- 1Murdoch Children’s Research Institute, Melbourne, VIC, Australia
- 2Australian Genomics, Melbourne, VIC, Australia
- 3Department of Paediatrics, University of Melbourne, Parkville, VIC, Australia
- 4Melbourne Law School, University of Melbourne, Parkville, VIC, Australia
Background: Newborn bloodspot screening (NBS) programs have improved neonatal healthcare since the 1960s. Genomic sequencing now offers potential to generate polygenic risk score (PRS) that could be incorporated into NBS programs, shifting the focus from treatment to prevention of future noncommunicable disease (NCD). However, Australian parents’ knowledge and attitudes regarding PRS for NBS is currently unknown.
Methods: Parents with at least one Australian-born child under 18 years were invited via social media platforms to complete an online questionnaire aimed at examining parents’ knowledge of NCDs, PRS, and precision medicine, their opinions on receiving PRS for their child, and considerations of early-intervention strategies to prevent the onset of disease.
Results: Of 126 participants, 90.5% had heard the term “non-communicable disease or chronic condition,” but only 31.8% and 34.4% were aware of the terms “polygenic risk score” and “precision medicine” respectively. A large proportion of participants said they would consider screening their newborn to receive a PRS for allergies (77.9%), asthma (81.0%), cancer (64.8%), cardiovascular disease (65.7%), mental illness (56.7%), obesity (49.5%), and type 2 diabetes (66.7%). Additionally, participants would primarily consider diet and exercise as interventions for specific NCDs.
Discussion: The results from this study will inform future policy for genomic NBS, including expected rate of uptake and interventions that parents would consider employing to prevent the onset of disease.
Introduction
Newborn Bloodspot Screening (NBS) programs have been an essential part of neonatal healthcare globally for more than 50 years (Guthrie and Susi, 1963; Woerner et al., 2021). As of 2015, NBS programs globally can screen for up to 50 conditions, commonly metabolic and/or genetic in nature (Therrell et al., 2015). In Australia, NBS programs employ tandem mass spectrometry as the standard screening approach (Metternick-Jones et al., 2015), with an average uptake rate of over 99% (Australian Government Department of Health and Aged Care, 2023; White et al., 2023). However, newer technologies such as genomic sequencing may be a feasible option for screening in the future as their reliability increases and costs decrease (Preston et al., 2021). Genomic sequencing technologies–encompassing exome and genome sequencing–can sequence a large number of genes implicated in health in parallel (Behjati and Tarpey, 2013), making them the logical next step in such population screening programs.
In cases requiring a diagnosis for critically ill infants, genomic sequencing has resulted in higher diagnostic rates than traditional clinical investigations (Donoghue et al., 2017; Farnaes et al., 2018; Stark et al., 2018). However, sequencing healthy newborns (rather than those already unwell) on a population-wide scale shifts the focus from diagnosis to risk assessment, and from treatment to prevention of onset of conditions. Additionally, sequencing a newborn’s genome can support a precision medicine approach. Precision medicine takes into account various genetic variations and environmental factors of subpopulations to facilitate more effective disease management, and is an alternative to the “one-size-fits-all” approach commonly used in therapeutic medicine (Gameiro et al., 2018).
Many studies globally have assessed the feasibility of introducing genomic sequencing into NBS programs–either as a first-line test or as a follow-up test after initial screening–and have stated its potential clinical utility for a range of metabolic and genetic conditions (Bodian et al., 2016; Park et al., 2016; Smon et al., 2018; Holm et al., 2019; van Campen et al., 2019). However, the feasibility of utilising genomic sequencing in NBS programs to screen for risk of non-communicable diseases (NCDs) has not been investigated. NCDs such as cancer, cardiovascular disease, diabetes, and chronic respiratory conditions, contribute to as much as 74% of deaths worldwide (World Health Organisation, 2022). Most NCDs are characterised as multifactorial or complex diseases, meaning many genes, as well as certain environmental factors such as diet, physical activity, and other socioeconomic factors, can contribute to the onset of disease (Chatterjee et al., 2016). Many studies have shown that early life events can impact an individual’s risk of developing various NCDs. For example, stressors such as malnutrition, physical trauma, air pollution, and an increasing sedentary lifestyle in early childhood can lead to a higher prevalence of NCDs in adulthood (Merrick et al., 2019; Rodriguez-Ayllon et al., 2019; Jaddoe et al., 2020; Nelson et al., 2020). Aside from environmental factors, genetic variants associated with various NCDs continue to be identified through genome-wide association studies (GWAS; Ejtahed et al., 2018; Goodarzi, 2018; Xue et al., 2018), demonstrating that some individuals may have a higher genetic predisposition to developing NCDs compared to others who do not have these associated variants. Implementing preventative measures from early in life may reduce the burden of NCDs in the future (Balbus et al., 2013; Ronto et al., 2018; Juan and Yang, 2020), with additional studies suggesting that early health interventions for children, such as improving diet, increasing physical activity, and reducing stress, can reduce an individual’s risk of developing NCDs (Rodriguez-Ayllon et al., 2019; Jacob et al., 2021; Curioni et al., 2022; Lioret et al., 2023).
Nonetheless, the feasibility of utilizing genomic sequencing on a population-wide level to screen for common NCDs remains a contentious debate. Decisions about which conditions should be included in NBS programs are generally guided by the Wilson and Junger criteria (Wilson et al., 1968) which, among other recommendations, suggest that a condition should only be added to a screening program if it is clinically actionable (Wilson et al., 1968; Johnston et al., 2018). While recent research has suggested the potential clinical and personal utility of genomic NBS (Downie et al., 2021; Armstrong et al., 2022), ethical concerns, such as conflicting interpretations of a child’s best interest (Ross and Clayton, 2019), remain around returning risk information about adult-onset conditions when sequencing children. Some countries, including Australia, have published guidelines for genetic and genomic testing that do not generally recommend testing children and/or disclosing risk information for adult-onset conditions in childhood (Botkin et al., 2015; Vears et al., 2020; Moore and Richer, 2022). However, the introduction of polygenic risk scores to predict an individual’s likelihood of developing certain NCDs in adulthood, could circumvent these guidelines by enabling families to adopt early intervention strategies to reduce their child’s risk of developing NCDs later in life (Khera et al., 2018).
Polygenic risk scores (PRS) are considered to be unchanging across an individual’s lifetime, and may therefore be used to estimate an individual’s lifetime genetic risk of disease. The prevailing view is that early identification of those of highest genetic risk, enables effective targeting of limited resources for disease monitoring, intervention and even prevention. However, for most common conditions, the current discriminative ability is low and the prognostic utility for most conditions remains unclear (Lewis and Vassos, 2020). It is likely that clinical implementation of PRS will be most useful where there is an established approach for intervention, either to treat a condition or even prevent its occurrence. The use of genetic screening in this regard is well established in adult cancers (Antoniou et al., 2008) and is further enhanced through a PRS approach (Mavaddat et al., 2019). However, the utility of PRS in newborn screening remains unclear, as it is likely highly dependent on severity of the disease of interest, age-of-onset, and various environmental and socioeconomic factors. Despite this, PRS-based pre-implantation genetic screening is already available internationally (LifeView, 2023; Virginia Center for Reproductive Medicine, 2023) and newborn PRS screening is being explored in several settings, particularly for type 1 diabetes (Winkler et al., 2019; Sims et al., 2022), giving rise to the possibility of utilising this sort of technology in clinical practice.
Research indicates that Australians have expressed some interest in health-related genomic testing in adulthood (Metcalfe et al., 2018; Metcalfe et al., 2019; Savard et al., 2019). Additionally, studies have shown that Australian parents offered genomic NBS have expressed interest in receiving broader findings relating to the health of their child (Downie et al., 2020). However, despite parents being proxy decision makers for their children, Australian parents’ perceptions of genomic NBS for NCDs have not been explicitly investigated. In this study, we therefore aimed to examine Australian parents’ knowledge and awareness of key concepts such as NCDs, PRS, and precision medicine. Furthermore, we aim to identify the proportion of Australian parents who would consider utilising genomic NBS to predict their child’s susceptibility of developing common NCDs. Finally, we explored the types of intervention strategies that Australian parents would consider in childhood to prevent the onset of these NCDs.
Materials and methods
Questionnaire design
The online questionnaire comprised five sections: demographics; knowledge and awareness of NCDs and PRS; personal experience with genetic testing; family history of NCDs; and considerations of genomic NBS for NCDs. Seven NCDs were chosen for the questionnaire and included allergies, asthma, cancer, cardiovascular disease, mental illness, obesity, and type 2 diabetes. These are a subset of the most common NCDs that represent the highest burden of disease internationally, as reported by the World Health Organisation (World Health Organisation, 2022). The questionnaire was modelled on a previous study assessing public perceptions of epigenetic concepts (Lynch et al., 2022), and the questions were adapted to inform opinions of genomic NBS for NCDs. Between 29 and 57 questions were shown to participants, depending on answers provided throughout the questionnaire. The full questionnaire can be found in the Supplementary Material.
A draft version of the questionnaire was piloted with researchers and students with a background in genetics, who provided feedback on questionnaire content, layout, and readability. Pilot responses were not included in the final analysis.
Recruitment
Individuals over the age of 18 who were parents of at least one Australian-born child under the age of 18, who could be reached via social media, and were able to read and understand written English, were eligible to participate in this study. Participants were recruited via an advertisement on various social media pages including the Facebook and Twitter pages of the Murdoch Children’s Research Institute, and online parenting forums. The study team chose to exclude non-parents from this study, and parents whose children were over the age of 18, as they were unlikely to have had participated in recent NBS programs. Eligibility was determined by a set of screening questions at the beginning of the questionnaire. Participation in the questionnaire was anonymous.
Data collection and analysis
The questionnaire was developed and administered using REDCap electronic data capture tools (RRID:SCR_003445; Harris et al., 2009). Responses were collected from July 31st to 23 September 2019. Responses that were incomplete beyond the screening questions were excluded from the final analysis.
Data were analysed using STATA version 14 (RRID:SCR_012763; Statacorp, 2015). Demographic and categorical data underwent basic descriptive analyses to obtain frequencies and percentages of responses. Participant postcodes were re-coded in STATA to their Index of Relative Socio-economic Advantage and Disadvantage (IRSAD) percentage and then grouped into quartiles to reflect their overall socioeconomic ratings as determined by average household income`, economic resources`, education`, and employment opportunities according to the Socio-Economic Index for Areas (SEIFA; Australian Bureau of Statistics, 2018). For all personal and family history questions for NCDs or a genetic condition, participants who had reported a personal or familial diagnosis of an NCD or genetic condition were re-coded as having a family history for the respective condition. Participants with no personal or familial diagnosis, or an unknown diagnostic history, were re-coded as having no diagnostic history of the condition. Responses regarding prior awareness and experience of various genetic tests were similarly categorised as “heard of the test and have had it”, “heard of the test but not had it”, and “not heard of this test”. Responses which reported having genetic testing comprised of participants who stated that either themselves or their child had undergone the test. Any “prefer not to say” responses for each question were excluded from analysis. For the purposes of statistical analysis, responses regarding education, marital status, yearly income, and employment status were collapsed into fewer categories.
Two-tailed z-tests of proportion were undertaken to analyse differences in awareness of NCDs, PRS, and precision medicine. Chi-squared tests of independence were performed to investigate any associations between genomic NBS preferences for each NCD and demographic groups. Statistical significance was defined as p < 0.05.
Results
Participant demographics
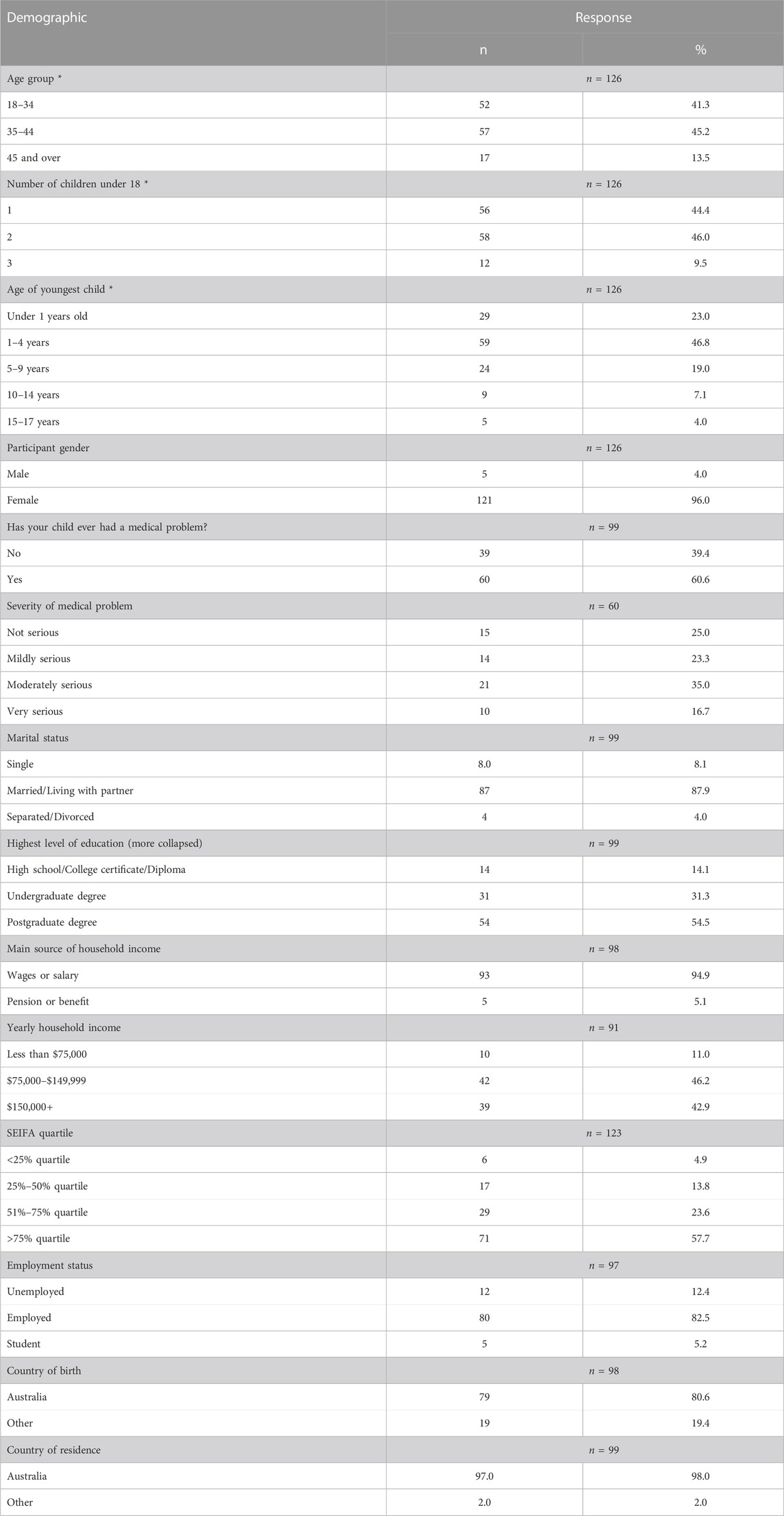
The questionnaire received a total of 145 responses. Of those, 19 responses were discarded due to incompleteness, leaving 126 responses for analysis.
The majority of participants identified as female (n = 121, 96.0%). The largest proportion of participants were aged between 35 and 44 (n = 57, 45.2%), and the mean age of participants was 37.1 years old. The mean number of children under 18 years of age was 1.7, and the average age of participants’ youngest child was 3.6 years, with the largest proportion of youngest children between 1 and 4 years of age (n = 59, 46.8%). Sixty participants stated that at least one of their children had a medical problem (n = 60, 60.6%), with 21 indicating that this was “moderately serious” (35.0%).
Most participants indicated a postgraduate degree was their highest form of education (n = 54, 54.5%) and were employed (n = 80, 82.5%). Most were married or living with their partner (n = 87, 87.9%) and the largest proportion of participants had a yearly household income of between AUD$75,000-$149,999 (n = 42, 46.2%), with the main source of income for coming from wages or a salary (n = 93, 94.9%). The majority of participants resided in postal areas within the highest SEIFA quartile (n = 71, 57.7%).
Most participants were born in Australia (n = 79, 79.8%) and currently resided in the country (n = 97, 98.0%). Participant demographics are summarised in Table 1.
Participants’ awareness of non-communicable diseases, polygenic risk scores and precision medicine
The first section of the questionnaire examined the level of awareness amongst participants of three terms chosen by the study team that reflected the rationale of genomic NBS for NCDs: “non-communicable condition or chronic condition”, “polygenic risk score”, and “precision medicine”. The term “non-communicable disease or chronic condition” was more well-known amongst participants (n = 114, 90.5%) compared to “polygenic risk score” (n = 40, 31.1%; z = 9.56, p < 0.001) and “precision medicine” (n = 43, 33.9%; z = 9.18, p < 0.001).
Participants’ prior experience with genetic testing
Participants were asked about whether they had heard of various genetic tests, and to recount prior experience with these tests either for themselves or for their child (Supplementary Figure S1). Genetic testing during pregnancy was the most well-recognised test as 98.3% (n = 118) of participants had heard of the genetic test, and a further 66.7% (n = 80) of participants or their children had undertaken the test. Pharmacogenomic testing was the least-known test amongst participants, with only 49.6% (n = 59) of participants having heard of it, and only one participant (0.8%) stated that they or their children had had a pharmacogenomic test.
Participants’ personal or familial history of non-communicable diseases or genetic conditions
Participants were asked to recall any personal or family medical history relating to genetic conditions or common NCDs (allergies, asthma, cancer, cardiovascular disease, mental illness, obesity, type 2 diabetes). The most reported condition that participants or their relatives had received a diagnosis for was cancer (n = 76, 66.7%). This was closely followed by allergies (n = 72, 64.9%) and asthma (n = 72, 63.7%). Obesity had the lowest reported diagnostic history, with 28.8% of participants (n = 32) reporting a personal or familial diagnosis. Responses are outlined in Figure 1.
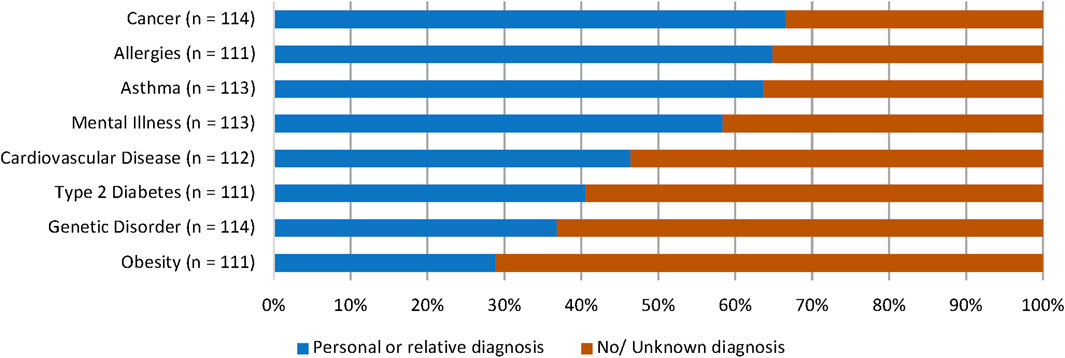
FIGURE 1. Participants’ personal or familial diagnostic history of NCDs and genetic conditions. Most participants stated that they or a relative had received a diagnosis for cancer (66.7%), allergies (64.9%), asthma (63.7%), and mental illness (58.4%).
Consideration of genomic newborn screening for common non-communicable diseases and implementation of health interventions
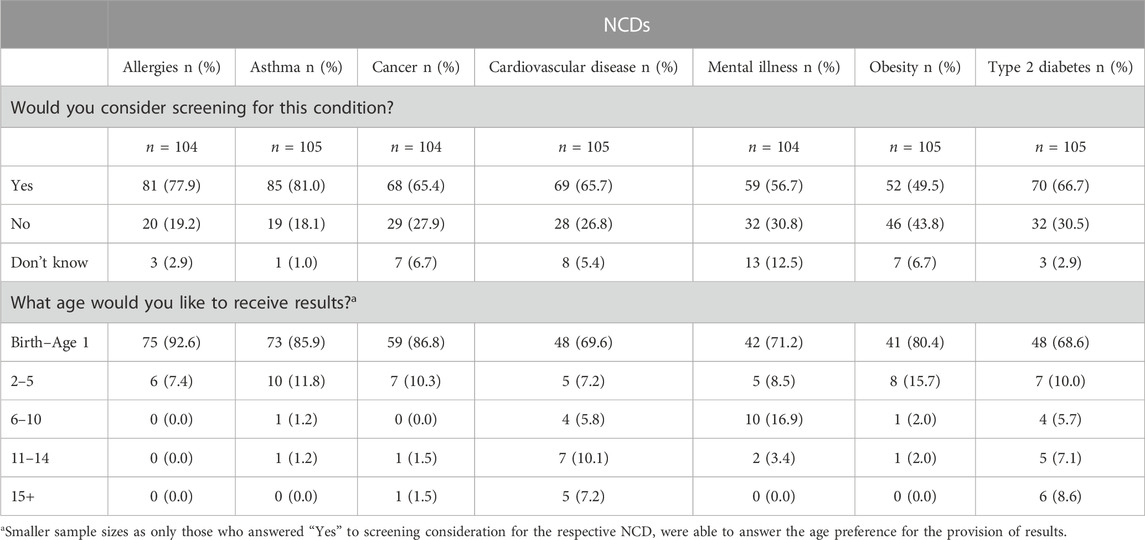
The third section of the questionnaire asked participants if they would consider genomic NBS for their children to determine their child’s risk of developing seven common NCDs. The largest proportion of participants indicated that they would consider genomic NBS for their child for each respective NCD. Participants’ preferences for genomic NBS across all NCDs are summarised in Table 2.
Participants who said they would consider genomic NBS for the NCDs presented were then asked about the age at which they would like to receive their child’s result for each specific condition (Table 2). Across all NCDs, most participants preferred to receive results between birth and age 1, irrespective of each condition’s indicative age of onset (as provided to participants in the survey question).
For each NCD, participants were asked who they would want to discuss the screening test with prior to ordering it, as well as who they would want to discuss the results with once received (Figure 2). Participants primarily chose to discuss both pre- and post-test information with a General Practitioner (GP) for asthma (n = 46, 54.8% and n = 50, 59.5% respectively), cardiovascular disease (n = 38, 55.9%; n = 37, 54.4%), mental illness (n = 31, 53.4%; n = 28, 48.3%), obesity (n = 29, 58.0%; n = 28, 56.0%), and type 2 diabetes (n = 40, 58.0%; n = 40, 58.0%). For allergies, pre-test discussions were also preferred to be held with a GP (n = 46, 57.5%), but most participants preferred post-test discussions to be held with a paediatrician (n = 46, 57.5%). For cancer, pre-test discussions were preferred to be held with a GP (n = 35, 51.5%), however for post-test discussions, participants equally preferred to speak with a GP or a paediatrician (n = 33, 48.5% each). One participant chose “other” for all NCDs when asked who to discuss the post-test results with and explained in a free-text comment that their choice depended on the severity of the NCD.

FIGURE 2. Who participants would prefer to discuss a genomic newborn screening test with (A) prior to ordering it and (B) who they would discuss results with post-screening test. Participants were able to select more than one answer for each condition, therefore the sum of answer frequencies for each NCD equate to over 100%.
At the end of this section of the questionnaire, participants had the opportunity to specify what common interventions they may employ to prevent the onset of each NCD for their children (Figure 3). Increased physical activity was the most common intervention chosen for obesity (n = 86, 89.6%), type 2 diabetes (n = 74, 78.7%), cardiovascular disease (n = 72, 83.7%), cancer (n = 48, 57.1%) and asthma (n = 47, 56.6%). Most participants chose mindfulness and meditation as an intervention for mental illness (n = 77, 82.8%), and a strict prescribed diet was the most commonly intervention strategy for allergies (n = 52, 58.4%).
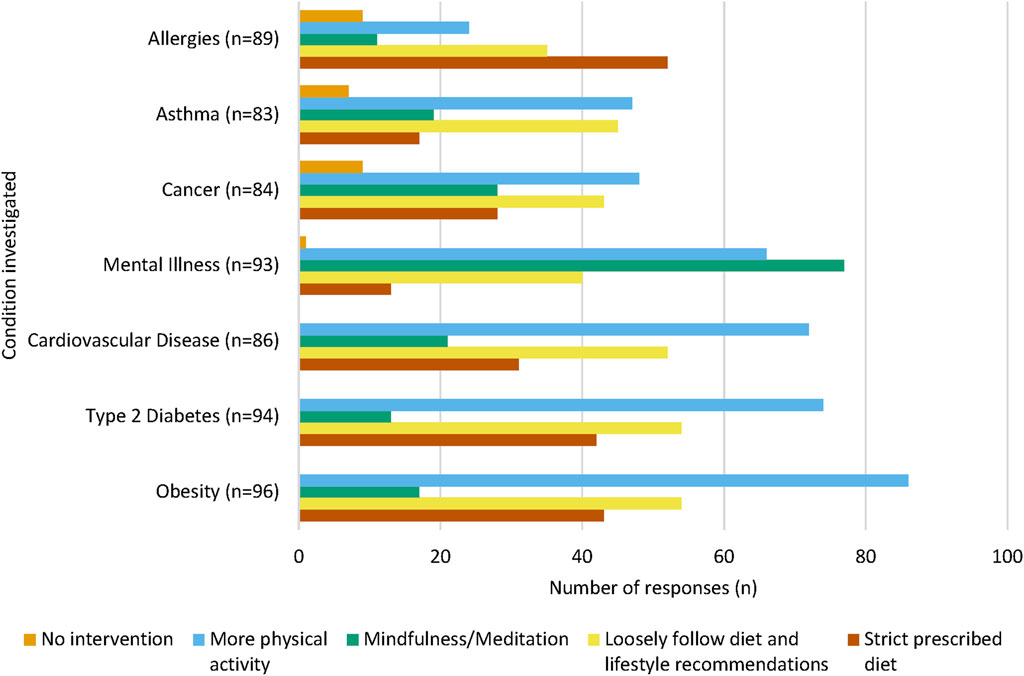
FIGURE 3. Intervention methods chosen for each NCD. For each NCD, a large proportion of participants chose “More physical activity” as a proposed intervention method. “Mindfulness and meditation” were chosen by the majority of participants to prevent the onset of mental illness. Participants could select multiple interventions for each condition, therefore the sum of each intervention per NCD will equate to over 100%.
Effect of participant characteristics on genomic newborn screening preferences for non-communicable diseases
Participants’ personal or familial history of each NCD were not evidenced to influence their preference of genomic NBS for the respective condition (Supplementary Table S1).
Whether or not participants had a child who experienced a medical problem did not show evidence of an effect on their screening preference for any of the NCDs except cancer [(χ2 (2) = 6.388, p = 0.041] (Supplementary Table S2), where parents with children who had not experienced a medical problem were more likely to prefer screening for cancer.
Participants’ age showed evidence of an effect on their preferences for screening for allergies [χ2 (4) = 11.485, p = 0.022], asthma [χ2 (4) = 15.929, p = 0.003], cancer [χ2 (4) = 10.382, p = 0.034], mental illness [χ2 (4) = 10.958, p = 0.027], and type 2 diabetes [χ2 (4) = 15.942, p = 0.003], but did not seem to influence their preferences for screening for cardiovascular disease [χ2 (4) = 9.480, p = 0.050] and obesity [χ2 (4) = 1.837, p = 0.766] (Supplementary Table S3).
Participants’ education showed evidence of influencing their screening questions for allergies [χ2 (4) = 12.473, p = 0.014], asthma [χ2 (2) = 10.736, p = 0.005], cancer [χ2 (4) = 14.076, p = 0.007], obesity [χ2 (4) = 10.938, p = 0.027], and type 2 diabetes [χ2 (4) = 9.568, p = 0.048] but did not show evidence of having an effect on their screening decisions for cardiovascular disease [χ2 (4) = 5.387, p = 0.250], and mental illness [χ2 (4) = 8.101, p = 0.088]. (Supplementary Table S4).
Participants’ SEIFA score based on their residential postcode did not show evidence of influencing their genomic NBS preferences for all NCDs investigated (Supplementary Table S5).
Discussion
This is the first study to explore Australian parents’ perceptions of genomic NBS, and their awareness of polygenic risk and precision medicine in the context of the NCDs presented. We found that most parents had previously heard about NCDs, but fewer knew about polygenic risk and precision medicine. However, despite the lack of awareness, most parents would consider genomic NBS to provide a PRS at birth for seven common NCDs if it were offered in addition to the current NBS program. Furthermore, the data highlights that parents would prefer to know their child’s risk of developing common NCDs earlier in their child’s life rather than later and would consider undertaking early intervention strategies to reduce their child’s risk of developing NCDs, therefore highlighting the personal and clinical utility of such a screening test.
Parents know about non-communicable diseases, but not polygenic risk scores or precision medicine
Most participants expressed familiarity with the concept of NCDs, which is comparable to research investigating public awareness and experiences with common NCDs including cancer, diabetes, obesity, and heart disease (Waters et al., 2014). In contrast, participants were largely unaware of PRS and the concept of “precision medicine”, which aligns with the lack of awareness and experience of health-related genomic testing, where PRS and precision medicine concepts may be more commonly discussed. The latter is consistent with a previous Australian study, showing that fewer individuals had undergone genomic tests (such as nutrigenomics and pharmacogenomics tests) compared to more common tests such as genetic carrier screening (Savard et al., 2019).
Parents value genomic newborn screening
This study showed that a large proportion of participants were in favour of expanding the current NBS program to include seven proposed common NCDs: allergies, asthma, cancer, cardiovascular disease, mental illness, obesity, and type 2 diabetes. Additionally, we found that parents would prefer to know about their child’s risk of developing NCDs earlier in life rather than later, irrespective of the average age of onset for the NCDs investigated.
There is conflicting literature examining parental attitudes around receiving genomic test results for children. Some literature suggests parents want to know their child’s risk of developing a disease in both childhood and adulthood (Tercyak et al., 2011; Kulchak Rahm et al., 2018; Holm et al., 2019; Armstrong et al., 2022) as it is in the child’s–and the family’s–best interest to begin clinical management as early as possible. On the other hand, other studies have demonstrated that parents are hesitant to know about genomic results for later-onset conditions (Sapp et al., 2014; Anderson et al., 2017), with the most common justification being that this type of information could cause additional worry and distress for the family. Nevertheless, our study suggests that parents still find value in genomic NBS for NCDs, highlighting their beliefs of personal utility of this kind of testing, irrespective of the level of information, whether perceived or actual, that a PRS could provide about a child’s health. We found that parents were willing to have their child undergo genomic NBS even if they stated that they had not previously heard of precision medicine or polygenic risk, suggesting that a lack of awareness around the nuances of utilising genomic sequencing to calculate polygenic risk could potentially drive overenthusiasm for its implementation into clinical practice. The notion of the general public’s enthusiasm for genomic sequencing is supported by a similar study, where parents hypothetically opted for genomic newborn sequencing due to the belief that newer sequencing technologies would be more accurate than traditional testing (Dodson et al., 2015). On the other hand, simply expressing interest in genomic NBS to obtain knowledge of their child’s risk status may not reflect the rate of actual participation in an expanded program (Genetti et al., 2018). Further research will be required prior to the implementation of such a program into the Australian healthcare system to investigate any potential discrepancy in anticipated versus actual uptake of genomic NBS.
Furthermore, while research has highlighted the clinical utility of PRS to assess an individual’s genetic predisposition to certain NCDs (Torkamani et al., 2018; Lambert et al., 2019), the observed lack of knowledge around PRS in this study suggests individuals may find PRS information difficult to understand (Naik et al., 2012; Folkersen et al., 2020). How the concept of PRS is communicated to patients will be crucial when considering their implementation. Information regarding the rationale and process of genomic NBS could be given to prospective parents in a similar fashion to the provision of current NBS information. Specifically in Victoria, Australia, parents are not contacted if their child’s screening results are normal, whereas positive screens are followed up immediately with the parents and hospitals (Victorian Government Department of Health and Human Services, 2018). However, further genomics education may be warranted for a parental audience prior to offering any genomic NBS. Additional research may also be required to assess the acceptability of genomic NBS more generally both before and after providing relevant information about the implications of PRS and precision medicine in the neonatal period for later-onset NCDs.
Influence of perceived risk and severity of non-communicable diseases on parents’ genomic newborn screening preferences
Participants’ opinions of genomic NBS for the seven proposed NCDs in our study did not appear to be significantly influenced by any personal or family medical history of each NCD respectively. This contrasts with previous literature showing that having a family history of a condition can impact parents’ decisions to consider NBS for that condition (Lipstein et al., 2010). The influence of personal or family history of a health condition on an individual’s preference for screening for the condition is explained by the Health Belief Model, which theorises that decision making in healthcare is determined by an individual’s perceived susceptibility and severity of a condition; perceived benefits of a health intervention (such as genomic screening); and perceived barriers to undertaking health interventions (Glanz et al., 2008). These factors combined may provide adequate cues to action that prompt an individual to seek out health interventions (Glanz et al., 2008). Despite this, our findings suggest that an individual’s perceived risk of disease did not significantly influence their decisions to consider genomic NBS for their children. Rather, other factors, not explored in this study, such as perceived severity of disease and perceived benefits of undergoing screening, may have an influence on participants’ considerations for screening.
Perceived severity of disease may also influence an individual’s motivation to adopt early intervention strategies and can therefore inform the development of policies and public health programs to promote healthy lifestyle choices (Azadi et al., 2021). Research has highlighted the importance of NCD prevention and early intervention, noting that consumption of high-calorie foods and a sedentary lifestyle are the two most common risk factors for obesity amongst school-aged children (Motlagh et al., 2017). In the context of polygenic risk, early health intervention strategies have been shown to reduce the risk of developing cardiovascular disease and diabetes in individuals with a higher genetic predisposition to these conditions (Wang et al., 2021). In contrast, other research has shown that genetic risk of NCDs is not necessarily the cause of adopting healthier lifestyle choices for individuals who have undertaken direct-to-consumer genomic testing (Nielsen et al., 2017), rather suggesting the act of undertaking the testing is influencing health behaviour change in individuals (Stewart et al., 2018). In accordance with the latter notion, our study found that Australian parents would largely choose diet and exercise interventions to prevent the onset of all presented NCDs for their child. The idea that the perceived benefits of testing overall are a key influence for parents rather than the actual perceived severity of each disease is important to consider when implementing genomic screening for NCDs into a NBS program. Further exploration is required to determine whether a nationwide genomic NBS program for NCDs (i.e., understanding the genetic predictors of the onset of NCDs), or other health intervention programs that don’t necessarily highlight genetic risk, would be more efficient to promote healthy lifestyle behaviours at an early childhood level.
Study limitations and future research
Our small study population consisted primarily of well-educated mothers, and many had a child with a medical condition. The questionnaire was shared across social media pages whose audience was mostly well-educated parents, likely with an interest in health research. Therefore, our findings about the attitudes towards genomic NBS are not representative of a diverse population. It is important to be aware of demographic differences amongst health beliefs when providing adequate information for expanded screening programs. For example, knowledge of NCDs and their risk factors, including genetic risk, tends to increase in more highly educated populations (Allen et al., 2017).
While the questionnaire was piloted primarily for the purposes of readability and layout, it was not piloted with a lay audience. This may further limit the transferability of the study findings to the general population. It is crucial that the attitudes of diverse populations regarding genomic newborn screening be appropriately explored before such screening is implemented (Dodson et al., 2015; Genetti et al., 2018). Additional research is needed utilising a more targeted approach to recruit diverse individuals and adequately capture the attitudes of parents from various cultural and socioeconomic backgrounds regarding genomic NBS and its potential outcomes.
Finally, this study aimed to provide an overview of the Australian public’s recognition of terms and phrases related to NBS for NCDs. The limited information in the questionnaire related to this terminology allowed us to examine the public’s superficial understanding of such terms and phrases, however it did not allow us to explore in-depth their accurate comprehension. Further research is needed to examine which terms are best understood by the public, and therefore which are best to use in public health communications.
Conclusion
This is the first study to examine Australian parental perceptions of genomic NBS for NCDs in the context of polygenic risk and precision medicine. While not representative of the general population, our findings establish a foundation for further research into the Australian public’s perceptions of polygenic risk and precision medicine in the context of NCDs, and considerations of genomic NBS should this type of testing be added to the current NBS program. Future research should focus on expanding the investigation to a more diverse population. Understanding different perspectives around the use of genomic technology in NBS programs will provide valuable information to researchers and policy makers about how to implement an expanded NBS program into the Australian public healthcare system.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by the Royal Children’s Hospital Human Research Ethics Committee. The patients/participants provided their written informed consent to participate in this study.
Author contributions
All authors contributed to the article and approved the submitted version. SC created the questionnaire, analysed the data, and drafted and revised the manuscript.
Acknowledgments
This research was completed in partial fulfilment of the requirements for SC’s Master of Genomics and Health from the University of Melbourne. The authors thank the Masters of Genomics and Health students for reviewing and piloting the original questionnaire. The authors thank all survey respondents for their participation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1209762/full#supplementary-material
References
Allen, L., Williams, J., Townsend, N., Mikkelsen, B., Roberts, N., Foster, C., et al. (2017). Socioeconomic status and non-communicable disease behavioural risk factors in low-income and lower-middle-income countries: A systematic review. Lancet Glob. Health 5, e277–e289. doi:10.1016/S2214-109X(17)30058-X
Anderson, J. A., Meyn, M. S., Shuman, C., Zlotnik Shaul, R., Mantella, L. E., Szego, M. J., et al. (2017). Parents perspectives on whole genome sequencing for their children: Qualified enthusiasm? J. Med. Ethics 43, 535–539. doi:10.1136/medethics-2016-103564
Antoniou, A. C., Cunningham, A. P., Peto, J., Evans, D. G., Lalloo, F., Narod, S. A., et al. (2008). The BOADICEA model of genetic susceptibility to breast and ovarian cancers: Updates and extensions. Br. J. Cancer 98, 1457–1466. doi:10.1038/sj.bjc.6604305
Armstrong, B., Christensen, K. D., Genetti, C. A., Parad, R. B., Robinson, J. O., Blout Zawatsky, C. L., et al. (2022). Parental attitudes toward standard newborn screening and newborn genomic sequencing: Findings from the BabySeq study. Front. Genet. 13, 867371. doi:10.3389/fgene.2022.867371
AUSTRALIAN BUREAU OF STATISTICS 2018. Socio-economic indexes for areas, In AUSTRALIAN bureau of statistics (Australia: SEIFA).
AUSTRALIAN GOVERNMENT DEPARTMENT OF HEALTH AND AGED CARE (2023). About newborn bloodspot screening. [Online]. Available: https://www.health.gov.au/our-work/newborn-bloodspot-screening/about-newborn-bloodspot-screening (Accessed June 4, 2023).
Azadi, N. A., Ziapour, A., Lebni, J. Y., Irandoost, S. F., Abbas, J., and Chaboksavar, F. (2021). The effect of education based on health belief model on promoting preventive behaviors of hypertensive disease in staff of the Iran University of Medical Sciences. Archives Public Health 79, 69. doi:10.1186/s13690-021-00594-4
Balbus, J. M., Barouki, R., Birnbaum, L. S., Etzel, R. A., Gluckman, P. D., Grandjean, P., et al. (2013). Early-life prevention of non-communicable diseases. Lancet 381, 3–4. doi:10.1016/S0140-6736(12)61609-2
Behjati, S., and Tarpey, P. S. (2013). What is next generation sequencing? Arch. Dis. Child. Educ. Pract. Ed. 98, 236–238. doi:10.1136/archdischild-2013-304340
Bodian, D. L., Klein, E., Iyer, R. K., Wong, W. S., Kothiyal, P., Stauffer, D., et al. (2016). Utility of whole-genome sequencing for detection of newborn screening disorders in a population cohort of 1,696 neonates. Genet. Med. 18, 221–230. doi:10.1038/gim.2015.111
Botkin, J. R., Belmont, J. W., Berg, J. S., Berkman, B. E., Bombard, Y., Holm, I. A., et al. (2015). Points to consider: Ethical, legal, and psychosocial implications of genetic testing in children and adolescents. Am. J. Hum. Genet. 97, 6–21. doi:10.1016/j.ajhg.2015.05.022
Chatterjee, N., Shi, J., and García-Closas, M. (2016). Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 17, 392–406. doi:10.1038/nrg.2016.27
Curioni, C. C., Da Silva, A. C. F., Da Silva Pereira, A., and Mocellin, M. C. (2022). “The role of dietary habits on development and progress of risk factors of chronic non-communicable diseases,” in Healthy lifestyle: From pediatrics to geriatrics. Editor R. KELISHADI (Cham: Springer International Publishing).
Dodson, D. S., Goldenberg, A. J., Davis, M. M., Singer, D. C., and Tarini, B. A. (2015). Parent and public interest in whole-genome sequencing. Public Health Genomics 18, 151–159. doi:10.1159/000375115
Donoghue, S., Downie, L., and Stutterd, C. (2017). Advances in genomic testing. Aust. Fam. Physician 46, 200–205.
Downie, L., Halliday, J., Lewis, S., and Amor, D. J. (2021). Principles of genomic newborn screening programs: A systematic review. JAMA Netw. Open 4, e2114336. doi:10.1001/jamanetworkopen.2021.14336
Downie, L., Halliday, J., Lewis, S., Lunke, S., Lynch, E., Martyn, M., et al. (2020). Exome sequencing in newborns with congenital deafness as a model for genomic newborn screening: The baby beyond hearing project. Genet. Med. 22, 937–944. doi:10.1038/s41436-019-0745-1
Ejtahed, H. S., Heshmat, R., Motlagh, M. E., Hasani-Ranjbar, S., Ziaodini, H., Taheri, M., et al. (2018). Association of parental obesity with cardiometabolic risk factors in their children: The CASPIAN-V study. PLoS One 13, e0193978. doi:10.1371/journal.pone.0193978
Farnaes, L., Hildreth, A., Sweeney, N. M., Clark, M. M., Chowdhury, S., Nahas, S., et al. (2018). Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. npj Genomic Med. 3, 10. doi:10.1038/s41525-018-0049-4
Folkersen, L., Pain, O., Ingason, A., Werge, T., Lewis, C. M., and Austin, J. (2020). Impute.me: An open-source, non-profit tool for using data from direct-to-consumer genetic testing to calculate and interpret polygenic risk scores. Front. Genet. 11, 578. doi:10.3389/fgene.2020.00578
Gameiro, G. R., Sinkunas, V., Liguori, G. R., and Auler-Júnior, J. O. C. (2018). Precision Medicine: Changing the way we think about healthcare. Clin. (Sao Paulo, Braz. 73, e723. doi:10.6061/clinics/2017/e723
Genetti, C. A., Schwartz, T. S., Robinson, J. O., Vannoy, G. E., Petersen, D., Pereira, S., et al. (2018). Parental interest in genomic sequencing of newborns: Enrollment experience from the BabySeq project. Genet. Med. 21, 622–630. doi:10.1038/s41436-018-0105-6
Glanz, K., Rimer, B. K., and Viswanath, K. (2008). Health behavior and health education: Theory, research, and practice. 4th ed. San Francisco, CA: Jossey-Bass.
Goodarzi, M. O. (2018). Genetics of obesity: What genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes Endocrinol. 6, 223–236. doi:10.1016/S2213-8587(17)30200-0
Guthrie, R., and Susi, A. (1963). A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics 32, 338–343. doi:10.1542/peds.32.3.338
Harris, P. A., Taylor, R., Thielke, R., Payne, J., Gonzalez, N., and Conde, J. G. (2009). Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inf. 42, 377–381. doi:10.1016/j.jbi.2008.08.010
Holm, I. A., Mcguire, A., Pereira, S., Rehm, H., Green, R. C., Beggs, A. H., et al. (2019). Returning a genomic result for an adult-onset condition to the parents of a newborn: Insights from the BabySeq project. Pediatrics 143, S37–s43. doi:10.1542/peds.2018-1099H
Jacob, C. M., Hardy-Johnson, P. L., Inskip, H. M., Morris, T., Parsons, C. M., Barrett, M., et al. (2021). A systematic review and meta-analysis of school-based interventions with health education to reduce body mass index in adolescents aged 10 to 19 years. Int. J. Behav. Nutr. Phys. Activity 18, 1. doi:10.1186/s12966-020-01065-9
Jaddoe, V. W. V., Felix, J. F., Andersen, A.-M. N., Charles, M.-A., Chatzi, L., Corpeleijn, E., et al. (2020). The LifeCycle project-EU child cohort network: A federated analysis infrastructure and harmonized data of more than 250,000 children and parents. Eur. J. Epidemiol. 35, 709–724. doi:10.1007/s10654-020-00662-z
Johnston, J., Lantos, J. D., Goldenberg, A., Chen, F., Parens, E., Koenig, B. A., et al. (2018). Sequencing newborns: A call for nuanced use of genomic technologies. Hastings Cent. Rep. 48 (2), S2–S6. doi:10.1002/hast.874
Juan, J., and Yang, H. (2020). Prevalence, prevention, and lifestyle intervention of gestational diabetes mellitus in China. Int. J. Environ. Res. Public Health 17, 9517. doi:10.3390/ijerph17249517
Khera, A. V., Chaffin, M., Aragam, K. G., Haas, M. E., Roselli, C., Choi, S. H., et al. (2018). Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 50, 1219–1224. doi:10.1038/s41588-018-0183-z
Kulchak Rahm, A., Bailey, L., Fultz, K., Fan, A., Williams, J. L., Buchanan, A., et al. (2018). Parental attitudes and expectations towards receiving genomic test results in healthy children. Transl. Behav. Med. 8, 44–53. doi:10.1093/tbm/ibx044
Lambert, S. A., Abraham, G., and Inouye, M. (2019). Towards clinical utility of polygenic risk scores. Hum. Mol. Genet. 28, R133-R142. doi:10.1093/hmg/ddz187
Lewis, C. M., and Vassos, E. (2020). Polygenic risk scores: From research tools to clinical instruments. Genome Med. 12, 44. doi:10.1186/s13073-020-00742-5
LIFEVIEW (2023). Embryo health score (PGT-P). [Online]. Available: https://lifeview.com/tests_pgtp.html (Accessed June 4, 2023).
Lioret, S., Harrar, F., Boccia, D., Hesketh, K. D., Kuswara, K., Van Baaren, C., et al. (2023). The effectiveness of interventions during the first 1,000 days to improve energy balance-related behaviors or prevent overweight/obesity in children from socio-economically disadvantaged families of high-income countries: A systematic review. Obes. Rev. 24, e13524. doi:10.1111/obr.13524
Lipstein, E. A., Nabi, E., Perrin, J. M., Luff, D., Browning, M. F., and Kuhlthau, K. A. (2010). Parents' decision-making in newborn screening: Opinions, choices, and information needs. Pediatrics 126, 696–704. doi:10.1542/peds.2010-0217
Lynch, F., Lewis, S., Macciocca, I., and Craig, J. M. (2022). Public knowledge and opinion of epigenetics and epigenetic concepts. J. Dev. Orig. Health Dis. 13, 431–440. doi:10.1017/S2040174421000520
Mavaddat, N., Michailidou, K., Dennis, J., Lush, M., Fachal, L., Lee, A., et al. (2019). Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet. 104, 21–34. doi:10.1016/j.ajhg.2018.11.002
Merrick, M. T., Ford, D. C., Ports, K. A., Guinn, A. S., Chen, J., Klevens, J., et al. (2019). Vital signs: Estimated proportion of adult health problems attributable to adverse childhood experiences and implications for prevention - 25 states, 2015-2017. MMWR Morb. Mortal. Wkly. Rep. 68, 999–1005. doi:10.15585/mmwr.mm6844e1
Metcalfe, S. A., Hickerton, C., Savard, J., Stackpoole, E., Tytherleigh, R., Tutty, E., et al. (2019). Australians’ perspectives on support around use of personal genomic testing: Findings from the Genioz study. Eur. J. Med. Genet. 62, 290–299. doi:10.1016/j.ejmg.2018.11.002
Metcalfe, S. A., Hickerton, C., Savard, J., Terrill, B., Turbitt, E., Gaff, C., et al. (2018). Australians' views on personal genomic testing: Focus group findings from the genioz study. Eur. J. Hum. Genet. 26, 1101–1112. doi:10.1038/s41431-018-0151-1
Metternick-Jones, S. C., Lister, K. J., Dawkins, H. J. S., White, C. A., and Weeramanthri, T. S. (2015). Review of current international decision-making processes for newborn screening: Lessons for Australia. Front. Public Health 3, 214. doi:10.3389/fpubh.2015.00214
Moore, A. M., and Richer, J. (2022). Genetic testing and screening in children. Paediatr. Child Health 27, 243–253. doi:10.1093/pch/pxac028
Motlagh, M. E., Ziaodini, H., Qorbani, M., Taheri, M., Aminaei, T., Goodarzi, A., et al. (2017). Methodology and early findings of the fifth survey of childhood and adolescence surveillance and prevention of adult noncommunicable disease: The CASPIAN-V study. Int. J. Prev. Med. 8, 4. doi:10.4103/2008-7802.198915
Naik, G., Ahmed, H., and Edwards, A. G. (2012). Communicating risk to patients and the public. Br. J. Gen. Pract. 62, 213–216. doi:10.3399/bjgp12X636236
Nelson, C. A., Bhutta, Z. A., Harris, N. B., Danese, A., and Samara, M. (2020). Adversity in childhood is linked to mental and physical health throughout life. bmj, 371. doi:10.1136/bmj.m3048
Nielsen, D. E., Carere, D. A., Wang, C., Roberts, J. S., Green, R. C., Green, R. C., et al. (2017). Diet and exercise changes following direct-to-consumer personal genomic testing. BMC Med. Genomics 10, 24. doi:10.1186/s12920-017-0258-1
Park, K. J., Park, S., Lee, E., Park, J. H., Park, J. H., Park, H. D., et al. (2016). A population-based genomic study of inherited metabolic diseases detected through newborn screening. Ann. Lab. Med. 36, 561–572. doi:10.3343/alm.2016.36.6.561
Preston, J., Vanzeeland, A., and Peiffer, D. (2021). Innovation at Illumina: The road to the $600 human genome. Nat. Portf.
Rodriguez-Ayllon, M., Cadenas-Sánchez, C., Estévez-López, F., Muñoz, N. E., Mora-Gonzalez, J., Migueles, J. H., et al. (2019). Role of physical activity and sedentary behavior in the mental health of preschoolers, children and adolescents: A systematic review and meta-analysis. Sports Med. 49, 1383–1410. doi:10.1007/s40279-019-01099-5
Ronto, R., Wu, J. H. Y., and Singh, G. M. (2018). The global nutrition transition: Trends, disease burdens and policy interventions. Public Health Nutr. 21, 2267–2270. doi:10.1017/S1368980018000423
Ross, L. F., and Clayton, E. W. (2019). Ethical issues in newborn sequencing research: The case study of BabySeq. Pediatrics 144, e20191031. doi:10.1542/peds.2019-1031
Sapp, J. C., Dong, D., Stark, C., Ivey, L. E., Hooker, G., Biesecker, L. G., et al. (2014). Parental attitudes, values, and beliefs toward the return of results from exome sequencing in children. Clin. Genet. 85, 120–126. doi:10.1111/cge.12254
Savard, J., Hickerton, C., Tytherleigh, R., Terrill, B., Turbitt, E., Newson, A. J., et al. (2019). Australians’ views and experience of personal genomic testing: Survey findings from the genioz study. Eur. J. Hum. Genet. 27, 711–720. doi:10.1038/s41431-018-0325-x
Sims, E. K., Besser, R. E. J., Dayan, C., Geno Rasmussen, C., Greenbaum, C., Griffin, K. J., et al. (2022). Screening for type 1 diabetes in the general population: A status report and perspective. Diabetes 71, 610–623. doi:10.2337/dbi20-0054
Smon, A., Repic Lampret, B., Groselj, U., Zerjav Tansek, M., Kovac, J., Perko, D., et al. (2018). Next generation sequencing as a follow-up test in an expanded newborn screening programme. Clin. Biochem. 52, 48–55. doi:10.1016/j.clinbiochem.2017.10.016
Stark, Z., Lunke, S., Brett, G. R., Tan, N. B., Stapleton, R., Kumble, S., et al. (2018). Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet. Med. 20, 1554–1563. doi:10.1038/gim.2018.37
STATACORP (2015). Stata statistical software: Release, 14. College Station, Texas, USA: StataCorp LP.
Stewart, K. F. J., Wesselius, A., Schreurs, M. A. C., Schols, A. M. W. J., and Zeegers, M. P. (2018). Behavioural changes, sharing behaviour and psychological responses after receiving direct-to-consumer genetic test results: A systematic review and meta-analysis. J. Community Genet. 9, 1–18. doi:10.1007/s12687-017-0310-z
Tercyak, K. P., Hensley Alford, S., Emmons, K. M., Lipkus, I. M., Wilfond, B. S., and Mcbride, C. M. (2011). Parents' attitudes toward pediatric genetic testing for common disease risk. Pediatrics 127, e1288–e1295. doi:10.1542/peds.2010-0938
Therrell, B. L., Padilla, C. D., Loeber, J. G., Kneisser, I., Saadallah, A., Borrajo, G. J., et al. (2015). Seminars in perinatology. Elsevier, 171–187.Current status of newborn screening worldwide: 2015
Torkamani, A., Wineinger, N. E., and Topol, E. J. (2018). The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590. doi:10.1038/s41576-018-0018-x
Van Campen, J. C., Sollars, E. S. A., Thomas, R. C., Bartlett, C. M., Milano, A., Parker, M. D., et al. (2019). Next generation sequencing in newborn screening in the United Kingdom national health service. Int. J. Neonatal Screen 5, 40. doi:10.3390/ijns5040040
Vears, D. F., Ayres, S., Boyle, J., Mansour, J., and Newson, A. J.Education, Ethics and Social Issues Committee of the Human Genetics Society of Australasia (2020). Human genetics society of australasia position statement: Predictive and presymptomatic genetic testing in adults and children. Twin Res. Hum. Genet. 23, 184–189. doi:10.1017/thg.2020.51
VICTORIAN GOVERNMENT DEPARTMENT OF HEALTH AND HUMAN SERVICES (2018). in Newborn bloospot screening: Policy and guidelines. Editor P. A. P. H. BRANCH (Victoria, Australia: Victorian Government Department of Health and Human Services).
VIRGINIA CENTER FOR REPRODUCTIVE MEDICINE (2023). Testing for polygenic disorders (PGT-P). [Online]. Available: https://www.vcrmed.com/genetic-screening/testing-for-polygenic-disorders-pgt-p/ (Accessed June 4, 2023).
Wang, C. A., Attia, J. R., Lye, S. J., Oddy, W. H., Beilin, L., Mori, T. A., et al. (2021). The interactions between genetics and early childhood nutrition influence adult cardiometabolic risk factors. Sci. Rep. 11, 14826. doi:10.1038/s41598-021-94206-4
Waters, E. A., Muff, J., and Hamilton, J. G. (2014). Multifactorial beliefs about the role of genetics and behavior in common health conditions: Prevalence and associations with participant characteristics and engagement in health behaviors. Genet. Med. 16, 913–921. doi:10.1038/gim.2014.49
White, S., Mossfield, T., Fleming, J., Barlow-Stewart, K., Ghedia, S., Dickson, R., et al. (2023). Expanding the Australian newborn blood spot screening program using genomic sequencing: Do we want it and are we ready? Eur. J. Hum. Genet. 31, 703–711. doi:10.1038/s41431-023-01311-1
Wilson, J. M. G., Jungner, G., and Organization, W. H. (1968). Principles and practice of screening for disease.
Winkler, C., Haupt, F., Heigermoser, M., Zapardiel-Gonzalo, J., Ohli, J., Faure, T., et al. (2019). Identification of infants with increased type 1 diabetes genetic risk for enrollment into primary prevention trials—GPPAD-02 study design and first results. Pediatr. Diabetes 20, 720–727. doi:10.1111/pedi.12870
Woerner, A. C., Gallagher, R. C., Vockley, J., and Adhikari, A. N. (2021). The use of whole genome and exome sequencing for newborn screening: Challenges and opportunities for population health. Front. Pediatr. 9, 663752. doi:10.3389/fped.2021.663752
WORLD HEALTH ORGANISATION (2022). Noncommunicable diseases. [Online]. Available: http://www.who.int/news-room/fact-sheets/detail/noncommunicable-diseases (Accessed June 4, 2023).
Keywords: genomic sequencing, newborn screening, non-communicable disease, polygenic risk score, quantitative
Citation: Casauria S, Lewis S, Lynch F and Saffery R (2023) Australian parental perceptions of genomic newborn screening for non-communicable diseases. Front. Genet. 14:1209762. doi: 10.3389/fgene.2023.1209762
Received: 21 April 2023; Accepted: 15 June 2023;
Published: 26 June 2023.
Edited by:
Manuel Corpas, University of Westminster, United KingdomReviewed by:
Sowmiya Moorthie, PHG Foundation, United KingdomAlice Virani, Provincial Health Services Authority, Canada
Copyright © 2023 Casauria, Lewis, Lynch and Saffery. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard Saffery, cmljaGFyZC5zYWZmZXJ5QG1jcmkuZWR1LmF1
 Sarah Casauria
Sarah Casauria Sharon Lewis
Sharon Lewis Fiona Lynch
Fiona Lynch Richard Saffery
Richard Saffery