
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 21 April 2023
Sec. Genomics of Plants and the Phytoecosystem
Volume 14 - 2023 | https://doi.org/10.3389/fgene.2023.1135577
This article is part of the Research Topic Genetic Mechanisms of Stress Tolerance in Crops View all 7 articles
 Yingfeng Wang1
Yingfeng Wang1 Yubo Wang1
Yubo Wang1 Wenjuan Chen1
Wenjuan Chen1 Yating Dong1
Yating Dong1 Guilian Zhang1
Guilian Zhang1 Huabing Deng1
Huabing Deng1 Xiong Liu1
Xiong Liu1 Xuedan Lu1
Xuedan Lu1 Feng Wang1
Feng Wang1 Guihua Chen1
Guihua Chen1 Yunhua Xiao1*
Yunhua Xiao1* Wenbang Tang1,2*
Wenbang Tang1,2*Heat stress (HS) has become a major abiotic stress in rice, considering the frequency and intensity of extreme hot weather. There is an urgent need to explore the differences in molecular mechanisms of HS tolerance in different cultivars, especially in indica and japonica. In this study, we investigated the transcriptome information of IR64 (indica, IR) and Koshihikari (japonica, Kos) in response to HS at the seedling stage. From the differentially expressed genes (DEGs) consistently expressed at six time points, 599 DEGs were identified that were co-expressed in both cultivars, as well as 945 and 1,180 DEGs that were specifically expressed in IR and Kos, respectively. The results of GO and KEGG analysis showed two different HS response pathways for IR and Kos. IR specifically expressed DEGs were mainly enriched in chloroplast-related pathways, whereas Kos specifically expressed DEGs were mainly enriched in endoplasmic reticulum and mitochondria-related pathways. Meanwhile, we highlighted the importance of NO biosynthesis genes, especially nitrate reductase genes, in the HS response of IR based on protein-protein interaction networks. In addition, we found that heat shock proteins and heat shock factors play very important roles in both cultivars. This study not only provides new insights into the differences in HS responses between different subspecies of rice, but also lays the foundation for future research on molecular mechanisms and breeding of heat-tolerant cultivars.
Agriculture production is threatened by the frequent change of the climate, which directly influences food security. In the second half of the 20th century, the average global temperature increased by 0.74°C, and it is expected that the global temperature will increase at an average rate of 0.2°C/10a in the next 20–30 years (Jaagus, 2006). Climate warming has led to an increase in the intensity and frequency of extreme hot weather, and results in heat stress (HS), which causes irreversible damage to the growth and development of plants (Xu et al., 2021). As a widely planted crop, rice (Oryza sativa L.) is a staple food for over half of the population and faces a multitude of biotic and abiotic stresses continuously during their lifespan, including salt, temperature, drought and so on. HS is one of the major abiotic stresses that affects rice growth and development, which may occur in all the growing stages of rice (Tripathi et al., 2016; Shahzad et al., 2021). For every 1.0°C increase in average daily temperature during the growing season, the yields of rice will approximately be reduced by 14%, which poses a great threat to the cultivation of rice. Therefore, exploring the HS molecular response mechanism and mining beneficial genes to cultivate heat-tolerant cultivars represent an attractive approach to cope with this challenge (Kan et al., 2022).
HS influences physiological and metabolic processes of rice, which cause growth and development impaired. In terms of physiological effect, it is manifested as membrane damage, reactive oxygen species (ROS) accumulation, photosynthesis damage, carbohydrate metabolism and partition chaos, and phytohormone imbalance. As the most sensitive component of plant cells, the structure and function of membranes are easily damaged by HS. HS alters the ratio of saturated to unsaturated fatty acids by decreasing the activities of fatty acid desaturases, thereby disrupting the integrity of the plasma membrane, which in turn increases permeability and leads to leakage of organic and inorganic ions from cells (Fukao et al., 2019; Yang et al., 2019). HS makes the content of superoxide dismutase (SOD), catalase (CAT) and intracellular malondialdehyde (MDA) abnormal and impairs the activity of antioxidant enzymes and protein function. HS destroys the permeability of thylakoid membrane and even disintegrates thylakoid grana, triggering a decrease in chlorophyll content, which further leads to a decrease in the ratio of variable fluorescence to maximum fluorescence (Fv/Fm) and photosynthetic rate (Hu et al., 2014; Wang et al., 2017). HS affects the expression of carbohydrate metabolism-related genes such as carbon starved anthers (CSA), monosaccharide transporter (MST8), anther-specific cell wall invertase (INV4) and sucrose transporter (OsSUT1), which disturb the energy production, transport and distribution. Jasmonates (JAs), brassinosteroids (BRs), ethylene and abscisic acid (ABA) are major phytohormones that can act either synergistically or antagonistically to mediate HS through activating the defense system and enhancing osmotic regulation (Yang et al., 2021).
In the long-term co-evolutionary with HS, plants have evolved different coping mechanisms. On the one hand, plants can improve their ability to scavenge unfolded proteins by “actively response”, thereby maintaining protein homeostasis for heat resistance. On the other hand, plants can also be insensitive by “static braking” to reduce the consumption of thermal responses, maintain normal physiological activity, and rapidly rebuild after HS to improve survival under HS. Mining heat-resistance genes or quantitative trait loci (QTL) and elucidating the molecular mechanism are the key points for HS tolerance improvement in crops. So far, more than 80 rice HS-related QTLs have been mapped, 25 genes have been cloned, including 12 genes (OsTT1、OsHTAS、TOGR1、ER、OsHCI1、OsHIRP1、OsSIZ1、OsNSUN2、MSD1、OsMDHAR4、OsUBP21、TT2) encoding proteases, 10 genes (OsHSP90、HSP101、OsANN1、OsRGB1、OsRab7、OsCNGC14、OsCNGC16、OsHSBP1、OsHSBP2、OsFBN1) encoding heat shock proteins (HSPs), channel proteins and other proteins, and 3 genes (OsMYB55、SNAC3、OsNTL3) are TFs (Rana et al., 2012; Lim et al., 2013; Wei et al., 2013; Li et al., 2015; Wang et al., 2015; Wang et al., 2016; Kilasi et al., 2018; Zhou et al., 2019; Chen et al., 2021). These genes through complex molecular mechanism to HS response, including classic cognition (reactive oxygen species scavenging, cytotoxic protein elimination and unfolded protein renaturation), and newly regulatory pathway (G protein regulation, Ca2+ signal transduction and decoding and wax metabolism) (Penke et al., 2018).
During the seedling stage, the optimum growth temperature of rice is 26°C–32°C, HS (above 35°C) results in growth retardation, accelerated aging, decreased absorption capacity, and maybe even death (Liu et al., 2021). There is a pathway correlation between the basic heat tolerance at seedling stage and later growth stage of rice, which makes the research on the molecular mechanism of HS in seedling stage of great application valuable. Different genetic backgrounds have different responses to HS, especially between rice subspecies (Indica and Japonica) which show a great significant difference in enduring tolerance.
High-throughput RNA-sequencing technology (RNA-seq) provides highly accurate and cost-effective tools to unravel the transcriptome dynamics and molecular mechanisms of rice response to HS (Vitoriano and Calixto, 2021). The process of natural and artificial selection has endowed indica rice with greater heat tolerance than japonica rice (Xu et al., 2020). The indica cultivar IR64 (IR) is the parent of modern indica rice cultivars (Maeda et al., 2019). The japonica cultivar Koshihikari (Kos) is an elite Japanese cultivar. Both IR and Kos are the most popular indica and japonica rice cultivars in the world and are widely grown in various rice producing areas (Ebitani et al., 2005; Nonoue et al., 2019). In this study, we analyzed the leaf transcriptomes of IR and Kos at 0.5, 1, 2, 4, 8, 24 h of HS treatment and performed Illumina RNA-seq, transcriptome expression analysis and qRT-PCR to identify differentially expressed genes (DEGs) and pathway details that are responds to HS. The results of this study will not only enhance our understanding of the mechanisms by which plants respond to HS, but also provide new insights into the key genes responsible for the differences in HS tolerance between indica and japonica.
IR64 (IR) and Koshihikari (Kos) were used in this study. Seeds of both cultivars were cultured at 37°C for 2 d for germination. Germinated seeds were transferred to 96-well plates and seedlings were cultured in distilled water for 4 days, and then replaced with Kimura B nutrient solution. Before the stage of three-leaf, the temperature was 28°C. After the stage of three-leaf of two cultivars, the seedlings were transferred to HS treatment with a temperature of 45°C. Seedlings of both species were grown in an artificial incubator with a relative humidity of 70%. After HS treatment, the seedlings were recovered at 28°C.
Here, we selected seven time points (0, 0.5, 1, 2, 4, 8, and 24 h) to collect leaves of seedling for RNA-seq. A total of 42 samples were obtained in this study, with 0 h as the control group and 0.5, 1, 2, 4, 8 and 24 h as the treatment groups. There were three biological replicates for each group, and one biological replicate with 10 plants sampled. For each biological replicate, samples from each treatment were harvested separately and immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction for transcriptome sequencing.
RNA extraction using the TRIzol method. RNA degradation and contamination was monitored on 1% agarose gels. Reverse transcription was performed using the PrimeScript RT kit (TakaRa, Dalian, China).
After treatment at 45°C for 40 h and recovery for 3 days, the seedlings were photographed and the survival rate was counted, using seedlings grown under normal conditions in another incubator as a control. Electrolyte leakage from seedlings treated with HS for 24 h was measured using a previously described method (Dai et al., 2020).
HTseq (1.99.2) was used to statistically calculate the read count values of each gene, which represent the basic expression of gene, and standardize each gene expression was using fragments per kilobase of transcript per million mapped reads (FPKM), which makes the genes of two cultivars and groups comparable. DESeq (1.34.0) was using to screen the differential expression of genes with the conditions: an expression difference of |log2 Fold Change|>0 and a Significant padj<0.05.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of DEG were performed by the PlantNGSTools R package. Terms or entries with corrected p-values less than 0.05 were considered significantly enriched for DEGs. The sequences of transcription factors (TFs), transcriptional regulators (TRs) and protein kinases (PKs) were searched in the iTAK database (http://itak.feilab.net/cgi-bin/itak/index.cgi, accessed on 1 July 2022) and compared with differential gene sequences. SIRING (https://string-db.org/, accessed on 1 July 2022) is used to analyze protein-protein interaction (PPI) networks. Cytoscape v.3.9.0 was used to drawn the regulatory network of target genes.
Total RNA extraction and reverse transcription as described in 2.1. Ten DEGs were randomly selected for the verification of the RNA-seq result. The primers for these genes are listed in Supplementary Table S1. The qRT-PCR was performed in 96-well plates in 10 µL reaction using ChamQ™ SYBR® Color qPCR Master Mix (Vazyme, Shanghai, China), UBQ gene was used as an internal control. Relative gene expression levels were determined from equation 2−△△CT. Three biological replicates were used for each gene under each time point.
To assess the different responses of the two cultivars (IR and Kos) to HS, we transferred rice seedlings at the three-leaf stage to an artificial incubator for a HS treatment (45°C). After 40 h of treatment and 3 days of recovery at 28°C, all Kos seedlings died with 0% survival rate, whereas all IR seedlings survived (Figure 1A, B). Moreover, the electrolyte leakage of Kos seedlings after HS treatment was significantly higher than that of IR seedlings (Figure 1C). These results demonstrated that the indica rice cultivar IR from the tropics has stronger HS tolerance than the japonica rice cultivar Kos from the temperate zone, and showed extremely significant difference between rice subspecies.
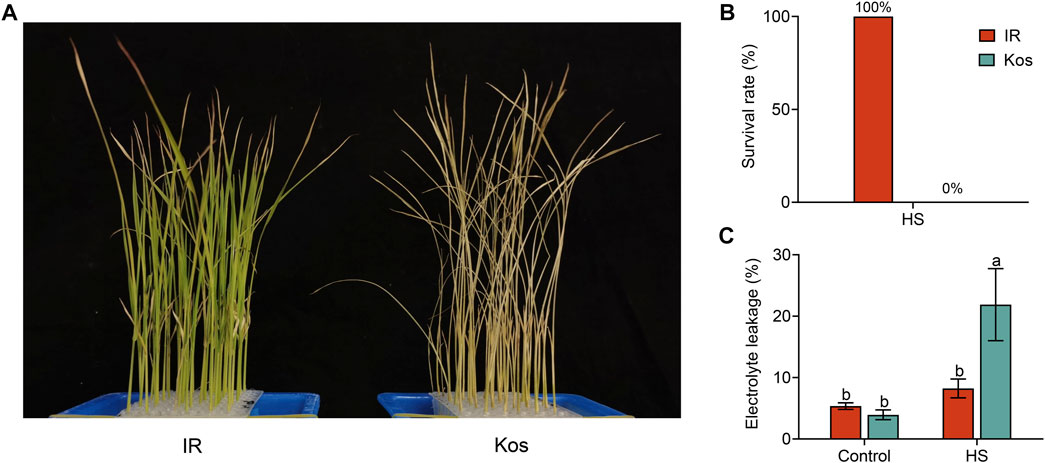
FIGURE 1. Phenotypes of two rice varieties subjected to a HS treatment. IR and Kos seedlings treated at 45°C for 40 h and recovered for 3 days (A), and the survival rate of two rice varieties (B). (C) Electrolyte leakage rate of IR and Kos seedlings after 24 h of high temperature treatment at 45°C. The corresponding seedlings grown under normal conditions in another chamber were used as controls. Survival rate in (B) is the proportion of surviving seedlings per 12 plants, and data are presented as mean ± SD, n = 4. The data shown in (C) are means ± SD of three independent experiments, and different letters indicate significant differences using ANOVA test, p < 0.05.
To study the gene expression changes in IR and Kos leaves under HS as described above, we used next-generation sequencing technology to measure the transcription in rice leaves after 45°C HS treatment. RNA sequencing data were generated from rice leaf samples at different time points. The transcriptome of each cultivar was analyzed at seven time points with three biological replicates for each time point, and Illumina RNA-seq analysis of 42 samples yielded 355.25G bp of data and 2,425,820,766 read pairs (obtained reads) (Supplementary Table S2A). After re-processing of the reads, we obtained 2,368,378,416 single-end clean reads (total records) (Supplementary Table S2B). The total map ratio ranged from 89.82% to 95.98%, and the unique map ratio ranged from 87.45% to 93.16%, few reads could not be mapped to the reference genomes (4.02%–10.18%) (Supplementary Table S2B).
We used sample-to-sample correlation analysis for the data exploration analysis. The overall relatedness of the transcriptome at different times was determined by a correlation heatmaps generated for the IR and Kos samples (Supplementary Figure S1). For all samples, the three biological replicates (IR and Kos) showed good correlation, and the transcriptome data were closely related at each time point. These results indicate that the samples and transcriptome sequencing results from this study are available and can be analyzed further.
We analyzed the cultivar DEGs at different time points in order to identify DEGs involved in the regulation of HS in rice (Table 1). Comparing the DEGs of each cultivar to their control at different time points, the number of upregulated genes were greater than the downregulated genes. IR and Kos had the largest number of DEGs at 24 h after HS, with 13,724 (7,526 upregulated, 6,198 downregulated) and 15,808 (8,364 upregulated, 7,444 downregulated) DEGs, respectively. In IR and Kos, the lowest number of DEGs were identified 4 h after HS, with 5,199 and 5,470, respectively.
To verify the reliability and accuracy of the transcriptome results in this study, 10 DEGs were randomly selected and the expression levels of the two cultivars were verified by qRT-PCR at each time point after HS treatment. The qRT-PCR results for the 10 genes are shown in Supplementary Figure S2. The qRT-PCR results showed that the trends of these 10 genes after HS treatment were consistent with the transcriptome results, which indicated that our transcriptome results were reliable.
We first analyzed all time point DEGs for the two cultivars IR and Kos separately. After HS treatment, 1,544 and 1,779 DEGs were shown to be continuously expressed within six time points in IR and Kos leaves, respectively (Figure 2A). We then compared the continuously expressed DEGs in the two cultivars, and as shown in Figure 2B these DEGs were classified into three categories. There are 599 common (non-cultivar-specific) HS-responsive (CHR) DEGs, 945 IR HS-responsive (IHR) DEGs and 1,180 Kos HS-responsive (KHR) DEGs.
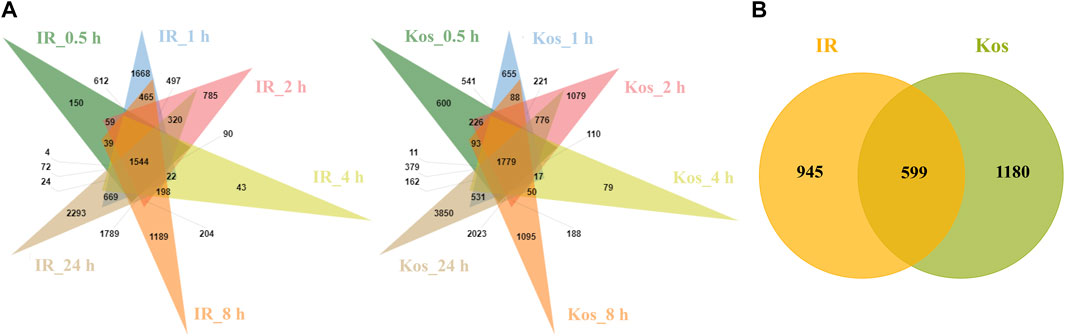
FIGURE 2. (A) Venn diagram of DEGs in both two cultivars at different time points after HS. IR_0.5 h, IR_1 h, IR_2 h, IR_4 h, IR_8 h, IR_24 h denote differentially expressed gene sets obtained by comparing IR samples at 0.5, 1, 2, 4, 8, and 24 h after HS, respectively, with the control sample IR_0 h. Kos_0.5 h, Kos_1 h, Kos_2 h, Kos_4 h, Kos_8 h, Kos_24 h denote differentially expressed gene sets obtained by comparing Kos samples at 0.5, 1, 2, 4, 8, and 24 h after HS, respectively, with the control sample Kos0. (B) Venn diagram of DEGs shared between both cultivars at all the time points after HS.
As shown in Figures 3A, B, the IHR and KHR DEGs annotated in the GO database were classified into three major categories including biological processes (BP), cellular component (CC), and molecular functions (MF), and the top ten terms with the smallest p-values in each category are shown by us in figure. The results revealed that the HS-response mechanisms were different in IR and Kos. For the selected 945 DEGs that were IHR, 714 had GO annotations and were involved in 132 terms. IHR DEGs are involve in transmembrane transport, component of chloroplast, heat acclimation, secondary active transmembrane transporter activity, nitric oxide biosynthetic process and others. For the selected 1,180 DEGs that were KHR, 832 had GO annotations and were involved in 94 terms. KHR DEGs involve response to heat, component of mitochondrial, beta-glucosidase and others.
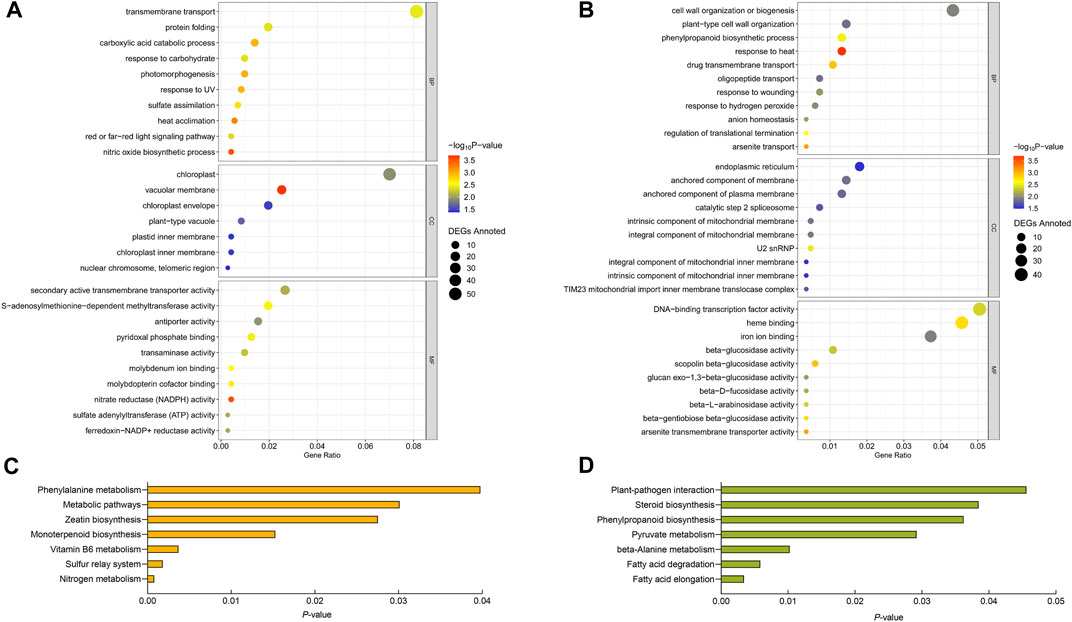
FIGURE 3. Analysis of GO and KEGG of IHR and KHR DEGs. GO analysis of IHR (A) and KHR (B) DEGs. Y-axis shows the GO terms. X-axis shows the Gene Ration. The size and color of each point represents the number of genes enriched in a particular GO term and the -log10 (p-value), respectively. KEGG pathway analysis of IHR (C) and KHR (D) DEGs. Y-axis shows the name of KEGG pathway. X-axis shows the p-values.
To further investigate the DEGs involved in various pathways in IHR and KHR, KEGG enrichment analysis was performed for these two categories of DEGs. DEGs of IHR and KHR were enriched into 86 and 74 KEGG pathways, respectively, and we showed the KEGG pathways with p-values <0.05 in Figures 3C, D. The DEGs of IHR are involved in metabolic pathways, nitrogen metabolism and zeatin biosynthesis, among others, with the largest number of DEGs in metabolic pathways at 66. The DEGs of KHR are involved in fatty acid elongation and degradation, pyruvate and beta-alanine metabolism, etc. In addition, both categories contain pathways associated with phenylalanine.
To better analyze the possible key genes of rice seedlings in response to HS, we identified TFs, TRs and PKs in IHR and KHR DEGs (Figure 4). As shown in Figure 4A, 53 and 77 TF genes were identified in the DEGs of IHR and KHR, respectively, which are contained in 36 TF families. In IHR, the TF family with the highest number of genes is MYB, while in KHR it is AP2/ERF-ERF. In addition, 10 TR families and 7 PK subfamilies were identified (Figure 4B). Among PKs, the majority of subfamilies belonged to the RLK (Figure 4B).
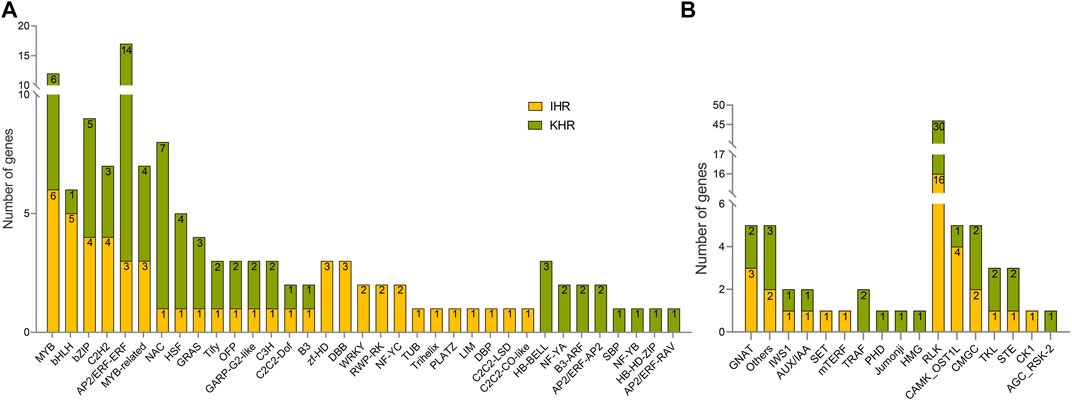
FIGURE 4. TF, TR and PK classification of IHR and KHR DEGs. (A) TF identification based on the iTAK database for DEGs of IHR and KHR, the numbers in the figure indicate the number of genes contained in each type of TF family. (B) The first 10 columns represent the TR identified in the IHR and the KHR. The latter 7 columns represent the PK identified in the IHR and the KHR. The numbers in the figure indicate the number of genes in TR and PK.
We constructed PPI networks of IHR and KHR DEGs to facilitate further identification of genes located in core positions. The top 20 DEGs with the highest degree of protein interactions were shown in the PPI network of IHR and KHR (Figures 5A,C), while the expression patterns of these genes were depicted in the heat map (Figures 5B,D). There were 11 genes showing downregulation in IHR, while only 3 genes were downregulated in KHR. Three HSPs gene (Os02g0710900, Os01g0840100, Os08g0500700) were included in these core genes, all of which were upregulated relative to the control. In addition, we identified five genes related to nitrogen metabolism in the IHR, including Os08g0468100 (OsNIA1), Os02g0770800 (OsNR2), Os08g0468700 (OsNIA2), Os02g0765900 (OsNIR2) and Os03g0223400 (OsGSr).
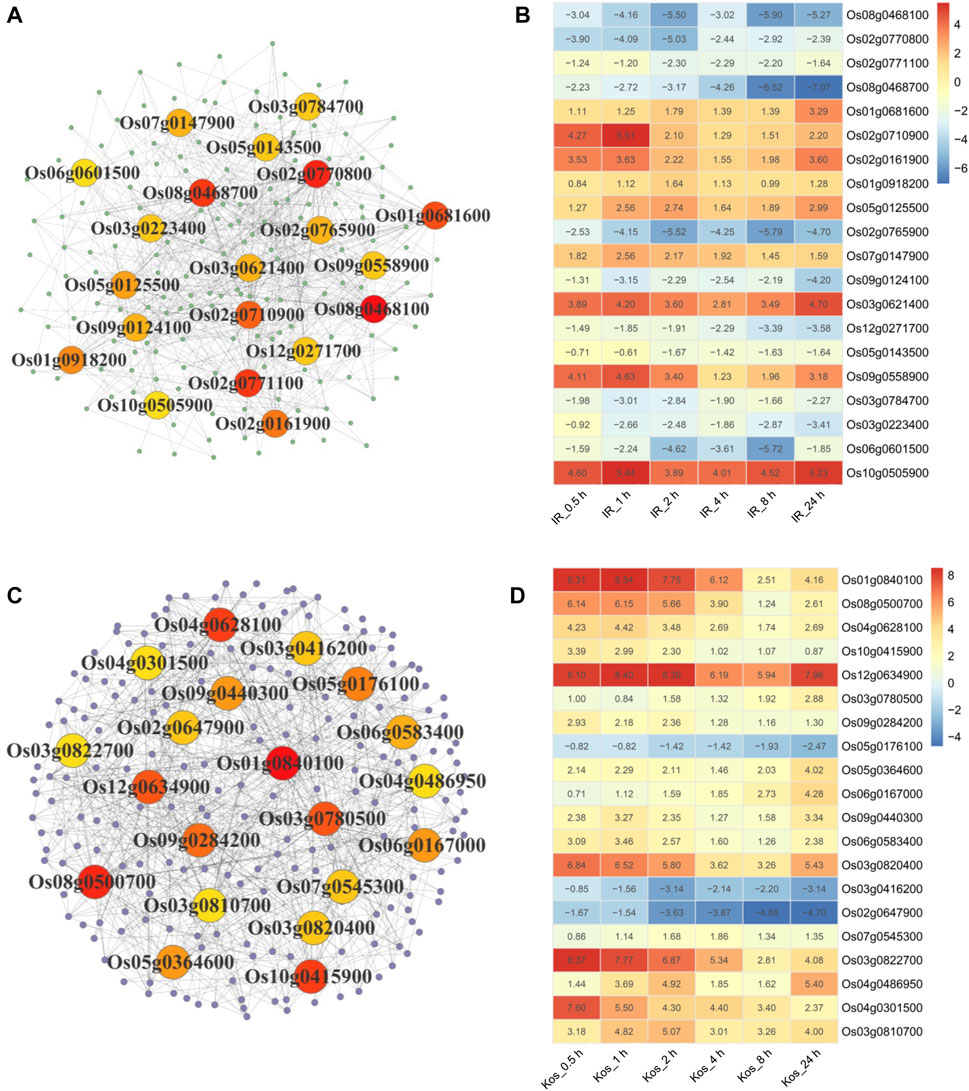
FIGURE 5. PPI analysis of IHR and KHR DEGs. Protein-protein interaction networks of IHR (A) and KHR (C) DEGs were constructed by STRING database. Each dot in the graph represents a gene, and the line indicates the relationship between the genes. The top 20 DEGs in the interaction network are calculated according to the degree, and the darker the color indicates the larger the degree unDir. The log2 Fold Change (log2 FC) of the top 20 DEGs in the IHR (B) and KHR (D) PPI network are represented in the heat map. The number indicates the log2 FC value of the DEGs at the corresponding time point.
Analysis of the DEGs of the two cultivars showed that there were 599 common DEGs, then the log2 FC value of these DEGs were shown in Supplementary Figure S3. After HS treatment, the CHR DEGs of the two cultivars showed similar expression patterns, implying that the response pattern of these genes to HS are the same in both cultivars. In addition, the number of upregulated genes were significantly higher than the number of downregulated genes in CHR DEGs.
As shown in Figure 6A, HSPs were the most abundant in the PPI network. Within the PPI network, it is worth noting that some key proteins may play key roles in the HS response, such as HSP81-2, CLPB1 (HSP100), OS11T0703900-01, OsJ_09939 (HSP70), OsJ_10337, HSP26.7, OsJ_09152, OS03T0271400-01, HSFA3, HSP23.2. In addition, some non-HSPs may also play important role in HS tolerance at seedlings, such as CIPK19, CBL1, APX1, a1, PP2A3, PDL1-3, HOX24, etc.
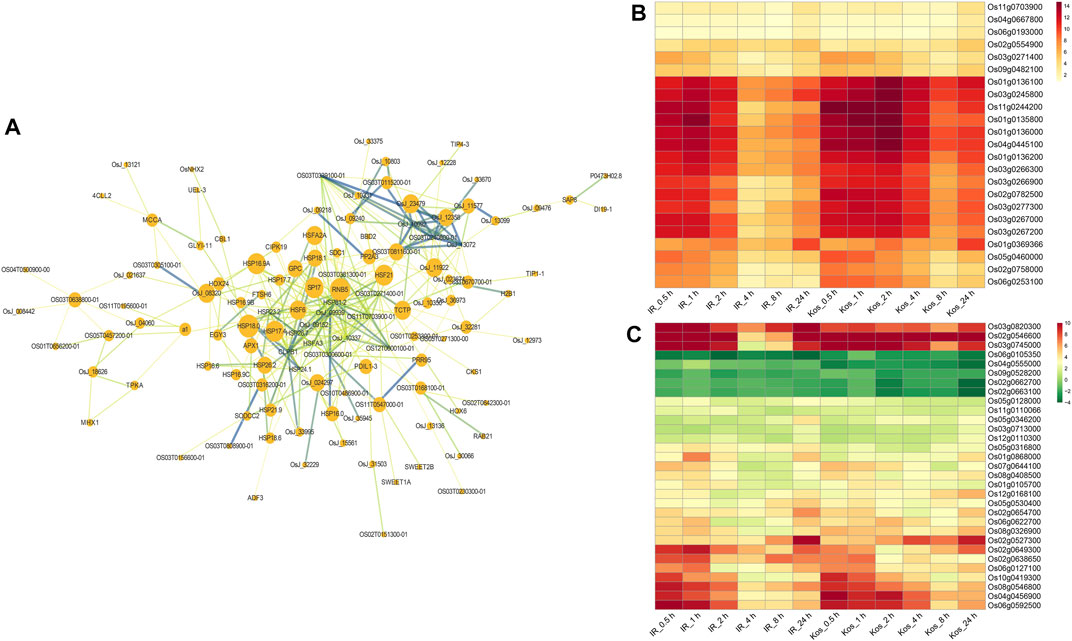
FIGURE 6. Analysis of CHR DEGs in two cultivars. (A) A PPI network for the CHR DEGs using the STRING database. Lines stand for relation of the target nodes. Color of lines stand for the distance of relationship between the nodes. Node size of nodes is proportional to the importance of protein. Heat map show the log2 FC values of genes in CHR DEGs belonging to the protein processing in endoplasmic reticulum (B) and DNA-binding transcription factor activity (C), respectively.
We performed GO and KEGG enrichment analysis of CHR DEGs. The results of GO enrichment analysis indicated that the DEGs of CHR were involved in heat acclimation, response to alcohol, regulation of programmed cell death, DNA-binding transcription factor activity, chaperone binding and Hsp90 protein binding, and so on (Supplementary Figure S4). Analysis of the KEGG pathway showed that the DEG of CHR were significantly involved in protein processing in endoplasmic reticulum, spliceosome, ubiquitin mediated proteolysis and RNA degradation (Supplementary Figure S5). We focused on the expression of DEGs in two pathways, protein processing in endoplasmic reticulum and DNA-binding transcription factor activity, which were significantly enriched. As shown in Figure 6B, there were 23 genes were enriched in the protein processing in endoplasmic reticulum pathway, and these included 18 HSP genes and 2 ubiquitin-conjugating enzyme genes, among others. Interestingly all genes that were enriched in the protein processing in endoplasmic reticulum pathway exhibit upregulation. A total of 31 DEGs were identified as belonging to the DNA-binding transcriptional activity pathway, of which 5 DEGs were downregulated (Figure 6C). Heat shock factors (Hsfs Os08g0546800, Os02g0527300, Os03g0745000, Os10g0419300), basic leucine zipper (bZIP) transcription factor (Os06g0622700, Os07g0644100), AP2/EREBP transcription factor gene (Os02g0546600, Os02g0654700, Os02g0638650, Os05g0346200, Os05g0316800, Os01g0868000, Os06g0127100, Os08g0408500), and TFIIIA-type zinc finger protein gene (Os03g0820300) were upregulated in both cultivars after HS treatment.
Temperature increased, due to global warming, has become one of the most important factors constraining rice yield and quality (Zhao et al., 2017). Therefore, the identification of more beneficial genes involved in HS tolerance in rice is necessary for adaptation to extremely high temperature. In this study, we used two different subspecies of rice, and analyzed the 2,044 cultivars-difference core genes as well as 599 co-expressed core DEGs. The results may facilitate the discovery and annotation of important genes to rice under HS and provided a comprehensive responsive expression profile to HS. Further studies on these differentially regulated genes will provided insights into the differential HS responses of different rice subspecies.
It is commonly recognized that domestication and selection process led cultivated rice to different subspecies, namely indica and japonica, which represents two partially isolated gene pools and major source of genetic diversity of the rice germplasm (Smýkal et al., 2018). Indica and japonica rice have amount of genotypic variation, which makes them behave differently in face of stress. Variation of nitrate-transporter gene (NRT 1.1B) has led to the divergence in nitrate use (Hu et al., 2015). bZIP73, OsAPX1, CTB4a and Ctb1 have a superimposed effect of dominant allelic variants that are important for adaptation to cold regions (Liu et al., 2018; Guo et al., 2020). Sequence variation in the OsHMA3 promoter contributes to differential grain Cd accumulation between the two-rice subspecies (Liu et al., 2020). Historically, different rice production areas favored different subspecies, and the different climatic conditions in each production areas also lead to different adaptations to temperature among different subspecies (Raza et al., 2019). Many studies have shown that seedlings of indica and japonica rice have genotypic differences in response to HS, with indica rice having a higher temperature threshold than japonica rice, showing a basal or inherent thermal tolerance to HS (Khan et al., 2019; Dar et al., 2021). Our results show that tropical indica (IR64) exhibits greater HS tolerance relative to temperate japonica (Koshihikari). In addition, the number of consistently expressed DEGs in Kos was greater than the number of consistently expressed DEGs in IR, which suggests a greater effect of HS on Kos gene expression.
In the present study, the results of GO analysis of IHR and KHR DEGs exhibited two distinct HS response pathways. IHR DEGs were significantly enriched in chloroplast-related pathways, such as chloroplast envelope, chloroplast inner membrane, plastid inner membrane, photomorphogenesis, response to UV and red or far-red light signaling pathway, etc. Numerous studies have concluded that chloroplasts are essential for activating cellular HS signals and have also been proposed as sensors of HS. Processes associated with photosynthesis are susceptible to HS, including chlorophyll biosynthesis, CO2 assimilation, electron transport, photophosphorylation, and chloroplast membrane fluidity, which are the main metabolic processes that optimize carbon fixation and growth (Wahid et al., 2007; Allakhverdiev et al., 2008; Kmiecik et al., 2016; Sun and Guo, 2016; Hu et al., 2020). In addition, several confirmations have been made that retrograde signaling from chloroplasts can regulate the response of the nucleus to HS (Sun and Guo, 2016). To verify this opinion, we measured the contents of chlorophyll a and chlorophyll b in the leaves of the two rice cultivars after 24 h of HS treatment, and the results showed that the contents of chlorophyll a and chlorophyll b were significantly lower in Kos after HS treatment, while there was no change in IR (Supplementary Figure S7). Unlike IHR, HS mainly affects DEGs of KHR in the endoplasmic reticulum (ER) and mitochondria-related pathway. Both ER and mitochondria are associated with the HS tolerance in plants. The accumulation of unfolded or misfolded proteins due to HS can activate ER stress, which in turn affects redox metabolism and ROS production (Cao et al., 2022). Mitochondria-mediated lipid homeostasis is a buffer for maintaining flower development in rice under HS (Zhang et al., 2016). In conclusion, the unique HS response pathways of IHR and KHR may confer different HS tolerance in IR and Kos.
Notably, among the core genes of the PPI network constructed by IHR DEGs, we identified three nitrate reductase (NR) genes (OsNIA1, OsNR2, OsNIA2), one nitrite reductase (NiR) gene (OsNIR2) and one glutamine synthetase gene (OsGSr). Meanwhile, the pathways in which IRH DEGs are significantly enriched in GO and KEGG also contain the nitric oxide (NO) biosynthetic pathway and the nitrogen metabolism pathway, respectively. Furthermore, our data showed that exogenous application of 1 mmol/L nitrate reductase inhibitor (sodium tungstate Na2WO4) significantly reduced the heat stress tolerance of IR seedlings after 46 h of high temperature treatment at 45°C (Supplementary Figure S6). Sodium tungstate can inhibit NR activity in plants, which in turn reduces the production of NO (Freschi et al., 2010; Kolbert et al., 2010; Kaya et al., 2021). NO has been shown to be a key signaling molecule during plant development and different stress responses, NR is the most important source of NO, and controlling the balance of NO signaling molecules (Chamizo-Ampudia et al., 2017). The NR-deficient double-knockout mutants has reduced NO levels and nitrite concentration is one of the rates limiting factors (Planchet et al., 2005). NO synthesis in guard cells mediated by NIR1 and NIR2 in Arabidopsis mediates ABA-induced stomatal closure (Desikan et al., 2002). In addition, tobacco plants expressing the antisense NiR gene showed a significant accumulation of nitrite and a significant 100-fold increase in NO emission rate compared to the wild type (Morot-Gaudry-Talarmain et al., 2002). Salt stress induces the expression of the nitrate-responsive transcription factor OsNLP2, which directly or indirectly activates the expression of OsNR1 and the ABA catabolism gene OsABA8ox1, leading to increased salt tolerance during rice seed germination (Yi et al., 2022). NO is actively involved in plant HS response by mitigating heat-induced oxidative stress, maintaining membrane integrity, and improving photosynthetic productivity (Parankusam et al., 2017). Xuan et al. (2010) showed that the nitrate reductase-deficient double mutant nia1nia2 had lower survival ratio and NO levels under HS treatment compared with the wild type, which could be reversible by treatment with exogenous NO. NO can act synergistically with hydrogen sulfide at high temperatures to reduce H2O2-induced oxidative stress through the ascorbate-glutathione cycle and enhance HS tolerance in wheat (Iqbal et al., 2021). Overall, these results strongly suggest that NO biosynthetic pathway-related genes, especially nitrate reductase genes, may play a very important role in the IR64 response to HS.
Disruption of protein homeostasis is an important damage caused by HS to plants, which will lead to cell death and cytotoxicity. Plants threatened by HS will result in unfolding, misfolding, or denaturation of proteins, and plants will produce enough HSPs to counteract this. As important molecular chaperones, HSPs stabilize and degrade damaged proteins and promote HS-induced refolding of aggregated proteins (Müller and Rieu, 2016). Purified OsHsp17.4 and OsHsp17.9A proteins have chaperone activity that prevents their substrates from forming large aggregates, and heterologously expressed transgenic tobacco plants have better HS tolerance (Sarkar et al., 2020). In addition to HSPs, Hsfs are also important components of plants in response to HS and are responsible for triggering transcriptional cascades and activating downstream genes such as ROS scavenging enzymes, metabolic enzymes and HSPs. Rice OsHSFA2dI participated in the unfolded protein response under HS by alternative splicing (Cheng et al., 2015). HsfA6a mediates transcriptional regulation of the HSP101 gene in rice (Singh et al., 2021). We identified a large number of genes encoding HSPs and Hsfs among the IHR KHR and CHR DEGs, including hsp82A, HSP70, HSP26.7, OsHsfB2b, hsp82B, OsHSP24.1, OsHSP17.7, HSA32, HSP100, OsHsfA2a and some uncloned genes containing heat shock protein typical domains, many of which are core genes in the PPI network. These genes have sustained expression for plants in response to HS and may be important candidate genes for HS tolerant breeding. Lin et al. (2014) showed that HSP100 and HSA32 interact and a positive feedback loop formed at the post-transcriptional level could extend the effect of heat acclimation at the seedling stage of rice. OsHsfB2b expression was strongly induced by heat treatment, but was barely affected by cold stress (Xiang et al., 2013). Overexpression of the small heat shock protein, OsHSP17.7, confers greater HS tolerance in rice (Murakami et al., 2004). Therefore, HSP and Hsfs play a central role in plants responding to HS, and may be good candidate genes for HS breeding in rice.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA917024.
Conceptualization of research (WT and YX), design of the experiment (YX, GZ, HD, and XuL), execution of experiments and data collection (YiW, WC, YD, and XiL), analysis of data and interpretation (YiW, FW, and GC), and preparation of the manuscript (YX, YiW, and YuW). All authors contributed to manuscript revision, read, and approved the submitted version.
This research was funded by the National Natural Science Foundation of China (No. 31901528, 31871599, and 32172078), the Major Project of Agricultural Biological Breeding (2022ZD0400402), and the Science and Technology Innovation Program of Hunan Province (No. 2022RC1147, 2021NK1001, 2022JJ30285, and 2021JC0007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1135577/full#supplementary-material
Allakhverdiev, S. I., Kreslavski, V. D., Klimov, V. V., Los, D. A., Carpentier, R., and Mohanty, P. (2008). Heat stress: An overview of molecular responses in photosynthesis. Photosynth. Res. 98 (1), 541–550. doi:10.1007/s11120-008-9331-0
Cao, J., Wang, C., Hao, N., Fujiwara, T., and Wu, T. (2022). Endoplasmic reticulum stress and reactive oxygen species in plants. Antioxidants 11 (7), 1240. doi:10.3390/antiox11071240
Chamizo-Ampudia, A., Sanz-Luque, E., Llamas, A., Galvan, A., and Fernandez, E. (2017). Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 22 (2), 163–174. doi:10.1016/j.tplants.2016.12.001
Chen, M., Fu, Y., Mou, Q., An, J., Zhu, X., Ahmed, T., et al. (2021). Spermidine induces expression of stress associated proteins (SAPs) genes and protects rice seed from heat stress-induced damage during grain-filling. Antioxidants 10 (10), 1544. doi:10.3390/antiox10101544
Cheng, Q., Zhou, Y., Liu, Z., Zhang, L., Song, G., Guo, Z., et al. (2015). An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol. 17 (2), 419–429. doi:10.1111/plb.12267
Dai, X., Wang, Y., Chen, Y., Li, H., Xu, S., Yang, T., et al. (2020). Overexpression of NtDOG1L-T improves heat stress tolerance by modulation of antioxidant capability and defense-heat-and ABA-related gene expression in tobacco. Front. Plant Sci. 11, 568489. doi:10.3389/fpls.2020.568489
Dar, M. H., Bano, D. A., Waza, S. A., Zaidi, N. W., Majid, A., Shikari, A. B., et al. (2021). Abiotic stress tolerance-progress and pathways of sustainable rice production. Sustainability 13 (4), 2078. doi:10.3390/su13042078
Desikan, R., Griffiths, R., Hancock, J., and Neill, S. (2002). A new role for an old enzyme: Nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 99 (25), 16314–16318. doi:10.1073/pnas.252461999
Ebitani, T., Takeuchi, Y., Nonoue, Y., Yamamoto, T., Takeuchi, K., and Yano, M. (2005). Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari. Breed. Sci. 55 (1), 65–73. doi:10.1270/jsbbs.55.65
Freschi, L., Rodrigues, M. A., Domingues, D. S., Purgatto, E., Van Sluys, M. A., Magalhaes, J. R., et al. (2010). Nitric oxide mediates the hormonal control of crassulacean acid metabolism expression in young pineapple plants. Plant physiol. 152 (4), 1971–1985. doi:10.1104/pp.109.151613
Fukao, T., Barrera-Figueroa, B. E., Juntawong, P., and Peña-Castro, J. M. (2019). Submergence and waterlogging stress in plants: A review highlighting research opportunities and understudied aspects. Front. Plant Sci. 10, 340. doi:10.3389/fpls.2019.00340
Guo, H., Zeng, Y., Li, J., Ma, X., Zhang, Z., Lou, Q., et al. (2020). Differentiation, evolution and utilization of natural alleles for cold adaptability at the reproductive stage in rice. Plant Biotechnol. J. 18 (12), 2491–2503. doi:10.1111/pbi.13424
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1. 1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47 (7), 834–838. doi:10.1038/ng.3337
Hu, H., Wang, L., Wang, Q., Jiao, L., Hua, W., Zhou, Q., et al. (2014). Photosynthesis, chlorophyll fluorescence characteristics, and chlorophyll content of soybean seedlings under combined stress of bisphenol A and cadmium. Environ. Toxicol. Chem. 33 (11), 2455–2462. doi:10.1002/etc.2720
Hu, S., Ding, Y., and Zhu, C. (2020). Sensitivity and responses of chloroplasts to heat stress in plants. Front. Plant Sci. 11, 375. doi:10.3389/fpls.2020.00375
Iqbal, N., Umar, S., Khan, N. A., and Corpas, F. J. (2021). Nitric oxide and hydrogen sulfide coordinately reduce glucose sensitivity and decrease oxidative stress via ascorbate-glutathione cycle in heat-stressed wheat (Triticum aestivum L.) plants. Antioxidants 10 (1), 108. doi:10.3390/antiox10010108
Jaagus, J. (2006). Climatic changes in Estonia during the second half of the 20th century in relationship with changes in large-scale atmospheric circulation. Theor. Appl. Climatol. 83 (1), 77–88. doi:10.1007/s00704-005-0161-0
Kan, Y., Mu, X. R., Zhang, H., Gao, J., Shan, J. X., Ye, W. W., et al. (2022). TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nat. Plants 8 (1), 53–67. doi:10.1038/s41477-021-01039-0
Kaya, C., Polat, T., Ashraf, M., Kaushik, P., Alyemeni, M. N., and Ahmad, P. (2021). Endogenous nitric oxide and its potential sources regulate glutathione-induced cadmium stress tolerance in maize plants. Plant Physiology Biochem. 167, 723–737. doi:10.1016/j.plaphy.2021.08.030
Khan, S., Anwar, S., Ashraf, M. Y., Khaliq, B., Sun, M., Hussain, S., et al. (2019). Mechanisms and adaptation strategies to improve heat tolerance in rice. A review. A Rev. Plants 8 (11), 508. doi:10.3390/plants8110508
Kilasi, N. L., Singh, J., Vallejos, C. E., Ye, C., Jagadish, S. K., Kusolwa, P., et al. (2018). Heat stress tolerance in rice (Oryza sativa L.): Identification of quantitative trait loci and candidate genes for seedling growth under heat stress. Front. Plant Sci. 9, 1578. doi:10.3389/fpls.2018.01578
Kmiecik, P., Leonardelli, M., and Teige, M. (2016). Novel connections in plant organellar signalling link different stress responses and signalling pathways. J. Exp. Bot. 67 (13), 3793–3807. doi:10.1093/jxb/erw136
Kolbert, Z., Ortega, L., and Erdei, L. (2010). Involvement of nitrate reductase (NR) in osmotic stress-induced NO generation of Arabidopsis thaliana L. roots. J. Plant Physiology 167 (1), 77–80. doi:10.1016/j.jplph.2009.08.013
Li, X. M., Chao, D. Y., Wu, Y., Huang, X., Chen, K., Cui, L. G., et al. (2015). Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 47 (7), 827–833. doi:10.1038/ng.3305
Lim, S. D., Cho, H. Y., Park, Y. C., Ham, D. J., Lee, J. K., and Jang, C. S. (2013). The rice RING finger E3 ligase, OsHCI1, drives nuclear export of multiple substrate proteins and its heterogeneous overexpression enhances acquired thermotolerance. J. Exp. Bot. 64 (10), 2899–2914. doi:10.1093/jxb/ert143
Lin, M. Y., Chai, K. H., Ko, S. S., Kuang, L. Y., Lur, H. S., and Charng, Y. Y. (2014). A positive feedback loop between HEAT SHOCK PROTEIN101 and HEAT STRESS-ASSOCIATED 32-KD PROTEIN modulates long-term acquired thermotolerance illustrating diverse heat stress responses in rice varieties. Plant Physiol. 164 (4), 2045–2053. doi:10.1104/pp.113.229609
Liu, C. L., Gao, Z. Y., Shang, L. G., Yang, C. H., Ruan, B. P., Zeng, D. L., et al. (2020). Natural variation in the promoter of OsHMA3 contributes to differential grain cadmium accumulation between Indica and Japonica rice. J. Integr. Plant Biol. 62 (3), 314–329. doi:10.1111/jipb.12794
Liu, C., Ou, S., Mao, B., Tang, J., Wang, W., Wang, H., et al. (2018). Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat. Commun. 9 (1), 3302–3312. doi:10.1038/s41467-018-05753-w
Liu, J., Meng, Q., Xiang, H., Shi, F., Ma, L., Li, Y., et al. (2021). Genome-wide analysis of Dof transcription factors and their response to cold stress in rice (Oryza sativa L.). BMC Genomics 22 (1), 800–812. doi:10.1186/s12864-021-08104-0
Maeda, H., Murata, K., Sakuma, N., Takei, S., Yamazaki, A., Karim, M. R., et al. (2019). A rice gene that confers broad-spectrum resistance to β-triketone herbicides. Science 365 (6451), 393–396. doi:10.1126/science.aax0379
Morot-Gaudry-Talarmain, Y., Rockel, P., Moureaux, T., Quillere, I., Leydecker, M., Kaiser, W., et al. (2002). Nitrite accumulation and nitric oxide emission in relation to cellular signaling in nitrite reductase antisense tobacco. Planta 215 (5), 708–715. doi:10.1007/s00425-002-0816-3
Müller, F., and Rieu, I. (2016). Acclimation to high temperature during pollen development. Plant Reprod. 29 (1), 107–118. doi:10.1007/s00497-016-0282-x
Murakami, T., Matsuba, S., Funatsuki, H., Kawaguchi, K., Saruyama, H., Tanida, M., et al. (2004). Over-expression of a small heat shock protein, sHSP17.7, confers both heat tolerance and UV-B resistance to rice plants. Mol. Breed. 13 (2), 165–175. doi:10.1023/b:molb.0000018764.30795.c1
Nonoue, Y., Hori, K., Ono, N., Shibaya, T., Ogiso-Tanaka, E., Mizobuchi, R., et al. (2019). Detection of heading date QTLs in advanced-backcross populations of an elite indica rice cultivar, IR64. Breed. Sci. 69 (2), 352–358. doi:10.1270/jsbbs.18172
Parankusam, S., Adimulam, S. S., Bhatnagar-Mathur, P., and Sharma, K. K. (2017). Nitric oxide (NO) in plant heat stress tolerance: Current knowledge and perspectives. Front. Plant Sci. 8, 1582. doi:10.3389/fpls.2017.01582
Penke, B., Bogár, F., Crul, T., Sántha, M., Tóth, M. E., and Vígh, L. (2018). Heat shock proteins and autophagy pathways in neuroprotection: From molecular bases to pharmacological interventions. Int. J. Mol. Sci. 19 (1), 325. doi:10.3390/ijms19010325
Planchet, E., Jagadis Gupta, K., Sonoda, M., and Kaiser, W. M. (2005). Nitric oxide emission from tobacco leaves and cell suspensions: Rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J. 41 (5), 732–743. doi:10.1111/j.1365-313X.2005.02335.x
Rana, R. M., Dong, S., Tang, H., Ahmad, F., and Zhang, H. (2012). Functional analysis of OsHSBP1 and OsHSBP2 revealed their involvement in the heat shock response in rice (Oryza sativa L.). J. Exp. Bot. 63 (16), 6003–6016. doi:10.1093/jxb/ers245
Raza, A., Razzaq, A., Mehmood, S. S., Zou, X., Zhang, X., Lv, Y., et al. (2019). Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 8 (2), 34. doi:10.3390/plants8020034
Sarkar, N. K., Kotak, S., Agarwal, M., Kim, Y. K., and Grover, A. (2020). Silencing of class I small heat shock proteins affects seed-related attributes and thermotolerance in rice seedlings. Planta 251 (1), 26–16. doi:10.1007/s00425-019-03318-9
Shahzad, A., Ullah, S., Dar, A. A., Sardar, M. F., Mehmood, T., Tufail, M. A., et al. (2021). Nexus on climate change: Agriculture and possible solution to cope future climate change stresses. Environ. Sci. Pollut. Res. 28 (12), 14211–14232. doi:10.1007/s11356-021-12649-8
Singh, G., Sarkar, N. K., and Grover, A. (2021). Hsp70, sHsps and ubiquitin proteins modulate HsfA6a-mediated Hsp101 transcript expression in rice (Oryza sativa L.). Physiol. Plant. 173 (4), 2055–2067. doi:10.1111/ppl.13552
Smýkal, P., Nelson, M. N., Berger, J. D., and Von Wettberg, E. J. (2018). The impact of genetic changes during crop domestication. Agronomy 8 (7), 119. doi:10.3390/agronomy8070119
Sun, A. Z., and Guo, F. Q. (2016). Chloroplast retrograde regulation of heat stress responses in plants. Front. Plant Sci. 7, 398. doi:10.3389/fpls.2016.00398
Tripathi, A., Tripathi, D. K., Chauhan, D. K., Kumar, N., and Singh, G. S. (2016). Paradigms of climate change impacts on some major food sources of the world: A review on current knowledge and future prospects. Agric. Ecosyst. Environ. 216, 356–373. doi:10.1016/j.agee.2015.09.034
Vitoriano, C. B., and Calixto, C. P. G. (2021). Reading between the lines: RNA-seq data mining reveals the alternative message of the rice leaf transcriptome in response to heat stress. Plants 10 (8), 1647. doi:10.3390/plants10081647
Wahid, A., Gelani, S., Ashraf, M., and Foolad, M. R. (2007). Heat tolerance in plants: An overview. Environ. Exp. Bot. 61 (3), 199–223. doi:10.1016/j.envexpbot.2007.05.011
Wang, D., Qin, B., Li, X., Tang, D., Zhang, Y. E., Cheng, Z., et al. (2016). Nucleolar DEAD-box RNA helicase TOGR1 regulates thermotolerant growth as a pre-rRNA chaperone in rice. PLoS Genet. 12 (2), e1005844. doi:10.1371/journal.pgen.1005844
Wang, H., Sun, R., Cao, Y., Pei, W., Sun, Y., Zhou, H., et al. (2015). OsSIZ1, a SUMO E3 ligase gene, is involved in the regulation of the responses to phosphate and nitrogen in rice. Plant Cell Physiology 56 (12), 2381–2395. doi:10.1093/pcp/pcv162
Wang, X., Xu, C., Cai, X., Wang, Q., and Dai, S. (2017). Heat-responsive photosynthetic and signaling pathways in plants: Insight from proteomics. Int. J. Mol. Sci. 18 (10), 2191. doi:10.3390/ijms18102191
Wei, H., Liu, J., Wang, Y., Huang, N., Zhang, X., Wang, L., et al. (2013). A dominant major locus in chromosome 9 of rice (Oryza sativa L.) confers tolerance to 48°C high temperature at seedling stage. J. Hered. 104 (2), 287–294. doi:10.1093/jhered/ess103
Xiang, J., Ran, J., Zou, J., Zhou, X., Liu, A., Zhang, X., et al. (2013). Heat shock factor OsHsfB2b negatively regulates drought and salt tolerance in rice. Plant Cell Rep. 32 (11), 1795–1806. doi:10.1007/s00299-013-1492-4
Xu, Y., Chu, C., and Yao, S. (2021). The impact of high-temperature stress on rice: Challenges and solutions. Crop J. 9 (5), 963–976. doi:10.1016/j.cj.2021.02.011
Xu, Y., Zhang, L., Ou, S., Wang, R., Wang, Y., Chu, C., et al. (2020). Natural variations of SLG1 confer high-temperature tolerance in indica rice. Nat. Commun. 11 (1), 5441. doi:10.1038/s41467-020-19320-9
Xuan, Y., Zhou, S., Wang, L., Cheng, Y., and Zhao, L. (2010). Nitric oxide functions as a signal and acts upstream of AtCaM3 in thermotolerance in Arabidopsis seedlings. Plant Physiol. 153 (4), 1895–1906. doi:10.1104/pp.110.160424
Yang, B., Tang, J., Yu, Z., Khare, T., Srivastav, A., Datir, S., et al. (2019). Light stress responses and prospects for engineering light stress tolerance in crop plants. J. Plant Growth Regul. 38 (4), 1489–1506. doi:10.1007/s00344-019-09951-8
Yang, J., Miao, W., and Chen, J. (2021). Roles of jasmonates and brassinosteroids in rice responses to high temperature stress-A review. Crop J. 9 (5), 977–985. doi:10.1016/j.cj.2021.02.007
Yi, Y., Peng, Y., Song, T., Lu, S., Teng, Z., Zheng, Q., et al. (2022). NLP2-NR module associated NO is involved in regulating seed germination in rice under salt stress. Plants 11 (6), 795. doi:10.3390/plants11060795
Zhang, B., Wu, S., Zhang, Y. E., Xu, T., Guo, F., Tang, H., et al. (2016). A high temperature-dependent mitochondrial lipase EXTRA GLUME1 promotes floral phenotypic robustness against temperature fluctuation in rice (Oryza sativa L.). PLoS Genet. 12 (7), e1006152. doi:10.1371/journal.pgen.1006152
Zhao, C., Liu, B., Piao, S., Wang, X., Lobell, D. B., Huang, Y., et al. (2017). Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. U. S. A. 114 (35), 9326–9331. doi:10.1073/pnas.1701762114
Zhou, H., Wang, X., Huo, C., Wang, H., An, Z., Sun, D., et al. (2019). A quantitative proteomics study of early heat-regulated proteins by two-dimensional difference gel electrophoresis identified OsUBP21 as a negative regulator of heat stress responses in rice. Proteomics 19 (20), 1900153. doi:10.1002/pmic.201900153
Keywords: rice (Oryza sativa L.), indica, japonica, heat stress, RNA-seq, DEGs
Citation: Wang Y, Wang Y, Chen W, Dong Y, Zhang G, Deng H, Liu X, Lu X, Wang F, Chen G, Xiao Y and Tang W (2023) Comparative transcriptome analysis of the mechanism difference in heat stress response between indica rice cultivar “IR64” and japonica cultivar “Koshihikari” at the seedling stage. Front. Genet. 14:1135577. doi: 10.3389/fgene.2023.1135577
Received: 01 January 2023; Accepted: 12 April 2023;
Published: 21 April 2023.
Edited by:
Ahmed Sallam, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), GermanyReviewed by:
Hua Wang, Zhejiang Academy of Agricultural Sciences, ChinaCopyright © 2023 Wang, Wang, Chen, Dong, Zhang, Deng, Liu, Lu, Wang, Chen, Xiao and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunhua Xiao, eHloa2VteUAxNjMuY29t; Wenbang Tang, dGFuZ3dlbmJhbmdAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.