
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 18 September 2023
Sec. Applied Genetic Epidemiology
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1248492
This article is part of the Research TopicWomen in Applied Genetic EpidemiologyView all 7 articles
 Maria Zanti1
Maria Zanti1 Maria A. Loizidou2
Maria A. Loizidou2 Denise G. O’Mahony1
Denise G. O’Mahony1 Leila Dorling3
Leila Dorling3 Joe Dennis3
Joe Dennis3 Peter Devilee4,5
Peter Devilee4,5 Douglas F. Easton3,6
Douglas F. Easton3,6 Mihalis I. Panayiotidis2
Mihalis I. Panayiotidis2 Andreas Hadjisavvas2
Andreas Hadjisavvas2 Kyriaki Michailidou1,3*
Kyriaki Michailidou1,3*Introduction: It is estimated that around 5% of breast cancer cases carry pathogenic variants in established breast cancer susceptibility genes. However, the underlying prevalence and gene-specific population risk estimates in Cyprus are currently unknown.
Methods: We performed sequencing on a population-based case-control study of 990 breast cancer cases and 1094 controls from Cyprus using the BRIDGES sequencing panel. Analyses were conducted separately for protein-truncating and rare missense variants.
Results: Protein-truncating variants in established breast cancer susceptibility genes were detected in 3.54% of cases and 0.37% of controls. Protein-truncating variants in BRCA2 and ATM were associated with a high risk of breast cancer, whereas PTVs in BRCA1 and PALB2 were associated with a high risk of estrogen receptor (ER)-negative disease. Among participants with a family history of breast cancer, PTVs in ATM, BRCA2, BRCA1, PALB2 and RAD50 were associated with an increased risk of breast cancer. Furthermore, an additional 19.70% of cases and 17.18% of controls had at least one rare missense variant in established breast cancer susceptibility genes. For BRCA1 and PALB2, rare missense variants were associated with an increased risk of overall and triple-negative breast cancer, respectively. Rare missense variants in BRCA1, ATM, CHEK2 and PALB2 domains, were associated with increased risk of disease subtypes.
Conclusion: This study provides population-based prevalence and gene-specific risk estimates for protein-truncating and rare missense variants. These results may have important clinical implications for women who undergo genetic testing and be pivotal for a substantial proportion of breast cancer patients in Cyprus.
Germline pathogenic variants (PVs) in cancer susceptibility genes have been associated with a significant risk of breast cancer (Dorling et al., 2021). Following the National Comprehensive Cancer Network (NCCN) guidelines, medical management recommendations are provided for certain genes contributing to increased risk of breast cancer (Daly et al., 2021). Genetic testing allows patients carrying PVs, to benefit from risk-reducing strategies including closer surveillance at an early age, prophylactic surgeries and chemoprevention, as well as targeted therapies (Pilie et al., 2019).
Gene panel testing of large population-based case-control studies has recently provided improved estimates of the prevalence of PVs and the respective magnitude of breast cancer risk associated with these PVs (Dorling et al., 2021; Hu et al., 2021). It is estimated that around 5% of breast cancer cases harbor PVs in established breast cancer susceptibility genes (ATM, BARD1, BRCA1, BRCA2, CHEK2, PALB2, RAD51C, RAD51D, TP53), of which the most prevalent occur in BRCA2, CHEK2 and BRCA1 (Dorling et al., 2021; Hu et al., 2021). Recently, it has been reported that 13% of high-risk Cypriot breast cancer patients are positive for PVs in BRCA1 and BRCA2 (Loizidou et al., 2017). However, the aggregate prevalence of cancer susceptibility genes among breast cancer cases or controls unselected for family history or age at diagnosis is not yet determined.
Here we used panel sequencing data generated as part of the Breast Cancer Association Consortium (BCAC) BRIDGES project (Dorling et al., 2021), to investigate the prevalence of PVs and rare missense variants in samples from the MASTOS study (Loizidou et al., 2008; Hadjisavvas et al., 2010), a population-based case-control study of breast cancer in Cyprus. We also estimated the risks of breast cancer associated with protein-truncating and rare missense variants in the Cypriot population.
This study included case-control samples from the MASTOS study, a population-based case-control study of breast cancer in Cyprus (Loizidou et al., 2008; Hadjisavvas et al., 2010). The study was approved by the National Bioethics Committee of Cyprus, and all participants provided signed informed consent (Approval No, ΕΕΒΚ/ΕΠ/2016/38) (Loizidou et al., 2008). The study includes 990 breast cancer cases and 1,094 age-matched healthy controls. All study participants were over 18 years of age. The average age of diagnosis for cases was 51.5 ± 9.3 standard deviation (sd) years, while the average age at the time of study enrolment for age-matched controls, was 55.7 ± 6.9 sd. Positive family history of breast cancer was reported for 16.2% (n = 158/973) and 8.0% (n = 87/1,091) for cases and controls, respectively. The majority of tumors were invasive carcinomas (92.5%, n = 656/709), estrogen receptor (ER)-positive (75.8%, n = 476/628), progesterone receptor (PR)-positive (61.6%, n = 381/619) and human epidermal growth factor receptor 2 (HER2)-negative (83.9%, n = 490/584). Out of the 990 tumors, 576 (58.2%) had available data for all ER, PR and HER2, of which 11.5% (n = 66/576) had a triple-negative phenotype, 76.6% (n = 441/576) had an ER + phenotype, 62.8% (n = 362/576) had a PR + phenotype and 16.1% (n = 93/576) had a HER2+ phenotype. All study sample characteristics are summarized in Table 1.
We analyzed targeted panel sequencing data on 35 actionable and suspected breast cancer susceptibility genes (Supplementary Table S1) using the BRIDGES panel (Dorling et al., 2021). Results are presented for variants in 34 genes since PPM1D was shown to be associated with breast or ovarian cancer risk, but in low allelic fractions (“somatic mosaicism”) (Ruark et al., 2013). These are potentially due to treatment, thus excluded from further analysis. Details on library preparation, sequencing and bioinformatics analysis including variant calling and quality control were previously documented (Dorling et al., 2021). In-frame insertions/deletions, intronic variants and variants in untranslated regions (UTRs) were not considered in the analysis.
Protein-truncating variants (PTVs) were defined as frameshift insertions/deletions, splicing variants (±2 positions) and nonsense variants as annotated by ANNOVAR (Wang et al., 2010). Splice variants affecting the penultimate exon (except variants in ATM, BARD1, BRCA1, RAD51C, RAD51C and PALB2 for which the truncated protein might still be pathogenic, irrespective of exon skipping; evidence previously documented (Dorling et al., 2021)), as well as PTVs in the last exon of each gene, were excluded from the analysis (Dorling et al., 2021). In addition, six canonical splice variants in BRCA1 (c.594-2A>C, c.4096 + 1G>A, c.4096 + 2T>C, c.4186-2A>G, c.4358-1G>C and c.4358-2del) were excluded, since they are of uncertain clinical significance according to ENIGMA (Evidence-based Network for the Interpretation of Germline Mutant Alleles) classification schemes (Spurdle et al., 2012).
Classifications for rare missense variants (allele frequency of less than 0.001 in gnomAD v2.1.1 non-Finnish European exome samples) were retrieved from ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, last accessed 11/12/2022), or the ENIGMA BRCA1 and BRCA2 expert panel (https://enigmaconsortium.org/) (Spurdle et al., 2012), along with missense TP53 variant classification based on ACMG/AMP guidelines (Richards et al., 2015), augmented with classifications made using a quantitative classification model which utilizes bioinformatics prediction tools alongside germline to somatic frequency ratios (Fortuno et al., 2019; Fortuno et al., 2021). Rare missense variants were further annotated for functional protein domain location defined by the UniProt database (https://www.uniprot.org/, release 2022_04). Rare missense variants classified as (likely) benign were not considered in the analysis.
Prevalence of protein-truncating and rare missense variants in each gene was tabulated for breast cancer patients and controls. Odds ratios (ORs) with 95% CIs were estimated using Firth’s bias-reduced penalized-likelihood logistic regression for overall, ER-positive and ER-negative breast cancer. For ER-negative cases, we also evaluated the associations for triple-negative breast cancer. Associations were adjusted for age at diagnosis or interview and first-degree family history of breast cancer. Logistic regression p values were estimated using Wald’s test. p values smaller than 0.05 were considered to be statistically significant. Association analysis was performed separately for PTVs and rare missense variants. Associations for rare missense variants were also evaluated according to gene and gene domain for the established breast cancer susceptibility genes (Dorling et al., 2021). Heterozygous and homozygous carriers of variants in a gene were not distinguished as it was not always possible to do so with certainty, and the number of homozygotes was too small for separate analysis.
Cumulative risks of breast cancer in the absence of other events were calculated by combining age-specific odds ratios with population incidence rates for Cyprus (2020) as a baseline, as previously described (Schmidt et al., 2016).
All the protein-truncating and rare missense variants are provided in Supplementary Table S2.
Among 990 breast cancer cases and 1,094 controls, we identified 27 unique PTVs in 4.55% (n = 45/990) of cases and 1.46% (n = 16/1,094) of controls (Table 2, Supplementary Table S2). Of these, 3.54% of cases (n = 35/990) and 0.37% of controls (n = 4/1,094), harbored PTVs in the established breast cancer susceptibility genes ATM, BARD1, BRCA1, BRCA2, CHEK2, PALB2, RAD51C, RAD51D and TP53 (Dorling et al., 2021).
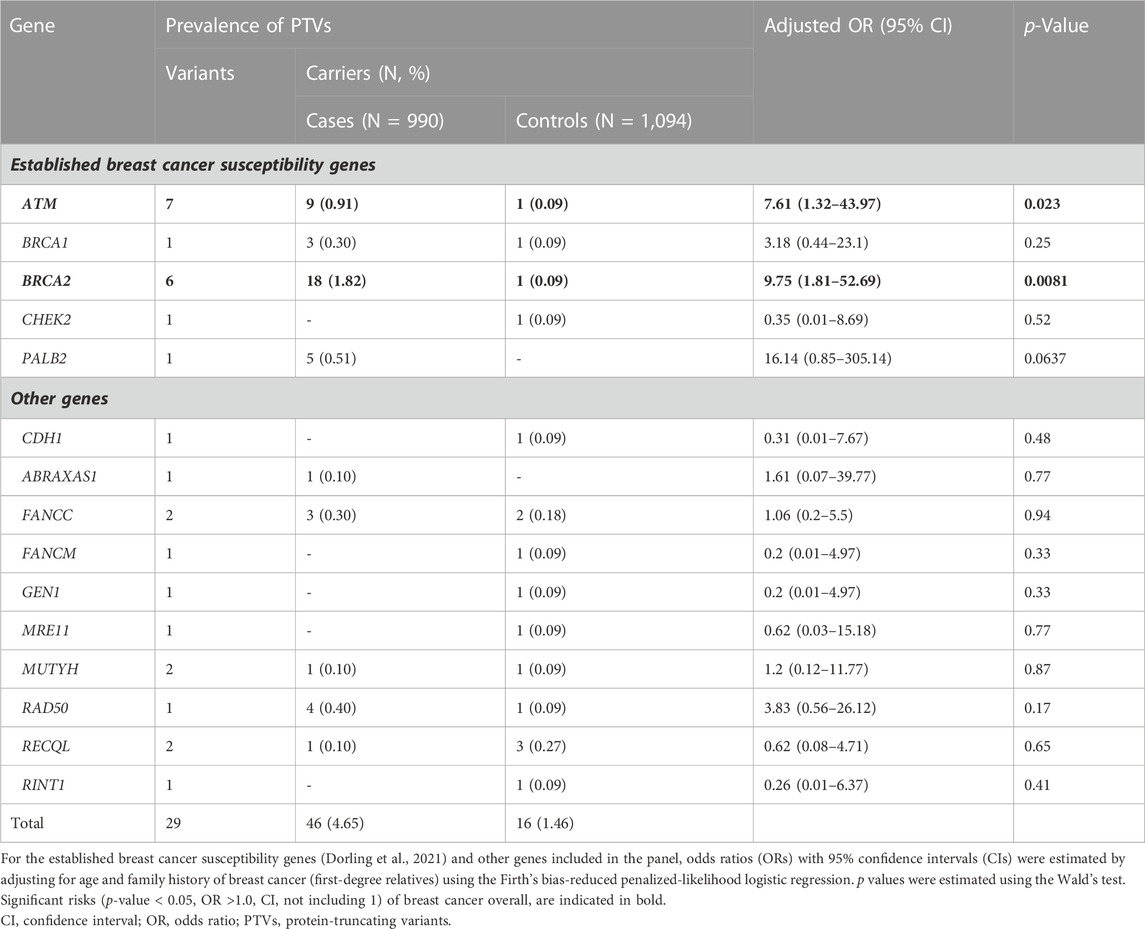
TABLE 2. Prevalence of protein-truncating variants and associations with overall breast cancer risk.
Among the case patients, the highest prevalence of PTVs was observed for BRCA2 (n = 18/990, 1.82%), ATM (n = 9/990, 0.91%), PALB2 (n = 5/990, 0.51%), RAD50 (n = 4/990, 0.40%), BRCA1 (n = 3/990, 0.3%) and FANCC (n = 3/990, 0.3%). We also identified one PTV in each of RECQL, ABRAXAS1 and MUTYH. Among controls, the highest prevalence was observed for RECQL (n = 3/1,094, 0.27%) and FANCC (n = 2/1,094, 0.18%). We also detected one PTV in each of ATM, BRCA1, BRCA2, CDH1, CHEK2, FANCM, GEN1, MRE11, MUTYH, RAD50 and RINT1 (Table 2). The founder BRCA2 c.8756delG PTV was detected in 1.01% of breast cancer cases (n = 10/990) and 0.09% of controls (n = 1/1,094) which corresponds to more than half of the BRCA2 PTVs observed among cases (55.56%, n = 10/18) and the only BRCA2 PTV identified among controls.
We report evidence of PTV association with overall breast cancer risk for BRCA2 and ATM with adjusted ORs of 9.75 (1.81-52.69 95% CI, p-value = 0.0081) and 7.61 (1.32-43.97 95% CI, p-value = 0.023) (Figure 1; Table 2, Supplementary Table S3). Association with overall breast cancer is also depicted for PTVs in PALB2 with adjusted ORs of 16.14 (0.85-305.14 95% CI, p-value = 0.064), although not statistically significant (Figure 1; Table 2, Supplementary Table S3). For other genes included in the panel, the evidence for an association between overall breast cancer and PTVs did not reach statistical significance (Figure 1; Table 2, Supplementary Table S3).
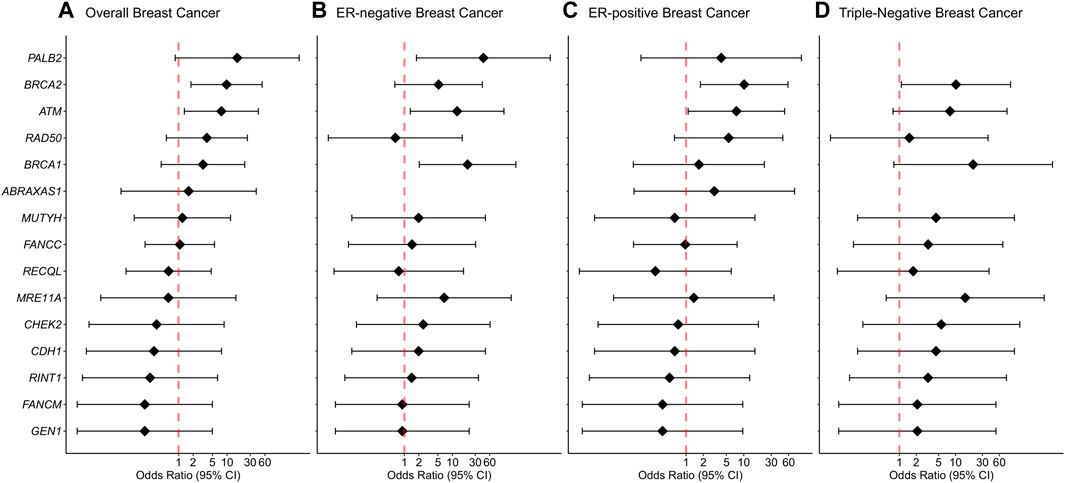
FIGURE 1. Associations of protein-truncating variants with breast cancer risk. (A) Overall breast cancer, (B) ER-negative breast cancer, (C) ER-positive breast cancer, (D) Triple-negative breast cancer. Odds ratios (ORs) and 95% confidence intervals (CIs) for overall breast cancer are represented only for genes with at least one protein-truncating variant detected in either cases or controls. Logistic regression analysis was conducted by adjusting for age and family history of breast cancer (first-degree relatives). The genes are listed in order of decreasing estimated odds ratios for overall breast cancer risk.
Breast cancer cases with PTVs in BRCA1 and BRCA2 had a relatively younger age at diagnosis (46.7 ± 10.4 sd) compared to cases without PTVs in BRCA1 and BRCA2 (51.6 ± 9.3 sd, Wilcoxon rank sum test p-value = 0.046) or cases with PTVs in genes other than BRCA1 and BRCA2 (53 ± 10.9 sd, two-sample t-test p-value = 0.051). The associations with breast cancer overall for both participants diagnosed or recruited at an age earlier or later than 50 years did not reach statistical significance, except for BRCA2 PTV-carriers who were at an increased risk of the disease if diagnosed at an age later than 50 years (Supplementary Table S4). Analysis by family history of breast cancer (first-degree relatives) revealed that the prevalence of PTVs among patients with a family history of breast cancer (8.23%, n = 13/158) was about two times the frequency reported among patients with no family history of breast cancer (3.56%, n = 29/815; p-value = 0.015). To investigate the influence of a family history of breast cancer, analysis was conducted separately for cases with and without a first-degree relative with breast cancer. Among participants with family history of breast cancer, PTVs in ATM (p-value = 0.002), BRCA2 (p-value = 0.005), BRCA1 (p-value = 0.046), PALB2 (p-value = 0.039) and RAD50 (p-value = 0.044) were significantly associated with increased risk of breast cancer. The exclusion of breast cancer patients with a family history of breast cancer had a minor impact on PTV association with overall breast cancer risk (i.e., BRCA2 adjusted OR of 8.74; 1.57-48.75 95% CI, p-value = 0.013) (Supplementary Table S5).
Protein-truncating variants in BRCA2 were strongly associated with an increased risk of ER-positive and triple-negative breast cancer with adjusted ORs of 10.18 (1.77-58.55 95% CI, p-value = 0.009) and 10.14 (1.08-94.84 95% CI, p-value = 0.042), respectively. Protein-truncating variants in BRCA1 were strongly associated with an increased risk of ER-negative breast cancer with adjusted ORs of 20.73 (2.04-210.69 95% CI, p-value = 0.0104). Protein-truncating variants in ATM were associated with an increased risk of both ER-negative and ER-positive disease with adjusted ORs of 12.55 (1.33-118.68 95% CI, p-value = 0.027) and 7.47 (1.09-51.23 95% CI, p-value = 0.041), respectively. Finally, PTVs in the PALB2 gene were associated with an increased risk of ER-negative breast cancer with adjusted ORs of 44.23 (1.78-1098.24 95% CI, p-value = 0.021) (Supplementary Table S3).
Odds ratios decreased significantly with increasing age for BRCA2 with ORs of 0.94 (0.89-0.99 95% CI, p-value = 0.013) (Supplementary Table S6). Estimated cumulative risks of breast cancer in the absence of other events were calculated by combining age-specific odds ratios with population incidence rates for Cyprus (2020) (Figure 2). For carriers of PTVs in BRCA2 and ATM the estimated cumulative risks by 80 years of age exceeded the 30% threshold for high risk, as defined by the NICE (National Institute for Health and Care Excellence) surveillance screening guidelines (NICE, 2019).
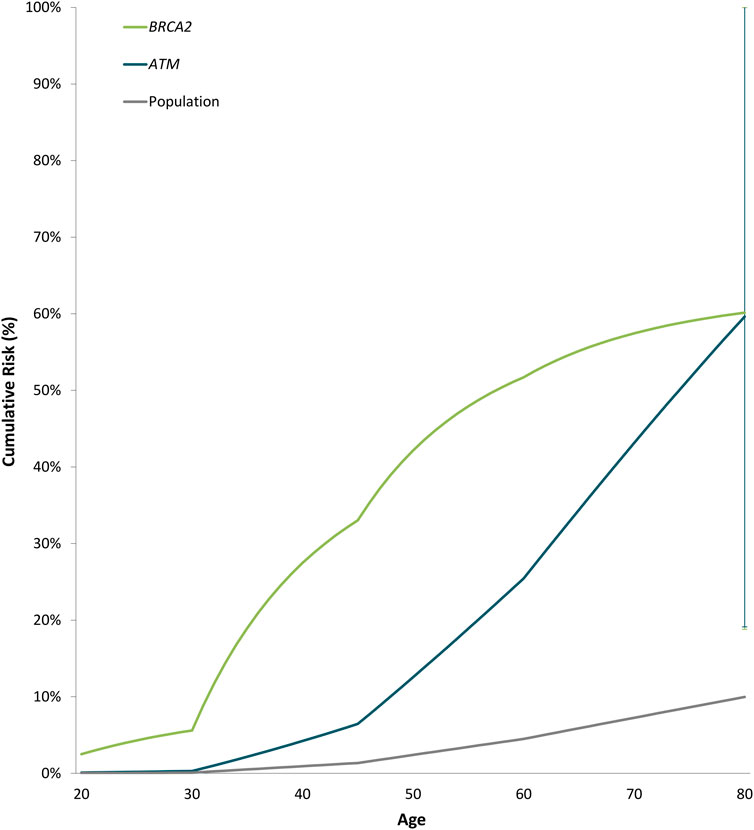
FIGURE 2. Estimated cumulative risk of breast cancer associated with protein-truncating variants in genes with significant evidence of an association with breast cancer overall. Cumulative risks of breast cancer in the absence of other events were calculated by combining age-specific odds ratios with population incidence rates for Cyprus (2020) as a baseline. The error bars indicate 95% confidence intervals.
Among 990 breast cancer cases and 1,094 controls, we identified 380 unique rare missense variants in 50.40% of cases (n = 499/990) and 41.77% of controls (n = 457/1,094) (Supplementary Table S2). Of these, 147 unique rare missense variants were detected in the established breast cancer susceptibility genes, among 195 out of 990 cases (19.70%) and 188 out of 1,094 controls (17.18%). None of the detected rare missense variants were classified as pathogenic.
Rare missense variants in BRCA1 were associated with an increased risk of overall breast cancer with an adjusted OR of 2.4 (1.14-5.08 95% CI, p-value = 0.022) (Figure 3). In the subtype-stratified analyses, rare missense variants in BRCA1 were associated with an increased risk of both ER-negative and ER-positive disease with adjusted ORs of 3.56 (1.15-11.01 95% CI, p-value = 0.028) and 2.77 (1.21-6.38 95% CI, p-value = 0.016), respectively (Supplementary Table S7, Figure 3). Increased risk of triple-negative breast cancer was observed for carriers of rare missense variants in PALB2 with an adjusted OR of 5.79 (1.32-25.29 95% CI, p-value = 0.020, Figure 3).
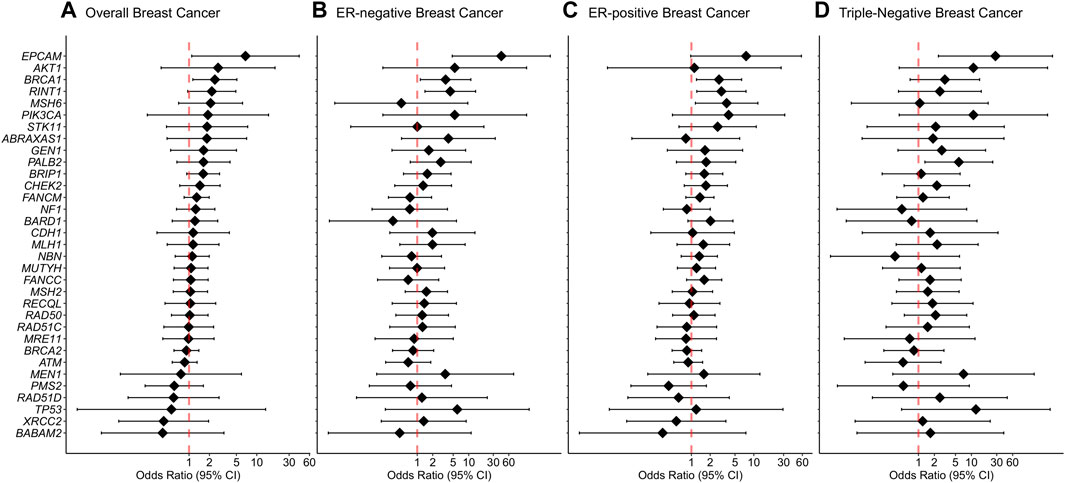
FIGURE 3. Associations of rare missense variants with breast cancer risk. (A) Overall breast cancer, (B) ER-negative breast cancer, (C) ER-positive breast cancer, (D) Triple-negative breast cancer. Odds ratios (ORs) and 95% confidence intervals (CIs) for overall breast cancer are represented only for genes with at least one missense variant detected in either cases or controls. Logistic regression analysis was conducted by adjusting for age and family history of breast cancer (first-degree relatives). The genes are listed in order of decreasing estimated odds ratios for overall breast cancer risk.
For missense variants in aggregate, the evidence for an association between overall breast cancer and any protein domain did not reach statistical significance. However, there was a significantly increased risk of ER-negative breast cancer associated with variants located within the BRCA1 Zinc-Finger, ATM FAT, PALB2 WD1 and CHEK2 FHA domains. In addition, rare missense variants within the ATM PI3L/PI4K catalytic domain demonstrated an association with an increased risk of ER-positive disease. Furthermore, rare missense variants within PALB2 domains WD1, domain for stimulation of POLH DNA synthesis and domain for interaction with RAD51, BRCA2 and POLH were significantly associated with increased risk of triple-negative breast cancer. Detailed results from the domain-specific analysis are shown in Supplementary Table S8.
This study evaluates the prevalence of protein-truncating and rare missense variants in putative breast cancer susceptibility genes and estimates population-specific risks of breast cancer in a large case-control dataset of breast cancer in Cyprus. We used panel sequencing data from 990 breast cancer cases unselected for family history of breast and/or ovarian cancer or age at disease diagnosis and 1,094 age-matched controls, generated as part of the Breast Cancer Association Consortium (BCAC) BRIDGES project (Dorling et al., 2021).
It is generally estimated that around 5%–6% of breast cancer cases and 1%–2% of the general population, harbor PVs in established breast cancer susceptibility genes (Dorling et al., 2021; Hu et al., 2021; Southey et al., 2021). Among breast cancer patients the most prevalent occur in BRCA2, CHEK2, BRCA1 and ATM (Dorling et al., 2021; Hu et al., 2021). Here, we report that 3.54% of breast cancer cases and 0.37% of controls in Cyprus are positive for PTVs in the established breast cancer susceptibility genes (Dorling et al., 2021). Among patients and the established breast cancer susceptibility genes, PTVs in BRCA2 and ATM, followed by PALB2, and BRCA1, were the most prevalent.
The prevalence of PVs in established breast cancer susceptibility genes possesses substantial ethnic and geographic disparities (Yadav et al., 2021). In Cyprus, unique founder PVs and more frequent BRCA2 PVs have been observed compared to other countries (Hadjisavvas et al., 2004; Loizidou et al., 2017). Protein-truncating variants in BRCA2 were the most prevalent in our study (1.82%), with the founder BRCA2 c.8756delG PTV being detected in ten breast cancer cases (1.01%) which corresponds to more than half of the BRCA2 PTVs observed among cases. Furthermore, BRCA2 PTVs were associated with a high risk of breast cancer overall, ER-positive and triple-negative disease and exceeded the 80-year cumulative risk threshold for high risk (30%), as defined by the NICE surveillance screening guidelines ((NICE) 2019). The cumulative risk estimates for BRCA2 PTVs in Cyprus are higher compared to risks reported by population- (Dorling et al., 2021; Hu et al., 2021) and family-based studies (Kuchenbaecker et al., 2017). However, it is consistent with cumulative risk estimates published by Kuchenbaecker et al. (2017).
Protein-truncating variants in BRCA1 generally confer around 8 to 10-fold increased risk of breast cancer (Antoniou et al., 2003; Kuchenbaecker et al., 2017; Dorling et al., 2021; Hu et al., 2021). In our study, BRCA1 PTVs showed weak associations with overall breast cancer risk (p > 0.05). However, BRCA1 was associated with an increased risk of ER-negative disease and was identified as a high-risk gene among case patients who had a first-degree relative with breast cancer. Moreover, breast cancer patients with PTVs in BRCA1 and BRCA2 had a relatively younger age at diagnosis compared to cases without PTVs in BRCA1 and BRCA2 or cases with PTVs in genes other than BRCA1 and BRCA2. These findings are consistent with recent data suggesting that the prevalence of PVs in genes other than BRCA1 and BRCA2 does not depend on age at diagnosis (Tung et al., 2016; Buys et al., 2017).
Among the established breast cancer susceptibility genes, ATM yielded an odds ratio of approximately 8 for breast cancer overall and its estimated cumulative risks by 80 years of age exceeded the 30% threshold for high risk [(NICE) 2019], compared to published moderate-risk estimates for ATM PTVs (Dorling et al., 2021). Although, if validated in a larger case-control series of breast cancer in Cyprus, it will be of clinical importance.
Pathogenic variants in PALB2 were previously identified as high- and moderate-risk in the large-scale population-based BRIDGES (Dorling et al., 2021) and CARRIERS projects (Hu et al., 2021). Here we report a possible association of PALB2 PTVs with high risk of breast cancer overall (p = 0.06) and ER-negative disease (p < 0.05). However, among cases with a family history of breast cancer, PTVs in PALB2 were significantly associated with a high risk of disease (p < 0.05), a finding consistent with reported associations (Yang et al., 2020; Hu et al., 2021). Furthermore, according to Cybulski et al. (2015) it is estimated that 34% of breast cancer patients with a germline PALB2 PV have a triple-negative phenotype. We have recently reported that among 163 BRCA-negative triple-negative breast cancer patients in Cyprus, 4.3% are positive for PVs in PALB2 (Zanti et al., 2020), whereas PALB2 PVs consisted 87.5% of the PVs detected using a panel of 94 cancer susceptibility genes. In the analysis presented here, triple-negative breast cancer patients did not carry any PTVs in PALB2. This may be due to the limited number of triple-negative breast cancer cases in our dataset. Hence, additional studies are required to draw more definite conclusions.
It is estimated that variants of uncertain clinical significance (VUS) account for around 30%–40% of the total number of variants identified in gene-panel sequencing studies (Tung et al., 2016; Buys et al., 2017; Federici and Soddu, 2020). Among all women and genes tested, we report a 45.65% prevalence of unclassified rare missense variants. Of these, 19.70% of breast cancer cases and 17.18% of controls had at least one rare missense variant detected in the established breast cancer susceptibility genes (Dorling et al., 2021). This is consistent with the frequency reported in the large-scale CARRIERS project (Hu et al., 2021). We further demonstrate that rare missense variants in BRCA1 were associated with an increased risk of breast cancer overall. In addition, rare missense variants in PALB2 were associated with a moderate risk of triple-negative breast cancer. For missense variants in aggregate, rare missense variants in specific domains in BRCA1, ATM, CHEK2, and PALB2 were significantly associated with increased risk of certain breast cancer subtypes. However, a possible caveat that should be recognized is the potential presence of missense pathogenic variants at an allele frequency higher than 0.001 which were not included in the current analyses.
Overall, we report that 3.54% of breast cancer cases in Cyprus are positive for PTVs in the established breast cancer susceptibility genes. We further provide population-specific evidence for the association of BRCA2 and ATM PTVs with overall breast cancer risk, and ER-negative breast cancer for PALB2 PTVs. Among the established breast cancer susceptibility genes, the most prevalent PTVs occurred in the BRCA2 and ATM, followed by PALB2, and BRCA1. Finally, we confirm the effect of family history, age at diagnosis and tumor subtype as critical factors important for risk stratification of women with breast cancer in the general population of Cyprus. These results, in combination with other risk factors, may have important clinical implications for women who undergo genetic testing for breast cancer susceptibility and be beneficial for a substantial proportion of breast cancer patients in Cyprus.
Carriers of variants included in the BRIDGES project can be found here: https://databases.lovd.nl/shared/screenings#order=id%2CASC&search_owned_by_=BRIDGES%20consortium&page_size=100&page=1 https://databases.lovd.nl/shared/view/BRCA1#object_id=VariantOnTranscript%2CVariantOnGenome%2CScreening%2CIndividual&id=BRCA1#=VariantOnTranscript%2FDNA%2CASC&search_transcriptid=00003478&search_owned_by_=BRIDGES%20consortium&page_size=100&page=1 https://bcac.ccge.medschl.cam.ac.uk/bcacdata/bridges/. Requests for raw data can be made to the Data Access Coordination Committee (DACC) of BCAC (http://bcac.ccge.medschl.cam.ac.uk/).
The study was approved by the National Bioethics Committee of Cyprus, and all participants provided signed informed consent (Approval No, ΕΕΒΚ/ΕΠ/2016/38).
MZ and KM drafted the manuscript. ML, MP, and AH recruited samples and collected clinical information. DE and PD obtained funding for the BRIDGES project. MZ, DM, LD, and JD performed statistical, bioinformatics and variant annotation analysis. All authors contributed to the article and approved the submitted version.
BCAC is funded by the European Union’s Horizon 2020 Research and Innovation Programme (grant numbers 634935 and 633784 for BRIDGES and B-CAST respectively), and the PERSPECTIVE I&I project, funded by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the Ministère de l’Économie et de l'Innovation du Québec through Genome Québec, the Quebec Breast Cancer Foundation. The EU Horizon 2020 Research and Innovation Programme funding source had no role in study design, data collection, data analysis, data interpretation or writing of the report. Additional funding for BCAC is provided via the Confluence project which is funded with intramural funds from the National Cancer Institute Intramural Research Program, National Institutes of Health. The BRIDGES panel sequencing was supported by the European Union Horizon 2020 research and innovation program BRIDGES (grant number, 634935) and the Wellcome Trust (v203477/Z/16/Z). The MASTOS study was supported by “Cyprus Research Promotion Foundation” grants 0104/13 and 0104/17, and the Cyprus Institute of Neurology and Genetics.
We thank all the study participants and express appreciation to the doctors: Yiola Marcou, Eleni Kakouri, Panayiotis Papadopoulos, Simon Malas and Maria Daniel, as well as to all the nurses and volunteers who provided valuable help towards the recruitment of the study participants.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1248492/full#supplementary-material
Antoniou, A., Pharoah, P. D., Narod, S., Risch, H. A., Eyfjord, J. E., Hopper, J. L., et al. (2003). Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: A combined analysis of 22 studies. Am. J. Hum. Genet. 72 (5), 1117–1130. doi:10.1086/375033
Buys, S. S., Sandbach, J. F., Gammon, A., Patel, G., Kidd, J., Brown, K. L., et al. (2017). A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 123 (10), 1721–1730. doi:10.1002/cncr.30498
Cybulski, C., Kluzniak, W., Huzarski, T., Wokolorczyk, D., Kashyap, A., Jakubowska, A., et al. (2015). Clinical outcomes in women with breast cancer and a PALB2 mutation: A prospective cohort analysis. Lancet Oncol. 16 (6), 638–644. doi:10.1016/S1470-2045(15)70142-7
Daly, M. B., Pal, T., Berry, M. P., Buys, S. S., Dickson, P., Domchek, S. M., et al. (2021). Genetic/familial high-risk assessment: breast, ovarian, and pancreatic, version 2.2021, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw. 19 (1), 77–102. doi:10.6004/jnccn.2021.0001
Dorling, L., Carvalho, S., Allen, J., Gonzalez-Neira, A., Luccarini, C., Wahlstrom, C., et al. (2021). Breast cancer risk genes - association analysis in more than 113,000 women. N. Engl. J. Med. 384 (5), 428–439. doi:10.1056/NEJMoa1913948
Federici, G., and Soddu, S. (2020). Variants of uncertain significance in the era of high-throughput genome sequencing: A lesson from breast and ovary cancers. J. Exp. Clin. Cancer Res. 39 (1), 46. doi:10.1186/s13046-020-01554-6
Fortuno, C., Cipponi, A., Ballinger, M. L., Tavtigian, S. V., Olivier, M., Ruparel, V., et al. (2019). A quantitative model to predict pathogenicity of missense variants in the TP53 gene. Hum. Mutat. 40 (6), 788–800. doi:10.1002/humu.23739
Fortuno, C., Pesaran, T., Dolinsky, J., Yussuf, A., McGoldrick, K., Tavtigian, S. V., et al. (2021). An updated quantitative model to classify missense variants in the TP53 gene: A novel multifactorial strategy. Hum. Mutat. 42 (10), 1351–1361. doi:10.1002/humu.24264
Hadjisavvas, A., Charalambous, E., Adamou, A., Neuhausen, S. L., Christodoulou, C. G., and Kyriacou, K. (2004). Hereditary breast and ovarian cancer in Cyprus: identification of a founder BRCA2 mutation. Cancer Genet. Cytogenet 151 (2), 152–156. doi:10.1016/j.cancergencyto.2003.09.020
Hadjisavvas, A., Loizidou, M. A., Middleton, N., Michael, T., Papachristoforou, R., Kakouri, E., et al. (2010). An investigation of breast cancer risk factors in Cyprus: A case control study. BMC Cancer 10, 447. doi:10.1186/1471-2407-10-447
Hu, C., Hart, S. N., Gnanaolivu, R., Huang, H., Lee, K. Y., Na, J., et al. (2021). A population-based study of genes previously implicated in breast cancer. N. Engl. J. Med. 384 (5), 440–451. doi:10.1056/NEJMoa2005936
Kuchenbaecker, K. B., Hopper, J. L., Barnes, D. R., Phillips, K. A., Mooij, T. M., Roos-Blom, M. J., et al. (2017). Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317 (23), 2402–2416. doi:10.1001/jama.2017.7112
Loizidou, M. A., Michael, T., Neuhausen, S. L., Newbold, R. F., Marcou, Y., Kakouri, E., et al. (2008). Genetic polymorphisms in the DNA repair genes XRCC1, XRCC2 and XRCC3 and risk of breast cancer in Cyprus. Breast Cancer Res. Treat. 112 (3), 575–579. doi:10.1007/s10549-007-9881-4
Loizidou, M. A., Hadjisavvas, A., Pirpa, P., Spanou, E., Delikurt, T., Tanteles, G. A., et al. (2017). BRCA1 and BRCA2 mutation testing in Cyprus; a population based study. Clin. Genet. 91 (4), 611–615. doi:10.1111/cge.12886
National Institute for Health and Care Excellence (NICE) (2019). Familial breast cancer: Classification, care and managing breast cancer and related risks in people with a family history of breast cancer. London: National Institute for Health and Care Excellence.
Pilie, P. G., Tang, C., Mills, G. B., and Yap, T. A. (2019). State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol. 16 (2), 81–104. doi:10.1038/s41571-018-0114-z
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 17 (5), 405–424. doi:10.1038/gim.2015.30
Ruark, E., Snape, K., Humburg, P., Loveday, C., Bajrami, I., Brough, R., et al. (2013). Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature 493 (7432), 406–410. doi:10.1038/nature11725
Schmidt, M. K., Hogervorst, F., van Hien, R., Cornelissen, S., Broeks, A., Adank, M. A., et al. (2016). Age- and tumor subtype-specific breast cancer risk estimates for CHEK2*1100delC carriers. J. Clin. Oncol. 34 (23), 2750–2760. doi:10.1200/JCO.2016.66.5844
Southey, M. C., Dowty, J. G., Riaz, M., Steen, J. A., Renault, A. L., Tucker, K., et al. (2021). Population-based estimates of breast cancer risk for carriers of pathogenic variants identified by gene-panel testing. NPJ Breast Cancer 7 (1), 153. doi:10.1038/s41523-021-00360-3
Spurdle, A. B., Healey, S., Devereau, A., Hogervorst, F. B., Monteiro, A. N., Nathanson, K. L., et al. (2012). ENIGMA--evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum. Mutat. 33 (1), 2–7. doi:10.1002/humu.21628
Tung, N., Lin, N. U., Kidd, J., Allen, B. A., Singh, N., Wenstrup, R. J., et al. (2016). Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J. Clin. Oncol. 34 (13), 1460–1468. doi:10.1200/JCO.2015.65.0747
Wang, K., Li, M., and Hakonarson, H. (2010). Annovar: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38 (16), e164. doi:10.1093/nar/gkq603
Yadav, S., LaDuca, H., Polley, E. C., Hu, C., Niguidula, N., Shimelis, H., et al. (2021). Racial and ethnic differences in multigene hereditary cancer panel test results for women with breast cancer. J. Natl. Cancer Inst. 113 (10), 1429–1433. doi:10.1093/jnci/djaa167
Yang, X., Leslie, G., Doroszuk, A., Schneider, S., Allen, J., Decker, B., et al. (2020). Cancer risks associated with germline PALB2 pathogenic variants: an international study of 524 families. J. Clin. Oncol. 38 (7), 674–685. doi:10.1200/JCO.19.01907
Keywords: breast cancer susceptibility, panel sequencing, MASTOS study, case-control, next-generation sequencing
Citation: Zanti M, Loizidou MA, O’Mahony DG, Dorling L, Dennis J, Devilee P, Easton DF, Panayiotidis MI, Hadjisavvas A and Michailidou K (2023) Multi-gene panel testing and association analysis in Cypriot breast cancer cases and controls. Front. Genet. 14:1248492. doi: 10.3389/fgene.2023.1248492
Received: 27 June 2023; Accepted: 28 August 2023;
Published: 18 September 2023.
Edited by:
Marta Olszewska, Polish Academy of Sciences, PolandReviewed by:
Zhi Ling Teo, Peter MacCallum Cancer Centre, AustraliaCopyright © 2023 Zanti, Loizidou, O’Mahony, Dorling, Dennis, Devilee, Easton, Panayiotidis, Hadjisavvas and Michailidou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyriaki Michailidou, a3lyaWFraW1pQGNpbmcuYWMuY3k=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.