- 1Jilin Province Engineering Laboratory of Plant Genetic Improvement, College of Plant Science, Jilin University, Changchun, China
- 2Developmental Biology, Laboratory of Plant Molecular and Zhejiang A & F University, Hangzhou, China
Nucleosome is the basic subunit of chromatin, consisting of approximately 147bp DNA wrapped around a histone octamer, containing two copies of H2A, H2B, H3 and H4. A linker histone H1 can bind nucleosomes through its conserved GH1 domain, which may promote chromatin folding into higher-order structures. Therefore, the complexity of histones act importantly for specifying chromatin and gene activities. Histone variants, encoded by separate genes and characterized by only a few amino acids differences, can affect nucleosome packaging and stability, and then modify the chromatin properties. Serving as carriers of pivotal genetic and epigenetic information, histone variants have profound significance in regulating plant growth and development, response to both biotic and abiotic stresses. At present, the biological functions of histone variants in plant have become a research hotspot. Here, we summarize recent researches on the biological functions, molecular chaperons and regulatory mechanisms of histone variants in plant, and propose some novel research directions for further study of plant histone variants research field. Our study will provide some enlightens for studying and understanding the epigenetic regulation and chromatin specialization mediated by histone variant in plant.
1 Introduction
Over the long evolutionary process, nucleosome has always been a defining unique feature of eukaryotes. Nucleosome, the most basic structural unit of chromatin, is consisting of ∼147 bp DNA wrapped around a histone octamer, two copies of H2A, H2B, H3, and H4. Besides these core histones, the linker histones H1, which binds to the nucleosome core at a ratio of one per nucleosome to form the chromosomes and play a role in compacting chromatin into higher order structures. This integral regulatory unit is indispensable in almost every DNA template involving processes, and the complexity, involving modifications and variants, of these histones code unquestionably has important significances on genome architecture and gene regulation (Foroozani et al., 2022).
The chromatin of plants exhibits a wide range of sequence variants of the core and linker histones. In plants, except for H4, the remaining histones H2A, H2B, H3 and linker H1 all exist in multiple variants forms that are encoded by different genes and distinguished by different protein sequences. Histone variants, acting as non-allelic protein isoforms of canonical histones, can antagonize deposition of canonical histones at specific chromatin loci to maintain stability of nucleosome and confer chromatin structure more diversity and flexibility (Martire and Banaszynski, 2020; Kurumizaka et al., 2021; Nunez-Vazquez et al., 2022; Probst, 2022). Additionally, variant protein can be modified by specific post-translational modifications (PTMs), defines distinct chromatin states that impact specific chromatin functions (Foroozani et al., 2022; Jiang and Berger, 2023). Increasing studies have indicated that histone variants influence in transcription and epigenetic states, chromosome segregation, DNA damage repair (Foroozani et al., 2022; Jiang and Berger, 2023). Overall, histone variants exhibit important transcriptional programming functions during developmental process and stress responses in plants.
In this review, we concisely summarize the researches about plant histone variants and mainly focus on their biological functions and regulatory mechanisms in transcriptional regulation during plant growth and development, environmental stress responses. Moreover, we also discuss the limitation in current researches, and propose new directions for researches in the field of plant histone variants.
2 Histone variants in plants
In plants, except for histone H4, all other core histones (H2A, H2B, and H3) and linker H1 have various variant forms, and most researches focus on variants of H2A and H3. H3.3 and H2A.Z are evolutionarily conserved throughout eukaryotes (Yelagandula et al., 2014; Giaimo et al., 2019). In addition, there are some lineage-specific and tissue-specific variants, such as H2A.W variants that only function in flowering plants (Bourguet et al., 2021; Lei et al., 2021), while the H3.10 and H2B.8 only existing in sperm cells of Arabidopsis (Borg et al., 2020; Jiang et al., 2020; Borg et al., 2021; Buttress et al., 2022). Species and tissues specificity endow histone variants with distinct functions. Cause of distinct amino acid sequences from the canonical histones, variant proteins exhibit distinct chromatin deposition pattern. For example, H2A.Z and H2A.X are more biased to be concentrated in the euchromatic region (Lei and Berger, 2020; Borg et al., 2021), while H2A.W is mainly located in the concentrated heterochromatin region, and different histone variants can endow nucleosomes and chromatin with unique properties. The combination of different histone variants can form hundreds or thousands different types of nucleosomes, and this flexible pattern confer great potential for regulating various physiological processes in plants by the epigenetic code. Histone variants can influence nucleosome stability, histone modification, DNA repair and methylation, transcriptional activity, epigenetic states, and other nuclear processes that profoundly involve in plant growth and development, environmental stress responses.
There is a wide variety of histone variants in plants and their physiological functions are sophisticated, therefore, in the next paragraph, histone variants will be classified according to their family, and the similarities and differences between different variants in the same family will be summarized, which will provide reference for further research on the biological functions of histone variants in plant. In Arabidopsis, the majority of histones variants are organized in Table 1 discussed below.
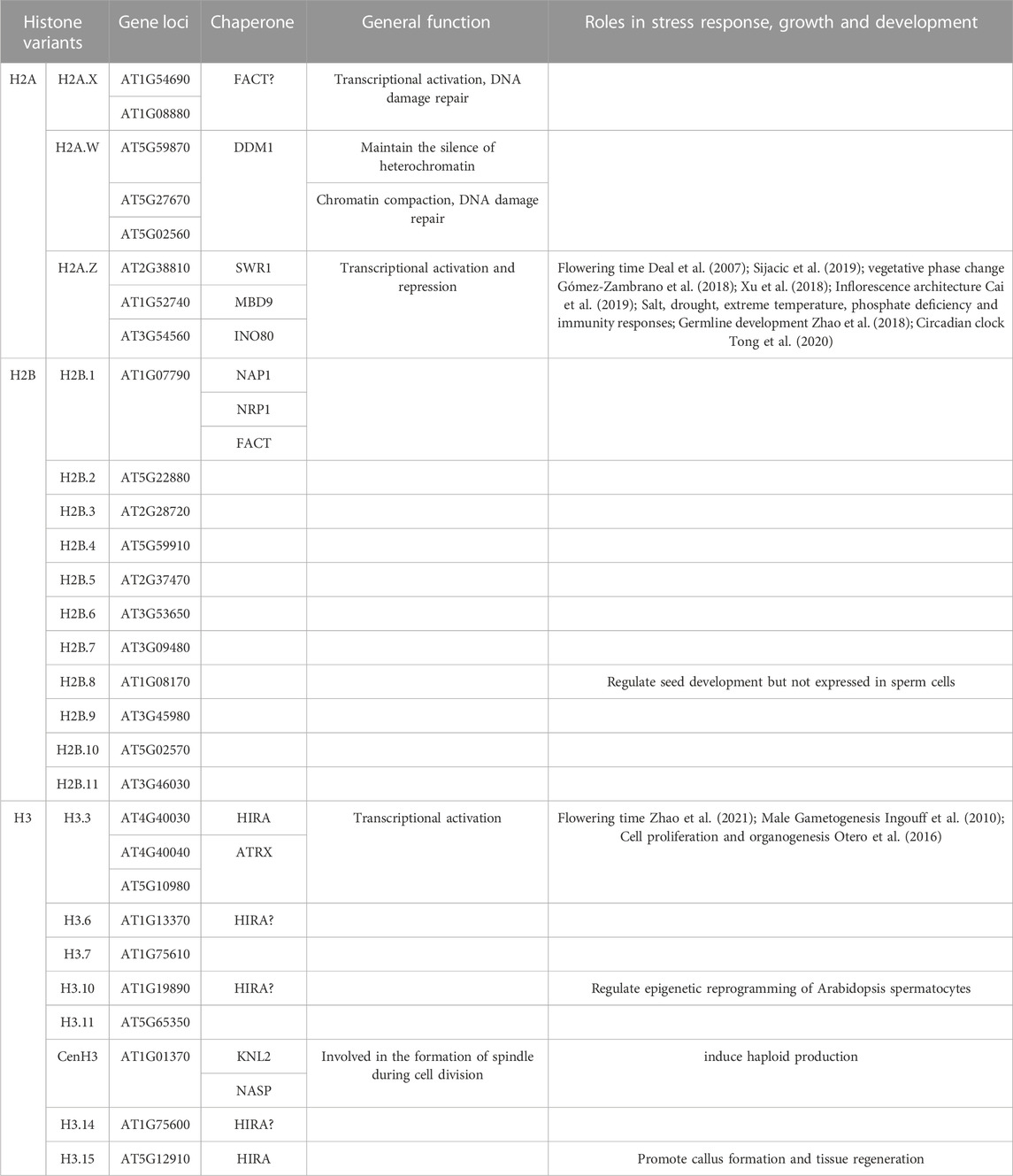
TABLE 1. Classification of histone variant family, including coding gene loci, chaperone, related generally physiological functions and roles in development and stress responses in Arabidopsis.
2.1 H2A variants
The H2A histone variants, playing vital roles in numerous biological processes, are widely studied. The three most widely studied histone variants of H2A are H2A.Z, H2A.X, and H2A.W, among them, H2A.W is specific in flowering plant, whereas H2A.X and H2A.Z are evolutionarily conserved among different species. Different H2A variants are distinguished mainly by three main features: L1 loop, docking domain and C-terminal tail.
The H2A.Z variant has a shorter C-terminal tail, which is a typical feature that differs from canonical H2A. The C-terminal tail of H2A.Z contains a KD/E conservative motif, and L1 loop contains a conservative S/TAHG motif, and the H2A.Z docking domain differs from other H2A histone variants. In Arabidopsis, there are a total of three genes encoding H2A proteins. The HTA8/9/11, which are redundant but exhibit different expression patterns, are responsible for encoding H2A.Z (Yi et al., 2006; Sijacic et al., 2019). In Arabidopsis, most active genes contain a prominent H2A.Z peak at the +1 nucleosome beyond the transcription start site (TSS) (Foroozani et al., 2022), while H2A.Z is mainly enriched in the gene body region of genes with low expression. H2A.Z is deposited into nucleosome by the ATP dependent SWR1 complex (SWI2/SNF2 related 1 complex) (Verbsky and Richards, 2001; Kobor et al., 2004; Mizuguchi et al., 2004), and the core subunits of SWR1 include PIE1 (PHOTOPERIOD-INDE PENDENT EARLY FLOWERING 1), ARP6 (ACTIN RELATED PROTEIN 6), SWC4 (SWR COMPLEX SUBUNIT 4) and SWC6 (SWR COMPLEX SUBUNIT 6) (Kobor et al., 2004; Mizuguchi et al., 2004; March-Díaz et al., 2007; Carter et al., 2018; Potok et al., 2019). Recent studies have shown that MBD9 (methy-CpG-binding domain 9) (Sijacic et al., 2019), YAF9 (yeast all1-fused gene from chromosome 9) (Crevillén et al., 2019) also participated in the assembly of H2A.Z into nucleosomes. On the contrary, NRP1 (NAP1 related protein 1) and NRP2 perform opposite function. Arabidopsis NRP1 and NRP2 proteins interacting with H2A.Z are also involved in the removal of H2A.Z from nucleosome in a SWR1 complex dependent manner, thus preventing the excessive accumulation of H2A.Z in nucleosome (Wang et al., 2020). INO80 control sliding and displacing of the canonical histone H2A and the variant H2A (Eustermann et al., 2018).
H2A.Z occupied a high proportion of total H2A content in cells, and involved many chromatins mediated processes, including changes of chromatin state as well as transcriptional activation or repression. In terrestrial plants, most H2A.Z-substituted nucleosomes are involved in the repression of transcription. H2A.Z inhibits transcriptional activity by promoting the recruitment of H3K27me3—a repressive histone modification, and preventing the deposition of H3K4me3, an active histone modification, thereby repressing gene transcription (Sequeira-Mendes et al., 2014; Yelagandula et al., 2014). Similar to conventional histones, histone variant H2A. Z could also be post-translationally modified, and the mono-ubiquitin modification of 129th lysine residue in H2A.Z by the PRC1 is also related to transcriptional repression (Gómez-Zambrano et al., 2019). Overall, H2A is involved in regulating all aspects of plant growth and development, and mutants loss of H2A function cause pleiotropic phenotypes including early vegetative phase change, early flowering, reduced fertility and others. In Arabidopsis, the biological functions of H2A.Z are organized in Figure 1 below.
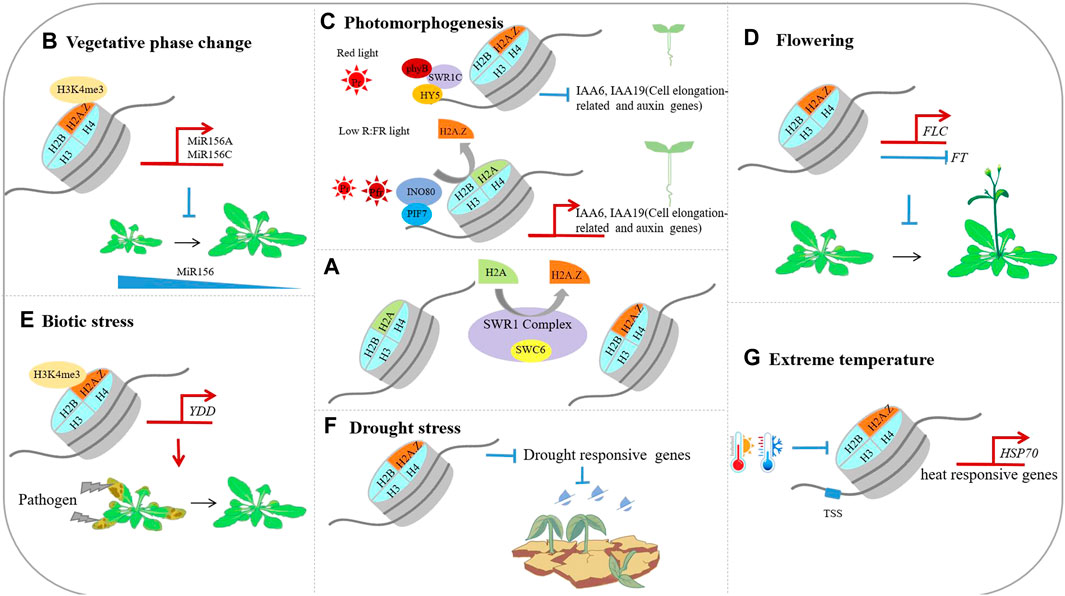
FIGURE 1. The biological functions of the histone variants in Arabidopsis thaliana. (A). H2A.Z is deposited into nucleosome by the ATP dependent SWR1 complex; (B). H2A.Z facilitates the deposition of H3K4me3 on MIR156A/MIR156C loci, and maintains a high level of MIR156A/MIR156C transcriptions; (C). Under red light conditions, the SWR1 complex deposit H2A.Z at auxin and cell elongation-related genes such as IAA6 and IAA19 to repress their expression, resulting in photomorphogenesis. Under low R∶FR light condition, the INO80 complex mediate H2A.Z eviction from PIF leading to shade avoidance response; (D). H2A.Z facilitates the transcription of FLC and inhibit the expression of FT, and prevent plants from entering reproductive stage early; (E). H2A.Z facilitates the deposition of H3K4me3 on YDD, thereby contributing the plants tolerance to Sclerotinia sclerotiorum; (F). H2A.Z facilitates the expression of drought responsive genes; (G). H2A.Z facilitates the expression of heat responsive genes such as HSP70.
Unlike H2A.Z, H2A.X variant has great similarity with H2A in terms of amino acid sequence, and the subtle feature that distinguishes H2A.X from canonical H2A is the presence of an SQEF motif at the C-terminal of H2A.X. H2A.X is enriched primarily in euchromatin and bodies of expressed genes (Yelagandula et al., 2014; Lorković et al., 2017). The histone chaperone of H2A.X is FACT (Facilitate Chromatin Transcription) complex in mammalian, while the assembly mechanism of H2A.X in plants has not been revealed. However, based on the fact that FACT chaperone is conserved in eukaryotes, it is reasonable to speculate that FACT may perform a similar function in plants (Foroozani et al., 2022). The most classic and characteristic function of H2A.X is to participate in the repair of DNA damage (Redon et al., 2011), which mainly depend on the phosphorylation of SQEF motif. When DNA is damaged, the serine residue of the SQEF motif in H2A.X is phosphorylated and then repair factors is recruited to repair the DNA damage (Bewersdorf et al., 2006).
The H2A.W variant is exclusive to the flowering plant (Kawashima et al., 2015). Particularly, the expression of H2A.W is restricted to S-phase, and its deposition is replication dependent (Gómez-Zambrano et al., 2019). H2A.W synergizes with the heterochromatin marker H3K9me2 and mainly distributed in the heterochromatin region, and H2A.W can promote heterochromatin condensation thus maintaining heterochromatin silence state (Yelagandula et al., 2014). The deposition of H2A.W alters the chromatin properties, therefore, preventing transposable elements (TEs) mobility mediated by DDM1 (DECREASE IN DNA METHYLATION 1) and leading to the silencing of TEs (Osakabe et al., 2021). At the same time, H2A.W can antagonize excessive H1 incorporation on heterochromatin, maintaining appropriate both CG and non-CG methylation level on the transposon (Bourguet et al., 2021). H2A.W has the same function as H2A.X in DNA damage repair. When DNA is damaged, the serine residue of the SQ motif in C-terminal of H2A.W. is phosphorylated by ATM (Ataxia-telangiectasia mutated kinase), thus playing an essential role in promoting DNA damage repair in the heterochromatin region (Lorković et al., 2017).
2.2 H2B variants
Compared with the well characterized histone variants of other core histones, the understanding of variants of H2B is relatively limited. The H2B variants of plants have undergone substantial evolutionary divergence, and the sequence between different H2B variants are diverse (Jiang et al., 2020). The H2B.S is highly expressed in sperm cells and mature embryo cells, both of which have highly concentrated chromatin. H2B.S variant specifically accumulates in dry seeds of several flowering plants. It indicates that the assembly of H2B.S may be beneficial to the concentration of chromatin. However, the study of H2B variants in plants is still in inception phase, and their specific functions need to be deeply studied in the future.
2.3 H3 variants
In plants, H3 has three main variants, H3.1, H3.3, and centromeric H3 variants (CenH3/CENP-A). Among them, H3.3 and CenH3 variants are most widely studied. Unlike other histone variant family, H3 family has high amino acid homology in convergent evolution. Canonical H3.1 and variants H3.3 are distinguished by four different amino acids: the 31st (Ala vs. Thr), 41st (Phe vs. Tyr), 87th (Ser vs. His) and 90th (Ala vs. Leu) (Talbert et al., 2012). Despite such tiny divergence, H3.1 and H3.3 have significant differences in modification and deposition manner. The distinction of these four different amino acids facilitates H3.1 to specifically recruit PRC2 to ensure the silence state of some developmental related genes. Recently, the combination of multiple technology methods such as immunofluorescence and genome-wide ChIP-seq data revealed that H3.1 and H3.3 have significant differences in deposition manner. H3.1 is predominantly associated with transposable elements, peri-centromeric heterochromatin and heterochromatin domains on chromosome arms, while H3.3 is enriched at the transcription end sites (TES) of genes (Stroud et al., 2012; Wollmann et al., 2012). A recent research shows that the variation of 41st amino acid (Phe vs. Tyr) partially contributes to the different distribution of H3.1 and H3.3 (Lu et al., 2018). Although H3.3 is predominantly enriched at the TES and associated with activated histone modification marks such as H3K4me3, H3K9me3, and H3K36me3, H3.3 does not affect global transcription but specifically affects the genes in response to biotic and abiotic stresses (Wollmann et al., 2017). H3.1 is mainly expressed in the S phase of the cell cycle, while H3.3 can be deposited through all stages of cell cycle in a DNA replication independent manner (Ahmad and Henikoff, 2002; Tagami et al., 2004). As the specific molecular chaperone of H3.1, CAF1 (chromatin assembly factor 1) can interact with DNA replication fork protein PCNA (PROLIFERATING CELL NUCLEAR ANTIGEN) to help H3.1 incorporate onto the newly synthesized nucleosome in S phase; whereas the H3.3 is transferred by ASF1 (Anti-Silencing Factor 1) to the typical chaperone HIRA (Histone regulator homolog A) complex, which can bind to 87th histidine and 90th leucine amino acids at C-terminal tail of H3.3 and get deposited through all stages of cell cycle (Daniel Ricketts et al., 2015). Since other less studied variants including H3.6, H3.14, and H3.10 all contain the 87th histidine and 90th leucine amino acids, whether HIRA also plays the role as molecular chaperone in their incorporations need to be demonstrated in future. Shorten breeding life.
The amino acid sequence and protein structure of CenH3 is very different from H3.1 and H3.3, especially at the N-terminal. CenH3 is located in the centromere region and plays key roles in centromere formation and kinetochore assembly, and the incorporation of CenH3 into nucleosomes occurs in the mitosis G2 stage (Lermontova et al., 2006; McKinley and Cheeseman, 2016). Unlike other H3 variants that have been thoroughly studied, there are few studies on the mechanism of CenH3 assembly into nucleosomes, and most studies of CenH3 are focused on its functions in shorting the breeding period. In recent years, scientists have exploited a series of techniques for haploid induction (HI) through manipulation of CenH3. HI can create true-breeding lines in a short period of time, which can greatly accelerate the pace of plant breeding (Lv et al., 2020; Aboobucker et al., 2023). At present, HI technology relying on CenH3/CENP-A transformation in wheat and maize has been reported (Lv et al., 2020; Wang N. et al., 2021).
In addition to the main variants of H3 described above, there are also some atypical H3 variants in Arabidopsis, for instance H3.15 and H3.10 while H3.15 plays an essential role in callus differentiation during plant regeneration (Yan et al., 2020), H3.10 is specially expressed in sperm and participates in the epigenetic reprogramming of spermatocytes (Borg et al., 2020).
2.4 Linker H1 variants
The linker H1 histone serves as a bridge connection for DNA and nucleosomes and facilitates chromatin compaction. H1 can bind to nucleosome through its binding sites in conserved GH1 domain and draw close to adjacent nucleosome to compress chromatin (Bednar et al., 2017). There are three variants of H1 in Arabidopsis: H1.1, H1.2, and H1.3. Among them, H1.1 and H1.2 are the main H1 variants donors. All of these H1 variants contain three conservative structures: a short N-terminus, GH1 central globular domain, and lysine rich C-terminus (Zhou et al., 2013). H1.3 is a histone variant induced by stresses such as low light, drought, or ABA (Rutowicz et al., 2015), and H1.3 with shorter N and C terminals and a lack of (S/T) PXK motifs that bind to DNA. All of these H1 variants are mainly enriched in the heterochromatin region and gene body, H1.1 and H1.2 are negatively correlated with gene expression and H3K4me3 deposition, while the negative correlation of H1.3 with gene expression and H3K4me3 deposition is weaker than H1.1 and H1.2 (Rutowicz et al., 2015). H1.3 is induced under stress and competes for the binding sites of H1.1 and H1.2, thus changing the accessibility of chromatin.
3 Biological functions of histone variants in plants
Plant growth and development is orchestrated by specific gene expression in a spatio-temporal manner. Additionally, plants also adjust their growth and metabolism in response to environmental stimuli by altering gene expression. Histone variants contribute importantly to specify the chromatin complexity and gene activities, and increasing findings on the variants reveal that they have essential functions during plant development in response to the external environment. Therefore, we summarize the important functions and the downstream mechanisms of histone variants redefining the plant chromatin landscape that plants perceive and transmit both developmental and environmental signals.
3.1 Roles of histone variants in plant growth and development
Flowering is an indispensable stage for higher plants, and histone variants play important roles in regulating plant flower.
H2A.Z affects flower through regulating the transcript levels of the floral repressor FLC. Reduction of H2A.Z enrichment near the TSS region on FLC chromatin repressing its expression and leading to premature flowering (Deal et al., 2007). In this process, FRI interacts with SWR1 complex to form a complex (FRI-C), which mediates H2A.Z enrichment at FLC chromatin loci and regulate FLC expression (Choi et al., 2011). Early flowering phenotypes were also observed in arp6, pie1, sef, yf9, mbd9 mutants, which were also associated with H2A.Z loading and deposition. The SWR1 complex (SWR1C) can catalyze the substitution of H2A to H2A.Z, and mutations in members of SWR1C, such as SWC6, SUF3 and PIE1, also lead to a series of development defective phenotypes, including leaf serration, apical dominance inhibition, flower deformity and accompanied with early flowering (Choi et al., 2007; Fan et al., 2022). Consistently, the early flowering phenomena of these mutants are all caused by the decrease of FLC expression caused by H2A.Z loading defects. Unexpectedly, nap1nap2 double mutants display an increase of H2A.Z enrichment at the TSS of FLC but a decrease of FLC expression, this finding indicated that H2A.Z enrichment at the TSS region of FLC is not prerequisite for FLC activation, and H2A.Z plays a dual role of activation and repression in gene transcription. The modification of H2A.Z also plays important roles in regulating the flower process, such as the acetylation of H2A.Z (Crevillén et al., 2019). Besides H2A.Z, H3.3 also participate in regulating floral transition in a FLC dependent pathway. H3.3 deposition and H3.3 mediated histone modifications are key controllers for flowering regulation through FLC transcription. FRI and HIRA chaperones collaborate to deposit H3.3 at the FLC locus and increase the activating histone modifications H3K4me3 and H3K36me3 at the FLC chromatin locus. Besides, the researchers discovered that H3.3 can facilitate the formation of 5′ to 3’ gene loop in FLC thereby promoting its transcriptional activation through chromosome conformation capture (3C) technology (Zhao et al., 2021). In addition to regulating flowering, H3.3 is also involved in regulating seed germination. H3.3 exhibits a seed specific 5′gene end distribution, and H3.3 is essential for proper gene transcriptional regulation during germination (Zhao et al., 2022).
In addition to regulating plant flowering in a FLC-dependent manner, H2A.Z could also regulate plant growth and development through some important microRNAs. H2A.Z facilitates the deposition of H3K4me3 on MIR156A/MIR156C loci, and maintains a high level of MIR156A/MIR156C transcriptions in the early stage of shoot development (Xu et al., 2018). In arp6 and pie1 mutants, reduced H2A.Z deposition and increased relative nucleosome possession of the MIR396A promoter significantly repressed miR396 expression, which accelerated plant leaf growth and vegetative phase change (Hou et al., 2019). These researches indicate that H2A.Z plays significant role in the process of plant growth and development regulated by miRNA.
In addition to regulate flower and vegetative transition, histone variants also involve in spermatocytes formation and callus differentiation. Unlike animals, specific histone variants package sperm chromatin of plants (Borg et al., 2020). H2B.8 performs a vital act in chromatin condensation of spermatocytes in flowering plants. H2B.8 (also known as H2B.S) specifically expressed in spermatocytes of flowering plants and is important for fertility. H2B.8 can specifically bind to the euchromatin transposon and the inter-gene region, condense the transcriptionally inactive region through phase separation, which facilitates nuclear compaction of the spermatocyte chromatin without affecting the gene expression (Buttress et al., 2022). In addition, variant of H3 family--H3.10 participate in the epigenetic reprogramming of sperm chromatin as well (Borg et al., 2020). H3.15 expression is rapidly induced to promote callus differentiation during plant regeneration. The replacement of lysine with histidine in the 27th amino acid of H3.15 leads to the failure of H3K27me3 deposition on the 27th amino acid mediated by the poly-comb repressive complex 2 (PRC2) on H3.15, and then the assembly of H3.15 will cause the removal of H3K27me3 from the chromatin, which will lead to transcriptional de-repression of genes related to callus formation and tissue regeneration (Yan et al., 2020).
Accurate modulation of hypocotyl cell extension is essential for plant growth and survival. Under the light, the plant presents the phenotype of the suppressed hypocotyl elongation and unfolded cotyledons, while under the dark condition, the hypocotyl elongates rapidly and cotyledons close. This process by which plants perceive the light and adjust their morphology is called photo-morphogenesis, a necessary developmental process from plant seedlings, which is jointly regulated by multiple pathways. Research has indicated that the deposition of H2A.Z is essential for the formation of photo-morphogenesis. NF-YCs (NUCLEAR FACTOR-Y) directly interact with ARP6 (ACTIN-RELATED PROTEIN6) to induce the deposition of H2A.Z in a light dependent manner. Enrichment of H2A.Z occupancy inhibits the expression of some auxin related genes such as IAA6 and IAA19, which then suppresses the growth of hypocotyls and promotes the photo-morphogenesis (Zhang et al., 2021).
PIFs direct interact with SWC6 and repress H2A.Z deposition at auxin-responsive genes such as IAA6 and IAA19, thus upregulate the expression of IAA6 and IAA19 in a red light dependent manner (Chen et al., 2023). Except to IAA6 and IAA19, H2A.Z deposited on gene body of auxin response genes HY5 to regulated hypocotyl elongation in the INO80 dependent manner (Yang et al., 2020). Compared with the deposition or eviction mechanism of H2A.Z, the transcriptional regulation of H2A.Z is still lacking in available research (Yin et al., 2023). Recently, Fang et al. shed a form of feedback regulation of auxin signaling through the transcription and deposition of H2A.Z, and ARF7/19-HB22/25-mediated H2A.Z transcription to modulate the activation of SAURs (SMALL AUXIN UP RNAs) and plant growth in Arabidopsis (Sun et al., 2022).
Similar to the photo-morphogenesis, plants will adjust their morphology and architecture under high temperature, such as elongations of hypocotyl and petiole as well as early flowering, which are collectively termed as thermal morphogenesis. Thermal morphogenesis can facilitate plant to reduce their own temperature and better adapt to high-temperature environment (Qin et al., 2022).
There is considerable overlap and crosstalk between plant thermal morphogenesis and photo-morphogenesis, and H2A.Z plays indispensable role in both these two physiological processes. Contrary to photo-morphogenesis, H2A.Z plays an inhibitory role in process of plant thermal morphogenesis. When the ambient temperature rises, INO80 chromatin remodeling complex interacts with PIF4 (PHYTOCHROME-INTERACTING FACTOR 4), and mediating the expulsion of H2A.Z from nucleosomes of PIF4 (Xue et al., 2021). INO80-C, a molecular chaperone, promotes H3K4me3 enrichment on PIF4 and enhances its transcription (Xue et al., 2021). INO80-C links H2A.Z eviction and transcription to co-modulate the expression of high temperature response associated genes, promoting the thermal morphogenesis of Arabidopsis. INO80 is an ATP-dependent chromatin remodeling factors that can establish and maintain the dynamic structure of chromatin employing the energy provided by ATP hydrolysis, and INO80 is necessary for H2A.Z loading to nucleosome of target genes. H2A.Z-INO80 module participates not only in photo morphogenesis and thermal morphogenesis, but also in the regulation of gibberellin pathway in rice. H2A.Z-INO80 module influences the GA biosynthesis pathway, and knockdown of INO80 causes decrease of H2A.Z enrichment at CPS1 and GA3ox2 loci, leading to a dwarf phenotype that can be partially restored by the spray of exogenous GA3 (Li et al., 2018).
3.2 Roles of histone variants in plant responses to biotic stress
In normal environments, plants maintain their growth and development accompanied by suppression of the functions of immune system. Microbial pathogens are the most significant biotic stresses to which plants are exposed, and therefore plants have evolved comprehensive immune response strategies to sense and deal with infection by viruses, bacteria, fungi, and other pathogens. With the refinement of the network between epigenetics and plant immunity, histone variants have been revealed to functions importantly in plants response to biotic stresses (Kim, 2021).
After perceiving pathogens, plants transduce the signals to alter phytohormone homeostasis and activate the transcriptions of resistance genes, ultimately initiating immune mechanisms. Puccinia striiformis f. sp. tritici (Pst) is a fungus that causes stripe rust in the host and reduces crop yield. Under high temperature, the TaCRK10 (Cysteine-rich receptor-like kinases 10) interacts with TaH2A.1, a variant of H2A.W, to mediate its phosphorylation and binding to related resistance genes, thereby enhancing resistance to stripe rust in wheat via the SA signaling pathway (Wang J. et al., 2021). Sclerotinia sclerotiorum causes white mold in plants, especially crucifers, whereas the critical element of SWR1C and the mutation of H2A.Z will affect the resistance of Arabidopsis to this fungus (Jakada et al., 2019; Cai et al., 2021). Further studies reveal that SWR1C promotes the expression of YDD (YODA DOWNSTREAM), a process that involves two epigenetic modifications, H2A.Z deposition and H3K4me3 modification, on YDD, thereby contributing to Arabidopsis tolerance to S. sclerotiorum (Cai et al., 2021). Similarly, arp6, pie1 and hta9hta11 mutants alter the disease resistance of Arabidopsis to the phytopathogenic bacteria Pseudomonas syringae (March-Díaz et al., 2008; Berriri et al., 2016). These results suggest that the SWR1C and histone variants regulate systemic acquired resistance in Arabidopsis. In addition, monoubiquitination of histone variants also affects plant disease resistance (Li et al., 2015). Under normal conditions, BRHIS1 (BIT-responsive Histone-interacting SNF2 ATPase 1) containing complex suppresses the expression of disease defense associated genes by interacting with histone variants H2A.Xa/H2A.Xb/H2A.3 and H2B.7, which are deposited in the promoter region of these genes. Upon pathogen infection in rice, decreased BRHIS1 expression is accompanied by increased deposition of monoubiquitinated histone variants to initiate the immune response (Li et al., 2015).
3.3 Roles of histone variants in plants responses to abiotic stresses
Plant growth and crop yield are co-regulated by both internal and external conditions, whereas the increased frequency and duration of extreme environments poses challenges to plants survival, so that plants have evolved multiple complex strategies to respond to abiotic stresses such as salt stress, phosphate deficiency, drought, high temperature, etc. Epigenetic regulation comes into action through histone variants in plant adaptation to adversities (Wollmann et al., 2017; Halder et al., 2022). Here, we review current researches on functions of histone variants in plant responses to abiotic stress and discuss the intricate mechanisms, with the hope of providing additional valuable references for crop breeding.
Excessive accumulation and deficiency of soil salinity alters plant photosynthesis, water uptake, and metabolism, which ultimately reduces crop production (Van Zelm et al., 2020; Liu, 2021). Plants have evolved multiple molecular mechanisms such as osmotic adjustment and ionic homeostasis to mitigate the damage caused by salt stress, where epigenetic modification of chromatin presents novel perspectives (Singroha et al., 2022). After salt treatment, a salt-tolerant grapevine rootstock exhibits three-fold decrease of H2A.X expression (Çakır Aydemir et al., 2020). Under salt stress, the reduction of H2A.Z deposition at the promoter and gene body region of AtMYB44 triggers the transcription of AtMYB44 and enhances Arabidopsis resistance, combined with unaffected levels of histone modifications, includingH3K4me3, H3 and H4 acetylation (Nguyen et al., 2018). Loss of function of ARP6, which affects H2A.Z deposition, causes inhibition of root development in Arabidopsis subjected to salt stress (Do et al., 2023). In rice, phosphate deficiency induces a decline in H2A.Z accumulation on the gene bodies of stress-responsive genes and facilitates the expression of these genes, allowing the plant to adapt to environmental changes (Zahraeifard et al., 2018).
Water deficit delays plant growth by causing a decline in biomass accumulation, while the drought response mechanisms are complex and unrefined. During response to mild water deficit, the higher stomatal density of h1.3 plants resulting in the enhanced photosynthetic rate and then in accelerated growth (Rutowicz et al., 2015). The increased transcription level of H1.3 contributes importantly to plant responses to drought and low light stresses (Rutowicz et al., 2015). Furthermore, H1.3 plays an irreplaceable role in maintaining redox homeostasis and stomatal development (Rutowicz et al., 2015; Probst et al., 2020). By studying the gene expression profile of Arabidopsis, it was revealed that the transcriptions of drought responsive genes are negatively correlated with the enrichments of H2A.Z in their gene body regions (Sura et al., 2017).
Extreme temperature affects the balance between development and reproduction of plants, which is also an important perspective for studying the functions of histone variants. Under high temperature, arp6 mutant exhibits hypocotyl and petiole elongation along with increased expression of heat responsive genes and HSP70 (Heat shock protein 70), and ChIP analysis reveals that the enrichment of H2A.Z on the HSP70 promoter is reduced, relieving its repressive effects on HSP70 expression (Kumar and Wigge, 2010). Further studies reveal that the INO80 complex mediates the eviction of H2A.Z and consequently activates the expressions of thermo-responsive genes, thereby contributing thermal morphogenesis of Arabidopsis (Xue et al., 2021). Heat stress regulates plant flowering time by altering the deposition of H2A.Z on FT gene, whereas this regulatory effect is specific in different plant species (Del Olmo et al., 2019; Abelenda et al., 2023). In conclusion, current researches on functions of histone variants in plants response to stresses have been focused on the H2A variants, and further investigations are needed to reveal the functions of other histone variants.
4 Conculsion
The dynamic properties and structural integrity of the nucleosome are crucial to maintain chromosome activity, and variants of the core canonical histone, profoundly affecting chromatin properties, play a prominent role in plant biological functions. At present, various researches are focusing on H2A.Z, especially on H2A.Z mediated transcriptional regulation, which has become a hot topic in the field of plant biology. The current research gives us a preliminary understanding of the biological functions and mechanisms of histones in plants, while there are still much explorations that need to be conducted for in-depth studies. Future researches can be explored in the following areas:
(1) As important epigenetic regulatory factors, histone variants play a key role in plant growth and development and stress responses. At present, most of the histone variant researches on plants is based on animal researches, and majority of researches in plant all focus on H2A.Z. There are many cell/tissues specific histone variants with low expression in plants, such as and H2B.S and CENH3/CENP-A, it is difficult to use conventional methods to reveal their functions. The cognitions of their biological functions are limited and still in primary exploration stage. With the continuous development of new technology such as single cell sequencing technology, CUT&Tag (Cleavage Under Target sand Tag mentation) can be applied to the study of histone variants. These technologies have the advantages of low sample input, high signal-to-noise ratio, and future research can use these technologies to clarify the molecular chaperone, depositing and evicting mechanism in distinct cell types;
(2) The assembly and deposition of histone variants undergo a dynamic process in a specific growth stage or receiving environmental stimuli signals such as salt stress, drought, extreme temperature, or nutrient deficit. And for plant cell, it is crucial to reprogram their chromosome landscape to respond to stimuli signals. What roles does epigenetic modifications play in these processes and whether there are tissue or time specific epigenetic modifications changes? The mechanism of how the molecular chaperone receive the internal or external environmental signals, thus assemble and unload the corresponding variants remain need to be intensively explored;
(3) In addition, related researches mainly focus on Arabidopsis, however, there are few studies relate on horticultural crops. With the deepening of research on the exploration of plant histone variants, the histone variants are expected to provide more important reference for crop breeding;
Author contributions
XW and XZ conceived the manuscript; XW and BH wrote the manuscript; HF and JH revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Jilin Province Development and Reform Commission (2021FGWCXNLJSSZ02) to JH, National Natural Science Foundation of China (32102367) and the Natural Science Foundation of Zhejiang Province (Grant No. Q21C060002) to HF.
Acknowledgments
We thank HF (Zhejiang A & F University) and JH (Jinlin University) for useful comments on this manuscript. We thank the reviewers for the constructive and positive comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abelenda, J. A., Trabanco, N., Del Olmo, I., Pozas, J., Martín-Trillo, M. d. M., Gómez-Garrido, J., et al. (2023). High ambient temperature impacts on flowering time in Brassica napus through both H2A. Z-dependent and independent mechanisms. Plant, Cell. Environ. 46 (5), 1427–1441. doi:10.1111/pce.14526
Aboobucker, S. I., Zhou, L., and Lübberstedt, T. (2023). Haploid male fertility is restored by parallel spindle genes in Arabidopsis thaliana. Nat. Plants 9, 214–218. doi:10.1038/s41477-022-01332-6
Ahmad, K., and Henikoff, S. (2002). The histone variant H3. 3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 9 (6), 1191–1200. doi:10.1016/s1097-2765(02)00542-7
Bednar, J., Garcia-Saez, I., Boopathi, R., Cutter, A. R., Papai, G., Reymer, A., et al. (2017). Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol. Cell. 66 (3), 729–397. doi:10.1016/j.molcel.2017.05.018
Berriri, S., Gangappa, S. N., and Kumar, S. V. (2016). SWR1 chromatin-remodeling complex subunits and H2A. Z have non-overlapping functions in immunity and gene regulation in Arabidopsis. Mol. plant 9 (7), 1051–1065. doi:10.1016/j.molp.2016.04.003
Bewersdorf, J., Bennett, B. T., and Knight, K. L. (2006). H2AX chromatin structures and their response to DNA damage revealed by 4Pi microscopy. Proc. Natl. Acad. Sci. 103 (48), 18137–18142. doi:10.1073/pnas.0608709103
Borg, M., Jacob, Y., Susaki, D., LeBlanc, C., Buendía, D., Axelsson, E., et al. (2020). Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Nat. Cell. Biol. 22 (6), 621–629. doi:10.1038/s41556-020-0515-y
Borg, M., Jiang, D., and Berger, F. J. (2021). Histone variants take center stage in shaping the epigenome. Curr. Opin. Plant Biol. 61, 101991. doi:10.1016/j.pbi.2020.101991
Bourguet, P., Picard, C. L., Yelagandula, R., Pélissier, T., Lorković, Z. J., Feng, S., et al. (2021). The histone variant H2A. W and linker histone H1 co-regulate heterochromatin accessibility and DNA methylation. Nat. Commun. 12 (1), 2683. doi:10.1038/s41467-021-22993-5
Buttress, T., He, S., Wang, L., Zhou, S., Saalbach, G., Vickers, M., et al. (2022). Histone H2B. 8 compacts flowering plant sperm through chromatin phase separation. Nature 611, 614–622. doi:10.1038/s41586-022-05386-6
Cai, H., Zhang, M., Chai, M., He, Q., Huang, X., Zhao, L., et al. (2019). Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A.Z and H3K4me3. Z and H3K4me3. New Phytol. 221 (1), 295–308. doi:10.1111/nph.15306
Cai, H., Huang, Y., Chen, F., Liu, L., Chai, M., Zhang, M., et al. (2021). ERECTA signaling regulates plant immune responses via chromatin-mediated promotion of WRKY33 binding to target genes. New Phytol. 230 (2), 737–756. doi:10.1111/nph.17200
Çakır Aydemir, B., Yüksel Özmen, C., Kibar, U., Mutaf, F., Büyük, P. B., Bakır, M., et al. (2020). Salt stress induces endoplasmic reticulum stress-responsive genes in a grapevine rootstock. PLoS One 15 (7), 0236424. doi:10.1371/journal.pone.0236424
Carter, B., Bishop, B., Ho, K. K., Huang, R., Jia, W., Zhang, H., et al. (2018). The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in Arabidopsis. Plant Cell. 30 (6), 1337–1352. doi:10.1105/tpc.17.00867
Chen, H., Wang, W., Chen, X., Niu, Y., Qi, Y., Yu, Z., et al. (2023). Phytochrome-interacting factors interact with SWI2/SNF2-related 1 complex subunit 6 to regulate H2A.Z deposition and photomorphogenesis in Arabidopsis. J. Genet. Genomics doi:10.1016/j.jgg.2023.04.008
Choi, K., Park, C., Lee, J., Oh, M., Noh, B., and Lee, I. (2007). Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134 (10), 1931–1941. doi:10.1242/dev.001891
Choi, K., Kim, J., Hwang, H.-J., Kim, S., Park, C., Kim, S. Y., et al. (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell. 23 (1), 289–303. doi:10.1105/tpc.110.075911
Crevillén, P., Gómez-Zambrano, Á., López, J. A., Vazquez, J., Piñeiro, M., and Jarillo, J. A. (2019). Arabidopsis YAF9 histone readers modulate flowering time through NuA4-complex-dependent H4 and H2A. Z histone acetylation at FLC chromatin. New Phytol. 222 (4), 1893–1908. doi:10.1111/nph.15737
Daniel Ricketts, M., Frederick, B., Hoff, H., Tang, Y., Schultz, D. C., Singh Rai, T., et al. (2015). Ubinuclein-1 confers histone H3 3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 6 (1), 7711. doi:10.1038/ncomms8711
Deal, R. B., Topp, C. N., McKinney, E. C., and Meagher, R. B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2AZ. Plant Cell. 19 (1), 74–83. doi:10.1105/tpc.106.048447
Del Olmo, I., Poza-Viejo, L., Piñeiro, M., Jarillo, J. A., and Crevillén, P. (2019). High ambient temperature leads to reduced FT expression and delayed flowering in Brassica rapa via a mechanism associated with H2A. Z dynamics. Plant J. 100 (2), 343–356. doi:10.1111/tpj.14446
Do, B. H., Hiep, N. T., Lao, T. D., and Nguyen, N. H. (2023). Loss-of-Function mutation of ACTIN-RELATED PROTEIN 6 (ARP6) impairs root growth in response to salinity stress. Mol. Biotechnol. 10, 1–7. doi:10.1007/s12033-023-00653-x
Eustermann, S., Schall, K., Kostrewa, D., Lakomek, K., Strauss, M., Moldt, M., et al. (2018). Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 556 (7701), 386–390. doi:10.1038/s41586-018-0029-y
Fan, J., Moreno, A. T., Baier, A. S., Loparo, J. J., and Peterson, C. L. (2022). H2A. Z deposition by SWR1C involves multiple ATP-dependent steps. Nat. Commun. 13 (1), 7052. doi:10.1038/s41467-022-34861-x
Foroozani, M., Holder, D. H., and Deal, R. B. (2022). Histone variants in the specialization of plant chromatin. Annu. Rev. plant Biol. 73, 149–172. doi:10.1146/annurev-arplant-070221-050044
Giaimo, B. D., Ferrante, F., Herchenröther, A., Hake, S. B., and Borggrefe, T. (2019). The histone variant H2A. Z in gene regulation. Epigenetics Chromatin 12 (1), 37–22. doi:10.1186/s13072-019-0274-9
Gómez-Zambrano, Á., Crevillén, P., Franco-Zorrilla, J. M., López, J. A., Moreno-Romero, J., Roszak, P., et al. (2018). Arabidopsis SWC4 binds DNA and recruits the SWR1 complex to modulate histone H2A. Z deposition at key regulatory genes. Mol. Plant 11 (6), 815–832. doi:10.1016/j.molp.2018.03.014
Gómez-Zambrano, Á., Merini, W., and Calonje, M. (2019). The repressive role of Arabidopsis H2A. Z in transcriptional regulation depends on AtBMI1 activity. Nat. Commun. 10 (1), 2828. doi:10.1038/s41467-019-10773-1
Halder, K., Chaudhuri, A., Abdin, M. Z., Majee, M., and Datta, A. (2022). Chromatin-based transcriptional reprogramming in plants under abiotic stresses. Plants 11 (11), 1449. doi:10.3390/plants11111449
Hou, N., Cao, Y., Li, F., Yuan, W., Bian, H., Wang, J., et al. (2019). Epigenetic regulation of miR396 expression by SWR1-C and the effect of miR396 on leaf growth and developmental phase transition in Arabidopsis. J. Exp. Bot. 70 (19), 5217–5229. doi:10.1093/jxb/erz285
Ingouff, M., Rademacher, S., Holec, S., Šoljić, L., Xin, N., Readshaw, A., et al. (2010). Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr. Biol. 20 (23), 2137–2143. doi:10.1016/j.cub.2010.11.012
Jakada, B. H., Aslam, M., Fakher, B., Greaves, J. G., Li, Z., Li, W., et al. (2019). Identification of SWI2/SNF2-related 1 chromatin remodeling complex (SWR1-C) subunits in pineapple and the role of pineapple SWR1 COMPLEX 6 (Ac SWC6) in biotic and abiotic stress response. Biomolecules 9 (8), 364. doi:10.3390/biom9080364
Jiang, D., and Berger, F. (2023). Variation is important: Warranting chromatin function and dynamics by histone variants. Curr. Opin. Plant Biol. 75, 102408. doi:10.1016/j.pbi.2023.102408
Jiang, D., Borg, M., Lorković, Z. J., Montgomery, S. A., Osakabe, A., Yelagandula, R., et al. (2020). The evolution and functional divergence of the histone H2B family in plants. PLoS Genet. 16 (7), e1008964. doi:10.1371/journal.pgen.1008964
Kawashima, T., Lorković, Z. J., Nishihama, R., Ishizaki, K., Axelsson, E., Yelagandula, R., et al. (2015). Diversification of histone H2A variants during plant evolution. Trends plant Sci. 20 (7), 419–425. doi:10.1016/j.tplants.2015.04.005
Kim, J.-H. (2021). Multifaceted chromatin structure and transcription changes in plant stress response. Int. J. Mol. Sci. 22 (4), 2013. doi:10.3390/ijms22042013
Kobor, M. S., Venkatasubrahmanyam, S., Meneghini, M. D., Gin, J. W., Jennings, J. L., Link, A. J., et al. (2004). A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A. Z into euchromatin. PLoS Biol. 2 (5), e131. doi:10.1371/journal.pbio.0020131
Kumar, S. V., and Wigge, P. A. (2010). H2A. Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 140 (1), 136–147. doi:10.1016/j.cell.2009.11.006
Kurumizaka, H., Kujirai, T., and Takizawa, Y. (2021). Contributions of histone variants in nucleosome structure and function. J. Mol. Biol. 433 (6), 166678. doi:10.1016/j.jmb.2020.10.012
Lei, B., and Berger, F. (2020). H2A variants in Arabidopsis: Versatile regulators of genome activity. Plant Commun. 1 (1), 100015. doi:10.1016/j.xplc.2019.100015
Lei, B., Capella, M., Montgomery, S. A., Borg, M., Osakabe, A., Goiser, M., et al. (2021). A synthetic approach to reconstruct the evolutionary and functional innovations of the plant histone variant H2A.W. W. Curr. Biol. 31 (1), 182–191.e5. doi:10.1016/j.cub.2020.09.080
Lermontova, I., Schubert, V., Fuchs, J., Klatte, S., Macas, J., and Schubert, I. (2006). Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell. 18 (10), 2443–2451. doi:10.1105/tpc.106.043174
Li, X., Jiang, Y., Ji, Z., Liu, Y., and Zhang, Q. (2015). BRHIS 1 suppresses rice innate immunity through binding to monoubiquitinated H2A and H2B variants. EMBO Rep. 16 (9), 1192–1202. doi:10.15252/embr.201440000
Li, C., Liu, Y., Shen, W. H., Yu, Y., and Dong, A. (2018). Chromatin-remodeling factor OsINO80 is involved in regulation of gibberellin biosynthesis and is crucial for rice plant growth and development. J. Integr. plant Biol. 60 (2), 144–159. doi:10.1111/jipb.12603
Liu, D. (2021). Root developmental responses to phosphorus nutrition. J. Integr. Plant Biol. 63 (6), 1065–1090. doi:10.1111/jipb.13090
Lorković, Z. J., Park, C., Goiser, M., Jiang, D., Kurzbauer, M.-T., Schlögelhofer, P., et al. (2017). Compartmentalization of DNA damage response between heterochromatin and euchromatin is mediated by distinct H2A histone variants. Curr. Biol. 27 (8), 1192–1199. doi:10.1016/j.cub.2017.03.002
Lu, L., Chen, X., Qian, S., and Zhong, X. (2018). The plant-specific histone residue Phe41 is important for genome-wide H3. 1 distribution. Nat. Commun. 9 (1), 630. doi:10.1038/s41467-018-02976-9
Lv, J., Yu, K., Wei, J., Gui, H., Liu, C., Liang, D., et al. (2020). Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat. Biotechnol. 38 (12), 1397–1401. doi:10.1038/s41587-020-0728-4
March-Díaz, R., García-Domínguez, M., Florencio, F. J., and Reyes, J. C. (2007). SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant physiol. 143 (2), 893–901. doi:10.1104/pp.106.092270
March-Díaz, R., García-Domínguez, M., Lozano-Juste, J., León, J., Florencio, F. J., and Reyes, J. C. (2008). Histone H2A. Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53 (3), 475–487. doi:10.1111/j.1365-313X.2007.03361.x
Martire, S., and Banaszynski, L. A. (2020). The roles of histone variants in fine-tuning chromatin organization and function. Nat. Rev. Mol. Cell. Biol. 21 (9), 522–541. doi:10.1038/s41580-020-0262-8
McKinley, K. L., and Cheeseman, I. M. (2016). The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell. Biol. 17 (1), 16–29. doi:10.1038/nrm.2015.5
Mizuguchi, G., Shen, X., Landry, J., Wu, W. H., Sen, S., and Wu, C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303 (5656), 343–348. doi:10.1126/science.1090701
Nguyen, N. H., Cheong, J.-J. J. B., and communications, b. r. (2018). H2A. Z-containing nucleosomes are evicted to activate AtMYB44 transcription in response to salt stress. Biochem. Biophys. Res. Commun. 499 (4), 1039–1043. doi:10.1016/j.bbrc.2018.04.048
Nunez-Vazquez, R., Desvoyes, B., and Gutierrez, C. (2022). Histone variants and modifications during abiotic stress response. Front. Plant Sci. 13, 984702. doi:10.3389/fpls.2022.984702
Osakabe, A., Jamge, B., Axelsson, E., Montgomery, S. A., Akimcheva, S., Kuehn, A. L., et al. (2021). The chromatin remodeler DDM1 prevents transposon mobility through deposition of histone variant H2A.W. W. Nat. Cell. Biol. 23 (4), 391–400. doi:10.1038/s41556-021-00658-1
Otero, S., Desvoyes, B., Peiró, R., and Gutierrez, C. (2016). Histone H3 dynamics reveal domains with distinct proliferation potential in the Arabidopsis root. Plant Cell. 28 (6), 1361–1371. doi:10.1105/tpc.15.01003
Potok, M. E., Wang, Y., Xu, L., Zhong, Z., Liu, W., Feng, S., et al. (2019). Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A. Z deposition. Nat. Commun. 10 (1), 3352. doi:10.1038/s41467-019-11291-w
Probst, A. V., Desvoyes, B., and Gutierrez, C. (2020). Similar yet critically different: The distribution, dynamics and function of histone variants. J. Exp. Bot. 71 (17), 5191–5204. doi:10.1093/jxb/eraa230
Probst, A. V. (2022). Deposition and eviction of histone variants define functional chromatin states in plants. Curr. Opin. Plant Biol. 69, 102266. doi:10.1016/j.pbi.2022.102266
Qin, W., Wang, N., Yin, Q., Li, H., Wu, A.-M., and Qin, G. (2022). Activation tagging identifies WRKY14 as a repressor of plant thermomorphogenesis in Arabidopsis. Mol. Plant 15 (11), 1725–1743. doi:10.1016/j.molp.2022.09.018
Redon, C. E., Nakamura, A. J., Martin, O. A., Parekh, P. R., Weyemi, U. S., and Bonner, W. M. (2011). Recent developments in the use of γ-H2AX as a quantitative DNA double-strand break biomarker. Aging 3 (2), 168–174. doi:10.18632/aging.100284
Rutowicz, K., Puzio, M., Halibart-Puzio, J., Lirski, M., Kotliński, M., Kroteń, M. A., et al. (2015). A specialized histone H1 variant is required for adaptive responses to complex abiotic stress and related DNA methylation in Arabidopsis. Plant Physiol. 169 (3), 2080–2101. doi:10.1104/pp.15.00493
Sequeira-Mendes, J., Aragüez, I., Peiró, R., Mendez-Giraldez, R., Zhang, X., Jacobsen, S. E., et al. (2014). The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell. 26 (6), 2351–2366. doi:10.1105/tpc.114.124578
Sijacic, P., Holder, D. H., Bajic, M., and Deal, R. B. (2019). Methyl-CpG-binding domain 9 (MBD9) is required for H2A. Z incorporation into chromatin at a subset of H2A. Z-enriched regions in the Arabidopsis genome. PLoS Genet. 15 (8), e1008326. doi:10.1371/journal.pgen.1008326
Singroha, G., Kumar, S., Gupta, O. P., Singh, G. P., and Sharma, P. (2022). Uncovering the epigenetic marks involved in mediating salt stress tolerance in plants. Front. Genet. 13, 811732. doi:10.3389/fgene.2022.811732
Stroud, H., Otero, S., Desvoyes, B., Ramírez-Parra, E., Jacobsen, S. E., and Gutierrez, C. (2012). Genome-wide analysis of histone H3. 1 and H3. 3 variants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 109 (14), 5370–5375. doi:10.1073/pnas.1203145109
Sun, A., Yin, C., Ma, M., Zhou, Y., Zheng, X., Tu, X., et al. (2022). Feedback regulation of auxin signaling through the transcription of H2A. Z and deposition of H2A. Z to SMALL AUXIN UP RNAs in Arabidopsis. New Phytol. 236 (5), 1721–1733. doi:10.1111/nph.18440
Sura, W., Kabza, M., Karlowski, W. M., Bieluszewski, T., Kus-Slowinska, M., Pawełoszek, Ł., et al. (2017). Dual role of the histone variant H2A. Z in transcriptional regulation of stress-response genes. Plant Cell. 29 (4), 791–807. doi:10.1105/tpc.16.00573
Tagami, H., Ray-Gallet, D., Almouzni, G., and Nakatani, Y. (2004). Histone H3. 1 and H3. 3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 116 (1), 51–61. doi:10.1016/s0092-8674(03)01064-x
Talbert, P. B., Ahmad, K., Almouzni, G., Ausió, J., Berger, F., Bhalla, P. L., et al. (2012). A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 5 (1), 7–19. doi:10.1186/1756-8935-5-7
Tong, M., Lee, K., Ezer, D., Cortijo, S., Jung, J., Charoensawan, V., et al. (2020). The evening complex establishes repressive chromatin domains via H2A. Z deposition. Plant Physiol. 182 (1), 612–625. doi:10.1104/pp.19.00881
Van Zelm, E., Zhang, Y., and Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. plant Biol. 71, 403–433. doi:10.1146/annurev-arplant-050718-100005
Verbsky, M. L., and Richards, E. J. (2001). Chromatin remodeling in plants. Curr. Opin. Plant Biol. 4 (6), 494–500. doi:10.1016/s1369-5266(00)00206-5
Wang, Y., Zhong, Z., Zhang, Y., Xu, L., Feng, S., Rayatpisheh, S., et al. (2020). NAP1-RELATED PROTEIN1 and 2 negatively regulate H2A. Z abundance in chromatin in Arabidopsis. Nat. Commun. 11 (1), 2887. doi:10.1038/s41467-020-16691-x
Wang, J., Wang, J., Li, J., Shang, H., Chen, X., and Hu, X. (2021a). The RLK protein TaCRK10 activates wheat high-temperature seedling-plant resistance to stripe rust through interacting with TaH2A. 1. Plant J. 108 (5), 1241–1255. doi:10.1111/tpj.15513
Wang, N., Gent, J. I., and Dawe, R. K. (2021b). Haploid induction by a maize cenh3 null mutant. Sci. Adv. 7 (4), eabe2299. doi:10.1126/sciadv.abe2299
Wollmann, H., Holec, S., Alden, K., Clarke, N. D., Jacques, P.-E., and Berger, F. (2012). Dynamic deposition of histone variant H3. 3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 8 (5), e1002658. doi:10.1371/journal.pgen.1002658
Wollmann, H., Stroud, H., Yelagandula, R., Tarutani, Y., Jiang, D., Jing, L., et al. (2017). The histone H3 variant H3. 3 regulates gene body DNA methylation in Arabidopsis thaliana. Genome Biol. 18, 94–10. doi:10.1186/s13059-017-1221-3
Xu, M., Leichty, A. R., Hu, T., and Poethig, R. S. (2018). H2A. Z promotes the transcription of MIR156A and MIR156C in Arabidopsis by facilitating the deposition of H3K4me3. Development 145 (2), 152868. doi:10.1242/dev.152868
Xue, M., Zhang, H., Zhao, F., Zhao, T., Li, H., and Jiang, D. (2021). The INO80 chromatin remodeling complex promotes thermomorphogenesis by connecting H2A. Z eviction and active transcription in Arabidopsis. Mol. Plant 14 (11), 1799–1813. doi:10.1016/j.molp.2021.07.001
Yan, A., Borg, M., Berger, F., and Chen, Z. (2020). The atypical histone variant H3 15 promotes callus formation in Arabidopsis thaliana. Development 147 (11), 184895. doi:10.1242/dev.184895
Yang, C., Yin, L., Xie, F., Ma, M., Huang, S., Zeng, Y., et al. (2020). AtINO80 represses photomorphogenesis by modulating nucleosome density and H2AZ incorporation in light-related genes. Proc. Natl. Acad. Sci. 117 (52), 33679–33688. doi:10.1073/pnas.2001976117
Yelagandula, R., Stroud, H., Holec, S., Zhou, K., Feng, S., Zhong, X., et al. (2014). The histone variant H2A. W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell. 158 (1), 98–109. doi:10.1016/j.cell.2014.06.006
Yi, H., Sardesai, N., Fujinuma, T., Chan, C.-W., and Gelvin, S. B. (2006). Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell. 18 (7), 1575–1589. doi:10.1105/tpc.105.039719
Yin, C., Sun, A., Zhou, Y., Liu, K., Wang, P., Ye, W., et al. (2023). The dynamics of H2A.Z on SMALL AUXIN UP RNAs regulate abscisic acid–auxin signaling crosstalk in Arabidopsis. J. Exp. Bot., erad131. doi:10.1093/jxb/erad131
Zahraeifard, S., Foroozani, M., Sepehri, A., Oh, D.-H., Wang, G., Mangu, V., et al. (2018). Rice H2A. Z negatively regulates genes responsive to nutrient starvation but promotes expression of key housekeeping genes. J. Exp. Bot. 69 (20), 4907–4919. doi:10.1093/jxb/ery244
Zhang, C., Qian, Q., Huang, X., Zhang, W., Liu, X., and Hou, X. (2021). NF-YCs modulate histone variant H2A. Z deposition to regulate photomorphogenic growth in Arabidopsis. J. Integr. Plant Biol. 63 (6), 1120–1132. doi:10.1111/jipb.13109
Zhao, L., Cai, H., Su, Z., Wang, L., Huang, X., Zhang, M., et al. (2018). KLU suppresses megasporocyte cell fate through SWR1-mediated activation of WRKY28 expression in Arabidopsis. Proc. Natl. Acad. Sci. 115 (3), 526–535. doi:10.1073/pnas.1716054115
Zhao, F., Zhang, H., Zhao, T., Li, Z., and Jiang, D. (2021). The histone variant H3. 3 promotes the active chromatin state to repress flowering in Arabidopsis. Plant Physiol. 186 (4), 2051–2063. doi:10.1093/plphys/kiab224
Zhao, T., Lu, J., Zhang, H., Xue, M., Pan, J., Ma, L., et al. (2022). Histone H3.3 deposition in seed is essential for the post-embryonic developmental competence in Arabidopsis. Nat. Commun. 13 (1), 7728. doi:10.1038/s41467-022-35509-6
Keywords: histone variants, epigenetic regulation, biological function, growth, development, stress response
Citation: Wu X, Zhang X, Huang B, Han J and Fang H (2023) Advances in biological functions and mechanisms of histone variants in plants. Front. Genet. 14:1229782. doi: 10.3389/fgene.2023.1229782
Received: 27 May 2023; Accepted: 21 July 2023;
Published: 31 July 2023.
Edited by:
Sarfraz Shafiq, Western University, CanadaReviewed by:
Suresh Kumar, Indian Agricultural Research Institute (ICAR), IndiaYue Lu, Yangzhou University, China
Giovanni Messina, Sapienza University of Rome, Italy
Copyright © 2023 Wu, Zhang, Huang, Han and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junyou Han, aGFuanlAamx1LmVkdS5jbg==; Huihui Fang, ZmFuZ2hoQHphZnUuZWR1LmNu
†These authors share first authorship
 Xi Wu1†
Xi Wu1† Huihui Fang
Huihui Fang