- 1Department of Pediatrics 3, George Emil Palade University of Medicine, Pharmacy, Sciences and Technology of Târgu Mureș, Târgu Mureș, Romania
- 2Genetics Department, Center for Advanced Medical and Pharmaceutical Research, George Emil Palade University of Medicine, Pharmacy, Science and Technology of Târgu Mureș, Targu Mures, Romania
MiRNAs are short, non-coding RNA molecules, which are involved in the regulation of gene expression and which play an important role in various biological processes, including inflammation and cell cycle regulation. The possibility of detecting their extracellular expression, within body fluids, represented the main background for their potential use as non-invasive biomarkers of various diseases. Salivary miRNAs particularly gained interest recently due to the facile collection of stimulated/unstimulated saliva and their stability among healthy subjects. Furthermore, miRNAs seem to represent biomarker candidates of gastrointestinal disorders, with miRNA-based therapeutics showing great potential in those conditions. This review aimed to highlight available evidence on the role of salivary miRNAs in different gastrointestinal conditions. Most salivary-based miRNA studies available in the literature that focused on pathologies of the gastrointestinal tract have so far been conducted on pancreatic cancer patients and delivered reliable results. A few studies also showed the diagnostic utility of salivary miRNAs in conditions such as esophagitis, esophageal cancer, colorectal cancer, or inflammatory bowel disease. Moreover, several authors showed that salivary miRNAs may confidently be used as biomarkers of gastric cancer, but the use of salivary miRNA candidates in gastric inflammation and pre-malignant lesions, essential stages of Correa’s cascade, is still put into question. On the other hand, besides miRNAs, other salivary omics have shown biomarker potential in gastro-intestinal conditions. The limited available data suggest that salivary miRNAs may represent reliable biomarker candidates for gastrointestinal conditions. However, their diagnostic potential requires validation through future research, performed on larger cohorts.
1 Introduction
MiRNAs represent short, non-coding RNA molecules that are able to regulate gene expression at post-transcriptional level, by targeting the 3′ untranslated region (3′-UTR) of messenger RNAs (mRNAs) (O’Brien et al., 2018; Dexheimer and Cochella, 2020). Specific, stable miRNA interactions with 5′-UTR have also been reported (Broughton et al., 2016). It is important though to acknowledge that miRNAs have multiple mRNA targets, detaining multiple binding sites and thus regulating gene expression in a complex manner (Bartel, 2009; Friedman et al., 2009). Most miRNAs are involved in gene expression suppression, also known as “gene silencing,” which is mediated by the interaction between the miRNA-induced silencing complex (miRISC) and the ribonucleoprotein (RNP) effector (Makarova et al., 2016). Given that miRISC-RNP interaction has been discovered in several cellular compartments, questions were raised about their function extension beyond gene expression modulation (Makarova et al., 2016). At the same time, miRISC-RNP complexes also seem to be able to stimulate gene expression at posttranscriptional levels, with miRNA-mediated effects being highly dependent upon factors such as cellular conditions, RNA sequence, or complementarity degree (Vasudevan, 2012). Thus, miRNAs represent fine tuners of gene expression, as schematically represented in Figure 1A.
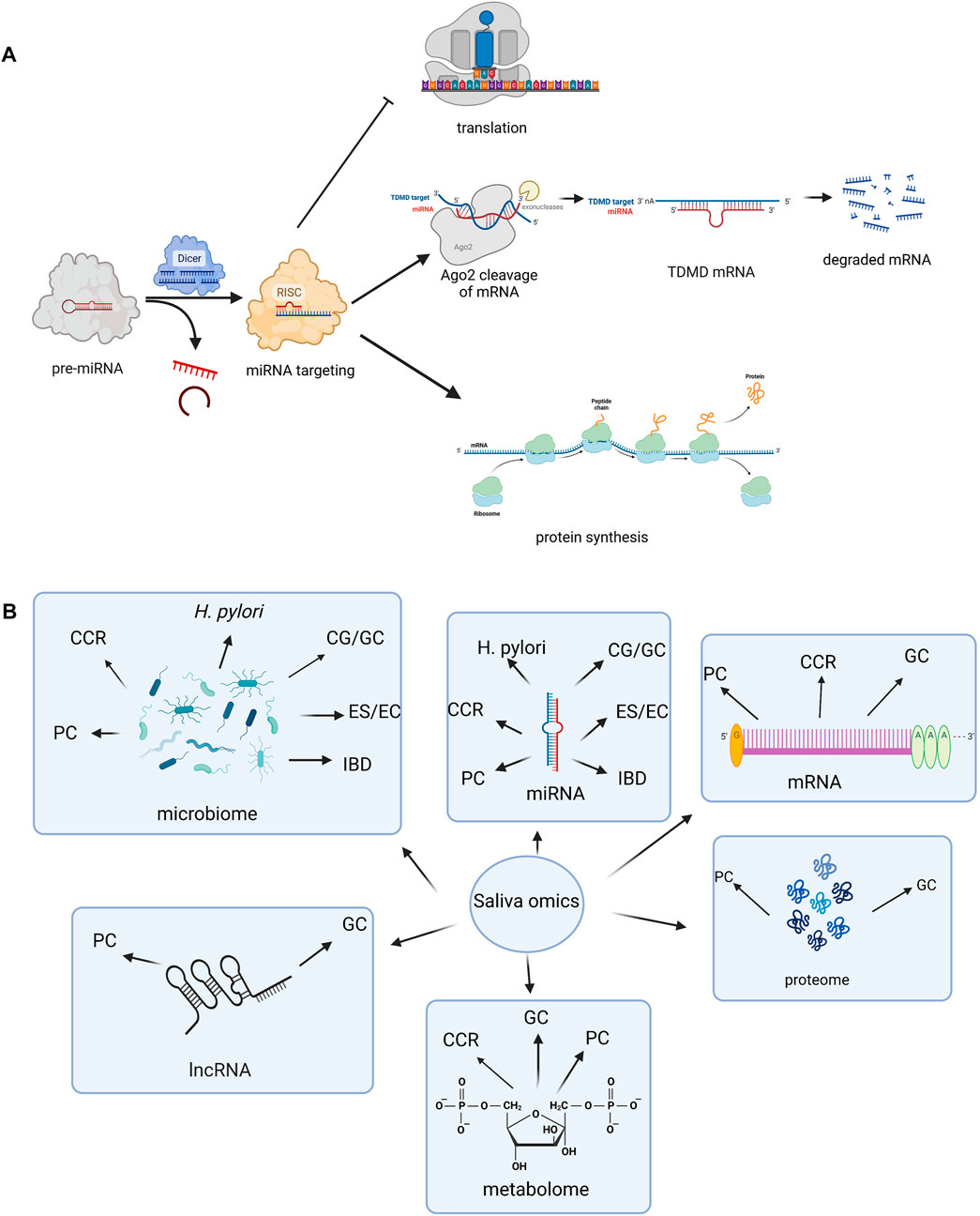
FIGURE 1. (A) Schematic representation of miRNA function (Created with BioRender.com). miRNAs take part in gene expression suppression, which is achieved through translation repression and/or mRNA degradation. Dicer (ribonuclease or helicase with RNase motif) is involved in processing of pre-miRNA (miRNA precursors) into mature miRNA. The main mediator in these processes is represented by the miRNA-induced silencing complex (miRISC) and the ribonucleoprotein (RNP) effector, and later its Ago2 ligand (an important component of RISC that facilitates binding of miRNA). TDMD represents a gene regulation mechanism by which the base-pairing between the miRNA and target RNA is achieved and afterwards their degradation is induced. miRISC-RNP interaction is also able to stimulate gene expression at a posttranscriptional level. Legend of figure: Ago2- Argonaut 2. RISC- RNA-induced silencing complex. TDMD- Target-directed microRNA (miRNA) degradation. (B) Salivary biomarkers of gastrointestinal disorders (Created with BioRender.com). Saliva omics are gaining more and more popularity due to their biomarker potential in gastro-intestinal disorders. Besides miRNAs, a few studies also sustain the diagnostic potential of other non-coding RNAs, such as mRNAs and lncRNAs. The prospective of salivary microbiome studies has been highlighted in a variety of gastro-intestinal disorders. Other omics, such as metabolomics and proteome are still scarcely studied, but might represent future non-invasive markers as well. Legend of figure: CCR-colorectal carcinoma. CG-chronic gastritis. EC- esophageal cancer. ES- esophagitis. GC- gastric cancer. H pylori- Helicobacter pylori. IBD-inflammatory bowel disease. lncRNA-long non-coding RNA. mRNA-messenger RNA. PC- pancreatic cancer.
MiRNAs are involved in various biological processes, including hematopoietic cell differentiation, cell growth, or epigenetic regulation of malignant transformation-associated pathways (Chen et al., 2004; Wang and Sen, 2011; Wang et al., 2016). Furthermore, miRNAs seem to substantially contribute to normal developmental processes, by modulating trophoblast cell proliferation and apoptosis. Thus, they function as part of the placental development, neuron, astrocyte, muscle cell development, or bone formation (Fu et al., 2013; Ivey and Srivastava, 2015). Their essential role in embryogenesis, cell development, hemostasis and differentiation has also been reported by several studies conducted on different animal species (He et al., 2022; Khanal et al., 2022; Luo et al., 2022). Given their involvement in numerous physiological processes, it is unsurprising that miRNAs and their aberrant expression have been regarded as potential diagnostic biomarkers in a variety of diseases (Hayes et al., 2014; Tüfekci et al., 2014; Huang, 2017). The quest for miRNA signatures is still ongoing, but multiple miRNAs might be validated as future biomarkers in malignant pathologies, viral infections, cardiovascular, renal or neurological disorders (Wang et al., 2016). The possibility of depicting stable, extracellular miRNAs in body fluids such as urine, blood or saliva, and their aberrant expression, which is sometimes similar to tissular analogues, gives the upperhand of potential non-invasiveness to the biomarker status (Huang, 2017; Fellizar et al., 2022).
2 Salivary miRNAs
As in all body fluids, circulating miRNAs are found in saliva in lipid/lipoprotein vesicular complexes such as exosomes (Leal-Galvan et al., 2022). Exosomes protect miRNA from degradation, ensuring adequate function of the transported molecular information (Théry et al., 2002; Théry et al., 2009). This translates into stability, circulating, exosomal plasma miRNAs being regarded as highly stable in healthy subjects (Sanz-Rubio et al., 2018). In similar fashion, exosomal salivary miRNAs seem to be easily isolated and quantified within the healthy population, constituting promising, easily available biomarkers (Michael et al., 2010). Some theories suggest that miRNAs could also be produced locally in oral fluids, arousing from apoptosis and cell necrosis. A theory based on the possible existence of transcellular and paracellular transport routes into salivary gland tissues has been issued (Yoshizawa et al., 2013). Moreover, salivary endogenous miRNAs seem to degrade at a slower rate than their exogenous homologues, according to a study which aimed to discover and validate miRNAs in the saliva of healthy subjects (Park et al., 2009).
Saliva collection, although characterized by non-invasiveness, is being subject to various debates. Type of saliva collection has been miscellaneous so far in various reports. Some authors have proposed collection of unstimulated saliva samples, while others have successfully isolated miRNA molecules from saliva collected after citric acid stimulation (Yoshizawa and Wong, 2013; Xie et al., 2015). Saliva composition seems to be influenced by various factors, including meal schedule, diet, time of the day, age or sex (Koopaie et al., 2021). Time of collection and amount of fasting time required prior to collection is still debated and varies from one study protocol to another. Furthermore, administration of agents such as citric acid for stimulation of saliva secretion, prior to its collection, still remains at the liberty of researchers (Setti et al., 2020). Saliva storage tubes vary in volume, with some subjects reporting difficulty in collecting saliva in smaller sized tubes (Urbizu et al., 2023). Isolation can be achieved both from whole saliva and supernatant, obtained after centrifugation, with identification of high resolution miRNA information (Patel et al., 2011). Centrifugation seems to be helpful in case of viscous saliva samples, but the performance rate should be adapted accordingly, in order to avoid mechanical rupture of cellular components (St John et al., 2004).
Due to their non-invasiveness, salivary miRNAs have emerged as promising biomarkers in the diagnosis and prognosis of various conditions, including neoplastic diseases, such as oral cancer, hepatic cancer, breast cancer or pancreatic cancer (Xie et al., 2015; Koopaie et al., 2021; Mariam et al., 2022; Scholtz et al., 2022), as well as inflammatory conditions, including endometriosis, inflammatory bowel disease, periodontitis or eosinophilic esophagitis (Schaefer et al., 2015; Bhardwaj et al., 2020; Lee et al., 2020; Bendifallah et al., 2022). Furthermore, salivary miRNAs seem to present altered expressions in relation to various types of injuries (Hicks et al., 2021; Hicks et al., 2022). Still, the question of similitude between the expression of miRNAs depicted in the saliva and their tissular or circulating homologues is raised. A few studies have tried to simultaneously assess the biomarker role of the same miRNA type in both saliva and blood or saliva and tissue (Nik Mohamed Kamal et al., 2020; Sembler-Møller et al., 2020). A clear view of the reliability of salivary miRNAs in comparison with miRNAs identified among tissue or other body fluids is still subject to future research.
3 Salivary miRNAs and gastrointestinal conditions
The quest for biomarkers in gastrointestinal disorders has gained more attention recently and research on the subject bloomed after miRNA-based therapeutics showed a great potential in treatment of various diseases of the gastrointestinal tract (Hossian et al., 2019). Most of the studies conducted so far have focused on the role of miRNA in the diagnosis of gastric cancer, as miRNA expression profile seems to be altered in different stages of Correa’s gastric precancerous cascade (Zheng et al., 2011; Deng et al., 2013; Sugiyama et al., 2016; Lario et al., 2018). However, miRNAs have been usually identified from gastric cellular lines, gastric or esophageal mucosa specimens or depicted in the serum or plasma (Matsushima et al., 2011; Sreedharan et al., 2017; Lario et al., 2018; Ohtsuka et al., 2021; Vasapolli et al., 2021). Studies analyzing the role of salivary miRNAs in gastrointestinal conditions are so far limited in number, but those available so far have shown promising results in terms of delivering non-invasive biomarkers. Therefore, this review aims to highlight available evidence on the role of salivary miRNAs in different gastrointestinal conditions.
3.1 Salivary miRNAs, esophagitis and esophageal cancer
Salivary miRNAs distinguished themselves as potential markers of eosinophilic esophagitis. In one pediatric study, in which 56 salivary miRNAs were detected, six different miRNAs (miR-26b-5p, miR-27b-3p, Let-7i-5p, miR-142-5p, miR-30a-5p and miR-205-5p) presented upregulation, quantified through variable importance projection (VIP) scores in subjects with eosinophilic esophagitis. The highest differences between the two study groups were obtained for miR-205-5p, but good sensitivity and specificity parameters were reported for each of the 6 miRNAs (Jhaveri et al., 2023). A similar outcome was described within an adult study, in which miR-4668 emerged as a biomarker candidate that can identify subjects with eosinophilic esophagitis and is correlated with the number of aeroallergens. According to the authors, the same miR-4668 can be used as a response predictor to topical corticosteroids, which suggests that this miRNA could be used as a marker of other allergic disorders as well (Bhardwaj et al., 2020).
More attention has been so far given to the role of miRNAs in the diagnosis and characterization of esophageal cancer. Saliva identifiable miRNAs such as miR-144, miR-451, miR-98, miR-10b and miR-363 might be able to modulate target genes involved in esophageal cancer, according to Du and Zhang (2017). Hoshino et al. describe the high diagnostic performance of miR-1246 in esophageal squamous cell carcinoma, when determined from blood, urine and saliva. Still, variations in salivary miR-1246 levels were found, in relation to saliva collection timing, and its expression differed among the three body fluid samples (Hoshino et al., 2021). On the other hand, plasmatic and salivary miR-21 expressions presented a positive, significant correlation, and comparable diagnostic value in esophageal cancer (Ye et al., 2014). Moreover, high expression of miR-144 was identified in the whole saliva and saliva supernatant of patients with esophageal cancer. Thus, Wu et al. (2013) proposed miR-144 as an early marker of esophageal cancer, with moderate diagnostic performance indices, based on sensitivity and specificity parameters. miRNA identification and expression might though differ between whole saliva and saliva supernatant samples. Hence, Xie et al. found that in subjects with esophageal cancer, three different miRNAs (miR-10b*, miR-144 and miR-451) were upregulated in the whole saliva samples, whereas four of the six identified miRNAs were upregulated (miR-10b*, miR-144, miR-21 and miR-451) within saliva supernatant (Xie et al., 2012; Xie et al., 2013). miR-196a has also been regarded as a suitable salivary biomarker for esophageal squamous cell carcinoma, as its salivary upregulation correlated with the expression augmentation seen in biopsy samples taken from neoplastic tissue, as opposed to the healthy adjacent mucosa (Fendereski et al., 2017).
Given the paucity of studies analyzing the role of salivary miRNAs in the diagnosis of esophageal cancer, and their miscellaneous sensitivity and specificity rates, one meta-analysis was performed and suggested that blood-based miRNAs are more accurate than those identified within saliva samples in detecting esophageal squamous cell carcinoma (Wang et al., 2014). The same conclusion was driven by Li et al., who suggested that both serum and plasma miRNAs are appropriate biomarkers for esophageal cancer (Li et al., 2017). However, Wan et al. claimed that type of body samples from which miRNAs were depicted did not constitute a source of heterogenicity among different miRNA-based studies of esophageal cancer (Wan et al., 2016).
3.2 Salivary miRNAs, Helicobacter pylori infection and gastric cancer
MiRNAs play a pivotal role in the modulation of host immune response and inflammation triggered by Helicobacter pylori (H. pylori) (Săsăran et al., 2021). A few studies have identified an upregulation of tissular and serum miRNAs such as miR-146a, miR-155, miR-127-5p, miR-181 in relation to H. pylori-induced chronic gastritis, while others failed to identify a significant variation from healthy subjects in the expression of miR-155 (Chung et al., 2017; Cortés-Márquez et al., 2018; Zabaglia et al., 2018; Mahboobi et al., 2022; Oana et al., 2022). However, only one study so far assessed the role of salivary miRNAs in aiding the identification of H. pylori infection, which included children with pulpitis. The authors found a downregulation of miR-204 in pulp tissue, serum and saliva of subjects who were concomitantly infected with H. pylori, as opposed to those in whom the bacterial infection was absent. At the same time, expression reduction of miR-204 was correlated with an upregulation of MMP-9, which is known to be influenced by inflammation, possibly caused by both pulpitis and H. pylori infection (Zhou and Xu, 2019). Similarly, downregulation of miR-204, in association with upregulation of MMP-9, was reported in patients with H. pylori-associated gastric ulcer (Li et al., 2016). Given the proven involvement of miRNAs in the regulation of inflammatory pathways related to H. pylori infection, as well as the alteration of miRNA expression in the setting of pre-neoplastic lesions, miRNAs were considered promising biomarkers for early gastric cancer detection, as well as potential therapeutic targets in this malignant setting (Shin and Chu, 2014; Vidal et al., 2016; Khayam et al., 2021). After tissular detection of miRNAs which showed biomarker potential, a search for non-invasive miRNAs, available in accessible body fluids was started (Adami et al., 2019; Zhang et al., 2019; Quirico and Orso, 2020; Lopes et al., 2023). Most of the studies available so far have assessed expression of multiple miRNAs among serum and plasma samples (Lopes et al., 2023). However, only two studies have investigated so far the biomarker potential of salivary miRNAs in gastric cancer individuals. Li et al. discovered and validated a panel of multiple extracellular, salivary miRNAs which were associated with gastric cancer. Among them, a significant downregulation of two miRNA candidates, namely, miR-140-5p and miR-301a, was noted in gastric cancer patients when compared to healthy controls (Li et al., 2018). miRNA panel of two whole saliva- identified miRNAs, miR-140 and miR-301a, might ensure early, non-invasive identification of gastric cancer subjects, according to Kaczor-Urbanowicz et al. However, the authors point out that the performance of these biomarkers, assessed with the help of receiver operating characteristic (ROC) curve, differs among ethnicities (Kaczor-Urbanowicz et al., 2022).
Besides salivary miRNAs, salivary microbiota starts to gain more and more popularity as potential biomarkers of H. pylori infection and gastric cancer. The alteration of the gastric microbiome trigerred by H. pylori impairs the oral-gut axis and leads to specific changes in salivary microbial community structure (Ji et al., 2020; Chen et al., 2022). In addition, an accumulation of pro-inflammatory bacteria has been noted in relation to gastric cancer, with a particular abundance of Neisseria species (Kageyama et al., 2019; Huang et al., 2021). Furthermore, salivary microbiome imbalance has been found in relation to chronic gastric inflammation, even in the absence of H. pylori infection (Contaldo et al., 2021).
3.3 Salivary miRNAs and differential diagnosis of inflammatory bowel disease (IBD)
The overlapping of clinical symptoms between Crohn’s disease and ulcerative colitis makes an accurate IBD diagnosis challenging, in the absence of endoscopic investigations and microscopic assessment of tissue biopsies. Peripheral blood miRNAs were firstly regarded as non-invasive biomarkers which can distinguish the two main IBD types, with 10 miRNAs presenting a significant upregulation and one miRNA an important expression decrease in the blood of active ulcerative colitis patients, when compared to Crohn’s disease patients (Wu et al., 2011). A panel of salivary miRNAs might be useful in the differential diagnosis of Chron’s disease and ulcerative colitis. In the study of Schaefer et al., expression of miR-101 was altered in the saliva of Chron’s disease patients, whereas miR-21, miR-31, miR-142-3p, and miR-142-5p presented particular expression variation patterns among patients with ulcerative colitis (Schaefer et al., 2015). No other studies have managed so far to evaluate differential expression of salivary miRNAs in IBD patients.
3.4 Salivary miRNAs and pancreatic cancer
The search for reliable biomarkers which can identify initial stages of pancreatic cancer has been of crucial importance in the last years, in light of the very low 1-year survival rates that characterize patients with this type of advanced malignancy (Burris et al., 1997). Humeau et al. identified 4 salivary miRNA candidates, namely, miR-21, miR-23a, miR-23b and miR-29c, which were upregulated in pancreatic cancer patients compared to controls, with these miRNAs presenting good sensitivity and excellent specificity. Moreover, miR-23a and miR-23b were already differentially expressed in precursor lesions of pancreatic cancer, while miR-216 was overexpressed in pancreatic cancer subjects when compared to pancreatitis patients (Humeau et al., 2015). Two miRNA candidates, miR-940 and miR-3679-5p, were upregulated and downregulated, respectively, in pancreatic cancer patients when compared to a study group consisting of subjects diagnosed with benign pancreatic tumors and healthy controls. Moreover, these two miRNAs could reliably identify individuals with resectable pancreatic cancer, according to ROC results (Xie et al., 2015). miR-1246 and miR-4644, detected among salivary exosomes, were other two miRNAs which exhibited a significant expression augmentation in pancreatobiliary tract cancer patients when compared to controls. The best area under the curve was calculated for the combined use of the two miRNAs, which suggested their concomitant panel utility for pancreatobiliary tract cancer diagnosis (Machida et al., 2016). On the other hand, another study concluded that whilst miR-1246 is upregulated within the serum and urine of pancreatic cancer patients, its salivary expression in the same individuals did not differ significantly from the one of healthy controls (Ishige et al., 2020).
3.5 Salivary miRNAs and colorectal cancer
MiRNAs constitute key players in the pathogenesis of colorectal adenocarcinoma, through their involvement in the regulation of mRNA expression, with eight different miRNAs which act on their mRNA targets being reported as having a differential expression in this malignant setting (miR-141, miR-19a, miR-20a, miR-19b-1, miR-19b-2, miR-16, miR-590 and miR-335) (Wang et al., 2017). Moreover, miR-21 has been particularly intensely studied in colorectal cancer. Circulating miR-21 seems to be a useful tool in the diagnosis of colorectal cancer, whereas tissular miR-21 emerged as a marker of prognosis in subjects with the same malignant condition (Peng et al., 2017). In both plasma and saliva, miR-21 has managed to accurately identify patients with colorectal cancer. Salivary miR-21 presented even higher sensitivity and specificity than plasma miR-21 in supporting colorectal cancer screening (Sazanov et al., 2017). Furthermore, Rapado-Gonzalez et al. proposed a panel of five salivary miRNAs (miR-186-5p, miR-29a-3p, miR-29c-3p, miR-766-3p, and miR-491-5p) for the non-invasive diagnosis of colorectal cancer, which presented significant upregulation in relation to this malignancy (Rapado-González et al., 2019).
4 Discussions
Salivaomics, which include proteome, transcriptome, metabolome, microbiome, as well as miRNAs, might represent revolutionary non-invasive tools for the diagnosis of multiple disorders, and especially for the early detection of neoplastic conditions (Wong, 2012). Salivary miRNAs first gained attention as biomarkers of oral pathologies, including periodontal disease or pre-neoplastic lesions or malignancies of the oral cavity (Martina et al., 2020; Kapoor et al., 2021; Manzano-Moreno et al., 2021). Non-coding RNAs, including miRNAs, circular RNAs (circRNAs) and long non-coding RNAs (lncRNAs) have been known to be involved in several important hallmarks of gastrointestinal cancers such as esophageal, gastric, pancreatic, colorectal cancer or hepatocellular carcinoma (Dragomir et al., 2020). Still, data on the biomarker utility of salivary miRNAs in conditions of the gastrointestinal tract is scarce, but research is continuously expanding. Igaz et al., who provided an update regarding the recent reported utility of fecal, biliary, salivary and urinary miRNAs, acknowledge the potential of these biomarkers in revolutionizing early diagnosis of malignancies of the gastrointestinal tract, but at the same out point out towards the need of spreading research on larger cohorts and using uniform methodological approaches (Igaz and Igaz, 2015). Through this review, we aimed to highlight current evidence available regarding the use of salivary miRNAs in various gastrointestinal pathologies. An overview of current studies adhering to our review objective, which suggested that miRNAs might serve as biomarker candidates in various types of conditions, has been provided through Table 1. In addition to salivary miRNAs, a synopsis of other, currently available salivary biomarkers in gastro-intestinal conditions has been provided through Figure 1B, based on recent literature data (Zhang et al., 2010; Xiao et al., 2016; Wang et al., 2017; Asai et al., 2018; Bel’skaya et al., 2020; Deutsch et al., 2020; Xu and Jiang, 2020; Huang et al., 2021; Eftekhari et al., 2022; Koopaie et al., 2022; Patel et al., 2022; Lopes et al., 2023).
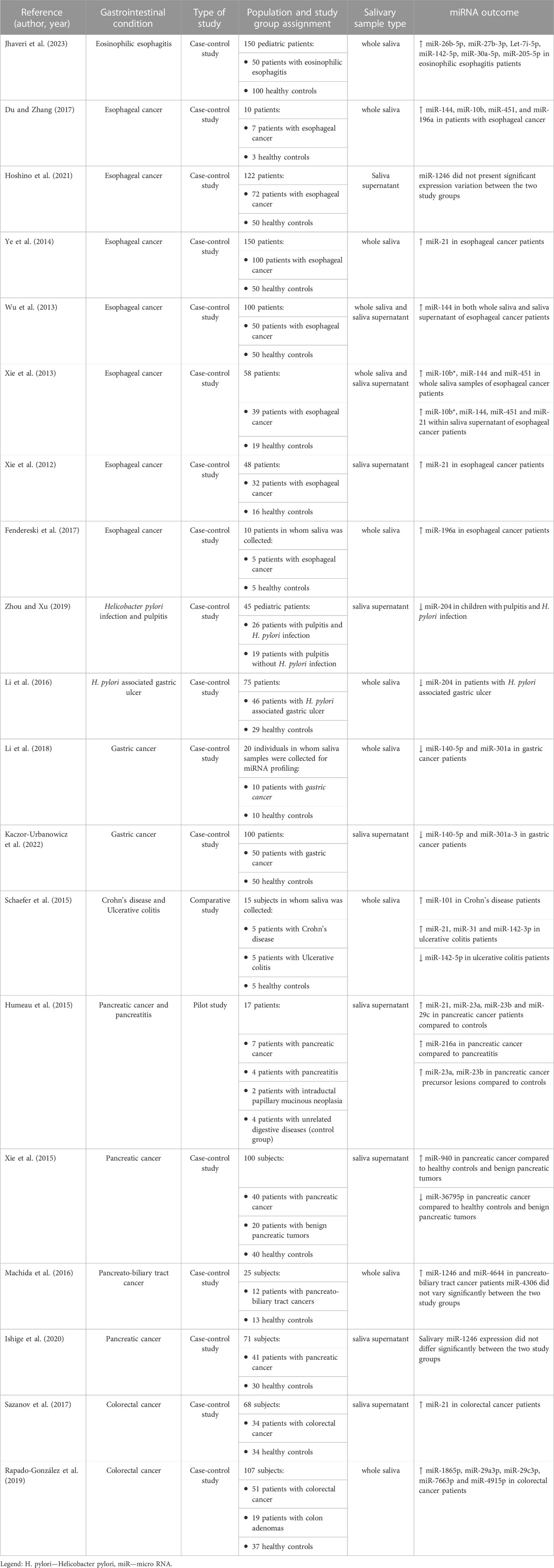
TABLE 1. Synopsis of studies that assessed the role of salivary miRNAs in gastrointestinal disorders.
As presented within the former chapters, most salivary-based miRNA studies available in the literature that focused on pathologies of the gastro-intestinal tract have been conducted on patients with pancreatic cancer. One meta-analysis which included the formerly described studies revealed that salivary miRNAs facilitate diagnosis of pancreatic cancer, with good performance indices such as pooled sensitivity, specificity, positive and negative likelihood ratios and diagnostic odds ratio. Hence, these have been proposed as biomarkers of early pancreatic cancer (Koopaie et al., 2021). The discriminatory power of salivary miRNAs in the detection of esophageal cancer cannot be foreseen, in spite of only a few available studies (Xie et al., 2012; Xie et al., 2013; Wu et al., 2013; Ye et al., 2014; Fendereski et al., 2017). The use of non-blood based liquid biopsies for timely detection of gastric cancer is another hot topic of current research, but the small number of available studies still raise the need for further research and validation (Lopes et al., 2023). Particular beneficial results could be obtained from the identification of initial stages of Correa’s cascade, which so far has been accomplished through plasmatic and tissular miRNAs (Lario et al., 2018; Kim et al., 2020). In colorectal cancer, the use of fecal miRNAs has so far been considered the ultimate non-invasive screening method, with only a few studies showing the biomarker characteristics of salivary miRNAs (Link et al., 2010; Sazanov et al., 2017; Rapado-González et al., 2019). Some studies offer though encouraging results, in spite of their unicity on the subject. For example, one recent study proved for the first time that salivary miRNA signatures could be used in the early identification of hepatocellular carcinoma (HCC). After identifying an impressive number of 4,565 precursor and mature miRNAs, the authors found that 283 of these were downregulated in patients with HCC (Mariam et al., 2022). Hence, abundance of miRNA candidates in the saliva calls for expansion of future research, in light of the limited data available.
In conclusion, salivary miRNAs may represent reliable biomarker candidates for gastrointestinal conditions. Their diagnostic potential has so far been underlined by a limited number of studies, but future research assessing salivary identifiable miRNAs, performed on larger cohorts, might validate or infirm their biomarker status in a multitude of gastrointestinal disorders, particularly in the early identification of malignancies.
Author contributions
MS and CB conceptualized and designed the study and conducted literature search and drafted the initial manuscript. MS and CB reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adami, B., Tabatabaeian, H., Ghaedi, K., Talebi, A., Azadeh, M., and Dehdashtian, E. (2019). miR-146a is deregulated in gastric cancer. J. Cancer Res. Ther. 15, 108–114. doi:10.4103/jcrt.JCRT_855_17
Asai, Y., Itoi, T., Sugimoto, M., Sofuni, A., Tsuchiya, T., Tanaka, R., et al. (2018). Elevated polyamines in saliva of pancreatic cancer. Cancers (Basel) 10, 43. doi:10.3390/cancers10020043
Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell 136, 215–233. doi:10.1016/j.cell.2009.01.002
Bel’skaya, L. V., Sarf, E. A., Shalygin, S. P., Postnova, T. V., and Kosenok, V. K. (2020). Identification of salivary volatile organic compounds as potential markers of stomach and colorectal cancer: A pilot study. J. Oral Biosci. 62, 212–221. doi:10.1016/j.job.2020.05.002
Bendifallah, S., Suisse, S., Puchar, A., Delbos, L., Poilblanc, M., Descamps, P., et al. (2022). Salivary MicroRNA signature for diagnosis of endometriosis. J. Clin. Med. 11, 612. doi:10.3390/jcm11030612
Bhardwaj, N., Sena, M., Ghaffari, G., and Ishmael, F. (2020). MiR-4668 as a novel potential biomarker for eosinophilic esophagitis. Allergy Rhinol. Provid. 11, 2152656720953378. doi:10.1177/2152656720953378
Broughton, J. P., Lovci, M. T., Huang, J. L., Yeo, G. W., and Pasquinelli, A. E. (2016). Pairing beyond the seed supports MicroRNA targeting specificity. Mol. Cell 64, 320–333. doi:10.1016/j.molcel.2016.09.004
Burris, H. A., Moore, M. J., Andersen, J., Green, M. R., Rothenberg, M. L., Modiano, M. R., et al. (1997). Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J. Clin. Oncol. 15, 2403–2413. doi:10.1200/JCO.1997.15.6.2403
Chen, C.-Z., Li, L., Lodish, H. F., and Bartel, D. P. (2004). MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86. doi:10.1126/science.1091903
Chen, X., Wang, N., Wang, J., Liao, B., Cheng, L., and Ren, B. (2022). The interactions between oral-gut axis microbiota and Helicobacter pylori. Front. Cell Infect. Microbiol. 12, 914418. doi:10.3389/fcimb.2022.914418
Chung, J.-W., Jeong, S. H., Lee, S. M., Pak, J. H., Lee, G. H., Jeong, J.-Y., et al. (2017). Expression of MicroRNA in host cells infected with Helicobacter pylori. Gut Liver 11, 392–400. doi:10.5009/gnl16265
Contaldo, M., Fusco, A., Stiuso, P., Lama, S., Gravina, A. G., Itro, A., et al. (2021). Oral microbiota and salivary levels of oral pathogens in gastro-intestinal diseases: Current knowledge and exploratory study. Microorganisms 9, 1064. doi:10.3390/microorganisms9051064
Cortés-Márquez, A. C., Mendoza-Elizalde, S., Arenas-Huertero, F., Trillo-Tinoco, J., Valencia-Mayoral, P., Consuelo-Sánchez, A., et al. (2018). Differential expression of miRNA-146a and miRNA-155 in gastritis induced by Helicobacter pylori infection in paediatric patients, adults, and an animal model. BMC Infect. Dis. 18, 463. doi:10.1186/s12879-018-3368-2
Deng, H., Guo, Y., Song, H., Xiao, B., Sun, W., Liu, Z., et al. (2013). MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene 518, 351–359. doi:10.1016/j.gene.2012.12.103
Deutsch, O., Haviv, Y., Krief, G., Keshet, N., Westreich, R., Stemmer, S. M., et al. (2020). Possible proteomic biomarkers for the detection of pancreatic cancer in oral fluids. Sci. Rep. 10, 21995. doi:10.1038/s41598-020-78922-x
Dexheimer, P. J., and Cochella, L. (2020). MicroRNAs: From mechanism to organism. Front. Cell Dev. Biol. 8, 409. doi:10.3389/fcell.2020.00409
Dragomir, M. P., Kopetz, S., Ajani, J. A., and Calin, G. A. (2020). Non-coding RNAs in GI cancers: From cancer hallmarks to clinical utility. Gut 69, 748–763. doi:10.1136/gutjnl-2019-318279
Du, J., and Zhang, L. (2017). Analysis of salivary microRNA expression profiles and identification of novel biomarkers in esophageal cancer. Oncol. Lett. 14, 1387–1394. doi:10.3892/ol.2017.6328
Eftekhari, A., Maleki Dizaj, S., Sharifi, S., Salatin, S., Khalilov, R., Samiei, M., et al. (2022). “Chapter six - salivary biomarkers in cancer,” in Advances in clinical chemistry. Editor G. S. Makowski (Elsevier), 171–192. doi:10.1016/bs.acc.2022.06.005
Fellizar, A., Refuerzo, V., Ramos, J. D., and Albano, P. M. (2022). Expression of specific microRNAs in tissue and plasma in colorectal cancer. J. Pathol. Transl. Med. 57, 147–157. doi:10.4132/jptm.2022.02.19
Fendereski, M., Zia, M. F., Shafiee, M., Safari, F., Saneie, M. H., and Tavassoli, M. (2017). MicroRNA-196a as a potential diagnostic biomarker for esophageal squamous cell carcinoma. Cancer Invest. 35, 78–84. doi:10.1080/07357907.2016.1254228
Friedman, R. C., Farh, K. K.-H., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi:10.1101/gr.082701.108
Fu, G., Brkić, J., Hayder, H., and Peng, C. (2013). MicroRNAs in human placental development and pregnancy complications. Int. J. Mol. Sci. 14, 5519–5544. doi:10.3390/ijms14035519
Hayes, J., Peruzzi, P. P., and Lawler, S. (2014). MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 20, 460–469. doi:10.1016/j.molmed.2014.06.005
He, T., Fan, Y., Wang, Y., Liu, M., and Zhu, A. J. (2022). Dissection of the microRNA network regulating hedgehog signaling in Drosophila. Front. Cell Dev. Biol. 10, 866491. doi:10.3389/fcell.2022.866491
Hicks, S. D., Onks, C., Kim, R. Y., Zhen, K. J., Loeffert, J., Loeffert, A. C., et al. (2021). Refinement of saliva microRNA biomarkers for sports-related concussion. J. Sport Health Sci. S2095-2546 (21), 369–378. doi:10.1016/j.jshs.2021.08.003
Hicks, S. D., Leddy, J., Lichak, B. P., Onks, C., Dretsch, M., Tennant, P., et al. (2022). Defining biological phenotypes of mild traumatic brain injury using saliva MicroRNA profiles. J. Neurotrauma 39, 923–934. doi:10.1089/neu.2022.0018
Hoshino, I., Ishige, F., Iwatate, Y., Gunji, H., Kuwayama, N., Nabeya, Y., et al. (2021). Cell-free microRNA-1246 in different body fluids as a diagnostic biomarker for esophageal squamous cell carcinoma. PLoS One 16, e0248016. doi:10.1371/journal.pone.0248016
Hossian, A. K. M. N., Mackenzie, G. G., and Mattheolabakis, G. (2019). miRNAs in gastrointestinal diseases: can we effectively deliver RNA-based therapeutics orally? Nanomedicine (Lond) 14, 2873–2889. doi:10.2217/nnm-2019-0180
Huang, K., Gao, X., Wu, L., Yan, B., Wang, Z., Zhang, X., et al. (2021). Salivary microbiota for gastric cancer prediction: An exploratory study. Front. Cell Infect. Microbiol. 11, 640309. doi:10.3389/fcimb.2021.640309
Huang, W. (2017). MicroRNAs: Biomarkers, diagnostics, and therapeutics. Methods Mol. Biol. 1617, 57–67. doi:10.1007/978-1-4939-7046-9_4
Humeau, M., Vignolle-Vidoni, A., Sicard, F., Martins, F., Bournet, B., Buscail, L., et al. (2015). Salivary MicroRNA in pancreatic cancer patients. PLoS One 10, e0130996. doi:10.1371/journal.pone.0130996
Igaz, I., and Igaz, P. (2015). Diagnostic relevance of microRNAs in other body fluids including urine, feces, and saliva. Exp. Suppl. 106, 245–252. doi:10.1007/978-3-0348-0955-9_11
Ishige, F., Hoshino, I., Iwatate, Y., Chiba, S., Arimitsu, H., Yanagibashi, H., et al. (2020). MIR1246 in body fluids as a biomarker for pancreatic cancer. Sci. Rep. 10, 8723. doi:10.1038/s41598-020-65695-6
Ivey, K. N., and Srivastava, D. (2015). microRNAs as developmental regulators. Cold Spring Harb. Perspect. Biol. 7, a008144. doi:10.1101/cshperspect.a008144
Jhaveri, P. B., Lambert, K. A., Bogale, K., Lehman, E., Alexander, C., Ishmael, F., et al. (2023). Salivary microRNAs in pediatric eosinophilic esophagitis. Allergy Asthma Proc. 44, 145–152. doi:10.2500/aap.2023.44.220102
Ji, Y., Liang, X., and Lu, H. (2020). Analysis of by high-throughput sequencing: Helicobacter pylori infection and salivary microbiome. BMC Oral Health 20, 84. doi:10.1186/s12903-020-01070-1
Kaczor-Urbanowicz, K. E., Saad, M., Grogan, T. R., Li, F., Heo, Y. J., Elashoff, D., et al. (2022). Performance of salivary extracellular RNA biomarker panels for gastric cancer differs between distinct populations. Cancers (Basel) 14, 3632. doi:10.3390/cancers14153632
Kageyama, S., Takeshita, T., Takeuchi, K., Asakawa, M., Matsumi, R., Furuta, M., et al. (2019). Characteristics of the salivary microbiota in patients with various digestive tract cancers. Front. Microbiol. 10, 1780. doi:10.3389/fmicb.2019.01780
Kapoor, P., Chowdhry, A., Bagga, D. K., Bhargava, D., and Aishwarya, S. (2021). MicroRNAs in oral fluids (saliva and gingival crevicular fluid) as biomarkers in orthodontics: Systematic review and integrated bioinformatic analysis. Prog. Orthod. 22, 31. doi:10.1186/s40510-021-00377-1
Khanal, S., Zancanela, B., Peter, J., and Flynt, A. (2022). The small RNA universe of Capitella teleta. FASEB J. 36 (Suppl. 1). doi:10.1096/fasebj.2022.36.S1.L7428
Khayam, N., Nejad, H. R., Ashrafi, F., and Abolhassani, M. (2021). Expression profile of miRNA-17-3p and miRNA-17-5p genes in gastric cancer patients with Helicobacter pylori infection. J. Gastrointest. Cancer 52, 130–137. doi:10.1007/s12029-019-00319-5
Kim, B., Jang, J., Heo, Y. J., Kang, S. Y., Yoo, H., Sohn, I., et al. (2020). Dysregulated miRNA in a cancer-prone environment: A study of gastric non-neoplastic mucosa. Sci. Rep. 10, 6600. doi:10.1038/s41598-020-63230-1
Koopaie, M., Abedinejad, F., Manifar, S., Mousavi, R., Kolahdooz, S., and Shamshiri, A. (2021). Salivary miRNA-21 expression as a potential non-invasive diagnostic biomarker in breast cancer. Gene Rep. 25, 101317. doi:10.1016/j.genrep.2021.101317
Koopaie, M., Kolahdooz, S., Fatahzadeh, M., and Aleedawi, Z. A. (2022). Salivary noncoding RNA in the diagnosis of pancreatic cancer: Systematic review and meta-analysis. Eur. J. Clin. Invest. 52, e13848. doi:10.1111/eci.13848
Lario, S., Brunet-Vega, A., Quílez, M. E., Ramírez-Lázaro, M. J., Lozano, J. J., García-Martínez, L., et al. (2018). Expression profile of circulating microRNAs in the Correa pathway of progression to gastric cancer. United Eur. Gastroenterol. J. 6, 691–701. doi:10.1177/2050640618759433
Leal-Galvan, B., Harvey, C., Thomas, D., Saelao, P., and Oliva Chavez, A. S. (2022). Isolation of microRNAs from tick ex vivo salivary gland cultures and extracellular vesicles. J. Vis. Exp. doi:10.3791/63618
Lee, N. H., Lee, E., Kim, Y. S., Kim, W. K., Lee, Y. K., and Kim, S. H. (2020). Differential expression of microRNAs in the saliva of patients with aggressive periodontitis: A pilot study of potential biomarkers for aggressive periodontitis. J. Periodontal Implant Sci. 50, 281–290. doi:10.5051/jpis.2000120006
Li, X.-J., Wang, L., Li, G., Zheng, X., and Duan, C. (2016). Expression of miR-204 and MMP-9 in Helicobacter pylori-associated gastric ulcer. Available at: https://www.semanticscholar.org/paper/Expression-of-miR-204-and-MMP-9-in-Helicobacter-Li-Wang/cb8f7e302b16639d0db3010bb82c1a1424253619 (Accessed May 20, 2023).
Li, M., Wu, F., Ji, Y., Yang, L., and Li, F. (2017). Meta-analysis of microRNAs as potential biomarkers for detecting esophageal carcinoma in Asian populations. Int. J. Biol. Markers 32, e375–e383. doi:10.5301/ijbm.5000296
Li, F., Yoshizawa, J. M., Kim, K.-M., Kanjanapangka, J., Grogan, T. R., Wang, X., et al. (2018). Discovery and validation of salivary extracellular RNA biomarkers for noninvasive detection of gastric cancer. Clin. Chem. 64, 1513–1521. doi:10.1373/clinchem.2018.290569
Link, A., Balaguer, F., Shen, Y., Nagasaka, T., Lozano, J. J., Boland, C. R., et al. (2010). Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol. Biomarkers Prev. 19, 1766–1774. doi:10.1158/1055-9965.EPI-10-0027
Lopes, C., Chaves, J., Ortigão, R., Dinis-Ribeiro, M., and Pereira, C. (2023). Gastric cancer detection by non-blood-based liquid biopsies: A systematic review looking into the last decade of research. United Eur. Gastroenterol. J. 11, 114–130. doi:10.1002/ueg2.12328
Luo, X., Chen, X., Lv, Y., Han, Y., Qu, X., Zhang, Y., et al. (2022). MicroRNA-101 regulates oocyte maturation in vitro via targeting HAS2 in porcine cumulus cells. Theriogenology 187, 119–126. doi:10.1016/j.theriogenology.2022.04.025
Machida, T., Tomofuji, T., Maruyama, T., Yoneda, T., Ekuni, D., Azuma, T., et al. (2016). miR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol. Rep. 36, 2375–2381. doi:10.3892/or.2016.5021
Mahboobi, R., Fallah, F., Yadegar, A., Dara, N., Kazemi Aghdam, M., Asgari, B., et al. (2022). Expression analysis of miRNA-155 level in Helicobacter pylori related inflammation and chronic gastritis. Iran. J. Microbiol. 14, 495–502. doi:10.18502/ijm.v14i4.10235
Makarova, J. A., Shkurnikov, M. U., Wicklein, D., Lange, T., Samatov, T. R., Turchinovich, A. A., et al. (2016). Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem Cytochem 51, 33–49. doi:10.1016/j.proghi.2016.06.001
Manzano-Moreno, F. J., Costela-Ruiz, V. J., García-Recio, E., Olmedo-Gaya, M. V., Ruiz, C., and Reyes-Botella, C. (2021). Role of salivary MicroRNA and cytokines in the diagnosis and prognosis of oral squamous cell carcinoma. Int. J. Mol. Sci. 22, 12215. doi:10.3390/ijms222212215
Mariam, A., Miller-Atkins, G., Moro, A., Rodarte, A. I., Siddiqi, S., Acevedo-Moreno, L.-A., et al. (2022). Salivary miRNAs as non-invasive biomarkers of hepatocellular carcinoma: A pilot study. PeerJ 10, e12715. doi:10.7717/peerj.12715
Martina, E., Campanati, A., Diotallevi, F., and Offidani, A. (2020). Saliva and oral diseases. J. Clin. Med. 9, 466. doi:10.3390/jcm9020466
Matsushima, K., Isomoto, H., Inoue, N., Nakayama, T., Hayashi, T., Nakayama, M., et al. (2011). MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int. J. Cancer 128, 361–370. doi:10.1002/ijc.25348
Michael, A., Bajracharya, S. D., Yuen, P. S. T., Zhou, H., Star, R. A., Illei, G. G., et al. (2010). Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 16, 34–38. doi:10.1111/j.1601-0825.2009.01604.x
Nik Mohamed Kamal, N. N. S., Awang, R. A. R., Mohamad, S., and Shahidan, W. N. S. (2020). Plasma- and saliva exosome profile reveals a distinct MicroRNA signature in chronic periodontitis. Front. Physiol. 11, 587381. doi:10.3389/fphys.2020.587381
O’Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 9, 402. doi:10.3389/fendo.2018.00402
Oana, S. M., Claudia, B., Lelia, R. A., Simona, M., Claudia, C., and Daniela, D. E. (2022). Differential expression of tissular miRNA-155 in pediatric gastritis. J. Clin. Med. 11, 3351. doi:10.3390/jcm11123351
Ohtsuka, M., Iwamoto, K., Naito, A., Imasato, M., Hyuga, S., Nakahara, Y., et al. (2021). Circulating MicroRNAs in gastrointestinal cancer. Cancers (Basel) 13, 3348. doi:10.3390/cancers13133348
Park, N. J., Zhou, H., Elashoff, D., Henson, B. S., Kastratovic, D. A., Abemayor, E., et al. (2009). Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 15, 5473–5477. doi:10.1158/1078-0432.CCR-09-0736
Patel, R. S., Jakymiw, A., Yao, B., Pauley, B. A., Carcamo, W. C., Katz, J., et al. (2011). High resolution of microRNA signatures in human whole saliva. Arch. Oral Biol. 56, 1506–1513. doi:10.1016/j.archoralbio.2011.05.015
Patel, V., Ma, S., and Yadlapati, R. (2022). Salivary biomarkers and esophageal disorders. Dis. Esophagus 35, doac018. doi:10.1093/dote/doac018
Peng, Q., Zhang, X., Min, M., Zou, L., Shen, P., and Zhu, Y. (2017). The clinical role of microRNA-21 as a promising biomarker in the diagnosis and prognosis of colorectal cancer: A systematic review and meta-analysis. Oncotarget 8, 44893–44909. doi:10.18632/oncotarget.16488
Quirico, L., and Orso, F. (2020). The power of microRNAs as diagnostic and prognostic biomarkers in liquid biopsies. Cancer Drug Resist 3, 117–139. doi:10.20517/cdr.2019.103
Rapado-González, Ó., Majem, B., Álvarez-Castro, A., Díaz-Peña, R., Abalo, A., Suárez-Cabrera, L., et al. (2019). A novel saliva-based miRNA signature for colorectal cancer diagnosis. J. Clin. Med. 8, 2029. doi:10.3390/jcm8122029
Sanz-Rubio, D., Martin-Burriel, I., Gil, A., Cubero, P., Forner, M., Khalyfa, A., et al. (2018). Stability of circulating exosomal miRNAs in healthy subjects. Sci. Rep. 8, 10306. doi:10.1038/s41598-018-28748-5
Săsăran, M. O., Meliț, L. E., and Dobru, E. D. (2021). MicroRNA modulation of host immune response and inflammation triggered by Helicobacter pylori. Int. J. Mol. Sci. 22, 1406. doi:10.3390/ijms22031406
Sazanov, A. A., Kiselyova, E. V., Zakharenko, A. A., Romanov, M. N., and Zaraysky, M. I. (2017). Plasma and saliva miR-21 expression in colorectal cancer patients. J. Appl. Genet. 58, 231–237. doi:10.1007/s13353-016-0379-9
Schaefer, J. S., Attumi, T., Opekun, A. R., Abraham, B., Hou, J., Shelby, H., et al. (2015). MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 16, 5. doi:10.1186/s12865-015-0069-0
Scholtz, B., Horváth, J., Tar, I., Kiss, C., and Márton, I. J. (2022). Salivary miR-31-5p, miR-345-3p, and miR-424-3p are reliable biomarkers in patients with oral squamous cell carcinoma. Pathogens 11, 229. doi:10.3390/pathogens11020229
Sembler-Møller, M. L., Belstrøm, D., Locht, H., and Pedersen, A. M. L. (2020). Distinct microRNA expression profiles in saliva and salivary gland tissue differentiate patients with primary Sjögren’s syndrome from non-Sjögren’s sicca patients. J. Oral Pathol. Med. 49, 1044–1052. doi:10.1111/jop.13099
Setti, G., Pezzi, M. E., Viani, M. V., Pertinhez, T. A., Cassi, D., Magnoni, C., et al. (2020). Salivary MicroRNA for diagnosis of cancer and systemic diseases: A systematic review. Int. J. Mol. Sci. 21, 907. doi:10.3390/ijms21030907
Shin, V. Y., and Chu, K.-M. (2014). MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J. Gastroenterol. 20, 10432–10439. doi:10.3748/wjg.v20.i30.10432
Sreedharan, L., Mayne, G. C., Watson, D. I., Bright, T., Lord, R. V., Ansar, A., et al. (2017). MicroRNA profile in neosquamous esophageal mucosa following ablation of Barrett’s esophagus. World J. Gastroenterol. 23, 5508–5518. doi:10.3748/wjg.v23.i30.5508
St John, M. A. R., Li, Y., Zhou, X., Denny, P., Ho, C.-M., Montemagno, C., et al. (2004). Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch. Otolaryngol. Head. Neck Surg. 130, 929–935. doi:10.1001/archotol.130.8.929
Sugiyama, T., Taniguchi, K., Matsuhashi, N., Tajirika, T., Futamura, M., Takai, T., et al. (2016). MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci. 107, 1767–1775. doi:10.1111/cas.13091
Théry, C., Zitvogel, L., and Amigorena, S. (2002). Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579. doi:10.1038/nri855
Théry, C., Ostrowski, M., and Segura, E. (2009). Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593. doi:10.1038/nri2567
Tüfekci, K. U., Oner, M. G., Meuwissen, R. L. J., and Genç, S. (2014). The role of microRNAs in human diseases. Methods Mol. Biol. 1107, 33–50. doi:10.1007/978-1-62703-748-8_3
Urbizu, A., Arnaldo, L., and Beyer, K. (2023). Obtaining miRNA from saliva-comparison of sampling and purification methods. Int. J. Mol. Sci. 24, 2386. doi:10.3390/ijms24032386
Vasapolli, R., Venerito, M., Schirrmeister, W., Thon, C., Weigt, J., Wex, T., et al. (2021). Inflammatory microRNAs in gastric mucosa are modulated by Helicobacter pylori infection and proton-pump inhibitors but not by aspirin or NSAIDs. PLoS One 16, e0249282. doi:10.1371/journal.pone.0249282
Vasudevan, S. (2012). Posttranscriptional upregulation by microRNAs. Wiley Interdiscip. Rev. RNA 3, 311–330. doi:10.1002/wrna.121
Vidal, A. F., Cruz, A. M. P., Magalhães, L., Pereira, A. L., Anaissi, A. K. M., Alves, N. C. F., et al. (2016). hsa-miR-29c and hsa-miR-135b differential expression as potential biomarker of gastric carcinogenesis. World J. Gastroenterol. 22, 2060–2070. doi:10.3748/wjg.v22.i6.2060
Wan, J., Wu, W., Che, Y., Kang, N., and Zhang, R. (2016). Insights into the potential use of microRNAs as a novel class of biomarkers in esophageal cancer. Dis. Esophagus 29, 412–420. doi:10.1111/dote.12338
Wang, J., and Sen, S. (2011). MicroRNA functional network in pancreatic cancer: From biology to biomarkers of disease. J. Biosci. 36, 481–491. doi:10.1007/s12038-011-9083-4
Wang, Y., Wang, Q., Zhang, N., Ma, H., Gu, Y., Tang, H., et al. (2014). Identification of microRNAs as novel biomarkers for detecting esophageal squamous cell carcinoma in asians: A meta-analysis. Tumour Biol. 35, 11595–11604. doi:10.1007/s13277-014-2350-x
Wang, J., Chen, J., and Sen, S. (2016). MicroRNA as biomarkers and diagnostics. J. Cell. Physiology 231, 25–30. doi:10.1002/jcp.25056
Wang, J.-Y., Wang, C.-L., Wang, X.-M., and Liu, F.-J. (2017). Comprehensive analysis of microRNA/mRNA signature in colon adenocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 21, 2114–2129.
Wong, D. T. W. (2012). Salivaomics. J. Am. Dent. Assoc. 143, 19S–24S. doi:10.14219/jada.archive.2012.0339
Wu, F., Guo, N. J., Tian, H., Marohn, M., Gearhart, S., Bayless, T. M., et al. (2011). Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm. Bowel Dis. 17, 241–250. doi:10.1002/ibd.21450
Wu, W., Hou, W., Wu, Z., Wang, Y., Yi, Y., and Lin, W. (2013). miRNA-144 in the saliva is a genetic marker for early diagnosis of esophageal cancer. Nan Fang. Yi Ke Da Xue Xue Bao 33, 1783–1786.
Xiao, H., Zhang, Y., Kim, Y., Kim, S., Kim, J. J., Kim, K. M., et al. (2016). Differential proteomic analysis of human saliva using tandem mass tags quantification for gastric cancer detection. Sci. Rep. 6, 22165. doi:10.1038/srep22165
Xie, Z.-J., Chen, G., Zhang, X.-C., Li, D.-F., Huang, J., and Li, Z.-J. (2012). Saliva supernatant miR-21: A novel potential biomarker for esophageal cancer detection. Asian Pac J. Cancer Prev. 13, 6145–6149. doi:10.7314/apjcp.2012.13.12.6145
Xie, Z., Chen, G., Zhang, X., Li, D., Huang, J., Yang, C., et al. (2013). Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS One 8, e57502. doi:10.1371/journal.pone.0057502
Xie, Z., Yin, X., Gong, B., Nie, W., Wu, B., Zhang, X., et al. (2015). Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev. Res. (Phila) 8, 165–173. doi:10.1158/1940-6207.CAPR-14-0192
Xu, F., and Jiang, M. (2020). Evaluation of predictive role of carcinoembryonic antigen and salivary mRNA biomarkers in gastric cancer detection. Medicine 99, e20419. doi:10.1097/MD.0000000000020419
Ye, M., Ye, P., Zhang, W., Rao, J., and Xie, Z. (2014). Diagnostic values of salivary versus and plasma microRNA-21 for early esophageal cancer. Nan Fang. Yi Ke Da Xue Xue Bao 34, 885–889.
Yoshizawa, J. M., and Wong, D. T. W. (2013). Salivary microRNAs and oral cancer detection. Methods Mol. Biol. 936, 313–324. doi:10.1007/978-1-62703-083-0_24
Yoshizawa, J. M., Schafer, C. A., Schafer, J. J., Farrell, J. J., Paster, B. J., and Wong, D. T. W. (2013). Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin. Microbiol. Rev. 26, 781–791. doi:10.1128/CMR.00021-13
Zabaglia, L. M., Sallas, M. L., Santos, M. P. D., Orcini, W. A., Peruquetti, R. L., Constantino, D. H., et al. (2018). Expression of miRNA-146a, miRNA-155, IL-2, and TNF-α in inflammatory response to Helicobacter pylori infection associated with cancer progression. Ann. Hum. Genet. 82, 135–142. doi:10.1111/ahg.12234
Zhang, L., Farrell, J. J., Zhou, H., Elashoff, D., Akin, D., Park, N.-H., et al. (2010). Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 138, 949–957.e1-7. doi:10.1053/j.gastro.2009.11.010
Zhang, Z., Dong, Y., Hua, J., Xue, H., Hu, J., Jiang, T., et al. (2019). A five-miRNA signature predicts survival in gastric cancer using bioinformatics analysis. Gene 699, 125–134. doi:10.1016/j.gene.2019.02.058
Zheng, B., Liang, L., Wang, C., Huang, S., Cao, X., Zha, R., et al. (2011). MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin. Cancer Res. 17, 7574–7583. doi:10.1158/1078-0432.CCR-11-1714
Keywords: salivary, miRNA, biomarker, gastrointestinal disorders, pancreatic cancer, gastric cancer
Citation: Săsăran MO and Bănescu C (2023) Role of salivary miRNAs in the diagnosis of gastrointestinal disorders: a mini-review of available evidence. Front. Genet. 14:1228482. doi: 10.3389/fgene.2023.1228482
Received: 30 May 2023; Accepted: 20 June 2023;
Published: 29 June 2023.
Edited by:
Peter Igaz, Semmelweis University, HungaryReviewed by:
Balint Nagy, University of Debrecen, HungaryCopyright © 2023 Săsăran and Bănescu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudia Bănescu, Y2xhdWRpYS5iYW5lc2N1QGdtYWlsLmNvbQ==
 Maria Oana Săsăran
Maria Oana Săsăran Claudia Bănescu
Claudia Bănescu