- 1Department of Pharmaceutical Sciences, University of Illinois Chicago College of Pharmacy, Chicago, IL, United States
- 2Comprehensive Sickle Cell Center, University of Illinois Chicago, Chicago, IL, United States
- 3Department of Biobehavioral Nursing Science, University of Florida College of Nursing, Gainesville, FL, United States
- 4Jesse Brown Veteran’s Administration Medical Center, Chicago, IL, United States
- 5Division of Hematology/Oncology, University of Illinois Chicago College of Medicine, Chicago, IL, United States
- 6Department of Neurology and Rehabilitation, University of Illinois Chicago College of Medicine, Chicago, IL, United States
- 7Department of Biomedical Engineering, University of Illinois Chicago College of Engineering, Chicago, IL, United States
Introduction: Pain is a lifelong companion of individuals with sickle cell disease (SCD) and has a severe impact on their quality of life. Both acute crisis pain and chronic non-crisis pain exhibit high variability between individuals, making it difficult to effectively manage sickle cell-related pain. We investigated the role of dopamine beta-hydroxylase (DBH) gene polymorphisms on pain variability in SCD. DBH is a key enzyme in the catecholamine biosynthesis pathway that catalyzes the conversion of dopamine to norepinephrine, both of which are known mediators of pain and pain-related behaviors.
Methods: Acute crisis pain-related utilization and non-crisis chronic pain scores of 131 African Americans with SCD were obtained.
Results and discussion: Association analyses revealed that the T allele of upstream variant rs1611115 and downstream variant rs129882 correlated with higher severity of chronic pain in an additive model. On the other hand, the A allele of missense variant rs5324 associated with lower risk of both acute crisis pain and chronic pain. Similarly, the C allele of intronic variant rs2797849 was associated with lower incidence of acute crisis pain in the additive model. In addition, tissue-specific eQTL revealed that the T allele of rs1611115 correlated with decreased expression of DBH in the frontal cortex and anterior cingulate cortex (GTEx), and decreased expression of DBH-AS1 in blood (eQTLGen). Bioinformatic approaches predicted that rs1611115 may be altering a transcription factor binding site, thereby, contributing to its potential effect. Taken together, findings from this study suggest that potential functional polymorphisms of DBH may modulate pain perception in SCD.
Introduction
The dopamine beta-hydroxylase (DBH) is a catalytic enzyme found in secretory vesicles of noradrenaline producing neurons in both the central and peripheral nervous systems (Cubells et al., 1998). It catalyzes the hydroxylation of dopamine to produce norepinephrine in these neurons (Diliberto and Allen, 1981) and is secreted along with the neurotransmitters during synaptic transmission (Weinshilboum et al., 1971). Therefore, DBH can be detected in the cerebrospinal fluid (CSF) and plasma, and polymorphisms in DBH have been found to alter plasma and CSF levels of the protein (Tunbridge et al., 2019). Altered DBH expression or activity would result in an imbalance of dopamine and norepinephrine. Since both of these neurotransmitters are key modulators in pain signaling (Jasmin et al., 2002; Leventhal et al., 2007; Jarcho et al., 2012), it was hypothesized that polymorphisms in DBH may be associated with variability in pain perception.
In that regard, we investigated the potential role of DBH variants on the heterogeneity of acute and chronic pain phenotypes in sickle cell disease (SCD). SCD is a debilitating genetic blood disorder in which patients suffer from pain throughout their lives, starting as early as 1 year after birth (Platt et al., 1991; Miller et al., 2000). Acute pain is caused by frequent and recurring episodes of ischemia-like events where deformed red blood cells agglomerate and block microvasculature in different organs (Manwani and Frenette, 2013). These events are known as painful vaso-occlusive crisis and are a leading cause for hospitalization among SCD patients (Shah et al., 2019). Chronic pain, on the other hand, is persistent daily pain experienced even on non-crisis days and believed to be caused by complex neuropathic pain mechanisms (Wang et al., 2010; He et al., 2016). It has been found that both pain phenotypes exhibit high variability between patients, thus rendering the assessment and management of pain in SCD a challenging task (Okpala and Tawil, 2002; Wilkie et al., 2010). A better understanding of the various genetic factors that modulate sickle cell pain variability can help us devise more effective pain management strategies.
Previously, we reported that genetic variants of other enzymes in the catecholamine biosynthesis pathway such as COMT, GCH1, PNMT can contribute to the inter-individual variability of pain in SCD (Jhun et al., 2014; Sadhu et al., 2018; Sadhu et al., 2020). In this study, we evaluate the role of genetic variants in another key monoaminergic enzyme DBH. The DBH gene is located on chromosome 9 and consists of 12 exons. Five single nucleotide polymorphisms (SNPs) rs1611115, rs2797849, rs5320, rs5324, and rs129882 spanning the gene were assessed in this study. rs1611115 is located upstream of the gene near its proximal promoter region. rs2797849 is an intronic SNP, whereas rs5320 and rs5324 are non-synonymous variants lying in exon 3 and 4, respectively. Finally, rs129882 is located in the 3’ untranslated region of DBH.
Methods
Study design
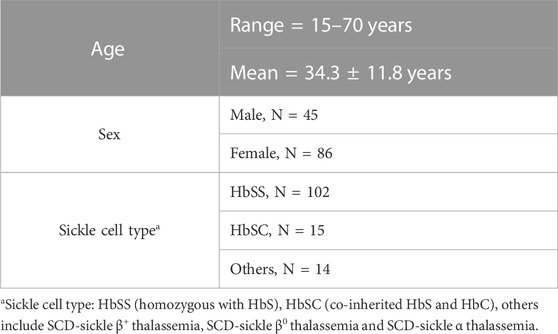
This study protocol was approved by the University of Illinois (UI) at Chicago’s Institutional Review Board. SCD patients were recruited at the Sickle Cell Clinic of the UI Hospital and Health Sciences System. Written informed consent of adult participants (aged 18 or older), and child assent and parental consent for participants under the age of 18 were obtained during subject recruitment. Eligibility criteria of subjects are provided in greater detail in previous publications (Ezenwa et al., 2014; Dyal et al., 2020). All subjects included in this study (N = 131) identified themselves as African Americans. Of the 131 subjects, 45 were male and 86 were female, their ages ranging from 15 to 70 years. A majority of the subjects (N = 102) had SS type sickle cell disease, i.e., homozygous for hemoglobin S. Subject demographics are outlined in Table 1.
Assessment of pain
Two types of pain phenotype data were collected—acute crisis pain and chronic non-crisis pain. Annual hospitalization counts of subjects due to crisis pain, referred to as utilization, was used as a marker for acute crisis pain measurement. Utilization data of patients were obtained mainly from the UI medical records of emergency department and acute care center visits (Ezenwa et al., 2014). Biweekly telephone calls to patients were made to record any out-of-UI-system visits. During their regular clinic visits, patients were asked to score various aspects of their baseline pain experience on a digital tool adapted from the McGill Pain Questionnaire. Scores from each category of questions, i.e., pain sites, pain intensity, pain pattern and quality, were aggregated and scaled to a range of 0–100 as described in earlier studies (Wilkie et al., 2010; Wilkie et al., 2015). This was reported as the Composite Pain Index (CPI) and used as a marker for chronic non-crisis pain measurement.
DNA isolation and genotyping
Blood or buccal swab samples were collected from subjects for genotyping. DNA was isolated from buccal swab samples using a modified phenol-chloroform method (Vandenbergh et al., 2003). DNA isolation from blood samples was performed using a modified salting-out procedure (Miller et al., 1988) or with the QuickGene DNA whole blood extraction kit (AutoGen, Holliston, MA). Candidate SNP genotyping was performed at the UIC Research Resource Center using the Agena Bioscience MassARRAY® System. SNPs included in this study had a call rate of at least 90% and were in Hardy Weinberg equilibrium (p > 0.05).
Statistical analysis
Based on the distribution of the pain phenotype data in our study sample, multiple linear regression was used to analyze CPI scores, whereas negative binomial regression was used to model utilization scores. Additive, and dominant genetic models of penetrance were used to assess the effect of the minor variant allele as compared to that of the major ancestral allele. All regression models were adjusted for covariates age, sex, and sickle type. R package ‘MASS’ was utilized for regression analysis.
Expression quantitative loci (eQTL) data were obtained from Genotype-Tissue Expression (GTEx) (version November 2019) for SNPs rs1611115 (chr9_133635393_T_C_b38), rs2797849 (chr9_133636819_G_C_b38), rs5320 (chr9_133642351_G_A_b38), and rs129882 (chr9_133658547_C_T_b38). The minor allele of rs5324 (chr9_133643536_G_A_b38) being less than 1% in occurrence was not included in the GTEx database. DBH gene expression in 11 different brain tissue types (preserved as fresh frozen tissue and whose main sampling site was at the Miami Brain Bank) were assessed. The tissue types included Amygdala, Anterior Cingulate Cortex, Caudate (basal ganglia), Cerebellar Hemisphere, Frontal Cortex, Hippocampus, Hypothalamus, Nucleus accumbens (basal ganglia), Putamen (basal ganglia), Spinal cord (cervical c-1), and Substantia nigra. Violin plots of significant associations between SNPs and tissue-specific gene expression levels were downloaded from the GTEx portal. We also looked up cis-eQTLs in the eQTLGen Consortium database (Võsa et al., 2021). Finally, functional analyses of rs1611115 was performed using web-based bioinformatic tools PROMO (http://alggen.lsi.upc.es/) and SNPinfo: FuncPred (https://snpinfo.niehs.nih.gov/), to predict putative transcription factor binding sites that may be affected by the SNP (Messeguer et al., 2002; Farré et al., 2003; Xu and Taylor, 2009).
Results
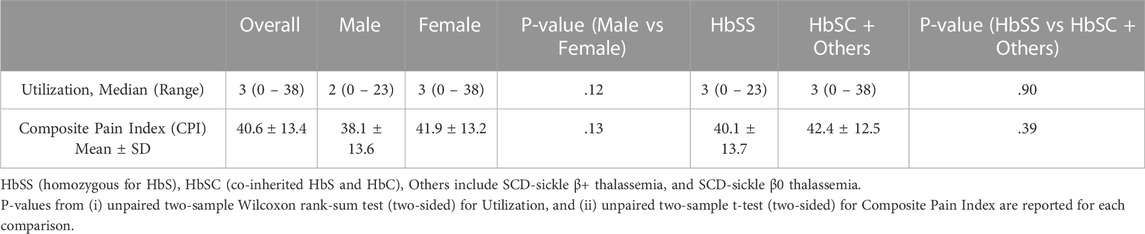
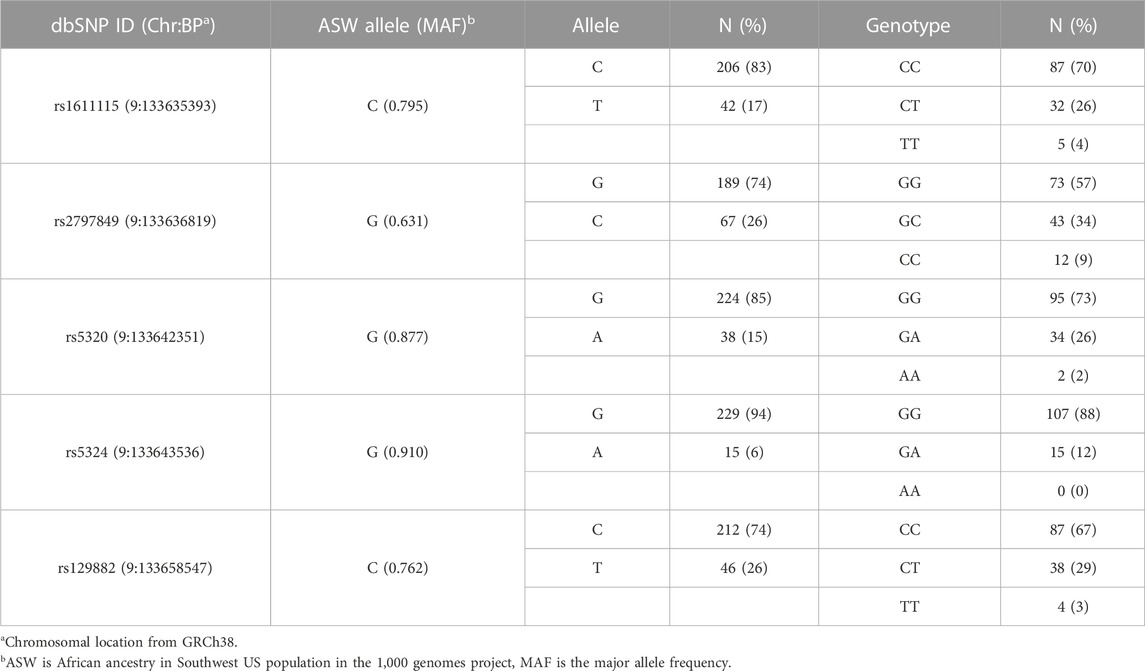
Acute and chronic pain phenotype data of study subjects have been summarized in Table 2. The heterogeneity of both pain phenotypes is reflected in the wide range of pain scores of the sample population. The 12-month utilization rate ranged from zero to 38, with a median of three utilizations per year. CPI scores on a scale of zero to 100 ranged from 14.8 to 86.5, with a mean score of 40.6. Neither utilization rate, nor CPI were found to be significantly different between males and females. Similarly, no significant differences in the pain phenotypes were observed based on sickle cell type. The distribution of the major and minor alleles and the corresponding genotypes in the study population is provided in Table 3. Comparison of allele frequencies with the African ancestry in Southwest US (ASW) population from the 1,000 genomes project showed that rs1611115, rs5320, rs5324, and rs129882 had comparable major allele frequencies. The major allele G of rs2797849 was, however, found to be more frequent in this study cohort (74%) than that in the ASW cohort (63%).
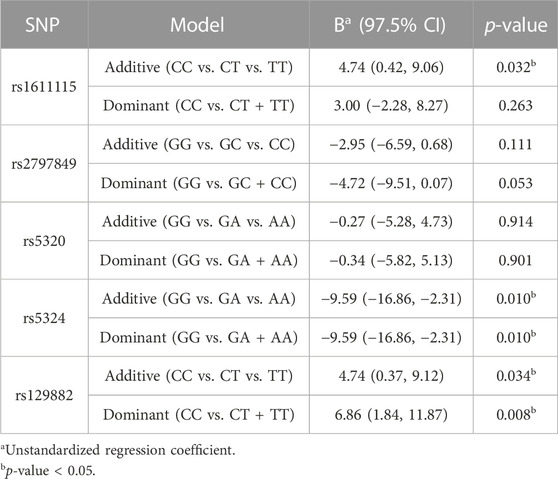
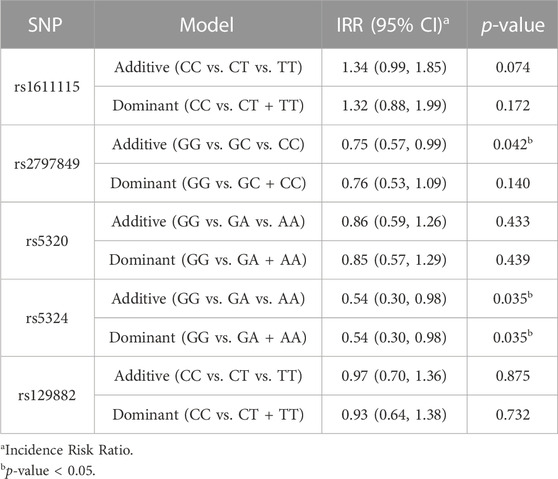
Multilinear regression models evaluating the association between these SNPs and CPI scores revealed that the minor alleles of rs1611115, rs5324, and rs129882 significantly associated with chronic pain (Table 4). The T allele of rs1611115 was found to significantly associate with higher CPI scores in the additive model (B = 4.74, p = 0.032). Similarly, the T allele of rs129882 also associated significantly with higher CPI scores (Additive: B = 4.74, p = 0.034; Dominant: B = 6.86, p = 0.008). Whereas the minor allele A of rs5324 associated with lower CPI scores (B = −9.59, p = 0.010). Since there were no subjects with homozygous recessive genotype for the SNP rs5324, the additive and dominant models were identical. As seen in Table 5, the rs5324 A allele associated significantly with decreased utilization rate of acute crisis pain (IRR = 0.54, p = 0.035) as well. The C allele of rs2797849 too was found to associate with decreased utilization scores (IRR = 0.75, p = 0.042).
To ascertain the potential functional role of these SNPs, the GTEx database was queried for tissue-specific association between expression levels of DBH and the SNPs. It was found that the T allele of rs1611115 associated with significantly lower expression of DBH in the frontal cortex and the anterior cingulate cortex in the GTEx database (Figure 1). Furthermore, rs1611115 was found to be a blood cis-eQTL of DBH-AS1 in eQTLGen database (p = 3.58 × 10−6), with the T allele being responsible for decreased gene expression.
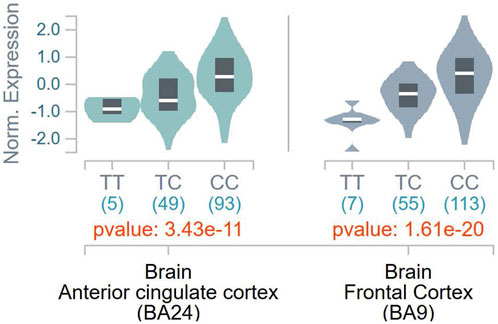
FIGURE 1. eQTL analysis of DBH-rs 1611115 from GTEx database. Violin plots of tissue-specific eQTLs of rs1611115 (chr9_133635393_T_C_b38) for DBH (ENSG00000123454.10) expression in Anterior Cingulate Cortex and Frontal Cortex obtained from the GTEx Analysis Release V8 (dbGap Accession phs000424.v8p2). The GTEX Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by NCL, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Since rs1611115 is located near the promoter region of DBH and was found to associate with altered expression level of DBH in specific regions of the brain, additional functional analysis was performed. Bioinformatic tools PROMO and SNPinfo:FuncPred were used to investigate the potential effect of this polymorphism on transcription factor binding sites (TFBS). It was found that the substitution of the allele C with variant T could lead to the loss of a putative transcription factor binding site, potentially that of E2F1 (Figure 2).
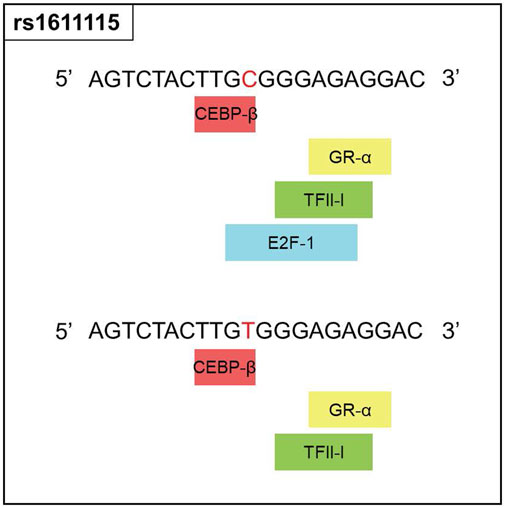
FIGURE 2. Functional prediction for rs1611115. Changes in the transcription factor blinding sites of rs1611115[C>T] as predicted by PROMO, Polymorphic variation in the input sequence is indicated by red letter. Input sequence was obtained from the dbSNP database.
Discussion
Polymorphisms on the DBH gene have been studied for their associations with several neurological disorders such as migraine (Fernandez et al., 2006; Fernandez et al., 2009; Ghosh et al., 2013), Parkinson’s disease (Ghosh et al., 2019), schizophrenia (Cubells et al., 2011; Sun et al., 2018), attention deficit hyperactivity disorder (Kieling et al., 2008), Alzheimer’s disease (Combarros et al., 2010), etc. In this study, we investigated the effect of DBH polymorphisms on pain in SCD and found that DBH rs5324 associated with both acute crisis pain and chronic pain, with the A allele being pain protective. On the other hand, rs1611115 and rs129882 were associated with chronic pain only, with the T alleles conferring a higher risk of pain. In addition, the C allele of rs2797849 was associated with decreased acute crisis pain.
As noted, rs5324 exhibited association with pain, while the other missense variant rs5320 did not associate with either of the pain phenotypes. In agreement with this observation, a previous study that developed an in-silico protein model of DBH predicted that the SNP rs5320 (Ala211Thr) may not alter enzyme activity since it did not lie close to the active site of the enzyme or its tetramer interface (Kapoor et al., 2011). The study also suggested that rs5324 (Asp290Asn) was situated close to the tetramer interface and could potentially interfere with tetramerization of the subunits into the enzyme’s quaternary structure. Thus, the exon SNP rs5324 may alter the enzymatic activity of DBH. It would be interesting to confirm such a hypothesis in future experiments.
Prior research examining the relationship between DBH variants and DBH levels in the plasma of African American found that while a few SNPs associated with altered plasma levels of DBH, rs1611115 accounted for the largest variation (30%–50%). The rs1611115 was also found to be in LD with other genetic variants and the T allele associated with significantly lower levels of DBH (Zabetian et al., 2001; Tang et al., 2007; Mustapic et al., 2014). Similarly, rs1611115 has been found to alter DBH proteins levels in the CSF (Weinshilboum et al., 1971). It has also been shown that the effect of DBH variants such as rs1611115 on gene expression can be tissue-specific (Barrie et al., 2014). With these studies in sight, the association of DBH variants (including rs1611115) with gene expression levels of DBH in different brain tissues were explored on the GTEx database. It should be noted that a majority of the samples in the GTEx database were obtained from white subjects, with African Americans accounting for approximately 13% of the overall cohort. It has been previously shown that the extent to which rs1611115 accounts for decreased levels of plasma DBH can vary between European Americans and African Americans (Zabetian et al., 2001). Nonetheless, it was found that the T allele of rs1611115 associated with decreased DBH expression levels in the frontal cortex tissue and the anterior cingulate cortex (ACC) tissue. This SNP-associated decrease in DBH expression may be explained by the loss of a putative transcriptional binding site for transcription factor E2F1. E2F1 belongs to a family of transcriptional factors that primarily regulate expression of genes involved in cellular proliferation (DeGregori et al., 1997; Wu et al., 2001). It has also been shown to mediate neurogenesis and dopamine-induced neuronal apoptosis (Hou et al., 2001; Cooper-Kuhn et al., 2002). However, the role of E2F1 is not limited to cell proliferation alone; thousands of genes contain the E2F1 binding site in their promoter region (Bieda et al., 2006; Iwanaga et al., 2006). E2F1 may activate or repress gene expression depending on the type of binding proteins it is programmed to recruit at that site (Denechaud et al., 2017).
Since DBH catalyzes the conversion of dopamine to noradrenaline, functional polymorphisms in DBH would alter the levels of both the neurotransmitters. Dopamine and norepinephrine can have pronociceptive or antinociceptive effects depending on the brain region, tissue type, as well as their release (Pertovaara, 2013; Taylor et al., 2016; Bravo et al., 2019). In one study, DBH knockout mice lacking noradrenaline production were found to develop chronic thermal hyperalgesia and exhibited diminished morphine efficacy (Jasmin et al., 2002). The frontal cortex, specifically the prefrontal cortex receives both dopaminergic and noradrenergic innervations from different regions in the midbrain, and their dysfunction have been implicated in pain conditions (Ong et al., 2019). The ACC is also believed to be critical in modulating pain and pain-related behavior (Fuchs et al., 2014; Bliss et al., 2016). Noxious stimuli have been found to activate the ACC in healthy individuals; ACC activation has also been noted in individuals with chronic pain (Talbot et al., 1991; Apkarian et al., 2005). However, the precise roles of dopaminergic and noradrenergic signaling in pain processing within the frontal cortex and ACC remains to be identified.
In summary, SNPs in the DBH gene were found to be associated with acute and chronic pain in SCD. The eQTL data revealed that the promoter region variant rs1611115-T also associated with DBH expression in specific regions of the brain known to be involved in pain signaling. Bioinformatic analysis indicated that this variant could be potentially involved in altered transcriptional regulation. Haplotype analysis would have been an interesting addition to this study but was not possible due to the relatively small sample size. While this study is exploratory in nature and limited by its sample size, the findings of this candidate-gene study suggest a possible role of DBH on pain variability in SCD. Larger genetic association studies and functional genomics experiments are needed to comprehensively understand how the identified SNPs may be altering DBH gene expression, its enzymatic activity, and consequently pain perception.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://data.mendeley.com/datasets/j9nsx69jyg.
Ethics statement
The studies involving human participants were reviewed and approved by University of Illinois Chicago Institutional Review Board. Written informed consent to participate in this study was provided by the participants or participantsʼ legal guardians.
Author contributions
ZW, YH, and NS contributed to conception and design of the study. NS and YY organized the database and performed the statistical analysis. NS wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported in part by grants from the Illinois Department of Public Health (IDPH) and the National Heart, Lung, and Blood Institute (R35 HL140031, 1R01 HL078536). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the IDPH, NIH, NHLBI, or Veteran’s Administration.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Apkarian, A. V., Bushnell, M. C., Treede, R. D., and Zubieta, J. K. (2005). Human brain mechanisms of pain perception and regulation in health and disease. Eur. J. Pain 9 (4), 463–484. doi:10.1016/j.ejpain.2004.11.001
Barrie, E. S., Weinshenker, D., Verma, A., Pendergrass, S. A., Lange, L. A., Ritchie, M. D., et al. (2014). Regulatory polymorphisms in human DBH affect peripheral gene expression and sympathetic activity. Circ. Res. 115 (12), 1017–1025. doi:10.1161/CIRCRESAHA.116.304398
Bieda, M., Xu, X., Singer, M. A., Green, R., and Farnham, P. J. (2006). Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16 (5), 595–605. doi:10.1101/gr.4887606
Bliss, T. V., Collingridge, G. L., Kaang, B. K., and Zhuo, M. (2016). Synaptic plasticity in the anterior cingulate cortex in acute and chronic pain. Nat. Rev. Neurosci. 17 (8), 485–496. doi:10.1038/nrn.2016.68
Bravo, L., Llorca-Torralba, M., Berrocoso, E., and Micó, J. A. (2019). Monoamines as drug targets in chronic pain: Focusing on neuropathic pain. Front. Neurosci. 13, 1268. doi:10.3389/fnins.2019.01268
Combarros, O., Warden, D. R., Hammond, N., Cortina-Borja, M., Belbin, O., Lehmann, M. G., et al. (2010). The dopamine β-hydroxylase -1021C/T polymorphism is associated with the risk of Alzheimer's disease in the Epistasis Project. BMC Med. Genet. 11, 162. doi:10.1186/1471-2350-11-162
Cooper-Kuhn, C. M., Vroemen, M., Brown, J., Ye, H., Thompson, M. A., Winkler, J., et al. (2002). Impaired adult neurogenesis in mice lacking the transcription factor E2F1. Mol. Cell. Neurosci. 21 (2), 312–323. doi:10.1006/mcne.2002.1176
Cubells, J. F., Sun, X., Li, W., Bonsall, R. W., McGrath, J. A., Avramopoulos, D., et al. (2011). Linkage analysis of plasma dopamine β-hydroxylase activity in families of patients with schizophrenia. Hum. Genet. 130 (5), 635–643. doi:10.1007/s00439-011-0989-6
Cubells, J. F., van Kammen, D. P., Kelley, M. E., Anderson, G. M., O'Connor, D. T., Price, L. H., et al. (1998). Dopamine β-hydroxylase: Two polymorphisms in linkage disequilibrium at the structural gene DBH associate with biochemical phenotypic variation. Hum. Genet. 102 (5), 533–540. doi:10.1007/s004390050736
DeGregori, J., Leone, G., Miron, A., Jakoi, L., and Nevins, J. R. (1997). Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. 94 (14), 7245–7250. doi:10.1073/pnas.94.14.7245
Denechaud, P. D., Fajas, L., and Giralt, A. (2017). E2F1, a novel regulator of metabolism. Front. Endocrinol. (Lausanne) 8, 311. doi:10.3389/fendo.2017.00311
Diliberto, E. J., and Allen, P. L. (1981). Mechanism of dopamine-beta-hydroxylation. Semidehydroascorbate as the enzyme oxidation product of ascorbate. J. Biol. Chem. 256 (7), 3385–3393. doi:10.1016/s0021-9258(19)69620-1
Dyal, B. W., Ezenwa, M. O., Yoon, S. L., Fillingim, R. B., Yao, Y., Schlaeger, J. M., et al. (2020). A QST-based pain phenotype in adults with sickle cell disease: Sensitivity and specificity of quality descriptors. Pain Pract. 20 (2), 168–178. doi:10.1111/papr.12841
Ezenwa, M. O., Molokie, R. E., Wang, Z. J., Yao, Y., Suarez, M. L., Angulo, V., et al. (2014). Outpatient pain predicts subsequent one-year acute health care utilization among adults with sickle cell disease. J. Pain Symptom Manage 48 (1), 65–74. doi:10.1016/j.jpainsymman.2013.08.020
Farré, D., Roset, R., Huerta, M., Adsuara, J. E., Roselló, L., Albà, M. M., et al. (2003). Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31 (13), 3651–3653. doi:10.1093/nar/gkg605
Fernandez, F., Colson, N., Quinlan, S., MacMillan, J., Lea, R. A., and Griffiths, L. R. (2009). Association between migraine and a functional polymorphism at the dopamine beta-hydroxylase locus. Neurogenetics 10 (3), 199–208. doi:10.1007/s10048-009-0176-2
Fernandez, F., Lea, R. A., Colson, N. J., Bellis, C., Quinlan, S., and Griffiths, L. R. (2006). Association between a 19 bp deletion polymorphism at the dopamine beta-hydroxylase (DBH) locus and migraine with aura. J. Neurol. Sci. 251 (1-2), 118–123. doi:10.1016/j.jns.2006.09.013
Fuchs, P. N., Peng, Y. B., Boyette-Davis, J. A., and Uhelski, M. L. (2014). The anterior cingulate cortex and pain processing. Front. Integr. Neurosci. 8, 35. doi:10.3389/fnint.2014.00035
Ghosh, A., Sadhukhan, T., Giri, S., Biswas, A., Das, S. K., Ray, K., et al. (2019). Dopamine β Hydroxylase (DBH) is a potential modifier gene associated with Parkinson's disease in Eastern India. Neurosci. Lett. 706, 75–80. doi:10.1016/j.neulet.2019.05.015
Ghosh, J., Pradhan, S., and Mittal, B. (2013). Identification of a novel ANKK1 and other dopaminergic (DRD2 and DBH) gene variants in migraine susceptibility. Neuromolecular Med. 15 (1), 61–73. doi:10.1007/s12017-012-8195-9
He, Y., Wilkie, D. J., Nazari, J., Wang, R., Messing, R. O., DeSimone, J., et al. (2016). PKCδ-targeted intervention relieves chronic pain in a murine sickle cell disease model. J. Clin. Investigation 126 (8), 3053–3057. doi:10.1172/JCI86165
Hou, S. T., Cowan, E., Walker, T., Ohan, N., Dove, M., Rasqinha, I., et al. (2001). The transcription factor E2F1 promotes dopamine-evoked neuronal apoptosis by a mechanism independent of transcriptional activation. J. Neurochem. 78 (2), 287–297. doi:10.1046/j.1471-4159.2001.00402.x
Iwanaga, R., Komori, H., Ishida, S., Okamura, N., Nakayama, K., Nakayama, K. I., et al. (2006). Identification of novel E2F1 target genes regulated in cell cycle-dependent and independent manners. Oncogene 25 (12), 1786–1798. doi:10.1038/sj.onc.1209210
Jarcho, J. M., Mayer, E. A., Jiang, Z. K., Feier, N. A., and London, E. D. (2012). Pain, affective symptoms, and cognitive deficits in patients with cerebral dopamine dysfunction. Pain 153 (4), 744–754. doi:10.1016/j.pain.2012.01.002
Jasmin, L., Tien, D., Weinshenker, D., Palmiter, R. D., Green, P. G., Janni, G., et al. (2002). The NK1 receptor mediates both the hyperalgesia and the resistance to morphine in mice lacking noradrenaline. Proc. Natl. Acad. Sci. U. S. A. 99 (2), 1029–1034. doi:10.1073/pnas.012598599
Jhun, E. H., He, Y., Yao, Y., Molokie, R. E., Wilkie, D. J., and Wang, Z. J. (2014). Dopamine D3 receptor Ser9Gly and catechol-o-methyltransferase Val158Met polymorphisms and acute pain in sickle cell disease. Anesth. Analg. 119 (5), 1201–1207. doi:10.1213/ANE.0000000000000382
Kapoor, A., Shandilya, M., and Kundu, S. (2011). Structural insight of dopamine β-hydroxylase, a drug target for complex traits, and functional significance of exonic single nucleotide polymorphisms. PLOS ONE 6 (10), e26509. doi:10.1371/journal.pone.0026509
Kieling, C., Genro, J. P., Hutz, M. H., and Rohde, L. A. (2008). The -1021 C/T DBH polymorphism is associated with neuropsychological performance among children and adolescents with ADHD. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B (4), 485–490. doi:10.1002/ajmg.b.30636
Leventhal, L., Smith, V., Hornby, G., Andree, T. H., Brandt, M. R., and Rogers, K. E. (2007). Differential and synergistic effects of selective norepinephrine and serotonin reuptake inhibitors in rodent models of pain. J. Pharmacol. Exp. Ther. 320 (3), 1178–1185. doi:10.1124/jpet.106.109728
Manwani, D., and Frenette, P. S. (2013). Vaso-occlusion in sickle cell disease: Pathophysiology and novel targeted therapies. Blood 122 (24), 362–369. doi:10.1182/asheducation-2013.1.362
Messeguer, X., Escudero, R., Farré, D., Núñez, O., Martínez, J., and Albà, M. M. (2002). Promo: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18 (2), 333–334. doi:10.1093/bioinformatics/18.2.333
Miller, S. A., Dykes, D. D., and Polesky, H. F. (1988). A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16 (3), 1215. doi:10.1093/nar/16.3.1215
Miller, S. T., Sleeper, L. A., Pegelow, C. H., Enos, L. E., Wang, W. C., Weiner, S. J., et al. (2000). Prediction of adverse outcomes in children with sickle cell disease. N. Engl. J. Med. 342 (2), 83–89. doi:10.1056/NEJM200001133420203
Mustapic, M., Maihofer, A. X., Mahata, M., Chen, Y., Baker, D. G., O'Connor, D. T., et al. (2014). The catecholamine biosynthetic enzyme dopamine β-hydroxylase (DBH): First genome-wide search positions trait-determining variants acting additively in the proximal promoter. Hum. Mol. Genet. 23 (23), 6375–6384. doi:10.1093/hmg/ddu332
Okpala, I., and Tawil, A. (2002). Management of pain in sickle-cell disease. J. R. Soc. Med. 95 (9), 456–458. doi:10.1258/jrsm.95.9.456
Ong, W. Y., Stohler, C. S., and Herr, D. R. (2019). Role of the prefrontal cortex in pain processing. Mol. Neurobiol. 56 (2), 1137–1166. doi:10.1007/s12035-018-1130-9
Pertovaara, A. (2013). The noradrenergic pain regulation system: A potential target for pain therapy. Eur. J. Pharmacol. 716 (1), 2–7. doi:10.1016/j.ejphar.2013.01.067
Platt, O. S., Thorington, B. D., Brambilla, D. J., Milner, P. F., Rosse, W. F., Vichinsky, E., et al. (1991). Pain in sickle cell disease. Rates and risk factors. N. Engl. J. Med. 325 (1), 11–16. doi:10.1056/NEJM199107043250103
Sadhu, N., Jhun, E. H., Posen, A., Yao, Y., He, Y., Molokie, R. E., et al. (2020). Phenylethanolamine N-methyltransferase gene polymorphisms associate with crisis pain in sickle cell disease patients. Pharmacogenomics 21 (4), 269–278. doi:10.2217/pgs-2019-0096
Sadhu, N., Jhun, E. H., Yao, Y., He, Y., Molokie, R. E., Wilkie, D. J., et al. (2018). Genetic variants of GCH1 associate with chronic and acute crisis pain in African Americans with sickle cell disease. Exp. Hematol. 66, 42–49. doi:10.1016/j.exphem.2018.07.004
Shah, N., Bhor, M., Xie, L., Halloway, R., Arcona, S., Paulose, J., et al. (2019). Treatment patterns and economic burden of sickle-cell disease patients prescribed hydroxyurea: A retrospective claims-based study. Health Qual. Life Outcomes 17 (1), 155. doi:10.1186/s12955-019-1225-7
Sun, Z., Ma, Y., Li, W., He, J., Li, J., Yang, X., et al. (2018). Associations between the DBH gene, plasma dopamine β-hydroxylase activity and cognitive measures in Han Chinese patients with schizophrenia. Schizophr. Res. 193, 58–63. doi:10.1016/j.schres.2017.06.028
Talbot, J. D., Marrett, S., Evans, A. C., Meyer, E., Bushnell, M. C., and Duncan, G. H. (1991). Multiple representations of pain in human cerebral cortex. Science 251 (4999), 1355–1358. doi:10.1126/science.2003220
Tang, Y.-l., Epstein, M. P., Anderson, G. M., Zabetian, C. P., and Cubells, J. F. (2007). Genotypic and haplotypic associations of the DBH gene with plasma dopamine β-hydroxylase activity in African Americans. Eur. J. Hum. Genet. 15 (8), 878–883. doi:10.1038/sj.ejhg.5201838
Taylor, A. M., Becker, S., Schweinhardt, P., and Cahill, C. (2016). Mesolimbic dopamine signaling in acute and chronic pain: Implications for motivation, analgesia, and addiction. Pain 157 (6), 1194–1198. doi:10.1097/j.pain.0000000000000494
Tunbridge, E. M., Narajos, M., Harrison, C. H., Beresford, C., Cipriani, A., and Harrison, P. J. (2019). Which dopamine polymorphisms are functional? Systematic Review and meta-analysis of COMT, DAT, DBH, DDC, DRD1-5, MAOA, MAOB, TH, VMAT1, and VMAT2. Biol. Psychiatry 86 (8), 608–620. doi:10.1016/j.biopsych.2019.05.014
Vandenbergh, D. J., Anthony, K., and Whitfield, K. E. (2003). Optimizing DNA yield from buccal swabs in the elderly: Attempts to promote buccal cell growth in culture. Am. J. Hum. Biol. 15 (5), 637–642. doi:10.1002/ajhb.10177
Võsa, U., Claringbould, A., Westra, H. J., Bonder, M. J., Deelen, P., Zeng, B., et al. (2021). Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 53 (9), 1300–1310. doi:10.1038/s41588-021-00913-z
Wang, Z. J., Wilkie, D. J., and Molokie, R. (2010). Neurobiological mechanisms of pain in sickle cell disease. Hematol. Am. Soc. Hematol. Educ. Program 2010, 403–408. doi:10.1182/asheducation-2010.1.403
Weinshilboum, R. M., Thoa, N. B., Johnson, D. G., Kopin, I. J., and Axelrod, J. (1971). Proportional release of norepinephrine and dopamine- -hydroxylase from sympathetic nerves. Science 174 (4016), 1349–1351. doi:10.1126/science.174.4016.1349
Wilkie, D. J., Molokie, R., Boyd-Seal, D., Suarez, M. L., Kim, Y. O., Zong, S., et al. (2010). Patient-reported outcomes: Descriptors of nociceptive and neuropathic pain and barriers to effective pain management in adult outpatients with sickle cell disease. J. Natl. Med. Assoc. 102 (1), 18–27. doi:10.1016/s0027-9684(15)30471-5
Wilkie, D. J., Molokie, R. E., Suarez, M. L., Ezenwa, M. O., and Wang, Z. J. (2015). Composite pain Index: Reliability, validity, and sensitivity of a patient-reported outcome for research. Pain Med. 16 (7), 1341–1348. doi:10.1111/pme.12703
Wu, L., Timmers, C., Maiti, B., Saavedra, H. I., Sang, L., Chong, G. T., et al. (2001). The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414 (6862), 457–462. doi:10.1038/35106593
Xu, Z., and Taylor, J. A. (2009). SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 37, W600–W605. doi:10.1093/nar/gkp290
Zabetian, C. P., Anderson, G. M., Buxbaum, S. G., Elston, R. C., Ichinose, H., Nagatsu, T., et al. (2001). A quantitative-trait analysis of human plasma-dopamine beta-hydroxylase activity: Evidence for a major functional polymorphism at the DBH locus. Am. J. Hum. Genet. 68 (2), 515–522. doi:10.1086/318198
Keywords: pain, sickle cell disease, chronic pain, acute pain, pharmacogenenomics and personalised medicine
Citation: Sadhu N, He Y, Yao Y, Wilkie DJ, Molokie RE and Wang ZJ (2023) Candidate gene association study suggests potential role of dopamine beta-hydroxylase in pain heterogeneity in sickle cell disease. Front. Genet. 14:1193603. doi: 10.3389/fgene.2023.1193603
Received: 27 March 2023; Accepted: 23 May 2023;
Published: 13 June 2023.
Edited by:
Marcelo Rizzatti Luizon, Federal University of Minas Gerais, BrazilReviewed by:
David J. Vandenbergh, The Pennsylvania State University (PSU), United StatesAthena Starlard-Davenport, University of Tennessee Health Science Center (UTHSC), United States
Copyright © 2023 Sadhu, He, Yao, Wilkie, Molokie and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zaijie Jim Wang, zjwang@uic.edu
 Nilanjana Sadhu
Nilanjana Sadhu Ying He
Ying He Yingwei Yao3
Yingwei Yao3 Diana J. Wilkie
Diana J. Wilkie Robert E. Molokie
Robert E. Molokie Zaijie Jim Wang
Zaijie Jim Wang