- 1College of Advanced Agriculture and Ecological Environment, Heilongjiang University, Harbin, China
- 2Laboratory of Genetic Breeding in Tomato, College of Horticulture and Landscape Architecture, Northeast Agricultural University, Harbin, China
- 3Horticultural Sub-Academy, Heilongjiang Academy of Agricultural Sciences, Harbin, China
Tomato (Solanum lycopersicum) is widely cultivated and consumed worldwide. Tomato leaf mold, caused by Cladosporium fulvum, is one of the most devastating diseases in tomato production. At present, some tomato leaf mold resistance (Cf series) genes used in production gradually lose resistance due to the continuous and rapid differentiation of C. fulvum physiological races. The Cf-16 gene derived from the “Ontario7816” tomato cultivar has shown effective resistance in field trials for many years, but few studies have reported on the mapping of the Cf-16 gene, which has not been cloned, limiting its utilization in tomato breeding. Here, we mapped Cf-16 using a novel comprehensive strategy including bulk segregation analysis (BSA), genome resequencing and SSR molecular markers. A genetic analysis revealed that Cf-16 resistance in “Ontario7816” is controlled by one major dominant locus. The Cf-16 gene was mapped in a region of 2.6 cM at chromosome 6 between two markers, namely, TGS447 and TES312, by using an F2 population from a cross between the resistant cultivar “Ontario7816” and susceptible line “Moneymaker.” Two nucleotide-binding-site-leucine-rich repeat (NBS-LRR) resistance genes, namely, XM_004240667.3 and XM_010323727.1, were identified in this interval. They are strong candidates for the Cf-16 gene. The mapping of Cf-16 may speed up its utilization for breeding resistant tomato varieties and represents an important step forward in our understanding of the mechanism underlying resistance to tomato leaf mold.
Introduction
Tomato (Solanum lycopersicum L.) is a global vegetable cultivated and consumed worldside that has outstanding nutritional. Because of its rich flavor, it is also often used as a fruit (Kim et al., 2017). Leaf mold disease caused by Cladosporium fulvum is considered to be one of the most devastating diseases in tomato crops and has threatened tomato-growing areas worldwide, especially for tomato grown in protected areas. This pathogen reduces both fruit yield and quality and sometimes even kills tomato plants (Thomma et al., 2005; Mesarich et al., 2014). In the early stages, tomato leaf mold disease symptoms appear as irregular or oval light yellow chlorotic spots on the backs of the leaves. As the disease progresses, the affected leaves grow gray‒brown or dark-brown velvety mold layers. When the disease is severe, the leaves senesce, wither and fall off prematurely (Zhao et al., 2022). However, it is increasingly difficult to control this disease in tomato-growing regions around the world. Thus, breeding for tomato leaf mold resistance will provide an effective and environmentally friendly alternative to chemical control. C. fulvum is the causal organism of tomato leaf mold, a fungal disease first described by Cooke (1883). The differentiation rate of C. fulvum physiological races is very rapid, and new races are continuously isolated and identified (Enya et al., 2009; Iida et al., 2010; Li et al., 2015). Thus, the resistance effectiveness of some tomato leaf mold resistance (Cf series) genes used in breeding production has gradually been undermined. Therefore, the mining, cloning and application of new Cf genes have become particularly critical.
Cf genes are a class of functional genes with typical AVR recognition ability (Rivas and Thomas, 2005). At least 24 Cf genes, named Cf-1 ∼ Cf-24, have been identified by Dr. Kerr’s laboratory. The susceptible cultivar ‘Moneymaker’ is known as a Cf-0 cultivar what means that it lack resistance genes. These Cf genes are derived from tomato and its wild relatives (Kanwar et al., 1980). At present, some Cf genes have been cloned using methods such as transposon tagging and map-based cloning, including Cf-2 (Hcr2-2B, Hcr2-2C), Cf-5, Cf-4 (Hcr9-4D), Cf-4E (Hcr9-9B), Cf-9 (Hcr9-9C), and 9DC (Hcr9-M205) (Zhao et al., 2019). Some Avr genes (Avr2, Avr4, Avr4E, Avr5, Avr9, Ecp1, Ecp2, Ecp4, Ecp5, Ecp6, and Ecp7) corresponding to Cf genes have also been isolated and identified (Zhao et al., 2022). Cf genes encode transmembrane glycoproteins with typical extracellular leucine-rich repeats (LRRs). The different LRR numbers in different Cf genes determine the ability of different Cf genes to recognize different physiological races of the pathogen (Kruijt et al., 2005).
DNA markers have been widely used in identifying and mapping plant disease resistance genes. In recent years, single-nucleotide polymorphism (SNP) markers have been widely used to identify markers associated with important traits, including tomato disease resistance (Yang et al., 2017; Liu et al., 2019). Bulked segregant analysis (BSA) can rapidly detect specific genes or genomic regions. In particular, the combination of BSA and genome resequencing accelerates the cloning of genes responsible for important traits (Michelmore et al., 1991; Zou et al., 2016). Moreover, Kompetitive allele-specific PCR (KASP) markers converted from special SNPs are more flexible and efficient than the original SNP markers for use in genotyping and marker-assisted selection (MAS) (Liu et al., 2019; Yang et al., 2022). However, simple sequence repeat (SSR) markers are still used to study traits of interest by linkage analysis (Rui et al., 2017).
In this study, we explored the genetic characteristics of the Cf-16 gene by developing a large F2 population and BC1 populations. Notably, this is the first time that BSA combined with genome resequencing technology and molecular markers has been used for mapping the tomato Cladosporium fulvum resistance gene Cf-16. This study will be valuable for Cf-16 cloning and resistance breeding in tomato.
Materials and methods
Plant materials and C. fulvum inoculation
Resistant female “Ontario7816” (P1, kindly provided by the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Science), whose genome contain the Cf-16 gene, was crossed with the susceptible male “Moneymaker” (P2, kindly provided by the Tomato Genetic Resource Center, LA2706). The F1 individuals were then self-crossed to harvest F2 seeds, and the BC1 plants were generated by backcrossing the F1 plants with “Ontario7816” and “Moneymaker.” All plants were grown in the greenhouse under favorable conditions.
Ten C. fulvum physiological races (1.2, 1.2.3, 1.2.3.4, 1.2.4, 1.3.4, 2.3, 1.3, 1.4, 1.2.3.4.5, and 1.2.3.4.9) provided by the tomato research group of Horticulture College, Northeast Agricultural University, were used to test to characterize the resistance responses of “Ontario7816” and “Moneymaker”.
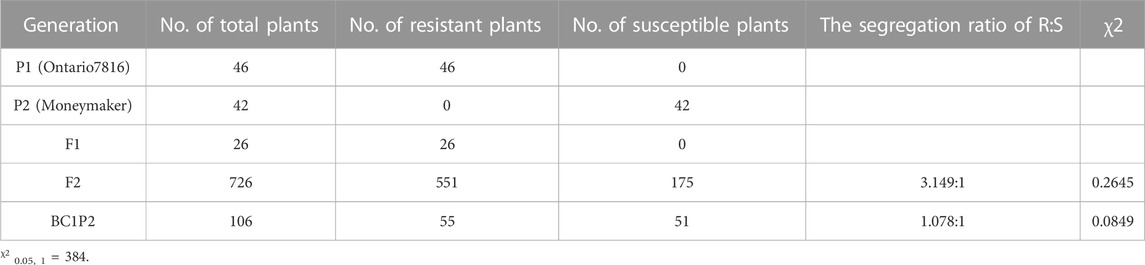
At the 5–6 leaf stage, all P1, P2, F1, F2, and BC1P2 plants were inoculated with C. fulvum race 1.2.3.4., which is a predominant physiological race in Heilongjiang Province, China. The plants were assessed for disease severity at 15 days postinoculation. Inoculation and assessment of disease severity ratings were performed as described by Xue et al. (2017). The plants were visually assessed for the severity of symptoms on a scale of 0–9 points. Plants with a disease index of 0–3 were regarded as resistant, and those with a score of 5–9 were regarded as susceptible.
DNA extraction, resequencing and association analysis
After inoculation, the F2 plants were used for genetic analysis and bulked segregant analysis (BSA). The parents (P1 and P2) and the F2 lines were prepared for genome resequencing and molecular marker detection, respectively. The resistant pool (F2R-pool, 25 resistant plants) and the susceptible pool (F2S-pool, 25 susceptible plants) were built by screening resistant and susceptible plants from the 726 F2 individuals. The cetyl-trimethylammonium bromide (CTAB) method was used for DNA extraction from young leaves, including those of the parents and the F2 lines. Bulked DNA samples were also subjected to the CTAB method by mixing equal amounts of DNA at a final concentration of 200 mg (Allen et al., 2006).
The resequencing and associated analysis were performed by BGI Tech (Shenzhen, China). The samples were constructed to generate 200–300 bp small fragment libraries for upsequencing on BGISEQ. The raw data were filtered to obtain clean data by the BGI in-house filter SOAPnuke. Subsequently, the clean data were aligned to the S. lycopersicum genome sequence (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/188/115/GCF_000188115.3_SL2.50) using Burrows-Wheeler Aligner (Li and Durbin, 2010). SNP and InDel detection processes were performed with Genome Analysis Toolkit (GATK) (Li et al., 2009), and we used an analysis tool developed by BGI to perform annotation and classification. The SNP-index method was used for the association analysis. The SNP index values of the resistant and susceptible pools were calculated using the resistant female “Ontario7816” as a reference.
SSR molecular marker analysis
Based on the candidate region of the association analysis, a total of 58 SSR markers from the Sol Genomics Network (SGN, http://solgenomics.net/) database were screened (Supplementary Table S1). SSR molecular markers were performed using the parents, F1 and 303 F2 individuals. Genetic linkage mapping was performed by JoinMap4 software (Ooijen and Van, 2006). Based on the electrophoresis results of 8% non-denaturing polyacrylamide gels.
General strategy used for mapping the Cf-16 gene
The general strategy of mapping the Cf-16 gene in “Ontario7816” is illustrated in Supplementary Figure S1. To determine the inheritance pattern of Cf-16 resistance in “Ontario7816” first, 726 F2 individuals and 106 BC1P2 individuals were planted and inoculated with C. fulvum race 1.2.3.4. The segregation ratios of resistant and susceptible phenotypes were recorded and further examined by the chi-square test. Next, to explore the rough position of the Cf-16 gene, bulked segregation analysis (BSA) combined with parental resequencing was performed. To further map the Cf-16 gene, 303 F2 individuals were planted and further screened with 58 SSR markers from the SGN (Supplementary Figure S1).
Results
Resistance and genetic analysis of the Cf-16 gene
To identify the resistance potential of the Cf-16 gene, we tested the resistance response of “Ontario7816” and “Moneymaker” to ten different races of C. fulvum (Table 1). “Moneymaker” showed susceptibility to all these physiological races of C. fulvum, while “Ontario7816” demonstrated resistance to all these races. These findings indicate that “Ontario7816” has highly resistant to C. fulvum and is suitable for future breeding work for disease resistance.
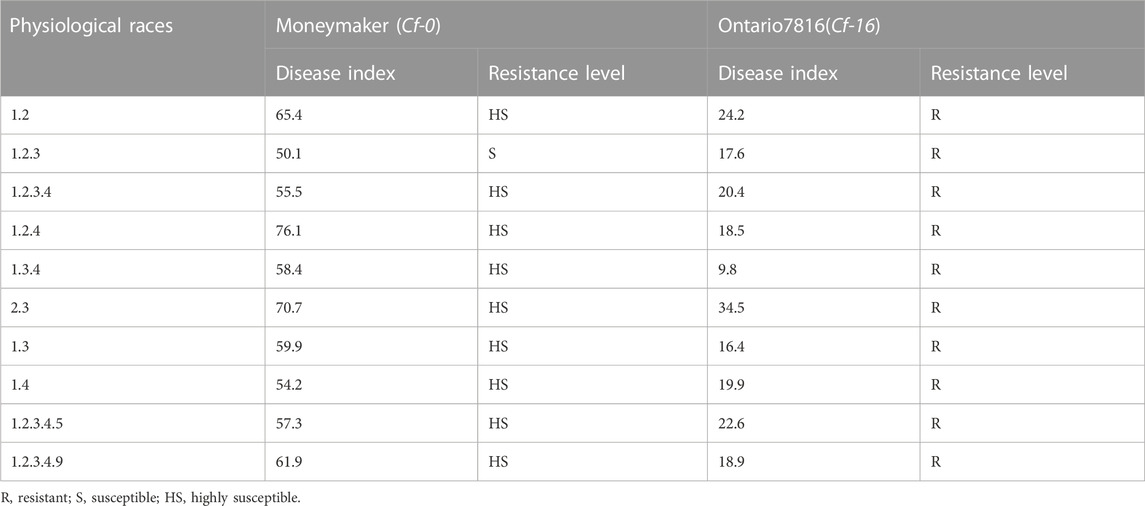
TABLE 1. The resistance response of “Ontario7816” and “Moneymaker” to ten different physiological races of C. fulvum.
Then, we performed a disease assay on the two parents, F1, F2 and BC1P2 individuals at 15 days postinoculation with the predominant physiological race 1.2.3.4 in Heilongjiang Province (Figure 1). “Ontario7816” and F1 plants were resistant to C. fulvum race 1.2.3.4, while “Moneymaker” plants were susceptible. Chi-square analysis showed that the segregation ratio of resistant and susceptible individuals of the F2 population was 3:1. The resistant and susceptible BC1P2 plants segregated according to the expected ratio of 1:1 (Table 2). This result confirms that the Cf-16 gene confers resistance via a single dominant gene.
Parental genome sequencing and SNP-index association analysis
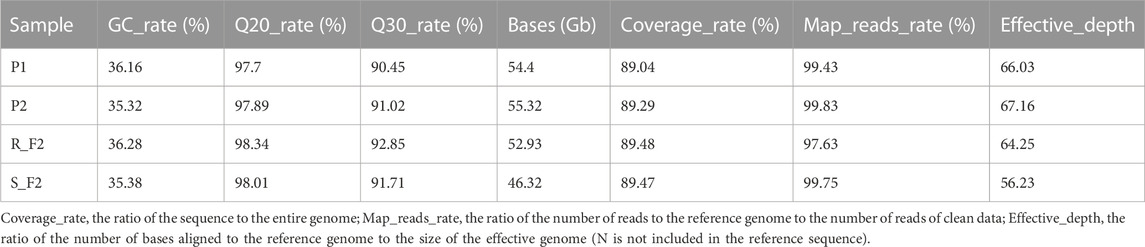
A total of 208.96 Gb clean data were obtained by Beijing Genomics Institute (BGI) sequencing, including 109.72 Gb from the parents and 99.26 Gb from the resistant and susceptible pools, all reads were of high quality (92.85% > Q30 > 90.45%) and with a stable GC content (36.28% > GC > 35.32%). The effective depths for the parents and the two F2 pools were between 56.23 and 67.16. The quality of the sequencing is high and the data are reliable and could be used for subsequent analysis (Table 3).
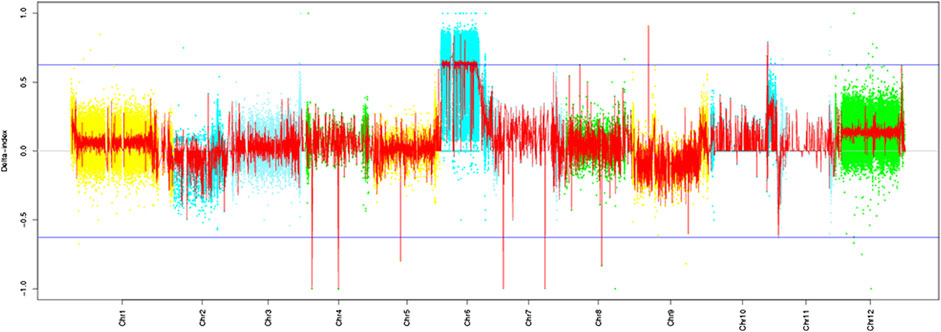
Based on the resequencing results, we calculated the SNP index values of the two mixed pools using the resistant parent “Ontario7816” as the reference. The Cf-16 gene was localized on tomato chromosome 6 by ΔSNP index analysis (Figure 2). Further analysis revealed an SNP imbalance between 11 and 38 Mb on chromosome 6, while the ΔSNP index of this region was greater than the threshold value at the 99% confidence level. Therefore, 11–38 Mb of chromosome 6 is the preliminary localization interval of the Cf-16 gene for leaf mold resistance in tomato.
Further mapping of the Cf-16 gene
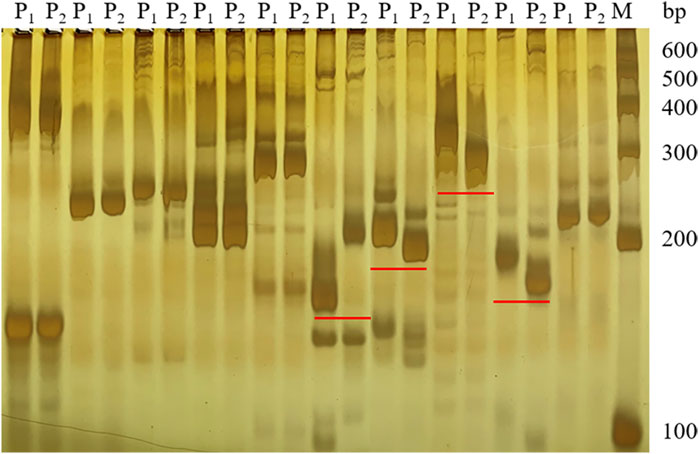
Based on the SNP index results, 58 SSR markers were used for polymorphic screening of the two parents and F1 generation to further map the Cf-16 gene (Figure 3). Subsequently, SSR marker analysis was performed on 303 F2 generations. Linkage mapping was performed using JoinMap 4.0 software based on the phenotypes and marker types of the F2 generation (Figure 4). The results showed that the most closely linked markers to the Cf-16 gene were TGS447 and TES312, both with a genetic distance of 1.3 cM. The positions of these two markers on chromosome 6 were 25.21 Mb and 28.84 Mb, respectively, and the physical distance of this candidate interval was 3.63 Mb.
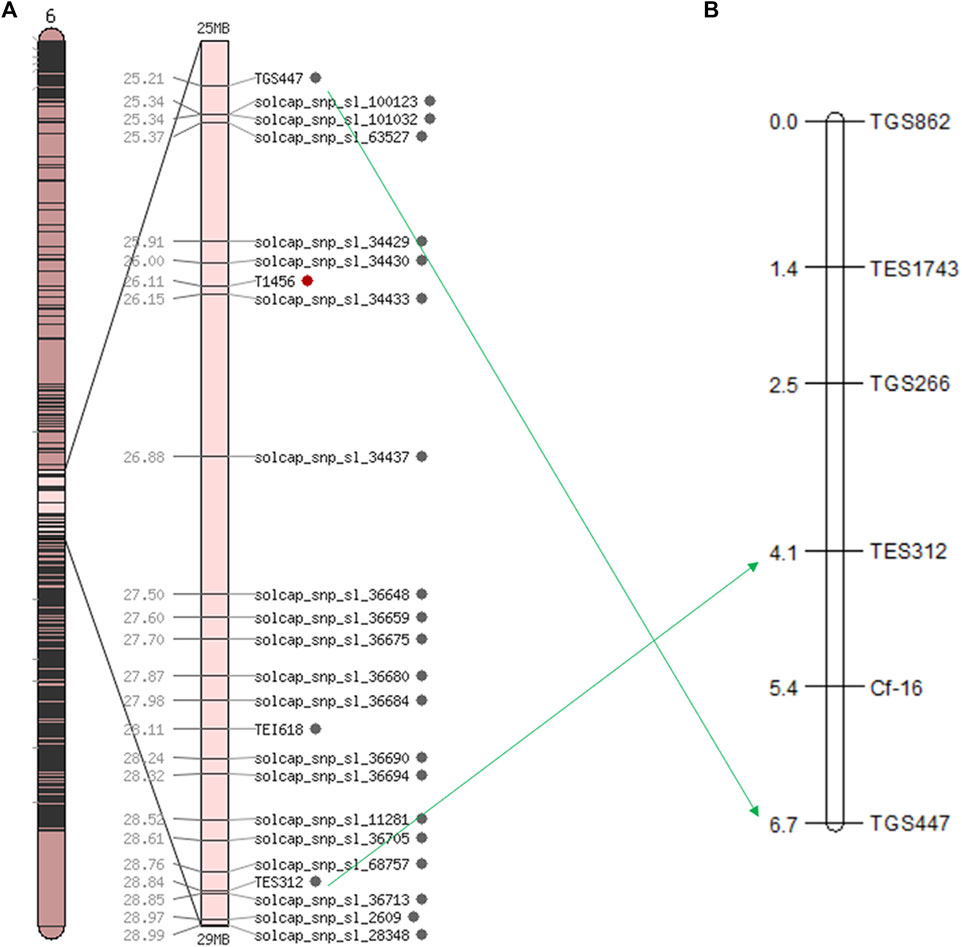
FIGURE 4. Linkage and chromosomal mapping of the leaf mold resistance gene Cf-16 in tomato. (A) Genetic map of chromosome 6 in tomato; (B) SSR linkage of the Cf-16 gene.
Analysis of candidate genes
Based on the candidate interval of the SNP index and SSR marker analysis, we found that 71 genes were distributed in the DNA fragment associated to the resistance gene Cf-16. Meanwhile, the number of nonsynonymous SNPs between the two parents in this region was 139. The 71 genes in the candidate interval were subjected to further functional annotation and structural analysis. Based on the typical structural features of the cloned tomato leaf mold resistance Cf gene encoding LRR-TM, 2 candidate genes were screened out, XM_004240667.3 and XM_010323727.1.
Discussion
In recent years, tomato leaf mold has become probably the disease that damage tomato production the most. An effective way to control the disease is to breed resistance to leaf mold. However, some tomato leaf mold resistance (Cf series) genes used available hybrids using currently in farms have gradually lost resistance, due to the continuous and rapid differentiation of C. fulvum physiological races. It was found that the application of the Cf-4 gene in production was overcome by C. fulvum physiological races such as 1.2.4 and 1.2.3.4. Subsequently, the application of Cf-5 and Cf-9, which are commonly used in breeding, was gradually limited by the appearance and identification of physiological races 1.2.3.4.5, 1.2.3.4.9, 2.5, and 2.4.5 (Li et al., 2015; Zhao et al., 2016). It is crucial to understand the differentiation of C. fulvum physiological races and to characterize the resistance range of new Cf genes before undertaking breeding efforts for disease resistance. The Cf-16 gene derived from the resistant material “Ontario7816” has shown effective resistance in field trials for many years. This study shows that “Ontario7816” has highly resistant to all the identified physiological races of C. fulvum. This result also indicates that this gene is a valuable resource against C. fulvum breeding. Therefore, mapping, cloning, and characterization of this gene may speed up its use in marker-assisted selection for resistance breeding and may have critical implications for our understanding of the resistance mechanism against tomato leaf mold disease.
Previous studies have shown that most Cf genes are inherited dominant single genes. In this study, the genetic analysis indicated that the segregation ratios between resistant and susceptible plants in both the F2 and BC1P2 populations were in accordance with Mendel’s law of segregation. The present results clarify that the Cf-16 gene confers resistance via a single dominant gene. At the same time, it also paves the way for the next localization of Cf-16.
In this study, we mapped the Cf-16 gene for the first time into a 2.6 cM region on tomato chromosome 6 between two markers, TGS447 and TES312, using BSA combined with parental resequencing and SSR molecular markers. By functional annotation and structural analysis of the genes in the candidate region, we ultimately predicted two candidate genes, XM_004240667.3 (LRR receptor-like serine-/threonine-protein kinase At1g51880) and XM_010323727.1 (LRR receptor-like serine-/threonine-protein kinase RFK1 isoform X1. LRR receptor-like serine-/threonine-protein kinase), the largest class of receptor-like protein kinase (RLK), has an LRR structure and transmembrane region composed of conserved amino acid sequences (Man et al., 2020). The protein structures encoded by the cloned Cf genes Cf-2, Cf-4, Cf-5, and Cf-9 all have the typical extracellular LRR domain, transmembrane domain and a short cytoplasmic domain, and the different numbers of LRRs are responsible for different Cf genes recognizing different pathogen physiological races (Kruijt et al., 2005). The two genes that were ultimately predicted in this study may have similar structures to Cf genes that have been cloned, so they are identified as candidate genes for Cf-16. However, it is essential to conduct expression pattern analysis and further functional verification of these two candidate genes.
Conclusion
In this study, we mapped the Cf-16 gene for the first time into a 2.6 cM region on tomato chromosome 6 between two markers, TGS447 and TES312, using F2 bulked segregant analysis combined with genome resequencing and SSR molecular markers. Furthermore, we screened 2 possible candidate genes, XM_004240667.3 and XM_010323727.1, with LRR-TM structures similar to those of the cloned Cf genes according to their annotation information. The location and candidate gene screening of Cf-16 could lay a robust foundation for later cloning of the Cf-16 gene and applications in MAS selection programs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra PRJNA937877.
Author contributions
XX, GF, and DZ designed the research. DZ and HL conducted the experiments. DZ, HL, and GL conducted data curation and formal analysis. XX, GF, and LX revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Postdoctoral Program Foundation of Heilongjiang Province (LBH-Z21026), the Basic Scientific Research Foundation of Heilongjiang Provincial Universities (HDRCCX-2020-KYYWF-1047) and the Natural Science Outstanding Youth Foundation of Heilongjiang Province (YQ 2020C038).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1219898/full#supplementary-material
References
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi:10.1038/nprot.2006.384
Enya, J., Ikeda, K., Takeuchi, T., Horikoshi, N., Higashi, T., Sakai, T., et al. (2009). The first occurrence of leaf mold of tomato caused by races 4.9 and 4.9.11 of Passalora fulva (syn. Fulvia fulva) in Japan. J. general plant pathology 75, 76–79. doi:10.1007/s10327-008-0134-0
Iida, Y., Iwadate, Y., Kubota, M., and Terami, F. (2010). Occurrence of a new race 2.9 of leaf mold of tomato in Japan. J. Gen. Plant Pathol. 76, 84–86. doi:10.1007/s10327-009-0207-8
Kanwar, J. S., Harney, P. M., and Kerr, E. A. (1980). Allelic relationships of genes for resistance to tomato leaf mold. Cladosporium fulvum cke. hortscience. 15, 418. doi:10.1007/BF00023230
Kim, H., Kim, B. S., Shim, J. E., Hwang, S., Yang, S., Kim, E., et al. (2017). TomatoNet: A genome-wide Co-functional Network for unveiling complex traits of tomato, a model crop for fleshy fruits. Mol. Plant 10, 652–655. doi:10.1016/j.molp.2016.11.010
Kruijt, M., De Kock, M. J., and de Wit, P. J. (2005). Receptor-like proteins involved in plant disease resistance. Mol. Plant Pathol. 6, 85–97. doi:10.1111/j.1364-3703.2004.00264.x
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. doi:10.1093/bioinformatics/btp698
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi:10.1093/bioinformatics/btp352
Li, S., Zhao, T., Li, H., Xu, X., and Li, J. (2015). First report of races 2.5 and 2.4.5 of cladosporium fulvum (syn. passalora fulva), causal fungus of tomato leaf mold disease in China. J. Gen. Plant Pathol. 81, 162–165. doi:10.1007/s10327-015-0577-z
Liu, G., Zhao, T., You, X., Jiang, J., Li, J., and Xu, X. (2019). Molecular mapping of the Cf-10 gene by combining SNP/InDel-index and linkage analysis in tomato (Solanum lycopersicum). BMC Plant Biol. 19, 15. doi:10.1186/s12870-018-1616-7
Man, J., Gallagher, J. P., and Bartlett, M. (2020). Structural evolution drives diversification of the large LRR-RLK gene family. New Phytol. 226, 1492–1505. doi:10.1111/nph.16455
Mesarich, C. H., Griffiths, S. A., van der Burgt, A., Okmen, B., Beenen, H. G., Etalo, D. W., et al. (2014). Transcriptome sequencing uncovers the Avr5 avirulence gene of the tomato leaf mold pathogen Cladosporium fulvum. Mol. Plant Microbe Interact. 27, 846–857. doi:10.1094/MPMI-02-14-0050-R
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U. S. A. 88, 9828–9832. doi:10.1073/pnas.88.21.9828
Ooijen, J., and Van, J. W. (2006). JoinMap 4, Software for the calculation of genetic linkage maps inexperimental populations. Wageningen, Netherlands: Kyazma B.V.
Rivas, S., and Thomas, C. M. (2005). Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu. Rev. Phytopathol. 43, 395–436. doi:10.1146/annurev.phyto.43.040204.140224
Rui, R., Liu, S., Karthikeyan, A., Wang, T., Niu, H., Yin, J., et al. (2017). Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to Soybean mosaic virus. Theor. Appl. Genet. 130, 2395–2410. doi:10.1007/s00122-017-2966-5
Thomma, B. P., Van Esse, H. P., Crous, P. W., and De Wit, P. J. (2005). Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 6, 379–393. doi:10.1111/j.1364-3703.2005.00292.x
Xue, D. Q., Chen, X. L., Zhang, H., Chai, X. F., Jiang, J. B., Xu, X. Y., et al. (2017). Transcriptome analysis of the Cf-12-Mediated resistance response to Cladosporium fulvum in tomato. Front. Plant Sci. 7, 2012. doi:10.3389/fpls.2016.02012
Yang, H., Wang, H., Jiang, J., Liu, M., Liu, Z., Tan, Y., et al. (2022). The Sm gene conferring resistance to gray leaf spot disease encodes an NBS-LRR (nucleotide-binding site-leucine-rich repeat) plant resistance protein in tomato. Theor. Appl. Genet. 135, 1467–1476. doi:10.1007/s00122-022-04047-6
Yang, H., Zhao, T., Jiang, J., Wang, S., Wang, A., Li, J., et al. (2017). Mapping and screening of the tomato Stemphylium lycopersici resistance gene, Sm, based on bulked segregant analysis in combination with genome resequencing. BMC Plant Biol. 17, 266. doi:10.1186/s12870-017-1215-z
Zhao, T., Jiang, J., Liu, G., He, S., Zhang, H., Chen, X., et al. (2016). Mapping and candidate gene screening of tomato Cladosporium fulvum-resistant gene Cf-19, based on high-throughput sequencing technology. BMC Plant Biol. 16, 51. doi:10.1186/s12870-016-0737-0
Zhao, T., Liu, W., Zhao, Z., Yang, H., Bao, Y., Zhang, D., et al. (2019). Transcriptome profiling reveals the response process of tomato carrying Cf-19 and Cladosporium fulvum interaction. BMC Plant Biol. 19, 572. doi:10.1186/s12870-019-2150-y
Zhao, T., Pei, T., Jiang, J., Yang, H., Zhang, H., Li, J., et al. (2022). Understanding the mechanisms of resistance to tomato leaf mold: A review. Hortic. Plant J. 6, 667–675. doi:10.1016/j.hpj.2022.04.008
Keywords: tomato, mapping, Cladosporium fulvum, resistance gene Cf-16, BSA, SSR markers
Citation: Zhang D, Li H, Liu G, Xie L, Feng G and Xu X (2023) Mapping of the Cladosporium fulvum resistance gene Cf-16, a major gene involved in leaf mold disease in tomato. Front. Genet. 14:1219898. doi: 10.3389/fgene.2023.1219898
Received: 09 May 2023; Accepted: 18 July 2023;
Published: 27 July 2023.
Edited by:
Zefeng Yang, Yangzhou University, ChinaReviewed by:
Junliang Yin, Yangtze University, ChinaPedro Alberto Balatti, National University of La Plata, Argentina
Copyright © 2023 Zhang, Li, Liu, Xie, Feng and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guojun Feng, ZmVuZ2d1b2p1bkBobGp1LmVkdS5jbg==; Xiangyang Xu, eHh5NzA5QDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
 Dongye Zhang
Dongye Zhang Huijia Li2†
Huijia Li2† Guan Liu
Guan Liu Xiangyang Xu
Xiangyang Xu