- 1Department of Molecular Biology and Genetic Engineering, Bihar Agricultural University, Bhagalpur, Bihar, India
- 2Department of Plant Pathology, Bihar Agricultural University, Bhagalpur, Bihar, India
- 3Department of Seed Science and Technology, Bihar Agricultural University, Bhagalpur, Bihar, India
- 4Deputy Director Research, Bihar Agricultural University, Bhagalpur, Bihar, India
- 5Department of Horticulture, Bihar Agricultural University, Bhagalpur, Bihar, India
Genome packaging is the crucial step for maturation of plant viruses containing an RNA genome. Viruses exhibit a remarkable degree of packaging specificity, despite the probability of co-packaging cellular RNAs. Three different types of viral genome packaging systems are reported so far. The recently upgraded type I genome packaging system involves nucleation and encapsidation of RNA genomes in an energy-dependent manner, which have been observed in most of the plant RNA viruses with a smaller genome size, while type II and III packaging systems, majorly discovered in bacteriophages and large eukaryotic DNA viruses, involve genome translocation and packaging inside the prohead in an energy-dependent manner, i.e., utilizing ATP. Although ATP is essential for all three packaging systems, each machinery system employs a unique mode of ATP hydrolysis and genome packaging mechanism. Plant RNA viruses are serious threats to agricultural and horticultural crops and account for huge economic losses. Developing control strategies against plant RNA viruses requires a deep understanding of their genome assembly and packaging mechanism. On the basis of our previous studies and meticulously planned experiments, we have revealed their molecular mechanisms and proposed a hypothetical model for the type I packaging system with an emphasis on smaller plant RNA viruses. Here, in this review, we apprise researchers the technical breakthroughs that have facilitated the dissection of genome packaging and virion assembly processes in plant RNA viruses.
1 Introduction
Genome packaging and assembly is a crucial step during the life cycle of RNA viruses infecting plants (Rao, 2006; Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). Three different approaches have been reported by which viruses condense their genome inside the empty prohead (Chelikani et al., 2014a; Ranjan et al., 2021; Mohanty et al., 2023). According to the recently upgraded type I classification system present in many of the smaller plant RNA viruses of genome size <20 kb, the processes of assembly and genome packaging involve the coating of the nucleic acid with viral capsid proteins (CPs) in the presence of ATP (Ranjan et al., 2021; Mohanty et al., 2023). On the contrary, a powerful ATP-fueled packaging motor, to package the genome in already assembled proheads of larger viruses with the genome size >20 kb, is involved in both type II and III systems (Burroughs et al., 2007; Chelikani et al., 2014a; Chelikani et al., 20014b). Both type II and III packaging systems have been reasonably well studied so far, but the details of the molecular mechanism of genome packaging, translocation, and assembly processes in plant RNA viruses are rather limited (Ranjan et al., 2021; Mohanty et al., 2023). Therefore, this review aims to update readers on the current knowledge of the molecular mechanism of genome packaging and the steps involved in viral maturation of plant RNA viruses, with a special emphasis on Polerovirus and Potexvirus.
After its entry into a host plant cell, an RNA virus carries out the following steps in a highly coordinated manner to complete a successful infection: 1) uncoating of virions; 2) immediate translation of proteins related to replication; 3) synthesis of new (−) strand and (+) strand RNAs; 4) translation of the movement protein (MP) and capsid protein from newly synthesized viral mRNAs; and finally, 5) the assembly of infectious virions capable of moving from one cell to neighbor cells through plasmodesmata or ultimately interacting with insect vectors for their spreading to healthy plants (Bol, 2005; Catalano, 2005; Rao, 2006; Kumari et al., 2020; Garmann et al., 2022). The entire process of the replication event has been explored in depth in several plant RNA viruses and tremendous reviews have been published worldwide on this subject (Annamalai and Rao, 2005; Bol, 2005; Rao, 2006; Annamalai et al., 2008; Rao et al., 2014). The RNA genome of viruses ranges from monopartite to multipartite, and RNA strands could be of positive or negative senses (Table 1). Table 1 represents the detailed characteristics of RNA viruses infecting plants with suitable examples (Table 1). The plant RNA virus replicates within a cytoplasmic compartment, which is heavily populated by cellular RNAs (Burd and Dreyfuss, 1994; Rao, 2006). Thus, CPs, the RNA-binding proteins, exhibit a remarkable degree of packaging specificity, despite their probability of co-packaging cellular RNAs (Burd and Dreyfuss, 1994; Ranjan et al., 2021; Kumar et al., 2022).
2 CPs with a novel ATPase domain: a new regime in the genome packaging of plant RNA viruses
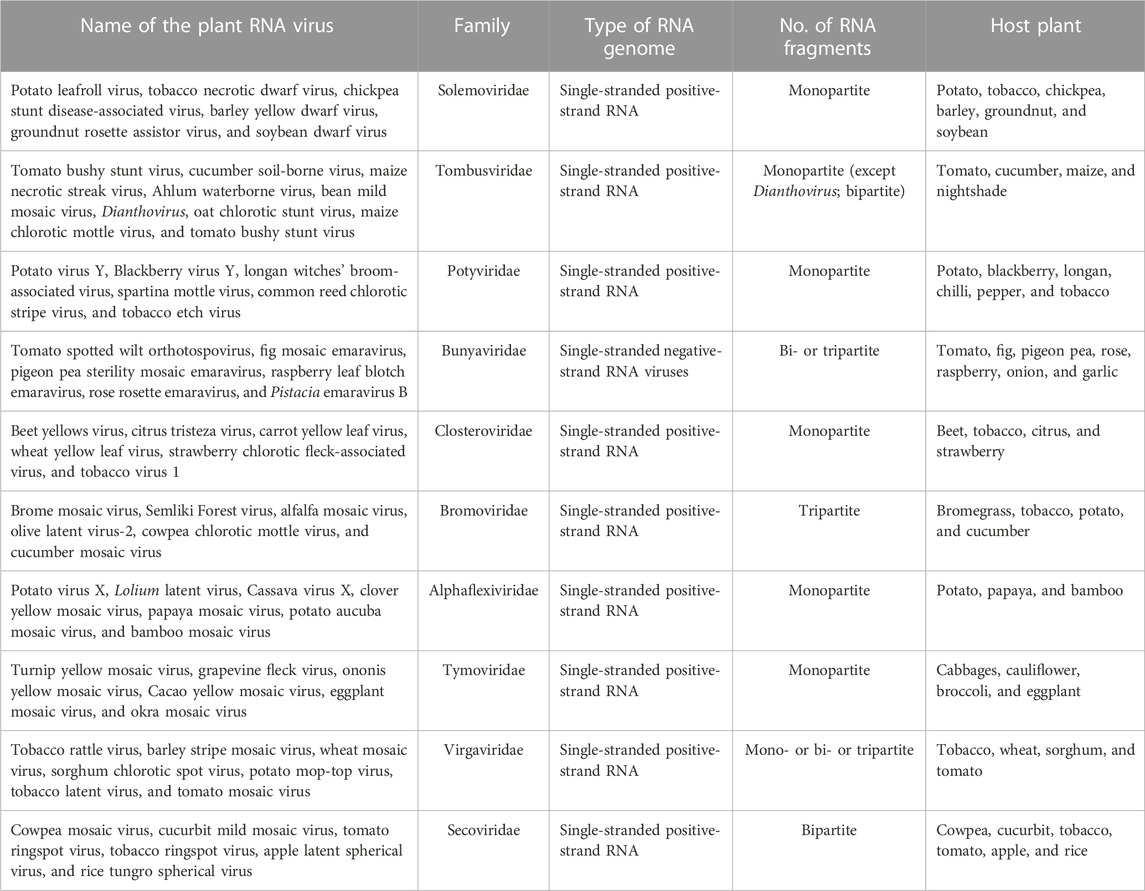
In our previous study, an attempt was made to expand and sub-classify type I packaging machinery for several plant RNA viruses, viz., Polerovirus and Potexvirus. These smaller RNA viruses infecting plants fall under an ATP-dependent sub-type IA packaging system, which possess a classical P-loop containing an ATPase domain situated over linear polypeptide chains of CPs (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). The viral encoded CP recognizes the genomic end, nucleates over the viral genome in the presence of ATP, and ultimately, encapsidates them to a mature virion (Ranjan et al., 2021; Mohanty et al., 2023). On the other hand, CPs of viruses belong to type IB, and IC does not possess ATPase folds over it and employs slightly different mechanisms for genome packaging (Ranjan et al., 2021). The type IB virus takes the help of viral- or host-encoded ATPase, whereas the type IC virus prefers an ATP-independent fashion to package their genome inside capsid coats (Ranjan et al., 2021). The role of packaging ATPase in genome packaging is well-known for viruses of type II and III systems, but our recent discovery of the presence of novel ATPase domains on polypeptide chains of CPs has completely changed our perception of genome packaging in plant RNA viruses and raised many interesting facts (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). An ATPase domain comprising different motifs responsible for ATP hydrolysis is situated on the linear primary structure of the CP of almost all plant RNA viruses (Figures 1A, B). Interestingly, apart from RNA viruses, the same ATPase fold consisting of Walker A, Walker B, sensors, and arginine fingers was also found to be present on the polypeptide chain of CPs of DNA viruses infecting plants, such as members of Nanoviridae and Geminiviridae families. Intriguingly, the presence of very rare patterns of a novel ATPase domain with multiple Walker A, sensor motifs, Walker B, and arginine motifs fetched during our thorough comprehensive analysis has indicated a variation within the ATPase superfamily (Figure 1A) (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). Such variations could be responsible for the evolution of different architectures of an ATPase domain with a single function of ATP hydrolysis during evolution time (Iyer et al., 2004; Chelikani et al., 2014b). These duplicated Walker A, sensors, Walker B, and arginine motifs together form an active site or a pocket for ATP binding and catalysis (Kumar et al., 2022). ATP hydrolysis is coordinated by Walker-A “P-loop” motifs during genome packaging. The Walker A motif with highly conserved sequences of RGRGSSET (lavender color) interacts with β and terminal γ-phosphates of ATP bound at the active site of CPs. On the other hand, the highly conserved Asp residue of Walker B motifs (consensus “hhhhDG”) situated at the tip of the β-strand forms a coordinate bond with the metal ion and further assists in the hydrolysis of ATP (Figures 1A, B) (Kumar et al., 2022). While a conserved Asp residue (blue) coordinates the Mg2+ cation, the other conserved Gly residue of Walker-B motifs (blue) involve in a nucleophilic attack toward the terminal γ-phosphate of the ATP molecule bound at active sites by priming a water molecule (Figure 1B) (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023).
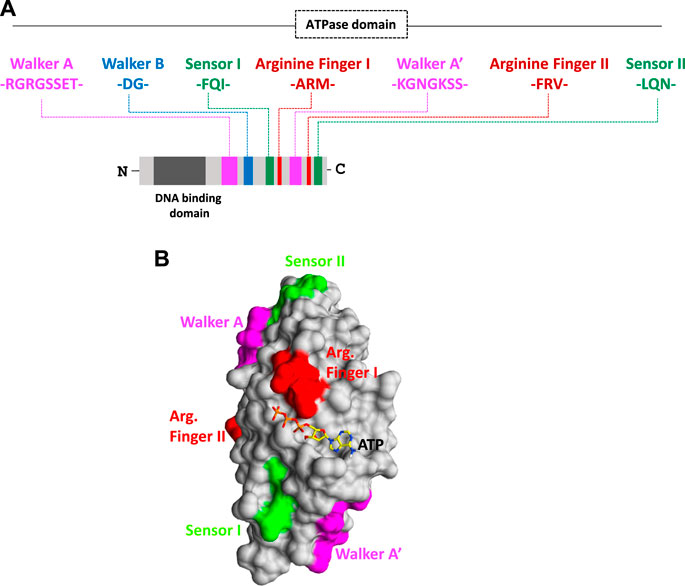
FIGURE 1. (A) Organization of functional motifs on the polypeptide chain of CPs of the Solemoviridae and Alphaflexiviridae family. The functional motifs at the N-terminal ATPase domain are represented in different colors, whereas the C-terminal end occupies the putative DNA-binding domain. Schematics are drawn approximately to the scale and represent the approximate consensus of representative homologs of RNA viruses infecting plants. (B) Interaction of the ATP molecule with the I-TASSER-predicted atomic model of CP. All the putative ATP catalysis motifs are highlighted in different colors. For details, refer to Kumar et al., 2022.
Intriguingly, all the motifs responsible for binding and catalysis of ATP are either part of the loop or are situated at the top of the β-strand in the generated 3D atomic model of CPs. This is a classical hallmark for the ASCE P-loop ATPase superfamily and makes these critical motifs very flexible for nucleophilic attack and catalysis inside active sites (Figure 1B) (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). The arginine finger I with a consensus sequence “ARM” and the arginine finger II with a consensus sequence “FRV” (in red) are situated at 9–10 amino acid residues next to the sensor I (“FQI” in green) motif and 5–6 amino acid residues downstream of another Walker-A-like motif (represented as Walker A’ in lavender color). Another sensor motif (designated as sensor II with the consensus sequence “LQN” in green color) is present almost after five residues of the arginine finger II. The arginine fingers I and II (red) and sensor motif II (green) are strictly conserved across plant RNA viruses (Figure 1A) (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). The ATPase fold of CPs of plant RNA viruses abides by a unique arrangement of motifs, where arginine fingers I and II and Walker A’ are found to be flanked with sensor I and II motifs from both the sides, and sensor motif I is situated next to Walker B almost after 48 amino acid residues downstream (Figure 1A) (Ranjan et al., 2021; Kumar et al., 2022). This permutation and different combinations of motifs has led to the origin and evolution of different homologs of ATPase folds across the domain of life (Iyer et al., 2004). Atomic structure prediction using I-TASSER revealed that the active site CP comprises all motifs necessary for ATP interaction, except arginine finger I, which is situated far from the rest of the motifs in their folded structure (highlighted in red; Figure 1B). Arginine finger is a classical hallmark of ATPases and is conserved across many ATPases. It completes the active site from a distinct location, forming contacts with the γ-phosphate of the nucleotide (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023).
The unique arrangement of ATP-binding motifs in the primary and tertiary structures of CPs indicates the structural similarity of CPs with the members of the well-known classical P-loop ATPase superfamily (Figures 1A, B) (Ranjan et al., 2021; Kumar et al., 2022). The functions of more than one Walker-A-like motifs in ATP binding and catalysis need to be explored. The use of site-directed mutagenesis (SDM) to replace critical amino acid residues in ATPase motifs could provide better insights into understanding the function of these motifs in genome translocation, packaging, assembly, and viral maturation (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). The P-loop was found to play a crucial role in directing ATP binding and hydrolysis with genome packaging, translocation, and assembly in all RNA/DNA viruses (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). Our preliminary experimental data suggest that the recombinant CPs of the potato virus X and potato leafroll virus, overexpressed in the bacterial system, showed enhanced ATP hydrolysis activity in the presence of DNA (T. Ranjan, unpublished data). The replacement of critical amino acid residues of Walker-A and Walker-B motifs obtained using SDM resulted in loss of ATPase function of recombinant CPs, indicating the role of these motifs in ATP binding and hydrolysis (T. Ranjan, unpublished data). Interestingly, we also observed the importance of the Walker A’ motif in ATP hydrolysis, the removal of which led to the loss of ATPase activity of recombinant CPs (T. Ranjan, unpublished data). Thus, our comprehensive sequence analysis and preliminary experimental data indicate the direct role of ATPase folds of CPs in the genome packaging of RNA viruses infecting plants (Kumari et al., 2020; Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023).
3 A model for genome translocation, packaging, and assembly in plant RNA viruses
Plant RNA viruses exhibit a wide range of virion symmetry including rods (e.g., Potyvirus), icosahedral (e.g., Cucumovirus), and bacilliform shape (e.g., alfalfa mosaic virus) (Ford et al., 2013; Rao et al., 2014). Despite these diversities among virus families, mature virions of a particular species often exhibit structural homogeneity and thus share the common mechanism of genome packaging and assembly (Chelikani et al., 2014a; Guo et al., 29019; Larson et al., 2005). Nucleation/oligomerization of capsid proteins around viral RNA genomes and virion assembly involves two primary molecular interactions: 1) protein–protein interactions (viz., capsid–capsid interaction) and ii) RNA–protein interactions (viz., RNA–capsid interaction) (Qu and Morris, 1997; Basnayake et al., 2009; Bunka et al., 2011; Ford et al., 2013). CPs recognize specific packaging signals (sequences) situated usually at the genomic end instead of nucleating at some random sites on a viral genome with the help of the N-terminal DNA-binding domain (Figure 2A) (Garoff et al., 1980; Bink et al., 2003; Zlotnick et al., 2020; Li et al., 2021). Our bioinformatics analysis also revealed the presence of helix-turn-helix and basic amino acid residues at the N-terminal nucleic acid-binding domain of CPs and indicated their importance during interactions with viral genomes (T. Ranjan, unpublished data). The thoroughly characterized packaging signals, viz., assembly sequence, tRNA-like sequence, hairpin-like structures, and some untranslated regions at the 5’- or 3’-end are recognized by CPs to discriminate viral RNA genomes from cellular RNAs (Choi and Rao, 2000; Choi et al., 2002; Bink et al., 2003; Choi and Rao, 2003). Selective packaging of the genomic RNA (gRNA) is mediated by high-affinity binding between the CP and the packaging signal (Figure 2A) (Wang et al., 1999; Choi and Rao, 2000; Choi et al., 2002; Choi and Rao, 2003; Kwon et al., 2005; Wang et al., 2005). Binding of one molecule of CP to the packaging signal changes the conformation of the CP from weak CP–CP interactions to strong interactions in a processive manner (Yang et al., 2017; Comas-Garcia, 2019). In almost all plant RNA viruses, the event of replication is functionally coupled with genome translocation, packaging, and virion assembly (Malik et al., 2005; Annamalai and Rao, 2006). A physical interaction with a virus-encoded replicase (p2a) expedites the CP to smoothly nucleate over the RNA genome and further increases the packaging specificity in an energy-dependent step (Figure 2B) (Annamalai and Rao, 2005; Annamalai and Rao, 2006; Annamalai et al., 2008; Chaturvedi and Rao, 2014; Comas-Garcia, 2019). Intriguingly, an interaction between host-encoded HSP70 and CP is critical during genome packaging and virion encapsidation in few plant RNA viruses, indicating variations within the type I genome packaging apparatus (Vriend et al., 1986; Alzhanova et al., 2001; Verchot, 2012; Gorovits et al., 2013). The disruption of such physical interactions leads to the packaging of cellular RNAs along with the viral RNA genome in a mature virion particle (Weiss et al., 1994; Wu and Shaw, 1998; Annamalai and Rao, 2005). Similarly, the inactivation of the ATPase domain of CPs employing the RNAi approach further inhibits the entire process of RNA genome packaging (Rakitina et al., 2005; Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). This confirms that genome packaging and assembly in a plant RNA virus is an ATP-dependent process (Figure 2B) (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023).
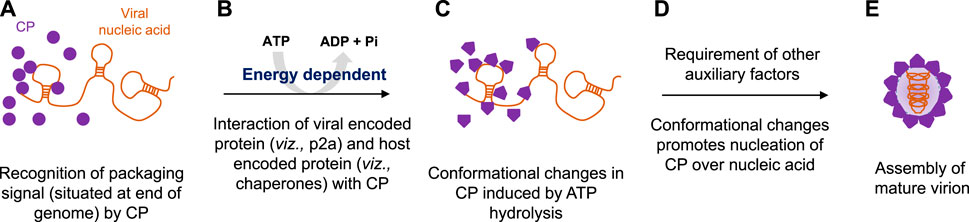
FIGURE 2. Proposed model for energy-dependent genome packaging and assembly in the plant RNA virus. (A) CPs first recognize the packaging signal or pac site, situated at the termini of viral genomes with the help of its RNA-binding domain. (B) Replication and genome packaging events in plant viruses are concurrent. Viral-encoded replicase protein (such as p2a in the case of Bromovirus) and sometimes host-encoded proteins (such as chaperonins in the case of Potyvirus) interact with CPs in an energy-dependent manner. (C) Binding of ATP molecules to the ATPase domain induces conformational changes in CPs. (D) Furthermore, ATP hydrolysis facilitates the nucleation of CPs over viral genomic RNAs, and (E) with the help of other auxiliary factors, CP encapsidates the genomic RNA, ultimately leading to the production of mature virion particles.
Intriguingly, the superimposed structure of the docked CP–ATP complex showed that each monomer has an active site at the interface (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). Binding of ATP molecules to the ATPase domain of CPs brings conformational changes in the tertiary structure of proteins (Figure 2C), and furthermore, their hydrolysis favors the nucleation of CPs onto viral RNA genomes (Figure 2D). ATP hydrolysis also promotes oligomerization (monomer-to-trimer-to-pentamer) of CPs into a large capsomere-like complex structure and ultimately packages RNA genomes into mature virions (Figure 2E) (Kumari et al., 2020; Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023).
4 Strategies to combat plant RNA viruses by targeting crucial steps of genome packaging
After entry into the host cytoplasm with the assistance of a vector (Figure 3A), viruses first increase their copy number of genomes via the replication process (Figure 3B). Furthermore, transcription (Figure 3C) and translation (Figure 3D) produce CPs, which ultimately help in the encapsidation of multiplied genomes into virion particles (Figure 3E). Targeting genome packaging and virus assembly processes could be one of the efficient approaches toward the development of virus resistance in plants (Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023) (Figures 3a–e). One of the most effective approaches to build plant resistance has been to introduce a part of the viral gene into the plant either to induce RNA silencing against viral RNAs or to express intact or modified viral proteins or RNAs that disturb the viral infection cycle (Goldbach et al., 2003). The RNA interference (RNAi) technology has emerged as a potential tool to target virus assembly for developing resistant crops (Lodge et al., 1993; Goldbach et al., 2003) (Figure 3). RNA silencing, a conserved regulatory mechanism of gene expression in eukaryotes, is triggered by dsRNA-provoking gene silencing through sequence-specific degradation of complementary mRNA transcripts (post-transcriptional gene silencing) (Waterhouse et al., 2001). The degradation of these target RNAs occurs in a sequence-specific manner via the formation of double-stranded RNAs, which further processed into small interfering RNAs (siRNAs) by the Dicer-like (DCL) proteins and the RNA-induced silencing complex (RISC) (Lodge et al., 1993; Waterhouse et al., 2001; Goldbach et al., 2003) (Figures 3a–c). Since it is well-acknowledged that the CP is essential for plant RNA virus genome packaging (Duggal and Hall, 1993; Hacker, 1995; Kumari et al., 2020; Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023), in our previous studies, an attempt was made to explore the conserved functional motifs situated across a polypeptide chain of CPs for developing strategies toward virus resistance in plants (Kumari et al., 2020; Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023) (Figures 3d–e).
FIGURE 3. Strategy for controlling plant viruses by targeting genome packaging events during their life cycle. (A) Plant virus enters inside the host cytoplasm with the help of a vector. (B) After uncoating the genome, the virus increases its copy number by replication, and further transcription (C) and translation (D) events enable the production of CP ATPase. (E) Ultimately, CP encapsidates the RNA genome with the input of ATP molecules and forms mature virion particles. The introduction of exogenous siRNA constructs specific to ATPase domains suppresses the expression of CP mRNAs (a–d) and ultimately inhibits the production of functional virions (e).
In our previous study, the efficiency of siRNA constructs of the CP_ATPase domain to silence and inhibit virus assembly was assessed in detail (Kumar et al., 2022). To further validate this efficient methodology, siRNAs designed against ATPase folds (comprising all motifs) of the CP were agroinfiltrated into virus-infected plants. Figure 4A depicts the details of the generation of siRNA constructs against the ATPase domain of CPs (Figure 4A). Agroinfiltrated plants did not show any symptoms of a virus infection. The suppression of viral infection could be attributed to the reduced expression of CPs due to its silencing by siRNA constructs (Figures 3, 4) (Kumar et al., 2022). To understand whether the knockdown of CPs, which possess a classical ATPase domain, can disrupt RNA virus genome packaging, we agroinfiltrated the pART27–CP_ATPase domain–siRNA constructs into potato plants (Kumar et al., 2022). Control plants (naturally virus-infected plants) (Figure 4B; lane I) and those agroinfiltrated with the empty vector (Figure 4B; lane II) showed rolling symptoms of potato leafroll virus (PLRV) infection in the upper leaves (Kumar et al., 2022). Intriguingly, the agroinfiltration of plantlets individually with sense (pART27–sense CP_ATPase) and antisense (pART27–antisense CP_ATPase) constructs of the CP_ATPase domain also displayed symptoms of leaf rolling in the upper leaves (Figure 4B; lane III and IV, respectively). This is obvious because these constructs were not able to form dsRNAs after agroinfiltration and Dicer cannot recognize them anymore. Plants agroinfiltrated with the siRNA construct of CP_ATPase (pART27–CP_ATPase) showed no symptoms of a viral infection (Figure 4B; lane V). These results demonstrated that an siRNA construct, specific to the ATPase domain (comprising all critical motifs) of CPs, leads to the suppression of viral genome packaging and assembly in plants (Kumar et al., 2022).
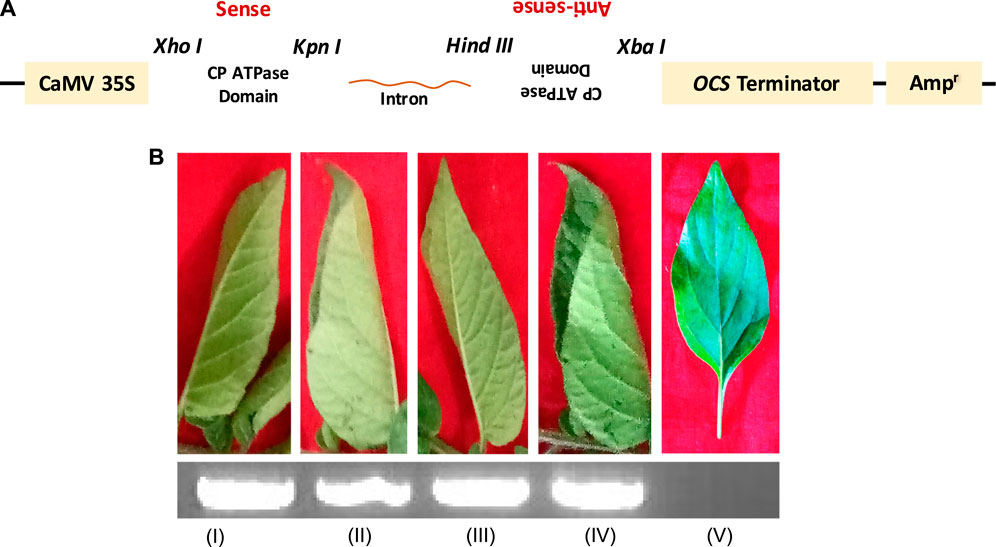
FIGURE 4. Genome packaging is an ATP-dependent process in plant viruses. (A) Schematic representation of the generation of siRNA construct specific to the ATPase domain of CP. (B) Symptoms observed in the tertiary leaves of the infected potato plants after 2 weeks of PLRV inoculation. (I) PLRV-infected control without agroinfiltration; (II) agroinfiltrated with the empty vector pART27; (III) agroinfiltrated with the plasmid containing only the antisense sequence (pART27–antisense CP_ATPase domain); (IV) agroinfiltrated with the plasmid containing only the sense sequence (pART27–sense CP_ATPase domain); and (V) agroinfiltrated with the pART27–CP_ATPase siRNA construct. The detection of the PLRV RNA using RT-PCR (inset; agarose gel). For details, refer to Kumar et al., 2022.
Our study showed that the transient expression of CP constructs specifically and efficiently inhibits genome packaging of plant RNA viruses (Kumar et al., 2022). Interestingly, CP mRNAs were found to be very high in the tertiary leaflet of control plants (natural infection of virus) and agroinfiltrated plants with an empty vector without any construct, sense construct, and antisense construct as well (Figure 4B I–IV; lower inset), whereas mRNAs of CPs were not even found in the new leaflets of agroinfiltrated plants with CP siRNAs targeted against ATPase folds (Figure 4B V; lower inset) (Kumar et al., 2022). These findings support the previous conjecture and strongly suggest a direct role of CP_ATPase in genome recognition and its packaging, the suppression of which may result in immature, non-functional, and genome-deficient viral particle production (Callaway et al., 2001; Rakitina et al., 2005; Hesketh et al., 2015).
5 Variations among type I genome packaging systems
Albeit the differences in the basic architecture and hosts of plant viruses, the process of assembly is coordinated by specific interactions of the nucleic acid-binding domain of capsid proteins with the viral genomic RNA followed by their oligomerization and nucleation in the genome. Linear polypeptide chains of CPs of type IA viruses encrypt a classical novel ATPase domain (Ranjan et al., 2021; Mohanty et al., 2023). On the other hand, CPs encoded by viruses in the type IB packaging and type IC packaging apparatus lack the ATPase domain and represent a variation within type I genome packaging (Ranjan et al., 2021). Our recent bioinformatics analysis revealed the relatedness of the type IB to type IC genome packaging system and the very early evolution of the type IA system. Interestingly, viruses from the type IA packaging system clustered into a separate clade and seemed to be diverged early from the rest of the two genome packaging systems during the evolution time (Kumari et al., 2020; Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). This represents a typical example of divergent evolution of the primordial viral genome packaging apparatus, and perhaps it could be the reminiscent genome packaging system encoded by the last universal common ancestor (LUCA). It seems that CPs of plant RNA viruses appear to derive from a common ancestor, regardless of their host (Ranjan et al., 2021; Mohanty et al., 2023). It could also represent a new variation and the adaptive evolution of the ancient viral genome packaging apparatus that likely existed in the LUCA (Burroughs et al., 2007; Chelikani et al., 2014b; Iyer et al., 2004; Mohanty et al., 2023; Ranjan et al., 2021).
6 Conclusion
RNA viruses cause serious diseases in crops, leading to significant reductions in annual yields worldwide. Innovative and state-of-the-art strategies are very much required to control/combat with plant viruses in order to meet the demands of the population growth worldwide (Hill et al., 2021; Ye et al., 2021). Among several advances in viral control measures, aiming for the steps of the viral assembly process has the potential to save crops from such destructive diseases (Burroughs et al., 2007; Dai et al., 2021; Kumar et al., 2022). Understanding the mechanism of genome packaging and assembly would be helpful in developing novel approaches to control plant viruses and ultimately to meet the demands of a growing world population (Kumar et al., 2020; Ranjan et al., 2021; Kumar et al., 2022; Mohanty et al., 2023). Deciphering the detailed machinery of ATP binding and catalysis by the ATPase fold would be helpful in designing novel inhibitors, which may hinder the entire process of genome packaging and virus assembly (Twarock and Stockley, 2019). Information collected from the limited available literature worldwide on Polerovirus and Potexvirus as model systems has provided novel insights into understanding the mechanism of ATP-dependent genome packaging and virus assembly. The extension of similar studies to other plant RNA viruses would be helpful in determining whether ATP-dependent genome packaging is universally conserved among different RNA viruses infecting plants. Still, important questions regarding the mechanisms regulating the process of genome packaging and assembly of uniformly sized virions remain unanswered and a challenge for future investigation.
Author contributions
TR and RR conceived the study concept. TR, MA, SD, and MFA further conceptualized the idea and contributed to visualization. JK, AM, AK, KJ, and KR wrote and edited the original draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India, New Delhi (India) (File Nos SRG/2019/002223 and CRG/2022/000478) to TR.
Acknowledgments
The authors would like to thank Bihar Agricultural University (BAU), Sabour, for providing the basic infrastructure for conducting the research works. This article bears BAU Communication No. 1328/221118.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alzhanova, D. V., Napuli, A. J., Creamer, R., and Dolja, V. V. (2001). Cell-to-cell movement and assembly of a plant closterovirus: Roles for the capsid proteins and Hsp70 homolog. EMBO J. 20, 6997–7007. doi:10.1093/emboj/20.24.6997
Annamalai, P., and Rao, A. L. N. (2006). Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J. Virology 80 (20), 10096–10108. doi:10.1128/JVI.01186-06
Annamalai, P., and Rao, A. L. N. (2005). Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: A functional role for viral replicase in RNA packaging. Virology 338 (1), 96–111. doi:10.1016/j.virol.2005.05.013
Annamalai, P., Rofail, F., DeMason, D. A., and Rao, A. L. N. (2008). Replication-coupled packaging mechanism in positive-strand RNA viruses: Synchronized coexpression of functional multigenome RNA components of an animal and a plant virus in nicotiana benthamiana cells by agroinfiltration. J. virology 82 (3), 1484–1495. doi:10.1128/JVI.01540-07
Basnayake, V. R., Sit, T. L., and Lommel, S. A. (2009). The Red clover necrotic mosaic virus origin of assembly is delimited to the RNA-2 trans-activator. Virology 384 (1), 169–178. doi:10.1016/j.virol.2008.11.005
Bink, H. H., Schirawski, J., Haenni, A. L., and Pleij, C. W. (2003). The 5′-proximal hairpin of turnip yellow mosaic virus RNA: Its role in translation and encapsidation. J. Virology 77 (13), 7452–7458. doi:10.1128/jvi.77.13.7452-7458.2003
Bol, J. F. (2005). Replication of alfamo-and ilarviruses: Role of the coat protein. Annu. Rev. Phytopathology 43, 39–62. doi:10.1146/annurev.phyto.43.101804.120505
Bunka, D. H., Lane, S. W., Lane, C. L., Dykeman, E. C., Ford, R. J., Barker, A. M., et al. (2011). Degenerate RNA packaging signals in the genome of satellite tobacco necrosis virus: Implications for the assembly of a T= 1 capsid. J. Mol. Biol. 413 (1), 51–65. doi:10.1016/j.jmb.2011.07.063
Burd, C. G., and Dreyfuss, G. (1994). Conserved structures and diversity of functions of RNA-binding proteins. Science 265 (5172), 615–621. doi:10.1126/science.8036511
Burroughs, A. M., Iyer, L. M., and Aravind, L. (2007). Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Gene Protein Evol. 3, 48–65. doi:10.1159/000107603
Callaway, A., Giesman-Cookmeyer, D., Gillock, E. T., Sit, T. L., and Lommel, S. A. (2001). The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathology 39 (1), 419–460. doi:10.1146/annurev.phyto.39.1.419
Catalano, C. E. (2005). Viral genome packaging machines: Genetics, structure, and mechanism. Springer US, 1–4.Viral genome packaging machines
Chaturvedi, S., and Rao, A. L. N. (2014). Live cell imaging of interactions between replicase and capsid protein of Brome mosaic virus using Bimolecular Fluorescence Complementation: Implications for replication and genome packaging. Virology 464, 67–75. doi:10.1016/j.virol.2014.06.030
Chelikani, V., Ranjan, T., and Kondabagil, K. (2014a). Revisiting the genome packaging in viruses with lessons from the “Giants”. Virology 466, 15–26. doi:10.1016/j.virol.2014.06.022
Chelikani, V., Ranjan, T., Zade, A., Shukla, A., and Kondabagil, K. (2014b). Genome segregation and packaging machinery in Acanthamoeba polyphaga mimivirus is reminiscent of bacterial apparatus. J. Virology 88 (11), 6069–6075. doi:10.1128/JVI.03199-13
Choi, Y. G., Dreher, T. W., and Rao, A. L. N. (2002). tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. 99 (2), 655–660. doi:10.1073/pnas.022618199
Choi, Y. G., and Rao, A. L. N. (2000). Molecular studies on bromovirus capsid protein: VII. Selective packaging of BMV RNA4 by specific N-terminal arginine residues. Virology 275 (1), 207–217. doi:10.1006/viro.2000.0513
Choi, Y. G., and Rao, A. L. N. (2003). Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virology 77 (18), 9750–9757. doi:10.1128/jvi.77.18.9750-9757.2003
Comas-Garcia, M. (2019). Packaging of genomic RNA in positive-sense single-stranded RNA viruses: A complex story. Viruses 11 (3), 253. doi:10.3390/v11030253
Dai, L., Singh, D., Lu, S., Kottadiel, V. I., Vafabakhsh, R., Mahalingam, M., et al. (2021). A viral genome packaging ring-ATPase is a flexibly coordinated pentamer. Nat. Commun. 12 (1), 6548. doi:10.1038/s41467-021-26800-z
Duggal, R., and Hall, T. C. (1993). Identification of domains in brome mosaic virus RNA-1 and coat protein necessary for specific interaction and encapsidation. J. Virology 67 (11), 6406–6412. doi:10.1128/JVI.67.11.6406-6412.1993
Ford, R. J., Barker, A. M., Bakker, S. E., Coutts, R. H., Ranson, N. A., Phillips, S. E., et al. (2013). Sequence-specific, RNA–protein interactions overcome electrostatic barriers preventing assembly of satellite tobacco necrosis virus coat protein. J. Mol. Biol. 425 (6), 1050–1064. doi:10.1016/j.jmb.2013.01.004
Garmann, R. F., Aaron, M., Goldfain, C. R., Tanimoto, C. E., Beren, F. F., Vasquez, D. A., et al. (2022). Single-particle studies of the effects of RNA–protein interactions on the self-assembly of RNA virus particles. Proc. Natl. Acad. Sci. 119 (39), e2206292119. doi:10.1073/pnas.2206292119
Garoff, H., Frischauf, A. M., Simons, K., Lehrach, H., and Delius, H. (1980). The capsid protein of Semliki Forest virus has clusters of basic amino acids and prolines in its amino-terminal region. Proc. Natl. Acad. Sci. 77 (11), 6376–6380. doi:10.1073/pnas.77.11.6376
Goldbach, R., Bucher, E., and Prins, M. (2003). Resistance mechanisms to plant viruses: An overview. Virus Res. 92 (2), 207–212. doi:10.1016/s0168-1702(02)00353-2
Gorovits, R., Moshe, A., Ghanim, M., and Czosnek, H. (2013). Recruitment of the host plant heat shock protein 70 by Tomato yellow leaf curl virus coat protein is required for virus infection. PLoS One 8 (7), e70280. doi:10.1371/journal.pone.0070280
Guo, P., Driver, D., Zhao, Z., Zheng, Z., Chan, C., and Cheng, X. (2019). Controlling the revolving and rotating motion direction of asymmetric hexameric nanomotor by arginine finger and channel chirality. ACS Nano 13 (6), 6207–6223. doi:10.1021/acsnano.8b08849
Hacker, D. L. (1995). Identification of a coat protein binding site on southern bean mosaic virus RNA. Virology 207 (2), 562–565. doi:10.1006/viro.1995.1117
Hesketh, E. L., Meshcheriakova, Y., Dent, K. C., Saxena, P., Thompson, R. F., Cockburn, J. J., et al. (2015). Mechanisms of assembly and genome packaging in an RNA virus revealed by high-resolution cryo-EM. Nat. Commun. 6 (1), 10113–10210. doi:10.1038/ncomms10113
Hill, S. R., Twarock, R., and Dykeman, E. C. (2021). The impact of local assembly rules on RNA packaging in a T= 1 satellite plant virus. PLoS Comput. Biol. 17 (8), e1009306. doi:10.1371/journal.pcbi.1009306
Iyer, L. M., Makarova, K. S., Koonin, E. V., and Aravind, L. (2004). Comparative genomics of the FtsK–HerA superfamily of pumping ATPases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 32 (17), 5260–5279. doi:10.1093/nar/gkh828
Kumar, J., Kumar, R. R., Das, D. K., Mohanty, A., Rajani, K., Kumari, N., et al. (2022). Knockdown of capsid protein encoding novel ATPase domain inhibits genome packaging in potato leafroll virus. 3Biotech 12 (3), 66–13. doi:10.1007/s13205-021-03085-z
Kumar, R. R., Ansar, M., Rajani, K., Kumar, J., and Ranjan, T. (2020). First report on molecular basis of potato leaf roll virus (PLRV) aggravation by combined effect of tuber and prevailing aphid. BMC Res. Notes 13 (1), 523–524. doi:10.1186/s13104-020-05370-1
Kumari, P., Kumar, J., Kumar, R. R., Ansar, M., Rajani, K., Kumar, S., et al. (2020). Inhibition of potato leafroll virus multiplication and systemic translocation by siRNA constructs against putative ATPase fold of movement protein. Sci. Rep. 10 (1), 22016–22111. doi:10.1038/s41598-020-78791-4
Kwon, S. J., Park, M. R., Kim, K. W., Plante, C. A., Hemenway, C. L., and Kim, K. H. (2005). cis-Acting sequences required for coat protein binding and in vitro assembly of Potato virus X. Virology 334 (1), 83–97. doi:10.1016/j.virol.2005.01.018
Larson, S. B., Lucas, R. W., Greenwood, A., and McPherson, A. (2005). The RNA of turnip yellow mosaic virus exhibits icosahedral order. Virology 334 (2), 245–254. doi:10.1016/j.virol.2005.01.036
Li, X., Gu, M., Zheng, Q., Gao, R., and Liu, X. (2021). Packaging signal of influenza A virus. Virology J. 18 (1), 36–10. doi:10.1186/s12985-021-01504-4
Lodge, J. K., Kaniewski, W. K., and Tumer, N. E. (1993). Broad-spectrum virus resistance in transgenic plants expressing pokeweed antiviral protein. Proc. Natl. Acad. Sci. 90 (15), 7089–7093. doi:10.1073/pnas.90.15.7089
Malik, P. S., Kumar, V., Bagewadi, B., and Mukherjee, S. K. (2005). Interaction between coat protein and replication initiation protein of Mung bean yellow mosaic India virus might lead to control of viral DNA replication. Virology 337 (2), 273–283. doi:10.1016/j.virol.2005.04.030
Mohanty, A., Ranjan, T., Kumar, J., Kumar, R. R., and Rajani, K. (2023). Is genome packaging in plant viruses energy independent? J. Animal Plant Sci. 33, 2023.
Qu, F., and Morris, T. J. (1997). Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J. Virology 71 (2), 1428–1435. doi:10.1128/JVI.71.2.1428-1435.1997
Rakitina, D. V., Kantidze, O. L., Leshchiner, A. D., Solovyev, A. G., Novikov, V. K., Morozov, S. Y., et al. (2005). Coat proteins of two filamentous plant viruses display NTPase activity in vitro. FEBS Lett. 579 (22), 4955–4960. doi:10.1016/j.febslet.2005.07.083
Ranjan, T., Pal, A. K., Prasad, B. D., Kumar, R. R., Kumar, M., Shamim, M., et al. (2021). Reassessing the mechanism of genome packaging in plant viruses with lessons from ATPase fold. Australas. Plant Pathol. 50 (3), 253–266. doi:10.1007/s13313-020-00772-y
Rao, A. L. N., Chaturvedi, S., and Garmann, R. F. (2014). Integration of replication and assembly of infectious virions in plant RNA viruses. Curr. Opin. Virology 9, 61–66. doi:10.1016/j.coviro.2014.09.008
Rao, A. L. N. (2006). Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathology 44, 61–87. doi:10.1146/annurev.phyto.44.070505.143334
Twarock, R., and Stockley, P. G. (2019). RNA-Mediated virus assembly: Mechanisms and consequences for viral evolution and therapy. Annu. Rev. Biophysics 48, 495–514. doi:10.1146/annurev-biophys-052118-115611
Verchot, J. (2012). Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front. Plant Sci. 3, 275. doi:10.3389/fpls.2012.00275
Vriend, G., Verduin, B. J. M., and Hemminga, M. A. (1986). Role of the N-terminal part of the coat protein in the assembly of cowpea chlorotic mottle virus: A 500 MHz proton nuclear magnetic resonance study and structural calculations. J. Mol. Biol. 191 (3), 453–460. doi:10.1016/0022-2836(86)90140-3
Wang, J., Carpenter, C. D., and Simon, A. E. (1999). Minimal sequence and structural requirements of a subgenomic RNA promoter for turnip crinkle virus. Virology 253 (2), 327–336. doi:10.1006/viro.1998.9538
Wang, X., Lee, W. M., Watanabe, T., Schwartz, M., Janda, M., and Ahlquist, P. (2005). Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virology 79 (21), 13747–13758. doi:10.1128/JVI.79.21.13747-13758.2005
Waterhouse, P. M., Wang, M. B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411 (6839), 834–842. doi:10.1038/35081168
Weiss, B., Geigenmüller-Gnirke, U., and Schlesinger, S. (1994). Interactions between Sindbis virus RNAs and a 68 amino acid derivative of the viral capsid protein further defines the capsid binding site. Nucleic Acids Res. 22 (5), 780–786. doi:10.1093/nar/22.5.780
Wu, X., and Shaw, J. G. (1998). Evidence that assembly of a potyvirus begins near the 5'terminus of the viral RNA. J. General Virology 79 (6), 1525–1529. doi:10.1099/0022-1317-79-6-1525
Yang, J., Qian, Q., Li, T. F., Yang, X., Won, S. J., and Zhou, X. (2017). Cypovirus capsid protein VP5 has nucleoside triphosphatase activity. Virol. Sin. 32 (4), 328–330. doi:10.1007/s12250-017-3999-2
Ye, L., Ambi, U. B., Olguin-Nava, M., Gribling-Burrer, A. S., Ahmad, S., Bohn, P., et al. (2021). RNA structures and their role in selective genome packaging. Viruses 13 (9), 1788. doi:10.3390/v13091788
Keywords: viral genome packaging, energy-dependent, capsid protein, ATPase fold, virus assembly, RNA virus
Citation: Ranjan T, Ranjan Kumar R, Ansar M, Kumar J, Mohanty A, Kumari A, Jain K, Rajani K, Dei S and Ahmad MF (2023) The curious case of genome packaging and assembly in RNA viruses infecting plants. Front. Genet. 14:1198647. doi: 10.3389/fgene.2023.1198647
Received: 01 April 2023; Accepted: 22 May 2023;
Published: 08 June 2023.
Edited by:
Davoud Koolivand, University of Zanjan, Zanjan, IranReviewed by:
Himanshu Tak, Bhabha Atomic Research Centre (BARC), IndiaBajarang Kumbhar, SVKM’s Narsee Monjee Institute of Management Studies, India
Copyright © 2023 Ranjan, Ranjan Kumar, Ansar, Kumar, Mohanty, Kumari, Jain, Rajani, Dei and Ahmad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tushar Ranjan, bWFpbDJ0dXNoYXJyYW5qYW5AZ21haWwuY29t
†These authors share first authorship
 Tushar Ranjan
Tushar Ranjan Ravi Ranjan Kumar1†
Ravi Ranjan Kumar1†