- 1Allergy Department, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Allergy Department, Beijing Key Laboratory of Precision Medicine for Diagnosis and Treatment of Allergic Diseases, National Clinical Research Center for Dermatologic and Immunologic Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 3Eight-Year Program of Clinical Medicine, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing, China
Background: It has been suggested that genetic factors may be substantially linked to allergy disorders. This study aims to investigate the relationship between the genetic susceptibility of Chinese patients with allergy disorders and the polymorphisms of plasminogen activator inhibitor 1 gene (PAI-1) rs1799762, cysteinyl leukotriene receptor 1 gene (CYSLTR1) rs320995, gasdermin B gene (GSDMB) rs7216389, glycoprotein IIIa gene (GPIIIa) rs5918, glycoprotein Ib alpha gene (GP1BA) rs6065, platelet endothelial aggregation receptor 1 gene (PEAR1) rs12041331, and tumor necrosis factor alpha gene (TNF-α) rs1800629.
Methods: From the Peking Union Medical College Hospital, this study enrolled 60 healthy participants and 286 participants with allergic diseases. TaqMan-minor groove binder (MGB) quantitative polymerase chain reaction (qPCR) was used to examine the gene polymorphisms in each group.
Results: The TaqMan-MGB qPCR results were completely consistent with the DNA sequencing results, according to other studies in this medical center (Kappa = 1, p < .001). Only the distribution of PAI-1 rs1799762 was different between patients with allergic cough and healthy people (χ2 = 7.48, p = .0238). With regard to cough patients, the 4G4G and 5G5G genotypes were more frequent (allergic cough vs. healthy individuals: 4G4G 57.9% vs. 26.7%; 5G5G 20.0% vs. 13.3%), but the 4G5G genotype was more frequent in healthy people (allergic cough vs. healthy individuals: 45.7% vs. 60.0%). The CYSLTR1 rs320995, GSDMB rs7216389, GPIIIa rs5918, GP1BA rs6065, PEAR1 rs12041331, and TNF-α rs1800629 polymorphisms, however, did not show any of such relationships.
Conclusion: The PAI-1 rs1799762 polymorphisms may be associated with the genetic susceptibility of Chinese allergic disease patients with cough performance.
1 Introduction
Due to a recent spike in prevalence, allergic disorders have become a considerable social burden (Simon, 2019). The prevalence of allergic diseases in children ranges from 7% to 10% for asthma, 15%–20% for allergic rhinitis and conjunctivitis, and 15%–20% for atopic dermatitis worldwide (Eigenmann, 2005). These can have an impact on a patient’s quality of life and increase healthcare costs. The prevalence of allergy illnesses has emerged as one of the major issues facing China’s healthcare system as a result of the country’s enormous population. Asthma, allergic and non-allergic rhinitis, and food allergies can all be diagnosed and treated thanks to the molecular and cellular basis of fundamental and clinical immunology research, which has considerably enhanced our understanding of allergic illnesses in the last 10 years (Han et al., 2020).
Allergy and associated illnesses are strongly heritable. Genetic vulnerability to allergy illnesses may be considerably influenced by disease-related single nucleotide polymorphisms (SNPs), according to genome-wide association studies (GWAS) (Tamari and Hirota, 2014; Bønnelykke et al., 2015; Haider et al., 2022). Studies of monogenic disorders have uncovered important cellular processes and protein roles in allergies. These complementary techniques suggest genetic underpinnings for T helper 2 cell (Th2) immunity, T-cell differentiation, transforming growth factor beta (TGFβ) signaling, regulatory T-cell activity, and skin/mucosal function, as well as as-yet-unknown mechanisms linked to newly discovered genes (Bønnelykke et al., 2015).
As a result, it may be sensible to find SNPs that are crucial to the development of the allergic condition. Numerous potential genes were grabbing the attention of researchers among the pathophysiological mediators involved in allergic rhinitis, asthma, severe anaphylactic reactions, atopic dermatitis, and other allergic illnesses. Cysteiny1 leukotrienes (CYSLTs) have been suggested to aid in promoting bronchoconstriction, vascular hypermeability, and mucous hypersecretion (Zhang et al., 2006). The CYSLTs receptor cysteiny1 leukotriene receptor 1 (CYSLTR1) has become a key target for the treatment of asthma and rhinitis. Antagonists against CYSLTR1, including montelukast, pranlukast and zafirlukast, have been widely prescribed in clinical practices (Trinh et al., 2019). There was also research reporting that polymorphism of rs321029 on CYSLTR1 gene was not related to the susceptibility and severity of allergic rhinitis in children, but it is closely related with the efficacy of montelukast on allergic rhinitis (Zhao et al., 2021). Additionally, it was observed that persistent mucosal inflammation may be associated to gasdermin B (GSDMB) (Moffatt et al., 2007). Moderate evidence exists for associations of the GSDMB rs7216389 variants with asthma (Zhao et al., 2015). GSDMB is a functional gene for both asthma susceptibility and severity. Single nucleotide polymorphisms in GSDMB associated with asthma severity, exacerbations, and GSDMB expression levels (Li et al., 2021).
The integrin, beta 3 (ITGB3) gene codes for glycoprotein IIIa (GPIIIa), the beta subunit of the platelet membrane adhesive protein receptor complex, which is a surface protein present in many tissues and involved in cell adhesion and cell-surface mediated signaling (Bennett, 2005). The leucine/proline polymorphism at position 33 (PlA1/A2) of the polypeptide chain correlates to the common ITGB3 polymorphism at codon 33 of exon 2 (rs5918). This SNP has been implicated as an asthma risk factor (Zimrin et al., 1990; Mikkelsson et al., 2000). The blocking of GPIIIa receptor with tirofiban can attenuate airway hyperresponsiveness and airway inflammation through the inhibition of platelet-eosinophil aggregation and activation (Kim et al., 2021).
The glycoprotein Ib alpha gene (GP1BA), which codes for the complex’s GP1ba transmembrane subunit, carries the SNPs rs2243093 and rs6065 (also known as the human platelet antigen-2, which leads to a threonine to methionine substitution at position 145). In an earlier investigation, GP1ba was linked to allergic airway disorders, especially severe, treatment-refractory asthma (Wu et al., 2021). Cardiovascular illness was found to be highly correlated with the SNPs of platelet endothelial aggregation receptor 1 (PEAR1), particularly rs12041331 and rs12566888 (Pi et al., 2019). The unique relationship between IgE-mediated allergy and cardiovascular disease was discovered to be PEAR1 (Sun et al., 2015). The omalizumab might be able to relieve the IgE-mediated inhibition of the FcεR1α-PEAR1 interaction, suggesting that omalizumab treatment could lead to alterations in the regulation of PEAR1 signaling (Sun et al., 2015).
The fibrinolytic system is significantly inhibited by plasminogen activator inhibitor 1 (PAI-1). According to recent research, PAI-1 controls airway remodeling, hyperresponsiveness, and allergic inflammation, which may contribute to the onset of asthma (Ma et al., 2009). There was a report showing that PAI-1 as a marker of esophageal functional changed in pediatric eosinophilic esophagitis (Williamson et al., 2022). A meta-analysis suggested that the −675 4G/5G polymorphism of PAI-1 gene was a risk factor of asthma (Nie et al., 2012). In a latest report, tumor necrosis factor alpha gene (TNF-α)-308G/A polymorphism in asthma patients was found to be associated with their metabolic profile (Yang et al., 2015). A large-scale meta-analysis supports a strong association between the TNF-α gene promoter polymorphism (−308G/A) and the development to asthma in both children and adults (Kang et al., 2019).
The association between allergy disorders and SNPs in the CYSLTR1 rs320995, GSDMB rs7216389, GPIIIa rs5918, GP1BA rs6065, PEAR1 rs12041331, PAI-1 rs1799762, and TNF-α rs1800629 gene has, regrettably, only been the subject of a few reports. As a result, it is a subject worth researching and may be applied to clinical detection.
The purpose of this study is to determine if gene polymorphisms in the CYSLTR1 rs320995, GSDMB rs7216389, GPIIIa rs5918, GP1BA rs6065, PEAR1 rs12041331, PAI-1 rs1799762, and TNF-α rs1800629 gene are related to various allergy diseases in Chinese patients.
2 Materials and methods
2.1 Ethics
The 1964 Helsinki declaration and its later amendments or analogous ethical standards were followed in all procedures carried out in studies involving human participants, as determined by the institutional and/or national research committee. The Chinese Academy of Medical Sciences and Peking Union Medical College Hospital Drug Clinical Trial Ethics Committee gave their clearance for this study, registration information: No. 002062, ethics approval No. KS2019282.
2.2 Study design and participants
We carried out a prospective study at the Peking Union Medical College Hospital (PUMCH) in Beijing, China, between July 2019 and December 2019.
In accordance with the patients’ presentations and the outcomes of auxiliary tests, patients underwent normal diagnostic workups. Including criteria for allergy patients (Simon, 2019): diagnosed with allergy diseases by clinical doctors, including allergic rhinitis, asthma, urticaria, atopic dermatitis, cough, atopic conjunctivitis, eczema, or a history of severe anaphylactic reaction (Eigenmann, 2005); positive serum specific IgE, positive skin prick test or intradermal test. Excluding criteria for allergy patients (Simon, 2019): patients with serious other diseases, such as diabetes, liver disease, kidney disease, etc., (Eigenmann, 2005); Immunocompromised patients.
Including criteria for healthy participants (Simon, 2019): No symptoms of any allergic diseases, including allergic rhinitis, allergic asthma, atopic dermatitis, allergic conjunctivitis, etc., (Eigenmann, 2005) No history of allergic diseases, family history (Han et al., 2020). No other immune system diseases (Bønnelykke et al., 2015). No organic disease (Tamari and Hirota, 2014). Voluntary acceptance of disease-related questionnaires (Haider et al., 2022). No participation in any drug clinical trials within 3 months. Excluding criteria for healthy participants (Simon, 2019): history of allergic diseases or chronic medical conditions associated with allergy diseases in this study (Eigenmann, 2005); history of significant allergen exposure (Han et al., 2020); patients with serious other diseases, such as diabetes, liver disease, kidney disease, etc., (Bønnelykke et al., 2015); Immunocompromised patients.
2.3 Clinical information collecting
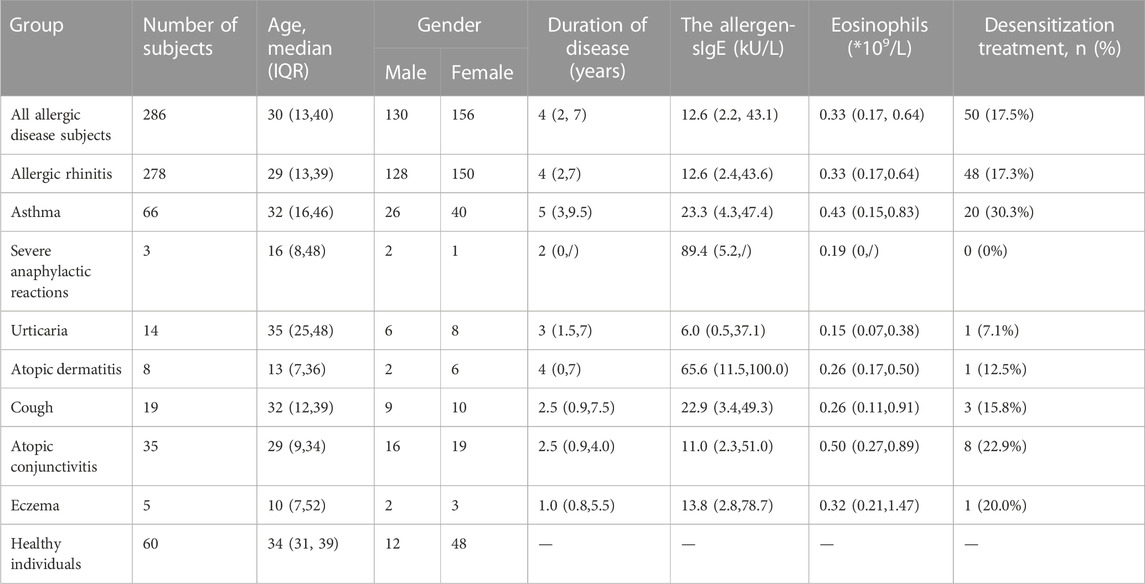
After informed consent, we collected information on the patients’ plasma allergen sIgE levels, disease duration (years), blood eosinophil counts, and whether desensitization was used. The demographic information about the study’s participants is displayed in Table 1.
2.4 TaqMan-MGB qPCR method
With the use of DNA extraction kits from Tianlong Technology Co. LTD. in Xi’an, China, genomic DNA was extracted from peripheral blood sample. Using a gene polymorphisms RT-PCR detection kit, the CYSLTR1 rs320995, GSDMB rs7216389, GPIIIa rs5918, GP1BA rs6065, PEAR1 rs12041331, PAI-1 rs1799762, and TNF-α rs1800629 genes were genotyped (Wuhan Healthcare Biotechnology Co., Ltd., Wuhan, China). According to the Applied Biosystems methodology, genotyping was accomplished by TaqMan chemistry utilizing the real-time Prism 3730XL Sequence Detection System (ABI Inc. CA, United States). In our prior research, we have demonstrated the effectiveness of the TaqMan-MGB qPCR kit for the detection of gene polymorphisms (Wang et al., 2021; Jin et al., 2022; Wang et al., 2022). The Kappa test was used to examine the agreement between DNA sequencing and TaqMan-MGB qPCR, with a Kappa value of 1 and a p-value <.001.
2.5 Data statistics and analysis
Excel 2022 (Microsoft Inc., United States), SPSS 26.0 (SPSS Inc., Chicago, IL, United States), R Project (version 4.2.0), and RStudio (Open-Source Edition) software were all used to analyze the data. To determine if the frequency distribution of polymorphisms across genomes was representative, the Hardy-Weinberg equilibrium (HWE) test was utilized. To determine if there was a significant difference in SNP between the illness group and the healthy control group, the Wilcoxon or Chi-square test was utilized. The “pwr” package in RStudio was used to do a statistical power analysis. Statistics were judged significant at p < .05.
3 Results
3.1 Demographic characteristics of participants
Table 1 shows the demographic information about the study’s participants. With a median age of 30, the 286 patients with allergic diseases included 130 men and 156 women. There were 178 patients with allergic rhinitis, 66 with asthma, 3 with severe anaphylactic reactions, 14 with urticaria, 8 with atopic dermatitis, 19 with cough, 35 with atopic conjunctivitis, and 5 with eczema, according to their clinical performance. Additionally, 60 healthy adults with a median age of 34 and a gender split of 12 men and 48 women were as controls in the study.
The median duration of allergic diseases for all patients in this study was 4 (IQR, 2, 7) years. The median allergen-sIgE of all patients was 12.6 (IQR, 2.2, 43.1) kU/L. The median count of blood eosinophils in all patients was .33 (IQR, .17, .64)/L. And 17.5% allergy patients received desensitization treatment. Compared to other allergic diseases, more asthma patients received desensitization treatment (30.3%, p < .05).
The amplification plots for genotypes of CYSLTR1 rs320995, GSDMB rs7216389, GPIIIa rs5918, GP1BA rs6065, PEAR1 rs12041331, PAI-1 rs1799762, and TNF-α rs1800629 are shown in Figure 1. The HWE equilibrium law was followed by the frequency distributions of the following polymorphisms in allergy patients: CYSLTR1 rs320995, GSDMB rs7216389, GPIIIa rs5918, GP1BA rs6065, PEAR1 rs12041331, PAI-1 rs1799762, and TNF-α rs1800629 (p > .05).
FIGURE 1. Amplification plots for genotypes of CYSLTR1 rs320995, GSDMB rs7216389, GPIIIa rs5918, GP1BA rs6065, PEAR1 rs12041331, PAI-1 rs1799762 and TNF-α rs1800629. (A–C) amplification plots of CYSLTR1 rs320995 TT, TC and CC genotypes in female. (D–F) amplification plots of GSDMB rs7216389 AA, GA, and GG genotypes. (G,H) amplification plots of GPIIIa rs5918 TT and TC genotypes. (I,J) amplification plots of GP1BA rs6065 CC and CT genotypes. (K–M) amplification plots of PEAR1 rs12041331 GG, GA and AA genotypes. (N–P) amplification plots of PAI-1 rs1799762 4G4G, 4G5G, and 5G5G genotypes. (Q–S) Amplification plots of TNF-α rs1800629 GG, GA and AA genotypes.
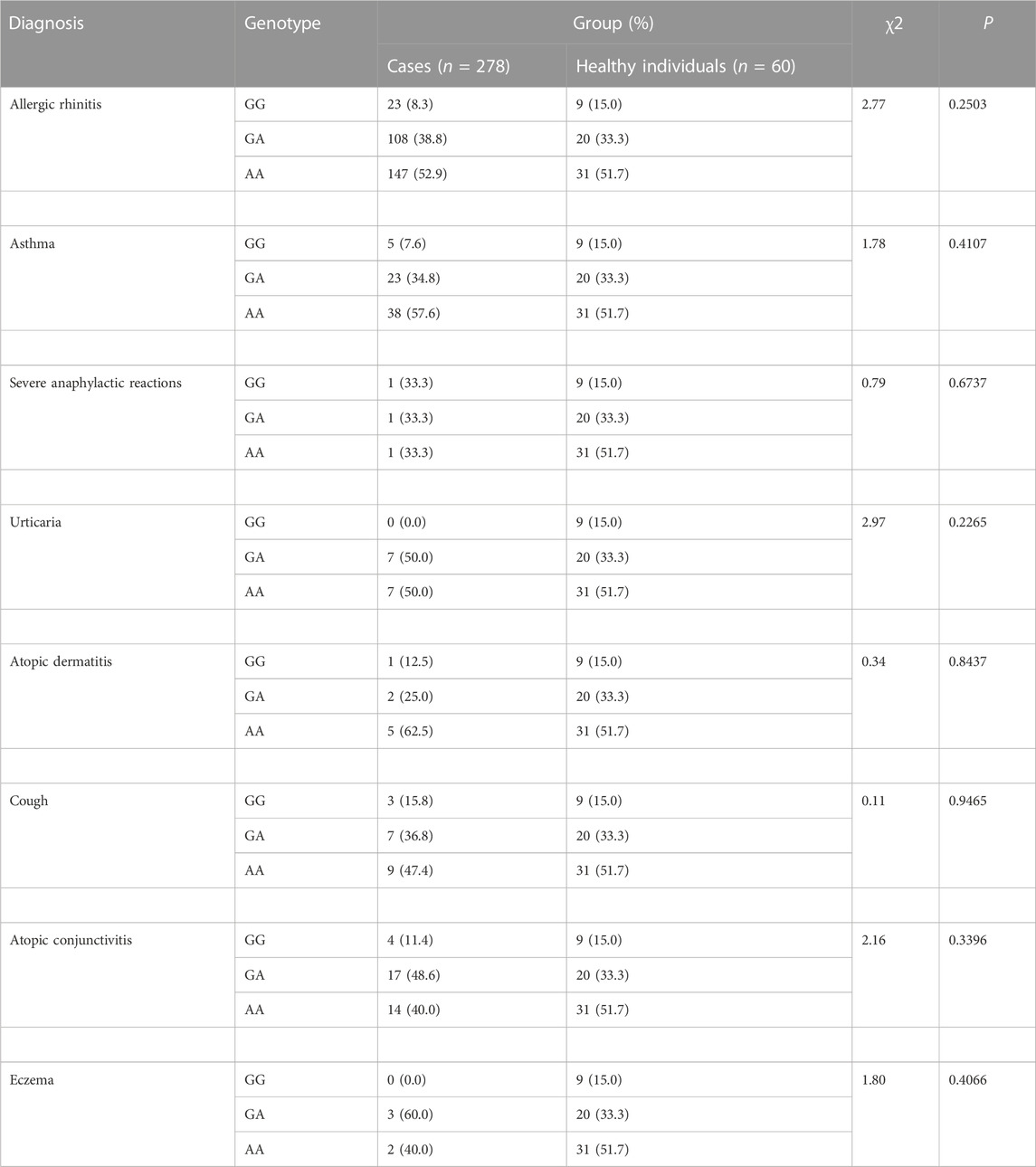
3.2 Analysis of association of PAI-1 rs1799762 with genetic susceptibility to allergic disease
Only distribution of PAI-1 rs1799762 differed from healthy controls in patients with allergic cough (χ2 = 7.48, p = .0238) (Table 2). The genotypes 4G4G and 5G5G were more common in cough patients (allergic cough vs. healthy individuals: 4G4G 57.9% vs. 26.7%; 5G5G 20.0% vs. 13.3%), but the 4G5G genotype was more common in healthy individuals (allergic cough vs. healthy individuals: 45.7% vs. 60.0%). Yet in other subgroups, no correlations between PAI-1 rs1799762 and allergic rhinitis, asthma, urticaria, atopic dermatitis, atopic conjunctivitis, eczema, or severe anaphylactic reaction were found.
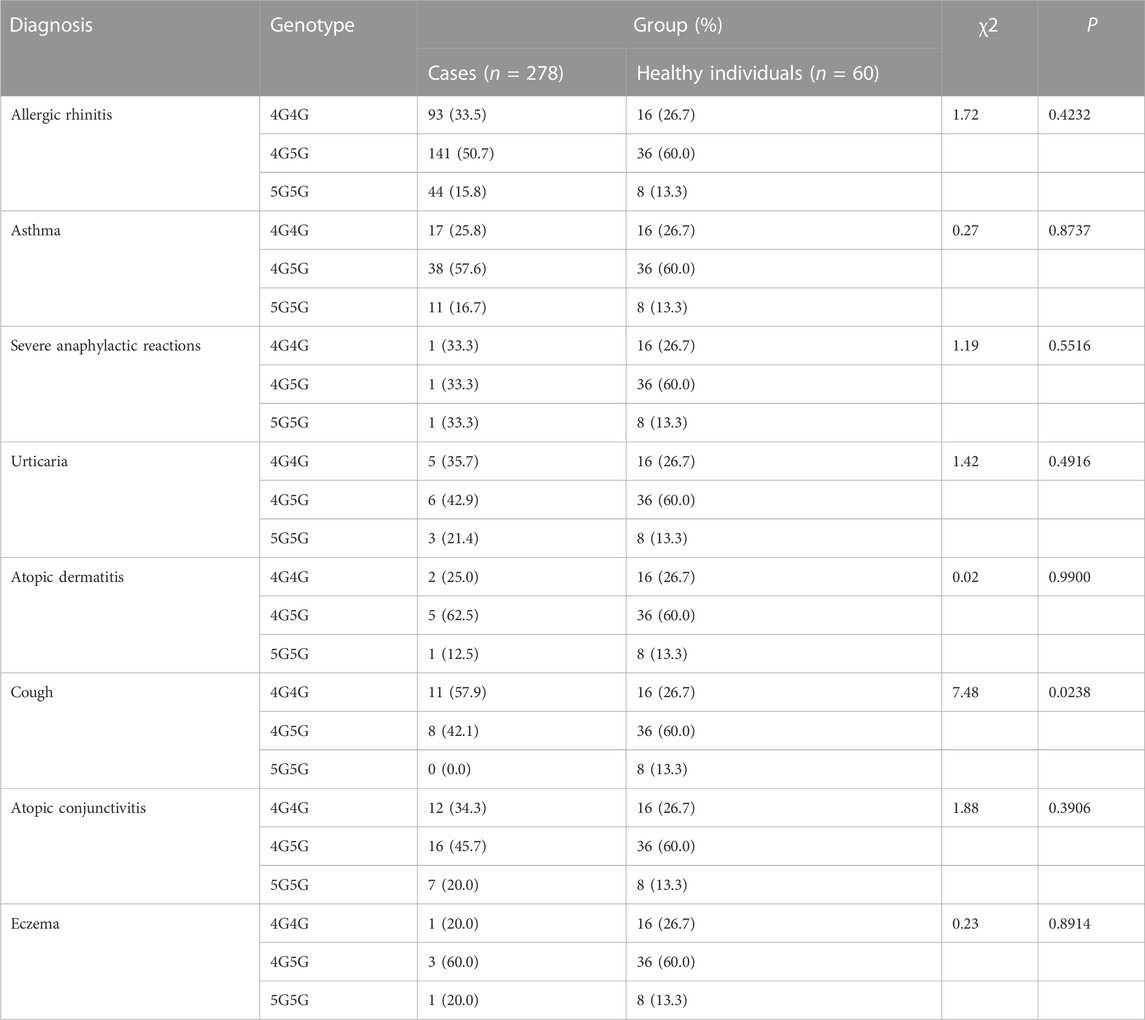
TABLE 2. Analysis of Association of PAI-1 rs1799762 (−675,4G5G) with Genetic Susceptibility to Diagnosis.
3.3 Analysis of association of other gene with genetic susceptibility to allergic disease
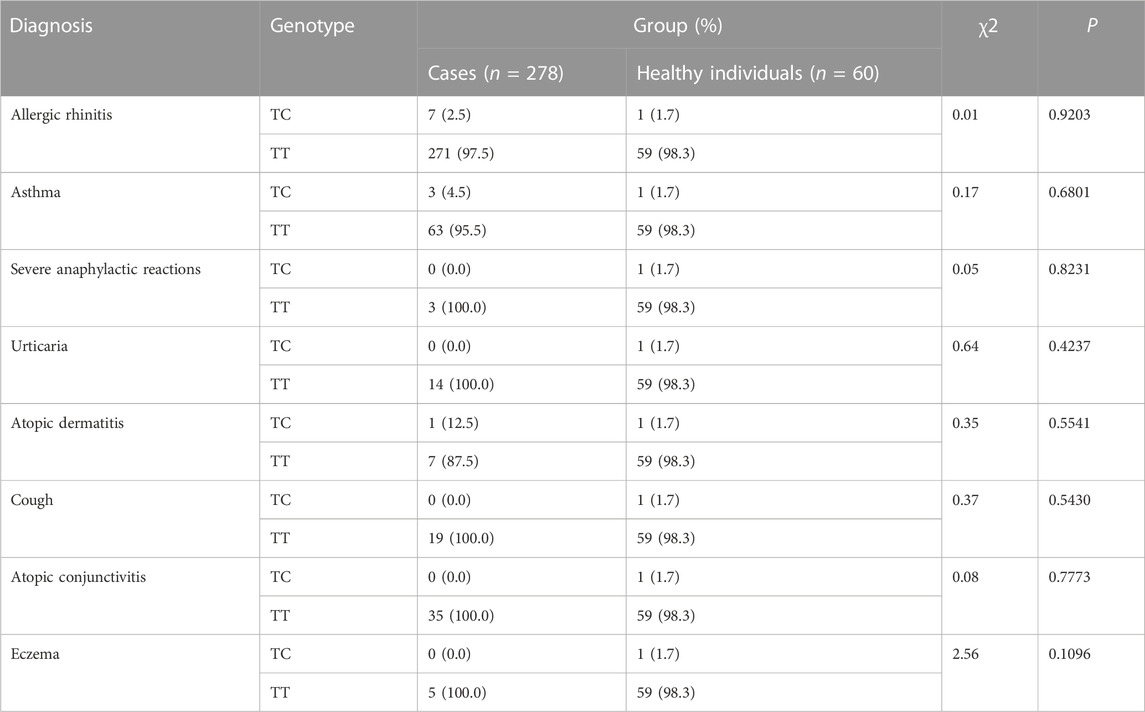
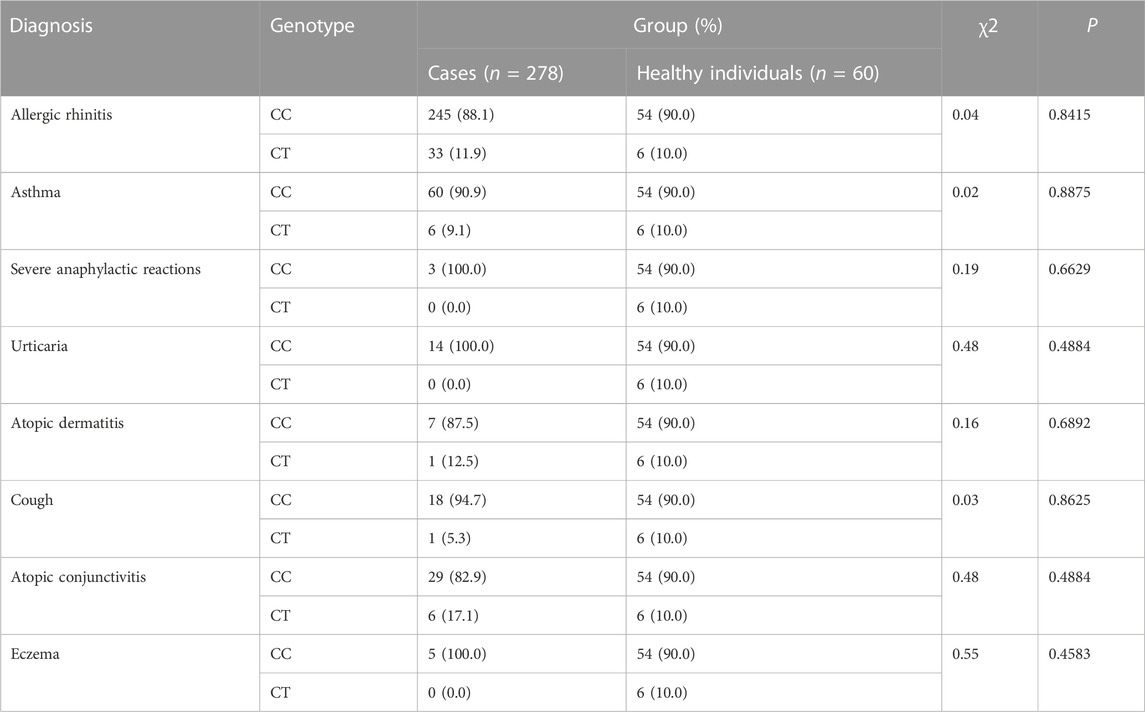
The CYSLTR1 rs320995, GSDMB rs7216389, GPIIIa rs5918, GP1BA rs6065, PEAR1 rs12041331, and TNF-α rs1800629 polymorphisms did not show any of such relationships (Table 3; Table 4; Table 5; Table 6; Table 7; Table 8).
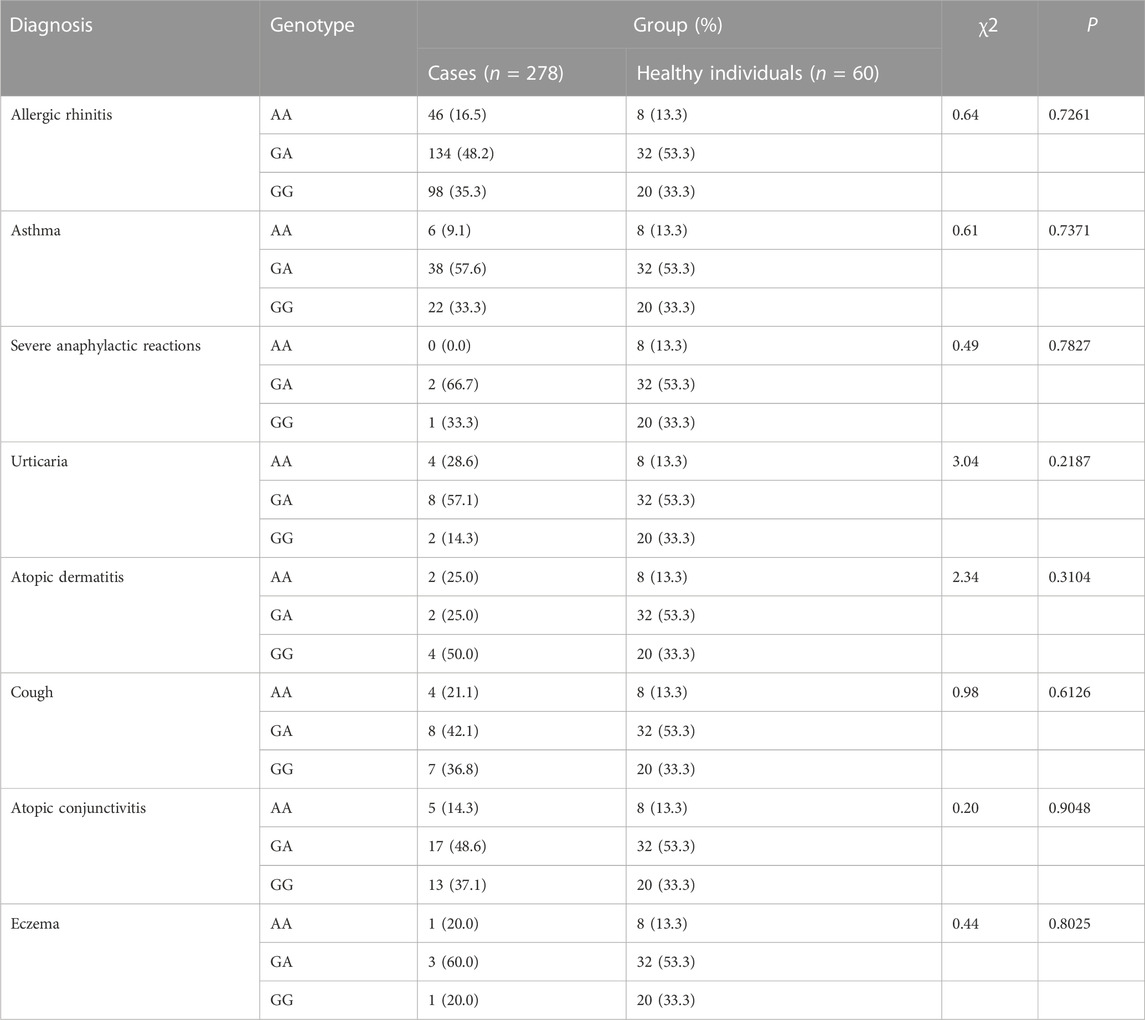
TABLE 7. Analysis of Association of PEAR1 rs12041331 (G2266A) with Genetic Susceptibility to Diagnosis.
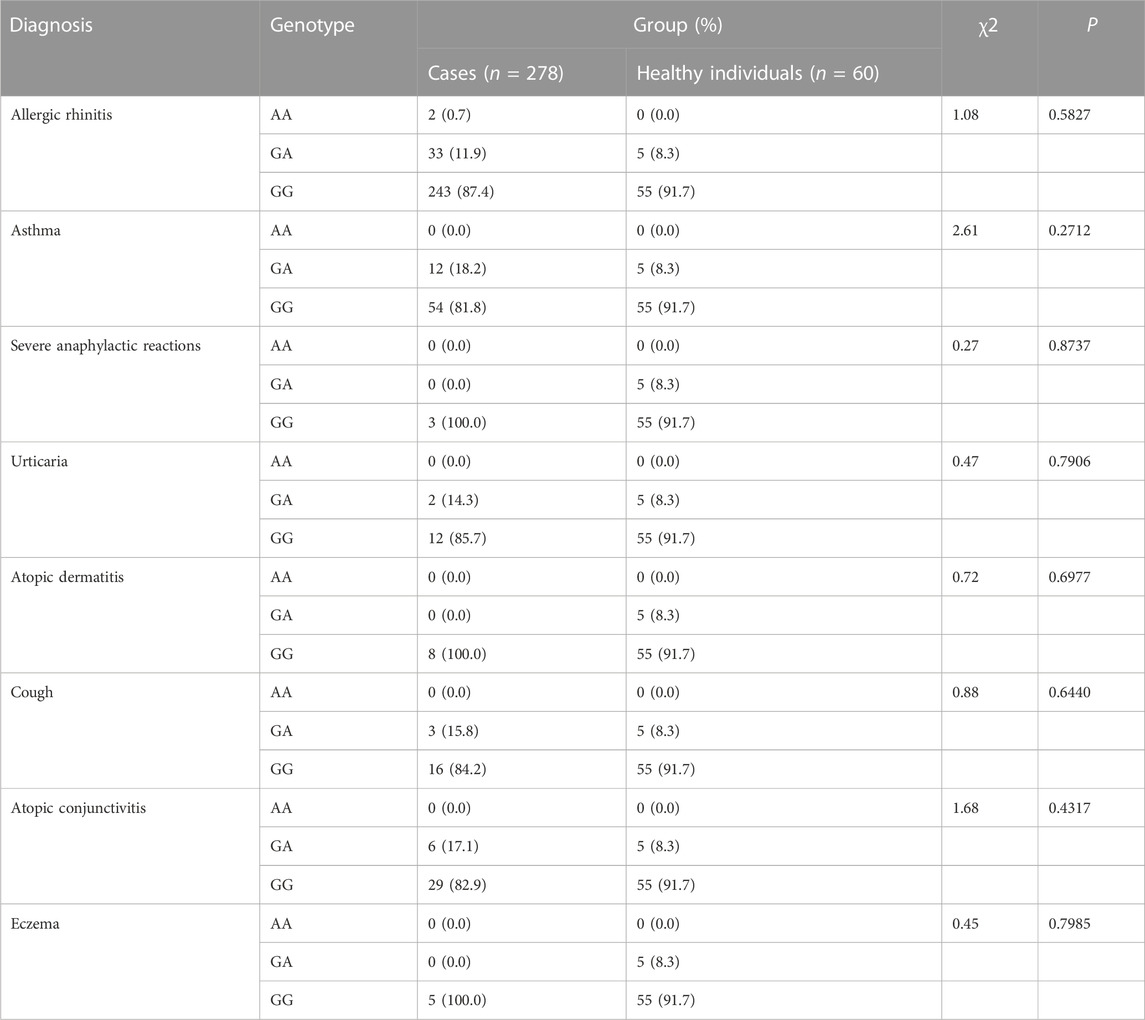
TABLE 8. Analysis of Association of TNF-α rs1800629 (−308 G>A) with Genetic Susceptibility to Diagnosis.
4 Discussion
Allergic disorders are frequent, hazardous, and occasionally fatal to patients. Growing research has emphasized how SNPs are linked to illness risk and prognosis, which is crucial for personalized medicine. Nevertheless, the field of allergic diseases had few corresponding reports. TaqMan-MGB qPCR offered a reliable and affordable method for investigating SNPs with allergy disorders (Wang et al., 2021; Jin et al., 2022; Wang et al., 2022). Using this technology, we initially discovered that patients with allergic cough had a different distribution of the PAI-1 rs1799762 gene than healthy people.
A key inhibitor of the fibrinolytic system is PAI-1. According to recent research, PAI-1 controls airway remodeling, hyperresponsiveness, and allergic inflammation, which may contribute to the onset of asthma (Ma et al., 2009). Many research are looking into the relationship between PAI-1 4G/5G polymorphisms and a variety of disorders, such as thrombosis (Zhang et al., 2020a), systemic lupus erythematosus (Anaya-Macias et al., 2020), thyroid eye disease (Katko et al., 2021), metabolic syndrome (Zhang et al., 2020b) and many more. Previous research from our center has shown that the PAI-1 4G/5G polymorphism may be useful for determining the prognosis of Chinese patients with venous thromboembolism (Wang et al., 2022).
The −675 4G/5G polymorphism of the PAI-1 gene was implicated as an asthma risk factor in a 2012 meta-analysis encompassing 1817 cases and 2327 controls (Nie et al., 2012). The 4G allele of the 4G/5G polymorphism in the PAI-1 gene may be a risk factor for IgE-mediated asthma and allergy disorders, according to comparative research published in 2002 (Bucková et al., 2002). Patients with asthma who carry at least one 4G allele of PAI-1 gene are more likely to experience allergic rhinitis symptoms (Lampalo et al., 2017). Turkish children with asthma or allergic rhinitis had a higher prevalence of the PAI-1 4G variant than their healthy classmates, according to a Turkish study (Ozbek et al., 2009). Additionally, it was discovered that the genes FCER1B and PAI-1 interact synergistically to increase asthma susceptibility (Hizawa et al., 2006).
The mechanism between asthma and PAI-1 4G/5G was explored by Ma et al. (2008). They discovered that upstream stimulatory factor-1’s binding to the E−4G/5G controls the production of PAI-1 in mast cells, a large source of PAI-1 and essential effector cells in asthma, in a manner, that is, reliant on the 4G/5G polymorphism. The primary genetic determinant of PAI-1 expression is the single guanosine nucleotide deletion/insertion polymorphism (4G/5G) at −675 base pairs of the PAI-1 gene. People with the 4G/4G genotype have plasma PAI-1 levels that are higher than people with the 5G/5G genotype.
The 4G/5G polymorphism of PAI-1 did not significantly differ between asthma patients and healthy individuals, according to this study. In contrast to healthy participants in this study, allergic patients with cough performance had higher levels of 4G4G and 5G5G. Additionally, a prior study found no statistically significant difference between the distribution of 4G/4G, 4G/5G, and 5G/5G between the group of asthmatic patients and the control group for the PAI-1 gene’s 4G/5G polymorphism (Lampalo et al., 2018). As a result, we initially discovered that in allergic individuals, the 4G/5G polymorphism of the PAI-1 gene related to cough performance rather than asthma. Future mechanistic components of the study might need to pay greater attention to the correlation between a cough and this gene in allergy patients rather than only asthma patients.
Nevertheless, there are some limitations in our study. Because of the small sample size, we had to be cautious when interpreting our findings. The small sample size of some group may cause lower power value. Since the primary result is a positive result, it is not affected by low power values. However, negative results may have some possibility of committing type II errors. In addition, it is essential to conduct future multi-center, large-scale, long-dimensional research to clarify the function of PAI-1 4G/5G polymorphisms in allergic patients with cough. Furthermore, we have not looked at how allergy individuals’ PAI-1 4G/5G polymorphisms relate to PAI-1. For the treatment and prevention of cough and other PAI-1-associated disorders, additional research examining the mechanisms of PAI-1 activity and regulation may result in the creation of a unique prognostic factor and therapeutic target. The strength of this study lies in the cross-sectional study of multiple loci polymorphisms in multiple allergic diseases. The rich variety of allergic diseases points to the direction for subsequent studies.
5 Conclusion
In conclusion, we discovered that allergic individuals with cough rather than asthma had a distinct distribution of PAI-1 rs1799762 genotypes. Allergic cough patients were more likely to have the 4G4G and 5G5G genotypes, whereas healthy people were more likely to have the 4G5G genotype.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by The Chinese Academy of Medical Sciences and Peking Union Medical College Hospital Drug Clinical Trial Ethics Committee gave their clearance for this study, ethics approval No. KS2019282. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This study was supported by grants from the National Natural Science Foundation of China (grant numbers 31972189). This study was supported by grants from the Central High Level Hospital Peking Union Medical College Hospital Clinical Research Special Youth Training Program (grant numbers 2022-PUMCH-A-222).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anaya-Macias, B. U., De la Cruz-Mosso, U., Palafox-Sánchez, C. A., Parra-Rojas, I., Martínez-Bonilla, G., González-López, L., et al. (2020). The -675 4G/5G PAI-1 polymorphism confers genetic susceptibility to systemic lupus erythematosus, its clinical manifestations, and comorbidities in Mexican-Mestizo population. Autoimmunity 53 (2), 71–77. doi:10.1080/08916934.2019.1700957
Bennett, J. S. (2005). Structure and function of the platelet integrin alphaIIbbeta3. J. Clin. investigation 115 (12), 3363–3369. doi:10.1172/JCI26989
Bønnelykke, K., Sparks, R., Waage, J., and Milner, J. D. (2015). Genetics of allergy and allergic sensitization: Common variants, rare mutations. Curr. Opin. Immunol. 36, 115–126. doi:10.1016/j.coi.2015.08.002
Bucková, D., Izakovicová Hollá, L., and Vácha, J. (2002). Polymorphism 4G/5G in the plasminogen activator inhibitor-1 (PAI-1) gene is associated with IgE-mediated allergic diseases and asthma in the Czech population. Allergy 57 (5), 446–448. doi:10.1034/j.1398-9995.2002.03582.x
Eigenmann, P. A. (2005). Diagnosis of allergy syndromes: Do symptoms always mean allergy? Allergy 60 (79), 6–9. doi:10.1111/j.1398-9995.2005.00850.x
Haider, S., Simpson, A., and Custovic, A. (2022). Genetics of asthma and allergic diseases. Handb. Exp. Pharmacol. 268, 313–329. doi:10.1007/164_2021_484
Han, X., Krempski, J. W., and Nadeau, K. (2020). Advances and novel developments in mechanisms of allergic inflammation. Allergy 75 (12), 3100–3111. doi:10.1111/all.14632
Hizawa, N., Maeda, Y., Konno, S., Fukui, Y., Takahashi, D., and Nishimura, M. (2006). Genetic polymorphisms at FCER1B and PAI-1 and asthma susceptibility. Clin. Exp. allergy J. Br. Soc. Allergy Clin. Immunol. 36 (7), 872–876. doi:10.1111/j.1365-2222.2006.02413.x
Jin, Z., Pan, Z., Wang, Z., Kong, L., Zhong, M., Yang, Y., et al. (2022). CYSLTR1 rs320995 (T927C) and GSDMB rs7216389 (G1199A) gene polymorphisms in asthma and allergic rhinitis: A proof-of-concept study. J. Asthma Allergy 15, 1105–1113. doi:10.2147/JAA.S371120
Kang, S. W., Kim, S. K., Han, Y. R., Hong, D., Chon, J., Chung, J. H., et al. (2019). Promoter polymorphism (-308G/A) of tumor necrosis factor-alpha (TNF-α) gene and asthma risk: An updated meta-analysis. Genet. Test. Mol. Biomarkers 23 (6), 363–372. doi:10.1089/gtmb.2018.0238
Katko, M., Galgoczi, E., Erdei, A., Gazdag, A., Berta, E., Bodor, M., et al. (2021). The 4G/5G polymorphism of plasminogen activator inhibitor type 1 is a predictor of moderate-to-severe thyroid eye disease. J. Inflamm. Res. 14, 1883–1890. doi:10.2147/JIR.S307046
Kim, S. H., Trinh, H. K. T., Park, H. S., and Shin, Y. S. (2021). The blocking effect of the glycoprotein IIb/IIIa receptor in the mouse model of asthma. Clin. Mol. Allergy 19 (1), 11. doi:10.1186/s12948-021-00149-6
Lampalo, M., Jukic, I., Bingulac-Popovic, J., Marunica, I., Petlevski, R., Pavlisa, G., et al. (2017). Polymorphism 4G/5G of the plasminogen activator inhibitor 1 gene as a risk factor for the development of allergic rhinitis symptoms in patients with asthma. Eur. archives oto-rhino-laryngology official J. Eur. Fed. Oto-Rhino-Laryngological Soc. (EUFOS) Affil. Ger. Soc. Oto-Rhino-Laryngology - Head Neck Surg. 274 (6), 2613–2619. doi:10.1007/s00405-017-4502-2
Lampalo, M., Jukić, I., Bingulac-Popović, J., Safić, H., Ferara, N., and Popović-Grle, S. (2018). The role of pai-1 gene 4g/5g polymorphism and diagnostic value of biomarkers in allergic and non-allergic asthma phenotype. Acta Clin. Croat. 57 (1), 96–102. doi:10.20471/acc.2018.57.01.11
Li, X., Christenson, S. A., Modena, B., Li, H., Busse, W. W., Castro, M., et al. (2021). Genetic analyses identify GSDMB associated with asthma severity, exacerbations, and antiviral pathways. J. allergy Clin. Immunol. 147 (3), 894–909. doi:10.1016/j.jaci.2020.07.030
Ma, Z., Jhun, B., Jung, S. Y., and Oh, C. K. (2008). Binding of upstream stimulatory factor 1 to the E-box regulates the 4G/5G polymorphism-dependent plasminogen activator inhibitor 1 expression in mast cells. J. allergy Clin. Immunol. 121 (4), 1006–1012.e2. doi:10.1016/j.jaci.2007.11.015
Ma, Z., Paek, D., and Oh, C. K. (2009). Plasminogen activator inhibitor-1 and asthma: Role in the pathogenesis and molecular regulation. Clin. Exp. allergy J. Br. Soc. Allergy Clin. Immunol. 39 (8), 1136–1144. doi:10.1111/j.1365-2222.2009.03272.x
Mikkelsson, J., Perola, M., Laippala, P., Penttilä, A., and Karhunen, P. J. (2000). Glycoprotein IIIa Pl(A1/A2) polymorphism and sudden cardiac death. J. Am. Coll. Cardiol. 36 (4), 1317–1323. doi:10.1016/s0735-1097(00)00871-8
Moffatt, M. F., Kabesch, M., Liang, L., Dixon, A. L., Strachan, D., Heath, S., et al. (2007). Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448 (7152), 470–473. doi:10.1038/nature06014
Nie, W., Li, B., and Xiu, Q. Y. (2012). The -675 4G/5G polymorphism in plasminogen activator inhibitor-1 gene is associated with risk of asthma: A meta-analysis. PloS one 7 (3), e34385. doi:10.1371/journal.pone.0034385
Ozbek, O. Y., Ataç, F. B., Ogus, E., and Ozbek, N. (2009). Plasminogen activator inhibitor-1 gene 4G/5G polymorphism in Turkish children with asthma and allergic rhinitis. Allergy asthma Proc. 30 (1), 41–46. doi:10.2500/aap.2009.30.3183
Pi, L., Xu, Y., Fu, L., Zhang, L., Liu, Y., Zhou, H., et al. (2019). A PEAR1 polymorphism (rs12041331) is associated with risk of coronary artery aneurysm in Kawasaki disease. Ann. Hum. Genet. 83 (1), 54–62. doi:10.1111/ahg.12285
Simon, D. (2019). Recent advances in clinical allergy and immunology 2019. Int. archives allergy Immunol. 180 (4), 291–305. doi:10.1159/000504364
Sun, Y., Vandenbriele, C., Kauskot, A., Verhamme, P., Hoylaerts, M. F., and Wright, G. J. (2015). A human platelet receptor protein microarray identifies the High affinity immunoglobulin E receptor subunit α (FcεR1α) as an activating platelet endothelium aggregation receptor 1 (PEAR1) ligand. Mol. Cell. Proteomics 14, 1265–1274. doi:10.1074/mcp.M114.046946
Tamari, M., and Hirota, T. (2014). Genome-wide association studies of atopic dermatitis. J. Dermatol 41 (3), 213–220. doi:10.1111/1346-8138.12321
Trinh, H. K. T., Lee, S. H., Cao, T. B. T., and Park, H. S. (2019). Asthma pharmacotherapy: An update on leukotriene treatments. Expert Rev. Respir. Med. 13 (12), 1169–1178. doi:10.1080/17476348.2019.1670640
Wang, Z., Kong, L., Luo, G., Zhang, H., Sun, F., Liang, W., et al. (2022). Clinical impact of the PAI-1 4G/5G polymorphism in Chinese patients with venous thromboembolism. Thromb. J. 20 (1), 68. doi:10.1186/s12959-022-00430-x
Wang, Z., Kong, L., Zhang, H., Sun, F., Guo, Z., Zhang, R., et al. (2021). Tumor necrosis factor alpha -308G/A gene polymorphisms combined with neutrophil-to-lymphocyte and platelet-to-lymphocyte ratio predicts the efficacy and safety of anti-TNF-α therapy in patients with ankylosing spondylitis, rheumatoid arthritis, and psoriasis arthritis. Front. Pharmacol. 12, 811719. doi:10.3389/fphar.2021.811719
Williamson, P., Proudfoot, J., Gharibans, A., Dohil, L., Newbury, R., Barsamian, J., et al. (2022). Plasminogen activator inhibitor-1 as a marker of esophageal functional changes in pediatric eosinophilic esophagitis. Clin. Gastroenterol. Hepatol. 20 (1), 57–64.e3. doi:10.1016/j.cgh.2020.09.040
Wu, Y., Zeng, Z., Guo, Y., Song, L., Weatherhead, J. E., Huang, X., et al. (2021). Candida albicans elicits protective allergic responses via platelet mediated T helper 2 and T helper 17 cell polarization. Immunity 54 (11), 2595–2610.e7. doi:10.1016/j.immuni.2021.08.009
Yang, Y. H., Liu, Y. Q., Zhang, L., Li, H., Li, X. B., Ouyang, Q., et al. (2015). Genetic polymorphisms of the TNF-α-308G/A are associated with metabolic syndrome in asthmatic patients from Hebei province, China. Int. J. Clin. Exp. Pathol. 8 (10), 13739–13746.
Zhang, J., Migita, O., Koga, M., Shibasaki, M., Arinami, T., and Noguchi, E. (2006). Determination of structure and transcriptional regulation of CYSLTR1 and an association study with asthma and rhinitis. Pediatr. Allergy Immunol. 17 (4), 242–249. doi:10.1111/j.1399-3038.2005.00347.x
Zhang, X., Cai, X., and Pan, J. (2020). Correlation between PAI-1 gene 4G/5G polymorphism and the risk of thrombosis in ph chromosome-negative myeloproliferative neoplasms. Clin. Appl. Thromb. Hemost. 26, 1076029620935207. doi:10.1177/1076029620935207
Zhang, X., Gao, B., and Xu, B. (2020). Association between plasminogen activator inhibitor-1 (PAI-1) 4G/5G polymorphism and risk of alzheimer's disease, metabolic syndrome, and female infertility: A meta-analysis. Medicine 99 (50), e23660. doi:10.1097/MD.0000000000023660
Zhao, C. N., Fan, Y., Huang, J. J., Zhang, H. X., Gao, T., Wang, C., et al. (2015). The association of GSDMB and ORMDL3 gene polymorphisms with asthma: A meta-analysis. Allergy Asthma Immunol. Res. 7 (2), 175–185. doi:10.4168/aair.2015.7.2.175
Zhao, M., Li, H., and Li, H. (2021). The relevance of CYSLTR1 gene polymorphism to the severity of allergic rhinitis and clinical responsiveness of montelukast in children. Eur. archives oto-rhino-laryngology official J. Eur. Fed. Oto-Rhino-Laryngological Soc. (EUFOS) Affil. Ger. Soc. Oto-Rhino-Laryngology - Head Neck Surg. 278 (12), 4847–4853. doi:10.1007/s00405-021-06771-z
Keywords: allergy and immunology, gene polymorphism, plasminogen activator inhibitor 1, cough, Hypersensitivity
Citation: Tang R, Lyu X, Li H and Sun J (2023) The 4G/5G polymorphism of plasminogen activator inhibitor type 1 is a predictor of allergic cough. Front. Genet. 14:1139813. doi: 10.3389/fgene.2023.1139813
Received: 07 January 2023; Accepted: 16 February 2023;
Published: 24 February 2023.
Edited by:
Beniamin Oskar Grabarek, University of Technology in Katowice, PolandReviewed by:
Anita Boron, Medical University of Silesia, PolandDariusz Boron, Academy of Silesia, Poland
Nikola Zmarzły, Katowice School of Technology, Poland
Copyright © 2023 Tang, Lyu, Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Li, bGlob25nMTIyOHNlY0AxNjMuY29t; Jinlyu Sun, amlubHl1c3VuQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Rui Tang1,2†
Rui Tang1,2† Xiaohong Lyu
Xiaohong Lyu