- 1Anhui University of Chinese Medicine, Hefei, China
- 2Key Laboratory of Xin’An Medicine, Ministry of Education, Hefei, China
- 3The First Affiliated Hospital of Anhui University of Chinese Medicine, Hefei, China
- 4The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 5Key Laboratory of Anhui Provincial Department of Education, Hefei, China
Background: Chronic obstructive pulmonary disease (COPD) affects approximately 400 million people worldwide and is associated with high mortality and morbidity. The effect of EPHX1 and GSTP1 gene polymorphisms on COPD risk has not been fully characterized.
Objective: To investigate the association of EPHX1 and GSTP1 gene polymorphisms with COPD risk.
Methods: A systematic search was conducted on 9 databases to identify studies published in English and Chinese. The analysis was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses reporting guidelines (PRISMA). The pooled OR and 95% CI were calculated to evaluate the association of EPHX1 and GSTP1 gene polymorphisms with COPD risk. The I2 test, Q test, Egger’s test, and Begg’s test were conducted to determine the level of heterogeneity and publication bias of the included studies.
Results: In total, 857 articles were retrieved, among which 59 met the inclusion criteria. The EPHX1 rs1051740 polymorphism (homozygote, heterozygote, dominant, recessives, and allele model) was significantly associated with high risk of COPD risk. Subgroup analysis revealed that the EPHX1 rs1051740 polymorphism was significantly associated with COPD risk among Asians (homozygote, heterozygote, dominant, and allele model) and Caucasians (homozygote, dominant, recessives, and allele model). The EPHX1 rs2234922 polymorphism (heterozygote, dominant, and allele model) was significantly associated with a low risk of COPD. Subgroup analysis showed that the EPHX1 rs2234922 polymorphism (heterozygote, dominant, and allele model) was significantly associated with COPD risk among Asians. The GSTP1 rs1695 polymorphism (homozygote and recessives model) was significantly associated with COPD risk. Subgroup analysis showed that the GSTP1 rs1695 polymorphism (homozygote and recessives model) was significantly associated with COPD risk among Caucasians. The GSTP1 rs1138272 polymorphism (heterozygote and dominant model) was significantly associated with COPD risk. Subgroup analysis suggested that the GSTP1 rs1138272 polymorphism (heterozygote, dominant, and allele model) was significantly associated with COPD risk among Caucasians.
Conclusion: The C allele in EPHX1 rs1051740 among Asians and the CC genotype among Caucasians may be risk factors for COPD. However, the GA genotype in EPHX1 rs2234922 may be a protective factor against COPD in Asians. The GG genotype in GSTP1 rs1695 and the TC genotype in GSTP1 rs1138272 may be risk factors for COPD, especially among Caucasians.
1 Introduction
Chronic obstructive pulmonary disease (COPD) is a common disease that is characterized by persistent airflow limitation and the associated respiratory symptoms. Oxidative stress and chronic inflammation are important components of the mechanism contributing to COPD (Cheng et al., 2004; Global Initiative for Chronic Obstruc, 2021). COPD is the leading cause of lung disease-associated morbidity and mortality, and its incidence has been increasing globally (Singh et al., 2019). It has been predicted that by 2030, COPD will be the third leading cause of death worldwide, imposing a heavy socioeconomic burden (Nikolaou et al., 2020). COPD onset is closely correlated with airway and lung inflammation caused by harmful particles and smoke (Lareau et al., 2019; Global Initiative for Chronic Obstruc, 2021). However, only 10%–20% of chronic smokers exhibit COPD-associated severe lung dysfunction and the risk of airflow limitation varies greatly among smokers (Bascom, 1991; Pillai et al., 2009). Furthermore, patients with early-onset COPD exhibit familial aggregation (Silverman et al., 1998), indicating that COPD is a complex disease that is caused by interactions between genetic and environmental factors 9). Genome-wide association studies (GWAS) have reported many COPD-associated susceptibility genes (Sakornsakolpat et al., 2019; Shrine et al., 2019). Several studies have also shown that gene polymorphisms play a key role in COPD pathogenesis (Xiao et al., 2004; An et al., 2016a; Cho et al., 2022). Thus, the key genetic variations associated with COPD susceptibility need to be identified to improve COPD prevention and treatment.
Cigarette smoke contains many toxic constituents which stimulate the release of vast amounts of reactive oxygen species (ROS) and reactive nitrogen species (RNS) by airway epithelial cells, granulocytes, and macrophages, leading to oxidative stress, oxidative inactivation of antiproteases, alveolar epithelial damage, increased neutrophils in pulmonary microvessels, and enhanced proinflammatory gene expression. Some of the genes involved in the metabolism of toxic substances found in cigarette smoke are thought to participate in COPD pathogenesis (Joos et al., 2002; Silverman, 2020). For instance, glutathione S-transferase P1 (GSTP1) and microsomal epoxide hydrolase (EPHX1) are typical detoxification enzyme genes known to be highly expressed in the lungs and are closely associated with oxidative stress and inflammatory responses in COPD (Tomaki et al., 2007).
EPHX1, which is involved in the metabolism and detoxification of exogenous chemicals, plays a key role in general oxidative defense in the lungs (Sandford and Silverman, 2002). The EPHX1 gene (≈35.48 kb long) is located on chromosome 1q42.1 and contains 9 exons and 8 introns (Akparova et al., 2017). Mutations of exon 3 Tyr113His (rs1051740) and exon 4 His139Arg (rs2234922) are the most common polymorphisms that influence the EPHX1 enzyme activities (Kiyohara et al., 2006). Genetic correlation case-control studies have shown that EPHX1 rs1051740 genetic variations increased the risk of COPD (Hersh et al., 2005; Hersh et al., 2006; Hersh et al., 2007), whereas EPHX1 rs2234922 variation decrease the risk of COPD (Smith and Harrison, 1997). However, these observations are controversial because some studies did not find any correlations between these polymorphisms and COPD risk (Brøgger et al., 2006; Chappell et al., 2008). GSTP1 (≈3 kb long), a member of the GST superfamily, is located on chromosome 11q13 and contains 6 introns and 7 exons. Compared to other GSTs, it is highly expressed in respiratory tissues including the alveoli, alveolar macrophages as well as bronchioles (Cantlay et al., 1994). GSTP1 catalyzes various electrophiles and glutathione, and serves to eliminate the products in tobacco smoke that cause toxicity associated with electrophiles and oxidative stress (Rodriguez et al., 2005; Du et al., 2019). Exon 5 Ile105Val (rs1695) and exon 6 Ala114Val (rs1138272) are the main GSTP1 polymorphisms (Cheng et al., 2004). Although studies have investigated the association of EPHX1 and GSTP1 gene polymorphisms with the risk of COPD in different ethnic groups, findings from such studies have been inconsistent which may be attributed to the small sample sizes in the studies. Here, we conducted a systematic review and meta-analysis to determine the association of EPHX1 (rs1051740 and rs2234922) and GSTP1 (rs1695 and rs1138272) polymorphisms with the risk of COPD risk, with the aim of providing evidence-based information on COPD pathogenesis which can be used to develop potential strategies for its diagnosis, prevention and treatment. The analysis was conducted in line with the PRISMA 2020 (Supplementary Table S1).
2 Materials and methods
2.1 Search strategy
A search was conducted on the PubMed, Embase, Web of Science, Cochrane Library, SCOPUS, CENTRAL, CINAHL, CNKI, and WANFANG DATA to identify relevant studies published up to 31 September 2022. Search terms included “COPD”, “gene”, “gene variation”, “single nucleotide polymorphism”, “EPHX1”, and “GSTP1”. Detailed retrieval strategies are provided in Supplementary Table S2. References in the retrieved literature were also reviewed.
2.2 Inclusion and exclusion criteria
The inclusion criteria for studies were: ①Explored the association of EPHX1 rs1051740, rs2234922, and GSTP1 rs1695, rs1138272 polymorphisms with the risk of COPD; ②case-control studies; ③ Reported specific genotype and allele counts or significant allele frequency (MAF) between groups; and ④ Involved human subjects.
The exclusion criteria for studies were: ① Were repeat published studies; ②Case reports, comments, or expert opinions; ③ Included other genetic polymorphisms; ④ Involved a non-healthy (with other diseases, such as lung cancer) control group; and ⑤ Lacked data that could be extracted from text, tables, or charts, or that could be obtained from the authors upon request.
2.3 Data extraction and literature quality evaluation
Two researchers (QY and WH) independently searched for the articles, assessed the inclusion/exclusion parameters, and conducted data extraction. Any discrepancies were resolved through discussions between two reviewers (JT and ZL). The extracted data included the first author’s name, year of publication, sample size, ethnicity, genotype, genotyping method, genotype count, allele count, and whether it met the Hardy Weinberger equilibrium (HWE). Using the Newcastle-Ottawa Scale (NOS) (Stang, 2010), the quality of included studies was evaluated based on the following criteria: selection of research subjects, comparability between groups, and outcome measurements. Studies with NOS scores ≥6 were considered high-quality studies (Mirzakhani et al., 2020).
2.4 Statistical analysis
The association of EPHX1 (rs1051740 and rs2234922) and GSTP1 (rs1695 and rs1138272) polymorphisms with the risk of COPD was analyzed using homozygote, heterozygote, dominant, recessive, and allele models. Higgin’s I2 and Cochran’s Q tests were used to evaluate heterogeneity between studies. Where heterogeneity was significant (I2 >50%, PQ<0.10), the random-effects model (DerSimonian–Lloyd method) was used, and in contrast (I2 ≤ 50%, PQ≥0.10), the fixed-effects model (Mantel‒Haenszel method) was used. The pooled odds ratio (OR) and 95% confidence interval (95% CI) were used as effect measurement indicators for each result. Furthermore, we conducted subgroup analyses according to different ethnic information. Egger’s and Begg’s tests were used to assess the publication bias, with p > 0.05 indicating no significant publication bias. If publication bias was found, the trim-and-fill method was used to assess the stability of the pooled results, and a funnel plot was drawn. A non-significant change in p values indicated that publication bias had little influence on the results. Sensitivity analysis was performed by excluding the studies one by one to determine the impact of each study on the total effects value and to assess the stability of results. All statistical analyses were done using Stata 17.0 (Stata Corp, College Station, TX, United States).
3 Results
3.1 Document screening process and results
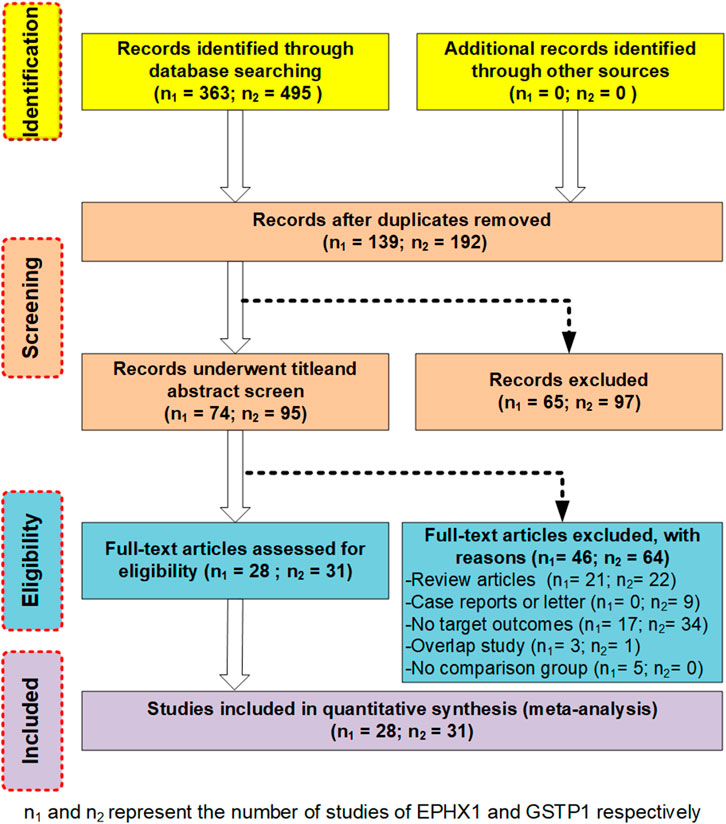
We employed an unrestricted literature search and a repeated search review procedure. Based on our inclusion and exclusion criteria, 59 articles containing genetic data were selected for analysis. Among them, 28 investigated EPHX1 rs1051740 and involved 5007 cases and 5476 controls, 26 explored EPHX1 rs2234922 and involved 4840 cases and 5326 controls, 31 investigated GSTP1 rs1695 and comprised 3975 cases and 4301 controls, while 7 explored GSTP1 rs1138272 and involved 1170 cases and 1455 controls. A schematic presentation of the literature screening process is shown in Figure 1. Based on the NOS scoring analysis of case controls, 16 studies had NOS scores of 6, while 43 had NOS scores of≥7, indicating that the included studies were of high quality. The basic characteristics of the included studies including the authors, publication years, NOS scores, ethnicity, genotype distributions of the case and control groups, and Hardy–Weinberg equilibrium analysis of the control group gene distribution are shown in Table 1 and Table 2.
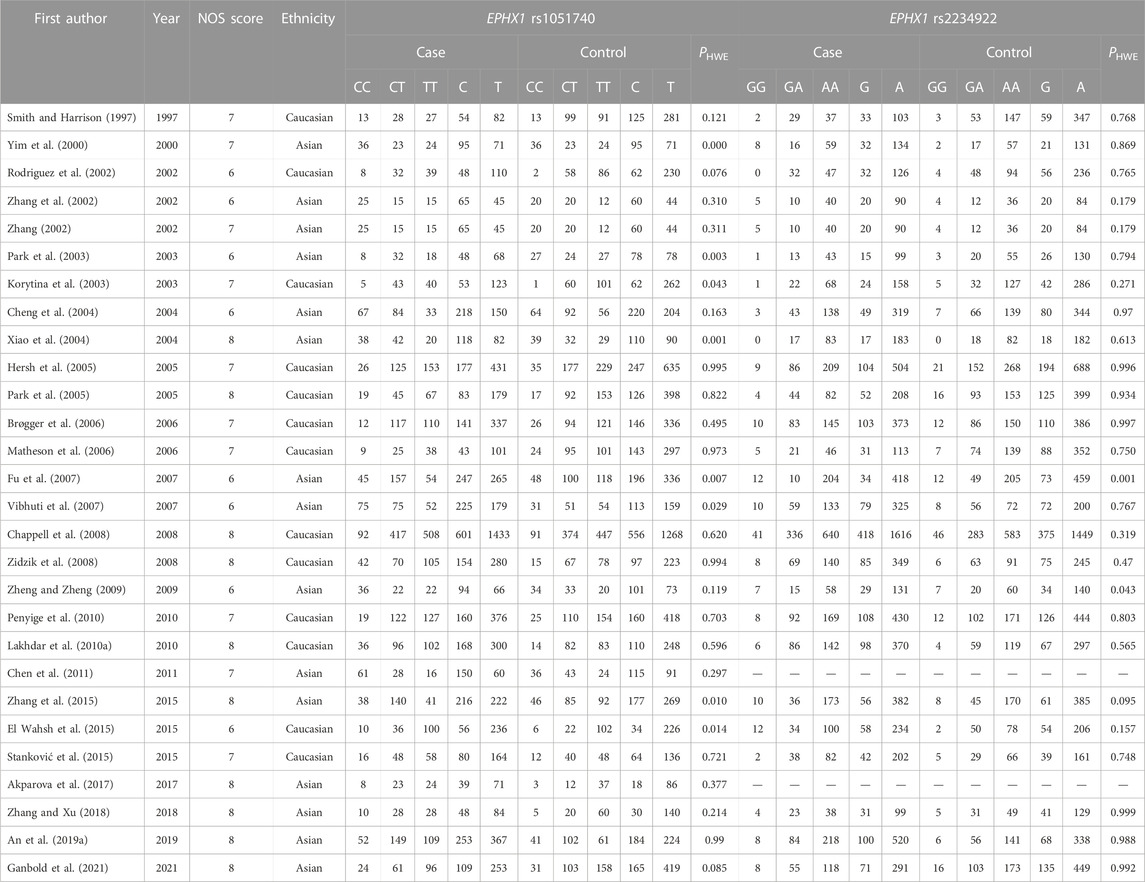
TABLE 1. Essential characteristics of EPHX1 (rs1051740 and rs2234922) polymorphism in the included studies
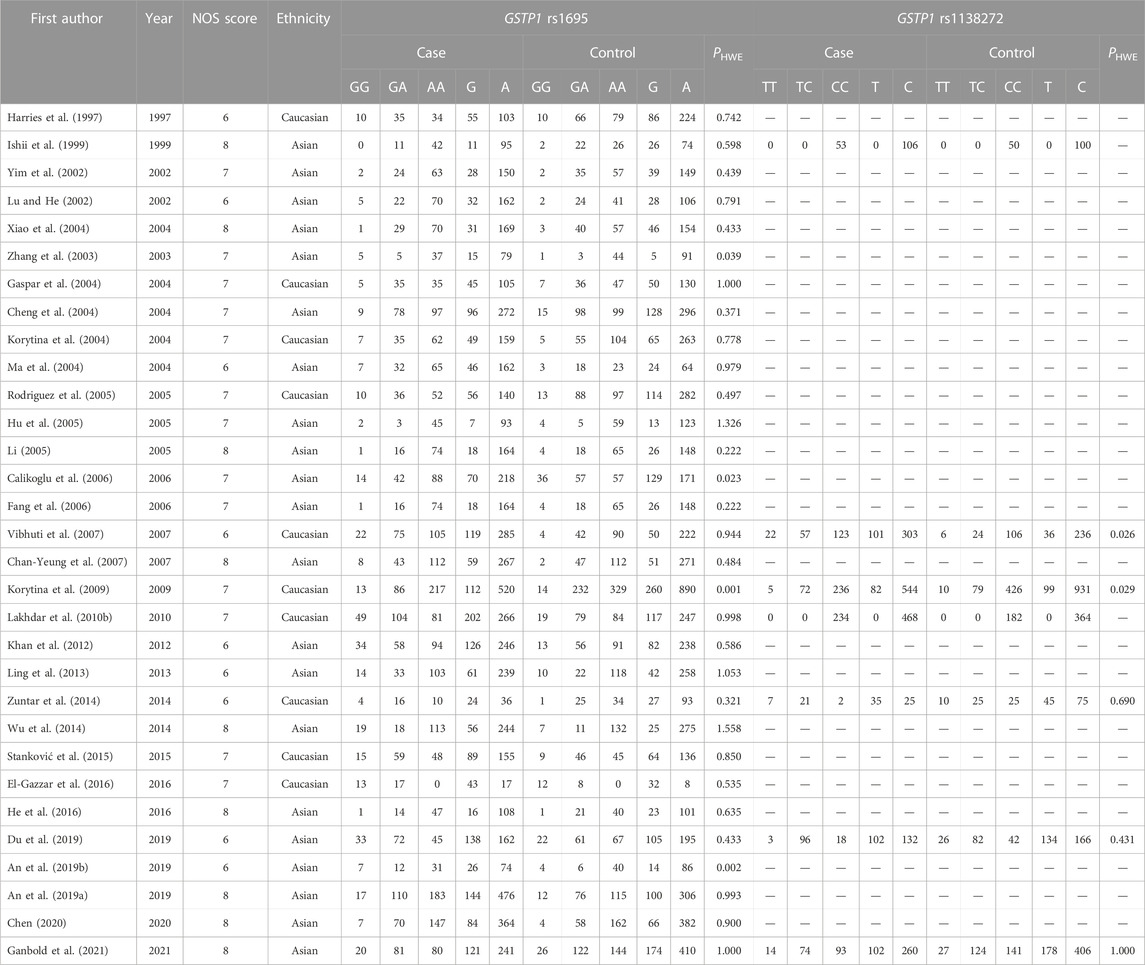
TABLE 2. Essential characteristics of GSTP1 (rs1695 and rs1138272) polymorphism in the included studies.
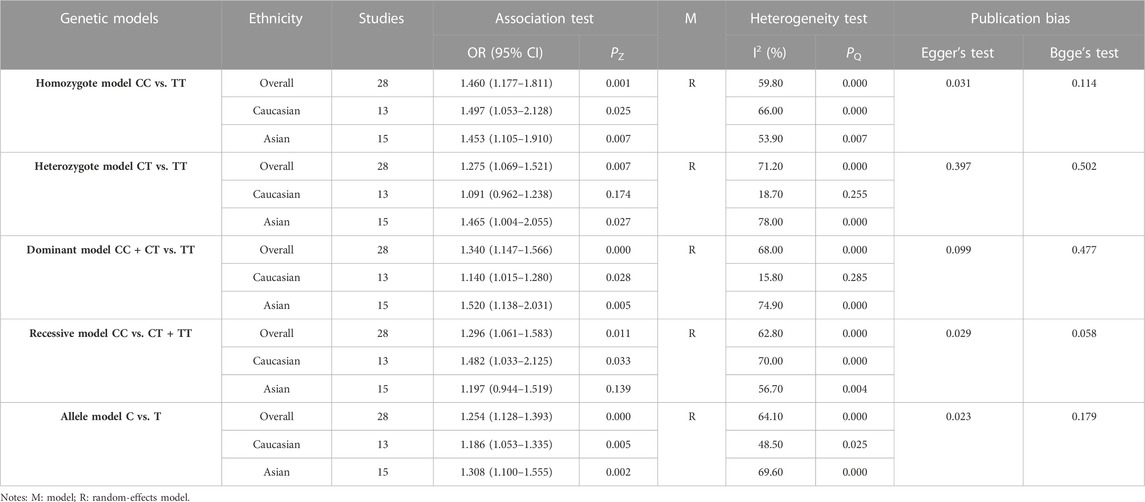
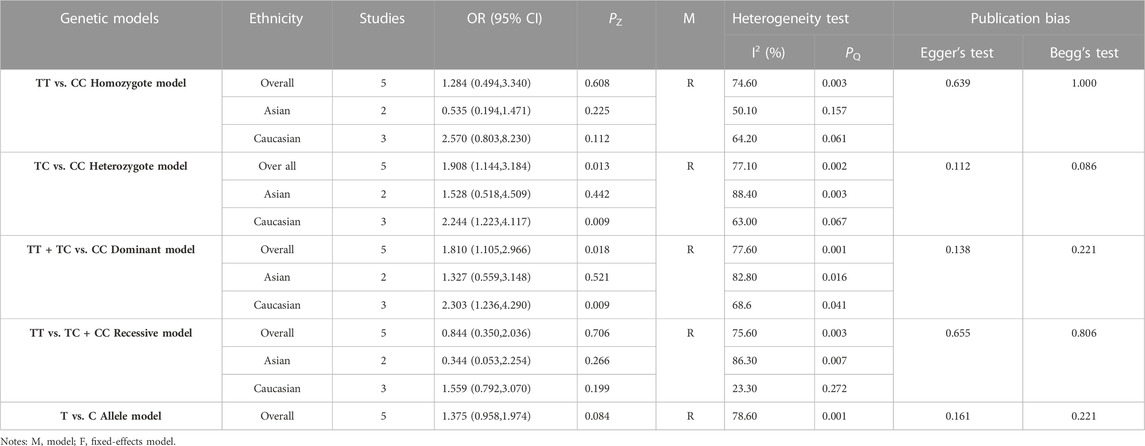
3.3 Association between the EPHX1 rs1051740 polymorphism and COPD risk
The results of meta-analyses of various models and subgroup analyses were utilized to explore the association of EPHX1 rs1051740 polymorphism with the COPD risk as shown in Table 3. In the overall analysis, heterogeneity among the five genetic models was high (I2>50%, PQ<0.10). Therefore, the random-effects model was used to determine the pooled OR and its 95% CI. For the homozygote model (OR = 1.460, PZ = 0.001), heterozygote model (OR = 1.275, PZ = 0.007), dominant model (OR = 1.340, PZ = 0.000), recessive model (OR = 1.296, PZ = 0.011) and allele model (OR = 1.254, PZ = 0.000), EPHX1 rs1051740 was significantly associated with COPD risk, indicating that the C allele is a risk factor for COPD. Subgroup analysis based on ethnicity showed that in the homozygote model (OR = 1.453, PZ = 0.007), heterozygote model (OR = 1.465, PZ = 0.027), dominant model (OR = 1.520, PZ = 0.005), and allele model (OR = 1.308, PZ = 0.002), EPHX1 rs1051740 was significantly associated with COPD risk among Asians, suggesting that the C allele is a risk factor for COPD in Asians. In addition, we also found that in the homozygote model (OR = 1.497, PZ = 0.025), dominant model (OR = 1.14, PZ = 0.028), recessive model (OR = 1.482, PZ = 0.033) and allele model (OR = 1.186, PZ = 0.005), EPHX1 rs1051740 was significantly associated with COPD risk among Caucasians, indicating that the CC genotype is a risk factor for COPD in Caucasians. Since 8 studies had PHWE<0.05 (Yim et al., 2000; Korytina et al., 2003; Park et al., 2003; Xiao et al., 2003; Xiao et al., 2004; Fu et al., 2007; Vibhuti et al., 2007; El Wahsh et al., 2015; Zhang et al., 2015), we conducted a subgroup analysis involving 20 studies with PHWE≥0.05. It was found that EPHX1 rs1051740 [homozygote model (OR = 1.409, 95% CI = 1.093–1.817, PZ = 0.008), recessive model (OR = 1.411, 95% CI = 1.116–1.784, PZ = 0.004) and allele model (OR = 1.203, 95% CI = 1.071–1.352, PZ = 0.002)] was significantly associated with COPD risk, consistent with findings from the overall analysis. Thus, studies with PHWE<0.05 did not affect the overall results, and the findings were generally reliable.
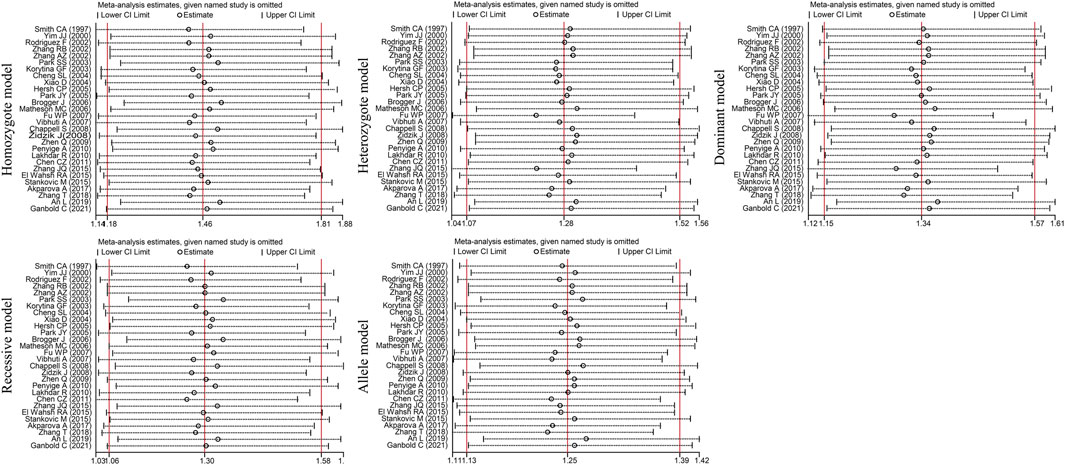
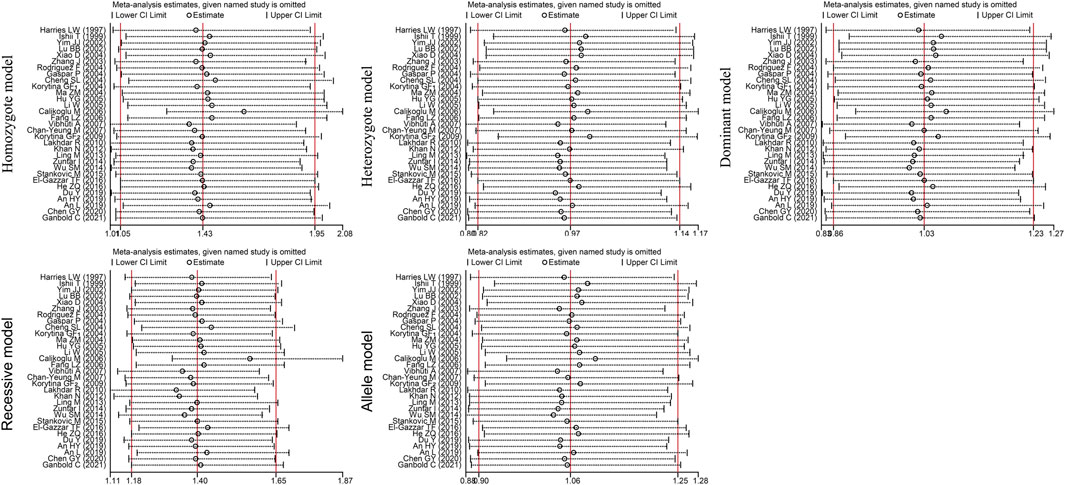
Egger’s test [homozygote model (PEgger = 0.031), recessive model (PEgger = 0.029) and allele model (PEgger = 0.023)] showed that publication bias might have been present, but Begg’s test did not reveal any publication bias (PBgge>0.05) (Table 3). The nonparametric trim-and-fill method was used to reduce the deviation in the combined effects. The trim-and-fill method is used to correct the impact of publication bias on the combined effect of meta-analysis. If the result of funnel plots show symmetry and Pz > 0.05 based on the trim-and-fill method results, the publication bias is not considered to significantly affect the reliability of the pooled results (Luo et al., 2022). We found that there were no significant changes in amounts of effects before and after pruning in the homozygote model (4 supplementary studies, OR = 1.331, 95% CI = 1.065–1.664, PZ = 0.012) and the allele model (3 excluded studies, OR = 1.254, 95% CI = 1.128–1.393, PZ = 0.000), indicating that the publication bias was too small to affect the stability of subgroup analysis results (Figures 2A, C). However, the shapes of the funnel plots were not symmetrical in the recessive models and Pz>0.05, indicating that the pooled results of the recessive model might not be stable (4 supplementary studies, OR = 1.254, 95% CI = 1.128–1.393, PZ = 0.567) (Figure 2B). Given the high heterogeneity (I2>50%, PQ<0.05) among the five genetic models, we conducted sensitivity analyses by excluding the studies one by one. The OR values of all studies fell within the 95% CI (Figure 3), which indicated that the overall estimates were stable and the results were reliable.

FIGURE 2. Funnel diagram of the trim-and-fill method of the EPHX1 rs1051740 polymorphism (Red points represent the supplementary studies). (A) Homozygote model. (B) Reccssive model. (C) Allele model.
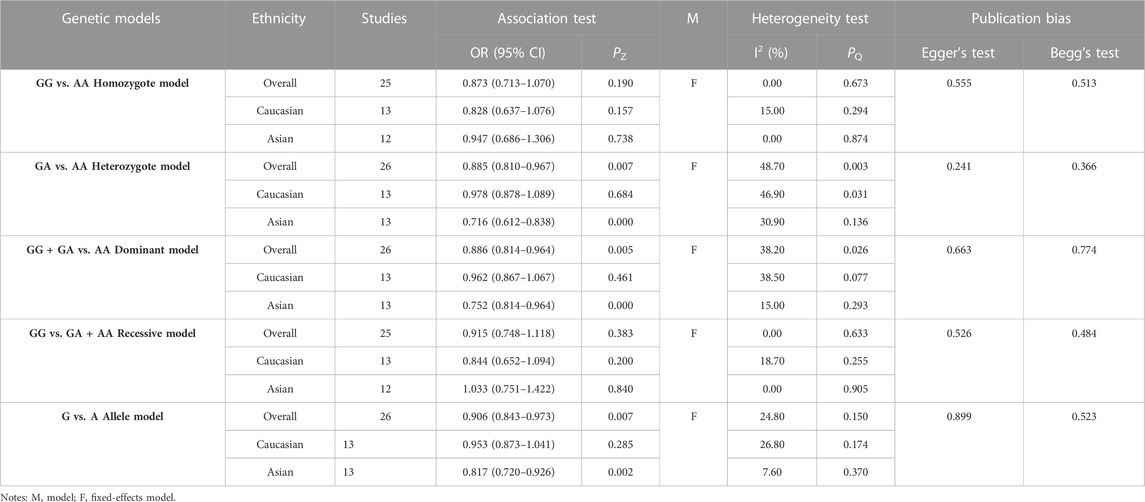
3.4 Association between EPHX1 rs2234922 polymorphism and COPD risk
The results of meta-analyses of various models and the subgroups used to explore the association between EPHX1 rs2234922 polymorphism and COPD risk are summarized in Table 4. In the overall analysis, heterogeneity among the five genetic models was low (I2<50%, PQ>0.10), and thus the fixed-effects model was used to determine the pooled OR and it is 95% CI. For the heterozygote model (OR = 0.885, PZ = 0.007), dominant model (OR = 0.886, PZ = 0.005) and allele model (OR = 0.906, PZ = 0.007), EPHX1 rs2234922 was found to be significantly associated with a lower risk of COPD, indicating that the G allele may be a protective factor for COPD. Subgroup analyses based on ethnicity showed that in the heterozygote model (OR = 0.716, PZ = 0.000), dominant model (OR = 0.752, PZ = 0.000) and allele model (OR = 0.817, PZ = 0.002), EPHX1 rs2234922 was significantly associated with reduced COPD risk among Asians, indicating that the GA genotype may be a protective factor for COPD among Asians. Considering that 2 studies had PHWE<0.05 (Fu et al., 2007; Zheng and Zheng, 2009), we conducted a subgroup analysis of 24 studies with PHWE≥0.05. Results showed that EPHX1 rs2234922 [dominant model (OR = 0.912, 95% CI = 0.836–0.994, PZ = 0.037) and allele model (OR = 0.922, 95% CI = 0.857–0.933, PZ = 0.032)] were associated with a lower risk of COPD, consistent with findings from the overall analysis, and indicating that some studies with PHWE <0.05 did not affect the overall results, which were reliable.
Egger’s and Begg’s tests of the included studies did not reveal any publication bias (PEgger>0.05, PBgge>0.05) (Table 4). Sensitivity analysis was conducted by excluding the studies one by one. The OR values of all studies fell within the 95% CI (Figure 4). These results suggested that the overall estimates were stable and the results were reliable.
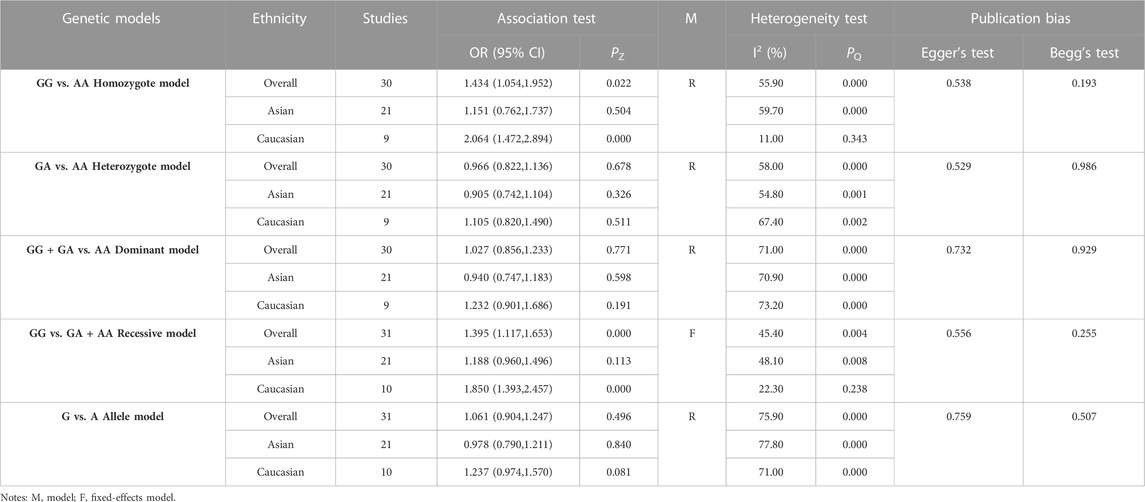
3.5 Association between the GSTP1 rs1695 polymorphism and COPD risk
The results of the meta-analysis among various models and subgroups used to explore the association between GSTP1 rs1695 and COPD risk are summarized in Table 5. In the overall analysis, the heterogeneity of the recessive model was low (I2 = 45.40%, PQ = 0.004), and therefore, the fixed-effects model was used to determine the pooled OR and its 95% CI. The other models had significant heterogeneity (I2>50%, PQ<0.10). Thus, the random-effects model was adopted. For the homozygote model (OR = 1.434, PZ = 0.022) and recessive model (OR = 1.395, PZ = 0.000), GSTP1 rs2234922 was found to be significantly associated with COPD risk, indicating that the GG genotype is a risk factor for COPD. Subgroup analysis based on ethnicity showed that in the homozygote model (OR = 2.064, PZ = 0.000) and recessive model (OR = 1.850, PZ = 0.000), GSTP1 rs1695 was significantly associated with COPD risk among Caucasians, while the association among Asians was not significant. Since 4 studies had PHWE <0.05 (Zhang et al., 2003; Calikoglu et al., 2006; Korytina et al., 2009; An et al., 2019b), we conducted a subgroup analysis of 27 studies with PHWE≥0.05 and established that GSTP1 rs1695 [homozygote model (OR = 1.586, 95% CI = 1.210–2.080, PZ = 0.001) and recessive model (OR = 1.533, 95% CI = 1.273–1.846, PZ = 0.000)] was significantly associated with COPD risk, consistent with findings in the overall analysis, indicating that some studies with PHWE <0.05 did not affect the overall results, which were reliable.
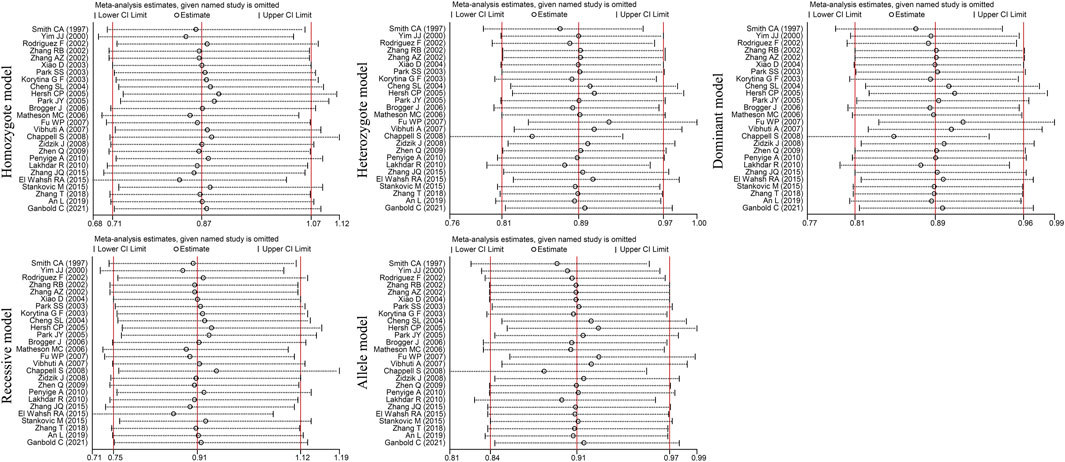
Egger’s and Begg’s tests did not reveal any publication bias (PEgger>0.05, PBgge>0.05) among the included studies (Table 5). In addition, sensitivity analysis was performed by excluding the studies one by one to determine the impact of each study on heterogeneity (I2 >50%, PQ <0.05). The OR values for all studies were within the 95% CI (Figure 5). These results indicate that the overall estimate was stable and the results were reliable.
3.6 Association between GSTP1 rs1138272 polymorphism and COPD risk
The results of the meta-analysis of various models and subgroups used to explore the association between GSTP1 rs1138272 and COPD risk are summarized in Table 6. In the overall analysis, significant heterogeneity was identified among the five genetic models (I2>50%, PQ<0.10). Therefore, the random-effects model was used to determine the pooled OR and its 95% CI. For the heterozygote model (OR = 1.908, PZ = 0.013) and dominant model (OR = 1.81, PZ = 0.018), GSTP1 rs1138272 was significantly associated with COPD risk, indicating that the TC genotype is a risk factor for COPD. Subgroup analysis by ethnicity revealed a significant association of GSTP1 rs1138272 with COPD risk among Caucasians for the homozygote model (OR = 2.570, PZ = 0.112), heterozygote model (OR = 2.244, PZ = 0.009) dominant model (OR = 2.303, PZ = 0.009) and allele model (OR = 1.822, PZ = 0.000). These results indicate that the TC genotype might be a risk factor for COPD among Caucasians. Since 2 studies had PHWE <0.05 (Vibhuti et al., 2007; Korytina et al., 2009), we conducted a subgroup analysis of 5 studies with PHWE ≥0.05 and observed that GSTP1 rs1138272 was not significantly correlated with COPD risk [homozygote model (OR = 1.085, 95% CI = 0.768–7.656, PZ = 0.918), heterozygote model (OR = 2.425, 95% CI = 0.768–7.656, PZ = 0.131), dominant model (OR = 2.112, 95% CI = 0.743–6.006, PZ = 0.161), recessive model (OR = 0.567, 95% CI = 0.158–2.038, PZ = 0.385), and allele model (OR = 1.155, 95% CI = 0.742–1.797, PZ = 0.524)], inconsistent with findings from the overall analysis.
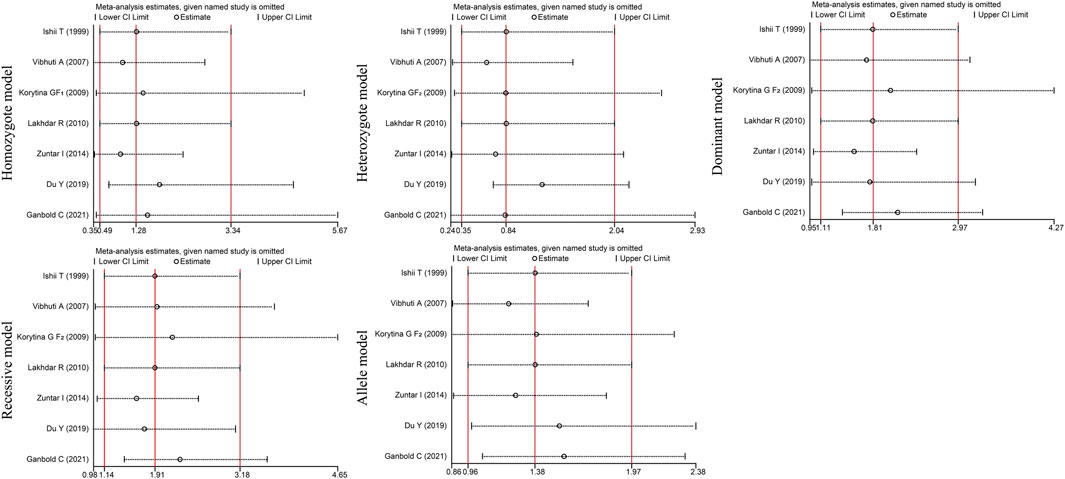
Egger’s and Begg’s tests did not reveal any publication bias among the included studies (PEgger>0.05, PBgge>0.05) (Table 6). Given the significant heterogeneity (I2>50%, PQ<0.05) and inconsistent results caused by articles with PHWE<0.05, we performed a sensitivity analysis by excluding the studies one by one. The OR values of all studies were within the 95% CI (Figure 6). These results indicate that the overall estimate was stable and the results were reliable.
4 Discussion
Chronic obstructive pulmonary disease (COPD) is a polygenic disease that is caused by various environmental factors and genetic factors (Silverman, 2020). The genes implicated in COPD occurrence are those that participate in anti-proteolysis, metabolism of toxic cigarette substances, airway hyperresponsiveness, inflammatory responses to smoking, and oxidative stress (Bossé, 2012). Among them, oxidative stress contributes to the upregulation of proinflammatory cytokine genes expression, enhances inflammatory responses, and damages the airway epithelium as well as the pulmonary interstitium and is a key driver of COPD development (Wu et al., 2014; Zhang et al., 2015). EPHX1 and GSTP1 are key oxidation-inhibiting enzymes which are xenobiotic metabolism enzyme genes. They are mainly involved in the first metabolism of foreign xenobiotic substances in the lungs, including cigarette smoke, oxides, and intermediate products of reactive oxides (An et al., 2019a). Although they have been extensively studied, the relationship between their mutations and the pathogenesis as well as risk of COPD is controversial. Thus, there is a need to explore the roles of EPHX1 and GSTP1 gene polymorphisms in COPD development to inform the development of diagnostic and therapeutic strategies for the disease. This meta-analysis investigated the association of EPHX1 (rs1051740 and rs2234922) and GSTP1 (rs1695 and rs1138272) polymorphisms with the risk of COPD. Since the ethnic background of gene-gene and gene-environment interactions affect SNP and disease risk, we conducted subgroup analysis based on ethnicity to determine the association between EPHX1 and GSTP1 gene polymorphisms and COPD risk in different ethnic groups.
Our meta-analysis of EPHX1 rs1051740 showed that the C allele of EPHX1 rs1051740 may be a risk factor for COPD. It was reported that a T/C mutation on codon 113 of exon 3 of EPHX1, substituting Tyr113 with His113, reduces the enzymatic activities of EPHX1 by 40%–50%, which increases the oxidation rate beyond the antioxidative capacity, resulting in the accumulation of reactive oxygen species in the organism and eventually, cell and lung tissue damage (Hassett et al., 1994). Clinical studies have demonstrated that the C allele is associated with lung dysfunction, reduced glutathione levels, and elevated malondialdehyde (MDA) levels in COPD patients (Vibhuti et al., 2007). In the subgroup analysis of EPHX1 rs1051740, it was found that the C allele is a risk factor for COPD in Asians. Furthermore, compared with genotype TT, genotype CC is a risk factor for COPD in Caucasians. These results are consistent with the observations that EPHX1 rs1051740 mutations decrease EPHX1 enzyme activity leading to an increase in the risk of COPD. A previous meta-analysis of 19 studies published in 2016 by An L et al. did not find any association between EPHX1 rs1051740 and COPD risk in Asians (OR = 0.92, PZ=0.31) and Caucasians (OR = 1.01, PZ=0.65) (An et al., 2016b). However, this study only used the allele model to pool analysis, which has greater limitations and needs further in-depth mining and research. Differences among the meta-analyses may also be attributed to the number of included studies that used the allelic model to perform the meta-analysis and publication bias. Here, we included all available studies that met our inclusion criteria (28 studies) to study the association between different genotypes of EPHX1 and COPD risk and used Egger’s and Begg’s tests to rule out any publication bias. And we also used the trim-and-fill method and sensitivity analysis to enhance the study’s rigor and reliability.
Furthermore, when a G/A mutation on codon 139 of EPHX1 exon 4 replaces Hisl39 with Arg139, it increases the activities of EPHX1 by 25% (Luo et al., 2019). Our meta-analysis findings indicate that the G allele of the EPHX1 rs2234922 gene may confer protection against COPD. Furthermore, in the subgroup analysis, we observed that Asians with the GA genotype had a decreased risk of COPD compared to individuals with the AA genotype. However, in the case of Caucasians, we did not identify any statistically significant associations. In contrast, studies of An L found that EPHX1 rs2234922 is not associated with COPD pathogenesis (OR = 1.01, PZ = 0.65) (An et al., 2016b). Lee J et al. concluded that although a statistically significant correlation was not observed, the presence of EPHX1 rs2234922 correlated with reduced COPD risk, consistent with the theory that the GA genotype of EPHX1 can increase the detoxification ability of the EPHX1 enzyme (Lee et al., 2011). We postulated that (rs1051740 and rs2234922) polymorphisms affect its enzymatic activities, which further involve the oxidative/antioxidative balance. Further investigations are required to establish if the polymorphisms of EPHX1 (rs1051740 and rs2234922) are significantly associated with COPD risk.
The tightly linked gene-gene interactions were observed between EPHX1 and GSTP1 gene polymorphisms, and alterations in the combined EPHX1-GSTP1 detoxification activity may affect COPD development (Ganbold et al., 2021). In view of the relationship between GSTP1 and EPHX1, we simultaneously investigated the correlation between GSTP1 polymorphisms and COPD risk. It was found that the GG genotype on GSTP1 rs1695 may be a risk factor for COPD. Subgroup analysis showed that Caucasians with the GG genotype were more likely to develop COPD than those with GA or AA genotypes, but the GG genotype was not associated with an increased risk of COPD among Asians. These findings are consistent with those of a previous meta-analysis published in 2010 that GSTP1 rs1695 is associated with increased COPD risk among Caucasians in the recessive model (OR = 1.59, PZ=0.001) but not among Asians (OR = 0.93, PZ = 0.64) (Zhong et al., 2010). However, another meta-analysis of 17 studies published in 2015 by Yang L found that there is no significant correlation between GSTP1 rs1695 polymorphism and COPD risk in any genetic model (Yang et al., 2015). The association between GSTP1 rs1695 polymorphism and COPD risk remains controversial, and no meta-analysis update has been conducted recently. Thus, we conducted a meta-analysis. In our study, we included 31 articles, including 5 newly published studies in the recent 7 years, and the analysis was more comprehensive and rigorous. Our results are more reliable than those of the other meta-analyses. It has been shown that the A/G mutations on GSTP1’s exon-5, which replace Ile105 with Val105, result in changing the volume and hydrophobicity of amino acids and inhibiting the enzyme’s activities as well as thermal stability, thereby reducing its detoxification capacity (Watson et al., 1998). It results in excess amounts of oxidants and free radicals in lung tissues and promotes airway tissue inflammation, which can cause bronchitis, emphysema, and COPD (Ganbold et al., 2021). This is consistent with our finding that the GSTP1 rs1695 GG genotype increases COPD risk. Large-sample clinical research showed that GSTP1 rs1695 was related to the rapid decline of lung function, which indirectly supported this evidence (He et al., 2004).
Our meta-analysis of GSTP1 rs1138272 showed that the TC genotype is a risk factor for COPD, and subgroup analysis showed that Caucasians carrying the TC genotype are more likely to suffer from COPD. It has been shown that the frequency of GSTP1 rs1138272 TC genotype in COPD patients was significantly higher than in normal people (28.57% vs. 14.45%), indicating that the GSTP1 rs1138272 polymorphism may be associated with COPD risk (Korytina et al., 2009). In contrast, Ganbold C et al. (Ganbold et al., 2021) concluded that GSTP1 rs1138272 polymorphisms are not correlated with COPD risk (OR = 1.38, p = 0.381). It is reported that the T/C variant of GSTP1 on exon 6 replaces Ala114 with Val114 without changing enzymatic activities (Watson et al., 1998). However, which is inconsistent with our findings. Our conclusion should be further validated because the number of studies involving the rs1138272 polymorphism of GSTP1 and the risk for COPD is small. We found that GSTP1 (rs1695 and rs1138272) mutations are associated with an increased risk of COPD. Given the correlation between GSTP and EPHX1, further studies should be performed to establish if EPHX1-GSTP1 interactions influence COPD development.
The PHWE<0.05 of the control group in the original article indicates that there may be a potential deviation in the study during control selection or genotyping errors (Lee et al., 2014). To avoid such deviations, we conducted subgroup analysis on studies with PHWE≥0.05 and found that only subgroup analysis results from GSTP1 rs1138272 differed from the original results. Our analysis found that data from 2 of the 5 studies involving GSTP1 rs1138272 (PHWE≥0.05) could not be calculated using the factor model and another 3 studies had a high heterogeneity (I2 >50%, PQ<0.05), indicating that the deviation may be caused by the high heterogeneity between studies and the small number of studies. Sensitivity analysis showed that OR values for all of the studies were within the 95% CI, indicating that the results were stable and reliable. After subgroup analysis according to PHWE≥0.05, the heterogeneity of EPHX1 1051740 (heterozygote model, dominant model and recessive model), EPHX1 rs2234922 (heterozygote model, dominant model and allele model), and GSTP1 rs1695 (homozygote model, heterozygote model, dominant model, recessive model and allele model) were decreased, so whether the control group included in the study conformed to PHWE ≥0.05 may be one of the sources of partial result heterogeneity.
5 Strengths and limitations of the study
The key strengths of this study are as follows. Firstly, strict inclusion and exclusion criteria were used to comprehensively assess the association of the polymorphisms of EPHX1 (rs1051740 and rs2234922) and GSTP1 (rs1695 and rs1138272) with the risk of COPD. Moreover, subgroup analysis was performed on different ethnicities to determine the effects of these polymorphisms on COPD susceptibility in diverse populations. However, this study has some limitations. For instance, our results are based on individual unadjusted estimates, and therefore, a more accurate prediction model need to be established after adjusting for potential confounding factors, such as sex, age, body mass index, lung functions, smoking status, and other environmental factors. However, subgroup analysis did not reveal whether these factors are associated with gene polymorphisms. Secondly, the results obtained from the subgroup analyses may be limited by the small number of studies involving African populations. Thirdly, although genetic and environmental factors may increase COPD risk, gene-gene and gene-environment interactions could not be assessed because of the limited data available. Finally, some of the studies included in this meta-analysis had significant heterogeneity which decreases the reliability of the final results.
6 Conclusion
EPHX1 (rs1051740 and rs2234922) and GSTP1 (rs1695 and rs1138272) polymorphisms are associated with the risk of COPD. The C allele of EPHX1 rs1051740 may increase the risk of COPD, especially among Asians, whereas the CC genotype may be a risk factor for COPD among Caucasians. In contrast, the G allele of EPHX1 rs2234922 may protect against COPD, especially the GA genotype significantly reducing COPD risk in Asians. The G allele of GSTP1 rs1695 may increase COPD risk, especially among Africans, whereas the TC genotype of GSTP1 rs1138272 may increase COPD risk, especially among Caucasians. These results indicate that EPHX1 and GSTP1 gene polymorphisms play key roles in COPD pathogenesis. Therefore, they are potential diagnostic and therapeutic targets in COPD. However, our conclusions should be validated in larger studies. Moreover, further analysis of gene-gene and gene-environment interactions should be performed to elucidate the mechanisms of COPD pathogenesis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
QY and WH conceptualization, methodology, writing-original draft, reviewing and editing. DY and LZ methodology, investigation, data curation, software and data analysis. YG methodology, writing review and editing. JT and ZL methodology, writing-review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was jointly funded by the joint key project of the National Natural Science Foundation of China (U20A20398), the major science and technology project of Anhui Province (201903a0702015), and the collaborative innovation project of public health in provincial medical colleges (GXXT-2020-025).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1128985/full#supplementary-material
References
Akparova, A., Abdrakhmanova, B., Banerjee, N., and Bersimbaev, R. (2017). EPHX1 Y113H polymorphism is associated with increased risk of chronic obstructive pulmonary disease in Kazakhstan population. Genet. Toxicol. Environ. Mutagen. 816-817, 1–6. doi:10.1016/j.mrgentox.2017.02.004
An, H. Y., Yang, P. Y., Zhu, X. W., Zhang, Y. P., Zhao, X. F., Ding, X. F., et al. (2019). Association of GSTP1 Ile (105) val polymorphisms diagnosed as chronic obstructive pulmonary disease in hui nationality population of henan. J. Mod. Med. Health 35, 1167–1173. doi:10.3969/j.issn.1009-5519.2019.08.014
An, L., Lin, Y., Yang, T., and Hua, L. (2016). Exploring the interaction among EPHX1, GSTP1, SERPINE2, and TGFB1 contributing to the quantitative traits of chronic obstructive pulmonary disease in Chinese Han population. Hum. genomics 10, 13. doi:10.1186/s40246-016-0076-0
An, L., Lin, Y., Zhang, H., Lu, Y., Zhang, S., and Hua, L. (2019). Genetic association of EPHX1, GSTP1 and susceptibility of chronic obstructive pulmonary disease and lung function in north Chinese Han population. Chin. J. Gerontology 39, 1860–1865. doi:10.3969/j.issn.1005-9202.2019.08.024
An, L., Xia, H., Zhou, P., and Hua, L. (2016). Exploration of association between EPHX1 and chronic obstructive pulmonary disease on the basis of combined data mining. Genet. Mol. Res. 15, 15. doi:10.4238/gmr.15028639
Bascom, R. (1991). Differential susceptibility to tobacco smoke: Possible mechanisms. Pharmacogenetics 1, 102–106. doi:10.1097/00008571-199111000-00008
Bossé, Y. (2012). Updates on the COPD gene list. Int. J. chronic Obstr. Pulm. Dis. 7, 607–631. doi:10.2147/COPD.S35294
Brøgger, J., Steen, V. M., Eiken, H. G., Gulsvik, A., and Bakke, P. (2006). Genetic association between COPD and polymorphisms in TNF, ADRB2 and EPHX1. Eur. Respir. J. 27, 682–688. doi:10.1183/09031936.06.00057005
Calikoglu, M., Tamer, L., Ates Aras, N., Karakaş, S., and Ercan, B. (2006). The association between polymorphic genotypes of glutathione S-transferases and COPD in the Turkish population. Biochem. Genet. 44, 307–319. doi:10.1007/s10528-006-9031-4
Cantlay, A. M., Smith, C. A., Wallace, W. A., Yap, P. L., Lamb, D., and Harrison, D. J. (1994). Heterogeneous expression and polymorphic genotype of glutathione S-transferases in human lung. Thorax 49, 1010–1014. doi:10.1136/thx.49.10.1010
Chan-Yeung, M., Ho, S. P., Cheung, A. H. K., So, L. K. Y., Wong, P. C., Chan, K. K., et al. (2007). Polymorphisms of glutathione S-transferase genes and functional activity in smokers with or without COPD. Int. J. Tuberc. Lung Dis. 11, 508–514. Available at: https://pubmed.ncbi.nlm.nih.gov/17439673/.
Chappell, S., Daly, L., Morgan, K., Guetta-Baranes, T., Roca, J., Rabinovich, R., et al. (2008). Genetic variants of microsomal epoxide hydrolase and glutamate-cysteine ligase in COPD. Eur. Respir. J. 32, 931–937. doi:10.1183/09031936.00065308
Chen, C. Z., Wang, R. H., Lee, C. H., Lin, C. C., Chang, H. Y., and Hsiue, T. R. (2011). Polymorphism of microsomal epoxide hydrolase is associated with chronic obstructive pulmonary disease and bronchodilator response. J. Formos. Med. Assoc. = Taiwan yi zhi 110, 685–689. doi:10.1016/j.jfma.2011.09.003
Chen, G. (2020). Relationship between GSTP1 gene polymorphism and susceptibility to chronic obstructive pulmonary disease. Kaifeng: Henan University.
Cheng, S., Yu, C., Chen, C., and Yang, P. (2004). Genetic polymorphism of epoxide hydrolase and glutathione S-transferase in COPD. Eur. Respir. J. 23, 818–824. doi:10.1183/09031936.04.00104904
Cho, M. H., Hobbs, B. D., and Silverman, E. K. (2022). Genetics of chronic obstructive pulmonary disease: Understanding the pathobiology and heterogeneity of a complex disorder. Lancet Respir. Med. 10, 485–496. doi:10.1016/S2213-2600(21)00510-5
Du, Y., Zhang, H., Xu, Y., Ding, Y., Chen, X., Mei, Z., et al. (2019). Association among genetic polymorphisms of GSTP1, HO-1, and SOD-3 and chronic obstructive pulmonary disease susceptibility. Int. J. COPD 14, 2081–2088. doi:10.2147/COPD.S213364
El Wahsh, R. A., Essa, E. S., Bakr, R. M., Zamzam, M. A., and Abozeid, S. M. (2015). GSTM1, GSTT1 and EPHX1 gene polymorphisms and susceptibility to COPD in a sample of Egyptian population. Egypt. J. Chest Dis. Tuberc. 64, 829–836. doi:10.1016/j.ejcdt.2015.05.005
El-Gazzar, T. F., El-dahdouh, S. S., and El-Mahalawy, (2016). Role of glutathione S-transferase P-1 (GSTP-1) gene polymorphism in COPD patients. Egypt. J. Chest Dis. Tuberc. 65, 739–744. doi:10.1016/j.ejcdt.2015.06.008
Fu, W., Sun, C., Dai, L., Yang, L., and Zhang, Y. (2007). Relationship between COPD and polymorphisms of HOX-1 and mEPH in a Chinese population. Oncol. Rep. 17, 483–488. doi:10.3892/or.17.2.483
Ganbold, C., Jamiyansuren, J., Tumurbaatar, A., Bayarmaa, A., Enebish, T., Dashtseren, I., et al. (2021). The cumulative effect of gene-gene interactions between GSTM1, CHRNA3, CHRNA5 and SOD3 gene polymorphisms combined with smoking on COPD risk. Int. J. chronic Obstr. Pulm. Dis. 16, 2857–2868. doi:10.2147/COPD.S320841
Gaspar, P., Moreira, J., Kvitko, K., Torres, M., and Moreira, A. (2004). CYP1A1, CYP2E1, GSTM1, GSTT1, GSTP1, and TP53 polymorphisms: Do they indicate susceptibility to chronic obstructive pulmonary disease and non-small-cell lung cancer? Genet. Mol. Biol. 27, 133–138. doi:10.1590/s1415-47572004000200001
Hassett, C., Aicher, L., Sidhu, J. S., and Omiecinski, C. J. (1994). Human microsomal epoxide hydrolase: Genetic polymorphism and functional expression in vitro of amino acid variants. Hum. Mol. Genet. 3, 421–428. doi:10.1093/hmg/3.3.421
He, J. Q., Connett, J. E., Anthonisen, N. R., Paré, P. D., and Sandford, A. J. (2004). Glutathione S-transferase variants and their interaction with smoking on lung function. Am. J. Respir. Crit. care Med. 170, 388–394. doi:10.1164/rccm.200312-1763OC
He, Z. Q., Fu, Y., and Liu, S. G. (2016). Correlation between gene polymorphisms of glutathione S-transferase M1 and glutathione S-transferase P1 and phenotypes of chronic obstructive pulmonary disease combined with obstructive sleep apnea hypopnea syndrome. Shanghai Med. J. 39, 400–406+452. CNKI: SUN:SHYX.0.2016-07-004.
Hersh, C. P., Demeo, D. L., Lange, C., Litonjua, A. A., Reilly, J. J., Kwiatkowski, D., et al. (2005). Attempted replication of reported chronic obstructive pulmonary disease candidate gene associations. Am. J. Respir. Cell. Mol. Biol. 33, 71–78. doi:10.1165/rcmb.2005-0073OC
Hersh, C. P., Demeo, D. L., Lazarus, R., Celedón, J. C., Raby, B. A., Benditt, J. O., et al. (2006). Genetic association analysis of functional impairment in chronic obstructive pulmonary disease. Am. J. Respir. Crit. care Med. 173, 977–984. doi:10.1164/rccm.200509-1452OC
Hersh, C. P., DeMeo, D. L., Reilly, J. J., and Silverman, E. K. (2007). Xenobiotic metabolizing enzyme gene polymorphisms predict response to lung volume reduction surgery. Respir. Res. 8, 59. doi:10.1186/1465-9921-8-59
Khan, N., Daga, M., Kamble, N., Mawari, G., Ahmed, I., and Husain, S. A. (2012). Association of xenobiotic metabolizing gene polymorphisms and chronic obstructive pulmonary disease in Indian population. Eur. Respir. J. 40, P4804. doi:10.1111/j.1744-313X.2010.00918.x
Kiyohara, C., Yoshimasu, K., Takayama, K., and Nakanishi, Y. (2006). EPHX1 polymorphisms and the risk of lung cancer: A HuGE review. Epidemiology 17, 89–99. doi:10.1097/01.ede.0000187627.70026.23
Korytina, G. F., Akhmadishina, L. Z., Cilousova, O. S., Zagidullin, S. Z., and Victorova, T. V. (2009). Polymorphism of the genes for antioxidant defense enzymes and their association with the development of chronic obstructive pulmonary disease in the population of Bashkortostan. Russ. J. Genet. 45, 848–856. doi:10.1134/s1022795409070138
Korytina, G. F., Yanbaeva, D. G., and Victorova, T. V. (2004). [Polymorphism of glutathione-S-transferase M1 and P1 genes in patients with cystic fibrosis and chronic respiratory tract diseases]. Genetika 40, 401–408. doi:10.1023/B:RUGE.0000021633.91036.92
Korytina, G. F., Yanbaeva, D. G., and Viktorova, T. V. (2003). [Role of polymorphic variants of cytochrome P450 genes (CYP1A1, CYP2E1) and microsomal epoxide hydrolase (mEPHX) in pathogenesis of cystic fibrosis and chronic respiratory tract diseases]. Mol. Biol. 37, 784–792. Available at: https://www.embase.com/search/results?subaction=viewrecord&id=L44253003&from=export.
Lakhdar, R., Denden, S., Knani, J., Leban, N., Daimi, H., Hassine, M., et al. (2010). Microsomal epoxide hydrolase gene polymorphisms and susceptibility to chronic obstructive pulmonary disease in the Tunisian population. Genet. Test. Mol. biomarkers 14, 857–863. doi:10.1089/gtmb.2009.0168
Lakhdar, R., Denden, S., Knani, J., Leban, N., Daimi, H., Hassine, M., et al. (2010). Relationship between glutathione S-transferase P1 polymorphisms and chronic obstructive pulmonary disease in a Tunisian population. GMR 9, 897–907. doi:10.4238/vol9-2gmr770
Lareau, S. C., Fahy, B., Meek, P., and Wang, A. (2019). Chronic obstructive pulmonary disease (COPD). Am. J. Respir. Crit. care Med. 199, P1–P2. doi:10.1164/rccm.1991P1
Lee, J., Nordestgaard, B. G., and Dahl, M. (2011). EPHX1 polymorphisms, COPD and asthma in 47,000 individuals and in meta-analysis. Eur. Respir. J. 37, 18–25. doi:10.1183/09031936.00012110
Lee, Y. H., Kim, J. H., and Song, G. G. (2014). Meta-analysis of associations between interleukin-10 polymorphisms and susceptibility to pre-eclampsia. Eur. J. Obstet. Gynecol. Reprod. Biol. 182, 202–207. doi:10.1016/j.ejogrb.2014.09.030
Li, W. (2005). A preliminary study on the relationship between GSTP1 exon 5 polymorphism and chronic obstructive pulmonary disease (COPD) and clinical phenotype in the han population of southwest China, kunming medical college. doi:10.3969/j.issn.1004-745X.2011.01.100
Ling, M., An, H. Y., Rong, Y., Wang, H., Niu, L., Zhu, J., et al. (2013). The relationship between glutathione S-transferase P1 gene polymorphism and the susceptibility of Kazakh patients with chronic obstructive pulmonary disease. Chin. J. Tuberc. Respir. Dis. 36, 459–460. doi:10.3760/cma.j.issn.1001-0939.2012.06.016
Lu, B. B., and He, Q. Y. (2002). Correlation between exon5 polymorphism of glutathione S-transferase P1 gene and susceptibility to chronic obstructive pulmonary disease in northern Chinese population of Han nationality living in Beijing, China. Zhonghua nei ke za zhi 41, 678–681. CNKI: SUN:ZHNK.0.2002-10-017.
Luo, C., Marks-Anglin, A., Duan, R., Lin, L., Hong, C., Chu, H., et al. (2022). Accounting for publication bias using a bivariate trim and fill meta-analysis procedure. Stat. Med. 41, 3466–3478. doi:10.1002/sim.9428
Luo, Y., Dai, L. M., Jia, M., Zhao, Z. H., Hu, C. M., Qi, W. Y., et al. (2019). Study on the relationship between EPHX1 gene polymorphism and antioxidant capacity in patients with chronic obstructive pulmonary disease. Zhonghua Jie He He Hu Xi Za Zhi 42, 760–764. doi:10.3760/cma.j.issn.1001-0939.2019.10.009
Ma, Z. M., Zhang, Z. X., Han, Z. M., and Xu, Y. J. (2004). A study on the correlation between the polymorphism at the gutathione S transferase P 1 Locus and the susceptibility to the deveopmentof COPD. J. Pract. Med., 741–743. doi:10.3969/j.issn.1006-5725.2004.07.008
Matheson, M. C., Raven, J., Walters, E. H., Abramson, M. J., and Ellis, J. A. (2006). Microsomal epoxide hydrolase is not associated with COPD in a community-based sample. Hum. Biol. 78, 705–717. doi:10.1353/hub.2007.0015
Mirzakhani, M., Shahbazi, M., Akbari, R., Dedinská, I., Nemati, E., and Mohammadnia-Afrouzi, M. (2020). Soluble CD30, the immune response, and acute rejection in human kidney transplantation: A systematic review and meta-analysis. Front. Immunol. 11, 295. doi:10.3389/fimmu.2020.00295
Nikolaou, V., Massaro, S., Fakhimi, M., Stergioulas, L., and Price, D. (2020). COPD phenotypes and machine learning cluster analysis: A systematic review and future research agenda. Respir. Med. 171, 106093. doi:10.1016/j.rmed.2020.106093
Park, J. Y., Chen, L., Wadhwa, N., and Tockman, M. S. (2005). Polymorphisms for microsomal epoxide hydrolase and genetic susceptibility to COPD. Int. J. Mol. Med. 15, 443–448. doi:10.3892/ijmm.15.3.443
Park, S. S., Kim, E. J., Son, C. Y., Wi, J. O., Park, K. H., Cho, G. J., et al. (2003). Genetic polymorphism of epoxide hydrolase and GSTM1 in chronic obstructive pulmonary disease. Tuberc. Respir. Dis. 55, 88–97. doi:10.4046/trd.2003.55.1.88
Penyige, A., Poliska, S., Csanky, E., Scholtz, B., Dezso, B., Schmelczer, I., et al. (2010). Analyses of association between PPAR gamma and EPHX1 polymorphisms and susceptibility to COPD in a Hungarian cohort, a case-control study. BMC Med. Genet. 11, 152. doi:10.1186/1471-2350-11-152
Pillai, S. G., Ge, D., Zhu, G., Kong, X., Shianna, K. V., Need, A. C., et al. (2009). A genome-wide association study in chronic obstructive pulmonary disease (COPD): Identification of two major susceptibility loci. PLoS Genet. 5, e1000421. doi:10.1371/journal.pgen.1000421
Rodriguez, F., de la Roza, C., Jardi, R., Schaper, M., Vidal, R., and Miravitlles, M. (2005). Glutathione S-transferase P1 and lung function in patients with alpha1-antitrypsin deficiency and COPD. Chest 127, 1537–1543. doi:10.1378/chest.127.5.1537
Rodriguez, F., Jardi, R., Costa, X., Juan, D., Galimany, R., Vidal, R., et al. (2002). Detection of polymorphisms at exons 3 (Tyr113-->His) and 4 (His139-->Arg) of the microsomal epoxide hydrolase gene using fluorescence PCR method combined with melting curves analysis. Anal. Biochem. 308, 120–126. doi:10.1016/s0003-2697(02)00219-1
Sakornsakolpat, P., Prokopenko, D., Lamontagne, M., Reeve, N. F., Guyatt, A. L., Jackson, V. E., et al. (2019). Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat. Genet. 51, 494–505. doi:10.1038/s41588-018-0342-2
Sandford, A. J., and Silverman, E. K. (2002). Chronic obstructive pulmonary disease. 1: Susceptibility factors for COPD the genotype-environment interaction. Thorax 57, 736–741. doi:10.1136/thorax.57.8.736
Shrine, N., Guyatt, A. L., Erzurumluoglu, A. M., Jackson, V. E., Hobbs, B. D., Melbourne, C. A., et al. (2019). New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat. Genet. 51, 481–493. doi:10.1038/s41588-018-0321-7
Silverman, E. K., Chapman, H. A., Drazen, J. M., Weiss, S. T., Rosner, B., Campbell, E. J., et al. (1998). Genetic epidemiology of severe, early-onset chronic obstructive pulmonary disease. Risk to relatives for airflow obstruction and chronic bronchitis. Am. J. Respir. Crit. care Med. 157, 1770–1778. doi:10.1164/ajrccm.157.6.9706014
Silverman, E. K. (2020). Genetics of COPD. Annu. Rev. Physiol. 82, 413–431. doi:10.1146/annurev-physiol-021317-121224
Singh, D., Agusti, A., Anzueto, A., Barnes, P. J., Bourbeau, J., Celli, B. R., et al. (2019). Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease: The GOLD science committee report 2019. Eur. Respir. J. 53, 1900164. doi:10.1183/13993003.00164-2019
Smith, C. A., and Harrison, D. J. (1997). Association between polymorphism in gene for microsomal epoxide hydrolase and susceptibility to emphysema. Lancet (London, Engl. 350, 630–633. doi:10.1016/S0140-6736(96)08061-0
Stang, A. (2010). Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605. doi:10.1007/s10654-010-9491-z
Stanković, M., Nikolić, A., Tomović, A., Mitić-Milikić, M., Nagorni-Obradović, L., Petrović-Stanojević, N., et al. (2015). Association of functional variants of phase I and II genes with chronic obstructive pulmonary disease in a Serbian population. J. Med. Biochem. 34, 207–214. doi:10.2478/jomb-2014-0024
Tomaki, M., Sugiura, H., Koarai, A., Komaki, Y., Akita, T., Matsumoto, T., et al. (2007). Decreased expression of antioxidant enzymes and increased expression of chemokines in COPD lung. Pulm. Pharmacol. Ther. 20, 596–605. doi:10.1016/j.pupt.2006.06.006
Vibhuti, A., Arif, E., Deepak, D., and Singh, B. (2007). Genetic polymorphisms of GSTP1 and mEPHX correlate with oxidative stress markers and lung function in COPD. Biochem. biophysical Res. Commun. 359, 136–142. doi:10.1016/j.bbrc.2007.05.076
Watson, M. A., Stewart, R. K., Smith, G. B., Massey, T. E., and Bell, D. A. (1998). Human glutathione S-transferase P1 polymorphisms: Relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis 19, 275–280. doi:10.1093/carcin/19.2.275
Wu, S. M., Wang, F. E., Guo, S. J., and Ling, M. (2014). Association between GSTP1 polymorphisms and susceptibility to chronic obstructive pulmonary disease in Xinjiang Uighur population. Xi'an, China: Journal of Xi'an Jiaotong University, 509–512. (Medical Sciences). doi:10.7652/jdyxb201404019
Xiao, D., Wang, C., Du, M., Pang, B., Zhang, H., Xiao, B., et al. (2003). Association between polymorphisms in the microsomal epoxide hydrolase (mEH) gene and chronic obstructive pulmonary disease. Zhonghua yi xue za zhi 83, 1782–1786.
Xiao, D., Wang, C., Du, M., Pang, B., Zhang, H., Xiao, B., et al. (2004). Relationship between polymorphisms of genes encoding microsomal epoxide hydrolase and glutathione S-transferase P1 and chronic obstructive pulmonary disease. Chin. Med. J. 117, 661–667. doi:10.3760/cma.j.issn.0366-6999.2004.05.107
Yang, L., Li, X., Tong, X., and Fan, H. (2015). Association between glutathione S-transferase P1 Ile (105) val gene polymorphism and chronic obstructive pulmonary disease: A meta-analysis based on seventeen case-control studies. Meta Gene 6, 59–64. doi:10.1016/j.mgene.2015.08.007
Yim, J. J., Park, G. Y., Lee, C. T., Kim, Y. W., Han, S. K., Shim, Y. S., et al. (2000). Genetic susceptibility to chronic obstructive pulmonary disease in Koreans: Combined analysis of polymorphic genotypes for microsomal epoxide hydrolase and glutathione S-transferase M1 and T1. Thorax 55, 121–125. doi:10.1136/thorax.55.2.121
Yim, J. J., Yoo, C. G., Lee, C. T., Kim, Y. W., Han, S. K., and Shim, Y. S. (2002). Lack of association between glutathione S-transferase P1 polymorphism and COPD in Koreans. Lung 180, 119–125. doi:10.1007/s004080000086
Zhang, A. Z. (2002). Association between in gene for microsomal epoxide hydrolase and susceptibility to the chronic obstructive pulmonary disease. Jinzhong: Shanxi Medical University.
Zhang, J., Wu, Y. L., Liu, X., and Shi, R. F. (2003). Study on the relationship between glutathione S-transferase P1 gene polymorphism and COPD in Chinese Han nationality. Chin. J. Tuberc. Respir. Dis. 14, 57–60. CNKI: SUN:ZHJH.0.2003-05-028.
Zhang, J. Q., Zhang, J. Q., Liu, H., Zhao, Z. H., Fang, L. Z., Liu, L., et al. (2015). Effect of N-acetylcysteine in COPD patients with different microsomal epoxide hydrolase genotypes. Int. J. chronic Obstr. Pulm. Dis. 10, 917–923. doi:10.2147/COPD.S79710
Zhang, R. B., Zhang, A. Z., He, Q. Y., and Lu, B. B. (2002). Microsomal epoxide hydrolase gene polymorphism and susceptibility to chronic obstructive pulmonary disease in Han nationality of North China. Chinese Journal of Internal Medicine. 14–17. Available at: http://rs.yiigle.com/CN112138200201/1142005.htm.
Zhang, T., and Xu, Z. B. (2018). Association of EPHX1 gene polymorphism with the risk of COPD and PH. Zhejiang Clin. Med. J. 20, 1346–1348. Available at: https://d.wanfangdata.com.cn/periodical/zjlcyx201808008.
Zheng, Q., and Zheng, W. (2009). Association between microsomal epoxide hydrolase gene polymorphism and susceptibility to chronic obstructive pulmonary disease. Chin. J. Prim. Med. Pharm., 1779–1780. doi:10.3760/cma.j.issn.1008-6706.2009.10.029
Zhong, L., Zhang, Y., Fu, W., Dai, L., Sun, C., and Wang, Y. (2010). The relationship between GSTP1 I105V polymorphism and COPD: A reappraisal. Am. J. Respir. Crit. care Med. 181, 763–765. doi:10.1164/ajrccm.181.7.763
Zidzik, J., Slabá, E., Joppa, P., Kluchová, Z., Dorková, Z., Skyba, P., et al. (2008). Glutathione S-transferase and microsomal epoxide hydrolase gene polymorphisms and risk of chronic obstructive pulmonary disease in Slovak population. Croat. Med. J. 49, 182–191. doi:10.3325/cmj.2008.2.182
Keywords: COPD risk, EPHX1, GSTP1, gene polymorphism, meta-analysis
Citation: Yang Q, Huang W, Yin D, Zhang L, Gao Y, Tong J and Li Z (2023) EPHX1 and GSTP1 polymorphisms are associated with COPD risk: a systematic review and meta-analysis. Front. Genet. 14:1128985. doi: 10.3389/fgene.2023.1128985
Received: 21 December 2022; Accepted: 10 May 2023;
Published: 22 May 2023.
Edited by:
Claudia Banescu, University of Medicine, RomaniaReviewed by:
Ingrid Fricke-Galindo, Instituto Nacional de Enfermedades Respiratorias-México (INER), MexicoStoian Adina, George Emil Palade University of Medicine, Romania
Copyright © 2023 Yang, Huang, Yin, Zhang, Gao, Tong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zegeng Li, YWh6eWZiQHNpbmEuY29t
†These authors have contributed equally to this work and share first authorship
 Qinjun Yang
Qinjun Yang Wanqiu Huang
Wanqiu Huang Dandan Yin
Dandan Yin Lu Zhang
Lu Zhang Yating Gao
Yating Gao Jiabing Tong1,3,5
Jiabing Tong1,3,5 Zegeng Li
Zegeng Li