
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Genet., 10 April 2023
Sec. Genetics of Common and Rare Diseases
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1140427
Introduction: Studies on the association between gene polymorphisms of various inflammatory factors and liver cirrhosis have been inconsistent. The purpose of this study was to comprehensively summarize the available evidence on the association between gene polymorphisms of inflammatory factors and liver cirrhosis through a systematic review.
Methods: We searched databases of PubMed, EMBASE, Web of Science, and the Cochrane Library for relevant articles published from building databases to 25 September 2022. A systematic review and meta-analysis were performed to investigate the association between gene polymorphisms of various inflammatory factors and liver cirrhosis. Odds ratios (OR) and 95% confidence intervals (CI) were used to assess the strength of association.
Results: A total of 43 articles were included in the systematic review and of them, 22 articles were included in the meta-analysis. The gene polymorphisms of IL-10–1082 GA + AA vs. GG (OR = 1.43, 95% CI = 1.12–1.83), IL-10–1082 AA vs. GG (OR = 2.03, 95% CI = 1.36–3.02), IL-18 -137 GG vs. CC (OR = 3.84, 95% CI = 1.29–11.40), TGF-β1 -509 T vs. C (OR = 2.52, 95% CI = 1.42–4.48), and IFN-γ +874 T vs. A (OR = 1.98, 95% CI = 1.32–2.98) were associated with liver cirrhosis significantly and no association was observed in other gene polymorphisms included in the meta-analysis. The review of inflammatory factors gene polymorphisms that were only reported by a single study indicated 19 gene polymorphisms were risk factors and 4 gene polymorphisms were protective factors for liver cirrhosis, while the association between other 27 gene polymorphisms and liver cirrhosis were not statistically significant.
Discussion: This study suggests that IL-10 -1082G/A, IL-18 -137G/C, TGF-β1 -509T/C, and IFN-γ +874T/A were potentially associated with the risk of liver cirrhosis susceptibility. These findings may provide comprehensive evidence for genetic susceptibility and immunogenetic pathology of liver cirrhosis.
Liver cirrhosis (LC) is a common chronic progressive liver disease, characterized by diffuse fibrosis, pseudo lobule formation, and eventually to portal hypertension, liver failure, or hepatocellular carcinoma (HCC) (Parola and Pinzani, 2019; Gines et al., 2021). It is reported that more than 1.32 million deaths are attributable to liver cirrhosis worldwide, accounting for 2.4% of global total deaths (G. B. D. C. Collaborators, 2020), and liver cirrhosis is the 11th most common cause of death and the third leading cause of death in people aged 45–64 years (Asrani et al., 2019). Nevertheless, there is no effective treatment for liver cirrhosis. Although antifibrotic or pro-regenerative drug therapies for cirrhosis have been approved, they have been in final stage of clinical trials and the effect has not been definitively determined (Fallowfield et al., 2021). A detailed understanding of cirrhosis pathogenesis may contribute to the development of new therapies.
The main causes of liver cirrhosis were chronic infection of hepatitis B virus (HBV) or hepatitis C virus (HCV), alcoholic liver disease (ALD), and non-alcoholic fatty liver disease (NAFLD). Other causes include primary biliary cirrhosis (PBC), primary sclerosing cholangitis (PSC), autoimmune hepatitis (AIH), and genetic and metabolic diseases, such as Wilson’s disease and hemochromatosis (Gines et al., 2021). These pathogenic processes involve a sequence of inflammation, during which various inflammatory factors play a key role (Bernsmeier et al., 2020). They can induce the immune system to secrete inflammatory factors or directly mediate liver cell damage. In the process of regulatory immune responses, the recruitment of both pro- and anti-inflammatory cells such as monocytes and macrophages amplify the response through the production of other inflammatory factors, which increase the stimulus of hepatic stellate cells (HSC) by activating proinflammatory cells (Altamirano-Barrera et al., 2017).
Furthermore, inflammatory factors themselves are manipulated by polymorphisms in their genes. Single nucleotide polymorphisms (SNPs) are the commonest form of genetic variants in human being and they may affect the level of inflammatory factors production and the intensity of their response. There is increasing evidence on the association between individual genetic polymorphisms of inflammatory factors and liver diseases, including chronic hepatitis, liver cirrhosis, and HCC (Aleagha et al., 2020; Wei et al., 2021). The genes mainly include Interleukin genes (IL), interferon-γ genes (IFN-γ), transforming growth factor-β genes (TGF-β), and tumor necrosis factor-α genes (TNF-α). Among them, IL-10, TGF-β and TNF-α have been studied the most. Previous studies have reported various inflammatory factors gene polymorphisms in liver cirrhosis, but the results were not consistent. Identification of genetic factors related to susceptibility to liver cirrhosis would help to elucidate the complex process of the disease and improve the scientific basis for therapeutics or preventive interventions.
So far, several common inflammatory factors gene polymorphisms in liver cirrhosis have been investigated by meta-analysis (Chen et al., 2011; Guo et al., 2015; Guo et al., 2018; Zheng et al., 2021; Liu and Yang, 2022). However, to our knowledge, there has not been a systematic review and meta-analysis that has comprehensively assessed the association of inflammatory factors gene polymorphisms with liver cirrhosis. Therefore, the purpose of this systematic review and meta-analysis was to comprehensively summarize the available evidence on the association between various different inflammatory factors gene polymorphisms and liver cirrhosis.
This systematic review and meta-analysis were performed according to the Preferred Re-porting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Moher et al., 2009). We systematically searched PubMed, EMBASE, Web of Science, and the Cochrane Library databases for relevant articles published from building databases to 25 September 2022. The search strategy included all possible combinations of keywords related to liver cirrhosis, inflammatory factors, and their associated outcomes. The search keywords were as follows: Liver Cirrhosis OR Hepatic Cirrhosis OR Cirrhosis, Hepatic OR Cirrhosis, Liver OR Fibrosis, Liver OR Liver Fibrosis AND Interleukin OR Cytokine OR Interferon OR Lymphocyte OR Macrophage OR Microglia OR Tumor Necrosis Factor OR C-Reactive Protein OR Transforming growth factor OR IFN* OR IL* OR CRP* OR TGF* OR TNF* OR CSF* OR GF* OR MCP* OR CCL* OR CXCL* OR Inflammatory factor* OR Pro-inflammatory cytokine* OR colony stimulating factor OR Chemokine OR Inflammatory cytokine.
Studies that met all of the following criteria were included: 1) the study design was case-control, cohort or cross-sectional; 2) the study investigated the association between inflammatory factors gene polymorphisms and liver cirrhosis; 3) the study established liver cirrhosis group and control group without liver cirrhosis; 4) diagnoses for liver cirrhosis were reported; 5) genotype frequencies were in line with Hardy–Weinberg equilibrium (HWE). Studies that met any of the following criteria were excluded from the study: 1) duplicated studies retrieved from various databases; 2) reviews, meta-analyses, conference papers, comments or case reports; 3) animal or in vitro cell experiments; 4) no full text; 5) no complete odds ratio (OR) and 95% confidence interval (95% CI).
Endnote 20 was used to manage articles and three investigators assessed articles and extracted relevant data from eligible articles independently, then a customized data extraction Excel spreadsheet was used to extract the data for this study. Differences among the three independent reviewers were resolved by another reviewer. The following data were extracted from the eligible studies: 1) the last name of the first author and year of publication; 2) the SNP of various inflammatory factors; 3) study design and sample size; 4) OR and 95% CI; 5) country where the research was conducted and race of participants; 6) type of case and control and diagnostic criteria; 7) material of sample, storage temperature, and genotyping method; 8) patients characteristics such as Mean age, Male gender, Body Mass Index (BMI), Aspartate aminotransferase (AST), Alanine transaminase (ALT) and γ-Glutamyl transpeptidase (GGT). Finally, the Newcastle-Ottawa Quality Assessment Scale (NOS) was used to assess the quality of eligible studies.
RevMan5.3 software was used for meta-analysis. OR and 95% CI were used to assess the strength of associations. Using Q test and I2 statistic to test the degree of heterogeneity. If p > 0.05 and I2 <50%, the results are homogeneous, and the fixed effect model is used for combined analysis; otherwise, the random effect model is used for combined analysis. Because the number of included studies of single SNP was less than 10, no formal assessment of publication bias was performed. Sensitivity analysis were conducted when I2 ≥ 50%. The sensitivity analysis adopted the method of eliminating individual studies item by item to explore the source of heterogeneity.
The search strategy retrieved a total of 2,622 studies, of which 1,305 were from PubMed, 522 from Embase, 340 from Web of Science, and 445 from Cochrane Library. The assessment of full texts of the 395 articles found that 318 articles examined the association between inflammatory factors concentration and liver cirrhosis and 34 provided incomplete OR and 95% CI, and were hence excluded, leaving 43 articles for the systematic review. Among them, 22 were included in the meta-analysis (Figure 1).
Characteristics of the 22 included articles in the meta-analysis are presented in Table 1. These studies were published between 2003 and 2022 and 7 studies were conducted in China, 5 in Egypt, 2 in Germany, 2 in Tunisian, 2 in Italy, 1 in Mexico, 1 in Turkey, 1 in Spain, and 1 in Thailand. Among 2,500 cases, 516 were HCV-induced liver cirrhosis, 803 were HBV-induced, 105 were alcohol-induced, 200 were Opisthorchis viverrini infection-induced, and 876 were mixed. Among 2,984 controls, 2,789 were healthy controls and 195 were chronic hepatitis C infection patients. PCR was used in 4 studies, PCR-RFLP in 12 studies, AS-PCR in 3 studies, ARMS-PCR in 2 studies, and TaqMan in 2 studies. Seven inflammatory factors and 12 SNPs were identified. In addition, the quality scores of study quality assessment varied between 6 and 8, with 8 studies classified as moderate quality and 14 as high quality. The assessment sheet is shown in Supplementary Appendix S1.
Gene polymorphisms of IL-10–1082 GA + AA vs. GG (OR = 1.43, 95% CI = 1.12–1.83), IL-10–1082 AA vs. GG (OR = 2.03, 95% CI = 1.36–3.02), IL-18 -137 GG vs. CC (OR = 3.84, 95% CI = 1.29–11.40), and TGF-β1 -509 T vs. C (OR = 2.52, 95% CI = 1.42–4.48) were significantly associated with liver cirrhosis. The sensitivity analysis showed that in the study of IFN-γ +874 T vs. A, the heterogeneity of other studies was significantly reduced and the result was reversed (OR = 1.98, 95% CI = 1.32–2.98) after excluding the article by Sun et al. (2015). Figure 2 displays the forest plot of these results. No association was observed in other SNPs included in the meta-analysis (Table 2) and the forest plots are shown in Supplementary Appendix S2.
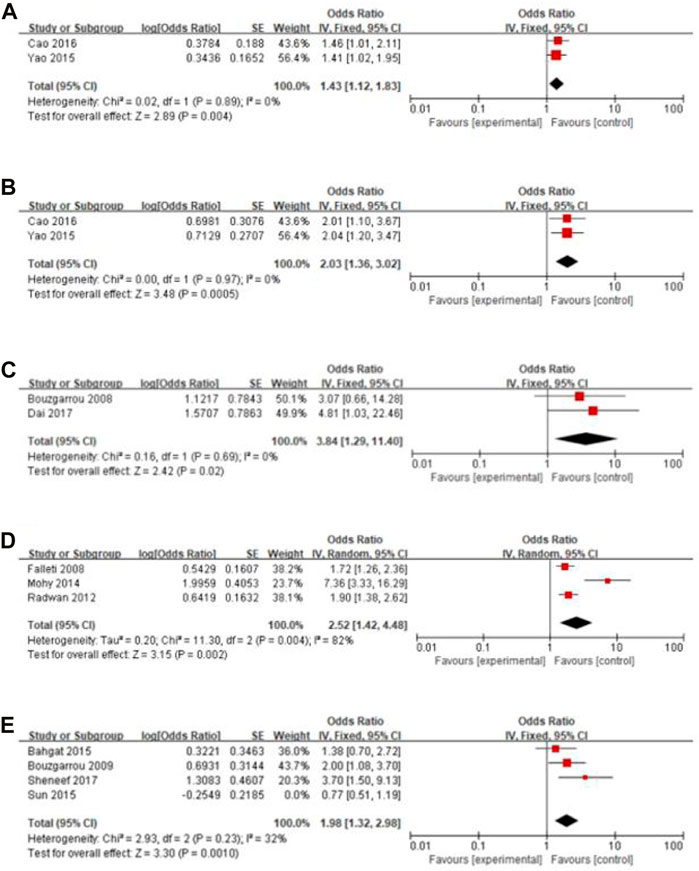
FIGURE 2. Forest plot of the significant results. (A) IL-10–1082 GA + AA vs. GG; (B) IL-10–1082 AA vs. GG; (C) IL-18 -137 GG vs. CC; (D) TGF-β1 -509 T vs. C; (E) IFN-γ +874 T vs. A.
Considering that liver cirrhosis has different etiologies, we performed subgroup analyses by classification of study subjects into HBV-LC, HCV-LC, Alcohol-LC, O viverrini-LC, and Mixed-LC. These analyses showed that there was no difference in gene polymorphisms of TGF-β1 -509 T vs. C between Mixed-LC and HCV-LC groups (I2 = 0%, p = 0.660), while there was a significant difference of IFN-γ + 874 T vs. A between HCV-LC and HBV-LC groups (I2 = 88%, p = 0.004) and heterogeneity was reduced in the subgroup of the HCV-LC group (Figure 3). Subgroup analyses for IL-10–1082 GA + AA vs. GG, IL-10–1082 AA vs. GG, and IL-18 -137 GG vs. CC were not conducted for each etiology of cirrhosis reported by only one study. Forest plots of subgroup analyses of other insignificant SNPs are shown in Supplementary Appendix S3.
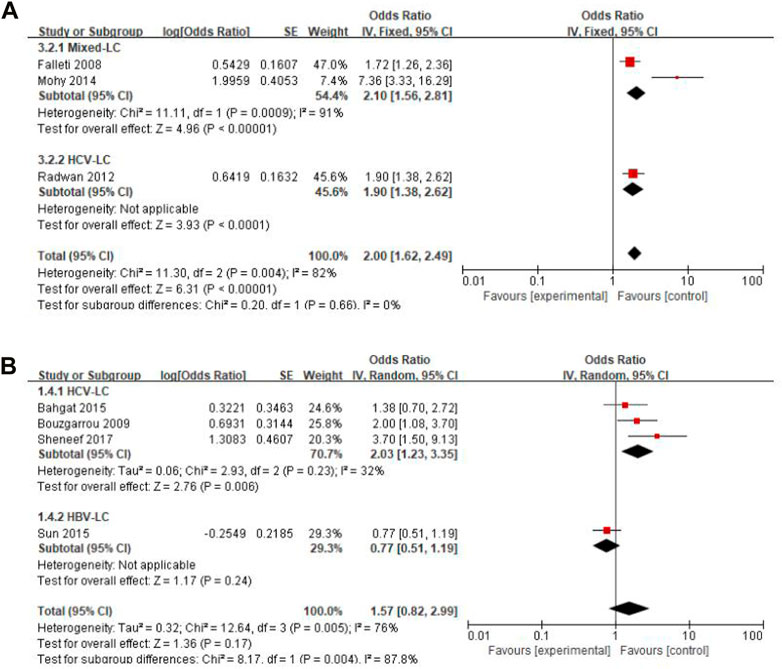
FIGURE 3. Forest plot of subgroup analyses by different etiologies of liver cirrhosis. (A) TGF-β1 -509 T vs. C; (B) IFN-γ +874 T vs. A.
In addition to the above inflammatory factors included in the meta-analysis, some inflammatory factors gene polymorphisms that were only reported by a single study. As part of the systematic review, their findings on gene polymorphisms are summarized in Supplementary Appendix S4. The results indicated that gene polymorphisms of CXCL-1 (rs4074G/A), IFR5(rs13242262A/T, rs10488630A/G), IL-13 (rs1800925A/T), IL-17 (rs4711998A/G), IL-1α(rs3783553del/ins), IL-22 (rs1026788A/G, rs1179249A/C, rs2227472A/G, rs2227485C/T, rs2227491A/G), IL-33 (rs1048274A/G, rs10975519T/C, rs4742170T/C), IL-6 (rs10499563T/C, rs2069837A/G), MIF(-173G/C), TNF-α(-857C/T, −863C/A) were risk factors for liver cirrhosis and gene polymorphisms of IFN-γ(+2109A/G, IL-2 (-330T/G), IL-6 (rs1474347A/C), IL-8 (-251A/T) were protective factors for liver cirrhosis. No significant association of gene polymorphisms of IL-13 (rs20541A/T), IL-17 (rs4711998A/G, rs763780C/T), IL-1α(-889A2A2), IL-1β(-3953A2A2, -31C/T), IL-2 (+114T/G, −384T/G), IL-22 (rs2046068A/C, rs2227473A/G, rs7314777T/C), IL-27 (-964A/G, 2905T/G), IL-28 (rs8099917T/G), IL-4 (-589C/T, −33C/T), IL-6 (rs1800796G/C, rs1800795G/C), IL-8 (+781C/T), IRF3(-925A/G), LT-α(+252A/G), TGF-β1 (-800G/A, Leu10ProC/T), TNFAIP3(rs148314165T/T del, rs200820567T/A, rs2230926G/T), VEGFA (+936C/T) with liver cirrhosis were found.
Only a few people develop cirrhosis after chronic liver injury such as viral infection, alcohol consumption, and metabolic, cholestatic or autoimmune damages, indicating that the modifiers determine the progress of these individuals towards liver cirrhosis. In addition to the recognized progression factors such as age, gender, race and exposure time window or duration, unknown individual genetic factors may affect disease progression. Inflammation is primarily responsible for the pathological progression of liver fibrosis and cirrhosis (Alegre et al., 2017; Koyama and Brenner, 2017). Inflammatory factors are a kind of endogenous polypeptides, produced mainly by immune system cells and are capable of mediating a variety of immune responses. Hence, we performed this systematic review and meta-analysis to explore the association between various inflammatory factors gene polymorphisms and liver cirrhosis. We found that IL-10 (−1082 GA + AA and AA), IL-18 (−137 GG), TGF-β1 (−509 T) and IFN-γ (+874 T) were potentially associated with the risk of liver cirrhosis susceptibility. Subgroup analyses showed that the results of TGF-β1 -509 T vs. C were consistent with the total analysis, indicating that the result was stable and not affected by different etiologies. While the associations of IFN-γ +874 T with liver cirrhosis vs. A and liver cirrhosis were affected by different etiologies, and the results were consistent with the sensitivity analysis.
HSC is the main receptor cell of inflammatory signal and the main effector cell of promoting fibrosis (Parola and Pinzani, 2019). In the process of inflammation, various immune cells (i.e., T cells B cells, macrophages mast cells and specifically Kupffer cells) are recruited in the liver tissue. These immune cells further release more inflammatory cytokines (i.e., IL-10, IL-18, TGF-β1 and IFN-γ), which, together with damage-associated molecular pattern (DAMPS), pathogen-associated molecular patterns (PAMPS) or inflammasomes released by injured or dead liver cells, can induce more secretion of inflammatory factors and stimulate HSC by activating various inflammatory pathways. The nuclear factor kappa B (NF-kB) pathway (Ramos-Tovar and Muriel, 2020), the c-Jun N terminal kinase (JNK) pathway (Seki et al., 2012) and the Toll-like receptor (TLR) signaling pathway (Leifer and Medvedev, 2016) were well-known pathways. The active HSC secrete much extracellular matrix (ECM) and cause deposition, so that chronic liver inflammation persists and leads to the formation of liver fibrosis. These processes are controlled by related genes (Figure 4). On the other hand, the degeneration and necrosis of liver cells cause liver cell regeneration and connective fiber tissue proliferation. The mechanism of liver cirrhosis is complex and has not been clearly understood yet. These processes may be primarily responsible for the progression of liver cirrhosis.
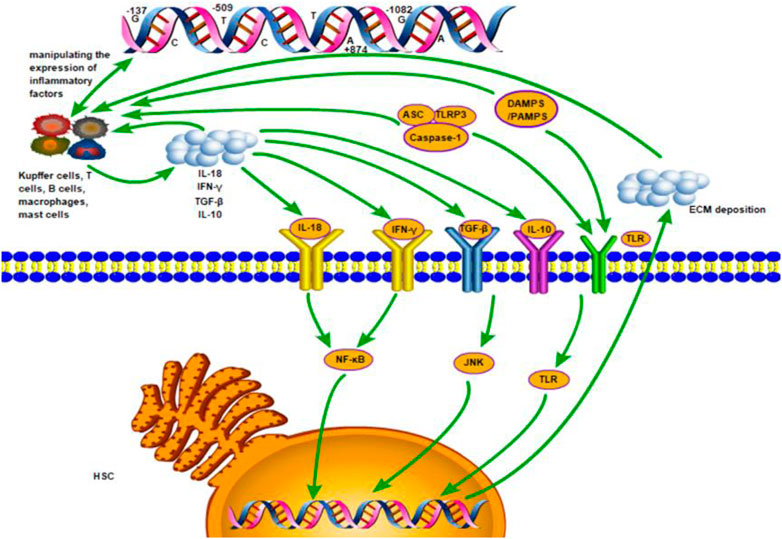
FIGURE 4. The inflammation mechanism of liver fibrosis mediated by inflammatory factors. Various immune cells (i.e., T cells B cells, macrophages mast cells and specifically Kupffer cells) are recruited in the liver tissue. These immune cells further release more inflammatory cytokines (i.e., IL-10, IL-18, TGF-β1 and IFN-γ), which, together with damage-associated molecular pattern (DAMPS), pathogen-associated molecular patterns (PAMPS) or inflammasomes released by injured or dead liver cells, induce more secretion of inflammatory factors and stimulate HSC by activating the nuclear factor kappa B (NF-kB) pathway, the c-Jun N terminal kinase (JNK) pathway and the Toll-like receptor (TLR) signaling pathway. The active HSC secrete much extracellular matrix (ECM) and cause deposition, so that chronic liver inflammation persists and leads to the formation of liver fibrosis. These processes are controlled by related genes.
TGF-β1 has been proven to be the most effective major cytokine to promote liver fibrosis by variable paths (Jeong et al., 2011; Fabregat et al., 2016; Tsuchida and Friedman, 2017). This study suggests that TGF-β1 -509T allele carriers maybe more likely to develop cirrhosis than C carriers. This was supported by the results of a meta-analysis (Guo et al., 2018) although the included literature was different. Our research did not include in other gene loci or comparative models due to the availability of original studies. IL-10 is an effective anti-inflammatory cytokine, mainly produced by Th2 cells, macrophages, and mast cells. IL-10 therapy has been reported to reduce the severity of liver inflammation and fibrosis (Nelson et al., 2000). In the promoter polymorphisms influencing the level of IL-10, the −1082G/A polymorphism is of some importance. This study indicates that IL-10–1082GA + AA and AA genotype carriers are more likely to develop cirrhosis than GG carriers. We included 7 pairs of comparison and no association was observed in other five comparison models. IL-18 plays a role in inflammation through the induction of inflammatory cytokines such as IFN-γ (Yoshimoto et al., 1998). This study indicates that IL-18 -137GG genotype carriers were more likely to develop cirrhosis than CC carriers. We included 3 pairs of comparison and no association was observed in other 2 comparison models. IFN-γ is one of the most important Th1 cytokines. Significant correlation was observed between expression level of IFN-γ mRNA and stage of fibrosis in chronic hepatitis C (Gigi et al., 2008). This study suggests that IFN-γ +874T allele carriers were more likely to develop cirrhosis than A carriers. This is the result of sensitivity analysis, and the sensitivity analysis of the other 4 comparative models also shows no correlation. We found that liver cirrhosis in excluded studies was caused by HBV, while other studies were caused by HCV, which suggested that the cause of liver cirrhosis was the source of heterogeneity. Considering the complex mechanisms of genetic action, further studies are needed to confirm the effects of these polymorphisms on inflammatory factors expression.
Our results showed that there were no significant associations of IL-10 (-592C/A, −819C/T), IL-18 (-607A/C), IL-1β −511C/T, IL-28 rs12979860C/T, TGF-β1(Arg25ProC/G), TNF-α (−308G/A, −238G/A) with liver cirrhosis. One of the reasons for this result may be the limited number of eligible studies. Gene polymorphisms of IL-10, IL-18, TGF-β1 have been confirmed to be related to liver cirrhosis by our study, however, it is accepted that IL-1β, IL-28, especially TNF-α plays an important role in the occurrence and development of liver cirrhosis. More research is needed to explore the relationship between these inflammatory factor gene polymorphisms and liver cirrhosis.
IL-6, a primary immunomodulatory cytokine, has been shown related to liver cirrhosis in a few studies. Zheng et al. (2021) identified three polymorphisms in IL-6 gene and perform a meta-analysis to explore their association with liver cirrhosis. Five polymorphisms (rs1474347, rs10499563, rs2069837, rs1800795, rs1800796) in IL-6 gene were also identified in our systematic review. It worths noting that the polymorphisms (rs1800795, rs1800796) identified simultaneously in both studies had the same results. However, we could not perform a meta-analysis to summarize the association because of the limitation of number of studies. Broader searches may be able to identify more studies on the association between IL-6 gene polymorphisms and liver cirrhosis. Besides, there were other inflammatory factors gene polymorphisms were only reported by single original study. We identified a few genetic polymorphisms this way and more studies are needed to further explore their association with liver cirrhosis.
This study has some limitations that should pay attention to. Firstly, only four databases were searched and some relevant original studies may be missed. Secondly, there were many predisposing factors such as age and race for liver cirrhosis, but we did not carry out further subgroup analysis on factors due to the limited number of relevant studies. Thirdly, we did not conduct a further analysis due to the lack of data on the severity of liver fibrosis, these may influence the precision of the association. Fourthly, only SNP was considered in this review due to the dearth of studies on the combined impact of gene-gene polymorphisms and haplotype gene polymorphisms. As liver cirrhosis is a multi-factors disease, the combined effects of factors like gene-gene and gene-environment should be considered. Moreover, liver cirrhosis and inflammatory diseases of the intestinal tract like inflammatory bowel diseases may share susceptibility gene loci, and expanded search of more databases may unveal different biomarkers.
Our systematic review and meta-analysis suggest that IL-10 -1082G/A, IL-18 -137G/C, TGF-β1 -509T/C and IFN-γ +874T/A are potentially associated with the risk of liver cirrhosis susceptibility. These findings may provide comprehensive evidence for genetic susceptibility and immunogenetic pathology of liver cirrhosis.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
XP and AL contributed to the study design, while XP, SX, XH, and YL contributed to the data collection. Statistical analyses and interpretation of results were performed by SX. SX, XP, and SW drafted the manuscript and edited the language. All the authors participated in the critical revisions, and approved the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1140427/full#supplementary-material
Aleagha, O. E., Oltulu, P., and Sadeghi, M. (2020). Association between interleukin 6 polymorphisms (rs1800796, rs1800795, rs2069837, rs17147230, and rs1800797) and hepatocellular carcinoma susceptibility: A meta-analysis. Clin. Exp. Hepatol. 6 (4), 359–366. doi:10.5114/ceh.2020.102171
Alegre, F., Pelegrin, P., and Feldstein, A. E. (2017). Inflammasomes in liver fibrosis. Semin. Liver Dis. 37 (2), 119–127. doi:10.1055/s-0037-1601350
Altamirano-Barrera, A., Barranco-Fragoso, B., and Mendez-Sanchez, N. (2017). Management strategies for liver fibrosis. Ann. Hepatol. 16 (1), 48–56. doi:10.5604/16652681.1226814
Armendáriz-Borunda, J., Rincón, A. R., Muñoz-Valle, J. F., Bueno-Topete, M., Oregón-Romero, E., Islas-Carbajal, M. C., et al. (2008). Fibrogenic polymorphisms (TGF-beta, PAI-1, AT) in Mexican patients with established liver fibrosis. Potential correlation with pirfenidone treatment. J. Investig. Med. 56 (7), 944–953. doi:10.2310/JIM.0b013e3181891512
Asrani, S. K., Devarbhavi, H., Eaton, J., and Kamath, P. S. (2019). Burden of liver diseases in the world. J. Hepatol. 70 (1), 151–171. doi:10.1016/j.jhep.2018.09.014
Bahgat, N. A., Kamal, M. M., Abdelaziz, A. O., Mohye, M. A., Shousha, H. I., ahmed, M. M., et al. (2015). Interferon-γ and interleukin-10 gene polymorphisms are not predictors of chronic hepatitis C (Genotype-4) disease progression. Asian Pac J. Cancer Prev. 16 (12), 5025–5030. doi:10.7314/apjcp.2015.16.12.5025
Bahr, M. J., el Menuawy, M., Boeker, K. H. W., Musholt, P. B., Manns, M. P., and Lichtinghagen, R. (2003). Cytokine gene polymorphisms and the susceptibility to liver cirrhosis in patients with chronic hepatitis C. Liver Int. 23 (6), 420–425. doi:10.1111/j.1478-3231.2003.00873.x
Bernsmeier, C., van der Merwe, S., and Perianin, A. (2020). Innate immune cells in cirrhosis. J. Hepatol. 73 (1), 186–201. doi:10.1016/j.jhep.2020.03.027
Bouzgarrou, N., Hassen, E., Farhat, K., Bahri, O., Gabbouj, S., Maamouri, N., et al. (2009). Combined analysis of interferon-gamma and interleukin-10 gene polymorphisms and chronic hepatitis C severity. Hum. Immunol. 70 (4), 230–236. doi:10.1016/j.humimm.2009.01.019
Bouzgarrou, N., Hassen, E., Schvoerer, E., Stoll-Keller, F., Bahri, O., Gabbouj, S., et al. (2008). Association of interleukin-18 polymorphisms and plasma level with the outcome of chronic HCV infection. J. Med. Virol. 80 (4), 607–614. doi:10.1002/jmv.21079
Cao, L. N., Cheng, S. L., and Liu, W. (2016). IL10 rs1800896 polymorphism is associated with liver cirrhosis and chronic Hepatitis B. Genet. Mol. Res. 15 (1). doi:10.4238/gmr.15017256
Chen, D., Liu, J. L., Liu, Y., Zhu, J., and Wang, S. W. (2011). Lack of an association between -308G>A polymorphism of the TNF-α gene and liver cirrhosis risk based on a meta-analysis. Genet. Mol. Res. 10 (4), 2765–2774. doi:10.4238/2011.November.8.2
Dai, Z. J., Liu, X. H., Wang, M., Guo, Y., Zhu, W., Li, X., et al. (2017). IL-18 polymorphisms contribute to Hepatitis B virus-related cirrhosis and hepatocellular carcinoma susceptibility in Chinese population: A case-control study. Oncotarget 8 (46), 81350–81360. doi:10.18632/oncotarget.18531
Fabregat, I., Moreno-Càceres, J., Sánchez, A., Dooley, S., Dewidar, B., Giannelli, G., et al. (2016). TGF-beta signalling and liver disease. FEBS J. 283 (12), 2219–2232. doi:10.1111/febs.13665
Fabris, C., Falleti, E., Cussigh, A., Bitetto, D., Fontanini, E., Bignulin, S., et al. (2011). IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: Role in the course of chronic viral hepatitis and the development of HCC. J. Hepatology 54 (4), 716–722. doi:10.1016/j.jhep.2010.07.019
Falleti, E., Fabris, C., Toniutto, P., Fontanini, E., Cussigh, A., Bitetto, D., et al. (2008). TGF-beta1 genotypes in cirrhosis: Relationship with the occurrence of liver cancer. Cytokine 44 (2), 256–261. doi:10.1016/j.cyto.2008.08.008
Fallowfield, J. A., Jimenez-Ramos, M., and Robertson, A. (2021). Emerging synthetic drugs for the treatment of liver cirrhosis. Expert Opin. Emerg. Drugs 26 (2), 149–163. doi:10.1080/14728214.2021.1918099
G. B. D. C. Collaborators (2020). The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990-2017: A systematic analysis for the global Burden of disease study 2017. Lancet Gastroenterol. Hepatol. 5 (3), 245–266. doi:10.1016/S2468-1253(19)30349-8
Gigi, E., Raptopoulou-Gigi, M., Kalogeridis, A., Masiou, S., Orphanou, E., Vrettou, E., et al. (2008). Cytokine mRNA expression in hepatitis C virus infection: TH1 predominance in patients with chronic hepatitis C and TH1-TH2 cytokine profile in subjects with self-limited disease. J. Viral Hepat. 15 (2), 145–154. doi:10.1111/j.1365-2893.2007.00908.x
Gines, P., Krag, A., Abraldes, J. G., Solà, E., Fabrellas, N., and Kamath, P. S. (2021). Liver cirrhosis. Lancet 398 (10308), 1359–1376. doi:10.1016/S0140-6736(21)01374-X
Guo, P. F., Jin, J., and Sun, X. (2015). Influence of IL10 gene polymorphisms on the severity of liver fibrosis and susceptibility to liver cirrhosis in HBV/HCV-infected patients. Infect. Genet. Evol. 30, 89–95. doi:10.1016/j.meegid.2014.12.011
Guo, P., Sun, X., Feng, X., and Zhang, C. (2018). Transforming growth factor-β1 gene polymorphisms with liver cirrhosis risk: A meta-analysis. Infect. Genet. Evol. 58, 164–170. doi:10.1016/j.meegid.2017.12.019
Jeong, W. I., Park, O., Suh, Y. G., Byun, J. S., Park, S. Y., Choi, E., et al. (2011). Suppression of innate immunity (natural killer cell/interferon-gamma) in the advanced stages of liver fibrosis in mice. Hepatology 53 (4), 1342–1351. doi:10.1002/hep.24190
Koyama, Y., and Brenner, D. A. (2017). Liver inflammation and fibrosis. J. Clin. Invest. 127 (1), 55–64. doi:10.1172/JCI88881
Leifer, C. A., and Medvedev, A. E. (2016). Molecular mechanisms of regulation of Toll-like receptor signaling. J. Leukoc. Biol. 100 (5), 927–941. doi:10.1189/jlb.2MR0316-117RR
Liu, C., and Yang, S. (2022). A meta-analysis of the influence of tumor necrosis factor-α-308 gene polymorphism on liver cirrhosis. J. Healthc. Eng. 2022, 9764770. doi:10.1155/2022/9764770
Liu, Y., Yu, M. C., Zhang, A. Q., Wang, Y. B., Jiang, K., and Dong, J. H. (2015). Interleukin-10 gene promoter polymorphism and risk of liver cirrhosis. Genet. Mol. Res. 14 (1), 1229–1234. doi:10.4238/2015.February.13.1
Lu, Y., Bao, J. G., Deng, Y., Rong, C-Z., Liu, Y-Q., Huang, X. L., et al. (2015). Role of IL-18 gene promoter polymorphisms, serum IL-18 levels, and risk of hepatitis B virus-related liver disease in the Guangxi Zhuang population: A Retrospective case-control study. Asian Pac J. Cancer Prev. 16 (14), 6019–6026. doi:10.7314/apjcp.2015.16.14.6019
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G.PRISMA Group (2009). Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ 339, b2535. doi:10.1136/bmj.b2535
Mohy, A., and Fouad, A. (2014). Role of transforming growth factor-β1 in serum and - 509 C>T promoter gene polymorphism in development of liver cirrhosis in Egyptian patients. Meta gene 2, 631–637. doi:10.1016/j.mgene.2014.08.002
Nelson, D. R., Lauwers, G. Y., Lau, J. Y., and Davis, G. L. (2000). Interleukin 10 treatment reduces fibrosis in patients with chronic hepatitis C: A pilot trial of interferon nonresponders. Gastroenterology 118 (4), 655–660. doi:10.1016/s0016-5085(00)70134-x
Nomair, A. M., Kandil, L. S., Nomeir, H. M., and Kandil, N. S. (2021). TGF-Β1 & PNPLA3 genetic variants and the risk of hepatic fibrosis and HCC in Egyptian patients with HCV-related liver cirrhosis. Asian Pac J. Cancer Prev. 22 (10), 3317–3326. doi:10.31557/APJCP.2021.22.10.3317
Öksüz, Z., Üçbilek, E., Serin, M. S., Yaraş, S., Temel, G. Ö., and Sezgin, O. (2022). Possible relationship between IL28B rs12979860 and TLR2 -196 to -174 del/ins polymorphisms and the liver fibrosis stage in hepatitis C patients. Arch. Virol. 167 (1), 153–161. doi:10.1007/s00705-021-05302-9
Parola, M., and Pinzani, M. (2019). Liver fibrosis: Pathophysiology, pathogenetic targets and clinical issues. Mol. Asp. Med. 65, 37–55. doi:10.1016/j.mam.2018.09.002
Pastor, I. J., Romero, A., and González-Sarmiento, R. (2005). 238 G>A polymorphism of tumor necrosis factor alpha gene (TNFA) is associated with alcoholic liver cirrhosis in alcoholic Spanish men. Alcohol Clin. Exp. Res. 29 (11), 1928–1931. doi:10.1097/01.alc.0000187595.19324.ca
Petrásek, J., Hubácek, J. A., Stickel, F., Sperl, J., Berg, T., Ruf, E., et al. (2009). Do common genetic variants in endotoxin signaling pathway contribute to predisposition to alcoholic liver cirrhosis? Clin. Chem. Lab. Med. 47 (4), 398–404. doi:10.1515/CCLM.2009.112
Radwan, M. I., Pasha, H. F., Mohamed, R. H., Hussien, H. I. M., and El-Khshab, M. N. (2012). Influence of transforming growth factor-β1 and tumor necrosis factor-α genes polymorphisms on the development of cirrhosis and hepatocellular carcinoma in chronic hepatitis C patients. Cytokine 60 (1), 271–276. doi:10.1016/j.cyto.2012.05.010
Ramos-Tovar, E., and Muriel, P. (2020). Molecular mechanisms that Link oxidative stress, inflammation, and fibrosis in the liver. Antioxidants 9 (12), 1279. doi:10.3390/antiox9121279
Seki, E., Brenner, D. A., and Karin, M. (2012). A liver full of JNK: Signaling in regulation of cell function and disease pathogenesis, and clinical approaches. Gastroenterology 143 (2), 307–320. doi:10.1053/j.gastro.2012.06.004
Sheneef, A., Esmat, M. M., Mohammad, A. N., Mahmoud, A. A., Moghazy, H. M., and Noureldin, A. K. (2017). Interleukin-10 and interferon gamma gene polymorphisms and hepatitis C virus-related liver cirrhosis risk. J. Interferon Cytokine Res. 37 (4), 175–180. doi:10.1089/jir.2016.0106
Sun, Y., Lu, Y., Xie, L., Deng, Y., Li, S., and Qin, X. (2015). Interferon gamma polymorphisms and Hepatitis B virus-related liver cirrhosis risk in a Chinese population. Cancer Cell Int. 15, 35. doi:10.1186/s12935-015-0184-2
Surapaitoon, A., Suttiprapa, S., Mairiang, E., Khuntikeo, N., Pairojkul, C., Bethony, J., et al. (2017). Subsets of inflammatory cytokine gene polymorphisms are associated with risk of carcinogenic liver Fluke Opisthorchis viverrini-associated advanced periductal fibrosis and cholangiocarcinoma. Korean J. Parasitol. 55 (3), 295–304. doi:10.3347/kjp.2017.55.3.295
Tsuchida, T., and Friedman, S. L. (2017). Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 14 (7), 397–411. doi:10.1038/nrgastro.2017.38
Wei, Y., Zhao, Z., Wang, Z., Zhang, K., Tang, Z., and Tao, C. (2021). Relationships between IL-1β, TNF-α genetic polymorphisms and HBV infection: A meta-analytical study. Gene 791, 145617. doi:10.1016/j.gene.2021.145617
Yang, A. M., Wen, L. L., Yang, C. S., Wang, S. C., Chen, C. S., and Bair, M. J. (2014). Interleukin 10 promoter haplotype is associated with alcoholic liver cirrhosis in Taiwanese patients. Kaohsiung J. Med. Sci. 30 (6), 291–298. doi:10.1016/j.kjms.2014.02.016
Yao, L., Xing, S., Fu, X., Song, H., Wang, Z., Tang, J., et al. (2015). Association between interleukin-10 gene promoter polymorphisms and susceptibility to liver cirrhosis. Int. J. Clin. Exp. Pathol. 8 (9), 11680–11684.
Yoshimoto, T., Takeda, K., Tanaka, T., Ohkusu, K., Kashiwamura, S., Okamura, H., et al. (1998). IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: Synergism with IL-18 for IFN-gamma production. J. Immunol. 161 (7), 3400–3407. doi:10.4049/jimmunol.161.7.3400
Keywords: inflammatory factors, gene polymorphism, liver cirrhosis, systematic review, meta-analysis
Citation: Xiao S, Pan X, Huang X, Liu Y, Wen SW and Liu A (2023) Gene polymorphisms of inflammatory factors in liver cirrhosis. Front. Genet. 14:1140427. doi: 10.3389/fgene.2023.1140427
Received: 09 January 2023; Accepted: 27 March 2023;
Published: 10 April 2023.
Edited by:
Jing Chen, Cincinnati Children’s Hospital Medical Center, United StatesReviewed by:
Jamshid Roozbeh, Shiraz University of Medical Sciences, IranCopyright © 2023 Xiao, Pan, Huang, Liu, Wen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aizhong Liu, bGF6cm95QGxpdmUuY24=
†Equal contribution
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.