- 1Hainan Academy of Ocean and Fisheries Sciences, Haikou, China
- 2Hainan Tropical Ocean University Yazhou Bay Innovation Institute, Sanya, China
- 3Hainan Provincial Engineering Research Center for Tropical Sea-Farming, Haikou, China
Introduction: Plectropomus leopardus, a commercially significant marine fish, is primarily found in the Western Pacific regions and along the coast of Southeast Asia. A thorough analysis of the molecular mechanisms involved in sex differentiation is crucial for gaining a comprehensive understanding of gonadal development and improving sex control breeding. However, the relevant fundamental studies of P. leopardus are relatively lacking.
Methods: In this study, a genome-wide association study (GWAS) was conducted to investigate the genetic basis mechanism of sex differentiation and gonadal developmental traits in P. leopardus utilizing about 6,850,000 high-quality single-nucleotide polymorphisms (SNPs) derived from 168 individuals (including 126 females and 42 males) by the genome-wide efficient mixed-model association (GEMMA) algorithm.
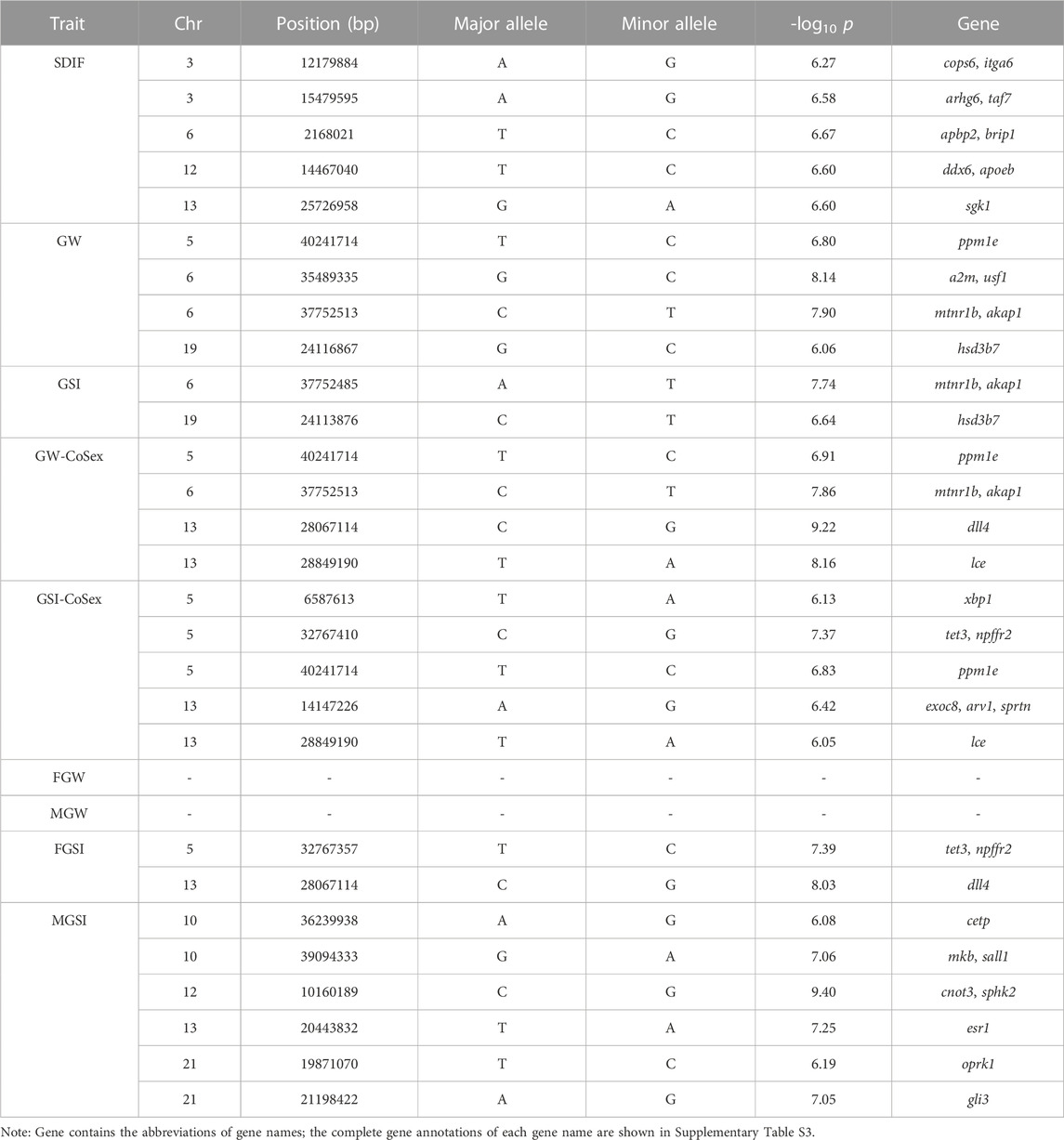
Results: The results of these single-trait GWASs showed that 46 SNP loci (-log10 p > 7) significantly associated with sex differentiation, and gonadal development traits were distributed in multiple different chromosomes, which suggested the analyzed traits were all complex traits under multi-locus control. A total of 1,838 potential candidate genes were obtained by considering a less-stringent threshold (-log10 p > 6) and ±100 kb regions surrounding the significant genomic loci. Moreover, 31 candidate genes were identified through a comprehensive analysis of significant GWAS peaks, gene ontology (GO) annotations, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, including taf7, ddx6, apoeb, sgk1, a2m, usf1, hsd3b7, dll4, xbp1, tet3, esr1, and gli3. These trait-associated genes have been shown to be involved in germline development, male sex differentiation, gonad morphogenesis, hormone receptor binding, oocyte development, male gonad development, steroidogenesis, estrogen-synthetic pathway, etc.
Discussion: In the present study, multiple genomic loci of P. leopardus associated with sex differentiation and gonadal development traits were identified for the first time by using GWAS, providing a valuable resource for further research on the molecular genetic mechanism and sex control in P. leopardus. Our results also can contribute to understanding the genetic basis of the sex differentiation mechanism and gonadal development process in grouper fish.
1 Introduction
Sex differentiation in aquatic fish is complex and elusive due to the interactive influence of heredity and environment. In teleost, a vast range of reproductive systems gives rise to an enormous plasticity of sex differentiation and change (Devlin and Nagahama, 2002). Elucidating the sex differentiation mechanisms and gonadal transformation into either a testis or an ovary in fish is important for both theoretical study and practical production (Xu et al., 2021). Various prominent genetic and environmental factors are involved during sex differentiation. At the genomic level, some candidate genes have been characterized as well-known factors and major determinants in sex differentiation. On the other hand, multiple physical and chemical environmental factors, such as water temperature, PH, salinity, and social interaction, have been demonstrated to influence sex (Baroiller et al., 2009; Rajendiran et al., 2021). Significantly, the fundamental and molecular mechanisms of sex differentiation in fish show high variability (Piferrer and Guiguen, 2008). To a certain extent, sex differentiation is usually visualized by gonadal differentiation that can take many forms in fish (Devlin and Nagahama, 2002); for example, some gonochoristic species develop only testis or ovaries and remain the same sex throughout their lifespans, some hermaphroditic species possess both sexes of immature gametes in a single gonad or functional sex reversal, and some species show various patterns and an extremely flexible process of sex differentiation. Revealing the molecular mechanisms of sex differentiation in fish will have important implications for research on reproductive development, sex-ratio control, and selective breeding. The gonad development process is an important manifestation of sex differentiation, which requires the involvement of genes and hormones (Mustapha et al., 2021). Accordingly, to comprehensively understand the mechanisms of sex differentiation (SDIF), some indicators of gonadal development, such as gonadal weight (GW) and gonadosomatic index (GSI), are usually considered together for relevant studies of sex-related traits in fish (Wang et al., 2021).
Many research ideas and basic methods with different formats have been used to solve the scientific problem and explore the fundamental mechanisms involved in the genetic information of sex-related traits in aquatic animals. Sex differentiation is a very important part of sex-related traits, which drives the primitive gonad to the testis or ovary (Stévant and Nef, 2019). Histological observation has been a conventional research methodology employed to investigate sex differentiation in gonochoristic fishes (Takashima et al., 1980). Using histological studies to inspect the different stages involved in the development of gonads can provide more detail about the morphological process of sex differentiation (Patil and Nayak, 2021). Moreover, model fish with a deficiency of an important sex-related gene were used to elucidate genetic mechanisms in the development of sexual traits (Zhai et al., 2018). With the development of molecular biology and high throughput sequencing technology, the study of sex differentiation has entered the era of omics. RNA-Seq has undoubtedly become the most popular and inexpensive option for operating omics data analysis. By using gonad transcriptome analysis of male and female channel catfish gonads during the critical period of sex differentiation, a few candidate genes and potential pathways underlying male-preferential cell development were revealed in catfish (Zeng et al., 2016). To uncover the genetic framework underlying sex differentiation of tilapia, a dynamic co-expression network analysis with RNA-Seq was carried out to detect differently expressed genes that were involved in sex differentiation (Tao et al., 2018). A high-quality gonadal transcriptome analysis of Siberian sturgeon was used to search for sex-differentiation genes at undifferentiated stages, and multiple genes related to the stem-cell niche and sex-specific nerve development were screened out (Vizziano-Cantonnet et al., 2018). However, we can only estimate the expression level for each gene with RNA-Seq without regard for the exact relationship between genes and observable characteristics of each individual across the population.
Genetic studies based on genome-wide scans have provided a new approach to sex differentiation in fish. Whole-genome wide association study (GWAS), which identifies associations of genotypes with phenotypes by using statistical inference for differences in the allele frequency of candidate genomic loci among objective populations (Uffelmann et al., 2021), is the most popular method of whole-genome scanning. Compared to previous linkage-based genome-wide studies, the GWAS approach is more powerful and sensitive for locating genomic loci associated with phenotypes (Talukder et al., 2019). To date, GWASs have been widely used to detect genetic loci or genes of sex-related traits in fish. For instance, several sex-related candidate genes within a sex-associated region were identified in Takifugu bimaculatus using GWAS (Patil and Nayak, 2021). An approximate GWAS result by ddRAD sequencing has also been obtained in a random breeding population of Larimichthys crocea, some candidate genes (i.e., dmrt1, dmrt3, piwil2, fam102a, and odf2) and some others (i.e., axl, cyp2a10, and cyp2g1) were detected separately to be involved in sex determination and GSI traits (Lin et al., 2021). Furthermore, a GWAS was conducted to reveal novel sex-related candidate genes (i.e., trap1 and fryl) involved in sex determination and differentiation in Mesocentrotus nudus using the genotyping-by-sequencing method (Wang et al., 2022). In addition, similar studies have been carried out on Cynoglossus semilaevis (Zhang et al., 2020), Salmo salar (Gutierrez et al., 2015; Gabián et al., 2019), and Scophthalmus maximus (Martínez et al., 2021). All these GWAS results provide potential genomic candidate loci and genes for dissecting molecular genetic mechanisms of sex-related traits in fish; they could be used to improve reproductive strategy and breeding efficiency.
The leopard coral grouper, Plectropomus leopardus, is a high-value species with bright external coloration that belongs to the Serranidae family. According to the available information, P. leopardus, the characteristic group of grouper species, exhibits a model of sex differentiation that undergoes a transition from ovary to intersexual gonad and then to testis (Zhou and Gui, 2010). Earlier studies have shown that the sex ratio, sex-specific size, and age structure of P. leopardus are variable with geographical location (Adams et al., 2000). However, the mechanism governing sex differentiation and gonadal development of P. leopardus is still poorly understood. Therefore, it is of interest to study the underlying genetic architecture of the sex differentiation mechanism in P. leopardus. Here, we performed GWAS of sex differentiation and gonadal development traits (GW and GSI) to identify relevant candidate genomic loci and genes.
2 Materials and methods
2.1 Experimental fish and library construction
All individuals of P. leopardus used in this work were obtained from the Hainan Ocean and Fisheries Sciences Research Base in Qionghai City, Hainan Province, P.R. China. The study population was the 4-year-old P. leopardus, which is the second filial generation of a breeding population collected from different regions throughout Hainan Island in 2009. A total of 168 individuals were sampled from the population for genomic analysis. Genomic DNA of each sample was extracted and purified from caudal fin samples using the standard phenol–chloroform protocol (Sambrook and Russell, 2006). Then, the quality of isolated genomic DNA was verified by 1% agarose gel electrophoresis and a Qubit™ dsDNA HS Assay Kit (Life Technologies, CA, United States of America). The sequencing libraries were constructed using a Nano DNA HT Sample Prep Kit (Illumina United States of America) with a total amount of 0.3 μg DNA per sample.
2.2 Whole genome resequencing and SNP calling
The fragments with 350 bp in size of genomic DNA samples were isolated by sonication, then end-polished, A-tailed, and ligated with the full-length adaptor for Illumina sequencing with further PCR amplification. Pair-end sequencing was performed to generate raw sequencing reads, followed by quality control using the fastp v0.20.1 (Chen et al., 2018). Then, the GATK v4.1.5.0 (Mckenna et al., 2010) was applied to split the pooling sequencing data and genotyping. For SNP calling, the sequencing reads were aligned to the P. leopardus genome (CNGBdb Accession No. CNP0000859), and SNPs were identified by HaplotypeCaller of the software package GATK. We used QC-Chain to filter the raw reads by discarding the adapter sequences, low-quality reads, and duplicated reads, and then all indels and non-biallelic SNPs were removed.
2.3 Statistics of phenotype data
In this study, 168 experimental fish were euthanized to measure body weight (BWE), after which they were dissected to determine gonadal weight (GW) and assess physiological sex. We used the GSI to indicate sexual maturity, which represented the gonadal development of individuals, by the formula below (Lin et al., 2021):
To distinctly perform a GWAS of GW and GSI for investigating the gonadal development process and to avoid sex differences affecting the results of the association analysis, a statistical analysis of male and female phenotypes, including female gonadal weight (FGW), male gonadal weight (MGW), female gonadosomatic index (FGSI), and male gonadosomatic index (MGSI), was conducted. Wilcox tests were implemented in GW and GSI between female and male individuals by R to ensure there were significant differences between the physiological sexes.
2.4 Statistical model
GWASs were conducted for each of the SDIF, GW, GSI, FGW, MGW, FGSI, and MGSI traits by univariate linear models (ULMs) using a genome-wide efficient mixed model association (GEMMA) algorithm. For the phenotypic traits influenced by gender (GW and GSI), sex difference (female or male) was considered as a fixed covariate or not in the statistical methods. Each phenotype could be explained by the following linear mixed model:
where y is the n × 1 vector of phenotypic values of each trait, n is the number of individuals, W is the n × c matrix of covariates (fixed effects) such as a column vector of 1 and sex difference for GW and GSI, α is the c × 1 vector of corresponding coefficients of fixed factors, c is the number of covariates, x is the n × 1 vector of SNP marker genotypes, β is the effect size of the marker, Z is an n×m incidence matrix, u is an m × 1 vector of random polygenic effects while excluding the tested SNP, and ε is the n × 1 vector of random residual errors. Manhattan and quantile–quantile (QQ) plots were used to visualize potential regions of interest that are associated with the phenotype and assess whether or not the dataset matches a specified probability distribution. All Manhattan and QQ plots in this study were drawn using R v4.0.5.
2.5 Potential candidate genomic loci and gene searching
To identify potential candidate genomic loci significantly associated with sex differentiation and gonadal development traits, the threshold p-value (-log10 p > 7) was used to calculate the genome-wide significance of each SNP marker. After the candidate genomic loci were screened out, we searched candidate genes based on the 100-kb regions surrounding the most significant SNPs according to their locations and functions. The less-stringent threshold p-value (-log10 p > 6) was also used for significant SNP identification. Subsequently, the candidate genes inferred from these SNPs were applied to explore the potential association with each objective trait by GO function annotation and KEGG pathway enrichment. All SNPs associated with the candidate genes involved in the aforementioned GO term were selected to compare with GWAS peaks on regions (±100 kb) with multiple SNPs exceeding the threshold in each trait. Finally, coincident SNPs were screened out as the most valuable genomic loci that were explicitly associated with sex differentiation and gonad development.
3 Results
3.1 Sequencing data and genotyping
Genomic DNA of 168 P. leopardus samples were sequenced by Illumina NovaSeq 6000 platform (Illumina, CA, United States) to generate 4.35 billion raw reads (∼1304.04 Gb in length). After the data filtering was conducted, 4.28 billion clean reads (∼1284.05 Gb in length) were obtained. A total of 14,653,768 SNPs of 168 samples were identified after filtering by VCFtools v0.1.15 (Danecek et al., 2011). Moreover, SNPs were further filtered by PLINK1.90 v1.90b4 (Purcell et al., 2007) with the parameters of “--mind 0.15 || --geno 0.10 || --maf 0.05 || --hwe 0.0001.” In addition, we used Beagle v5.4 (Browning et al., 2021) to impute all missing SNPs with default parameters. Finally, 168 samples with more than six million SNPs (6,890,254 SNPs for SDIF, GW, and GSI; 6,823,720 SNPs for FGW and FGSI; and 6,431,681 SNPs for MGW and MGSI) remained for the GWAS research.
3.2 Statistics of phenotypic analysis
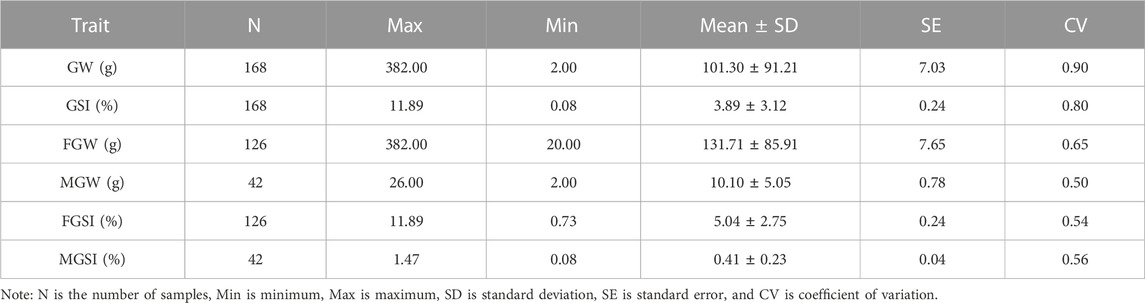
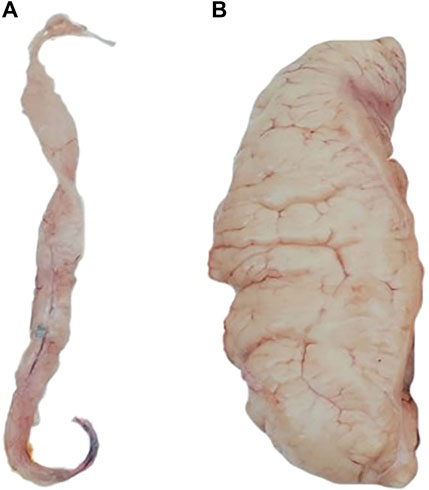
The phenotypic records of 168 individuals for each trait were counted in Table 1. According to the statistical analysis, we found a significant difference between the phenotype of FGW, which is 131.71 ± 85.91 g, and the phenotype of MGW, which is 10.10 ± 5.05 g. The visual difference between male and female gonads is shown in Figure 1. A significant difference was also presented between the phenotype of FGSI (5.04% ± 2.75%) and MGSI (0.41% ± 0.23%). By using the Wilcoxon test, the significant differences (p < 0.0001) were proved between the female and male groups of the GW and GSI phenotypes (Supplementary Figure S1). The coefficient of variation (CV) for each phenotypic trait value was greater than or equal to 0.5, which indicated that all traits possessed relatively discrete phenotypes. However, gender-specific statistics for GW and GSI might help decrease the CV of the phenotypes (Table 1).
3.3 Genome-wide association study
To investigate the genomic loci and genes significantly associated with sex differentiation and gonadal development process, GWAS on SDIF and a total of eight quantitative traits, including GW, GSI, FGW, MGW, FGSI, and MGSI, were conducted by using a linear mixed-model. Furthermore, in order to clarify the impact of gender on GW and GSI, the physiological sex of each individual was taken as the covariable of the GWAS model. Therefore, GW and GSI with physiological sex as covariates (GW-CoSex and GSI-CoSex) were added to the analysis. Manhattan and quantile–quantile (QQ) plots of nine specific traits are shown in Figure 2 and Supplementary Figure S2. In addition, genomic control (GC) values of each analysis were estimated to evaluate false negatives or false positives in GWAS (Supplementary Table S1). Based on the genome-wide threshold of -log10 p > 7, we identified 46 SNPs that significantly correlated with sex differentiation and gonadal development traits (Supplementary Table S2). Here, four SNPs associated with the SDIF trait were located on chromosomes 1, 6, 7, and 18 (Figure 2). For GW and GSI traits, six (located on chromosomes 6 and 13) and three (located on chromosomes 6 and 12) SNPs showed a significant correlation, respectively (Figure 2). To consider the effect of physiological sex on gonadal development, GWASs of GW-CoSex and GSI-CoSex were implemented to identify five (located on chromosomes 5, 8, and 13) and eight (located on chromosomes 2, 5, 6, and 13) significant SNPs (Supplementary Figure S2). To avoid sex differentiation affecting the associated results of GW and GSI, we also separately conducted the statistical analysis of male and female phenotypes, whereafter four significantly associated SNPs that exceeded the threshold were found in FGW (located on chromosome 13), two were found in MGW (located on chromosome 7), seven were found in FGSI (located on chromosomes 1, 2, 5, and 13), and 17 were found in MGSI (located on chromosomes 1, 2, 6, 9, 12, 13, 15, 16, 17, and 21) traits (Supplementary Figure S2).

FIGURE 2. Manhattan and QQ plots of genome-wide SNP significance for the traits of SDIF, GW, and GSI in P. leopardus. The X-axis shows all SNPs across chromosomes 1–24, and the Y-axis represents the -log10 p. The gray dotted line indicates the genome-wide significant thresholds (p < 10E-7).
3.4 Identification and functional characterization of genomic loci and candidate genes
To conduct a less-stringent threshold of -log10 p > 6 and ±100 kb regions surrounding the significantly associated loci, we obtained 595 potential candidate genes for SDIF, GW, and GSI traits that play an important role in illuminating the molecular mechanisms of SDIF and 1,243 potential candidate genes for GW-CoSex, GSI-CoSex, FGW, MGW, FGSI, and MGSI that have an effect on the gonadal development process. Subsequently, gene ontology (GO) analyses of the two groups of candidate genes were applied to excavate the functional association with the objective traits. In detail, 88 GO terms (SDIF, GW, and GSI) and 143 GO terms (GW-CoSex, GSI-CoSex, FGW, MGW, FGSI, and MGSI) were annotated as related to sex differentiation and gonad development, such as female/male sex differentiation, female/male gonad development, gonad morphogenesis, steroid biosynthetic process, hormone biosynthetic process, oocyte construction, spermatid differentiation, etc., whereafter 156 candidate genes probably associated with multiple biological processes of sex differentiation and gonad development were selected for GO and KEGG signaling pathway (KEGG) analyses (Figure 3). Accordingly, 23 coincident SNPs have been identified as the most valuable genomic loci from 156 candidate genes with related functional characterization, and GWAS peaks on regions (±100 kb) with at least two SNPs exceeding the threshold (-log10 p > 6) (Table 2).
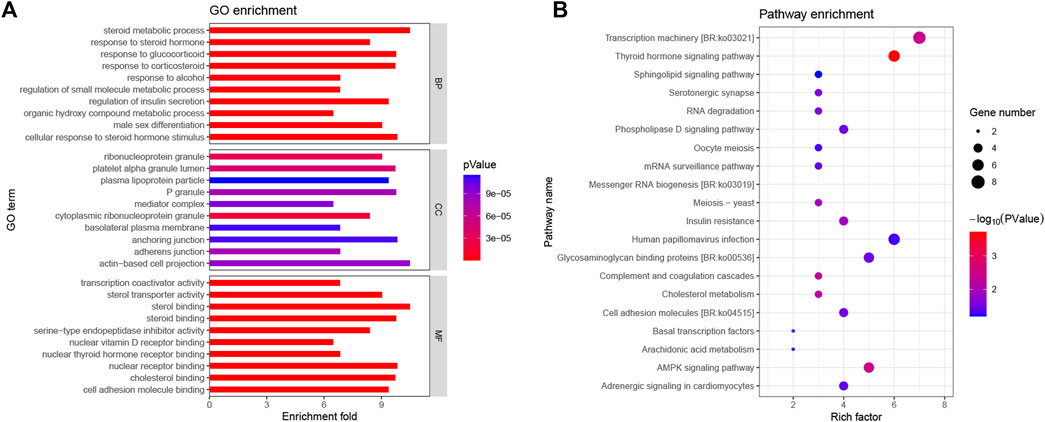
FIGURE 3. GO and KEGG analysis of the 156 potential candidate genes associated with sex differentiation and gonad development of P. leopardus. (A) Top 10 most significant GO terms (BP, biological process; CC, cellular component; and MF, molecular function) associated with the candidate genes; (B) top 20 most significant KEGG pathways associated with the candidate genes.
4 Discussion
The formation mechanism of sex differentiation and transition of P. leopardus is a complex subject. By examining the gonads of physio-sexual dimorphic sex-changing individuals, P. leopardus first differentiated as an immature female, and then a male-type gonad would be derived from a female-type mature gonad through sex change surgery (Adams, 2003). Early sex change is considered a latent defect for P. leopardus, as it shortens the female reproductive period while affecting the male sperm quality. Late sex change, on the other hand, is an untapped potential economic trait in P. leopardus because it would bring about higher female fecundity and efficient utilization of energy for reproduction. The sex determination, sex differentiation, or gonadal development mechanisms have been broadly applied in the explanation of the genetic mechanism of aquatic animals by using GWAS. It is interesting to note that existing GWASs on sex-related traits of fish were implemented for various phenotypes, such as sex determination, gonadal development, and sex reversal. Several sex-associated genomic regions and genetic loci related to the sex-determination system and putative sex-determination genes were detected in Protosalanx hyalocranius (Li et al., 2020) and Thunnus orientalis (Nakamura et al., 2021). Moreover, a single locus with a large controlling effect was identified through GWAS to elucidate the mechanism of sex reversal in Cynoglossus semilaevis (Jiang and Li, 2017). Gonadal development is a significant feature when elaborating sex differentiation, which is triggered by sex-determination signals. Gonadal weight and gonadosomatic index were representative phenotypes for identifying genomic loci of sex-related traits in fish (Gao et al., 2018; Lin et al., 2021; Patil and Nayak, 2021). The male heterogametic sex-determination system as a special sex-linked inheritance was also interpreted using GWAS in Oncorhynchus mykiss (Fraslin et al., 2020) and Clarias macrocephalus (Nguyen et al., 2021).
Plectropomus leopardus, a grouper from the subfamily Epinephelinae, has commanded high market prices locally and overseas. Since the rapid increase in market demand, its culture area continues to grow, and breeding technology continues to improve. However, knowledge of the sex differentiation mechanism of this species is still unclear. Therefore, to investigate the molecular mechanisms of sex differentiation and gonadal development traits of P. leopardus, we performed GWASs using 168 sexually mature individuals (126 females and 42 males) and more than six million sequencing-based SNPs in this study. The traits to be analyzed were divided into three categories, such as sex differentiation, gonadal weight, and gonadosomatic index, for thoroughly exploring molecular genetic mechanisms of sex development in P. leopardus. Especially, phenotypic sex was considered as a covariate, and sex-specific phenotypes were separately conducted for the GW and GSI traits to avoid sex differences affecting the results of association analysis.
In order to identify more genomic loci associated with the analyzed traits, the stringent threshold p-value (-log10 p > 7) and less-stringent threshold p-value (-log10 p > 6) were used for screening significant loci. Afterward, 46 significant SNPs exceeded the stringent threshold, and 250 significant SNPs were positioned above the less-stringent threshold. Moreover, we identified 156 candidate genes within or very close (±100 kb) to the 250 significant SNPs. GO and KEGG enrichment analysis of these candidate genes suggested that GO terms and pathways related to steroid and thyroid hormones were important for sex differentiation and gonadal development. Furthermore, 23 intersecting loci and 31 nearby genes on multiple chromosomes were extracted from functional enrichment results and GWAS peaks. These results indicated that sex differentiation and gonadal development of P. leopardus are complex traits under multi-locus control.
For the GWAS of the SDIF trait in this study, we identified nine trait-associated genes that were mainly involved in such germline development, male sex differentiation, gonad morphogenesis, hormone receptor binding, oocyte development, male gonad development, the steroid metabolic process, and the response to glucocorticoid. itga6 (integrin alpha 6) has been identified as a surface marker of stem spermatogonia for fertility preservation, and its expression in the male germinal epithelium has been confirmed (Sá et al., 2013). It has also been proved that taf7l, as a paralogue of taf7 (transcription initiation factor TFIID subunit 7), cooperates with trf2 to regulate spermiogenesis (Zhou et al., 2013), and the variation of taf7 affects the genetic etiology of idiopathic male infertility (Pal et al., 2021). As reported, ddx6 (probable ATP-dependent RNA helicase DDX6) belongs to a family of DEAD-box RNA helicases that was highly expressed during various stages of gonad development in the Chinese mitten crab (Eriocheir sinensis) (Li et al., 2015). Based on the ontogenic gonadal transcriptomes, the mechanisms of sex change of the ricefield eel (Monopterus albus) have been elucidated (Fan et al., 2022), and the expression of apoeb (apolipoprotein Eb) detected was dramatically increased and peaked at the early intersexual stage. Another study has suggested that the expression of apoeb was significantly different between old and young female brains in zebrafish (Danio rerio) (Zhai et al., 2022). Furthermore, SGK1 (integrin alpha 6) is responsible for regulating foxo1 (forkhead box O1) (Ding et al., 2017), and foxo1 knockdown suppressed expression of sox9 (sex-determining region Y cox 9) (Kurakazu et al., 2019). Previous studies reported that sox9 promoted testis cord formation during testicular differentiation (Li et al., 2014) and played an important role as a master regulator of Sertoli cell differentiation during testis development (Jakob and Lovel-Bafge, 2011). In addition, sox9 was cloned in the lambari fish (Astyanax altiparanae) and evidenced its role in sex determination, sex differentiation, and the male reproductive cycle (Adolfi et al., 2015). Thus, we assume there is a complex regulatory mechanism of sgk1 for male sex differentiation in P. leopardus.
Gonadal weight and gonadosomatic index are important indicators that reflect the degree of reproductive ability and gonadal development of fish and are useful to the mechanism research of reproductive biology and breeding. The GWASs on GW and GSI traits in this study showed that 22 candidate genes associated with sex-related traits were identified on chromosomes 5, 6, 10, 12, 13, 19, and 21. Moreover, sex as a covariate and unisex analysis were performed to avoid sex differences affecting the GW and GSI GWASs. It is noteworthy that several genomic loci and genes were identified simultaneously in different traits. All genes detected through the GWASs might play important roles in the gonadal development and differentiation of P. leopardus. a2m (alpha-2-macroglobulin) has been shown to be the regulatory gene in the development of the oviduct, which is stimulated in response to estrogen in chickens (Gallus gallus) (Lim et al., 2011; Lim and Song, 2014). Usf1 (upstream stimulatory factor 1) is critical for the maintenance of germline stem cells in male mice (Mus musculus) (Faisal et al., 2019) and the control of gene expression during the onset of rat Sertoli cell differentiation (Wood et al., 2011). hsd3b7 (3 beta-hydroxysteroid dehydrogenase type 7) was suggested to be involved in the estrogen-synthetic pathway in the zebra finch and considered primarily as a bile-synthesizing enzyme in mammals (London and Clayton, 2010). Furthermore, significant upregulation of hsd3b7 in female clownfish (Amphiprion clarkii) provided obvious evidence for the importance of steroidogenesis in ovarian development and maintenance (Wang et al., 2022). dll4 (delta-like protein 4) is a canonical Notch ligand in the Notch signaling pathway involved in mammalian sex determination and gonad development (Windley and Wilhelm, 2015; Whiteley et al., 2021). To investigate the effect of xbp1 (x-box-binding protein 1) on steroidogenesis, knockdown of xbp1 by RNAi was conducted in mouse granulosa cells and showed that xbp1 depletion significantly affected the concentrations of estradiol (Wang et al., 2017). tet3 (methylcytosine dioxygenase TET3) is involved in primordial germ-cell development, and interruption of the tet3 gene could reduce germ cell numbers in zebrafish (Wang et al., 2021). Interestingly, primordial germ cells undergo significant DNA methylation reprogramming that is similar to the reprogramming during the adult zebrafish female-to-male sex transition (Wang et al., 2021). esr1 (estrogen receptor alpha) is dispensable for sexual development and reproduction in medaka (Oryzias latipes) (Tohyama et al., 2017), and it can also modulate estrogen-induced sex reversal in the American alligator (Alligator mississippiensis) (Kohno et al., 2015). gli3 (zinc finger protein GLI3) plays a key role in regulation of dmrt3 (doublesex and Mab-3 related transcription factor 3), which was related to male sexual development in mice (Mus musculus) (Bellefroid et al., 2013). In addition, gli3, which resides at the intersection of hedgehog and androgen action, has been considered to be associated with male sex differentiation (Kothandapani et al., 2020). In our study, these associated genes were suggested to play an important role in sex differentiation and gonadal development.
5 Conclusion
Identification of reliable associated genomic and potentially interesting candidate genes affecting the sex differentiation and gonadal development process are important for parent selection and controlled breeding in P. leopardus. In the present GWAS analysis, multiple genomic loci and genes were identified as being significantly associated with the sex differentiation and gonadal development traits (including SDIF, GW, GSI, GW-CoSex, GSI-CoSex, FGW, MGW, FGSI, and MGSI) of P. leopardus. By comprehensively considering the associated peaks, GO annotation, KEGG pathway enrichment analyses, and biological information of genes, several candidate genes, such as taf7, ddx6, apoeb, sgk1, a2m, usf1, hsd3b7, dll4, xbp1, tet3, esr1, and gli3, provided insight into the investigation of not only the genetic mechanism of sex differentiation and gonadal development but also the sex control technologies in P. leopardus.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by the Laboratory Animal Ethics Committee at Hainan Academy of Ocean and Fisheries Sciences (EAEC-HAOFS-No.2022002).
Author contributions
JG and SF conceived and designed the experiments. JG, YW, JL, YG, FC, and XW collected the experimental fish and measured the phenotype. JG, YW, JL, and SF performed the experiments. JG analyzed the data. JG and YW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Major Science and Technology Plan of Hainan Province (ZDKJ2021017), the Hainan Provincial Departmental Budget Project (KLY-2022-08), the Hainan Provincial Joint Project of Sanya Yazhou Bay Science and Technology City (320LH030), and the China Agriculture Research System (CARS-47-Z18).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1229242/full#supplementary-material
References
Adams, S., Mapstone, B. D., Russ, G. R., and Davies, C. R. (2000). Geographic variation in the sex ratio, sex specific size, and age structure of Plectropomus leopardus (Serranidae) between reefs open and closed to fishing on the Great Barrier Reef. Can. J. Fish. Aquatic Sci. 57, 1448–1458. doi:10.1139/f00-076
Adams, S. (2003). Morphological ontogeny of the gonad of three plectropomid species through sex differentiation and transition. J. Fish. Biol. 63, 22–36. doi:10.1046/j.1095-8649.2003.00098.x
Adolfi, M. C., Carreira, A. C., Jesus, L. W., Bogerd, J., Funes, R. M., Schartl, M., et al. (2015). Molecular cloning and expression analysis of dmrt1 and sox9 during gonad development and male reproductive cycle in the lambari fish, Astyanax altiparanae. Reprod. Biol. Endocrinol. 13, 2. doi:10.1186/1477-7827-13-2
Baroiller, J. F., D’Cotta, H., and Saillant, E. (2009). Environmental effects on fish sex determination and differentiation. Sex. Dev. 3, 118–135. doi:10.1159/000223077
Bellefroid, E. J., Leclère, L., Saulnier, A., Keruzore, M., Sirakov, M., Vervoort, M., et al. (2013). Expanding roles for the evolutionarily conserved Dmrt sex transcriptional regulators during embryogenesis. Cell Mol. Life Sci. 70, 3829–3845. doi:10.1007/s00018-013-1288-2
Browning, B. L., Tian, X., Zhou, Y., Browning, S. R., et al. (2021). Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 108, 1880–1890. doi:10.1016/j.ajhg.2021.08.005
Chen, S. F., Zhou, Y. Q., Chen, Y. R., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi:10.1093/bioinformatics/bty560
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi:10.1093/bioinformatics/btr330
Devlin, R. H., and Nagahama, Y. (2002). Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208, 191–364. doi:10.1016/S0044-8486(02)00057-1
Ding, L., Zhang, L., Biswas, S., Schugar, R. C., Brown, J. M., Byzova, T., et al. (2017). Akt3 inhibits adipogenesis and protects from diet-induced obesity via WNK1/SGK1 signaling. JCI Insight 2, e95687. doi:10.1172/jci.insight.95687
Faisal, I., Cisneros-Montalvo, S., Hamer, G., Tuominen, M. M., Laurila, P. P., Tumiati, M., et al. (2019). Transcription factor USF1 is required for maintenance of germline stem cells in male mice. Endocrinology 160, 1119–1136. doi:10.1210/en.2018-01088
Fan, M., Yang, W., Zhang, W., and Zhang, L. (2022). The ontogenic gonadal transcriptomes provide insights into sex change in the ricefield eel Monopterus albus. BMC Zool. 7, 56. doi:10.1186/s40850-022-00155-4
Fraslin, C., Phocas, F., Bestin, A., Charles, M., Bernard, M., Krieg, F., et al. (2020). Genetic determinism of spontaneous masculinisation in XX female rainbow trout: new insights using medium throughput genotyping and whole-genome sequencing. Sci. Rep. 10, 17693. doi:10.1038/s41598-020-74757-8
Gabián, M., Morán, P., Fernández, A. I., Villanueva, B., Chtioui, A., Kent, M. P., et al. (2019). Identification of genomic regions regulating sex determination in Atlantic salmon using high density SNP data. BMC Genomics 20, 764. doi:10.1186/s12864-019-6104-4
Gao, Y., Dong, L., Xu, S., Xiao, S., Fang, M., Wang, Z., et al. (2018). Role of alternative splicing in hematopoietic stem cells during development. Aquaculture 486, 26–30. doi:10.21037/sci.2018.08.02
Gutierrez, A. P., Yáñez, J. M., Fukui, S., Swift, B., and Davidson, W. S. (2015). Genome-wide association study (GWAS) for growth rate and age at sexual maturation in atlantic salmon (Salmo salar). PLoS ONE 10, e0119730. doi:10.1371/journal.pone.0119730
Jakob, S., and Lovell-Badge, R. (2011). Sex determination and the control of Sox9 expression in mammals. FEBS J. 278, 1002–1009. doi:10.1111/j.1742-4658.2011.08029.x
Jiang, L., and Li, H. (2017). Single locus maintains large variation of sex reversal in half-smooth tongue sole (Cynoglossus semilaevis). G3 (Bethesda) 7, 583–589. doi:10.1534/g3.116.036822
Kohno, S., Bernhard, M. C., Katsu, Y., Zhu, J., Bryan, T. A., Doheny, B. M., et al. (2015). Estrogen receptor 1 (ESR1; ERα), not ESR2 (ERβ), modulates estrogen-induced sex reversal in the American Alligator, a species with temperature-dependent sex determination. Endocrinology 156, 1887–1899. doi:10.1210/en.2014-1852
Kothandapani, A., Lewis, S. R., Noel, J. L., Zacharski, A., Krellwitz, K., Baines, A., et al. (2020). GLI3 resides at the intersection of hedgehog and androgen action to promote male sex differentiation. PLoS Genet. 16, e1008810. doi:10.1371/journal.pgen.1008810
Kurakazu, I., Akasaki, Y., Hayashida, M., Tsushima, H., Goto, N., Sueishi, T., et al. (2019). FOXO1 transcription factor regulates chondrogenic differentiation through transforming growth factor β1 signaling. J. Biol. Chem. 294, 17555–17569. doi:10.1074/jbc.RA119.009409
Li, Q., Wang, Y., Xie, J., Sun, W., Zhu, M., He, L., et al. (2015). Characterization and expression of DDX6 during gametogenesis in the Chinese mitten crab Eriocheir sinensis. Genet. Mol. Res. 14, 4420–4437. doi:10.4238/2015.April.30.15
Li, Y., Xing, T., and Liu, J. (2020). Genome-wide association analyses based on whole-genome sequencing of Protosalanx hyalocranius provide insights into sex determination of Salangid fishes. Mol. Ecol. Resour. 20, 1038–1049. doi:10.1111/1755-0998.13172
Li, Y., Zheng, M., and Lau, Y. C. (2014). The sex-determining factors SRY and SOX9 regulate similar target genes and promote testis cord formation during testicular differentiation. Cell Rep. 8, 723–733. doi:10.1016/j.celrep.2014.06.055
Lim, W., Jeong, W., Kim, J., Lee, J., Kim, J., Bazer, F. W., et al. (2011). Differential expression of alpha 2 macroglobulin in response to dietylstilbestrol and in ovarian carcinomas in chickens. Reprod. Biol. Endocrinol. 9, 137. doi:10.1186/1477-7827-9-137
Lim, W., and Song, G. (2014). Identification of novel regulatory genes in development of the avian reproductive tracts. PLoS ONE 9, e96175. doi:10.1371/journal.pone.0096175
Lin, H., Zhou, Z., Zhao, J., Zhou, T., Bai, H., Ke, Q., et al. (2021). Genome-wide association study identifies genomic loci of sex determination and gonadosomatic index traits in large yellow croaker (Larimichthys crocea). Mar. Biotechnol. 23, 127–139. doi:10.1007/s10126-020-10007-2
London, S. E., and Clayton, D. F. (2010). Genomic and neural analysis of the estradiol-synthetic pathway in the zebra finch. BMC Neurosci. 11, 46. doi:10.1186/1471-2202-11-46
Martínez, P., Robledo, D., Taboada, X., Blanco, A., Moser, M., Maroso, F., et al. (2021). A genome-wide association study, supported by a new chromosome-level genome assembly, suggests sox2 as a main driver of the undifferentiatiated ZZ/ZW sex determination of turbot (Scophthalmus maximus). Genomics 113, 1705–1718. doi:10.1016/j.ygeno.2021.04.007
Mckenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi:10.1101/gr.107524.110
Mustapha, U. F., Huang, Y., Huang, Y., Assan, D., Shi, H., Jiang, M., et al. (2021). Gonadal development and molecular analysis revealed the critical window for sex differentiation, and E2 reversibility of XY-male spotted scat, Scatophagus argus. Aquaculture 544, 737147. doi:10.1016/j.aquaculture.2021.737147
Nakamura, Y., Higuchi, K., Kumon, K., Yasuike, M., Takashi, T., Gen, K., et al. (2021). Prediction of the sex-associated genomic region in tunas (Thunnus fishes). Int. J. Genomics 2021, 7226353. doi:10.1155/2021/7226353
Nguyen, D. H. M., Ponjarat, J., Laopichienpong, N., Kraichak, E., Panthum, T., Singchat, W., et al. (2021). Genome-wide SNP analysis suggests male heterogamety in bighead catfish (Clarias macrocephalus, Günther, 1864). Aquaculture 543, 737005. doi:10.1016/j.aquaculture.2021.737005
Pal, S., Paladhi, P., Dutta, S., Bose, G., Ghosh, P., Chattopadhyay, R., et al. (2021). Novel variations in spermatogenic transcription regulators RFX2 and TAF7 increase risk of azoospermia. J. Assist. Reprod. Genet. 38, 3195–3212. doi:10.1007/s10815-021-02352-5
Patil, S. S., and Nayak, M. S. (2021). Gonadal sex differentiation in the fish, Oreochromis mossambicus. Discovery 57, 632–639.
Piferrer, F., and Guiguen, Y. (2008). Fish gonadogenesis. Part II: molecular biology and genomics of sex differentiation. Rev. Fish. Sci. 16, 35–55. doi:10.1080/10641260802324644
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi:10.1086/519795
Rajendiran, P., Jaafar, F., Kar, S., Sudhakumari, C., Senthilkumaran, B., and Parhar, I. S. (2021). Sex determination and differentiation in teleost: roles of genetics, environment, and brain. Biology 10, 973. doi:10.3390/biology10100973
Sá, R., Miranda, C., Carvalho, F., Barros, A., and Sousa, M. (2013). Expression of stem cell markers: OCT4, KIT, ITGA6, and ITGB1 in the male germinal epithelium. Syst. Biol. Reprod. Med. 59, 233–243. doi:10.3109/19396368.2013.804964
Sambrook, J., and Russell, D. W. (2006). Purification of nucleic acids by extraction with phenol: chloroform. CSH Protoc. 1, pdb.prot4455. pdb.prot4455. doi:10.1101/pdb.prot4455
Stévant, I., and Nef, S. (2019). Genetic control of gonadal sex determination and development. Trends Genet. 35, 346–358. doi:10.1016/j.tig.2019.02.004
Takashima, F., Patino, R., and Nomura, M. (1980). Histological studies on the sex differentiation in Rainbow Trout. Nippon. Suisan Gakkaishi 46, 1317–1322. doi:10.2331/SUISAN.46.1317
Talukder, Z. I., Ma, G., Hulke, B. S., Jan, C., and Qi, L. (2019). Linkage mapping and Genome-wide association studies of the Rf gene cluster in Sunflower (Helianthus annuus L.) and their distribution in world Sunflower collections. Front. Genet. 10, 216. doi:10.3389/fgene.2019.00216
Tao, W., Chen, J., Tan, D., Yang, J., Sun, L., Wei, J., et al. (2018). Transcriptome display during tilapia sex determination and differentiation as revealed by RNA-Seq analysis. BMC Genom. 19, 363. doi:10.1186/s12864-018-4756-0
Tohyama, S., Ogino, Y., Lange, A., Myosho, T., Kobayashi, T., Hirano, Y., et al. (2017). Establishment of estrogen receptor 1 (ESR1)-knockout medaka: ESR1 is dispensable for sexual development and reproduction in medaka, Oryzias latipes. Oryzias Latipes. Dev. Growth Differ. 59, 552–561. doi:10.1111/dgd.12386
Uffelmann, E., Huang, Q., Munung, N. S., Vries, J. D., Okada, Y., Martin, A. R., et al. (2021). Genome-wide association studies. Nat. Rev. Methods Prim. 59, 59–20. doi:10.1038/s43586-021-00056-9
Vizziano-Cantonnet, D., Lasalle, A., Landro, S. D., Klopp, C., and Genthon, C. (2018). De novo transcriptome analysis to search for sex-differentiation genes in the Siberian sturgeon. Gen. Comp. Endocrinol. 268, 96–109. doi:10.1016/j.ygcen.2018.08.007
Wang, H., Qu, M., Tang, W., Liu, S., and Ding, S. (2022). Transcriptome profiling and expression localization of key sex-related genes in a socially-controlled hermaphroditic clownfish, Amphiprion clarkia. Int. J. Mol. Sci. 23, 9085. doi:10.3390/ijms23169085
Wang, M., Li, L., Lin, H., Zhou, Z., Liu, B., Zhong, J., et al. (2022). Genome-wide association study identifies genomic loci of sex determination, gonadal weight and gonadosomatic index traits in Takifugu bimaculatus. Aquaculture 546, 737389. doi:10.1016/j.aquaculture.2021.737389
Wang, N., Zhao, F., Lin, P. F., Zhang, G., Tang, K., Wang, A., et al. (2017). Knockdown of XBP1 by RNAi in mouse granulosa cells promotes apoptosis, inhibits cell cycle, and decreases estradiol synthesis. Int. J. Mol. Sci. 18, 1152. doi:10.3390/ijms18061152
Wang, Q., Liu, Y., Wang, Y., Jiang, S., Zhang, C., and Li, B. (2022). GWAS reveal novel sex-related markers and candidate genes in Sea urchin Mesocentrotus nudus. Mar. Biotechnol. 24, 32–39. doi:10.1007/s10126-021-10084-x
Wang, X., Ma, X., Wei, G., Ma, W., Zhang, Z., Chen, X., et al. (2021). The role of DNA methylation reprogramming during sex determination and transition in zebrafish. Genomics Proteomics Bioinforma. 19, 48–63. doi:10.1016/j.gpb.2020.10.004
Whiteley, S. L., Holleley, C. E., Wagner, S., Blackburn, J., Deveson, I. W., Marshall Graves, J. A., et al. (2021). Two transcriptionally distinct pathways drive female development in a reptile with both genetic and temperature dependent sex determination. PLoS Genet. 17, e1009465. doi:10.1371/journal.pgen.1009465
Windley, S. P., and Wilhelm, D. (2015). Signaling pathways involved in mammalian sex determination and gonad development. Sex. Dev. 9, 297–315. doi:10.1159/000444065
Wood, M. A., Mukherjee, P., Toocheck, C. A., and Walker, W. H. (2011). Upstream stimulatory factor induces Nr5a1 and shbg gene expression during the onset of rat sertoli cell differentiation. Biol. Reprod. 85, 965–976. doi:10.1095/biolreprod.111.093013
Xu, W., Cui, Z., Wang, N., Zhang, M., Wang, J., Xu, X., et al. (2021). Transcriptomic analysis revealed gene expression profiles during the sex differentiation of Chinese tongue sole (Cynoglossus semilaevis). Comp. Biochem. 40, 100919. doi:10.1016/j.cbd.2021.100919
Zeng, Q. F., Liu, S. K., Yao, J., Zhang, Y., Yuan, Z., Jiang, C., et al. (2016). Transcriptome display during testicular differentiation of Channel Catfish (Ictalurus punctatus) as revealed by RNA-Seq analysis. Biol. Reprod. 95, 19. doi:10.1095/biolreprod.116.138818
Zhai, G., Jia, Y., Bereketoglu, C., Yin, Z., and Pradhan, A. (2022). Sex-specific differences in zebrafish brains. Biol. Sex. Differ. 13, 31. doi:10.1186/s13293-022-00442-2
Zhai, G., Shu, T., Xia, Y., Lu, Y., Shang, G., Jin, X., et al. (2018). Characterization of sexual trait development in cyp17a1-deficient Zebrafish. Endocrinology 159, 3549–3562. doi:10.1210/en.2018-00551
Zhang, B., Zhao, N., Peng, K., He, X., Chen, C., Liu, H., et al. (2020). A combination of genome-wide association study screening and SNaPshot for detecting sex-related SNPs and genes in Cynoglossus semilaevis. Comp. Biochem. Physiol. Part D. Genomics Proteomics 35, 100711. doi:10.1016/j.cbd.2020.100711
Zhou, H., Grubisic, I., Zheng, K., He, Y., Wang, P. J., Kaplan, T., et al. (2013). Taf7l cooperates with Trf2 to regulate spermiogenesis. Proc. Natl. Acad. Sci. U. S. A. 110, 16886–16891. doi:10.1073/pnas.1317034110
Keywords: Plectropomus leopardus, genome-wide association study, sex differentiation, gonadal development, genomic loci, candidate genes
Citation: Gao J, Wang Y, Liu J, Chen F, Guo Y, Ke H, Wang X, Luo M and Fu S (2023) Genome-wide association study reveals genomic loci of sex differentiation and gonadal development in Plectropomus leopardus. Front. Genet. 14:1229242. doi: 10.3389/fgene.2023.1229242
Received: 12 June 2023; Accepted: 17 July 2023;
Published: 14 August 2023.
Edited by:
Jie Mei, Huazhong Agricultural University, ChinaReviewed by:
Zhongdian Dong, Guangdong Ocean University, ChinaZhenhua Ma, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2023 Gao, Wang, Liu, Chen, Guo, Ke, Wang, Luo and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuyuan Fu, ZnVzaHV5dWFuQGhuaGt5LmNu
†These authors have contributed equally to this work
 Jin Gao
Jin Gao Yongbo Wang
Yongbo Wang Jinye Liu1,3
Jinye Liu1,3