
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet., 08 January 2024
Sec. Evolutionary and Population Genetics
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1174584
This article is part of the Research TopicComparative and Evolutionary Analyses of Organelle GenomesView all 5 articles
 Jiaqiao Li1,2,3
Jiaqiao Li1,2,3 Enjiong Huang4
Enjiong Huang4 Yifan Wu1
Yifan Wu1 Changqiang Zhu1
Changqiang Zhu1 Wenhao Li1
Wenhao Li1 Lele Ai1
Lele Ai1 Qinghua Xie3
Qinghua Xie3 Zhi Tian1
Zhi Tian1 Weiwen Zhong5
Weiwen Zhong5 Gang Sun2*
Gang Sun2* Lingling Zhang3
Lingling Zhang3 Weilong Tan1*
Weilong Tan1*Background: The invasive brownrat (Rattus norvegicus) and the Oriental rats (Rattus tanezumi) are common commensal murid that are important hosts for rodent-borne diseases in southeast Asia. Understanding their population structure and genetic diversity is essential to uncover their invasion biology and distribution dynamics that are essential for controlling rodent-borne diseases.
Methods: TA total of 103 R. norvegicus and 85 R. tanezumi were collected from 13 to 9 coastal areas of six provincial monitoring sentinel sites, respectivelyto assess patterns in their microsatellite loci and their mitochondrial coxl gene region.
Results: Eleven sampled populations of R. norvegicus were divided into two major clusters by region. The observed heterozygosity values of all regional populations were smaller than expected genetic diversity heterozygosity values and deviated from Hardy-Weinberg equilibrium Nine sample populations of R. tanezumi were divided into three clusters; two that included sample from Hainan and Fujian provinces, and one that included samples from the other provinces and cities. The genetic diversity of R. tanezumi was highest in samples from Jiangsu and Guangdong provinces.
Conclusion: The data in this paper confirm the two invasive rodent species from the southeastern coastal region of China may have relied on maritime transport to spread from the southern region of China to the Yangtze River basin. R. tanezumi may then hanve migrated unidirectionally, along the southeastern provinces of China towards the north, while R. norvegicus spread in a complex and multidirectional manner in Hainan, Fujian, Zhejiang and Jiangsu Provinces of the country.
Rats are the most adaptable and widespread mammals, that live close to humans and are associated with human migrations (Matisoo-Smith and Robins, 2004; Suzuki et al., 2013). Generally, four major murid rodent species are invasive and commensal, namely, Rattus rattus, Rattus tanezumi, Rattus exulans, and Rattus norvegicus (Puckett et al., 2020). Rattus tanezumi and R. norvegicus serve as hosts for numerous pathogens, representing a significant threat to human health due to their presence. (Himsworth et al., 2013). They are also major pests in farmlands in southern and southwestern China, causing damage to crops during spring and autumn before harvest (Htwe et al., 2012).
In China, the distribution range of R. tanezumi and R. norvegicus has expanded extensively over the past decades (Wang, 2003; Mora et al., 2010). In the 1950s, R. tanezumi in China was mainly distributed in the Yangtze River basin, eastern and southern regions (Robins et al., 2008; Deinum et al., 2015). Due to the rapid changes in transportation and the landscape, the geographic distribution of R. tanezumi has increased significantly in recent years (Guo et al., 2019). So far, R. tanezumi has been reported in Qinghai, Tibet Autonomous Region, west of Xinjiang Uyghur Autonomous Region, north of Hebei and Liaoning (Hou and Jiang, 2008; Ma et al., 2011; Yang et al., 2011). Rattus norvegicus is widely distributed in all provinces except Tibet due to its excellent long-distance transmission ability (Wang et al., 2021).
The origin and genetic evolution of the R. norvegicus has been controversial. The regions of Eastern Central Asia, including present-day China and Mongolia, have long been regarded as its birthplace. Concurrently, an alternative hypothesis proposes that R. norvegicus originated in the realms of southeastern Siberia and northeastern China, as suggested by Lin et al. (2012) (Lin et al., 2012), subsequently embarking on a westward journey across the vast Eurasian grasslands and ultimately reaching Europe. (Gibbs et al., 2004). Recent advancements in fossil dating have unveiled a more illuminating perspective. These cutting-edge analyses contribute to a more robust narrative, indicating that the species originated in southwestern China approximately 1.2–1.6 million years ago. (Jin et al., 2008; Wu and Wang, 2012).
Emerging and re-emerging infectious diseases endanger human health and public safety, and their infections are influenced by the rate by transmission of their natural hosts between geographic areas (Morens et al., 2004). Rats as typical zoonotic hosts, harbour ectoparasites such as ticks, lice and mites, and a variety of internal parasites (Puckett et al., 2020). In addition, they harbourrange of bacteria, rickettsiae and pathogens (Huang et al., 2013; Yin et al., 2021). Currently, they are known to be vectors of pathogens of plague (caused by Yersinia pestis), leptospirosis (Leptospira), hemorrhagic fever with renal syndrome (Hantavirus), COVID-19 (SARS-CoV-2), and scrub typhus (Plyusnina et al., 2009; Blasdell et al., 2011). Among these, plague has had three major outbreaks in human history. Europe was devastated by Justinian’s plague (541–767 AD) and the Black Death (1346–18th Century) (Pollitzer, 1951). In China, tens of people of individuals died of plague in the 19th Century (Morelli et al., 2010). Plague is carried by rodents and spread by the bite of infected fleas, and owing to the endeavors of rodents, notably those thriving in close proximity to human habitats, such as R. tanezumi and R. norvegicus, ubiquitous in the human environment and frequently interacting with humans in their daily routines, there is a significant escalation in the potential for ticks harbored on these rodents to transmit diseases within human populations. Rodent population dynamics and distribution may also affect the risk of human hantavirus infections in both (Xiao et al., 2018) developing or developed countries (Pimentel et al., 2000). To prevent and monitor the risk posed by the migration of these invasive and commensal rodents, their population genetic structure can be assessed by commonly used molecular genetic markerssuch as among which microsatellite loci and mtDNA as indicators of genetic diversity and phylogenetic relationships (Brown et al., 1979; Montagutelli, 1991). For instance, Zhao et al. (2014) illuminated, via mitochondrial DNA sequencing of the brown house mouse (R. norvegicus), that rivers had negligible impact on the genetic patterns of the species (Zhao et al., 2016). In a similar vein, Guo et al. (2019) demonstrated, utilizing microsatellite markers and mitochondrial DNA (COI and D-loop) sequence analyses, that populations of the yellow marmot from Tibet and Sichuan exhibited a linkage transmitted via the Sichuan-Tibet Motorway, as opposed to being introduced via the Tea Horse Road (Guo et al., 2019).
To investigate dispersal patterns and population structure of R. tanezumi and R. norvegicus along the southeastern coastal areas of China, their populations from six provinces were sampled and screened using microsatellite loci and mtDNA sequences, respectively. The microsatellite loci and mtDNA employed in this study have been previously documented for their informative genetic polymorphism in various studies. These markers have been utilized to analyze the genetic structure and information of mouse populations in regions such as the United States and Asia. (Kunieda et al., 1992; Lack et al., 2012; Guo et al., 2019). Based on the genetic diversity and population structure among each populations, the dispersal patterns of populations were evaluated.
As challenges like mounting industrialization and global warming persist, a growing impact is observed on diverse commensal rodent species. Consequently, we posit that the genetic structure and migration patterns of these two commensal rodents in the coastal regions of China are probably influenced by human activities or transportation mechanisms, reflecting the dynamics within the framework of contemporary society. Our results will provide insights into genetic diversity, genetic differentiation, gene flow patterns, and possible hybridization among populations of different regions, with implications on the potential control of these problem invasive and commensal murid rodents.
In this study, R. norvegicus were sample from 13 localities and R. tanezumi from 9 localities in six provinces along the southeastern coast of China (Table 1; Figure 1). Given that it was necessary to avoid factors of kinship among samples, we randomly plac trapping cages and sticky boards in residential areas, ports and other areas at a distance of more than 2 km between sample sites (All captured rodent samples were executed by spinal subluxation (Ethics Committee Batch No: 2018001003)). Muscle and liver tissues were dissected out and frozen at −80 °C. DNA was extracted from mouse samples using the TaKaRa MiniBEST Universal Genomic DNA kit according to the manufacturer’s requirements. OD260/OD280 values and nucleic acid concentrations were recorded in a spectrophotometer (KAIAO,K5600,Beijing, China)to determine the concentration and purity of the DNA. The species were genetically-identified based on the screening of the cytochrome (cytb)gene region (S1 Table), leading to 88 samples being identified as Rtanezumi and 103 as R. norvegicus.
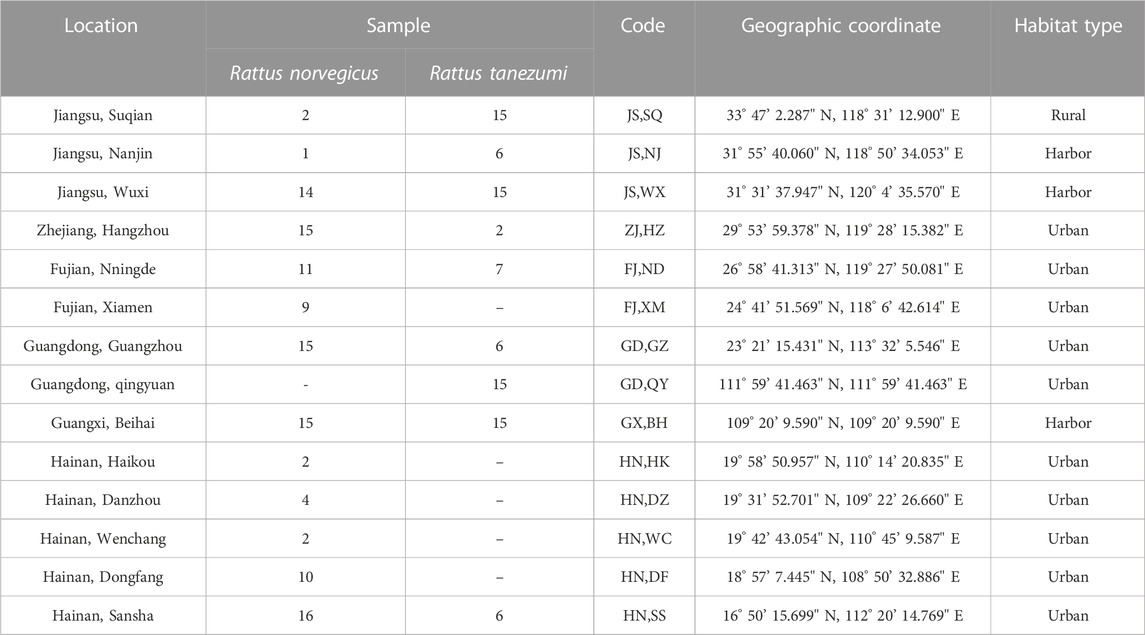
TABLE 1. Sampling localities and their geographic coordinates and habitat types of rat samples from the southeastern coast of China.
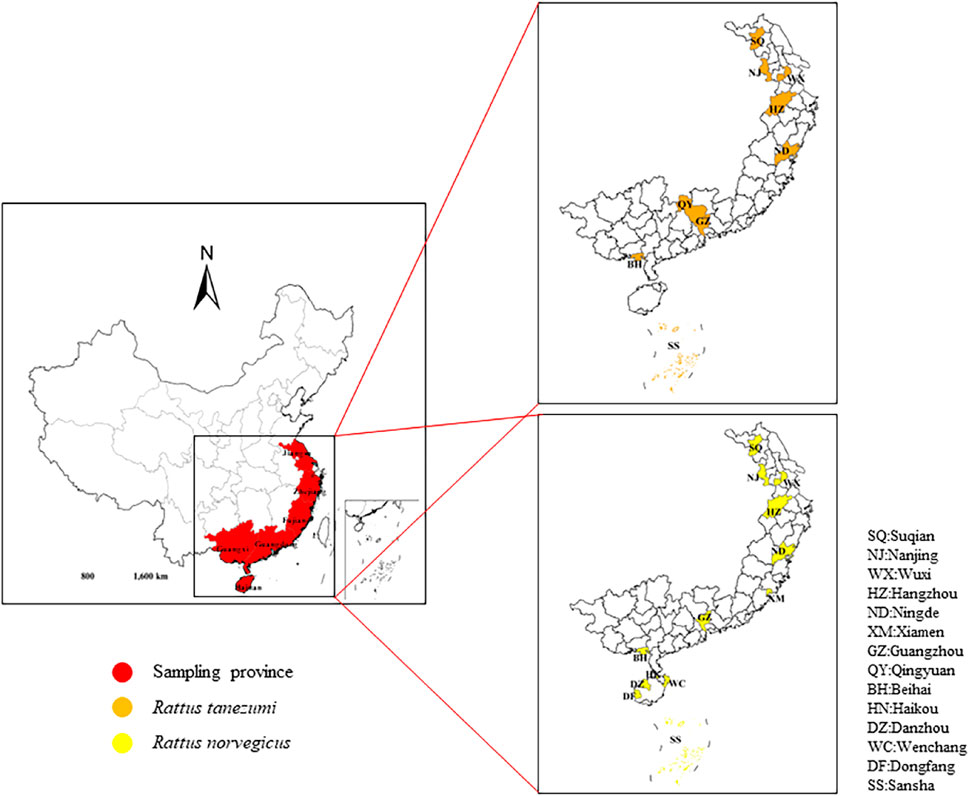
FIGURE 1. Sampling loca lities. Sampling sites of Rattus norvegicus and R. tanezumi from different regions southeastern coast of China. The sampling areas are marked with different colors.
According to the principles of high conservation and polymorphism, 9 and 10 microsatellite loci for R. tanezumi and R. norvegicus were screened. Sequences and primers were obtained from Genebank and the literature (Supplementary Table S1), and were synthesized by Biotech Bioengineering (Shanghai) Co.,shanghai, China. The reaction system included 50 μL and contained 10 µL PCR buffer, 10 ng DNA, 0.25 U Taq enzyme (10 p.m./µL, TaKaRa), 10 μM dNTPs (2.5 mM, TaKaRa) and approximately 20 μM of each primer. The 5′ends of the forward primers were labeled with fluorescent dyes (FAM, HEX and ROX). The cycling conditions were as follows: 5 min at 94°C; 10 cycles of 95°C for 30 s, 60°C for 40 s, and 72°C for 50 s; 27 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 50 s, 72°C for 10 min. Formamide was then mixed with the internal lane standards at a volume ratio of 100:1, 15 uL was taken and mixed with 10-fold diluted PCR products. Capillary electrophoresis was then performed using a 3730XL sequencer, and the raw data obtained from the sequencer were analyzed using Fragment (Plant) analysis software in Genemarker to obtain fragment sizes.
Mitochondrial sequences of R. tanezumi and R. norvegicus were amplified using coxl, D-LOOP and ND4 primersand. the detailed primer information is provided in Supplementary Table S1. The amplification system was configured in a thermal cycler with a total volume of 50 μL using genomic DNA as template: 5 μL of template DNA, 2.5 μL of each primer (10 μM) and 40 μL of Ex Taq (TaKaRa Taq). The PCR amplification program was set as follows: predenaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C (coxl/ND4/D-LOOP) for 30 s and elongation at 72°C for 1 min, with a final extension at 72°C for 10 min. The PCR reaction products were separated by 1.2% agarose gel electrophoresis, and then imaged by UV gel imager (ClinX, GenoSens 2150,shanghai, China). Samples exhibiting clear, singular positive bands were forwarded to the Institute of Bioengineering in Shanghai, China, for bidirectional sequencing. Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to confirm whether they were the target sequences.
PIC Calc 0.6 was used to calculate polymorphic information content (PIC) values per microsatellite locus for each sample of R. tanezumi and R. norvegicus based on the microsatellite data (Nagy et al., 2012). Popgene 1.32 (Rousset, 2008) was used to calculate diversity indices including number of different alleles (Na), number of effective alleles (Ne), expected genetic diversity (He), observed heterozygosity (Ho), Shannon Wiener Diversity Index I) and inbreeding coefficient (FIS) for each microsatellite locus in both species rat. Based on the mitochondrial data, nucleotide diversity indices that included number of samples S), number of haplotypes H), haplotype diversity index (Hd), and nucleotide diversity (Pi) were calculated for all populations using DNAsp 6.0 software (Rozas et al., 2017).
Fisher’s exact test with Bonferroni correction based on microsatellite data was used to determine possible deviations from Hardy-Weinberg equilibrium (HWE) and heterozygosity defects (Cristescu et al., 2009). Exact p-values were estimated using the Markov chain algorithm with 10,000 dememorizations, 500 batches, and 5,000 iterations per batch (Slatkin, 1995). Pairwise differences between populations and AMOVA analyses based on allele frequencies were analysed using Arlequin software 3.5.1.3 (Slatkin, 1995; Excoffier and Lischer, 2010). Genetic variation was evaluated through Analysis of Molecular Variance (AMOVA) at three distinct levels, aiming to capture genetic diversity and structure among various positional groups within the same rat species. Additionally, the fixation index (FST) for each population was computed, relying on the mean of pairwise differences between populations. (Goudet, 2001). The Unweighted Pair Group Method with Arithmetic Mean (UPGMA) phenogram was built using NTsys software 2.10e (Piry et al., 2017). Alternatively, the discriminant analysis of principal components (DAPC) analyses were performed by the R package “adegenet 1.3" (Jombart, 2008).
Simultaneously, potential migration routes were reconstructed utilizing the R package “divMigrate,” configuring the number of bootstrap replicates to 3, setting the alpha value to 0.05, opting for the Nm method for the migration statistic, and establishing the filter threshold value at 0.25. (Kharzinova et al., 2016). Statistical assessments for neutral detection, encompassing Fu’s Fs and Tajima’s D, were executed utilizing mitochondrial data with Arlequin 3.5.2.2. The analysis involved plotting observed pairwise nucleotide differences between mtDNA haplotypes against their corresponding expected frequencies. Furthermore, the construction of a TCS network, elucidating haplotype relationships among rodent populations, was accomplished using popART software and was grounded in mitochondrial data. (Leigh and Bryant, 2015).
A total of 14 rat populations were sampled from urban, harbor, and rural areas of southeastern coast of China (Table 1; Figure 1). Each of the breeding site buffer zones was isolated from each other, indicating the independence of each population. In addition, 16 pairs of microsatellite markers were obtained from previous studies, all of which were highly polymorphic, and were therefore selected for microsatellite genotyping (Supplementary Table S1). The PIC values for each locus ranged from 0.707 to 0.939, and nearly all selected markers were highly informative (PIC values > 0.5) according to (Ditta et al., 2018) definition of PIC values. Microsatellite results showed average number of alleles (Na) per R. tanezumi population ranged from 3.000 ± 1.323 to 14.200 ± 3.645, and the allele numbers of Jiangsu and Guangdong regions were significantly higher (Table 2). Shannon index of the two provinces was the highest. The observed heterozygosity (Ho) values of all areas ranged from 0.685 ± 0.243 to 0.343 ± 0.340, and were lower than the expected heterozygosity (He) values (ranging from 0.869 ± 0.044 to 0.500 ± 0.258). The FIS values for each population ranged from −0.026 to 0.218, and rat populations in both the Jiangsu and Guangdong regions showed significant deviations from HWE, indicating that these populations contain varying degrees of inbreeding and heterozygote deficiency. The observed Na value interval in the R. norvegicus population ranged from 2.917 ± 0.996 to 11.333 ± 2.570, with the largest Na values in the Hainan and Guangdong populations and the smallest Na values in the Zhejiang population. The mean Ho value (0.552 ± 0.179) was significantly lower than the mean He value (0.827 ± 0.067) in the five regional rat populations except Zhejiang, and the Ho value (0.599 ± 0.387) was greater than the He value (0.555 ± 0.218) in the Zhejiang population. The FIS values for each population ranged from −0.026 to 0.218, and the FIS values of Hainan, Guangdong and Guangxi populations were significantly different from HWE values.
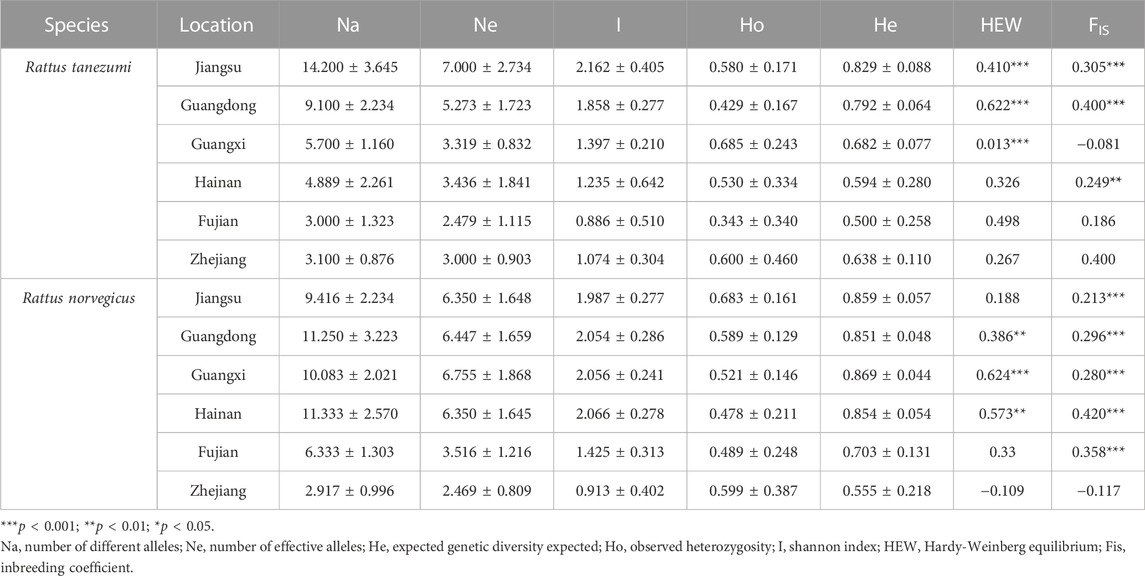
TABLE 2. Genetic variation of microsatellite loci between Rattus norvegicus and Rattus tanezumi from coastal cities in the southeastern coast of China.
Given the significant deviation from Hardy-Weinberg equilibrium (HWE) and the heterozygote defect, it was decidedthat there was further structural refinement among all samples. This study stratified all populations of R. tanezumi and R. norvegicus into three and two genetically distinct clades, respectively, employing UPGMA cluster tree analysis. (Figures 2B, E). In the cluster analysis of R. tanezumi populations, Guangxi and Hainan each constituted distinct branches, while Jiangsu, Guangdong, Fujian, and Zhejiang clustered together, forming another branch. In the Discriminant Analysis of Principal Components (DAPC), all populations exhibited proximity, except for the Guangxi population. Conversely, R. norvegicus populations displayed distinct genetic structures, with Guangdong, Hainan, Fujian, Jiangsu, and Zhejiang forming one cohesive group, while Guangxi stood as an isolated group.
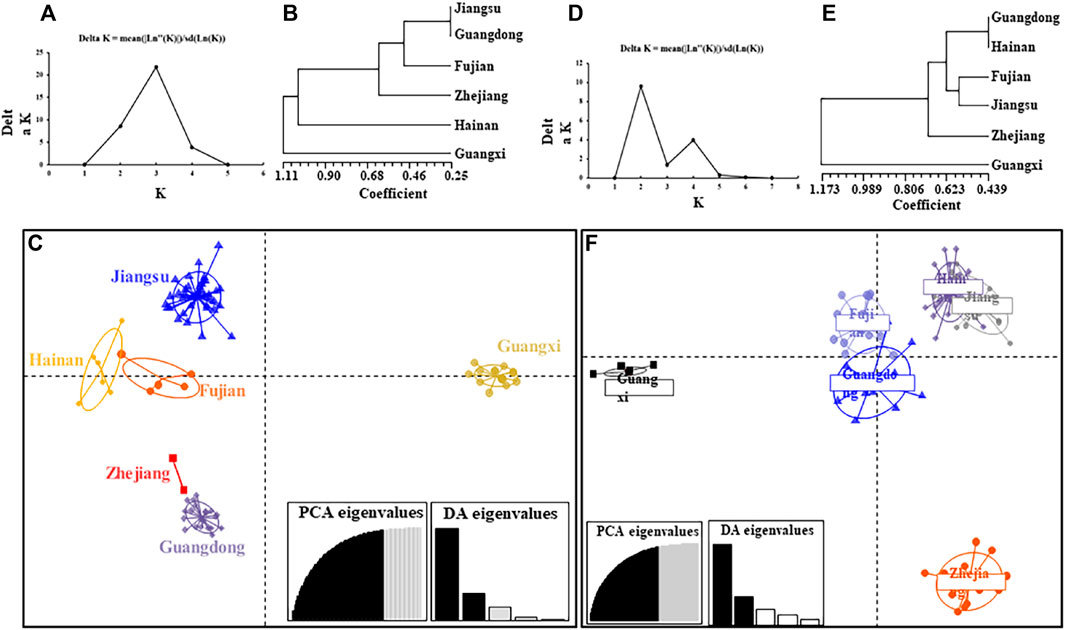
FIGURE 2. The population structure of Rattus norvegicus and Rattus tanezumi from six regions in southeastern coast of China based on microsatellite loci. (A) Best molecular evolutionary model identified by Bayesian information criteria; (B) UPGMA cluster tree analysis of all R. norvegicus populations; (C) principal components (DAPC) analysis of all R. norvegicus populations, and 86.4% of variation was explained by 50 PCs; (D) Best molecular evolutionary model identified by Bayesian information criteria; (E) UPGMA cluster tree analysis of all Rattus tanezumi populations; (F) DAPC analysis of all Rattus tanezumi populations, and 86.4% of variation was explained by 50 PCs.
The AMOVA results (Table 3) showed that among the different levels of genetic variation in the R. tanezumi population, 23.40% of the variation came from differences among individuals within the population; 23.86% of the variation in the R. norvegicus population came from differences among individuals within the population. Paired FST analysis among various regions revealed significant (p < 0.05) genetic differentiation of R. tanezumi samples from Fujian and Zhejiang compared to populations from other regions, with values ranging from 0.006 to 0.154. Rattus norvegicus populations showed moderate to high genetic differentiation in all regional populations (ranging from 0.040 to 0.306) (Table 4).
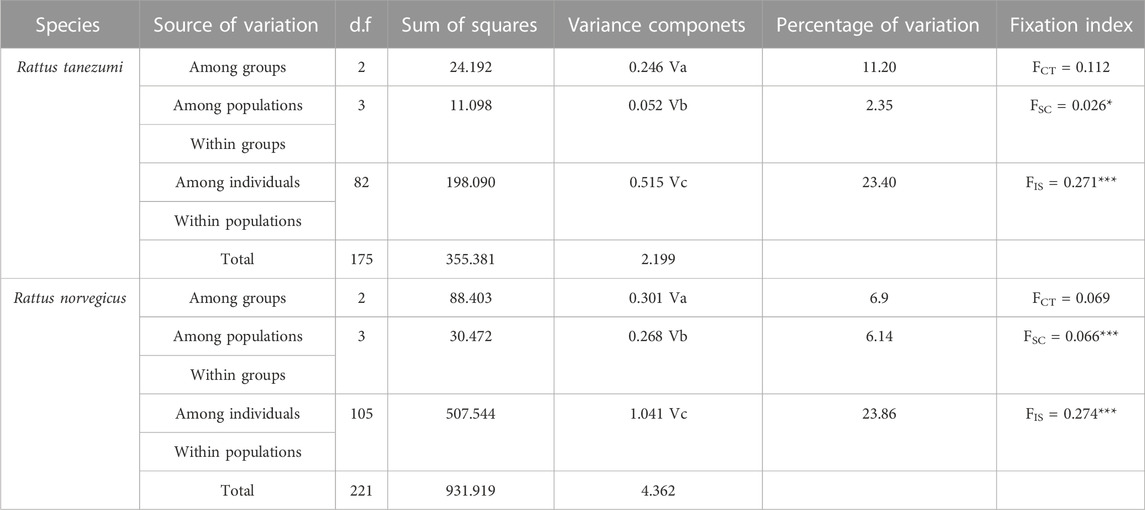
TABLE 3. Analysis of molecular variance (AMOVA) results of Rattus norvegicus and Rattus tanezumi from the southeastern coast of China.
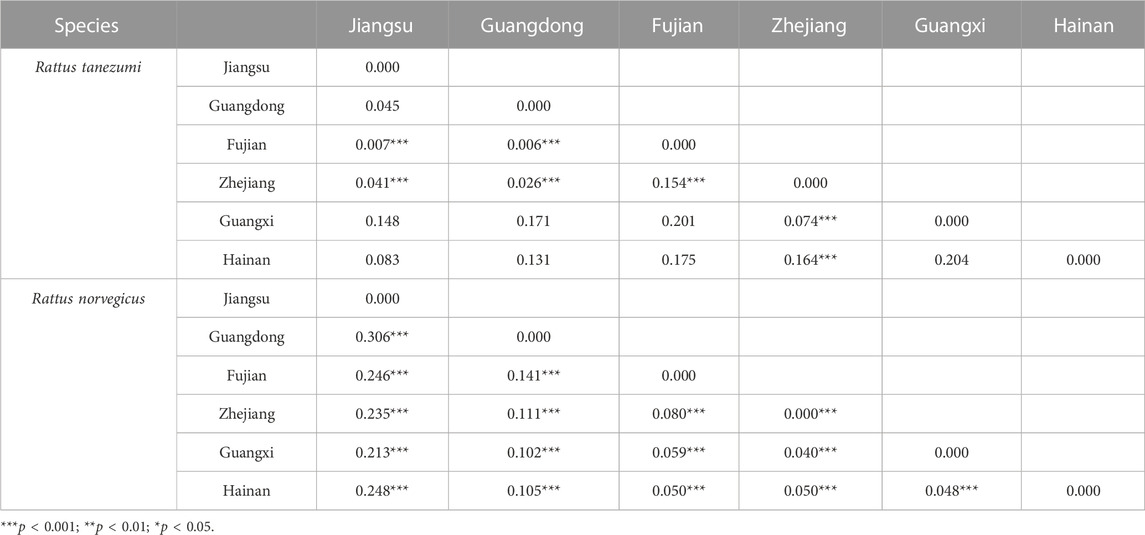
TABLE 4. Differentiation coefficient (FST) analysis results of Rattus norvegicus and Rattus tanezumi populations from different regions in southeastern coast of China based on mtDNA pairing.
Three mitochondrial sequences of coxl (750bp), D-LOOP (950bp) and ND4 (450bp) were obtained from 188 samples. The polymorphism of the control region of D-LOOP was significantly higher than that of coxl and ND4, but the rapid evolutionary rate may lead to unstable results and a large number of mutations. The coxl sequence with stable evolutionary rate was selected for analysis, and a total of 16 haplotypes were found in each species of R. tanezumi and R. norvegicus. The highest genetic diversity of the R. tanezumi was found in the Jiangsu populations, and the lowest genetic diversity was found in the Fujian populations (Table 5). The nucleotide diversity of R. norvegicus population in Fujian was the highest (Pi = 0.01214), while that of R. norvegicus population from Guangxi was the lowest (Pi = 0.00200) (Table 5). In the neutral test of Tajima’s D and Fu’s F, R. norvegicus was positive in all regions, while R. tanezumi was significantly negative (p < 0.05) except Guangxi and Zhejiang. Migration patterns were assessed using a divMigrate network representing all rat populations. For gene flow analyses, although both R. norvegicus and R. tanezumi had migration routes from other regions to the Jiangsu region, their gene flow mobility levels were at low values (Figure 3).
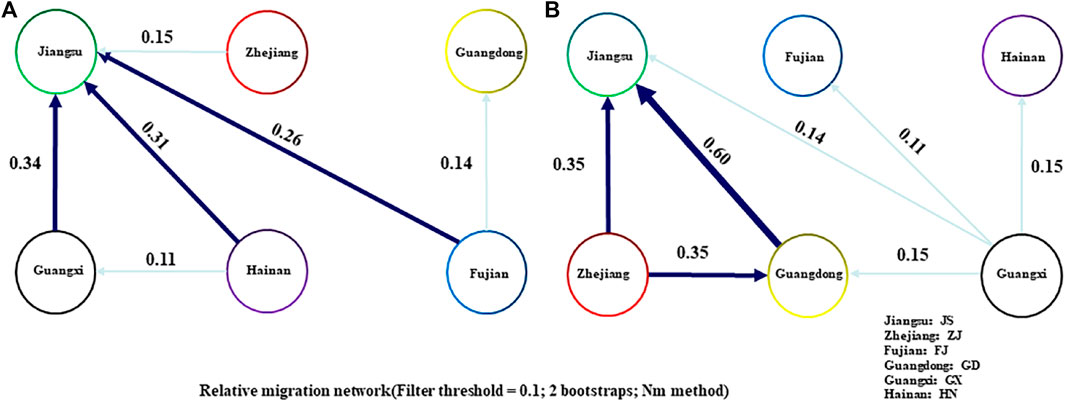
FIGURE 3. Migration patterns of Rattus tanezumi and Rattus norvegicus from southeastern coast of. Migration routes are marked with different types of arrows (dark blue: gene flow >0.3; light blue: gene flow <0.3). Diffusion diagrams of the R. tanezumi (A) and Rattus norvegicus (B).
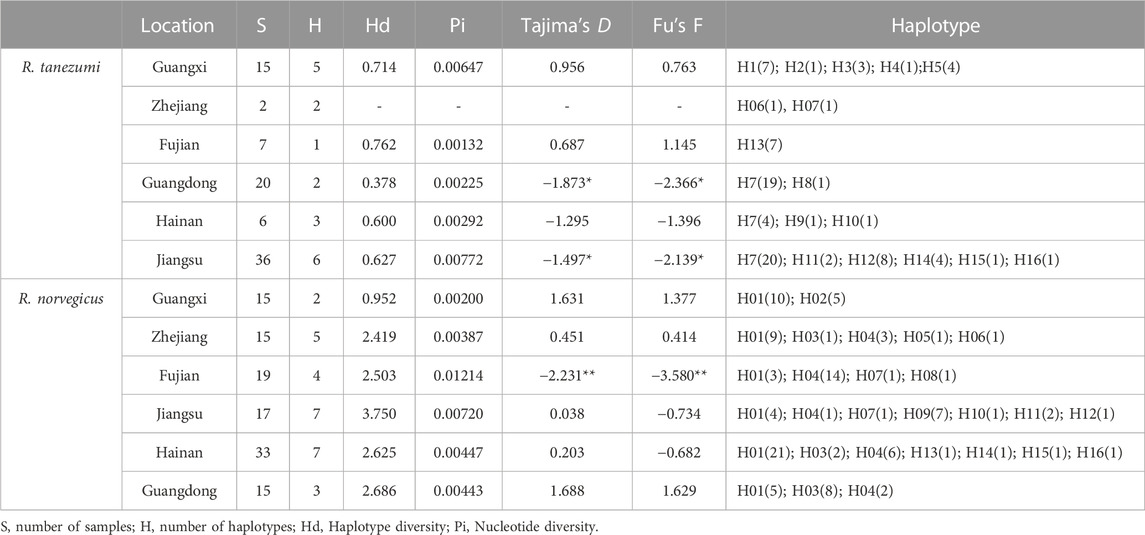
TABLE 5. Mitochondrial polymorphism indicators and neutral detection cytochrome c oxidase subunit I (coxl) Rattus norvegicus and Rattus tanezumi populations from different regions in southeastern coast of China.
A TCS network was reconstructed using all cox1 sequences (Figure 4). Within the R. tanezumi haplotype networks, haplotype 7 (H7), the most prevalent haplotype, predominantly occurred in Jiangsu, Hainan, and Guangdong, shaping a star-like network. The number of haplotypes exhibited a decline in southern Jiangsu and eastern Guangdong, with haplotype 13 (H13) exclusively present in Fujian. The TCS network map of R. norvegicus portrayed a star-shaped network structure, predominantly centered on haplotype 1 (H1) and haplotype 4 (H4), prevalent in Guangxi, Zhejiang, Fujian, and Jiangsu. Meanwhile, haplotype 9 (H9), haplotype 10 (H10), haplotype 11 (H11), and haplotype 12 (H12) were confined to distribution solely in Jiangsu.
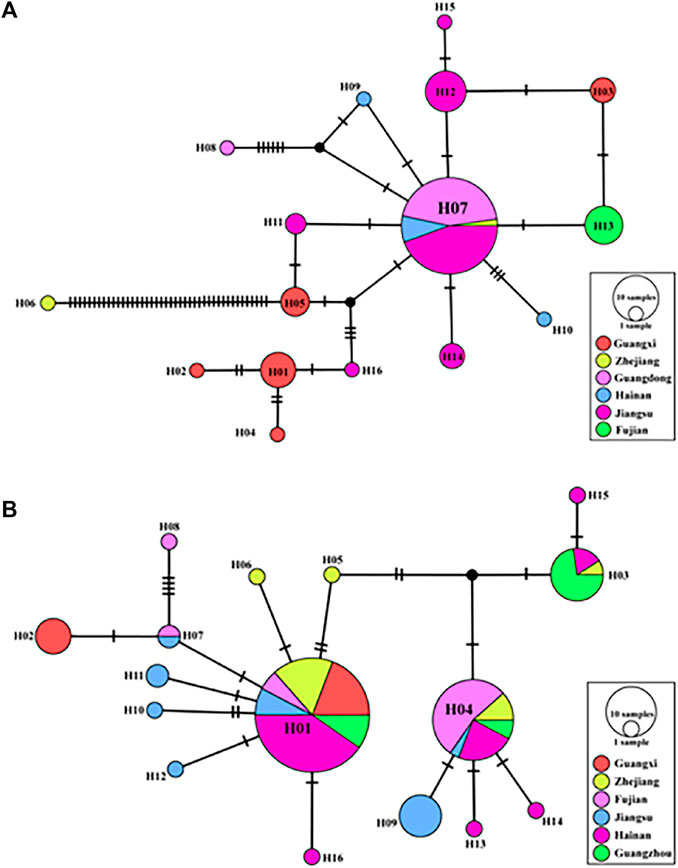
FIGURE 4. A topography of haplotype Haplotype network based on mitochondrial cytochrome c oxidase subunit I (coxl) sequences in Rattus norvegicus and R. tanezumi from southeastern coast of China. Haplotype network diagrams of the Rattus tanezumi (A) R. norvegicus (B). The solid pie chart represents the observed haplotypes, the circle size represents the number of individuals possessing the haplotype, and the colors represent the different sampling locations. Each black connecting line represents a mutant base step between different haplotypes.
The southeastern coast features a warm and humid subtropical marine monsoon climate, transitioning into a tropical monsoon climate. Characterized by mild weather and consistently high annual temperatures in most regions, this climatic profile historically designated it as the primary distribution area for R. tanezumi and R. norvegicus. Its climatic condition is a suitable breeding ground for rodents, and the main channel of economic and cultural exchange between China and continental Europe is seaport shipping. Therefore, seaports are considered introduction epicentres for invasive alien species (Drake and Lodge, 2004; Early et al., 2016). The combination of adequate shelter, food and water supply has led to a high population of rat in seaports (Himsworth et al., 2013; Johnson and Munshi-South, 2017). The economic development brought by the well-developed trade in coastal cities accelerated local urbanization, making it an important area for the export and migration of rats to other areas.
Of our mitochondrial data showed that R. tanezumi populations from Jiangsu and Guangxi possess higher genetic diversity than populationsfrom other regions. Genetic diversity has a significant impact on the evolutionary process can play an important catalytic and driving role at all levels, including population, community and ecosystem levels (Hughes et al., 2008). The same results were obtained from allele (Na) and heterozygosity (He and Ho) analyses in microsatellite analysis. It is therefore hypothesized that there may be an invasion of exotic populations of R. tanezumi in Hainan, Zhejiang and Fujian, which may also account for its lack of significant deviation from HEW. This conclusion was also supported by the neutral detection of Tajima’s D and Fu’s F values, which were negative and significant, which may be caused by the recent expansion of the population in R. tanezumi.
The findings pertaining to R. norvegicus revealed elevated haplotype diversity and nucleotide diversity within the Jiangsu and Hainan populations. This observation suggests the possibility of secondary evolution or interpopulation contact influencing the genetic dynamics in this particular group. (Wenzel et al., 2015). The values of He across different populations varied from 0.555 to 0.869, unequivocally signifying the presence of high polymorphism. Additionally, the Ho values for all populations were consistently lower than their corresponding He values. Only the Zhejiang population had a higher Ho value, indicating that there was no heterozygous deficiency in this population. This may be means that diversity is high in that site. Conversely, the departure from Hardy-Weinberg equilibrium (HWE) observed in both R. tanezumi and R. norvegicus populations in Guangxi and Guangdong may be attributed to the hot and humid climate, as well as the complex geological environment in southern China. Additionally, the impact of population migration and expansion driven by social development emerges as a crucial influencing factor.
The results of the three methods of analysis based on microsatellites data showed slightly different results. All samples of the R. tanezumi population were divided into three clusters in UPGMA. Hainan and Guangxi were divided into one branch each, and Jiangsu, Guangdong, Fujian and Zhejiang were unified into one branch. The DPCA analysis showed the same results as the cluster analysis, and interestingly, the Jiangsu and Guangdong populations clustered together in the genetic structure of both species of house rat (Figures 2C, F). This suggests that there are some factors that led to the exchange and spread of species between the two areas. The close affiliation between Jiangsu and Guangdong populations was further evident in the haplotype analysis of their mitochondrial sequences. In the cox1 haplotype network of R. tanezumi’s mtDNA sequences, the network structure centered around the principal haplotypes in Jiangsu and Guangdong, radiating in a star-shaped pattern. This pattern indicates population expansion in these two regions, aligning with the results of neutral detection. The widespread distribution of H7 haplotypes across populations in all provinces along the southeastern coast suggests that, despite the geographical distance between Jiangsu and Guangdong, they are positioned at the extremities of the southeastern coast, fostering genetic exchange or shared ancestry. Additionally, haplotype analysis of mitochondrial sequences in R. norvegicus identified two predominant haplotypes (H1 and H4), with H4 being prominent in Fujian and H1 in Hainan regions Fujian exhibited an expansion in the outcomes of its neutrality test, accompanied by a noteworthy richness in haplotype diversity. The haplotype network diagram further illustrates a connection among rat populations in Fujian, Hainan, and Jiangsu. The coastal regions, particularly in Fujian, are distinguished by mountainous terrains and seascapes, featuring high forest cover and favorable temperatures, creating ideal environments for rat breeding and dissemination. Jiangsu Province, situated in the Yangtze River Economic Zone and east of the Yellow Sea, is intersected by the Yangtze River and Huai River, encompassing two major water systems. With a geographical expanse from north to south, the province displays a hierarchically diverse climate and vegetation. The intricate river trade and railroad transportation network further contribute to the complexity, potentially facilitating the spread of rodents.
Analyzing the history of segregation and gene flow during species differentiation provides valuable insights into the process of their formation. Notably, a substantial level of gene exchange is evident among the populations of R. norvegicus in Guangdong, Zhejiang, and Jiangsu, as depicted in the gene flow dispersal map. The discernible trend indicates a movement from Guangdong and Zhejiang towards the Jiangsu region, aligning with the overarching northern expansion pattern observed in R. norvegicus. In contrast, the Fujian population is notably geographically isolated from several other regions. Despite this isolation, its high genetic diversity suggests the absence of a significant founder effect, hinting at potential transmission exchanges with Southeast Asia. While R. norvegicus was initially considered to have originated from northern China and Mongolia, the intricate genetic dynamics observed in the Fujian population present a more nuanced perspective (Lin et al., 2012; Song et al., 2014). Subsequent analysis of dated fossils discovered in Guangxi Province, China, believed to represent the ancestors of R. norvegicus, proposed an origin for the species in present-day southwestern China, around 1.2–1.6 million years ago (Jin et al., 2008). This finding provides an explanation for the observed high genetic diversity of R. norvegicus in Fujian and Hainan provinces in southern China. Previous studies have indicated that a significant portion of the genetic structure in R. norvegicus is attributed to geographic and environmental heterogeneity (Puckett et al., 2016; Zhao et al., 2020).
Rattus tanezumi populations in the Yangtze River basin diverged from rats in central regions such as Yunnan, and Aplin et al. (2011). Reported an expansion of R. tanezumi through human shipping 4,000 years ago (Aplin et al., 2011). Guo et al. (2019) reported that R. tanezumi may have originated from Yunnan and neighboring Southeast Asia, and relied primarily on maritime shipping for early dispersal (Guo et al., 2019). This lends support to the notion that the ancestors of R. tanezumi were transported along the road to the Yangtze River basin during that period, expanding towards the warmer inland and southeastern coastal regions. Simultaneously, population migration also occurred in neighboring provinces such as Guangxi and Guangdong, influenced by anthropogenic factors. It can therefore be speculated that the Guangdong and Jiangsu populations may have originated from the same ancestor. On the flip side, the populations of R. tanezumi in Fujian, Hainan, and Zhejiang regions appear to have propagated from both Guangdong and Jiangsu through trade routes. The reduced population genetic diversity in these regions may be attributed to the accumulation of deleterious alleles or mutations linked to invasion and post-colonial expansion (Sakai et al., 2001). Nevertheless, invasive populations, even with reduced genetic diversity, can still successfully colonize and expand in new environments (Tsutsui et al., 2000; Carlson et al., 2014).
Within China, R. norvegicus and R. tanezumi emerge as key commensal rodent species. This study reveals a significant resemblance in their distribution patterns with respect to overall genetic structure. Jiangsu province, serving as a crucial pathway within the Yangtze River basin, displays remarkably high genetic diversity. Nevertheless, the observed contrast in genetic diversity between the compared populations of R. norvegicus and R. tanezumi in Fujian suggests potential distinctions in their dispersal capabilities and pathways. R. norvegicus populations exhibit lower dispersal ability over long distances when competing with R. tanezumi in the United States (Lack et al., 2013). Over the past half century, R. norvegicus has invaded the Xinjiang region with the aid of rail transport and other modern forms of transportation (Liao et al., 2014). Compared to the northwestern expansion of R. norvegicus, R. tanezumi has expanded northward at a lower rate in recent years. This suggests that R. tanezumi is less suitable than the R. norvegicus to use modern transportation systems for long-distance dispersal. This may be due to the different daily routines of these two parasitic rodents around humans. Although they both inhabit human housing and various types of buildings, the primary range of R. tanezumi is in high-rise spaces, as opposed to the R. norvegicus, which results in easier migration and dispersal via transportation.
Worth noting is the impact of the migration of commensal rodents, including the spread of rodent-borne diseases carried by these two species of rodents that accompany the migration of populations. Urban environments have proven to be favorable for rat population growth and associated transmission of zoonoses (Tian et al., 2018; Clement et al., 2019). Rattus tanezumi and R. norvegicus are natural hosts of hantaviruses, and they are far more mobile than other hantavirus hosts. This has led in many cases to correlations between host taxa and their associated hantaviruses (Plyusnin and Sironen, 2014; Zuo et al., 2018). Through phylogenetic analysis of the high concordance between hantavirus and its host (R. norvegicus), Lin et al. (2012) found that the global spread of Orthohantavirus was a consequence of the outward expansion of R. norvegicus in China (Lin et al., 2012). Domestic reports have also described a high similarity of 98.0%–99.0% between the Hantaan strain from Ningde port and the strain from Shandong (Lin et al., 2012). It is interesting to note that the increase in mild HFRS coincides with the rapid socioeconomic development that began in 1978 (Lin et al., 2012). In addition to the risk of hantavirus transmission, plague deserves attention as an important rodent-borne disease, with bubonic pest epidemics occurring from western Yunnan to the eastern part of the country in the 26 years from 1986 to 2005 (Li et al., 2008). This is similar to our findings that the two symbiotic rats expanded from coastal areas to central China through shipping and modern transportation, respectively. Collectively this shows that the virus creates a broader risk of transmission during its activity with the host, and according to previous studies the hosts of hantavirus are mostly R. norvegicus, but the virus sequences were found to be highly similar in R. tanezumi and R. norvegicus, so there is also the possibility of cross-species transmission in bothrat species (He et al., 2021). Currently, fast transportation has started to provide these house rat more opportunities for long-distance transmission and various rodent-borne diseases have been transmitted by rodents over relatively long distances. Previously this expansion of the geographic range of commensal rat species was considered to be the result of human activities, therefore detailed biological and anthropological investigations are necessary to provide traceable information for future infectious diseases.
The results of this study further support the dispersal trend of R. tanezumi populations in the southeastern coastal areas of China from the southern regions or coastal routes to the inland Yangtze River basin. Historically, the population spread towards the warmer interior and the south, and gradually to the southeast. Subsequently, with the development of global warming and rail transport, the overall spread was toward the north. Rattus norvegicus has a similar trend of invasion from south to north, but its rate of spreadhigher. Many specific dispersal mechanisms still need to be further studied. This study provides a more refined population structure of R. tanezumi and R. norvegicus populations in the southeastern coastal region, which can provide new perspectives and assist in the prevention and control of zoonotic disease transmission to assist in the development of more effective rodent control programs.
The datasets presented in this article are not readily available because of policy and privacy restrictions. Requests to access the datasets should be directed to the corresponding authors.
JL and WT participated in manuscript writing design. JL and EH did the literature search and wrote the first draft of the manuscript. GS and LZ revised the manuscript, and provided important intellectual content. YW, WL, ZT, CZ, LA, QX, WZ and JL organized the field work and participated in data collection. EH and JL participated in data collection and developed the illustrations. All authors contributed to the article and approved the submitted version.
This study was supported by grants from YQ Grants (Number: 2023YQFH08), Jiangsu Social Development Plan (Grants numbers BE2022682), Jiangsu Natural Science Foundation (Grants numbers BK20221196).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1174584/full#supplementary-material
R. norvegicus, Rattus norvegicus; R. tanezumi, Rattus tanezumi; HFRS, hemorrhagic fever with renal syndrome; PIC, PIC polymorphism information content; Na, Number of allele; Ne, effective number of allele; Ho, observed heterozygosity; He, expected heterozygosity; I, shannon index; FIS, inbreeding coefficient; FST, genetic differences among populations; HWE, hardy-weinberg equilibrium; AMOVA, analysis of molecular variance; H, haplotype number; Hd, haplotype diversity index; Nm, number of migrants; DAPC, discriminant analysis of principal components; π, nucleotide diversity; mtDNA, mitochondrial DNA; PCR, polymerase chain reaction.
Aplin, K. P., Suzuki, H., Chinen, A. A., Chesser, R. T., Ten Have, J., Donnellan, S. C., et al. (2011). Multiple geographic origins of commensalism and complex dispersal history of Black Rats. PLoS One 6 (11), e26357. doi:10.1371/journal.pone.0026357
Blasdell, K., Cosson, J. F., Chaval, Y., Herbreteau, V., Douangboupha, B., Jittapalapong, S., et al. (2011). Rodent-borne hantaviruses in Cambodia, Lao PDR, and Thailand. Ecohealth 8 (4), 432–443. doi:10.1007/s10393-011-0725-7
Brown, W. M., George, M., and Wilson, A. C. (1979). Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. U. S. A. 76 (4), 1967–1971. doi:10.1073/pnas.76.4.1967
Carlson, S. M., Cunningham, C. J., and Westley, P. A. (2014). Evolutionary rescue in a changing world. Trends Ecol. Evol. 29 (9), 521–530. doi:10.1016/j.tree.2014.06.005
Clement, J., LeDuc, J. W., Lloyd, G., Reynes, J. M., McElhinney, L., Van Ranst, M., et al. (2019). Wild rats, laboratory rats, pet rats: global seoul hantavirus disease revisited. Viruses 11 (7), 652. doi:10.3390/v11070652
Cristescu, R., Sherwin, W. B., Handasyde, K., Cahill, V., and Cooper, D. W. (2009). Detecting bottlenecks using BOTTLENECK 1.2.02 in wild populations: the importance of the microsatellite structure. Conserv. Genet. 11, 1043–1049. doi:10.1007/s10592-009-9949-2
Deinum, E. E., Halligan, D. L., Ness, R. W., Zhang, Y. H., Cong, L., Zhang, J. X., et al. (2015). Recent evolution in Rattus norvegicus is shaped by declining effective population size. Mol. Biol. Evol. 32 (10), 2547–2558. doi:10.1093/molbev/msv126
Ditta, A., Zhou, Z., Cai, X., Shehzad, M., Wang, X., Okubazghi, K., et al. (2018). Genome-wide mining and characterization of SSR markers for gene mapping and gene diversity in Gossypium barbadense L. and Gossypium darwinii. G. Watt Access. Agron. 8, 1–12. doi:10.3390/agronomy8090181
Drake, J. M., and Lodge, D. M. (2004). Global hot spots of biological invasions: evaluating options for ballast-water management. Proc. Biol. Sci. 271 (1539), 575–580. doi:10.1098/rspb.2003.2629
Early, R., Bradley, B. A., Dukes, J. S., Lawler, J. J., Olden, J. D., Blumenthal, D. M., et al. (2016). Global threats from invasive alien species in the twenty-first century and national response capacities. Nat. Commun. 7, 12485. doi:10.1038/ncomms12485
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10 (3), 564–567. doi:10.1111/j.1755-0998.2010.02847.x
Gibbs, R. A., Weinstock, G. M., Metzker, M. L., Muzny, D. M., Sodergren, E. J., Scherer, S., et al. (2004). Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428 (6982), 493–521. doi:10.1038/nature02426
Goudet, J. (2001). FSTAT, Version. 1.2: a computer program to calculate F-statistics. Heredity 86, 485–486. doi:10.1093/oxfordjournals.jhered.a111627
Guo, S., Li, G., Liu, J., Wang, J., Lu, L., and Liu, Q. (2019). Dispersal route of the Asian house rat (Rattus tanezumi) on mainland China: insights from microsatellite and mitochondrial DNA. BMC Genet. 20 (1), 11. doi:10.1186/s12863-019-0714-3
He, W., Fu, J., Wen, Y., Cheng, M., Mo, Y., and Chen, Q. (2021). Detection and genetic characterization of seoul virus in liver tissue samples from Rattus norvegicus and Rattus tanezumi in urban areas of southern China. Front. Vet. Sci. 8, 748232. doi:10.3389/fvets.2021.748232
Himsworth, C. G., Parsons, K. L., Jardine, C., and Patrick, D. M. (2013). Rats, cities, people, and pathogens: a systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers. Vector Borne Zoonotic Dis. 13 (6), 349–359. doi:10.1089/vbz.2012.1195
Hou, Y. F., and Jiang, X. C. (2008). Survey of yellow-breasted mice in Shijiazhuang in 2004-2007. Chin. J. Vector Biol. Control 19 (2), 125. doi:10.3969/j.issn.1003-4692.2008.02.033
Htwe, N. M., Singleton, G. R., Hinds, L. A., Propper, C. R., and Sluydts, V. (2012). Breeding ecology of rice field rats, Rattus argentiventer and R. tanezumi in lowland irrigated rice systems in the Philippines. Agric. Ecosyst. Environ. 161, 39–45. doi:10.1016/j.agee.2012.07.023
Huang, L. Q., Guo, X. G., Speakman, J. R., and Dong, W. G. (2013). Analysis of gamasid mites (Acari: mesostigmata) associated with the Asian house rat, Rattus tanezumi (Rodentia: muridae) in Yunnan Province, southwest China. Parasitol. Res. 112 (5), 1967–1972. doi:10.1007/s00436-013-3354-y
Hughes, A. R., Inouye, B. D., Johnson, M. T., Underwood, N., and Vellend, M. (2008). Ecological consequences of genetic diversity. Ecol. Lett. 11 (6), 609–623. doi:10.1111/j.1461-0248.2008.01179.x
Jin, C. Z., Qin, D. G., Pan, W. S., Wang, Y., and Zhang, Y. Q. (2008). Micromammals of the giantopithecus fauna from sanhe cave, chongzuo, Guangxi. Quat. Sci. 28, 1129–1137. doi:10.3321/j.issn:1001-7410.2008.06.018
Johnson, M. T. J., and Munshi-South, J. (2017). Evolution of life in urban environments. Science 358 (6363), eaam8327. doi:10.1126/science.aam8327
Jombart, T. (2008). adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24 (11), 1403–1405. doi:10.1093/bioinformatics/btn129
Kharzinova, V. R., Dotsev, A. V., Kramarenko, A. S., Layshev, V., and Zinovieva, N. A. (2016). Study of the allele pool and the degree of genetic introgression of semi-domesticated and wild populations of reindeer (Rangifer tarandus L., 1758) using microsatellites. Sel'skokhozyaistvennaya Biol. 51, 811–823. doi:10.15389/agrobiology.2016.6.811eng
Kunieda, T., Kobayashi, E., Tachibana, M., Ikadai, H., and Imamichi, T. (1992). Polymorphic microsatellite loci of the rat (Rattus norvegicus). Mamm. Genome 3 (10), 564–567. doi:10.1007/BF00350622
Lack, J. B., Greene, D. U., Conroy, C. J., Hamilton, M. J., Braun, J. K., Mares, M. A., et al. (2012). Invasion facilitates hybridization with introgression in the Rattus rattus species complex. Mol. Ecol. 21 (14), 3545–3561. doi:10.1111/j.1365-294x.2012.05620.x
Lack, J. B., Hamilton, J., Braun, J. K., Mares, M. A., and Bussche, D. (2013). Comparative phylogeography of invasive Rattus rattus and Rattus norvegicus in the U.S. reveals distinct colonization histories and dispersal. Biol. Invasions 15 (5), 1067–1087. doi:10.1007/s10530-012-0351-5
Leigh, J. W., and Bryant, D. (2015). PopART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6 (9), 1110–1116. doi:10.1111/2041-210X.12410
Li, J. Y., Dong, X. Q., Zhao, W. H., and Liang, Y. (2008). Analysis on epidemiologic characteristics of human plague case form Rattus flavipectus plague natural foci in Yunnan Province. Chinese Jouranl of Endemiology 27 (2), 3. doi:10.3760/cma.j.issn.1000-4955.2008.02.028
Liao, L. F., Wu-So, B., and Yan, S. S. (2014). Migration patterns of Rattus norvegicus in desert area. China. Chin. J. Vector Biol. Control. 7 (02), 76–78.
Lin, X. D., Guo, W. P., Wang, W., Zou, Y., Hao, Z. Y., Zhou, D. J., et al. (2012). Migration of Norway rats resulted in the worldwide distribution of Seoul hantavirus today. J. Virol. 86 (2), 972–981. doi:10.1128/jvi.00725-11
Ma, Y., Wei, Y. W., and Luo, J. (2011). Spatial niche analysis of the small mammals in haidong region of Qinghai province,China. Chin. J. Zoology 46 (4), 126–130. doi:10.1111/j.1749-4877.2010.00232.x
Matisoo-Smith, E., and Robins, J. H. (2004). Origins and dispersals of Pacific peoples: evidence from mtDNA phylogenies of the Pacific rat. Proc. Natl. Acad. Sci. U. S. A. 101 (24), 9167–9172. doi:10.1073/pnas.0403120101
Montagutelli, X., Serikawa, T., and Guénet, J. L. (1991). PCR-analyzed microsatellites: data concerning laboratory and wild-derived mouse inbred strains. Mamm. Genome 1, 255–259. doi:10.1007/BF00352333
Mora, M. S., Mapelli, F. J., Gaggiotti, O. E., Kittlein, M. J., and Lessa, E. P. (2010). Dispersal and population structure at different spatial scales in the subterranean rodent Ctenomys australis. BMC Genet. 11, 9. doi:10.1186/1471-2156-11-9
Morelli, G., Song, Y., Mazzoni, C. J., Eppinger, M., Roumagnac, P., Wagner, D. M., et al. (2010). Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42 (12), 1140–1143. doi:10.1038/ng.705
Morens, D. M., Folkers, G. K., and Fauci, A. S. (2004). The challenge of emerging and re-emerging infectious diseases. Nature 430 (6996), 242–249. doi:10.1038/nature02759
Nagy, S., Poczai, P., Cernák, I., Gorji, A. M., Hegedűs, G., and Taller, J. (2012). PICcalc: an online program to calculate polymorphic information content for molecular genetic studies. Biochem. Genet. 50 (9-10), 670–672. doi:10.1007/s10528-012-9509-1
Pimentel, D., Lach, L., Zuniga, R., and Morrison, D. (2000). Environmental and economic costs of nonindigenous species in the United States. BioScience 50 (1), 53–65. doi:10.1641/0006-3568(2000)050[0053:EAECON]2.3.CO;2
Piry, S., Luikart, G., and Cornuet, J. M. (2017). Bottleneck: a computer program for detecting recent reductions in the effective population size using allele frequency data. APS Meet. Abstr. 90, 502–503. doi:10.1039/cc9960002269
Plyusnin, A., and Sironen, T. (2014). Evolution of hantaviruses: Co-speciation with reservoir hosts for more than 100 MYR. Virus Res. 187, 22–26. doi:10.1016/j.virusres.2014.01.008
Plyusnina, A., Ibrahim, I. N., and Plyusnin, A. (2009). A newly recognized hantavirus in the Asian house rat (Rattus tanezumi) in Indonesia. J. Gen. Virol. 90 (Pt 1), 205–209. doi:10.1099/vir.0.006155-0
Pollitzer, R. (1951). Plague studies. 1. A summary of the history and survey of the present distribution of the disease. Bull. World Health Organ 4 (4), 475–533. doi:10.1016/0165-1765(79)90152-6
Puckett, E. E., Orton, D., and Munshi-South, J. (2020). Commensal rats and humans: integrating rodent phylogeography and zooarchaeology to highlight connections between human societies. Bioessays 42 (5), e1900160. doi:10.1002/bies.201900160
Puckett, E. E., Park, J., Combs, M., Blum, M. J., Bryant, J. E., Caccone, A., et al. (2016). Global population divergence and admixture of the brown rat (Rattus norvegicus). Proc. Biol. Sci. 283 (1841), 20161762. doi:10.1098/rspb.2016.1762
Robins, J. H., McLenachan, P. A., Phillips, M. J., Craig, L., Ross, H. A., and Matisoo-Smith, E. (2008). Dating of divergences within the Rattus genus phylogeny using whole mitochondrial genomes. Mol. Phylogenet Evol. 49 (2), 460–466. doi:10.1016/j.ympev.2008.08.001
Rousset, F. (2008). genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8 (1), 103–106. doi:10.1111/j.1471-8286.2007.01931.x
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34 (12), 3299–3302. doi:10.1093/molbev/msx248
Sakai, A. K., Allendorf, F. W., Holt, J. S., Lodge, D. M., Molofsky, J., With, K. A., et al. (2001). The population biology of invasive species. Annu. Rev. Ecol. Syst. 32 (1), 305–332. doi:10.1146/annurev.ecolsys.32.081501.114037
Slatkin, M. (1995). A measure of population subdivision based on microsatellite allele frequencies. Genetics 139 (1), 457–462. doi:10.1093/genetics/139.1.457
Song, Y., Lan, Z., and Kohn, M. H. (2014). Mitochondrial DNA phylogeography of the Norway rat. PLoS One 9 (2), e88425. doi:10.1371/journal.pone.0088425
Suzuki, H., Nunome, M., Kinoshita, G., Aplin, K. P., Vogel, P., Kryukov, A. P., et al. (2013). Evolutionary and dispersal history of Eurasian house mice Mus musculus clarified by more extensive geographic sampling of mitochondrial DNA. Hered. (Edinb). 111 (5), 375–390. doi:10.1038/hdy.2013.60
Tian, H., Hu, S., Cazelles, B., Chowell, G., Gao, L., Laine, M., et al. (2018). Urbanization prolongs hantavirus epidemics in cities. Proc. Natl. Acad. Sci. U. S. A. 115 (18), 4707–4712. doi:10.1073/pnas.1712767115
Tsutsui, N. D., Suarez, A. V., Holway, D. A., and Case, T. J. (2000). Reduced genetic variation and the success of an invasive species. Proc. Natl. Acad. Sci. 97 (11), 5948–5953. doi:10.1073/pnas.100110397
Wang, Q. W., Huang, P., Yue, M., Huang, E. J., Cai, Y. S., Liang, P. D., et al. (2021). Genetic characterization of rodent hantavirus in the summer of 2020 in Ningde port, fujian province. Chin. J. Epidemiol. 42, 1266–1273. doi:10.3760/cma.j.cn112338-20210126-00065
Wang, Y. X. (2003). Taxonomic list and distribution of mammalian species and subspecies in China. China: China Forestry Press.
Wenzel, M. A., Webster, L. M. I., Blanco, G., Burgess, M. D., Kerbiriou, C., Segelbacher, G., et al. (2015). Erratum to: pronounced genetic structure and low genetic diversity in European red-billed chough (Pyrrhocorax pyrrhocorax) populations. Conserv. Genet. 16 (4), 1011–1012. doi:10.1007/s10592-014-0681-1
Wu, X. Z., and Wang, Y. F. (2012). Fossil materials and migrations of Mus musculus and Rattus norvegicus. Res. China's Front. Archaeol. 1, 1–9.
Xiao, H., Tong, X., Huang, R., Gao, L., Hu, S., Li, Y., et al. (2018). Landscape and rodent community composition are associated with risk of hemorrhagic fever with renal syndrome in two cities in China, 2006-2013. BMC Infect. Dis. 18 (1), 37. doi:10.1186/s12879-017-2827-5
Yang, X. G., Wang, T. L., and Ning, Z. D. (2011). Distribution characteristics and development trend of the invasive yellow-breasted mouse (Rattus flavipectus) in Shanxi Province. Shanxi Agric. Sci. 39 (5), 462–464. doi:10.3969/j.issn.1002-2481.2011.05.24
Yin, P. W., Guo, X. G., Jin, D. C., Fan, R., Zhao, C. F., Zhang, Z. W., et al. (2021). Distribution and host selection of tropical rat mite, Ornithonyssus bacoti, in yunnan province of southwest China. Anim. (Basel) 11 (1), 110. doi:10.3390/ani11010110
Zhao, F., Lin, G. H., and Wu, A. G. (2016). Effects of rivers on genetic patterns of the yellow-breasted mouse, Rhinopithecus roxellana. Sichuan Anim. 2, 6. doi:10.11984/j.issn.1000-7083.20150286
Zhao, L., Zhang, J. X., and Zhang, Y. H. (2020). Genetic boundary and gene flow between 2 parapatric subspecies of brown rats. Curr. Zool. 66 (6), 677–688. doi:10.1093/cz/zoaa027
Zhao, S., Guo, Y., Sheng, Q., and Shyr, Y. (2014). Advanced heat map and clustering analysis using heatmap3. Biomed. Res. Int. 2014, 986048. doi:10.1155/2014/986048
Keywords: R. norvegicus, R. tanezumi, population Genetics, haplotype, microsatellite loci, southeast coast of China
Citation: Li J, Huang E, Wu Y, Zhu C, Li W, Ai L, Xie Q, Tian Z, Zhong W, Sun G, Zhang L and Tan W (2024) Population structure, dispersion patterns and genetic diversity of two major invasive and commensal zoonotic disease hosts (Rattus norvegicus and Rattus tanezumi) from the southeastern coast of China. Front. Genet. 14:1174584. doi: 10.3389/fgene.2023.1174584
Received: 26 February 2023; Accepted: 11 December 2023;
Published: 08 January 2024.
Edited by:
Thales Renato Ochotorena De Freitas, Federal University of Rio Grande do Sul, BrazilReviewed by:
Christian Timothy Chimimba, University of Pretoria, South AfricaCopyright © 2024 Li, Huang, Wu, Zhu, Li, Ai, Xie, Tian, Zhong, Sun, Zhang and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weilong Tan, bmpjZGNAMTYzLmNvbQ==; Gang Sun, c3VuZ2FuZ0BuZW51LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.