto, J., and Abubaker, J. (2021). Editorial: diabetes in the Middle East. Front. Endocrinol. (Lausanne) 12, 638653. doi:10.3389/fendo.2021.638653
Agardh, E., Lundstig, A., Perfilyev, A., Volkov, P., Freiburghaus, T., Lindholm, E., et al. (2015) Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy. BMC Med. 13, Article number: 182. doi:10.1186/s12916-015-0421-5
Akalin, A., Kormaksson, M., Li, S., Garrett-Bakelman, F. E., Figueroa, M. E., Melnick, A., et al. (2012). methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13 (10), R87. doi:10.1186/gb-2012-13-10-r87
Al-Awadi, S. A., Moussa, M. A., Naguib, K. K., Farag, T. I., Teebi, A. S., el-Khalifa, M., et al. (1985). Consanguinity among the Kuwaiti population. Clin. Genet. 27 (5), 483–486. doi:10.1111/j.1399-0004.1985.tb00236.x
Al-Gazali, L., and Hamamy, H. (2014). Consanguinity and dysmorphology in Arabs. Hum. Hered. 77 (1-4), 93–107. doi:10.1159/000360421
Al-Kandari, H., Al-Abdulrazzaq, D., Davidsson, L., Nizam, R., Jacob, S., Melhem, M., et al. (2021). Identification of Maturity-Onset-Diabetes of the Young (MODY) mutations in a country where diabetes is endemic. Sci. Rep. 11 (1), 16060. doi:10.1038/s41598-021-95552-z
Andersen, M. K., Sterner, M., Forsén, T., Käräjämäki, A., Rolandsson, O., Forsblom, C., et al. (2014). Type 2 diabetes susceptibility gene variants predispose to adult-onset autoimmune diabetes. Diabetologia 57 (9), 1859–1868. doi:10.1007/s00125-014-3287-8
Bittles, A. H., and Black, M. L. (2010). Evolution in health and medicine Sackler colloquium: consanguinity, human evolution, and complex diseases. Proc. Natl. Acad. Sci. U. S. A., 107 (1), 1779–1786. doi:10.1073/pnas.0906079106
Bonnefond, A., Unnikrishnan, R., Doria, A., Vaxillaire, M., Kulkarni, R. N., Mohan, V., et al. (2023). Monogenic diabetes. Nat. Rev. Dis. Prim. 9 (1), 12. doi:10.1038/s41572-023-00421-w
Channanath, A. M., Farran, B., Behbehani, K., and Thanaraj, T. A. (2013). State of diabetes, hypertension, and comorbidity in Kuwait: showcasing the trends as seen in native versus expatriate populations. Diabetes Care 36 (6), e75. doi:10.2337/dc12-2451
Dashti, M., Nizam, R., Hebbar, P., Jacob, S., John, S. E., Channanath, A., et al. (2022). Differentially methylated and expressed genes in familial type 1 diabetes. Sci. Rep. 12 (1), 11045. doi:10.1038/s41598-022-15304-5
Del Bosque-Plata, L., Hernández-Cortés, E. P., and Gragnoli, C. (2022). The broad pathogenetic role of TCF7L2 in human diseases beyond type 2 diabetes. J. Cell Physiol. 237 (1), 301–312. doi:10.1002/jcp.30581
Disanto, G., Veelakova, J., Pakpoor, J., Elangovan, R. M., Sumnik, Z., Ulmannova, T., et al. (2013). DNA methylation in monozygotic quadruplets affected by type 1 diabetes. Diabetologia 56, 2093–2095. doi:10.1007/s00125-013-2972-3
Eftychi, C., Howson, J. M., Barratt, B. J., Vella, A., Payne, F., Smyth, D. J., et al. (2004). Analysis of the type 2 diabetes-associated single nucleotide polymorphisms in the genes IRS1, KCNJ11, and PPARG2 in type 1 diabetes. Diabetes 53 (3), 870–873. doi:10.2337/diabetes.53.3.870
Elashi, A. A., Toor, S. M., Diboun, I., Al-Sarraj, Y., Taheri, S., Suhre, K., et al. (2022). The genetic spectrum of maturity-onset diabetes of the young (MODY) in Qatar, a population-based study. Int. J. Mol. Sci. 24 (1), 130. doi:10.3390/ijms24010130
Gaunt, T. R., Shihab, H. A., Hemani, G., Min, J. L., Woodward, G., Lyttleton, O., et al. (2016). Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 17, 61. doi:10.1186/s13059-016-0926-z
Gillies, C. E., Putler, R., Menon, R., Otto, E., Yasutake, K., Nair, V., et al. (2018). An eQTL landscape of kidney tissue in human nephrotic syndrome. Am. J. Hum. Genet. 103 (2), 232–244. doi:10.1016/j.ajhg.2018.07.004
GTEx Consortium (2017). Genetic effects on gene expression across human tissues. Nature 550 (7675), 204–213. doi:10.1038/nature24277
Heller, S., Li, Z., Lin, Q., Geusz, R., Breunig, M., Hohwieler, M., et al. (2021). Transcriptional changes and the role of ONECUT1 in hPSC pancreatic differentiation. Commun. Biol. 4 (1), 1298. doi:10.1038/s42003-021-02818-3
John, S. E., Antony, D., Eaaswarkhanth, M., Hebbar, P., Channanath, A. M., Thomas, D., et al. (2018). Assessment of coding region variants in Kuwaiti population: implications for medical genetics and population genomics. Sci. Rep. 8 (1), 16583. doi:10.1038/s41598-018-34815-8
Johnson, R. K., Vanderlinden, L. A., Don, F., Carry, P. M., Seifert, J., Waugh, K., et al. (2020). Longitudinal DNA methylation differences precede type 1 diabetes. Sci. Rep. 10, 3721. doi:10.1038/s41598-020-60758-0
Juszczak, A., Pryse, R., Schuman, A., and Owen, K. R. (2016). When to consider a diagnosis of MODY at the presentation of diabetes: aetiology matters for correct management. Br. J. Gen. Pract. 66 (647), e457–e459. doi:10.3399/bjgp16X685537
Kheradpour, P., and Kellis, M. (2014). Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 42 (5), 2976–2987. doi:10.1093/nar/gkt1249
Krueger, F., and Andrews, S. R. (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27 (11), 1571–1572. doi:10.1093/bioinformatics/btr167
Kudtarkar, P., Costanzo, M. C., Sun, Y., Jang, D., Koesterer, R., Mychaleckyj, J. C., et al. (2023). Leveraging type 1 diabetes human genetic and genomic data in the T1D knowledge portal. PLoS Biol. 21 (8), e3002233. doi:10.1371/journal.pbio.3002233
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. doi:10.1038/nmeth.1923
Mahajan, A., Go, M. J., Zhang, W., Below, J. E., Gaulton, K. J., Ferreira, T., et al. (2014). Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 46 (3), 234–244. doi:10.1038/ng.2897
O'Beirne, S. L., Salit, J., Rodriguez-Flores, J. L., Staudt, M. R., Abi Khalil, C., Fakhro, K. A., et al. (2018). Exome sequencing-based identification of novel type 2 diabetes risk allele loci in the Qatari population. PLoS One 13 (9), e0199837. doi:10.1371/journal.pone.0199837
Patel, S., and Santani, D. (2009). Role of NF-kappa B in the pathogenesis of diabetes and its associated complications. Pharmacol. Rep. 61 (4), 595–603. doi:10.1016/s1734-1140(09)70111-2
Philippi, A., Heller, S., Costa, I. G., Senée, V., Breunig, M., Li, Z., et al. (2021). Mutations and variants of ONECUT1 in diabetes. Nat. Med. 27 (11), 1928–1940. doi:10.1038/s41591-021-01502-7
Prudente, S., Andreozzi, F., Mercuri, L., Alberico, F., Di Giamberardino, A., Mannino, G. C., et al. (2022). Contribution of ONECUT1 variants to different forms of non-autoimmune diabetes mellitus in Italian patients. Acta Diabetol. 59 (8), 1113–1116. doi:10.1007/s00592-022-01889-w
Raj, S. M., Howson, J. M., Walker, N. M., Cooper, J. D., Smyth, D. J., Field, S. F., et al. (2009). No association of multiple type 2 diabetes loci with type 1 diabetes. Diabetologia 52 (10), 2109–2116. doi:10.1007/s00125-009-1391-y
Rimoldi, M., Wang, N., Zhang, J., Villar, D., Odom, D. T., Taipale, J., et al. (2022). DNA methylation patterns of transcription factor binding regions characterize their functional and evolutionary contexts. bioRxiv - Prepr. Serv. Biol. doi:10.1101/2022.07.21.500978
Rosselot, C., Baumel-Alterzon, S., Li, Y., Brill, G., Lambertini, L., Katz, L. S., et al. (2021). The many lives of Myc in the pancreatic β-cell. J. Biol. Chem. 296, 100122. doi:10.1074/jbc.REV120.011149
Rudan, I., Campbell, H., Carothers, A. D., Hastie, N. D., and Wright, A. F. (2006). Contribution of consanguinuity to polygenic and multifactorial diseases. Nat. Genet. 38 (11), 1224–1225. doi:10.1038/ng1106-1224
Russ-Silsby, J., Patel, K. A., Laver, T. W., Hawkes, G., Johnson, M. B., Wakeling, M. N., et al. (2023). The role of ONECUT1 variants in monogenic and type 2 diabetes mellitus. Diabetes, db230498. doi:10.2337/db23-0498
Shaltout, A. A., Wake, D., Thanaraj, T. A., Omar, D. M., Al-AbdulRazzaq, D., Channanath, A., et al. (2017). Incidence of type 1 diabetes has doubled in Kuwaiti children 0-14 years over the last 20 years. Pediatr. Diabetes 18 (8), 761–766. doi:10.1111/pedi.12480
Tadmouri, G. O., Nair, P., Obeid, T., Al Ali, M. T., Al Khaja, N., and Hamamy, H. A. (2009). Consanguinity and reproductive health among Arabs. Reprod. Health 6, 17. doi:10.1186/1742-4755-6-17
Thompson, M. J., Rubbi, L., Dawson, D. W., Donahue, T. R., and Pellegrini, M. (2015). Pancreatic cancer patient survival correlates with DNA methylation of pancreas development genes. PLoS One 10 (6), e0128814. doi:10.1371/journal.pone.0128814
Ward, L. D., and Kellis, M. (2012). HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934. doi:10.1093/nar/gkr917
Keywords: ONECUT1, SNP, MODY, T1D, T2D, Arab ethnicity, differential methylation profiles, different types of diabetes
Citation: Dashti M, Nizam R, John SE, Melhem M, Channanath A, Alkandari H, Thanaraj TA and Al-Mulla F (2023) ONECUT1 variants beyond type 1 and type 2 diabetes: exploring clinical diversity and epigenetic associations in Arab cohorts. Front. Genet. 14:1254833. doi: 10.3389/fgene.2023.1254833
Received: 08 August 2023; Accepted: 11 October 2023;
Published: 24 October 2023.
Edited by:
Ahmed Rebai, Centre of Biotechnology of Sfax, TunisiaReviewed by:
Francisco Martin Barajas-Olmos, National Institute of Genomic Medicine (INMEGEN), MexicoCopyright © 2023 Dashti, Nizam, John, Melhem, Channanath, Alkandari, Thanaraj and Al-Mulla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fahd Al-Mulla, ZmFoZC5hbG11bGxhQGRhc21hbmluc3RpdHV0ZS5vcmc=; Thangavel Alphonse Thanaraj, YWxwaG9uc2UudGhhbmdhdmVsQGRhc21hbmluc3RpdHV0ZS5vcmc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.