- School of Computer Science, Qufu Normal University, Rizhao, China
Identification of disease-associated long non-coding RNAs (lncRNAs) is crucial for unveiling the underlying genetic mechanisms of complex diseases. Multiple types of similarity networks of lncRNAs (or diseases) can complementary and comprehensively characterize their similarities. Hence, in this study, we presented a computational model iLncDA-RSN based on reliable similarity networks for identifying potential lncRNA-disease associations (LDAs). Specifically, for constructing reliable similarity networks of lncRNAs and diseases, miRNA heuristic information with lncRNAs and diseases is firstly introduced to construct their respective Jaccard similarity networks; then Gaussian interaction profile (GIP) kernel similarity networks and Jaccard similarity networks of lncRNAs and diseases are provided based on the lncRNA-disease association network; a random walk with restart strategy is finally applied on Jaccard similarity networks, GIP kernel similarity networks, as well as lncRNA functional similarity network and disease semantic similarity network to construct reliable similarity networks. Depending on the lncRNA-disease association network and the reliable similarity networks, feature vectors of lncRNA-disease pairs are integrated from lncRNA and disease perspectives respectively, and then dimensionality reduced by the elastic net. Two random forests are at last used together on different lncRNA-disease association feature sets to identify potential LDAs. The iLncDA-RSN is evaluated by five-fold cross-validation to analyse its prediction performance, results of which show that the iLncDA-RSN outperforms the compared models. Furthermore, case studies of different complex diseases demonstrate the effectiveness of the iLncDA-RSN in identifying potential LDAs.
1 Introduction
Evidences from many studies suggest that the complex process of cancer development is regulated not only by protein-coding RNAs but also by long non-coding RNAs (lncRNAs), a class of RNAs larger than 200 bp with no coding potential (Schmitt and Chang, 2016; Wong et al., 2018). With in-depth research on associations between diseases and lncRNAs, lots of lncRNAs have been identified to have oncogenic potential and cancer-suppressive effects (Taniue and Akimitsu, 2021). For example, the expression of lncRNA HOTAIR is significantly associated with poor prognosis in lung, colon and primary breast cancers, which implies that it may be used as biomarkers for cancer diagnosis and prognosis, as well as potential treatment targets for various cancer types (Gupta et al., 2010; Aprile et al., 2020b). The lncRNA NORAD facilitates cancer development, whose expression is upregulated and associated with poor prognosis in several cancers, including bladder, squamous cell, breast, colorectal, esophageal, and pancreatic cancers (Li et al., 2017; Li et al., 2018; Tan et al., 2019; Zhou et al., 2019; Aprile et al., 2020a; Soghli et al., 2021). Besides, some lncRNAs play essential roles in the regulation of tumor suppressor functions. For instance, the expression of lncRNA GAS5 is negatively related to tumor size, metastasis and stage in prostate, pancreatic, colon, bladder and breast cancer (Goustin et al., 2019). Therefore, identifying potential disease-associated lncRNAs will be helpful for understanding the disease pathogenesis, and facilitating the diagnosis and therapeutics of complex diseases.
Nowadays, more and more biologically validated lncRNA-disease associations (LDAs) are reported, which make it possible to use computational models to predict potential LDAs (Chen and Yan, 2013). Introduced a semi-supervised framework LRLSLDA to identify LDAs, in which the hypothesis of similar diseases normally being associated with similar lncRNAs was proposed. Based on this hypothesis, a series of computational models were developed, which can be mainly divided into three categories, including matrix decomposition, random walk, and machine learning. For the matrix decomposition category (Lu et al., 2018), proposed the SIMCLDA, which uses the principal feature vectors in the constructed feature matrices to complement the association matrix based on an inductive matrix complementation framework. (Wang et al., 2021) regarded as the association prediction problem as the problem of recommendation system, and presented the LDGRNMF to employ graph-regularized nonnegative matrix decomposition to identify potential LDAs. (Liu et al., 2021) proposed the DSCMF to predict potential LDAs, which deals with the sparsity by adding
Although these models show promising results, there are still several limitations. For instance, some of them only used one type of similarity network of lncRNAs or diseases, which only describe their biological characteristics in a single perspective. It is confirmed that multiple types of similarity networks of lncRNAs (or diseases) can complementary and comprehensively characterize their similarities. However, it is a challenge to properly integrate them without bringing in redundancy and noises. Besides, heuristic information or priori knowledge of other biomolecules that associated with lncRNAs and/or diseases should be considered in the model to fully identifying potential LDAs. Taking the lncRNA-miRNA interaction as an example, the lncRNA MALAT1 has been proven to act as a sponge for miRNA miR-129-5p promoting the development of triple-negative breast cancer (Volovat et al., 2020).
In this study, we proposed a computational model, namely, iLncDA-RSN in short, to identify potential LDAs, which based on reliable similarity networks for integrating multiple types of similarity networks and utilizing miRNA heuristic information. Specifically, for constructing reliable similarity networks of lncRNAs and diseases, miRNA heuristic information with lncRNAs and diseases is firstly introduced to construct their respective Jaccard similarity networks; then GIP kernel similarity networks and Jaccard similarity networks of lncRNAs and diseases are provided based on the lncRNA-disease association network; a random walk with restart strategy is finally applied on Jaccard similarity networks, GIP kernel similarity networks, as well as lncRNA functional similarity network and disease semantic similarity network to construct reliable similarity networks. Depending on the lncRNA-disease association network and the reliable similarity networks, feature vectors of lncRNA-disease pairs are integrated from lncRNA and disease perspectives respectively, and then dimensionality reduced by the elastic net. Two random forests are at last used together on different lncRNA-disease association feature sets to identify potential LDAs. The iLncDA-RSN is evaluated by five-fold cross-validation to analyse its prediction performance, results of which show that the iLncDA-RSN outperforms the compared models. Furthermore, case studies of different complex diseases demonstrate the effectiveness of the iLncDA-RSN in identifying potential LDAs.
2 Methods
2.1 Disease similarity networks
2.1.1 Disease semantic similarity network and GIP kernel similarity network
The disease semantic similarity network is constructed using disease ontology information containing multiple directed acyclic graphs (Schriml et al., 2012). The disease
where
where the semantic contribution factor
Under the assumption that diseases with similar phenotypes tend to be more associated with similar lncRNAs, and vice versa, based on the lncRNA-disease association network, the GIP kernel similarity value
where
where
2.1.2 Disease Jaccard similarity network based on the lncRNA-disease association network
Jaccard similarity is a common statistic used to describe the degree of similarity between two groups of items and has been widely applied in the calculation of biological data (Luo et al., 2017; Zhou et al., 2021). Based on the lncRNA-disease association network, the disease Jaccard similarity value
where
2.1.3 Disease Jaccard similarity network based on the miRNA-disease association network
It is believed that heuristic information of other biomolecules that associated with diseases can help to provide supplementary prior knowledge for accurately identifying potential LDAs. In this study, miRNA-disease association network is introduced for calculating the disease Jaccard similarity value
where
2.2 LncRNA similarity networks
2.2.1 LncRNA functional similarity network and GIP kernel similarity network
The computation of functional similarity between two lncRNAs is based on the assumption that lncRNAs with shared functions are more probable correlated with diseases with similar phenotypes (Chen et al., 2015). Suppose the disease set
According to the definition of the semantic similarity value
Similar with the computational process of the GIP kernel similarity value between two diseases, based on the lncRNA-disease association network, the GIP kernel similarity value
where
where
2.2.2 LncRNA Jaccard similarity network based on the lncRNA-disease association network
Based on the lncRNA-disease association network, the lncRNA Jaccard similarity value
where
2.2.3 LncRNA Jaccard similarity network based on the lncRNA-miRNA association network
Likewise, lncRNA-miRNA association network is also introduced for calculating the lncRNA Jaccard similarity value
where
2.3 iLncDA-RSN
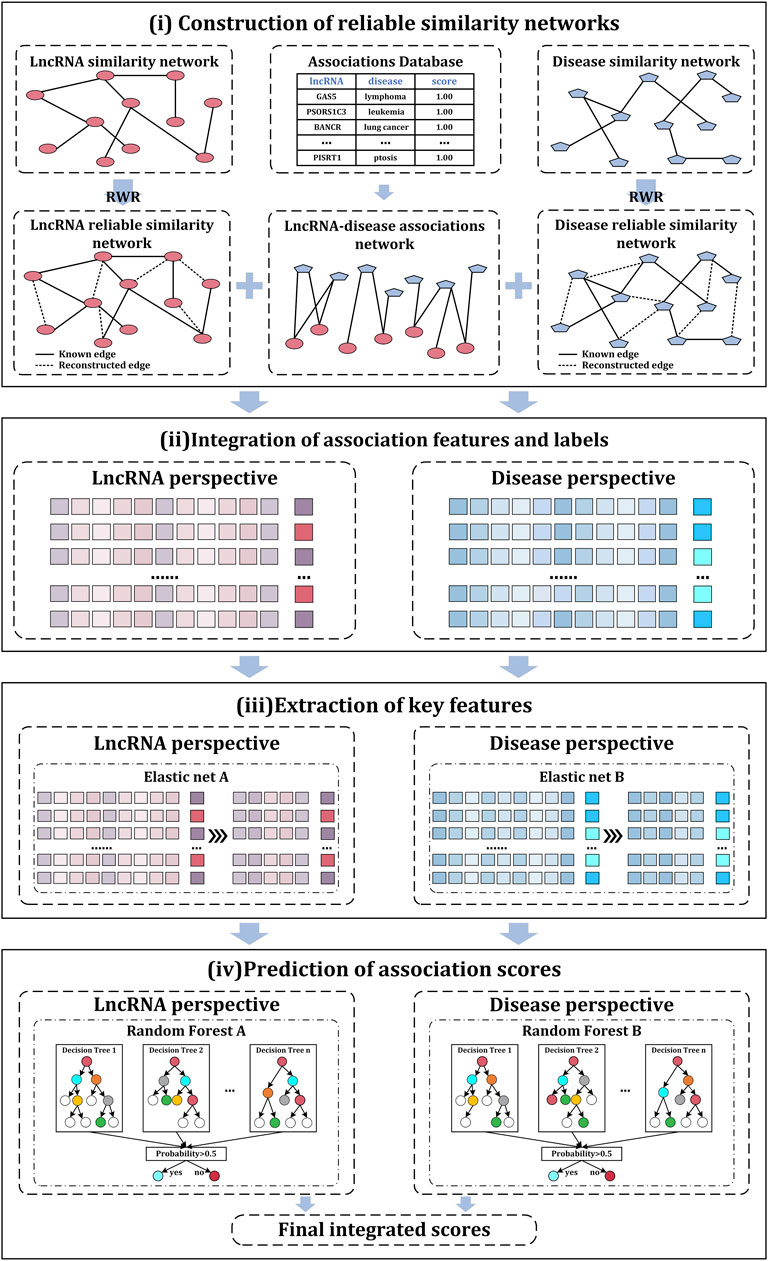
In this study, a computational model iLncDA-RSN is proposed for the Identification of LncRNA-Disease Associations based on Reliable Similarity Networks. Figure 1 shows its flowchart, from which it is seen that the iLncDA-RSN mainly has four steps, i.e., construction of reliable similarity networks, integration of association features and labels, extraction of key features, and prediction of association scores.
2.3.1 Construction of reliable similarity networks
One type of similarity network of lncRNAs or diseases only describe their biological characteristics in a single perspective and multiple types of similarity networks of lncRNAs (or diseases) can complementary and comprehensively characterize their similarities. Hence, it is a challenge to properly integrate them without bringing in redundancy and noises. In this study, a random walk with restart (RWR) strategy is applied to construct reliable similarity networks, rather than directly fuse similarity networks together, since RWR can take into account the topological connectivity patterns globally and locally within the network by introducing predefined restart probabilities at the initial nodes of each iteration to exploit potential relationships between nodes, either directly or indirectly (Liao et al., 2009; Cao et al., 2014). Specifically,
Then,
where
2.3.2 Integration of association features and labels
Depending on the lncRNA-disease association network
Labels of samples in these two association feature sets are marked as known LDAs, i.e., if the lncRNA-disease pair between the disease
2.3.3 Extraction of key features
To remove redundant features from the association feature sets to improve the prediction accuracy of LDAs, a feature extraction method, i.e., elastic net (Liu et al., 2020) is employed in this study. The elastic net is a regularization and variable selection method that has been widely used for processing data (Yu et al., 2021). The elastic net employs two penalty terms (
where the penalty degree of the model is controlled by adjusting the weight terms
2.3.4 Prediction of association scores
The random forest is based on the idea of Bagging ensemble learning, which introduces sample randomness and attributes randomness. With strong robustness and generalization, the random forest is extensively applied in the field of bioinformatics (Chen et al., 2018; Wei et al., 2021). In this study, we also apply the random forest to the iLncDA-RSN as its classifier to predict the scores of LDAs. Since there are two lncRNA-disease association feature sets constructed from lncRNA and disease perspectives respectively, two random forests are used together on them to identify potential LDAs. The final predicted association score
where
3 Results
In the study, a lncRNA-disease association network is downloaded from the Lnc2Cancer (Ning et al., 2016), GeneRIF (Lu et al., 2007) and LncRNADisease (Chen et al., 2013) databases, which includes 412 diseases, 240 lncRNAs, and 2,697 known LDAs. For a fair experimental comparison, we divided 80% of the samples into the benchmark dataset and the remaining 20% into the independent validation set (Zhang et al., 2022). The benchmark dataset is employed to select optimal parameters as well as to train the iLncDA-RSN, while the independent validation set is employed to compare the iLncDA-RSN with other computational models. To provide prior knowledge for accurately identifying potential LDAs, a miRNA-disease association network is introduced from the HMDD 2.0 database (Li et al., 2014), in which includes 13,562 experimentally validated miRNA-disease associations, and a lncRNA-miRNA association network is also introduced from the starBase database (Li et al., 2014), in which includes 1,002 experimentally validated lncRNA-miRNA associations.
We performed the 5-fold cross-validation on the benchmark dataset and used five evaluation metrics to evaluate the iLncDA-RSN, i.e., area under the receiver operating characteristic curve (AUC), Accuracy (Acc), Sensitivity (Sen), Matthews correlation coefficient (MCC) and F1-score (F1), which are defined as,
where
3.1 Evaluation of prediction ability
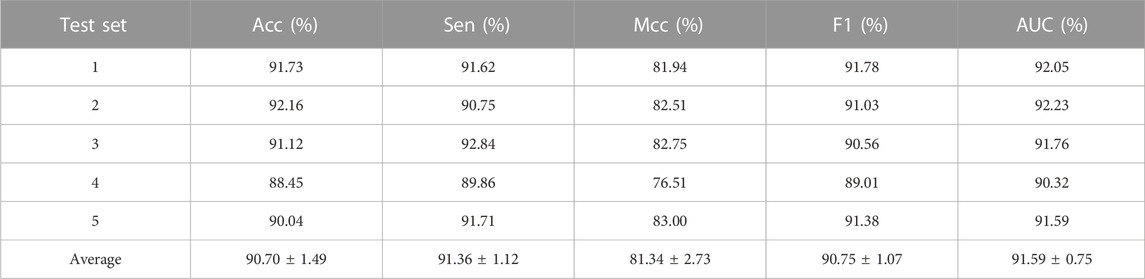
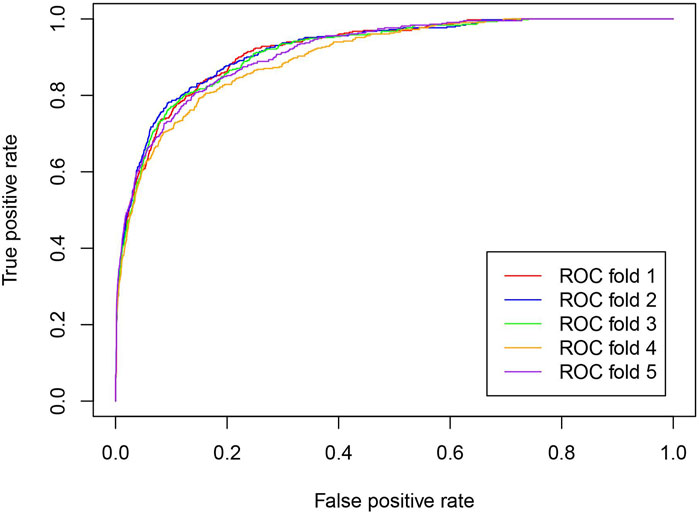
To comprehensively evaluate the prediction ability of the iLncDA-RSN, this study performed experiments on the benchmark dataset using the 5-fold cross-validation, and evaluated experimental results using 5 metrics, including AUC, Acc, Sen, MCC, and F1. Table 1 lists its experimental results, from which it is seen that the iLncDA-RSN obtained an average AUC of 91.59%, Acc of 90.70%, Sen of 91.36%, MCC of 81.34% and F1 of 90.75%, respectively. These results demonstrate that the iLncDA-RSN has high prediction ability and can play an important role in identifying potential LDAs. Besides, it is also seen that the prediction ability of the iLncDA-RSN is stable since the standard deviations are small in terms of 5 metrics. Figure 2 shows receiver operating characteristic (ROC) curves of the iLncDA-RSN on the benchmark dataset under the 5-fold cross-validation. It is seen that the ROC curves on different test sets are very similar, implying that its high stability and reliability.
3.2 Evaluation of the reliable similarity network
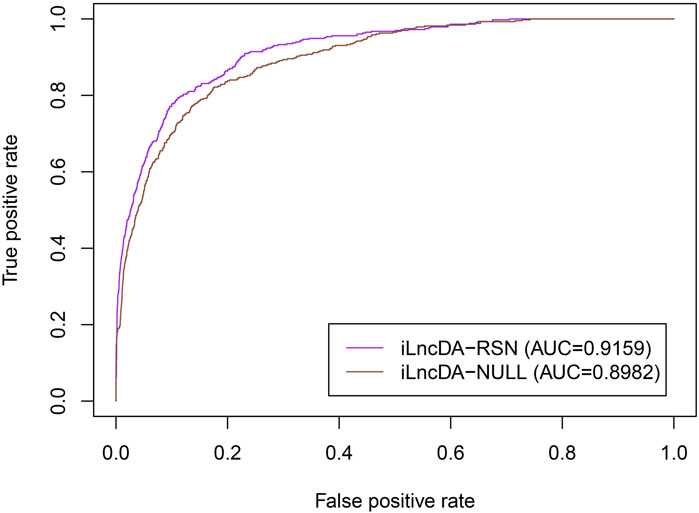
To demonstrate that the reliable similarity network is important for the iLncDA-RSN to improve the prediction ability, we performed a comparison experiment between the iLncDA-RSN and the iLncDA-NULL. Compared with the iLncDA-RSN, the iLncDA-NULL uses the directly integrated similarity networks of lncRNAs and diseases, rather than reliable similarity networks. For a fair comparison, all experimental steps and parameter settings are the same. Figure 3 shows ROC curves of the iLncDA-RSN and the iLncDA-NULL under the 5-fold cross-validation on the benchmark dataset. It is seen that the iLncDA-RSN significantly outperforms the iLncDA-NULL with their respective AUC values being 0.9159 and 0.8982, implying that the reliable similarity network is indeed important for improving the prediction ability.
3.3 Evaluation of the miRNA heuristic information
To validate that the iLncDA-RSN is advantageous by introducing the miRNA heuristic information to construct reliable similarity network, we performed a comparison experiment between the iLncDA-RSN and the same model that does not introduce the miRNA heuristic information. Figure 4 shows ROC curves of the iLncDA-RSN with and without miRNA heuristic information on the benchmark dataset. It is seen that the iLncDA-RSN is significantly superior to the model without introducing the miRNA heuristic information in terms of AUC, implying that the introduced miRNA heuristic information can help to provide supplementary prior knowledge for accurately identifying potential LDAs.
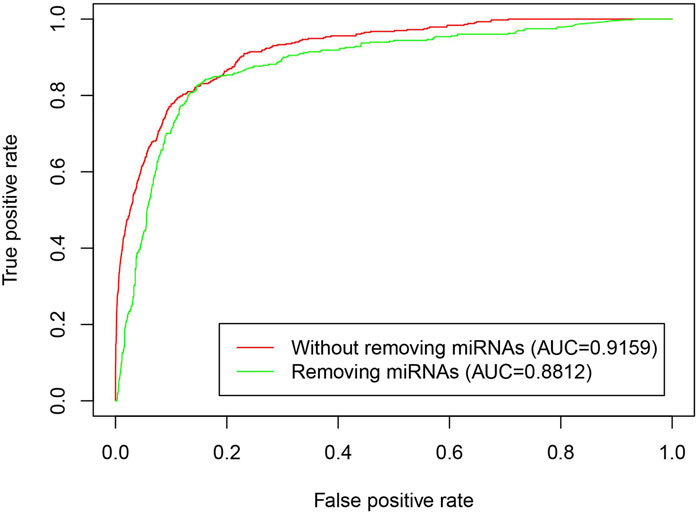
FIGURE 4. ROC curves of the iLncDA-RSN with and without miRNA heuristic information on the benchmark dataset.
3.4 Comparison with other dimensionality reduction methods
To test the performance of the elastic net for dimensionality reduction in the iLncDA-RSN, we compared it with other three dimensionality reduction methods, including extra-trees (ETS) (Liu et al., 2020), LASSO (Ranstam and Cook, 2018) and SVD (Zeng et al., 2020). The feature extraction part of the iLncDA-RSN is replaced by these three dimensionality reduction methods and other parts are the same to ensure a fair comparison. Figure 5 shows ROC curves of the iLncDA-RSN with different dimensionality reduction methods on the benchmark dataset. It is seen that their AUC values are 0.9025, 0.8982, 0.8838, and 0.9159 corresponding to LASSO, SVD, ETS and the elastic net, respectively. Hence, in the iLncDA-RSN, the elastic net method is employed to remove redundant features from the association feature sets to improve the prediction accuracy of LDAs.
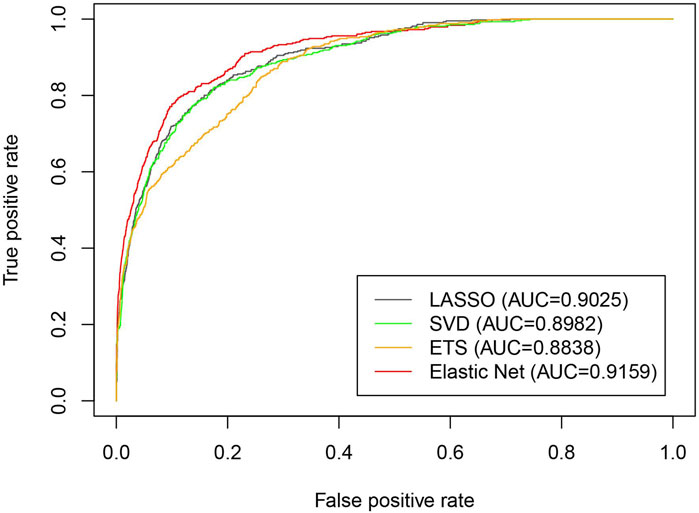
FIGURE 5. ROC curves of the iLncDA-RSN with different dimensionality reduction methods on the benchmark dataset.
3.5 Comparison with other classifiers
To find the most suitable classifier for the iLncDA-RSN, multiple classic classifiers, including random forest (RF), XGBoost (XGB) (Chen and Guestrin, 2016), k-nearest neighbor (KNN) (Liu et al., 2020), AdaBoost (Zhao et al., 2019) and Bayesian network (BN) (Marcot and Penman, 2019), were tested. Figure 6 shows ROC curves of the iLncDA-RSN with different classifiers on the benchmark dataset. It is seen that AUC values of RF, XGB, KNN, AdaBoost, and BN are 0.9159, 0.8962, 0.9042, 0.8762, and 0.8222, respectively, implying that the winner random forest is the most suitable classifier among them.
3.6 Comparison with other computational models
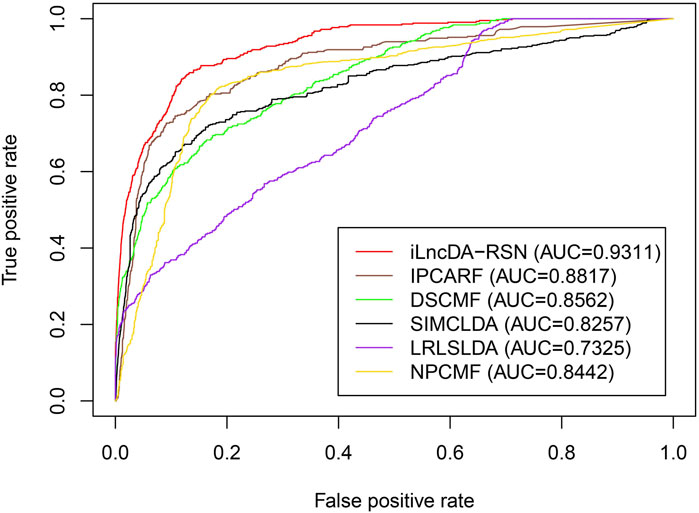
To further evaluate the prediction ability of the iLncDA-RSN, 5-fold cross-validation was performed to compare the iLncDA-RSN and other five state-of-the-art models, including IPCARF (Zhu et al., 2021), DSCMF (Liu et al., 2021), SIMCLDA (Lu et al., 2018), LRLSLDA (Chen and Yan, 2013) and NPCMF (Gao et al., 2019) on the independent validation set. Figure 7 shows ROC curves of all compared computational models. It is seen that the iLncDA-RSN has the largest area under the ROC curve, achieving an AUC value of 0.9311, while the other five computational models have AUC values of 0.8817, 0.8562, 0.8257, 0.7325, and 0.8442, respectively. This indicates that the iLncDA-RSN has better prediction ability and can predict potential LDAs more accurately.
3.7 Case study
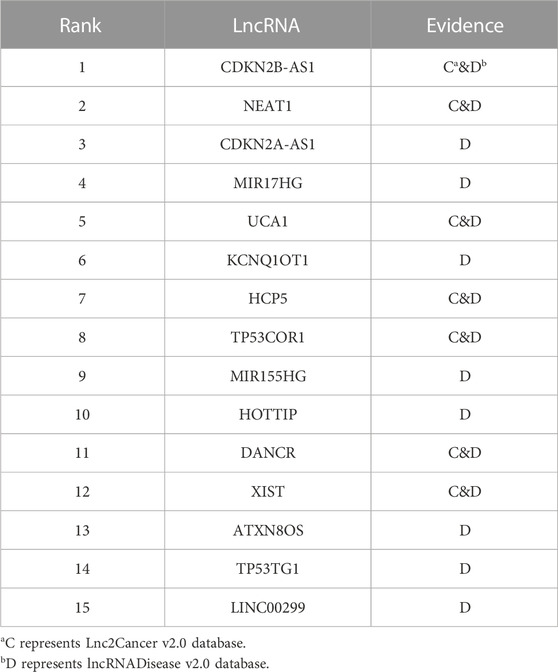
To validate the ability of the iLncDA-RSN in predicting potential LDAs, we performed case studies for cervical cancer, colon cancer and gastric cancer. All known LDAs and miRNA-disease associations were employed to train the iLncDA-RSN, which then predicts lncRNAs associated with each disease, and gives their association scores. The predicted lncRNAs were ranked based on their association scores and the top 15 lncRNAs would be verified through the databases Lnc2Cancer v2.0 (Ning et al., 2016) and lncRNADisease v2.0 (Chen et al., 2013).
Cervical cancer is diagnosed in more than 500,000 women, which causes more than 300,000 deaths worldwide (Jiang et al., 2021). Top 15 lncRNAs predicted by the iLncRNA-RSN for the cervical cancer is recorded in Table 2. Through a series of experiments, Zhang et al. (2017) demonstrated that the expression of lncRNA CDKN2B-AS1 is remarkably high in both cervical cancer tissues and cell lines, and the CDKN2B-AS1 may take an essential part in the progression of cervical cancer, implying that CDKN2B-AS1 may work as a new cervical cancer therapeutic target and prognostic biomarker. Wang and Zhu (2018) demonstrated that lncRNA NEAT1 serves as a miR-101 sponge in cervical cancer and its upregulated level is associated with poor prognosis and poor clinical-pathological factors, implying that NEAT1 might be a target for the treatment of cervical cancer. Yan et al. (2018) performed a luciferase reporter gene analysis, which showed that there is a binding site between the UCA1 lncRNA and miR-206, and the UCA1 is upregulated in the tissues of cervical cancer patients.
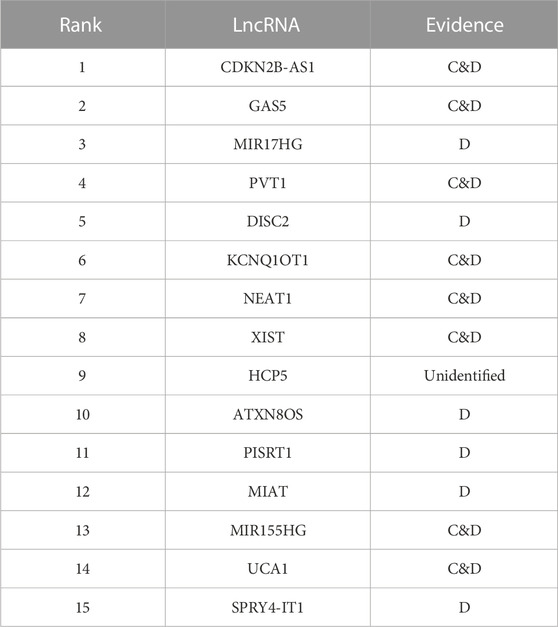
Colon cancer, a common preventable cancer, has been increasing in incidence and mortality among young people under the age of 50 in the past 25 years (Ahmed, 2020). Top 15 lncRNAs predicted by the iLncRNA-RSN for the colon cancer is recorded in Table 3. Of them, 14 lncRNAs are verified in databases C and D. (Tseng et al., 2014) found that lncRNA PVT1 increases MYC protein level, which in turn increases the cancer rate of colon cancer. (Li et al., 2019) showed that lncRNA KCNQ1OT1 fosters chemoresistance in colon cancer via sponging miR-34a and may act as a possible target for the therapy of colon cancer. (Sun et al., 2018) used qRT-PCR to measure the expression of lncRNA XIST in colon cancer tissues as well as in adjacent normal tissues, and showed that XIST expression is upregulated remarkably in tissues of colon cancer, thus indicating that XIST plays an oncogenic role in colon cancer.
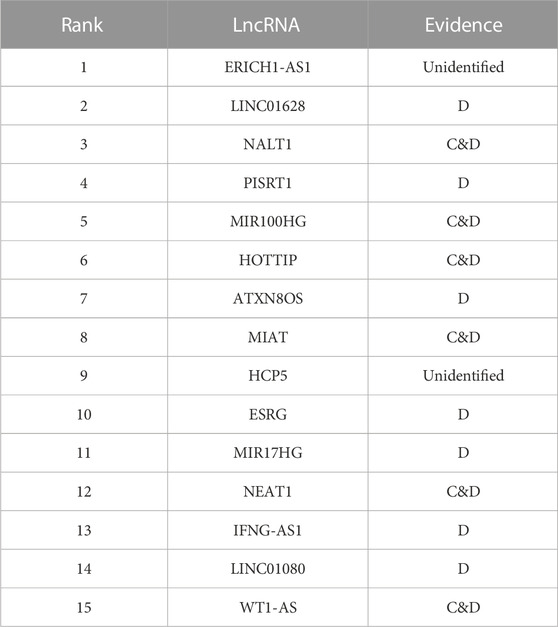
Most patients with gastric cancer are diagnosed at an advanced phase and suffer from a poor prognosis (Lian et al., 2016). Top 15 lncRNAs predicted by the iLncRNA-RSN for the gastric cancer is recorded in Table 4. Several studies (Chang et al., 2016; Wang et al., 2016; Ye et al., 2016) found that lncRNA HOTTIP may play a significant part in the initiation and progression of gastric cancer, and may be both a new prognostic marker and a prospective target for the therapy of gastric cancer. Sha et al. (2018) conducted real-time PCR with gastric cancer specimens and adjacent matched regular tissues, and showed that the level of lncRNA MIAT in gastric cancer tissues is elevated. (Tan et al., 2019b) found that the downregulation of lncRNA NEAT1 significantly inhibited gastric cancer progression, while overexpression of NEAT1 induced gastric cancer development. (Du et al., 2016) showed that the expression of lncRNA WT1-AS is downregulated in the tissues and cells of gastric cancer, and demonstrated that WT1-AS may be associated with gastric cancer of tumor progression.
4 Conclusion
In this study, we presented a computational model iLncDA-RSN based on reliable similarity networks for identifying potential LDAs. Specifically, for constructing reliable similarity networks of lncRNAs and diseases, miRNA heuristic information with lncRNAs and diseases is firstly introduced to construct their respective Jaccard similarity networks; then GIP kernel similarity networks and Jaccard similarity networks of lncRNAs and diseases are provided based on the lncRNA-disease association network; a random walk with restart strategy is finally applied on Jaccard similarity networks, GIP kernel similarity networks, as well as lncRNA functional similarity network and disease semantic similarity network to construct reliable similarity networks. Depending on the lncRNA-disease association network and the reliable similarity networks, feature vectors of lncRNA-disease pairs are integrated from lncRNA and disease perspectives respectively, and then dimensionality reduced by the elastic net. Two random forests are at last used together on different lncRNA-disease association feature sets to identify potential LDAs. The iLncDA-RSN is evaluated by five-fold cross-validation and five experiments were performed, including evaluation of prediction ability, evaluation of the reliable similarity network, evaluation of the miRNA heuristic information, comparison with other dimensionality reduction methods, comparison with other classifiers, and comparison with other computational models. Experimental results show that the iLncDA-RSN outperforms the compared models. Furthermore, case studies of different complex diseases demonstrate the effectiveness of the iLncDA-RSN in identifying potential LDAs.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
YL and MZ designed the iLncDA-RSN. YL and JS implemented and performed the experiments. YL, FL, QR, and J-XL analysed the experiment results and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Science Foundation of China (61972226 and 62172254). The funder played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Acknowledgments
The authors thank the referees for suggestions that helped improved the paper substantially.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, M. (2020). Colon cancer: A clinician’s perspective in 2019. Gastroenterology Res. 13 (1), 1–10. doi:10.14740/gr1239
Aprile, M., Katopodi, V., Leucci, E., and Costa, V. (2020). LncRNAs in cancer: From garbage to junk. Cancers (Basel) 12 (11), 3220. doi:10.3390/cancers12113220
Cao, M., Pietras, C. M., Feng, X., Doroschak, K. J., Schaffner, T., Park, J., et al. (2014). New directions for diffusion-based network prediction of protein function: Incorporating pathways with confidence. Bioinformatics 30 (12), i219–i227. doi:10.1093/bioinformatics/btu263
Chang, S., Liu, J., Guo, S., He, S., Qiu, G., Lu, J., et al. (2016). HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression. Oncol. Rep. 35 (6), 3577–3585. doi:10.3892/or.2016.4743
Chen, G., Wang, Z., Wang, D., Qiu, C., Liu, M., Chen, X., et al. (2013). LncRNADisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 41 (D1), D983–D986. doi:10.1093/nar/gks1099
Chen, T., and Guestrin, C. (2016). “Xgboost: A scalable tree boosting system,” San Francisco California USA, August 2016, 785–794. doi:10.1038/s41598-017-12763-zProc. 22nd acm sigkdd Int. Conf. Knowl. Discov. data Min.
Chen, X., Clarence Yan, C., Luo, C., Ji, W., Zhang, Y., and Dai, Q. (2015). Constructing lncRNA functional similarity network based on lncRNA-disease associations and disease semantic similarity. Sci. Rep. 5 (1), 11338–11412. doi:10.1038/srep11338
Chen, X., Wang, C. C., Yin, J., and You, Z. H. (2018). Novel human miRNA-disease association inference based on random forest. Molecuar Ther. Nucleic Acids 13, 568–579. doi:10.1016/j.omtn.2018.10.005
Chen, X., and Yan, G. Y. (2013). Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics 29 (20), 2617–2624. doi:10.1093/bioinformatics/btt426
Du, T., Zhang, B., Zhang, S., Jiang, X., Zheng, P., Li, J., et al. (2016). Decreased expression of long non-coding RNA WT1-AS promotes cell proliferation and invasion in gastric cancer. Biochimica Biophysica Acta-Molecular Basis Dis. 1862 (1), 12–19. doi:10.1016/j.bbadis.2015.10.001
Gao, Y. L., Cui, Z., Liu, J. X., Wang, J., and Zheng, C. H. (2019). Npcmf: Nearest profile-based collaborative matrix factorization method for predicting miRNA-disease associations. BMC Bioinforma. 20 (1), 353. doi:10.1186/s12859-019-2956-5
Goustin, A. S., Thepsuwan, P., Kosir, M. A., and Lipovich, L. (2019). The growth-arrest-specific (GAS)-5 long non-coding rna: A fascinating lncRNA widely expressed in cancers. Noncoding RNA 5 (3), 46. doi:10.3390/ncrna5030046
Gu, C., Liao, B., Li, X., Cai, L., Li, Z., Li, K., et al. (2017). Global network random walk for predicting potential human lncRNA-disease associations. Sci. Rep. 7 (1), 12442. doi:10.1038/s41598-017-12763-z
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464 (7291), 1071–1076. doi:10.1038/nature08975
Jiang, H.-J., Wang, Y.-B., and Huang, Y. (2021). “Prediction of drug-disease associations based on long short-term memory network and Gaussian interaction profile kernel,” in Bio-inspired computing: Theories and applications (Berlin, Germany: Springer), 432–444.
Li, H., Wang, X., Wen, C., Huo, Z., Wang, W., Zhan, Q., et al. (2017). Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol. Cancer 16 (1), 169. doi:10.1186/s12943-017-0738-0
Li, J. H., Liu, S., Zhou, H., Qu, L. H., and Yang, J. H. (2014a). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42 (D1), D92–D97. doi:10.1093/nar/gkt1248
Li, J., Zhao, H., Xuan, Z., Yu, J., Feng, X., Liao, B., et al. (2021). A novel approach for potential human LncRNA-disease association prediction based on local random walk. IEEE/ACM Trans. Comput. Biol. Bioinforma. 18 (3), 1049–1059. doi:10.1109/TCBB.2019.2934958
Li, Q., Li, C., Chen, J., Liu, P., Cui, Y., Zhou, X., et al. (2018). High expression of long noncoding RNA NORAD indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Urol. Oncol. 36 (6), e315–e310. doi:10.1016/j.urolonc.2018.02.019
Li, Y., Li, C., Li, D., Yang, L., Jin, J., and Zhang, B. (2019). lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. Oncotargets Ther. 12, 2649–2660. doi:10.2147/OTT.S188054
Li, Y., Qiu, C., Tu, J., Geng, B., Yang, J., Jiang, T., et al. (2014b). HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 42 (D1), D1070–D1074. doi:10.1093/nar/gkt1023
Lian, Y., Cai, Z., Gong, H., Xue, S., Wu, D., and Wang, K. (2016). Hottip: A critical oncogenic long non-coding RNA in human cancers. Mol. Biosyst. 12 (11), 3247–3253. doi:10.1039/c6mb00475j
Liao, C. S., Lu, K., Baym, M., Singh, R., and Berger, B. (2009). IsoRankN: Spectral methods for global alignment of multiple protein networks. Bioinformatics 25 (12), i253–i258. doi:10.1093/bioinformatics/btp203
Liu, J. X., Gao, M. M., Cui, Z., Gao, Y. L., and Li, F. (2021). Dscmf: Prediction of LncRNA-disease associations based on dual sparse collaborative matrix factorization. BMC Bioinforma. 22 (3), 241. doi:10.1186/s12859-020-03868-w
Liu, W., Lin, H., Huang, L., Peng, L., Tang, T., Zhao, Q., et al. (2022). Identification of miRNA-disease associations via deep forest ensemble learning based on autoencoder. Briefings Bioinforma. 23 (3), bbac104. doi:10.1093/bib/bbac104
Liu, Y., Yu, Z., Chen, C., Han, Y., and Yu, B. (2020). Prediction of protein crotonylation sites through LightGBM classifier based on SMOTE and elastic net. Anal. Biochem. 609, 113903. doi:10.1016/j.ab.2020.113903
Lu, C., Yang, M., Luo, F., Wu, F. X., Li, M., Pan, Y., et al. (2018). Prediction of lncRNA-disease associations based on inductive matrix completion. Bioinformatics 34 (19), 3357–3364. doi:10.1093/bioinformatics/bty327
Lu, Z., Cohen, K. B., and Hunter, L. (2007). GeneRIF quality assurance as summary revision. Pac. Symposium Biocomput., 269–280. doi:10.1142/9789812772435_0026
Luo, Y., Zhao, X., Zhou, J., Yang, J., Zhang, Y., Kuang, W., et al. (2017). A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 8 (1), 573. doi:10.1038/s41467-017-00680-8
Marcot, B. G., and Penman, T. D. (2019). Advances in Bayesian network modelling: Integration of modelling technologies. Environ. Model. Softw. 111, 386–393. doi:10.1016/j.envsoft.2018.09.016
Ning, S., Zhang, J., Wang, P., Zhi, H., Wang, J., Liu, Y., et al. (2016). Lnc2Cancer: A manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 44 (D1), D980–D985. doi:10.1093/nar/gkv1094
Ranstam, J., and Cook, J. (2018). LASSO regression. J. Br. Surg. 105 (10), 1348. doi:10.1002/bjs.10895
Schmitt, A. M., and Chang, H. Y. (2016). Long noncoding RNAs in cancer pathways. Cancer Cell. 29 (4), 452–463. doi:10.1016/j.ccell.2016.03.010
Schriml, L. M., Arze, C., Nadendla, S., Chang, Y. W., Mazaitis, M., Felix, V., et al. (2012). Disease ontology: A backbone for disease semantic integration. Nucleic Acids Res. 40 (D1), D940–D946. doi:10.1093/nar/gkr972
Sha, M., Lin, M., Wang, J., Ye, J., Xu, J., Xu, N., et al. (2018). Long non-coding RNA MIAT promotes gastric cancer growth and metastasis through regulation of miR-141/DDX5 pathway. J. Exp. Clin. Cancer Res. 37 (1), 58. doi:10.1186/s13046-018-0725-3
Soghli, N., Yousefi, T., Abolghasemi, M., and Qujeq, D. (2021). NORAD, a critical long non-coding RNA in human cancers. Life Sci. 264, 118665. doi:10.1016/j.lfs.2020.118665
Sun, J., Shi, H., Wang, Z., Zhang, C., Liu, L., Wang, L., et al. (2014). Inferring novel lncRNA-disease associations based on a random walk model of a lncRNA functional similarity network. Mol. Biosyst. 10 (8), 2074–2081. doi:10.1039/c3mb70608g
Sun, N., Zhang, G., and Liu, Y. (2018). Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/β-catenin signaling pathway. Gene 665, 141–148. doi:10.1016/j.gene.2018.04.014
Tan, B. S., Yang, M. C., Singh, S., Chou, Y. C., Chen, H. Y., Wang, M. Y., et al. (2019a). LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene 38 (28), 5612–5626. doi:10.1038/s41388-019-0812-8
Tan, H. Y., Wang, C., Liu, G., and Zhou, X. (2019b). Long noncoding RNA NEAT1-modulated miR-506 regulates gastric cancer development through targeting STAT3. J. Cell. Biochem. 120 (4), 4827–4836. doi:10.1002/jcb.26691
Taniue, K., and Akimitsu, N. (2021). The functions and unique features of LncRNAs in cancer development and tumorigenesis. Int. J. Mol. Sci. 22 (2), 632. doi:10.3390/ijms22020632
Tseng, Y. Y., Moriarity, B. S., Gong, W., Akiyama, R., Tiwari, A., Kawakami, H., et al. (2014). PVT1 dependence in cancer with MYC copy-number increase. Nature 512 (7512), 82–86. doi:10.1038/nature13311
Volovat, S. R., Volovat, C., Hordila, I., Hordila, D.-A., Mirestean, C. C., Miron, O. T., et al. (2020). MiRNA and LncRNA as potential biomarkers in triple-negative breast cancer: A review. Front. Oncol. 10, 526850. doi:10.3389/fonc.2020.526850
Wang, D., Wang, J., Lu, M., Song, F., and Cui, Q. (2010). Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics 26 (13), 1644–1650. doi:10.1093/bioinformatics/btq241
Wang, L., and Zhu, H. (2018). Long non-coding nuclear paraspeckle assembly transcript 1 acts as prognosis biomarker and increases cell growth and invasion in cervical cancer by sequestering microRNA-101. Mol. Med. Rep. 17 (2), 2771–2777. doi:10.3892/mmr.2017.8186
Wang, M.-N., You, Z.-H., Wang, L., Li, L.-P., and Zheng, K. (2021). Ldgrnmf: LncRNA-disease associations prediction based on graph regularized non-negative matrix factorization. Neurocomputing 424, 236–245. doi:10.1016/j.neucom.2020.02.062
Wang, S. S., Wuputra, K., Liu, C. J., Lin, Y. C., Chen, Y. T., Chai, C. Y., et al. (2016). Oncogenic function of the homeobox A13-long noncoding RNA HOTTIP-insulin growth factor-binding protein 3 axis in human gastric cancer. Oncotarget 7 (24), 36049–36064. doi:10.18632/oncotarget.9102
Wei, H., Xu, Y., and Liu, B. (2021). iPiDi-PUL: identifying Piwi-interacting RNA-disease associations based on positive unlabeled learning. Briefings Bioinforma. 22 (3), bbaa058. doi:10.1093/bib/bbaa058
Wong, C. M., Tsang, F. H., and Ng, I. O. (2018). Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 15 (3), 137–151. doi:10.1038/nrgastro.2017.169
Yan, Q., Tian, Y., and Hao, F. (2018). Downregulation of lncRNA UCA1 inhibits proliferation and invasion of cervical cancer cells through miR-206 expression. Oncol. Res. doi:10.3727/096504018X15185714083446
Ye, H., Liu, K., and Qian, K. (2016). Overexpression of long noncoding RNA HOTTIP promotes tumor invasion and predicts poor prognosis in gastric cancer. Oncotargets Ther. 9, 2081–2088. doi:10.2147/OTT.S95414
Yu, B., Chen, C., Wang, X., Yu, Z., Ma, A., and Liu, B. (2021). Prediction of protein–protein interactions based on elastic net and deep forest. Expert Syst. Appl. 176, 114876. doi:10.1016/j.eswa.2021.114876
Zeng, M., Lu, C., Zhang, F., Li, Y., Wu, F. X., Li, Y., et al. (2020). Sdlda: lncRNA-disease association prediction based on singular value decomposition and deep learning. Methods 179, 73–80. doi:10.1016/j.ymeth.2020.05.002
Zhang, D., Sun, G., Zhang, H., Tian, J., and Li, Y. (2017). Long non-coding RNA ANRIL indicates a poor prognosis of cervical cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed. Pharmacother. 85, 511–516. doi:10.1016/j.biopha.2016.11.058
Zhang, W., Wei, H., and Liu, B. (2022). idenMD-NRF: a ranking framework for miRNA-disease association identification. Briefings Bioinforma. 23 (4), bbac224. doi:10.1093/bib/bbac224
Zhao, Y., Chen, X., and Yin, J. (2019). Adaptive boosting-based computational model for predicting potential miRNA-disease associations. Bioinformatics 35 (22), 4730–4738. doi:10.1093/bioinformatics/btz297
Zhou, F., Yin, M. M., Jiao, C. N., Zhao, J. X., Zheng, C. H., and Liu, J. X. (2021). Predicting miRNA-disease associations through deep autoencoder with multiple kernel learning. IEEE Trans. Neural Netw. Learn. Syst., 1–10. doi:10.1109/TNNLS.2021.3129772
Zhou, K., Ou, Q., Wang, G., Zhang, W., Hao, Y., and Li, W. (2019). High long non-coding RNA NORAD expression predicts poor prognosis and promotes breast cancer progression by regulating TGF-beta pathway. Cancer Cell. Int. 19, 63. doi:10.1186/s12935-019-0781-6
Keywords: lncRNA-disease association, reliable similarity network, random forest, random walk with restart, elastic net
Citation: Li Y, Zhang M, Shang J, Li F, Ren Q and Liu J-X (2023) iLncDA-RSN: identification of lncRNA-disease associations based on reliable similarity networks. Front. Genet. 14:1249171. doi: 10.3389/fgene.2023.1249171
Received: 28 June 2023; Accepted: 27 July 2023;
Published: 08 August 2023.
Edited by:
Min Zeng, Central South University, ChinaCopyright © 2023 Li, Zhang, Shang, Li, Ren and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junliang Shang, c2hhbmdqdW5saWFuZzExMEAxNjMuY29t
†These authors have contributed equally to this work
 Yahan Li
Yahan Li Mingrui Zhang
Mingrui Zhang Junliang Shang
Junliang Shang Feng Li
Feng Li Qianqian Ren
Qianqian Ren Jin-Xing Liu
Jin-Xing Liu